- 1Department of Biology and Center for Molecular Signaling, Wake Forest University, Winston-Salem, NC, United States
- 2Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, United States
- 3Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI, United States
The inhibition of hypocotyl elongation by ethylene in dark-grown seedlings was the basis of elegant screens that identified ethylene-insensitive Arabidopsis mutants, which remained tall even when treated with high concentrations of ethylene. This simple approach proved invaluable for identification and molecular characterization of major players in the ethylene signaling and response pathway, including receptors and downstream signaling proteins, as well as transcription factors that mediate the extensive transcriptional remodeling observed in response to elevated ethylene. However, the dark-adapted early developmental stage used in these experiments represents only a small segment of a plant’s life cycle. After a seedling’s emergence from the soil, light signaling pathways elicit a switch in developmental programming and the hormonal circuitry that controls it. Accordingly, ethylene levels and responses diverge under these different environmental conditions. In this review, we compare and contrast ethylene synthesis, perception, and response in light and dark contexts, including the molecular mechanisms linking light responses to ethylene biology. One powerful method to identify similarities and differences in these important regulatory processes is through comparison of transcriptomic datasets resulting from manipulation of ethylene levels or signaling under varying light conditions. We performed a meta-analysis of multiple transcriptomic datasets to uncover transcriptional responses to ethylene that are both light-dependent and light-independent. We identified a core set of 139 transcripts with robust and consistent responses to elevated ethylene across three root-specific datasets. This “gold standard” group of ethylene-regulated transcripts includes mRNAs encoding numerous proteins that function in ethylene signaling and synthesis, but also reveals a number of previously uncharacterized gene products that may contribute to ethylene response phenotypes. Understanding these light-dependent differences in ethylene signaling and synthesis will provide greater insight into the roles of ethylene in growth and development across the entire plant life cycle.
Introduction
Plant responses to the gaseous hormone ethylene are an excellent model for studying the relationships between hormone synthesis, signaling, transcriptional changes, and development. The identification of ethylene-insensitive mutants in Arabidopsis using molecular genetics opened a new era in dissecting plant hormone signaling (Bleecker et al., 1988; Guzman and Ecker, 1990). Ethylene-insensitive mutants were identified as lacking the ethylene “triple response” in dark-grown seedlings (short, thick hypocotyl and exaggerated apical hook), remaining tall in the presence of excess ethylene (Alonso et al., 2003; Guo and Ecker, 2003; Yanagisawa et al., 2003). This approach enabled the isolation of mutations affecting the activities of core ethylene response machinery, including receptors, signaling proteins, and transcription factors. The functions of these signaling components, as well as the pathways for ethylene synthesis, have subsequently been assayed in additional tissues beyond dark-grown hypocotyls, demonstrating that many of these proteins function in all tissues and growth conditions, but also revealing branches of the ethylene signaling and synthesis pathways that have distinct roles in light-grown plants and in other developmental stages. In particular, ethylene-responsive transcriptional networks and regulatory controls of ethylene biosynthesis show profound differences between light- and dark-grown tissues. Although some of these differences have been reviewed previously (Rodrigues et al., 2014; Booker and DeLong, 2015; Yoon, 2015; Yu and Huang, 2017), recent studies have identified new mechanisms and yielded insight into light-dependent differences. This review highlights the similarities and differences in light-dependent regulation of ethylene synthesis and response in seedlings grown at a range of light levels, focusing on recent publications establishing that the genetic redundancy in ethylene biosynthetic machinery, ethylene receptors, and transcriptional machinery may allow a complex suite of light-dependent developmental responses to this important hormone.
Basics of the Ethylene Signaling Pathway
The triple response of dark-grown seedlings was exploited in elegant genetic screens that identified mutants exhibiting either ethylene-insensitivity (ein or etr mutants) (Bleecker et al., 1988; Guzman and Ecker, 1990; Chang et al., 1993), enhanced ethylene signaling in the constitutive triple response (ctr) (Kieber et al., 1993; Huang et al., 2003), or synthesis in the ethylene overproducer (eto) mutants (Guzman and Ecker, 1990). The genes responsible for these phenotypes have been cloned and mapped to the ethylene signaling and biosynthetic pathways. The signaling pathway begins with ethylene binding to ER-localized receptor proteins (Kendrick and Chang, 2008), which act as negative regulators of the pathway (Hua and Meyerowitz, 1998). In Arabidopsis, these receptors are ETR1, ETR2, EIN4, ERS1, and ERS2 (Chang et al., 1993; Schaller and Bleecker, 1995; Hua and Meyerowitz, 1998; Sakai et al., 1998), which fall into two subfamilies based on sequence similarity of the ethylene binding domains and the presence of conserved histidine kinase domains (Kendrick and Chang, 2008; Stepanova and Alonso, 2009; Shakeel et al., 2013). When ethylene binds, the receptors are turned off, resulting in decreased activity of the inhibitory CTR1 protein kinase and increased EIN2 output (Kieber et al., 1993; Alonso et al., 1999; Huang et al., 2003; Qiao et al., 2009). C-terminal proteolytic cleavage of EIN2 promotes the nuclear localization of the EIN2 C-terminal proteolytic fragment (EIN2-CEND) (Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). EIN2-CEND-mediated targeting of EBF1/2 mRNA to the processing body further enhances signaling output (Li et al., 2015; Merchante et al., 2015). Nuclear EIN2-CEND alters transcription via activation of the EIN3 and EIN3-LIKE (EIL1 and EIL2) transcription factors (TFs), which then turn on expression of genes encoding other TFs, such as ERF1 and EDF1-EDF4 (Chao et al., 1997; Solano et al., 1998; Alonso et al., 2003; Chang et al., 2013). These core TFs likely work with other TFs as part of a gene regulatory network leading to a diversity of transcriptional responses, which have been characterized in multiple genome-wide transcriptional studies (Stepanova et al., 2007; Chang et al., 2013; Feng et al., 2017; Harkey et al., 2018). Ethylene signaling is also modulated by EIN2-mediated translational regulation (Merchante et al., 2015), as well as F-box dependent proteolysis of EIN2 and EIN3 via ETP1/2 and EBF1/2, respectively (Guo and Ecker, 2003; Potuschak et al., 2003; Qiao et al., 2009). EBF1/2 are also destabilized by ethylene in an EIN2-dependent manner, allowing increased accumulation of EIN3 (An et al., 2010).
Ethylene signaling proteins have roles that extend beyond their functions in dark-grown Arabidopsis hypocotyls. Genes encoding these proteins have been found across the plant kingdom (Wang et al., 2015), and the proteins have been shown to function in a diversity of tissues and under a range of light conditions (Lanahan et al., 1994; Binder et al., 2006; Plett et al., 2009; Wilson et al., 2014a). Both CTR1 and EIN2 are required for normal ethylene responsiveness in all light conditions in Arabidopsis, indicating that each of these gene products plays a central and non-redundant role in ethylene signaling, regardless of light conditions. Mutants lacking CTR1 show constitutive ethylene responses in roots and shoots grown in light or dark (Kieber et al., 1993). Mutations in EIN2 confer insensitivity to added ethylene in dark-grown hypocotyls (Alonso et al., 1999), light-grown rosettes (Kieber et al., 1993), light-grown hypocotyls (Smalle et al., 1997), and roots of dark-grown (Stepanova et al., 2005) and light-grown seedlings (Negi et al., 2008; Harkey et al., 2018).
Ethylene receptors are members of a conserved multi-gene family (Shakeel et al., 2013). As these receptors function as negative regulators, dominant gain-of-function (GOF) mutations, such as etr1-1 and etr1-3 in Arabidopsis (Bleecker et al., 1988; Guzman and Ecker, 1990; Chang et al., 1993) and Neverripe in tomato (Wilkinson et al., 1995), yield ethylene-insensitive plants. In contrast, null or loss-of-function (LOF) alleles can confer constitutive ethylene response phenotypes (Hua and Meyerowitz, 1998; Shakeel et al., 2013). In Arabidopsis, the five ethylene receptors have been shown to have distinct roles that are tied to specific developmental responses (Shakeel et al., 2013), some of which can be studied only in older plants, which are necessarily grown in light. Similarly, the tomato Neverripe gene belongs to a seven-member ethylene receptor gene family and the Neverripe mutant carries a GOF mutation that confers ethylene insensitivity in phenotypes observed in both light and dark conditions (e.g., fruit ripening, hypocotyl triple response, and root development) (Wilkinson et al., 1995; Negi et al., 2010; Klee and Giovannoni, 2011). Tomato plants with knockdown of mRNA encoding receptors have also revealed distinct functions for two tomato ethylene receptors (Kevany et al., 2007). In the sections below, we highlight studies that have revealed differences in ethylene responses that are influenced by light and developmental stage, and which require distinct ethylene signaling or synthesis machinery.
Basics of the Ethylene Biosynthesis Pathway
The enzymatic steps of the ethylene biosynthetic pathway were uncovered in fruit; subsequent work in fruit and in dark-grown Arabidopsis seedlings identified a conserved biosynthetic pathway and revealed important regulatory mechanisms that control pathway activity (Adams and Yang, 1979; Yang and Hoffman, 1984; Booker and DeLong, 2015; Yoon, 2015). The simple and highly conserved pathway has only two committed steps: conversion of S-adenosyl-l-methionine (SAM) to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS), followed by conversion of ACC to ethylene by ACC oxidase (ACO) (Houben and Van de Poel, 2019). ACS has been a primary target for researchers interested in understanding regulation of ethylene biosynthesis, as this enzyme catalyzes the first biosynthetic step, which is frequently described as the rate-limiting step (Adams and Yang, 1979; Yang and Hoffman, 1984). ACS gene families in land plants encode isozymes belonging to three classes, type-1, type-2, and type-3 (El-Sharkawy et al., 2008; Lin et al., 2009; Booker and DeLong, 2015; Zhu et al., 2015; Lee et al., 2019). The evolution and regulation of ACO, including consideration of conditions under which ACO activity is limiting for ethylene production, have been recently reviewed (Houben and Van de Poel, 2019). There are both transcriptional and post-translational mechanisms that control which ACS and ACO isozymes are expressed and active, leading to distinct enzyme populations in tissue- and developmental stage-specific contexts (Booker and DeLong, 2015; Houben and Van de Poel, 2019). Positive feedback loops, largely driven by transcriptional controls of these biosynthetic enzymes, drive dramatic increases in ethylene production to accelerate fruit ripening (Klee and Giovannoni, 2011). This review will examine new insight into the molecular mechanisms by which ethylene synthesis is modulated by light levels at both transcriptional and post-translational levels.
Light-Dependent and -Independent Ethylene Responses
Ethylene Effects in Hypocotyls Are Opposite in Light and Dark
The ethylene response in the hypocotyls of young seedlings is highly dependent on light level. The triple response of etiolated seedlings, including inhibited hypocotyl elongation, is the basis of much of the current molecular insight into ethylene signaling (Bleecker et al., 1988; Guzman and Ecker, 1990). Ethylene treatment under shade covering, rather than complete darkness, also leads to decreased hypocotyl growth (Das et al., 2016). The hypocotyl response to ethylene is coordinated with light-dependent hypocotyl elongation changes during photomorphogenesis (Yu and Huang, 2017). Light inhibits hypocotyl elongation, which is important as plants growing in soil transition to light (Montgomery, 2016). In opposition to the effect of ethylene in the dark, light-grown Arabidopsis seedlings show increased hypocotyl elongation in response to ethylene (Smalle et al., 1997; Le et al., 2005; Das et al., 2016; Seo and Yoon, 2019), as illustrated in Figure 1. In both light and dark, the ACC or ethylene response is tied to differences in cell expansion (Smalle et al., 1997; Seo and Yoon, 2019). These light-dependent differences have more frequently been reported in response to treatment with the ethylene precursor, ACC (Smalle et al., 1997; Le et al., 2005), but ethylene yields the same light-dependent increases in elongation (Figure 1), and ethylene-insensitive mutants are shorter than wild-type in the light (Le et al., 2005). Intriguingly, the nutrient content of the growth media affects the ethylene response in light-grown, but not dark-grown, seedlings (Smalle et al., 1997; Collett et al., 2000).
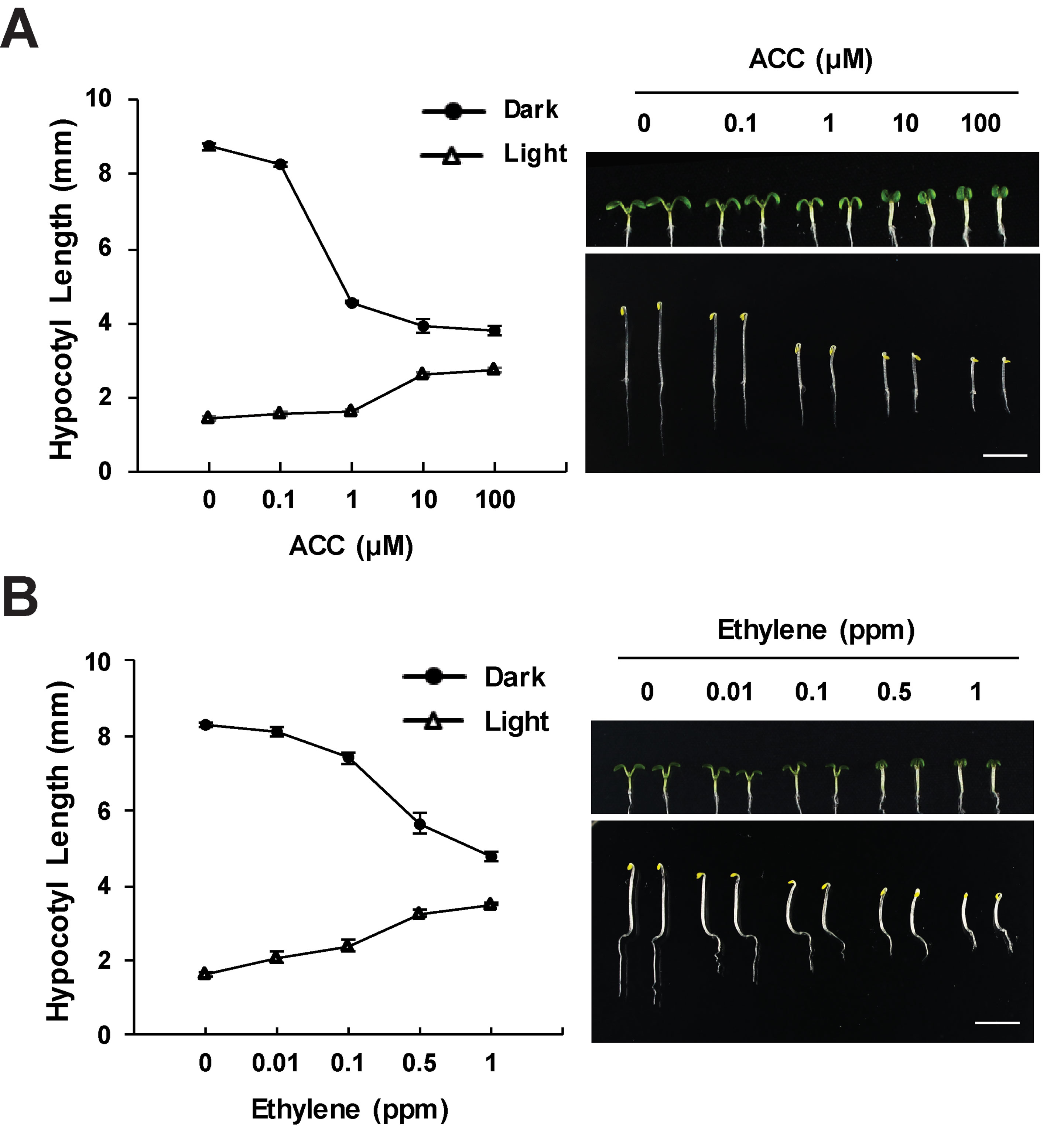
Figure 1 Ethylene and ACC inhibit hypocotyl elongation in the dark and increase elongation in the light. Wild-type seedlings were grown on media containing the indicated concentrations of ACC or on control media and treated with ethylene gas for 3 days in the dark or 5 days in light. The effects of (A) ACC or (B) ethylene on hypocotyl growth in dark and light conditions. Images generated by the Yoon lab (Seo and Yoon, 2019), recapitulating previous findings (Bleecker et al., 1988; Guzman and Ecker, 1990; Smalle et al., 1997; Le et al., 2005; Liang et al., 2012). Values shown are the average and SD of three replicates, each containing at least 20 seedlings.
Another striking feature of the ethylene triple response in etiolated seedlings is the accentuation of the apical hook. As part of photomorphogenesis, the apical hook opens and cotyledons expand, so it is important to ask whether this ethylene response, like hypocotyl elongation, is also light dependent (Bleecker et al., 1988; Raz and Ecker, 1999; Mazzella et al., 2014; Van de Poel et al., 2015). The formation of apical hooks in etiolated seedlings protects the shoot apical meristem during growth through soil, and ethylene build-up in denser soil exaggerates this hook to assist in emergence (Zhong et al., 2014; Shi et al., 2016a). Ethylene insensitive mutants with receptor and signaling defects show impaired hook formation, while the ctr1-1 null mutant has an exaggerated hook (Abbas et al., 2013). Localized accumulation of ACO across the hook may also contribute to hook maintenance in dark-grown seedlings (Peck et al., 1998; Raz and Ecker, 1999). Mutants with elevated ethylene synthesis show enhanced hook formation (Guzman and Ecker, 1990). A central feature of ethylene-accentuated hook formation is crosstalk with auxin. Asymmetries in auxin synthesis and auxin transport, which lead to accumulation of growth-inhibiting auxin levels on the inside of the hook, are enhanced by ethylene treatment (Vandenbussche et al., 2010; Zádníková et al., 2010). The process of hypocotyl hook opening in response to light is also ethylene regulated (Vandenbussche et al., 2010; Zádníková et al., 2010; Van de Poel et al., 2015). In dark-grown seedlings, the ein3-1 eil1-1 double mutant has enhanced hook opening, while an EIN3 overexpression line has a tightly closed, exaggerated hook like ctr1-1, and shows delayed hook opening in the light (Zhang et al., 2018), consistent with ethylene negatively regulating hook opening in both light and dark.
Ethylene Modulates Light-Dependent and Light-Independent Root Development
In seedling roots, ethylene and ACC inhibit elongation in both light and dark conditions (Rahman et al., 2001; Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007; Negi et al., 2008; Negi et al., 2010; Strader et al., 2010) while enhancing root hair initiation (Cutter, 1978; Tanimoto et al., 1995; Pitts et al., 1998; Dolan, 2001; Rahman et al., 2002; Strader et al., 2010). In both light- and dark-grown seedlings, these root responses to ethylene are lost in ethylene-insensitive etr1-3, a dominant gain of function (GOF) receptor mutant, and in the ein2-5 signaling mutant (Ruzicka et al., 2007; Swarup et al., 2007; Negi et al., 2008; Lewis et al., 2011a). These effects on root elongation are tied to auxin and ethylene cross-talk in a light-independent fashion. Ethylene enhances auxin synthesis, transport, and signaling to control root development (Stepanova et al., 2005; Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007; Negi et al., 2008; Stepanova et al., 2008; Lewis et al., 2011a; Muday et al., 2012).
In contrast, the inhibitory effect of ethylene and ACC on lateral root (LR) formation in Arabidopsis and tomato has been examined only in light-grown seedlings, as LRs do not form in roots of dark-grown seedlings (Ivanchenko et al., 2008; Negi et al., 2008; Negi et al., 2010; Lewis et al., 2011b; Lewis et al., 2011a). Ethylene and ACC block early stages of LR initiation (Ivanchenko et al., 2008). As with the inhibition of root elongation, ethylene inhibits LR formation by modulating auxin synthesis, signaling, and transport, which control this process (Stepanova et al., 2007; Muday et al., 2012). Similarly, the effects of ethylene and ACC on root gravitropism and root waving, which have been assayed only in light-grown seedlings, also are blocked in the ethylene signaling mutants ein2-5 and the GOF etr1-3 receptor mutant (Buer et al., 2003; Buer et al., 2006). Overall, published data support a light-independent function of the EIN2 protein in ethylene signaling in roots (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007; Negi et al., 2008; Lewis et al., 2011a). However, these data do not reveal which specific receptors function in the roots, because the use of GOF mutants (like etr1-1 and etr1-3) can perturb the functions of the entire receptor family (Chang et al., 1993; Shakeel et al., 2013). Using LOF alleles in each receptor subtype is a powerful strategy to resolve the specific function of the family of ethylene receptors; this approach has been used to understand ethylene-regulated growth and development in a light-dependent context, as discussed below.
Mechanistic Connections Between Light Response and Ethylene Biosynthesis
Changes in ethylene synthesis in response to changing light levels have been reported in many different species and under many different growth conditions, with dramatically varying results. The ability of light to modulate ethylene synthesis was reported half a century ago, when a single dose of red light was shown to decrease ethylene levels in etiolated pea seedlings in a far-red reversible manner, suggesting that phytochrome negatively controls ethylene biosynthesis (Goeschl et al., 1967). Conversely, high-intensity illumination of green seedlings induced an increase in ethylene synthesis, demonstrating a positive effect of light on ethylene production (Weckx and Van Poucke, 1989). Subsequent studies have confirmed that the effect of light on ethylene synthesis is complex and context-dependent (Foo et al., 2006; Khanna et al., 2007; Jeong et al., 2016; Song et al., 2018), and is also affected by crosstalk with other plant hormone response pathways (Vandenbussche et al., 2003; Arteca and Arteca, 2008; Muday et al., 2012; Lee et al., 2017). For instance, etiolated Arabidopsis seedlings show age- and light-dependent increases in ethylene biosynthesis with higher levels in the light; increased ethylene production is detectable as rapidly as 4 h after transfer to light, but becomes more dramatic with increasing time in light (Seo and Yoon, 2019). As discussed below, these effects are mediated at both the transcriptional and post-translational levels, and although much work has focused on regulation of ACS expression and activity, additional data reveal light-dependent effects on regulation of ACO function.
Light-Mediated Transcriptional Regulation of ACS and ACO
Regulation of ethylene synthesis via alteration of ACS and/or ACO gene expression is a primary mechanism through which differences in the quality, quantity, or periodicity of light modulate ethylene production and signaling outputs to coordinate plant growth and development (Yamagami et al., 2003; Tsuchisaka and Theologis, 2004; Wang et al., 2005). The combinatorial effects of light with phytohormones and biotic or abiotic stresses add further complexity to light-mediated control of ethylene biosynthesis. For example, IAA treatment induces expression of Arabidopsis ACS genes in seedlings grown in darkness or in constant light, but this induction is less dramatic in plants grown with a light/dark cycle (Rashotte et al., 2005). Furthermore, light differentially influences the transcript levels of various ACS genes, depending on the developmental stage and the length of light treatment (Seo and Yoon, 2019). The mRNA levels of a subset of type-1 and type-2 ACSs (ACS6 and ACS5, 8, and 9, respectively) declined rapidly and steeply after etiolated seedlings were transferred to light, and these transcript levels remained low for 5 days. Meanwhile, ACS2 (type-1) and ACS4 (type-2) showed gradual increases in their transcript levels after light exposure (Seo and Yoon, 2019). Together, these data suggest distinct roles for ACS isozymes depending on the light conditions, with ACS5, 6, 8, and 9 playing the primary roles in dark-grown seedlings, while expression of ACS2 and ACS4 is implicated in controlling ethylene production in the light.
Analysis of light signaling mutants and transgenic lines expressing light signaling components has also provided insight into the light-mediated regulation of ethylene biosynthesis. Mutations in the phytochrome genes PHYA and PHYB increased ethylene biosynthesis in pea, consistent with a negative effect of light on ethylene synthesis, with a more profound effect observed in the phyA mutant (Foo et al., 2006). Intriguingly, in Arabidopsis and sorghum, phyA mutants show less profound increases in ethylene biosynthesis than do phyB mutants, indicating species-specific functions of these photoreceptors in controlling ethylene levels. Similarly, transgenic lines overexpressing Arabidopsis PHYTOCHROME-INTERACTING FACTOR5 (PIF5), a basic helix-loop-helix transcription factor that specifically interacts with the photoactivated form of PhyB, showed a marked increase in ethylene production in the dark that is correlated with increased abundance of ACS4, ACS8, and other ACS transcripts (Khanna et al., 2007). Although the pif1 pif3 pif4 pif5 (pifq) mutant initially produced less ethylene than wild-type seedlings, consistent with the higher ethylene levels in PIF5 overexpression lines, at later time-points the pifq mutant showed higher ethylene production (Jeong et al., 2016), indicating a developmental stage-dependent role of PIFs in controlling ethylene biosynthesis.
The regulation of ACO gene expression has received much less study than that of ACS (Houben and Van de Poel, 2019), yet the levels of ACO transcripts are also regulated by light and other factors that control pathway activity (Argueso et al., 2007; Rodrigues et al., 2014). In tomato fruits, ACO1 is upregulated by pulses of white light (Scott et al., 2018). Classic work demonstrated that ACO expression is both a driver of ethylene production and a reporter for ethylene response in etiolated tissues (Peck and Kende, 1995; Kim et al., 1997), creating a positive feedback loop. ACO transcript increases have also been reported after ACC treatment of aerial tissues of light-grown seedlings (Zhong and Burns, 2003). The meta-analysis discussed below provides strong support for this feed-forward mechanism. Furthermore, when ACS activity is elevated during climacteric ripening in tomato or banana fruits (and during flooding stress), ACO activity becomes rate limiting, and ACO expression is up-regulated (Ruduś et al., 2013; Xiao et al., 2013; Houben and Van de Poel, 2019). This suggests that one role of the feed-forward mechanism is to “clear” excess ACC when ACO activity limits ethylene production.
Light-Mediated Post-Translational Control of ACS and ACO Activity
An early study suggested that light regulates ethylene biosynthesis by altering stability/activity of ACS isozymes (Rohwer and Schierle, 1982). More recent work has confirmed that light modulates ethylene biosynthesis via post-translational mechanisms including reversible phosphorylation and protein turnover (Steed et al., 2004; Chae and Kieber, 2005; Yoon and Kieber, 2013b; Zdarska et al., 2015; Seo and Yoon, 2019). Post-translational regulation of ACS is largely dependent on the regulatory motifs located in the C-terminus of ACS proteins (Chae and Kieber, 2005). All three ACS types contain a well-conserved N-terminal catalytic domain, whereas the C-termini vary among ACS isoforms. Type-1 ACSs (ACS1, 2, and 6 in Arabidopsis) possess phosphorylation target sites for mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) (Tatsuki and Mori, 2001; Hernández Sebastià et al., 2004; Liu and Zhang, 2004). Type-2 ACSs (ACS4, 5, 8, 9, and 11 in Arabidopsis) contain a phosphorylation site for CDPKs and a unique regulatory motif called Target of ETO1 (TOE) in the C-terminus. The TOE motif is the binding site for ETHYLENE OVERPRODUCER1 (ETO1) and its two paralogs, ETO1-LIKE1 and 2 (EOL1 and EOL2). ETO1/EOL1/EOL2 are BTB/TRP-containing E3 ligases that control the degradation of type-2 ACS proteins via the 26S proteasome (Yoshida et al., 2005). In contrast to both type-1 and type-2 ACSs, the single type-3 ACSs does not contain known regulatory motifs in the C-terminus, but as discussed below, an N-terminal motif may control the stability of Arabidopsis ACS7 (Xiong et al., 2014), a sole type 3 isozyme in Arabidopsis.
The protein stability of all three ACS isozyme types is regulated by 14-3-3 proteins (Yoon and Kieber, 2013a). 14-3-3 proteins are an evolutionarily well-conserved family of regulatory proteins involved in numerous cellular processes such as cell cycle regulation, cell division, cell metabolism, proliferation, and protein oligomerization and localization (Dougherty and Morrison, 2004; Darling et al., 2005; Oecking and Jaspert, 2009; Freeman and Morrison, 2011). 14-3-3 activity influences ethylene biosynthesis by destabilizing ETO/EOL proteins and by stabilizing ACS proteins in an ETO/EOL-independent manner (Yoon and Kieber, 2013a). The range of light-dependent developmental phenotypes observed in 14-3-3 LOF mutants (Pnueli et al., 2001; Mayfield et al., 2007; Tseng et al., 2012; Adams et al., 2014) suggests interaction with multiple light signaling components. Although there is no direct evidence that light regulates interactions between 14-3-3 proteins, ACS isozymes, and ETO/EOLs, the 14-3-3s proteins are logical candidates to mediate crosstalk between light signaling and ethylene biosynthesis pathways.
Light-dependent post-translational control of ACS5 (and perhaps other type-2 ACSs) and the associated increase in ethylene production are critical for regulating hypocotyl elongation during the dark-to-light transition. Intriguingly, PIF3 may be involved in this process (Seo and Yoon, 2019). As described above, PIF3 is required for ethylene-induced stimulation of hypocotyl elongation in the light, and ethylene treatment specifically antagonizes light-induced degradation of PIF3 (Zhong et al., 2012). Light-induced stabilization of type-2 ACS enzymes should lead to increased ethylene production, which may play a role in PIF3 stabilization, thereby driving ethylene-induced hypocotyl elongation in the light (Seo and Yoon, 2019). PP2A is another regulatory component that contributes to post-translational regulation of ACS stability. Genetic analysis indicated that PP2A-mediated dephosphorylation negatively controls the protein stability of ACS6 in the dark, but has a much weaker effect on ethylene production in the light (Skottke et al., 2011). Paradoxically, the stability of ACS5, a type-2 isozyme, is positively regulated by PP2A; differential effects on the two isozyme types likely accounts for the lesser effect of PP2A inhibition in light-grown plants (Muday et al., 2006; Skottke et al., 2011).
Compared to type-1 and type-2 ACS isozymes, the sole Arabidopsis type-3 isozyme, ACS7, has unique protein stability characteristics; regulation of ACS7 turnover remains somewhat controversial (Lyzenga et al., 2012; Xiong et al., 2014; Lee et al., 2017). Because of the lack of C-terminal regulatory motifs in type-3 ACS, it was thought that these isozymes might be generally stable compared to other ACS isozymes. However, recent work showed that the stability of type-3 ACS is negatively regulated by ubiquitin-dependent turnover mediated by XBAT32, a RING-type E3 ligase (Lyzenga et al., 2012). Moreover, a putative N-terminal degron of ACS7 is active only in light-grown plants (Xiong et al., 2014) and is poorly conserved (Booker and DeLong, 2015). This light-dependent regulation of ACS7 stability may be similar to the turnover regulation of type-2 ACS, allowing the fine-tuning of ethylene production to impose transient growth control under changing conditions. Considering the regulatory role of the N-terminal domain in ACS7, it may be important to revisit the question of N-terminal motifs that could be involved in regulating the stability of other ACS proteins in response to various stimuli, including light.
The post-translational modifications of ACO have been examined in less detail than those that regulate ACS activity. However, recent work has identified several post-translational mechanisms for controlling ACO activity, including glutathionylation (Dixon et al., 2005) and sulfhydration of cysteine residues on ACO (Friso and van Wijk, 2015). While the effect of glutathionylation on ACO activity has not been reported, S-sulfhydration of LeACO1 and LeACO2 results in a decrease in ACO activity and ethylene production (Jia et al., 2018), establishing an in vivo role for post-translational control of ACO. Determining whether these modifications contribute to light-dependent regulation of ethylene production is an open question for future research.
Mechanistic Connections Between Light Response and the Ethylene Signaling Pathway
Ethylene Receptor Function Is Dependent on Light and Developmental Context
The five ethylene receptors in Arabidopsis are not functionally equivalent, with sub-functionalization observed for responses in different tissues and developmental stages (as reviewed by Shakeel et al., 2013). This subfunctionalization was revealed though detailed phenotypic analysis of LOF receptor mutants (Wang et al., 2003; Binder et al., 2004; Binder et al., 2006; Qu et al., 2007; Liu et al., 2010; McDaniel and Binder, 2012; Wilson et al., 2014b; Bakshi et al., 2015; Harkey et al., 2018). This sub-functionalization is likely due to diversity in receptor structure and signaling capabilities (O’Malley et al., 2005; Wang et al., 2006; Shakeel et al., 2013; Bakshi et al., 2015). Like the central signaling mutant ein2-1, a GOF etr1-3 mutant was insensitive to ethylene or ACC in seedlings growth in light or dark (Guzman and Ecker, 1990; Roman et al., 1994; Negi et al., 2008). In an examination of nutation of etiolated hypocotyls, ethylene-dependent nutations were lost in the etr1-7 LOF mutant no other single receptor LOF mutations affected this process (Binder et al., 2004; Binder et al., 2006). In contrast, the function of EIN4 was light-dependent. In dark-grown seedlings the ein4-1 receptor GOF mutant showed no ethylene response (Roman et al., 1994). When grown in the light, however, ein4-1 seedlings show a partial response to ACC (Smalle et al., 1997), suggesting differences in this receptor’s role in dark vs. light.
The functional role of the five ethylene receptors has been explored in roots of light-grown Arabidopsis seedlings (Harkey et al., 2018). Transcripts encoding all five ethylene receptors are expressed in roots, and the abundance of transcripts encoding three receptors, ETR2, ERS1, and ERS2, is increased by treatments that elevated ethylene (Hua et al., 1998; Harkey et al., 2018). The GOF ETR1 mutant (etr1-3) is insensitive to the effects of ethylene on root elongation, LR development, and root hair initiation (Negi et al., 2008; Lewis et al., 2011a). Using null mutants in each of the five receptors, the major role of ETR1 in controlling root responses to ACC was reported, with subtle changes in development in null mutants in any of the other receptors (Harkey et al., 2018). Using multiple LOF mutants in two or three receptor genes, minor and redundant roles for ETR2 and EIN4 were identified, especially in root hair formation. A triple mutant carrying etr1-6, etr2-3, and ein4-4 LOF mutations has short roots, with no LRs and with extreme proliferation of root hairs. All three phenotypes are largely complemented with a genomic copy of ETR1 (Harkey et al., 2018). These results argue that the ETR1 receptor has a predominant role in controlling ethylene-inhibited LR formation, and ethylene-stimulated root hair initiation in light-grown roots, similar to the major role of this receptor in controlling nutations and responses to silver ions (Shakeel et al., 2013). Two specific receptors regulate the size of the root apical meristem, however (Street et al., 2015). In contrast with findings in LRs and root hairs, LOF etr1-9 or ers1-3 single mutants showed wild-type meristem size, but the LOF etr1-9 ers1-3 double mutant exhibited a substantially reduced root apical meristem size, similar to that found in the ctr1-2 mutant, consistent with multiple receptors controlling this aspect of root development (Street et al., 2015).
The role of specific ethylene receptors in root elongation in dark-grown seedlings has also been reported. Images of dominant GOF mutants in ETR1, ERS1, ERS2, and EIN4 show roots that appear to be ethylene-insensitive (Hua et al., 1995; Hua et al., 1998). Responses to added ACC were quantified for several etr1 and ers1 mutant alleles, which showed reduced sensitivity (Hua et al., 1995). In comparison, the GOF etr2-1 mutant appears to have an intermediate phenotype, with roots shorter in ethylene than in air, but not as short as wild-type roots in ethylene (Sakai et al., 1998). One study observed that subfamily 2 receptors (ETR2, ERS2, and EIN4) are not required for ethylene root response, as the etr1-9 ers1-3 double mutant which carries strong LOF alleles has constitutive ethylene signaling, suggesting that the remaining receptors were not sufficient to repress ethylene signaling (Hall et al., 2012). Additionally, complementation with a wild-type copy of ETR1 was adequate to restore ethylene sensitivity (Hall et al., 2012). Another group assayed phenotypes of receptor mutants in both light and dark (Adams and Turner, 2010), but in the absence of sucrose, which is also known to influence ethylene response (Zhou et al., 1998; Gibson et al., 2001; Yanagisawa et al., 2003; Haydon et al., 2017). Root length in the GOF etr1-1 mutant was unchanged in response to ethylene under conditions of continuous darkness, but not continuous light. Some differences in the responses of other receptor LOF mutants were observed in dark- versus light-grown seedlings, but all receptors were at least partially required under both conditions (Adams and Turner, 2010). Together, these results demonstrate that ethylene receptors in Arabidopsis have distinct functions, dependent on tissue and light context.
The EIN3 and EBFs Mediate Light-Dependent Transcriptional Responses to Ethylene
The EIN3 TF is an essential mediator of ethylene response in hypocotyls of dark-grown seedlings, but its role is more complex in light-grown seedlings. The ein3-1 mutant has ethylene-insensitive hypocotyl elongation in either light- or dark-grown hypocotyls (Chao et al., 1997; Smalle et al., 1997), suggesting that elongation responses to ethylene in the hypocotyl require EIN3 in a light-independent manner. EIN3 also regulates chlorophyll biosynthesis during the dark-to-light transition (Liu et al., 2017). However, the function of EIN3 in roots is light-dependent. In roots of dark-grown seedlings, double mutants between ein3-1 and either eil1-1 or eil1-2 show no response to added ACC, while single mutants in ein3 and eil1 show partial response to this treatment (Alonso et al., 2003). In contrast, in roots of light-grown seedlings, ein3-1, eil1-1, and the double mutant all exhibit ACC-inhibition of root elongation and LR formation, and ACC stimulation of root hair formation (Harkey et al., 2018). A subset of ethylene-responsive transcripts from light-grown roots were identified as binding targets of EIN3 (Harkey et al., 2018) as reported by a DAP-Seq dataset (O’Malley et al., 2016), but many other transcripts were not direct EIN3 targets. These results are consistent with EIN3 and EIL1 controlling only a subset of ethylene responses in roots of light-grown seedlings. One example where there is light-dependent function of EIN3 is in regulation of ACO2 transcript abundance. Upregulation of ACO2 after ethylene treatment was lost in dark-grown ein3-1 mutant seedlings (and EIN3 has been shown to bind to ACO2 via ChIP-Seq) (Chang et al., 2013). In contrast, in light-grown plants, that upregulation was present in the ein3-1 single mutant, but was lost in both ein3-1 eil1-1 and ein3-1 eil1-2 double mutants (Lee et al., 2006), suggesting that EIL1 can compensate for EIN3 in regulating ACO2 only in light-grown plants.
Recent results have suggested that differences in EIN3 function in the light and dark may be controlled at the level of turnover of this protein. Although EIN3 transcript accumulation is not regulated by ethylene (Chao et al., 1997; Harkey et al., 2018), EIN3 and EIL1 protein accumulation is tightly controlled via ethylene-regulated turnover. In the absence of ethylene, EIN3 and EIL1 are ubiquitinated by EIN3-BINDING F-BOX PROTEIN1 and 2 (EBF1 and 2), two F-box proteins that act in SCF complexes, leading to EIN3 degradation. When ethylene levels rise, EBF1 and 2 are targeted for degradation in an EIN2-dependent manner, stabilizing EIN3 (Guo and Ecker, 2003; Gagne et al., 2004; Binder et al., 2007; An et al., 2010). EIN3 and EIL1 protein turnover is also regulated by crosstalk with light signaling via cryptochromes and HY5. The stimulation of hypocotyl elongation by ethylene in light-grown plants requires CRY1 or CRY2 (Vandenbussche et al., 2007), as well as HY5 (Yu et al., 2013). In darkness, CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1), an integrator of light signaling, targets EBF1/2 and HY5 for ubiquitination and degradation, allowing EIN3 accumulation (Shi et al., 2016a), and preventing HY5-mediated inhibition of hypocotyl elongation. Movement of COP1 from the nucleus to the cytoplasm in light conditions allows HY5 to accumulate and inhibit growth. If ethylene signaling is activated in light conditions, EIN3 antagonizes HY5 and stimulates elongation by promoting nuclear localization of COP1, leading to HY5 degradation (Yu et al., 2013). The red light receptor PhyB also directly interacts with EIN3 and EBF1/2 after exposure to red light and enhances degradation of EIN3 (Shi et al., 2016b).
EIN3 regulation of PIF3 and ERF1, which have antagonistic roles in regulating growth, constitutes one of the primary mechanisms driving the inverse hypocotyl responses to ethylene in light versus dark (Zhong et al., 2012). Both PIF3 and ERF1 are direct transcriptional targets of EIN3 (Chang et al., 2013). ERFs are stabilized by light, and they generally inhibit growth. EIN3 upregulates ERF1 both in darkness and in light, but ERF1 effects on hypocotyl growth are only measurable under darkness, where other ERFs are absent. Conversely, pif3 mutants are insensitive to ethylene-induced hypocotyl elongation in light, but not to hypocotyl inhibition in the dark (Zhong et al., 2012). PIFs generally promote elongation, and are destabilized in light, contributing to reduced elongation in light-grown seedlings. Transcriptional regulation of PIF3 by ethylene via EIN3 is inconsequential in darkness, where many other PIFs are also active, but becomes significant under light, where other PIFs are degraded, and PIF3 activation leads to increased hypocotyl growth. EBF1/2 also mediate red light-dependent degradation of PIF3 (Dong et al., 2017). EBFs can synergistically reduce PIF3 levels both directly, by promoting PIF3 degradation, and indirectly, by targeting EIN3 for degradation and thus reducing PIF3 mRNA. This modulation of EIN3 and its targets by light enables complex responses to ethylene under different light contexts, such as opposite response in hypocotyl elongation. As discussed above light-dependent ethylene synthesis may also contribute to PIF3 stabilization and amplification of ethylene responses.
Downstream transcriptional effects of EIN3 and light signaling pathways cannot be completely disentangled. Recent work revealed that an ein3 eil1 double mutant retains shade response, although ethylene-stimulated hypocotyl elongation is abolished (Das et al., 2016), suggesting that shade does not induce hypocotyl elongation by acting directly through the EIN3/EIL1 response pathway. The similar growth effects of ethylene and light are accompanied by many common transcriptional responses (Das et al., 2016). The COP1 effects on EIN3 targets are also complex. COP1 has been shown to increase EIN3 protein levels by targeting EBF1/2 for degradation in the dark (Shi et al., 2016a). In the light, ACC treatment and EIN3 overexpression lead to increased transcript levels of growth-promoting genes such as YUCCA1 and 5. This effect is lost in the dark but is restored in the cop1-4 null mutant (Liang et al., 2012). This suggests that COP1 works by some mechanism downstream of EIN3 to fine tune expression of these particular genes so that they promote elongation in the light, but not in the dark. EIN3 and PIF1 transcriptionally regulate many of the same gene targets independently from one another, but mostly in the same direction (Jeong et al., 2016), and EIN3 and PIF1 pathways are each sufficient to maintain skotomorphogenesis (Shi et al., 2018). Overlapping transcriptional responses are also involved in EIN3/EIL1- and PIF3-mediated regulation of hypocotyl hook opening (Zhang et al., 2018). Downstream transcription factors, such as ERF72, may also have activity that is modulated by light to influence developmental responses (Liu et al., 2018). As described above, differential regulation of specific proteins, such as HY5, contributes to the opposing ethylene effects observed in light and dark (Smalle et al., 1997).
Downstream Ethylene Transcriptional Effects Are Influenced by Light
A number of ethylene transcriptome studies have been performed with plants grown under a range of light conditions, revealing distinct transcriptional networks downstream of ethylene perception. We previously compared a dataset from dark-grown seedlings treated with ethylene (Chang et al., 2013) with another dataset from light-grown roots treated with ACC (Harkey et al., 2018). Both datasets used similar time points across a 24-h period after treatment, and we used the same statistical analysis of both datasets. However, we found limited overlap in differentially expressed (DE) genes (71 common genes out of 449 in the light-grown root dataset and out of 971 in the dark-grown seedling dataset). In principle, these changes could be explained by differences in light condition, tissue type, and/or method of elevating ethylene levels (ACC treatment vs. ethylene gas). This last possibility seems unlikely because all ACC responses were lost in the ethylene-insensitive etr1-3 and ein2-5 mutants (Harkey et al., 2018). Comparing a larger number of transcriptomic data sets is essential for more complete understanding of the light-dependent effects of ethylene on transcript accumulation.
To identify transcriptional responses to ethylene that are light- and tissue-specific, we looked for datasets that were suitable for a meta-analysis that could resolve differences and similarities in ethylene-responsive transcriptomes in the light and dark. We searched the Gene Expression Omnibus (GEO) for the term “ethylene.” Twenty-five datasets were identified in the original search based on treatment with ACC, ethylene, or with compounds that block ethylene synthesis (such as AVG), and/or mutations or transgenes that alter ethylene production or response. Many of these datasets were not usable because of dissimilar approaches or incomplete information. Five datasets were excluded due to insufficient information on experimental methods; another five used specific mutants or transgenic lines that were not found in any other dataset and did not include wild-type seedlings treated with ACC or ethylene. Although there were many datasets utilizing Col-0 and/or ein2, ein3, and eil1 mutants in light and dark conditions, they used experimental methods, tissue types, or plants that were not developmentally matched. Seven additional datasets used 3- or 4-day-old whole dark-grown seedlings, while the remaining five datasets came from light-grown material using a variety of ages and tissue types. This highlights the need for future work that directly compares ethylene effects in light versus dark.
Ultimately, we identified three datasets with highly similar experimental methods and plant age in which transcript abundance was quantified after 4 h of ethylene or ACC treatment in roots (Stepanova et al., 2007; Feng et al., 2017; Harkey et al., 2018), and a fourth that provided an interesting comparison between ethylene treatment and shade treatment in hypocotyls or in cotyledons (Das et al., 2016). The most relevant differences between the three root datasets can be found in Figure 3, and further details on the process of identifying these datasets can be found in Supplemental Datasheet 1, along with a description of the experimental conditions used in each study. The fourth dataset was of particular interest because the authors compared the transcriptional effects of shade and ethylene in experimental conditions that were otherwise identical (Das et al., 2016). The authors noted that the effect of combined shade and ethylene on hypocotyl elongation was intermediate between the two individual treatments, consistent with ethylene and light signaling pathways sharing downstream signaling and/or effector components. However, samples treated with both ethylene and shade were not included in the transcriptomic analysis. Among genes that responded to ethylene and shade consistently and with the same direction of change, the authors found enrichments for annotations including hormone signaling, cell wall, and photomorphogenesis, among others, as well as two TFs, AtHB28 and IBL1. Analysis of mutant and overexpression lines showed that AtHB28 and IBL1 are important for both shade and ethylene response (Das et al., 2016).
We developed a statistical pipeline to apply to all datasets used in our analysis to avoid discrepancies that might arise from differences in data analysis methods. We generated lists of DE genes that could more properly be compared to one another. (Note that this re-analysis results in DE lists that differ from those derived in the original publications.) For the three root datasets, we combined expression data from all three experiments into one master dataframe; both this dataframe and the Das et al. dataset were analyzed for differential expression using limma and other packages in R (Davis and Meltzer, 2007; R Core Team, 2014; Ritchie et al., 2015; Gu et al., 2016). Additional details of these analyses can be found in Supplemental Datasheet 1.
To identify the entire overlap between ethylene and shade transcriptional responses in the Das et al. (2016) dataset, we used this data analysis pipeline. First, we identified the complete set of ethylene-responsive genes, and then queried their expression responses in the shade dataset. Compared to cotyledons, hypocotyls showed a greater response to ethylene, which is expected given the changes in hypocotyl growth that occur in etiolated seedlings treated with ethylene, described above, so we focused on that tissue type. Not surprisingly, of the 7,248 hypocotyl transcripts that showed a significant response to ethylene, more than half of those genes also showed a shade response (4,239; Figure 2A). The majority of these gene expression changes occurred in the same direction and with similar kinetics. Full results for all ethylene-responsive transcripts can be found in Supplemental Datasheet 2.
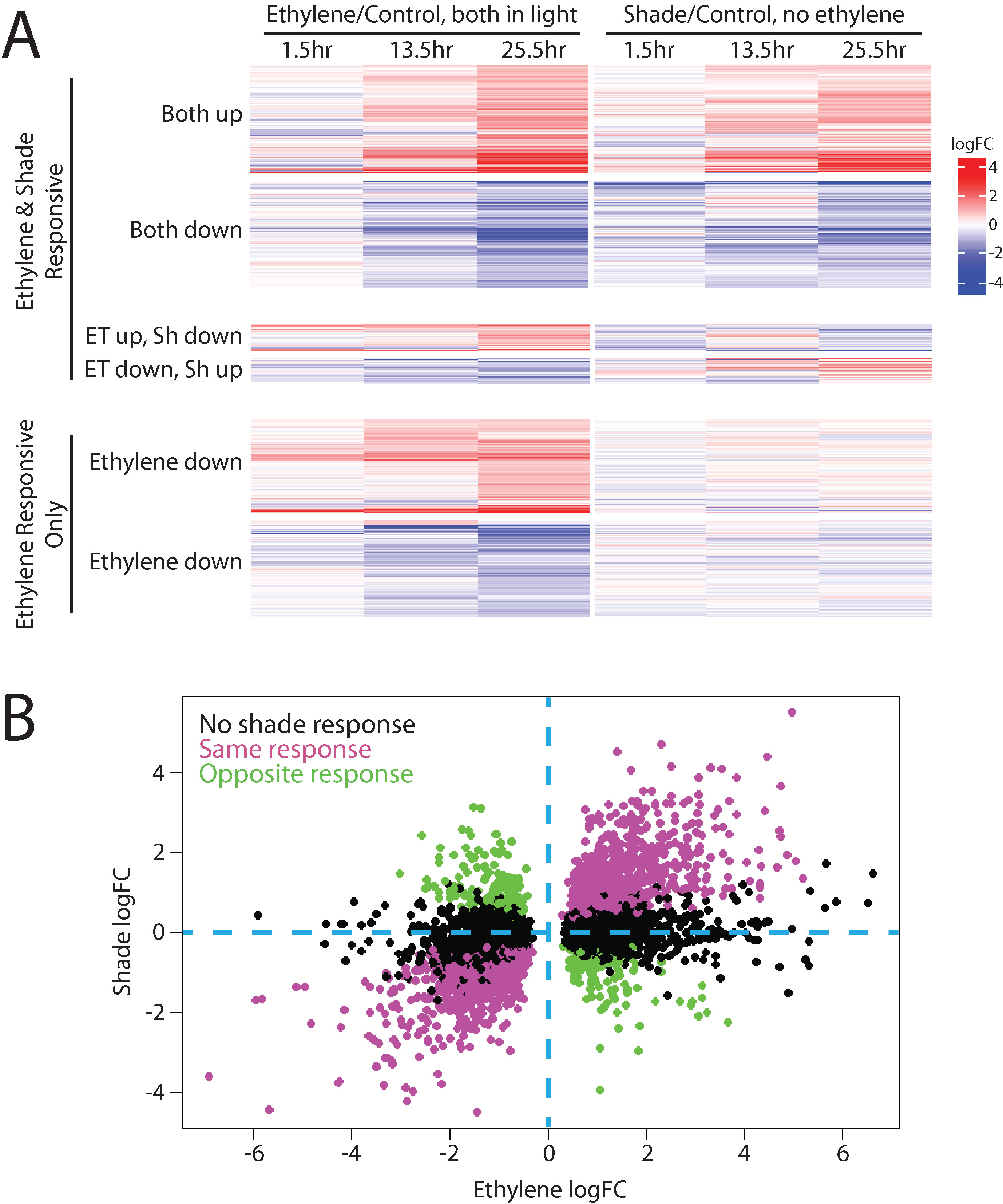
Figure 2 Ethylene and shade regulate many of the same genes in Arabidopsis hypocotyls. A transcriptional dataset in which seedlings were grown in the light and then either treated with ethylene or moved to shade (Das et al., 2016) and were refiltered as described in Supplemental Datasheet 1, revealing that many transcripts share both ethylene and shade regulation. (A) A heat map, generated using the Complex Heatmaps package in R (Gu et al., 2016), shows transcripts that had statistically significant responses to ethylene in at least one time point and how those transcripts responded to shade treatment. Most genes regulated by ethylene were also regulated by shade, with the majority changing in the same direction and a smaller subset changing in opposite directions, and with a limited number of transcripts showing no response to shade. (B) To better define the relationship between magnitude change in response to ethylene and light, the transcripts that showed significant changes in abundance with ethylene treatment in the 25.5 h sample (which showed most dramatic ethylene-induced abundance changes) were plotted as a function of their change in response to shading. Genes that were also regulated by shade in this dataset showed strong statistical correlations between ethylene logFC and shade logFC (positive for genes with the same direction of regulation (Pearson’s correlation, r = 0.89, p < 0.001), and negative for genes with the opposite direction of regulation (Pearson’s correlation, r = −0.87, p < 0.001).
To better illustrate the relationship between ethylene and shade response, we plotted the log2 fold-changes in transcripts in response to ethylene against the fold-change in response to transition to shade (using the 25.5-h time point, which showed the most striking changes from the control) using the previously published transcript abundance values from Das et al. (2016). This graph highlights the strong correlation between ethylene response and shade response (Figure 2B). The correlation between the magnitude of change in response to ethylene and shade is statistically significant both for genes with the same direction of response (Pearson’s correlation, r = 0.89, p < 0.001) and in genes with the opposite direction of response (Pearson’s correlation, r = −0.87, p < 0.001). Dark- or shade-grown plants exhibit a different transcriptional landscape than their light-grown counterparts. Our analysis illustrates that many transcripts show similar responses to ethylene and shade; thus, studies that use dark-grown tissues to examine ethylene response will likely miss changes that occur only in light-grown plants.
We performed a meta-analysis using the three root-specific ethylene-response datasets identified as sufficiently matched for comparison (Stepanova et al., 2007; Feng et al., 2017; Harkey et al., 2018) to screen for light-dependent and light-independent changes in ethylene-regulated transcript abundance. We used our new pipeline to reanalyze the root-specific transcriptomes to identify differences that are linked to the light environment of seedling growth. This analysis yielded interesting patterns of light-dependent and light-independent changes in transcript abundance that are summarized in a Venn Diagram in Figure 3. A list of all transcripts that showed significant responses to ethylene or ACC in at least one dataset and their magnitude of change can be found in Supplemental Datasheet 2. As expected, many more DE genes were identified in the RNA-seq dataset (Feng et al., 2017) than in the microarray-based datasets (Stepanova et al., 2007; Harkey et al., 2018), because RNA-Seq has a greater dynamic range. Although only 3% of the DE genes identified responded to ethylene in all three datasets, nearly a third (32%) were DE in two datasets. A number of genes were DE in the two datasets from light-grown seedlings (Feng et al., 2017; Harkey et al., 2018), but not in the dark (Stepanova et al., 2007), suggesting light-dependent regulation by ethylene. There was also substantial overlap (433 transcripts) between the two datasets that used ethylene treatment but differed in the presence of light during growth. We identified 169 transcripts in the overlap between the dark-grown ethylene dataset (Stepanova et al., 2007) and light-grown ACC dataset (Harkey et al., 2018). This number is greater than in our previously reported comparison of these two datasets (80, transcripts; Harkey et al., 2018), due to the common filtering used for both datasets in this meta-analysis. A surprising number of genes, however, were specifically regulated in one dataset, and not in the other two, despite the similarity of experimental techniques. These differences may be related to other conditions such as plant age (3, 5, or 6 days), light cycle (continuous light vs. 16 h light 8 h dark), or differences in media (e.g., sucrose concentration, which is also known to influence ethylene response; Gibson et al., 2001; Haydon et al., 2017; Yanagisawa et al., 2003). These results demonstrate the need for direct comparisons of ethylene effects under experimental conditions that vary only by light level.
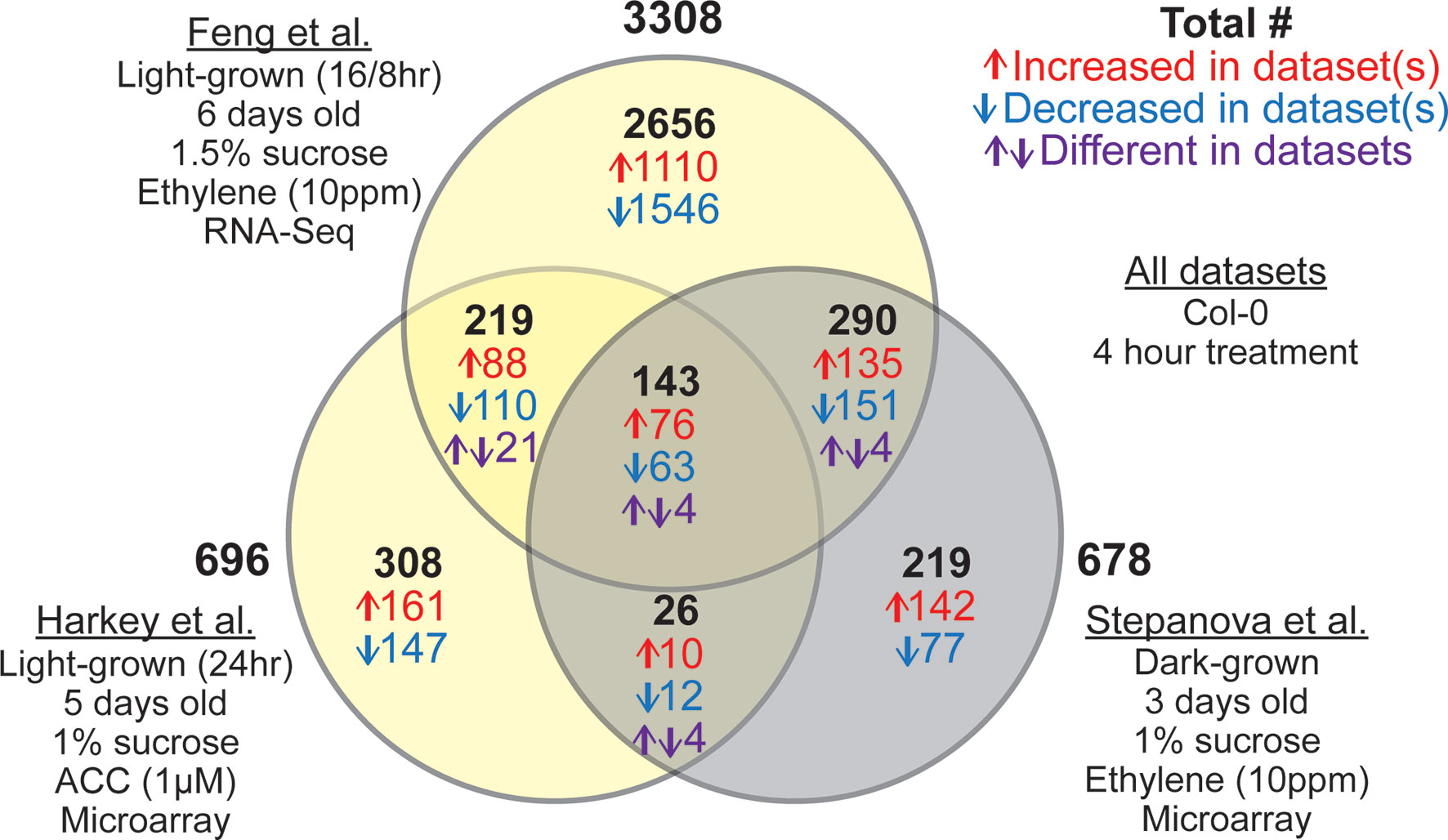
Figure 3 Three root-specific ethylene response datasets show light-dependent and light-independent overlaps. Venn diagram represents number of overlapping and non-overlapping DE genes between three root-specific transcriptomic datasets: Stepanova et al. (2007), Harkey et al. (2018), and Feng et al. (2017). Differences in experimental conditions are summarized under each dataset name. Details of the analysis can be found in Supplemental Datasheet 1. Once DE lists were generated for individual datasets, we compared the lists to find overlapping and non-overlapping genes. In the Venn diagram, the two light-grown datasets are represented in yellow, and the dark-grown dataset is represented in gray. The number of transcripts within each overlap are color coded, with the total in black, the number increasing in both or all three in red, the number decreasing in blue, and purple indicating transcripts that changed in different directions between datasets.
In addition to the light-specific transcripts described above, this analysis identified a core set of 143 transcripts that responded to ethylene or ACC in all three datasets, regardless of light. Of these transcripts, 139 (97%) changed in the same direction in all treatments (Figure 3). This set of 139 genes with consistent direction of change should be considered the “gold standard,” for root ethylene response, much like a previously identified set of cytokinin-responsive genes from another meta-analysis (Bhargava et al., 2013). The full list of ethylene- or ACC-responsive genes from any dataset can be found in Supplemental Datasheet 2 , with “gold standard” genes indicated.
A subset of the “gold standard” genes is summarized in Table 1. This group of 44 genes was chosen based on three criteria: the largest logFC values (in the positive or negative direction), known roles in ethylene synthesis or signaling (highlighted in red in Table 1), and/or known EIN3 targets based on DAP-Seq (O’Malley et al., 2016) and/or CHiP-Seq (Chang et al., 2013) analysis. Interestingly, most of the upregulated “gold” genes were identified as EIN3 targets by at least one method (72.4%), but very few downregulated “gold” genes were bound by EIN3 (6.3%). “Gold standard” genes also included a number of auxin-related genes (e.g., SAUR76, SAUR8, IAA2, and IAA4/AUX2-11), and genes involved in cell wall regulation (e.g., a pectin methylesterase inhibitor). Not surprisingly, the 139 transcripts were also enriched in gene annotations for cellular response to ethylene stimulus and negative regulation of the ethylene pathway.
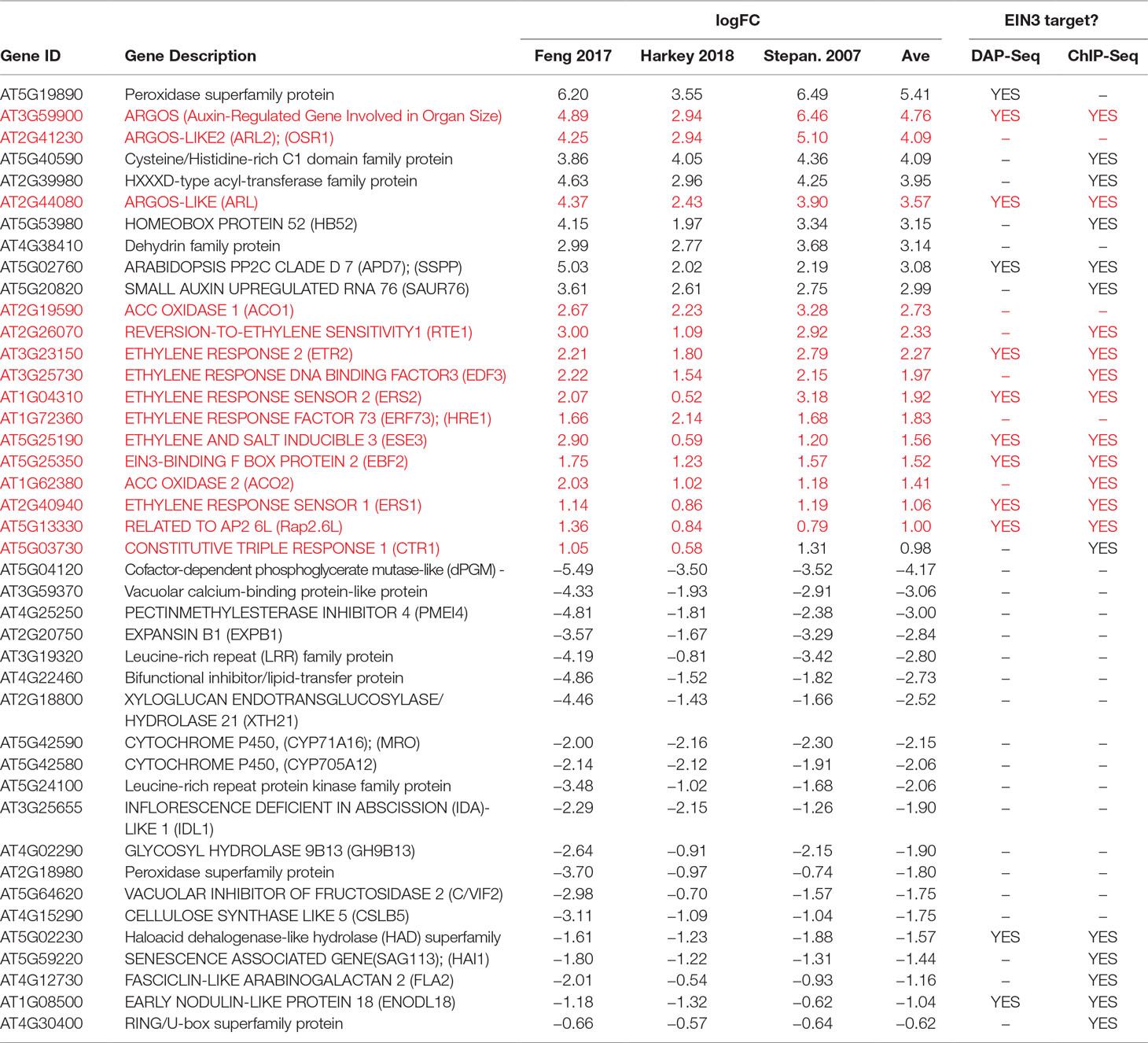
Table 1 Selected gold standard transcripts regulated in all three datasets. The transcripts in red are all implicated in ethylene signaling or synthesis.
Within this group of 139 transcripts, we identified 13 core genes in ethylene signaling or synthesis whose levels increased in all three datasets (and in Das et al., 2016). This core gene set includes genes encoding TFs that participate in ethylene signaling (for example, EDF1, EDF3, EDF4, and several ERFs), negative regulators of the signaling pathway CTR1, EBF2 and ARGOS, and the ethylene receptors ETR2, ERS1, and ERS2. Thus, a core output of the ethylene response is upregulation of its own signaling pathway components including both positive and negative regulators of ethylene responses. The core set also includes transcripts encoding ethylene biosynthetic proteins. There is consistent upregulation of transcripts encoding the ACO enzymes, with ACO1 and ACO2 upregulated in all three datasets and ACO3, ACO4, and ACO5 upregulated in two of the three datasets. ACO2 was also upregulated by ethylene, although down-regulated in shade in Das et al. (2016). Interestingly, ACS transcript levels show less consistent positive regulation, showing no changes for any ACS gene in two datasets (Stepanova et al., 2007; Harkey et al., 2018) and changes in only two to four ACS transcripts (out of 11 family members) in two other data sets (Das et al., 2016; Feng et al., 2017). These results indicate that a positive feedback loop drives ethylene synthesis via upregulation of ACO expression, while ACS mRNA levels appear to be subject to a more complex control network, as discussed above (see Light-Mediated Transcriptional Regulation of ACS and ACO).
Finally, included in this comparison is an annotation of genes that are regulated by ethylene in dark-grown whole seedlings as detected by RNA-Seq (Chang et al., 2013) (as found in a separate column in the Supplemental Datasheet 2). Of the 77 up-regulated genes in the gold-standard list, 40 were also found to be sites of EIN3 binding while only 2 of the 62 down-regulated genes showed ethylene-regulated expression. Therefore, one can further refine these genes into root-specific and tissue-independent transcripts, using the detailed annotations in Supplemental Datasheet 2. Together, this meta-analysis reveals many candidate genes for conserved ethylene responses that are also induced by the ethylene precursor, ACC, and transcripts whose responses depend on light or tissue type. This information can allow formulation of a wealth of hypotheses that can be tested to further refine our understanding of ethylene signaling across plant development.
Conclusions
As seedlings germinate, elongate through soil, and then emerge into light, they undergo profound changes in development. The importance of ethylene levels in controlling development is best understood in the early dark phases, but new studies that examine the role of ethylene during developmental transitions from dark to light or in light-dependent development are providing new insight into the functions of ethylene during seedling development. Recent studies have revealed novel mechanisms that modulate ethylene biosynthesis, including important transcriptional and post-translational regulatory strategies that control production of this hormone. The pathways that control ethylene response include central signaling proteins that function in ethylene response under all conditions, but also receptors and transcription factors with light- and developmental stage-specific functions. Comparison of genome-wide transcriptional datasets allows identification of candidate genes that contribute to all ethylene responses and other genes that may contribute to developmental outputs that are specific to the light environment. Together, light regulation of ethylene biosynthesis, signaling, and developmental response have far-reaching effects on a plant’s ability to adapt to the environment in early stages of development and throughout the life cycle. Understanding the mechanisms by which light and ethylene interact at the molecular and organismal levels is an important goal of future research.
Author Contributions
AH performed the meta-analysis, drafted text, prepared figures, and edited the manuscript; GY drafted text, prepared figures, and edited the manuscript; DS prepared figures; AD and GM drafted text and edited the manuscript.
Funding
This work was supported by grants from the US National Science Foundation to GM (MCB-1716279) and GY (MCB-1817286).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the assistance of Joëlle Mühlemann with the meta-analysis and the helpful comments on the manuscript from Emily Martin.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01094/full#supplementary-material
Supplemental Datasheet 1 | Extended methods for meta-analysis of ethylene-regulated transcripts and ethylene- and shade-regulated transcripts
Supplemental Datasheet 2 | Summary of differentially expressed transcripts from roots in response to ethylene or ACC and overlap of shade-and ethylene-regulated transcriptomes
References
Abbas, M., Alabadí, D., Blázquez, M. A. (2013). Differential growth at the apical hook: all roads lead to auxin. Front. Plant Sci. 4, 441. doi: 10.3389/fpls.2013.00441
Adams, D. O., Yang, S. F. (1979). Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. 76, 170–174. doi: 10.1073/pnas.76.1.170
Adams, E., Diaz, C., Hong, J.-P., Shin, R. (2014). 14-3-3 proteins participate in light signaling through association with PHYTOCHROME INTERACTING FACTORs. Int. J. Mol. Sci. 15, 22801–22814. doi: 10.3390/ijms151222801
Adams, E., Turner, J. (2010). COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J. Exp. Bot. 61, 4373–4386. doi: 10.1093/jxb/erq240
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S. D., Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. doi: 10.1126/science.284.5423.2148
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. 100, 2992–2997. doi: 10.1073/pnas.0438070100
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Argueso, C. T., Hansen, M., Kieber, J. J. (2007). Regulation of ethylene biosynthesis. J. Plant Growth Regul. 26, 92–105. doi: 10.1007/s00344-007-0013-5
Arteca, R. N., Arteca, J. M. (2008). Effects of brassinosteroid, auxin, and cytokinin on ethylene production in Arabidopsis thaliana plants. J. Exp. Bot. 59, 3019–3026. doi: 10.1093/jxb/ern159
Bakshi, A., Wilson, R. L., Lacey, R. F., Kim, H., Wuppalapati, S. K., Binder, B. M. (2015). Identification of regions in the receiver domain of the ETHYLENE RESPONSE1 ethylene receptor of Arabidopsis important for functional divergence. Plant Physiol. 169, 219–232. doi: 10.1104/pp.15.00626
Bhargava, A., Clabaugh, I., To, J. P., Maxwell, B. B., Chiang, Y.-H., Schaller, G. E., et al. (2013). Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis1[C][W][OA]. Plant Physiol. 162, 272–294. doi: 10.1104/pp.113.217026
Binder, B. M., Malley, R. C. O., Wang, W., Moore, J. M., Parks, B. M., Spalding, E. P., et al. (2004). Arabidopsis seedling growth response and recovery to ethylene. A Kinetic Analysis. Plant Physiol. 136, 2913–2920. doi: 10.1104/pp.104.050369
Binder, B. M., O’Malley, R. C., Wang, W., Zutz, T. C., Bleecker, A. B. (2006). Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol. 142, 1690–1700. doi: 10.1104/pp.106.087858
Binder, B. M., Walker, J. M., Gagne, J. M., Emborg, T. J., Hemmann, G., Bleecker, A. B., et al. (2007). The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell Online 19, 509–523. doi: 10.1105/tpc.106.048140
Bleecker, A. B., Estelle, M. A., Somerville, C., Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. doi: 10.1126/science.241.4869.1086
Booker, M. A., DeLong, A. (2015). Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes1. Plant Physiol. 169, 42–50. doi: 10.1104/pp.15.00672
Buer, C. S., Sukumar, P., Muday, G. K. (2006). Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140, 1384–1396. doi: 10.1104/pp.105.075671
Buer, C. S., Wasteneys, G. O., Masle, J. (2003). Ethylene modulates root-wave responses in Arabidopsis. Plant Physiol. 132, 1085–1096. doi: 10.1104/pp.102.019182
Chae, H. S., Kieber, J. J. (2005). Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10, 291–296. doi: 10.1016/j.tplants.2005.04.006
Chang, C., Kwok, S., Bleecker, A. B., Meyerowitz, E. M. (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262, 539–544. doi: 10.1126/science.8211181
Chang, K. N., Zhong, S., Weirauch, M. T., Hon, G., Pelizzola, M., Li, H., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2, e00675. doi: 10.7554/eLife.00675
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
Collett, C. E., Harberd, N. P., Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124, 553–562. doi: 10.1104/pp.124.2.553
Darling, D. L., Yingling, J., Wynshaw-Boris, A. (2005). Role of 14–3–3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68, 281–315. doi: 10.1016/S0070-2153(05)68010-6
Das, D., Onge, K. R. S., Voesenek, L. A. C. J., Pierik, R., Sasidharan, R. (2016). Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172, 718–733. doi: 10.1104/pp.16.00725
Davis, S., Meltzer, P. S. (2007). GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinforma. Oxf. Engl. 23, 1846–1847. doi: 10.1093/bioinformatics/btm254
Dixon, D. P., Skipsey, M., Grundy, N. M., Edwards, R. (2005). Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138, 2233–2244. doi: 10.1104/pp.104.058917
Dolan, L. (2001). The role of ethylene in root hair growth in Arabidopsis. J. Plant Nutr. Soil Sci. 164, 141–145. doi: 10.1002/1522-2624(200104)164:2<141::AID-JPLN141>3.0.CO;2-Z
Dong, J., Ni, W., Yu, R., Deng, X. W., Chen, H., Wei, N. (2017). Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr. Biol. 27, 2420+. doi: 10.1016/j.cub.2017.06.062
Dougherty, M. K., Morrison, D. K. (2004). Unlocking the code of 14-3-3. J. Cell Sci. 117, 1875–1884. doi: 10.1242/jcs.01171
El-Sharkawy, I., Kim, W. S., Jayasankar, S., Svircev, A. M., Brown, D. C. W. (2008). Differential regulation of four members of the ACC synthase gene family in plum. J. Exp. Bot. 59, 2009–2027. doi: 10.1093/jxb/ern056
Feng, Y., Xu, P., Li, B., Li, P., Wen, X., An, F., et al. (2017). Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. 114, 13834–13839. doi: 10.1073/pnas.1711723115
Foo, E., Ross, J. J., Davies, N. W., Reid, J. B., Weller, J. L. (2006). A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J. 46, 911–921. doi: 10.1111/j.1365-313X.2006.02754.x
Freeman, A. K., Morrison, D. K. (2011). 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 22, 681–687. doi: 10.1016/j.semcdb.2011.08.009
Friso, G., van Wijk, K. J. (2015). Posttranslational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487. doi: 10.1104/pp.15.01378
Gagne, J. M., Smalle, J., Gingerich, D. J., Walker, J. M., Yoo, S.-D., Yanagisawa, S., et al. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 6803–6808. doi: 10.1073/pnas.0401698101
Gibson, S. I., Laby, R. J., Kim, D. (2001). The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem. Biophys. Res. Commun. 280, 196–203. doi: 10.1006/bbrc.2000.4062
Goeschl, J., Pratt, H., Bonner, B. (1967). An effect of light on production of ethylene and growth of plumular portion of etiolated pea seedlings. Plant Physiol. 42, 1077–. doi: 10.1104/pp.42.8.1077
Gu, Z., Eils, R., Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma. Oxf. Engl. 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Guo, H., Ecker, J. R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2- dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. doi: 10.1016/S0092-8674(03)00969-3
Guzman, P., Ecker, J. R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. doi: 10.1105/tpc.2.6.513
Hall, B. P., Shakeel, S. N., Amir, M., Ul Haq, N., Qu, X., Schaller, G. E. (2012). Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 159, 682–695. doi: 10.1104/pp.112.196790
Harkey, A. F., Watkins, J. M., Olex, A. L., DiNapoli, K. T., Lewis, D. R., Fetrow, J. S., et al. (2018). Identification of transcriptional and receptor networks that control root responses to ethylene. Plant Physiol. 176, 2095–2118. doi: 10.1104/pp.17.00907
Haydon, M. J., Mielczarek, O., Frank, A., Roman, A., Webb, A. A. R. (2017). Sucrose and ethylene signaling interact to modulate the circadian clock. Plant Physiol. 175, 947–958. doi: 10.1104/pp.17.00592
Hernández Sebastià, C., Hardin, S. C., Clouse, S. D., Kieber, J. J., Huber, S. C. (2004). Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 428, 81–91. doi: 10.1016/j.abb.2004.04.025
Houben, M., Van de Poel, B. (2019). 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 10, 695. doi: 10.3389/fpls.2019.00695
Hua, J., Chang, C., Sun, Q., Meyerowitz, E. M. (1995). Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269, 1712–1714. doi: 10.1126/science.7569898
Hua, J., Meyerowitz, E. M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. doi: 10.1016/S0092-8674(00)81425-7
Hua, J., Sakai, H., Nourizadeh, S., Chen, Q. G., Bleecker, A. B., Ecker, J. R., et al. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332. doi: 10.1105/tpc.10.8.1321
Huang, Y., Li, H., Hutchison, C. E., Laskey, J., Kieber, J. J. (2003). Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 33, 221–233. doi: 10.1046/j.1365-313X.2003.01620.x
Ivanchenko, M. G., Muday, G. K., Dubrovsky, J. G. (2008). Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 55, 335–347. doi: 10.1111/j.1365-313X.2008.03528.x
Jeong, J., Kim, K., Kim, M. E., Kim, H. G., Heo, G. S., Parka, O. K., et al. (2016). Phytochrome and ethylene signaling integration in Arabidopsis occurs via the transcriptional regulation of genes co-targeted by PIFs and EIN3. Front. Plant Sci. 7, 1055. doi: 10.3389/fpls.2016.01055
Jia, H., Chen, S., Liu, D., Liesche, J., Shi, C., Wang, J., et al. (2018). Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 9, 1517. doi: 10.3389/fpls.2018.01517
Ju, C., Mee, G., Marie, J., Lin, D. Y., Ying, Z. I., Chang, J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis, PNAS 109, 19486–19491. doi: 10.1073/pnas.1214848109
Kendrick, M. D., Chang, C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485. doi: 10.1016/j.pbi.2008.06.011
Kevany, B. M., Tieman, D. M., Taylor, M. G., Cin, V. D., Klee, H. J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467. doi: 10.1111/j.1365-313X.2007.03170.x
Khanna, R., Shen, Y., Marion, C. M., Tsuchisaka, A., Theologis, A., Schäfer, E., et al. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929. doi: 10.1105/tpc.107.051508
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., Ecker, J. R., Kieber, J. J., et al. (1993). CTRI, a negative regulator of the ethylene pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. doi: 10.1016/0092-8674(93)90119-B
Kim, J. H., Kim, W. T., Kang, B. G., Yang, S. F. (1997). Induction of 1-aminocyclopropane-1-carboxylate oxidase mRNA by ethylene in mung bean hypocotyls: involvement of both protein phosphorylation and dephosphorylation in ethylene signaling. Plant J. 11, 399–405. doi: 10.1046/j.1365-313X.1997.11030399.x
Klee, H. J., Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. doi: 10.1146/annurev-genet-110410-132507
Lanahan, M. B., Yen, H. C., Giovannoni, J. J., Klee, H. J. (1994). The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6, 521–530. doi: 10.1105/tpc.6.4.521
Le, J., Vandenbussche, F., De Cnodder, T., Van Der Straeten, D., Verbelen, J.-P. (2005). Cell Elongation and Microtubule Behavior in the Arabidopsis Hypocotyl: responses to Ethylene and Auxin. J. Plant Growth Regul. 24, 166–178. doi: 10.1007/s00344-005-0044-8
Lee, H. Y., Chen, Y.-C., Kieber, J. J., Yoon, G. M. (2017). Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J. 91, 491–504. doi: 10.1111/tpj.13585
Lee, H. Y., Chen, Z., Zhang, C., Yoon, G. M. (2019). Editing of the OsACS locus alters phosphate deficiency-induced adaptive responses in rice seedlings. J. Exp. Bot. 70, 1927–1940. doi: 10.1093/jxb/erz074
Lee, J.-H., Deng, X. W., Kim, W. T. (2006). Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem. Biophys. Res. Commun. 350, 484–491. doi: 10.1016/j.bbrc.2006.09.074
Lewis, D. R., Negi, S., Sukumar, P., Muday, G. K. (2011a). Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138, 3485–3495. doi: 10.1242/dev.065102
Lewis, D. R., Ramirez, M. V., Miller, N. D., Vallabhaneni, P., Ray, W. K., Helm, R. F., et al. (2011b). Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156, 144–164. doi: 10.1104/pp.111.172502
Li, W., Ma, M., Feng, Y., Li, H., Wang, Y., Ma, Y., et al. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163, 670–683. doi: 10.1016/j.cell.2015.09.037
Liang, X., Wang, H., Mao, L., Hu, Y., Dong, T., Zhang, Y., et al. (2012). Involvement of COP1 in ethylene- and light-regulated hypocotyl elongation. Planta 236, 1791–1802. doi: 10.1007/s00425-012-1730-y
Lin, Z., Zhong, S., Grierson, D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. doi: 10.1093/jxb/erp204
Liu, K., Li, Y., Chen, X., Li, L., Liu, K., Zhao, H., et al. (2018). ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J. Exp. Bot. 69, 3933–3947. doi: 10.1093/jxb/ery220
Liu, Q., Xu, C., Wen, C.-K. (2010). Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 10, 60. doi: 10.1186/1471-2229-10-60
Liu, X., Liu, R., Li, Y., Shen, X., Zhong, S., Shi, H. (2017). EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried Seedlings. Plant Cell 29, 3051–3067. doi: 10.1105/tpc.17.00508
Liu, Y., Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Lyzenga, W. J., Booth, J. K., Stone, S. L. (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71, 23–34. doi: 10.1111/j.1365-313X.2012.04965.x
Mayfield, J. D., Folta, K. M., Paul, A.-L., Ferl, R. J. (2007). The 14-3-3 proteins μ and υ influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702. doi: 10.1104/pp.107.108654
Mazzella, M. A., Casal, J. J., Muschietti, J. P., Fox, A. R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5, 52. doi: 10.3389/fpls.2014.00052
McDaniel, B. K., Binder, B. M. (2012). Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J. Biol. Chem. 287, 26094–26103. doi: 10.1074/jbc.M112.383034
Merchante, C., Binder, B. M., Heber, S. (2015). Gene-specific translation regulation mediated by the Hormone-signaling molecule EIN2 article gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684–697. doi: 10.1016/j.cell.2015.09.036
Muday, G. K., Brady, S. R., Argueso, C., Deruere, J., Kieber, J. J., DeLong, A. (2006). RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol. 141, 1617–1629. doi: 10.1104/pp.106.083212
Muday, G. K., Rahman, A., Binder, B. M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195. doi: 10.1016/j.tplants.2012.02.001
Negi, S., Ivanchenko, M. G., Muday, G. K. (2008). Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 55, 175–187. doi: 10.1111/j.1365-313X.2008.03495.x
Negi, S., Sukumar, P., Liu, X., Cohen, J. D., Muday, G. K. (2010). Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 61, 3–15. doi: 10.1111/j.1365-313X.2009.04027.x
Oecking, C., Jaspert, N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12, 760–765. doi: 10.1016/j.pbi.2009.08.003
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
O’Malley, R. C., Rodriguez, F. I., Esch, J. J., Binder, B. M., O’Donnell, P., Klee, H. J., et al. (2005). Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41, 651–659. doi: 10.1111/j.1365-313X.2004.02331.x
Peck, S. C., Kende, H. (1995). Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in etiolated peas. Plant Mol. Biol. 28, 293–301. doi: 10.1007/BF00020248
Peck, S. C., Pawlowski, K., Kende, H. (1998). Asymmetric responsiveness to ethylene mediates cell elongation in the apical hook of peas. Plant Cell 10, 713–719. doi: 10.1105/tpc.10.5.713
Pitts, R. J., Cernac, A., Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. doi: 10.1046/j.1365-313x.1998.00321.x
Plett, J. M., Cvetkovska, M., Makenson, P., Xing, T., Regan, S. (2009). Arabidopsis ethylene receptors have different roles in Fumonisin B1-induced cell death. Physiol. Mol. Plant Pathol. 74, 18–26. doi: 10.1016/j.pmpp.2009.08.004
Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N., et al. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702. doi: 10.1105/tpc.010293
Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., et al. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. doi: 10.1016/S0092-8674(03)00968-1
Qiao, H., Chang, K. N., Yazaki, J., Ecker, J. R. (2009). Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521. doi: 10.1101/gad.1765709
Qiao, H., Shen, Z., Huang, S. C., Schmitz, R. J., Mark, A., Briggs, S. P., et al. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338, 390–393. doi: 10.1126/science.1225974
Qu, X., Hall, B. P., Gao, Z., Schaller, G. E. (2007). A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 7, 3. doi: 10.1186/1471-2229-7-3
R Core Team (2014), R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: http://www.r-project.org/.
Rahman, A., Amakawa, T., Goto, N., Tsurumi, S. (2001). Auxin is a positive regulator for ethylene-mediated response in the growth of arabidopsis roots. Plant Cell Physiol. 42, 301–307. doi: 10.1093/pcp/pce035
Rahman, A., Hosokawa, S., Oono, Y., Amakawa, T., Goto, N., Tsurumi, S. (2002). Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 130, 1908–1917. doi: 10.1104/pp.010546
Rashotte, A. M., Chae, H. S., Maxwell, B. B., Kieber, J. J. (2005). The interaction of cytokinin with other signals. Physiol. Plant. 123, 184–194. doi: 10.1111/j.1399-3054.2005.00445.x
Raz, V., Ecker, J. R. (1999). Regulation of differential growth in the apical hook of Arabidopsis. Dev. Camb. Engl. 126, 3661–3668.
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Rodrigues, M. A., Bianchetti, R. E., Freschi, L. (2014). Shedding light on ethylene metabolism in higher plants. Front. Plant Sci. 5, 665. doi: 10.3389/fpls.2014.00665
Rohwer, F., Schierle, J. (1982). Effect of light on ethylene production: red light enhancement of 1-aminocyclopropane-1-carboxylic acid concentration in etiolated pea shoots. Z. Pflanzenphysiol. 107, 295–300. doi: 10.1016/S0044-328X(82)80195-5
Roman, G., Lubarsky, B., Kieber, J. J., Rothenberg, M., Ecker, J. R. (1994). Genetic analysis of ethylene signal transduction in arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409
Ruduś, I., Sasiak, M., Kępczyński, J. (2013). Regulation of ethylene biosynthesis at the level of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiol. Plant. 35, 295–307. doi: 10.1007/s11738-012-1096-6
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Sakai, H., Hua, J., Chen, Q. G., Chang, C., Medrano, L. J., Bleecker, A. B., et al. (1998). ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. 95, 5812–5817. doi: 10.1073/pnas.95.10.5812
Schaller, G. E., Bleecker, A. B. (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811. doi: 10.1126/science.270.5243.1809
Scott, G., Dickinson, M., Shama, G., Rupar, M. (2018). A comparison of the molecular mechanisms underpinning high-intensity, pulsed polychromatic light and low-intensity UV-C hormesis in tomato fruit. Postharvest Biol. Technol. 137, 46–55. doi: 10.1016/j.postharvbio.2017.10.017
Seo, D. H., Yoon, G. M. (2019). Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. Cell Mol. Biol. 98, 898–911 doi: 10.1111/tpj.14289
Shakeel, S. N., Wang, X., Binder, B. M., Schaller, G. E. (2013). Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants 5, plt010–plt010. doi: 10.1093/aobpla/plt010
Shi, H., Liu, R., Xue, C., Shen, X., Wei, N., Deng, X. W., et al. (2016a). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26, 139–149. doi: 10.1016/j.cub.2015.11.053
Shi, H., Lyu, M., Luo, Y., Liu, S., Li, Y., He, H., et al. (2018). Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. 115, 6482–6487. doi: 10.1073/pnas.1803861115
Shi, H., Shen, X., liu, R., Xue, C., Wei, N., Deng, X. W., et al. (2016b). The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39, 597–610. doi: 10.1016/j.devcel.2016.10.020
Skottke, K. R., Yoon, G. M., Kieber, J. J., DeLong, A. (2011). Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 7, e1001370. doi: 10.1371/journal.pgen.1001370
Smalle, J., Haegman, M., Kurepa, J., Van Montagu, M., Straeten, D. V. D. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. 94, 2756–2761. doi: 10.1073/pnas.94.6.2756
Solano, R., Stepanova, A. N., Chao, Q., Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Song, Q., Ando, A., Xu, D., Fang, L., Zhang, T., Huq, E., et al. (2018). Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis. Proc. Natl. Acad. Sci. 115, 5606–5611. doi: 10.1073/pnas.1722068115
Steed, C. L., Taylor, L. K., Harrison, M. A. (2004). Red light regulation of ethylene biosynthesis and gravitropism in etiolated pea stems. Plant Growth Regul. 43, 117–125. doi: 10.1023/B:GROW.0000040116.10016.c3
Stepanova, A. N., Alonso, J. M. (2009). Ethylene signaling and response: where different regulatory modules meet. Curr. Opin. Plant Biol. 12, 548–555. doi: 10.1016/j.pbi.2009.07.009
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A., Alonso, J. M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242. doi: 10.1105/tpc.105.033365
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D.-Y., Doležal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Stepanova, A. N., Yun, J., Likhacheva, A. V., Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. doi: 10.1105/tpc.107.052068
Strader, L. C., Chen, G. L., Bartel, B. (2010). Ethylene directs auxin to control root cell expansion. Plant J. Cell Mol. Biol. 64, 874–884. doi: 10.1111/j.1365-313X.2010.04373.x
Street, I. H., Aman, S., Zubo, Y., Ramzan, A., Wang, X., Shakeel, S. N., et al. (2015). Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 169, 338–350. doi: 10.1104/pp.15.00415
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T. S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Tanimoto, M., Roberts, K., Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 8, 943–948. doi: 10.1046/j.1365-313X.1995.8060943.x
Tatsuki, M., Mori, H. (2001). Phosphorylation of Tomato 1-aminocyclopropane-1-carboxylic Acid Synthase, LE-ACS2, at the C-terminal Region. J. Biol. Chem. 276, 28051–28057. doi: 10.1074/jbc.M101543200
Tseng, T.-S., Whippo, C., Hangarter, R. P., Briggs, W. R. (2012). The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24, 1114–1126. doi: 10.1105/tpc.111.092130
Tsuchisaka, A., Theologis, A. (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136, 2982–3000. doi: 10.1104/pp.104.049999
Van de Poel, B., Smet, D., Van Der Straeten, D. (2015). Ethylene and hormonal cross talk in vegetative growth and development1. Plant Physiol. 169, 61–72. doi: 10.1104/pp.15.00724
Vandenbussche, F., Petrásek, J., Zádníková, P., Hoyerová, K., Pesek, B., Raz, V., et al. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Dev. Camb. Engl. 137, 597–606. doi: 10.1242/dev.040790
Vandenbussche, F., Vancompernolle, B., Rieu, I., Ahmad, M., Phillips, A., Moritz, T., et al. (2007). Ethylene-induced Arabidopsis hypocotyl elongation is dependent on but not mediated by gibberellins. J. Exp. Bot. 58, 4269–4281. doi: 10.1093/jxb/erm288
Vandenbussche, F., Vriezen, W. H., Smalle, J., Laarhoven, L. J. J., Harren, F. J. M., Straeten, D. V. D. (2003). Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 133, 517–527. doi: 10.1104/pp.103.022665
Wang, C., Liu, Y., Li, S.-S., Han, G.-Z. (2015). Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886. doi: 10.1104/pp.114.247403
Wang, N. N., Shih, M.-C., Li, N. (2005). The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 56, 909–920. doi: 10.1093/jxb/eri083
Wang, W., Esch, J. J., Shiu, S.-H., Agula, H., Binder, B. M., Chang, C., et al. (2006). Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18, 3429–3442. doi: 10.1105/tpc.106.044537
Wang, W., Hall, A. E., O’Malley, R., Bleecker, A. B. (2003). Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. 100, 352–357. doi: 10.1073/pnas.0237085100
Weckx, J., Van Poucke, M. (1989). “The effect of white light on the ethylene biosynthesis of intact green seedlings,” in Biochemical and Physiological Aspects of Ethylene Production in Lower and Higher Plants: proceedings of a Conference held at the Limburgs Universitair Centrum, Diepenbeek, Belgium, 22–27 August 1988 Advances in Agricultural Biotechnology. Eds. Clijsters, H., De Proft, M., Marcelle, R., Van Poucke, M. (Dordrecht: Springer Netherlands), 279–290. doi: 10.1007/978-94-009-1271-7_32
Wen, X., Zhang, C., Ji, Y., Zhao, Q., He, W., An, F., et al. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22, 1613–1616. doi: 10.1038/cr.2012.145
Wilkinson, J. Q., Lanahan, M. B., Yen, H.-C., Giovannoni, J. J., Klee, H. J. (1995). An ethylene-inducible component of signal transduction encoded by Never-ripe. Science 270, 1807–1809. doi: 10.1126/science.270.5243.1807
Wilson, R. L., Bakshi, A., Binder, B. M. (2014a). Loss of the ETR1 ethylene receptor reduces the inhibitory effect of far-red light and darkness on seed germination of Arabidopsis thaliana. Front. Plant Sci. 5, 1–13. doi: 10.3389/fpls.2014.00433
Wilson, R. L., Kim, H., Bakshi, A., Binder, B. M. (2014b). The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 165, 1353–1366. doi: 10.1104/pp.114.241695
Xiao, Y., Chen, J., Kuang, J., Shan, W., Xie, H., Jiang, Y., et al. (2013). Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 64, 2499–2510. doi: 10.1093/jxb/ert108
Xiong, L., Xiao, D., Xu, X., Guo, Z., Wang, N. N. (2014). The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis. J. Exp. Bot. 65, 4397–4408. doi: 10.1093/jxb/eru211
Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W. F., Harden, L. A., Theologis, A. (2003). Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. doi: 10.1074/jbc.M308297200
Yanagisawa, S., Yoo, S.-D., Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. doi: 10.1038/nature01984
Yang, S. F., Hoffman, N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. doi: 10.1146/annurev.pp.35.060184.001103
Yoon, G. M. (2015). New insights into the protein turnover regulation in ethylene biosynthesis. Mol. Cells 38, 597–603. doi: 10.14348/molcells.2015.0152
Yoon, G. M., Kieber, J. J. (2013a). 14-3-3 Regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028. doi: 10.1105/tpc.113.110106
Yoon, G. M., Kieber, J. J. (2013b). ACC synthase and its cognate E3 ligase are inversely regulated by light. Plant Signal. Behav. 8, e26478. doi: 10.4161/psb.26478
Yoshida, H., Nagata, M., Saito, K., Wang, K. L., Ecker, J. R. (2005). Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 5, 14. doi: 10.1186/1471-2229-5-14
Yu, Y., Huang, R. (2017). Integration of ethylene and light signaling affects hypocotyl growth in Arabidopsis. Front. Plant Sci. 8, 57. doi: 10.3389/fpls.2017.00057
Yu, Y., Wang, J., Zhang, Z., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLOS Genet. 9, e1004025. doi: 10.1371/journal.pgen.1004025
Zádníková, P., Petrásek, J., Marhavy, P., Raz, V., Vandenbussche, F., Ding, Z., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Dev. Camb. Engl. 137, 607–617. doi: 10.1242/dev.041277
Zdarska, M., Dobisová, T., Gelová, Z., Pernisová, M., Dabravolski, S., Hejátko, J. (2015). Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J. Exp. Bot. 66, 4913–4931. doi: 10.1093/jxb/erv261
Zhang, X., Ji, Y., Xue, C., Ma, H., Xi, Y., Huang, P., et al. (2018). Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 30, 1971–1988. doi: 10.1105/tpc.18.00018
Zhong, G. Y., Burns, J. K. (2003). Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 53, 117–131. doi: 10.1023/B:PLAN.0000009270.81977.ef
Zhong, S., Shi, H., Xue, C., Wang, L., Xi, Y., Li, J., et al. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22, 1530–1535. doi: 10.1016/j.cub.2012.06.039
Zhong, S., Shi, H., Xue, C., Wei, N., Guo, H., Deng, X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. 111, 3913–3920. doi: 10.1073/pnas.1402491111
Zhou, L., Jang, J., Jones, T. L., Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. 95, 10294–10299. doi: 10.1073/pnas.95.17.10294
Keywords: ethylene, light, transcriptomic meta-analysis, ethylene response, ethylene biosynthesis, hypocotyl, root
Citation: Harkey AF, Yoon GM, Seo DH, DeLong A and Muday GK (2019) Light Modulates Ethylene Synthesis, Signaling, and Downstream Transcriptional Networks to Control Plant Development. Front. Plant Sci. 10:1094. doi: 10.3389/fpls.2019.01094
Received: 02 June 2019; Accepted: 09 August 2019;
Published: 12 September 2019.
Edited by:
Dominique Van Der Straeten, Ghent University, BelgiumReviewed by:
Jan Smalle, University of Kentucky, United StatesAnna N. Stepanova, North Carolina State University, United States
Copyright © 2019 Harkey, Yoon, Seo, DeLong and Muday. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gloria K. Muday, muday@wfu.edu
 Alexandria F. Harkey
Alexandria F. Harkey Gyeong Mee Yoon
Gyeong Mee Yoon Dong Hye Seo
Dong Hye Seo Alison DeLong
Alison DeLong Gloria K. Muday
Gloria K. Muday