- 1Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, Hebei, China
- 2Institute of Biotechnology and Food Science, Hebei Academy of Agriculture and Forestry Science/the Key Laboratory of Plant Genetic Engineering of Hebei Province, Shijiazhuang, Hebei, China
- 3The National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 4The State Key Laboratory for Biology of Plant Disease and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 5The Innovative Academy for Seed Design, Chinese Academy of Sciences, Beijing, China
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a destructive fungal disease of wheat throughout the world. Utilization of effective powdery mildew resistance genes and cultivars is considered as the most economic, efficient, and environmental-friendly method to control this disease. Synthetic hexaploid wheat (SHW), which was developed through hybridization of diploid Aegilops and tetraploid wheat, is a valuable genetic resource for resistance to powdery mildew. SHW line YAV249 showed high levels of resistance to powdery mildew at both the seedling and adult stages. Genetic analysis indicated that the resistance was controlled by a single dominant gene, temporarily designated PmYAV. Bulked segregant analysis with wheat 660K single nucleotide polymorphism (SNP) array scanning and marker analysis showed that PmYAV was located on chromosome 2AL and flanked by markers Xgdm93 and Xwgrc763, respectively, with genetic distances of 0.8 cM and 1.2 cM corresponding to a physic interval of 1.89 Mb on the Chinese Spring reference genome sequence v1.0. Sequence alignment analysis demonstrated that the sequence of PmYAV was consistent with that of Pm4a but generated an extra splicing event. When inoculated with different Bgt isolates, PmYAV showed a significantly different spectrum from Pm4a, hence it might be a new resistant resource for improvement of powdery mildew resistance. The flanked markers GDM93 and WGRC763, and the co-segregated markers BCD1231 and JS717/JS718 were confirmed to be easily performed in marker-assisted selection (MAS) of PmYAV. Using MAS strategy, PmYAV was transferred into the commercial cultivar Kenong 199 (KN199) and a wheat line YK13 was derived at generation BC3F3 from the population of YAV249/4*KN199 due to its excellent agronomic traits and resistance to powdery mildew. In conclusion, an alternative splicing variant of Pm4 was identified in this study, which informed the regulation of Pm4 gene function.
Introduction
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a destructive fungal disease of common wheat throughout the world, accounting for approximately 5% of the total yield losses caused by wheat pathogens and pests (Serge et al., 2019). Utilization of effective powdery mildew resistance genes (Pm genes) and resistant cultivars is the most economic, efficient, and environmental-friendly method to control this disease.
So far, more than 100 Pm genes/alleles from wheat and its relatives have been reported, including formally named Pm1 to Pm68 at 63 loci (noting that Pm8=Pm17, Pm18=Pm1c, Pm22=Pm1e, Pm23=Pm4c, and Pm31=Pm21) and 30 temporarily named genes (He et al., 2020; McIntosh et al., 2020). With the development of sequencing technologies and releases of reference genomes in succession, 13 Pm genes, including Pm1a, Pm2, Pm3, Pm4b, Pm5e, Pm8, Pm17, Pm21, Pm24, Pm38, Pm41, Pm46 and Pm60 have been cloned (Krattinger et al., 2009; Brunner et al., 2011; Hurni et al., 2013; Moore et al., 2015; Sánchez-Martín et al., 2016; Singh et al., 2018; Xing et al., 2018; Zou et al., 2018; Li et al., 2020; Lu et al., 2020; Xie et al., 2020; Hewitt et al., 2021; Sánchez-Martín et al., 2021). Because of the large-scale deployment of single resistance genes and the evolution of corresponding pathogens, many resistance genes have lost their effectiveness, such as Pm1, Pm3, Pm7 and Pm8,either regionally or across the world (Jin et al., 2021). Therefore, it is essential to explore novel resistance genes/alleles of powdery mildew and transfer them into wheat cultivars to enrich the diversity of resistance sources and enhance the durability of disease resistance.
Gene Pm4 has been one of the most widely used powdery mildew resistance genes and can be found in many cultivars all over the world (Ullah et al., 2018; Jin et al., 2021). Since Pm4 allele Pm4a was first mapped on the long arm of wheat chromosome 2A in Khapli (The et al., 1979), five alleles (Pm4a-Pm4e) have been reported at that locus (Ullah et al., 2018). Recently, Pm4b, encoded a putative chimeric protein of a serine/threonine kinase with multiple C2 domains and transmembrane regions, has been cloned through Mutant Chromosome Flow Sorting and Sequencing (Sánchez-Martín et al., 2021). Pm4b consists of seven exons and contains two alternative transcripts, denoted Pm4b_V1 and Pm4b_V2. Subsequently, three new Pm4 alleles, tentatively denoted as Pm4f, Pm4g and Pm4h were discovered by the diagnostic functional markers JS717/JS718 (Sánchez-Martín et al., 2021). It is noted that other loci, for example, Pm1, Pm2, and Pm5 also have multiple allelic variations because of the long term interactions with the co-evolving pathogen. It is reported that the allelic variations of the documented Pm genes played important roles in resistance breeding, for their different spectra of effectiveness to pathogen isolates. Therefore, identification of the allelic variations in detail is important, not only for increasing the diversity of resistance resource, but also for understanding the host-pathogen interaction mechanism and applying in resistance gene pyramiding breeding (Srichumpa et al., 2005; Koller et al., 2018).
Synthetic hexaploid wheat (SHW), artificially created hexaploid wheat, is usually developed from crosses of durum wheat, Triticum turgidum ssp. durum (2n = 4x = 28, AABB), and Aegilops tauschii (2n = 2x = 14, DD), followed by chromosome doubling of the F1 hybrids (Kamali et al., 2014). SHW possesses valuable genes for wheat improvement including disease resistance, abiotic-stress tolerance and yield related genes, which can be transferred from tetraploid wheat and Ae. tauschii to common wheat cultivars as a bridge resource (Trethowan and Mujeeb-Kazi, 2008; Li et al., 2011). Since the late 1980s, the International Maize and Wheat Improvement Center (CIMMYT) has developed more than 1000 SHW lines (Das et al., 2016), these SHW lines are widely used to improve wheat quality, yield, and other important agronomic traits all over the world (Li et al., 2018). Some powdery mildew resistance genes from SHW have been reported. The SHW line “XX 194” was derived from the cross of susceptible cultivar T. durum “Moroccos 182” and the selected resistant accession Ae. squarrosa “AE 457/78”. Lutz et al. (1995) revealed “XX 194” conferred the same response pattern to Bgt isolates as Pm2. The SHW line “XX 186” which was produced from a cross between susceptible cultivar T. durum “Santa Marta” and Ae. squarrosa “BGRC 1458” showed a different response pattern from all other named Pm genes, being designated Pm19 (Lutz et al., 1995). Hu et al. (2001) found that SHW line M81 (68.111/RGB-U//WARD/3/Ae. tauschii 452) showed resistance to powdery mildew at both the seedling and adult plant stages. Genetic analyses indicated that the resistance gene originated from Ae. tauschii (Hu et al., 2001).
YAV249 is a SHW line that showed effective resistance to powdery mildew in the field over multiple years. To make better use of its Pm gene(s), the major objectives of this study were to: (i) determine the inheritance of powdery mildew resistance and identify the Pm gene in YAV249; (ii) investigate the relationship between PmYAV and Pm4; (iii) evaluate diagnostic markers for marker-assisted selection (MAS); (iv) apply PmYAV in wheat powdery mildew resistance breeding.
Materials and methods
Plant materials
SHW line YAV249 was derived from the cross YAV-2/TEZ//Ae. squarrosa 249 (T. durum and Ae. tauschii) and is highly resistant to powdery mildew at all growth stages. Wheat cultivar Shixin 828 (SX828) is susceptible to the tested Bgt isolates and used as the susceptible parent to produce F2 and F2:3 populations for genetic analysis and mapping of PmYAV in this study. Wheat cultivar Mingxian 169, which does not carry any known Pm genes, was used as a susceptible control for phenotypic evaluation (Qie et al., 2019). The 47 wheat accessions with known Pm genes or gene combinations were tested with 24 different Bgt isolates collected from diseased wheat fields in different wheat growing areas in China to make comparison with the Pm gene(s) in YAV249 (Table 1) (An et al., 2019). Additionally, 192 wheat accessions including cultivars, landraces, advanced lines and introduced cultivars were determined by using the flanked or co-segregated markers of Pm gene(s) in YAV249 to validate the applicability for MAS (Supplementary Table 1). The susceptible commercial cultivar Kenong 199 (KN199) was used as the recurrent parent to cross and backcross with YAV249 to produce progenies.
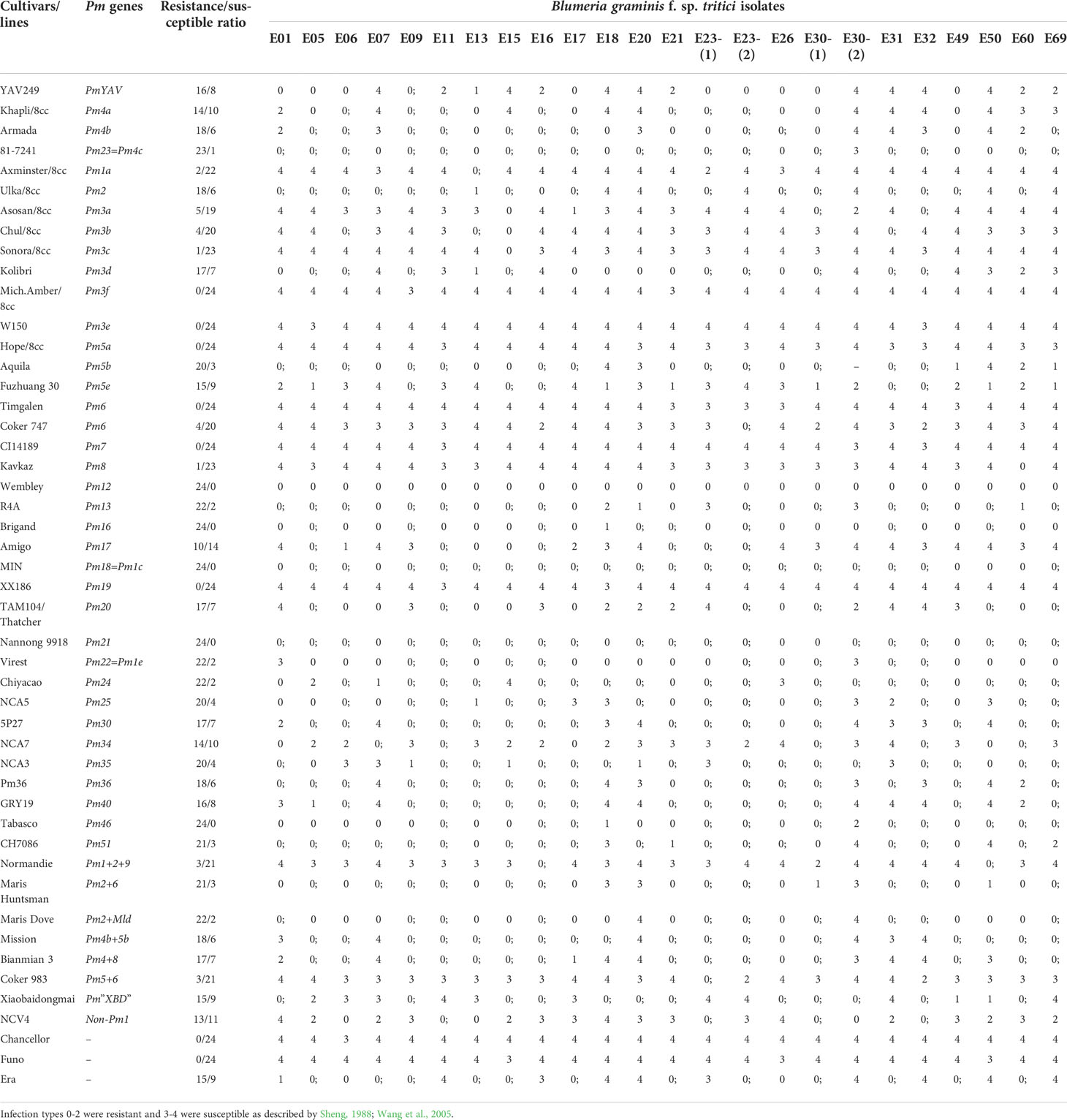
Table 1 Response spectra of YAV249 and wheat accessions with known powdery mildew (Pm) resistance genes to 24 different Blumeria graminis f. sp. tritici (Bgt) isolates at the seedling stage.
Reaction-phenotyping to Bgt isolates
At the adult stage, YAV249 was inoculated with Bgt isolates mixture of E09, E11, E18 and E20 in the field nurseries with three replicates. The assessments were performed from 2019 to 2021 at Luancheng Agro-Ecological Experimental Station (37° 53′ 15″ N, 114° 40′ 47″ E), Chinese Academy of Sciences, Shijiazhuang, Hebei Province, China. YAV249 was planted with four rows, 30 seeds per row (1.5 m), and wheat cultivar Mingxian 169 was planted around the plot as susceptible control for each replicate. When Mingxian 169 showed severe disease symptoms, the phenotype reaction of YAV249 was assessed at least two times in weekly intervals with a 0-9 scale for ITs, ITs 0-2 were considered as highly resistant, ITs 3-4 as moderately resistant, ITs 5-6 as moderately susceptible and ITs 7-9 as highly susceptible (Sheng and Duan, 1991).At the seedling stage, YAV249 and 47 wheat accessions with known Pm genes or gene combinations were tested for response to 24 single-pustule-derived Bgt isolates to compare their resistance spectra (Table 1). Bgt isolate E09 prevalent in the main wheat producing regions of China (Zhou et al., 2005) was used to inoculate YAV249, SX828, KN199 and progenies of YAV249/SX828 and YAV249/KN199. At least 20 plants of each variety/progeny were used for phenotyping of Bgt isolates in three replicates. Response to powdery mildew of all materials was executed in a greenhouse. Materials were planted in the rectangular trays with 128 wells and inoculated at the one-leaf stage by dusting fresh conidia of a Bgt isolate, and Mingxian 169 was planted randomly in the trays as the susceptible control. After inoculation, the trays were treated in a high humidity environment at 22 ± 2°C and 10 h of darkness at 18°C. When the first leaf of Mingxian 169 showed full development of pustules about 14-15 days after inoculation, infection types (ITs) for each plant were scored using a 0-4 scale, and plants with ITs 0-2 were regarded as resistant and those with ITs 3 and 4 susceptible (Sheng, 1988; Wang et al., 2005).
Microscopic analyses of powdery mildew resistance reaction
Microscopic analyses of the plants were performed as previously described (Wang et al., 2014). Two cm leaf segments of YAV249 and SX828 at 7 days post-inoculation (dpi) inoculated with Bgt isolate E09 were fixed at 37°C for 24 h in 2 ml of Carnoy’s Fluid (ethanol: acetic acid, 3:1, v/v), then stained with 2 ml of 0.6% (w/v) Coomassie blue solution for 3 min. Excess dye was rinsed off carefully with distilled water. Samples were observed under an Olympus BX-53 microscope (Olympus, Tokyo, Japan).
Bulked segregant analysis with the wheat 660K SNP array
Genomic DNA was extracted from young leaf tissues after evaluation of their powdery mildew reactions following the method of cetyletrimethylammonium bromide (CTAB) (Sharp et al., 1988). Resistant and susceptible bulks were made from equal amounts of DNA from 20 homozygous resistant and 20 homozygous susceptible F2:3 families of YAV249/SX828 for Illumina wheat 660K single nucleotide polymorphism (SNP) array scanning by China Golden Marker Company (Beijing, China). Single nucleotide polymorphism genotype calling and clustering was performed with software Genome Studio Polyploid Clustering v1.0 (Illumina, http://www.illumina.com). The monomorphic and poor quality SNP markers were excluded from further analyses. Then, the distribution of polymorphic SNPs between the resistant and susceptible DNA bulks on wheat chromosomes was analyzed. An enriched peak of differential SNPs was considered as the candidate interval of the Pm gene(s) in YAV249.
Molecular markers analysis
Based on the predicted interval of 660K SNP array scanning, 52 documented SSR molecular markers in the interval, including 40 SSR markers published on the GrainGenes website (http://wheat.pw.usda.gov) and 12 SSR markers developed by Ullah et al. (2018), were used to test the polymorphisms between the resistant and susceptible parents and bulks. The polymorphic markers between the parents and the bulks were used to genotype the F2:3 families of YAV249/SX828. Then, the markers were aligned on the Chinese Spring reference genome sequence v1.0 (International Wheat Genome Sequencing Consortium (IWGSC), 2018) to confirm their physical locations.
PCR procedure was performed as previously described (Ma et al., 2015). The amplification products of marker BCD1231 were detected by agarose gel electrophoresis with a concentration of 1.5%, and stained with Super GelRed, while the remaining amplification products were separated in 8% non-denaturing polyacrylamide gels with 1×TBE buffer, and visualized by silver staining (Santos et al., 1993).
Map construction
A Chi squared (χ2) test was carried out to investigate deviations of the observed phenotypic data of the F2:3 families from the theoretically expected segregation ratios for analyzing the goodness-of-fit. Then, the linkage map of PmYAV was constructed using MAPMAKER 3.0 and the Kosambi function as reported previously (Kosambi, 1944; Lander et al., 1987).
Cloning and analysis of the Pm4 homologous sequence in YAV249
Since PmYAV was mapped to the Pm4 interval, the homologous sequence of YAV249 was cloned and assembled by using a homology-based cloning strategy according to the previous report on cloning Pm4 (Sánchez-Martín et al., 2021). Then, the homologous sequence of YAV249 was compared with the cloned Pm4 sequence.
RNA extraction and gene expression analysis
The wheat leaf samples of YAV249 were collected at 0, 2, 4, 8, 12, 24, 48 and 72 hours post inoculation (hpi) after inoculating Bgt isolate E09 and with three biological replicates. Total RNA was extracted using RNAiso Plus (TaKaRa, Shiga, Japan). For each sample, 2 μg of RNA was used for reverse transcription with a FastQuant RT Kit (Tiangen, Beijing, China). The transcript levels were detected by qRT-PCR on Bio-Rad CFX 96 with TB Green Premix Ex Taq™ II (TaKaRa, Shiga, Japan). The procedure included an initial step at 95°C for 30 s followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 10 s. The expression levels of target genes were normalized to that of TaActin. All qRT-PCR assays were performed in three independent replications.
Evaluation of the markers for MAS
The 192 wheat accessions including cultivars, landraces, advanced lines and introduced cultivars were tested by using the flanked or co-segregated markers of PmYAV and a specific marker of Pm4 to evaluate their applicability for MAS breeding (Supplementary Table 1). If the polymorphic band(s) of a marker were same in both YAV249 and the tested accessions, it was considered not suitable for MAS. However, when the markers that amplified products in YAV249 that were different from the products in the tested accessions, this indicated that the marker could be used to detect PmYAV when it was transferred into these accessions and considered to be applicable for MAS breeding.
Application of PmYAV in wheat powdery mildew breeding
To transfer PmYAV into susceptible wheat cultivars to improve the resistance to powdery mildew, YAV249 was crossed and backcrossed with wheat cultivar KN199 that is widely grown in Huanghuai region (Zhang et al., 2011). Combined with identification of powdery mildew resistance and evaluation of agronomic traits, the flanked markers and co-segregated marker were used to trace PmYAV in the BC1F1, BC2F1, BC3F1, BC3F2 and BC3F3 populations.
The homozygous resistant progeny line YK13 derived from BC3F3 generation was selected to evaluate agronomic performance along with the parents YAV249 and KN199. These materials were planted at Luancheng Agro-ecosystem Experimental Station (37° 53′ 15″ N, 114° 40′ 47″ E) from 2019 to 2021 with three replicates. Each line was planted in five rows (1.5 m length and 0.25 m between rows) and 20 seeds per row. For each parent/line, 10 plants in the middle three internal rows were randomly sampled to investigate the plant height (PH), spike numbers per plant (SNPP), spike length (SL), spikelet numbers per spike (SNS), sterile spikelet numbers per spike (SSNS), kernel numbers per spike (KNS), thousand-kernel weight (TKW) and kernel related traits. PH and SNPP were assessed based on the mean of 10 plants. SL, SNS, SSNS and KNS were identified based on the mean of the main spike of 10 plants. TKW and kernel related traits (kernel length (KL), kernel width (KW), kernel length/kernel width (KL/W)) of at least 500 kernels randomly were measured three times using the rapid SC-G grain appearance quality image analysis system (WSeen Detection, Hangzhou, China). Analysis of variance (ANOVA) and least significant difference (LSD) test were performed with SPSS statistics v20.0 software (SPSS, Chicago, USA) to test the significance of differences between YK13 and its parents YAV249 and KN199 for each agronomic trait.
Results
Inheritance of powdery mildew resistance in YAV249
From 2019 to 2021, YAV249 was highly resistant with ITs 0-1 to the Bgt isolate mixture of E09, E11, E18 and E20 at the adult stage. When inoculated with Bgt isolate E09 at the seedling stage, YAV249 was highly resistant with IT 0, while SX828 was highly susceptible with IT 4 (Figure 1A). The results of microscopic observation indicated that many mycelia were produced in SX828 but not in YAV249 (Figure 1B). Twenty F1 plants of the cross YAV249/SX828 were resistant with IT 0-1, indicating that the resistance of YAV249 to Bgt isolate E09 was controlled by dominant Pm gene(s). The F2 population from the cross of YAV249/SX828 segregated with 231 resistant and 88 susceptible plants, which fit the theoretical ratio of 3:1 for monogenic segregation of a dominant gene (χ2 = 1.14; P = 0.29). All 319 F2 plants were transplanted in the field to produce F2:3 families. The 250 F2:3 families segregated with 64 homozygous resistant (RR), 119 segregating (Rr) and 67 homozygous susceptible (rr). The result further confirmed the ratio of monogenic inheritance of a single dominant powdery mildew resistance gene (χ2 = 0.65; P = 0.72). Therefore, it was concluded that the resistance to Bgt isolate E09 in YAV249 was controlled by a single dominant gene, tentatively designated PmYAV.
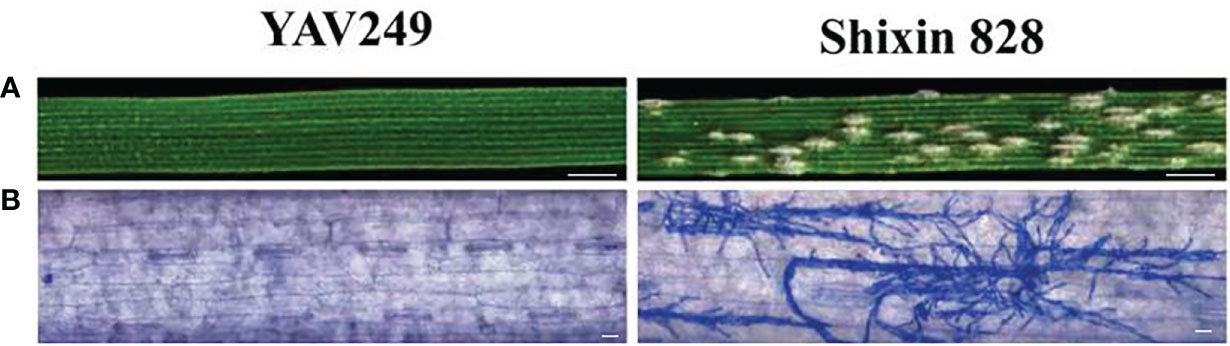
Figure 1 Macroscopic and microscopic characterization of YAV249 and Shixin 828 after inoculating isolate E09 of Blumeria graminis (f) sp. tritici (Bgt). (A) Macroscopic view of the infected representative leaf segments from YAV249 and Shixin 828 at 10 d post-inoculation (dpi). Bar, 5 mm. (B) Microscopic view of leaves from YAV249 and Shixin 828 at 7 dpi to visualize fungal mycelia. Bar, 100 μm.
Bulked segregant analysis with wheat 660K SNP array
To map PmYAV, the resistant and susceptible DNA bulks were genotyped with the 660K SNP array. A total of 1,901 SNPs which was distributed across 21 wheat chromosomes were homozygous polymorphic between the two bulks. Among them, 1,653 SNPs were enriched on chromosome 2A, and 248 SNPs were distributed on other chromosomes (Figure 2A and Supplementary Table 2). Of the 1,653 SNPs located on chromosome 2A, 1,179 SNPs were enriched in an interval of 29 Mb (734 Mb to 763 Mb) on chromosome 2AL (Figure 2B), indicating that PmYAV was mapped in or near this region.
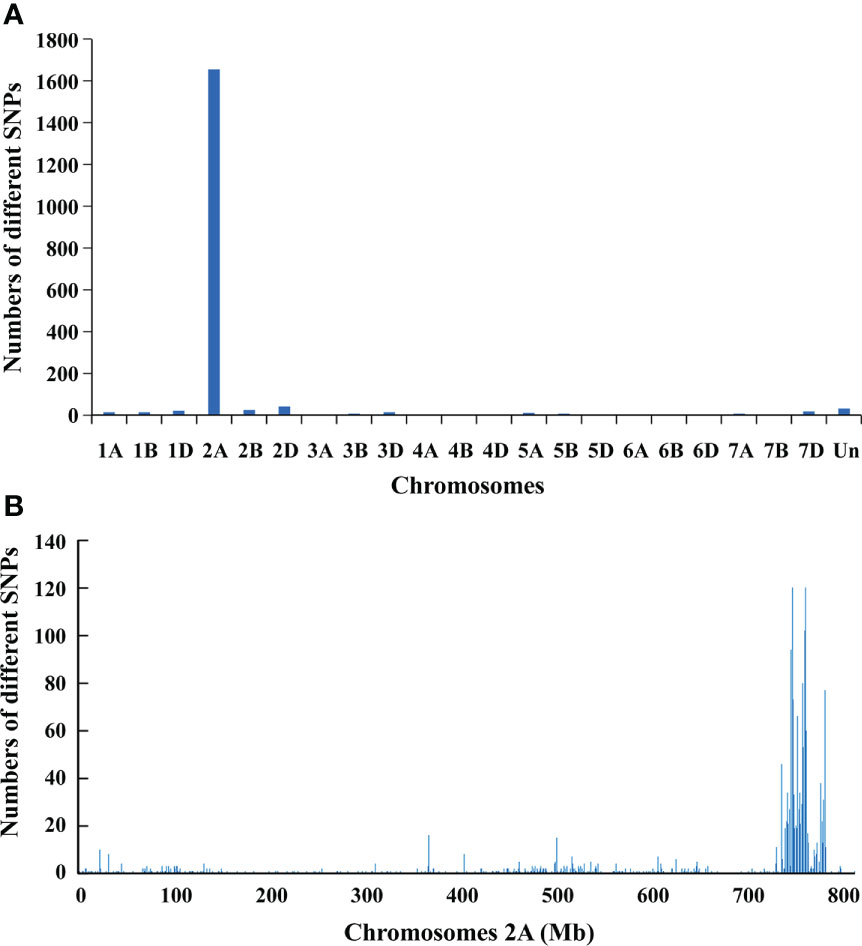
Figure 2 (A) Distribution of polymorphic SNP loci between the resistant and susceptible DNA bulks from the F2:3 families of YAV249/Shixin 828 on 21 chromosomes. (B) Density of polymorphism SNP loci on chromosome 2A with 1 Mb intervals.
Molecular mapping of PmYAV
Based on the mapping interval of the 660K SNP array, 52 SSR molecular markers located on chromosome 2AL were used to screen between resistant parent YAV249, susceptible parent SX828, resistant bulk and susceptible bulk. Among them, 14 markers PSP3039, GPW4474, GWM356, GWM311, GWM382, GDM93, GWM526, GPW4456, BCD1231, WGRC763, WGRC872, WGRC1096, WGRC869 and WGRC883 showed consistent polymorphism between parents and bulks. Subsequently, these markers were used to genotype the 250 F2:3 families and confirm linkage with PmYAV. The result demonstrated that PmYAV co-segregated with five markers Xwgrc883, Xwgrc969, Xwgrc1096, Xwgrc872 and Xbcd1231 and was flanked by Xwgrc763 and Xgdm93 with genetic distances 0.8 cM and 1.2 cM corresponding to a physic interval of 1.89 Mb (760.58-762.47 Mb) on the Chinese Spring reference genome sequence v1.0 (Figure 3). There were four reported and formally designated Pm genes including Pm4, Pm33, Pm50 and Pm65 on chromosome 2AL (Li et al., 2019), and PmYAV mapped closed to Pm4.
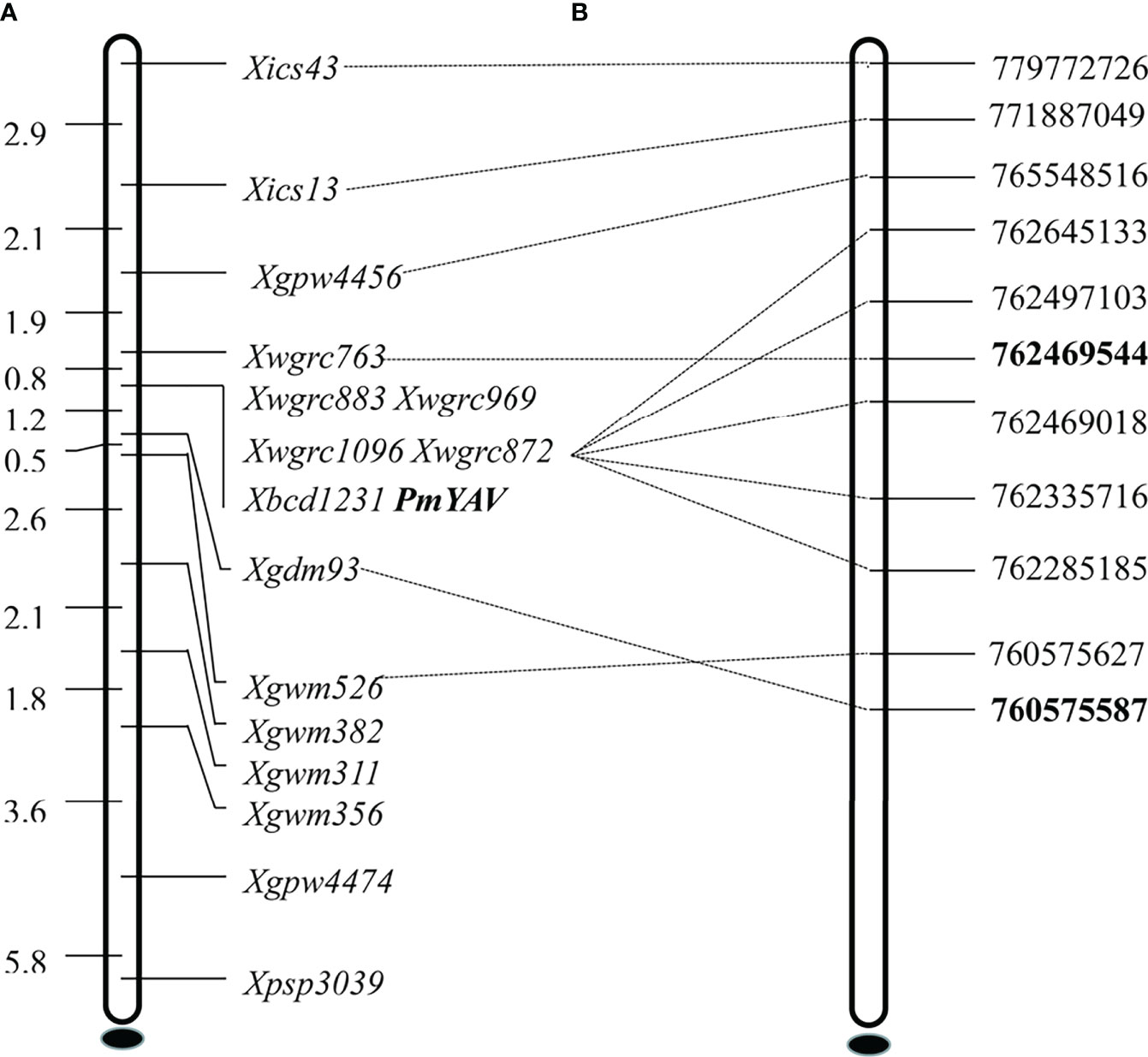
Figure 3 Genetic linkage map (A) and physical map (B) of powdery mildew resistance gene PmYAV using the F2:3 families of YAV249/Shixin 828.
Cloning and analysis of the Pm4 homologous sequence in YAV249
To further explore the relationship between PmYAV and Pm4, we genotyped all the 250 F2:3 families from YAV249/SX828 with the functional molecular marker JS717/JS718 of Pm4. JS717/JS718 showed positive amplification in YAV249 and co-segregated with PmYAV in the F2:3 families. Then, the Pm4 homologous coding sequence in YAV249 was cloned using nested PCR, and the result showed that Pm4 homologous sequence in YAV249 was totally consistent with the Pm4a sequence. However, PmYAV generated an extra splicing event compared to the previously reported Pm4a_V1 and Pm4a_V2 (Sánchez-Martín et al., 2021), suggesting that there was an extra 67 bp-intron in PmYAV. The intron was located on the second exon in the serine/threonine kinase domain of Pm4a_V1 (Figure 4).
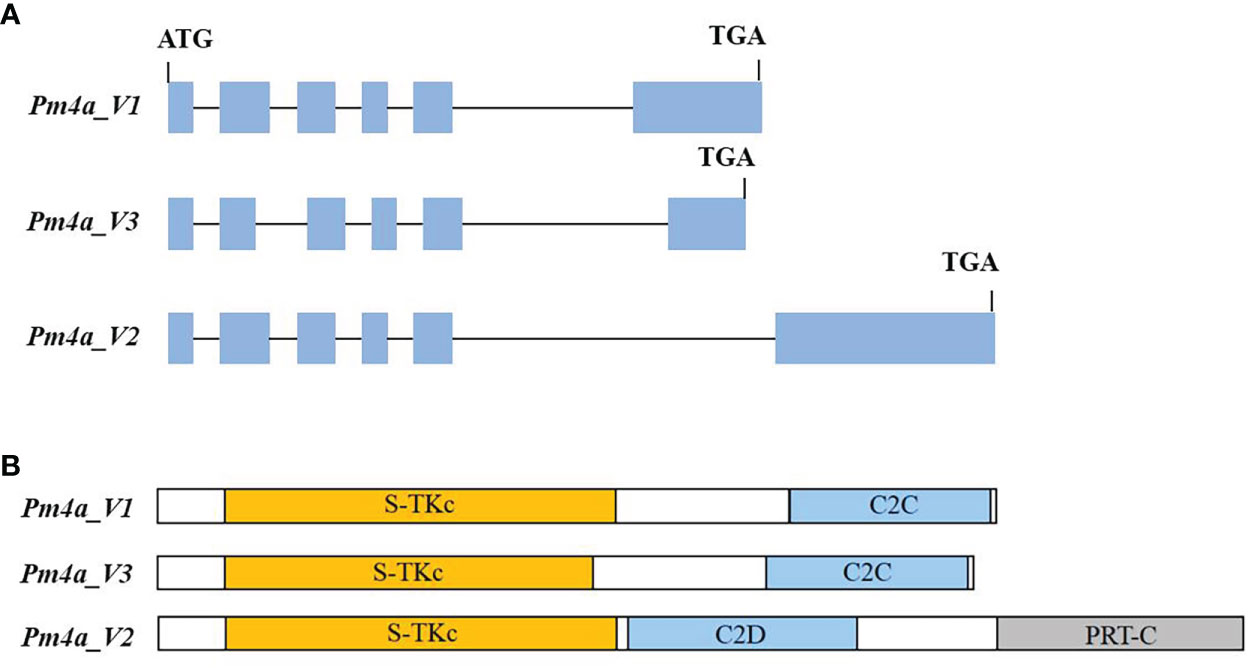
Figure 4 (A) Schematic representation of gene structures of PmYAV splicing variants. Gene exons and introns are shown by blue boxes and black lines, respectively. (B) Pm4a_V1, Pm4a_V2 and Pm4a_V3 protein isoforms with domains indicated by colours: yellow, serine/threonine kinase (S-TKc); light blue, C2; grey, phosphoribosyl transferase C-terminal (PRT-C).
Gene expression analysis of PmYAV
To investigate the expression patterns of PmYAV, we performed qRT-PCR analysis. Pm4a_V1 and Pm4a_V2 in YAV249 showed similar expression patterns and were upregulated after Bgt isolate E09 infection, peaked at 24 hpi, and then reduced at later stages between 48 and 72 hpi. The level of Pm4a_V2 was higher significantly than that of Pm4a_V1 at most of the time points (Figure 5). We speculated that Pm4a_V2 had a greater contribution than Pm4a_V1 to resistance in YAV249.
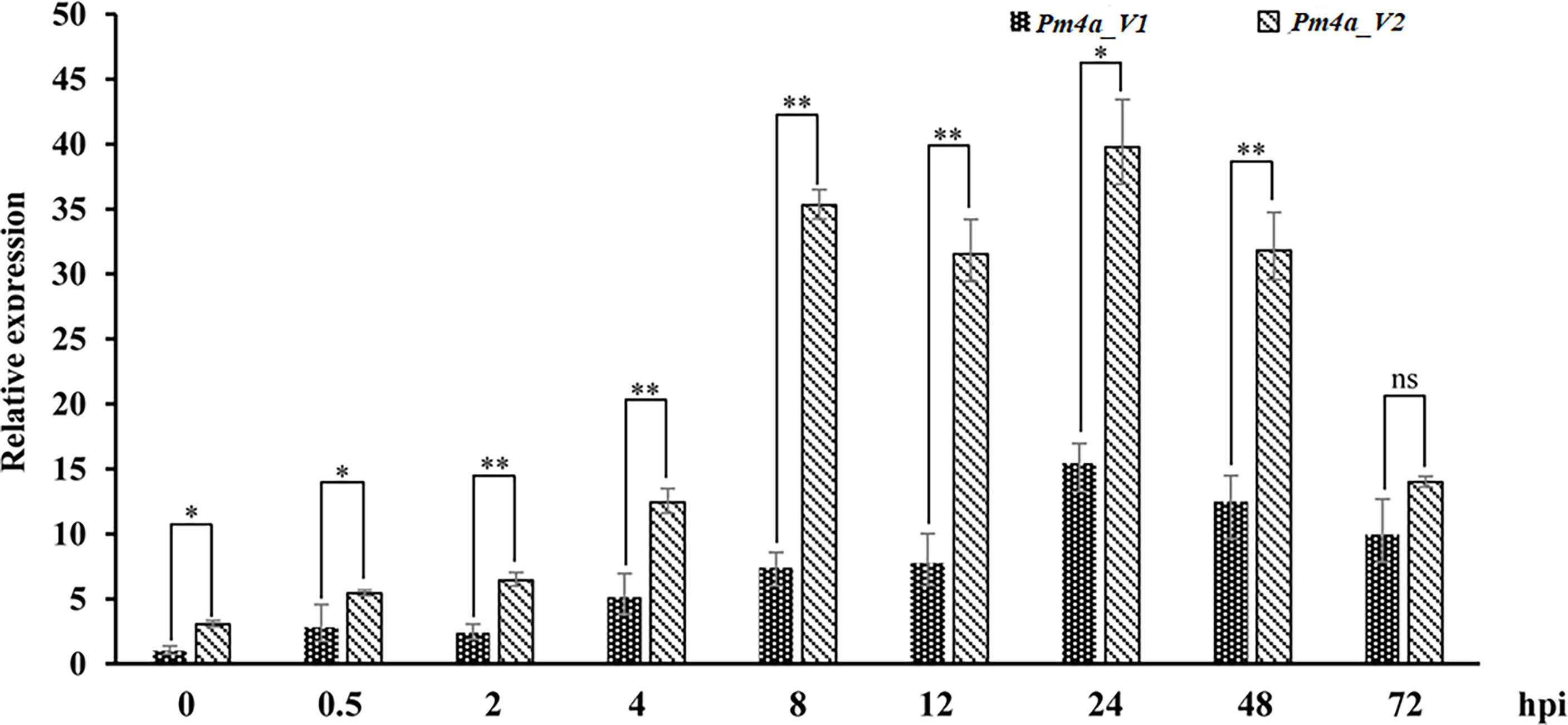
Figure 5 Expression patterns of the Pm4a_V1 and Pm4a_V2 splice variants in YAV249 during a 72-hour time course. Error bars represent SD based on three independent repeats. Asterisks indicate significant differences (t-tests) between Pm4a_V1 and Pm4a_V2 at each time point (*P < 0.05, **P < 0.01, ns: not significant). TaActin was used as the internal control.
The difference of powdery mildew resistance spectrum between YAV249 and documented Pm4a stock
When inoculated with 24 Bgt isolates, Pm4a was resistant to 14 out of 24 Bgt isolates, including E01, E05, E06, E09, E11, E13, E16, E17, E21, E23-1, E23-2, E26, E30-1 and E49, while PmYAV was also resistant to E60 and E69 besides those 14 Bgt isolates (Table 1). Therefore, PmYAV showed a different reaction to two Bgt isolates compared to Pm4a.
Evaluation of the markers of PmYAV for MAS
To investigate the applicability of the markers linked or co-segregated with PmYAV in MAS, the flanked markers GDM93 and WGRC763 and the co-segregating markers BCD1231 and JS717/JS718 of PmYAV were used to test 192 wheat accessions (Supplementary Table 1). Markers GDM93 and WGRC763 amplified specific bands that differed from YAV249 in 147, and 140 out of 192 accessions respectively. The same as JS717/JS718, BCD1231 amplified different bands between YAV249 and 137 out of 192 accessions. The results indicated that these four markers could be used in MAS for detecting PmYAV in those wheat genetic backgrounds (Figure 6 and Supplementary Table 1). However, this was not the case for all the marker alleles in wheat cultivars Aikang 58, Xinmai 1998, ROANE, Safha 3 and Taixue 12 due to the same bands between YAV249 and them (Figure 6 and Supplementary Table 1).
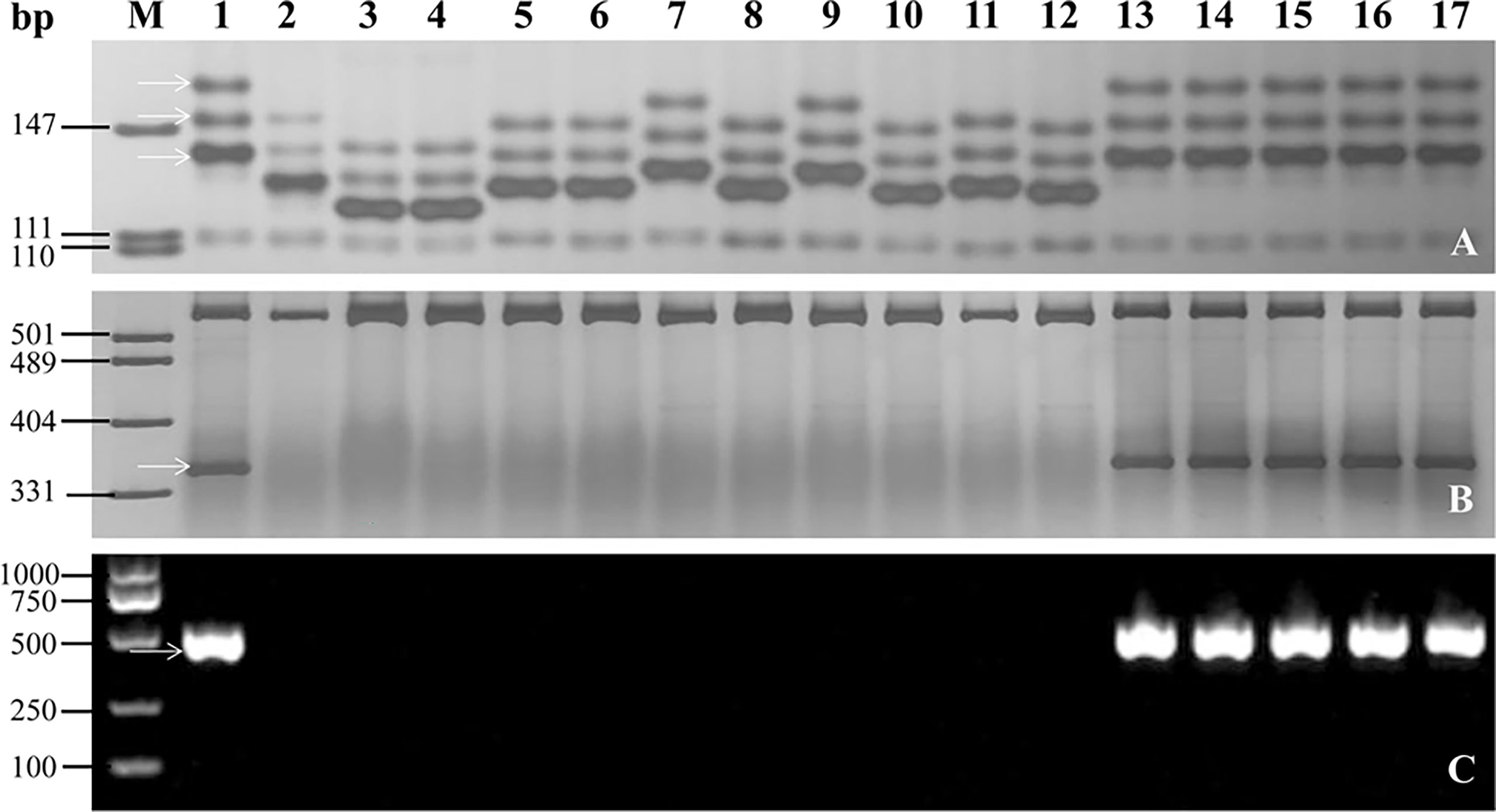
Figure 6 Amplification patterns of the flanked markers GDM93 (A) and WGRC763 (B) and the co-segregated marker BCD1231 (C) of PmYAV in YAV249, Shixin 828 and selected wheat cultivars/lines. M: pUC19/MspI; 1: YAV249; 2: Shixin 828; 3: Shi 4185; 4: Shiyou 17; 5: Shimai 18; 6: Shixin 633; 7: Ji 5265; 8: Kenong 199; 9: Zhengyumai 9989; 10: Zhou 16; 11: Jinmai 41; 12: Gaoyou 503; 13: Xinmai 1998; 14: ROANE; 15: Safha 3; 16: Aikang 58; 17: Taixue 12. The white arrows indicate the specific amplified fragments.
Application of PmYAV in wheat powdery mildew breeding
To develop a resistance pre-breeding resource, we transferred PmYAV into the commercial susceptible cultivar KN199 by MAS, combined with evaluation of powdery mildew resistance and analysis of agronomic traits. An advanced breeding line YK13 was selected from a BC3F3 population of YAV249/4*KN199 due to its desirable agronomics traits and resistance to powdery mildew. During 2019-2021, analysis of agronomic traits indicated that the PmYAV donor parent YAV249 had a greater SL, TKW and KL but a poorer PH than the recurrent parent KN199. Compared with KN199, YK13 showed improved SL, TKW and KL in one or two years. Moreover, there was no significant difference on PH, SNPP, SNS, KNS and KW between YK13 and its recurrent parent KN199 (Table 2).
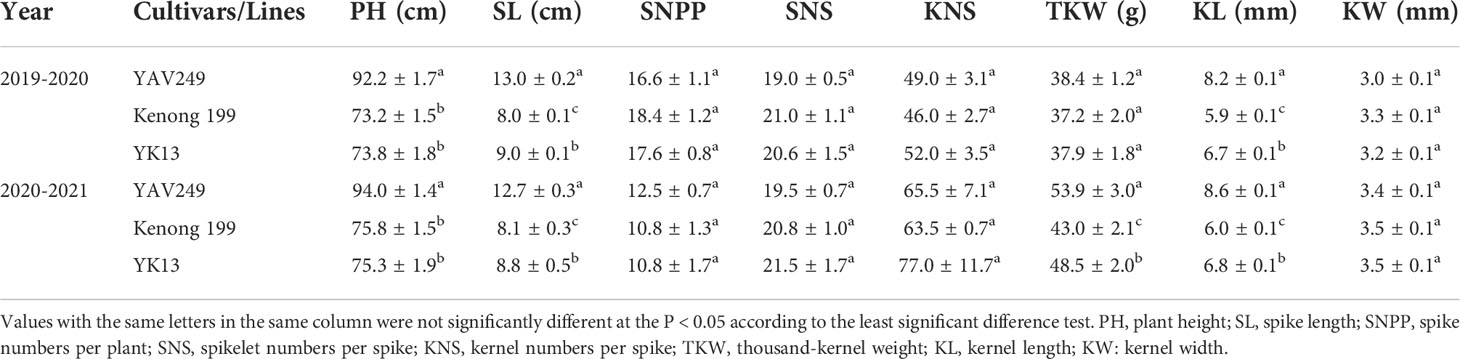
Table 2 The agronomic trait evaluation of the advanced breeding line YK13 with its donor parent YAV249 and recurrent parent Kenong 199.
Discussion
SHW lines possess important resistance genes effective against various diseases, therefore, regular screening of SHW genotypes is important for identifying new sources of resistance to powdery mildew that could eventually be used in wheat disease resistance breeding programs. In this study, the SHW line YAV249 derived from the cross YAV-2/TEZ//Ae. squarrosa 249 was identified to be resistant to powdery mildew at all stages. Genetic analysis indicated that its resistance was conferred by a single dominant gene, temporarily designated PmYAV. Bulked segregant analysis by using wheat 660K SNP array scanning and marker analysis showed that PmYAV was mapped in the Pm4 interval on chromosome 2AL.
Up to now, eight alleles (Pm4a-Pm4h) have been reported at the Pm4 locus (Sánchez-Martín et al., 2021), which makes it multiple allelic loci as previously known Pm1 (Hsam et al., 1998), Pm2 (Jin et al., 2018), Pm3 (Bhullar et al., 2010) and Pm5 (Xie et al., 2020) loci. Pm4a was derived from tetraploid wheat (T. turgidum) cultivars Yuma and Khapli (Briggle, 1966). However, Pm4b was originally reported in three bread wheat cultivars Weihenstephan M1, TP29 and ELS (Wolfe, 1965; The et al., 1979). Subsequently, Pm4b was postulated in French cultivar VPM1 (Bariana and McIntosh, 1994). Three alleles at Pm4 locus, including Pm4c (Pm4c = Pm23), Pm4d, and Pm4e, were identified in wheat line 81-7241, Tm27d2, and landrace D29, respectively (Hao et al., 2008; Schmolke et al., 2012; Li et al., 2017). Pm4f, Pm4g and Pm4h which were susceptible to most of the tested Bgt isolates were discovered by using the haplotype-specific marker JS717/JS718 in a global wheat collection of 512 accessions (Sánchez-Martín et al., 2021). In this study, PmYAV was resistant to 16 of 24 Bgt isolates at the seedling stage which was different from the tested Pm4a and Pm4b (Table 1). Meanwhile, the donor of PmYAV was also different from those of Pm4a-Pm4h. Therefore, PmYAV might be a new allele at the Pm4 locus. The identification of PmYAV enriched the genetic diversity of Pm4 locus, which was available for increasing the durable resistance conferred by Pm4 locus in wheat powdery mildew resistance breeding since it has a resistance spectrum different from that of Pm4a, Pm4b and Pm4c, but unknown with that of Pm4d to Pm4h which were not tested in the phenotyping study.
Recently, Pm4b, which consisted of seven exons and contained two alternative transcripts, was cloned by using the strategy of MutChromSeq (Sánchez-Martín et al., 2021). Subsequently, all the alleles at Pm4 locus were cloned with the full-length amplification and Sanger sequencing and indicated Pm4 alleles contain SNPs and/or combinations of shared SNPs which have main effects on the kinase domain. What is even more interesting is that most of the SNP cause amino acid changes in the transmembrane and S_TKc domains. Pm4 generated two isoforms which were both required for resistance (Sánchez-Martín et al., 2021). In this study, using homology-based cloning, we found that the Pm4 homologous sequence in YAV249 was consistent with Pm4a sequence, while PmYAV generated an extra splicing event for the reported Pm4a_V1 and Pm4a_V2, suggesting that there was an extra 67 bp-intron in PmYAV (Figure 4). It may cause different resistance reactions between PmYAV and other alleles. Then, the function of the extra splicing needs to be further investigated in future by other methods, such as virus-induced gene silencing and transgenic assay analysis. On the other hand, Sánchez-Martín et al. (2021) reported that the expression of Pm4b_V1 and Pm4b_V2 on the wild-type Pm4b wheat genotype Fed-Pm4b after infection with powdery mildew did not significantly differ from each other. Similar expression levels of both transcripts indicate that Pm4b_V1 and Pm4b_V2 have a similar contribution to powdery mildew resistance. However, in this study, when inoculated Bgt isolate E09, the expression level of Pm4b_V2 in YAV249 was higher significantly than that of Pm4b_V1 at most of the time points (Figure 5) which might cause the different resistance spectra of PmYAV from other alleles at Pm4 locus.
When a valuable resistance gene or accession was identified, its rational utilization was the first factor to be considered in wheat disease resistance breeding. MAS is a high priority in wheat breeding programs to efficiently transfer R gene to susceptible cultivars or pyramid with other R gene(s). Sánchez-Martín et al. (2021) reported a dominant diagnostic marker JS717/JS718 which could detect the Pm4 in different genetic background. In this study, YAV249 showed a high level of resistance to powdery mildew and possessed desirable agronomic traits, such as higher TKW and KL/KW. Two dominant markers WGRC763 closely linked to PmYAV and BCD1231 and JS717/718 co-segregated with PmYAV and can be used for MAS in many genetic backgrounds. More importantly, the co-dominant marker GDM93 which was closely linked to PmYAV could be used for selection of homozygotes during MAS breeding. In fact, an advanced breeding line YK13, which possessed not only desirable agronomic traits but excellent resistance to powdery mildew, was selected in BC3F3 population of YAV249/KN199 by MAS.
SHW is a valuable genetic resource for improving disease resistance in wheat cultivars. In our study, we identified an extra splicing variant of Pm4a from SHW line YAV249 and selected a new wheat breeding line YK13 with high resistance to powdery mildew and excellent agronomic traits. The results will add diversity to the available genes for powdery mildew resistance and provide the foundation for analyzing alternative splicing mechanism in regulating the Pm4 gene function.
Conclusion
To sum up, in the present study, we identified a dominant powdery mildew resistance gene PmYAV in synthetic hexaploid wheat line YAV249 using bulked segregant analysis with wheat 660K single nucleotide polymorphism (SNP) array scanning and marker analysis. Sequence alignment analysis demonstrated that the sequence of PmYAV was totally consistent with that of Pm4a but generated an extra splicing. The flanked markers GDM93 and WGRC763, and the co-segregated markers BCD1231 and JS717/JS718 were confirmed to be easily performed in marker-assisted selection (MAS) of PmYAV. Using MAS strategy, PmYAV was transferred into the commercial cultivar Kenong 199 (KN199) and a wheat line YK13 was selected in BC3F3 from the population of YAV249/KN199 due to its excellent agronomic traits and resistance to powdery mildew. Our study can be valuable for enhancing the genetic diversity of powdery mildew resistance in breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
HZ and DA conceived the research. YJ, TG, XL, GH and ZS performed the experiments. TG, XL, YZ, JF, JW and HL developed the experimental materials. YZ, JF, JW and WL performed the phenotypic assessment. YJ and TG wrote the manuscript. DA and HZ supervised and revised the writing of the article. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key Research and Development Program of China No. 2021YFD1200600, and the National Natural Science Foundation of China No. 32272105.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1048252/full#supplementary-material
Supplementary Table 1 | Validation of the flanked markers GDM93 and WGRC763 and the co-segregated markers BCD1231 and JS717/JS718 of PmYAV on 192 wheat accessions in marker-assisted selection (MAS). ‘+’ amplified marker allele differs from that in YAV249, indicating that the marker might be useful in MAS, and ‘−’ marker is similar to that in YAV249, indicating that the marker could not be used in MAS.
References
An, D. G., Ma, P. T., Zheng, Q., Fu, S. L., Li, L. H., Han, F. P., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Bariana, H. S., McIntosh, R. A. (1994). Characterization and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 76, 53–61. doi: 10.1007/bf00024020
Bhullar, N. K., Zhang, Z. Q., Wicker, T., Keller, B. (2010). Wheat gene banks as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol. 10, 88. doi: 10.1186/1471-2229-10-88
Briggle, L. W. (1966). Transfer of resistance to Erysiphe graminis f. sp. tritici from khapli emmer and yuma durum to hexaploid wheat. Crop Sci. 6, 459–461. doi: 10.2135/cropsci1966.0011183x000600050020x
Brunner, S., Hurni, S., Herren, G., Kalinina, O., von Burg, S., Zeller, S. L., et al. (2011). Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol. J. 9, 897–910. doi: 10.1111/j.1467-7652.2011.00603.x
Das, M. K., Bai, G., Mujeeb-Kazi, A., Rajaram, S. (2016). Genetic diversity among synthetic hexaploid wheat accessions (Triticum aestivum) with resistance to several fungal diseases. Genet. Resour. Crop Evol. 63, 1285–1296. doi: 10.1007/s10722-015-0312-9
Hao, Y. F., Liu, A. F., Wang, Y. H., Feng, D. S., Gao, J. R., Li, X. F., et al. (2008). Pm23: a new allele of Pm4 located on chromosome 2AL in wheat. Theor. Appl. Genet. 117, 1205–1212. doi: 10.1007/s00122-008-0827-y
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., Wu, Q. H., et al. (2020). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
Hsam, S. L. K., Huang, X. Q., Ernst, F., Hartl, L., Zeller, F. J. (1998). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum l. em thell.). 5. alleles at the Pm1 locus. Theor. Appl. Genet. 96, 1129–1134. doi: 10.1007/s001220050848
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969. doi: 10.1111/tpj.12345
Hu, Y. K., Xin, Z. Y., Chen, X., Zhang, Z. Y., Duan, X. Y. (2001). Genetic analysis and gene deduction of powdery mildew resistance in T. durum-ae. squarrosa amphiploid. Acta Genet Sin. 28, 152–157. doi: 10.1088/0256-307x/18/11/313
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jin, Y. L., Shi, F. Y., Liu, W. H., Fu, X. Y., Gu, T. T., Han, G. H., et al. (2021). Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2,978 wheat accessions. Plant Dis. 105, 3900–3908. doi: 10.1094/pdis-03-21-0532-re
Jin, Y. L., Xu, H. X., Ma, P. T., Fu, X. Y., Song, L. P., Xu, Y. F., et al. (2018). Characterization of a new Pm2 allele associated with broad-spectrum powdery mildew resistance in wheat line subtil. Sci. Rep. 8, 475. doi: 10.1038/s41598-017-18827-4
Kamali, S., Majidi, E., Khodarahmi, M., Mehrabi, R. (2014). Evaluation of CIMMYT synthetic hexaploid wheats for resistance to septoria tritici blotch. Crop Breed. J. 4, 23–33. doi: 10.22092/cbj.2014.109669
Koller, T., Brunner, S., Herren, G., Hurni, S., Keller, B. (2018). Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor. Appl. Genet. 131, 861–871. doi: 10.1007/s00122-017-3043-9
Kosambi, D. D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1007/978-81-322-3676-4_16
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daley, M., Lincoln, S., et al. (1987). Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Li, G. Q., Cowger, C., Wang, X. W., Carver, B. F., Xu, X. Y. (2019). Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor. Appl. Genet. 132, 2625–2632. doi: 10.1007/s00122-019-03377-2
Li, M. M., Dong, L. L., Li, B. B., Wang, Z. Z., Xie, J. Z., Qiu, D., et al. (2020). A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228, 1027–1037. doi: 10.1111/nph.16761
Li, N., Jia, H. Y., Kong, Z. X., Tang, W. B., Ding, Y. X., Liang, J. C., et al. (2017). Identification and marker-assisted transfer of a new powdery mildew resistance gene at the Pm4 locus in common wheat. Mol. Breed. 37, 79. doi: 10.1007/s11032-017-0670-4
Li, A., Liu, D., Yang, W., Kishii, M., Mao, L. (2018). Synthetic hexaploid wheat: yesterday, today, and tomorrow. Engineering 4, 552–558. doi: 10.1016/j.eng.2018.07.001
Li, J., Wei, H. T., Hu, X. R., Li, C. S., Tang, Y. L., Liu, D. C., et al. (2011). Identification of a high-yield introgression locus from synthetic hexaploid wheat in chuanmai 42. Acta Agronom. Sin. 37, 255–262. doi: 10.3724/sp.j.1006.2011.00263
Lu, P., Guo, L., Wang, Z. Z., Li, B. B., Li, J., Li, Y. H., et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680. doi: 10.1038/s41467-020-14294-0
Lutz, J., Hsam, S., Limpert, E., Zeller, F. J. (1995). Chromosomal location of powdery mildew resistance genes in Triticum aestivum l. (common wheat). 2. genes Pm2 and Pm19 from Aegilops squarrosa l. Heredity 74, 152–156. doi: 10.1038/hdy.1995.22
Ma, P. T., Xu, H. X., Xu, Y. F., Li, L. H., Qie, Y. M., Luo, Q. L., et al. (2015). Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 128, 613–622. doi: 10.1007/s00122-015-2457-5
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., Raupp, W. J. (2020). Catalogue of gene symbols for wheat: 2020 supplement. Available at: https://wheat.pw.usda.gov/GG3/WGC.
Moore, J. W., Herrera-Foessel, S., Lan, C. X., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Qie, Y. M., Sheng, Y., Xu, H. X., Jin, Y. L., Ma, F. F., Li, L. H., et al. (2019). Identification of a new powdery mildew resistance gene PmDHT at or closely linked to the Pm5 locus in the Chinese wheat landrace dahongtou. Plant Dis. 103, 2645–2651. doi: 10.1094/pdis-02-19-0401-re
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. doi: 10.1186/s13059-016-1082-1
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants. 7, 327–341. doi: 10.1038/s41477-021-00869-2
Santos, F. R., Pena, S. D. J., Epplen, J. T. (1993). Genetic and population study of a y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 90, 655–656. doi: 10.1007/bf00202486
Schmolke, M., Mohler, V., Hartl, L., Zeller, F. J., Hsam, S. L. K. (2012). A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol. Breed. 29, 449–456. doi: 10.1007/s11032-011-9561-2
Sharp, P. J., Kreis, M., Shewry, P. R., Gale, M. D. (1988). Location of β-amylase sequences in wheat and its relatives. Theor. Appl. Genet. 75, 286–290. doi: 10.1007/bf00303966
Sheng, B. Q. (1988). Wheat powdery mildew was recorded using infection type in seedling stage. Plant Prot. 14, 49. Available at: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CJFD&dbname=CJFD8589&filename=ZWBH198801035&uniplatform=NZKPT&v=EleQOgszeUXu-PqypFjom3Acylv32Ikwf0r2_3oTXJpIMQ4n8xGbEPM7PZ-6Sx1q
Sheng, B. Q., Duan, X. Y. (1991). Improvement of scale 0-9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing. Agr. Sci. 1, 38–39. Available at: https://kns.cnki.net/kcms/detail/detail.aspx?filename=BJNK199101011&dbcode=CJFD&dbname=CJFD1991&v=Rxg8UkEIGV3B5TxBLD9dSq7XlRDsCXbtegphAfhso0TfBjG8a6XZCKAo6ZUWwcFN
Singh, S. P., Hurni, S., Ruinelli, M., Brunner, S., Sanchez-Martin, J., Krukowski, P., et al. (2018). Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98, 249–260. doi: 10.1007/s11103-018-0780-3
Srichumpa, P., Brunner, S., Keller, B., Yahiaoui, N. (2005). Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895. doi: 10.1104/pp.105.062406
The, T. T., McIntosh, R. A., Bennett, F. G. A. (1979). Cytogenetical studies in wheat. IX. monosomic analyses, telocentric mapping and linkage relationships of genes Sr21, Pm4, and Mle. Aust. J. Biol. Sci. 32, 115–125. doi: 10.1071/bi9790115
Trethowan, R. M., Mujeeb-Kazi, A. (2008). Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 48, 1255–1265. doi: 10.2135/cropsci2007.08.0477
Ullah, K. N., Li, N., Shen, T., Wang, P. S., Tang, W. B., Ma, S. W., et al. (2018). Fine mapping of powdery mildew resistance gene Pm4e in bread wheat (Triticum aestivum l.). Planta 248, 1329–1329. doi: 10.1007/s00425-018-2990-y
Wang, Y. P., Cheng, X., Shan, Q. W., Zhang, Y., Liu, J. X., Gao, C. X., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/pd-89-0457
Wolfe, M. S. (1965). Physiologic specialization of Erysiphe graminis f. sp. tritici in the united kingdom. Trans. Bril. Mycol. Soc 48, 315–326. doi: 10.1016/S0007-1536(65)80099-7
Xie, J. Z., Guo, G. H., Wang, Y., Hu, T. Z., Wang, L. L., Li, J. T., et al. (2020). A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228, 1011–1026. doi: 10.1111/nph.16762
Xing, L. P., Hu, P., Liu, J. Q., Witek, K., Zhou, S., Xu, J. F., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS- LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878. doi: 10.1016/j.molp.2018.02.013
Zhang, W., Wang, J., Ji, J., Wang, Z. G., An, D. G., Zhang, X. Q., et al. (2011). ). development of “Kn199” new winter wheat variety and its cultivation in China. Chin. J. Eco. Agric. 19, 1215–1219. doi: 10.3724/sp.j.101l.2011.01215
Zhou, R. H., Zhu, Z. D., Kong, X. Y., Huo, N. X., Tian, Q. Z., Li, P., et al. (2005). Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 110, 640–648. doi: 10.1007/s00122-004-1889-0
Keywords: Blumeria graminis f. sp. tritici, PmYAV, Pm4a, markers, marker-assisted selection
Citation: Jin Y, Gu T, Li X, Liu H, Han G, Shi Z, Zhou Y, Fan J, Wang J, Liu W, Zhao H and An D (2022) Characterization of a new splicing variant of powdery mildew resistance gene Pm4 in synthetic hexaploid wheat YAV249. Front. Plant Sci. 13:1048252. doi: 10.3389/fpls.2022.1048252
Received: 19 September 2022; Accepted: 10 October 2022;
Published: 26 October 2022.
Edited by:
Xiaoli Fan, Chengdu Institute of Biology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Huagang He, Jiangsu University, ChinaCheng Liu, Crop Research Institute, Shandong Academy of Agricultural Sciences, China
Copyright © 2022 Jin, Gu, Li, Liu, Han, Shi, Zhou, Fan, Wang, Liu, Zhao and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: He Zhao, hezhao311@163.com; Diaoguo An, dgan@sjziam.ac.cn
†These authors have contributed equally to this work
 Yuli Jin
Yuli Jin Tiantian Gu1†
Tiantian Gu1† Hong Liu
Hong Liu Yilin Zhou
Yilin Zhou Jieru Fan
Jieru Fan Jing Wang
Jing Wang Wei Liu
Wei Liu Diaoguo An
Diaoguo An