- 1Department of Life Sciences, Yeungnam University, Gyeongsan, Gyeongsan-buk, Republic of Korea
- 2Plants Resource Division, Biological Resources Research Department, National Institute of Biological Resources, Seo-gu, Incheon, Republic of Korea
Introduction: The chloroplast (cp) is an autonomous plant organelle with an individual genome that encodes essential cellular functions. The genome architecture and gene content of the cp is highly conserved in angiosperms. The plastome of Corydalis belongs to the Papaveraceae family, and the genome is comprised of unusual rearrangements and gene content. Thus far, no extensive comparative studies have been carried out to understand the evolution of Corydalis chloroplast genomes.
Methods: Therefore, the Corydalis platycarpa cp genome was sequenced, and wide-scale comparative studies were conducted using publicly available twenty Corydalis plastomes.
Results: Comparative analyses showed that an extensive genome rearrangement and IR expansion occurred, and these events evolved independently in the Corydalis species. By contrast, the plastomes of its closely related subfamily Papaveroideae and other Ranunculales taxa are highly conserved. On the other hand, the synapomorphy characteristics of both accD and the ndh gene loss events happened in the common ancestor of the Corydalis and sub-clade of the Corydalis lineage, respectively. The Corydalis-sub clade species (ndh lost) are distributed predominantly in the Qinghai-Tibetan plateau (QTP) region. The phylogenetic analysis and divergence time estimation were also employed for the Corydalis species.
Discussion: The divergence time of the ndh gene in the Corydalis sub-clade species (44.31 – 15.71 mya) coincides very well with the uplift of the Qinghai-Tibet Plateau in Oligocene and Miocene periods, and maybe during this period, it has probably triggered the radiation of the Corydalis species.
Conclusion: To the best of the authors’ knowledge, this is the first large-scale comparative study of Corydalis plastomes and their evolution. The present study may provide insights into the plastome architecture and the molecular evolution of Corydalis species.
Introduction
In angiosperms, chloroplast (cp) genomes are highly conserved in terms of their structure, gene content, and gene arrangement contains a pair of inverted repeats (IRs) that separate with a large single-copy (LSC) and a small single-copy (SSC) region (Palmer, 1983; Palmer, 1985; Wicke et al., 2011; Maliga, 2014; Lee et al., 2016; Mower and Vickrey, 2018). The cp genome of angiosperms comprises roughly 80 protein-coding genes, which play a role in essential cellular functions and photosynthesis, along with 30 transfer and four ribosomal RNA genes (Bock, 2007). Among these, approximately seventeen genes were duplicated in the IR region. In addition, most of the land plant cp genome size varies from 110 to 170 kb, and the difference in cp size is frequently ascribed to extension, reduction, or loss of the IR region (Chumley et al., 2006; Wicke et al., 2011). The large-scale IR expansion is identified in the Pelargonium transvallense (Geraniaceae) (Weng et al., 2014), in which the IR enlarged higher than tripled (87.7 kb) compared to the typical size of the IR region (~25 kb). On the other hand, the IR region of two lineages of Erodium (Geraniaceae) (Blazier et al., 2016; Ruhlman et al., 2017), Carnegiea gigantean (Cactaceae) (Sanderson et al., 2015), Tahina spectabilis (Arecaceae) (Choi et al., 2019), the Putranjivoid clade of Malpighiales (Jin et al., 2020a), and IR-lacking clade (IRLC) of Papilionoideae (Fabaceae) (Palmer and Thompson, 1982) plastome size reduced significantly. Generally, gene arrangement is not often in most angiosperms plastomes (Frailey et al., 2018). If so, the plastome rearrangement is relatively small (Xu and Wang, 2021). Nevertheless, a large amount of rearrangement is rare, but it is infrequently present in a few lineages, namely Asteraceae (Jansen and Palmer, 1987; Kim et al., 2005; Sablok et al., 2019), Campanulaceae (Knox et al., 1993; Cosner et al., 2004; Knox, 2014; Knox and Li, 2017; Uribe-Convers et al., 2017), Fabaceae (Kolodner and Tewari, 1979; Palmer and Thompson, 1981; Lavin et al., 1990; Doyle et al., 1996; Cai et al., 2008; Martin et al., 2014; Schwarz et al., 2015; Wang et al., 2017), Geraniaceae (Palmer et al., 1987; Chumley et al., 2006; Guisinger et al., 2011; Weng et al., 2014; Roschenbleck et al., 2017; Weng et al., 2017), Oleaceae (Lee et al., 2007), Plantaginaceae (Zhu et al., 2016; Kwon et al., 2019; Asaf et al., 2020), and Poaceae (Palmer and Thompson, 1982; Doyle et al., 1992; Michelangeli et al., 2003; Burke et al., 2016; Liu et al., 2020).
In the family Papaveraceae, Corydalis belongs to the Fumarioideae subfamily. It comprises more than 465 species and is the largest genus in the Papaveraceae family (Zhang et al., 2008). Some Corydalis species have medicinal properties and have tremendous potential against hepatitis, tumor, muscular pain, and cardiovascular diseases (Luo et al., 1984; Zhang et al., 2016). The morphological characteristics of the Corydalis species are diversified and adapt to various habitats, such as grasslands, forests, riversides, shrubs, and cliffs. In addition, the Corydalis species can grow from sea level to more than 6,000 meters in elevation, which is of great interest to ecologists and evolutionary biologists (Niu et al., 2014; Niu et al., 2017). Moreover, these species are distributed widely from the north temperate regions, specifically Qinghai–Tibet Plateau (QTP), to southeast China, Myanmar, the Korean peninsula, and Japan. Xu and Wang (2021) reported that the Corydalis species had undergone severe and rapid differentiation (Xu and Wang, 2021). The Corydalis plastomes must have also experienced a sequence of genetic shifts to adapt to the radically altered environment. Therefore, ecology biologists are interested in understanding how the plastome structure and its content have fluctuated on a fine scale in the evolutionary period and when rare plastome rearrangements were derived, which is why these modifications occurred (Xu and Wang, 2021). Nevertheless, few Corydalis species have been reported since 2019, and a large-scale plastome rearrangement in their structure has been observed. Thus far, twenty Corydalis cp genomes have been sequenced, but most studies have reported only briefly. Among these, two research articles explained the genomes in detail. Xu and Wang (2021) used six, and Ren et al. (2021) used two Corydalis plastomes for comparative studies (Ren et al., 2021; Xu and Wang, 2021). On the other hand, there are no extensive comparative studies of Corydalis plastomes to understand their genome rearrangement patterns, such as inversion, relocation, expansion, and contraction of IR regions, and their molecular evolution patterns in detail. In addition, divergent time-related molecular studies of the Corydalis species could not be found. Therefore, a new plastome of C. platycarpa was sequenced, and detailed comparative genomic analyses of all the publicly available twenty Corydalis plastomes were conducted. Based on this, this study examined the complexity of the genome structure and rearrangement, gene content, gain/loss of genes and introns, repeats, RNA editing, nucleotide diversity, and adaptive evolution of the Corydalis plastomes. Furthermore, the phylogenetic position and divergent time of the Corydalis lineages were estimated.
Materials and methods
Genomic DNA isolation and Corydalis genome sequencing
Fresh leaves of C. platycarpa were sampled from Cheongok mountain, Bonghwa-gun, South Korea (geospatial coordinates: N37°4′9″, E128°57′47″). A voucher specimen (YNUH22C183) was deposited at Yeungnam University Plant Herbarium, Gyeongsan, South Korea. The total gDNA was extracted from the fresh Corydalis leaves by a modified CTAB method (Doyle, 1990). Next-generation sequencing was performed with an Illumina HiSeq2500 by Phyzen Ltd., South Korea. The paired-end (PE) library (2 × 150 bp) was constructed using TruSeq PCR free kit and then paired reads with 550 bp insert size were sequenced and ~3 GB of raw data were obtained. FastQC v0.11 (Andrews, 2010) was used to check the low-quality reads, which were removed using Trimmomatic 0.39 (Bolger et al., 2014).
Assembly and annotation of the Corydalis chloroplast genome
For the de novo chloroplast (cp) genome assembly, the plastid-like reads were obtained from clean reads using the GetOrganelle pipeline v1.7.6.1 (Jin et al., 2020b). The filtered reads were then assembled using SPAdes v3.15.2 (Nurk et al., 2013) for the circular cp genome assembly in the paired-end mode. The assembled C. platycarpa cp genome coverage is 26,922×. The complete cp genome sequence and gene annotation were made using the online DOGMA program (Wyman et al., 2004) along with the cp genome annotations of Nicotiana tabacum (NCBI Reference sequence: NC_001879). Manual curation was carried out to adjust the start and stop codons of protein and ribosomal coding genes. The Corydalis cp genome circular map was drawn using OGDRAW v1.3.1 (Lohse et al., 2007). The annotated genome sequence was submitted to GenBank and assigned the accession number OP142703.
Chloroplast genome sequence divergence and comparison
The newly sequenced C. platycarpa cp genome and other 20 publicly available Corydalis plastomes (Supplementary Table S1) were compared to determine the cp genome structure synteny and identify the possible rearrangements with the plastome of N. tabacum as a reference using Mauve v1.1.3 with the progressiveMauve algorithm (Katoh et al., 2019). A single IR region was used in this analysis. The schematic diagram was drawn manually based on their plastome structure to access the expansion/contraction of the LSC, IR, and SSC junctions of the 21 Corydalis plastomes. The entire plastome sequences of all the Corydalis were used to visualize the sequence similarity using mVISTA in Shuffle-LAGAN mode (Frazer et al., 2004), with the default parameters and C. platycarpa plastome used as a reference.
Analyses of repetitive sequences
The simple sequence repeats (SSR) motifs were analyzed in the 21 plastomes of Corydalis using MISA v2.1 (Thiel et al., 2003) with the smallest number of repeats set to ten repetitions for mononucleotide SSRs, six repeat units for dinucleotide SSRs, and five repeat units for tri, tetra, penta, and hexanucleotide SSRs. The tandem repeats were searched using the Phobos Tandem Repeats Finder v1.0.6 with the parameters 1 for the match, −5 for mismatch and gap, and 0 for N positions (Mock et al., 2017). In addition, the forward, reverse, complement, and palindromic repeats were detected using REPuter with a Hamming distance of 3, 90% minimum sequence identity, and 30 bp of a minimal repeat size (Kurtz et al., 2001). For all these analyses, one copy of the IR region was used.
Analyses of the genetic divergence
All 59 protein-coding genes were extracted and aligned individually using Geneious Prime (Biomatters, New Zealand) to evaluate the genetic divergence of the 21 Corydalis plastomes. All gaps and missing data were excluded before the analysis. The genetic divergence of 21 Corydalis plastomes was calculated by applying nucleotide diversity (π) and the total number of polymorphic sites in the DnaSP v6.12.03 (Librado and Rozas, 2009).
Analysis of RNA editing sites in the protein-coding genes
The predictive RNA Editor for Plants (PREP) suite was applied to analyze the potential RNA editing sites in the protein-coding genes of the 21 Corydalis plastomes. The PREP-cp program has 35 reference genes explaining the RNA editing sites in the cp genomes (Mower, 2009). Therefore, 35 protein-coding genes of the Corydalis plastomes were utilized. In the present analysis, the cut-off value was set to 0.8.
Analysis of substitution rate
The complete cp genome of C. platycarpa was compared with the other 20 Corydalis plastomes. The synonymous (KS) and non-synonymous (KA) substitution rates were analyzed by extracting the identical specific 59 functional protein-coding DNA sequences and translating them into protein sequences and aligning them independently using Geneious Prime (Biomatters, New Zealand). The synonymous and non-synonymous substitution rates were assessed in DnaSP v6.12.03 (Librado and Rozas, 2009). Similarly, the substitution analyses for all the 37 Ranunculales and all the Ranunculales (16 taxa) excluding the genus Corydalis cp genomes were compared.
Analysis of positive selection
Positive selection analysis was performed based on substitution rate analyses of the 21 Corydalis plastomes. The site-specific model was employed using EasyCodeML v1.4 (Gao et al., 2019) to investigate the positive selection analysis. The 24 protein-coding gene sequence was aligned individually using the MAFFT alignment v1.5 (Katoh et al., 2019), and the maximum likelihood phylogenetic tree was built using RAxML v. 7.2.6 (Stamatakis et al., 2008). The codon substitution models, likelihood ratio test and the Bayes Empirical Bayes (BEB) analysis were conducted as described earlier.
Analysis of phylogenetic tree
Thirty-seven cp genomes from the order Ranunculales were selected to construct a phylogenetic tree with N. tabacum selected as an outgroup, determine the location of the C. platycarpa in the order Ranunculales, and analyze the phylogenetic correlation of the Corydalis genus. The cp genome sequences of 29 species across the Papaveraceae family corresponding to two subfamilies (Fumarioideae and Papaveroideae) were downloaded. In addition, two chloroplast genomes of each subfamily of Berberidaceae, Ranunculaceae, Menispermaceae, and Ciraeasteraceae were included in this analysis (Supplementary Table S1). The 59 protein-coding genes shared by 38 plastomes were concatenated, aligned and saved in PHYLIP format using Clustal X v2.1 (Larkin et al., 2007). The Maximum-Likelihood (ML) tree was built using RAxML v7.2.6 with a General Time Reversible + Proportion Invariant model. One thousand non-parametric bootstrap replicates were performed to estimate the support of the data for each internal branch of the phylogeny (Stamatakis et al., 2008).
Analysis of the evolutionary rate
Molecular divergence analysis of the Ranunculales lineages was performed with the Bayesian inference through Bayesian Markov chain Monte Carlo (MCMC) sampling implemented in BEAST v1.4 (Drummond and Rambaut, 2007) with a few modifications, as described earlier (Raman et al., 2021). A relaxed-clock log-normal model was applied using MCMC (500 million steps, sampled every 1000 generations, burn-in of 10%). A maximum clade credibility (MCC) tree was analyzed using TreeAnnotator v2.1.2 (Center for Computational Evolution, University of Auckland, New Zealand). Multiple calibration points were set for the divergence of the Berberidaceae subfamily, such as Berberis bealei at 88.94 mya (71.13–100.39 million years ago (mya), 78.22 mya (62.05–90.8 mya) for Jeffersonia diphylla, 62 mya (46.9–75.65 mya) for Epimedium koreanum and Diphylleia cymosa, 55.79 mya (38.65–73.14 mya) for Nandina domestica and 13.68 mya (8.05–21.75 mya) for Caulophyllum robustum and Gymnospermium microrrhynchum, which were employed with a log-normal distribution (Wang et al., 2016a).
Results
General features of the Corydalis chloroplast genome
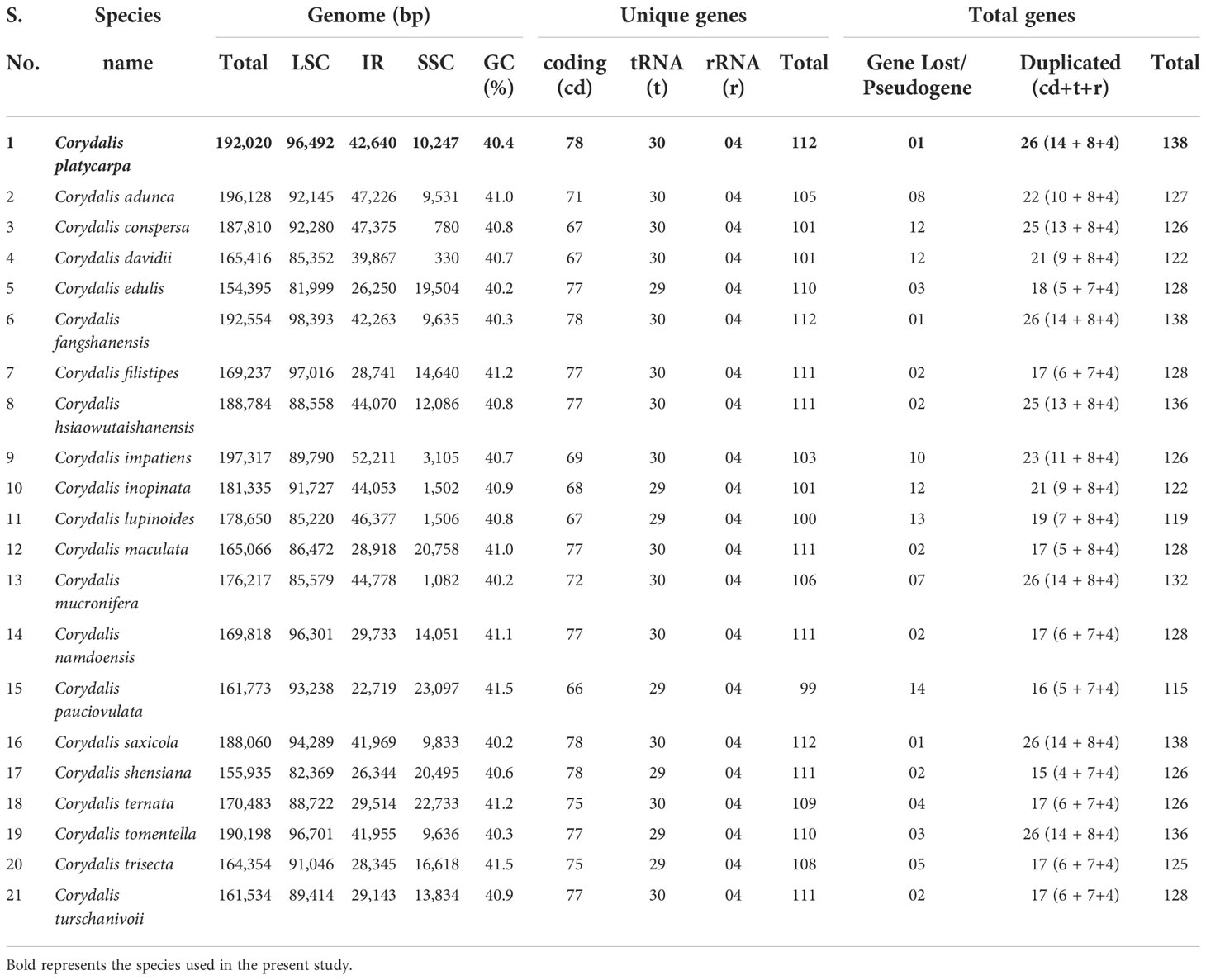
The complete chloroplast (cp) genome sequence of Corydalis platycarpa (GenBank: OP142703) is 192,20 bp, with an inverted repeat (IR) of 42,640 bp separating a large single-copy (LSC) region of 96,492 bp and a small single-copy region of 10,247 bp (Figure 1). The average G+C content of the cp genome was 40.4%. The C. platycarpa cp genome includes 112 unique genes, such as 78 protein-coding, 30 tRNA, and four rRNA genes. In 112 genes, nine protein-coding and six tRNA genes contained a single intron, and ycf3 and rps12 encoded two introns, whereas clpP coded for three introns. Moreover, 26 genes were replicated in IR regions, fourteen involving protein-coding, eight tRNA, and four rRNA genes (Supplementary Table S2). The gene accD was entirely lost in the cp genome of C. platycarpa. In addition, the C. platycarpa was compared with other Corydalis species and other Fumarioideae (Table 1) and Papaveroideae plastomes (Figure 2; Supplementary Table S3). The average plastome size of the Fumarioideae was 177 kb, whereas the Papaveroideae was only 156.5 kb (Figure 2A). Similarly, the GC content of Fumarioideae and Papaveroideae was 40.7 and 38.7%, respectively (Figure 2B). In addition, the average length of the LSC region of Fumarioideae and Papaveroideae was 90.3 kb and 85.5 kb, and 10.78 and 18.2 kb of the SSC and 38 and 26.3 kb of the IR regions, respectively (Figures 2C–E).

Figure 1 Circular chloroplast genome map of Corydalis platycarpa. Genes drawn outside the circle are transcribed clockwise, and those inside are counterclockwise. Genes belonging to different functional groups are color-coded. The darker grey in the inner circle shows the GC content, while the lighter grey shows the AT content.
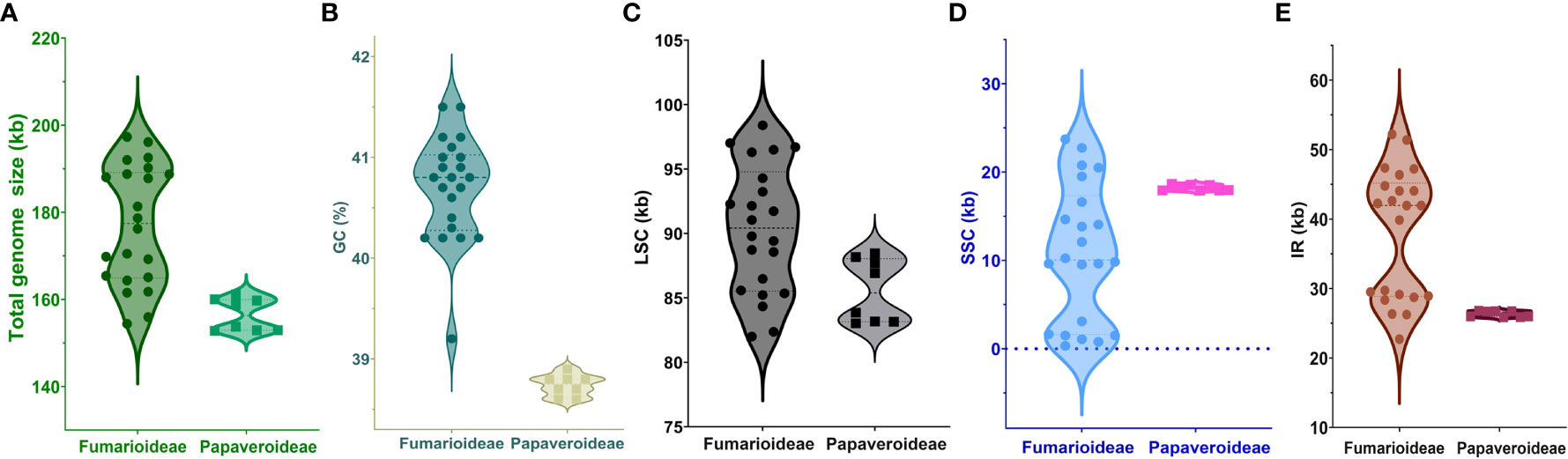
Figure 2 Visualization of (A) total genome size, (B) GC content, (C) LSC, (D) SSC, and (E) IR size of the Fumarioideae plastomes relative to Papaveroideae using a Violin plot.
Comparative analysis of the Corydalis chloroplast genome structure
The mauve alignment revealed many rearrangements in the cp genome of the C. platycarpa and their relatives of Corydalis species (Supplementary Figure S1). Therefore, many events, namely, inversion, translocation, expansion, and contraction duplication, occurred in the SC and IR regions in the cp genomes of Corydalis. In the LSC region, the rps16 gene was relocated within the LSC region of C. adunca; rbcL – trnV-UAC inversion and relocation occur in all the cp genomes of Corydalis except the species of C. edulis and C. shensiana. Similarly, the ndhB – trnR-ACG inversion was found in all the IR regions of the Corydalis cp genomes except the C. edulis and C. shensiana plastomes. In addition, a ndhI – ycf1 inversion occurs in the C. pauciovulata. In addition, the expansion and contraction of the SC and IR boundaries of the 21 Corydalis cp genomes were evaluated using comparative analyses of the genes across the boundary regions (Figure 3). The rps19 gene straddled the boundary of the LSC/IRB regions of the C. shensiana, C. lupinoides, and C. edulis cp genomes, whereas the rpl2 gene straddled the LSC/IRB regions of the remaining 18 Corydalis cp genomes that lead to the length of LSC regions varies from 82 kb to 98.4 kb (Figure 2). In contrast, the IR regions are highly expanded in most Corydalis cp genomes ranging from 22.7 to 52.2 kb. The ndhF gene is spanned in the IRB/SSC region in the C. shensiana and C. edulis cp genomes. Nevertheless, ndhI, ycf1, rps15, rpl32, trnN, and ndhH genes traversed the remaining Corydalis cp genomes due to the relocation, inversion, and expansion of the IR regions. Correspondingly, contraction occurs in the SSC region in most of the Corydalis cp genomes (Figures 2, 3) that affect the shuffling of the boundary genes (ndhA, ndhI, rps15, trnfM, ycf1, trnN, and ndhA) in the SSC/IRA regions. Similarly, most of the Corydalis genome encodes the rpl2 pseudogene in the IRA/LSC boundary regions.
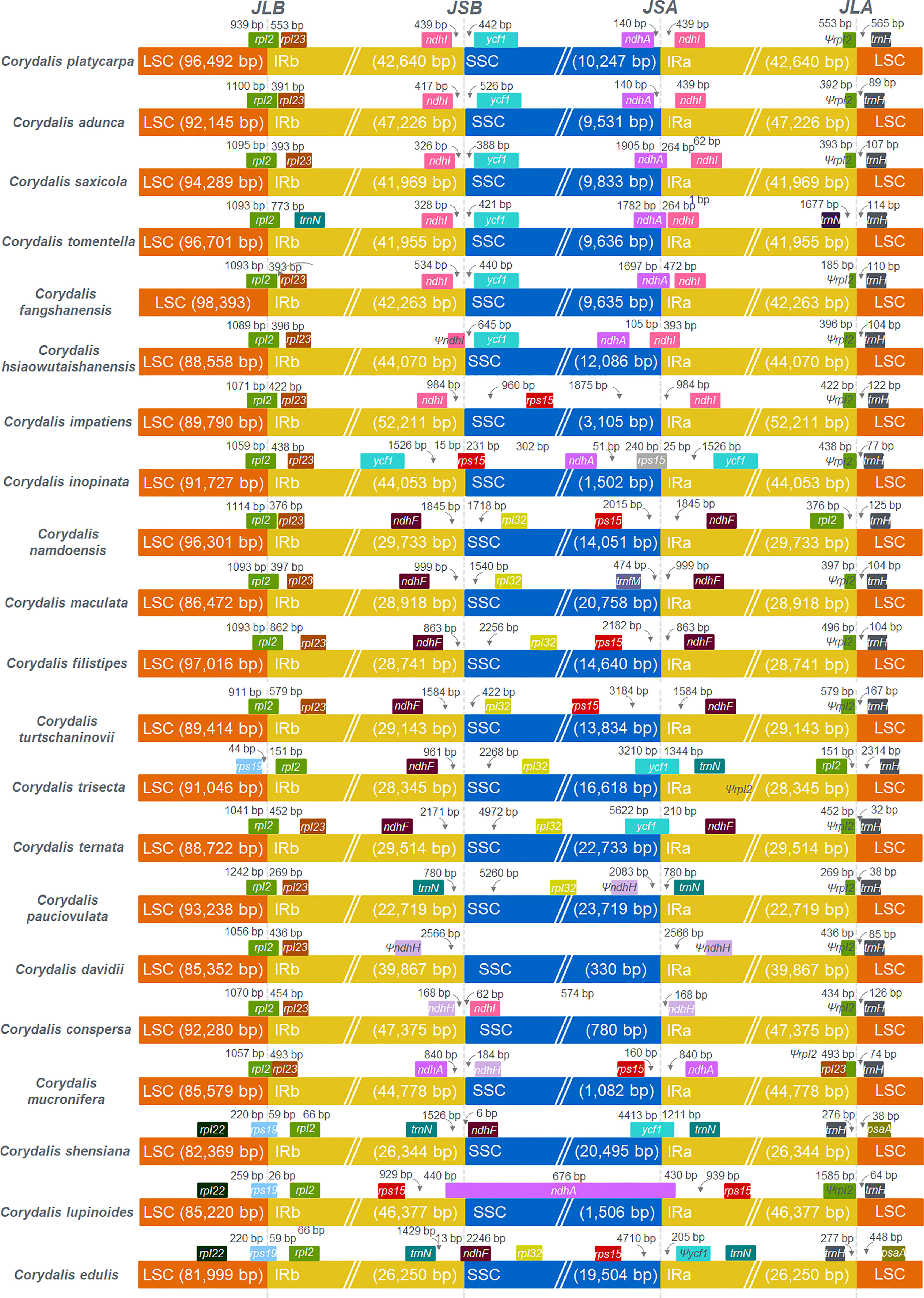
Figure 3 Comparison of the borders of LSC, SSC, and IR regions among 21 Corydalis chloroplast genomes. JLB indicates the junction line between LSC and IRb; JSB indicates the junction line between SSC and IRb; JSA indicates the junction line between SSC and IRa; JLA indicates the junction between LSC and IRa.
Comparative analysis of the repeat sequences in the Corydalis cp genomes
The results show that the total number of simple sequence repeats (SSRs) ranges from 19 (C. ternata) to 51 (C. pauciovulata), and the distribution of SSRs differs among the 21 plastomes of Corydalis (Figure 4A). Mononucleotides are the most frequent in the SSRs, distributing 88%, followed by dinucleotide and trinucleotides at 11% and 1%, respectively, in the Corydalis plastomes (Figure 4B). Among the mononucleotides, all the cp genomes occupy 96% of A and T type SSRs in their genomes, while most of the species lack dinucleotides, such as AG and CT trinucleotides, namely ATG, ATT, CAA, TTA, and TTG (Supplementary Table S4). Similarly, the distribution of tandem repeats in the Corydalis cp genomes ranges from 18 to 71. In addition to SSRs and tandem repeats, 1038 dispersed repeats are identified using REPuter (Figure 4A; Supplementary Table S4). Among the Corydalis cp genomes, the forward (76%), palindrome (21%) and reverse (3%) repeats are observed (Figure 4C).
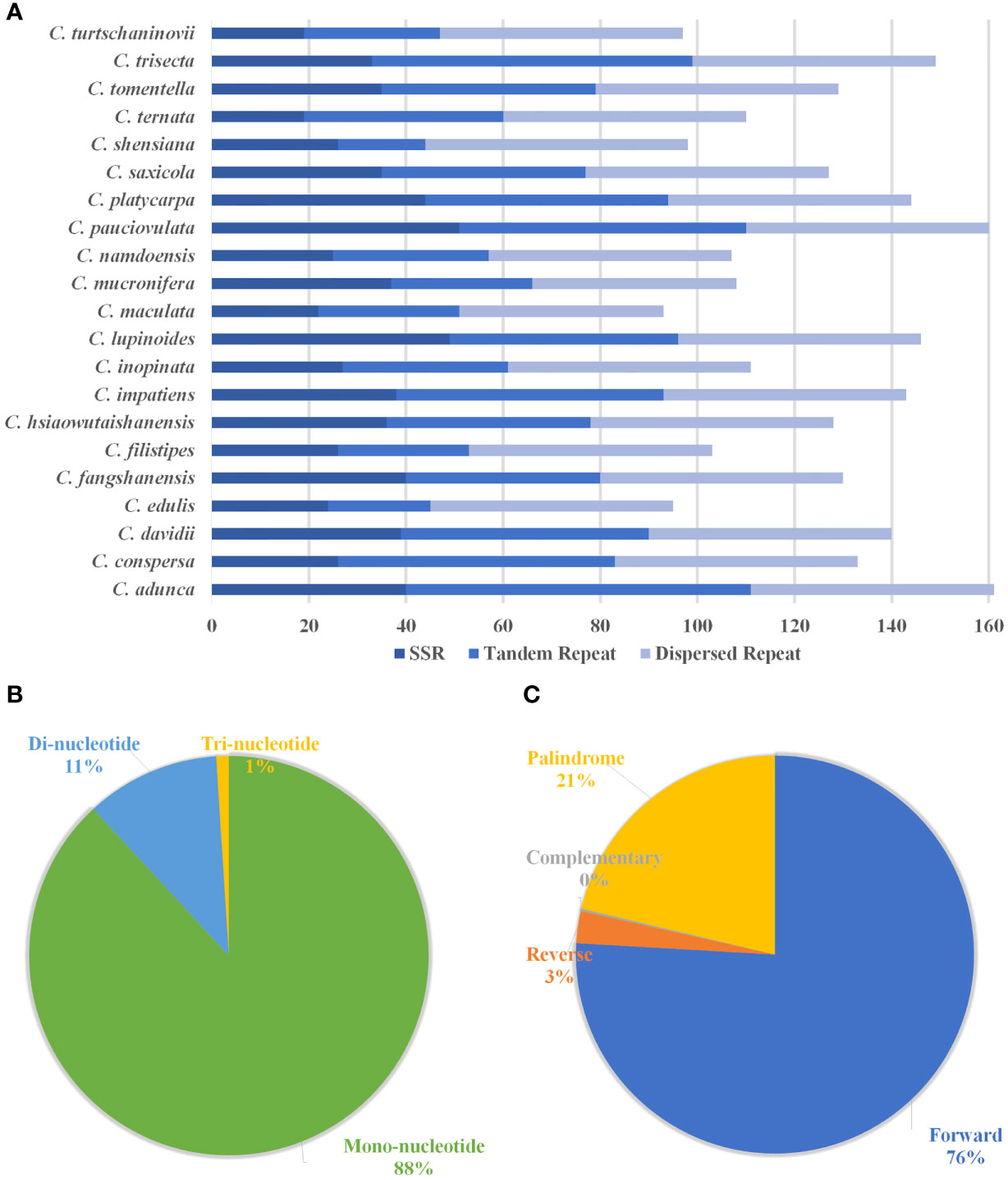
Figure 4 Histogram shows the number of repeats in 21 Corydalis chloroplast genomes. (A) The distribution of simple sequence repeats (SSRs), tandem repeats, and dispersed repeats in the 21 Corydalis plastomes. (B) Proportion of different SSR repeat types in the 21 plastomes of Corydalis. (C) The number of different types of dispersed repeats in the 21 Corydalis plastomes.
RNA editing site analysis in the Corydalis cp genomes
The possible RNA editing sites for 35 protein-coding genes were predicted by the PREP suite in the 21 Corydalis cp genomes. One thousand and seventy RNA editing sites were detected in their coding genes (Figure 5A; Supplementary Table S5). The number of editing sites varied from the 46 (C. mucronifera) to 57 (C. adunca) (Figure 5B; Supplementary Table S5). Among the 35 protein-coding genes of the Corydalis plastomes, the rpoB gene encoded the highest RNA editing sites (143), followed by rpoC2 (135), rpoC1 (122), atpA (84), matK (80), rps2 (71), ycf3 (59), rpl2 (51), rpl20 (44), and petD (42) (Figure 5A; Supplementary Table S5). In the RNA editing sites, 31% of sites converted serine to leucine, followed by 14% of proline to leucine, 9% of histidine to tyrosine, 8% of proline to serine, 7% of serine to phenylalanine, and 7% of arginine to tryptophane amino acids (Figure 5C; Supplementary Table S5). All predictable RNA editing sites are cytosine to uracil (C–U) transitions, the maximum of which are situated at the second codon position (66%), followed by the first codon position (30%), first and second codon position (4%), besides no transitions at the third codon position (Figure 5D; Supplementary Table S5).
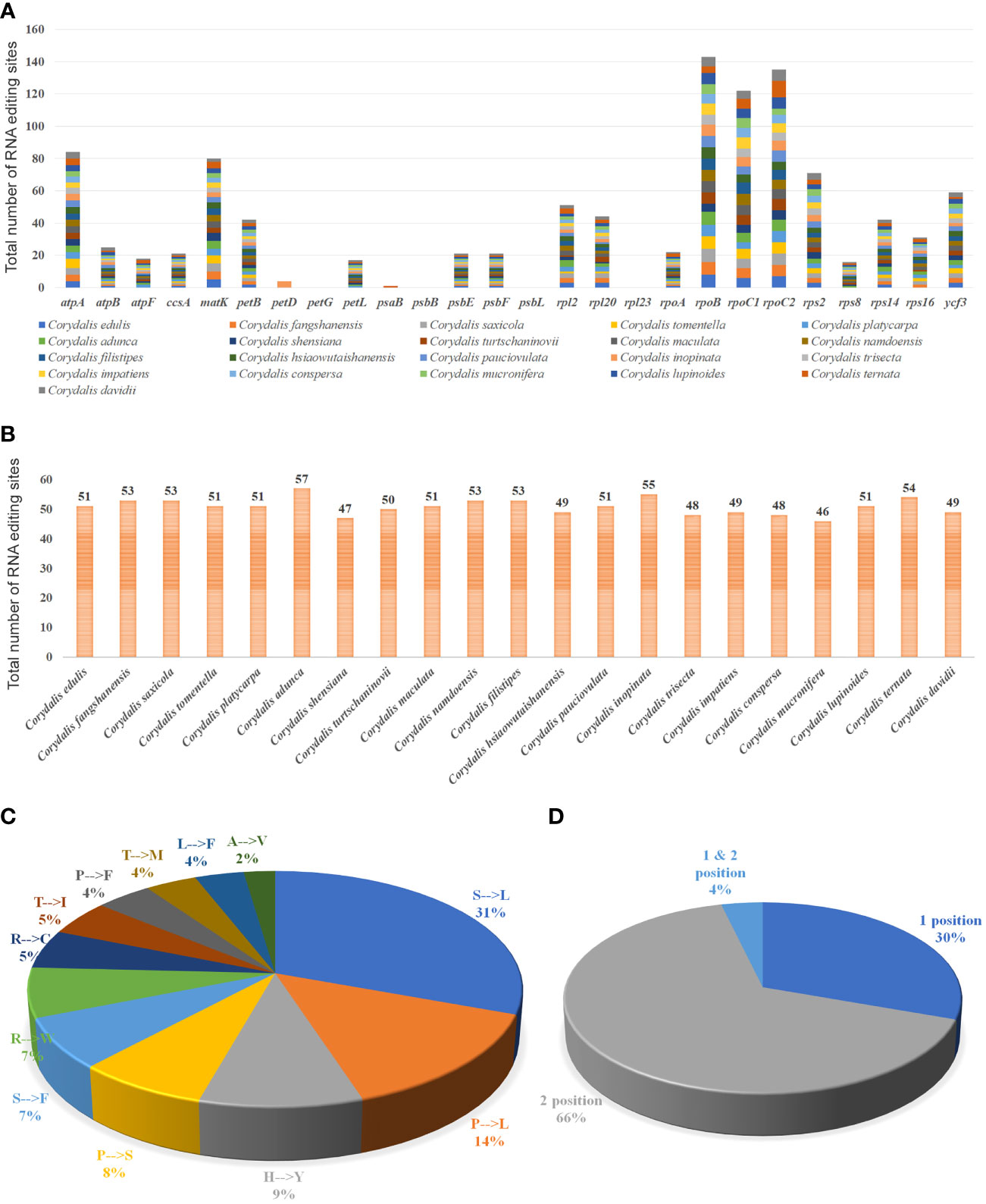
Figure 5 Analyses of RNA editing in the 35 protein-coding genes of the 21 Corydalis plastomes. (A) the distribution of RNA editing sites in the protein-coding genes of each Corydalis genome. (B) The number of RNA editing sites in each Corydalis cp genome. (C) Pie diagram represents the conversion percentage of amino acids in the RNA editing sites. (D) Represents the RNA editing site in the triplet codon of the nucleotide. S, serine; L, leucine; P, proline; H, histidine; Y, tyrosine; F, phenylalanine; R, arginine; W, tryptophan; C, cysteine; T, threonine; I, isoleucine; M, methionine; A, alanine; V, valine.
Sequence divergence analysis in the Corydalis cp genomes
The sequence divergence of all the 21 plastomes of Corydalis was analyzed using mVISTA and sequence identity plots constructed (Figure 6) with the annotated cp genome of C. platycarpa as the reference. The results showed that the ribosomal RNA genes in the IR regions were highly conserved and less divergent than the other coding and non-coding sequences in the LSC, SSC, and IR regions. In addition, the nucleotide diversity (Pi) of 59 protein genes in the Corydalis cp genomes was calculated. All 59 genes were highly variable regions (>0.03) that are associated with photosynthetic, transcription, and translational processes (Figure 7). Among these 59 genes, psaC has the lowest Pi value (0.041), and rps16 has the highest Pi value 0.642.
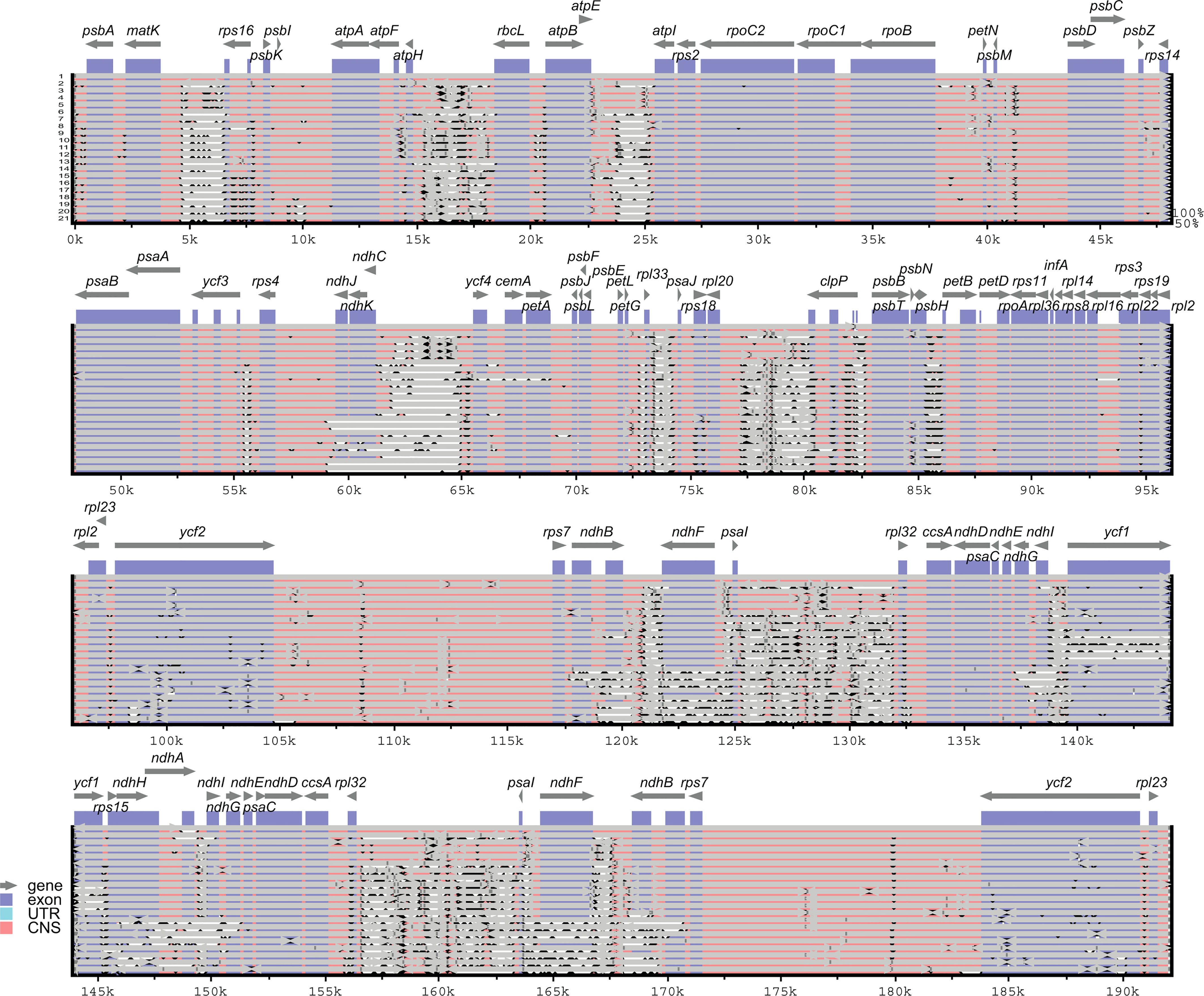
Figure 6 mVISTA-based sequence identity plot of 21 Corydalis plastomes with C. platycarpa as a reference. The gray arrows indicate the direction of the gene transcription. The y-axis represents the percent identity ranging from 50 to 100% is represented by the vertical scale. Coding and non-coding regions are colored purple and pink, respectively. 1. Corydalis platycarpa; 2. Corydalis adunca; 3. Corydalis saxicola; 4. Corydalis tomentella; 5. Corydalis fangshanensis; 6. Corydalis hsiaowutaishanensis; 7. Corydalis impatiens; 8. Corydalis inopinata; 9. Corydalis namdoensis; 10. Corydalis maculata; 11. Corydalis filistipes; 12. Corydalis turtschaninovii; 13. Corydalis trisecta; 14. Corydalis ternata; 15. Corydalis pauciovulata; 16. Corydalis davidii; 17. Corydalis conspersa; 18. Corydalis mucronifera; 19. Corydalis shensiana; 20. Corydalis lupinoides; 21. Corydalis edulis.
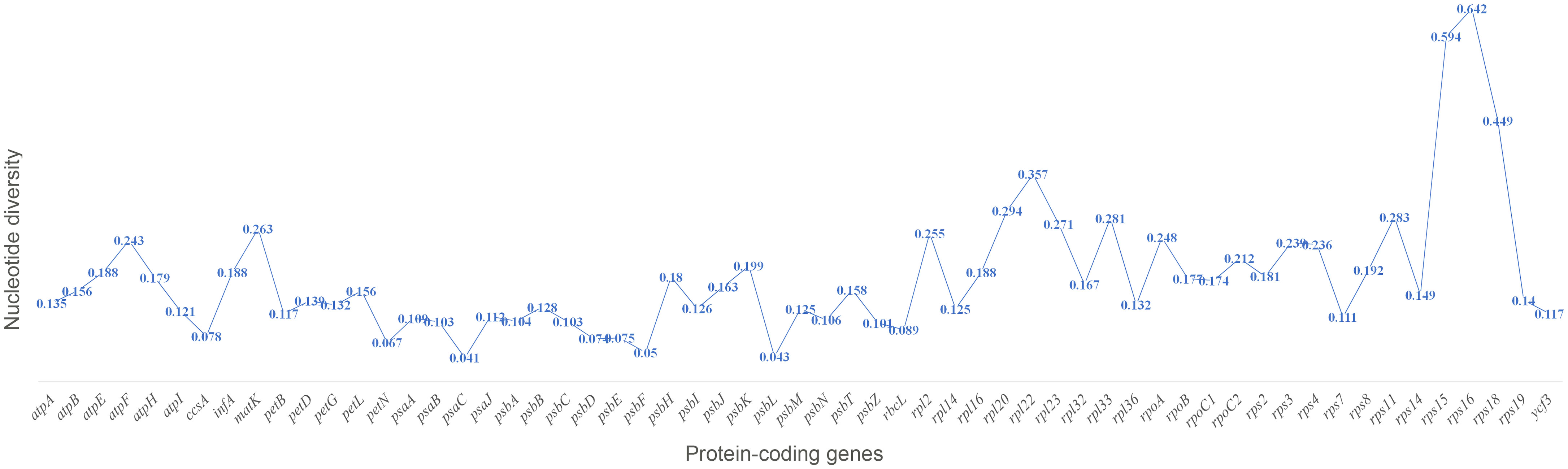
Figure 7 Percentage of variable characters (SNPs) in the protein-coding genes of 21 Corydalis plastomes.
Adaptive evolution analysis in the Corydalis cp genomes
Fifty-nine shared protein-coding genes of all the 21 Corydalis plastomes were used for synonymous (KS) and non-synonymous (KA) substitution rates. The results showed that most protein-coding genes have relatively high average KS values (>0.05) except the ccsA, petN, psaJ, psbE, psbF, psbL, psbM, psbZ, rpl32, rpl36, and rps7 genes (Figure 8A; Supplementary Table S6). In the same way, most of the protein-coding genes are comparatively high average KA values (>0.02) except atpA, atpB, atpI, ccsA, infA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbL, psbM, psbN, psbZ, rbcL, rpl14, rpl16, rpl36, rps7, rps19, and ycf3 genes (Figure 8B; Supplementary Table S6). The protein-coding genes, rps16 and rps18, show the highest average KA/KS ratio of 1.42 and 1.37, respectively. On the other hand, the KA/KS ratios of all the protein-coding genes ranged from 0 to 1.42, with an average ratio of only 0.28 (Figure 8A; Supplementary Table S6). Similarly, all the 37 Ranunculales taxa were analyzed for substitution analysis and revealed that the KA/KS ratios of all the protein-coding genes differed from 0 to 6.83, with an average ratio of 0.21 (Supplementary Figure S2; Supplementary Table S7). Furthermore, the substitution analysis of all the 59 protein-coding genes of Ranunculales, excluding the genus Corydalis (16 taxa), revealed that the KA/KS rate varied from 0 to 0.89, with the average rate of 0.13 (Supplementary Figure S3; Supplementary Table S8).
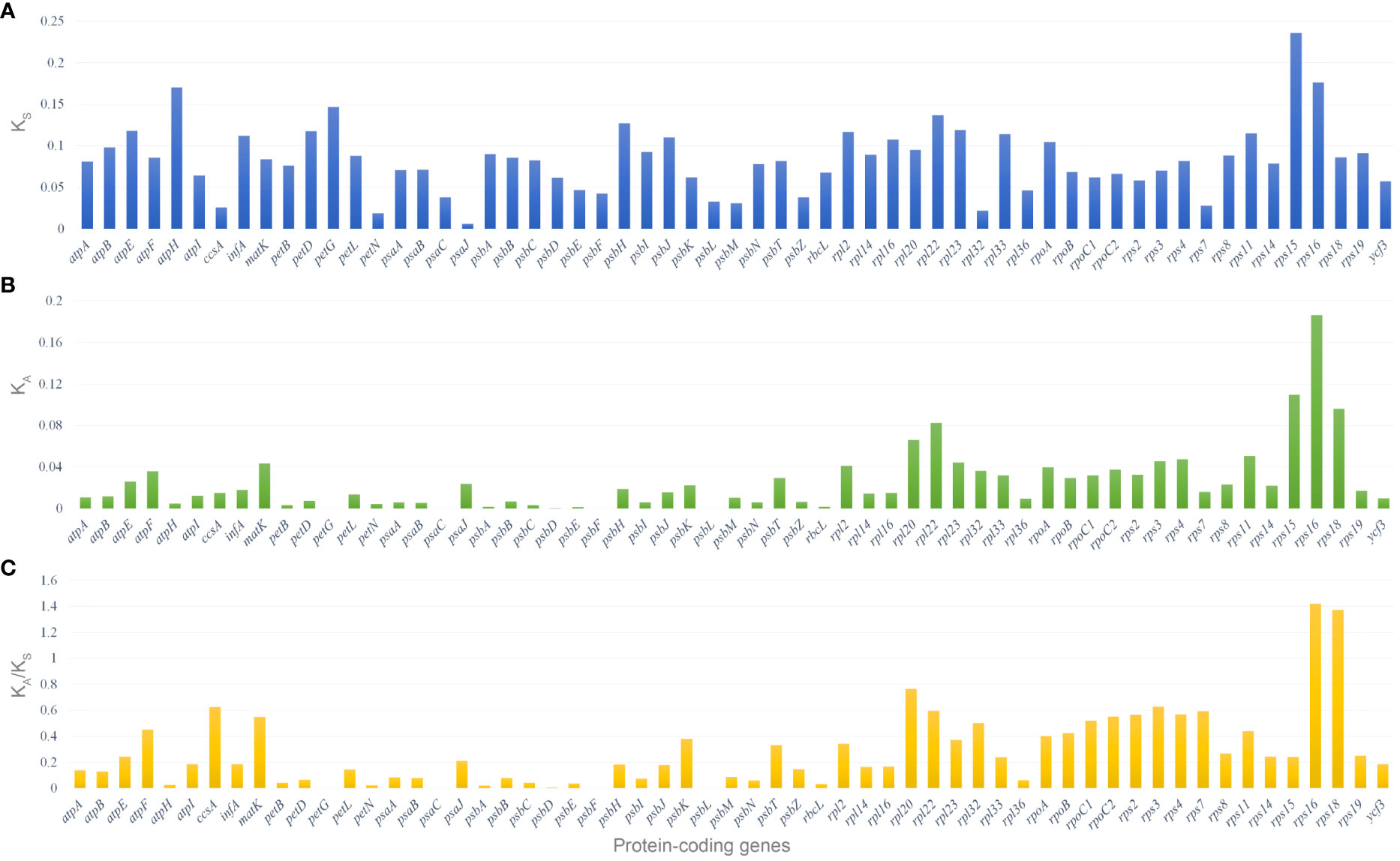
Figure 8 Selective pressure of 59 protein-coding genes in the 21 Corydalis plastomes. (A) KS, rate of synonymous substitution; (B) KA, rate of non-synonymous substitution; (C) KA/KS, rate of non-synonymous vs. synonymous substitution.
Suppose the substitution ratio of the specific protein-coding genes among two cp genomes or the whole genomes is > 1.0. In that case, these genes are considered to be under positive selection. Therefore, the KA/KS (ω) ratio of 24 protein-coding genes is > 1.0, and they were analyzed for selective pressure events. The ω2 ratio of 24 protein-coding genes ranges from 1.0 – 107.1382 in the M2a model (Supplementary Table S9). Bayes empirical Bayes (BEB) analysis was employed to assess the position of coherent selective sites in the 24 protein-coding genes utilizing the M7 vs. M8 model and determine that seventeen sites under possibly positive selection in the four protein-coding genes (rpl20 – 2; rpl22 – 3; rpl23 – 1; rps2 – 2; rps3 – 1; rps4 – 3; rps11 – 1; rps14 – 2 and rps18 – 2) with posterior probabilities >0.95 and 21 sites (ccsA – 1; psbJ – 1; psbK – 1; rpl20 – 1; rpl22 – 3; rps3 – 3; rps4 – 5; rps7 – 1; rps8 – 1; rps11 – 1 and rps16 – 3) >0.99 (Supplementary Table S9). The positive selection models’ likelihood ratio test (LRT) statistics against their null models (2ΔLnL) for all 59 genes of 21 Corydalis species were evaluated. The 2ΔLnL value ranged from 1.450266 – 115.737378 (Table 2). In contrast, the protein-coding genes atpB, atpE, atpF, matK, psbH, psbT, rpl16, rpl33, and rps15 did not positively the encode selected sites in their genes.

Table 2 Comparison of the likelihood ratio test (LRT) statistics of positive selection models against their null models (2ΔLnL) for across all Corydalis species.
Phylogenetic analysis of the Ranunculales
In the present study, 59 concatenated protein-coding genes were used to investigate the phylogenetic relationship of Ranunculales. All the Ranunculales species were clustered into two lineages (clade I and II). In the Papaveraceae lineage, it was grouped into Fumarioideae and Papaveroideae clades. All the Corydalis species were clustered into three clades, and C. adunca is the basal group in the tree (Figure 9). C. platycarpa is the sister to C. Saxicola, C. tomentella, C. fangshanensis, and C. edulis and formed one clade. C. ternata, C. turtschaninovii, C. flistipes, C. maculata, and C. namdoensis formed another clade, whereas C. davidii, C. lupinoides, C. pauciovulata, C. inopinata, C. trisecta, C. impatiens, C. conspersa, and C. mucronifera formed the third clade. All the Corydalis species were supported with strong bootstrap values.
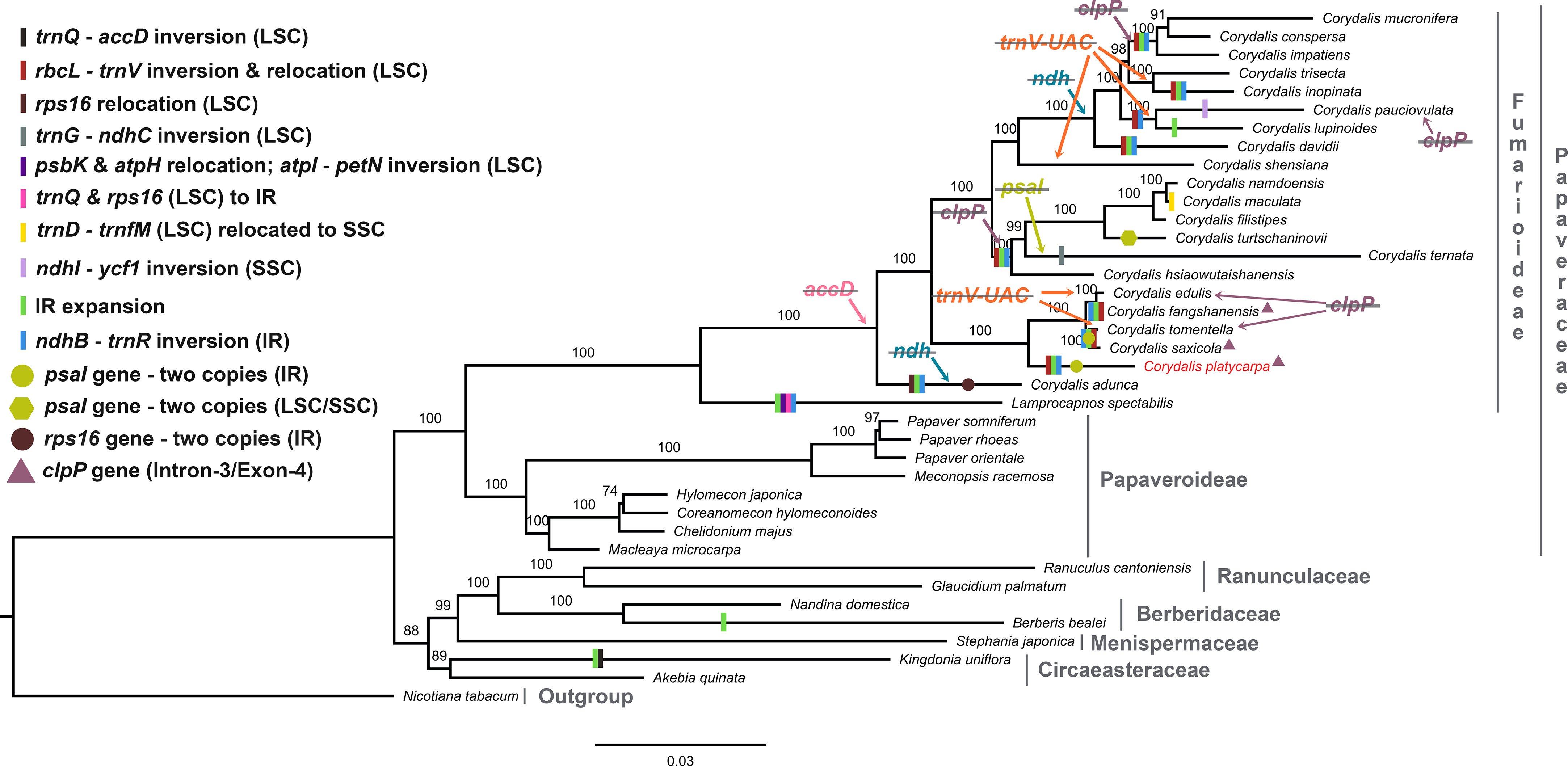
Figure 9 Maximum likelihood (ML) tree for 38 taxa based on 59 common plastid protein-coding genes. Values above the branches represent the maximum likelihood bootstrap value.
Molecular clock analysis of the Ranunculales
The dataset for 59 protein-coding genes of 37 Ranunculales species was used to estimate the divergent time for the Corydalis species. Owing to a lack of calibration points, five other species of Ranunculales were also included. The divergent time was estimated using the previous data of the Ranunculales, which are similar to those obtained in the present study. Among order Ranunculales, the families Circaesteraceae, Menispermaceae, Ranunculaceae, and Berberidaceae diverged 138.13 million years ago (mya) (95% highest posterior density [HPD]: 198.81–90.25 mya). In the Papaveraceae family, Fumarioideae and Papaveroideae diverged 181.08 mya (95% HPD: 272.02–112.44 mya) and 155.65 mya (95% HPD: 223.22–98.61 mya), respectively (Figure 10). The chronogram resulting from a BEAST analysis showed that whole speciation events within Corydalis occurred from 98.6 to 1.51 mya. The C. adunca diverged from the ancestor of other remaining members of the Corydalis species at 98.6 mya (95% HPD: 154.44–56.86 mya). Among the Corydalis, the C. platycarpa diverged in the early Oligocene period (31.15 mya [95% HPD: 62.15–11.97].
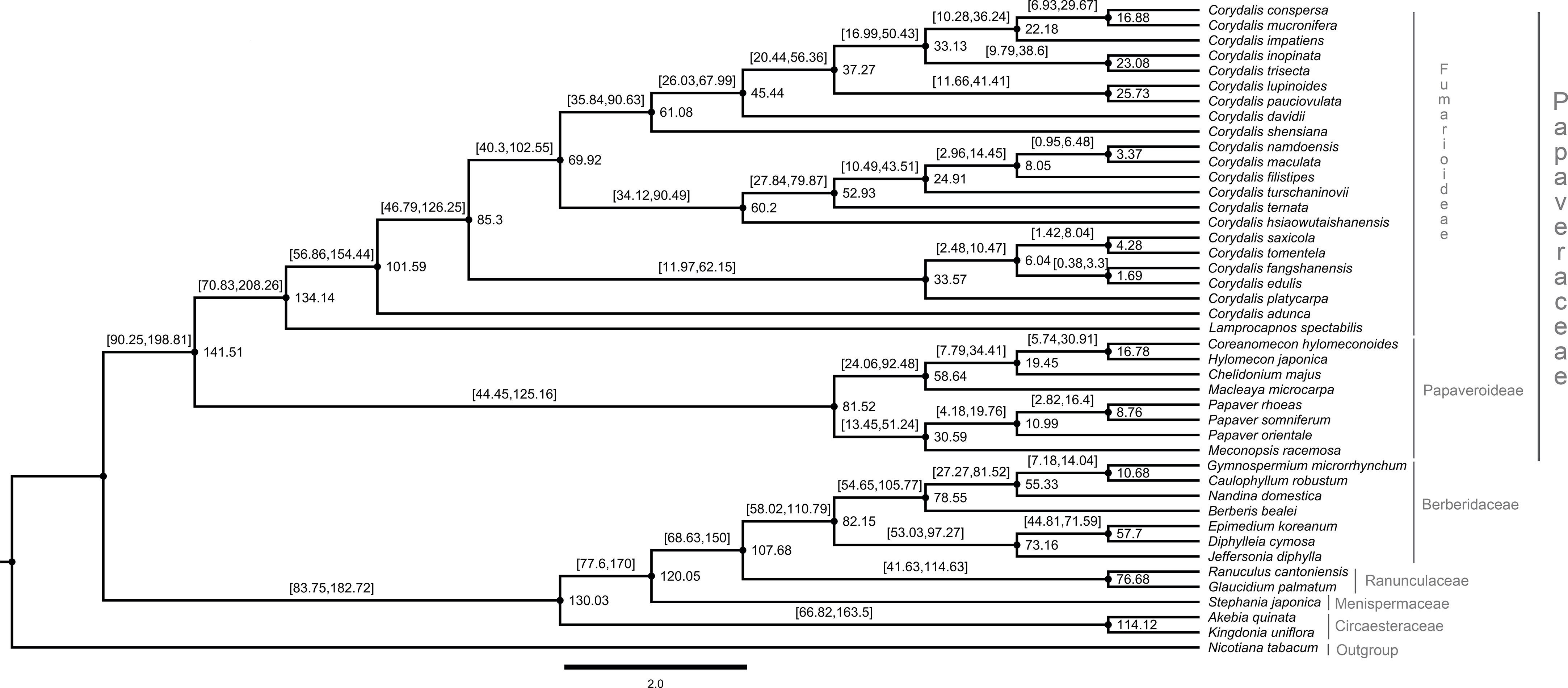
Figure 10 Estimation of the divergence time of Corydalis species using BEAST based on 59 plastid protein-coding genes of 42 Ranunculales and one outgroup species. The estimated mean ages are shown near the nodes, and blue bars represent 95% high posterior density.
Discussion
Corydalis is the largest genus within the Papaveraceae family and contains more than 465 species (Zhang et al., 2008). Thus far, 20 chloroplast genomes have been sequenced and analyzed. No extensive or comparative studies of the Corydalis plastomes have been conducted thus far. Therefore, in the current study, the cp genome of C. platycarpa was sequenced and characterized, and comparative studies were carried out with twenty other species of the Corydalis genus. The results showed that the cp genomes of Corydalis displayed the total genome and size of the LSC, SSC, and IR regions (Figure 2; Supplementary Table S3). The gene order and its contents and the GC% content varied and were unusually greater or less than other Papaveraceae cp genomes. The total cp genome size of the Corydalis ranged from 154.4 kb (C. edulis) to 197.3 kb (C. impatiens), and C. platycarpa was the fourth largest cp genome (192 kb) in the Corydalis and the fifth largest among the Ranunculales cp genomes (Figure 2; Supplementary Table S3). The average size of the Corydalis cp genomes is 176.5 kb. In addition, the typical size of the cp genomes of Fumarioideae (Corydalis + Lamprocapnos) is 177 kb. In contrast, among the Papaveraceae family, the average genome size of the Papaveroideae is only 156.5 kb. Similarly, the average length of the LSC, SSC, and IR of the Fumarioideae was found to be 90.3, 10.78, and 38 kb, respectively. By contrast, the average lengths of the LSC, SSC and IR regions of Papaveroideae were 85.5 kb, 18.2 kb, and 26.3 kb, respectively. The variation in the Furmarioideae was attributed to the expansion of IR regions in their genomes, leading to a shift of the SC genes into the IR regions (Figure 3). In particular, the IR region was extended into the SSC region in most Corydalis cp genomes. C. impatiens encodes the largest IR region (52.2 kb), and C. davidii contains the smallest SSC region (330 bp). This variation in the size of Fumarioideae cp genomes also affects their GC content. The GC content of the C. pauciovulata and C. trisecta is the highest (41.5%) in the Ranunuculales plastomes. Commonly, a high GC content imparts more stability to the genome than the AT. In addition, the larger amount of GC base pairs in the genome might impact their adaptation to various adverse environments. On the other hand, the high percentage of AT affects the gene order and its content in the Fumarioideae cp genomes.
In Ranunculales, many relocations, inversions, and rearrangements occurred with all the Fumarioideae plastomes except for C. edulis, C. shensiana, and C. trisecta. Moreover, at least eight events have occurred in all three LSC, SSC, and IR regions (Figure 9; Supplementary Figure S1) and the following events occurred in the LSC region: (i) a ~ 10 kb of rbcL – trnV-UAC inverted and relocated into the upstream of atpH and downstream of the atpI gene in the LSC region (all the Fumarioideae genomes except Lamprocapnos spectabilis, C. adunca, C. edulis, C. shensiana and C. trisecta); (ii) the rps16 gene (LSC) relocated into the IR region (C. adunca) and trnQ -UUG – rps16 (LSC) into the IR (L. spectabilis); (iii) ~16 kb of trnG-GCC – ndhC inverted and relocated into the upstream of psbZ and downstream of psbJ in the LSC region (C. ternata); (iv) ~7 kb of psbK – atpH relocated and ~15 kb of atpI - petN inverted in the LSC region (L. spectabilis) (Park et al., 2018); (v) ~7.5 kb of trnD-GUC – trnfM relocated into the SSC region (C. maculata). The following occurred in the SSC region: (vi) ~10.5 kb of ndhI – ycf1 inverted in the IR region (C. pauciovulata): (vii) ~14.4 kb of ndhB – trnR-ACG inverted (except C. edulis, C. shensiana and C. trisecta in the Fumarioideae); (viii) IR expanded (except C. edulis, C. shensiana, C. pauciovulata and C. trisecta in the Fumarioideae). Among these eight events in Fumarioideae, seven events occurred in at least one species of the Corydalis cp genome (Figure 9). Furthermore, the translocation and IR expansion analyses were extended in the Ranunculales cp genomes. In the Berberis bealei (Berberidaceae) (Ma et al., 2013), the ~13 kb of the LSC region (rps19 – psbB) was transferred to the IR region (Figure 9). Similarly, ~6 kb of the SSC region moved to the IR region, and ~50 kb inversion (trnQ-UUG – accD) occurred within the LSC region of the Kingdonia uniflora (Circaeasteraceae) (Sun et al., 2020) (Figure 9). In contrast, Xu and Wang (2021) reported that the ndhB – trnR-ACG inversion event occurred in the IR region of the common ancestor of Fumarioideae plastomes (Xu and Wang, 2021). This is due to the limitation of the six plastomes used in their comparative studies. In the present study, 21 Corydalis species and one L. spectabilis (Park et al., 2018) cp genome (Fumarioideae) were used for comparative analyses, suggesting that the ndhB – trnR-ACG event did not occur in C. edulis, C. shensiana, and C. trisecta in the Fumarioideae (Figure 9). Therefore, single or all the genome rearrangement/relocation events did not take place in the common ancestor of either the Corydalis genera or Fumarioideae clade. Earlier studies reported that several cp genome rearrangements and relocation are feasibly lowest common in angiosperms. To support the present study, previous studies reported similar rearrangement events, such as inversion and relocation of trnV-UAC – rbcL event in the Oleaceae (Lee et al., 2007) and Campanulaceae (Knox, 2014; Knox and Li, 2017; Uribe-Convers et al., 2017) and trnQ -UUG – rbcL in the Circaeaster agrestis and K. uniflora of Ranunculaceae (Sun et al., 2017; Sun et al., 2020) occurred independently in their plastomes rather than there being a common ancestor.
The gene content in the Corydalis plastomes was compared. Usually, the cp genome comprises 79 protein-coding genes (excluding ycf15 and ycf68 genes), 30 transfer and four ribosomal RNA genes (Raman et al., 2019; Raman and Park, 2022). On the other hand, the genus Corydalis varied from 66 (C. pauciovulata) to 78 protein-coding genes (C. platycarypa, C. fangshanensis, C. saxicola, and C. shensiana) in their plastomes (Table 1). All the Corydalis species lost accD, and some of the Corydalis lost clpP, ndh, rps16, psaI, and trnV-UAC genes in their plastomes (Figure 9; Supplementary Table S10). The tRNA and rRNA contents were the same in almost all species except for the loss of the trnV-UAC gene in a few Corydalis cp genomes (C. edulis, C. inopinata, C. lupinoides, C. pauciovulata, C. shensiana, C. tomentella, and C. trisecta). In contrast, trnV-UAC was reported to be highly conserved in all monocot plants, whereas tRNALys, tRNAAla, tRNAIle, tRNASec, tRNAPyl and suppressor tRNA were absent in most of the monocot cp genomes (Mohanta and Bae, 2017; Mohanta et al., 2019; Mohanta et al., 2020b). By contrast, the absence of all the tRNA genes in the monocots was highly conserved in the Corydalis and other closely related cp genomes. Overall, losses of a minimum of one gene to a maximum of fourteen genes occurred in the Corydalis plastomes (Table 1; Supplementary Table S10). Among the protein-coding genes, the accD gene was lost in all Corydalis plastomes. This event happened in the common ancestor of the Corydalis lineages (Figure 9). The accD encodes one of four subunits of the acetyl-CoA carboxylase enzyme (ACC), which is necessary for fatty acid biosynthesis (Elborough et al., 1996; Sasaki and Nagano, 2004). Moreover, this enzyme is involved in the first process. Kode et al., 2005 suggested that the loss of the accD is detrimental to the plants, as observed in a study of tobacco (Kode et al., 2005). Earlier studies confirmed that the missing accD gene in the plastome is relocated in the nucleus of angiosperms species, such as Trifolium repens (Magee et al., 2010), Campanulaceae (Rousseau-Gueutin et al., 2013), and Platycodon grandiflorum (Hong et al., 2017). Because there were no transcriptome data, this study could not confirm whether the cp-encoded accD gene was lost wholly or functionally, which was relocated to the nucleus in the genus Corydalis. Therefore, further transcriptome studies will be needed to confirm whether the plastid copy is relocated in the Corydalis nuclear genome.
Typically, the plastid DNA of most of the higher plants encodes eleven ndh genes (Maier et al., 1995; Yukawa et al., 2005) that produce ndh polypeptides, forming a thylakoid ndh complex (Sazanov et al., 1998; Casano et al., 2000). This ndh complex is similar to the mitochondrial complex I, which catalyzes the transfer of electrons from NADH to plastoquinone (Martin and Sabater, 2010). Among the eleven ndh genes in the plastomes, the ndhC, ndhK, and ndhJ genes are situated in one transcriptional unit (ndhC-J operon) in the LSC region of the plastome (Serrot et al., 2008). The genes ndhH, ndhA, ndhI, ndhG, ndhE, and ndhD are located in the SSC region (ndhH-D operon), which also includes the gene psaC (encodes a polypeptide of the photosystem I complex, PSI) between the genes ndhE and ndhD (Del Campo et al., 2000). In addition, the ndhF gene is represented in the SSC region, and two identical copies of the ndhB gene exist in IR regions (one on each). The ndhF gene and the two ndhB genes are possibly transcribed autonomously as monocistronic mRNAs (Martin and Sabater, 2010). In the present study, some Corydalis lineages that displayed a wide-ranging pseudogenization or absence of the ndh genes in their plastomes were identified (Figure 9; Supplementary Table S10). A similar plastome rearrangement event accompanied by either pseudogenization or a loss of ndh genes was also identified in other Ranunculales species, K. uniflora (Sun et al., 2017) and Orchidaceae species (Lin et al., 2015). These two events occured independently in these species. In addition, comparative analyses of 2511 cp genomes showed that anyone of the ndh gene losses occurred commonly in at least one species of all lineages, such as algae, bryophytes, eudicots, gymnosperms, magnoliids, monocots, protists and pteridophytes (Mohanta et al., 2020a). However, in the Corydalis plastomes, at least three to all (eleven genes) ndh genes were either pseudogenized or lost in the plastomes of C. adunca, C. conspersa, C. davidii, C. impatiens, C. inopinata, C. lupinoides, C. mucronifera, C. pauciovulata, and C. trisecta plastomes (Figure 9; Supplementary Table S10). Among the loss of ndh genes in the nine plastomes, the ndhC and ndhF loss occurred in the plastomes of all nine species (Figure 9; Supplementary Table S10). The C. adunca is basal for the remaining Corydalis species, and the ndh gene loss occurred in their genome. The remaining eight species formed a single clade in the phylogenetic tree, and the ndh gene loss occurred in this clade, suggesting that after divergence from the C. shensiana, it probably occurred in the common ancestor of this clade plastomes (Figure 9). This event appears to be a synapomorphy that occurred at the subgenus level in the Corydalis clade. It is probably associated with the rearrangement and relocation of SC and IR genes, boundary shift, and expansion of the IR regions in the Corydalis plastomes. Interestingly, all nine species (except C. pauciovulata) were distributed predominantly on the Qinghai–Tibet Plateau (QTP) regions. The photosynthetic systems (ndh genes) in these plants, might have been lost due to the high altitude conditions, such as low temperatures, strong winds, and low atmospheric pressure, and adapted to their ecological environment.
The clpP gene is a proteolytic subunit of the ATP-dependent Clp protease found in higher plant chloroplasts (Shikanai et al., 2001). Usually, the clpP encoded three exons spliced by two type II introns in the cp genome (Raman et al., 2019; Raman and Park, 2022). Earlier studies stated that the loss of introns of the clpP gene had been determined in the Geranium, legume, Silene, and Hypericum cp genomes (Erixon and Oxelman, 2008; Dugas et al., 2015; Park et al., 2017; Claude et al., 2022). In this study, the clpP gene loss also took place in some of the Corydalis species, such as C. conspersa, C. mucronifera, C. impatiens, C. namdoensis, C. maculata, C. filistipes, C.turtschaninovii, C. ternata, C. hsiaowutaishanensis, C. edulis and C. tomentalla (Supplementary Table S10). Among these, C. conspersa, C. mucnifera, and C. impatiens formed one clade in the phylogenetic tree using 59 protein-coding concatenated datasets and C. namdoensis, C. maculata, C. filistipes, C.turtschaninovii, C. ternata, and C. hsiaowutaishanensis formed another (Figure 9). Except for the C. edulis and C. tomentella, the clpP gene loss may have occurred in the common ancestor of these two clades at the subgenera level (Figure 9). On the other hand, it is essential to use additional species to understand the clpP loss in the genomes of Corydalis. This is supported by a similar type of clpP loss that occurred in the common ancestor of the Actinidiaceae family (Clematoclethra, Actinidia, and Saurauia) (Wang et al., 2016b). In contrast, a few Corydalis plastomes (C. platycarpa, C. saxicola, and C. fangshanensis) encoded four exons and three introns in the clpP gene (Supplementary Figure S3). In the clpP, there was a ~115 bp insertion after exon 1 in the gene leading to the formation of an additional intron in their plastomes. The inserted nucleotide sequence similarity between the three species was 73.5%. In addition, this insertion sequence in the clpP gene was analyzed using BLASTN, but reliable results could not be obtained. The acquisition of one extra intron in the clpP gene may be due to the selective pressure in their genome. This could play a role in the evolutionary maintenance of the group II introns and provide more stability to the genome (Petersen et al., 2011). Selective pressures may have been significant and undervalued in the evolution of spliceosomal introns from group II intron progenitors (Chalamcharla et al., 2010). To the best of the authors’ knowledge, this rare event has not been identified in any other plastomes. Therefore, further studies will be needed to understand the molecular mechanisms of the clpP gene in their plastomes.
In addition, gene duplication also occurred in the Corydalis plastomes because of genome rearrangements and boundary shifts. The two copies of the rps16 gene in the IR regions of C. adunca are replaced with rrn16 in the LSC region, suggesting that at least two rearrangements might have occurred in their plastome simultaneously or independently (Xu and Wang, 2021). At the same time, the rps16 gene is a pseudogene in the C. ternata. Similarly, two copies of the psaI gene were found in the IR regions of C. platycarpa, C. saxicola, and C. tomentella: one copy from LSC and another from SSC in the C. turtschaninovii cp genomes (Figure 9; Supplementary Figure S4). Typically the psaI gene is located upstream of accD and downstream of the ycf4 gene in the plastomes. The psaI gene duplication might have occurred in their cp genomes and was inserted into the IR region. The psaI was copied into another IR region due to the copy correction mechanism. In addition, the pseudogenization of the psaI gene in the LSC region was identified. A double-strand break might have occurred between the ndhK and psaI region (which contains ndhK, trnV-UAC, trnM-CAU, atpE, atpB, rbcL, accD, and psaI). This leads to the excision and inversion of this fragment inserted between the LSC regions in atpH and atpI. During this process, accD gene loss may have occurred in these three species, but this hypothesis could not be concluded for the remaining Corydalis plastomes, which might have played a role in the transposon activity. On the other hand, there is no direct evidence of transposable elements with the Corydalis cp genome, even though they may have been present transiently.
The SSRs a significant role during genome rearrangements and the recombination process (Ogihara et al., 1988; Milligan et al., 1989; Cole et al., 2018). Therefore, this study analyzed the presence of SSRs in the Corydalis plastomes. The distribution of the SSRs in the plastomes of Corydalis was quite different from 19 to 51. In addition, the Corydalis plastomes distributed many repeats in their genome ranging from 93 to 161 (Figure 4; Supplementary Table S4). Moreover, the presence of repeat sequences does not correlate with their genome rearrangements and relocation events in Corydalis. The C. edulis has 95 repeat sequences and does not encode major rearrangement events in its genome (Figures 4, 6, 9; Supplementary Table S4). On the other hand, the significant events (inversion, relocation, gene loss, and IR expansion) occurred in the C. maculata, C. turtschaninovii, and C. shensiana plastomes that encoded similar numbers of repeats (~95) regions in their genomes (Figures 4, 9; Supplementary Table S4). Generally, the RNA editing process arises in the mitochondrial genomes but is less common in the plastomes (Chen et al., 2011; Raman and Park, 2015; Raman et al., 2016). In addition, the seed plant has ~30–40 RNA editing sites in its plastomes (Stern et al., 2010). Nevertheless, all the Corydalis have similar numbers (~51) of RNA editing sites in their genomes (Figures 5A–D). This process mainly occurred in the second position, followed by the first position of the triplet codon (Figure 5D). In addition, ~45% of the amino acids were converted to leucine (Figure 5C). Previous studies also reported that C to U RNA editing in the second codon position occurred mainly in plant organelles to enhance the hydrophobic amino acid leucine frequency. Chen et al. (2011) also reported that the closely associated taxa usually contribute to more RNA editing sites due to the evolutionary process but not in this study.
The mVISTA and nucleotide diversity analysis results showed a high degree of variation in both coding and non-coding regions in the Corydalis plastomes (Figures 6, 7). The KA/KS rate is associated with gene adaptive evolution, such as the positive and purification selection effects (Raman et al., 2020; Raman and Park, 2020). The genes under positive selection might result from natural selection and adaptation to the living environment (Raven et al., 2013; Raman et al., 2020; Raman and Park, 2020; Scobeyeva et al., 2021). Therefore, the substitution rates of all the independent protein-coding genes of 21 Corydalis species are averaged. The results showed that the photosynthetic, transcription and transcription-related genes show accelerated non-synonymous rates (Figures 8A, B). Furthermore, the ratio of KA/KS (ω) showed that the majority of the protein-coding genes were less than 1, excluding rps16 and rps18 genes (Figure 8C). A separate analysis of synonymous and non-synonymous substitution rates was also conducted for all protein-coding genes. Similarly, the substitution analysis of 59 protein-coding genes of all Ranunculales taxa (37 taxa) showed that the KA/KS ratio varies from 6.83, with an average ratio of 0.21 (Supplementary Figure S2; Supplementary Table S7). In contrast, the substitution analysis of all the Ranunculales except Corydalis taxa (16 taxa) revealed that the KA/KS ratio of all these protein-coding genes varies from 0 to 0.89, with an average ratio of 0.13 (Supplementary Figure S3; Supplementary Table S8). This result indicates that all the Ranunculales cp genomes, excluding Corydalis taxa, are highly conserved. Therefore, if the ω value is more than 1.0 of the particular protein-coding genes between two plastomes, or the whole genomes of Corydalis taxa, these genes are considered to be under positive selection. Therefore, in the present study, 24 protein-coding genes were identified in the Corydalis plastomes under positive selection pressure events (Table 2; Supplementary Table S9). In the selective pressure events, six forms of photosynthesis, transcription and translation-related genes were characterized: (i) subunits of ATP synthase (atpB, atpE, and atpF); (ii) C-type cytochrome synthesis gene (ccsA); (iii) maturase (matK); (iv) subunits of photosystem II (psbH, psbJ, psbK, and psbT); (v) large subunits of the ribosome (rpl16, rpl20, rpl22, rpl23, and rpl33); (vi) small subunit of the ribosome (rps2, rps3, rps4, rps7, rps8, rps11, rps14, rps15, rps16, and rps18). Among these, fourteen genes (ccsA, psbJ, psbK, rpl20, rpl22, rpl23, rps2, rps3, rps4, rps8, rps11, rps14, rps16, and rps18) have positively selected sites, providing evidence of the adaptive evolution of proteins (Supplementary Table S9). Genes with various functions, such as genetic and photosynthetic systems, might play a crucial role in the adaptation to the terrestrial ecological environment (Xu et al., 2015; Xu et al., 2020) because most of the Corydalis species live at QTP high altitudes and various North, Central, and East Asia terrestrial regions (Supplementary Table S11) and must adapt to high rates of UV radiation, oxygen depletion conditions, temperature fluctuations, and drought stress conditions. Such genes can be a significant genetic foundation for evolutionary adaptation at the chloroplast level (Xu et al., 2020).
The cp genomes are significant genomic resources for reconstructing precise and high-resolution phylogenetic relationships and taxonomic positions in angiosperms (Jansen et al., 2005). In addition to the whole cp genomes, protein-coding genes have been used widely to determine the phylogenetic relationships at every taxonomic level (Li et al., 2017). The phylogenomic analysis showed two distinct clades, such as Papaveraceae and the rest of the Ranunuculales. These results are consistent with the previous results. All the Corydalis lineages are highly supported with a >97% bootstrap value in the phylogenetic tree, and C. adunca is an early divergence species for the remaining Corydalis species (Figure 9). No molecular age studies for Corydalis species have been reported. Therefore, the divergent times for the genus Corydalis were analyzed. The Corydalis is estimated to have originated at 98.6 mya (95% HPD: 154.44–56.86 mya) in the early upper Cretaceous period and diverged. It took approximately 16 mya to form the rest of the Corydalis species (Figure 10). C. platycarpa, C. edulis, C. fangshanensis, C. saxicola, C. hsiaowutaishanensis, C. ternata , C. turschaninovii, C. filistipes, C. maculata, C. namdoensis, and C. shensiana, distributed in east Asia evolved from 82.86 to 1.51 mya. The remaining eight species are C. davidii, C. pauciovulata, C. lupoinoides, C. trisecta, C. inopinata, C. impatiens, and C. mucronifera and C. conspersa, mainly distributed in the QTP regions. The uplift of the QTP from the period of 25 to 17 mya (Li and Fang, 1999; Wang et al., 2012) changed the environment of East Asia dramatically. The molecular age results of all the eight QTP region Corydalis species (44.31 mya [95% HPD: 67.99–26.03 mya] – 15.71 mya [95% HPD: 29.67–6.93 mya]) correlated very well with the uplift of the Qinghai–Tibet Plateau period. This may have caused the radiation of Corydalis species during this period. Nevertheless, more taxa will be needed to understand the genome architecture, evolution, and divergence of the Corydalis species.
Conclusion
The complete chloroplast genome sequence of Corydalis platycarpa species was determined using a de novo assembly approach. This is the first comprehensive systematic analysis comparing the plastome rearrangement features and adaptive evolution and inferring phylogenetic and molecular clock relationships using the plastome data of Corydalis and its relatives in detail. The comparative analysis showed that Fumarioideae species exhibited high rearrangements, translocation, inversion, duplication, and loss of several protein-coding genes in their genomes. The remaining cp genomes (Papaveroideae, Ranunculaceae, Berberidaceae, Menispermaceae, and Cicaeasteraceae) in the Ranunculales are highly conserved. The accD and ndh gene loss likely provides a prominent synapomorphic characteristic of the genus Corydalis. Phylogenetic and molecular clock studies offer new insights into the systematic relationships between Corydalis and will serve as a basis for future research on the phylogenetic, evolution, and biogeography relationships of Corydalis species.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number OP142703.
Author contributions
GR, SP, and G-HN conceived the project. G-HN provided plant sources. GR designed the experiments. SP and G-HN supervised the project. GR performed the experiments, and analyzed the data, interpreted the results, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Institute of Biological Resources of Korea (NBR201731201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1043740/full#supplementary-material
Supplementary Table 1 | List of taxa and GenBank accession numbers used in the phylogenetic and molecular clock analyses.
Supplementary Table 2 | List of genes present in the chloroplast genome of Corydalis platycarpa.
Supplementary Table 3 | Summary of the total genome size, GC content, LSC, SSC, and IR regions length, and gene content of 21 Corydalis cp genomes.
Supplementary Table 4 | Distribution of distinct types of repeats in the 21 Corydalis cp genomes.
Supplementary Table 5 | Presence of RNA editing sites, codon position, and amino acid conversion in the protein-coding genes of the 21 Corydalis plastomes.
Supplementary Table 6 | Amount of synonymous and non-synonymous substitution rates present in the 59 protein-coding genes of the 21 Corydalis plastomes.
Supplementary Table 7 | Amount of synonymous and non-synonymous substitution rates present in the 59 protein-coding genes of all the Ranunculales plastomes (37 plastomes).
Supplementary Table 8 | Amount of synonymous and non-synonymous substitution rates present in the 59 protein-coding genes of all the Ranunculales (16 taxa) except the genus Corydalis plastomes.
Supplementary Table 9 | Comparison of site models, positive selective amino acid loci, and estimation of parameters for 24 protein-coding genes in the Corydalis species.
Supplementary Table 10 | List of pseudogenes and lost genes in the 21 Corydalis plastomes.
Supplementary Table 11 | List of Corydalis plants distribution areas.
Supplementary Figure 1 | MAUVE alignment of Ranunculales plastomes using Geneious Prime. Local collinear blocks are represented by blocks of the same color and linked within each of the alignments.
Supplementary Figure 2 | Selective pressure analysis for 59 protein-coding genes of all the Ranunculales plastomes (37 taxa). (A) KS: rate of synonymous substitution; (B) KA: rate of non-synonymous substitution; (C) KA/KS: rate of non-synonymous vs. synonymous substitution.
Supplementary Figure 3 | Selective pressure analysis for 59 protein-coding genes of all the Ranunculales plastomes (16 taxa) except Corydalis taxa. (A) KS: rate of synonymous substitution; (B) KA: rate of non-synonymous substitution; (C) KA/KS: rate of non-synonymous vs. synonymous substitution.
Supplementary Figure 4 | Comparison of clpP gene in the Corydalis platycarpa, C. fangshanensis, C. saxicola with C. adunca plastome.
Supplementary Figure 5 | Comparison of psaI gene in the LSC and IR region of Corydalis platycarpa plastome with LSC copy of C. tomentela psaI.
References
Andrews, S. (2010). FASTQC. A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Asaf, S., Khan, A. L., Lubna, K. A., Khan, A., Khan, G., Lee, I. J., et al. (2020). Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci. Rep. 10, 3881. doi: 10.1038/s41598-020-60803-y.
Blazier, J. C., Jansen, R. K., Mower, J. P., Govindu, M., Zhang, J., Weng, M. L., et al. (2016). Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 117, 1209–1220. doi: 10.1093/aob/mcw065
Bock, R. (2007). “Structure, function, and inheritance of plastid genomes,” in Cell and molecular biology of plastids. Ed. Bock, R. (Berlin, Heidelberg: Springer Berlin Heidelberg), 29–63.
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burke, S. V., Lin, C. S., Wysocki, W. P., Clark, L. G., Duvall, M. R. (2016). Phylogenomics and plastome evolution of tropical forest grasses (Leptaspis, streptochaeta: Poaceae). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01993
Cai, Z. Q., Guisinger, M., Kim, H. G., Ruck, E., Blazier, J. C., Mcmurtry, V., et al. (2008). Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67, 696–704. doi: 10.1007/s00239-008-9180-7
Casano, L. M., Zapata, J. M., Martin, M., Sabater, B. (2000). Chlororespiration and poising of cyclic electron transport - plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J. Biol. Chem. 275, 942–948. doi: 10.1074/jbc.275.2.942
Chalamcharla, V. R., Curcio, M. J., Belfort, M. (2010). Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 24, 827–836. doi: 10.1101/gad.1905010
Chen, H., Deng, L., Jiang, Y., Lu, P., Yu, J. (2011). RNA Editing sites exist in protein-coding genes in the chloroplast genome of Cycas taitungensis. J. Integr. Plant Biol. 53, 961–970. doi: 10.1111/j.1744-7909.2011.01082.x
Choi, I. S., Jansen, R., Ruhlman, T. (2019). Lost and found: Return of the inverted repeat in the legume clade defined by its absence. Genome Biol. Evol. 11, 1321–1333. doi: 10.1093/gbe/evz076
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190. doi: 10.1093/molbev/msl089
Claude, S.-J., Park, S., Park, S. (2022). Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae). BMC Plant Biol. 22, 135. doi: 10.1186/s12870-022-03515-x
Cole, L. W., Guo, W., Mower, J. P., Palmer, J. D. (2018). High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Mol. Biol. Evol. 35, 2773–2785. doi: 10.1093/molbev/msy176
Cosner, M. E., Raubeson, L. A., Jansen, R. K. (2004). Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evolutionary Biol. 4, 27. doi: 10.1186/1471-2148-4-27
Del Campo, E. M., Sabater, B., Martin, M. (2000). Transcripts of the ndhH-D operon of barley plastids: possible role of unedited site III in splicing of the ndhA intron. Nucleic Acids Res. 28, 1092–1098. doi: 10.1093/nar/28.5.1092
Doyle, J. J., Davis, J. I., Soreng, R. J., Garvin, D., Anderson, M. J. (1992). Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. U.S.A. 89, 7722–7726. doi: 10.1073/pnas.89.16.7722
Doyle, J. J., Doyle, J. L., Ballenger, J. A., Palmer, J. D. (1996). The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet Evol. 5, 429–438. doi: 10.1006/mpev.1996.0038
Drummond, A. J., Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Dugas, D. V., Hernandez, D., Koenen, E. J., Schwarz, E., Straub, S., Hughes, C. E., et al. (2015). Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep. 5, 16958. doi: 10.1038/srep16958
Elborough, K. M., Winz, R., Deka, R. K., Markham, J. E., White, A. J., Rawsthorne, S., et al. (1996). Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem. J. 315 (Pt 1), 103–112. doi: 10.1042/bj3150103
Erixon, P., Oxelman, B. (2008). Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One 3, e1386. doi: 10.1371/journal.pone.0001386
Frailey, D. C., Chaluvadi, S. R., Vaughn, J. N., Coatney, C. G., Bennetzen, J. L. (2018). Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol. 18, 30. doi: 10.1186/s12870-018-1249-x
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Gao, F. L., Chen, C. J., Arab, D. A., Du, Z. G., He, Y. H., Ho, S. Y. W. (2019). EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Guisinger, M. M., Kuehl, J. V., Boore, J. L., Jansen, R. K. (2011). Extreme Reconfiguration of Plastid Genomes in the Angiosperm Family Geraniaceae: Rearrangements, Repeats, and Codon Usage (vol 28, pg 583, 2011). Mol. Biol. Evol. 28, 1543–1543. doi: 10.1093/molbev/msq229
Hong, C. P., Park, J., Lee, Y., Lee, M., Park, S. G., Uhm, Y., et al. (2017). accD nuclear transfer of Platycodon grandiflorum and the plastid of early Campanulaceae. BMC Genomics 18, 607. doi: 10.1186/s12864-017-4014-x
Jansen, R. K., Palmer, J. D. (1987). A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci. U.S.A. 84, 5818–5822. doi: 10.1073/pnas.84.16.5818
Jansen, R. K., Raubeson, L. A., Boore, J. L., Depamphilis, C. W., Chumley, T. W., Haberle, R. C., et al. (2005). Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 395, 348–384. doi: 10.1016/S0076-6879(05)95020-9
Jin, D. M., Wicke, S., Gan, L., Yang, J. B., Jin, J. J., Yi, T. S. (2020a). The loss of the inverted repeat in the putranjivoid clade of Malpighiales. Front. Plant Sci. 11, 942. doi: 10.3389/fpls.2020.00942
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., Depamphilis, C. W., Yi, T. S., et al. (2020b). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kim, K. J., Choi, K. S., Jansen, R. K. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792. doi: 10.1093/molbev/msi174
Knox, E. B. (2014). The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. United States America 111, 11097–11102. doi: 10.1073/pnas.140336311
Knox, E., Downie, S., Palmer, J. (1993). Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol. Biol. Evol. 10, 414–414. doi: 10.1093/oxfordjournals.molbev.a040017
Knox, E. B., Li, C. J. (2017). The East Asian origin of the giant lobelias. Am. J. Bot. 104, 924–938. doi: 10.3732/ajb.1700025
Kode, V., Mudd, E. A., Iamtham, S., Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Kolodner, R., Tewari, K. K. (1979). Inverted repeats in chloroplast DNA from higher plants. Proc. Natl. Acad. Sci. U.S.A. 76, 41–45. doi: 10.1073/pnas.76.1.41
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Kwon, W., Kim, Y., Park, C. H., Park, J. (2019). The complete chloroplast genome sequence of traditional medical herb, Plantago depressa willd. (Plantaginaceae). Mitochondrial DNA Part B-Resources 4, 437–438. doi: 10.1080/23802359.2018.1553530
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lavin, M., Doyle, J. J., Palmer, J. D. (1990). Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 44, 390–402. doi: 10.1111/j.1558-5646.1990.tb05207.x
Lee, J., Cho, C. H., Park, S. I., Choi, J. W., Song, H. S., West, J. A., et al. (2016). Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 14, 75. doi: 10.1186/s12915-016-0299-5
Lee, H. L., Jansen, R. K., Chumley, T. W., Kim, K. J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Li, J., Fang, X. (1999). Uplift of the Tibetan plateau and environmental changes. Chin. Sci. Bull. 44, 2117–2124. doi: 10.1007/BF03182692
Lin, C. S., Chen, J. J., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5, 9040. doi: 10.1038/srep09040
Liu, Q., Li, X., Li, M., Xu, W., Schwarzacher, T., Heslop-Harrison, J. S. (2020). Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 20, 406. doi: 10.1186/s12870-020-02621-y
Li, Y., Zhou, J. G., Chen, X. L., Cui, Y. X., Xu, Z. C., Li, Y. H., et al. (2017). Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two Hemiparasitic Taxillus species. Sci. Rep. 7, 12834. doi: 10.1038/s41598-017-13401-4
Lohse, M., Drechsel, O., Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52, 267–274.
Luo, D. S., Feng, C. H., Xia, G. C. (1984). The resources of the Tibetan drugs in Qinghai-Xizang plateau — preliminary studies on the plants of Corydalis. Zhong Cao Yao 15, 33–36.
Magee, A. M., Aspinall, S., Rice, D. W., Cusack, B. P., Semon, M., Perry, A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710. doi: 10.1101/gr.111955.110
Maier, R. M., Neckermann, K., Igloi, G. L., Kossel, H. (1995). Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. doi: 10.1006/jmbi.1995.0460
Martin, G. E., Rousseau-Gueutin, M., Cordonnier, S., Lima, O., Michon-Coudouel, S., Naquin, D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210. doi: 10.1093/aob/mcu050
Martin, M., Sabater, B. (2010). Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645. doi: 10.1016/j.plaphy.2010.04.009
Ma, J., Yang, B., Zhu, W., Sun, L., Tian, J., Wang, X. (2013). The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 528, 120–131. doi: 10.1016/j.gene.2013.07.037
Michelangeli, F. A., Davis, J. I., Stevenson, D. W. (2003). Phylogenetic relationships among Poaceae and related families as inferred from morphology, inversions in the plastid genome, and sequence data from the mitochondrial and plastid genomes. Am. J. Bot. 90, 93–106. doi: 10.3732/ajb.90.1.93
Milligan, B. G., Hampton, J. N., Palmer, J. D. (1989). Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol. Biol. Evol. 6, 355–368. doi: 10.1093/oxfordjournals.molbev.a040558
Mock, T., Otillar, R. P., Strauss, J., Mcmullan, M., Paajanen, P., Schmutz, J., et al. (2017). Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540. doi: 10.1038/nature20803
Mohanta, T. K., Bae, H. (2017). Analyses of genomic tRNA reveal presence of novel tRNAs in Oryza sativa. Front. Genet. 8, 90. doi: 10.3389/fgene.2017.00090
Mohanta, T. K., Khan, A. L., Hashem, A., Abd Allah, E. F., Yadav, D., Al-Harrasi, A. (2019). Genomic and evolutionary aspects of chloroplast tRNA in monocot plants. BMC Plant Biol. 19, 39. doi: 10.1186/s12870-018-1625-6
Mohanta, T. K., Mishra, A. K., Khan, A., Hashem, A., Abd Allah, E. F., Al-Harrasi, A. (2020a). Gene loss and evolution of the plastome. Genes 11, 1133. doi: 10.21203/rs.2.16576/v2
Mohanta, T. K., Yadav, D., Khan, A., Hashem, A., Abd Allah, E. F., Al-Harrasi, A. (2020b). Analysis of genomic tRNA revealed presence of novel genomic features in cyanobacterial tRNA. Saudi J. Biol. Sci. 27, 124–133. doi: 10.1016/j.sjbs.2019.06.004
Mower, J. P. (2009). The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 37, W253–W259. doi: 10.1093/nar/gkp337
Mower, J. P., Vickrey, T. L. (2018). Structural diversity among plastid genomes of land plants. Plastid Genome Evol. 85, 263–292. doi: 10.1016/bs.abr.2017.11.013
Niu, Y., Chen, G., Peng, D. L., Song, B., Yang, Y., Li, Z. M., et al. (2014). Grey leaves in an alpine plant: a cryptic colouration to avoid attack? New Phytol. 203, 953–963. doi: 10.1111/nph.12834
Niu, Y., Chen, Z., Stevens, M., Sun, H. (2017). Divergence in cryptic leaf colour provides local camouflage in an alpine plant. Proc. Biol. Sci. 284, 20171654. doi: 10.1098/rspb.2017.1654
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Ogihara, Y., Terachi, T., Sasakuma, T. (1988). Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. U.S.A. 85, 8573–8577. doi: 10.1073/pnas.85.22.8573
Palmer, J. D. (1983). Chloroplast DNA exists in two orientations. Nature 301, 92–93. doi: 10.1038/301092a0
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Palmer, J. D., Nugent, J. M., Herbon, L. A. (1987). Unusual structure of Geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. U. S. A. 84, 769–773. doi: 10.1073/pnas.84.3.769
Palmer, J. D., Thompson, W. F. (1981). Rearrangements in the chloroplast genomes of mung bean and pea. Proc. Natl. Acad. Sci. U.S.A. 78, 5533–5537. doi: 10.1073/pnas.78.9.5533
Palmer, J. D., Thompson, W. F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550. doi: 10.1016/0092-8674(82)90170-2
Park, S., An, B., Park, S. (2018). Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 8, 13568. doi: 10.1038/s41598-018-31938-w
Park, S., Ruhlman, T. A., Weng, M.-L., Hajrah, N. H., Sabir, J. S. M., Jansen, R. K. (2017). Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of Geranium. Genome Biol. Evol. 9, 1766–1780. doi: 10.1093/gbe/evx124
Petersen, K., Schöttler, M. A., Karcher, D., Thiele, W., Bock, R. (2011). Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucleic Acids Res. 39, 5181–5192. doi: 10.1093/nar/gkr105
Raman, G., Choi, K. S., Park, S. (2016). Phylogenetic relationships of the fern Cyrtomium falcatum (Dryopteridaceae) from dokdo island based on chloroplast genome sequencing. Genes 7, 115. doi: 10.3390/genes7120115
Raman, G., Lee, E. M., Park, S. (2021). Intracellular DNA transfer events restricted to the genus Convallaria within the asparagaceae family: Possible mechanisms and potential as genetic markers for biographical studies. Genomics 113, 2906–2918. doi: 10.1016/j.ygeno.2021.06.033
Raman, G., Park, S. (2015). Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS One 10, e0141329. doi: 10.1371/journal.pone.0141329.
Raman, G., Park, S. (2020). The complete chloroplast genome sequence of the Speirantha gardenii: Comparative and adaptive evolutionary analysis. Agronomy. 10, 1405. doi: 10.3390/agronomy10091405
Raman, G., Park, S. (2022). Structural characterization and comparative analyses of the chloroplast genome of Eastern Asian species Cardamine occulta (Asian C. flexuosa with.) and other cardamine species. Front. Biosci. (Landmark Ed) 27, 124. doi: 10.31083/j.fbl2704124
Raman, G., Park, K. T., Kim, J.-H., Park, S. (2020). Characteristics of the completed chloroplast genome sequence of Xanthium spinosum: comparative analyses, identification of mutational hotspots and phylogenetic implications. BMC Genomics 21, 855. doi: 10.1186/s12864-020-07219-0
Raman, G., Park, S., Lee, E. M., Park, S. (2019). Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the asparagales. Sci. Rep. 9 5028. doi: 10.1038/s41598-019-41377-w
Raven, J. A., Beardall, J., Larkum, A. W., Sanchez-Baracaldo, P. (2013). Interactions of photosynthesis with genome size and function. Philos. Trans. R Soc. Lond B Biol. Sci. 368, 20120264. doi: 10.1098/rstb.2012.0264
Ren, F., Wang, L., Li, Y., Zhuo, W., Xu, Z., Guo, H., et al. (2021). Highly variable chloroplast genome from two endangered Papaveraceae lithophytes Corydalis tomentella and Corydalis saxicola. Ecol. Evol. 11, 4158–4171. doi: 10.1002/ece3.7312
Roschenbleck, J., Wicke, S., Weinl, S., Kudla, J., Muller, K. F. (2017). Genus-wide screening reveals four distinct types of structural plastid genome organization in Pelargonium (Geraniaceae). Genome Biol. Evol. 9, 64–76. doi: 10.1093/gbe/evw271
Rousseau-Gueutin, M., Huang, X., Higginson, E., Ayliffe, M., Day, A., Timmis, J. N. (2013). Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161, 1918–1929. doi: 10.1104/pp.113.214528
Ruhlman, T. A., Zhang, J., Blazier, J. C., Sabir, J. S. M., Jansen, R. K. (2017). Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 104, 559–572. doi: 10.3732/ajb.1600453
Sablok, G., Amiryousefi, A., He, X., Hyvonen, J., Poczai, P. (2019). Sequencing the plastid genome of giant ragweed (Ambrosia trifida, Asteraceae) from a herbarium specimen. Front. Plant Sci. 10, 218. doi: 10.3389/fpls.2019.00218
Sanderson, M. J., Copetti, D., Burquez, A., Bustamante, E., Charboneau, J. L. M., Eguiarte, L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102, 1115–1127. doi: 10.3732/ajb.1500184
Sasaki, Y., Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Sazanov, L. A., Burrows, P. A., Nixon, P. J. (1998). The plastid ndh genes code for an NADH-specific dehydrogenase: Isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 1319–1324. doi: 10.1073/pnas.95.3.1319
Schwarz, E. N., Ruhlman, T. A., Sabir, J. S. M., Hajrah, N. H., Alharbi, N. S., Al-Malki, A. L., et al. (2015). Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Systematics Evol. 53, 458–468. doi: 10.1111/jse.12179
Scobeyeva, V. A., Artyushin, I. V., Krinitsina, A. A., Nikitin, P. A., Antipin, M. I., Kuptsov, S. V., et al. (2021). Gene loss, pseudogenization in plastomes of genus Allium (Amaryllidaceae), and putative selection for adaptation to environmental conditions. Front. Genet. 12, 674783. doi: 10.3389/fgene.2021.674783
Serrot, P. H., Sabater, B., Martín, M. (2008). Expression of the ndhCKJ operon of barley and editing at the 13th base of the mRNA of the ndhC gene. Biol. Plantarum 52, 347–350. doi: 10.1007/s10535-008-0071-y
Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273. doi: 10.1093/pcp/pce031
Stamatakis, A., Hoover, P., Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stern, D. B., Goldschmidt-Clermont, M., Hanson, M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. doi: 10.1146/annurev-arplant-042809-112242
Sun, Y., Deng, T., Zhang, A., Moore, M. J., Landis, J. B., Lin, N., et al. (2020). Genome sequencing of the endangered Kingdonia uniflora (Circaeasteraceae, Ranunculales) reveals potential mechanisms of evolutionary specialization. iScience 23, 101124. doi: 10.1016/j.isci.2020.101124
Sun, Y. X., Moore, M. J., Lin, N., Adelalu, K. F., Meng, A. P., Jian, S. G., et al. (2017). Complete plastome sequencing of both living species of circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genomics 18, 592. doi: 10.1186/s12864-017-3956-3
Thiel, T., Michalek, W., Varshney, R., Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Uribe-Convers, S., Carlsen, M. M., Lagomarsino, L. P., Muchhala, N. (2017). Phylogenetic relationships of burmeistera (Campanulaceae: Lobelioideae): Combining whole plastome with targeted loci data in a recent radiation. Mol. Phylogenet. Evol. 107, 551–563. doi: 10.1016/j.ympev.2016.12.011
Wang, W. C., Chen, S. Y., Zhang, X. Z. (2016b). Chloroplast genome evolution in Actinidiaceae: clpP loss, heterogenous divergence and phylogenomic practice. PLoS One 11, e0162324. doi: 10.1371/journal.pone.0162324
Wang, W., Lin, L., Xiang, X. G., Ortiz, R. D., Liu, Y., Xiang, K. L., et al. (2016a). The rise of angiosperm-dominated herbaceous floras: Insights from Ranunculaceae. Sci. Rep. 6, 27259. doi: 10.1038/srep27259.
Wang, Y. H., Qu, X. J., Chen, S. Y., Li, D. Z., Yi, T. S. (2017). Plastomes of mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet. Genomes 13, 41. doi: 10.1007/s11295-017-1124-1.
Wang, Y., Zheng, J., Zhang, W., Li, S., Liu, X., Yang, X., et al. (2012). Cenozoic uplift of the Tibetan Plateau: Evidence from the tectonic–sedimentary evolution of the western Qaidam basin. Geosci. Front. 3, 175–187.
Weng, M. L., Blazier, J. C., Govindu, M., Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Weng, M. L., Ruhlman, T. A., Jansen, R. K. (2017). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214, 842–851. doi: 10.1111/nph.14375
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Muller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wyman, S. K., Jansen, R. K., Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Xu, Z., Jiang, Y., Zhou, G. (2015). Response and adaptation of photosynthesis, respiration, and antioxidant systems to elevated CO2 with environmental stress in plants. Front. Plant Sci. 6, 701. doi: 10.3389/fpls.2015.00701
Xu, X. D., Wang, D. (2021). Comparative chloroplast genomics of Corydalis species (Papaveraceae): Evolutionary perspectives on their unusual Large scale rearrangements. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.600354
Xu, S., Wang, J., Guo, Z., He, Z., Shi, S. (2020). Genomic convergence in the adaptation to extreme environments. Plant Commun. 1, 100117. doi: 10.1016/j.xplc.2020.100117
Yukawa, M., Tsudzuki, T., Sugiura, M. (2005). The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum). Plant Mol. Biol. Rep. 23, 359–365. doi: 10.1007/BF02788884
Zhang, B., Huang, R., Hua, J., Liang, H., Pan, Y., Dai, L., et al. (2016). Antitumor lignanamides from the aerial parts of Corydalis saxicola. Phytomedicine 23, 1599–1609. doi: 10.1016/j.phymed.2016.09.006
Keywords: Corydalis, Plastome rearrangement, relocation, IR expansion, accD, clpP, ndh, divergent time
Citation: Raman G, Nam G-H and Park S (2022) Extensive reorganization of the chloroplast genome of Corydalis platycarpa: A comparative analysis of their organization and evolution with other Corydalis plastomes. Front. Plant Sci. 13:1043740. doi: 10.3389/fpls.2022.1043740
Received: 14 September 2022; Accepted: 07 November 2022;
Published: 09 December 2022.
Edited by:
Tapan Kumar Mohanta, University of Nizwa, OmanReviewed by:
Gopal Pandi, Madurai Kamaraj University, IndiaKrishnaveni Muthan, Manonmaniam Sundaranar University, India
Ravendran Vasudevan, University of Cambridge, United Kingdom
Copyright © 2022 Raman, Nam and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: SeonJoo Park, sjpark01@ynu.ac.kr; Gi-Heum Nam, namgih@korea.kr
 Gurusamy Raman
Gurusamy Raman Gi-Heum Nam2*
Gi-Heum Nam2* SeonJoo Park
SeonJoo Park