- Hubei Province Key Laboratory of Traditional Chinese Medicine Resource and Chemistry, Hubei University of Chinese Medicine, Wuhan, China
Background: Evodiamine (EVO), an alkaloid extracted from the traditional Chinese medicine Euodia rutaecarpa, plays an important role in the treatment of cancer. This study was performed to clarify the effects of evodiamine in mice tumor model studies.
Methods: Electronic databases and search engines involved China Knowledge Resource Integrated Database (CNKI), Wanfang Database, Chinese Scientific Journal Database (CSJD-VIP), China Biomedical Literature Database (CBM), PubMed, Embase, Web of Science, and ClinicalTrials.gov databases, which were searched for literature related to the antitumor effects of evodiamine in animal tumor models (all until 1 October 2021). The evodiamine effects on the tumor volume and tumor weight were compared between the treatment and control groups using the standardized mean difference (SMD).
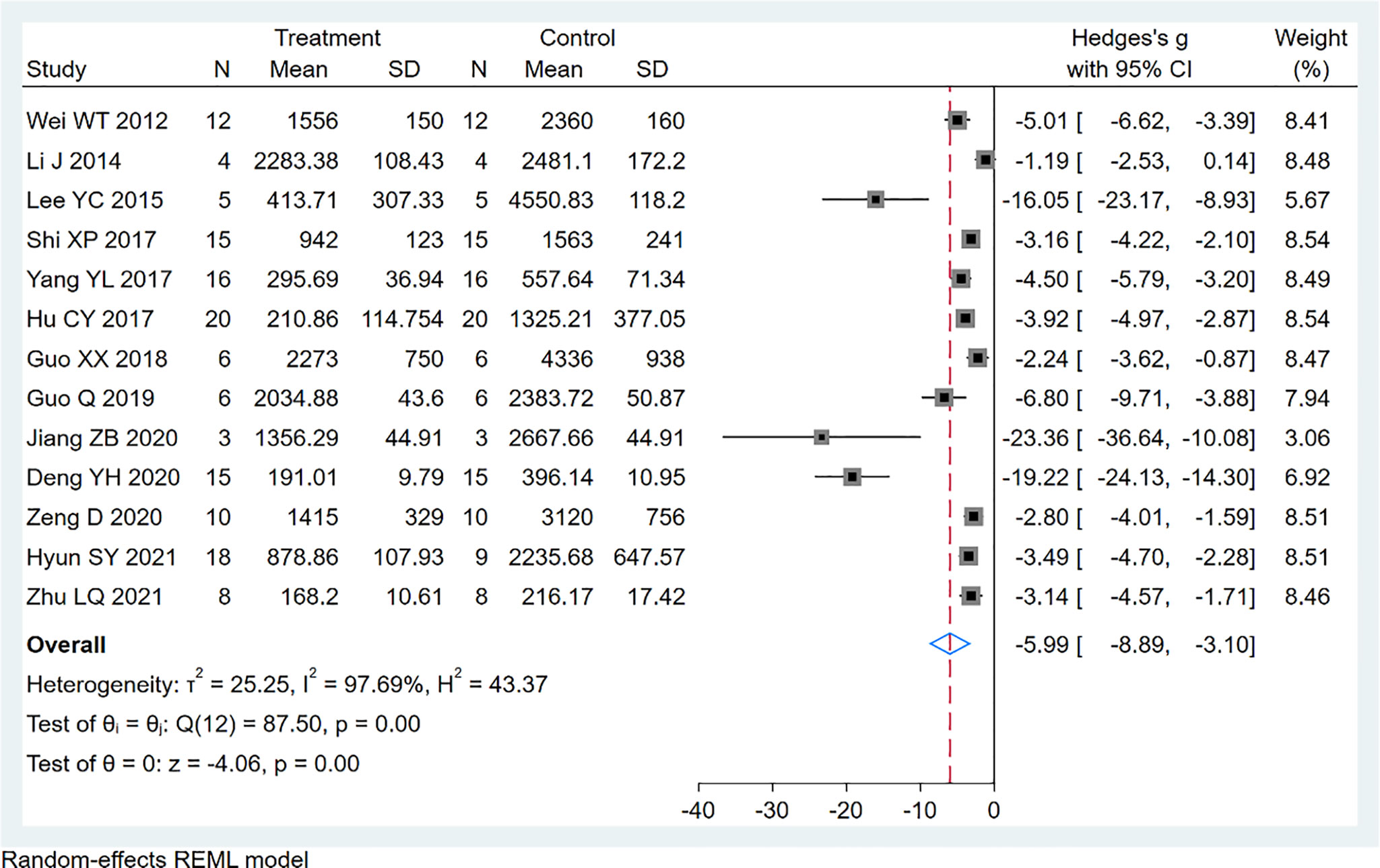
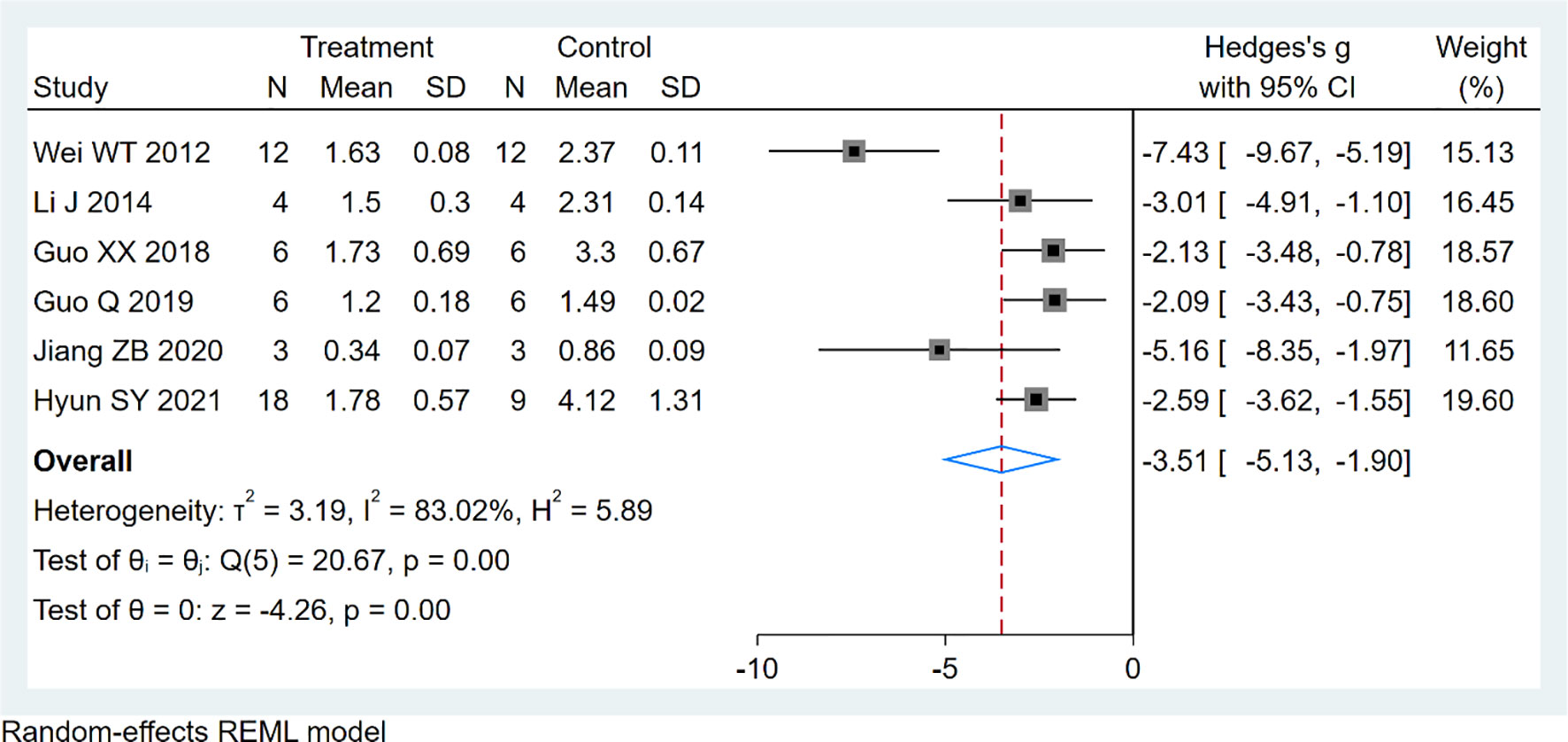
Results: Evodiamine significantly inhibited tumor growth in mice, as was assessed with tumor volume [13 studies, n=267; 138 for EVO and 129 for control; standard mean difference (SMD)= -5.99; 95% (CI): -8.89 to -3.10; I2 = 97.69%, p ≤ 0.00], tumor weight [6 studies, n=89; 49 for EVO and 40 for control; standard mean difference (SMD)= -3.51; 95% (CI): -5.13 to -3.90; I2 = 83.02%, p ≤ 0.00].
Conclusion: EVO significantly suppresses tumor growth in mice models, which would be beneficial for clinical transformation. However, due to the small number of studies included in this meta-analysis, the experimental design and experimental method limitations should be considered when interpreting the results. Significant clinical and animal studies are still required to evaluate whether EVO can be used in the adjuvant treatment of clinical tumor patients.
Introduction
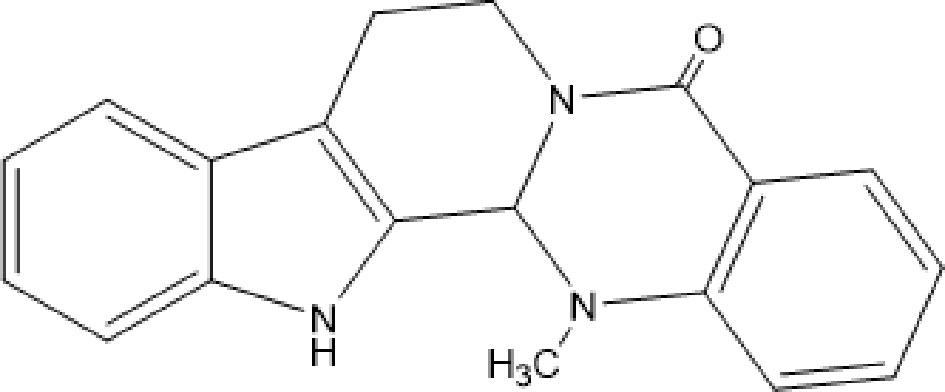
Evodiamine (EVO, Figure 1), a kind of quinazolinocarboline alkaloid, is one of the components isolated from a Chinese herbal medicine, called Wu-Chu-Yu (Evodia rutaecarpa) (1). It has been considered an effective Chinese medicine for the treatment of gastropathy, hypertension, and eczema (2). In recent years, the antitumor effect of EVO in vivo has become a focus for innovation and a hot topic in medical research. Recent work in this field suggests that the anticancer effects of EVO resist several high-risk cancers, including pancreatic cancer, lung cancer, colon cancer, lymphoma, and oral cancer (3–7). EVO, as a novel occurring indole alkaloid with attractive multitargeting antiproliferative activity, has been investigated as a leading compound that possesses multitargeting profiles (8, 9). Novel boron-containing EVO derivatives were designed, which have improved the antitumor potency of the EVO and showed a good antitumor activity in vitro and in vivo by reactive oxygen species (ROS) (8).
Although many preclinical experiments have undoubtedly shown that EVO exerts an antitumor effect, less evidence, however, has established clinical associations to guide healthcare decisions. Moreover, quality assessment of these animal studies reporting the antitumor effect of EVO is lacking. A systematic review of preclinical studies (especially animal experiments) can evaluate the efficacy and mechanism of treatment, provide reliable information for drug research, and lay the foundation for future clinical research (10). To accelerate the transformation of preclinical experiments to clinical studies, a systematic review and meta-analysis would be performed. The aim of this study is to investigate the impact of EVO on a mice tumor model.
Materials and Methods
This study was performed according to the systematic reviews of animal experiments (11) (Supplementary Table S1) and preferred reporting items for systematic reviews and meta-analyses (PRISMA) (12) guidelines (Supplementary Table S2).
Search Strategy
We searched China Knowledge Resource Integrated Database (CNKI), Wanfang Database, Chinese Scientific Journal Database (CSJD-VIP), China Biomedical Literature Database (CBM), PubMed, Embase, Web of Science, and ClinicalTrials.gov databases to find relevant articles up to 1 October 2021 in all languages using a combination of the main search terms “evodiamine” and “Neoplasms.” The search strategy was designed to be as broad as possible. Then, the reference lists of the relevant articles were checked for additional articles. The detailed search strategy is presented in Supplementary Table S3.
Two authors (YC and PHB) independently reviewed the titles and abstracts. Discrepancies were resolved through discussions between the two authors. The two authors then independently analyzed the full text of the remaining articles to determine the final inclusion.
Selection Criteria
The inclusion criteria were as follows—(1) types of animals: mice; (2) types of intervention: EVO only; (3) type of outcome measure: the effects of EVO in mice cancer models, including tumor volume and tumor weight; and (4) study design: experiments should be prospectively controlled. The exclusion criteria were as follows: (1) review, conference reports, or editorial articles; (2) repeatedly published studies; (3) articles with incomplete data; and (4) articles with no use of experimental animals.
Data Extraction
Two authors (YC and PHB) extracted data from the selected trials. This process was verified by another investigator (LJ). An Excel database was used to extract the following information from the included studies: (1) author name, year of publication, and country; (2) animal species, sex, and sample size; (3) methods of inducing tumor; and (4) EVO treatment conditions including administration route, dosage, frequency, duration, and types of tumor. To obtain the data not presented in the original articles, we sent two e-mails 1 week apart to the corresponding authors.
Data of all outcomes including sample size of animals (N), means, standard deviation (SD), or standard error of the mean (SEM) were extracted. If the data were presented in the form of graph, data extraction would be performed using digital image analysis software (Getdata.Ink). SEM was converted to SD by a mathematical formula: SD=SEM×√N.
Assessment of the Methodological Quality
Each study was evaluated according to the Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE) for animal model studies (13). The tool, which aims to assess methodological quality, was adapted to appraise bias in animal studies, including random sequence generation (selection bias), baseline characteristics (selection bias), allocation concealment (selection bias), random housing (performance bias), blinding of participants and personnel (performance bias), random selection of animals in the evaluation results (detection bias), blinding of outcome assessment (detection bias), incomplete outcome data (attrition bias), selective reporting (reporting bias), and other biases. Two trained researchers (YC and PHB) independently evaluated and cross-checked the inherent risk of bias in the included studies. Differences in opinion were negotiated or settled by a third party (LJ). Answers to the assessment questions (tools) were either “yes” to indicate a low risk of bias or “no” to indicate a high risk of bias. An answer of “unclear” was assigned to items for which a “yes” or “no” answer was not clear.
Statistical Analysis
We performed the analysis with use of STATAMP, version 16. For the outcome measures of tumor volume and tumor weight, we will use standard mean difference (SMD). Considering the variation between studies, the random-effects model was used at a 95% confidence level. In addition, the heterogeneity of the studies was assessed by forest plots using I2 statistics. Then, a subgroup analysis was performed based on gender, mode of administration, the types of tumor, and animal species. A p-value of less than 0.05 was considered significant. Funnel plots were performed to analyze publication bias. A sensitivity analysis was conducted only when three or more studies were included in the comparison. Sensitivity analysis was used to assess the robustness of meta-analysis results by sequentially removing individual included studies. The presence of the publication bias was checked for each outcome through funnel plots. Publication bias was assessed using the Egger regression asymmetry test when the comparison contained at least 10 studies (14).
Results
Selection of Studies
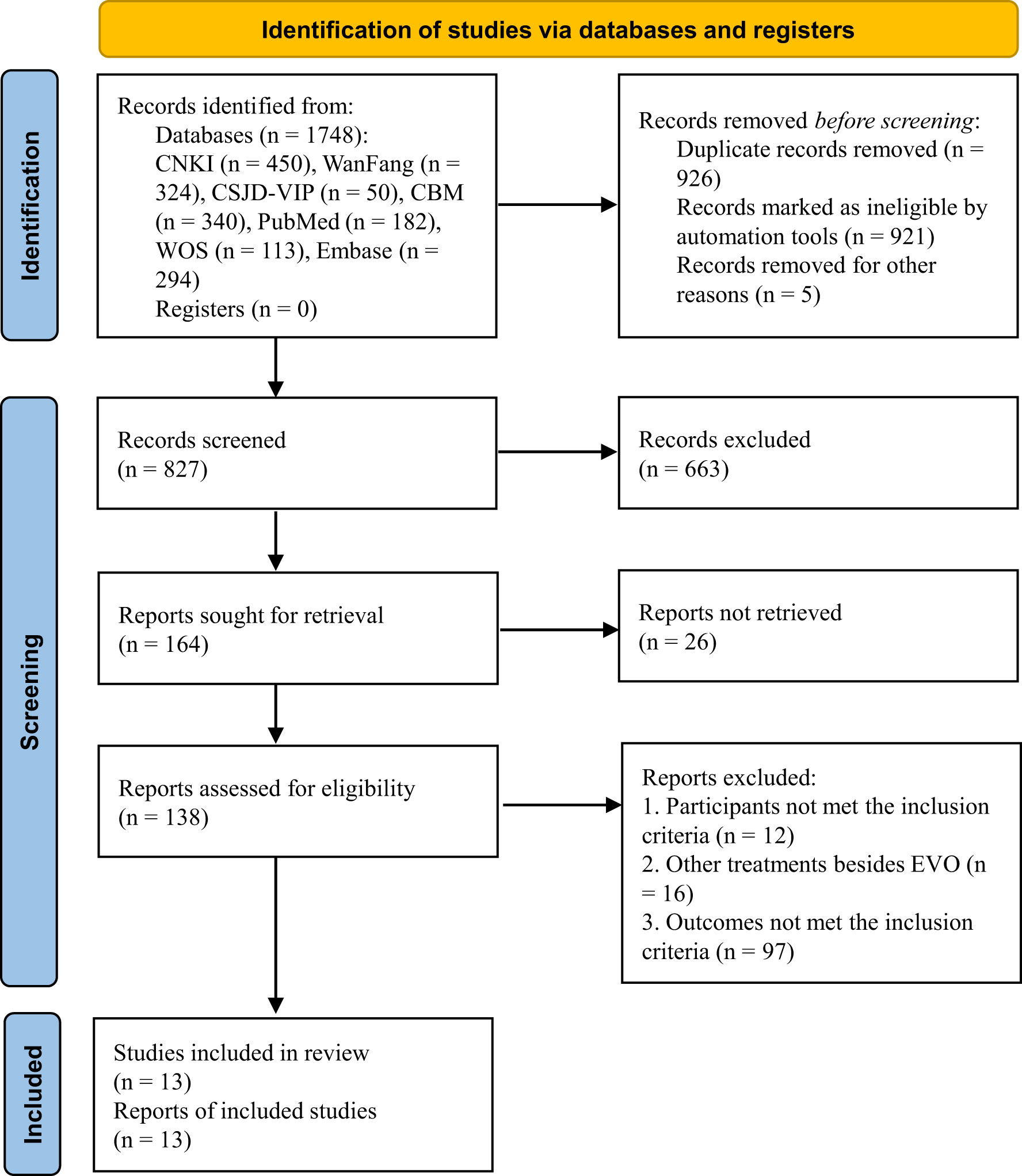
A total of 1,748 studies were identified in the electronic database search. After selection, 13 studies (1, 3, 4, 6, 15–23) met the inclusion criteria and were included in our meta-analysis. The flow of literature screening is shown in Figure 2.
Study Characteristics
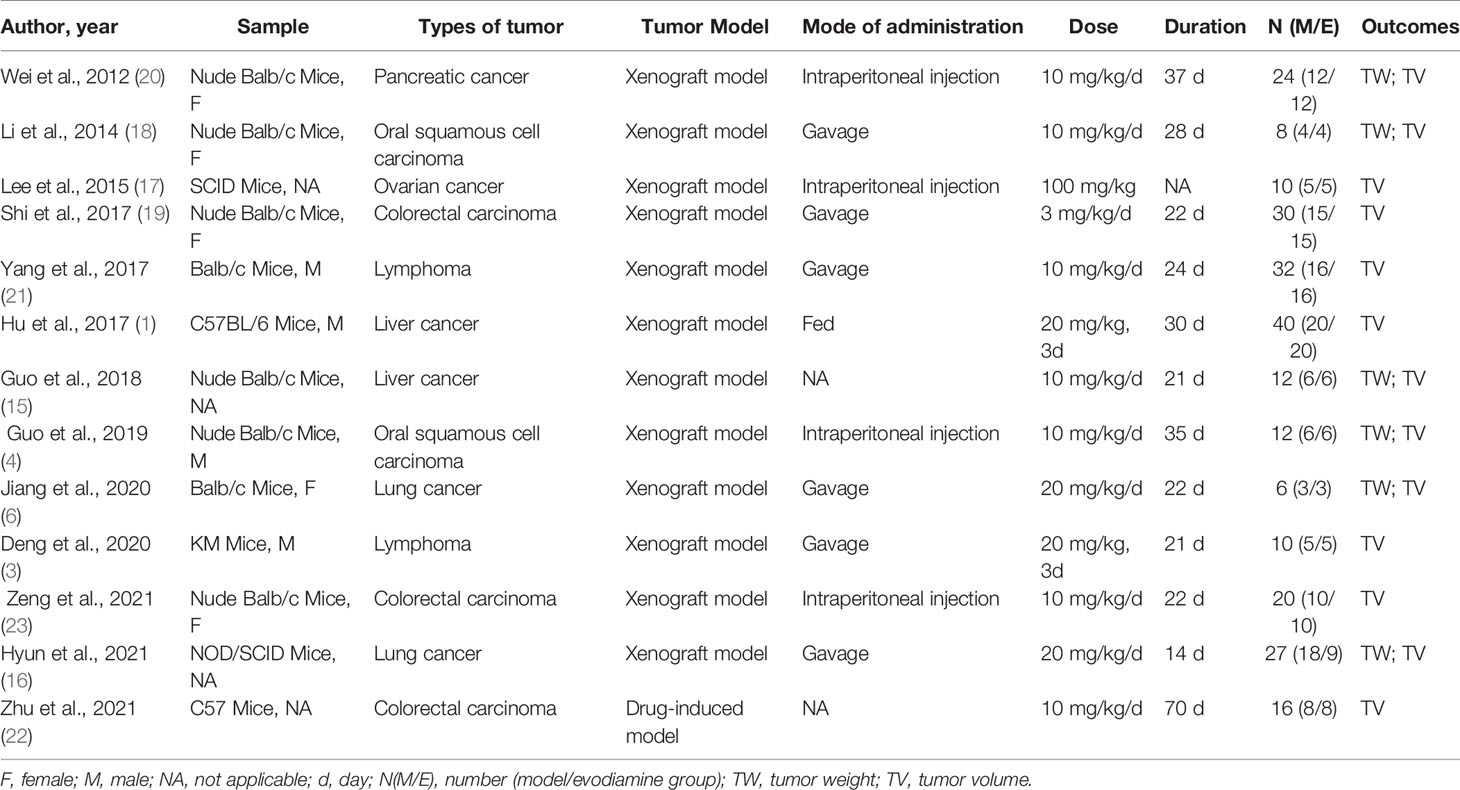
Study characteristics are presented in Table 1. A total of 13 studies from 2012 to 2021 met the inclusion criteria, containing various cancer types, including lymphoma, oral cancer, colon cancer, hepatocellular cancer, ovarian cancer, lung cancer, and pancreatic cancer. The studies all used mice, and the mice were modeled via subcutaneous tumor implantation or induced tumor formation. EVO was administered in doses ranging between 3 and 100 mg/kg body weight through subcutaneous injection, intraperitoneal injection, and gavage. The size of the study sample ranged from 4 to 18, while the course of treatment ranged from 14 to 70 days.
Quality Assessment
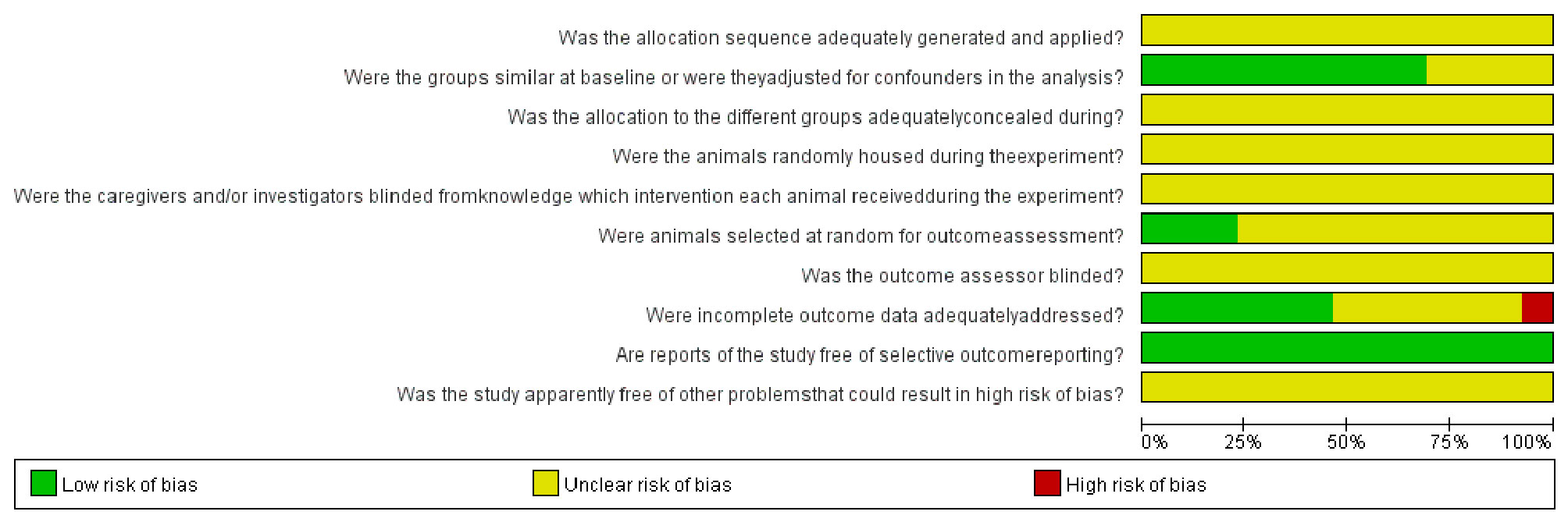
The quality assessment results of the 13 included studies are shown in Figure 3 and Supplementary Figure S1. All of the studies did not report the following aspects: random sequence generation, allocation concealment, random housing, blinding of participants and personnel, random selection of animals in the evaluation results, and blinding of outcome assessment. Baseline characteristics were not fully reported in most studies. One study showed high risk of bias on the attrition bias item of incomplete outcome data with the main problem of an unclear number of animals. Although the overall quality of the studies was poor, none of the papers were excluded because of their quality or risk of bias assessment.
Tumor Volume
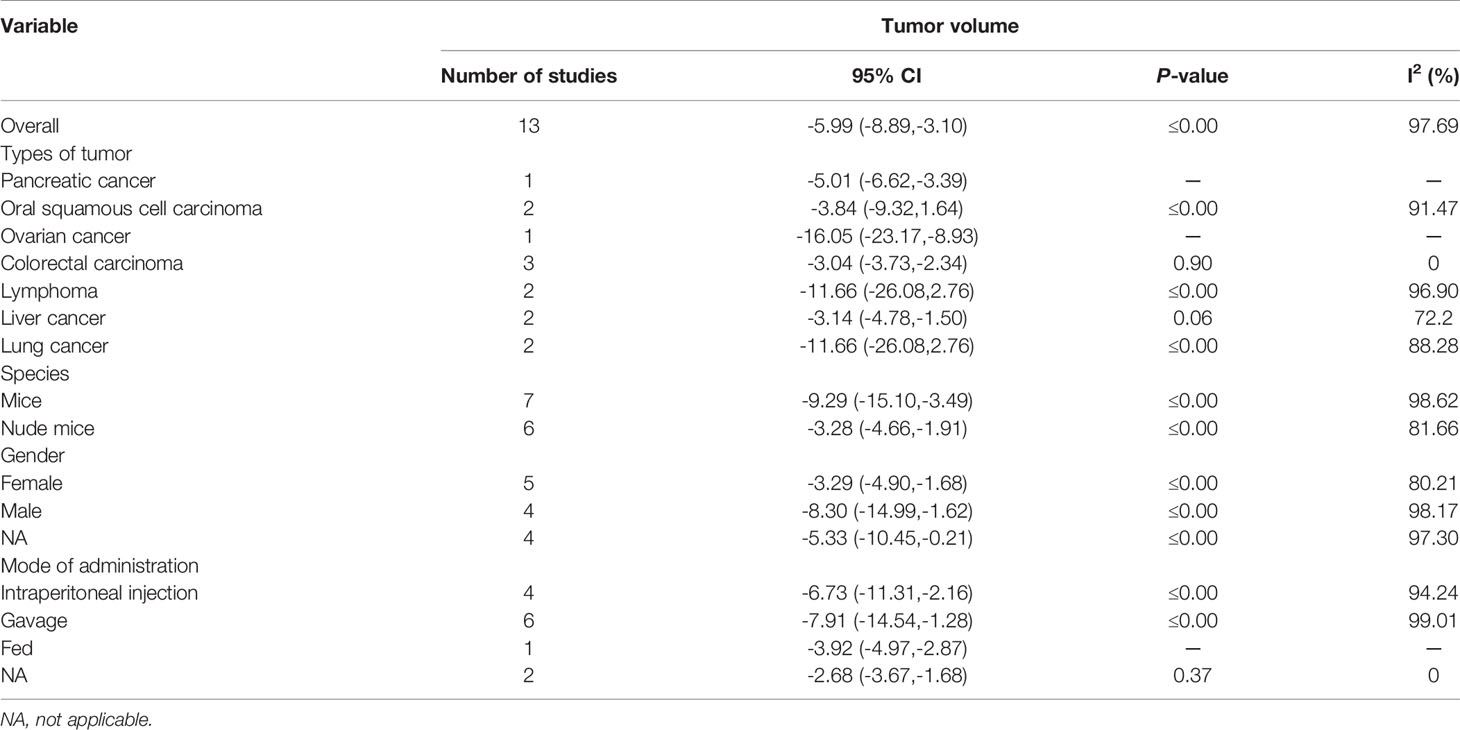
Thirteen studies using xenograft models or an induced tumor model were included to investigate the effect of EVO treatment on tumor volume (Figure 4). A total of 138 mice were in the intervention group, while 129 mice were in the control group. The pooled estimate suggested a significant inhibition of tumor volume [SMD -5.99; 95% CI -8.89, -3.10; P ≤ 0.00]. The heterogeneity among studies was relatively high (I2 = 97.69%, P ≤ 0.00). Subgroup analysis had not found the sources of the significant heterogeneity (Table 2).
Tumor Weight
Six studies using xenograft models were included to investigate the effect of EVO treatment on tumor weight (Figure 5). A total of 95 mice were in the intervention group, while 95 mice were in the control group. The pooled estimate suggested a significant inhibition of tumor weight [SMD -3.51; 95% CI -5.13, -1.90; P ≤ 0.00]. The heterogeneity among studies was relatively high (I2 = 83.02%, P ≤ 0.00). Subgroup analysis had not found the sources of the significant heterogeneity (Table 3).
Sensitivity Analysis
Supplementary Figures S2, S3 show that the results of tumor volume and tumor weight were robust.
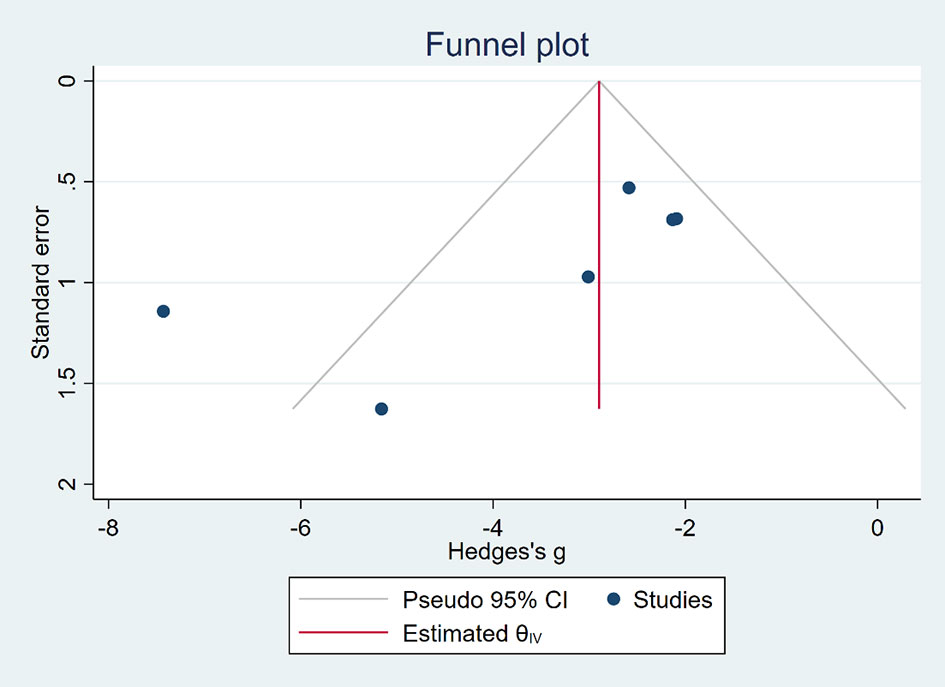
Publication Bias
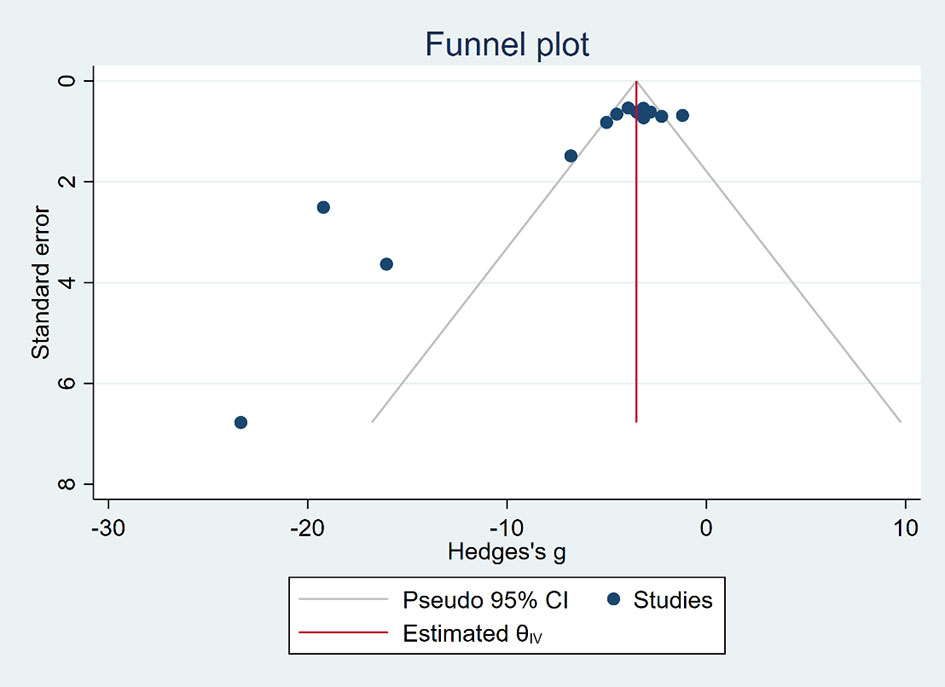
Funnel plots (Figures 6, 7) were used to express publication bias. The standard error was plotted against the tumor volume and tumor weight (13 publications evaluating the inhibitory effects of EVO on tumor volume and 6 on tumor weight). Visual inspection of the funnel plot test suggested that publication bias was possible. In this research, we were unable to evaluate publication bias because some positive results may be much easier to be published.
Discussion
This meta-analysis was conducted to evaluate the effects of EVO on animal tumor models. It suggested that EVO generates a significant antitumor activity in animal models of tumor. It has been shown that EVO mediates tumor apoptosis mainly through a mitochondrial pathway (24, 25). EVO could increase the ratio of BAX/BCL to sensitize cells to death signals (15, 20, 26). Then, it would activate apoptotic initiation protease caspase-9 and its downstream apoptotic execution protease caspase-3 (27–30). Also, the key rate-limiting step of apoptosis release of cyto-C was observed (28). Moreover, EVO suppresses cancer growing by eliminating stemness and inhibiting invasion and migration (7, 31). This is mainly due to the downregulation of epithelial-mesenchymal transition (EMT) (32–34). For instance, EVO elevated epithelial marker E-cadherin and reduced the expression of mesenchymal markers N-cadherin and vimentin (33), as well as matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) protein levels (32).
Many in vitro and in vivo studies had reported beneficial effects of EVO against tumor, and as far as we know, this is the first meta-analysis that assessed the anticancer effect of EVO in preclinical experiments. Our results showed that both the tumor volume and tumor weight results of mice animal models treated with EVO showed significant improvement, indicating that EVO can probably be used as a new treatment strategy in the treatment of clinical tumor patients.
Although this study achieved positive results, the possible shortcomings should not be ignored. First, our research only includes the available data; some negative results are less likely to have been published. Therefore, this meta-analysis may have exaggerated the effect size. Second, the heterogeneity among the various animal studies was quite considerable, as animal studies are often explorative and heterogeneous compared with clinical trials. As a consequence, a subgroup analysis involving cancer types, species, mode of administration, and gender was conducted; however, due to the limited sample size of each subgroup and insufficient statistical power, it did not find the source of significant heterogeneity. Heterogeneity might have originated from different animal ages, varied administration routes, and different initiation times for treatment. Third, our study still lacks the findings of the effect of EVO in tumor mice models with specific types of diseases. Clinical patients are more likely to present with different types of underlying diseases, so there is still a lot of work to be done in clinical translation. In addition, we could not get exact conclusion in this condition: the treatment methods in control arms did not unify to an extent; the numbers of animals varied from group to group.
Conclusions
EVO appears to have the highest efficacy in reducing tumor volume and weight, which increases our confidence in the results and their transformation to the clinical situation. This is a result of antiproliferative, anti-invasive, and apoptosis-inducing functions of EVO in animal experiments. However, due to the small number of studies included in this meta-analysis, the experimental design and experimental method limitations should be considered when interpreting the results. Significant clinical and animal studies are still required to evaluate whether EVO can be used in the adjuvant treatment of clinical tumor patients.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
YC, LJ, and CKL contributed to conception and design of the study. YC and CJ organized the database. YC, CJ, PHB, and YSJ performed the statistical analysis. YC, CJ, and LJ wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302); The Opening Project of Hubei Provincial Key Laboratory of Occurrence and Intervention of Rheumatic Diseases (Hubei Minzu University)(PT022002); The Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University (No. ZYAOXYB2019003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.774201/full#supplementary-material
References
1. Hu CY, Wu HT, Su YC, Lin CH, Chang CJ, Wu CL. Evodiamine Exerts an Anti-Hepatocellular Carcinoma Activity Through a WWOX-Dependent Pathway. Molecules (Basel Switzerland) (2017) 22(7):1175. doi: 10.3390/molecules22071175
2. Meng T, Fu S, He D, Hu G, Gao X, Zhang Y, et al. Evodiamine Inhibits Lipopolysaccharide (LPS)-Induced Inflammation in BV-2 Cells via Regulating AKT/Nrf2-HO-1/NF-κb Signaling Axis. Cell Mol Neurobiol (2021) 41(1):115–27. doi: 10.1007/s10571-020-00839-w
3. Deng YH, Dai YC, Tang B. Effect of Evodiamine on the Ki-67 Expression in Tumor Tissue of Lymphoma Transplanted Mice Model. Clin Basic Bridging Res (2020) 36(6):655–7. doi: 10.16368/j.issn.1674-8999.2017.06.240
4. Guo Q, Liu Y, Zhao J, Wang J, Li Y, Pang Y, et al. Evodiamine Inactivates NF-κb and Potentiates the Antitumor Effects of Gemcitabine on Tongue Cancer Both In Vitro and In Vivo. OncoTargets Ther (2019) 12:257–67. doi: 10.2147/OTT.S181062
5. Hong Z, Wang Z, Zhou B, Wang J, Tong H, Liao Y, et al. Effects of Evodiamine on PI3K/Akt and MAPK/ERK Signaling Pathways in Pancreatic Cancer Cells. Int J Oncol (2020) 56(3):783–93. doi: 10.3892/ijo.2020.4956
6. Jiang ZB, Huang JM, Xie YJ, Zhang YZ, Chang C, Lai HL, et al. Evodiamine Suppresses non-Small Cell Lung Cancer by Elevating CD8(+) T Cells and Downregulating the MUC1-C/PD-L1 Axis. J Exp Clin Cancer Res CR (2020) 39(1):249. doi: 10.1186/s13046-020-01741-5
7. Kim H, Yu Y, Choi S, Lee H, Yu J, Lee JH, et al. Evodiamine Eliminates Colon Cancer Stem Cells via Suppressing Notch and Wnt Signaling. Molecules (Basel Switzerland) (2019) 24(24):4520. doi: 10.3390/molecules24244520
8. Li X, Wu S, Dong G, Chen S, Ma Z, Liu D, et al. Natural Product Evodiamine With Borate Trigger Unit: Discovery of Potent Antitumor Agents Against Colon Cancer. ACS Medicinal Chem Lett (2020) 11(4):439–44. doi: 10.1021/acsmedchemlett.9b00513
9. Yu H, Jin H, Gong W, Wang Z, Liang H. Pharmacological Actions of Multi-Target-Directed Evodiamine. Molecules (Basel Switzerland) (2013) 18(2):1826–43. doi: 10.3390/molecules18021826
10. Sena ES, Currie GL, McCann SK, Macleod MR, Howells DW. Systematic Reviews and Meta-Analysis of Preclinical Studies: Why Perform Them and How to Appraise Them Critically. J Cereb Blood Flow Metab Off J Int Soc Cereb Blood Flow Metab (2014) 34(5):737–42. doi: 10.1038/jcbfm.2014.28
11. Peters JL, Sutton AJ, Jones DR, Rushton L, Abrams KR. A Systematic Review of Systematic Reviews and Meta-Analyses of Animal Experiments With Guidelines for Reporting. J Environ Sci Health Part B Pesticides Food Contaminants Agric Wastes (2006) 41(7):1245–58. doi: 10.1080/03601230600857130
12. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ (Clinical Res ed) (2021) 372:n71. doi: 10.1136/bmj.n71
13. Hooijmans CR, Rovers MM, de Vries RB, Leenaars M, Ritskes-Hoitinga M, Langendam MW. SYRCLE's Risk of Bias Tool for Animal Studies. BMC Med Res Method (2014) 14:43. doi: 10.1186/1471-2288-14-43
14. Yin C, Li Z, Xiang Y, Peng H, Yang P, Yuan S, et al. Effect of Intermittent Fasting on Non-Alcoholic Fatty Liver Disease: Systematic Review and Meta-Analysis. Front Nutr (2021) 8:709683. doi: 10.3389/fnut.2021.709683
15. Guo XX, Li XP, Zhou P, Li DY, Lyu XT, Chen Y, et al. Evodiamine Induces Apoptosis in SMMC-7721 and HepG2 Cells by Suppressing NOD1 Signal Pathway. Int J Mol Sci (2018) 19(11):3419. doi: 10.3390/ijms19113419
16. Hyun SY, Le HT, Min HY, Pei H, Lim Y, Song I, et al. Evodiamine Inhibits Both Stem Cell and non-Stem-Cell Populations in Human Cancer Cells by Targeting Heat Shock Protein 70. Theranostics (2021) 11(6):2932–52. doi: 10.7150/thno.49876
17. Lee YC, Lee CH, Tsai HP, An HW, Lee CM, Wu JC, et al. Targeting of Topoisomerase I for Prognoses and Therapeutics of Camptothecin-Resistant Ovarian Cancer. PloS One (2015) 10(7):e0132579. doi: 10.1371/journal.pone.0132579
18. Li J, Zhang KL, Hu CQ, Han YY, Kang ZQ, Sun QQ, et al. Inhibitory Effect of Evodiamine Combined With Radiotherapy on the Growth of Xenografts of Human Tongue Squamous-Cell Carcinoma Tca-8113 Cells in Nude Mice. TUMOR (2014) 34(2):108–12. doi: 10.3781/j.issn.1000-7431.2014.02.002
19. Shi XP, Li XP, Xiong W, Li H, Guo P, Wang F, et al. Evodiamine Suppresses Proliferation of Colon Cancer HCT-116 Cells in Mice. Basic Clin Med (2017) 37(10):1373–7. doi: 10.3969/j.issn.1001-6325.2017.10.005
20. Wei WT, Chen H, Wang ZH, Ni ZL, Liu HB, Tong HF, et al. Enhanced Antitumor Efficacy of Gemcitabine by Evodiamine on Pancreatic Cancer via Regulating PI3K/Akt Pathway. Int J Biol Sci (2012) 8(1):1–14. doi: 10.7150/ijbs.8.1
21. Yang YL. Effect of Evodiamine on the Expression of Bcl-2 mRNA and Ki-67 mRNA in the Model Mice With Lymphoma. China J Chin Med (2017) 32(6):921–4. doi: 10.16368/j.issn.1674-8999.2017.06.240
22. Zhu LQ, Zhang L, Zhang J, Chang GL, Liu G, Yu DD, et al. Evodiamine Inhibits High-Fat Diet-Induced Colitis-Associated Cancer in Mice Through Regulating the Gut Microbiota. J Integr Med (2021) 19(1):56–65. doi: 10.1016/j.joim.2020.11.001
23. Zeng D, Zhou P, Jiang R, Li XP, Huang SY, Li DY, et al. Evodiamine Inhibits Vasculogenic Mimicry in HCT116 Cells by Suppressing Hypoxia-Inducible Factor 1-Alpha-Mediated Angiogenesis. Anti Cancer Drugs (2021) 32(3):314–22. doi: 10.1097/CAD.0000000000001030
24. Adrain C, Martin SJ. The Mitochondrial Apoptosome: A Killer Unleashed by the Cytochrome Seas. Trends Biochem Sci (2001) 26(6):390–7. doi: 10.1016/S0968-0004(01)01844-8
25. Fang C, Zhang J, Qi D, Fan X, Luo J, Liu L, et al. Evodiamine Induces G2/M Arrest and Apoptosis via Mitochondrial and Endoplasmic Reticulum Pathways in H446 and H1688 Human Small-Cell Lung Cancer Cells. PloS One (2014) 9(12):e115204. doi: 10.1371/journal.pone.0115204
26. Lalier L, Cartron PF, Juin P, Nedelkina S, Manon S, Bechinger B, et al. Bax Activation and Mitochondrial Insertion During Apoptosis. Apoptosis an Int J Programmed Cell Death (2007) 12(5):887–96. doi: 10.1007/s10495-007-0749-1
27. Huang YC, Guh JH, Teng CM. Induction of Mitotic Arrest and Apoptosis by Evodiamine in Human Leukemic T-Lymphocytes. Life Sci (2004) 75(1):35–49. doi: 10.1016/j.lfs.2003.11.025
28. Mohan V, Agarwal R, Singh RP. A Novel Alkaloid, Evodiamine Causes Nuclear Localization of Cytochrome-C and Induces Apoptosis Independent of P53 in Human Lung Cancer Cells. Biochem Biophys Res Commun (2016) 477(4):1065–71. doi: 10.1016/j.bbrc.2016.07.037
29. Yang F, Shi L, Liang T, Ji L, Zhang G, Shen Y, et al. Anti-Tumor Effect of Evodiamine by Inducing Akt-Mediated Apoptosis in Hepatocellular Carcinoma. Biochem Biophys Res Commun (2017) 485(1):54–61. doi: 10.1016/j.bbrc.2017.02.017
30. Zhao N, Tian KT, Cheng KG, Han T, Hu X, Li DH, et al. Antiproliferative Activity and Apoptosis Inducing Effects of Nitric Oxide Donating Derivatives of Evodiamine. Bioorganic Medicinal Chem (2016) 24(13):2971–8. doi: 10.1016/j.bmc.2016.05.001
31. Wang D, Ge S, Chen Z, Song Y. Evodiamine Exerts Anticancer Effects via Induction of Apoptosis and Autophagy and Suppresses the Migration and Invasion of Human Colon Cancer Cells. J BUON Off J Balkan Union Oncol (2019) 24(5):1824–9.
32. Kim SH, Kang JG, Kim CS, Ihm SH, Choi MG, Lee SJ. Evodiamine Suppresses Survival, Proliferation, Migration and Epithelial-Mesenchymal Transition of Thyroid Carcinoma Cells. Anticancer Res (2018) 38(11):6339–52. doi: 10.21873/anticanres.12992
33. Yang S, Chen J, Tan T, Wang N, Huang Y, Wang Y, et al. Evodiamine Exerts Anticancer Effects Against 143B and MG63 Cells Through the Wnt/β-Catenin Signaling Pathway. Cancer Manage Res (2020) 12:2875–88. doi: 10.2147/CMAR.S238093
Keywords: evodiamine, tumor, systematic review, meta-analysis, mice model experiments
Citation: Yin C, Cheng J, Peng HB, Yuan SJ, Chen KL and Li J (2021) Antitumor Effects of Evodiamine in Mice Model Experiments: A Systematic Review and Meta-Analysis. Front. Oncol. 11:774201. doi: 10.3389/fonc.2021.774201
Received: 11 September 2021; Accepted: 18 October 2021;
Published: 09 November 2021.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Masoud Keikha, Mashhad University of Medical Sciences, IranYoubo Zhang, Peking University, China
Copyright © 2021 Yin, Cheng, Peng, Yuan, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Li, lz198207@126.com; Keli Chen, kelichen@126.com
†These authors have contributed equally to this work
 Cong Yin
Cong Yin Jing Cheng†
Jing Cheng†