- 1Henan International Joint Laboratory for Nuclear Protein Regulation, School of Basic Medical Sciences, Henan University, Kaifeng, China
- 2School of Life Sciences, Henan University, Kaifeng, China
- 3Faculty of Pharmacy, The University of Lahore, Lahore, Pakistan
- 4Key Laboratory of Natural Medicine and Immune Engineering, School of Medicine, Henan University, Kaifeng, China
- 5Department of Biological Sciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, Bangi, Malaysia
- 6Department of Bio-Sciences, University Wah, Rawalpindi, Pakistan
- 7Institute of Nursing and Health, Henan University, Kaifeng, China
- 8School of Stomatology, Henan University, Kaifeng, China
- 9Kaifeng Key Laboratory of Infection and Biological Safety, Henan University College of Medicine, Kaifeng, China
PEST-containing nuclear protein (PCNP), a novel nuclear protein, is involved in vital cellular processes like cell proliferation and mediates tumorigenesis. PCNP is a short-living, small nuclear protein of only 178 amino acids with two remarkable PEST sequences that are rich in proline (P), glutamic acid (E), serine (S), and threonine (T). The current understanding of PCNP reveals that PCNP has the ability to interact with cell cycle regulatory proteins; tumor suppressors (p53 and pRB), and promoters (cyclin E and cyclin D) to determine the fate of tissues to facilitate the process of either apoptosis or cell proliferation. In many preclinical studies, it has been evaluated that PCNP expression has associations with the development and progression of various cancers like neuroblastoma, lung adenocarcinoma, and ovarian cancer. Based on these depicted novel roles of PCNP in cell cycleregulation and of PCNP in tumorigenesis, it is logical to consider PCNP as a potential molecular target for cancer research. The aim of the current communication is to present an update on PCNP research and discussion on the potential role of PCNP in cancer development with challenges and opportunities perspectives. Considering the available evidence as a baseline for our statement, we anticipate that in the future, new research insights will strengthen the aim to develop PCNP-based diagnostic and therapeutic approaches that will move the PCNP from the laboratory to the cancer clinic.
Introduction
To maintain normal continuity of cell life, nuclear proteins (NPs) have central importance in regulating vital innate mechanisms. Molecular and biochemical evidences have attested to the notion that NPs with amazing features play important roles to regulate the cell division and stem cell generation, condense and repair the genomic contents, organize the nuclear metabolism, and maintain the architecture of cell machinery with the process of ubiquitination (1–6). Based on their nature and functions, a family of NPs, rich in proline (P), glutamic acid (E), serine (S), and threonine (T) sequences, known as PEST-proteins (PEST-NPs), is widely distributed and involved in various cellular functions. This class of PEST-NPs is known as the guardian of the cell and is believed to play roles in the ubiquitin–proteasome pathway, glycosylation of nuclear pores, and the hexosamine biosynthetic pathways (7, 8). Furthermore, these proteins are considered active players in cellular processes like cell cycle regulation, cell nutrient uptake and cellular metabolism regulation, nucleocytoplasmic transport and regulating cyclic nucleotide signaling pathways (9–12). Some of the PEST-NPs are primarily involved in regulating cancer metabolism by interfering with key signaling pathways and cancer-immune mechanisms via apoptosis and autophagy (7). In particular, there is a growing volume of evidence about PEST-NPs PEST sequence function as proteolytic signals to target the degradation of different proteins via the proteasome or by calpain proteolysis pathways (8, 13, 14).
In 2002, through database mining, Mori and his co-workers discovered a novel PEST-containing protein, named PCNP, localized in the nucleus. Rich in these PEST sequences, PCNP is a novel short-living NP and regulates many cellular activities in the form of a transcriptional factor or acting as cell cycle regulatory proteins (15).
Through the proteomic studies databases and in vitro–in vivo experiments, PCNP expression has been confirmed in several cancer cells, including HepG2 hepatoma cells, U-937 myeloid leukemia cells, and HT-1080 fibrosarcoma cells (15, 16). Furthermore, studies confirmed the potential mediating role of PCNP in the proliferation, migration, and invasion of human neuroblastoma, lung adenocarcinoma cells, and ovarian cancer cells. Findings demonstrate that PCNP is involved in disrupting key cellular signaling cascades like PI3K/AKT/mTOR and MAPK, which lead to the development of different cancers (17, 18). Results of a recent novel study depicted that PCNP is involved in promoting ovarian cancer progression through activating the Wnt/β-catenin signaling pathway and epithelial–mesenchymal transition (EMT) (19).
Herein, we have summarized the available literature on the biology of PCNP and its potential role as a molecular target in the different cellular activities through which it takes part in different cancer developments. Considering the available evidence of a candidate protein as a potential target in cancer research, we anticipate that in-depth further exploration into elucidating the precise associations of PCNP and mechanisms of cancer progression holds great promise for PCNP use as a diagnostic and therapeutic target for different cancers.
Ubiquitination of PCNP
Inside a cell, ubiquitination is the most dynamic and highly controlled biological phenomenon to mark proteins for degradation, control their activity, alter their localization, and reinforce or hinder their protein interactions (20, 21). These crucial phenomena regulate protein functions, happen via special types of protease, and determine cell survival, differentiation, and other physiological processes (22, 23). Owing to its PEST sequences, PCNP is likely to be very prone to the machinery of ubiquitination in cells (15). Through the mammalian two-hybrid system, GST pull-down assay, and nuclear co-localization studies, it has been evaluated that PCNP shows interaction with ubiquitin-like Np95/ICBP90‐like RING finger protein (UHRF2/NIRF) (16). NIRF is a ubiquitin multi-domain E3 ligase that plays a very important role in maintaining DNA methylation, regulating cell cycle via p53, and DNA damage (24–27). Later on, in a follow-up study through co-immunoprecipitation and Western blotting analysis, it has been found that NIRF is a potential ubiquitin ligase and is capable of ubiquitinating the PCNP (16). Going through these in vivo and in vitro validations of PCNP ubiquitination through NIRF highlights the importance of NIRF as the target to determine the potential role of PCNP in different cancers. In addition, these findings also open a new avenue to investigate the mutual roles of NIRF and PCNP in different signaling pathways concerned with cell cycle regulation and genome stability. Figure 1 shows the mechanisms of PCNP ubiquitination by NIRF.
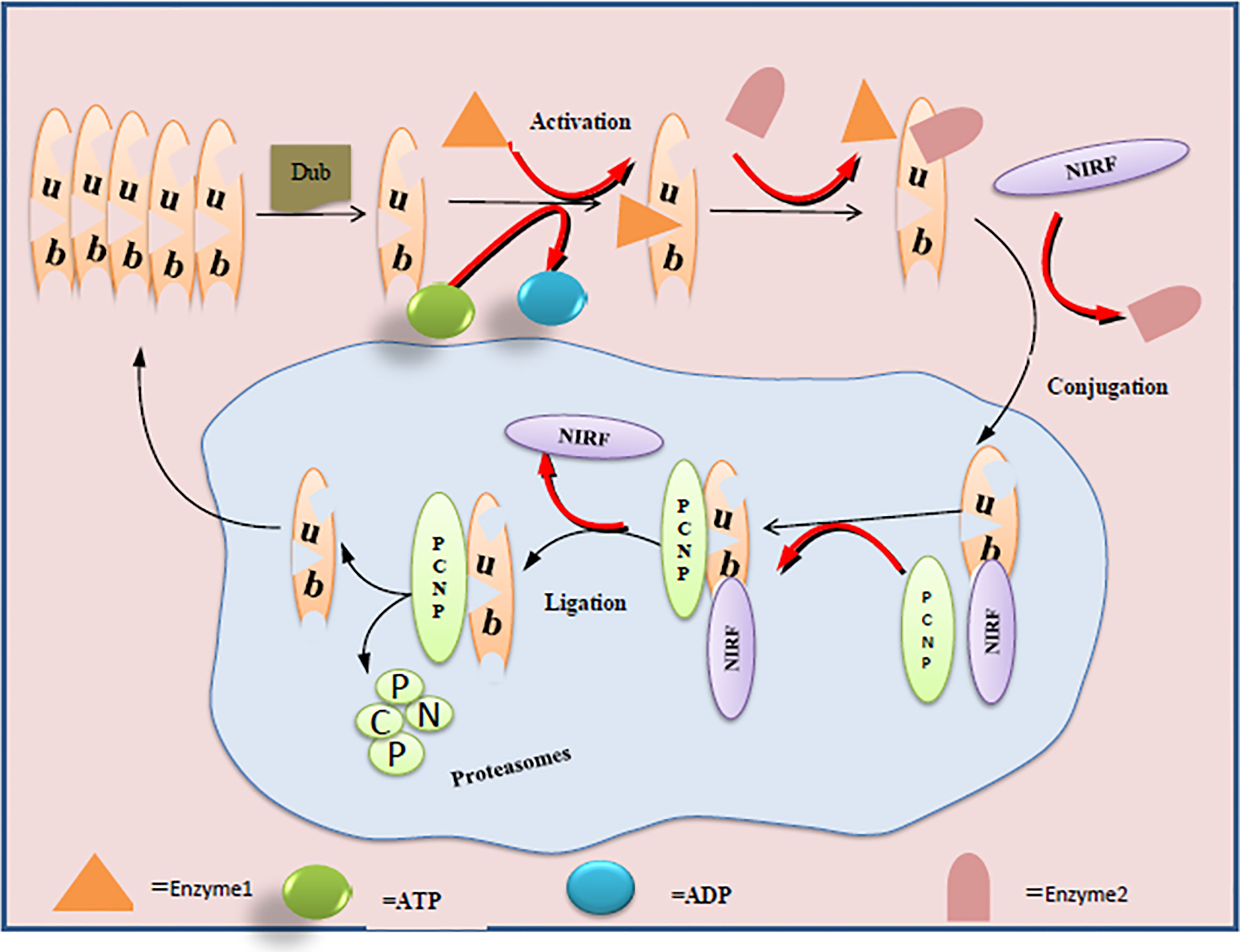
Figure 1 Ubiquitination of PCNP by NIRF in the nuclear cell: ligase enzymes (E1, E2) facilitate the conjugation and ligation of NIRF with PCNP. C-terminus of NIRF performs the ubiquitin ligase function and acts on the PCNP as substrate and mediates the proteasome activity.
Transcriptional Pattern of PCNP With Other Proteins
The help of new high-throughput technologies and advanced levels of bioinformatics tools has enabled the scientists to elucidate the functions of the large fraction of genes in the genome whose functions are currently unknown and help to discover how different genes interact to perform biological functions. Expression patterns of a gene determine the functionality of the protein encoded. Published literature reveals that genes that encode the proteins participating in the same pathway or part of the same protein complex are often co-regulated and exhibit the same expression patterns (28–30); however, co-regulation does not necessarily imply that genes are functionally related (28).
Co-Expression of PCNP and PPP1CC
Protein phosphatase 1 (PP1) is a major eukaryotic protein serine/threonine phosphatase that regulates an enormous variety of cellular functions through the interaction of its catalytic subunit (PP1c). It is an indispensable component in the process of cell division and apoptosis and participates in the regulation of glycogen metabolism, muscle contraction, and protein synthesis (31–33). Employing the weighted gene co-expression network analysis (WGCNA) in a large-scale blood gene expression study on amyotrophic lateral sclerosis (ALS) patients, it has been found that PCNP shows a similarity in pattern expression to PPP1CC (34). The findings give insights into developing the association of PCNP function with PPP1CC and demand further exploration of their co-expression and synchronization in functions.
Co-Expression of PCNP and CGREF1
Cell growth regulator with EF hand domain protein 1 (CGREF1) is involved in mediating cell adhesion by binding to calcium and inhibiting the proliferation of some cell lines (35, 36). It has been evaluated that PCNP shows mRNA transcription level similarity with CGREF1 in the regulation of the hepatoma cell cycle in the zebrafish hepatoma model (37). Considering the importance and evidence of expression pattering in genes, this mRNA transcription similarity of PCNP with a protein involved in cell adhesion and proliferation predicts the PCNP role in cancer cell metastasis.
Co-Expression of PCNP and SRP9
In 2016, de Jong and his team investigated attention-deficit hyperactivity disorder (ADHD) and major depressive disorder (MDD). In whole genome blood gene expression analysis and genetic risk scores of 318 individuals, they found that PCNP shows co-expression with Signal recognition particle 9-kDa protein (SRP9) (38).
SRP9 plays an important role in directing secretory proteins to the rough endoplasmic reticulum. SRP9, together with the Alu part of SRP14 and SRP RNA, constitutes the extended inhibition domain of SRP. The complex of SRP9 and SRP14 is necessary for the binding of SRP RNA (39, 40). Without any doubt, this co-expression of PCNP with SRP9 holds the promise of important association and PCNP functions on further exploration.
Co-Expression of PCNP and DPM1
Dolichol-phosphate mannosyltransferase subunit 1 (DPM1) is a mannose donor in various glycosylation reactions and a catalytic subunit of the polyol-mannose phosphate (DPM) synthesis complex (41–43). As with other proteins like PPP1CC, PCNP have the co-expression pattern with DPM1 at the post-translational modification level in patients with ALS (43). DPM1 and PCNP co-expression could be considered, as synchronization of PCNP functions with DPM1 and demands in-depth exploration.
PCNP Kinship With Other Proteins
As mentioned earlier, the expression patterns of a gene determine the functionality of the protein encoded. Transcription patterning analysis of a PCNP showed that it is co-expressed with MARCH7, BMI1, TMEM123, TRAM1, and PSMC6. The function of MARCH7 includes regulating DNA damage through the p53 gene, regulating cell migration, invasion, and autophagy induced by TGF-β (44, 45). BMI1 proto-oncogene, polycomb ring finger (BMI1) can form a PCG–PRC1 complex, which plays a role through chromatin remodeling and histone modification, and can mediate the ubiquitination of histones, resulting in hereditary changes in the expression rate of chromatin (46, 47). Moreover, co-expression of MARCH7, BMI1, and PCNP is associated with ALS, but the correlation is low (34). Transmembrane protein 123 (TMEM123) can cause cancer cell swelling, cancer cell organelle swelling, vacuolization, and increased membrane permeability, thus participating in the death of tumor cells (47). Translocation associated membrane protein 1 (TRAM1) stimulates secretory proteins through the endoplasmic reticulum membrane and promotes the translocation of endoplasmic reticulum membrane proteins (48). Proteasome 26S subunit (PSMC6), a component of 26S proteasome, is a multi-protein complex that participates in ubiquitin recognition of proteasome and plays a key role in maintaining protein dynamic balance by removing misfolded or damaged proteins that may damage cell function, and by removing proteins with functions that are no longer needed (49). However, PCNP and these three proteins are only co-expressed experimentally, and the role of their co-expression has not been reported in the literature.
Multifaceted Behavior PEST-Containing Nuclear Protein
PEST sequence proteins are considered guardians of the cell and perform vital physiological functions. These proteins execute the process of the ubiquitin–proteasome pathway, glycosylation of nuclear pores, and the hexosamine biosynthetic pathway (8). In initial research on PEST-containing protein PCNP, it has been found that, on the one hand, PCNP is ubiquitinated by NIRF as a tumor suppressor cell regulatory protein in neuroblastoma like p53 and pRB (17). On the other hand, it acts as a tumor-promoting cell regulatory protein in lung adenocarcinoma like cyclin D and cyclin E (18). Furthermore, overexpressed PCNP mediates apoptosis via mitochondrial-mediated pathway, which is evident with a higher apoptotic index and higher expressions of cleaved caspases 3, 8, and 9 as compared with control in Western blotting analysis. However, the exact mechanism of the role of PCNP in mediating cancer cell death is not clear. It has been supposed that PCNP may bind to the DNA directly inside the nucleus and start apoptosis via specialized proteins or after being localized in the cytosol and mediating the mitochondrial-dependent apoptotic pathway (7).
PCNP as Potential Therapeutic Target for Different Cancers
PCNP Promotes the Progression of Ovarian Cancer
Owing to its involvement in the regulation of cell proliferation, apoptosis, and EMT, the Wnt signal pathway has been considered an important pathway to investigate the development and progression of tumors (50, 51).
In particular, it has been found that any sort of aberration in Wnt pathway proteins imposes a significant impact on the ovarian tissues and may become worse in the form of ovarian cancer (52). Dong et al. found a novel association of PCNP with the Wnt pathway. Their results indicate that PCNP is overexpressed in both ovarian cancer tissues and cells more than those in para-cancerous tissues and ovarian epithelial cells (IOSE80), respectively. Through in vitro and in vivo experiments on ovarian cancer cells (SK-OV-3 and A2780) and xenografted nude mice models, it has been found that high expression of PCNP promotes the growth, migration, and invasion of ovarian cancer cells and inhibits apoptosis. They further evaluated that PCNP binds to β-catenin and promotes β-catenin nuclear translocation and then activates the Wnt/β-catenin signal pathway (19). The findings suggest that PCNP may be a regulatory protein upstream of the Wnt pathway involved in the occurrence and development of ovarian cancer and could act as a novel target for treating ovarian cancer. Figure 2 illustrates the pathway of PCNP and catenin β in ovarian cancer.
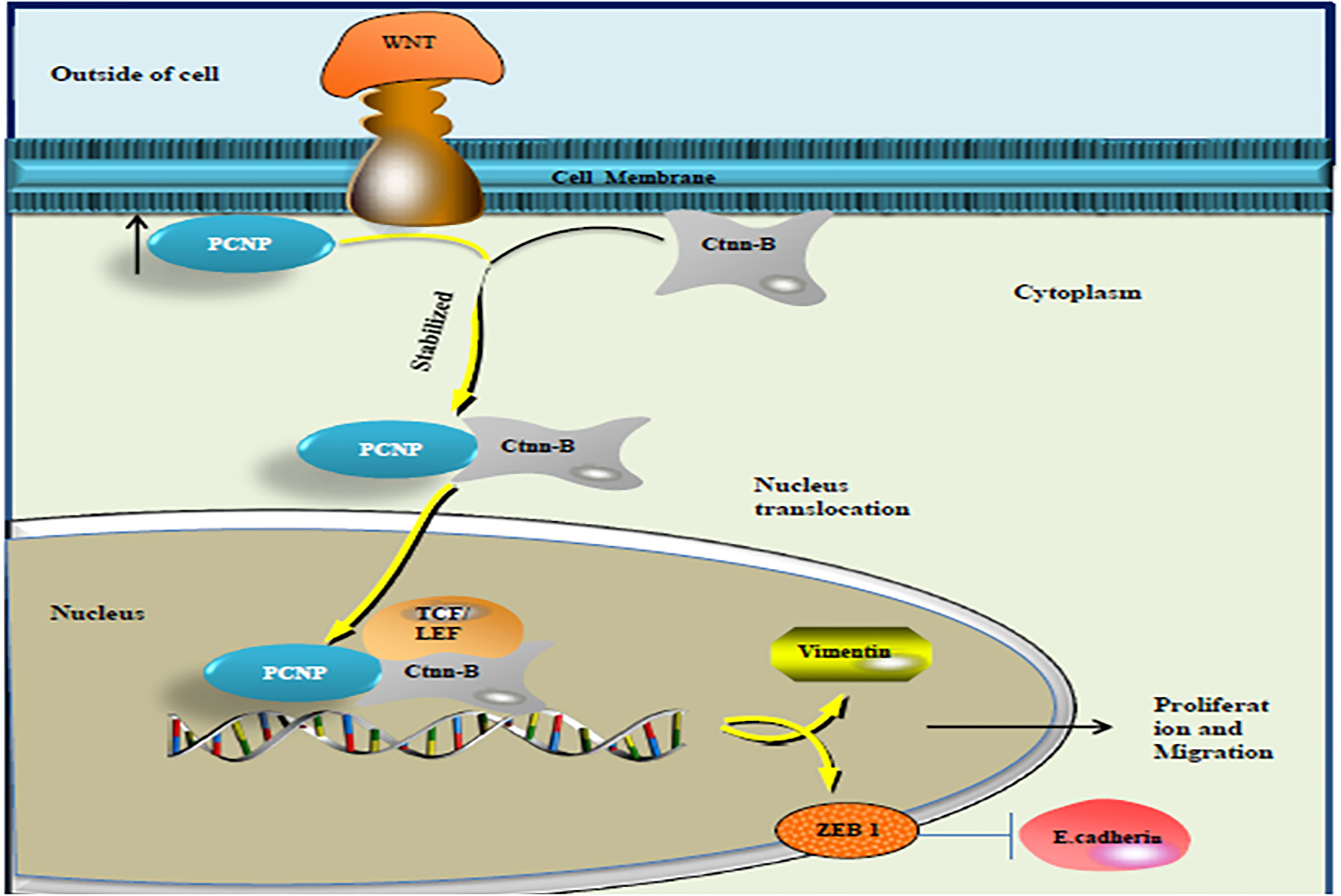
Figure 2 PCNP binding with β catenin: after binding with β catenin, PCNP becomes stabilized and accelerates the expression of β catenin in the nucleus, which further activates the Wnt signaling pathway and promotes ovarian cancer progression.
Overexpressed PCNP Promotes Adenocarcinoma
Lung cancer is the leading cause of cancer-related mortality worldwide. A study published in Oncogenesis observed that PCNP is overexpressed in the human lung adenocarcinoma tissues more than in the corresponding adjacent non-tumor tissues. Overexpression of PCNP promoted the proliferation, migration, and invasion of adenocarcinoma cells, while knockdown of PCNP exhibited the opposite effects. The authors further investigated that overexpressed PCNP mediates the upregulation of the protein expression of p-STAT3 and p-STAT5 and decreases apoptosis. Moreover, compared with the blank control group, it has also been found that in the subcutaneous tumor of nude mice established by A549 and H1299 cells, the growth of tumor cells in the PCNP overexpression group was significantly faster (volume and weight increased, and the tumor doubling time was shortened), and tumor microvessel density (TMD) was also increased (18). These results exploit clearly that PCNP could be considered as a promising biomarker for diagnosis and prognosis in patients with lung adenocarcinoma. Furthermore, designing a potent PCNP inhibitor strategy could be adopted for the treatment of lung adenocarcinoma.
Upregulated PCNP Inhibits Neuroblastoma
Novel findings on the expression association of PCNP with the development and progression of various cancers have made the PCNP a flashpoint of oncology research and urges the biologists for further exploration.
Recently, PCNP gene-expression (downregulation and upregulation) association with the progression of human neuroblastoma was evaluated by using the SH-SY5Y and SK-N-SH cell lines. Findings show that compared with the normal control group, the high expression of PCNP decreased the growth, migration, and invasion of human neuroblastoma through MAPK and PI3K/AKT/mTOR signaling pathways and promoted the apoptosis of neuroblastoma. Opposite results were observed by silencing the PCNP gene in human neuroblastoma cells. Results were further strengthened by xenograft tumor analysis in animal experiments for both PCNP overexpression and knockdown (17). These novel findings imply that PCNP is a potential therapeutic target for designing drugs to treat patients with neuroblastoma.
Future Outlook Under Cell-Biology Prospective
The occurrence and development of cancer are the result of the joint action of many factors, not only with the participation of oncogenes such as c-Jun, c-Fos, p53, and other oncogenes, but also by the regulation of signal transduction pathways such as PI3K/Akt, and Wnt (53–55).
Although the current findings have well established the PCNP participation in the regulation of cell cycle, and its role in affecting PI3K/Akt and Wnt signaling pathways in different cancers, however, there are still many dots that are unconnected. For example, there is no study on the direct association of PCNP with signaling molecules and on their impact on cell internal regulation.
In terms of protein–protein interaction, for the normal continuity of cells, each physiological function relies on thousands of important protein–protein interactions (56). PCNP interactions with reported proteins have been partially elucidated, and there are still many problems that have not been solved, such as: a) whether UHRF2 ubiquitination of PCNP leads to its degradation by the proteasome or whether UHRF2 can affect some cancers; and b) whether proteins containing PEST sequence through PCNP can be rapidly degraded and what stabilizes PCNP in tumor cells; c) whether PCNP can be ubiquitinated by UHRF2 and whether MARCH7 can also ubiquitinate PCNP; and d) whether CGREF1 can mediate cell adhesion and inhibit the growth of multiple cell lines and what the effect of co-expression of PCNP and CGREF is on the migration and proliferation of cancer cells. In particular to this study, we also believe that PCNP exploration in system biology perspective will account to gain a better understanding regarding its biology and potential roles in cancers. System-level understanding constructs the theme in cell biology to depict the anatomy and physiology of biomolecules (57). To date, PCNP lacked a three-dimensional (3D) structure on the protein data bank and there was no reported study on docking ability of PCNP with other oncogenic proteins. Through the use of breakthrough advancements in devices and analytical methods with high-throughput measurements in genomic studies, we look forward to examining the structure and cellular dynamics of PCNP functions in biochemical pathways.
Conclusion Remarks
In conclusion, the current published literature on PCNP represents the critical and clinical roles of PCNP in tumor development, and it highlights the need to develop new understandings of the role and interactions of NPs in regulating the cell cycle. The findings of all preclinical studies on PCNP have established the fact that PCNP is the key regulatory protein in tumorigenesis and has the potential to be considered as a novel molecular target for designing drugs to treat cancer. However, as an attractive molecular target for cancer, it demands exporting the underlying mechanisms of PCNP actions both in mediating apoptosis and in cell proliferation. We anticipate and look forward that in the future, new research insights that will strengthen the aim of developing PCNP-based diagnostic and therapeutic approaches that will move the PCNP from the laboratory to the cancer clinic.
Author Contributions
NHK, DDW, XYJ, and ESJ participated in the conception and manuscript drafting. NHK, HC, YF, MS, MFA, YQ, MS, RV, did the literature mining and wrote the draft. MS, RV did graphical work and draw the figure to explain pathways of candidate protein. DDW, XYJ, ESJ supervised the study put the intellectual input to the review. All authors read and approved the final manuscript.
Funding
This work was supported by Henan Provincial Science and Technology Research Project [No. 212102310147], the National Natural Science Foundation of China (Nos. 81900375, 81802718 and 81670088), the Foundation of Science & Technology Department of Henan Province, China (Nos. 202102310480 and 192102310151), and the Training Program for Young Backbone Teachers of Institutions of Higher Learning in Henan Province, China (No. 2020GGJS038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the expert reviewer for taking his time to kindly review the manuscript and provide his valuable suggestions to improve the manuscript. The authors gratefully acknowledge the assistance and motivation energy of Dr. Attia Afzal. NK( Hussain) would like to express his gratitude and love to his Lala for his presence and support in the PhD journey.
Abbreviations
PCNP, PEST‐containing nuclear protein; NPs, nuclear proteins; PEST, proline (P), glutamic acid (E), serine (S), threonine (T); EMT, epithelial–mesenchymal transition; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; CGREF1, cell growth regulator with EF hand domain protein 1; SRP9, signal recognition particle 9-kDa protein; DPM1, dolichol-phosphate mannosyltransferase subunit 1; PP1, protein phosphatase 1; NIRF, Np95/ICBP90‐like RING finger protein.
References
1. Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Alaluf I, et al. Dynamic Proteomics in Individual Human Cells Uncovers Widespread Cell-Cycle Dependence of Nuclear Proteins. Nature Methods (2006) 3(7):525–31. doi: 10.1038/nmeth892
2. Liu W, Long Q, Chen K, Li S, Xiang G, Chen S, et al. Mitochondrial Metabolism Transition Cooperates With Nuclear Reprogramming During Induced Pluripotent Stem Cell Generation. Biochem Biophys Res Commun (2013) 431(4):767–71. doi: 10.1016/j.bbrc.2012.12.148
3. Ramisetty SK, Langlete P, Lale R, Dias RS. In Vitro Studies of DNA Condensation by Bridging Protein in a Crowding Environment. Int J Biol Macromol (2017) 103:845–53. doi: 10.1016/j.ijbiomac.2017.05.079
4. Boon R, Silveira GG, Mostoslavsky RJNM. Nuclear Metabolism and the Regulation of the Epigenome. Nat Metabol (2020) 2(11):1190–203. doi: 10.1038/s42255-020-00285-4
5. Hurst V, Shimada K, Gasser S. Nuclear Actin and Actin-Binding Proteins in DNA Repair. Trends Cell Biol (2019) 29(6):462–76. doi: 10.1016/j.tcb.2019.02.010
6. Dissmeyer N, Rivas S, Graciet EJNP. Life and Death of Proteins After Protease Cleavage: Protein Degradation by the N-End Rule Pathway. New Phytol (2018) 218(3):929–35. doi: 10.1111/nph.14619
7. Sarfraz M, Afzal A, Khattak S, Saddozai UAK, Li H-M, Zhang Q-Q, et al. Multifaceted Behavior of PEST Sequence Enriched Nuclear Proteins in Cancer Biology and Role in Gene Therapy. J Cell Physiol (2021) 236(3):1658–76. doi: 10.1002/jcp.30011
8. Afzal A, Sarfraz M, Li G-L, Ji S-P, Duan S-F, Khan NH, et al. Taking a Holistic View of PEST-Containing Nuclear Protein (PCNP) in Cancer Biology. Cancer Med (2019) 8(14):6335–43. doi: 10.1002/cam4.2465
9. Rogers S, Wells R, Rechsteiner M. Amino Acid Sequences Common to Rapidly Degraded Proteins: The PEST Hypothesis. Science(1986) 234(4774):364–8. doi: 10.1126/science.2876518
10. Jackson PK. Cell Cycle: Cull and Destroy. Cell (1996) 6(10):1209–12. doi: 10.1016/S0960-9822(96)00697-5
11. Yan J, Wang J, Zhang H. An Ankyrin Repeat-Containing Protein Plays a Role in Both Disease Resistance and Antioxidation Metabolism. Plant J (2002) 29(2):193–202. doi: 10.1046/j.0960-7412.2001.01205.x
12. Sekhar KR, Freeman M, S. Transduction. PEST Sequences in Proteins Involved in Cyclic Nucleotide Signalling Pathways. J Recept Signal Transduct (1998) 18(2-3):113–32. doi: 10.3109/10799899809047740
13. Rechsteiner M, Rogers S. PEST Sequences and Regulation by Proteolysis. Trends Biochem Sci (1996) 21(7):267–71. doi: 10.1016/S0968-0004(96)10031-1
14. Salazar-Retana AL, Maruri-Lopez I, Hernández-Sánchez IE, Becerra-Flora A, de la Luz Guerrero-Gonzalez M, Jiménez-Bremont JF. PEST Sequences From a Cactus Dehydrin Regulate its Proteolytic Degradation. PeerJ (2019) 7:e6810. doi: 10.7717/peerj.6810
15. Mori T, Li Y, Hata H, Ono K, Kochi H. NIRF, A Novel RING Finger Protein, Is Involved in Cell-Cycle Regulation. Biochem Biophys Res Commun (2002) 296(3):530–6. doi: 10.1016/S0006-291X(02)00890-2
16. Mori T, Li Y, Hata H, Kochi H. NIRF is a Ubiquitin Ligase That Is Capable of Ubiquitinating PCNP, a PEST-Containing Nuclear Protein. FEBS Lett (2004) 557(1-3):209–14. doi: 10.1016/S0014-5793(03)01495-9
17. Wu D-D, Gao Y-R, Li T, Wang D-Y, Lu D, Liu S-Y, et al. PEST-Containing Nuclear Protein Mediates the Proliferation, Migration, and Invasion of Human Neuroblastoma Cells Through MAPK and PI3K/AKT/mTOR Signaling Pathways. BMC Cancer (2018) 18(1):1–15. doi: 10.1186/s12885-018-4391-9
18. Wang D-Y, Hong Y, Chen Y-G, Dong P-Z, Liu S-Y, Gao Y-R, et al. PEST-Containing Nuclear Protein Regulates Cell Proliferation, Migration, and Invasion in Lung Adenocarcinoma. Oncogenesis (2019) 8(3):1–14. doi: 10.1038/s41389-019-0132-4
19. Dong P, Hao F, Lin C, Shihui Z, Xin Z, Huimin L, et al. PCNP Promotes Ovarian Cancer Progression by Accelerating β-Catenin Nuclear Accumulation and Triggering EMT Transition. J Cell Mol Med (2020) 24(14):8221–35. doi: 10.1111/jcmm.15491
20. Mansour M, c. Ubiquitination: Friend and Foe in Cancer. Int J Biochem Cell Biol (2018) 101:80–93. doi: 10.1016/j.biocel.2018.06.001
21. Maxwell BA, Gwon Y, Mishra Y, Peng J, Zhang K, Kim HJ, et al. Ubiquitination is Essential for Recovery of Cellular Activities Following Heat Shock. bioRxiv (2021). doi: 10.1101/2021.04.22.440934
22. Gallo L, Ko J, Donoghue DJCC. The Importance of Regulatory Ubiquitination in Cancer and Metastasis. Cell Cycle (2017) 16(7):634–48. doi: 10.1080/15384101.2017.1288326
23. Young M-J, Hsu K-C, Lin TE, Chang W-C, Hung J-J. The Role of Ubiquitin-Specific Peptidases in Cancer Progression. J Biomed Sci (2019) 26(1):1–14. doi: 10.1186/s12929-019-0522-0
24. Vaughan RM, Dickson BM, Cornett EM, Harrison JS, Kuhlman B, Rothbart SB. Comparative Biochemical Analysis of UHRF Proteins Reveals Molecular Mechanisms That Uncouple UHRF2 From DNA Methylation Maintenance. Nucleic Acids Res (2018) 46(9):4405–16. doi: 10.1093/nar/gky151
25. Mori T, Ikeda DD, Yamaguchi Y, Unoki M. NIRF/UHRF2 Occupies a Central Position in the Cell Cycle Network and Allows Coupling With the Epigenetic Landscape. FEBS Lett (2012) 586(11):1570–83. doi: 10.1016/j.febslet.2012.04.038
26. Wang Y, Yan X, Zeng S, Zhang T, Cheng F, Chen R, et al. UHRF2 Promotes DNA Damage Response by Decreasing P21 via RING Finger Domain. Biotechnol Lett (2018) 40(8):1181–8. doi: 10.1007/s10529-018-2577-5
27. Motnenko A, Liang C-C, Yang D, Lopez-Martinez D, Yoshikawa Y, Zhan B, et al. Identification of UHRF2 as a Novel DNA Interstrand Crosslink Sensor Protein. PLoS Genet (2018) 14(10):e1007643. doi: 10.1371/journal.pgen.1007643
28. Stuart JM, Segal E, Koller D, Kim SK. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. (2003) 302(5643):249–55. doi: 10.1126/science.1087447
29. Bergmann S, Ihmels J, Barkai N. Similarities and Differences in Genome-Wide Expression Data of Six Organisms. PLoS Biol (2004) 2(1):e9. doi: 10.1371/journal.pbio.0020009
30. Mutwil M, Øbro J, Willats WGT, Persson S. GeneCAT—Novel Webtools That Combine BLAST and Co-Expression Analyses. Nucleic Acids Res (2008) 36(suppl_2):W320–6. doi: 10.1093/nar/gkn292
31. Rebelo S, Santos M, Martins F, da Cruz e Silva EF, da Cruz e Silva OAB. Protein Phosphatase 1 Is a Key Player in Nuclear Events. Cell Signal (2015) 27(12):2589–98. doi: 10.1016/j.cellsig.2015.08.007
32. Kumar GS, Choy MS, Koveal DM, Lorinsky MK, Lyons SP, Kettenbach AN, et al. Identification of the Substrate Recruitment Mechanism of the Muscle Glycogen Protein Phosphatase 1 Holoenzyme. Sci Adv (2018) 4(11):eaau6044. doi: 10.1126/sciadv.aau6044
33. Nicolaou P, Hajjar RJ, Kranias EG. Role of Protein Phosphatase-1 Inhibitor-1 in Cardiac Physiology and Pathophysiology. J Mol Cell Cardiol (2009) 47(3):365–71. doi: 10.1016/j.yjmcc.2009.05.010
34. Saris CG, et al. Weighted Gene Co-Expression Network Analysis of the Peripheral Blood From Amyotrophic Lateral Sclerosis Patients. BMC Genom (2009) 10(1):1–16. doi: 10.1186/1471-2164-10-405
35. Reddy VS, Day IS, Thomas T, Reddy ASN. KIC, A Novel Ca2+ Binding Protein With One EF-Hand Motif, Interacts With a Microtubule Motor Protein and Regulates Trichome Morphogenesis. Plant Cell (2004) 16(1):185–200. doi: 10.1105/tpc.016600
36. Deng W, Wang L, Xiong Y, Li J, Wang Y, Shi T, et al. The Novel Secretory Protein CGREF1 Inhibits the Activation of AP-1 Transcriptional Activity and Cell Proliferation. Int J Biochem Cell Biol (2015) 65:32–9. doi: 10.1016/j.biocel.2015.05.019
37. Li Z, Zheng W, Wang Z, Zeng Z, Zhan H, Li C, et al. A Transgenic Zebrafish Liver Tumor Model With Inducible Myc Expression Reveals Conserved Myc Signatures With Mammalian Liver Tumors. Dis Models Mech (2013) 6(2):414–23. doi: 10.1242/dmm.010462
38. de Jong S, Newhouse SJ, Patel H, Lee S, Dempster D, Curtis C, et al. Immune Signatures and Disorder-Specific Patterns in a Cross-Disorder Gene Expression Analysis. British J Psychiatry (2016) 209(3):202–8. doi: 10.1192/bjp.bp.115.175471
39. Berger A, Ivanova E, Gareau C, Scherrer A, Mazroui R, Strub K. Direct Binding of the Alu Binding Protein Dimer SRP9/14 to 40S Ribosomal Subunits Promotes Stress Granule Formation and Is Regulated by Alu RNA. Nucleic Acids Res (2014) 42(17):11203–17. doi: 10.1093/nar/gku822
40. Grudnik P, Bange G, Sinning I. Protein Targeting by the Signal Recognition Particle. Biol Chem (2009) 390(8):775–82. doi: 10.1515/BC.2009.102
41. Maeda Y, Tanaka S, Hino J, Kangawa K, Kinoshita T. Human Dolichol-Phosphate-Mannose Synthase Consists of Three Subunits, DPM1, DPM2 and DPM3. Biol Chem (2000) 19(11):2475–82. doi: 10.1093/emboj/19.11.2475
42. van Tol W, Ashikov A, Korsch E, Abu Bakar N, Willemsen MA, Thiel C, et al. A Mutation in Mannose-Phosphate-Dolichol Utilization Defect 1 Reveals Clinical Symptoms of Congenital Disorders of Glycosylation Type I and Dystroglycanopathy. JIMD Reps (2019) 50(1):31–9. doi: 10.1002/jmd2.12060
43. Kim S, Westphal V, Srikrishna G, Mehta DP, Peterson S, Filiano J, et al. Dolichol Phosphate Mannose Synthase (DPM1) Mutations Define Congenital Disorder of Glycosylation Ie (CDG-Ie). J Clin Invest (2000) 105(2):191–8. doi: 10.1172/JCI7302
44. Zhao K, Yang Y, Zhang G, Wang C, Wang D, Wu M, et al. Regulation of the Mdm2–p53 Pathway by the Ubiquitin E3 Ligase MARCH 7. EMBO Reps (2018) 19(2):305–19. doi: 10.15252/embr.201744465
45. Hu J, Zhang L, Mei Z, Jiang Y, Yi Y, Liu L. Interaction of E3 Ubiquitin Ligase MARCH7 With Long Noncoding RNA MALAT1 and Autophagy-Related Protein ATG7 Promotes Autophagy and Invasion in Ovarian Cancer. Cell Physiol Biochem (2018) 47(2):654–66. doi: 10.1159/000490020
46. Wang H, Lee H-W, Deng Y, Lu Z, Hsu P-C, Liu Y, et al. Bifunctional Non-Noble Metal Oxide Nanoparticle Electrocatalysts Through Lithium-Induced Conversion for Overall Water Splitting. Nat Commun (2015) 6(1):1–8. doi: 10.1038/ncomms8261
47. Matsuno Y, Torigoe H, Yoshioka K. Establishment of Cellular Quiescence Together With H2AX Downregulation and Genome Stability Maintenance. J Clin Exp Pathol (2018) 8(335):2161-0681.1000335. doi: 10.4172/2161-0681.1000335
48. Ng CL, Oresic K, Tortorella D. TRAM1 Is Involved in Disposal of ER Membrane Degradation Substrates. Exp Cell Res (2010) 316(13):2113–22. doi: 10.1016/j.yexcr.2010.04.010
49. Bard JA, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. Structure and Function of the 26S Proteasome. Ann Rev Biochem (2018) 87:697–724. doi: 10.1146/annurev-biochem-062917-011931
50. Clevers HJC. Wnt/β-Catenin Signaling in Development and Disease. Cell (2006) 127(3):469–80. doi: 10.1016/j.cell.2006.10.018
51. Ghahhari NM, Babashah S. Interplay Between microRNAs and WNT/β-Catenin Signalling Pathway Regulates Epithelial–Mesenchymal Transition in Cancer. Eur J Cancer (2015) 51(12):1638–49. doi: 10.1016/j.ejca.2015.04.021
52. Arend RC, et al. The Wnt/β-Catenin Pathway in Ovarian Cancer: A Review. Lancet Oncol (2013) 131(3):772–9. doi: 10.1016/j.ygyno.2013.09.034
53. Nia HT, Munn LL, Jain RKJS. Physical Traits of Cancer. Science (2020) 370(6516):eaaz0868. doi: 10.1126/science.aaz0868
54. Bugter JM, Fenderico N, Maurice MM. Mutations and Mechanisms of WNT Pathway Tumour Suppressors in Cancer. Nat Rev Cancer (2021) 21(1):5–21. doi: 10.1038/s41568-020-00307-z
55. Hoxhaj G, Manning BD. The PI3K–AKT Network at the Interface of Oncogenic Signalling and Cancer Metabolism. Nat Rev Cancer (2020) 20(2):74–88. doi: 10.1038/s41568-019-0216-7
56. Zhang Y, Fernie ARJB. On the Detection and Functional Significance of the Protein–Protein Interactions of Mitochondrial Transport Proteins. Biomolecules (2020) 10(8):1107. doi: 10.3390/biom10081107
Keywords: cancer, PEST-sequence, nuclear proteins, molecular research target, PCNP, therapeutics
Citation: Khan NH, Chen H-J, Fan Y, Surfaraz M, Ahammad M, Qin Y-Z, Shahid M, Virk R, Jiang E, Wu D-D and Ji X-Y (2022) Biology of PEST‐Containing Nuclear Protein: A Potential Molecular Target for Cancer Research. Front. Oncol. 12:784597. doi: 10.3389/fonc.2022.784597
Received: 28 September 2021; Accepted: 06 January 2022;
Published: 04 February 2022.
Edited by:
Shiv K. Gupta, Mayo Clinic, United StatesCopyright © 2022 Khan, Chen, Fan, Surfaraz, Ahammad, Qin, Shahid, Virk, Jiang, Wu and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enshe Jiang, esjiang@gmail.com; Dong-Dong Wu, ddwubiomed2010@163.com; Xin-Ying Ji, 10190096@vip.henu.edu.cn
 Nazeer Hussain Khan
Nazeer Hussain Khan Hao-Jie Chen
Hao-Jie Chen Yuanyuan Fan
Yuanyuan Fan Muhammad Surfaraz3
Muhammad Surfaraz3 Enshe Jiang
Enshe Jiang Dong-Dong Wu
Dong-Dong Wu