- State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China
Mulberry (genus Morus) is an economically important woody plant with an altered ploidy level. The variable number of Morus species recognized by different studies indicates that the genus is in need of revision. In this study, the chloroplast (CP) genomes of 123 Morus varieties were de novo assembled and systematically analyzed. The 123 varieties represented six Morus species, namely, Morus alba, Morus nigra, Morus notabilis, Morus rubra, Morus celtidifolia, and Morus serrata. The Morus CP genome was found to be 158,969~159,548 bp in size with 125 genes, including 81 protein coding, 36 tRNA, and 8 rRNA genes. The 87 out of 123 mulberry accessions were assigned to 14 diverse groups with identical CP genome, which indicated that they are maternally inherited and share 14 common ancestors. Then 50 diverse CP genomes occurred in 123 mulberry accessions for further study. The CP genomes of the Morus genus with a quadripartite structure have two inverted repeat (IR) regions (25,654~25,702 bp) dividing the circular genome into a large single-copy (LSC) region (87,873~88,243 bp) and small single-copy (SSC) region (19,740~19,994 bp). Analysis of the phylogenetic tree constructed using the complete CP genome sequences of Morus revealed a monophyletic genus and that M. alba consisted of two clades, M. alba var. alba and M. alba var. multicaulis. The Japanese cultivated germplasms were derived from M. alba var. multicaulis. We propose that the Morus genus be classified into six species, M. nigra, M. notabilis, M. serrata, M. celtidifolia, M. rubra, and M. alba with two subspecies, M. alba var. alba and M. alba var. multicaulis. Our findings provide a valuable resource for the classification, domestication, and breeding improvement of mulberry.
Introduction
Mulberry (Morus L., Moraceae) (Group, 2009) comprises a variable number of species, with the first 7 described by Linnaeus (1753). The traditional taxonomy of Morus is often based on minor morphological differences (Gardner et al., 2021). Various researchers identified 5, 8, 13, 16, 24, and 35 Morus species using morphological and/or molecular methods (Bureau, 1873; Koidzumi, 1930; Hotta, 1954; Zhou and Gilbert, 2003; Zeng et al., 2015; Jain et al., 2022). The classification of the Morus genus based on morphology did not truly reflect the phylogenetic relationships (Jiao et al., 2020). In 2015, we proposed eight species in the Morus genus on the basis of ITS (internal transcribed spacer) sequences, which were recently verified by an analysis of population genetics (Jiao et al., 2020). A recent investigation of Moraceae revealed that the genus Morus is a monophyletic group without M. mesozygia and M. insignis, and the delimitation of M. alba may be worth further investigating (Gardner et al., 2021). However, the genome-based taxonomy of the genus Morus remains unexplored (Jiao et al., 2020).
Since the first mulberry genome (M. notabilis) was published (He et al., 2013), six other mulberry genomes, including chromosome-level genomes, have been reported (Jiao et al., 2020; Muhonja et al., 2020; Jain et al., 2022; Xia et al., 2022; Xuan et al., 2022), which represent excellent reference genomes for genomic and population analyses of mulberry resources. Different chromosome numbers (14, 28, 35, 42, 49, 56, 84, 112, 126, and 308) with various ploidy levels have been reported in mulberry (Tikader and Kamble, 2008; Xuan et al., 2022). For example, black mulberry (M. nigra) is a polyploid with 308 chromosomes (Agaev and Fedorova, 1970; SB et al., 1990), and Morus serrata is a natural polyploid with 56 or 84 chromosomes (SB and Rajan, 1989). Variable ploidy levels are often observed in white mulberry (M. alba) (Xuan et al., 2019). The variable ploidy levels in Morus adversely affect population genetic analyses for taxonomic purposes. Mulberry has been cultivated by farmers for over 5000 years (He et al., 2013), and many varieties or cultivars have been generated by natural and artificial breeding selection. More than 2600, 1500, and 1120 mulberry germplasm resources were recorded in China, Japan, and India, respectively (Vijayan et al., 2011; Vijayan and B.da Silva, 2011). The evolutionary relationships of these cultivars or varieties remain unclear.
Chloroplast, a plant cell organelle with its own genome, is essential for the growth and development of plants. Compared with the large nuclear genome, chloroplast genomes are smaller. CP genomes with their numerous advantages for plant phylogeny reconstruction, including a relatively conserved rate of evolution and usually uniparental inheritance, provide an important resource for elucidating morphological evolution (Gitzendanner et al., 2018; Li et al., 2021; Hua et al., 2022). CP genomes also provide critical insights into historically difficult relationships of the major angiosperm subclades (Moore et al., 2007; Moore et al., 2010; Stull et al., 2015; Sun et al., 2016; Li H-T. et al., 2019). We proposed that the CP genomes possibly provide insights into the evolution and taxonomy of the Morus genus. Since the CP genome of M. indica var. K2 was first obtained (Ravi et al., 2006), those of M. mongolica (Kong and Yang, 2016), M. alba var. atropurpurea (Li et al., 2016), M. notabilis (Chen et al., 2016), M. alba var. multicaulis and M. cathayana (Kong and Yang, 2017), M. alba (Luo et al., 2019; He et al., 2020), etc., have been reported. However, a large-scale comparative analysis of the CP genome across the Morus genus has not yet been conducted.
Therefore, the purpose of this study is to conduct a large-scale comparative genomic analysis of Morus CP genomes and reconstruct phylogenetic tree based on CP genomes to explore the taxonomy of genus Morus. The evolutionary relationships of mulberry accessions were also explored based on their CP genomes. These results provide an important information for the classification, domestication, and breeding improvement of mulberry.
Material and methods
Sample collection and sequencing
Morus serrata was collected from Jilong, Tibet Autonomous Region, China, and propagated at the Mulberry Germplasm Nursery at Southwest University. Morus celtidifolia was identified by Professor Elizabeth Makings from Arizona State University, USA. Morus notabilis was collected from a pristine forest in Ya’an, Sichuan Province, China. Morus yunnanensis was obtained from the Institute of Sericulture and Apiculture, Yunnan Academy of Agricultural Sciences, Mengzi, Yunnan Province, China. Morus nigra was collected from Yutian County, Xinjiang Uygur Autonomous Region, China. Other samples were obtained from the Mulberry Germplasm Nursery at Southwest University, China. For each sample, 10 µg genomic DNA was extracted from young leaves according to a standard cetyltrimethylammonium bromide protocol for the subsequent construction of sequencing libraries. Specifically, sequencing libraries with an average insert size of 350 bp were constructed according to the Illumina standard protocol, after which they were sequenced by BGI-Shenzhen (Shenzhen, China) using the Illumina HiSeq XTen or MGISEQ-2000 platform (Illumina, San Diego, CA, United States) to generate 150-bp paired-end reads. The raw data of 35 samples have been deposited in the CNGB Sequence Archive of the China National GeneBank Database (CNGBdb) under accession number CNP0001407. Using these data and publicly available genomic data downloaded from the NCBI or CNGB database, the Morus CP genomes were studied (Supplementary 1). The adapters and low-quality sequences were removed using the program fastp (Chen et al., 2018) from the raw reads to obtain clean reads for the subsequent analyses.
De novo assembly and annotation of the chloroplast genome
NOVOPLasty (version 4.3) (Dierckxsens et al., 2017) and GetOrganelle (v1.7.6.1) (Jin et al., 2020), which were developed for the de novo assembly of organelle genomes, were used for assembling CP genomes. For NOVOPLasty, default parameters were applied, with the following exceptions: read length (100 or 150), genome range (150,000–170,000), and K-mer optimized. For GetOrganelle, default parameters were applied, with the following exception: the heyebai chloroplast genome sequence (KU981119) as a reference sequence. Because the two haplotypes are present in the same proportion in a cell (Wang and Lanfear, 2019), we then selected the haplotype with the same SSC orientation as that in the CP genome sequences for further analyses.
The complete CP genomes were annotated in CPGAVAS2 (Shi et al., 2019) with default parameters. The ambiguous gene positions were manually corrected by NCBI BLASTN searches. All transfer RNA genes were confirmed on the tRNAscan-SE 2.0 web server with default settings (Lowe and Chan, 2016). Their high-quality graphical maps were drawn by OGDRAW (Lohse et al., 2013) with default parameters. All annotated chloroplast genome sequences were submitted to GenBank through BankIt (https://www.ncbi.nlm.nih.gov/WebSub/index.cgi).
Comparative chloroplast genome analysis
The mVISTA program (http://genome.lbl.gov/vista/mvista/about.shtml) was employed to determine the differences in the whole chloroplast genomes of M. notabilis (MK211167), M. serrata (MT154044), M. celtidifolia (MT154045), M. alba var. multicaulis (OP153908), M. alba var. alba (OP153917), M. nigra (OP153918), M. alba var. indica (OP153922), and M. rubra (OP161259), in the Shuffle-LAGAN mode with M. notabilis as the reference genome.
Sequence divergence analysis
MAFFT v7.455 software (Katoh and Standley, 2013) was employed to align the CP genomes of 50 Morus accessions. DnaSP v5.10 software (Librado and Rozas, 2009) was used to identify rapidly evolving molecular markers with a sliding window analysis (window length and step size set as 500 and 250 bp, respectively). The R package ggmsa (Zhou et al., 2022) was used to visualize multiple sequence alignments of target regions from 50 CP genomes.
Phylogenetic analysis
We assigned mulberry accessions with the identical CP genomes to a group, therefore 87 out of 123 mulberry accessions were assigned to 14 different groups and other 36 accessions were classified to an unclassified group. In 14 different groups, each group randomly selected a representative CP genome, together with other 36 CP genomes from the unclassified group, to form 50 CP genomes for phylogenetic analysis. The CP genome sequences of Broussonetia papyrifera (Broussonetia genus, GenBank: MZ662865), Ficus carica (Ficus genus, GenBank: KY635880) and Morus mesozygia (Afromorus genus, GenBank: MZ274134) (Gardner et al., 2021) were selected as outgroups. All complete CP genome sequences were aligned using MAFFT v7.455 (Katoh and Standley, 2013), and the alignments were trimmed with trimAl v1.4.rev15 (Capella-Gutiérrez et al., 2009). IQ-TREE 2.0 (Minh et al., 2020) was employed to construct a maximum likelihood (ML) phylogenetic tree with 1000 bootstrap replicates. Finally, the phylogenetic tree was edited using iTOL 5.0 (https://itol.embl.de/) (Letunic and Bork, 2019) and Adobe Photoshop® CC (Adobe Systems Inc., California, U.S.A.).
Results
Features of the Morus species chloroplast (CP) genomes
We de novo assembled 123 complete CP genomes of Morus species with sizes ranging from 158,969 to 159,548 bp (Figure 1; Supplementary 1). These CP genomes display a typical circular quadripartite architecture, with an LSC region (87,873~88,243 bp) and an SSC region (19,740~19,994 bp) separated by two inverted repeat (IR) regions (25,654~25,702 bp) (Supplementary 1). All CP genomes showed similar total GC contents (ranging from 36.13% to 36.21%). The largest length change in CP genome sequences occurred upstream of psbA of M. alba var. indica with a 135 bp missing sequence, which led M. alba var. indica to have the shortest CP genome (Supplementary 1).
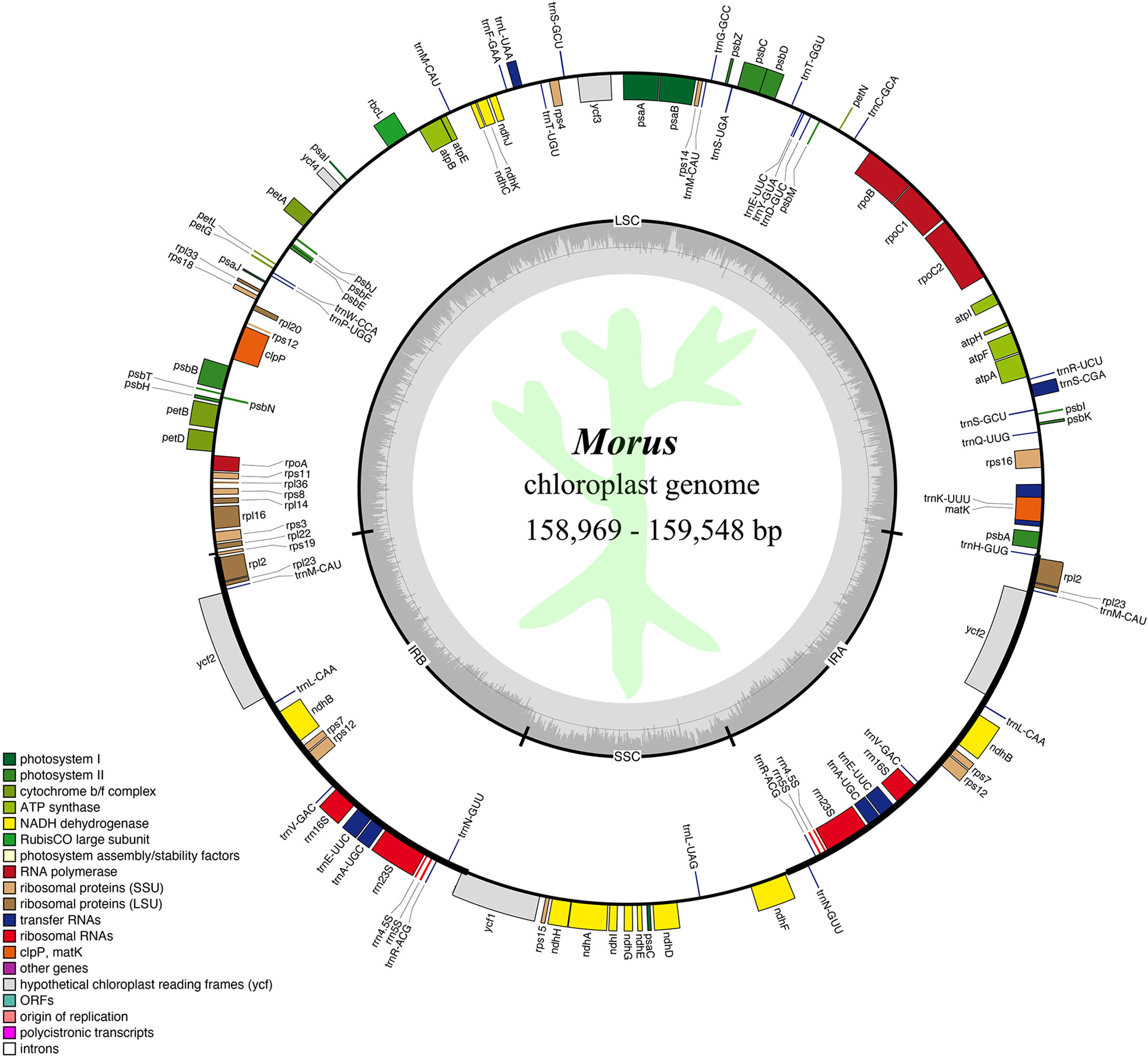
Figure 1 Genome map of the average Morus CP genome studied in this work. The inner circle represents the quadripartite structure, with an LSC, an SSC, and inverted repeat regions (IRA and IRB) and GC content shown in dark gray and AT content shown in light gray. The external circle represents gene content, with those outside the circle transcribed counterclockwise, while those located inside are transcribed clockwise. Genes are colored according to functional groups defined in the legend shown in the bottom left. The script in light green represents Jiaguwen mulberry.
The mulberry accessions with same CP genomes were assigned to a group, 87 out of 123 accessions were assigned to 14 diverse groups and other 36 accessions with different CP genomes were assigned to an unclassified group (Table 1 and Supplementary 1), which indicated that these 87 mulberry accessions were inherited maternally and shared 14 common ancestors. Of them, the largest group, ONE, comprises 28 Morus accessions, including 9 Japanese mulberry accessions and 9 Husang (M. alba var. multicaulis) accessions (Table 1). The nine Husang germplasms include heyebai (Husang-32), Jiantouheyebai, Husang-103, Husang-26, Husang-37, Husang-10, and Husang-60. Group TWO consists of 13 Morus accessions, including 12 Indian mulberry samples. Group THREE consists of 10 different Chuansang (M. notabilis) samples collected in a pristine forest around Ya’an, Sichuan Province. Group SIX comprises 4 samples, including one maternal parent and its three hybrid progenies. Group EIGHT comprises 4 Japanese samples, Yamasou-159, Ichinose, Fengwei-Ichinose, and Shinichinose. In 14 different groups, each group randomly selected a representative CP genome, together with other 36 CP genomes from the unclassified group Table 1, to form 50 diverse CP genomes occurred in 123 mulberry accessions for further analysis. And these 50 CP genomes were deposited in NCBI with GenBank accession numbers OP153890-OP153924, OP161257-OP161267, OP380682-OP380686, MK211167, MT154044, MT154045, and OP142713 for further analysis (Supplementary 1).
We annotated 125 genes in each Morus CP genome, consisting of 81 protein-coding genes, 8 rRNAs, and 36 tRNAs (Figure 1). In detail, 17 duplicated genes in the IR region were identified, including 6 protein-coding genes, 4 rRNA genes, and 7 tRNA genes. Ten protein-coding genes and 1 tRNA gene exist in the SSC region, while 59 protein-coding genes and 21 tRNA genes are present in the LSC region. All annotated genes were concatenated into a supermatrix, whose sizes in the 50 CP genomes ranged from 109,073 to 109,159 bp (Supplementary 1). Twenty-six out of 50 supermatrices showed identical sequences in ten separate groups (Supplementary 2). Of them, the largest three groups contained five, four, and three members, respectively, indicating high conservation.
There were 22 genes containing introns in all Morus CP genomes (Supplementary 3). Among these genes, 7 are tRNA genes, and 15 are protein-coding genes. Most genes have only a single intron, whereas the clpP and ycf3 genes contain two introns (Supplementary 3). rps12 is a trans-splicing gene composed of three exons, containing one 5’ exon located in the LSC region and two 3’ exons located in the IR region (Figure 1). Compared to other intron-containing genes, the trnK-UUU gene embodied the matK genes and has the largest intron (2,553-2,563 bp) (Supplementary 3).
Similarity analysis and nucleotide diversity of CP genomes
The sequence homology of the Morus species was investigated with M. notabilis as a reference using mVISTA software (Figure 2). The nucleotide variability (Pi) was calculated to further confirm the sequence variations (Figure 3). The Morus CP genomes were highly conserved and displayed similar structures and gene orders (Figure 2). The divergence level of the noncoding regions was higher than that of the coding regions. The protein-coding regions were highly conserved, and the ndhF genes displayed obvious polymorphism (Figure 2). The Pi values were rather low, ranging from 0 to 0.00442 among the 50 CP genomes, and 2 hotspot regions were identified with Pi >0.003 (rps16-trnQ-UUG and trnL-UAG-ndhF) (Figure 3). No highly variable loci were detected in the IR regions, and the nucleotide diversity values were significantly lower than those in the single-copy regions (Figure 3). Because of the highly conserved sequences, structure, and size of the CP genomes of Morus, no obvious hypervariable regions were noted (Figures 2, 3). As a result, the complete CP genomes were considered to distinguish Morus species.
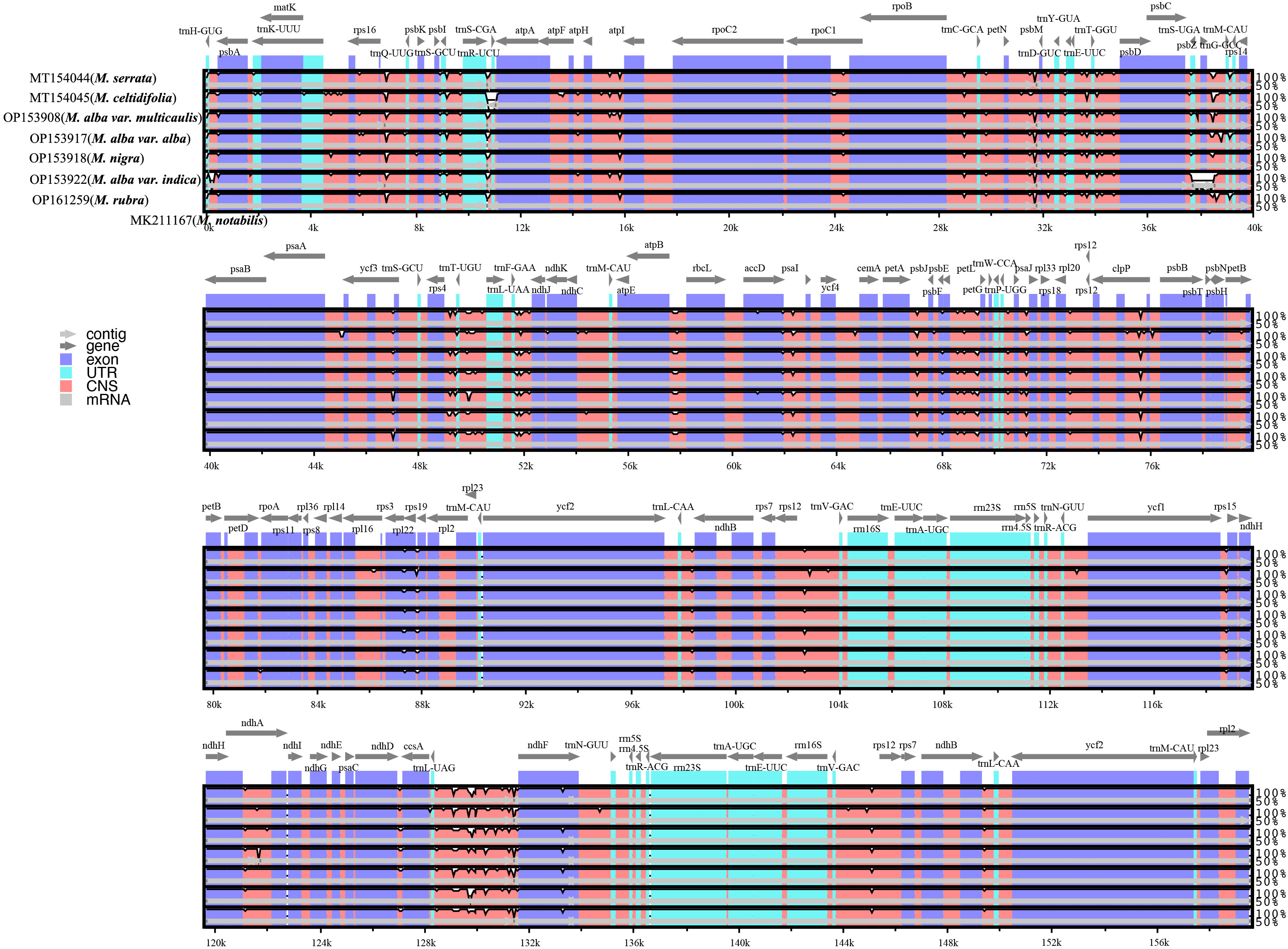
Figure 2 Sequence identity plot comparing the chloroplast genomes of six Morus species with M. notabilis as a reference. The vertical scale indicates the percentage of identity, ranging from 50 to 100%. The horizontal axis indicates the coordinates within the CP genome.
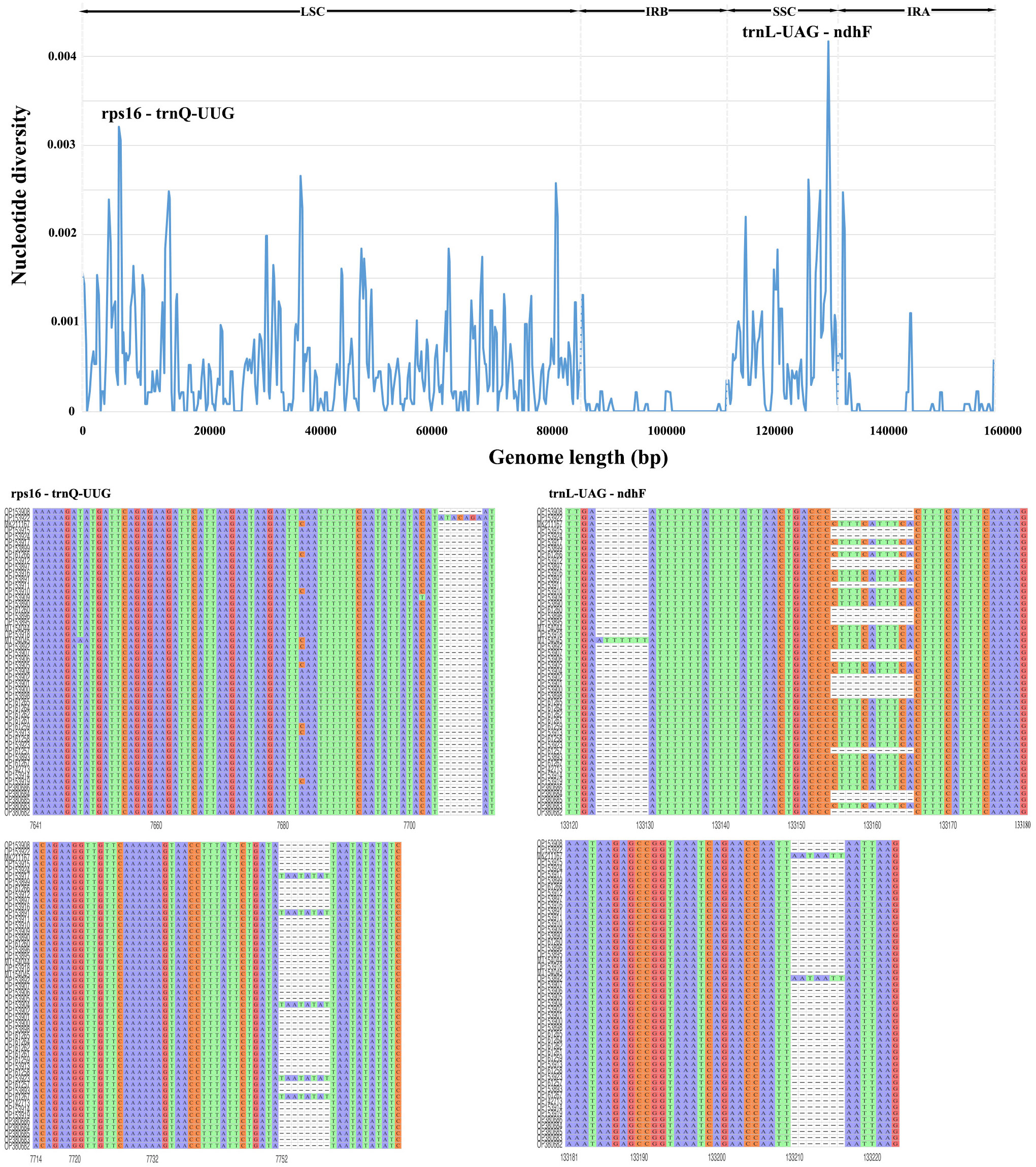
Figure 3 Comparative analysis of nucleotide variability by Pi values of the 50 CP genomes presented in a sliding window (window length: 500 bp; step size: 250 bp). X-axis: position of the midpoint of a window; Y-axis: nucleotide diversity in each window. The R package ggmsa was used to visualize the sequence alignment of two hotspot regions in the 50 CP genomes.
Phylogenetic analysis
In this study, the 50 CP genomes representing 123 mulberry samples were utilized to explore the phylogenetic positions of Morus species. Because of the highly conserved coding-region sequences in the Morus CP genomes, the complete genomes were used to construct the maximum-likelihood (ML) tree. As illustrated in Figure 4, the phylogenetic tree was divided into five clades. Among them, Outgroup is a clade containing three different genera (Ficus, Broussonetia, and Afromorus) at the root. Ficus carica was clustered with Broussonetia papyrifera, which was a sister genus of Morus, indicating a close relationship between Ficus and Broussonetia. Morus mesozygia, an outgroup member, was recognized as a Morus species native to Africa and belongs to the Afromorus genus. M. celtidifolia, a species native to America, was an independent clade. M. notabilis, native to Sichuan Province, and M. yunnanensis, native to Yunnan Province, formed a clade. Black mulberry (M. nigra) was a clade. White mulberry (M. alba) was the most complex and largest clade and was further divided into two subclades, M. alba var. alba and M. alba var. multicaulis, indicating that there were two subspecies of M. alba species. The M. alba var. multicaulis subclade comprised three subgroups containing all Husang, M. alba var. indica, and Japanese mulberry accessions. The M. alba var. alba subclade contained three subgroups, including the red mulberry (M. rubra), M. serrata, and a wild mulberry collected in Tibet, China. In addition, the mulberry resources (M. alba var. Taiwanchangguosang, M. alba var. shuisang, M. wittiorum, M. alba var. Yun7 and M. alba var. Yun6) with long fruits (over 4 cm) were clustered in the M. alba var. alba subclade. At the same time, two M. alba var. atropurpurea germplasms (M. alba var. Lunjiao109 and M. alba var. Kanqin283) were placed in the M. alba var. alba subclade.
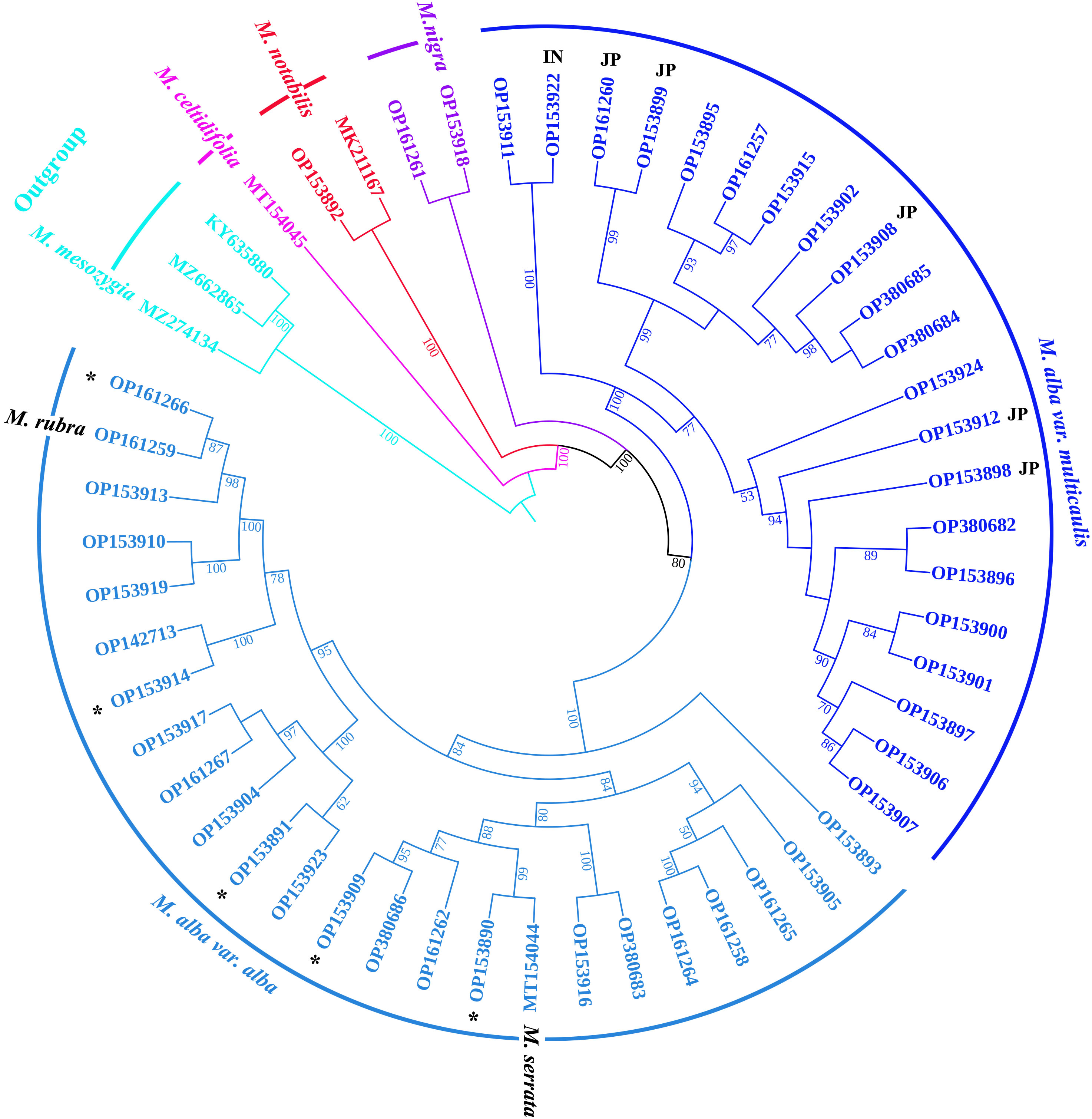
Figure 4 Phylogenetic relationships among Morus species based on their CP genomes with 50 Morus accessions and three outgroup genera (Ficus, Broussonetia, and Afromorus). The maximum-likelihood tree constructed with IQ-TREE2 is presented with complete CP genomes. The percentage of statistical support for the nodes is based on 1,000 bootstrap replicates. The black asterisks represent Morus accessions with mulberry fruit lengths over 4 cm. JP (Japan) and IN (India) in bold black represent mulberry accessions from Japan and India, respectively.
Discussion
Morus CP genome characterization
Maternally inherited CP genomes provide useful information for phylogenetic reconstruction (Bruun-Lund et al., 2017; Gitzendanner et al., 2018; Li et al., 2021; Wang et al., 2021; Zhang et al., 2022). Although some Morus CP genomes (Chen et al., 2016; Kong and Yang, 2016; Li et al., 2016; Kong and Yang, 2017; Luo et al., 2019; He et al., 2020) have previously been reported since the first was reported in 2006 (Ravi et al., 2006), there is a lack of large-scale comparative analysis of these genomes. Some Morus CP genomes deposited in the NCBI database were reference-based assemblies (Chen et al., 2016; Kong and Yang, 2016; Li et al., 2016), which may lack some useful information. For example, the CP genome of M. notabilis showed length differences between de novo assembly (GenBank: MK211167, size: 159,548 bp) and reference-based assembly (GenBank: KP939360, size: 158,680 bp). In addition, two indels over 40 bp were detected in the CP genome of the de novo assembly (GenBank: OP153912, size: 159,200 bp) compared with the reference-based assembly (GenBank: KU981119, size: 159,103 bp) using the same raw data. The performance of the reference-based assembly was dependent on the references employed (Scheunert et al., 2020). As a result, all Morus CP genomes were de novo assembled in this study, and Morus CP genomes of the reference-based assembly were not included. The size of Morus CP genomes ranged from 158,969 to 159,548 bp, which was larger than the first Morus CP genome (158,484 bp) (Ravi et al., 2006) and suggested that CP genome length in Morus was highly conserved. GC content is often considered an important indicator of species affinity (Chen et al., 2021), and the GC content of Morus CP genomes showed slight differences, ranging from 36.13% to 36.21%, which indicated high conservation in the Morus CP genomes. Twenty-two intron-containing genes out of 125 genes were detected in these CP genomes. Among them, the trnK-UUU gene embodied the matK genes and had the largest intron (over 2,500 bp), which has been reported in previous studies (Li X. et al., 2019; Souza et al., 2020; Ren et al., 2022). matK is a well-known gene that is often used for molecular identification and analysis of genetic relationships in plants (Hilu and Liang, 1997; Ramesh et al., 2022), including Morus (Venkateswarlu et al., 2012). As a result, over one hundred matK genes of the Morus genus have been deposited in the GenBank of the NCBI. Intron-containing genes often have important physiological functions; for example, the clpP gene is relevant to proteolysis (Shikanai et al., 2001), and the ndhB gene has an important role in mediating photosystem I cyclic electron transport (Shen et al., 2022). Therefore, introns in Morus CP genomes may be useful in terms of physiological function.
The largest group with identical CP genome contained 28 mulberry germplasm accessions, including 9 Husang accessions and 9 Japanese mulberry accessions. Husang (or Hu mulberry, M. alba var. multicaulis, with multicaulis meaning many stalks or branches), a well-known cultivar of domesticated mulberry, is widely distributed worldwide (Kenrick, 1839; Jiao et al., 2020). Heyebai, named Husang-32, is a control cultivar in the National Mulberry New Cultivar Identification Test (Jiao et al., 2020). The selection of Husang germplasms was mainly performed for open-pollinated seedlings after the Song Dynasty, and the excellent traits were retained by the asexual method (Jiao et al., 2020). Tens of Husang germplasms were obtained and recorded after selection over hundreds of years. Most of the cultivated mulberry varieties in Japan are derived from the three original species, namely, Yamaguwa (Morus bombycis), Karayumaguwa (M. alba), and Roguwa (Morus lhou) (Minamizawa, 1997; Muhonja et al., 2020), of which M. lhou and M. bombycis (Koidzumi, 1930) belong to M. alba (Zeng et al., 2015). Additionally, Karayumaguwa and Roguwa were introduced to Japan from Chinese M. alba species around A.D. 677 and A.D. 1873, respectively (Hotta, 1958). Therefore, those three original species in Japan belong to the M. alba species. Ichibei, Kenmochi, and Gunmaakagi are related to Yamaguwa, whereas Ichinose, Kairyonezumigaeshi, Nezumigaeshi, and Kairyowasejumonji are related to Karayumaguwa (Minamizawa, 1997). In this study, 9 Japanese samples shared the same progenitor with Husang, indicating that the 9 Japanese samples belonged to M. alba var. multicaulis. Kairyonezumigaeshi was selected from among Nezumigaeshi plants in 1907 (Muhonja et al., 2020), which is consistent with their identical CP genomes (Table 1), whereas Ichinose (group EIGHT) was isolated from Nezumigaeshi (group ONE) seedlings in 1901 (Yamanouchi et al., 2010; Smethurst, 2014; Muhonja et al., 2020) and showed a different CP genome (Supplementary 1), which implied that hybridization may increase the genetic diversity of CP genomes (Van Droogenbroeck et al., 2006; Daniell et al., 2016). In addition, in group EIGHT, 4 Japanese samples, Yamasou-159, Ichinose, Fengwei-Ichinose and Shinichinose, had common ancestors. Shinichinose, a hybrid variety derived from Ichinose (Muhonja et al., 2020), showed an identical CP genome with Ichinose, which is a typical character of maternal inheritance. In addition, group SIX showed another case of maternal inheritance because the maternal parent exhibited the same CP genome as its three hybrid progenies (Table 1 and Supplementary 1). The autotriploid cultivar Shaansang305 (group NINE, 159,200 bp) induced from the diploid cultivar Shinichinose using colchicine (Liu et al., 2021) showed obvious differences from the CP genome of Shinichinose (group EIGHT, 159,219 bp) (Supplementary 1), which suggested that polyploidizations affected the DNA of the nucleus and chloroplast (Choopeng et al., 2019; Zhai et al., 2021).
Group TWO consisted of 13 different accessions, including 12 M. alba var. indica, which indicated that they had common ancestors. Group THREE consisted of 9 different M. notabilis trees located in regions with an approximately 10 km radius of the pristine forest in Ya’an, Sichuan Province, Southwest China, and one seedling germinated from an M. notabilis seed. Group ELEVEN contained two different M. yunnanensis trees collected on Dawei Mountain, Yunnan Province, Southwest China.
The genome size (158,969-159,548 bp) and GC content (36.13%-36.21%) of the Morus CP genomes exhibited slight differences, which indicated high conservation of Morus CP genomes. DnaSP (Librado and Rozas, 2009) and mVISTA software were employed to investigate the divergence of CP genomes of the Morus genus. The results showed high conservation of gene order and rather low Pi values (0-0.00442), and noncoding regions were more variable than coding regions. At the same time, over half of the supermatrices showed identical sequences (Supplementary 2), which further supported that the coding regions were highly conserved. Therefore, the complete CP genomes of Morus were considered to construct the phylogenetic tree for identifying Morus species.
Phylogenetic analysis and taxonomical review of Morus
The 50 complete CP genomes of Morus were used to construct the ML tree for exploring the phylogenetic positions of the Morus species. It is clear that the genus Morus is monophyletic and is divided into five clades (Figure 4). Recently, M. mesozygia and M. insignis, native to Africa, which used to belong to the Morus genus, were eliminated from the Morus genus on the basis of phylogenetic analyses of supercontig sequences from 246 Moraceae samples (Gardner et al., 2021). Here, we also found that M. mesozygia did not belong to Morus because M. mesozygia was clustered with Ficus and Broussonetia (Figure 4).
Morus yunnanensis, similar to M. notabilis found in Southwest China with the same chromosome number, is a wild mulberry native to Yunnan Province, China (Xia et al., 2022). Morus yunnanensis was clustered with M. notabilis into a clade, indicating a close phylogenetic relationship (Figure 4). This close phylogenetic relationship between M. yunnanensis and M. notabilis was strongly supported by a phylogenomic tree (Xia et al., 2022). Combined with evidence from the nuclear genome and CP genome, we classify M. yunnanensis as belonging to M. notabilis.
Black mulberry (M. nigra), native to western Asia, is a Morus species with 308 chromosomes, which hinders the exploration of phylogenetic relationships based on the nuclear genome. It has been reported that M. nigra originated from M. alba (Agaev and Fedorova, 1970; Browicz, 2000; Lim., 2012), but molecular evidence is lacking. Fortunately, the CP genome is independent of the nuclear genome and is commonly used in phylogenetic studies. In our phylogenetic tree inferred from complete CP genomes, M. nigra displayed a close phylogenetic relationship with M. alba (Figure 4), which indicates that M. nigra originated from M. alba.
The largest clade in the phylogenetic tree of the Morus genus is the M. alba clade, comprising two subclades, M. alba var. alba and M. alba var. multicaulis, which is consistent with the taxonomy presented in the Flora of China (Zhou and Gilbert, 2003) and the phylogenetic tree based on domesticated mulberry accessions (Jiao et al., 2020). The M. alba var. multicaulis clade was divided into three subclades, including all Husang (M. alba var. multicaulis) and 16 Japanese mulberry samples, M. alba var. indica and other samples. Nine Japanese samples sharing the same CP genomes with Husang were clustered with seven other Japanese samples into the M. alba var. multicaulis clade, showing a close phylogenetic relationship, which indicated that these Japanese samples may have been derived from M. alba var. multicaulis through maternal inheritance. Recently, gene flow between Husang and Japanese samples was observed in the population structure analysis of mulberry accessions (Xia et al., 2022). Sixteen Japanese samples were clustered into two subclades, which was consistent with the findings of a previous report (Muhonja et al., 2020). Here, we provided molecular evidence that Japanese cultivated mulberry was derived from Chinese M. alba (Hotta, 1958; Jiao et al., 2020); therefore, we conclude that Japanese cultivated mulberry belongs to M. alba var. multicaulis. The subclade of M. alba var. alba contained red mulberry (M. rubra), M. serrata, M. alba var. atropurpurea, long-fruited mulberry germplasms, and other samples. Among them, M. rubra, a Morus species native to America, was clustered into M. alba var. alba, which may be triggered by common hybridization with M. alba (Burgess et al., 2008). It has been reported that M. rubra commonly hybridizes with M. alba and that M. alba potentially poses a threat to the existence of M. rubra, which leads to the endangerment of native M. rubra in America (Nepal and Wichern, 2013). In field observations, the direction of introgression of hybrids between M. rubra and M. alba was biased toward M. alba as the maternal parent (Nepal and Wichern, 2013). Mulberry germplasms with long fruits include M. wittiorum and M. macroura, which have been recognized as M. alba (Zeng et al., 2015). Here, we supplied new molecular evidence at the genome level. Morus serrata was classified as a species based on morphological taxonomy and molecular marker genes (Hotta, 1958; Zhou and Gilbert, 2003; Nepal and Ferguson, 2012; Zeng et al., 2015). However, in this study, M. serrata was clustered with M. alba var. alba in the phylogenetic tree based on the CP genome (Figure 4). The classification of M. serrata requires more samples and further investigation using molecular evidence.
Indian mulberry (M. indica or M. alba var. indica) is recognized as a variety of M. alba (Datta, 1954; Rao and Jarvis, 1986; Zeng et al., 2015; Muhonja et al., 2020). The first Morus CP genome (GenBank: DQ226511, 158,484 bp) was identified in M. indica with the reference genome method using plastid genomic DNA (Ravi et al., 2006). In this study, the CP genomes of 12 different samples, including M. indica var. kanva2, were de novo assembled and found to have identical sequences (size: 158,969 bp). Sample SL2 in group TWO, native to Sri Lanka (Xia et al., 2022), may be a hybrid progeny of M. alba var. indica. M. indica was clustered with M. alba var. multicaulis, indicating a close phylogenetic relationship with this taxon (Figure 4). Additional molecular evidence of the phylogenetic relationships of M. indica was provided (Muhonja et al., 2020). Therefore, M. indica may be derived from M. alba var. multicaulis.
In the present study, we de novo assembled 123 Morus CP genomes and found that they are highly conserved. Many Morus CP genomes displayed identical sequences, which indicated that they shared common maternal ancestors. We propose that the Morus genus includes six species, namely, M. notabilis, M. celtidifolia, M. nigra, M. rubra, M. serrata, and M. alba comprising two subspecies, M. alba var. alba and M. alba var. multicaulis. The Japanese cultivated germplasms were derived from M. alba var. multicaulis. Our findings provide valuable information for studies on the classification, domestication, and breeding improvement of mulberry.
Data availability statement
The raw sequence data were deposited in the CNGB Sequence Archive of the China National GeneBank Database (CNGBdb) under accession number CNP 0001407.
Author contributions
QZ, NH, and ZX conceived the project and designed the experiments. QZ assembled, annotated, and analyzed the genomes, QZ wrote the manuscript. MC, SW and XX contributed materials and isolated DNA and analyzed the data. TL helped QZ to analyze the data. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Key Research and Development Program (No. 2018YFD1000602) and the Chongqing Research Program of Basic Research and Frontier Technology (cstc2021yszx-jcyj0004).
Acknowledgments
We are grateful to Professor Elizabeth Makings from Arizona State University for kindly providing leaves of M. celtidifolia. We thank for the help of Ni Yang and Li Jingling from the Institute of Medicinal Plant Development for their suggestion on the annotation and analysis of chloroplast genomes. We also thank the reviewers for helpful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1047592/full#supplementary-material
References
Agaev, Y. M., Fedorova, H. E. (1970). Investigation of meiosis in the diploid species moras alba l., the 22-ploid m. nigra l. and their cross in relation to the origin of the species m. nigra l. Pak. J. Bot. 2 (1), 65–76.
Browicz, K. (2000). Where is the place of origin of morus nigra (Moraceae) ? Fragm. Flor. Geobot. 45 (1/2), 273–280.
Bruun-Lund, S., Clement, W. L., Kjellberg, F., Rønsted, N.. (2017). First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of ficus L.(Moraceae). Mol. Phylogenet. Evol. 109, 93–104. doi: 10.1016/j.ympev.2016.12.031
Bureau, L.É. (1873). “Moraceae,” in Prodromus systematis naturalis regni vegetabilis. Ed. DeCandolle, A. P. (Paris, France: Tuettel and Wurtz), 211–288.
Burgess, K. S., Martin, M., Husband, B. C. (2008). Interspecific seed discounting and the fertility cost of hybridization in an endangered species . New Phytol. 177 (1), 276–283. doi: 10.1111/j.1469-8137.2007.02244.x
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinf. (Oxford England) 25 (15), 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, C., Zhou, W., Huang, Y., Wang, Z. Z. (2016). The complete chloroplast genome sequence of the mulberry morus notabilis (Moreae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27 (4), 2856–2857. doi: 10.3109/19401736.2015.1053127
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, J., Zang, Y., Shang, S., Liang, S., Zhu, M., Wang, Y., et al. (2021). Comparative chloroplast genomes of zosteraceae species provide adaptive evolution insights into seagrass. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.741152
Choopeng, S., Te-Chato, S., Khawnium, T. (2019). Effect of colchicine on survival rate and ploidy level of hydrid between dendrobium santana x d. friedericksianum orchid. Int. J. Agric. Technol. 15 (2), 249–260.
Daniell, H., Lin, C. -S., Yu, M., Chang, W. -J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17 (1), 134. doi: 10.1186/s13059-016-1004-2
Datta, M. (1954). Cytogenetical studies on two species of morus. Cytologia 19 (1), 86–95. doi: 10.1508/cytologia.19.86
Dierckxsens, N., Mardulyn, P., Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45 (4), e18. doi: 10.1093/nar/gkw955
Gardner, E. M., Garner, M., Cowan, R, Dodsworth, S, Epitawalage, N, Arifiani, D, et al. (2021). Repeated parallel losses of inflexed stamens in moraceae: Phylogenomics and generic revision of the tribe moreae and the reinstatement of the tribe olmedieae (Moraceae). Taxon. 70, 946 988. doi: 10.1002/tax.12526
Gitzendanner, M. A., Soltis, P. S., Wong, G. K. S., Ruhfel, B. R., Soltis, D. E., et al. (2018). Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am. J. Bot. 105 (3), 291–301. doi: 10.1002/ajb2.1048
Group A. P. (2009). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161 (2), 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
He, N., Zhang, C., Qi, X., Zhao, S., Tao, Y., Yang, G., et al. (2013). Draft genome sequence of the mulberry tree morus notabilis. Nat. Commun. 4, 2445. doi: 10.1038/ncomms3445
He, S.-L., Tian, Y., Yang, Y., Shi, C. -Y. (2020). Chloroplast genome and phylogenetic analyses of morus alba (Moraceae) . Mitochondrial DNA Part B 5 (3), 2203–2204. doi: 10.1080/23802359.2019.1673242
Hilu, K. W., Liang, G. (1997). The matK gene: sequence variation and application in plant systematics. Am. J. Bot. 84 (6), 830–839. doi: 10.2307/2445819
Hotta, T. (1958). Taxonomical studies on the morus plants and their distributions in Japan and its vicinities (Tokyo: Japan Society for the Promotion of Science).
Hua, Z., Tian, D., Jiang, C., Song, S., Chen, Z., Zhao, Y., et al. (2022). Towards comprehensive integration and curation of chloroplast genomes . Plant Biotechnol. J. doi: 10.1111/pbi.13923
Jain, M., Bansal, J., Rajkumar, M. S., Sharma, N., Khurana, J. P., Khurana, P. (2022). Draft genome sequence of Indian mulberry (Morus indica) provides a resource for functional and translational genomics. Genomics 114 (3), 110346. doi: 10.1016/j.ygeno.2022.110346
Jiao, F., Luo, R., Dai, X., Lui, H., Yu, G., Han, S., et al. (2020). Chromosome-level reference genome and population genomic analysis provide insights into the evolution and improvement of domesticated mulberry (Morus alba). Mol. Plant 13 (7), 1001–1012. doi: 10.1016/j.molp.2020.05.005
Jin, J.-J., Yu, W. -B., Yang, J. B., Song, Y., Depamphilis, C. W., Yi, Y. -S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21 (1), 1–31. doi: 10.1186/s13059-020-02154-5
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kenrick, W. (1839). The American silk grower's guide : or, the art of raising the mulberry and silk, and the system of successive crops in each season. 2d ed (Boston: Weeks, Jordan & co).
Koidzumi, G. (1930). Florae symbolae orientali-asiaticae: Sive, contributions to the knowledge of the flora of Eastern Asia (Kyoto, Japan) 115 pp. doi: 10.11501/167884
Kong, W., Yang, J. (2016). The complete chloroplast genome sequence of morus mongolica and a comparative analysis within the fabidae clade. Curr. Genet. 62 (1), 165–172. doi: 10.1007/s00294-015-0507-9
Kong, W. Q., Yang, J. H. (2017). The complete chloroplast genome sequence of morus cathayana and morus multicaulis, and comparative analysis within genus morus l. PeerJ 5, e3037. doi: 10.7717/peerj.3037
Letunic, I., Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments . Nucleic Acids Res. 47 (W1), W256–W259. doi: 10.1093/nar/gkz239
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 (11), 1451–1452. doi: 10.1093/bioinformatics/btp187
Li, Q. L., Guo, J. Z., Yan, N., Li, C. C. (2016). Complete chloroplast genome sequence of cultivated morus l. species. Genet. Mol. Res. 15 (4), 1–13. doi: 10.4238/gmr15048906
Li, H.-T., Yi, T. -S., Gao, L. -M., Ma, P. -F., Zhang, T., Yang, J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5 (5), 461–470. doi: 10.1038/s41477-019-0421-0
Li, X., Zuo, Y., Zhu, X., Liao, S., Ma, J. (2019). Complete chloroplast genomes and comparative analysis of sequences evolution among seven aristolochia (Aristolochiaceae) medicinal species. Int. J. Mol. Sci. 20 (5), 1045. doi: 10.3390/ijms20051045
Li, H.-T., Luo, Y., Gan, L., Ma, P. -F., Gao, L. -M., Yang, J. -B., et al. (2021). Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 19 (1), 1–13. doi: 10.1186/s12915-021-01166-2
Lim., T. K. (2012). “Morus nigra, in edible medicinal and non-medicinal plants,” in Fruits, vol. Volume 3. (Netherlands: Springer), 430–438.
Linnaeus, C. “Morus,” in Species plantarum (1753). (Stockholm, Impensis Laurentii Salvii) 968. doi: 10.5962/bhl.title.669
Liu, H., Sun, H., Bao, L., Han, S., Hui, T., Zhang, R., et al. (2021). Secondary metabolism and hormone response reveal the molecular mechanism of triploid mulberry (Morus alba l.) trees against drought. Front. Plant Sci. 12, 720452. doi: 10.3389/fpls.2021.720452
Lohse, M., Drechsel, O., Kahlau, S., Bock, P.. (2013). OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets . Nucleic Acids Res. 41 (Web Server issue), W575. doi: 10.1093/nar/gkt289
Lowe, T. M., Chan, P. P. (2016). tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes . Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Luo, J., Wang, Y., Zhao, A. Z. (2019). The complete chloroplast genome of morus alba (Moraceae: Morus), the herbal medicine species in china . Mitochondrial DNA Part B 4 (2), 2467–2468. doi: 10.1080/23802359.2019.1638328
Minamizawa, K. (1997). Moriculture: Science of mulberry cultivation. 2 ed (Boca Raton, Florida, USA: CRC Press).
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–34. doi: 10.1093/molbev/msaa015
Moore, M. J., Bell, C. D., Soltis, P. S., Soltis, D. E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. 104 (49), 19363–19368. doi: 10.1073/pnas.0708072104
Moore, M. J., Soltis, P. S, Bell, C. D., Burleigh, J. D., Soltis, D. E., et al. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl.Acad. Sci. U.S.A 107 (10), 4623–4628. doi: 10.1073/pnas.0907801107
Muhonja, L., Yamanouchi, H., Yang, C. -C., Kuwazaki, S., Yokoi, K., Kameda, T., et al. (2020). Genome-wide SNP marker discovery and phylogenetic analysis of mulberry varieties using double-digest restriction site-associated DNA sequencing. Gene. 726, 144162. doi: 10.1016/j.gene.2019.144162
Nepal, M. P., Ferguson, C. J. (2012). Phylogenetics of morus (Moraceae) inferred from ITS and trnL-trnF sequence data. Syst. Bot. 37 (2), 442–450. doi: 10.1600/036364412X635485
Nepal, M. P., Wichern, D. J. (2013). Taxonomic status of red mulberry (morus rubra, moraceae) At its northwestern boundar. Proc. South Dakota Acad. Sci. 92, 19–29.
Ramesh, G. A., Mathew, D., John, K. J., Ravisankar, V. (2022). Chloroplast gene matK holds the barcodes for identification of momordica (Cucurbitaceae) species from Indian subcontinent. Hortic. Plant J. 8 (1), 89–98. doi: 10.1016/j.hpj.2021.04.001
Rao, C. K., Jarvis, C. (1986). Lectotypification, taxonomy and nomenclature of morus alba, m. tatarica and m. indica (Moraceae). Taxon. 35 (4), 705–708. doi: 10.2307/1221619
Ravi, V., Khurana, J. P., Tyagi, A. K., Khurana, P. (2006). The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet. Genomes 3 (1), 49–59. doi: 10.1007/s11295-006-0051-3
Ren, J., Tian, J., Jiang, H., Zhu, X. -X., Mutie, F. M., Wang, F. M., et al. (2022). Comparative and phylogenetic analysis based on the chloroplast genome of coleanthus subtilis (Tratt.) Seidel, a protected rare species of monotypic genus. Front. Plant Sci. 13, 828467. doi: 10.3389/fpls.2022.828467
SB, D., Dhar, A., Sengupta, K. (1990). Meiosis in natural decosaploid (22x) morus nigra l. Cytologia 55 (3), 505–509.
SB, D., Rajan, M. V. (1989). Microsporogenesis in hexaploid morus serrata roxb. Cytologia 54 (4), 747–751.
Scheunert, A., Dorfner, M., Lingl, T., Oberprieler, C. (2020). Can we use it? on the utility of de novo and reference-based assembly of nanopore data for plant plastome sequencing. PloS One 15 (3), e0226234. doi: 10.1371/journal.pone.0226234
Shen, L., Tang, K., Wang, W., Wang, C., Wu, H., Mao, Z., et al. (2022). Architecture of the chloroplast PSI–NDH supercomplex in hordeum vulgare. Nature 601 (7894), 649–654. doi: 10.1038/s41586-021-04277-6
Shi, L., Chen, H., Jiang, M., Wang, M, Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer . Nucleic Acids Res. 47 (W1), W65–W73. doi: 10.1093/nar/gkz345
Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42 (3), 264–273. doi: 10.1093/pcp/pce031
Smethurst, R. J. (2014). Agricultural development and tenancy disputes in Japan (Princeton, NJ: Princeton University Press) 1870–1940.
Souza, U. J. B. D., Vitorino, L. C., Bessa, L. A., Silva, F. G., et al. (2020). The complete plastid genome of artocarpus camansi: a high degree of conservation of the plastome structure in the family moraceae. Forests 11 (11), 1179. doi: 10.3390/f11111179
Stull, G. W., Duno De Stefano, R., Soltis, D. E., Soltis, P. S. (2015). Resolving basal lamiid phylogeny and the circumscription of icacinaceae with a plastome-scale data set. Am. J. Bot. 102 (11), 1794–1813. doi: 10.3732/ajb.1500298
Sun, Y., Moore, M. J., Zhang, S., Soltis, P. S., Soltis, D. E., Zhao, T., et al. (2016). Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol. Phylogenet. Evol. 96, 93–101. doi: 10.1016/j.ympev.2015.12.006
Tikader, A., Kamble, C. K. (2008). Mulberry wild species in India and their use in crop improvement–a review. Aust. J. Crop Sci. 2 (2), 64–72.
Van Droogenbroeck, B., Kyndt, T., Romeijn-Peeters, E., Van Thuyne, W, Goetghebeur, P., Romero-Motochi, J., et al. (2006). Evidence of natural hybridization and introgression between vasconcellea species (Caricaceae) from southern Ecuador revealed by chloroplast, mitochondrial and nuclear DNA markers. Ann. Bot. 97 (5), 793–805. doi: 10.1093/aob/mcl038
Venkateswarlu, M., Ravikumar, G., Vijayaprakash, N., Rao, C., Kamble, C., Tikadar, A. (2012). Molecular phylogeny of morus species differentiation based on chloroplast matK sequences. Indian J. Sericulture 51 (1), 16–19.
Vijayan, K. S., B.da Silva, J. A. T. (2011). Germplasm conservation in mulberry (Morus spp.). Scientia Hortic. 128 (4), 371–379. doi: 10.1016/j.scienta.2010.11.012
Vijayan, K., Tikader, A., Weiguo, Z., Nair, C. V., Ercisli, S., Tsou, C. -H. (2011). “Morus,” in Wild crop relatives: Genomic and breeding resources (Tropical and subtropical fruits). Ed. Chittaranjan, K. (Berlin Heidelberg: Springer-Verlag), 75–95.
Wang, G., Zhang, X., Herre, E. A., Mckey, D., Machado, C. A., Yu, W. -B., et al. (2021). Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nat. Commun. 12 (1), 1–14. doi: 10.1038/s41467-021-20957-3
Wang, W., Lanfear, R. (2019). Long-reads reveal that the chloroplast genome exists in two distinct versions in most plants. Genome Biol. Evol. 11 (12), 3372–3381. doi: 10.1093/gbe/evz256
Xia, Z., Dai, X., Fan, W., Liu, Z., Zhang, M., Bian, P., et al. (2022). Chromosome-level genomes reveal the genetic basis of descending dysploidy and sex determination in morus plants. Genomics Proteomics Bioinf. doi: 10.1016/j.gpb.2022.08.005
Xuan, Y., Wu, Y., Li, P., Liu, R., Luo, Y., Yuan, J., et al. (2019). Molecular phylogeny of mulberries reconstructed from ITS and two cpDNA sequences. PeerJ. 7, e8158. doi: 10.7717/peerj.8158
Xuan, Y., Ma, B., Li, D., Tian, Y., Zeng, Q., He, N. (2022). Chromosome restructuring and number change during the evolution of Morus notabilis and Morus alba. Hortic. Res. 9, uhab030. doi: 10.1093/hr/uhab030
Yamanouchi, H., Koyama, A., Takyu, T., Muramatsu, N. (2010). Nuclear DNA amounts in diploid mulberry speceis (Morus. spp.) J. Insect Biotechnol. Sericology 79, 1–8.
Zeng, Q., Chen, H., Zhang, C., Han, M., Li, T., Qi, X., et al. (2015). Definition of eight mulberry species in the genus morus by internal transcribed spacer-based phylogeny. PloS One 10 (8), e0135411. doi: 10.1371/journal.pone.0135411
Zhai, Y., Yu, X., Zhou, J., Li, J., Tian, Z., Wang, P. (2021). Complete chloroplast genome sequencing and comparative analysis reveals changes to the chloroplast genome after allopolyploidization in Cucumis. Genome 64 (6), 627–638. doi: 10.1139/gen-2020-0134
Zhang, Z.-R., Yang, X., Li, W. -Y., Peng, Y. -Q., Gao, J. (2022). Comparative chloroplast genome analysis of ficus (Moraceae): Insight into adaptive evolution and mutational hotspot regions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.965335
Zhou, L., Feng, T., Xu, S., Gao, F., Lam, T. T., Wang, Q., et al. (2022). Ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Briefings Bioinf. doi: 10.1093/bib/bbac222
Keywords: Mulberry, Chloroplast genome, Phylogenetic tree, Taxonomy, Morus alba
Citation: Zeng Q, Chen M, Wang S, Xu X, Li T, Xiang Z and He N (2022) Comparative and phylogenetic analyses of the chloroplast genome reveal the taxonomy of the Morus genus. Front. Plant Sci. 13:1047592. doi: 10.3389/fpls.2022.1047592
Received: 18 September 2022; Accepted: 24 October 2022;
Published: 24 November 2022.
Edited by:
Wenpan Dong, Beijing Forestry University, ChinaReviewed by:
Xiwen Li, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, ChinaLihong Xiao, Zhejiang Agriculture and Forestry University, China
Copyright © 2022 Zeng, Chen, Wang, Xu, Li, Xiang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ningjia He, hejia@swu.edu.cn
 Qiwei Zeng
Qiwei Zeng Miao Chen
Miao Chen Shouchang Wang
Shouchang Wang Xiaoxiang Xu
Xiaoxiang Xu Tian Li
Tian Li Zhonghuai Xiang
Zhonghuai Xiang Ningjia He
Ningjia He