- 1Plant Molecular Biology Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
- 2Department of Botany and Forestry, Vidyasagar University, Midnapore, West Bengal, India
Mature seeds of most of the higher plants harbor dormant embryos and go through the complex process of germination under favorable environmental conditions. The germination process involves dynamic physiological, cellular and metabolic events that are controlled by the interplay of several gene products and different phytohormones. The small non-coding RNAs comprise key regulatory modules in the process of seed dormancy and germination. Recent studies have implicated the small RNAs in plant growth in correlation with various plant physiological processes including hormone signaling and stress response. In this review we provide a brief overview of the regulation of seed germination or dormancy while emphasizing on the current understanding of the role of small RNAs in this regard. We have also highlighted specific examples of stress responsive small RNAs in seed germination and discussed their future potential.
Introduction
The seeds of higher plants contain the dormant embryos, as miniature new plants, along with adequate food reserves to sustain the growing seedlings until they establish themselves as self-sufficient, autotrophic organisms (Figure 1A). Germination is one of the most important physiological process of a seed which begins with the uptake of water by the quiescent dry seed and is completed when a part of the embryo, usually a radical oozes out of the seed coat (Bewley, 1997). Seed dormancy is regarded as the temporary failure or block of a viable seed to complete germination under seemingly unfavorable conditions and is an adaptive feature for optimizing the timing of germination (Bewley, 1997). The dynamic process of seed germination is triphasic (Figure 1B) and involves a complex coordination of many physiological, cellular and metabolic events (Bewley, 1997; Weitbrecht et al., 2011). Phase-I includes rapid leakage of solutes which paves the way for respiration and protein synthesis. Phase-II represents a plateau stage where new mRNAs and proteins are synthesized. There is also an accumulation of the mitochondrion to support the energy requirements at this stage. During phase-III, radicle cells elongate and divide. This is also the stage of rapid DNA synthesis and replication together with the mobilization of stored reserves (Bewley, 1997).
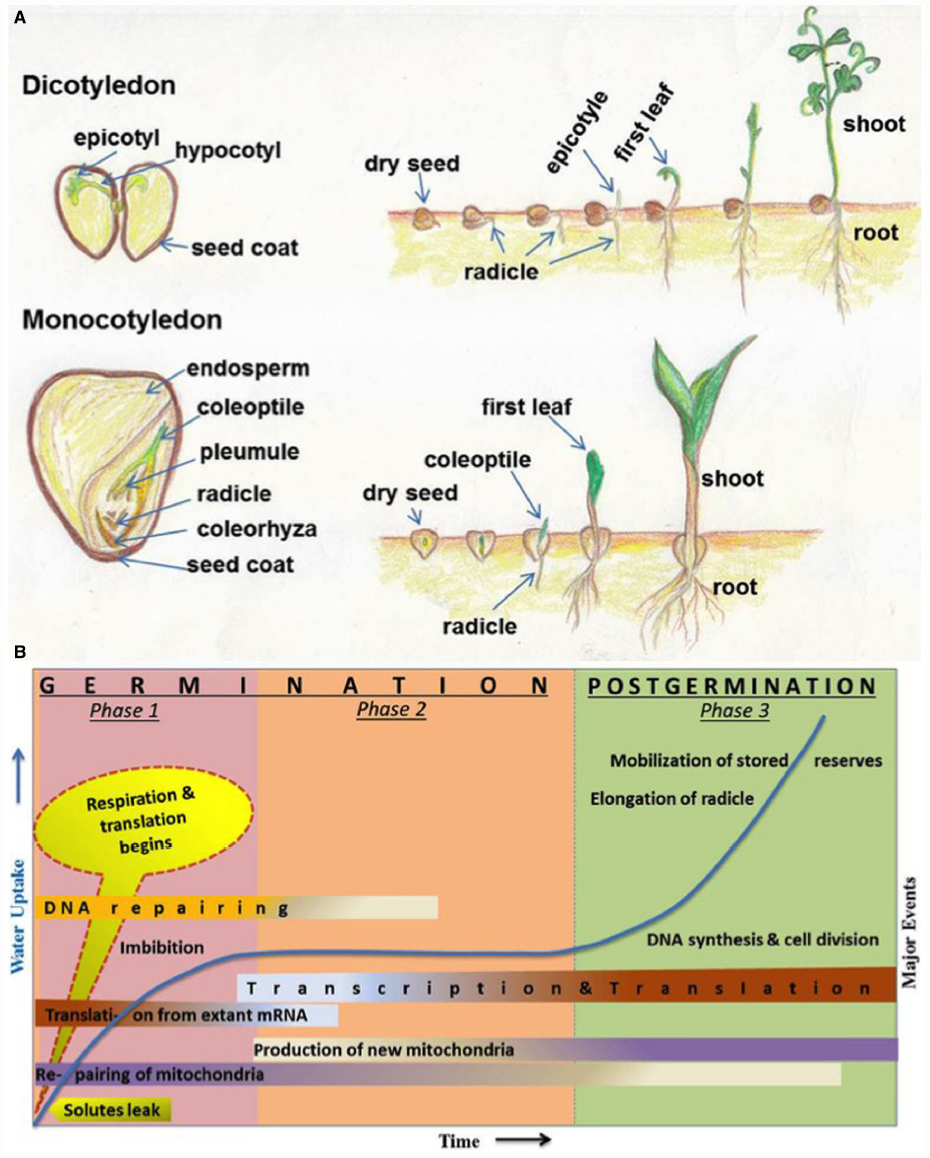
Figure 1. (A) Schematic representation (hand drawn by SS) of different parts of seeds and seed germination stages. Seeds and germination stages of dicotyledonous (chickpea) and monocotyledonous (maize) plants have been shown in upper and lower panels, respectively. (B) Major events associated with seed germination and post-germinative growth phases. Germination stages are represented by phase 1 and phase 2; postgermination events includes phase 3. The time (x-axis) for events varies from several hours to many weeks, depending on different plant species and germination conditions. Uptake of water and related increase in biomass is indicated in y-axis and shown in line graph during three phases. Some events (such as DNA repairing, transcription, translation, and mitochondria production etc.) are spread over more than one phases and indicated with shaded color; dark colors indicate more activity and light colors indicate less activity. This figure has been reproduced with modification, after written permission of the corresponding author (Prof. J. D. Bewley) and the original publisher, American Society of Plant Biologists (ASPB).
Various environmental factors such as light, temperature, moisture, oxygen, soil, humidity, stress etc and some physiological factors such as viability of seeds, thickness of seed coat, dormancy period etc also play vital role in seed germination stages (Martin et al., 2010; Weitbrecht et al., 2011). Several studies have implicated that the interactions between different phytohormones such as abscisic acid (ABA), gibberellins (GAs), ethylene, brassinosteroids (BRs), auxin, and cytokinins (CKs) play a key role in regulating the interconnected molecular processes that control dormancy release and activation of the stages of seed germination (Liu et al., 2007; Finkelstein et al., 2008). The activity of plant hormones needs to be precisely regulated, since some phytohormones exert crucial but contrasting influence on the process. ABA is a positive regulator of dormancy and its maintenance, while it is a negative regulator of germination (Finkelstein et al., 2008). The absence of, or insensitivity to ABA during seed development results in the production of viviparous or precociously germinating seeds as exemplified by maize viviparous (vp), tomato sitiens (sit), and Arabidopsis ABA-deficient (aba), and ABA-insensitive (abi) mutants (Finkelstein et al., 2008). GA releases dormancy and its role is analogous to that of Ethylene and BR in promoting germination by counteracting ABA effects. Recently, the crosstalk between ABA and auxin has also been highlighted (Finkelstein et al., 2008).
The discovery of small non-coding RNAs (of 19–24 nucleotides length) has added a new dimension to the understanding of the regulation of cellular environment (Bartel, 2004; Axtell et al., 2007). They have been shown to play diverse roles in growth, development, morphogenesis and stress responses of both plants and animals (Chen, 2012; Kamthan et al., 2015). The functional small RNAs are produced from double stranded RNA precursors through the activity of RNA-dependent RNA Polymerase (RDR), DICER-like (DCL), and ARGONAUTE (AGO) proteins (Allen et al., 2005; Mallory et al., 2008; Axtell, 2013). There are two major classes of small non-coding RNAs—short interfering RNAs (siRNAs) and microRNAs (miRNAs) that negatively regulate their target genes by binding to the complementary sequences. At the transcriptional level, the small RNAs may be involved in chromatin remodeling (Huettel et al., 2007; Pontier et al., 2012; Xie and Yu, 2015) while at the post-transcriptional level, depending upon the nature of homology they can bring about the cleavage of the target mRNA (Rajagopalan et al., 2006; Vaucheret, 2006) or block their translation (Poethig et al., 2006; Vaucheret, 2006; Bartel, 2009). The biosynthesis and function of many of these small RNA genes are also regulated by different plant hormones and environmental stress (Mallory et al., 2005; Reyes and Chua, 2007; Sunkar et al., 2007; Shukla et al., 2008; Martin et al., 2010; Khraiwesh et al., 2011; Sanan-Mishra et al., 2013).
Biogenesis of miRNA and ta-siRNA
miRNA biogenesis is a multistep process that is mainly resistricted to the nucleus in plants. Briefly, the miRNA gene is transcribed into a capped and poly-adenylated primary miRNA (pri-miRNA) by enzyme RNA polymerase II (Chen, 2012; Axtell, 2013). The pri-miRNA is processed to precursor miRNA (pre-miRNA), of around 70–100 nt long, by DCL protein (Axtell, 2013). The pre-miRNA is further processed to form miRNA and miRNA* duplex by the activity of DCL protein. The duplex is then methylated at the 2′OH of the 3′nucleotide end by HEN1 (Allen et al., 2005; Chen, 2009) and transported to the cytoplasm.
One strand of the duplex is loaded into RISC (RNA-induced silencing complex) containing AGO1. The strand selection widely depends on the relative stability of the two ends of the duplex (Axtell, 2013). It is observed that generally the strand whose 5′ end is comparatively loose, gets incorporated into the RISC (Allen et al., 2005; Axtell, 2013). The RISC complex containing the miRNA identifies its target transcripts based on perfect or nearly perfect sequence complementarity. In plants the stringency of target recognition is very high and the target transcripts are normally cleaved, however, the central mismatches in the miRNA:mRNA pair direct the inhibition of translation (Allen et al., 2005; Axtell, 2013).
Recently, the ta-siRNA (trans-acting small interferring RNAs) have also been implicated in plant development thereby attracting major research interest for many plant biologists (Nogueira et al., 2006; Axtell, 2013). ta-siRNAs are generated from TAS (Trans-Acting SiRNA locus) gene derived non-coding transcripts through specific miRNA guided cleavage. The cleaved precursors of ta-siRNAs are bounded and stabilized by SUPPRESSOR of GENE SILENCING3 (SGS3) and further synthesized into double stranded RNAs by RDR6 (Chen, 2009; Allen and Howell, 2010; Axtell, 2013). The double stranded RNAs are cleaved several times by DCL4 from the miRNA mediated cleavage sites, so that 21-nt long phased ta-siRNAs are produced. Similar to miRNAs, the ta-siRNAs are incorporated into RISCs, where they cleave the target mRNAs or repress translation (Allen et al., 2005; Allen and Howell, 2010). There are four families of TAS gene in Arabidopsis, namely TAS1, TAS2, TAS3, TAS4 (Rajagopalan et al., 2006; Allen and Howell, 2010). For the initial processing TAS1 and 2 require miR173 whereas TAS3 and TAS4 require miR390 and miR828, respectively for initial processing (Chen, 2009; Allen and Howell, 2010; Axtell, 2013).
The Role of miRNAs and ta-siRNAs in Plant Growth and Development
The miRNAs constitute a major class that play important and diverse roles in regulation of various aspects of plant development (Sanan-Mishra and Mukherjee, 2007; Chen, 2012; Sharma et al., 2015). The classical examples include regulation of CUC1/CUC2 and NAC1 transcripts by miR164 to affect reproductive and root development (Guo et al., 2005); determination of abaxial/adaxial leaf polarity and root development by miR166/165 mediated control of Class III HD-ZIP transcription factor mRNAs (Chen, 2012; Barik et al., 2014; Singh et al., 2014) and the regulation of flower development in Arabidopsis thaliana by miR172 targeted AP2 and other mRNAs (Wollmann et al., 2010). The function of miRs has been shown to be affected by hormones and stress responses (Mallory et al., 2005; Liu et al., 2007; Reyes and Chua, 2007)
The miRNA mediated, ta-siRNA production is also significantly altered in drought, salinity and hypoxia stresses, besides their regulation by auxin and other hormones (Moldovan et al., 2009; Matsui et al., 2014). This is evident by TAS3 derived ta-siR-ARF that target different AUXIN RESPONSE FACTOR2, 3 and 4 (ARF2,3,4) and regulate various aspects of plant development such as vegetative to reproductive phase changes, leaf polarity and lateral root development in Arabidopsis (Peragine et al., 2004; Chitwood et al., 2007; Allen and Howell, 2010; Marin et al., 2010). Mutations in ta-siRNA biogenesis pathway lead to the upregulation of target mRNAs and affect the aforesaid aspects of plant development. The rice and maize ta-siRNA biogenesis mutants have been shown to have severely affected shoot and leaf development (Itoh et al., 2006; Nogueira et al., 2006; Nagasaki et al., 2007; Douglas et al., 2010). DCL4 is suggested to redundantly regulate processing of some miRNAs, besides role in ta-siRNA biogenesis (Rajagopalan et al., 2006). ABA signaling is shown to be, at least partially, affect RDR6 accumulation (Zhang et al., 2013). Although ta-siRNA has not directly been implicated in seed germination, their cross talk with miRNA and hormone signaling in feed-back loops (Marin et al., 2010; Chen, 2012) as well as role in seed development (Zhang et al., 2013) indicate their potential function in seed maturation and germination. This remains to be an interesting area to be explored in the complex process of seed germination.
Molecular Network of Small RNAs in Seed Germination and Dormancy
Throughout the life cycle of an angiosperm plant, there are two major developmental phase transitional periods. One is germination (from seed to seedling stage; Huang et al., 2013) and another is flowering (from vegetative to reproductive stage; Wu et al., 2009). Recent studies indicate that genes, regulating phase transition to flowering are also involved in transition from dormancy to germination (Huang et al., 2013). The genes that regulate cellular phase transitions from embryo to seedling growth also play important role in the process. In addition, phytohormones and environmental factors affect expression of seed germination (Liu et al., 2007; Finkelstein et al., 2008). Recently, a role for small RNAs has been indicated in this process by characterizing the mutants of small RNA biogenesis pathway genes, such as DCL1, HYL1, HEN1, and AGO1 that display severe defects in embryogenesis and seed development (Willmann et al., 2011). This can be illustrated by the dcl1 mutant, which shows early seed maturation phenotype than the normal wild type seeds. The positive regulators of DCL1 gene are leafy cotyledon (LEC) genes like LEC2 and FUS3. Whereas the negative regulators or repressors of early embryo maturation are ASIL1, ASIL2, and HDA6/SIL1 (Willmann et al., 2011).
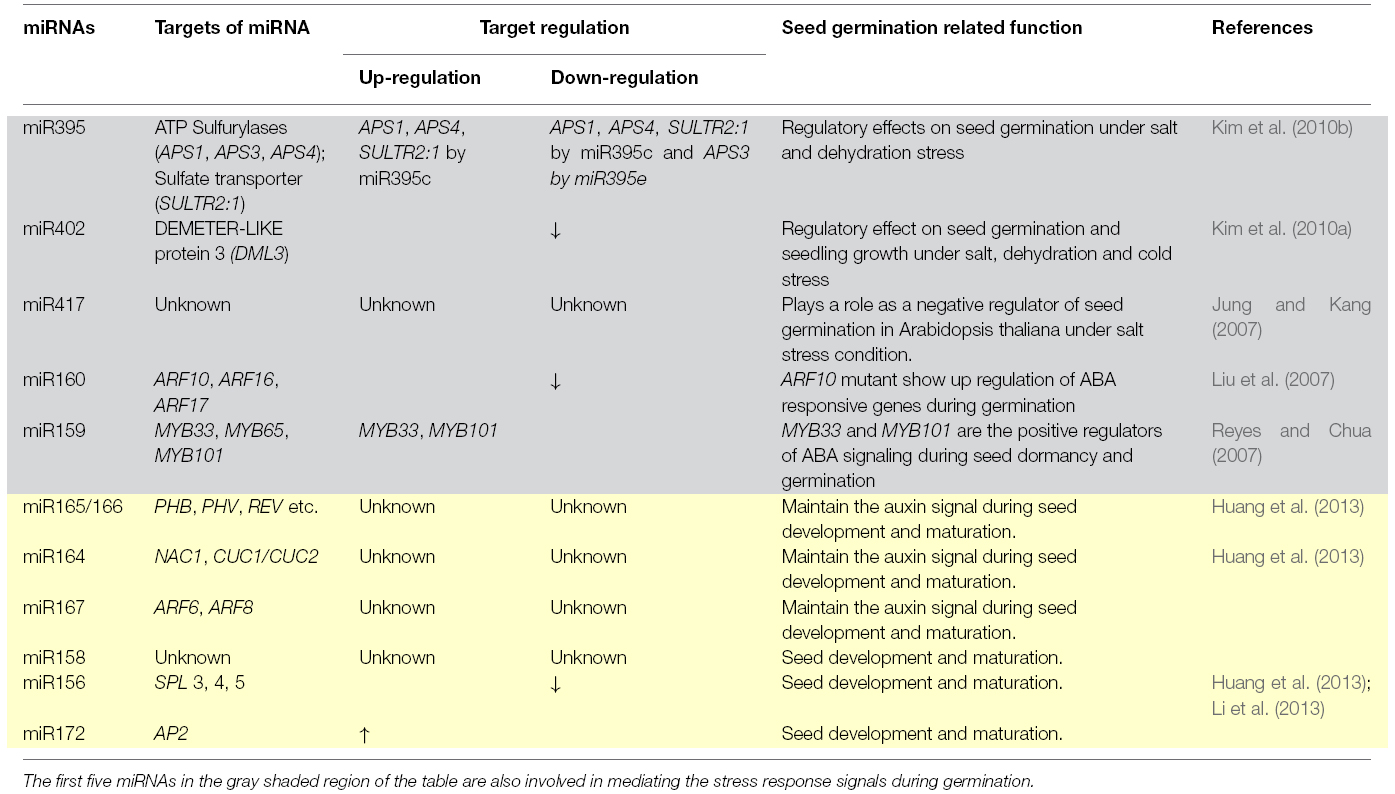
Different miRNAs like are miR160, miR159, miR417, miR395, miR402, mir165/166, miR164, miR167, miR156, miR172, and miR158 (Table 1) are known to control both the activators and repressors of seed germination and dormancy (Jung and Kang, 2007; Liu et al., 2007; Reyes and Chua, 2007; Kim et al., 2010a,b; Martin et al., 2010; Huang et al., 2013). Increased level of miR156 and reduced level of SPLs and miR172 (Table 1) in the mature embryo could down regulate the developmental transition and keep seeds in dormant stages (Martin et al., 2010; Huang et al., 2013). The imbibition step itself has been shown to differentially down-regulate twelve miRNA families, miR156, miR159, miR164, miR166, miR167, miR168, miR169, miR172, miR319, miR393, miR394, and miR397; while four families, miR398, miR408, miR528, and miR529 were up-regulated during the seed germination (Li et al., 2013). Interestingly, miR156 and miR157 have also been implicated in vegetative to reproductive phase change (Wu et al., 2009), indicating their functional diversification.
The complex regulatory cross-talk between the hormones and the small RNAs, was evident by the identification of two ABA supersensitive mutants for germination viz. absg1 and absg2 as the alleles of dcl1 and hen1. The absg1 and absg2 mutants show up regulation of the expression of ABA responsive genes (Zhang et al., 2008). An important role for miR159 has been demonstrated in regulating the dynamic seed germination procedure by modulating GA and ABA hormone signaling (Table 1). The expression of miR159 is controlled by both GA and ABA (Martin et al., 2010). The GAMYB proteins act as the positive regulators, whereas DELLA proteins act as the negative regulators of the GA signaling cascade (Peng and Harberd, 2002; Finkelstein et al., 2008; Weitbrecht et al., 2011). The GAMYB mRNAs are regulated by miR159 during floral development, fertility and seed germination (Reyes and Chua, 2007). Recently, it was shown that alurone vacuolation, a GA-mediated (GAMYB protein) programmed cell death (PCD) process in alurone is required for seed germination (Peng and Harberd, 2002; Finkelstein et al., 2008; Alonso-Peral et al., 2010). The miR159 also regulates transcription factors MYB33 and MYB101, which are the positive regulators of ABA signaling during seed dormancy and germination (Reyes and Chua, 2007; Martin et al., 2010). miR159 expression is upregulated in rdr2 and dcl2 dcl3 dcl4 triple mutants. Interestingly, RDR2, DCL2, DCL3, DCL4 are the essential factors in case of siRNA biogenesis, especially heterochromatic siRNA biogenesis pathway (Allen and Howell, 2010; Axtell, 2013). This suggests that different kinds of small RNAs, besides miRNAs, could essentially play significant role in seed germination and dormancy.
The role of phytohormone Auxin in seed germination, became evident when Liu et al. (2007) showed that miR160 mediated down regulation of ARF10 plays crucial roles in seed germination (Table 1; Liu et al., 2007). ARFs are transcription factors involved in auxin signaling pathway during many plant growth and developmental stages. The miR160 also appears to be the converging point of Auxin and ABA mediated cross-talk during seed germination, since mutation in ARF10 results in developmental defects and overexpression of ABA responsive genes (Liu et al., 2007). Similarly, it was shown that over expression of miR160 caused hyposensitivity to ABA during germination (Liu et al., 2007). Auxin homeostasis is vital for embryo development and is mediated by the action of miR165/166, miR167, miR164, miR158, and miR160 (Martin et al., 2010). This suggests an important role for the miRNAs in mediating suitable auxin signaling during embryo and seed development (Table 1). Thus, it could be concluded that these miRNAs play important roles in maintaining dormancy and breaking of dormancy to promote embryo into seedling stage through seed germination (Martin et al., 2010; Huang et al., 2013; Zhang et al., 2013).
Gaseous hormone ethylene promotes seed germination through interaction with ABA signaling (Finkelstein et al., 2008) The two mutants namely ethylene resistant1 (etr1) and ethylene insensitive2 (ein2) or, enhanced response to aba3 (era3) show upregulation of ABA responsive genes and delay in seed germination (Finkelstein et al., 2008).Whereas wild type seeds treated with ethylene precursor ACC (1-aminocyclopropane -1-carboxylic acid) show downregulation of ABA response factors (Finkelstein et al., 2008). Again, etr1-2 mutant show the over accumulation of GA content, which could be a compensation to over accumulation of ABA (Finkelstein et al., 2008). Since miR160 and miR159 both have regulatory effects on ABA and GA, and ethylene has a cross talk with ABA and GA, therefore, it is hypothesized that these miRNAs may have direct or indirect control over ethylene mediated regulation during seed germination and dormancy.
Plant steroid hormone BRs that mainly effect stem elongation and leaf unfurling also effect seed germination. The mutants for BR biosynthetic and signaling pathway are sensitive to ABA leading to decrease in the germination potential (Finkelstein et al., 2008). The possibility of a cross talk between BR and ABA signaling cannot be ruled out in the activation of the miR160 regulatory pathway in seed germination (Liu et al., 2007). Also, BRs induce the expression of distinct EXPANSIN (EXP) family members, which are cell wall loosening proteins that can indirectly influence seed germination (Bewley, 1997).
Parallel studies have shown that the small RNA biogenesis pathway mutants, that show high expression of ABA, are highly sensitive to salt and osmotic stresses (Zhang et al., 2008), thereby indicating the overlap with the environmental cues. Under abiotic stress conditions, miR395 (Table 1) acts both as a positive and negative regulator of seed germination (Kim et al., 2010b). miR395 has six family members in Arabidopsis genome, that target the proteins APS1, APS3, APS4, and SULTR, involved in sulfate assimilation and transport. It was shown that miR395e that differs from miR395c in a single nucleotide cannot target APS1 and APS4. These miRNAs have different effects on the seed germination of Arabidopsis under high salt or dehydration stress conditions (Kim et al., 2010b). Over expression of miR395c reduces the germination potential under high salt or dehydration stress condition; whereas over expression of miR395e enhances the germination potential under the same stress condition in Arabidopsis thaliana (Kim et al., 2010b). Similarly over expression of miR402 (Table 1) enhances the seed germination potential in Arabidopsis under salt, dehydration and cold stress conditions (Kim et al., 2010a). miR402 downregulates its target gene DML3 (DEMETER-LIKE protein3), which is involved in DNA demethylation, an epigenetic regulatory process of plants in various stress conditions (Kim et al., 2010a). miR417 (Table 1) also exhibits a negative regulation over seed germination under salt stress condition (Jung and Kang, 2007). However, its mechanism of molecular action is not yet clear.
Conclusion and Future Perspective
Agriculture exclusively depends on growing crops; so the success of cultivation as well as productivity largely depends on seed viability, seed germination and efficiency of seed development. Small RNAs play critical roles in regulation of gene expression in developing and germinating seeds (Kamthan et al., 2015). In this review we describe that specific small RNAs, mainly miRNAs regulated nodes, play crucial roles in regulating seed germination in response to different phyto-hormones and abiotic stresses. But the mechanism of action and the interconnection of the various signaling cascades with their regulatory networks remain largely unknown till date. Thus, functional analysis of small RNAs expressed in seeds or during germination process will provide useful information for seed biology. Future studies are required to unravel the molecular details of small RNAs regulated pathways in seed germination and viability maintenance, and their association with the stress responses and hormonal signals, especially in crop plants. Expression and functional analysis using transgenic approach, proteomic analysis and the use of different bioinformatics tools could also help to throw light on this issue.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
SS acknowledges Women Scientist-A (Wos-A) fellowship from Department of Science and Technology (DST), India WOS-A/LS-1276/2014. We sincerely apologize to all colleagues whose relevant work could not be mentioned due to space restrictions.
References
Allen, E., and Howell, M. D. (2010). miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21, 798–804. doi: 10.1016/j.semcdb.2010.03.008
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Alonso-Peral, M. M., Li, J., Li, Y., Allen, R. S., Schnippenkoetter, W., Ohms, S., et al. (2010). The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 154, 757–771. doi: 10.1104/pp.110.160630
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
Axtell, M. J., Snyder, J. A., and Bartel, D. P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19, 1750–1769. doi: 10.1105/tpc.107.051706
Barik, S., Sarkardas, S., Singh, A., Gautam, V., Kumar, P., Majee, M., et al. (2014). Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 103, 114–121. doi: 10.1016/j.ygeno.2013.11.004
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bewley, J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. doi: 10.1105/tpc.9.7.1055
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/annurev.cellbio.042308.113417
Chen, X. (2012). Small RNAs in development—insights from plants. Curr. Opin. Genet. Dev. 22, 361–367. doi: 10.1016/j.gde.2012.04.004
Chitwood, D. H., Guo, M., Nogueira, F. T., and Timmermans, M. C. (2007). Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 134, 813–823. doi: 10.1242/dev.000497
Douglas, R. N., Wiley, D., Sarkar, A., Springer, N., Timmermans, M. C., and Scanlon, M. J. (2010). Ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22, 1441–1451. doi: 10.1105/tpc.109.071613
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Guo, H. S., Xie, Q., Fei, J. F., and Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Huang, D., Koh, C., Feurtado, J. A., Tsang, E. W., and Cutler, A. J. (2013). MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 14:140. doi: 10.1186/1471-2164-14-140
Huettel, B., Kanno, T., Daxinger, L., Bucher, E., Van Der Winden, J., Matzke, A. J., et al. (2007). RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta 1769, 358–374. doi: 10.1016/j.bbaexp.2007.03.001
Itoh, J., Sato, Y., Nagato, Y., and Matsuoka, M. (2006). Formation, maintenance and function of the shoot apical meristem in rice. Plant Mol. Biol. 60, 827–842. doi: 10.1007/s11103-005-5579-3
Jung, H. J., and Kang, H. (2007). Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol. Biochem. 45, 805–811. doi: 10.1016/j.plaphy.2007.07.015
Kamthan, A., Chaudhuri, A., Kamthan, M., and Datta, A. (2015). Small RNAs in plants: recent development and application for crop improvement. Front. Plant Sci. 6:208. doi: 10.3389/fpls.2015.00208
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2011). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kim, J. Y., Kwak, K. J., Jung, H. J., Lee, H. J., and Kang, H. (2010a). MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 51, 1079–1083. doi: 10.1093/pcp/pcq072
Kim, J. Y., Lee, H. J., Jung, H. J., Maruyama, K., Suzuki, N., and Kang, H. (2010b). Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232, 1447–1454. doi: 10.1007/s00425-010-1267-x
Li, D., Wang, L., Liu, X., Cui, D., Chen, T., Zhang, H., et al. (2013). Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS ONE 8:e55107. doi: 10.1371/journal.pone.0055107
Liu, P. P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H., and Carrington, J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146. doi: 10.1111/j.1365-313X.2007.03218.x
Mallory, A. C., Bartel, D. P., and Bartel, B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17, 1360–1375. doi: 10.1105/tpc.105.031716
Mallory, A. C., Elmayan, T., and Vaucheret, H. (2008). MicroRNA maturation and action–the expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol 11, 560–566. doi: 10.1016/j.pbi.2008.06.008
Marin, E., Jouannet, V., Herz, A., Lokerse, A. S., Weijers, D., Vaucheret, H., et al. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117. doi: 10.1105/tpc.109.072553
Martin, R. C., Liu, P. P., Goloviznina, N. A., and Nonogaki, H. (2010). microRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J. Exp. Bot. 61, 2229–2234. doi: 10.1093/jxb/erq063
Matsui, A., Mizunashi, K., Tanaka, M., Kaminuma, E., Nguyen, A. H., Nakajima, M., et al. (2014). tasiRNA-ARF pathway moderates floral architecture in Arabidopsis plants subjected to drought stress. Biomed. Res. Int. 2014, 303451. doi: 10.1155/2014/303451
Moldovan, D., Spriggs, A., Yang, J., Pogson, B. J., Dennis, E. S., and Wilson, I. W. (2009). Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 61, 165–177. doi: 10.1093/jxb/erp296
Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., et al. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 14867–14871. doi: 10.1073/pnas.0704339104
Nogueira, F. T., Sarkar, A. K., Chitwood, D. H., and Timmermans, M. C. (2006). Organ polarity in plants is specified through the opposing activity of two distinct small regulatory RNAs. Cold. Spring Harb. Symp. Quant. Biol. 71, 157–164. doi: 10.1101/sqb.2006.71.045
Peng, J., and Harberd, N. P. (2002). The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 5, 376–381. doi: 10.1016/S1369-5266(02)00279-0
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Poethig, R. S., Peragine, A., Yoshikawa, M., Hunter, C., Willmann, M., and Wu, G. (2006). The function of RNAi in plant development. Cold. Spring Harb. Symp. Quant. Biol. 71, 165–170. doi: 10.1101/sqb.2006.71.030
Pontier, D., Picart, C., Roudier, F., Garcia, D., Lahmy, S., Azevedo, J., et al. (2012). NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell. 48, 121–132. doi: 10.1016/j.molcel.2012.07.027
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Reyes, J. L., and Chua, N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Sanan-Mishra, N., and Mukherjee, S. K. (2007). A peep into the plant miRNA world. Open Plant J. 1, 1–9. doi: 10.2174/1874294700701010001
Sanan-Mishra, N., Varanasi, S. P., and Mukherjee, S. K. (2013). Micro-regulators of auxin action. Plant Cell Rep. 32, 733–740. doi: 10.1007/s00299-013-1425-2
Sharma, N., Tripathi, A., and Sanan-Mishra, N. (2015). Profiling the expression domains of a rice-specific microRNA under stress. Front. Plant Sci. 6:333. doi: 10.3389/fpls.2015.00333
Shukla, L. I., Chinnusamy, V., and Sunkar, R. (2008). The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta 1779, 743–748. doi: 10.1016/j.bbagrm.2008.04.004
Singh, A., Singh, S., Panigrahi, K. C., Reski, R., and Sarkar, A. K. (2014). Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 33, 945–953. doi: 10.1007/s00299-014-1573-z
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771. doi: 10.1101/gad.1410506
Weitbrecht, K., Muller, K., and Leubner-Metzger, G. (2011). First off the mark: early seed germination. J. Exp. Bot. 62, 3289–3309. doi: 10.1093/jxb/err030
Willmann, M. R., Mehalick, A. J., Packer, R. L., and Jenik, P. D. (2011). MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155, 1871–1884. doi: 10.1104/pp.110.171355
Wollmann, H., Mica, E., Todesco, M., Long, J. A., and Weigel, D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642. doi: 10.1242/dev.036673
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xie, M., and Yu, B. (2015). siRNA-directed DNA methylation in plants. Curr. Genomics 16, 23–31. doi: 10.2174/1389202915666141128002211
Zhang, J., Zhang, S., Han, S., Li, X., Tong, Z., and Qi, L. (2013). Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in Larches (Larix leptolepis). PLoS ONE 8:e81452. doi: 10.1371/journal.pone.0081452
Keywords: seed germination, seed, small RNA, miRNA, stress response, seed dormancy
Citation: Das SS, Karmakar P, Nandi AK and Sanan-Mishra N (2015) Small RNA mediated regulation of seed germination. Front. Plant Sci. 6:828. doi: 10.3389/fpls.2015.00828
Received: 03 August 2015; Accepted: 22 September 2015;
Published: 13 October 2015.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research, IndiaCopyright © 2015 Das, Karmakar, Nandi and Sanan-Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neeti Sanan-Mishra, neeti@icgeb.res.in
 Shabari Sarkar Das
Shabari Sarkar Das