- Beijing Key Laboratory for Forest Pest Control, Beijing Forestry University, Beijing, China
A special mutual relationship exists between the pine wood nematode (PWN) Bursaphelenchus xylophilus and its vector beetles of genus Monochamus, which enables PWN to spread, at the same time provides longhorned beetles with more weak hosts. PWN are attracted to the pupal chambers and then carried inside the trachea of beetle adults, which is a necessary part to complete the B. xylophilus infection cycle. The growth and immune responses of the vector beetle will affect this carrying process, however, they were rarely studied in Monochamus saltuarius. Real-time quantitative polymerase chain reaction (RT-qPCR), one of the most common methods for quantitative gene expression analysis, was performed to explore the key genes and pathways involved in the growth, development and immune responses of M. saltuarius at different developmental stages associated with infection of PWN and PWN treatment conditions. To enhance the accuracy of RT-qPCR data, the expression of target genes needs to be normalized with reference genes, which are stably expressed under varied experimental conditions. In our study, the stability of 14 candidate reference genes in M. saltuarius samples at different developmental stages associated with infection of PWN or PWN treatment conditions was evaluated using delta Ct, geNorm, NormFinder, BestKeeper and RefFinder algorithms. Moreover, KLF gene was used to validate the stability of the selected reference genes. Under experimental conditions of this study, RPL7 and TER were suitable reference genes at different developmental stages associated with infection of PWN. RPL7 and RPS5 were considered the most stable reference genes in the pupae treated with PWN. RPS5 and SNX6 could be used as reference genes in the adults treated with PWN. RPL7, EF1-γ, and RPS5 could be used as stable reference genes in all the samples. This work is the first to evaluate reference genes in M. saltuarius, laying a foundation for further gene expression experimental procedures and understanding the phoretic relationship between M. saltuarius and B. xylophilus.
Background
Pine wilt disease is one of the most dangerous and devastating diseases caused by Bursaphelenchus xylophilus (pine wood nematode; PWN) worldwide. B. xylophilus originated in North America, and was introduced to Japan, Korea, China, Portugal, and other countries, causing serious damage in these invasion areas (Dropkin and Foudin, 1979; Mamiya, 1988; Cheng et al., 1983; Han et al., 2008; Khan, 1991; Manuel et al., 1999; Abelleira et al., 2011). PWN is transmitted to dead or dying trees by its insect vector, the Monochamus beetles, during oviposition or maturation feeding (Akbulut and Stamps, 2012; Kim et al., 2020; Li et al., 2020).
In China, PWN was first widely spread and damaged in southern area, then spread to northern and other regions. In the southern area, Monochamus alternatus as a main vector of the PWN was widely studied. However, Monochamus saltuarius emerged as a new and unique vector in Liaoning Province, China (Yu and Wu, 2018), although it was regarded as a common vector in Korea and Japan (Sato, 1987; Kim et al., 2006). It greatly promotes the transmission of B. xylophilus to Larix spp., Pinus koraiensis, Pinus sylvestris var. mongolica, and Pinus tabuliformis (Yu and Wu, 2018; Yu et al., 2019; 2020). Therefore, the research about interaction between B. xylophilus and M. saltuarius is of significance to prevent and control the prevalence of pine wilt disease in north of China.
Gene expression is an important method to study the potential function of insect genes in different conditions. Real-time quantitative polymerase chain reaction (RT-qPCR), one of the most common methods for quantitative gene expression analysis, has the characteristics of high accuracy, specificity, sensitivity, and rapidity (Bustin, 2002; Bustin et al., 2005; Valasek and Repa, 2005). However, the quality and quantity of RNA extraction, polymerase amplification efficiency, and cDNA synthesis efficiency can all lead to systematic errors during RT-qPCR operation (Klein, 2002; Fleige and Pfaffl, 2006). To eliminate these errors, various strategies have been used to normalize RT-qPCR data, and using internal controls or reference genes has become the most reliable method (Silver et al., 2006; Borowski et al., 2014; Silveira et al., 2021). Nevertheless, there are no absolute stable reference genes because of spatio-temporal specificity of genes and variable experimental conditions. So, it is necessary to screen suitable reference genes according to specific experimental materials and conditions for RT-qPCR analysis.
Studies of reference genes in insects are common. In coleoptera, reference gene screening has been performed in approximately twenty insect species, such as Dendroctonus valens, Harmonia axyridis, and Cerambycidae species, Monochamus alternatus, Anoplophora glabripennis (Toutges et al., 2010; Rajarapu et al., 2012; Shi et al., 2013; Barros Rodrigues et al., 2014; Wang et al., 2014; Song et al., 2015; Tan et al., 2015; Feng et al., 2016; Rodrigues et al., 2017; Yang et al., 2018; Yang et al., 2020; Zhang et al., 2020; Zheng et al., 2020; Guo et al., 2021; Sellamuthu et al., 2021). And common reference genes for Coleoptera studies include ACT (actin), β-TUB (beta-tubulin), α-TUB (alpha-tubulin), RPs (ribosomal proteins), 18S rRNA (18S ribosomal RNA), 28S rRNA (28S ribosomal RNA), EF1-α (elongation factor1-α) and so forth (Toutges et al., 2010; Zhang et al., 2020; Guo et al., 2021). These genes are involved in normal cell metabolic processes. However, reference genes in M. saltuarius have not been reported. Therefore, we required to find the appropriate reference genes for gene expression analysis under different PWN treatments.
In this study, we aimed to identify the optimal reference genes in M. saltuarius at different developmental stages associated with infection of PWN or PWN treatment conditions. Based on prior experimental reports regarding reference genes in Coleoptera and other insects, 14 candidate reference genes including sorting nexin 6 (SNX6), phospholipid-transporting ATPase (ATPase), palmitoyltransferase ZDHHC15 isoform X2 (ZDhhc15), transcription factor A, mitochondrial-like (TFAM), 60S ribosomal protein L18 (RPL18), 60S ribosomal protein L7 (RPL7), 40S ribosomal protein S5 (RPS5), transitional endoplasmic reticulum ATPase TER94 (TER), transmembrane and ubiquitin-like domain-containing protein 1 (Tmub1), eukaryotic translation initiation factor 4B (EIF), elongation factor 1-gamma (EF1-γ), cytochrome c oxidase subunit 7C (COX7), tubulin alpha-1 chain (α-TUB), and triosephosphate isomerase (TPI) were selected from the genome and transcriptome data of M. saltuarius (unpublished data). Five algorithms were used to evaluate reference genes stability and perform a comprehensive ranking. In addition, the expression profile of the Krüppel-like factor luna (KLF) gene was used to verify our result. This study provides valuable information for further exploration on the growth and immune mechanism of M. saltuarius, and serves as a reference for exploring its phoretic relationship with B. xylophilus.
Materials and Methods
Insect
In August and December 2020, the fourth and fifth instar larvae of M. saltuarius were collected from Dahuofang Forest Farm, Fushun City, Liaoning Province, China. To ensure the absence of B. xylophilus all times, after sterilizing the larvae surface with 75% alcohol, the fifth instar larvae were incubated at artificial media at 25°C with 75% relative humidity. All procedures were performed at the Plant Quarantine Laboratory, Beijing Forestry University, Beijing, China.
Sample Treatment
Samples collected at different developmental stages associated with infection of PWN in M. saltuarius included two instar larval stages (L4 and L5), 1-day-age pupae (P1), 5-days-age pupae (P5), 10-days-age pupae (P10), and newly emerged adult (1-day-old) males (AM) and females (AF). Three independent biological replicates were performed at each stage, and each replicate was derived using an individual beetle. All samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
To test the effect of B. xylophilus on M. saltuarius, the artificial co-culture medium of B. xylophilus and M. saltuarius was prepared using the previous method (Li et al., 2021). Fifth instar larvae with weights ranging from 300 to 500 mg were selected from Dahuofang Forest Farm in December 2020. After sterilizing the larvae surface with 75% alcohol, the fifth instar larvae inoculated in the artificial co-culture medium were cultured with B. xylophilus at 25°C and 75% relative humidity. The growth and developmental stage of beetles was observed once every 24 h. After the beetle larvae pupated, 5-days-age pupae (BP5), 10-days-age pupae (BP10), and newly emerged adult (1-day-old) males (BAM) and females (BAF) were collected respectively. Three biological replicates were performed at each stage, and each replicate included one sample. All samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
RNA Extraction and cDNA Synthesis
Total RNA from all 33 samples at different developmental stages associated with infection of PWN and PWN treatment conditions were extracted using EASY Spin Plus Tissue/Cell RNA Extraction Kit (Aidlab, China). Quality and quantity of total RNA were evaluated using 1.2% (w/v) agarose gel electrophoresis and NanoDrop 2,000 spectrophotometry. The PrimeScript™ RT Reagent Kit (Takara, China) was used to synthesize the first strand cDNA of each sample according to the manufacturer’s protocol. Obtained cDNAs were diluted 5-fold and stored at −20°C for subsequent RT-qPCR experiments.
Selection of Candidate Reference Genes and Primer Design
Fourteen candidate reference genes with relatively high transcript abundance and stable expression [fragments per kilobase of transcript per million mapped reads (FPKM) value >20 and a fold change in expression <2] were selected based on the transcriptome and genome data (unpublished data) of M. saltuarius. These genes were SNX6, ATPase, ZDhhc15, TFAM, RPL18, RPL7, RPS5, TER, Tmub1, EIF, EF1-γ, COX7, α-TUB, and TPI (Supplementary Table S1). Their coding DNA sequences were obtained from M. saltuarius genome data (GeneBank: OM471799–OM471813), and primers were designed using the web software Primer 3.0 (https://bioinfo.ut.ee/primer3-0.4.0/) and IDT (https://sg.idtdna.com/pages). The primers used for amplification are listed in Table 1.
RT-qPCR Analysis
RT-qPCR was performed using Bio-Rad CFX Connect real-time PCR instrument (Bio-Rad, United States) with TB Green® Premix Ex Taq™ II (Takara, Japan). A 25-μl reaction volume consisted of 12.5 μl of TB Green Premix Ex Taq Ⅱ (2×), 1 μl of forward and reverse primers (10 μM), respectively, 1 μl of cDNA template, and 9.5 μl of RNA-free water. Amplification conditions were as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. Then, we performed a melt curve analysis using the default parameters with a steady increase in temperature from 65 to 95°C. All RT-qPCR assays were performed in three biological replicates, each biological replicate with three technical replicates. The amplification efficiency (E) and correlation coefficients (R2) were determined for each gene using the standard curves with a 5-fold dilution series of the template (1, 1/5, 1/25, 1/125, and 1/625), where R2 was the slope of the standard curve. Amplification efficiency was calculated according to the equation: E% = (10 [−1/slope]−1) × 100%.
Stability Analysis of Candidate Reference Genes
Five algorithms, including delta Ct, geNorm, NormFinder, BestKeeper and RefFinder, were used to analyze the expression stability of the candidate reference genes in different groups. The delta Ct algorithm (based on the delta Ct method) was used to calculate the mean standard deviation (SD) of the paired genes in each sample to assess the gene expression stability. Genes with lower SD value had more stable expression (Silver et al., 2006). The GeNorm was used to calculate the M value based on the pairwise variation between two reference genes. If the M value was less than 1.5, it could be considered a suitable reference gene. The smaller the M value, the higher the stability of gene. The optimal number of reference genes was determined by calculating pairwise variation (Vn/Vn+1) by geNorm. A value of Vn/Vn+1 less than 0.15 indicated that the most suitable reference gene number is n without introducing n + 1 (Vandesompele et al., 2002). In NormFinder, the S value of reference gene according to variance analysis decided the stability of candidate reference genes. The lower the S value, the more stable they were (Andersen et al., 2004). Before using geNorm or NormFinder analysis, the original Ct values were converted to 2−ΔCt values (ΔCt = original Ct value − lowest Ct value in each group). BestKeeper evaluated the expression stability of all candidate reference genes by calculating the SD and stability value (SV) based on the original Ct values. A gene could not be used as an internal reference gene if the SD value was more than 1 (Pfaffl et al., 2004). Similarly, genes with lower SD and SV values had more stable expression. Finally, the comprehensive ranking of candidate reference genes under different conditions was obtained according to RefFinder (Xie et al., 2012).
Validation of Reference Genes
Krüppel-like transcription factor luna (KLF) belongs to a family of 15 different zinc finger proteins of the C2H2 type that are involved invertebrate development, and which controls cell proliferation, growth and differentiation. De Graeve et al. (2003) proposed that KLF was a novel transcriptional determinant of Drosophila development (De Graeve et al., 2003). Therefore, the KLF gene was selected as target gene to validate the stability of the selected reference genes based on the transcriptome data. The primers used are shown in Table 1.
We used RT-qPCR (method same as above) to detect the KLF expression level in M. saltuarius samples at different developmental stages associated with infection of PWN and PWN treatment conditions. The relative quantification of the KLF gene was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). One-way analysis of variance (ANOVA) followed by post-hoc Tukey’s honestly significant difference (HSD) test on SPSS Statistics Software was used to determine the significance of KLF expression levels at different developmental stages associated with infection of PWN and PWN treatment conditions (Wu et al., 2021; Fu et al., 2022).
Results
Primer Performance Analysis of Candidate Reference Genes
A total of 14 candidate reference genes were selected for gene-normalization studies in different samples. RT-qPCR products showed a single peak in the melting curve analysis (Supplementary Figure S1) and 1.2% agarose gel electrophoresis showed a specific band for each gene (Supplementary Figure S2). The amplification efficiency (E) values of all candidate genes ranged from 90% (ATPase) to 109% (ZDhhc15), and regression analysis of all primer pairs showed a correlation coefficient (R2) greater than 0.99 (Table 1). These results indicated that all primer pairs designed for the candidate reference genes had good efficiency and specificity in RT-qPCR amplification. Therefore, the primers of these candidate reference genes were used for further analysis.
Expression Analysis of Selected Reference Genes
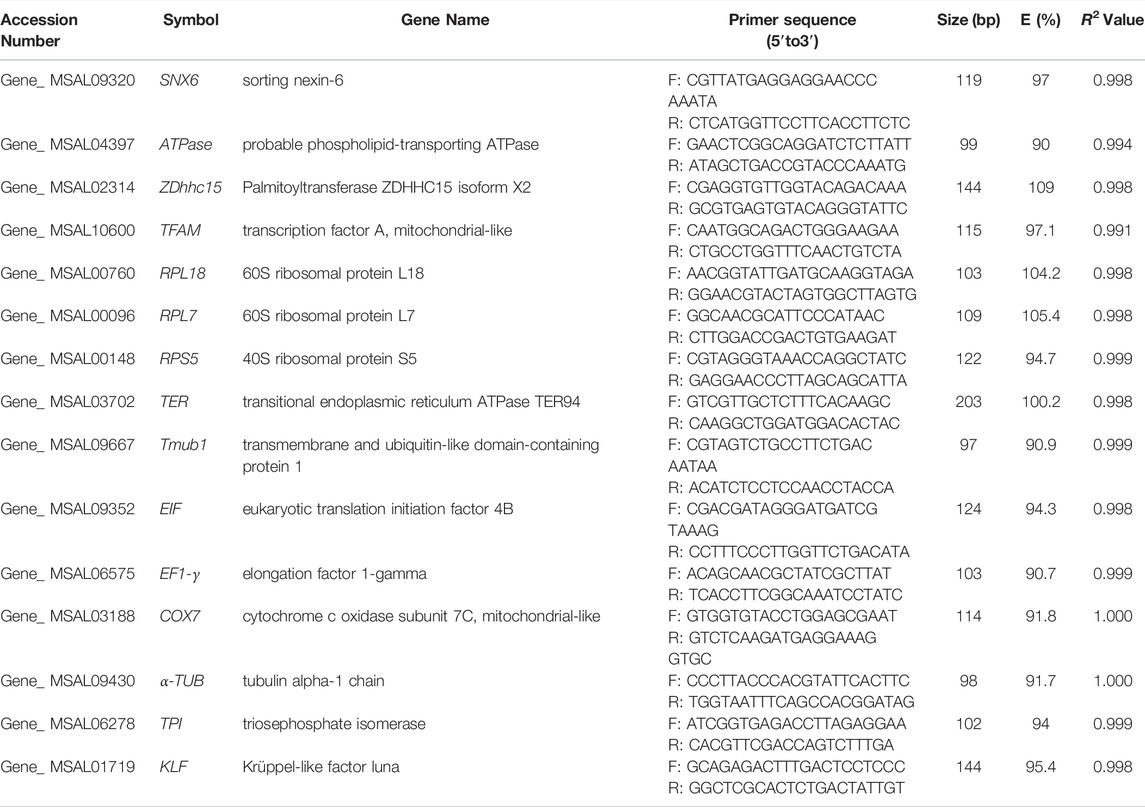
Transcript abundance and cycle threshold (Ct) variation are important parameters for screening reference genes. The Ct value of 14 candidate reference genes across 33 samples showed a wide range of expression levels and significant differences. Ct values of these candidate reference genes ranged from 14.98 to 27.14 for total samples. Among these, α-TUB, RPL7 and RPL18 were the most abundant transcripts (average Ct = 17.41, 18.34, 18.74, respectively). The least frequently expressed reference gene were Zdhhc15, Tmub1, and ATPase (average Ct = 24.80, 25.19, 26.21, respectively). According to the SD values, variance in Ct values increased in the following order: RPS5 < RPL18 < RPL7 < EF1-γ < Zdhhc15 < TER < Tmub1 < SNX6 < ATPase < TPI < TFAM < EIF < α-TUB < COX7 (Figure 1).
FIGURE 1. Cycle threshold (Ct) values of 14 candidate reference genes across total samples in M. saltuarius. Boxes indicate the 25th and 75th percentiles, and lines in the boxes represent the median value.
Expression Stability of Candidate Reference Genes in Different Developmental Stages Associated With Infection of PWN and PWN Treatments
To identify the most suitable reference genes of M. saltuarius under the three conditions—different developmental stages associated with infection of PWN, PWN treatment at the pupal stage, and PWN treatment at the adult stage, their expression stability was evaluated using five algorithms as elaborated below.
Delta Ct Analysis
For candidate reference genes in total samples, RPL7, EF1-γ, and TER were more stable than other reference genes (average SD = 0.73, 0.74, 0.76, respectively). For different treatments (Table 2), TER, Zdhhc15 and EF1-γ had the most stable expression levels (average SD = 0.71, 0.73, 0.74, respectively) at different developmental stages associated with infection of PWN. RPL7, RPS5, and TFAM were the most stable reference genes (average SD = 0.57, 0.60, 0.65, respectively) in PWN-treated pupal groups; while SNX6, RPS5, and RPL7 were the most stable reference genes (average SD = 0.40, 0.41, 0.43, respectively) in PWN-treated adult groups. On the whole α-TUB, COX7, and TPI were the least stable under most conditions.
GeNorm Analysis
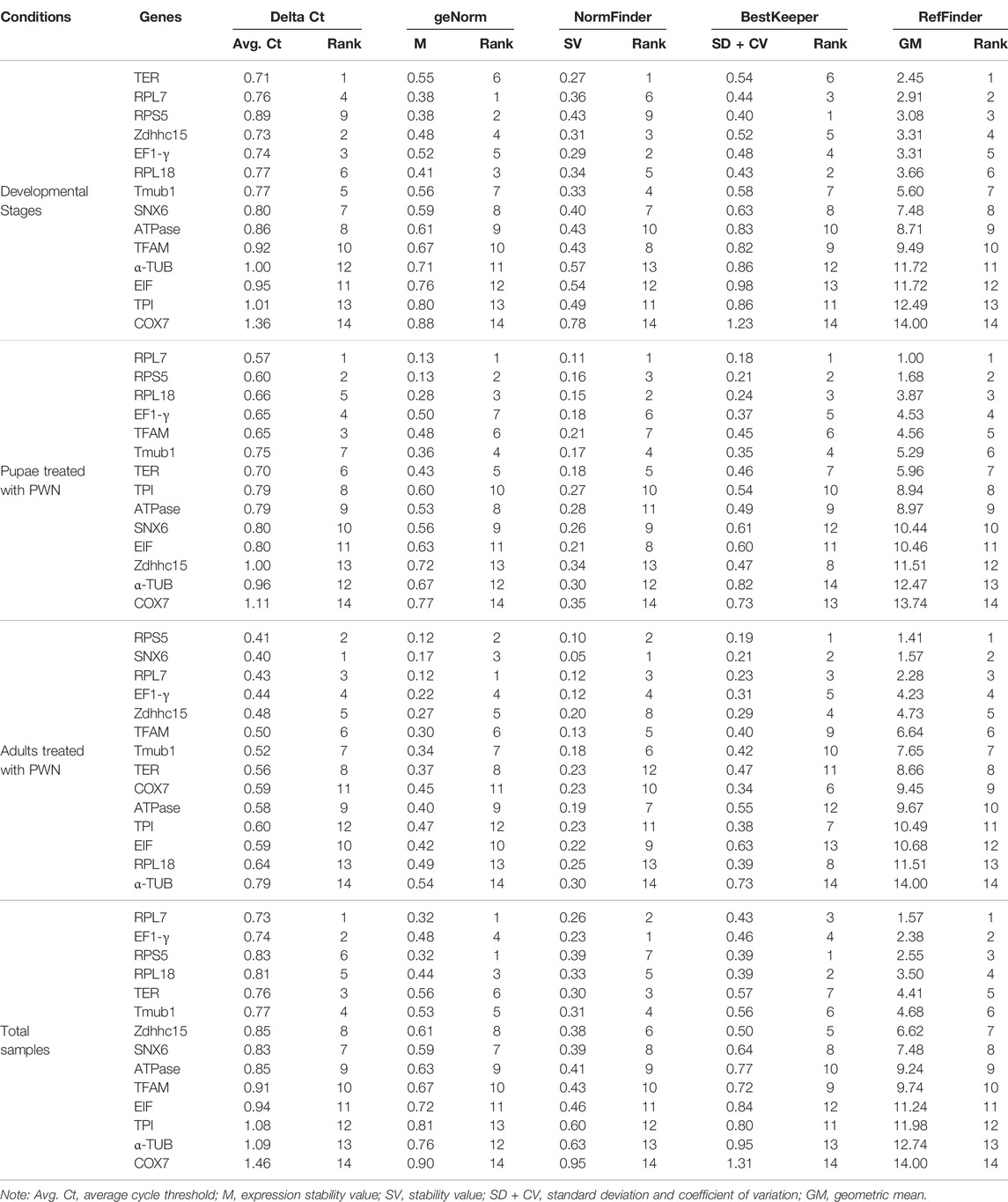
Among all samples, RPL7 and RPS5 were the most stable reference genes, similar with the results found in the sample sets of different developmental stages associated with infection of PWN and all PWN treatment conditions (Figure 2). In addition, we found that the most unstable genes greatly varied in different experimental conditions. In developmental stages associated with infection of PWN and total samples, the M values of COX7 and TPI were higher than other genes. In pupae treated with PWN, COX7 and Zdhhc15 had the least stability. In adults treated with PWN, α-TUB and RPL18 exhibited the most unstable expression levels.
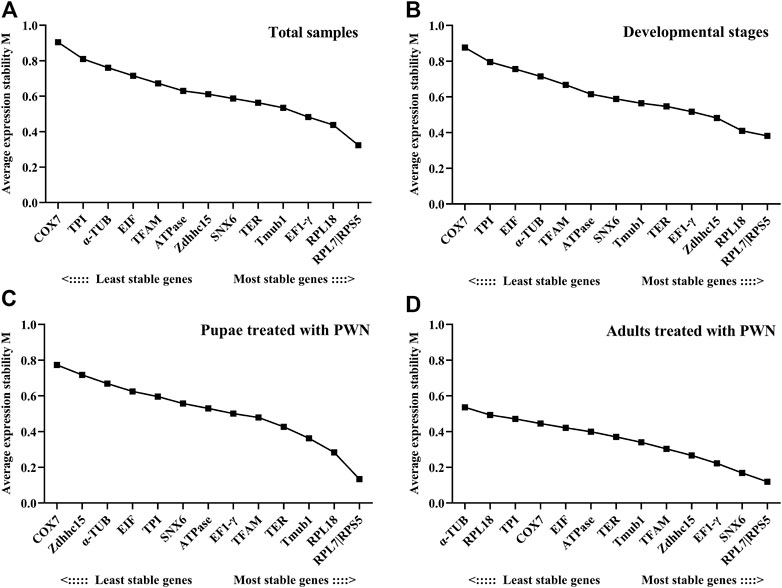
FIGURE 2. Average expression stability and ranking of candidate reference genes calculated by geNorm. Candidate reference genes with lower M values were more stable. The least stable genes are listed on the left, and the most stable genes are listed on the right. (A) Total samples. (B) Developmental stages associated with infection of PWN. (C) Pupae treated with PWN. (D) Adults treated with PWN.
The pairwise changes (Vn/Vn+1) were calculated using geNorm with a threshold value of 0.15 to assess the number of reference genes for all treatment conditions. Three groups including different developmental stages associated with infection of PWN, pupae treated with PWN, and adults treated with PWN, V2/3 values were all <0.15 (0.126, 0.118, and 0.062, respectively), indicating that two reference genes were sufficient for RT-qPCR normalization. In 33 total samples, the V2/3 value was 0.156, which was greater than the split-off value, and the V3/4 value was 0.115. Therefore, three reference genes were needed to normalize the expression of the target gene in all samples (Figure 3).
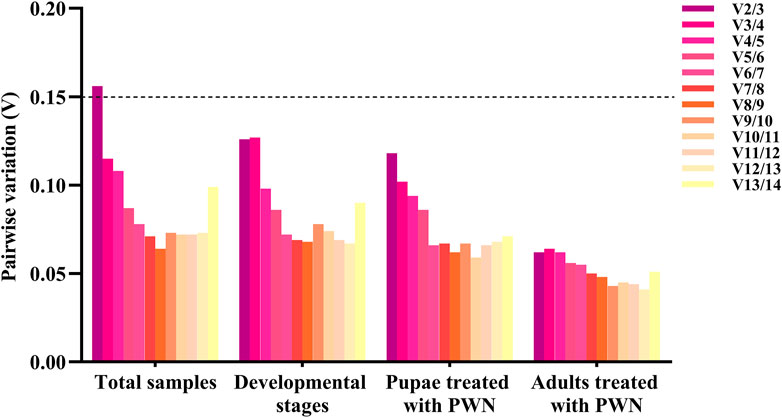
FIGURE 3. Pairwise variation (V) of 14 reference genes in different conditions calculated by geNorm. The threshold value for assessing the optimal number of reference genes for RT-qPCR normalization is 0.15.
NormFinder Analysis
At different developmental stages associated with infection of PWN, the most stable genes were EF1-γ and TER. In pupae treated with PWN, RPL7, and RPL18 had the strongest stability (Table 2). In adults treated with PWN, SNX6, and RPS5 were the most stable reference genes. In total samples, EF1-γ and RPL7 were the best reference genes combination. Unsurprisingly, α-TUB and COX7 were also the least stable genes in most cases.
BestKeeper Analysis
For BestKeeper algorithm, the most stable genes showed the lowest SD ± CV values, and genes with an SD value >1 were considered unstable. In total samples, RPS5, RPL18, and RPL7 were the most stable genes. For different developmental stages associated with infection of PWN and pupae treated with PWN, two conditions had similar results that RPS5, RPL18, and RPL7 were identified as the most stable genes, but α-TUB, COX7, EIF, and SNX6 were poor stable genes. Whereas the PWN-treated adult group was slightly different from the above. For adults treated with PWN, RPS5, SNX6, and RPL7 showed the highest stability; ATPase, α-TUB, and EIF had the least stable expression level (Figure 4). In some groups, COX7, whose SD value was greater than 1, was considered an unstable reference gene.
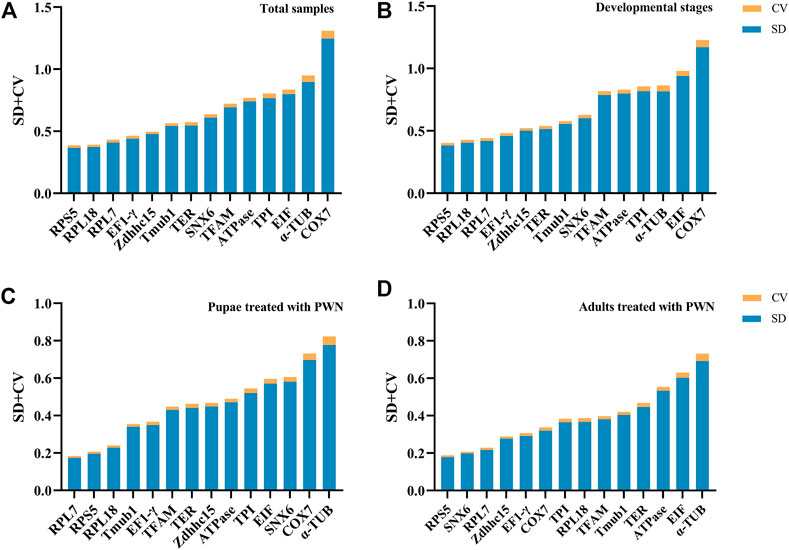
FIGURE 4. Stability rankings of 14 candidate reference genes by BestKeeper. Blue bars represent standard deviation (SD) of average Ct values, and yellow bars represent coefficients of variation (CV). (A) Total samples. (B) Developmental stages associated with infection of PWN. (C) Pupae treated with PWN. (D) Adults treated with PWN.
RefFinder Analysis: Comprehensive Stability Analysis of Reference Genes
The RefFinder program was used to obtain a comprehensive reference gene ranking based on the geometric mean of four algorithms. The expression stabilities of candidate reference genes in all samples decreased in the order: RPL7 > EF1-γ > RPS5 > RPL18 > TER > Tmub1 > Zdhhc15 > SNX6 > ATPase > TFAM > EIF > TPI > α-TUB > COX7. The stability ranking at different developmental stages associated with infection of PWN was as the following: TER > RPL7 > RPS5 > Zdhhc15 > EF1-γ > RPL18 > Tmub1 > SNX6 > ATPase > TFAM > α-TUB > EIF > TPI > COX7. The stability ranking at pupae treated with PWN was: RPL7 > RPS5 > RPL18 > EF1-γ > TFAM > Tmub1 > TER > TPI > ATPase > SNX6 > EIF > Zdhhc15 > α-TUB > COX7, and at adults treated with PWN was RPS5 > SNX6 > RPL7 > EF1-γ > Zdhhc15 > TFAM > Tmub1 > TER > COX7 > ATPase > TPI > EIF > RPL18 > α-TUB. The comprehensive analysis showed that RPL7, EF1-γ, and RPS5 genes were the most stable reference genes combination for total samples. RPL7, RPS5, and RPL18 were the most suitable reference genes in pupae treated with PWN. RPS5, SNX6, and RPL7 were the most suitable reference genes in adult treated with PWN (Table 2). TER, RPL7, and RPS5 were the optimal reference genes at different developmental stages associated with infection of PWN.
Validation of the Selected Reference Genes
To verify the reliability of the selected reference genes, KLF was used as the target gene for RT-qPCR analysis. We used the four most stable candidate reference genes (RPL7, RPS5, SNX6, and TER), the combination of these stable genes (RPL7 + RPS5 + EF1-γ, RPL7 + RPS5, RPS5+SNX6, and RPL7+TER), and two most unstable reference genes (α-TUB, COX7) in different treatment conditions to normalize the expression of KLF (Figure 5).
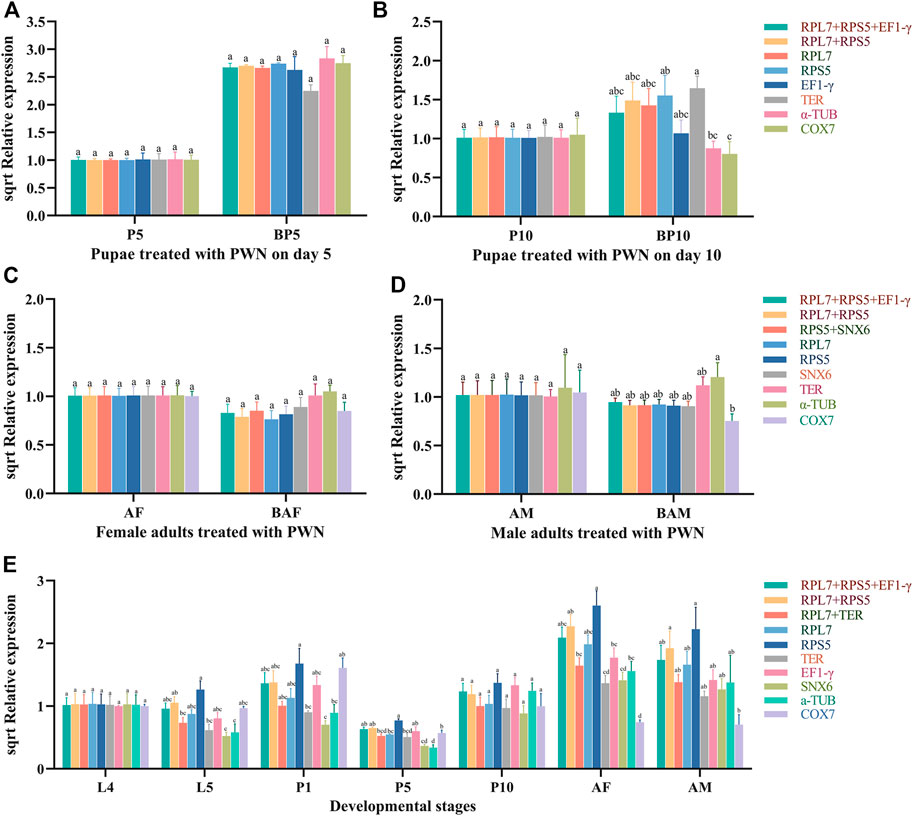
FIGURE 5. Relative expression levels of KLF normalized by candidate reference genes. Different letters indicate the significant differences in KLF expression levels (ANOVA, HSD, p < 0.05). Sqrt (Relative expression) represents the square root of the relative expression value. (A) Pupae treated with PWN on day 5. (B) Pupae treated with PWN on day 10. (C) Female adults treated with PWN. (D) Male adults treated with PWN. (E) Developmental stages associated with infection of PWN in M. saltuarius.
In the pupae treated with PWN, the relative expression level of KLF was significantly up-regulated in BP5 and BP10 groups compared to control groups (P5, P10), which was normalized by the top-ranked gene (RPL7, RPS5 or their combinations). Similar expression-profile changes were obtained by the combination of stable reference genes (RPL7 + RPS5 + EF1-γ), and there were no significant differences among those normalized by RPL7, RPS5 individually, and RPL7 + RPS5. However, the normalization by the least stable reference gene (TER, α-TUB, and COX7) led to a strong bias in the expression level of KLF in different treatments. α-TUB and COX7 significantly decreased the transcription of KLF in BP10, and TER decreased in BP5. In the adults treated with PWN, although the expression trends were very similar, normalization with the unstable reference gene TER and α-TUB increased the expression level of KLF in BAF and BAM, which resulted in larger standard deviation values. At the different developmental stages associated with infection of PWN, the expression levels of KLF normalized by RPL7 and TER individually, or RPL7 + TER were different with COX7 and RPS5. When normalized by RPS5, the expression of KLF increased in every developmental stage (L5, P1, P5, P10, AF and AM), and significantly decreased in AF and AM stages normalized by COX7.
Discussion
M. saltuarius is a unique vector of B. xylophilus in northeast China. Its molecular physiology and the function of genes has been actively explored with the unpublished genomes, and recent transcriptomic advances have provided an opportunity for exploring the interspecific interaction mechanism between M. saltuarius and B. xylophilus, which was in favour of controlling the spread of pine wilt disease to north China. Therefore, it is necessary to probe gene function and quantify gene expression in M. saltuarius. Due to high sensitivity, rapidity, specificity, and accuracy, RT-qPCR is an effective method to study this mechanism. To reduce some inter-sample errors, appropriate reference genes are needed to normalize target genes (Zhao et al., 2022). However, there is no research on the reference genes of M. saltuarius. We systematically selected the reliable inference genes for standardization of gene expression by using five assessment algorithms (delta Ct, geNorm, NormFinder, BestKeeper, and RefFinder) in M. saltuarius at different developmental stages associated with infection of PWN and treated with PWN at the pupal and adult stages.
In our results, some candidate reference genes varied with different algorithms. TER ranked first in delta Ct and NormFinder, whereas it ranked sixth in geNorm and BestKeeper at different developmental stages associated with infection of PWN in M. saltuarius. The ranking of genes by different software was diverse, probably because different programs have different algorithmics, and the differences in the scaling systems used by the algorithms can also lead to these variations (Zhai et al., 2014; Sagri et al., 2017). Although the ranking order varies depending on the analysis program used, the overall trend was similar. For instance, in the adults treated with PWN, RPS5, SNX6, RPL7, EF1-γ, and Zdhhc15 were all the top five stable genes in the delta Ct, geNorm, BestKeeper, and RefFinder. According to the geNorm, NormFinder, RefFinder, and BestKeeper, PWN, RPL7, RPS5, and RPL18 were all the top three most stable genes in the pupae treated with PWN. Therefore, in practical application, the results provided by these algorithms are required to be considered comprehensively.
Most studies have found that two or more reference genes rather than a single reference gene can increase the accuracy of relative quantification (Vandesompele et al., 2002; Haller et al., 2004; Veazey and Golding, 2011). In our study, the optimal number of reference genes was calculated by geNorm. Most experimental conditions showed values below the proposed 0.15 cut-off value at V2/3. This result indicated that combining the top two reference genes would be adequate for the normalization of gene expression data at developmental stages associated with infection of PWN and PWN treatment conditions.
In this study, the stability of reference genes in M. saltuarius could differ under various experimental conditions. TER and RPL7 were stable reference genes at different developmental stages associated with infection of PWN. At the same time, RPL7 + RPS5 and RPS5 + SNX6 were identified as optimal reference genes in pupal stage treated with PWN, adult stage treated with PWN, respectively. Previous studies have also shown that no reference gene has always been stably expressed under different experimental conditions, in which species, growth stage, tissue, temperature, strain, population, and pesticide varied. Sellamuthu et al. (2021) showed that β-TUB, Eef2 and RPS3 were the most stable gene under different developmental stages and sex, while UBQ and V-ATPase were the most stable genes after Juvenile Hormone III treatment in Ips sexdentatus (Sellamuthu et al., 2021). β-TUB expression was also stable in Aquatica leii at different developmental stages, but GST was the most stably gene under different temperatures (Yang et al., 2020). Similarly, RPS32 was stably expressed in different tissues of Agasicles hygrophila while showing lower stability under different nutritional conditions (Guo et al., 2021).
Besides, it was observed that B. xylophilus induced more variations in the Ct values in M. saltuarius. In M. alternatus, PWN caused significant changes at the physiological and molecular level. Zhao et al. (2016) found that ascarosides secreted by dispersal juveniles (LIII) of B. xylophilus could facilitate M. alternatus pupation by upregulating ecdysone-dependent gene expression. When dispersal juveniles (LⅣ) of B. xylophilus entered the vector beetle, PWN affected the gene expression of Toll signal pathway (Zhou et al., 2018). In this study, the stable reference genes at PWN treatment conditions were different from normal developmental stages. This result suggested that B. xylophilus can also cause variations in transcript levels in M. saltuarius.
Among the 14 reference genes studied in this study, ribosomal proteins exhibited more stability compared to other candidate genes in relation to different biotic (developmental stages and PWN treatment) factors. Ribosomal protein genes, which play an important role in ribosome biogenesis, protein translation, and cell development, were one of the most stable reference genes in diverse biotic and abiotic conditions in many insects (Zhou et al., 2015). In Tribolium castaneum and Coccinella septempunctata, ribosomal proteins exhibited a high level of stability at different developmental stages (Yang et al., 2016; Lü et al., 2018). In different sexes of M ylabris cichorii, and I. sexdentatus, RPL22 and RPS3, respectively, were the most suitable reference genes for RT-qRCR normalization (Wang et al., 2014; Sellamuthu et al., 2021). Our results demonstrated that the ribosomal proteins were also transcriptionally conserved in M. saltuarius under PWN treatment.
The genes of tubulin, a protein that maintains the cytoskeletal structure and morphology in eukaryotic cells, are also frequently used as reference genes (Caridi et al., 2019). For example, α-TUB was stably expressed in Drosophila melanogaster exposed to different temperatures (Fleur et al., 2011). In Antheraea pernyi, α-TUB was suitable reference gene for normalizing RT-qPCR data infected by multicapsid nucleopolyhedrovirus (Zhao et al., 2019). However, α-TUB was unstable as reference genes under certain conditions, such as in Spodoptera litura larvae treated with azadirachtin (Lu et al., 2018). In our research, α-TUB showed instability under PWN treatment conditions. COX responds to a wide variety of metabolic states and is also considered a novel reference gene for different tissues in A. hygrophila and Spodoptera frugiperda (Guo et al., 2021; Shu et al., 2021), while was inconsistent with our results. In M. saltuarius subjected to several experimental conditions (different developmental stages, adults treated with PWN, and all samples), COX7 was particularly unstable reference gene. These results suggested that reference genes differ from species to species.
KLF is a key DNA-binding transcriptional factor that regulates various pathways that pertain to insect metamorphosis, metabolism, and other cellular mechanisms, and was selected as the target gene (Weber et al., 2014). The overall transcription pattern of KLF normalized with the most stable internal reference genes was similar to the transcriptome data at different developmental stages associated with infection of PWN and PWN treatment. At different developmental stages, the expression level of KLF increased at emergence period (AF and AM) when normalized by the top ranked genes and their combinations. On the contrary, normalization with COX7 showed the lowest transcription of KLF in adults (AF and AM). Under certain conditions, normalizing with unsuitable reference genes affected the gene expression and resulted in more significant standard deviations (Lü et al., 2018). The expression level of KLF in the pupae treated with PWN (BP5 and BP10) was higher than pupae treated without PWN when normalized by the top ranked genes and their combinations, and the same expression pattern was also observed in M. alternatus (Zhao et al., 2016). However, normalization by the least stable reference gene resulted in a strong bias. The transcriptions of KLF significantly decreased in the pupae treated with PWN (BP10). Similar results were observed in the condition of adults treated with PWN. Consequently, our findings confirmed the importance of selecting and validated accurate reference genes for RT-qPCR analysis to avoid the misinterpretation of target gene transcription data.
Conclusion
This is the first study evaluating reference genes in M. saltuarius. We evaluated the stability of 14 candidate reference genes in samples from this beetle at different developmental stages associated with infection of PWN and PWN treatment conditions by delta Ct, geNorm, NormFinder, BestKeeper and RefFinder algorithms. We concluded that RPL7 and TER were suitable reference genes at different developmental stages associated with infection of PWN. RPL7 and RPS5 were considered the most stable reference genes in pupae treated with PWN. RPS5 and SNX6 could be used as reference genes in adults treated with PWN. RPL7, EF1-γ, and RPS5 could be used as stable reference genes in all the samples. Overall, RPL7 and RPS5 were the most stable reference genes for M. saltuarius under different conditions. Our results could provide stable reference genes for RT-qPCR gene expression analysis of M. saltuarius, also lay a foundation for the study of its phoretic relationship with B. xylophilus.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository. The names of the repository and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/genbank/; OM471799, OM471800, OM471801, OM471802, OM471803, OM471804, OM471805, OM471806, OM471807, OM471808, OM471809, OM471810, OM471811, OM471812, OM471813.
Author Contributions
JL carried out the majority of the bioinformatics studies and participated in performing the experiments. NF was involved in experimental data analysis. JL and NF wrote the manuscript. LR and YL participated in the design of the study and helped to draft the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Key Research and Development Program of China (2021YFD1400900) and the Chinese National Natural Science Foundation (31870642).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge Xiaoyu Xin, Sixun Ge, and Zhenxiao Li (Beijing Forestry University, Beijing), for kind assistance in specimen collection. We also thank the staff of Liaoning Station of Forest and Grassland Pest Management in Shenyang for their support in our collection work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.882792/full#supplementary-material
References
Abelleira A., Picoaga A., Mansilla J. P., Aguin O. (2011). Detection of Bursaphelenchus Xylophilus, Causal Agent of Pine Wilt Disease on Pinus pinaster in Northwestern Spain. Plant Dis. 95, 776. doi:10.1094/PDIS-12-10-0902
Akbulut S., Stamps W. T. (2012). Insect Vectors of the Pinewood Nematode: a Review of the Biology and Ecology of Monochamus Species. For. Pathol 42, 89–99. doi:10.1111/j.1439-0329.2011.00733.x
Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: a Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and colon Cancer Data Sets. Cancer Res. 64 (15), 5245–5250. doi:10.1158/0008-5472.CAN-04-0496
Barros Rodrigues T., Khajuria C., Wang H., Matz N., Cunha Cardoso D., Valicente F. H., et al. (2014). Validation of Reference Housekeeping Genes for Gene Expression Studies in Western Corn Rootworm (Diabrotica Virgifera Virgifera). Plos One 9 (10), e109825. doi:10.1371/journal.pone.0109825
Borowski J. M., Galli V., da Silva Messias R., Perin E. C., Buss J. H., dos Anjos e Silva S. D., et al. (2014). Selection of Candidate Reference Genes for Real-Time PCR Studies in Lettuce under Abiotic Stresses. Planta 239, 1187–1200. doi:10.1007/s00425-014-2041-2
Bustin S. A., Benes V., Nolan T., Pfaffl M. W. (2005). Quantitative Real-Time RT-PCR - a Perspective. J. Mol. Endocrinol. 34, 597–601. doi:10.1677/jme.1.01755
Bustin S. (2002). Quantification of mRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 29, 23–39. doi:10.1677/jme.0.0290023
Caridi C. P., Plessner M., Grosse R., Chiolo I. (2019). Nuclear Actin Filaments in DNA Repair Dynamics. Nat. Cell Biol. 21 (9), 1068–1077. doi:10.1038/s41556-019-0379-1
Cheng H. R., Lin M. S., Li W. Q., Fang Z. D. (1983). Pine Wilt Disease on Pinus Thunbergii Parl in Nanjing. Forest Pest and Disease 4, 1–5.
De Graeve F., Smaldone S., Laub F., Mlodzik M., Bhat M., Ramirez F. (2003). Identification of the Drosophila Progenitor of Mammalian Krüppel-like Factors 6 and 7 and a Determinant of Fly Development. Gene 314, 55–62. doi:10.1016/S0378-1119(03)00720-0
Dropkin B. H., Foudin A. S. (1979). Report of the Occurrence of Burasphelenchus lignicolus Induced Pine Wilt Disease in Missouri. Plant Dis. Rep. 63, 904–905.
Feng B., Guo Q. S., Mao B. P., Du Y. J. (2016). Identification and Validation of Reference Genes for qRT-PCR Analysis in Chemosensory Tissue of Monochamus Alternatus. Acta Entomologica Sinica 59, 427–437.
Fleige S., Pfaffl M. W. (2006). RNA Integrity and the Effect on the Real-Time qRT-PCR Performance. Mol. Aspects Med. 27, 126–139. doi:10.1016/j.mam.2005.12.003
Fu N., Li J., Wang M., Ren L., Zong S., Luo Y. (2022). Identification and Validation of Reference Genes for Gene Expression Analysis in Different Development Stages of Amylostereum Areolatum. Front. Microbiol. 12, 827241. doi:10.3389/fmicb.2021.827241
Guo Y.-Q., Yang Y., Chai Y., Gao L.-L., Ma R. (2021). Identification and Evaluation of Reference Genes for Quantitative PCR Normalization in Alligator Weed Flea Beetle (Coleoptera: Chrysomelidae). J. Insect Sci. 21 (5), 9. doi:10.1093/jisesa/ieab067
Haller F., Kulle B., Schwager S., Gunawan B., Heydebreck A. v., Sültmann H., et al. (2004). Equivalence Test in Quantitative Reverse Transcription Polymerase Chain Reaction: Confirmation of Reference Genes Suitable for Normalization. Anal. Biochem. 335, 1–9. doi:10.1016/j.ab.2004.08.024
Han H., Chung Y.-J., Shin S.-C. (2008). First Report of Pine Wilt Disease on Pinus Koraiensis in Korea. Plant Dis. 92, 1251. doi:10.1094/PDIS-92-8-1251A
Khan V. H. (1971). The Pine Wilt Disease Caused by Bursaphelenchus xylophilus in Nigeria. Pakistan J. Nematol. 9 (1), 57–59.
Kim B.-N., Kim J. H., Ahn J.-Y., Kim S., Cho B.-K., Kim Y.-H., et al. (2020). A Short Review of the Pinewood Nematode, Bursaphelenchus Xylophilus. Toxicol. Environ. Health Sci. 12, 297–304. doi:10.1007/s13530-020-00068-0
Kim M., Kim J., Han J., Kim Y., Yoon C., Kim G. (2006). Mating Behavior of Pine Sawyer, Monochamus Saltuarius Gebler (Coleoptera: Cerambycidae). J. Asia-pac. Entomol. 9 (3), 275–280. doi:10.1016/s1226-8615(08)60303-9
Klein D. (2002). Quantification Using Real-Time PCR Technology: Applications and Limitations. Trends Mol. Med. 8, 257–260. doi:10.1016/s1471-4914(02)02355-9
Li J. X., Ren L. L., Luo Y. Q., Xin X. Y. (2021). A Method and Application of a Culture Medium for Co-culture of Monochamus Saltuarius and Pine Wood Nematode. CN113475628A. Beijing, China: China National Intellectural Property Administration.
Li M., Li H., Sheng R.-C., Sun H., Sun S.-H., Chen F.-M. (2020). The First Record of Monochamus Saltuarius (Coleoptera; Cerambycidae) as Vector of Bursaphelenchus Xylophilus and its New Potential Hosts in China. Insects 11, 636. doi:10.3390/insects11090636
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lü J., Yang C., Zhang Y., Pan H. (2018). Selection of Reference Genes for the Normalization of RT-qPCR Data in Gene Expression Studies in Insects: A Systematic Review. Front. Physiol. 9, 1560. doi:10.3389/fphys.2018.01560
Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2018). Identification and Validation of Reference Genes for Gene Expression Analysis Using Quantitative PCR in Spodoptera Litura (Lepidoptera: Noctuidae). Plos One 8 (7), e68059. doi:10.1371/journal.pone.0068059
Manuel M. M., Helen B., Maria A. B., Ana C. P., Wolfgang B., Kai M. (1999). First Report of Bursaphelenchus Xylophilus in Portugal and in Europe. Nematology 1 (7), 727–734.
Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper - Excel-based Tool Using Pair-wise Correlations. Biotechnol. Lett. 26, 509–515. doi:10.1023/b:bile.0000019559.84305.47
Ponton F., Chapuis M.-P., Pernice M., Sword G. A., Simpson S. J. (2011). Evaluation of Potential Reference Genes for Reverse Transcription-qPCR Studies of Physiological Responses in Drosophila melanogaster. J. Insect Physiol. 57 (6), 840–850. doi:10.1016/j.jinsphys.2011.03.014
Rajarapu S. P., Mamidala P., Mittapalli O. (2012). Validation of Reference Genes for Gene Expression Studies in the Emerald Ash Borer (Agrilus Planipennis). Insect Sci. 19, 41–46. doi:10.1111/j.1744-7917.2011.01447.x
Rodrigues T. B., Dhandapani R. K., Duan J. J., Palli S. R. (2017). RNA Interference in the Asian Longhorned Beetle:Identification of Key RNAi Genes and Reference Genes for RT-qPCR. Sci. Rep. 7 (1), 1–10. doi:10.1038/s41598-017-08813-1
Sagri E., Koskinioti P., Gregoriou M.-E., Tsoumani K. T., Bassiakos Y. C., Mathiopoulos K. D. (2017). Housekeeping in Tephritid Insects: the Best Gene Choice for Expression Analyses in the Medfly and the Olive Fly. Sci. Rep. 7, 45634. doi:10.1038/srep45634
Sato H. S. T. A. (1987). Transmission of Bursaphelenchus Xylophilus Nickle (Nematoda, Aphelenchoididae) by Monochamus Saltuarius (Gebler) (Coleptera, Cerambycidae). J. Jpn. Soc. Hortic. Sci. 69, 492–496.
Sellamuthu G., Amin S., Bílý J., Synek J., Modlinger R., Sen M. K., et al. (2021). Reference Gene Selection for Normalizing Gene Expression in Ips Sexdentatus (Coleoptera: Curculionidae: Scolytinae) under Different Experimental Conditions. Front. Physiol. 12. doi:10.3389/fphys.2021.752768
Shi X.-Q., Guo W.-C., Wan P.-J., Zhou L.-T., Ren X.-L., Ahmat T., et al. (2013). Validation of Reference Genes for Expression Analysis by Quantitative Real-Time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 6, 93. doi:10.1186/1756-0500-6-93
Shu B.-s., Yu H.-k., Dai J.-h., Xie Z.-g., Qian W.-q., Lin J.-t. (2021). Stability Evaluation of Reference Genes for Real-Time Quantitative PCR Normalization in Spodoptera Frugiperda (Lepidoptera: Noctuidae). J. Integr. Agric. 20, 2471–2482. doi:10.1016/S2095-3119(20)63298-1
Silveira G. O., Amaral M. S., Coelho H. S., Maciel L. F., Pereira A. S. A., Olberg G. G. O., et al. (2021). Assessment of Reference Genes at Six Different Developmental Stages of Schistosoma Mansoni for Quantitative RT-PCR. Sci. Rep. 11, 16816. doi:10.1038/s41598-021-96055-7
Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol 7, 33. doi:10.1186/1471-2199-7-33
Song W., Wang X. J., Guo R. J., Zhang W., Zhang Z. Q., Li M. L. (2015). Selection of Reference Genes for qRT-PCR Analysis of Dastarcus Helophoroides. Acta Agriculturae Boreali-occidentalis Sinica. 24, 156–161.
Tan Q.-Q., Zhu L., Li Y., Liu W., Ma W.-H., Lei C.-L., et al. (2015). A De Novo Transcriptome and Valid Reference Genes for Quantitative Real-Time PCR in Colaphellus Bowringi. Plos One 10, e0118693. doi:10.1371/journal.pone.0118693
Toutges M. J., Hartzer K., Lord J., Oppert B. (2010). Evaluation of Reference Genes for Quantitative Polymerase Chain Reaction across Life Cycle Stages and Tissue Types of Tribolium castaneum. J. Agric. Food Chem. 58, 8948–8951. doi:10.1021/jf101603j
Valasek M. A., Repa J. J. (2005). The Power of Real-Time PCR. Adv. Physiol. Edu. 29, 151–159. doi:10.1152/advan.00019.2005
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 3 (7), research0034.1. doi:10.1186/gb-2002-3-7-research0034
Veazey K. J., Golding M. C. (2011). Selection of Stable Reference Genes for Quantitative Rt-PCR Comparisons of Mouse Embryonic and Extra-embryonic Stem Cells. Plos One 6, e27592. doi:10.1371/journal.pone.0027592
Wang Y., Wang Z.-K., Huang Y., Liao Y.-F., Yin Y.-P., Tu J. (2014). Identification of Suitable Reference Genes for Gene Expression Studies by qRT-PCR in the Blister Beetle Mylabris Cichorii. Mylabris Cichorii. J. Insect Sci. 14, 94. doi:10.1093/jis/14.1.94
Weber U., Rodriguez E., Martignetti J., Mlodzik M. (2014). Luna, a Drosophila KLF6/KLF7, Is Maternally Required for Synchronized Nuclear and Centrosome Cycles in the Preblastoderm Embryo. Plos One 9, e96933. doi:10.1371/journal.pone.0096933
Wu Y., Zhang C., Yang H., Lyu L., Li W., Wu W. (2021). Selection and Validation of Candidate Reference Genes for Gene Expression Analysis by RT-qPCR in Rubus. Ijms 22, 10533. doi:10.3390/ijms221910533
Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). miRDeepFinder: a miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 80, 75–84. doi:10.1007/s11103-012-9885-2
Yang C., Preisser E. L., Zhang H., Liu Y., Dai L., Pan H., et al. (2016). Selection of Reference Genes for RT-qPCR Analysis in Coccinella septempunctata to Assess Un-intended Effects of RNAi Transgenic Plants. Front. Plant Sci. 7, 1672. doi:10.3389/fpls.2016.01672
Yang X.-J., Zheng H.-L., Liu Y.-Y., Li H.-W., Jiang Y.-H., Lin L.-B., et al. (2020). Selection of Reference Genes for Quantitative Real-Time PCR in Aquatica Leii (Coleoptera: Lampyridae) under Five Different Experimental Conditions. Front. Physiol. 11. doi:10.3389/fphys.2020.555233
Yang X., Pan H., Yuan L., Zhou X. (2018). Reference Gene Selection for RT-qPCR Analysis in Harmonia axyridis, a Global Invasive Lady Beetle. Sci. Rep. 8. doi:10.1038/s41598-018-20612-w
Yu H. Y., Wu H., Huang R. F., Wang J., Zhang R. X., Song Y. S. (2020). Isolation and Identification of pine wood Nematode from Pinus Sylvestris in Fushun, Liaoning Province. For. Pest Dis. 39, 6–10.
Yu H. Y., Wu H. (2018). New Host Plants and Vectors of pine wood Nematodes Were Found in Liaoning Province. For. Pest Dis. 37, 61.
Yu H. Y., Wu H., Zhang X. D., Wang L. M., Zhang X. F., Song Y. S. (2019). Report on Infection of pine wood Nematode in Natural Condition of Larch. For. Pest Dis. 38, 7–10.
Zhai Y., Lin Q., Zhou X., Zhang X., Liu T., Yu Y. (2014). Identification and Validation of Reference Genes for Quantitative Real-Time PCR in Drosophila Suzukii (Diptera: Drosophilidae). Plos One 9, e106800. doi:10.1371/journal.pone.0106800
Zhang Y., Chen J., Chen G., Ma C., Chen H., Gao X., et al. (2020). Identification and Validation of Reference Genes for Quantitative Gene Expression Analysis in Ophraella communa. Front. Physiol. 11. doi:10.3389/fphys.2020.00355
Zhao L., Zhang X., Wei Y., Zhou J., Zhang W., Qin P., et al. (2016). Ascarosides Coordinate the Dispersal of a Plant-Parasitic Nematode with the Metamorphosis of its Vector Beetle. Nat. Commun. 7, 12341. doi:10.1038/ncomms12341
Zhao X., Geng Y., Hu T., Zhao Y., Yang S., Hao D. (2022). Evaluation of Optimal Reference Genes for qRT-PCR Analysis in Hyphantria cunea (Drury). Insects 13, 97. doi:10.3390/insects13010097
Zhao Z., Wang L., Yue D., Ye B., Li P., Zhang B., et al. (2019). Evaluation of Reference Genes for Normalization of RT-qPCR Gene Expression Data forTrichoplusia niCells DuringAntheraea pernyi(Lepidoptera: Saturniidae) Multicapsid Nucleopolyhedrovirus (AnpeNPV) Infection. J. Insect Sci. 19. doi:10.1093/jisesa/iey133
Zheng C., Zhao D., Xu Y., Shi F., Zong S., Tao J. (2020). Reference Gene Selection for Expression Analyses by qRT-PCR in Dendroctonus Valens. Insects 11, 328. doi:10.3390/insects11060328
Zhou J., Zhao L.-L., Yu H.-Y., Wang Y.-H., Zhang W., Hu S.-N., et al. (2018). Immune Tolerance of Vector Beetle to its Partner Plant Parasitic Nematode Modulated by its Insect Parasitic Nematode. FASEB j. 32, 4862–4877. doi:10.1096/fj.201800247R
Keywords: Monochamus saltuarius, Bursaphelenchus xylophilus, RT-qPCR, reference genes, developmental stages
Citation: Li J, Fu N, Ren L and Luo Y (2022) Identification and Validation of Reference Genes for Gene Expression Analysis in Monochamus saltuarius Under Bursaphelenchus xylophilus Treatment. Front. Physiol. 13:882792. doi: 10.3389/fphys.2022.882792
Received: 25 February 2022; Accepted: 21 March 2022;
Published: 25 April 2022.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation, BrazilReviewed by:
Benshui Shu, Zhongkai University of Agriculture and Engineering, ChinaLifeng Zhou, Zhejiang A & F University, China
Copyright © 2022 Li, Fu, Ren and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Ren, lily_ren@bjfu.edu.cn; Youqing Luo, youqingluo@126.com
 Jiaxing Li
Jiaxing Li Ningning Fu
Ningning Fu Lili Ren
Lili Ren Youqing Luo*
Youqing Luo*