- 1School of Grassland Science, Beijing Forestry University, Beijing, China
- 2Inner Mongolia M-Grass Ecology And Environment (Group) Co., Ltd, Hohhot, China
- 3The University of Western Australia (UWA) Institute of Agriculture, and University of Western Australia School of Agriculture and Environment, The University of Western Australia, Perth, WA, Australia
Introduction: Alfalfa (Medicago sativa) is a kind of high quality leguminous forage species, which was widely cultivated in the world. Leaf senescence is an essential process in plant development and life cycle. Here, we reported the isolation and functional analysis of an alfalfa SENESCENCE-ASSOCIATED GENE113 (MsSAG113), which belongs to the PP2C family and mainly plays a role in promoting plant senescence.
Methods: In the study, Agrobacterium-mediated, gene expression analysis, next generation sequencing, DNA pull-down, yeast single hybridization and transient expression were used to identify the function of MsSAG113 gene.
Results: The MsSAG113 gene was isolated from alfalfa, and the transgenic plants were obtained by Agrobacterium-mediated method. Compared with the wildtype, transgenic plants showed premature senescence in leaves, especially when cultivated under dark conditions. Meanwhile, application of exogenous hormones ABA, SA, MeJA, obviously acclerated leaf senescence of transgenic plants. Furthermore, the detached leaves from transgenic plants turned yellow earlier with lower chlorophyll content. Transcriptome analysis identified a total of 1,392 differentially expressed genes (DEGs), involving 13 transcription factor families. Of which, 234 genes were related to phytohormone synthesis, metabolism and transduction. Pull-down assay and yeast one-hybrid assay confirmed that alfalfa zinc finger CCCH domain-containing protein 39 (MsC3H-39) could directly bind the upstream of MsSAG113 gene. In conclusion, the MsSAG113 gene plays a crucial role in promoting leaf senescence in alfalfa via participating in the hormone regulatory network.
Discussion: This provides an essential basis for further analysis on the regulatory network involving senescence-associated genes in alfalfa.
Introduction
Leaf senescence is the late stage in plant development, during which leaves change from pale yellow to yellow. During senescence, nutrients from old leaves are diverted to other parts of the plant to initiate new tissue, such as new leaves or developing organs and seeds (Jing et al., 2002). As a result, plant leaf cells undergo a series of changes in morphology, structure, and metabolism leading to leaf abscission, a process known as senescence syndrome (Bleecker and Patterson, 1997). Although senescence is an age-dependent process, it is often triggered by a range of biological and abiotic stresses (Gan and Amasino, 1997). External factors, such as environmental stress and long-term darkness may also induce leaf senescence, which is considered as a self-protection mechanism to cope with the stress to prevent crop yield or quality reduction (Chao et al., 2018).
Recently, the study of plant leaf senescence has been favored by a large number of researchers, and the related molecular mechanisms have also made good progress. In senescent leaves, the senescence-associated genes (SAG) are expressed, which are considered as a marker of cellular senescence (Wistrom and Villeponteau, 1992). Senescence associated genes have been cloned and identified in many plants. OsSAG12-2 codes for a functional protease that negatively regulates stress-induced cell death in rice (Singh et al., 2016). A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis thaliana, chemically induced overexpression of SAG101 causing premature senescence in both attached and isolated leaves of transgenic Arabidopsis plants (He and Gan, 2002). The senescence associated gene AhSAG under aluminum stress promoted senescence in transgenic plants and further induced or promoted the occurrence of programmed cell death (PCD) (Zhan et al., 2013). Growing isolated leaves in the dark can induce leaf yellowing and reduce chlorophyll content. Therefore, isolated leaves are used as models to study leaf senescence (Liebsch and Keech, 2016).
In addition, plant hormones play an important role in plant growth, development and metabolism. The senescence process is also regulated by various plant hormones such as salicylic acid (SA), abscisic acid (ABA), ethylene, jasmonic acid (JA), cytokinins, and auxin (He et al., 2002). Studies have shown that the ABA receptor gene PLY9 has a role in aging old leaves and inhibiting the growth of young tissues (Zhao et al., 2016). There are many reports on the complex mechanisms of gene interactions regulating leaf senescence. RhHB1 is a gene strongly over-expressed in senescent petals, which can be induced expression by ABA or ethylene. It was shown that ABA or ethylene treatment could induce increased RhHB1 expression, while gibberellin GA3 treatment delayed this process (Lü et al., 2014). Leaf senescence is usually accompanied by a decrease in cytokinin content, but the increase of cytokinin content can alleviate leaf senescence (Balibrea Lara et al., 2004). In 2015, Surya et al. found that the expression of converted structural genes (IPT) encoding the cytokinin biosynthetic enzyme isoprenyl transferase delayed leaf senescence of the transgenic rapeseed plants under controlled environments (Kant et al., 2015).
Protein phosphatases of the PP2C type are monomeric enzymes present in prokaryotes and eukaryotes. PP2C is the largest family of protein phosphatases in plant species. Plant PP2C family proteins play important roles in transcriptional regulatory pathways such as stress response, ABA synthesis, and developmental signaling (Schweighofer et al., 2004). The AHG1 gene of the PP2C protein family has special functions during seed development and germination (Nishimura et al., 2007). Two phosphatases 2C genes, ABI1 and ABI2, negatively feedback the ABA signaling pathway (Merlot et al., 2001). SAG113 belongs to the PP2C family and was founded to localize to the cis-Golgi as a negative regulator of ABA signaling, specifically inhibiting stomatal closure, further leading to senescence and eventual desiccation (Zhang et al., 2012).
Alfalfa (Medicago sativa)is an important leguminous herbage speceis, which is rich in protein, trace elements and vitamins. It has been widely used in the world because of its important ecological and economic values (Hrbáčková et al., 2020). Therefore, it is important to study the senescence of alfalfa leaves to improve its application value.
Although senescence associated genes have been widely reported in many different plants, only a few studies involved in alfalfa. Therefore, in this study the role of a senescence associated gene, MsSAG113 was explored in the senescence regulatory network, aiming to lay a foundation for improving alfalfa quality by genetic engineering methods.
Materials and methods
Plant materials and growth conditions
Seeds of alfalfa (Medicago sativa) (cv. Zhongmu No. 1) were vernalized at 4°C for 4 d and cultivated under 16 h light (25°C) and 8 h dark (23°C) until root development. The seedlings were transferred to pots filled with mixed peat, vermiculite and perlite, and grown under 16 h light (25°C) and 8 h dark (23°C). It is difficult to control the developmental senescence, thus many senescence-related studies have been performed on isolated leaves in dark cultures or attached leaves in dark cultures alone (Pruzinská et al., 2005). For phenotype analysis of detached leaves, alfalfa leaves of 3-month-old wild type (WT) and MsSAG113 transgenic plants were collected and immersed in 2-(N-Morpholino) ethanesulfonic acid hydrate (MES) buffer (3mM, pH 5.8) in the dark or with different plant hormone treatments.
Subcellular localization
The SAG113-GFP-F/R primers (Supplementary Table S1) were used for the construction of subcellular localization vector 35S::MsSAG113:GFP and the 35S::GFP vector was used as a control. The recombinant plasmid was transferred into EHA105 Agrobacterium tummefaciens and injected into tobacco plants and cultured for subcellular localization. The position of green fluorescent protein (GFP) signal in the cells was observed with a confocal microscope (Leica TCS SP8, Leica Microsystems).
Exogenous hormone treatments and gene expression
Two-month-old alfalfa plants were selected and sprayed with hormones involving 50 μmol/L ABA, 0.5 mmol/L SA, 10 μmol/L MeJA, 10 μmol/L 6BAP, respectively. The leaves were collected at 0 min, 10 min, 20 min, 30 min, 1 h, 3 h, 6 h, 9 h, and 12 h for expression analysis. Meanwhile, leaves at different developmental stage (1 month-old, 2 month-old and 3 month-old) were harvested for MsC3H-39 gene expression analysis. Total RNA from the leaf samples of different treatments was extracted by a Plant RNA Kit (Omega Bio-tek, Inc., USA), and cDNA was synthesize using PrimeScript™ RT reagent Kit (TaKaRa, Japan) for real-time quantitative RT-PCR (qRT-PCR) with primer pair of SAG113-RT-F/R and C3H-39-RT-F/R (Supplementary Table S1). Expression of MsSAG113 and MsC3H-39 was calculated by the 2-ΔΔCT method with three biological replicates (Schmittgen and Livak, 2008).
Transformation of plants
The sequence of MsSAG113 (GenBank Access No.: KT592510) was obtained from NCBI (National Center for Biotechnology Information) and analyzed in MEGA version 6.0. Two primers for SAG113-F/R (Supplementary Table S1) were used to clone the coding domain sequence (CDS) from alfalfa. Primer 3302Y-SAG113-F/R (Supplementary Table S1) was used to obtain plant expression vector 35S::SAG113. The expression vector was then transformed into alfalfa mediated by Agrobacterium transformation (Hoekema et al., 1983). Three different lines (S1, S6 and S7) with high expression levels were selected for further experiments.
Measurement of endogenous hormones and chlorophyll contents
Two-month-old transgenic and WT leaves with similar state were selected and endogenous hormones were detected by enzyme-linked immunoassay (ELISA) as described previously (Engvall and Perlmann, 1971). Chloroform was used to extract the chlorophyll content as previously reported (Matile et al., 1988). The absorbance of the extract was measured by spectrophotometer, and the content of each pigment in the extract was calculated. All samples were repeated three times.
Next generation sequencing and analysis
Mature leaves of three independent lines were selected for transcriptiome sequencing and DEGs were identified by DESeq R package (version 1.18.0) (Robinson et al., 2009) with parameters: adjusted p-value <0.05 and |log2FC|≥1. Gene Ontology (GO) analysis of DEGs between samples was performed using GOseq R package (version: Release 2.12) (Young et al., 2010). KOBAS software (version: 2.0) was used for functional analysis of DEGs in the KEGG pathway (Mao et al., 2005).
DNA pull-down
The potential cis elements of upstream of MsSAG113 were investigated via the PlantCARE website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Based on the predicted result, one pair of primers SAG113-bition-F/R labeled with biotin (Supplementary Table S1) was synthesized and used for DNA pull-down. The alfalfa nuclear proteins were extracted by a nuclear protein extraction kit (Solarbio, China) and the extraction was incubated with the target sequence of MsSAG113 containing biotin. Compound was pulled down with streptavidin magnetic beads (Beyotime, China) according to the manual. The compound products were washed five times, separated by SDS-PAGE, and identified by mass spectrometry.
Yeast one-hybrid
Two pairs of primers, pAbAi-SAG113pro-F/R and pGADT7-C3H-39-F/R (Supplementary Table S1) were used for construction of vectors for yeast one-hybrid. Recombinant plasmids pAbAi-MsSAG113pro and pGADT7-MsC3H-39 were co-transformed into yeast Y1H strain. Different concentrations of aureobasidin (AbA) (TaKaRa, Japan) were used to screen a suitable concentration for yeast one-hybrid, and transformed yeast cells were grown on SD/-Leu/AbA medium for further investigation.
Transient expression of MsC3H-39
The MsC3H CDS was cloned with MsC3H-39-F/R primers (Supplementary Table S1) and ligased into 3302Y vector, resulting in plant expression vector. The recombinant plasmids were transiently transferred into 2 month-old alfalfa leaves with empty vector 3302Y as controls and the leaf phenotype was observed. After 48h of dark treatment, total RNA was isolated from each leaf sample, and the expression levels of MsSAG113 was analyzed with qRT-PCR as described above.
Results
Location and expression pattern of MsSAG113
Based on the Cell-PLoc package and UniProt analysis, MsSAG113 was predicted to be a nuclear-localized protein. The expression of MsSAG113-GFP fusion protein in tobacco leaves was captured showing strong GFP signal in the nucleus and cytoplasm (Figure 1), which indicated subcellular localization of MsSAG113 protein was in nucleus and cytoplasm.
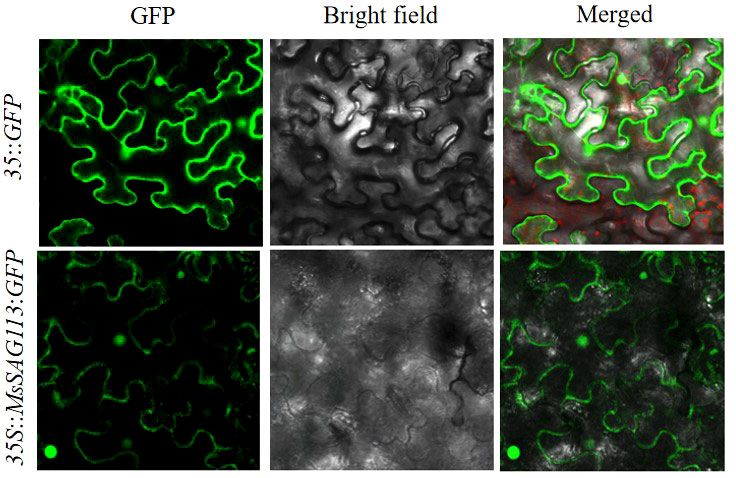
Figure 1 Subcellular localization of MsSAG113 in the nucleus and cytoplasm detected by GFP signal. Scale bars =25 μm.
All hormones used in this study had significant effects on the expression of MsSAG113,which was increased rapidly at first and then decreased with the passage of time in ABA,SA and MeJA treated plants. The expression of MsSAG113 reached its peak at 20 min after spraying with ABA and MeJA hormones (Figures 2A–C). It indicated that the expression of MsSAG113 gene was regulated by ABA, MeJA or SA in the short term. The expression of MsSAG113 in plants treated with 6BAP showed a down-regulated trend within 6 h and then recovered rapidly at 9 h (Figure 2D). These results indicated that MsSAG113 was involved in short-term regulatory of plant hormone network.
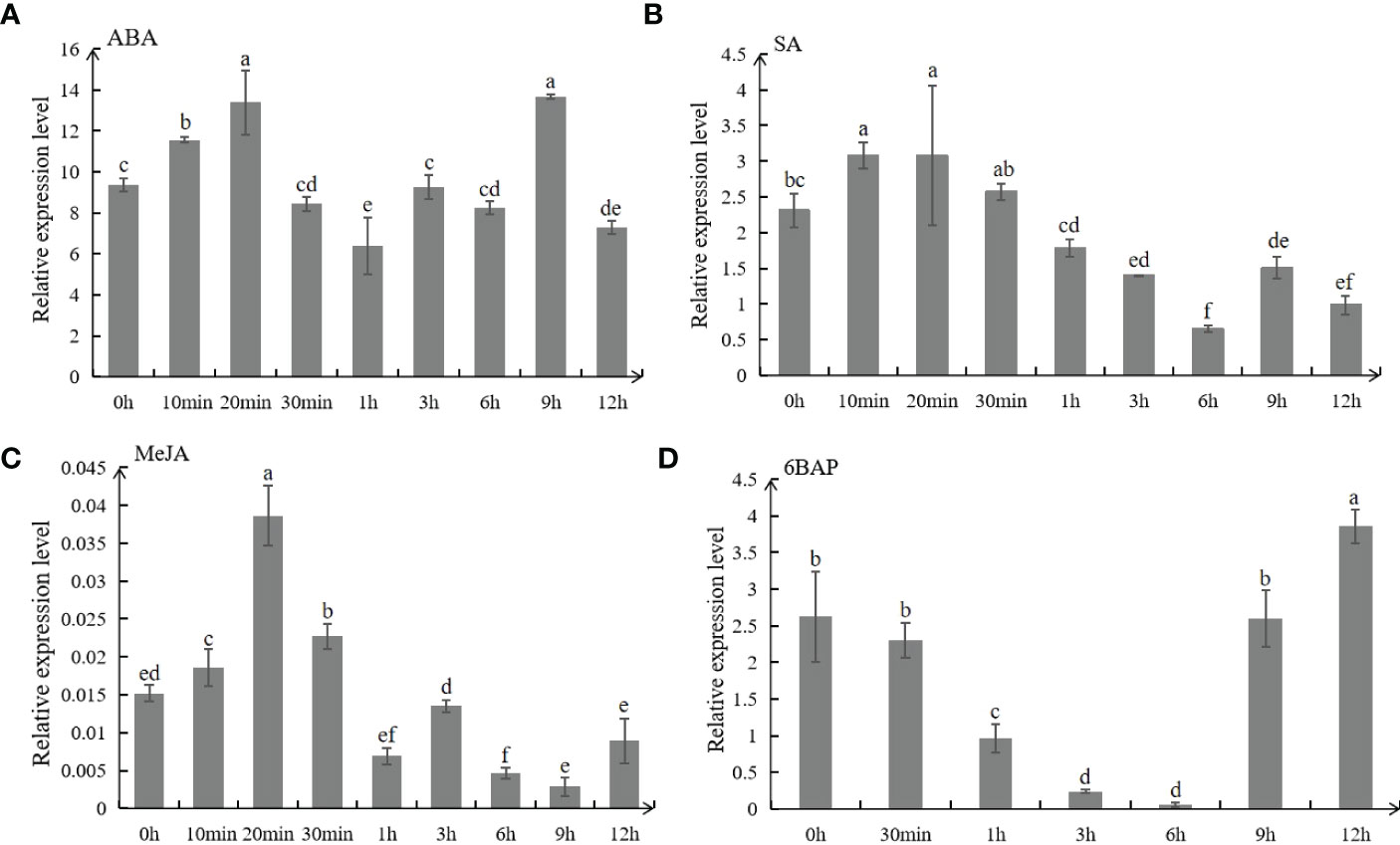
Figure 2 Expression analysis of MsSAG113 in alfalfa by qRT-PCR. MsSAG113 under different hormones treatments: 50 μM ABA (A), 0.5 mM SA (B), 10 μM MeJA (C), and 10 μM 6BAP (D). The values are means ± SD (n=3). In graphs, bars with different lowercase letters indicate significant difference (p<0.05).
The role of MsSAG113 in premature leaf senescence
Through the analysis of transgenic plants S1, S6 and S7, we found that under normal condition, transgenic plants exhibited early senescence phenotype and leaves turned yellow much earlier than those of WT plants. All three transgenic samples showed significantly higher expression levels of MsSAG113 than those in WT plants (Figure 3B). Meanwhile, the chlorophyll contents in transgenic plants were drastically lower than those in WT (Figure 3C). These results indicated that MsSAG113 promoted early senescence and chlorophyll degradation in alfalfa.
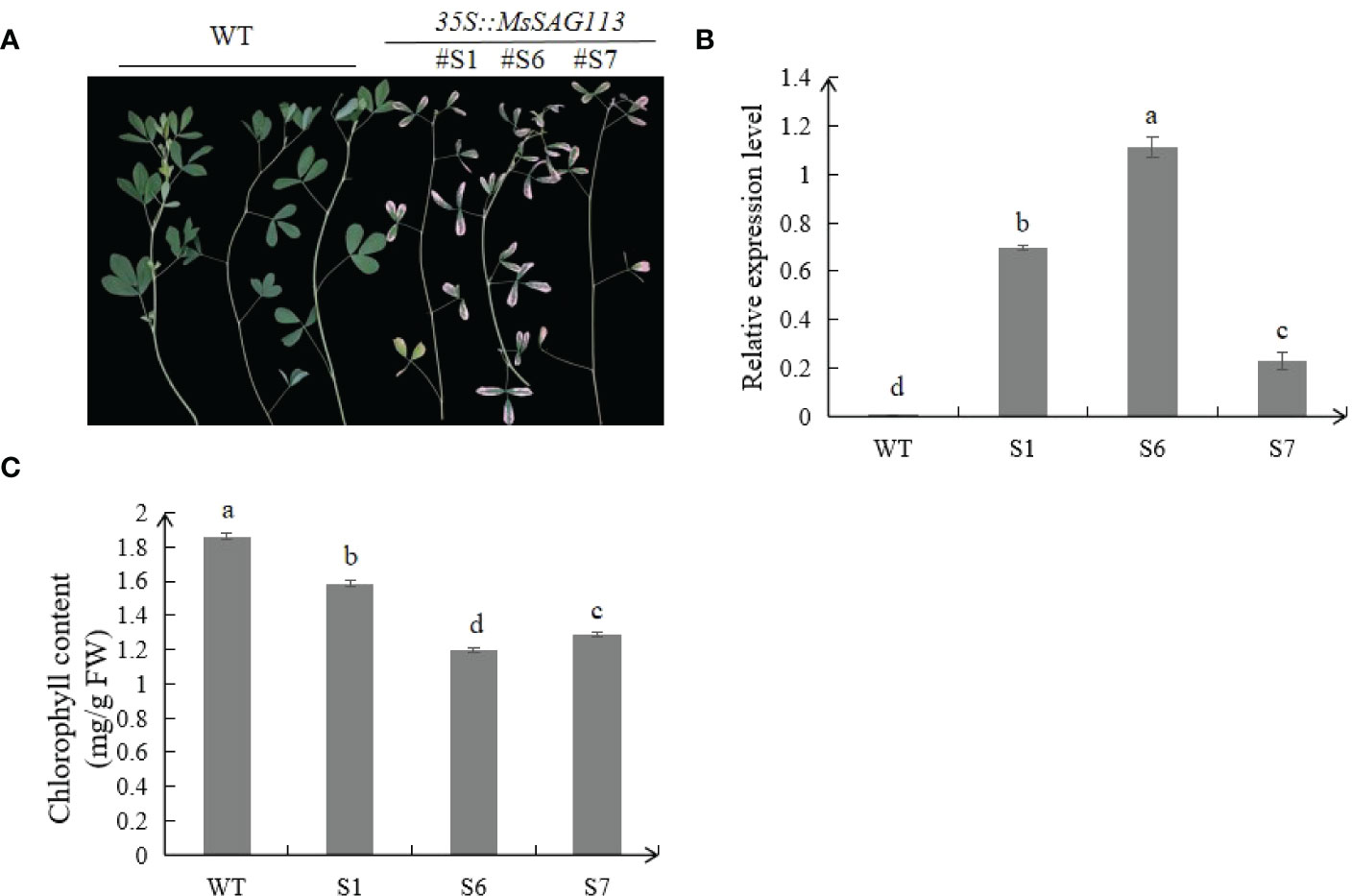
Figure 3 Phenotypes analysis of transgenic and WT alfalfa. Phenotype of three transgenic lines and WT alfalfa of 2 month-old (A). Relative expression levels of MsSAG113 in transgenic and WT plants (B). Chlorophyll contents of three transgenic and WT plants (C). The values are means ± SD (n = 3). In graphs, bars with different lowercase letters indicate significant difference at p<0.05.
Dark accelerates senescence of detached leaves of transgenic alfalfa
The detached leaves from transgenic plants turned yellow much earlier than WT under continuous dark for 7 days (Figure 4A), and chlorophyll contents were consistent with the senescence phenotype. The chlorophyll content of transgenic lines S1, S6 and S7 was 19.6%, 21.1% and 9.3% of that of wild-type lines, respectively (Figure 4B).
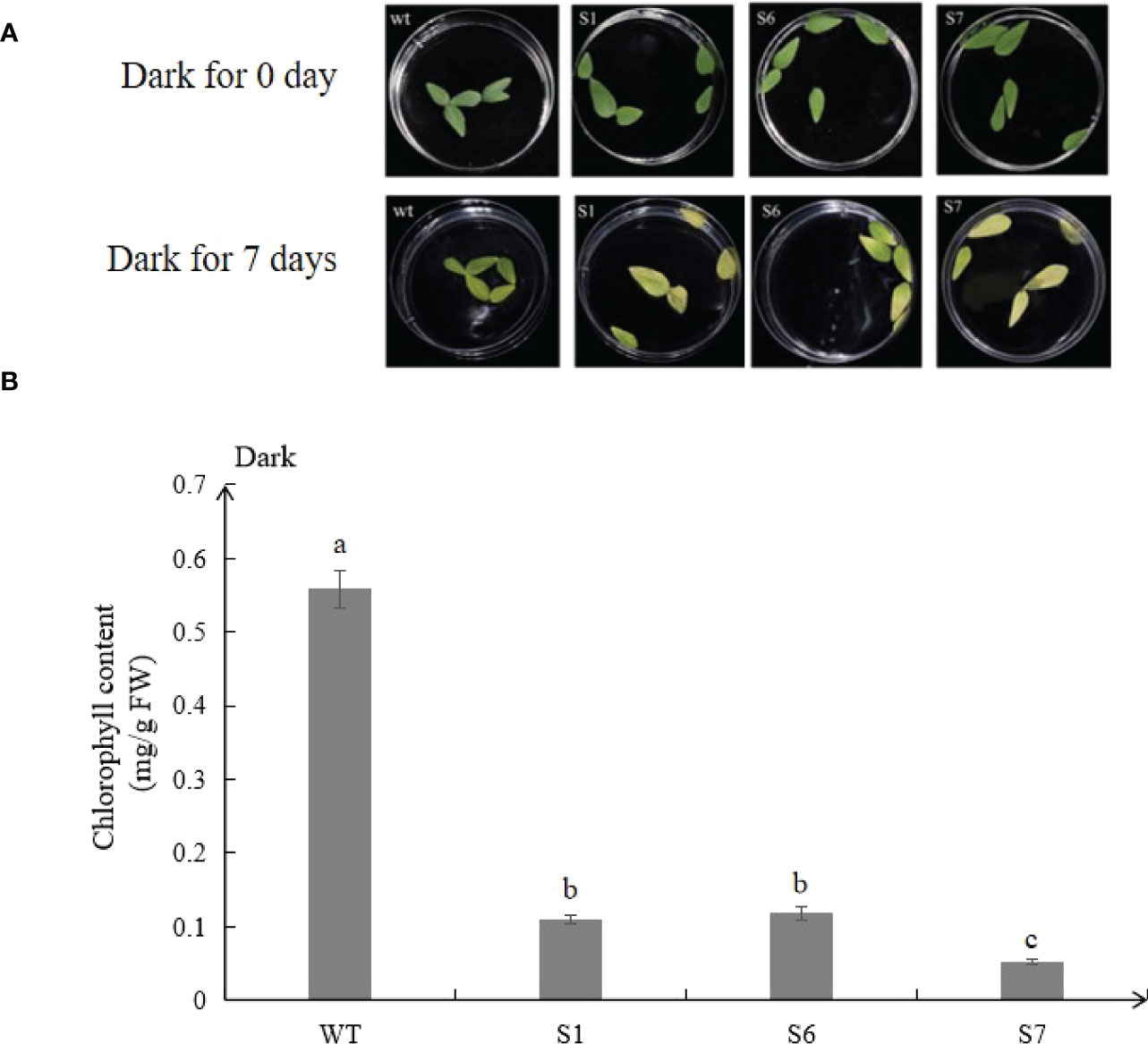
Figure 4 MsSAG113 is involved in dark-induced leaf senescence. Phenotypic analysis of 2 month-old wild type and transgenic isolated leaves after dark treatment (A). Chlorophyll contents of leaves under dark treatment (B). The values are means ± SD (n = 3). In graphs, bars with different lowercase letters indicate significant difference at p<0.05.
Detection of endogenous hormones in overexpressed plants
The contents of plant hormones in WT and transgenic plants were detected. Results showed that MeJA and ZR contents of transgenic plants were lower than those of WT (Figures 5A, B). In addition, ABA contents of the three MsSAG113 transgenic lines were all lower than those in WT. Compared with WT, ABA content in S1, S6 and S7 plants was decreased by 53.3%, 16.7% and 41.2%, respectively (Figure 5C).
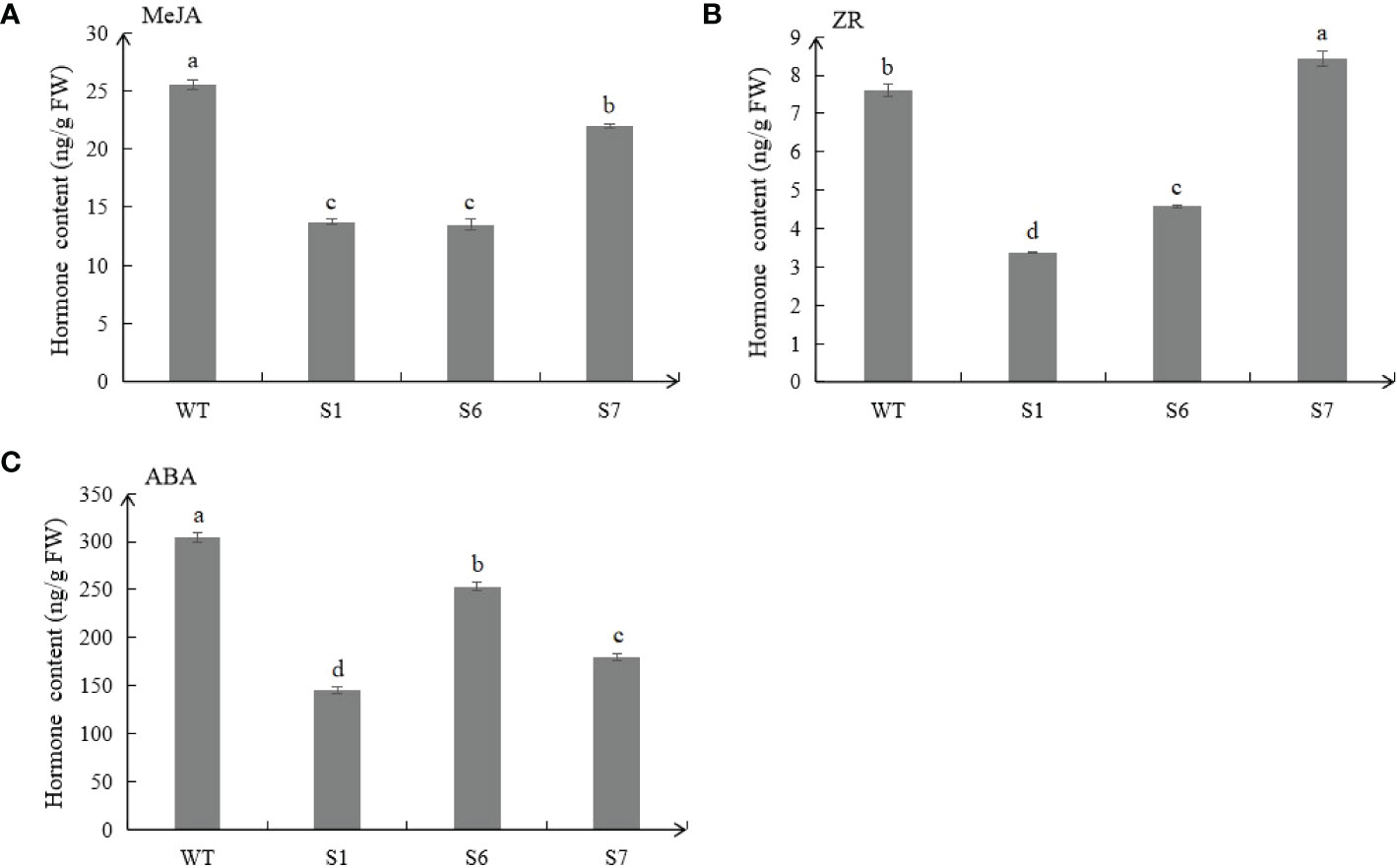
Figure 5 Endogenous hormone contents in transgenic and WT plants. Different hormones, including MeJA (A), ZR (B) and ABA (C) contents in transgenic and WT plants.The values are means ± SD (n = 3). In graphs, bars with different lowercase letters indicate significant difference at p<0.05.
Analysis of detached leaves of transgenic MsSAG113 treated with exogenous hormones
Detached leaves of S1, S6, and S7 treated with ABA, SA or MeJA showed obvious senescence on day 5, 8 or 12, respectively (Figure 6). Meanwhile, leaf senescence in S1, S6, and S7 leaves were also more pronounced than that in WT (Figures 6A–C). After 12 days of treatment with 10 μM 6BAP, the leaves of S1, S6 and S7 were significantly greener than those without MsSAG113 gene in plant senescence, indicating that MsSAG113 gene is involved in hormone network.
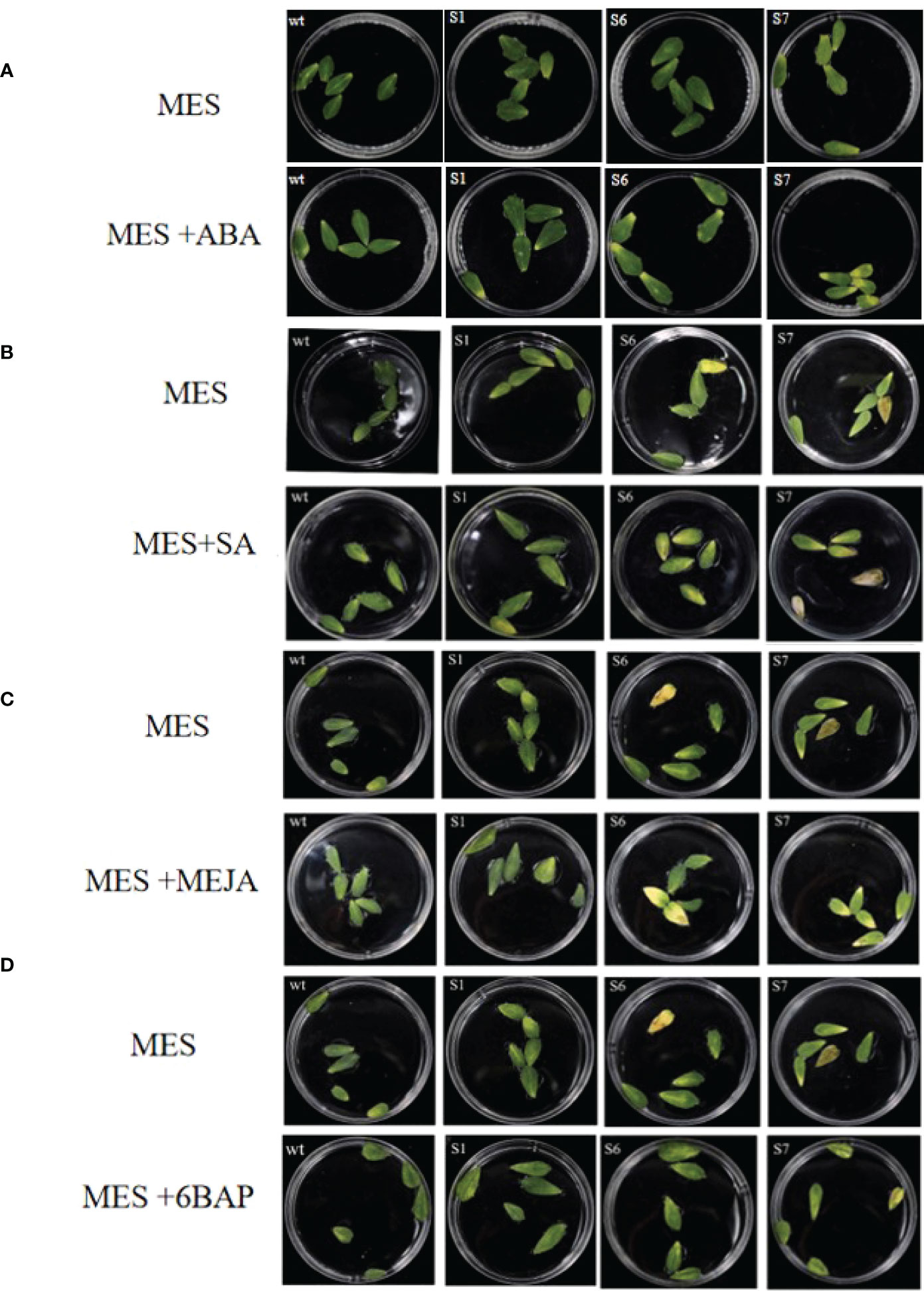
Figure 6 Detached leaves in MES with different hormones. Transgenic and WT detached leaves with or without ABA treatments for 5 days (A). Transgenic and WT detached leaves with or without SA treatments for 8 days (B). Transgenic and WT detached leaves with or without MeJA treatments for 12 days (C). Transgenic and WT detached leaves with or without 6BAP treatments for 12 days (D).
Under the treatments of ABA, SA and MeJA, the chlorophyll contents of transgenic plants, especially in S6 and S7 were lower than those from WT, which were consistent with the senescence phenotypes (Figures 7A–C). Under 6BAP treatments, the chlorophyll contents of WT leaves were higher than those in S6 and S7 (Figure 7D). However, samples of transgenic S1 showed no significant difference in chorophyll contents with WT under different hormone treatments except MeJA.
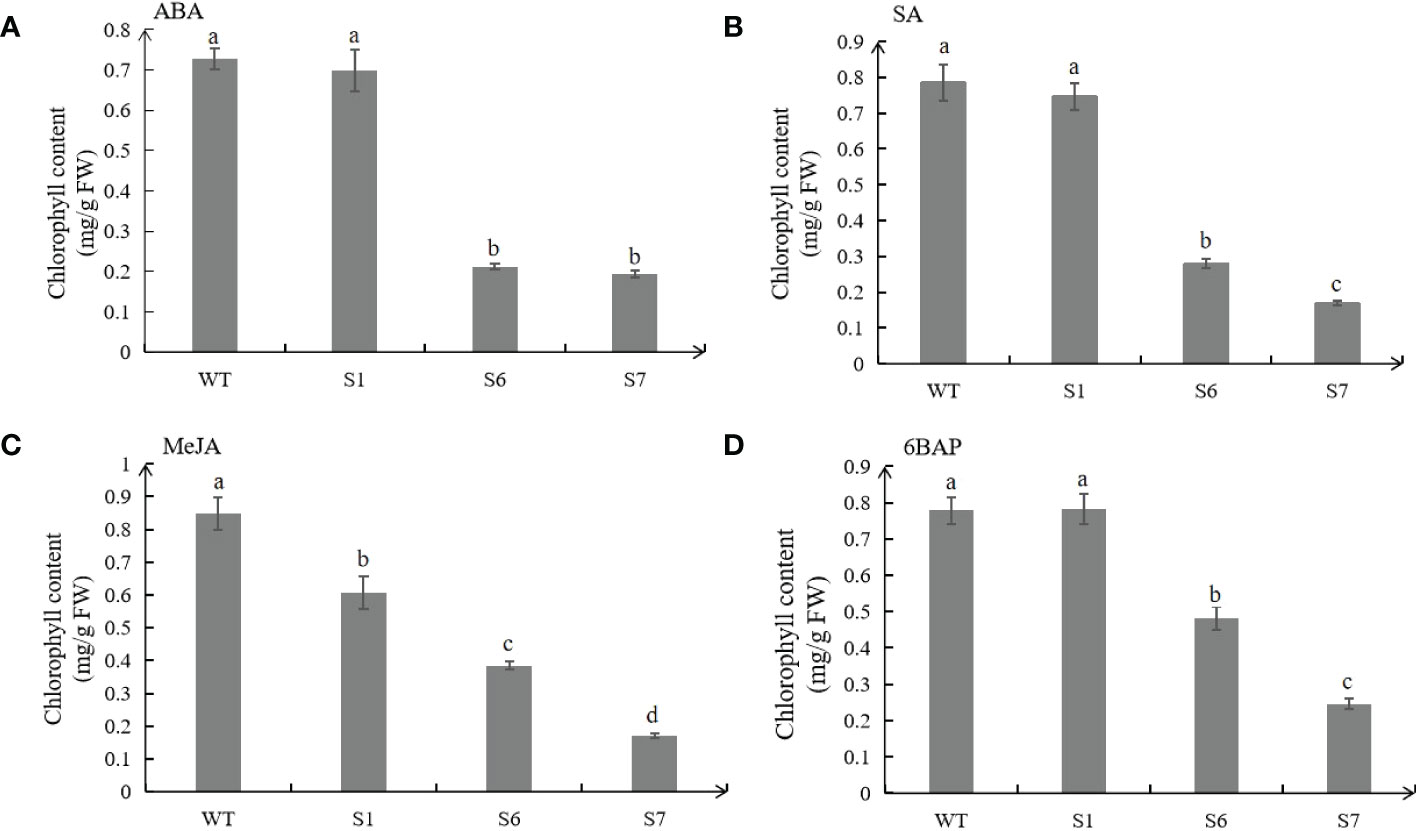
Figure 7 Chlorophyll contents in leaves of wild type (WT) and transgenic plants under hormone treatments. Chlorophyll contents of WT and transgenic leaves treated with 50 μM ABA for 5 days (A). Chlorophyll contents of WT and transgenic leaves treated with 0.5 mM SA for 8 days (B). Chlorophyll contents of WT and transgenic leaves treated with 10 μM MeJA for 10 days (C). Chlorophyll contents of WT and transgenic leaves treated with 10 μmol/L 6BAP for 12 days (D). The values are means ± SD (n = 3). In graphs, bars with different lowercase letters indicate significant difference at p<0.05.
DEGs and TFs were identified by transcriptome analysis
Transcriptome analysis identified a total of 1,392 DEGs in the transgenic and WT plants, of which 898 were up-regulated and 494 were down-regulated (Figure 8A; Supplementary Table S2). There were 42 differentially expressed TFs from 13 families identified, of which MYB (13), bHLH (7), Homeobox (5), ZBTB (4) and zf-CCCH (2) families were among the top 5 (Figure 8B; Supplementary Table S3). Meanwhile, 234 DEGs related to plant hormones were identified mainly involving ABA, Auxin, JA and SA, among which 145 were up-regulated and 77 were down-regulated (Figure 8C; Supplementary Table S4). These results showed that over-expression of MsSAG113 gene affected multiple senescence associated genes in alfalfa.
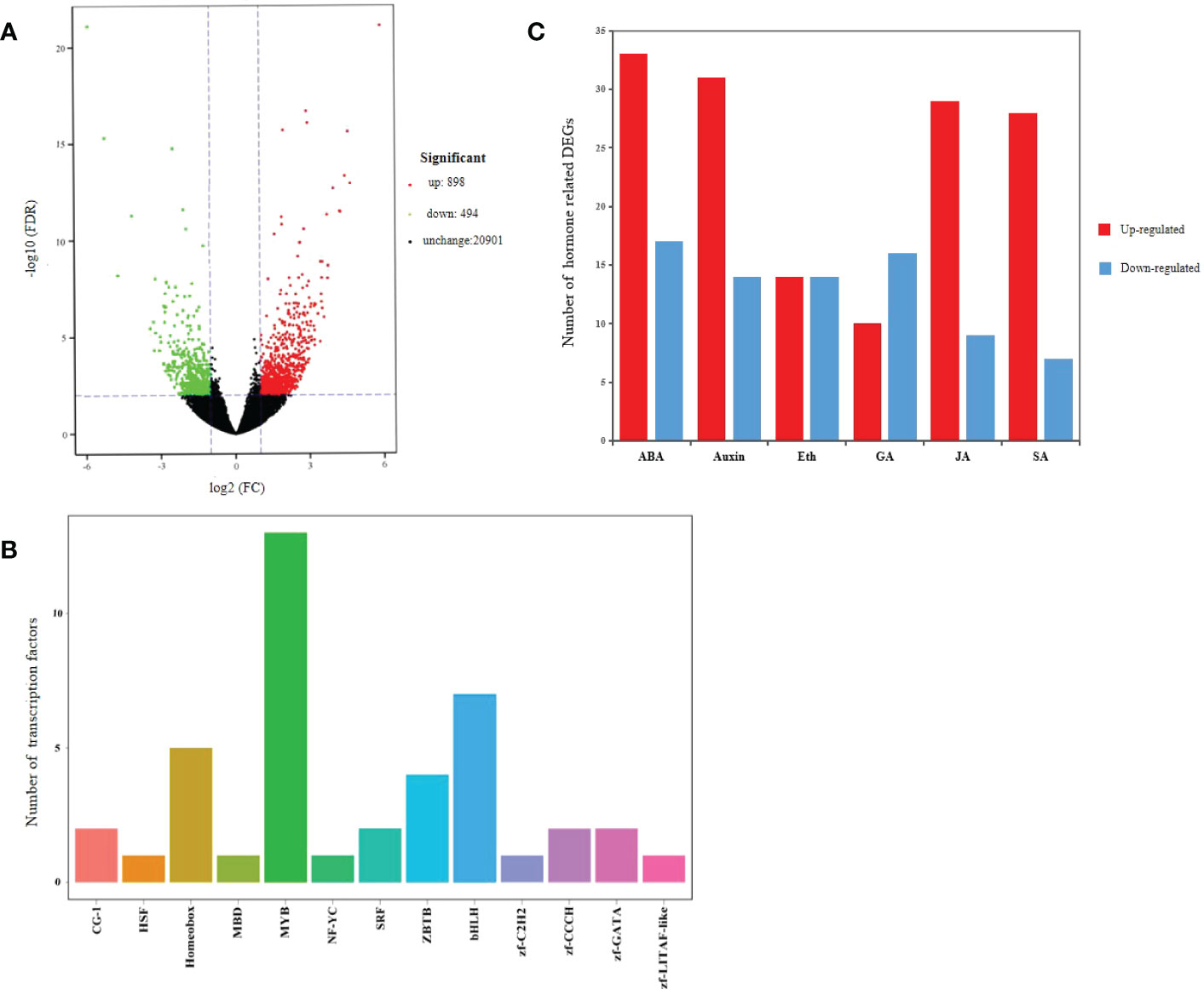
Figure 8 Transcriptome analysis of transgenic and WT alfalfa. Volcano plot of DEGs (A). In graphs, each plot represents one gene with three colors, including red (up-regulated), green (down-regulated) and blank (unchanged). The X-axis represents the value of log2 (Fold Change) in the two samples, and the Y-axis indicates the negative value of log10 (FDR). Differentially expressed transcription factor genes identified with MsSAG113 overexpression (B). In graphs, the X-axis represents transcription factor family, and the Y-axis represents the number of transcription factor genes. Hormone related DEGs (C). Eth indicates Ethylene. Red indicates up-regulated, and blue indicates down-regulated.
TF MsC3H-39 enhanced MsSAG113 expression
By DNA pull-down, 54 target proteins were collected, including alfalfa zinc finger protein MsC3H-39, which was also found in transcriptome analysis. To verify whether the TF can recognize and change MsSAG113 expression, we performed yeast one-hybrid and qRT-PCR analysis of alfalfa with transient expression of MsC3H-39 gene. Yeast one-hybrid assay showed that only those yeast clones containing MsSAG113 promoter and MsC3H-39 gene could survive on selection medium (SD/-leu/AbA300) (Figure 9A). These results suggest that MsC3H-39 transcription factor can bind directly to the promoter of MsSAG113.
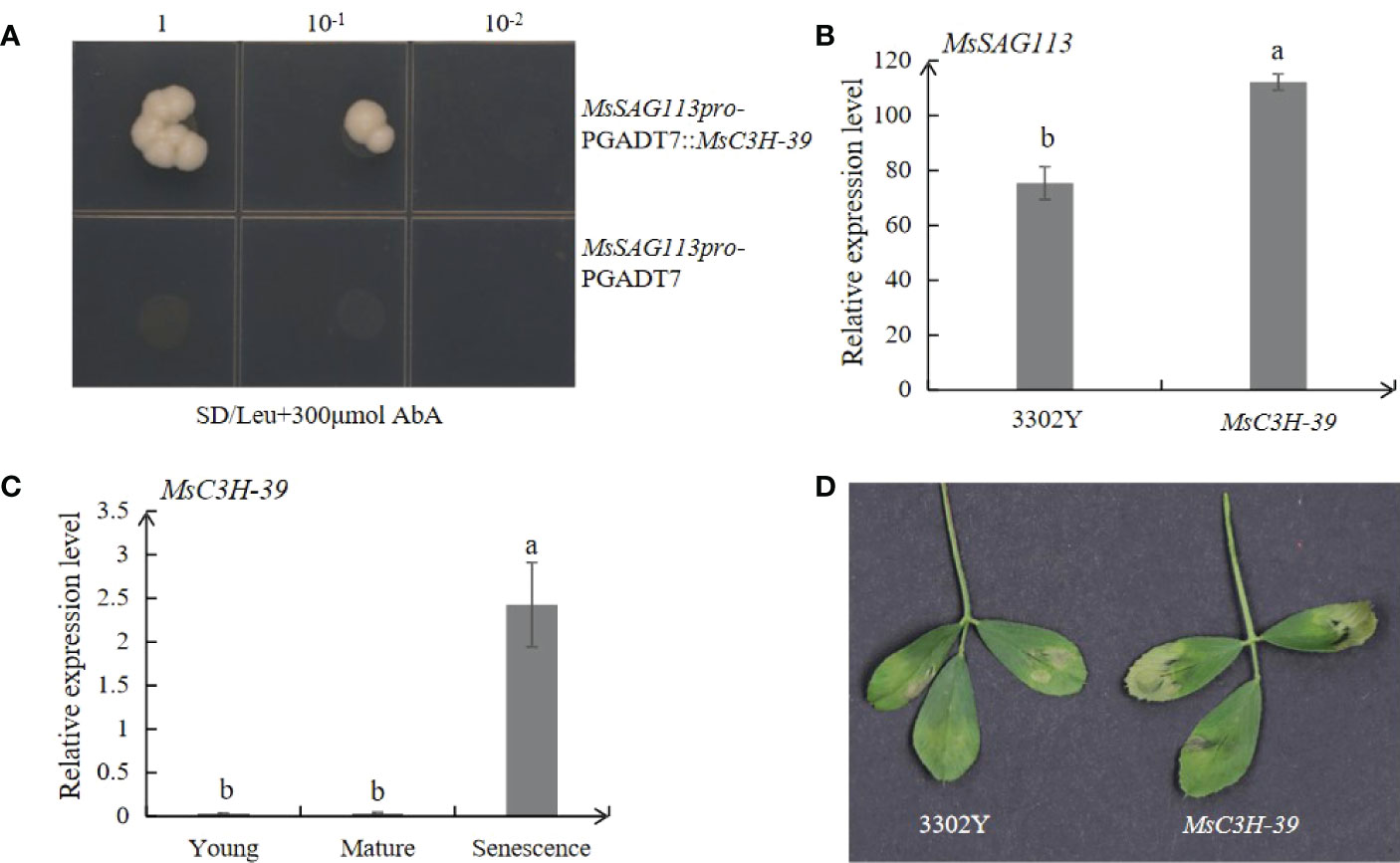
Figure 9 Interaction and expression analysis of MsSAG113 with TF MsC3H-39. (A) Interaction analysis of upstream of MsSAG113 and MsC3H-39 transcription factors. The number indicates dilution times of transformed yeast cells. (B) Relative expression levels of MsSAG113 gene in alfalfa with transcient expression of empty vector or MsC3H-39. (C) Expression of MsC3H-39 in young, mature and senescent leaves of alfalfa. (D) 3302Y empty vector and MsC3H-39 transiently express the senescent phenotype in alfalfa leaves. The values are means ± SD (n = 3). In graphs, bars with different lowercase letters indicate significant difference at p<0.05.
The expression of MsC3H-39 in alfalfa leaves was the highest in senescence leaves indicating the involvement of MsC3H-39 in plant senescence. Meanwhile, transiently expression of MsC3H-39 result in apparent senescence symptoms compared with empty vector 3302Y (Figures 9C, D). Transient expression of MsC3H-39 gene and qRT-PCR analysis of MsSAG113 showed that MsC3H-39 significantly increased the expression of MsSAG113, suggesting that MsC3H-39 transcription factor positively regulated the expression of MsSAG113 by binding the upstream of the gene (Figures 9B–D).
Discussion and conclusion
The regulatory network of senescence associated genes is constantly updated, and many SAGs were found being involved in gene expression regulation, signal transduction, macromolecular degradation and other senescence processes (He et al., 2001; Gepstein et al., 2003; Buchanan-Wollaston et al., 2005; Guo and Gan, 2012). SAG113 belongs to the PP2C superfamily and plays an indispensable role in plant senescence. Senescence can limit crop yield and normal plant growth and development (Gan and Amasino, 1997). In this study, we explored the senescence regulation mechanism of SAG113 gene in alfalfa, which could provide a reference for studying the growth and development mechanism of other legumes and improving crop yield.
Expression analysis showed that MsSAG113 gene was affected by ABA, SA, MeJA and 6BAP hormone treatments. These hormones are considered to be the main internal factors that control the aging process of leaves (Balibrea Lara et al., 2004; Liebsch and Keech, 2016). It was reported that application of ABA-treated daffodils promoted senescence, and the increase in leaf ABA content was visually consistent with leaf senescence (Hunter et al., 2004). In Arabidopsis thaliana mutants deficient in SA signaling, the expression patterns of many genes were altered, and the mutants delayed yellowing and reduced necrosis (Morris et al., 2000). Exogenous application of JA led to premature senescence in leaves of wild-type Arabidopsis thaliana, but such effect absented in JA mutant plants (He et al., 2002). In our study, exogenous ABA, SA or MeJA accerlated early senescence of detached leaves with MsSAG113 overexpression (Figure 7). These results indicated that ABA, SA and MeJA might participate in plant senescence regulation of MsSAG113 gene. Abscisic acid promoted aging, while 6BAP generally inhibited aging (Smart, 1994). Early physiological studies have shown that exogenous cytokinin treatment of most monocotyledonous and dicotyledonous plant species could delay leaf senescence (Richmond et al., 1957). Our study achieved similar results. Exogenous 6BAP hormone treatment of isolated leaves indicated that leaf senescence was delayed, and the delay in transgenic leaf senescence was more obvious (Figure 7). Therefore, MsSAG113 gene, as a senescence regulator, may have a role in regulating leaf senescence by participating in a hormonal regulatory network.
PP2C enzymes play an important role in signal transduction. Six groups of PP2C family protein phosphatases are considered to be negative regulators of ABA (Schweighofer et al., 2004). Previous studies showed that SAG113 is a negative regulator of ABA signaling pathway, which can inhibit stomatal closing in leaves and specifically involved in the control of water loss during leaf senescence (Wang et al., 2011; Zhang and Gan, 2012). Overexpression of FsPP2C1 in Arabidopsis reduced the degree of seed dormancy and was insensitive to ABA, so FsPP2C1 had a negative regulatory effect on ABA (Gonzalez-Garcia et al., 2003). This precisely corroborates our results that ABA contents were reduced in plants overexpressing MsSAG113. Therefore, it is reasonable to speculate that the increased transcription level of SAG113 may be involved in negatively regulating ABA signaling, leading to the inhibition of ABA synthesis.
Compared with the wild type, the leaves of MsSAG113 transgenic alfalfa had obvious yellowing, and the chlorophyll content was significantly reduced (Figure 3). These results are consistent with previous research. During senescence, chlorophyll was degraded, which was visually manifested as yellowing of leaves (Curty and Engel, 1996). Overexpression of MYBR1 could delay leaf senescence in Arabidopsis. Loss-of-function MYBR1 plants exhibited faster chlorophyll loss and senescence (Jaradat et al., 2013). Darkness was often thought to be a cause of senescence, in which the synthesis of chlorophyll was blocked during the reductive phase (Weaver and Amasino, 2001; Eckhardt et al., 2004). Our results showed that after 7 days of dark treatment, the color of the isolated leaves in wild type was greener than that of the transgenic plants, with about 10 times difference in chlorophyll content (Figures 4, 5). These results suggest that MsSAG113 gene be involved in the metabolic process of chlorophyll to regulate leaf senescence in alfalfa.
Our study showed that application of exogenous ABA, SA and MeJA significantly increased chlorophyll content in WT compared to transgenic plants. Previous studies on other plants observed the similar results (He et al., 2002; Lü et al., 2014; Zhao et al., 2016). However, we found that in isolated leaves treated with exogenous 6BAP, the chlorophyll content in S1 was increased compared with WT, while S6 and S7 were significantly reduced. Studies have shown that at the onset of senescence, under the activation of the SAG12 promoter, feedback regulation of cytokinins occurs, which can be inhibited in delaying leaf senescence (Kant et al., 2015). The SAG12 and SAG13 promoters retarded the degradation of chlorophyll in isolated tomato senescent leaves under dark induction, while only PSAG13::IPT plants showed delayed chlorophyll degradation in isolated leaves and florets of broccoli (Chen et al., 2001; Swartzberg et al., 2006). Therefore, we inferred that under exogenous 6BAP treatment, the SAG113 promoter was activated in S6 and S7 plants, resulting in feedback regulation of cytokinins, affecting chlorophyll synthesis, and thus reducing chlorophyll content of leaves. In addition, there was no significant difference in chlorophyll content between S1 plants and WT plants treated with exogenous hormones ABA, SA and 6BAP, which may be due to the different expression levels of MsSAG113 in different transgenic lines and a phenotype in the transgenic plant is affected by gene expression levels and environmental factors.
RNA-Seq has become one of the important techniques for analyzing plant leaf senescence. Transcription factors and plant hormones are central to the senescence regulatory networks. In Arabidopsis, NAC, WRKY, AP2-EREBP and bHLH family transcription factors are regulated (Balazadeh et al., 2008). In this study, a total of 1,164 aging-related genes and 42 TFs from 13 families were identified, mainly including MYB, bHLH, Homeobox, ZBTB and zf-CCCH. The studies of plant senescence in Arabidopsis thaliana showed that many hormones were involved in the regulation of senescence in different ways (van der Graaff et al., 2006). By transcriptome analysis, we identified a total of 234 genes involved in hormone biosynthesis, metabolism, signal transduction and response. These results provided further evidence that MsSAG113 gene plays important roles in regulating senescence in alfalfa and participating in hormone related regulatory network.
The identification of upstream regulators revealed the mechanism of MsSAG113 gene in regulating senescence. This study screened the upstream regulatory protein of MsC3H-39 by DNA pull-down and verified it by yeast one-hybrid and transient expression analysis. MsC3H-39 protein belongs to CCCH-like of zinc finger proteins, which can be classified into C2H2, C2C2, C2HC, C2C2C2C2, C2HCC2C2 and CCCH types according to the number and order of zinc ion-bound Cys and His residues in the secondary structure of fingers (Sanchez-Garcia and Rabbitts, 1994; Klug and Schwabe, 1995; Mackay and Crossley, 1998). CCCH-like zinc lipoproteins have been shown to be involved in senescence regulation in previous reports. As a CCCH-type protein, OsDOS can delay leaf senescence, partially through the JA pathway (Jan et al., 2013). Two CCCH zinc finger proteins, AtC3H49 and AtC3H20 can delay leaf senescence and participate in ABA and JA responses in Arabidopsis (Lee et al., 2012). KHZ1 and KHZ2, two proteins of CCCH, positively regulated leaf senescence in Arabidopsis thaliana (Yan et al., 2017). The present study identified MsSAG113 as a target gene of MsC3H-39 by DNA pull-down assay and confirmed by yeast one-hybrid and qRT-PCR analysis. Meanwhile, we found that expression levels of MsC3H-39 were the highest in senescence leaves, much higher than those in young and mature samples. Furthermore, MsC3H-39 can cause an earlier senescence phenotype and the senescence associated gene, MsSAG113 can be induced by MsC3H-39 (Figures 9A–D). These data suggest that MsC3H-39 directly regulates the expression of MsSAG113 and promotes leaf senescence.
In summary, MsSAG113 gene, as a senescence regulator, can be induced by hormones and regulates plant senescence in alfalfa by participating in the hormone regulatory network. MsC3H-39 directly recognizes and binds the upstream of MsSAG113 and enhances its expression (Figure 10). Meanwhile, A total of 42 TFs associated with plant senescence and 234 genes involved in hormone regulation was identified by transcriptome analysis, which further indicated the important role of MsSAG113 gene in leaf senescence and hormone regulation network in alfalfa. This study lays a foundation for studying the mechanism of alfalfa senescence and provides new insights for improving the growth and production in pasture legumes.
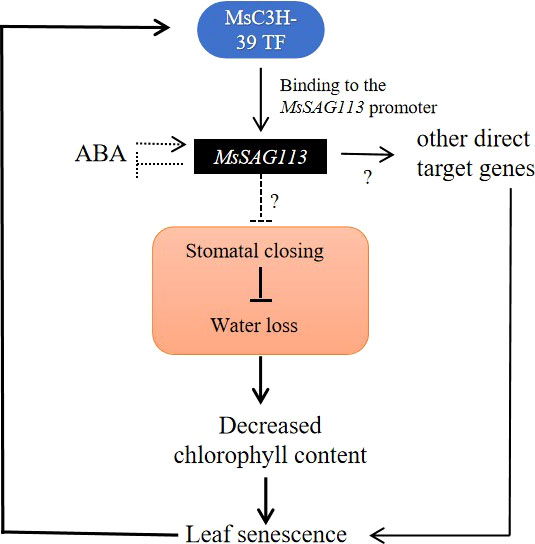
Figure 10 Potiential regulatory of MsSAG113 in senescence in alfalfa. MsC3H-39 TF can enhance the expression of MsSAG113 gene by recognizing and binding its promoter. ABA induces MsSAG113 expression and MsSAG113 negatively regulates ABA accumulation. MsSAG113 may negatively regulate stomatal closure, resulting in accelerated water loss, a decrease in chlorophyll content and triggering plant senescence in alfalfa.
Data availability statement
The data presented in the study are deposited in the NCBI repository. The accession numbers are PRJNA822614 and OP880214.
Author contributions
YHC, LH, and SL conceived and designed the experiment. SL and LZ conducted the experiments. DD and HX analyzed the data. CJ and YL contributed analysis tools. YHC provided financial support. SL wrote the manuscript. YHC and YLC edited and revised the manuscript. All authors made significant contributions to the manuscript and approved the final version for publication.
Funding
This work was supported by the Inner Mongolia Science and Technology Plan “Jiebangguashuai” Project (Breeding and Industrialization Demonstration of New Alfalfa Varieties with High Quality) and the Fundamental Research Funds for the Central Universities (Grant No. 2021ZY84).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1085497/full#supplementary-material
Supplementary Table 1 | List of primers.
Supplementary Table 2 | Differentially expressed genes.
Supplementary Table 3 | Involved in hormone regulation genes in DEGs.
Supplementary Table 4 | Transcriptional regulators in DEGs.
References
Balazadeh, S., Riano-Pachon, D. M., Mueller-Roeber, B. (2008). Transcription factors regulating leaf senescence in arabidopsis thaliana. Plant Biol. 10, 63–75. doi: 10.1111/j.1438-8677.2008.00088.x
Balibrea Lara, M. E., Gonzalez Garcia, M., Fatima, T., Ehneß, R., Lee, T. K., Proels, R., et al. (2004). Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16, 1276–1287. doi: 10.1105/tpc.018929
Bleecker, A. B., Patterson, S. E. (1997). Last exit: Senescence, abscission, and meristem arrest in arabidopsis. Plant Cell 9, 1169–1179. doi: 10.1105/tpc.9.7.1169
Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P. O., Nam, H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in arabidopsis. Plant J. 42, 567–585. doi: 10.1111/j.1365-313X.2005.02399.x
Chao, Y., Xie, L., Yuan, J., Guo, T., Li, Y., Liu, F., et al. (2018). Transcriptome analysis of leaf senescence in red clover (Trifolium pratense l.). Physiol. Mol. Biol. Plants 24, 753–765. doi: 10.1007/s12298-018-0562-z
Chen, L. F. O., Hwang, J. Y., Charng, Y. Y., Sun, C. W., And Yang, S. F. (2001). Transformation of broccoli (Brassica oleracea var. italica)with isopentenyltransferase gene via agrobacterium tumefaciens for post-harvest yellowing retardation. Mol. Breed. 7, 243–257. doi: 10.1023/A:1011357320259
Curty, C., Engel, N. (1996). Detection, isolation and structure elucidation of a chlorophyll a catabolite from autumnal senescent leaves of cercidiphyllum japonicum. Phytochemistry 42, 1531–1536. doi: 10.1016/0031-9422(96)00155-0
Eckhardt, U., Grimm, B., Hortensteiner, S. (2004). Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 56, 1–14. doi: 10.1007/s11103-004-2331-3
Engvall, E., Perlmann, P. (1971). Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. doi: 10.1016/0019-2791(71)90454-X
Gan, S., Amasino, R. M. (1997). Making sense of senescence. Plant Physiol. 113, 313–319. doi: 10.1104/pp.113.2.313
Gepstein, S., Sabehi, G., Carp, M., Hajouj, T., Nesher, M. F. O., Yariv, I., et al. (2003). Large-Scale identification of leaf senescence-associated genes. Plant J. 36, 629–642. doi: 10.1046/j.1365-313X.2003.01908.x
Gonzalez-Garcia, M. P., Rodriguez, D., Nicolas, C., Rodriguez, P. L., Nicolas, G., Lorenzo, O. (2003). Negative regulation of abscisic acid signaling by the fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 133, 135–144. doi: 10.1104/pp.103.025569
Guo, Y., Gan, S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35, 644–655. doi: 10.1111/j.1365-3040.2011.02442.x
He, Y., Fukushige, H., Hildebrand, D. F., Gan, S. (2002). Evidence supporting a role of jasmonic acid in arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
He, Y., Gan, S. (2002). A gene encoding an acyl hydrolase is involved in leaf senescence in arabidopsis. Plant Cell 14, 805–815. doi: 10.1105/tpc.010422
He, Y. Y., Tang, W. W., Swain, J. D. J. D., Green, A. L. A. L., Jack, T. P. T. P., Gan, S. S. (2001). Networking senescence-regulating pathways by using arabidopsis enhancer trap lines1. Plant Physiol. (Bethesda) 126, 707–716. doi: 10.1104/pp.126.2.707
Hoekema, A., Hirsch, P. R., Al, H. P. J. J. (1983). 20-a binary plant vector strategy based on separation of vir- and T-region of the agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Hrbáčková, M., Dvořák, P., Takáč, T., Tichá, M., Luptovčiak, I., Šamajová, O., et al. (2020). Biotechnological perspectives of omics and genetic engineering methods in alfalfa. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00592
Hunter, D. A., Ferrante, A., Vernieri, P., Reid, M. S. (2004). Role of abscisic acid in perianth senescence of daffodil (Narcissus pseudonarcissus " Dutch master "). Physiol. Plant. 121, 313–321. doi: 10.1111/j.1399-3054.2004.00311.x
Jan, A., Maruyama, K., Todaka, D., Kidokoro, S., Abo, M., Yoshimura, E., et al. (2013). OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216. doi: 10.1104/pp.112.205385
Jaradat, M. R., Feurtado, J. A., Huang, D., Lu, Y., Cutler, A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13, 192. doi: 10.1186/1471-2229-13-192
Jing, H. C., Sturre, M. J., Hille, J., Dijkwel, P. P. (2002). Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J. 32, 51–63. doi: 10.1046/j.1365-313x.2002.01400.x
Kant, S., Burch, D., Badenhorst, P., Palanisamy, R., Mason, J., Spangenberg, G. (2015). Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus l.). PloS One 10, e116349. doi: 10.1371/journal.pone.0116349
Klug, A., Schwabe, J. W. (1995). Protein motifs 5. zinc fingers. FASEB J. 9, 597–604. doi: 10.1096/fasebj.9.8.7768350
Lee, S., Jung, H. J., Kang, H., Kim, S. Y. (2012). Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 53, 673–686. doi: 10.1093/pcp/pcs023
Liebsch, D., Keech, O. (2016). Dark-induced leaf senescence. new insights into a complex light-dependent regulatory pathway. New Phytol. 212, 563–570. doi: 10.1111/nph.14217
Lü, P., Zhang, C., Liu, J., Liu, X., Jiang, G., Jiang, X., et al. (2014). RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J. 78, 578–590. doi: 10.1111/tpj.12494
Mackay, J. P., Crossley, M. (1998). Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4. doi: 10.1016/S0968-0004(97)01168-7
Mao, X., Cai, T., Olyarchuk, J. G., Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Matile, P., Ginsburg, S., Schellenberg, M., Thomas, H. (1988). Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc. Natl. Acad. Sci. United States America 85, 9529–9532. doi: 10.1073/pnas.85.24.9529
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Morris, K., Mackerness, S. A. H., Page, T., John, C. F., Murphy, A. M., Carr, J. P., et al. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. For Cell Mol. Biol. 23, 677–685. doi: 10.1046/j.1365-313x.2000.00836.x
Nishimura, N., Yoshida, T., Kitahata, N., Asami, T., Shinozaki, K., Hirayama, T. (2007). ABA-hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in arabidopsis seed. Plant J. 50, 935–949. doi: 10.1111/j.1365-313X.2007.03107.x
Pruzinská, A., Tanner, G., Aubry, S., Anders, I., Moser, S., Müller, T., et al. (2005). Chlorophyll breakdown in senescent arabidopsis leaves. characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139, 52–63. doi: 10.1104/pp.105.065870
Richmond, Amos, E., Lang, A. (1957). Effect of kinetin on protein content and survival of detached xanthium leaves. Science 125, 650–651. doi: 10.1126/science.125.3249.650
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2009). EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sanchez-Garcia, I., Rabbitts, T. H. (1994). The LIM domain a new structural motif found in zinc-finger-like proteins. Trends Genet. 10, 315–320. doi: 10.1016/0168-9525(94)90034-5
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schweighofer, A., Hirt, H., Meskiene, I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9, 236–243. doi: 10.1016/j.tplants.2004.03.007
Singh, S., Singh, A., Nandi, A. K. (2016). The rice OsSAG12-2 gene codes for a functional protease that negatively regulates stress-induced cell death. J. Biosci. 41, 445–453. doi: 10.1007/s12038-016-9626-9
Smart, C. M. (1994). Tansley review no. 64. gene expression during leaf senescence. New Phytol. 126, 419–448. doi: 10.1111/j.1469-8137.1994.tb04243.x
Swartzberg, D., Dai, N., Gan, S., Amasino, R., Granot, D. (2006). Effects of cytokinin production under two SAG promoters on senescence and development of tomato plants. Plant Biol. 8, 579–586. doi: 10.1055/s-2006-924240
van der Graaff, E. E., Schwacke, R. R., Schneider, A. A., Desimone, M. M., Flügge, U. U., Kunze, R. R. (2006). Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence1. Plant Physiol. (Bethesda) 141, 776–792. doi: 10.1104/pp.106.079293
Wang, R. S., Pandey, S., Li, S., Gookin, T. E., Zhao, Z., Albert, R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of arabidopsis guard cells. BMC Genomics 12, 216. doi: 10.1186/1471-2164-12-216
Weaver, L. M., Amasino, R. M. (2001). Senescence is induced in individually darkened arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127, 876–886. doi: 10.1104/pp.010312
Wistrom, C., Villeponteau, B. (1992). Cloning and expression of SAG: A novel marker of cellular senescence. Exp. Cell Res. 199, 355. doi: 10.1016/0014-4827(92)90445-E
Yan, Z., Jia, J., Yan, X., Shi, H., Han, Y. (2017). Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 95, 549–565. doi: 10.1007/s11103-017-0667-8
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, K., Gan, S. (2012). An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing arabidopsis leaves. Plant Physiol. 158, 961–969. doi: 10.1104/pp.111.190876
Zhang, K., Xia, X., Zhang, Y., Gan, S. (2012). An ABA-regulated and golgi-localized protein phosphatase controls water loss during leaf senescence in arabidopsis. Plant J. 69, 667–678. doi: 10.1111/j.1365-313X.2011.04821.x
Zhan, J., He, H., Wang, T., Wang, A., Li, C., He, L. (2013). Aluminum-induced programmed cell death promoted by AhSAG, a senescence-associated gene in arachis hypoganea l. Plant Sci. 210, 108–117. doi: 10.1016/j.plantsci.2013.05.012
Keywords: alfalfa, MsC3H-39, MsSAG113, RNA-seq, senescence
Citation: Li S, Xie H, Zhou L, Dong D, Liu Y, Jia C, Han L, Chao Y and Chen Y (2022) Overexpression of MsSAG113 gene promotes leaf senescence in alfalfa via participating in the hormone regulatory network. Front. Plant Sci. 13:1085497. doi: 10.3389/fpls.2022.1085497
Received: 31 October 2022; Accepted: 25 November 2022;
Published: 08 December 2022.
Edited by:
Shangang Jia, China Agricultural University, ChinaReviewed by:
Yuefei Xu, Northwest A&F University, ChinaJun Li, Inner Mongolia University, China
Xunzhong Zhang, Virginia Tech, United States
Copyright © 2022 Li, Xie, Zhou, Dong, Liu, Jia, Han, Chao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liebao Han, hanliebao@bjfu.edu.cn; Yuehui Chao, chaoyuehui@bjfu.edu.cn
 Shuwen Li
Shuwen Li Hong Xie1
Hong Xie1 Di Dong
Di Dong Yuehui Chao
Yuehui Chao Yinglong Chen
Yinglong Chen