- 1Research Institute of Pomology, Chinese Academy of Agricultural Sciences, Xingcheng, China
- 2Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Germplasm Resources Utilization), Research Institute of Pomology, Chinese Academy of Agricultural Sciences, Ministry of Agriculture, Xingcheng, China
Alternaria blotch disease, caused by Alternaria alternata apple pathotype (AAAP), is one of the major fungal diseases in apple. Early field observations revealed, the anther-derived homozygote Hanfu line (HFTH1) was highly susceptible to AAAP, whereas Hanfu (HF) exhibited resistance to AAAP. To understand the molecular mechanisms underlying the difference in sensitivity of HF and HFTH1 to AAAP, we performed allele-specific expression (ASE) analysis and comparative transcriptomic analysis before and after AAAP inoculation. We reported an important immune gene, namely, MdFLS2, which displayed strong ASE in HF with much lower expression levels of HFTH1-derived alleles. Transient overexpression of the dominant allele of MdFLS2-1 from HF in GL-3 apple leaves could enhance resistance to AAAP and induce expression of genes related to salicylic acid pathway. In addition, MdFLS2-1 was identified with an insertion of an 85-bp terminal-repeat retrotransposon in miniature (TRIM) element-like sequence in the upstream region of the nonreference allele. In contrast, only one terminal direct repeat (TDR) from TRIM-like sequence was present in the upstream region of the HFTH1-derived allele MdFLS2-2. Furthermore, the results of luciferase and β-glucuronidase reporter assays demonstrated that the intact TRIM-like sequence has enhancer activity. This suggested that insertion of the TRIM-like sequence regulates the expression level of the allele of MdFLS2, in turn, affecting the sensitivity of HF and HFTH1 to AAAP.
1 Introduction
Apple (Malus domestica Borkh.) is one of the significant fruit species that is widely consumed globally. However, apple is susceptible to fungal diseases during its cultivation. Alternaria blotch disease, caused by Alternaria alternata apple pathotype (AAAP), is one of the most serious fungal diseases of apples (Filajdic and Sutton, 1991; Zhang et al., 2015). As a pathogenic variant of Alternaria, AAAP mainly infects apple leaves and causes early defoliation of the tree, severely damaging the yield and quality of apples (Moriya et al., 2019). The most effective measure for overcoming fungal diseases in apple is the selective breeding of resistant cultivars (Zhang et al., 2018). However, the mechanism of resistance to AAAP in apples is still not clearly studied (Hou et al., 2021). Therefore, it is urgent to investigate the molecular mechanism underlying Alternaria blotch disease resistance in apple.
To defend themselves against pathogens, plants have evolved with strong immune mechanisms (Ngou et al., 2022). Pattern recognition receptors (PRRs) located on plant cell membranes recognize conserved molecular pathogen-molecular patterns (PAMPs) secreted by pathogens, thereby activating pattern-triggered immunity (PTI) to limit pathogenicity (Zipfel, 2008). In contrast, nucleotide-binding leucine-rich repeat receptors located inside plant cells can activate effector-triggered immune (ETI) responses by directly or indirectly sensing specific effectors secreted by pathogens (Nomura et al., 2011). PTI is an important component of basal resistance in plants. Recent studies have reported that defense responses triggered by PTI are equally essential for ETI activation (Ngou et al., 2021; Yuan et al., 2021). As PRRs trigger PTI responses, they have been at the forefront of plant immune research, with the receptor kinase FLS2 being the most extensively studied (Zipfel et al., 2004; Lu et al., 2011; Shi et al., 2016; Zhang et al., 2020b). FLS2 can form a complex with the co-receptor kinase BAK1 to recognize the conserved protein polypeptide flg22 in the flagellin of pathogenic bacteria to activate the immune response of plants (Sun et al., 2013). In addition, FLS2 may be involved in the recognition of other ligands, e.g., the sensitivity of Arabidopsis AtFLS2 mutants to the protein polypeptide Ax21 secreted by Xanthomonas oryzae pv. oryzae (Xoo) is significantly reduced by the action of FLS2 (Danna et al., 2011). PTI-mediated immune responses involve MAPK cascade responses and phytohormone signaling (Asai et al., 2002; Kong et al., 2016). Upon recognition of flg22 by FLS2, the MAPK cascade response in plants is activated to further phosphorylate WRKY transcription factors, thereby regulating the expression of immune-related genes (Sarowar et al., 2019). Previous studies in apple reported one FLS2 ortholog, namely, MdFLS2 (Chen-Hui et al., 2018). Overexpression of MdFLS2 in Arabidopsis increased its resistance to Botryosphaeria dothidea, whereas the expression levels of genes related to salicylic acid pathway were significantly enhanced (Liu et al., 2018). However, it is unclear whether MdFLS2 has a broad-spectrum resistance to various plant pathogens, similar to AtFLS2, and the mechanisms regulating its expression level should be further studied.
Apple is typically hetero-pollinated plant, and its highly heterozygous genotypes confer diversity in response to biotic (Reim et al., 2019) and abiotic (Ren et al., 2017) stresses. After 3 years of field observations, we observed that Hanfu (HF) exhibited resistance, whereas the anther-derived homozygote Hanfu line (HFTH1) was highly susceptible to AAAP (Figure 1A). The homozygosis of some important genes predisposes plants to exhibit extreme phenotypes (Wang et al., 2014; Zhang et al., 2020a). However, heterozygotes can overcome these adverse effects by selecting more favorable allele expression under specific conditions, i.e., allele-specific expression (ASE) (Shao et al., 2019). Recent studies have reported that ASE is involved in the regulation of many important traits in apples, such as fruit ripening (Sun et al., 2020) and flower color formation (Tian et al., 2022). To understand the molecular mechanisms underlying the difference in sensitivity of HF and HFTH1 to AAAP, we aimed to investigate the ASE patterns in HF under AAAP stress. Previously, we have obtained the HFTH1 haplotype reference genome (Zhang et al., 2019), providing a genomic resource for ASE analysis in HF. In this study, we integrated ASE analysis and comparative transcriptomic analysis of HF and HFTH1 before and after AAAP inoculation to identify key genes and regulatory patterns that influence differences in susceptibility to AAAP. These results can provide new insights and references for breeding with Alternaria blotch disease resistance in apple.
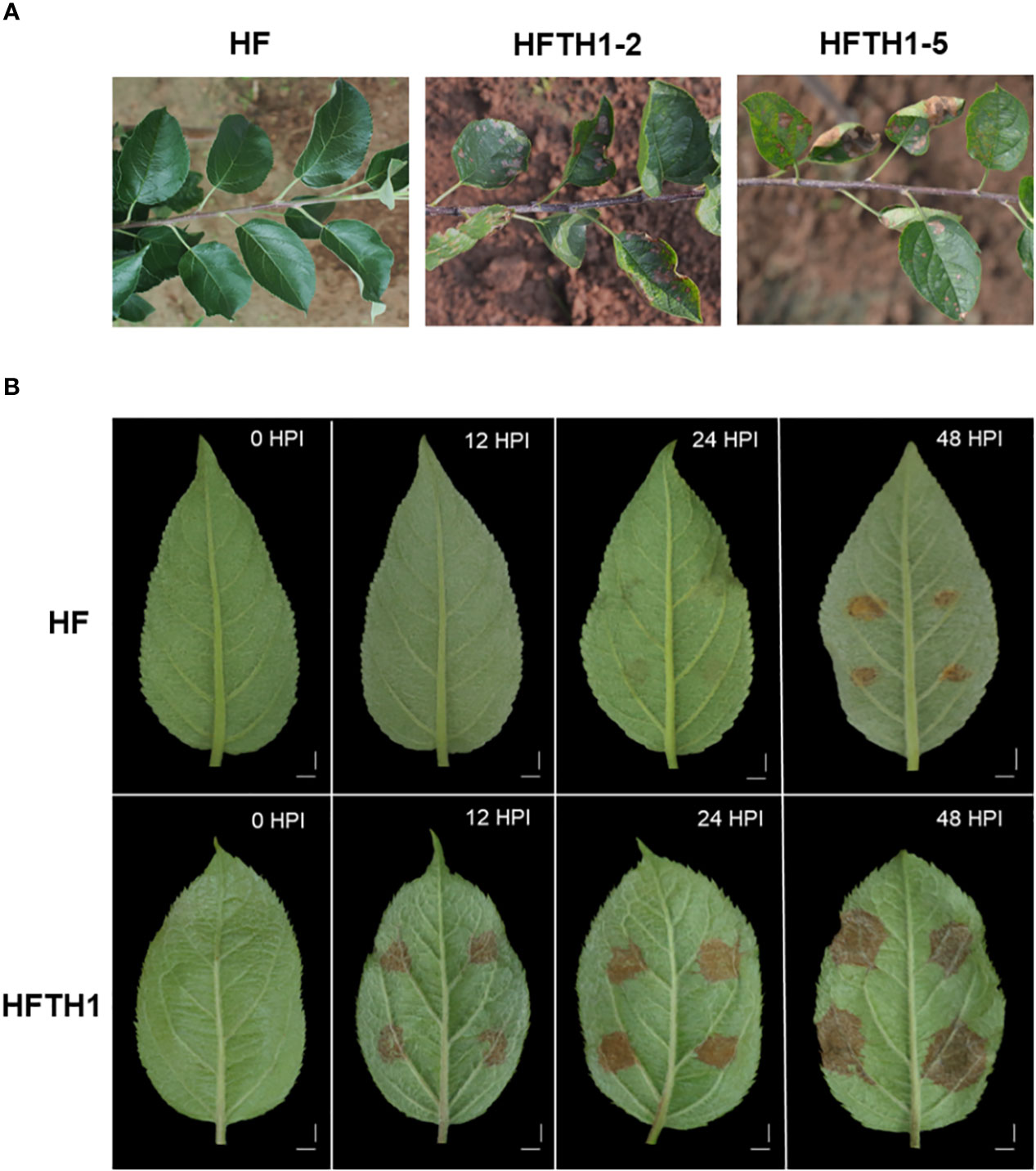
Figure 1 Different susceptibility of HF and HFTH1 to AAAP. (A) Investigation of the susceptibility of HF and HFTH1 to AAAP in the field experiments. (B) Changes in symptoms of HF and HFTH1 at 12, 24, and 48 h after inoculation with AAAP in vitro. Mock inoculation of HF and HFTH1 leaves without AAAP was used as the control (0HPI). Scale bar = 1 cm.
2 Materials and methods
2.1 Plant materials, AAAP culture, and leaf inoculation
To reduce the genetic difference between two apple species, we selected HF (exhibiting resistance to AAAP) and HFTH1 (an anther-derived homozygote Hanfu line susceptible to AAAP) as experimental materials. Both materials were grafted on GM256 rootstock in 2010 and grown in a research orchard at Research Institute of Pomology, Chinese Academy of Agricultural Sciences (CAAS). AAAP was obtained from Plant Protection Center of Institute of Pomology, CAAS. AAAP was cultured on potato dextrose agar (PDA; 200 g potato extract, 20 g dextrose, and 20 g agar in 1 L water) medium and incubated in dark until the mycelia spread to two-third of the plate. The mycelium was inoculated onto 20-day-old HF and HFTH1 leaves according to the experimental method described previously (Zhu et al., 2017). In each treatment, four cakes of mycelium were used to inoculate both sides of the midvein on the abaxial surface of the leaves. Six treatment groups were set up at 12-h intervals for both plant materials, and the control group was mock-inoculated using PDA medium cakes without mycelium. After inoculation, the apple leaves were incubated in a sterilized culture chamber at 25°C under a 14 h light/10 h dark conditions with moisture control. Further, 48 h after inoculation in the first group, samples were simultaneously collected from each group to observe the incidence of the two materials at three stages after inoculation (12 HPI, 24 HPI, and 48 HPI) compared with the control group (0 HPI). Apple leaves were collected from three HF plants and used for whole genome resequencing.
Leaf samples were quickly placed in liquid nitrogen and stored at -80°C for resequencing. Three parallel leaves on three trees were collected for each sample set, representing three independent biological replicates. The strand-specific transcriptome sequencing and whole genome resequencing was performed by BerryGenomics (Beijing, China).
2.2 Genome and strand-specific transcriptome sequencing
Total cellular DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany), as per the manufacturer’s instructions. The final libraries constructed above were sequenced using the Illumina Novaseq6000 platform, and 150-bp paired-end reads were generated with an insert size of approximately 350 bp. The raw reads obtained from sequencing were quality controlled, and the clean reads obtained after screening were compared with the HFTH1 reference genome (Zhang et al., 2019) using BWA (version 0.7.15; Li and Durbin, 2009) for variant calling.
As per the manufacturer’s instructions, total RNA was isolated using cetyltrimethyl ammonium bromide method (Pavy et al., 2008). The purity of RNA was measured using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and analyzed for RNA degradation and contamination using agarose gel electrophoresis. Strand-specific cDNA libraries were constructed using the dUTP method, as per a previous study (Parkhomchuk et al., 2009). Further, the 24 individual libraries were sequenced using the Illumina Novaseq6000 platform (BerryGenomics, Beijing, China).
2.3 Analysis of data and allele−specific expression
The raw reads of sequenced raw sequences were subjected to strict quality control to obtain valid and high-quality clean reads. The clean reads were compared with the HFTH1 apple reference genome using HISAT2 (v 2.20; Kim et al., 2015) to obtain information such as the position of the read on the compared genome and quality of the match; thus, the gene or transcript is annotated and quantified. Gene-level quantification was performed using featureCounts (Version 2.0.1; Liao et al., 2014), and gene expression levels were calculated as FPKM. Based on the comparison results, the differential expression of genes in each sample was analyzed using edgeR package (Robinson et al., 2010), and the pvalue and padj values of differential expression were calculated. The differential genes with padj < 0.05 and |log2FoldChange| > 1 were screened. GO enrichment analysis was performed using topGO software, and KEGG enrichment analysis was performed using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/). The heatmap was created using Tbtools (Chen et al., 2020).
To identify alleles, genome and transcriptome sequencing data compared with the reference genome were called for variants using GATK (version 3.8.0; Brouard et al., 2019). In this study, only SNPs were used to distinguish alleles using VCFTools (version 0.1.14) to filter SNPs and retaining SNP sites with read depths above 8×, the remaining SNP sites were annotated using snpEff (Cingolani et al., 2012). The SNP sites located in the CDS region of the genome were used for the identification of ASE. Genes that simultaneously satisfy the following conditions were considered ASE genes (ASEGs; Tian et al., 2022): (1) read counts of the reference allele divided by total read counts were >0.75 or <0.25, (2) different SNPs on the same gene exhibited same direction of significant bias, and (3) genes were identified as ASEGs in at least two out of three replicates.
2.4 Vector construction and transient overexpression in apple leaves
Specific primers were designed to amplify MdFLS2-1 and MdFLS2-2 in HF based on the coding sequence of MdFLS2 predicted from the HFTH1 reference genome (Supplementary Table S7). The CDS region of MdFLS2 allele was inserted into the Nde I digest site of PRI101 vector using 2× MultiF Seamless Assembly Mix (ABclonal, Wuhan, China) according to the in-fusion cloning method. Specific primers were designed to amplify the 2058-bp promoter sequence of MdFLS2 allele based on the HFTH1 reference genome (Supplementary Table S7) and ligated to the pESI-Blunt vector. The ligated vector was transformed into E. coli DH5α competent cells, and the monoclonal colonies were picked and sent to GENEWIZ (Tianjin, China) for sequencing. The allelic variants of the gene were analyzed using ClustalX 2.1.
A recombinant plasmid inserted with the CDS region of MdFLS2-1 was transformed into Agrobacterium tumefaciens strain GV3101 (Bai et al., 2013). The vector-transformed Agrobacterium was cultured in 10 mL YEP medium and shaken at 28°C and 130 rpm until OD600 reached to 1. The bacterial broth was centrifuged at 5000 rpm, resuspended in 10 mL of buffer (10 mM MgCl2,10 mM MES [pH 5.7], and 200 μM acetosyringone), and used after 3 h of activation at 28°C. To improve the transformation efficiency, we selected GL-3 apple plants that were favorable for transient transformation experiments. GL-3 apple plants cultured in vitro for 1 month were selected, and two spots on both sides of the leaf veins were selected to be infiltrated by injection. The infiltrated plants were placed in new MS medium and incubated for 4 days at 25°C in a light incubator under 16 h light/8 h dark conditions. Three Agrobacterium-infiltrated plants were randomly selected, and RNA from the injected leaves was extracted for gene expression analysis. After 4 days of infiltration, the remaining plants were inoculated with AAAP spore suspension at the location of the Agrobacterium injection, and the inoculated plants were placed back into MES medium and incubated in a light incubator for 48 h. In the control group, GL-3 apple leaves were treated in the same manner with sterile injection buffer and injection buffer containing empty vector, and the same batch of AAAP spore suspension was used to inoculate the infiltration site of the leaves. Plants with AAAP-infected leaves were examined at 48 HPI, and three independent biological replicates were collected and photographed. The differences in incidence levels were analyzed using student’s t test (Zhang et al., 2018).
2.5 Luciferase reporter assay
The TRIM-like sequence from the promoter of MdFLS2-1 and TDR2 sequence from the promoter of MdFLS2-2 were respectively fused to the cauliflower mosaic virus 35S minimal promoter (mpCaMV) and ligated to the HindIII-BamHI site of pGrernII0800-LUC vector, to generate the construct structures TRIM:mpCaMV : LUC and TDR2:mpCaMV : LUC. Sequences were synthesized (Supplementary Table S8) by GENEWIZ (Tianjin, China). Two reporter constructs were used to simultaneously transform Agrobacterium GV3101. Leaves of 8-week-old Nicotiana benthamiana plants were permeated with a needle-free syringe using the above injection buffer. The infiltrated plants were first incubated in a phytotron at 23°C for 12 h, followed by 60 h of growth under 16 h light/8 h dark conditions. The working solution (100 mM stock solution; 0.1% Triton X-100) was evenly spread onto the plant leaves using a high-pressure sprayer. The leaves were left in dark for 7 min, and the LUC images were captured using a low-light cooled CCD imaging apparatus (Tanon 5200Multi, China). The LUC/REN ratio was analyzed as described in a previous study (Liu et al., 2019). The experiment was independently repeated three times to obtain similar results.
2.6 Analysis of β-glucuronidase activity
The TRIM-like and TDR2 sequences fused with the 35S minimal promoter were respectively ligated to the HindIII-BamHI site of the pBI121 vector to generate reporter constructs and used to transform Agrobacterium GV3101. The GV3101 strain containing the recombinant plasmid was infiltrated into the abaxial leaf surface of N. benthamiana plants using the injection buffer described above. Three biological replicates were set for each infection. Infected plants were grown in a climatic chamber for 3 days, and β-glucuronidase (GUS) activity was detected as described in a previous study (Jefferson et al., 1987).
2.7 RNA extraction and RT-qPCR analysis
After freezing the plant leaves in liquid nitrogen, total RNA was extracted using Quick RNA Isolation Kit (Huayueyang Biotechnology, China) according to the manufacturer’s instructions. RNA quality was detected using agarose gel electrophoresis, and RNA concentration was determined using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The same concentration of RNA was reverse transcribed to cDNA using PrimeScript RT Master Mix (TakaRa, Tokyo, Japan). RT-qPCR was conducted using a TB-Green PCR kit (TakaRa, Tokyo, Japan) as per the manufacturer’s instructions. The relative expression of each gene was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001) using Tubulin as the reference gene to normalize the gene expression level. Three experimental replicates were set up for each sample, and the mean standard deviation was calculated. RT-qPCR primers (Supplementary Table S7) were designed using primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast).
3 Results
3.1 HF and HFTH1 have different sensitivity to AAAP
Based on field observation for 3 years, the homozygous Hanfu line HFTH1 exhibited symptoms of severe spotted defoliation disease (Figure 1A), causing early defoliation in the wet rainy season. Meanwhile, the heterozygote HF exhibited no symptoms of susceptibility to the disease.
We performed simultaneous infection experiments on isolated leaves of HF and HFTH1 using the AAAP strain and recorded the symptoms on leaves (Figure 1B). The leaves of HFTH1 started exhibiting tissue necrosis by 12 HPI, brown spots by 24 HPI, and dark brown spots with conspicuous white hyphae on the necrotic tissue by 48 HPI. In contrast, HF leaves exhibited a later onset and milder symptoms. By 12HPI, no signs of disease were observed on HF leaves. By 24HPI, slight symptoms of the disease appeared. By 48HPI, lesions expanded and deepened to brown.
3.2 Sequencing and identification of ASE
The leaves of HF and HFTH1 after 12, 24, and 48 h of AAAP inoculation were set up in three replicates. We mixed the mock-inoculated leaves of three time points into one sample as the control (0 HPI). A total of 24 samples were sequenced using the Illumina Novaseq6000 platform for Strand-specific transcriptome sequencing. In total, 20,379,378–36,174,753 clean reads were generated from 24 RNA-seq libraries, with Clean GC% of 45.43–46.52, CleanQ20 of 97.33–98.10, and CleanQ30 of 92.80– 94.50 (Supplementary Table S1). This indicated that the obtained RNA-Seq data were of high quality and suitable for further analysis. The transcriptome sequencing data were matched with the HFTH1 apple reference genome with 94.76%–97.19% similarity for the HFTH1 and 93.75%–95.19% similarity for the HF. The high matching rate ensured the utilization and accuracy of the transcriptome data.
Whole genome resequencing of heterozygous diploid HF was performed at a sequencing depth of 35×, and a total of 571,019 SNPs and 695,491 indels (Figure 2A) were identified using the HFTH1 genome as a reference. To improve the accuracy of ASE analysis, 52,765–46,854 SNP sites (Supplementary Table S2) located in the CDS region of genes and shared in both genome and transcriptome with transcriptome sequencing depths > 8 were retained in combination with transcriptome data of HF at each period post AAAP inoculation and control (Gao et al., 2021). These SNPs were used to analyze the expression levels between alleles. Alleles located in the HFTH1 reference genome with expression ratios > 0.75 or < 0.25 were considered to have ASE, whereas the presence of ASE in at least two of the three replicate samples with the same orientation was identified as an ASE gene (ASEG). Finally, a total of 2406 ASEGs were identified in the transcriptome, and 899–1086 ASEGs were identified in each of the four time periods (Figure 2B). Most ASEGs were biased toward reference genome expression during at each time period after AAAP inoculation and mock inoculation, and more genes exhibited ASE as the time of AAAP infection progressed (Supplementary Table S3). These results suggested that AAAP infection may have altered the allele expression pattern in HF leaves.
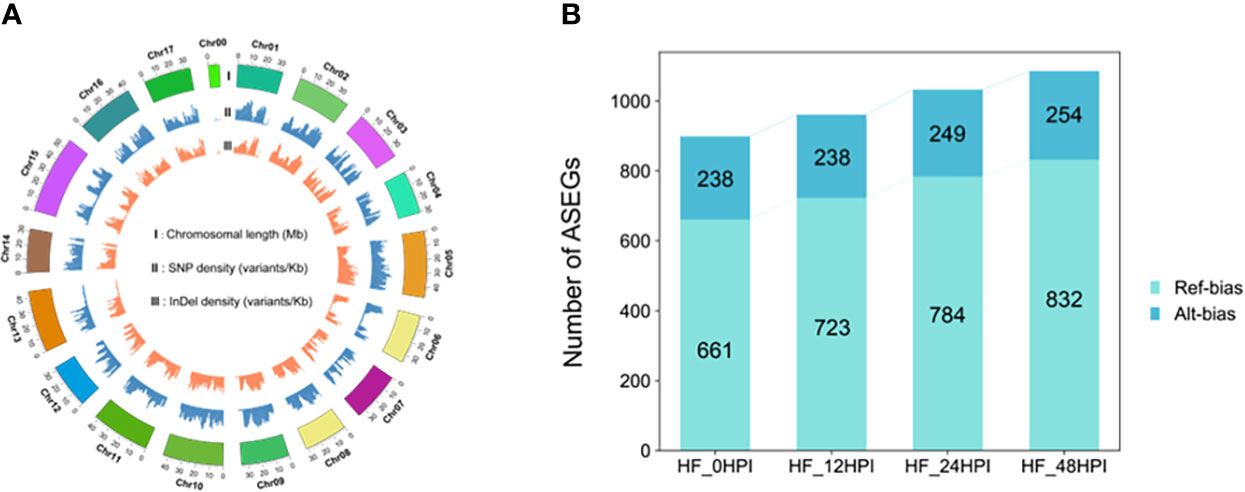
Figure 2 Identification of ASEGs. (A) Circos plot of HF resequencing data, chromosome length, SNP density, and indel density. (B) The number of ASEGs in HF leaves at 12, 24, and 48 h after AAAP inoculation and mock inoculation (0 HPI).
3.3 Two modes of ASEGs at different time periods before and after AAAP infection
A comparative analysis of ASEGs at different time periods before and after AAAP infection revealed that 132 genes exhibited ASE with the same direction of expression bias before and after the infection. Of them, 114 genes were biased toward the reference genome expression and 18 genes were biased toward the alternative genome expression (Figure 3A). Several studies have suggested that genes that exhibit consistent ASE across conditions may have dominant effects (Shao et al., 2019). GO enrichment analysis was used to analyze the biological functions of these ASEGs (Supplementary Table S4), and 89 genes were grouped into three main categories of GO: biological process, cellular component, and molecular function (Figure 3B). In the category of biological processes, more genes were involved in “oxidation-reduction process,” “protein phosphorylation,” and “proteolysis.” In the category of cellular components, most genes were classified as “integral component of membrane.” In the category of molecular functions, most genes were involved in “ATP binding.”
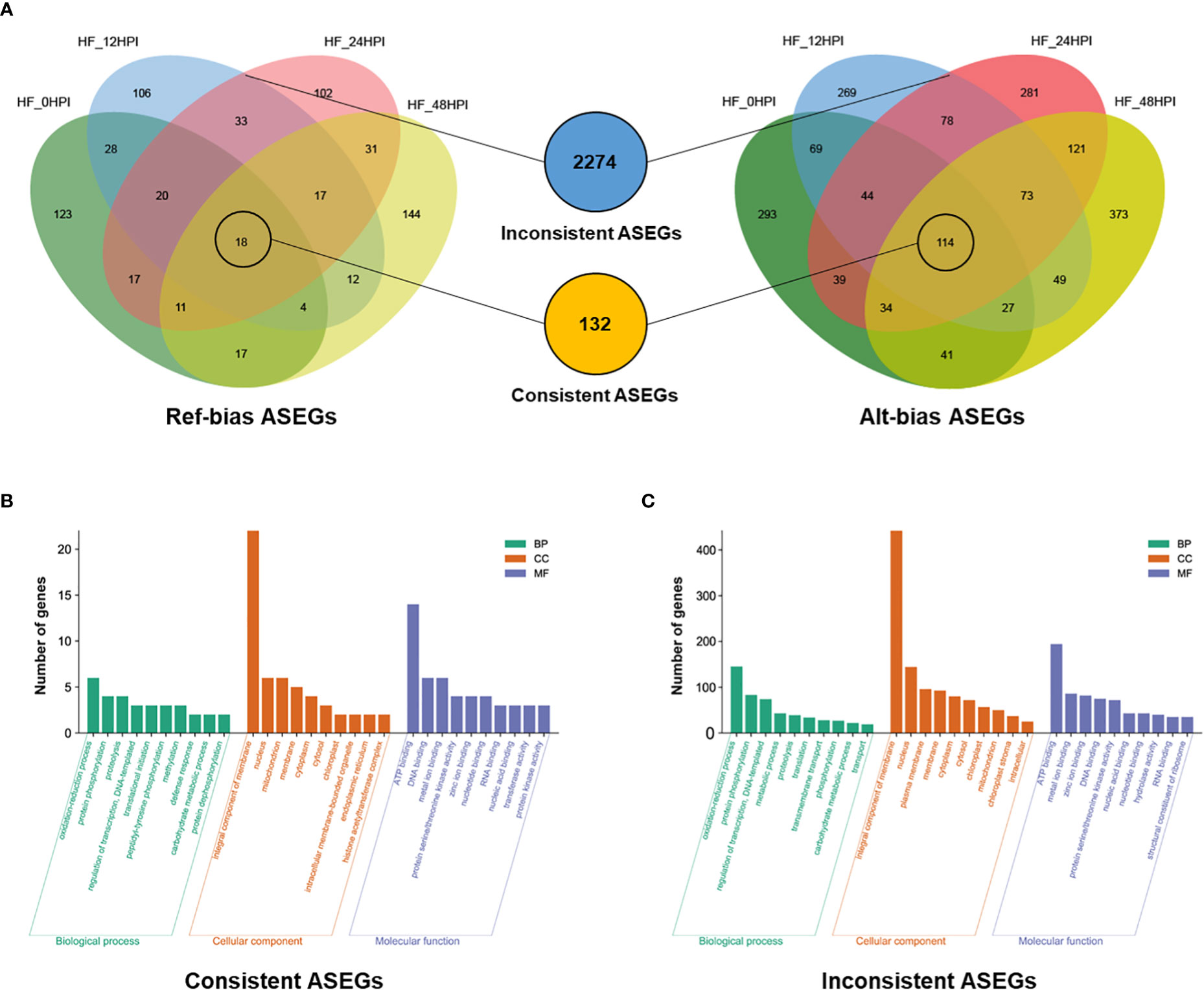
Figure 3 Characteristics of the two models of ASEGs. (A) Venn diagram analysis of reference genomic bias (Ref-bias) and alternative genomic bias (Alt-bias) at various times before and after AAAP infection. The middle blue and orange circles indicate inconsistent and consistent ASEGs, respectively. GO enrichment analysis of (B) consistent and (C) inconsistent ASEGs.
The remaining 2274 genes of the 2406 ASEGs exhibited inconsistent patterns of allelic bias expression at different time periods before and after the infection (Figure 3A). The expression pattern of this allele may cause an overdominance effect in heterozygotes (Zhang et al., 2021). GO enrichment analysis of these genes (Supplementary Table S5) revealed that in the category of biological processes, most of the genes were involved in “oxidation-reduction process,” “protein phosphorylation,” and “regulation of transcription, DNA-templates” (Figure 3C). In the category of cellular components, the vast majority of genes were classified as “integral component of membrane.” Among the category of molecular functions, most of the genes were enriched in “ATP binding,” “metal ion binding,” and “zinc ion binding.” These results were similar with the enrichment results of biased consistent ASEGs, suggesting that ASEGs may function mainly in some specific subcategories of cellular components and biological processes. These biological processes are closely related to plants’ resistance to pathogens (Turra et al., 2014; David et al., 2019), predicting that ASEGs may play an important role in the process of plant immune response.
3.4 Differential expression analysis and KEGG functional annotation
To identify defense genes that affect the difference in sensitivity of HF and HFTH1 to AAAP, we performed a comparative analysis of the transcriptomic data of HF and HFTH1 at 12, 24, and 48 h after the infection and mock inoculation (control; 0 HPI). Gene expression was compared between sample sets at different time periods after AAAP infection and mock inoculation, and differentially expressed genes (DEGs) that were up- and downregulated were counted. The results revealed that the up- and downregulated DEGs after AAAP infection in HF and HFTH1 increased with the advancement of infection time, compared with the control (Supplementary Figures S1A-D). This suggested that more genes were involved in the response to AAAP as the time of infection advanced. Meanwhile, the number of DEGs, both up- and downregulated, was higher in HF than in HFTH1 at each post-infection time compared with the control (Supplementary Figures S1A, B). This indicated that more genes were involved in disease-resistant varieties in response to AAAP stress. The transcriptome data of HF and HFTH1 in the same time after infection and mock inoculation were analyzed, and the DEGs that were up- and downregulated by HF compared with HFTH1 at each time period were separately counted. Due to the similar genetic background between the two species, relatively few DEGs were obtained. Moreover, and the number of DEGs upregulated in HF relative to HFTH1 was higher than that of DEGs downregulated in all four time periods after infection and mock inoculation (Figure 4A).
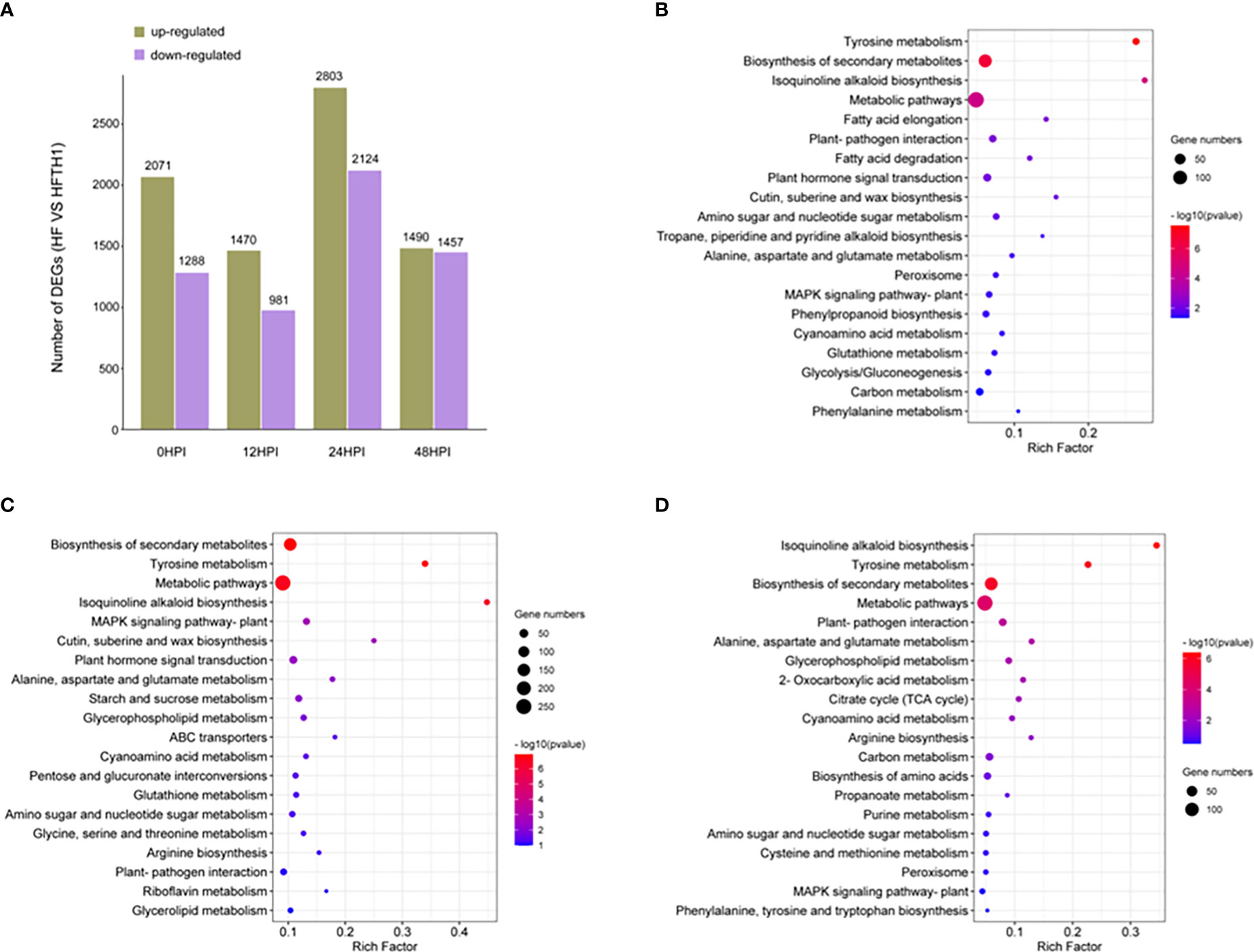
Figure 4 Comparative transcriptomic analysis of HFTH1 and HF before and after AAAP infection. (A) DEGs in HF compared with HFTH1 at the same time before and after AAAP infection. (B–D) Compared with HFTH1, HF up-regulated the KEGG pathway with significant enrichment of DEGs at 12 HPI (B), 24 HPI, and (C) and 48 HPI (D).
Based on orthology (KO) terminology mapping of the upregulated DEGs of HF compared with HFTH1 at the same period after pathogenic inoculation to the KEGG pathway, the top 20 significantly enriched pathways at each time period were selected for analysis (Figures 4B-D). Multiple KEGG pathways were significantly simultaneously enriched in the comparative combinations at each time period; they were “Tyrosine metabolism,” “Biosynthesis of secondary metabolites,” “Isoquinoline alkaloid biosynthesis,” “Metabolic pathways,” “Plant-pathogen interaction,” “Amino sugar and nucleotide sugar metabolism,” “Alanine, aspartate, and glutamate metabolism,” “MAPK signaling pathway-plant,” and “Cyanoamino acid metabolism.” Among them, “Plant-pathogen interaction” and “MAPK signaling pathway-plant” are directly related to the defense response of plants against pathogens (Dodds and Rathjen, 2010). MAPK signaling is a central pathway in immune response transduction and plays a crucial role in phytohormone-induced immune responses (Meng and Zhang, 2013). DEGs enriched in “Plant-pathogen interaction” and “MAPK signaling pathway-plant” at three time periods after pathogenic infection comprised a dataset containing 51 “core” immune genes (Supplementary Table S6). This dataset provides candidate genes for further analysis and functional identification.
3.5 Identification of MdFLS2 and expression analysis of related genes
Combining the above “core” immune genes dataset with the 2406 ASEGs, the Venn diagram indicated that six genes were simultaneously present in both datasets (Figure 5A). Heatmaps of the expression of these genes between HF and HFTH1 leaves indicated that MdFLS2 consistently exhibited significant differences in expression levels at three time periods after AAAP infection (Figure 5B). Meanwhile, MdFLS2 was consistently biased toward alternative genomic ASE after AAAP infection and mock inoculation. Previous studies have reported that overexpression of MdFLS2 in Arabidopsis enhances salicylic acid signaling and improves resistance to Botryosphaeria dothidea in Arabidopsis (Liu et al., 2018). There are some similarities in the disease resistance mechanisms of apples to AAAP and Botryosphaeria dothidea; for example, sorbitol can enhance the resistance of apples to both AAAP and Botryosphaeria dothidea (Meng et al., 2018; He et al., 2022).
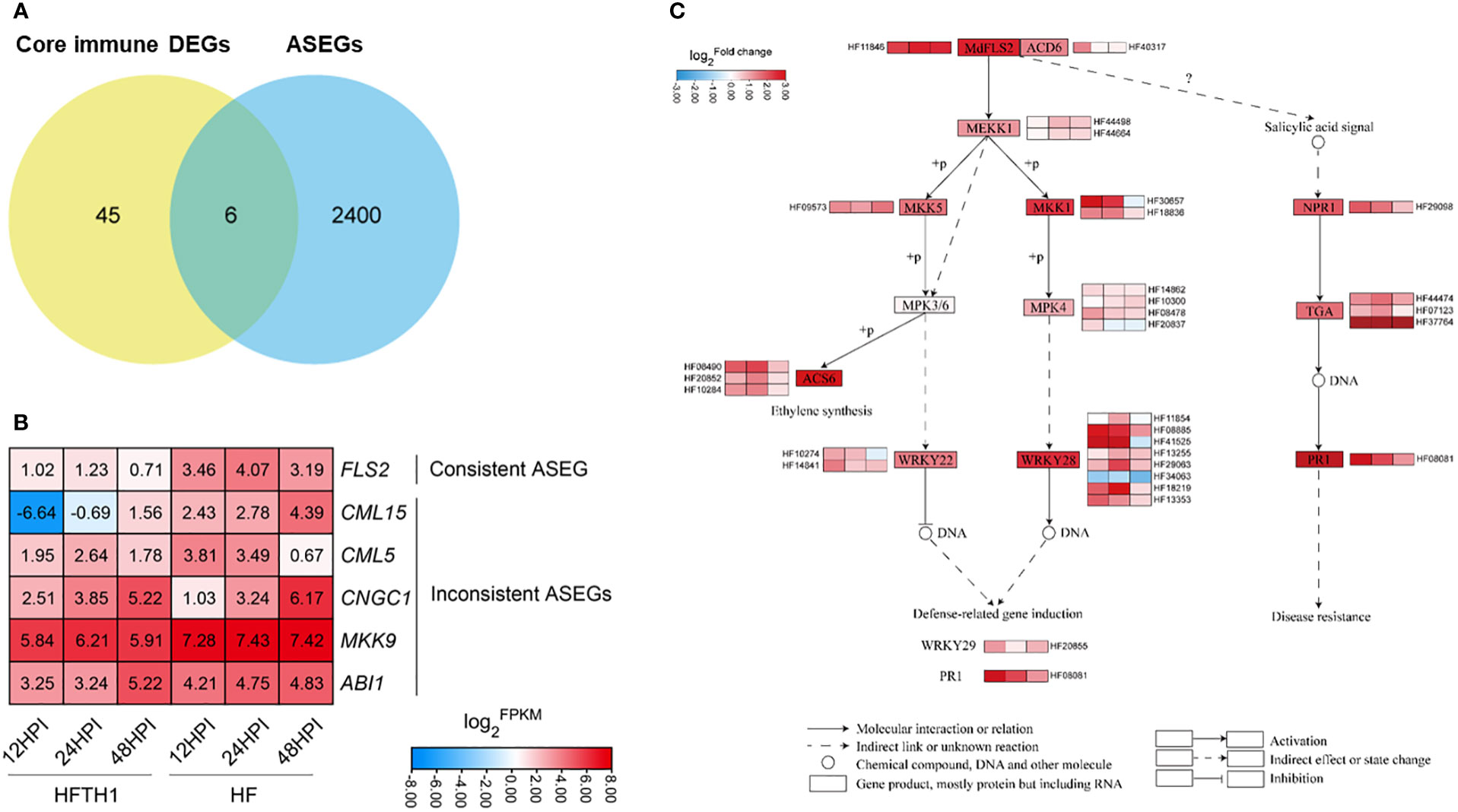
Figure 5 The ASE gene MdFLS2 may affect the defense response of apple leaves to AAAP. (A) Venn diagram analysis of “core” immune DEGs and ASEGs. (B) Heatmap showing log2FPKM of six “core” immune ASEGs in HFTH1 and HF at 12, 24, and 48 h after AAAP inoculation. (C) Heatmap sequentially shows the log2Fold change of genes from MdFLS2-related pathway in HF compared with HFTH1 at 12, 24, and 48 h after AAAP inoculation.
To further clarify the differential expression of genes from MdFLS2-related pathway between the two plant materials, we analyzed the differential expression of related pathway genes in HFTH1 and HF, including those from “Plant-pathogen interaction,” “MAPK signaling pathway-plant,” and salicylic acid pathway in plant hormone signal transduction. Heatmap analysis revealed that the expression levels of MdFLS2-related pathway genes were basically higher in HF than in HFTH1; particularly, expression levels of genes related to salicylic acid pathway were significantly higher in disease-resistant HF than disease-susceptible HFTH1 at the early stage of pathogenic infection, i.e., at 12 HPI and 24 HPI after AAAP inoculation (Figure 5C). These genes may play an essential role in the early defense against pathogenic infection.
3.6 MdFLS2-1 is a positive regulator of resistance to AAAP
The MdFLS2 allele located in the HF alternative genome was named MdFLS2-1, and that located in HFTH1 was named MdFLS2-2. ASE analysis revealed that MdFLS2-1 exhibited a significant expression advantage compared with MdFLS2-2 in HF after AAAP infection and mock inoculation. To identify the allelic variants located in the CDS region of the MdFLS2, we cloned MdFLS2-1 and MdFLS2-2 in HF leaves. MdFLS2-1 and MdFLS2-2 had 10 SNPs in the CDS region and caused difference in five amino acids. By comparing the protein sequence of FLS2 with that of A. thaliana (Gomez-Gomez and Boller, 2000), Citrus sinensis, Prunus dulcis, Rosa chinensis, Trifolium pratense, Medicago truncatula, Glycine max, and P. persica, we discovered that the amino acid sites of the variants were not conserved in several species (Supplementary Figure S2). However, these results are insufficient to indicate whether amino acid variants of MdFLS2-1 and MdFLS2-2 cause functional differences.
We observed that the previously reported MdFLS2 is actually MdFLS2-2 (Liu et al., 2018). To determine the function of MdFLS2-1 and its role in AAAP infection, we constructed the PRI01-MdFLS2-1 vector and performed transient expression experiments in GL-3 apple leaves. RT-qPCR analysis of transgenic leaves revealed that MdFLS2 expression levels were significantly increased in OE-MdFLS2-1 transgenic leaves compared with those in untransformed control leaves (WT) with empty vector (EV) (Figure 6A). AAAP inoculation tests on transgenic leaves and controls revealed that apple leaves overexpressing MdFLS2-1 had significantly less disease incidence than WT and EV plants at 48 HPI (Figures 6C, D). These results suggested that MdFLS2-1 positively regulates resistance to AAAP in apple. RT-qPCR analysis of genes related to salicylic acid pathway in transgenic leaves revealed that the expression levels of marker genes of salicylic acid signaling pathway (MdNPR1, MdTGA3, and MdPR1) were significantly higher in OE-MdFLS2-1 plants compared with WT and EV plants (Figure 6B). These results suggested that the expression of both MdFLS2-1 and MdFLS2-2 is positively correlated with the salicylic acid signaling pathway, and MdFLS2-2 positively regulates the resistance of apple leaves to AAAP. Therefore, we speculated that MdFLS2-1 and MdFLS2-2 play at least similar positive roles in the resistance of apples to fungi.
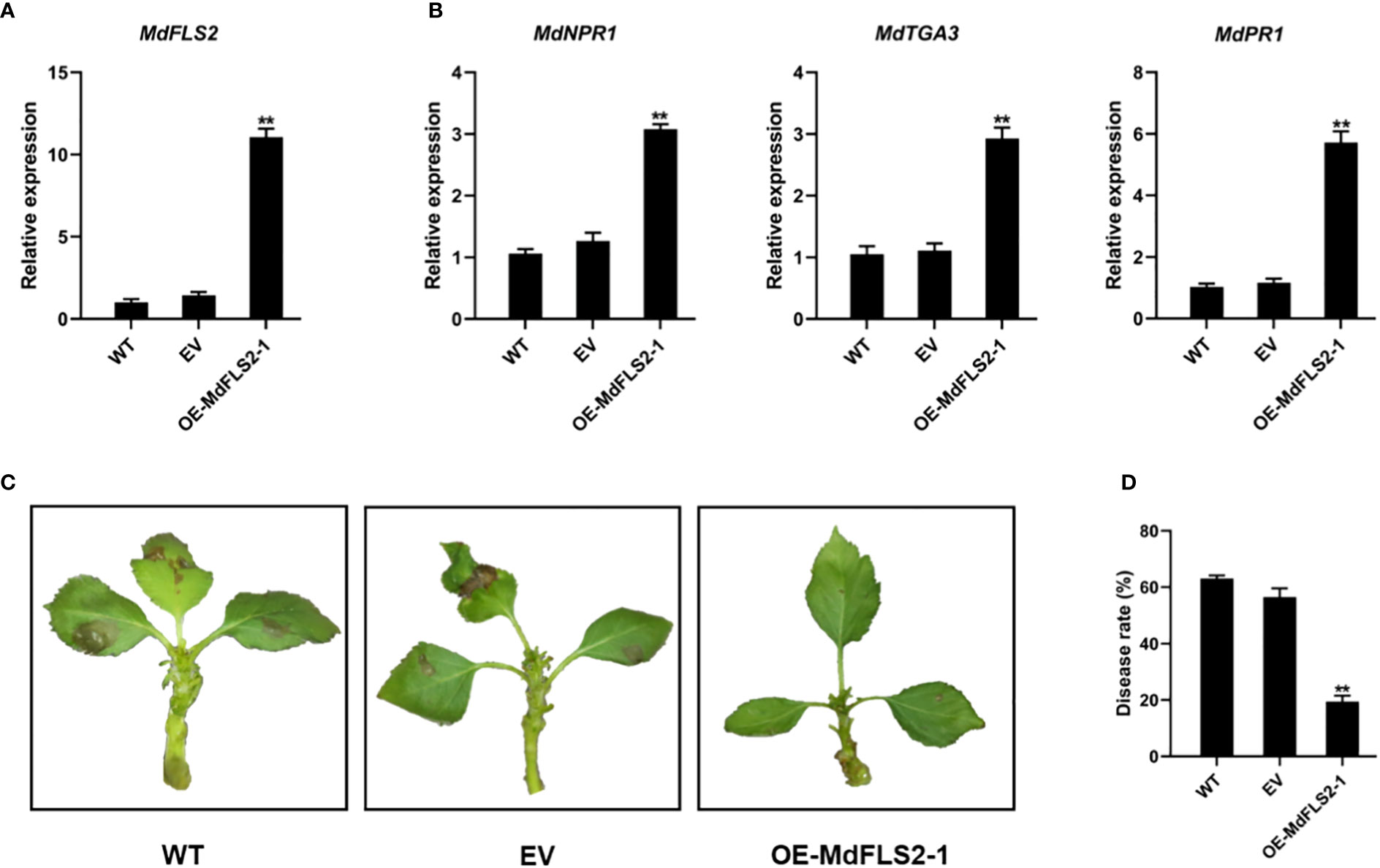
Figure 6 MdFLS2-1 positively regulates resistance of apple to AAAP. RT-qPCR analysis of the expression level of (A) MdFLS2 and (B) salicylate pathway marker genes MdNPR1, MdTGA3, and MdPR1 in leaves of WT, EV, and OE-MdFLS2-1 plants. Asterisks denote Student’s t-tests, significance: **P < 0.01. The mean ± SD of biological triplicates were taken for every value. (C) Symptoms of infection in leaves of WT, EV, and OE-MdFLS2-1 plants at 48 h after AAAP inoculation. (D) Incidence statistics of leaves of WT, EV, and OE-MdFLS2-1 plants at 48 h after AAAP inoculation. Values are the means ± SD of three biological replicates (Student’s t-tests, **P < 0.01).
3.7 TRIM-like sequence as an enhancer of MdFLS2-1 expression
Since in heterozygous diploids, alleles are placed in the same trans regulation background and cellular environment, the effect of trans regulation on allele expression can be temporarily shielded, thus prioritizing cis regulation of variants in the genome (Hill et al., 2021). To investigate the cis-regulatory mechanism causing specific expression of the MdFLS2 allele, we searched the promoter region of the MdFLS2 allele using whole-genome resequencing data of HF. Based on resequencing data and MdFLS2 allele promoter cloning, we identified an 85-bp TRIM-like sequence insertion in the promoter of MdFLS2-1, which was located 235-bp upstream of the translation start codon (Figure 7A). The TRIM-like sequence consists of two almost identical 32-bp TDRs and a 21-bp intermediate sequence (Figure 7A). The TRIM-like sequence is flanked by 3-bp target site duplication that was generated after insertion. The two TDRs of the TRIM-like element have two base differences, named TDR1 and TDR2, respectively. In the promoter region of MdFLS2-2, the TRIM-like sequence undergoes truncation, retaining only a TDR2 sequence (Figure 7A).
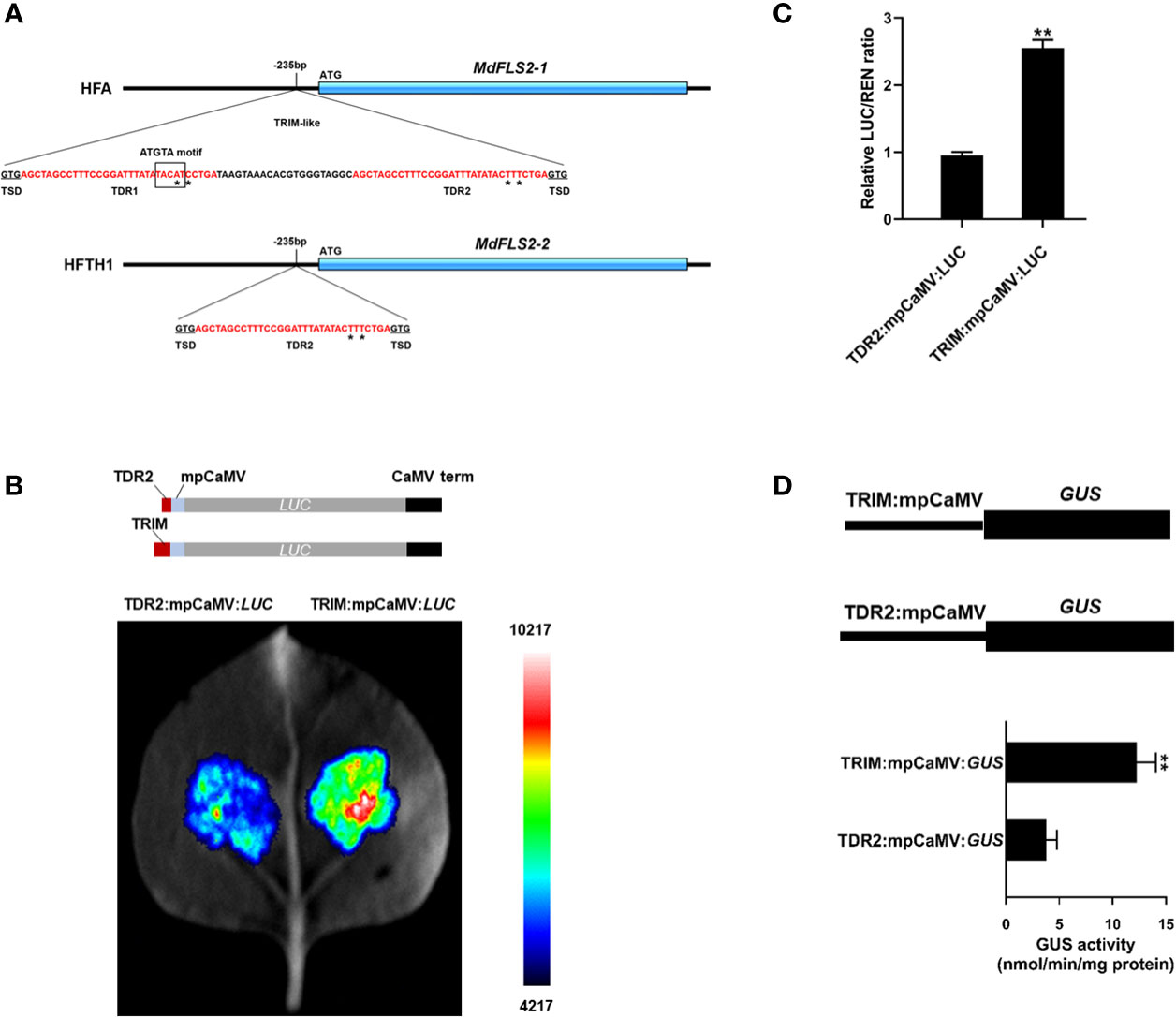
Figure 7 Insertion of the TRIM-like sequence affects the expression level of MdFLS2 allele. (A) Molecular structure of MdFLS2-1 from HF alternative genome (HFA) and of MdFLS2-2 from HFTH1 reference genome with flanking sequence. The TRIM-like sequence contains the following sequence features: TSD (black underlined), TDRs (red font), and intermediate sequence (black font). Base differences between TDR1 and TDR2 are marked with asterisks. The black box indicates the ATGTA motif. (B) Transient expression analysis revealed that TRIM-like sequence insertion could significantly increase the expression level of luciferase. Upper panel represents the TDR2:mpCaMV:LUC (up) and TRIM:mpCaMV:LUC (down) constructs, both containing the minimal promoter from the cauliflower mosaic virus (mpCaMV), luciferase ORF, and cauliflower mosaic virus terminator (CaMV term), to which the TDR2 sequence and TRIM-like sequence are respectively added. Lower panel indicates luciferase assay of Nicotiana benthamiana leaves 72 h after infection with Agrobacterium strains containing TDR2:mpCaMV:LUC (left) and TRIM:mpCaMV:LUC (right). (C) The relative ratio of LUC/REN for both TDR2:mpCaMV:LUC and TRIM:mpCaMV:LUC. Values are the means ± SD of three biological replicates (Student’s t-tests, **P < 0.01). (D) GUS activity in the transiently transformed N. benthamiana leaves with constructs TRIM:mpCaMV:GUS and TDR2:mpCaMV:GUS under normal conditions. The mean ± SD of biological triplicates were taken for every value. Asterisks denote significant differences by Student’s t-test (**P < 0.01).
Several studies have reported that transposon insertion in the promoter region can provide enhancer activity and thus increase the expression level of genes (Wei and Cao, 2016). MdFLS2 alleles with intact transposon insertion in the promoter region had higher expression levels, which is consistent with the pattern of transposon insertions enhancing gene expression. To determine whether the insertion of the TRIM-like element provided enhancer activity, we ligated an 85-bp TRIM-like sequence inserted in the promoter of MdFLS2-1 to the 35S minimal promoter and performed transient experiments in leaves of N. benthamiana to assess the effect of the TRIM-like element on the expression of firefly luciferase gene and β-glucuronidase gene. The results revealed that the TRIM-like sequence led to a significant increase in reporter gene expression, relative to the use of only a TDR2 sequence from the promoter of MdFLS2-2-containing structure (Figures 7B-D). This suggested that the insertion of the TRIM-like sequence in the promoter region of MdFLS2-1 increased the expression level of the gene, causing differential expression of the MdFLS2 allele in HF leaves. This natural variation in the promoter region of the MdFLS2 allele resulted in its significantly higher expression level in HF than in the homozygote HFTH1.
4 Discussion
ASE analysis in heterozygous organisms provides an unprecedented method to compare the effects of cis-regulatory variants on gene expression in the same trans-regulatory background and cellular environment (Bao et al., 2019). Genome-wide ASE analysis was performed by RNA-seq of leaves of HF, an important disease-resistant cultivar, after AAAP infection and mock infection. A large number of ASE genes were identified. These genes form a validated dataset to rapidly identify the cis-regulatory mechanisms affecting the expression levels of these alleles by comparing genomes.
4.1 MdFLS2 is involved in the resistance of apple leaves against AAAP
PTI is an important mode of plant immunity (Bigeard et al., 2015). Some receptor kinases are involved in the resistance of plants to fungi by linking hormone signals (Tang et al., 2017). By comparative transcriptomic analysis, we identified an important receptor kinase, MdFLS2, that exhibited significant differences in expression between HF and HFTH1 after AAAP inoculation. There was a consistent bias of ASE for MdFLS2 in HF after AAAP infection and mock inoculation, and the expression level of the HF-derived allele MdFLS2-1 was significantly higher than that of the HFTH1-derived allele MdFLS2-2. Previous studies have reported that MdFLS2-2 is involved in the defense response of apple to fungi by enhancing the expression levels of the genes related to salicylic acid pathway. In the transcriptome data, we observed an association of MdFLS2 with the genes related to salicylic acid pathway, with their higher expression levels in the AAAP-resistant HF compared with AAAP-susceptible HFTH1 at the early stage of AAAP infection; the genes down-regulated at a later stage. Related studies have indicated that an intact salicylic acid pathway is essential in the defense of Solanum tuberosum against Alternaria solani (Brouwer et al., 2020). Studies on the defense of Chrysanthemum morifolium against Alternaria (Liu et al., 2020; Zhao et al., 2020) have reported that the salicylic acid pathway plays an important role in the early response of plants against pathogenic attacks. Transient overexpression of MdFLS2-1 could improve resistance of apple leaves to AAAP (Figures 6C, D). At the same time, we observed that overexpression of MdFLS2-1 was accompanied by increased expression levels of genes related to salicylic acid pathway (Figures 6A, B). These results predicted a conserved and active role of salicylic acid pathway in the defense of various plants against various pathogenic variants of Alternaria. On the other hand, MdFLS2-1 and MdFLS2-2 could regulate the level of salicylic acid signaling in apple leaves during resistance to fungi.
Studies on Arabidopsis indicated that AtFLS2 receptor kinase can function in complex with various proteins, including AtACD6, which can form a complex with AtFLS2 to enhance salicylic acid signaling (Tateda et al., 2014), and AtACD6 and AtFLS2 can promote the activation of salicylic acid signaling even in the absence of PAMPs (Tateda et al., 2015). Significant differences in AtACD6 homolog MdACD6 were reflected at 12 HPI in susceptible varieties, which is consistent with the expression trend of genes related to salicylic acid pathway (Figure 5C). We speculated that a similar mechanism may exist in apple, where MdFLS2 regulates salicylic acid signaling through MdACD6. However, further studies are needed to elucidate this mechanism.
4.2 A TRIM-like insertion in MdFLS2 allele promoter causes its ASE
It is widely reported that insertion of transposable elements in the promoter region affects expression level of the genes (Mao et al., 2015; Xia et al., 2017; Niu et al., 2022). In this study, we identified a TRIM-like sequence inserted in the promoter region of MdFLS2-1, which was experimentally demonstrated to have enhancer activity. In contrast, promoter region of the other allele MdFLS2-2 only had a TDR2 sequence of the TRIM-like element; thus, the expression level of MdFLS2-2 was significantly lower than that of MdFLS2-1. Based on these results, we developed a model to explain the difference in the sensitivity of HF and HFTH1 to AAAP. After AAAP infection, HF containing MdFLS2-1 exhibited high expression levels of genes related to salicylic acid pathway, which in turn led to resistance of HF to AAAP. In contrast, HFTH1 with lower expression levels of MdFLS2-2 exhibited susceptibility to AAAP (Figure 8).
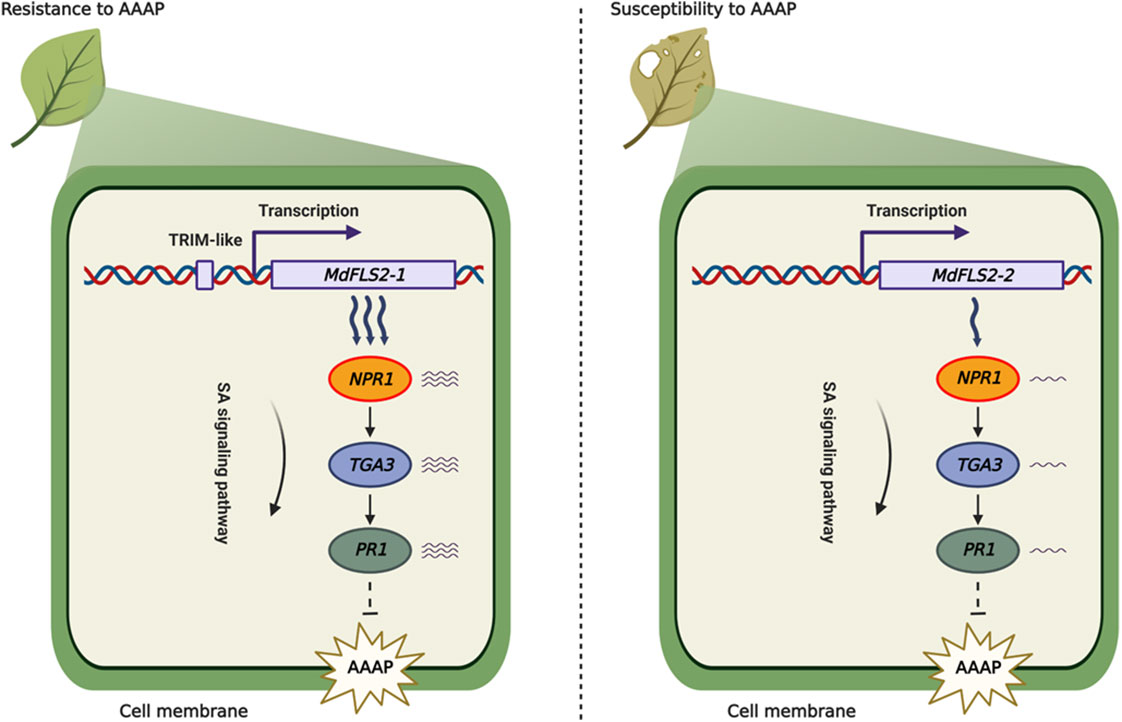
Figure 8 Model for the role of a TRIM-like sequence insertion in the MdFLS2-1 promoter region in regulating resistance of apple to Alternaria blotch disease. MdFLS2 allele is involved in the resistance of apple to Alternaria blotch disease via increasing the expression level of genes related to salicylic acid pathway (NPR1, TGA3, and PR1). Insertion of a TRIM-like sequence into the MdFLS2-1 promoter region enhances MdFLS2-1 expression to improve resistance of apple to Alternaria blotch disease. Arrow: activation; Bar: repression.
Some studies have reported that insertion of transposable elements can provide effective binding sites for transcription factors (Sahebi et al., 2018). An ATGTA motif is present in the TDR1 sequence of the TRIM-like element. Due to mutations during evolution, the ATGTA motif is not present in the TDR2 sequence. Previous studies have identified the ATGTA motif as a binding site for the MdEIL1 transcription factor in apple (An et al., 2018). Interestingly, AtEIL1, the homolog of MdEIL1 in Arabidopsis, can bind to the promoter region of AtFLS2 to regulate its expression level (Boutrot et al., 2010). In apples, this regulatory mechanism is expected to be further revealed in future studies. Overall, our study provided new insights into the molecular mechanism of resistance to AAAP by apple and provided a reference for breeding with disease resistance varieties.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under BioProject ID: PRJNA896728 and PRJNA897033.
Author contributions
LZ, PC and ZL initiated the project and designed the study. ZL, KL, CJ, AY, JY performed biological experiments. ZL performed data analysis and wrote the manuscript. XH, CZ optimized experimental protocol. LZ, and PC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2021-RIP-02) and the National Key Research and Development Program of China (2019YFD1001403).
Acknowledgments
We thank Dr. Zhihong Zhang from Shenyang Agricultural University for providing tissue-cultured GL-3 plants, and Dr. Jia-Long Yao from The New Zealand Institute for Plant & Food Research Limited for providing the pGreenII0800-LUC vector.
Conflict of interest
The handling editor LC declared a shared affiliation with the Author/s ZL & LZ at the time of review Chinese Academy of Agricultural Sciences CAAS, Beijing, China
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1090621/full#supplementary-material
References
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 178, 808–823. doi: 10.1104/pp.18.00068
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bai, T. T., Xie, W. B., Zhou, P. P., Wu, Z. L., Xiao, W. C., Zhou, L., et al. (2013). Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with fusarium oxysporum f. sp. cubense tropical race 4. PLoS One 8, e73945. doi: 10.1371/journal.pone.0073945
Bao, Y., Hu, G., Grover, C. E., Conover, J., Yuan, D., Wendel, J. F. (2019). Unraveling cis and trans regulatory evolution during cotton domestication. Nat. Commun. 10, 5399. doi: 10.1038/s41467-019-13386-w
Bigeard, J., Colcombet, J., Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Boutrot, F., Segonzac, C., Chang, K. N., Qiao, H., Ecker, J. R., Zipfel, C., et al. (2010). Direct transcriptional control of the arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. United States America 107, 14502–14507. doi: 10.1073/pnas.1003347107
Brouard, J. S., Schenkel, F., Marete, A., Bissonnette, N. (2019). The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J. Anim. Sci. Biotechnol. 10. doi: 10.1186/s40104-019-0359-0
Brouwer, S. M., Odilbekov, F., Burra, D. D., Lenman, M., Hedley, P. E., Grenville-Briggs, L., et al. (2020). Intact salicylic acid signalling is required for potato defence against the necrotrophic fungus alternaria solani. Plant Mol. Biol. 104, 1–19. doi: 10.1007/s11103-020-01019-6
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen-Hui, Q., Xian-Yan, Z., Han, J., Hai-Tao, L., Yong-Xu, W., Da-Gang, H., et al. (2018). Molecular cloning and functional identification of an apple flagellin receptor MdFLS2 gene. J. Integr. Agric. 17, 2694–2703. doi: 10.1016/S2095-3119(18)62009-X
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6, 80–92. doi: 10.4161/fly.19695
Danna, C. H., Millet, Y. A., Koller, T., Han, S. W., Bent, A. F., Ronald, P. C., et al. (2011). The arabidopsis flagellin receptor FLS2 mediates the perception of xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. U.S.A. 108, 9286–9291. doi: 10.1073/pnas.1106366108
David, L., Kang, J., Chen, S. (2019). Targeted metabolomics of plant hormones and redox metabolites in stomatal immunity. Methods Mol. Biol. 2085, 79–92. doi: 10.1007/978-1-0716-0142-6_6
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Filajdic, N., Sutton, T. B. (1991). Identification and distribution of alternaria mali on apples in north Carolina and susceptibility of different varieties of apples to alternaria blotch. Plant Dis. 75, 1045–1048. doi: 10.1094/PD-75-1045
Gao, Z. Y., Li, H., Yang, X. Y., Yang, P. F., Chen, J. M., Shi, T. (2021). Biased allelic expression in tissues of F1 hybrids between tropical and temperate lotus (Nelumbo nuicfera). Plant Mol. Biol. 106, 207–220. doi: 10.1007/s11103-021-01138-8
Gomez-Gomez, L., Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
He, X., Meng, H., Wang, H., He, P., Chang, Y., Wang, S., et al. (2022). Quantitative proteomic sequencing of f 1 hybrid populations reveals the function of sorbitol in apple resistance to botryosphaeria dothidea. Hortic. Res. 9, uhac115. doi: 10.1093/hr/uhac115
Hill, M. S., Vande Zande, P., Wittkopp, P. J. (2021). Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet. 22, 203–215. doi: 10.1038/s41576-020-00304-w
Hou, Y., Yu, X., Chen, W., Zhuang, W., Wang, S., Sun, C., et al. (2021). MdWRKY75e enhances resistance to alternaria alternata in malus domestica. Hortic. Res. 8, 225. doi: 10.1038/s41438-021-00701-0
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: betaâ glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kong, Q., Sun, T., Qu, N., Ma, J., Li, M., Cheng, Y. T., et al. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171, 1344–1354. doi: 10.1104/pp.15.01954
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Li, H., Durbin, R.. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, X., Liang, Y., Zhang, W., Hou, Y., Feng, S., Qiu, H., et al. (2018). MdFLS2 recognizes bacterial flagellin flg22 and enhances immune resistance against apple ring rot causal fungi in arabidopsis fls2 mutant. Acta Hortic. Sin. 45 (5), 827–844. doi: 16420/j.issn.0513-353x.2017-0687
Liu, Y., Xin, J., Liu, L., Song, A., Guan, Z., Fang, W., et al. (2020). A temporal gene expression map of chrysanthemum leaves infected with alternaria alternata reveals different stages of defense mechanisms. Hortic. Res. 7, 23. doi: 10.1038/s41438-020-0245-0
Liu, Y., Yang, T., Lin, Z., Gu, B., Xing, C., Zhao, L., et al. (2019). A WRKY transcription factor PbrWRKY53 from pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. doi: 10.1111/pbi.13099
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta biotechnology c) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, D., Lin, W., Gao, X., Wu, S., Cheng, C., Avila, J., et al. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442. doi: 10.1126/science.1204903
Mao, H. D., Wang, H. W., Liu, S. X., Li, Z., Yang, X. H., Yan, J. B., et al. (2015). A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6 8326. doi: 10.1038/ncomms9326
Meng, D., Li, C., Park, H. J., Gonzalez, J., Wang, J., Dandekar, A. M., et al. (2018). Sorbitol modulates resistance to alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 30, 1562–1581. doi: 10.1105/tpc.18.00231
Meng, X., Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Moriya, S., Terakami, S., Okada, K., Shimizu, T., Adachi, Y., Katayose, Y., et al. (2019). Identification of candidate genes responsible for the susceptibility of apple (Malus x domestica borkh.) to alternaria blotch. BMC Plant Biol. 19, 132. doi: 10.1186/s12870-019-1737-7
Ngou, B. P. M., Ahn, H. K., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Ngou, B. P. M., Jones, J. D. G., Ding, P. (2022). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Niu, C., Jiang, L., Cao, F., Liu, C., Guo, J., Zhang, Z., et al. (2022). Methylation of a MITE insertion in the MdRFNR1-1 promoter is positively associated with its allelic expression in apple in response to drought stress. Plant Cell 34, 3983–4006. doi: 10.1093/plcell/koac220
Nomura, K., Mecey, C., Lee, Y. N., Imboden, L. A., Chang, J. H., He, S. Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 10774–10779. doi: 10.1073/pnas.1103338108
Parkhomchuk, D., Borodina, T., Amstislavskiy, V., Banaru, M., Hallen, L., Krobitsch, S., et al. (2009). Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123. doi: 10.1093/nar/gkp596
Pavy, N., Boyle, B., Nelson, C., Paule, C., Giguere, I., Caron, S., et al. (2008). Identification of conserved core xylem gene sets: conifer cDNA microarray development, transcript profiling and computational analyses. New Phytol. 180, 766–786. doi: 10.1111/j.1469-8137.2008.02615.x
Reim, S., Rohr, A.-D., Winkelmann, T., Weiß, S., Liu, B., Beerhues, L., et al. (2019). Genes involved in stress response and especially in phytoalexin biosynthesis are upregulated in four malus genotypes in response to apple replant disease. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01724
Ren, J., Mao, J., Zuo, C., Calderón-Urrea, A., Dawuda, M. M., Zhao, X., et al. (2017). Significant and unique changes in phosphorylation levels of four phosphoproteins in two apple rootstock genotypes under drought stress. Mol. Genet. Genomics 292, 1307–1322. doi: 10.1007/s00438-017-1348-7
Robinson, M. D., Mccarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sahebi, M., Hanafi, M. M., Van Wijnen, A. J., Rice, D., Raffi, M. Y., Azizi, P., et al. (2018). Contribution of transposable elements in the plant's genome. Gene 665, 155–166. doi: 10.1016/j.gene.2018.04.050
Sarowar, S., Alam, S. T., Makandar, R., Lee, H., Trick, H. N., Dong, Y., et al. (2019). Targeting the pattern-triggered immunity pathway to enhance resistance to fusarium graminearum. Mol. Plant Pathol. 20, 626–640. doi: 10.1111/mpp.12781
Shao, L., Xing, F., Xu, C., Zhang, Q., Che, J., Wang, X., et al. (2019). Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. U.S.A. 116, 5653–5658. doi: 10.1073/pnas.1820513116
Shi, Q., Febres, V. J., Jones, J. B., Moore, G. A. (2016). A survey of FLS2 genes from multiple citrus species identifies candidates for enhancing disease resistance to xanthomonas citri ssp. citri. Hortic. Res. 3, 16022. doi: 10.1038/hortres.2016.22
Sun, X., Jiao, C., Schwaninger, H., Chao, C. T., Ma, Y., Duan, N., et al. (2020). Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 52, 1423–1432. doi: 10.1038/s41588-020-00723-9
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. doi: 10.1126/science.1243825
Tang, D., Wang, G., Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tateda, C., Zhang, Z. Q., Greenberg, J. T. (2015). Linking pattern recognition and salicylic acid responses in arabidopsis through ACCELERATED CELL DEATH6 and receptors. Plant Signaling Behav. 10. doi: 10.1080/15592324.2015.1010912
Tateda, C., Zhang, Z., Shrestha, J., Jelenska, J., Chinchilla, D., Greenberg, J. T. (2014). Salicylic acid regulates arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26, 4171–4187. doi: 10.1105/tpc.114.131938
Tian, Y., Thrimawithana, A., Ding, T., Guo, J., Gleave, A., Chagne, D., et al. (2022). Transposon insertions regulate genome-wide allele-specific expression and underpin flower colour variations in apple (Malus spp.). Plant Biotechnol. J. 20, 1285–1297. doi: 10.1111/pbi.13806
Turra, D., Segorbe, D., Di Pietro, A. (2014). Protein kinases in plant-pathogenic fungi: Conserved regulators of infection. Annu. Rev. Phytopathol. 52, 267–288. doi: 10.1146/annurev-phyto-102313-050143
Wang, H., Dong, B., Jiang, J., Fang, W., Guan, Z., Liao, Y., et al. (2014). Characterization of in vitro haploid and doubled haploid chrysanthemum morifolium plants via unfertilized ovule culture for phenotypical traits and DNA methylation pattern. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00738
Wei, L., Cao, X. (2016). The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci. China Life Sci. 59, 24–37. doi: 10.1007/s11427-015-4993-2
Xia, C., Zhang, L., Zou, C., Gu, Y., Duan, J., Zhao, G., et al. (2017). A TRIM insertion in the promoter of Ms2 causes male sterility in wheat. Nat. Commun. 8, 15407. doi: 10.1038/ncomms15407
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhang, X., Chen, S., Shi, L., Gong, D., Zhang, S., Zhao, Q., et al. (2021). Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant camellia sinensis. Nat. Genet. 53, 1250–1259. doi: 10.1038/s41588-021-00895-y
Zhang, M., Huang, S., Gao, Y., Fu, W., Qu, G., Zhao, Y., et al. (2020a). Fine mapping of a leaf flattening gene bralcm through BSR-seq in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Sci. Rep. 10, 13924. doi: 10.1038/s41598-020-70975-2
Zhang, L., Hu, J., Han, X., Li, J., Gao, Y., Richards, C. M., et al. (2019). A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 10, 1494. doi: 10.1038/s41467-019-09518-x
Zhang, Q., Ma, C., Zhang, Y., Gu, Z., Li, W., Duan, X., et al. (2018). A single-nucleotide polymorphism in the promoter of a hairpin RNA contributes to alternaria alternata leaf spot resistance in apple (Malus x domestica). Plant Cell 30, 1924–1942. doi: 10.1105/tpc.18.00042
Zhang, N., Pombo, M. A., Rosli, H. G., Martin, G. B. (2020b). Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to pseudomonas syringae. Plant Physiol. 183, 1869–1882. doi: 10.1104/pp.20.00144
Zhang, C. X., Tian, Y., Cong, P. H. (2015). Proteome analysis of pathogen-responsive proteins from apple leaves induced by the alternaria blotch alternaria alternata. PLoS One 10, e0122233. doi: 10.1371/journal.pone.0122233
Zhao, X., Song, L., Jiang, L., Zhu, Y., Gao, Q., Wang, D., et al. (2020). The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus alternaria sp. Hortic. Res. 7, 80. doi: 10.1038/s41438-020-0297-1
Zhu, L., Ni, W., Liu, S., Cai, B., Xing, H., Wang, S. (2017). Transcriptomics analysis of apple leaves in response to alternaria alternata apple pathotype infection. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00022
Zipfel, C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. doi: 10.1016/j.coi.2007.11.003
Keywords: alternaria alternata apple pathotype, malus domestica borkh., allele-specific expression, MdFLS2, TRIM element
Citation: Liang Z, Liu K, Jiang C, Yang A, Yan J, Han X, Zhang C, Cong P and Zhang L (2022) Insertion of a TRIM-like sequence in MdFLS2-1 promoter is associated with its allele-specific expression in response to Alternaria alternata in apple. Front. Plant Sci. 13:1090621. doi: 10.3389/fpls.2022.1090621
Received: 05 November 2022; Accepted: 09 December 2022;
Published: 29 December 2022.
Edited by:
Lin Chen, Institute of Animal Sciences (CAAS), ChinaReviewed by:
Mingjun Li, Northwest A&F University, ChinaLiangju Wang, Nanjing Agricultural University, China
Copyright © 2022 Liang, Liu, Jiang, Yang, Yan, Han, Zhang, Cong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyi Zhang, zhangliyi@caas.cn; Peihua Cong, congph@163.com
 Zhaolin Liang
Zhaolin Liang Kai Liu1,2
Kai Liu1,2 Liyi Zhang
Liyi Zhang