- 1Engineering Research Center of Agricultural Microbiology Technology, Ministry of Education, Heilongjiang University, Harbin, China
- 2Heilongjiang Provincial Key Laboratory of Plant Genetic Engineering and Biological Fermentation Engineering for Cold Region, School of Life Sciences, Heilongjiang University, Harbin, China
- 3State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 4College of Forest Resources and Environmental Science, Michigan Technological University, Houghton, MI, United States
WUSCHEL-related homeobox (WOX) genes are plant-specific transcription factors (TFs) involved in multiple processes of plant development. However, there have hitherto no studies on the WOX TFs involved in secondary cell wall (SCW) formation been reported. In this study, we identified a Populus trichocarpa WOX gene, PtrWOX13A, which was predominantly expressed in SCW, and then characterized its functions through generating PtrWOX13A overexpression poplar transgenic lines; these lines exhibited not only significantly enhanced growth potential, but also remarkably increased SCW thicknesses, fiber lengths, and lignin and hemicellulose contents. However, no obvious change in cellulose content was observed. We revealed that PtrWOX13A directly activated its target genes through binding to two cis-elements, ATTGATTG and TTAATSS, in their promoter regions. The fact that PtrWOX13A responded to the exogenous GAs implies that it is responsive to GA homeostasis caused by GA inactivation and activation genes (e.g., PtrGA20ox4, PtrGA2ox1, and PtrGA3ox1), which were regulated by PtrWOX13A directly or indirectly. Since the master switch gene of SCW formation, PtrWND6A, and lignin biosynthesis regulator, MYB28, significantly increased in PtrWOX13A transgenic lines, we proposed that PtrWOX13A, as a higher hierarchy TF, participated in SCW formation through controlling the genes that are components of the known hierarchical transcription regulation network of poplar SCW formation, and simultaneously triggering a gibberellin-mediated signaling cascade. The discovery of PtrWOX13A predominantly expressed in SCW and its regulatory functions in the poplar wood formation has important implications for improving the wood quality of trees via genetic engineering.
Introduction
WUSCHEL-related homeobox (WOX) family, a plant-specific clade of homeobox transcription factors (TFs) (Park et al., 2005), have been found to play an important role not only in plant developmental tissues and organs (Cheng et al., 2018; Hao et al., 2019) but also in plant response to abiotic stresses (Yang et al., 2017; Minh-Thu et al., 2018; Sajjad et al., 2021). The WOX proteins usually have a highly conserved homeodomain harboring a short stretch of amino acids (60–66 residues) with a helix–loop–helix–turn–helix structure (Yang et al., 2017), which is responsible for binding to a specific DNA sequence (Alvarez et al., 2018; Minh-Thu et al., 2018). To date, WOX genes have been identified in various plant species at the whole genome scale (Deveaux et al., 2008; Zhang et al., 2010; Gambino et al., 2011; Cao et al., 2017; Chang et al., 2020; Daude et al., 2020; Tang et al., 2020). The numbers of WOX genes in plants have been reported to be more related to the levels of ploidy and duplication events rather than genome sizes (Li et al., 2019; Wang et al., 2019; Wu et al., 2019). For example, although Brassica napus and Phyllostachys edulis both are tetraploid genome, B. napus has the largest WOX family consisting of 52 members (Li et al., 2019), while the WOX family P. edulis only has five members (Xu et al., 2019). The WOX genes have been clustered into three clades, known as WUS/modern clade, intermediate clade, and ancient clade (van der Graaff et al., 2009). For example, AtWOX1-7 and AtWUS of Arabidopsis thaliana belong to the WUS/modern clade, AtWOX8-9 and AtWOX11-12 belong to the intermediate clade, and AtWOX10 and AtWOX13-14 are of the ancient clade (Sarkar et al., 2007; Stahl et al., 2009). In addition, each clade of the WOX family has been found to contain specific conserved motifs (Kieffer et al., 2006; Deveaux et al., 2008; Ikeda et al., 2009), such as ancient clade having NVFYWFQNH motif and an ancient/basal motif (Wu et al., 2019).
It has been reported that WOX genes participate in multiple developmental processes in plants (Rathour et al., 2020; Han et al., 2021), such as stem cell maintenance in the shoot and root apical meristems, cambium and embryo apical–basal patterning, and lateral organ formation (Deveaux et al., 2008; Stahl et al., 2009; Ji et al., 2010; Zhang et al., 2010; Suer et al., 2011). Besides these, mounting evidence suggests that some members of the WOX ancient clade play a function in wood formation. For example, AtWOX13 has been reported to be expressed not only in meristematic tissues but also in replums, lateral roots, and root vasculature (Romera-Branchat et al., 2013). AtWOX14 overexpression induces radial growth with strong lignification of vascular stem tissues (Denis et al., 2017). GhWOX13 participates in cotton fiber elongation (He et al., 2019). In addition, there is evidence of the link between WOXs’ function and the gibberellin (GA) biosynthesis pathway. For example, DWT1, which encodes a WOX TF homologous to the Arabidopsis WOX8 and WOX9, regulates the internode elongation which is directly or indirectly associated with GA signaling (Wang et al., 2014). The expression levels of OsWOX12A and OsWOX12B were greatly upregulated by GAs after a 3-h treatment (Cheng et al., 2014). WOX14 overexpression upregulated GA3ox while repressing GA2ox, resulting in the accumulation of bioactive GA (Denis et al., 2017). However, up to now, there is a lack of reports regarding the involvement of WOX ancient clade members in the wood formation and GA homeostasis in woody plants such as trees, especially the exact roles they play during secondary cell wall (SCW) formation.
Being the most abundant biomass produced by land plants, wood, mainly comprising dead SCW, serves as raw material for multiple end uses by humans, and is considered to be one of the most environmentally cost-effective and renewable sources of bioenergy (Zhong et al., 2010). To genetically modify wood properties, it is essential to unveil the regulatory genes that control the various developmental processes of SCW in woody plants, especially in trees. Past studies have revealed that SCW biosynthesis is mainly controlled by a hierarchical transcription regulatory network, where the high-hierarchical regulators or switches activate the middle-level master hubs and then the lower-level TFs, and they together coordinately regulate the expression of bottom structural genes responsible for different SCW component biosynthesis (Zhou et al., 2009; Du and Groover, 2010; Yamaguchi et al., 2010; Zhong et al., 2010; Nakano et al., 2015; Kumar et al., 2016). This delicate network ensures a differential regulation of the formation of diverse SCW with varied components and thicknesses in secondary xylem under various conditions (Zhou et al., 2009; Zhong et al., 2010; Kumar et al., 2016). For example, PsnSHN2 (Liu et al., 2017) and AtSHN2 (Ambavaram et al., 2011), two higher hierarchical master switches of this network, significantly increase cellulose and hemicellulose contents and SCW thickness with significantly decreased lignin content through activating or repressing their downstream TFs and structural genes in overexpression of tobacco and rice, respectively. In addition, other higher hierarchical regulators that have been reported include PtrGATA12 and PtrHAT22 (Ren et al., 2021, 2022). For this reason, we need to continuously identify more higher hierarchical regulators that may be useful for leveraging the manipulation of important traits.
In this report, we revealed that overexpression of PtrWOX13A (Potri.005G101800.1), a member of the ancient clade of Populus trichocarpa WOX family, manifested not only the significantly enhanced growth potentials but also notably altered SCW and fiber characteristics, accompanied by the expression-level changes of the TFs in different hierarchical layers and the structural genes. The bioactive GA contents were elevated presumably due to the upregulation of three key genes involved in GA activation in transgenic poplar. PtrWOX13A was shown to be capable of directly activating some SCW-associated TFs, structural genes, PtrGA20ox4, and PtrGA3ox1 through binding to two cis-elements that are presented in the promoters of these genes. Collectively, our findings suggest that PtrWOX13A is a higher hierarchic regulator that regulates some wood-formation TFs, structural genes, and/or through triggering GA-mediated signaling cascade. The PtrWOX13A and its target genes identified in this study will be instrumental in developing new strategies for genetic improvement of wood quality.
Materials and Methods
Plant Materials
The plantlets of P. trichocarpa clone Nisqually-1 were obtained from the Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, and vegetatively propagated in our laboratory through tissue culture (Ren et al., 2021).
In the Northeast Forestry University greenhouse, 1-year-old P. trichocarpa trees were propagated and planted in a mixture of turfy peat and sand (2:1 v/v) and grown for 90 days under 16 h day/8 h night photoperiod at 25°C. The roots, primary leaves, transition leaves, secondary leaves, primary phloem, transition phloem, secondary phloem, primary xylem, transition xylem, and secondary xylem were collected from the above plants and immediately frozen in liquid nitrogen and stored at −80°C. Tissue culture seedlings that were 1-month old were treated with GA3 (1 μM), and the samples were collected after treatment for 0, 3, 6, 12, 24, 48, 72, and 96 h, respectively. The RNA was isolated according to the method described in a previous report (Liao et al., 2004) and subsequently treated with DNase I (Qiagen) to remove genomic DNA.
Sequence Comparisons, Protein Sequence, and Promoter Analysis
Multiple sequence alignments were carried out using ClustalW21 with default setting (Larkin et al., 2007). The homologous gene sequences from Populus trichocarpa [Potri.005G101800.1 (PtrWOX13A), Potri.002G008800.1 (PtrWOX13B1), and Potri.005G252800.1 (PtrWOX13B)] and Arabidopsis thaliana [AT4G35550.1 (AtWOX13)] were retrieved from Phytozome Database2 using TBLASTP with the AtWOX13 protein sequence as the query sequence.
The conserved motifs in the PtrWOX13 protein were analyzed by Multiple Expectation Maximization for Motif Elicitation (MEME) v. 5.33 with default parameters (Bailey et al., 2006). Conserved motifs were identified with the motif widths of 6-50 residues.
Sequences of 2,000 nucleotides before the transcription start codon were extracted from the genomic sequences of the PtrWOX13A, PtrWOX13B1, and PtrWOX13B from Phytozome (see the footnote 2), and the cis-regulatory elements were predicted by the program PlantCARE online4 (Lescot et al., 2002).
Cloning PtrWOX13A From P. trichocarpa
The full PtrWOX13A (Potri.005G101800.1) cDNA was amplified with gene-specific primers (Supplementary Table 1) using the method described in a previous report (Liu et al., 2017), and then transformed into Escherichia coli cells (DH5α, TsingKe) for validation by Sanger sequencing.
Subcellular Localization
The full-length coding region of PtrWOX13A without termination codon was amplified using specific primers (Supplementary Table 1) and then fused to the N-terminal of GFP under the control of CaMV 35S promoter in the pBI121 vector. The two fusion constructs were delivered into onion epidermal cells via particle bombardment. The GFP fluorescent images were photographed with confocal microscopy (Leica TCS SP5) 48 h after the bombardment with a gene gun (GJ-1000).
Transcriptional Activation Assay
The transcriptional activation of PtrWOX13A on putative target genes was tested using the yeast two–hybrid system. The complete CDS of PtrWOX13A was amplified using specifically designed primers (Supplementary Table 1). The amplified fragments were fused in-frame to the pGBKT7 vector to generate the pGBKT7-PtrWOX13A construct. The pGBKT7-PtrWOX13A and the pGBKT7 blank vector (as negative control) were transformed into Y2H yeast cells independently. The transformed Y2H yeast cells were plated onto SD/-Trp (growth control), SD/-Trp/-His/-Ade, and X-α-Gal media and incubated at 30°C for 3–5 days to identify the PtrWOX13A transcriptional activation.
Transformation of P. trichocarpa for Generating PtrWOX13A Overexpression Lines
The PtrWOX13A was amplified with specific primers (Supplementary Table 1) and then inserted into the pROKII vector. The pROKII-PtrWOX13A was first transferred into Agrobacterium tumefaciens GV3101 using the freeze–thaw method. The transgenic method was described as the previous report (Alvarez et al., 2018). The genomic DNA of all kanamycin-resistant shoots amplified by regular PCR using the PROKII sequencing primers is listed in Supplementary Table 1 to verify whether PtrWOX13A was integrated into the poplar genome.
The verified PtrWOX13A transgenic lines and wild-type (WT) poplars were propagated and planted in a mixture of turfy peat and sand (2:1 v/v), and grown under 16 h/8 h day/night photoperiod at 25°C in the greenhouse. After the PtrWOX13A transgenic lines were grown for 90 days, they were used for further characterization assays.
Phenotypic Trait Measurement
The phenotypic traits, which included lengths and widths of leaves, heights, base diameters, fresh and dry weights, and breaking forces of stems, of wild type (WT) and PtrWOX13A transgenic lines were measured following the method described in our previous publication (Ren et al., 2021).
Fiber Measurements
Pieces of outer xylem (approximately 1 mm × 1 cm × 0.5 mm) from Internode 8 (IN8) of the plants used for the anatomical characterization were macerated to separate the wood cells. The cells were observed under an Axioplan 2 microscope (Zeiss), and lengths and diameters of 100 fibers per plant were measured using AxioVision 4.6 software.
Determination of Gibberellin Contents
Fresh stems between IN5 and IN8 of PtrWOX13A transgenic lines and WT were used to measure GA1 and GA4 contents. The contents of GAs were measured with high-performance liquid chromatography (HPLC) by Suzhou Michy Biomedical Technology Co., Ltd., in China (Yang et al., 2014). Values are means ± SD of three biological replicates of three individual plants.
Scanning Electron Microscopy and Histological Analysis
Stem segments were prepared by freeze-drying for measuring SCW thickness using scanning electron microscopy (SEM) (S-4800, HITACHI) following the method described in a previous report (Liu et al., 2017). The SCW thicknesses of fibers in the SEM micrographs were quantified in a randomly selected area of 45 cells using ImageJ software5.
Histological Analysis and Determination of Cellulose, Hemicellulose, and Lignin Contents
The histological analysis was carried out with a Laser Scanning Confocal Microscope (LSM800) and the determinations of lignin, cellulose, and hemicellulose contents were conducted using the ANKOM 2000i Automatic Fiber Analyzer (ANKOM). Both of these analyses can be found in our previous studies (Liu et al., 2017).
Gene Expression Analysis of Poplar
RNA from the xylem of stems, spanning IN5 to IN8 of PtrWOX13A transgenic lines and WT, were used for synthesizing cDNA. Samples of cDNA were run in triplicate using the Applied Biosystems 7500 Real-Time PCR System to determine the critical threshold (Ct) with the SYBR Premix Ex Taq kit (TaKaRa). Then, the relative gene expression levels were calculated.
The primers used for the analysis of the expression levels of the PtrWOX13A, PtrWOX13B1, and PtrWOX13B2 by qRT-PCR are listed in Supplementary Table 2. The expression levels of genes involved in cellulose biosynthesis (PtrCESA4, PtrCESA7, and PtrCESA8) (Suzuki et al., 2006), xylan biosynthesis (PtrGT43A, PtrGT47C, and PtrGT8F) (Zhou et al., 2006, 2007), lignin biosynthesis (PtrPAL4, PtrC4H1, PtrC3H3, Ptr4CL5, PtrCCoAOMT3, PtrCOMT2, PtrCCR2, PtrCAld5H2, and PtrCAD1), lignin polymerization (PtrLAC19, PtrLAC21, PtrLAC26, and PtrLAC41) (Marjamaa et al., 2009; Lu et al., 2013), fiber elongation (PtrCSLD2, PtrXTH5, PtrEXPA8, and PtrFRA2) (Burk et al., 2001; Gray-Mitsumune et al., 2004; Samuga and Joshi, 2004; Esmon et al., 2006; Seale, 2020), the TFs belonging to well-known hierarchical transcription regulatory network governing poplar SCW formation (PtrWND6A, PtrWND6B, PtrMYB20, PtrMYB21, PtrMYB157, PtrMYB28, PtrMYB152, PtrMYB52, and PtrMYB54) (Zhong et al., 2010, 2011; Zhong and Ye, 2010; Hussey et al., 2013; Zhang et al., 2018; Cubria-Radio and Nowack, 2019; Zhang J. et al., 2020), and genes involved in homeostasis of bioactive GAs (GA4 and GA1) (PtrGA20ox4, PtrGA3ox1, and PtrGA2ox1) (Israelsson et al., 2005) were analyzed by qRT-PCR using specific primers (Supplementary Table 2). PtrActin was employed as internal control, and the delta-delta CT method was used to quantify gene expression levels relative to PtrActin as described in a previous report (Zhang Y. et al., 2020).
Transient Expression Assay
The full coding region of PtrWOX13A was cloned into pROKII under the control of the CaMV 35S promoter, which was used as the effector construct. The reporter construct contained the GUS reporter gene driven by the various 2-kb promoters of genes, which were chosen based on the changes in the expression levels in PtrWOX13A transgenic lines as compared with WT. The effector vector, reporter vector, and 35S-LUC-pGreenII 8000 vector transfected tobacco leaves together. GUS activity in tobacco leaves transfected with pROKII empty vector (without PtrWOX13A), and reporter vector (with one of the selected promoters), and 35S-LUC-pGreenII 8000 were used as a control. The 35S-LUC was an internal baseline control for the transient expression analysis to use for the normalization of GUS activity. Each promoter was amplified using the primers listed in Supplementary Table 3 designed from P. trichocarpa genomic DNA. The GUS activity was measured by the method described in our previous study (Ren et al., 2021). GUS activity for each promoter tested is expressed as the ratio of GUS/LUC obtained with the effector pROKII-PtrWOX13A to GUS/LUC obtained with the control effector pROKII empty vector.
Analysis of the Downstream Cis-Regulatory Elements of the PtrWOX13A
Three tandem copies of two cis-regulatory elements, ATTGATTG and TTAATSS (Supplementary Table 4), were fused to the multiple cloning site upstream of the HIS3 reporter gene in pHIS2. The full CDS of PtrWOX13A amplified with the primers in Supplementary Table 1 was inserted into pGADT7-Rec2 as the effector vector. Both constructs were co-transformed into Y187 yeast cells, which were plated onto DDO, TDO, and TDO plus 60 mM 3-aminotriazole to test the expression of the His3 gene at 30°C for 3–5 days. The pair of pHIS2-p53 and pGADT7-rec2-p53 was used as a positive co-transformation control, whereas the pair of pHIS2-p53 and pGADT7-rec2-PtrWOX13A was used as a negative control. The interactions of these sequences with PtrWOX13A were studied using the yeast one-hybrid analysis (Y187). The ExactSearch tool was employed to identify these motifs in the promoter regions of putative target genes we identified (Supplementary Table 5; Gunasekara et al., 2016).
ChIP-PCR
For immunoprecipitation analysis, 1-month-old 35-PtrWOX13A-Flag overexpressing transgenic seedlings that had been subjected to tissue culture were harvested and vacuumed for 15 min in 10 ml buffer solution, followed by the addition of 4 ml 2M glycine to quench cross-linking by and then vacuumed for 2 min. The treated plants were washed with H2O three times and ground in liquid nitrogen. Immunoprecipitation was performed with anti-FLAG antibody according to Zhao et al. (2020). PCR was used to determine the binding sites of PtrWOX13A in the promoter regions. Primers are listed in Supplementary Table 6. All experiments were performed in three biological replications with three technical replicates.
Statistical Analysis
The Dunnett’s test (SPSS 17.0) was used to test the statistical significance of the data. The difference between two groups of data for comparisons in this study was evaluated by significance (*p < 0.05) or high significance (**P < 0.01).
Results
Identification and Characterization of PtrWOX13
The previous phylogenetic analysis revealed that the ancient clade of Arabidopsis and poplar WOX families include three Arabidopsis and three poplar WOX genes (Liu et al., 2014). The ortholog counterparts of AtWOX10 and AtWOX14 in poplar were lost, while the ortholog counterpart of AtWOX13 in poplar was evolved into three paralogous genes that include PtrWOX13A (Potri.005G101800.1), PtrWOX13B1 (Potri.002G008800.1), and PtrWOX13B2 (Potri.005G252800.1) (Supplementary Figure 1A).
Multiple sequence alignment of AtWOX13 and three PtrWOX13 protein sequences showed that the three PtrWOX13 proteins contained not only the typic HD domain consisting of three α-helices separated by a loop and a turn but also the following conservative features unique to WOX ancient clade: (1) the N-terminal domain located in the upstream of the HD domain (Sakakibara et al., 2014); (2) the ESExE motif located in the downstream of the HD domain (Deveaux et al., 2008); (3) the WOX13 MOG motif consisting of 31 amino acids that are found upstream of the HD domain (Wu et al., 2019); (4) the YxDpl motif between the WOX13 MOG motif and the HD domain (Deveaux et al., 2008); and (5) the signature motif of the ancient clade consisting of NVYNWFQNR (Wu et al., 2019; Supplementary Figure 1B).
In addition, the results of the conserved motif analysis of the three PtrWOX13 proteins suggested that PtrWOX13B1 and PtrWOX13B2 proteins shared all five aforementioned domain/motifs while PtrWOX13A had no motif 5 (Supplementary Figure 2A). Moreover, the similarity of protein sequences was only 54.8% between the PtrWOX13A and PtrWOX13B1, 53.4% between PtrWOX13A and PtrWOX13B2, but 86.1% between PtrWOX13B1 and PtrWOX13B2. In summary, these data suggested that PtrWOX13A has evolved to be more distal to both PtrWOX13B1 and PtrWOX13B2, and thus may be functionally distinct from both of them.
To further determine if there were differential responses to environmental and intracellular cues among the three PtrWOX13 genes, we analyzed the cis-regulatory elements in their promoters. The results showed that there were 11 common cis-regulatory elements, for example, MYC, MYB, MBS, ARE, and ABRE among all three genes (Supplementary Figures 2B,C). In addition, there were 20 common cis-regulatory elements between PtrWOX13B1 and PtrWOX13B2 promoters, 13 between PtrWOX13A and PtrWOX13B1 promoters, and 14 between PtrWOX13A and PtrWOX13B1 promoters (Supplementary Figure 2C), which indicates that responses of PtrWOX13B1 and PtrWOX13B2 appear to be more similar among the three PtrWOX13 genes. Moreover, each promoter of these genes also had unique cis-regulatory elements. For example, there were P-box, TGA-element, and TCA-element that were only present in the PtrWOX13A promoter; G-Box, ACE, and AAGAA-motifs that were only present in the PtrWOX13B1 promote; and RY-element, TC-rich repeats, and LTR that were only present in the PtrWOX13B2 promoter (Supplementary Figure 2C). These data further suggested that these three PtrWOX13 genes in poplar have subfunctionalized after duplication.
To further determine which of the three PtrWOX13 genes participated in wood formation, we analyzed their expression levels in various tissues of poplar by quantitative RT-PCR (qRT-PCR). As shown in Figure 1A, the three PtrWOX13 genes were differently expressed at detectable levels in all examined tissues. Among them, the transcript levels of PtrWOX13A in the transition and secondary xylem were much higher than those in any other tissues and those of the other two PtrWOX13 transcripts in the same tissues, indicating that PtrWOX13A was the most likely gene to participate in wood formation among the three PtrWOX13 genes (Figure 1A). In addition, given that GAs have important roles in wood formation and PtrWOX13A has GAs response cis-element, a P-box, in its promoter, we further tested if the PtrWOX13 was inducible in the secondary tissues of poplar stems by exogenous GA treatment. The transcript levels of PtrWOX13A were evaluated by qRT-PCR after treatment with exogenous GAs using GA3. The results show that the expression of PtrWOX13A increased after treatment with exogenous GA3 for 24 h, and then decreased from 24 to 96 h, whereas the transcriptional levels of PtrWOX13B1 and PtrWOX13B2 had no obvious changes in response to exogenous GA treatment (Figure 1B). Thus, we chose PtrWOX13A for studying the functions of WOX genes in wood formation.
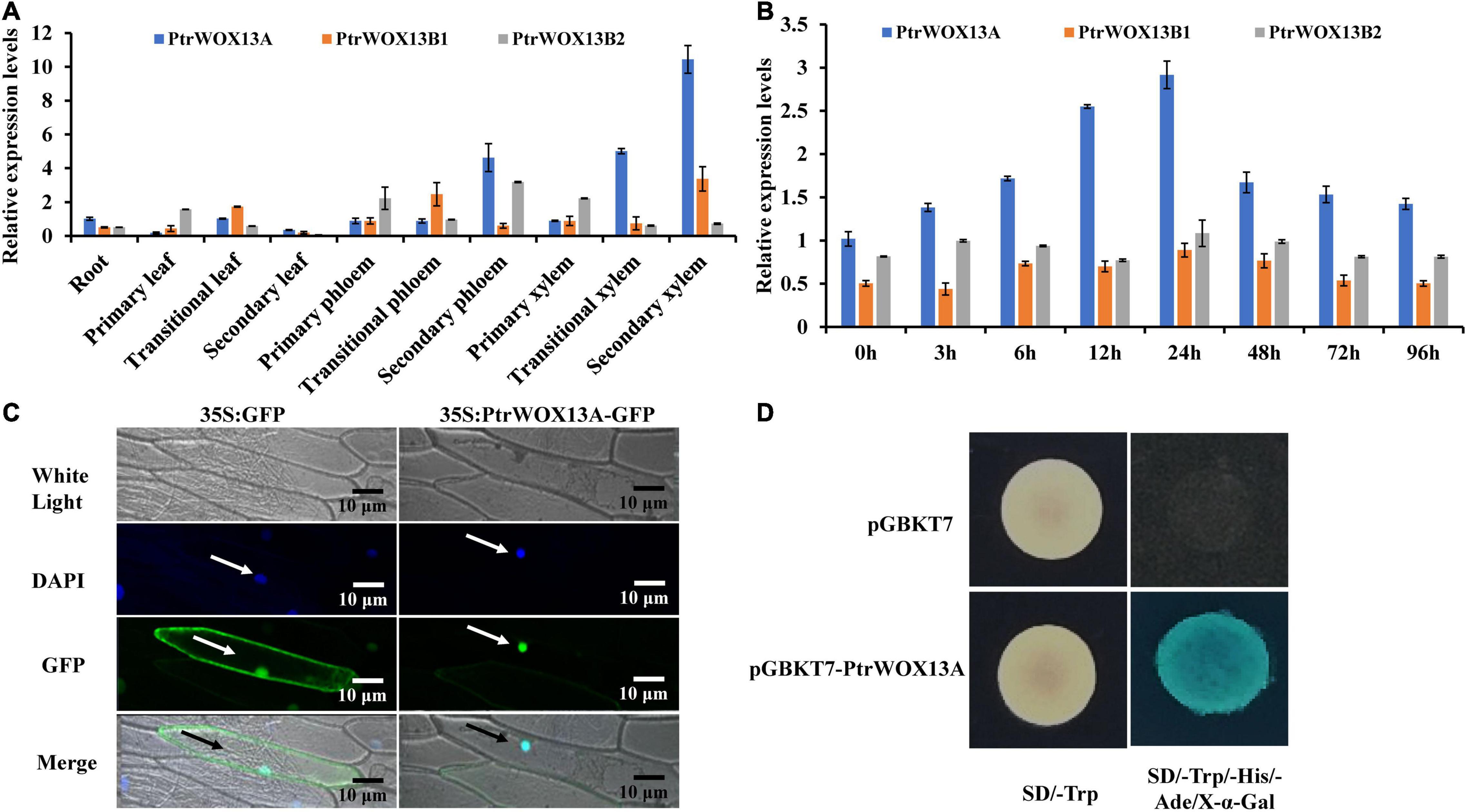
Figure 1. Tissue-specific expression patterns, subcellular localization, and transcriptional activity of PtrWOX13A. (A) The tissue-specific expression patterns of three PtrWOX13 genes as determined by qRT-PCR analysis. PtrActin was used as a reference gene. The expression of PtrWOX13A in root was set to 1 and expression in all other tissues is relative to roots. (B) The temporal expression patterns of three PtrWOX13 genes after GA treatments as determined by qRT-PCR analysis. PtrActin was used as a reference gene. The expression of PtrWOX13A at 0 h was set to 1, all other time points were relative to 0 h. (C) Subcellular localization of PtrWOX13A. Confocal images manifested the localization of PtrWOX13A: DAPI, a nuclear staining dye; GFP proteins in the nuclei of onion epidermal cells. Merge: The merged images of White light, DAPI, and GFP staining. Arrows indicate cells located in the epidermis of onions. (D) Transcriptional activation analysis of PtrWOX13A fused with the GAL4 DNA binding domain (GAL4DB) in yeast showed its potential to activate the expression of the His-3 and X-α-Gal reporter genes. Arrows indicate cells located in the epidermis of onions.
Subcellular Localization and Transcriptional Activation Activity of PtrWOX13A
To determine whether PtrWOX13A localized in the nuclei, we performed the transient expression of PtrWOX13A in onion epidermal cells using the particle gun bombardment method. As visualized with a fluorescence confocal microscope, the PtrWOX13A–GFP fusion protein was exclusively colocalized to DAPI-stained nuclei (Figure 1C), indicating that PtrWOX13A was a nuclear-localized TF.
In addition, the presence of an acidic amino-terminal domain next to an HD domain in the C-terminal region, as seen in Supplementary Figure 1B, suggested that PtrWOX13A was a transcriptional activator. To verify this, we fused PtrWOX13A with the GAL4 DNA-binding domain and tested its potential to activate the reporter gene expression in yeast. As shown in Figure 1D, PtrWOX13A activated the expression of His3 and X-α-Gal reporter genes, indicating that it was indeed a transcriptional activator.
Phenotypic Changes of the PtrWOX13A Overexpression Transgenic Lines
To investigate the functions of PtrWOX13A in poplar, we expressed PtrWOX13A under the control of the 35S promoter in WT. In total, eight transgenic lines were generated and corroborated to harbor the transformed PtrWOX13A by genomic PCR (Supplementary Figure 3A). Then, the expression levels of PtrWOX13A in these transgenic lines were quantitatively determined using qRT-PCR. Three transgenic lines, OE-2, OE-4, and OE-7, which had higher expression levels of PtrWOX13A compared with other transgenic lines, were chosen for further characterization of PtrWOX13A’s functions (Supplementary Figure 3B).
The three PtrWOX13A transgenic lines all exhibited significantly enhanced growth potentials compared with WT. For example, the lengths and widths of leaf, heights, base diameters, and fresh and dry weights of the three PtrWOX13A transgenic lines increased by 18.8, 18.1, 9.7, 12.6, 22.4, and 16.6% on average as compared with those of the WT, respectively (Figures 2A–D). In addition, the breaking forces of stems, which is the important physical property of wood, increased 17.8% on average in PtrWOX13A transgenic lines compared to WT (Figure 2D). These data indicate that PtrWOX13A overexpression results in enhanced vegetative growth and increased biomass productivity of transgenic lines.
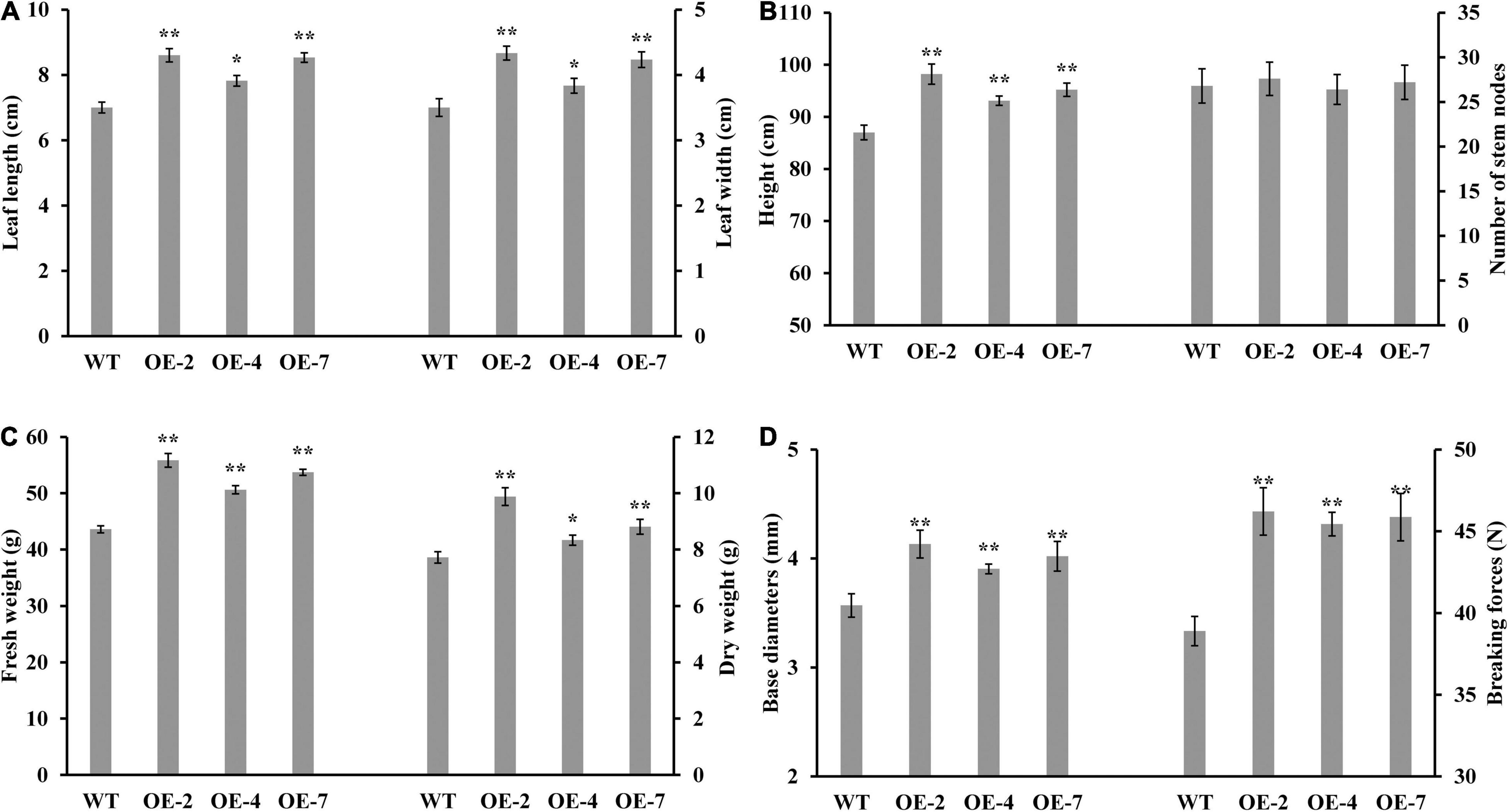
Figure 2. Effect of PtrWOX13A overexpression on growth-related traits in Populus trichocarpa transgenic lines. (A) Average leaf length and leaf width; (B) Average height and number of stem nodes and standard error of each transgenic line; (C) Average fresh weight and dry weight; (D) Average base diameters and breaking forces. All the data were measured on the 90-day-old wild type (WT) and PtrWOX13A transgenic lines (OE-2, OE-4, and OE-7). Each error bar represents the standard deviation of three biological replicates. Asterisks denote levels of significance (Dunnett’s test; *p < 0.05 significant, and **p < 0.01, highly significant).
Effects of PtrWOX13A Overexpression on Wood Characteristics and Bioactive Gibberellin Homeostasis in Transgenic Poplar
To investigate the effects of PtrWOX13A overexpression on poplar wood formation, we performed histochemical staining of the stem cross-sections of PtrWOX13A transgenic lines. The toluidine blue and phloroglucinol-HCl were used to stain cell morphology and lignin, respectively. As shown in Supplementary Figure 4, the staining intensities of the cross-sections from IN2 showed no obvious difference between the PtrWOX13A transgenic lines and WT, but the staining intensities of the cross-sections in the IN4, IN6, to IN8 exhibited increasingly strong staining signals in PtrWOX13A transgenic lines compared to those of WT, which implicates that PtrWOX13A enhances wood lignification during stem development in the transgenic lines.
Then, we further analyzed the wood characteristics in the IN8 of PtrWOX13A transgenic lines. The scanning electron microscope of stem cross-sections revealed that the average SCW thicknesses of fibers increased by 20.8% on average, while the average SCW thickness of vessels had little or no changes in the PtrWOX13A transgenic lines compared to that of the WT (Figures 3A–H). These results indicate that overexpression of PtrWOX13A has different effects on the fibers and vessel formation in the transgenic poplars.
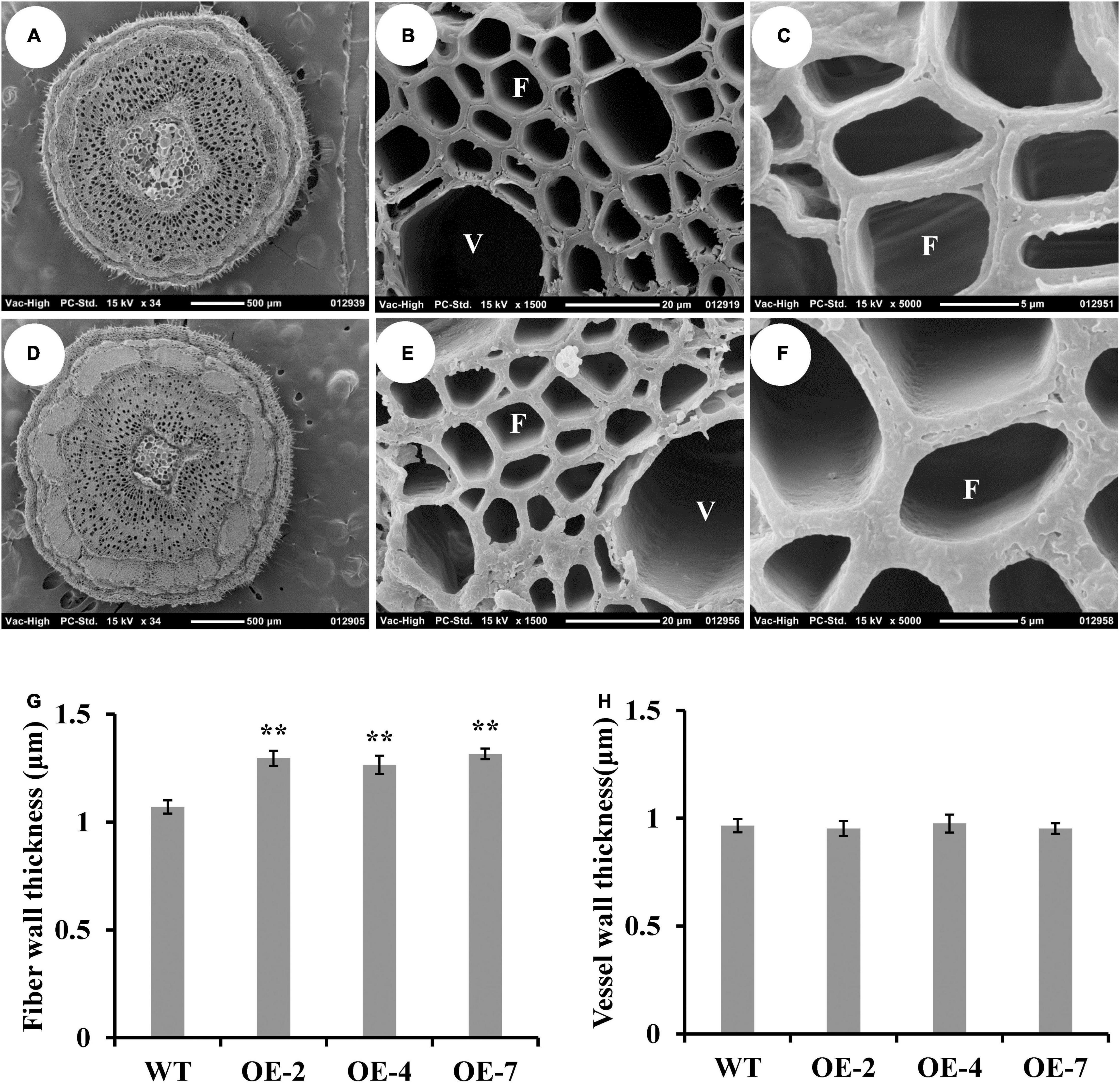
Figure 3. Effect of PtrWOX13A overexpression on the secondary wall thickness of stems in Populus trichocarpa. (A–F) The scanning electron microscope of cross stem sections of wild type (A–C) and PtrWOX13A overexpression transgenic lines (D–F). (A,D) represent 34×, (B,E) represent 1,500×, and (C,F) represent 5,000× scanning electron microscope of cross stem sections, respectively. V and F in (B,C,E,F) represent vessel cell and fiber cell, respectively. (G) The fiber wall thicknesses of cross stem sections in wild type (WT) and PtrWOX13A overexpression transgenic lines (OE-2, OE-4, and OE-7). (H) The vessel wall thicknesses of cross stem sections in WT and PtrWOX13A overexpression transgenic lines. Error bars represent the standard deviation of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; **p < 0.01). All data were measured on the 8th stem of 90-day-old poplars.
To determine which component (e.g., cellulose, hemicellulose, or lignin) contributed more to fiber SCW thickening, the phloroglucinol-HCl and calcofluor white were used to stain lignin and cellulose, respectively, and the monoclonal antibody LM10 was used to label hemicellulose (xylan) immunologically. As shown in Figure 4, the widths of staining zones of phloroglucinol-HCl (Figure 4A) and antibody LM10 calcofluor (Figure 4C) in PtrWOX13A transgenic lines were larger than those in WT, while the width of staining zone of calcofluor white in PtrWOX13A transgenic lines (Figure 4B) was not larger than that in WT (Figures 4A–F). In addition, the staining intensities of both dyes and antibodies had stronger intensities in transgenic lines than in WT. To further accurately assess these changes in SCW components, we analyzed the three main components of SCW using the automatic fiber analyzer. The results revealed that the lignin and hemicellulose contents significantly increased by 15.3 and 10.2%, respectively, on average, while the cellulose contents in PtrWOX13A transgenic lines had no significant changes compared with that in WT (Figures 5A–C). These results indicate that PtrWOX13A overexpression results in increased contents of lignin and hemicellulose but had no obvious effects on the cellulose deposition in transgenic lines compared with WT. In addition, PtrWOX13A transgenic lines exhibited notably increased fiber length and base diameter by 11.3 and 8.1% on average, respectively, as compared with WT (Figure 5D).
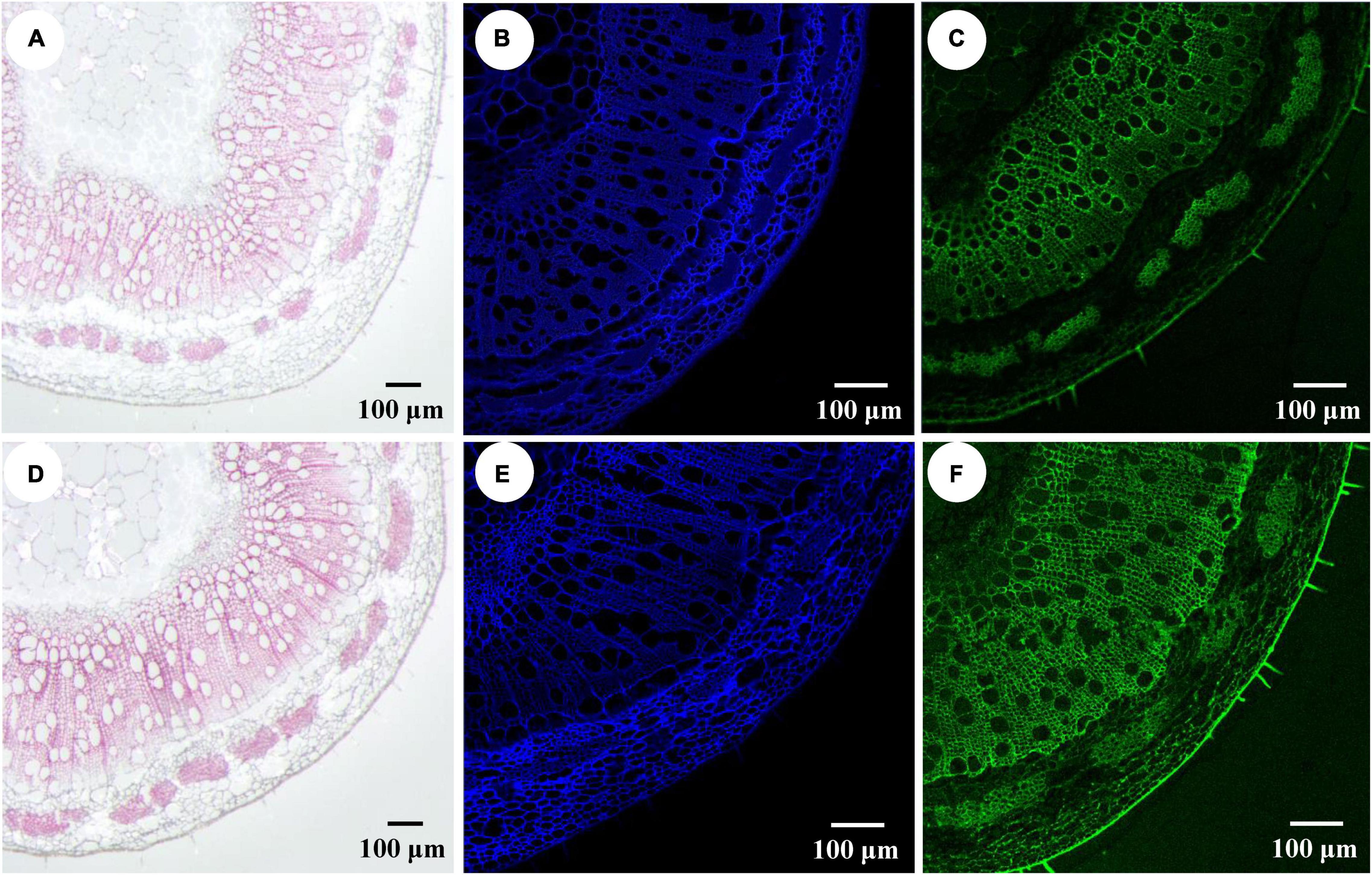
Figure 4. Impact of PtrWOX13A overexpression on components of the secondary cell wall of stems in Populus trichocarpa. (A) Stem section of wild type stained with Phloroglucinol-HCl (red color). (B) Stem section of wild type stained with Calcofluor white staining (blue color). (C) Stem section of wild type stained with Monoclonal Antibody LM10 (green color). (D) Stem section of PtrWOX13A overexpression transgenic lines stained with Phloroglucinol-HCl (red color). (E) Stem section of PtrWOX13A overexpression transgenic lines stained with Calcofluor white (blue color). (F) Stem section of PtrWOX13A overexpression transgenic lines stained with Monoclonal Antibody LM10 (green color). Scale bars = 100 μm. All data were measured on the 8th stem of 90-day-old poplars.
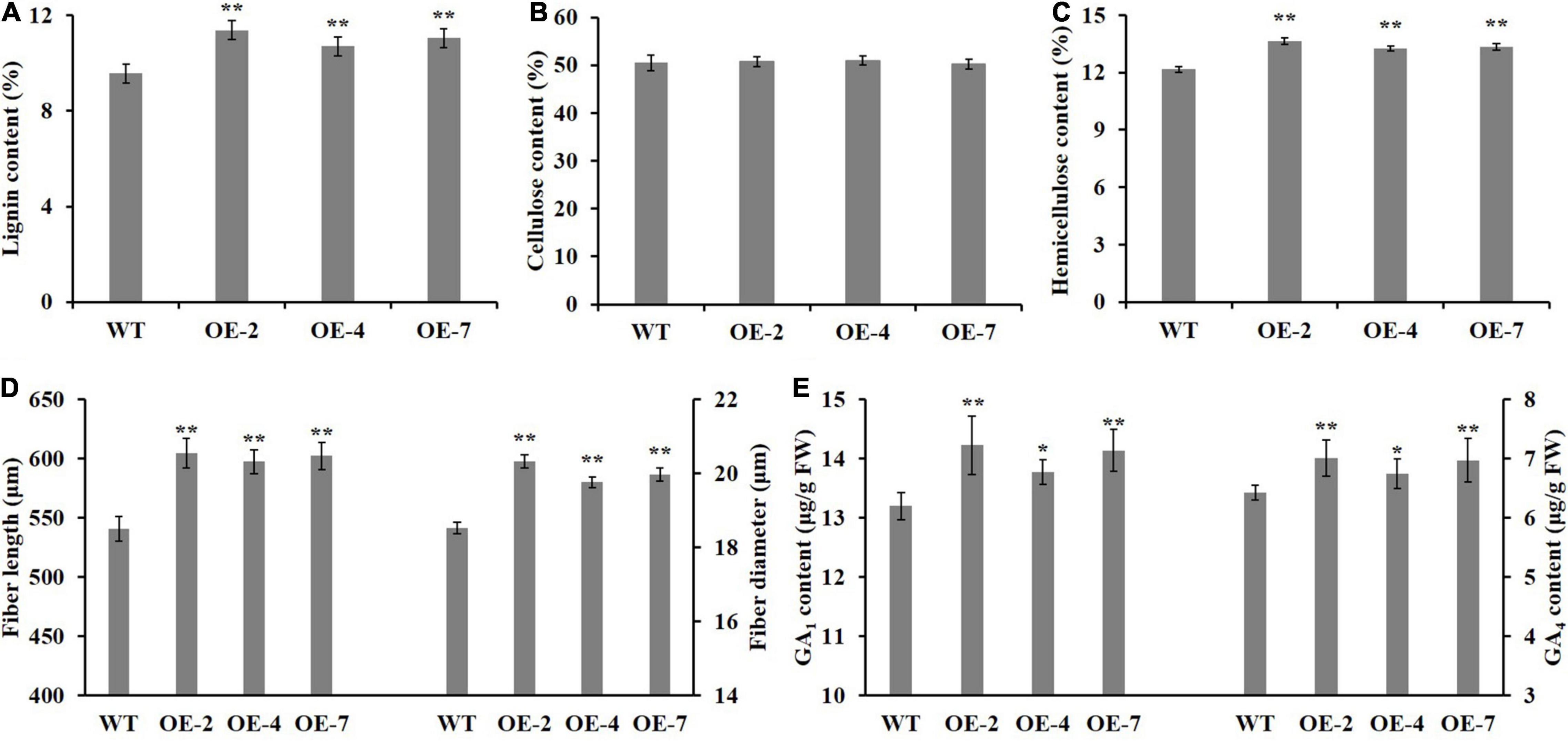
Figure 5. Effect of PtrWOX13A overexpression on secondary cell wall of stems in Populus trichocarpa. (A–E) Represent lignin content, cellulose content, hemicellulose content, fiber length, fiber diameter, GA1 content, and GA4 content, respectively. Error bars represent the standard deviation of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; *p < 0.05 significant, and **p < 0.01, highly significant).
Given that AtWOX14 is involved in bioactive GA homeostasis (Denis et al., 2017), we suspected that PtrWOX13A, a member of the WOX ancient clade that includes AtWOX14, also affected the bioactive GA homeostasis in its overexpression transgenic poplars. The analysis of the bioactive GAs showed that the contents of bioactive GA1 and GA4 increased by 6.39 and 7.52%, respectively, on average in the IN5-8 of PtrWOX13A transgenic lines compared with WT (Figure 5E). Taken together, these results implicate that PtrWOX13A participates in wood formation and bioactive GA homeostasis of transgenic poplar.
Overexpression of PtrWOX13A Altered the Expressions of Genes Involved in Secondary Cell Wall and Fiber Formation and Bioactive Gibberellin Homeostasis in Poplar
To uncover the molecular mechanism underlying the aforementioned changes in secondary growth and bioactive GA homeostasis, we analyzed the expression levels of the genes related to these changes in the PtrWOX13A transgenic lines. The qRT-PCR results demonstrated that the expression levels of lignin biosynthesis genes including PtrPAL4, PtrC4H1, PtrC3H3, Ptr4CL5, PtrCCoAOMT3, PtrCOMT2, PtrCCR2, PtrCAld5H2, and PtrCAD1, lignin polymerization genes including PtrLAC19, PtrLAC21, PtrLAC26, and PtrLAC41, and hemicellulose biosynthesis genes including PtrGT43A, PtrGT47C, and PtrGT8F, increased at extremely significant levels, while the expression levels of the cellulose biosynthesis genes such as PtrCESA4, PtrCESA7, and PtrCESA8, had no obvious increases in PtrWOX13A transgenic lines compared with WT (Figure 6A). In addition, the genes related to fiber cell elongation, such as PtrCSLD2, PtrXTH5, PtrEXPA8, and PtrFRA2, were notably upregulated in PtrWOX13A transgenic lines compared with WT. Moreover, we observed that the bioactive GA biosynthesis genes such as PtrGA20ox4 and PtrGA3ox1 were significantly upregulated, whereas the expression levels of PtrGA2ox1, a bioactive GA deactivation gene, significantly decreased in PtrWOX13A transgenic lines compared with WT (Figure 6B).
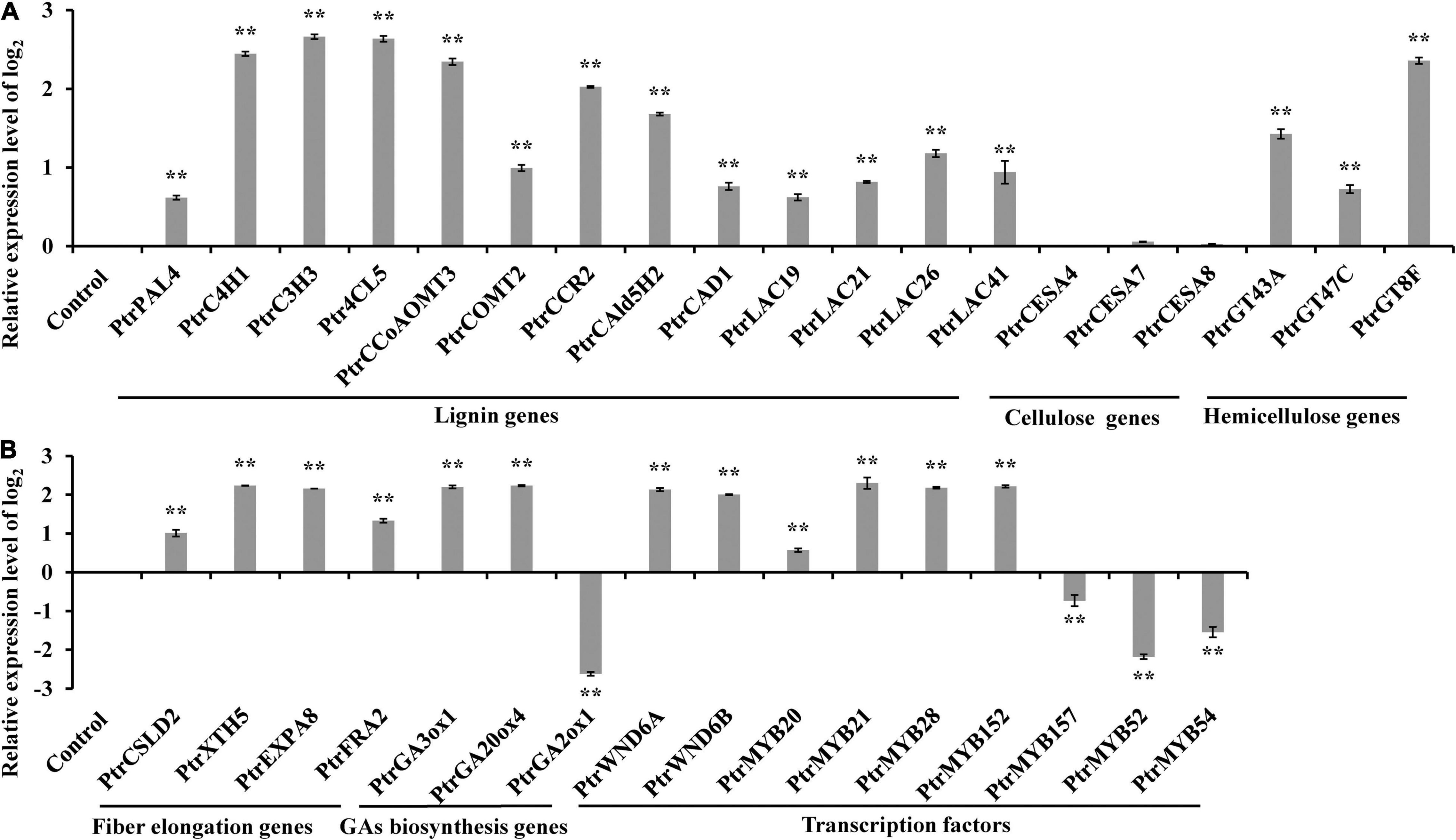
Figure 6. Expression analysis of wood formation pathway genes and their regulatory genes in 90 days old wild-type and PtrWOX13A transgenic lines. (A) Represents expression analysis of lignin biosynthesis genes, cellulose biosynthesis genes, and hemicellulose biosynthesis genes. (B) Represents expression of fiber cell elongation genes, GA biosynthesis genes, and TFs. PtrActin was used as a reference gene. Control represents the normalized expression level (namely 0 in this case) of each gene examined in wild type plants. Each error bar represents the standard deviation of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; **p < 0.01, highly significant). Those genes include lignin biosynthesis genes (PtrPAL4, PtrC4H1, PtrC3H3, Ptr4CL5, PtrCCoAOMT3, PtrCOMT2, PtrCCR2, PtrCAld5H2, and PtrCAD1), lignin polymerization genes (PtrLAC19, PtrLAC21, PtrLAC26, and PtrLAC41), cellulose biosynthesis genes (PtrCESA4, PtrCESA7, and PtrCESA8), hemicellulose biosynthesis genes (PtrGT43A, PtrGT47C, and PtrGT8F), fiber cell elongation genes (PtrCSLD2, PtrXTH5, PtrEXPA8, and PtrFRA2), GA biosynthesis genes (PtrGA20ox4, PtrGA3ox1, and PtrGA2ox1), and TFs (PtrWND6A, PtrWND6B, PtrMYB20, PtrMYB21, PtrMYB28, PtrMYB152, PtrMYB157, PtrMYB52, and PtrMYB54).
It was noteworthy that the expression levels of the master switches of SCW formation, such as PtrWND6A, PtrWND6B, PtrMYB20, and PtrMYB21, significantly increased in PtrWOX13A transgenic lines compared with WT (Figure 6B). In addition, the expression levels of TFs that are capable of activating lignin biosynthesis, such as PtrMYB28 and PtrMYB152, also increased significantly. On the contrary, the expression levels of TFs that are capable of repressing lignin biosynthesis, such as PtrMYB157, PtrMYB52, and PtrMYB54, significantly decreased in PtrWOX13A transgenic lines compared with WT (Figure 6B). These data aligned well with the changes in SCW and fiber characteristics observed in PtrWOX13A transgenic lines.
PtrWOX13A Activated the Promoters of the Genes Involved in Wood Formation and Bioactive Gibberellin Homeostasis
Based on the fact that overexpression of PtrWOX13A significantly altered the expression of several genes in transgenic poplar, we investigated whether it could activate these genes directly rather than indirectly, whose expression levels had a two-fold change in PtrWOX13A transgenic lines compared with WT, by conducting transient expression assays in tobacco leaves. The 2-kb proximal promoter regions of these genes, upon being amplified from P. trichocarpa genomic DNA, were linked to the GUS reporter gene to create the reporter constructs, and the full-length cDNA of PtrWOX13A was ligated to the immediately downstream of 35S promoter to generate the effector construct (Figures 7A,B). The reporter and effector constructs were co-transfected into tobacco leaves by the Agrobacterium-mediated method. The subsequent assay of the GUS activity demonstrated that PtrWOX13A significantly activated the lignin biosynthesis genes of PtrC4H1 and Ptr4CL5, the fiber cell elongation genes of PtrXTH5 and PtrEXPA8, GA biosynthesis genes of PtrGA20ox4 and PtrGA3ox1), and TFs of PtrWND6A, PtrWND6B, and PtrMYB28 albeit to different levels (Figure 7C), Contrastingly, PtrWOX13A did not activate or inhibit lignin biosynthesis genes of PtrC3H3, PtrCCoAOMT3, and PtrCCR2), hemicellulose biosynthesis genes of PtrGT8F, GA biosynthesis genes of PtrGA2ox1, and TFs of PtrMYB21, PtrMYB152, and PtrMYB52 (Figure 7C). These data suggest that PtrWOX13A directly regulates some genes involved in wood formation and bioactive GA homeostasis through binding to their promoters, while some other significantly changed genes were supposedly regulated indirectly.
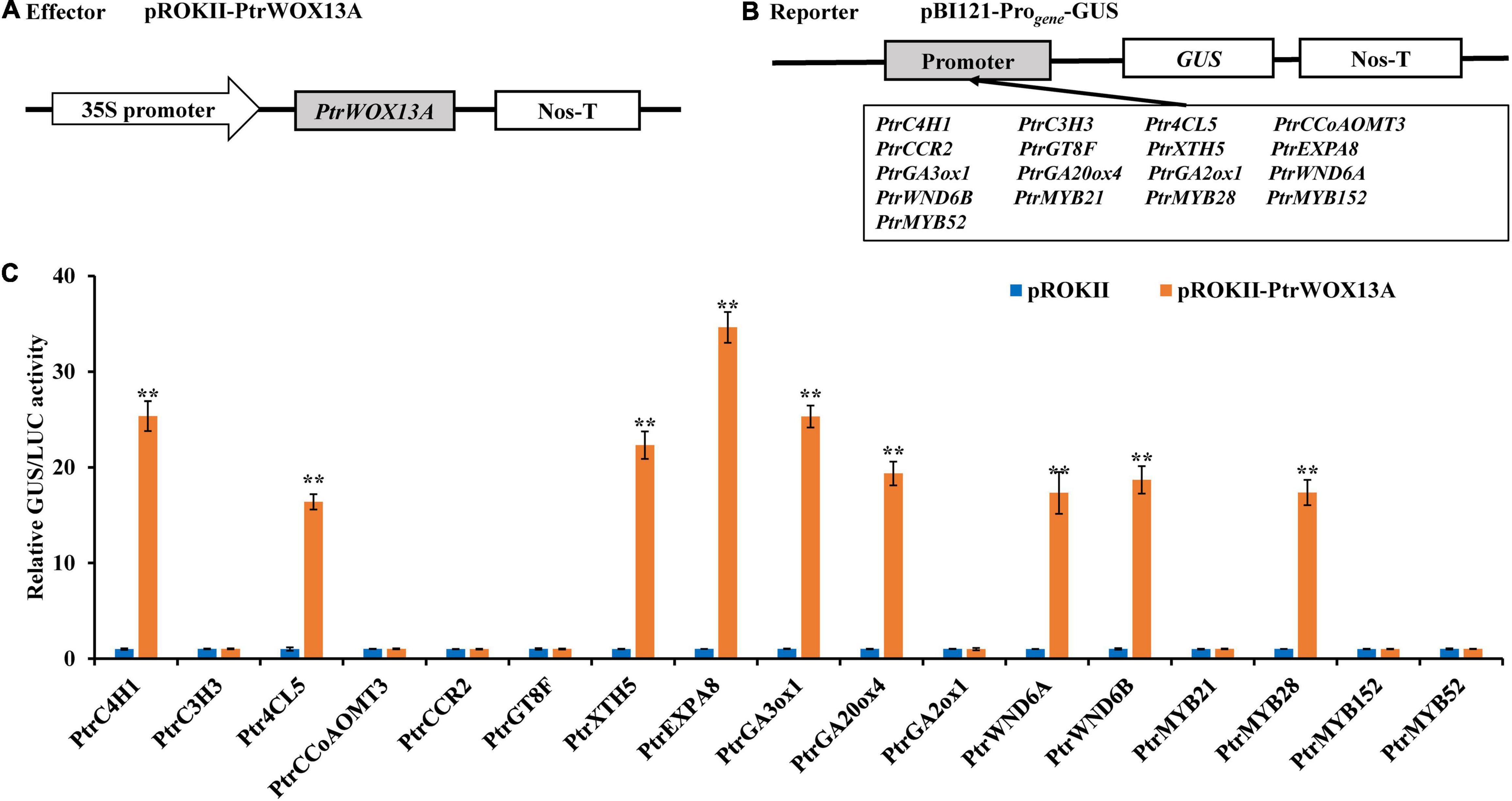
Figure 7. Activation of the promoters of poplar transcription factors and secondary cell wall biosynthesis pathway genes by PtrWOX13A. (A,B) Diagrams of the effector and the reporter constructs. (C) The expression of the GUS under the control of those promoters that were activated by PtrWOX13A. GUS activity in tobacco leaves transfected with pROKII empty vector, reporter vector, and 35S-LUC-pGreenII 8000 vector was used as a control and was set to 1. GUS activity for each promoter tested is expressed as the ratio of GUS/LUC obtained with the effector pROKII-PtrWOX13A to GUS/LUC obtained with the control effector pROKII empty vector. Error bars represent the standard deviation of three biological replicates. Asterisks indicate levels of significance of differential expression (Dunnett’s test; **p < 0.01, highly significant). Those genes include lignin biosynthesis genes (PtrC4H1, PtrC3H3, Ptr4CL5, PtrCCoAOMT3, and PtrCCR2), hemicellulose biosynthesis genes (PtrGT8F), fiber cell elongation genes (PtrXTH5 and PtrEXPA8), GA biosynthesis genes (PtrGA20ox4, PtrGA3ox1, and PtrGA2ox1), and TFs (PtrWND6A, PtrWND6B, PtrMYB21, PtrMYB28, PtrMYB152, and PtrMYB52).
PtrWOX13A Bound to Two Cis-Regulatory Elements in the Promoters of Its Directly Regulated Genes
The key mechanism for a TF to directly regulate downstream genes depends on the sequence-specific binding site to DNA motifs present in the promoters of its target genes. Thus, we further performed the yeast one-hybrid assay to test if PtrWOX13A could bind to the aforementioned cis-regulatory elements, ATTGATTG and TTAATSS (Minh-Thu et al., 2018; Ren et al., 2022). In addition, we found that promoter regions of the genes, such as PtrC4H1, Ptr4CL5, PtrXTH5, PtrEXPA8, PtrGA20ox4, PtrGA3ox1, PtrWND6A, PtrWND6B, and PtrMYB28, also contain these two cis-regulatory elements (Supplementary Table 5). As shown in Supplementary Figure 5, PtrWOX13A had obvious binding affinities to ATTGATTG (denoted by A in the figure) and TTAATSS (denoted by T in the figure) cis-regulatory elements.
To test the binding regions in the promoters of PtrC4H1, Ptr4CL5, PtrXTH5, PtrEXPA8, PtrGA20ox4, PtrGA3ox1, PtrWND6A, PtrWND6B, and PtrMYB28 by PtrWOX13A in vivo, chromatin immunoprecipitation (ChIP) assays were performed using transgenic poplars, which expressed Flag-tagged PtrWOX13A protein. PtrWOX13A–Flag-bound DNA fragments were immunoprecipitated using anti-Flag antibodies, and purified DNA fragments were used as templates in PCR analysis. The primer pair spanning DNA fragments of P1, P2, and P3 in PtrC4H1, P1 in Ptr4CL5, P1 and P2 in PtrXTH5 and PtrEXPA8, P1 in PtrGA20ox4, PtrGA3ox1, PtrWND6A, PtrWND6B and P1, P2, P3, and P4 in PtrMYB28 promoter sequences were designed (Figure 8A). The results showed that PtrWOX13A bound directly to the promoters of PtrC4H1, Ptr4CL5, PtrXTH5, PtrEXPA8, PtrGA20ox4, PtrGA3ox1, PtrWND6A, and PtrMYB28 to regulate their expression (Figure 8B). These results, together with other pieces of evidence shown above, suggested that PtrWOX13A acted as a higher hierarchical regulator to directly activate downstream middle-layered TFs and structural genes involved in SCW component biosynthesis and GA homeostasis.
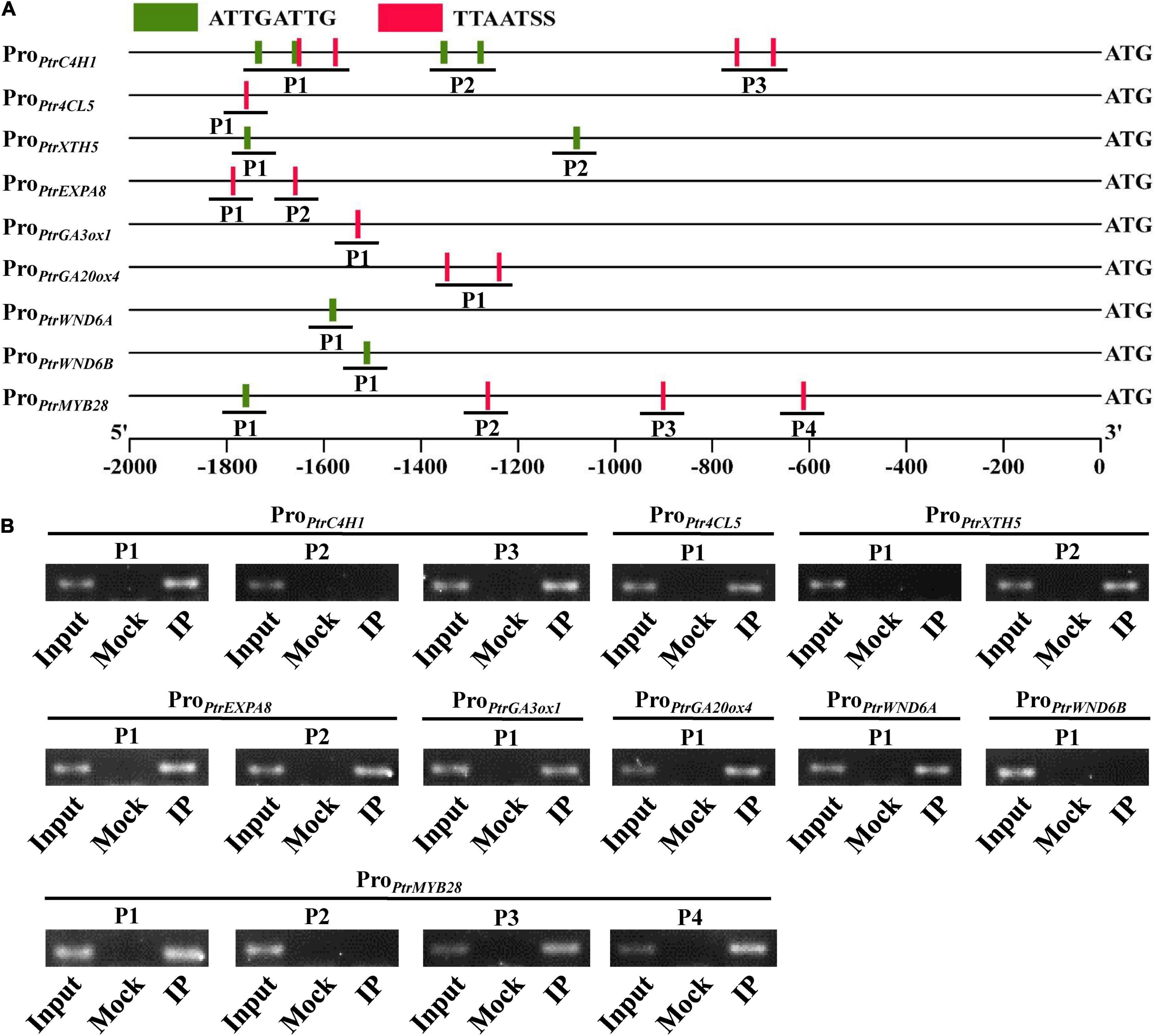
Figure 8. PtrWOX13A directly binds to the promoters of the poplar transcription factors and secondary cell wall biosynthesis pathway genes. (A) Schematic diagrams of the promoters showing the locations amplified by ChIP-PCR. (B) ChIP-PCR analysis of the promoter fragments enriched due to PtrWOX13A binding during immunoprecipitation; 30-day-old poplar plants overexpressing PtrWOX13A-Flag used for ChIP with anti-Flag antibodies. Input, Mock, and IP indicate the chromatin before immunoprecipitation (IP), IP with no antibodies, and IP with anti-FLAG antibodies, respectively. Those genes include lignin biosynthesis genes (PtrC4H1 and Ptr4CL5), fiber cell elongation genes (PtrXTH5 and PtrEXPA8), GA biosynthesis genes (PtrGA20ox4 and PtrGA3ox1), and TFs (PtrWND6A, PtrWND6B, and PtrMYB28).
Discussion
Multiple studies on herbaceous plants demonstrated that WOX genes participated in diverse development processes including shoot apical meristem (Nardmann and Werr, 2006), lateral organ development (Deveaux et al., 2008), plant stem cell maintenance (Sarkar et al., 2007; Stahl et al., 2009), and floral determinacy (Deveaux et al., 2008). In addition, although it has been reported that some members of the WOX ancient clade were involved in secondary growth (Romera-Branchat et al., 2013; Denis et al., 2017; He et al., 2019), none of the members of the woody plant WOX ancient clade gene family regulating wood formation has been reported. Therefore, PtrWOX13A is the first WOX ancient clade gene in tree that has been found to regulate SCW component biosynthesis.
PtrWOX13A Regulated Secondary Cell Wall and Fiber Formation Through Regulating Transcription Factors and Structural Genes Related to Wood Formation
Overexpression of PtrWOX13A increased lignin and hemicellulose contents, wall thickness, lengths, and diameter of the fiber, and did not alter cellulose content (Figures 3, 5). These changes aligned well with the changes in TFs and structural genes involved in the SCW component biosynthesis (Zhong et al., 2010; Nakano et al., 2015; Gunasekara et al., 2016). As a result, significant changes in PtrWOX13A transgenic lines were conspicuous (Figure 6). In addition, the transient expression assay analysis revealed that the activities of the promoters of some TFs and structural genes, including PtrC4H1, Ptr4CL5, PtrXTH5, PtrEXPA8, PtrWND6A, and PtrMYB28, were directly activated by PtrWOX13A (Figures 7C, 8B). In summary, these data suggested that PtrWOX13A participated in wood formation as a higher hierarchy TF in the transcriptional regulation network of poplar SCW formation.
PtrWOX13A Regulated Secondary Cell Wall and Fiber Formation Through Modulation of Gibberellin-Mediated Signaling Pathway
It has been recognized for a long time that bioactive GAs promoted SCW biosynthesis (Wang et al., 2013, 2017; Yuan et al., 2019), generally through promoting DELLA repressor degradation which leads to the activation of SCW master TFs (Biemelt et al., 2004). For example, GA is found to trigger the transactivation of SCW master switch NAC29/31, resulting in activating the expression of MYB61, which specifically upregulates CESAs and thereby enhances the cellulose biosynthesis in rice (Huang et al., 2015). Presumably, upregulation or downregulation of GA homeostasis key genes, such as GA20ox, GA3ox, and GA2ox (Yamaguchi, 2008), alters bioactive GA contents and affects SCW formation, especially lignin deposition, in the stems of several plants.
The previous studies have revealed that some WOX family members regulate plant development through modulating GA-homeostasis and GA-signaling. For example, WOX8 and WOX9 activities are associated with GA signaling in the internode elongation of Arabidopsis (Wang et al., 2014), while WOX14 overexpression promotes the bioactive GA accumulation through activating GA3ox and repressing GA2ox, and thus enhances lignification of transgenic plants (Denis et al., 2017). Moreover, OsWOX3A participates in diverse developmental processes through negative feedback that controls the GA biosynthetic pathway (Cho et al., 2016). Similarly, PtrWOX13A overexpression resulted in the increased expression of PtrGA20ox4 and PtrGA3ox1 and decreased expression of PtrGA2ox1 (Figure 6B), accompanied by the increased contents of bioactive GAs (Figure 5E). Thus, PtrWOX13A perhaps regulated wood formation partially through GA-mediated signaling cascade, such as DELLA-NAC (Huang et al., 2015).
Possibility for PtrWOX13A to Modulate Bioactive Gibberellin Homeostasis Through Negative Feedback Regulation Pathway
In this study, we found that both PtrGA20ox4 and PtrGA3ox1 were significantly upregulated, while the expression of the PtrGA2ox1 significantly decreased in PtrWOX13A overexpression lines (Figure 6B), which was also aligned well with the increased contents of bioactive GAs (Figure 5E). In addition, we showed that PtrWOX13A directly activated the promoter of PtrGA3ox1 by binding to the TTAATSS cis-regulatory element in PtrGA3ox1 and PtrGA20ox4 promoter, but not the promoter of PtrGA2ox1 (Figure 7C). This result resembles the previous report that OsWOX3A acts as both a positive regulator of auxin-related genes and a negative regulator of YABBY3 in leaf development (Dai et al., 2007; Cho et al., 2013). Moreover, PtrWOX13A had a GA-responsive cis-regulatory element, P-BOX, in its promoter, and its expression rapidly increased within 24 h and then decreased after 24 h under exogenous GA treatment (Figure 1B), which indicates that the high bioactive GAs beyond a threshold might repress PtrWOX13A. Thus, when the activation of PtrGA20ox4 and PtrGA3ox1 and the repression of PtrGA2ox1 resulting from PtrWOX13A overexpression caused the active GAs to increase beyond the threshold, the expression levels of PtrWOX13A decreased, resulting in the downregulation of the aforementioned genes regulating GA homeostasis, which then decreased the concentration of bioactive GAs. These results indicate that PtrWOX13A not only acts as a positive regulator and a negative regulator in GA homeostasis but also participates in GA homeostasis and signaling through the negative feedback regulation pathway in poplar. However, further studies are needed to test this.
The Putative Regulatory Model of PtrWOX13A on Wood Formation
Finally, we proposed the two putative regulation pathways of PtrWOX13A participating in wood formation (Figure 9): (1) As a higher hierarchical switch TF, PtrWOX13A directly and indirectly regulated SCW TFs and structural genes; (2) PtrWOX13A enhanced bioactive GA contents through regulating the key genes regulating bioactive GA homeostasis and subsequently promoted DELLA proteins degradation through GA-mediated signaling cascade to derepressed wood-associated NAC or MYB TFs. The PtrWOX13A could participate in poplar wood formation through one or two of these pathways. This study revealed a new higher hierarchical regulator and its multiple regulatory chains in the hierarchical network governing SCW biosynthesis in tree species. The findings are instrumental for the molecular breeding of tree species for high biomass productivity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YZ and YL conducted most of the experiments and data analysis. XW and RW contributed to histochemical staining. XC and SW contributed to the vegetative propagation of Populus trichocarpa plantlets and phenotype analysis. HW contributed to figure preparation and manuscript revision. ZW designed the experiments and wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
The experimental design and implementation, data preprocessing analysis, and interpretation were supported by the National Natural Science Foundation of China (31770640).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.835035/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis and protein sequence alignment of PtrWOX13. (A) Phylogenetic tree of WOX proteins from Populus trichocarpa and Arabidopsis thaliana. PtrWOX13 proteins were shown in an orange trapezoid. The tree showed three major phylogenetic subfamilies (subfamilies I–III) that are marked with different colors. (B) AtWOX13A (AT4G35550) protein of A. thaliana aligned with PtrWOX13 proteins from Populus trichocarpa (Potri.005G101800, Potri.002G008800, and Potri.005G252800). The conserved HD and MOG motifs represent by blue arrows and green arrows, respectively; yellow, red, brown, and black boxes represent the YxDpl motif, NVYNWFQNR motif, activation region, and ESExE motif, respectively.
Supplementary Figure 2 | Cis-regulatory elements in promoter regions of PtrWOX13 genes and motifs of PtrWOX13 proteins. (A) Motifs of PtrWOX13 proteins. Colors of boxes denote different motifs while lengths of boxes represent relative motif lengths. (B) Venn diagram of common and unique motifs present in promoter regions of PtrWOX13A, PtrWOX13B1, and PtrWOX13B2. (C) Cis-regulatory elements in the 2,000 bp promoter sequences upstream of transcription start codons of PtrWOX13A, PtrWOX13B1, and PtrWOX13B2 genes using the online tool PlantCARE. These elements have different functions, which are represented by different colors.
Supplementary Figure 3 | Identification of PtrWOX13A in Populus trichocarpa overexpression transgenic lines. (A) PCR detection of PtrWOX13A transgenic lines. Lane M, DNA marker DL2000; Lane 1, wild type; Lane 2–9. PCR products of PtrWOX13A transgenic lines. (B) qRT-PCR detection of PtrWOX13A transgenic lines. PtrActin was used as a control. Each error bar represents the standard deviation of three biological replicates. Asterisks represent levels of significance (Dunnett’s test; *p < 0.01 < p < 0.05, **p < 0.01). (C) 90-day-old wild type (WT) and PtrWOX13A transgenic lines (OE-2, OE-3, and OE-7).
Supplementary Figure 4 | Anatomical and histochemical analyses of stems of different developmental stages in Populus trichocarpa transgenic lines. (A–D) Represent phloroglucinol-HCl stained transverse sections of the internode 2 (IN2), 4 (IN4), 6 (IN6), and 8 (IN8), respectively. (E–H) Represent toluidine blue O-stained transverse sections of IN2, IN4, IN6, and IN8 in WT, respectively. (I–L) were phloroglucinol-HCl-stained transverse sections of IN2, IN4, IN6, and IN8 in PtrWOX13A transgenic lines, respectively. (M–P) Represent toluidine blue O-stained transverse sections of IN2, IN4, IN6, and IN8 in PtrWOX13A transgenic lines, respectively. Scale bars = 200 μm. The toluidine blue and phloroglucinol-HCl were used to stain cell morphology and lignin, respectively.
Supplementary Figure 5 | Analysis of PtrWOX13A binding to cis-acting elements by yeast one-hybrid assay. The pHIS2-p53/pGADT7-rec2-p53 and pHIS2-p53/pGADT7-rec2-PtrWOX13A were used as the positive and negative control, respectively. A represents ATTGATTG cis-regulatory element; AM1 and AM2 represent mutated ATTGATTG cis-regulatory elements; T represents TTAATSS cis-regulatory element; TM1 and TM2 represent mutated TTAATSS cis-regulatory elements; 10–1, 10–2, and 10–3 represent solution dilution ratio of transformed Y187 yeast cells.
Footnotes
- ^ http://www.ebi.ac.uk/Tools/msa/clustalw2/
- ^ https://phytozome-next.jgi.doe.gov/
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://rsbweb.nih.gov/ij/
References
Alvarez, J. M., Bueno, N., Canas, R. A., Avila, C., Canovas, F. M., and Ordas, R. J. (2018). Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: new insights into the gene family evolution. Plant Physiol. Biochem. 123, 304–318. doi: 10.1016/j.plaphy.2017.12.031
Ambavaram, M. M., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi: 10.1104/pp.110.168641
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Biemelt, S., Tschiersch, H., and Sonnewald, U. (2004). Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol. 135, 254–265. doi: 10.1104/pp.103.036988
Burk, D. H., Liu, B., Zhong, R., Morrison, W. H., and Ye, Z. H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827.
Cao, Y., Han, Y., Meng, D., Li, G., Li, D., Abdullah, M., et al. (2017). Genome-wide analysis suggests the relaxed purifying selection affect the evolution of WOX genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 8:78. doi: 10.3389/fgene.2017.00078
Chang, Y., Song, X., Zhang, Q., Liu, H., Bai, Y., Lei, X., et al. (2020). Genome-wide identification of WOX Gene family and expression analysis during rejuvenational rhizogenesis in walnut (Juglans regia L.). Forests 11:16.
Cheng, S., Huang, Y., Zhu, N., and Zhao, Y. (2014). The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 549, 266–274. doi: 10.1016/j.gene.2014.08.003
Cheng, S., Tan, F., Lu, Y., Liu, X., Li, T., Yuan, W., et al. (2018). WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 46, 2356–2369. doi: 10.1093/nar/gky017
Cho, S. H., Kang, K., Lee, S. H., Lee, I. J., and Paek, N. C. (2016). OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa). J. Exp. Bot. 67, 1677–1687. doi: 10.1093/jxb/erv559
Cho, S. H., Yoo, S. C., Zhang, H., Pandeya, D., Koh, H. J., Hwang, J. Y., et al. (2013). The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 198, 1071–1084. doi: 10.1111/nph.12231
Cubria-Radio, M., and Nowack, M. K. (2019). Transcriptional networks orchestrating programmed cell death during plant development. Curr. Top. Dev. Biol. 131, 161–184. doi: 10.1016/bs.ctdb.2018.10.006
Dai, M., Hu, Y., Zhao, Y., Liu, H., and Zhou, D. X. (2007). A Wuschel-Like Homeobox gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144, 380–390. doi: 10.1104/pp.107.095737
Daude, M. M., Dos Santos Silva, T. W., Freitas, N. C., Ságio, S. A., Paiva, L. V., and Barreto, H. G. (2020). Transcriptional analysis of WUSCHEL-related HOMEOBOX (WOX) genes in Coffea arabica L. Biologia 75, 1483–1495.
Denis, E., Kbiri, N., Mary, V., Claisse, G., Conde, E. S. N., Kreis, M., et al. (2017). WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 90, 560–572. doi: 10.1111/tpj.13513
Deveaux, Y., Toffano-Nioche, C., Claisse, G., Thareau, V., Morin, H., Laufs, P., et al. (2008). Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 8:291. doi: 10.1186/1471-2148-8-291
Du, J., and Groover, A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52, 17–27. doi: 10.1111/j.1744-7909.2010.00901.x
Esmon, C. A., Tinsley, A. G., Ljung, K., Sandberg, G., Hearne, L. B., and Liscum, E. (2006). A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. U.S.A. 103, 236–241. doi: 10.1073/pnas.0507127103
Gambino, G., Minuto, M., Boccacci, P., Perrone, I., Vallania, R., and Gribaudo, I. (2011). Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis vinifera. J. Exp. Bot. 62, 1089–1101. doi: 10.1093/jxb/erq349
Gray-Mitsumune, M., Mellerowicz, E. J., Abe, H., Schrader, J., Winzell, A., Sterky, F., et al. (2004). Expansins abundant in secondary xylem belong to subgroup A of the alpha-expansin gene family. Plant Physiol. 135, 1552–1564. doi: 10.1104/pp.104.039321
Gunasekara, C., Subramanian, A., Avvari, J. V., Li, B., Chen, S., and Wei, H. (2016). ExactSearch: a web-based plant motif search tool. Plant Methods 12:26. doi: 10.1186/s13007-016-0126-6
Han, N., Tang, R., Chen, X., Xu, Z., Ren, Z., and Wang, L. (2021). Genome-wide identification and characterization of WOX genes in Cucumis sativus. Genome 64, 761–776. doi: 10.1139/gen-2020-0029
Hao, Q., Zhang, L., Yang, Y., Shan, Z., and Zhou, X. A. (2019). Genome-wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants (Basel) 8:215. doi: 10.3390/plants8070215
He, P., Zhang, Y., Liu, H., Yuan, Y., Wang, C., Yu, J., et al. (2019). Comprehensive analysis of WOX genes uncovers that WOX13 is involved in phytohormone-mediated fiber development in cotton. BMC Plant Biol. 19:312. doi: 10.1186/s12870-019-1892-x
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681–1696. doi: 10.1105/tpc.15.00015
Hussey, S. G., Mizrachi, E., Creux, N. M., and Myburg, A. A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4:325. doi: 10.3389/fpls.2013.00325
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21, 3493–3505. doi: 10.1105/tpc.109.069997
Israelsson, M., Sundberg, B., and Moritz, T. (2005). Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. Plant J. 44, 494–504. doi: 10.1111/j.1365-313X.2005.02547.x
Ji, J., Shimizu, R., Sinha, N., and Scanlon, M. J. (2010). Analyses of WOX4 transgenics provide further evidence for the evolution of the WOX gene family during the regulation of diverse stem cell functions. Plant Signal. Behav. 5, 916–920. doi: 10.4161/psb.5.7.12104
Kieffer, M., Stern, Y., Cook, H., Clerici, E., Maulbetsch, C., Laux, T., et al. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18, 560–573. doi: 10.1105/tpc.105.039107
Kumar, M., Campbell, L., and Turner, S. (2016). Secondary cell walls: biosynthesis and manipulation. J. Exp. Bot. 67, 515–531. doi: 10.1093/jxb/erv533
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, M., Wang, R., Liu, Z., Wu, X., and Wang, J. (2019). Genome-wide identification and analysis of the WUSCHEL-related homeobox (WOX) gene family in allotetraploid Brassica napus reveals changes in WOX genes during polyploidization. BMC Genomics 20:317. doi: 10.1186/s12864-019-5684-3
Liao, Z., Chen, M., Guo, L., Gong, Y., Tang, F., Sun, X., et al. (2004). Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep. Biochem. Biotechnol. 34, 209–214. doi: 10.1081/PB-200026790
Liu, B., Wang, L., Zhang, J., Li, J., Zheng, H., Chen, J., et al. (2014). WUSCHEL-related homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics 15:296. doi: 10.1186/1471-2164-15-296
Liu, Y., Wei, M., Hou, C., Lu, T., Liu, L., Wei, H., et al. (2017). Functional characterization of Populus PsnSHN2 in coordinated regulation of secondary wall components in tobacco. Sci. Rep. 7:42. doi: 10.1038/s41598-017-00093-z
Lu, S., Li, Q., Wei, H., Chang, M. J., Tunlaya-Anukit, S., Kim, H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110, 10848–10853. doi: 10.1073/pnas.1308936110
Marjamaa, K., Kukkola, E. M., and Fagerstedt, K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60, 367–376. doi: 10.1093/jxb/ern278
Minh-Thu, P. T., Kim, J. S., Chae, S., Jun, K. M., Lee, G. S., Kim, D. E., et al. (2018). A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice. Mol. Cells 41, 781–798. doi: 10.14348/molcells.2018.0203
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Nardmann, J., and Werr, W. (2006). The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 23, 2492–2504. doi: 10.1093/molbev/msl125
Park, S. O., Zheng, Z., Oppenheimer, D. G., and Hauser, B. A. (2005). The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 132, 841–849. doi: 10.1242/dev.01654
Rathour, M., Sharma, A., Kaur, A., and Upadhyay, S. K. (2020). Genome-wide characterization and expression and co-expression analysis suggested diverse functions of WOX genes in bread wheat. Heliyon 6:e05762. doi: 10.1016/j.heliyon.2020.e05762
Ren, M., Zhang, Y., Liu, C., Liu, Y., Tian, S., Cheng, H., et al. (2021). Characterization of a High hierarchical regulator, PtrGATA12, functioning in differentially regulating secondary wall component biosynthesis in Populus trichocarpa. Front. Plant Sci. 12:657787. doi: 10.3389/fpls.2021.657787
Ren, M., Zhang, Y., Wang, R., Liu, Y., Li, M., Wang, X., et al. (2022). PtrHAT22, as a higher hierarchy regulator, coordinately regulates secondary cell wall component biosynthesis in Populus trichocarpa. Plant Sci. 316:111170. doi: 10.1016/j.plantsci.2021.111170
Romera-Branchat, M., Ripoll, J. J., Yanofsky, M. F., and Pelaz, S. (2013). The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 73, 37–49. doi: 10.1111/tpj.12010
Sajjad, M., Wei, X., Liu, L., Li, F., and Ge, X. (2021). Transcriptome analysis revealed GhWOX4 intercedes myriad regulatory pathways to modulate drought tolerance and vascular growth in cotton. Int. J. Mol. Sci. 22:898. doi: 10.3390/ijms22020898
Sakakibara, K., Reisewitz, P., Aoyama, T., Friedrich, T., Ando, S., Sato, Y., et al. (2014). WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 141, 1660–1670. doi: 10.1242/dev.097444
Samuga, A., and Joshi, C. P. (2004). Cloning and characterization of cellulose synthase-like gene, PtrCSLD2 from developing xylem of aspen trees. Physiol. Plant 120, 631–641. doi: 10.1111/j.0031-9317.2004.0271.x
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Seale, M. (2020). Cell wall remodeling during wood development. Plant Physiol. 182, 1800–1801. doi: 10.1104/pp.20.00260
Stahl, Y., Wink, R. H., Ingram, G. C., and Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909–914. doi: 10.1016/j.cub.2009.03.060
Suer, S., Agusti, J., Sanchez, P., Schwarz, M., and Greb, T. (2011). WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23, 3247–3259. doi: 10.1105/tpc.111.087874
Suzuki, S., Li, L., Sun, Y. H., and Chiang, V. L. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. doi: 10.1104/pp.106.086678
Tang, Y., Li, H., Guan, Y., Li, S., Xun, C., Dong, Y., et al. (2020). Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5. Front. Genet. 11:670. doi: 10.3389/fgene.2020.00670
van der Graaff, E., Laux, T., and Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10:248.
Wang, G. L., Que, F., Xu, Z. S., Wang, F., and Xiong, A. S. (2017). Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 254, 839–848. doi: 10.1007/s00709-016-0995-6
Wang, P., Guo, Y., Chen, X., Zheng, Y., Sun, Y., Yang, J., et al. (2019). Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 33, 1129–1142.
Wang, W., Li, G., Zhao, J., Chu, H., Lin, W., Zhang, D., et al. (2014). Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 10:e1004154. doi: 10.1371/journal.pgen.1004154
Wang, Z. G., Chai, G. H., Wang, Z. Y., Tang, X. F., Sun, C. J., Zhou, G. K., et al. (2013). [Molecular mechanism of AtGA3OX1 and AtGA3OX2 genes affecting secondary wall thickening in stems in Arabidopsis]. Yi Chuan 35, 655–665.
Wu, C. C., Li, F. W., and Kramer, E. M. (2019). Large-scale phylogenomic analysis suggests three ancient superclades of the wuschel-related homeobox transcription factor family in plants. PLoS One 14:e0223521.
Xu, X., Lou, Y., Yang, K., Shan, X., Zhu, C., and Gao, Z. (2019). Identification of homeobox genes associated with lignification and their expression patterns in bamboo shoots. Biomolecules 9:862. doi: 10.3390/biom9120862
Yamaguchi, M., Goue, N., Igarashi, H., Ohtani, M., Nakano, Y., Mortimer, J. C., et al. (2010). Vascular-related NAC-DOMAIN6 and vascular-related NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153, 906–914.
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251.
Yang, R., Yang, T., Zhang, H., Qi, Y., Xing, Y., Zhang, N., et al. (2014). Hormone profiling and transcription analysis reveal a major role of ABA in tomato salt tolerance. Plant Physiol. Biochem. 77, 23–34. doi: 10.1016/j.plaphy.2014.01.015
Yang, Z., Gong, Q., Qin, W., Yang, Z., Cheng, Y., Lu, L., et al. (2017). Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 17:113. doi: 10.1186/s12870-017-1065-8
Yuan, H., Zhao, L., Guo, W., Yu, Y., Tao, L., Zhang, L., et al. (2019). Exogenous application of phytohormones promotes growth and regulates expression of wood formation-related genes in Populus simonii x P. nigra. Int. J. Mol. Sci. 20:792.
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., and Chen, J. G. (2020). Transcriptional and post-transcriptional regulation of lignin biosynthesis pathway genes in Populus. Front. Plant Sci. 11:652. doi: 10.3389/fpls.2020.00652
Zhang, J., Xie, M., Tuskan, G. A., Muchero, W., and Chen, J. G. (2018). Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Front. Plant Sci. 9:1535. doi: 10.3389/fpls.2018.01535
Zhang, X., Zong, J., Liu, J., Yin, J., and Zhang, D. (2010). Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 52, 1016–1026. doi: 10.1111/j.1744-7909.2010.00982.x
Zhang, Y., Liu, C., Cheng, H., Tian, S., Liu, Y., Wang, S., et al. (2020). DNA methylation and its effects on gene expression during primary to secondary growth in poplar stems. BMC Genomics 21:498.
Zhao, H., Li, H., Jia, Y., Wen, X., Guo, H., Xu, H., et al. (2020). Building a Robust Chromatin Immunoprecipitation method with substantially improved efficiency. Plant Physiol. 183, 1026–1034. doi: 10.1104/pp.20.00392
Zhong, R., and Ye, Z. H. (2010). The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 5, 469–472. doi: 10.4161/psb.5.4.11400
Zhong, R., Lee, C., and Ye, Z. H. (2010). Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., McCarthy, R. L., Lee, C., and Ye, Z. H. (2011). Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. doi: 10.1104/pp.111.181354
Zhou, G. K., Zhong, R. Q., Himmelsbach, D. S., McPhail, B. T., and Ye, Z. H. (2007). Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiology 48, 689–699. doi: 10.1093/pcp/pcm037
Zhou, G. K., Zhong, R. Q., Richardson, E. A., Morrison, W. H., Nairn, C. J., Wood-Jones, A., et al. (2006). The poplar glycosyltransferase GT47C is functionally conserved with Arabidopsis Fragile fiber8. Plant and Cell Physiol. 47, 1229–1240. doi: 10.1093/pcp/pcj093
Keywords: PtrWOX13A, wood formation, bioactive gibberellins biosynthesis, Populus trichocarpa, coordinated regulation
Citation: Zhang Y, Liu Y, Wang X, Wang R, Chen X, Wang S, Wei H and Wei Z (2022) PtrWOX13A Promotes Wood Formation and Bioactive Gibberellins Biosynthesis in Populus trichocarpa. Front. Plant Sci. 13:835035. doi: 10.3389/fpls.2022.835035
Received: 14 December 2021; Accepted: 19 May 2022;
Published: 28 June 2022.
Edited by:
Chang Liu, University of Hohenheim, GermanyReviewed by:
Xingliang Hou, South China Botanical Garden (CAS), ChinaYves Deveaux, Université Paris-Sud, France
Copyright © 2022 Zhang, Liu, Wang, Wang, Chen, Wang, Wei and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Wei, zhigangwei1973@163.com
†These authors have contributed equally to this work
 Yang Zhang
Yang Zhang Yingying Liu3†
Yingying Liu3† Ruiqi Wang
Ruiqi Wang Zhigang Wei
Zhigang Wei