- State Key Laboratory of Subtropical Silviculture, Institute of Bamboo Research, Zhejiang A&F University, Hangzhou, China
Mariner-like elements (MLEs) are promising tools for gene cloning, gene expression, and gene tagging. We have characterized two MLE transposons from moso bamboo, Ppmar1 and Ppmar2. Ppmar2, is smaller in size and has higher natural activities, thus making it a more potential genomic tool compared to Ppmar1. Using a two-component system consisting of a transposase expression cassette and a non-autonomous transposon cotransformed in yeast, we investigated the transposition activity of Ppmar2 and created hyperactive transposases. Five out of 19 amino acid mutations in Ppmar2 outperformed the wild-type in terms of catalytic activities, especially with the S347R mutant having 6.7-fold higher transposition activity. Moreover, 36 yeast mutants with single-gene deletion were chosen to screen the effects of the host factors on Ppmar2NA transposition. Compared to the control strain (his3Δ), the mobility of Ppmar2 was greatly increased in 9 mutants and dramatically decreased in 7 mutants. The transposition ability in the efm1Δ mutant was 15-fold higher than in the control, while it was lowered to 1/66 in the rtt10Δ mutant. Transcriptomic analysis exhibited that EFM1 defection led to the significantly impaired DDR2, HSP70 expression and dramatically boosted JEN1 expression, whereas RTT10 defection resulted in significantly suppressed expression of UTP20, RPA190 and RRP5. Protein methylation, chromatin and RNA transcription may affect the Ppmar2NA transposition efficiency in yeast. Overall, the findings provided evidence for transposition regulation and offered an alternative genomic tool for moso bamboo and other plants.
Introduction
Transposons are found in a wide variety of species and migrate across the genome. There are two types of transposons: DNA transposons and RNA transposons. Mariner-like elements (MLEs) belong to the DNA transposon family. Mariner transposon was first characterized in the Drosophila mauritania mutant with white eyes, and later homologous elements were discovered in abundance in plant and animal genomes (Hartl et al., 1997). MLEs are typically comprised of an Open Reading Frame (ORF) that encodes the transposase (Tpase), which locates between two paired Terminal Inverted Repeats (TIRs) and Target Site Duplications (TSDs) (Plasterk et al., 1999). Transposase has three well-defined domains. The N-terminal domain contains helix-turn-helix (HTH) motifs that recognize and bind the TIRs. The C-terminus contains a catalytic domain with a DDE/D triad (Yuan and Wessler, 2011). The two aspartic acid residues (D) and a glutamic acid (E) or another aspartic acid (D) are commonly located 34 or 39 amino acids apart in animal and plant transposases (DD34E/D, DD39D, respectively) (Hartl et al., 1997; Feschotte and Wessler, 2002). The Linker region, harboring a conserved WVPHEL motif, connects the HTH motif and catalytic domain (Liu and Chalmers, 2014).
The conservation of the transposase sequence has a significant impact on transposition efficiency. The three most well-studied MLEs to date are Sleeping Beauty (SB) from fish (Ivics et al., 1997), Mos1 from Drosophila mauritiana (Bryan et al., 1990), as well as Himar1 from Haematobia irritans (Robertson and Lampe, 1995). In SB, a single amino acid mutation yielded around 2-3-fold transposition efficiency, and 9 amino acid mutations (K14R, K33A, R115H, RKEN214-217DAVQ, M243H, T314N) by DNA shuffling resulted in a hyperactive mutant SB100X with a 100-fold increase in transposition activity (Mátés et al., 2009). Germon et al. (2009) used systematic single amino acid substitutions to create two hyperactive Mos1 mutants (FETY and FET) that were 60- and 800-fold more active than the wild-type Mos1 version. The Linker region is also important for transposition efficiency. Almost all single mutations of the WVPHEL motif in Himar1 and Hsmar1 transposase led to highly active transposase variants, but which easily produce non-productive DNA double-strand breaks that can induce DNA damage and mutations (Butler et al., 2006; Lampe, 2010; Liu and Chalmers, 2014).
Besides the sequences of transposases, DNA methylation, chromatin status, over-production inhibition (OPI) and host proteins may influence the transposition frequency. It was shown that the methylated SB was at least 100-fold more active than the unmethylated version. CpG methylation of the SB region and heterochromatin formation facilitated the transposition reaction (Yusa et al., 2004). The MLE transposase activity was also inhibited by OPI (Lohe and Hartl, 1996). Overproduction of wild-type transposase enhanced the attachment of the transposase dimer and competition for free transposon ends, ultimately lowering the transposase activity. OPI started to occur when the number of transposase dimers was superior to the number of available TIRs (Claeys et al., 2013). High Mobility Group B1 (HMGB1) was a host-encoded cofactor of SB transposition and was involved in the formation of transposase-transposon complexes. Transposition of SB was severely suppressed in the HMGB1-deficient mouse cells (Zayed et al., 2003). In yeast (Saccharomyces cerevisiae), more than 200 host factors have been found to be associated with Ty1 and Ty3 retrotransposon (Irwin et al., 2005; Curcio et al., 2015). These factors were hypothesized of being involved in chromatin and transcript elongation, translation and cytoplasmic RNA processing, vesicular trafficking, nuclear transport, and DNA maintenance.
Two full-length MLEs named Ppmar1 and Ppmar2 were previously identified from the genome of moso bamboo (Phyllostachys edulis) (Zhou et al., 2015). Ppmar1 and Ppmar2 were shown to be transposable in Arabidopsis thaliana and yeast (Zhou et al., 2016; Zhou et al., 2017; Ramakrishnan et al., 2019a; Ramakrishnan et al., 2019b). Site-directed mutation boosted the activity of the Ppmar1 mutant (S171A) by more than 10-fold (Zhou et al., 2017). Ppmar2 is smaller in size (Zhou et al., 2016) and has a higher natural activity (Zhou et al., 2017), thus making it a more potential genomic tool compared to Ppmar1. Moso bamboo has a very lengthy vegetative growth cycle (~60 years), and hence rarely reproduces sexually, but reproduces via rhizomes during the vegetative period (Janzen, 1976; Watanabe et al., 1982). Breeding of moso bamboo via hybridization is extremely difficult due to its occasional sexual reproduction. So, a bamboo mutant library via an efficient transformation system will be a preference for future breeding. Currently, the most used genomic tools are T-DNA insertion and gene targeting (e.g., CRISPR/Cas). Insertional mutagenesis based on active transposons may be a promising tool to manipulate the genome of moso bamboo.
In the present study, we focused on the Ppmar2 transposon to develop hyperactive Ppmar2 mutants by rational mutagenesis. The host factors in yeast which regulated Ppmar2 transposition were also screened. The hyperactive Ppmar2 transposon system reported in this study with its outstanding features including its compact size and non-linked insertion sites could provide an alternative genomic tool for moso bamboo and other plants.
Methods
Construction of the Ppmar2 transposition system in yeast
The Ppmar2 transposition system was constructed using the transposon donor vector, pWL89A, and the transposase expression vector, pAG415gal-ccdB, as described by Ramakrishnan et al. (2019a; 2019b). Ppmar2 non-autonomous transposon (Ppmar2NA) was cloned by amplifying its 5’ and 3’ TIRs as well as the adjacent sequence, followed by an overlap PCR. Ppmar2NA was inserted into the Xho I site at the 5’ untranslated region (UTR) of the Ade2 gene in the vector pWL89A possessing two selectable markers, Ura3 and Ade2. The transposon donor vector was named pWL89A-Ppmar2NA. The Ppmar2 transposase sequences were amplified by adding Not I and EcoR V sites at both ends to fit the pAG415gal-ccdB vector. Both restriction enzymes cut the pAG415gal-ccdB vector. The transposase fragment and the backbone of pAG415gal-ccdB were then ligated by T4 DNA ligase to obtain the recombined vectors pAG415gal-transposase with Leu2 selectable marker. The transposase was promoted to be expressed under the gal promoter.
Yeast transposition assay
The two prepared plasmids (pWL89a-Ppmar2NA and pAG415gal-transposase) were co-transformed into S. cerevisiae strain DG2523. Transformed yeast strains were grown on medium lacking Leucine and uracil (SD-his-ura) but with 2% galactose at 30°C in the dark for 3 days, followed by the suspension of single colonies in 150 μl water and plating onto medium lacking adenine, Leucine, and uracil (SD-ade-his-ura) with 2% galactose as the carbon sources. The plates were incubated at 30°C in the dark for about 3 days to allow the growth of ADE2 revertant colonies.
Transposition footprints analysis
The fragments covered Ppmar2NA excision spots on pWL89a were amplified using primers of yeast T-5 (5’-CAC CCC AGG CTT TAC ACT TTA TG-3’) and T-3 (5’-GTT GCT TAT TTG TTT GGC AGG AG-3’). PCR products were cloned into the pUC18-T vector for sequencing.
Insertion sites and insertion bias analysis
Genomic DNA was extracted from the single ADE2 revertant colonies and then was sheared into 500-bp fragments using Covaris E220 ultrasonicator (Covaris, UK). The libraries were normalized, pooled, and sequenced via Illumina high-throughput sequencing platform NovaSeq 6000 (2×250 bp paired-end run). Low-quality sequences were filtered by Sickle (v1.33) with Q30 and 125bp minimal length (Joshi and Fass, 2011). Filtered reads were aligned to the Ppmar2NA sequence through the local blast. Subsequently, the reads containing the Ppmar2NA sequence and adjacent sequence were aligned to the S. cerevisiae reference genome (https://yeastgenome.org/) to identify the insertion sites. The 20bp upstream and downstream of the insertion sites were aligned to verify the nucleotide distribution characteristics of the Ppmar2NA insertion sites.
Site-directed mutagenesis of Ppmar2 transposase
To identify Ppmar2 non-conserved transposase sites, we downloaded 22 MLE transposase sequences from GenBank and aligned them with the Ppmar2 transposase using MEGA11.0.10 (Tamura et al., 2021). Mutagenesis was performed with the QuikChange Lightning Site-Directed Mutagenesis Kit using the primers listed in Supplementary Table S1. The selected sites of the Ppmar2 transposase were mutated into corresponding amino acids (Supplementary Table S2). The mutated Ppmar2 transposases then replaced the template on pAG415gal-transposase to examine the transposition activities. All plasmids were sequenced to confirm the presence of the targeted mutation. Homologous hyperactive mutation sites in the Mos1 and Himar1 transposase were mutated into the corresponding amino acid in the Ppmar2 transposase.
Transposition frequency of Ppmar2 mutants
The transposition frequencies of Ppmar2 mutants were evaluated by ADE2 revertant frequencies. Each galactose-induced colony was suspended in 50 μl of water and plated on media without adenine. The cell suspension was equally diluted to 1×10-5 volume and was plated on SD media lacking adenine, Leucine and uracil (SD-ade-his-ura) to obtain the total number of galactose-induced colonies. The assay of each mutant was performed with six biological replicates.
Transposition frequency analysis of Ppmar2 in yeast mutants
To examine the host factors’ effects on Ppmar2 transposition in yeast, 36 mutants with single-gene deletions were selected (Table S3) which were kindly provided by Charles Boone (Toronto University, Toronto, in Canada), and His3Δ was used as the control strain. These genes are involved in methylation, DNA replication, transcription, translation, as well as the regulation of retrotransposon (Ty1 and Ty3) (Irwin et al., 2005; Curcio et al., 2015).
Gene expression via RNA-seq
The efm1Δ, rtt10Δ mutants and the wild type yeast His3Δ were cotransformed with pWL89a-Ppmar2NA and pAG415gal-transposase plasmids with three colonies for each genotype. The mRNA of the single ADE2 revertant colonies was extracted, when strains grew in YPD liquid medium with the OD600 of 0.5, and was synthesized into cDNA strands with dNTPs and DNA polymerase I. The final cDNA library was obtained through the AMPure XP system and was sequenced on an Illumina Hiseq 2000 platform (Tianjin Nuohe Zhiyuan Bioinformatics Co., Ltd.) to generate 125 bp/150 bp paired-end reads. The adapter, ploy-N and low-quality reads from raw data were removed using Illumina PIPELINE software. Index of the yeast genome has been built using STAR (v2.5.1b) and paired-end clean reads were aligned to the yeast reference genome (https://yeastgenome.org/) with TopHat v2.0.12. The reads mapped to each gene were counted by HTSeq v0.6.0 and were normalized to fragments per kilobase of exon per million fragments mapped (FPKM) (Trapnell et al., 2010). The method of Benjamini and Hochberg was used to adjust the P-value to control the error detection rate (Haynes, 2013). In this experiment, the correction of p < 0.05, with | log2foldchange | > 1 was used for screening differentially expressed genes (DEGs).
Quantitative real-time PCR analysis
Eight DEGs were selected for qRT-PCR analysis. One μg total RNA of each sample was reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser. The qRT-PCR was performed using the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) on the iQ™5 Multi-channel Real-time PCR Detection System (Bio-Rad). The relative abundance of each gene expression was calculated from the 2-ΔΔCt values between the target gene and ALG9 (Protein amino acid glycosylase 9) (Teste et al., 2009). The results were analyzed by Bio-Rad CFX Manager 3.1 software. Each reaction was performed at least three times.
Results
Verification of the Ppmar2 transposition potential in yeast
To determine whether Ppmar2 can transpose in the yeast genome, we performed transposition assays in yeast utilizing the methods described previously (Ramakrishnan et al., 2019a; Ramakrishnan et al., 2019b). The assays used two constructs, one transposase expression vector and one transposon donor vector. In the transposase expression plasmid, the Ppmar2 transposase coding sequence was fused to the inducible GAL1 promoter, and Leu2 served as the selectable marker. In the transposon donor plasmid, the non-autonomous Ppmar2NA element was inserted in the 5’UTR of the ADE2 reporter gene, and Ura3 served as the selectable marker. Transformants containing both plasmids were selected on a medium with 2% galactose but lacking Leucine and uracil. Colonies of the double transformants were picked and regrown on agar plates without adenine for the selection of ADE2 revertants, which represented the excision of the Ppmar2NA. Notably, ADE2 revertant colonies were obtained in the presence of the Ppmar2 transposase, but none when the control plasmid, pAG415-ccdB, was used.
Plasmid DNA was prepared from the independent ADE2 revertants, and the excision products of Ppmar2NA in the ADE2 5’UTR region were PCR amplified using primers of yeast T-5 and T-3. Sequencing results revealed diverse footprints of excision were generated by Ppmar2NA in the donor plasmid. Between two Ppmar2NA TIRs, one to four bases were retained (Figure 1).
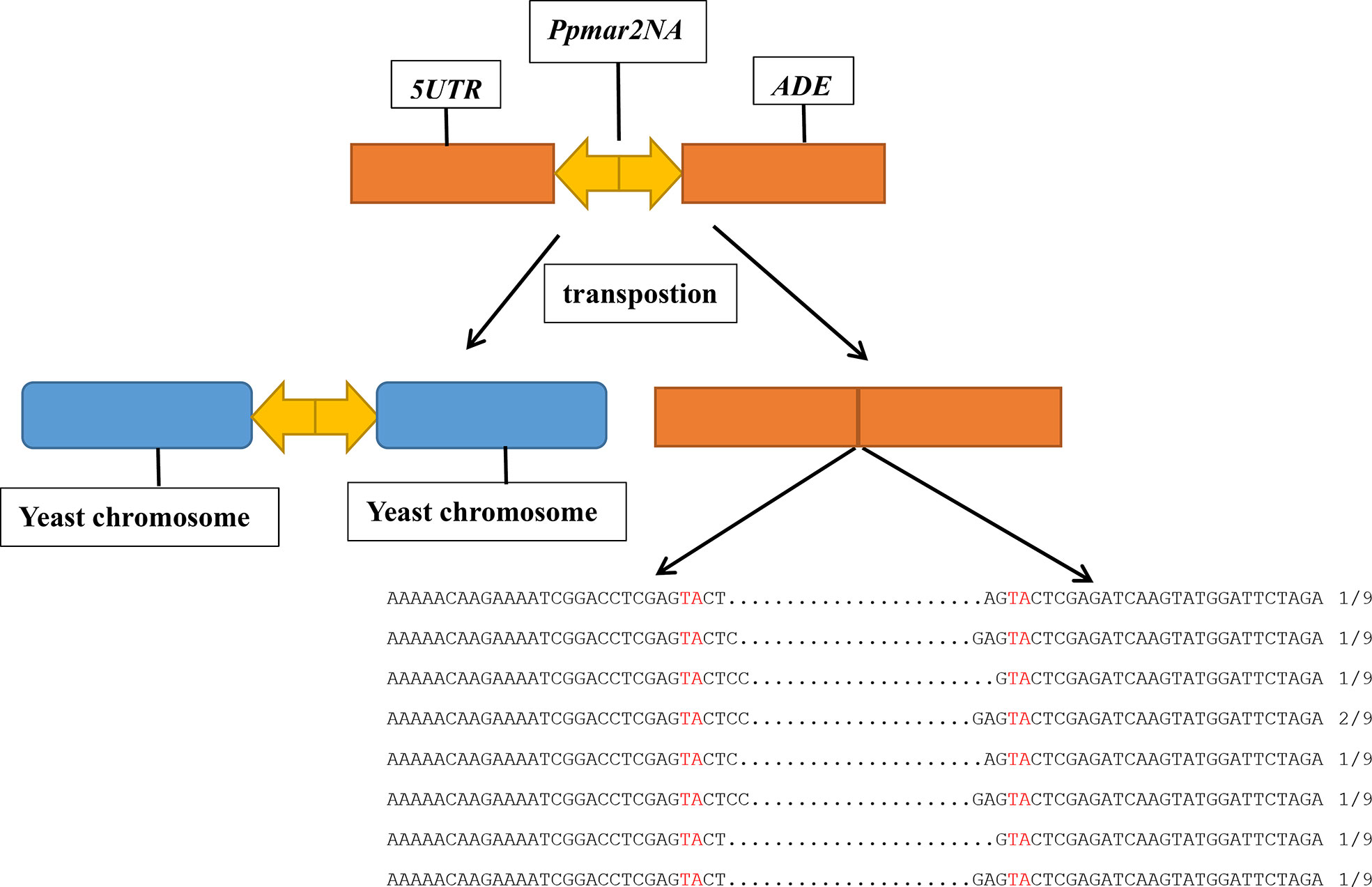
Figure 1 Sequence alignment of Ppmar2NA at excision spots on pWL89a. Sequences of Ppmar2NA at excision spots were amplified using primers yeast T-5 and T-3. The TA-target sites of TSDs are marked in red, and the interval between the red TA represents the footprints after cleavages.
Reinsertion preferences of the excised Ppmar2NA
To follow the fate of the excised Ppmar2NA and localize the reinsertion sites in the yeast genome, genomic DNA was extracted from the independent ADE2 revertants, and sequenced (with 30 times coverage). Following alignment, we identified 7 genetic integration events on 6 yeast chromosomes. All insertions occurred within ~500 bp of the coding regions, with one insertion occurring within the gene (Table 1). As expected, Ppmar2NA in all detected events was inserted into a TA dinucleotide where the AT content of 50 bp sequences nearby the insertion sites was more than 60% (Figure 2). To further validate the Ppmar2NA insertion bias, we analysed the 20 bp sequences flanking the TA on both sides. Alignment of the 7 integration sites revealed that the excised Ppmar2NA was preferentially inserted into AT-rich regions (Figure 2 and Table 1).

Figure 2 Reinsertion preference analysis of excised Ppmar2NA. The pictogram shows the relative nucleotide frequencies of the 7 detected insertion events in the yeast genome. On either side of the reinserted Ppmar2NA elements, 20 bases flank the conserved TA insertion spot. The nucleotide A is shown in green, C in red, G in blue, and T in yellow. The lightning symbol indicates the Ppmar2NA reinsertion site.
Targeted mutagenesis of the Ppmar2 transposase to enhance Ppmar2NA transposition in yeast
To improve the transposition activity of Ppmar2, the transposase was modified following the strategy as described (Zhou et al., 2017). We selected 22 MLE transposase sequences from animals and plants, including Mos1 from D. mauritiana (Bryan et al., 1990), Himar1 from H. irritans (Robertson and Lampe, 1995), Ppmar1 from Moso bamboo, and three MLE transposases from rice (Yang et al., 2006; Yang et al., 2009) (Figure 3). Other sequences were filtered in the NCBI database by blast using the Ppmar2 transposase sequence as a query, which showed high homology with the Ppmar2 transposase. We examined amino acid residues that were partially conserved in MLEs but not in the Ppmar2 transposase after alignment. Thirteen such sites scattered across the three conserved domains of the Ppmar1 transposase (the DNA binding domain, the Linker domain, and the catalytic domain) were identified (Figure 3). Moreover, we selected six homology sites in the Ppmar2 transposase that corresponded to the hyperactive mutation sites in the Mos1 and Himar1 transposase (D129A, D129R, G132A, A168R, L174K and K289A) (Figure 3) (Butler et al., 2006; Germon et al., 2009). The 18 candidate sites, including four in the HTH binding domain, three in the Linker region, and eleven in the catalytic domain, were systematically modified one by one, and the transposase variants were evaluated in the yeast excision assay (Table S2).
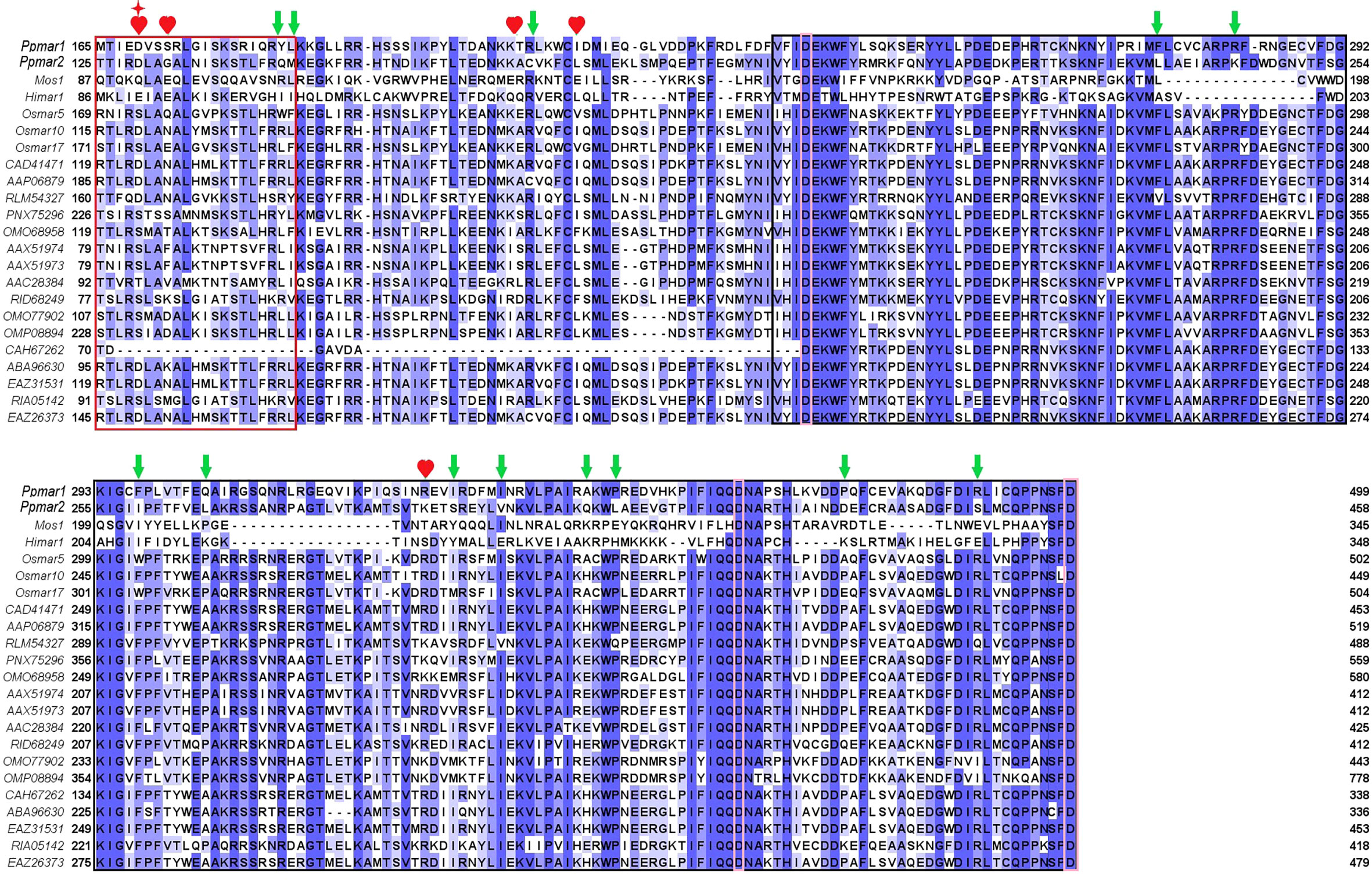
Figure 3 Homology alignment of 22 MLE transposases. The red box is the HTH domain, the black box is the DDD domain, and the Linker area is in the middle. Three aspartic acid residues (D) in the DDD domain were highlighted with pink boxes. Mos1 from D. mauritiana (Bryan et al., 1990), Himar1 from H. irritans (Robertson and Lampe, 1995), Ppmar1 from moso bamboo and three MLE transposases (Osmar5, Osmar10, Osmar17) from rice (Yang et al., 2009). Other entries are named according to the first letter of the genus name and the species name, followed by the GenBank accession number. The 12 non-conserved sites (marked by green arrows) of the Ppmar2 transposase were mutated into corresponding conserved amino acids. The 6 homologous hyperactive mutation sites in the Mos1 and Himar1 transposase were mutated into the corresponding amino acid in the Ppmar2 transposase (marked by a red heart shape). The amino acid at the 129th position was mutated into two different amino acids (marked by a red star).
Among the 19 transposase mutations (two mutations at the 129th amino acid), 5 of them (L235F, K243R, S347R, S292I, and L174K) dramatically enhanced the Ppmar2 excision activity by more than 2-fold (Figure 4). Four amino acid substitutions, including L266P, L309P, D333P, and A168R, suppressed the transposition. The remaining 10 amino acid substitutions had no significant effects (Figure 4). The S347R mutant exhibited the highest transposition activity, which was 6.7-fold that of the wild type (Figure 4).
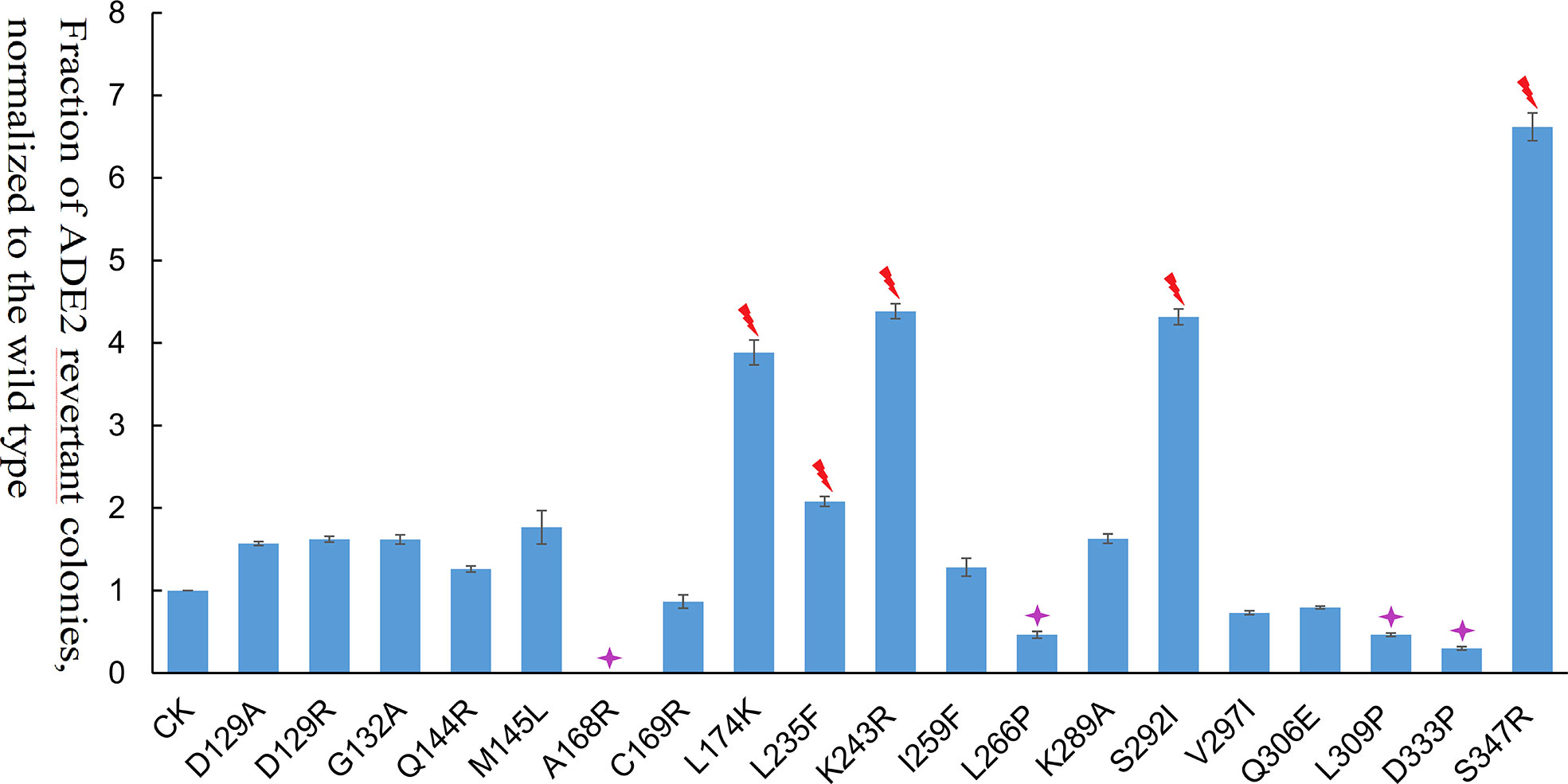
Figure 4 Transposition frequencies catalyzed by Ppmar2 transposase mutants (CK is the wild type Ppmar2 transposase). The mean excision frequency of the 19 Ppmar2 mutants was normalized by that of the wild type. Y-axis represents the fraction of ADE2 revertant colonies, normalized to the wild type. Each excision assay was conducted with six biological replicates. The transposase mutations were marked by red heart shapes which dramatically enhanced Ppmar2 transposase catalytic activity by more than 2-fold. The transposase mutations were marked by pink star shapes which dramatically reduced Ppmar2 transposase catalytic activity to less than 1/2-fold.
Host factors’ effects on Ppmar2 transposition in yeast
To investigate the effects of host factors on transposition efficiency, 36 yeast mutants with single-gene deletions were screened (Table S2). Of the 36 genes, 8 genes were involved in methylation, 7 genes in DNA replication, transcription, and translation, and 21 genes were host factors of retrotransposons (Ty1 and Ty3) (Irwin et al., 2005; Curcio et al., 2015). The 36 yeast mutants were double transformed with pWL89a-Ppmar2NA and pAG415gal-transposase. The positive colony assay in each yeast mutant revealed that the transposition frequency was significantly higher in 9 yeast mutants (tmt1Δ, efm1Δ, efm5Δ, mgt1Δ, pdr3Δ, rtt102Δ, chd1Δ, exg2Δ, and cur1Δ) than in the control strain (His3Δ), but significantly lower in 7 yeast mutants (set2Δ, rtt106Δ, rtt10Δ, mlh2Δ, vac7Δ, mrpl10Δ, and itr2Δ), with no obvious differences in the other 20 yeast mutants (Figure 5). Ppmar2NA showed the most active transposition in the efm1Δ mutant, which was 15-fold higher than in His3Δ. Ppmar2NA had the lowest transposition frequency in the rtt10Δ mutant, which was only 1/66 that in the His3Δ strain.
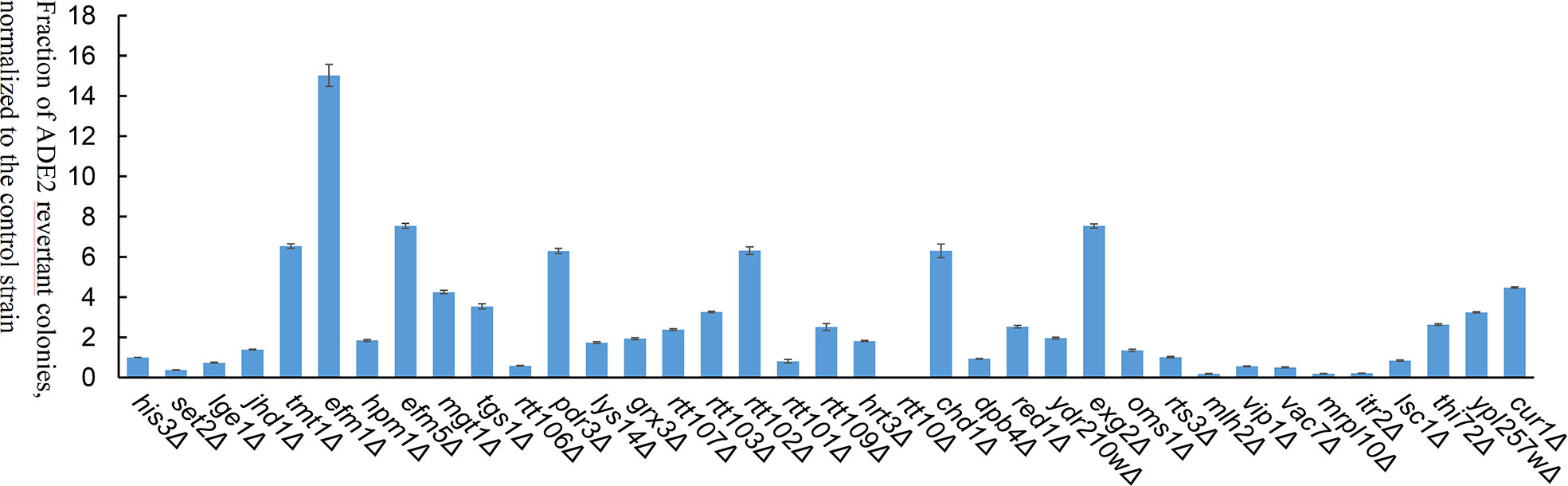
Figure 5 Transposition efficiency of Ppmar2NA in 36 yeast single-gene mutants. The mean excision frequency of Ppmar2NA in the 36 yeast mutants was normalized to that of the control strain (His3Δ). Y-axis represents the fraction of ADE2 revertant colonies, normalized to the control strain. Each excision assay was conducted with six biological replicates.
DEGs in the mutants with dramatically distinct transposition ability Ppmar2NA
To explore the host factors’ effects and investigate the DEGs involved in the transposition regulation, we performed the transcriptome analyses for the strains with distinct transposition competence Ppmar2NA (efm1Δ vs. His3Δ and rtt10Δ vs. His3Δ). Sixty-seven DEGs were validated with a P-value lower than 0.0001 (Figure 6) and enriched via GO and KEGG. These DEGs were related to ribosome biogenesis, RNA modification and DNA modifications which might be involved in the regulation the Ppmar2 transposition (Figures 1S, 2S). In the mutant efm1Δ with highly active Ppmar2NA, three genes linked with DNA repair and RNA transcription (DNA Damage Responsive 2 (DDR2), Heat Shock Protein 70 (HSP70) and Monocarboxylate/proton symporter of the plasma membrane (JEN1)), might be likely related to regulation of the Ppmar2NA transposition. DDR2 and HSP70 were highly expressed in the efm1Δ strain, while the expression of JEN1 was repressed. In the rtt10Δ null mutant, three genes (U3 snoRNA-associated protein 20 (UTP20), DNA-directed RNA polymerase I subunit (RPA190), U3 small nucleolar RNA-associated protein (RRP5)), involved in ribosome assembly and RNA polymerase synthesis, were markedly downregulated.
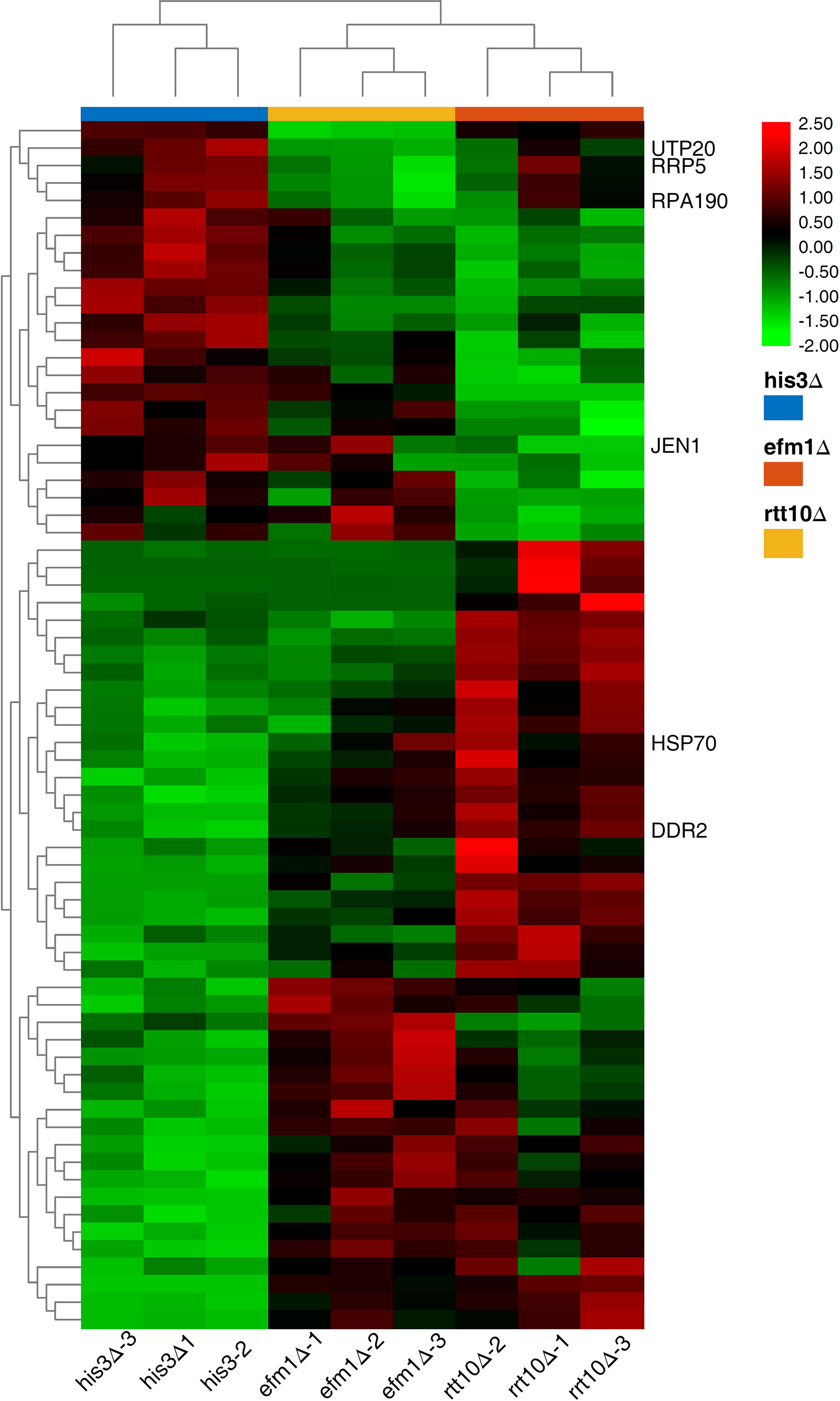
Figure 6 Heat map of DEGs expression in HisΔ3, efm1Δ, rtt10Δ strains. The expression patterns of DDR2, HSP70, JEN1, UTP20, RPA190 and RRP5 genes were marked. The color scale indicates the relative expression level.
qRT-PCR was performed to validate the RNA-seq data by checking the expression levels of Elongation Factor Methyltransferase (EFM1), Regulator of Ty1 Transposition (RTT10), DDR2, HSP70, JEN1, UTP20, RPA190 and RRP5. No expression of EFM1 was detected in efm1Δ, and a low-level expression of RTT10 in rtt10Δ, as predicted. The other gene expression patterns displayed via qRT-PCR were completely consistent with the RNA-seq data-derived gene expression pattern (Supplementary Figure 3).
Discussion
Yeast has been widely used as a heterologous host to evaluate the transposition activity of plant DNA transposons, such as Osmar5 and Stowaways MITE in rice (Yang et al., 2006; Yang et al., 2009), and Ac/Ds in maize (Lazarow et al., 2012). We have identified two MLE transposons in moso bamboo, Ppmar1 and Ppmar2, which both could jump in Arabidopsis thaliana and yeast genome (Zhou et al., 2016; Zhou et al., 2017; Ramakrishnan et al., 2019a; Ramakrishnan et al., 2019b). Ppmar2 has a relatively smaller size and naturally higher activity than Ppmar1, thus making it a more potential genomic tool. To improve the transposition efficiency and develop an alternative gene-tagging tool, we create a hyperactive Ppmar2 via mutagenesis and screen the host factors influencing transposition of Ppmar2 in yeast.
Ppmar2NA’s transposition footprints and insertion preferences resembled the other MLEs
Using a two-component system consisting of a transposase expression cassette and a non-autonomous transposon cotransformed in yeast, we examined the transposition activity of Ppmar2NA. The footprints of Ppmar2NA after excision revealed that Ppmar2NA was preferentially cut between the two TIRs, leaving 1-4 staggered nucleotides (Figure 1). Similarly, the Osmar5 transposase generated a staggered cut with one to four nucleotides at both ends of the transposon element (Yang et al., 2006), whereas the animal transposases Mos1, Sleeping Beauty, and Frog Prince were cleaved but left no more than three nucleotides (Luo et al., 1998; Dawson and Finnegan, 2003; Miskey et al., 2003). On the other hand, Ppmar2NA tended to insert into the TA-rich regions and generated AT TSDs (Figure 2). In short, as a member of the MLEs family transposons, Ppmar2NA shared a similar cleavage footprint and insertion preference.
Some amino acids in key sites are important for the catalytic activity of Ppmar2 transposase
Eighteen non-conversed amino acids in Ppmar2 transposase, including 4 in the HTH binding domain, 3 in the linker junction region, and 11 in the DDD catalytic domain, were selected for mutagenesis to improve the transposition activity (Figure 3). Another 6 homology sites in Ppmar2 corresponding to hyperactive mutation sites in Mos1 and Himar1 were also chosen (D129A, D129R, G132A, A168R, L174K and K289A) (Butler et al., 2006; Germon et al., 2009). The catalytic activities in five of the six mutants were improved, which was consistent with the mutation effect in Mos1 and Himar1 except for A168R (Figure 4) (Butler et al., 2006; Germon et al., 2009). Strangely, the five hyperactive mutation sites are not conserved in the sequences of Ppmar2 transposase (Figure 4).
The linker region connects the HTH motif and catalytic domain, which are not conserved as WVPHEL motif in Ppmar2 transposase. Three mutations (A168R, C169R, and L174K) located in the Linker region of the Ppmar2 transposase, which regulated the spatial structure of the transposase, exhibited divergent transposition actives (Butler et al., 2006; Liu and Chalmers, 2014). The catalytic activity of the C169R mutation did not change significantly, but the A168R mutant totally lost the catalytic activity. The L174K transposase mutant, on the other hand, showed considerably higher catalytic activity than the wild type (Figure 4). The catalytic domain featured by a conserved DDE/D motif played a vital role in transposition activity (Yang et al., 2006). It has been reported that substitution of the DDD to the DDE in the Tc1 family abolished the transposase activity (Lohe et al., 1997). The transposition efficiency of Ppmar2 transposase varied among the targeted mutation sites (L235F, K243R, I259F, L266P, K289A, S292I, V297I, Q306E, L309P, D333P, and S347R). The L235F, K243R, S347R and S292I mutation greatly boosted the transposition. Especially, the S347R mutation resulted in a 6.7-fold higher transposition frequency (Figure 4). The hyperactive Ppmar2 system would be used in plant genomic engineering as an alternative transposon-based technique.
It should be noted that the high catalytic activity of transposase mutations may not directly lead to high-frequency transposition due to OPI. Construction of transposase mutations which are usually less sensitive to OPI, e.g. low TIR binding affinity, and low stability of the transposase dimers will contribute to the high transposition frequency (Liu and Chalmers, 2014; Tellier and Chalmers, 2020). In the following, mutations that change in Ppmar2 transposition kinetics that shift the OPI equilibrium will be considered.
Host factors, including EFM1 and RTT10 genes, significantly affected the transposition frequency of Ppmar2NA in yeast
The yeast single-gene deletion mutant library was effectively used to investigate the impact of host factors on the activity of retrotransposons Ty1 and Ty3. More than 200 host factors, including those involved in chromatin and transcript elongation, translation and cytoplasmic RNA processing, vesicular trafficking, nuclear transport, and DNA maintenance, regulated the transposon activity of yeast Ty1 (Curcio et al., 2015) and Ty3 (Irwin et al., 2005). The 36 related yeast mutants with single gene deletions were selected in this study. The transposition efficiencies of Ppmar2NA in 9 yeast mutants (tmt1Δ, efm1Δ, efm5Δ, mgt1Δ, chd1Δ, pdr3Δ, cur1Δ, exg2Δ, and rtt102Δ) were 2-fold more active than that in the control yeast. Notably, in the efm1Δ mutant, the transposition efficiency increased by 15-fold. Among the 9 genes, four are related to protein methylation, one to DNA methylation, one to chromatin organization, and the remaining 4 were host factors of retrotransposon. In another 5 yeast mutants (set2Δ, rtt10Δ, mlh2Δ, mrpl10Δ, and itr2Δ), the transposition efficiency of Ppmar2NA was at least 1/2 less active than in the control strain His3Δ, especially in rtt10Δ mutant, where the transposition efficiency decreased by more than 65-fold. One of the 5 genes was associated with histone methylation, whereas the other 4 genes were host factors of retrotransposon. These findings indicated that DNA transposon and RNA transposon may share common host factors (Irwin et al., 2005; Curcio et al., 2015).
In yeast, Elongation Factor Methyltransferase (EFM1), which encoded lysine methyltransferase SET8, may characteristically methylate H4K20 (Zhang and Bruice, 2008) and function in protein modification, chromosome-protein binding regulation, and gene transcription and translation. Studies have shown that EFM1 methylates H4K20 into three proteins H4K20me1, H4K20me2 and H4K20me3 (Yang and Mizzen, 2009). H4K20me1 could further promote chromatin condensation with the help of Condensin II (Weirich et al., 2015). Also, H4K20me1 and H4K20me2 inhibited gene transcription and chromosomal shrinkage by binding to the Lethal (3) Malignant Brain Tumor-Like Protein 1 (L3MBTL1) (Li et al., 2007). H4K20me3 was generally regarded as a marker of transcriptional inhibition in heterochromatin (Schotta et al., 2004). H4K20 was not methylated when the EFM1 gene was knocked out, which might be related to the high transposition frequency of the Ppmar2 in the efm1Δ mutant yeast.
The DEGs identified with the comparison of efm1Δ and His3Δ revealed that the efm1Δ cell conditions were changed significantly. The DDR2 gene encoded a stress protein involved in DNA repair (Kobayashi et al., 1996). The expression of this gene was dramatically elevated in the efm1Δ mutant, suggesting that it may efficiently repair the damaged DNA. The HSP70, which encoded heat shock factor (Abrams et al., 2014), was also obviously upregulated in the efm1Δ mutant. Jardim et al. (2015) found evidence that mos1 may be co-activated with HSP70 genes. JEN1 mediated high-affinity uptake of carbon sources lactate, pyruvate, acetate, and micronutrient selenite, JEN1 expression and localization are tightly regulated, with transcription repression, mRNA degradation, and protein endocytosis and degradation all occurring in the presence of glucose (Haurie et al., 2001).
The RTT10 gene, which encoded a member of the WD40 protein family, was a retrotransposition-related gene. Ty1 was more active in the rtt10Δ mutant than in the His3Δ strain, and the RTT10 gene might prevent Ty1 RNA from being reversely transcribed into cDNA (Scholes et al., 2001). Moreover, RTT10 contributed to the synthesis of transcription factor TFIID, regulating the ribosome small subunit rRNA synthesis (Dragon et al., 2002). Three DEGs (UTP20, RPA190 and RRP5) in the rtt10Δ mutant might be related to the lowered transposition activity. The UTP20 protein was a component of the small-subunit (SSU) processome, which modulated the 18S rRNA synthesis (Gallagher et al., 2004). The expression level of this gene in the rtt10Δ mutant was significantly lowered. RRP5 was another component of the SSU processome and 90S preribosome related to the synthesis of 18S and 5.8S rRNAs (Venema and Tollervey, 1996). The expression level was much lower in the rtt10Δ mutant than in His3Δ. RPA190, encoding RNA polymerase A (Mémet et al., 1988), was also downregulated in the rtt10Δ mutant. The discovery of host factors associated with Ppmar2 mobility lays an important foundation for understanding the mechanism of Ppmar2 transposition in the heterologous host.
In the present study, we created hyperactive Ppmar2 transposons via site-direct mutagenesis and screened the heterologous host factors that influenced the transposition activity. Ppmar2 was proved to efficient to able to excision and reinserted into the TA-rich region, which was the same as the other MLEs in plants. EFM1 and RTT10 related to protein methylation, chromatin and RNA transcription greatly impact the heterologous transposition efficiency of Ppmar2 in yeast. The Ppmar2 transposon system is a promising tool for insertion mutagenesis in moso bamboo and might be used as an alternative to the existing transposon tagging systems in the other plants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MZ designed the experiments; XZ and JX performed the research; CX, XC and L-HZ participated in the research; MZ and XZ wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was funded by grants from the Zhejiang Provincial Natural Science Foundation of China (No. LZ19C160001 and LQ21C160003), the National Natural Science Foundation of China (No. 31870656, 31470615 and 32001326), the Talent Research Foundation of Zhejiang A&F University (No. 2019FR055) and Independent research project of state key laboratory of subtropical silviculture (ZY20200301).
Acknowledgments
We would like to extend our sincere gratitude and appreciation to all reviewers for their valuable comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1004732/full#supplementary-material
References
Abrams, J. L., Verghese, J., Gibney, P. A., Morano, K. A. (2014). Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289 (19), 13155–13167. doi: 10.1074/jbc.M113.530014
Bryan, G., Garza, D., Hartl, D. (1990). Insertion and excision of the transposable element mariner in Drosophila. Genetics 125 (1), 103–114. doi: 10.1093/genetics/125.1.103
Butler, M. G., Chakraborty, S. A., Lampe, D. J. (2006). The n-terminus of Himar1 mariner transposase mediates multiple activities during transposition. Genetica 127 (1-3), 351–366. doi: 10.1007/s10709-006-6250-x
Claeys, B., Lipkow, K., Andrews, S. S., Liu, D., Chalmers, R. (2013). The autoregulation of a eukaryotic DNA transposon. Elife 2, e00668. doi: 10.7554/eLife.00668
Curcio, M. J., Lutz, S., Lesage, P. (2015). The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 3 (2), MDNA3–0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014
Dawson, A., Finnegan, D. J. (2003). Excision of the Drosophila mariner transposon Mos1: Comparison with bacterial transposition and V(D)J recombination. Mol. Cell 11 (1), 225–235. doi: 10.1016/S1097-2765(02)00798-0
Dragon, F., Gallagher, J., Compagnone-Post, P. A., Mitchell, B. M., Porwancher, K. A., Wehner, K. A., et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417 (6892), 967–970. doi: 10.1038/nature00769
Feschotte, C., Wessler, S. R. (2002). Mariner-like transposases are widespread and diverse in flowering plants. Proc. Natl. Acad. Sci. U.S.A. 99 (1), 280–285. doi: 10.1073/pnas.022626699
Gallagher, J. E., Dunbar, D. A., Granneman, S., Mitchell, B. M., Osheim, Y., Beyer, A. L., et al. (2004). RNA Polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18 (20), 2506–2517. doi: 10.1101/gad.1226604
Germon, S., Bouchet, N., Casteret, S., Carpentier, G., Adet, J., Bigot, Y., et al. (2009). Mariner Mos1 transposase optimization by rational mutagenesis. Genetica 137 (3), 265–276. doi: 10.1007/s10709-009-9375-x
Hartl, D., Lohe, L., Lozovskaya, E. R. (1997). Modern thoughts of an ancyent marinere: Function, evolution, regulation. Annu. Rev. Genet. 31: 337–358. doi: 10.1146/annurev.genet.31.1.337
Haurie, V., Perrot, M., Mini, T., Jeno, P., Sagliocco, F., Boucherie, H. (2001). The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276 (1), 76–85. doi: 10.1074/jbc.M008752200
Haynes, W. (2013). “Benjamini–hochberg method,” in Encyclopedia of systems biology. Eds. Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H. (New York, NY: Springer New York), 78–78.
Irwin, B., Aye, M., Baldi, P., Beliakova-Bethell, N., Cheng, H., Dou, Y., et al. (2005). Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15 (5), 641–654. doi: 10.1101/gr.3739005
Ivics, Z., Hackett, P. B., Plasterk, R. H., Izsvák, Z. (1997). Molecular reconstruction of Sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91 (4), 501–510. doi: 10.1016/S0092-8674(00)80436-5
Janzen, D. H. (1976). Why bamboos wait so long to flower. Annu. Rev. Ecol. syst. 7 (1), 347–391. doi: 10.1146/annurev.es.07.110176.002023
Jardim, S. S., Schuch, A. P., Pereira, C. M., Loreto, E. L. (2015). Effects of heat and UV radiation on the mobilization of transposon mariner-Mos1. Cell Stress Chaperones 20 (5), 843–851. doi: 10.1007/s12192-015-0611-2
Joshi, N., Fass, J. (2011). “Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33)[Software]”. Available at:https://github.com/najoshi/sickle.
Kobayashi, N., Mcclanahan, T. K., Simon, J. R., Treger, J. M., Mcentee, K. (1996). Structure and functional analysis of the multistress response gene DDR2 from Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 229 (2), 540–547. doi: 10.1006/bbrc.1996.1840
Lampe, D. J. (2010). Bacterial genetic methods to explore the biology of mariner transposons. Genetica 138 (5), 499–508. doi: 10.1007/s10709-009-9401-z
Lazarow, K., Du, M. L., Weimer, R., Kunze, R. (2012). A hyperactive transposase of the maize transposable element activator (Ac). Genetics 191 (3), 747–756. doi: 10.1534/genetics.112.139642
Li, H., Fischle, W., Wang, W., Duncan, E. M., Liang, L., Murakami-Ishibe, S., et al. (2007). Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28 (4), 677–691. doi: 10.1016/j.molcel.2007.10.023
Liu, D., Chalmers, R. (2014). Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. Nucleic Acids Res. 42 (4), 2637–2645. doi: 10.1093/nar/gkt1218
Lohe, A. R., De Aguiar, D., Hartl, D. L. (1997). Mutations in the mariner transposase: the D,D(35)E consensus sequence is nonfunctional. Proc. Natl. Acad. Sci. 94 (4), 1293–1297. doi: 10.1073/pnas.94.4.1293
Lohe, A. R., Hartl, D. L. (1996). Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. Mol. Biol. Evol. 13 (4), 549–555. doi: 10.1093/oxfordjournals.molbev.a025615
Luo, G., Ivics, Z., Izsvák, Z., Bradley, A. (1998). Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. United States America 95, 10769–10773. doi: 10.1073/pnas.95.18.10769
Mátés, L., Chuah, M. K., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., et al. (2009). Molecular evolution of a novel hyperactive Sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41 (6), 753–761. doi: 10.1038/ng.343
Mémet, S., Gouy, M., Marck, C., Sentenac, A., Buhler, J. M. (1988). RPA190, the gene coding for the largest subunit of yeast RNA polymerase a. J. Biol. Chem. 263 (6), 2830–2839. doi: 10.1016/s0021-9258(18)69144-6
Miskey, C., Izsvak, Z., Plasterk, R. H., Ivics, Z. (2003). The Frog prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31 (23), 6873–6881. doi: 10.1093/nar/gkg910
Plasterk, R. H., Izsvák, Z., Ivics, Z. (1999). Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15 (8), 326–332. doi: 10.1016/s0168-9525(99)01777-1
Ramakrishnan, M., Zhou, M., Pan, C., Hanninen, H., Yrjala, K., Vinod, K. K., et al. (2019a). Affinities of terminal inverted repeats to DNA binding domain of transposase affect the transposition activity of bamboo Ppmar2 mariner-like element. Int. J. Mol. Sci. 20 (15), 3692. doi: 10.3390/ijms20153692
Ramakrishnan, M., Zhou, M. B., Pan, C. F., Hanninen, H., Tang, D. Q., Vinod, K. K. (2019b). Nuclear export signal (NES) of transposases affects the transposition activity of mariner-like elements Ppmar1 and Ppmar2 of moso bamboo. Mob DNA 10, 35. doi: 10.1186/s13100-019-0179-y
Robertson, H. M., Lampe, D. J. (1995). Recent horizontal transfer of a mariner transposable element among and between diptera and neuroptera. Mol. Biol. Evol. 12 (5), 850–862. doi: 10.1093/oxfordjournals.molbev.a040262
Scholes, D. T., Banerjee, M., Bowen, B., Curcio, M. J. (2001). Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159 (4), 1449–1465. doi: 10.1093/genetics/159.4.1449
Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., et al. (2004). A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18 (11), 1251–1262. doi: 10.1101/gad.300704
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Tellier, M., Chalmers, R. (2020). Compensating for over-production inhibition of the Hsmar1 transposon in escherichia coli using a series of constitutive promoters. Mobile DNA 11, 5. doi: 10.1186/s13100-020-0200-5
Teste, M. A., Duquenne, M., Francois, J. M., Parrou, J. L. (2009). Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 10, 99. doi: 10.1186/1471-2199-10-99
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–515. doi: 10.1038/nbt.1621
Venema, J., Tollervey, D. (1996). RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15 (20), 5701–5714. doi: 10.1002/j.1460-2075.1996.tb00954.x
Watanabe, M., Ueda, K., Manabe, I., Akai, T. (1982). Flowering, seeding, germination, and flowering periodicity of Phyllostachys pubescens. J. Japanese Forestry Soc. 64 (3), 107–111. doi: 10.11519/jjfs1953.64.3_107
Weirich, S., Kusevic, D., Kudithipudi, S., Jeltsch, A. (2015). Investigation of the methylation of numb by the SET8 protein lysine methyltransferase. Sci. Rep. 5, 13813. doi: 10.1038/srep13813
Yang, H., Mizzen, C. A. (2009). The multiple facets of histone H4-lysine 20 methylation. Biochem. Cell Biol. 87 (1), 151–161. doi: 10.1139/O08-131
Yang, G., Nagel, D. H., Feschotte, C., Hancock, C. N., Wessler, S. R. (2009). Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. science 325 (5946), 1391–1394. doi: 10.1126/science.1175688
Yang, G., Weil, C. F., Wessler, S. R. (2006). A rice Tc1/mariner-like element transposes in yeast. Plant Cell 18 (10), 2469–2478. doi: 10.1105/tpc.106.045906
Yuan, Y. W., Wessler, S. R. (2011). The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. U.S.A. 108 (19), 7884–7889. doi: 10.1073/pnas.1104208108
Yusa, K., Takeda, J., Horie, K. (2004). Enhancement of Sleeping beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol. Cell Biol. 24 (9), 4004–4018. doi: 10.1128/MCB.24.9.4004-4018.2004
Zayed, H., Izsvak, Z., Khare, D., Heinemann, U., Ivics, Z. (2003). The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping beauty transposition. Nucleic Acids Res. 31 (9), 2313–2322. doi: 10.1093/nar/gkg341
Zhang, X., Bruice, T. C. (2008). Product specificity and mechanism of protein lysine methyltransferases: Insights from the histone lysine methyltransferase SET8. Biochemistry 47 (25), 6671–6677. doi: 10.1021/bi800244s
Zhou, M. B., Zhong, H., Hu, J. L., Tang, D. Q. (2015). Ppmar1 and Ppmar2: the first two complete and intact full-length mariner-like elements isolated in Phyllostachys edulis. Acta Botanica Gallica 162 (2), 127–137. doi: 10.1080/12538078.2014.999117
Zhou, M., Hu, H., Liu, Z., Tang, D. (2016). Two active bamboo mariner-like transposable elements (Ppmar1 and Ppmar2) identified as the transposon-based genetic tools for mutagenesis. Mol. Breed. 36 (12), 163. doi: 10.1007/s11032-016-0588-2
Keywords: mariner-like element (MLE), artificial optimization, transposase, host factor, bamboo, yeast
Citation: Zhou X, Xie J, Xu C, Cao X, Zou L-H and Zhou M (2022) Artificial optimization of bamboo Ppmar2 transposase and host factors effects on Ppmar2 transposition in yeast. Front. Plant Sci. 13:1004732. doi: 10.3389/fpls.2022.1004732
Received: 27 July 2022; Accepted: 07 October 2022;
Published: 20 October 2022.
Edited by:
Daqiu Zhao, Yangzhou University, ChinaReviewed by:
Michael Tellier, University of Oxford, United KingdomSharmistha M, Indian Institute of Technology Gandhinagar, India
Corentin Claeys Bouuaert, Université catholique de Louvain, Belgium
Copyright © 2022 Zhou, Xie, Xu, Cao, Zou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingbing Zhou, zhoumingbing@zafu.edu.cn
 Xiaohong Zhou
Xiaohong Zhou Jiamin Xie
Jiamin Xie Xiuling Cao
Xiuling Cao Long-Hai Zou
Long-Hai Zou Mingbing Zhou
Mingbing Zhou