- 1Department of Hepato-Pancreato-Biliary Surgery and Liver Transplantation, University Medical Center Groningen, University of Groningen, Groningen, Netherlands
- 2Department of Genetics, University Medical Center Groningen, University of Groningen, Groningen, Netherlands
Background: The current prognostication of patient survival after surgery for colorectal liver metastases is based on clinical characteristics, but low accuracy makes it difficult to guide treatment for the individual patient. Rapidly evolving technologies have led to the expectation that biomarkers will be able to outperform the current clinical scoring systems and provide more effective personalised treatment. Two main topics prevail in cancer treatment, namely the role of the immune system and the prediction and prognostication by application of high-throughput methodology. The aim of this review is to examine the evidence for prognostic immunological and molecular markers studied in tumour tissue obtained at surgical resection for colorectal liver metastases.
Methods: First we analysed immunophenotypical protein markers, that are mainly studied by immunohistochemistry. Second, we review molecular markers by analysing high-throughput studies on tumour mRNA and microRNA expression.
Results: CD3, CD4, and CD8 are the most frequently studied protein markers. High intra-tumoural CD3+ T cell infiltration and low CXCR4 expression have the best association with favourable patient survival. Studies that analysed microRNA or mRNA expression data showed very little overlap in prognostic genes.
Conclusions: Patient prognostication after surgery for colorectal liver metastases by analysing the immune system remains difficult. Current data are based on diverse and heterogeneous patient populations which prohibits drawing firm conclusions.
Introduction
Rationale
In Europe, colorectal carcinoma (CRC) is the cancer with the third highest incidence and the second highest mortality rate (1). The liver is the most common site of metastases, and a curative resection of colorectal liver metastases (CRLM) is impossible in 75–80% of patients because of widespread liver involvement, extra-hepatic disease or comorbidity (2). Although recent advances in the treatment of CRLM have extended the possibilities to increase curability, disease will still recur in many patients undergoing a potentially curative resection (3). The mean 5-year survival of patients undergoing intentionally curative surgery varies from 15 to 60% (4).
Models that predict the outcome of treatment in patients with CRLM can support the therapeutical management of individual patients. To this end, several prognostic scoring systems have been developed to guide treatment decisions, predominantly based on clinicopathological characteristics. Unfortunately, these clinical models still have high variability and a review of these models found no common prognostic factor between them (5). Multiple prognostic factors have been identified in patients with CRLM, e.g., KRAS and BRAF mutational status and surgical resection margin (5–7). Rapidly evolving technologies have led to the expectation that immunological and molecular biomarkers will be able to outperform the current clinical scoring systems and provide more effective personalised treatment.
Objectives and Research Question
Two main topics prevail in cancer treatment, namely the role of the immune system (8) and the prediction and prognostication by application of high-throughput methodology. The latter has been shown to be of great prognostic value in for instance breast cancer (9). Both topics are not recently analysed in a systematic review on the treatment of CRLM. Therefore, the aim of this review is to examine the evidence for prognostic immunological and molecular markers studied in tumour tissue obtained at surgical resection for CRLM. To do this, we first review studies using tissue-based immunophenotypical protein markers. We then review tissue-based molecular markers by assessing tumour mRNA and microRNA expression, focusing on genes related to the immune system.
Methods
Study Design and Search Strategy
This systematic review is based on literature that analyses the effectiveness of tissue-based prognostic markers of patient survival and recurrence rate after surgery for CRLM. Our review examines papers published in English between January 1, 2005 until November 10, 2017. We chose 2005 as the starting year because the last systematic review on prognostic markers included papers until 2005 (10). Online publications ahead of print were also included. The databases of Pubmed and Web of Science were screened using the search terms: “colorectal liver metastases” OR “colorectal liver metastasis” AND “tumour biology” OR “tumour biology” OR “genetic” OR “genetics” OR “molecular” OR “markers” OR “expression” OR “mutation” OR “mirna” or “microRNA” OR “lncrna” OR “DNA” OR “RNA.” We first screened the studies for eligibility based on title and abstract, and then thereafter on reading the entire manuscript. References cited in the studies were also checked to identify other relevant papers not found in our initial search. After this initial search process, we also performed searches on all the individual immunological markers, e.g., using search terms like: “CD4” AND “colorectal liver metastases.” In all instances, the data used in this review was extracted from the original papers.
Data Collection and Analysis
The following variables were collected: immune markers, patient survival, administration of chemotherapy, scientific method, and statistics. Survival rates in the various studies were variable and were recorded as median survival (in months), overall survival (OS), cancer-specific survival (CSS) or disease-free survival (DFS). Additionally, disease specific survival (DSS) was recorded as CSS, and recurrence-free survival and progression-free survival were recorded as DFS. To assess the risk of bias in the selected studies, several study and patient characteristics were analysed, including administration of neoadjuvant and adjuvant chemotherapy, number of patients and the use of univariable/multivariable analyses. However, reporting bias in the original papers could not always be assessed, with non-significant findings in particular not always reported. The principal summary measures are the correlation of the immune-related markers with patient survival. The data was analysed by combining the results of the selected studies.
Results
Study Selection
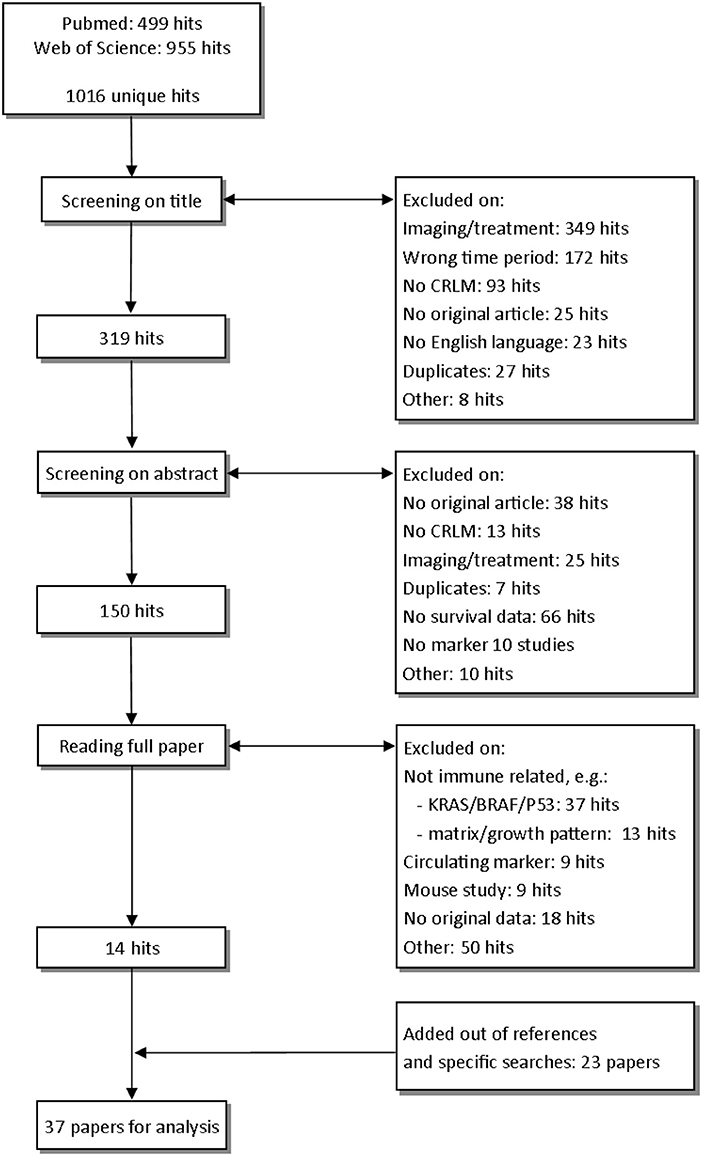
The studies examined in this review are shown in Figure 1. In total, after removing duplicates, we identified 1,016 unique hits using our search terms. After screening on title and abstract, we read 150 studies in full. After reading the full paper, 14 papers were eligible for analysis. In cited references and specific searches, we found another 23 eligible studies, resulting in the 37 studies that we discuss in this review (Figure 1).
Immunophenotypical Protein Markers
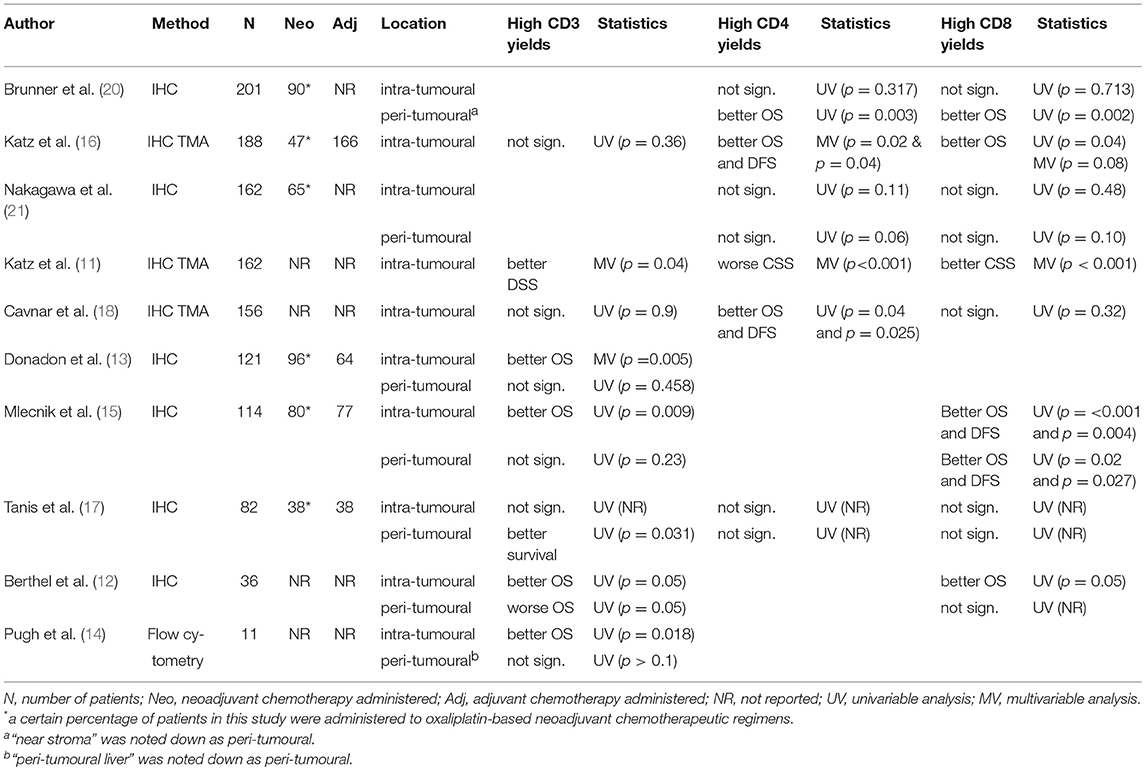
Phenotypes of tumour infiltrating inflammatory cells and profiles of cytokines were mostly studied by immunohistochemistry (IHC) on tissue sections, by scoring the presence of tumour infiltrating lymphocytes (TILs) in the intra-tumoural and peri-tumoural region. In contrast, studies that use tissue microarrays (TMA) for the IHC studies only analyse intra-tumoural areas. Distant liver parenchyma is seldom studied. The largest proportion of TILs in the tumour microenvironment consists of T cells, which are further subdivided according to their IHC staining pattern. The most commonly used markers are CD3 (all T cells), CD4 (T helper cells), CD8 (cytotoxic T cells), and FOXP3 (regulatory T cells). Supplementary Table 1 shows the summary of all studies that analyse immune-related protein and RNA expression.
General T Cells
In eight studies, CD3+ was used as a marker for T cells. All eight analysed intra-tumoural expression and five also analysed peri-tumoural regions (Table 1). An association between high infiltration of intra-tumoural CD3+ T cells and favourable patient survival was seen in 5/8 studies (11–15) analysing a total of 444 patients, while the remaining three studies (16–18), analysing 426 patients, yielded non-significant findings. The difference in prognostic value of CD3+ seems to be associated with neoadjuvant chemotherapy. That is, in the studies that show an association between high intra-tumoural CD3+ T cells and favourable patient survival, more patients received neoadjuvant chemotherapy (176/235; 74.9%) (13, 15) compared to studies that show no association with patient survival (85/270; 31.5%) (16, 17). Five studies analysed the peri-tumoural regions (12–15, 17), three showed no association with patient survival (13–15), one showed a favourable DFS (17), and one showed an unfavourable OS (12). In this latter paper, only 3 out of 36 patients had tumours with high numbers of CD3+ cells, yielding results that are difficult to interpret (12). In addition, one study analysed general T cells by quantification of CD45 and showed an association between a high peri-tumoural infiltration and a favourable survival (19).
T Helper Cells (CD4) and Cytotoxic T Cells (CD8)
Out of the eight papers that reported on CD4+ and CD8+ T cells, six reported on both markers (11, 16–18, 20, 21) and two only on CD8 (12, 15) (Table 1). Interestingly, out of the six papers that studied both CD4 and CD8, four described similar associations with patient survival related to these two markers (16, 17, 20, 21) (Table 1). An association between high intra-tumoural infiltration of CD4+ T cells and favourable patient survival was described in 2/6 papers (16, 18) analysing a total of 344 patients, while three other studies analysing 544 patients yielded non-significant results (17, 20, 21). In contrast, one study showed an association between a high intra-tumoural CD4 expression and unfavourable patient survival (11). Additionally, in the papers that study peri-tumoural regions (17, 20, 21), an association between high peri-tumoural infiltration of CD4+ T cells and favourable patient survival was seen in 1/3 papers (20). Four out of eight papers describe an association between high intra-tumoural CD8+ T cells and a favourable patient survival (11, 12, 15, 16), analysing a total of 500 patients, while the other 4 studies, analysing 601 patients, observed no such association (17, 18, 20, 21). In addition, two out of five studies describe an association between high CD8+ T cells in the peri-tumoural regions and favourable patient survival (15, 20), while this association was not observed in the other three studies (12, 17, 21) (Table 1). Of note, one study analysed granzyme B positive immune cells by IHC, which is primarily a marker for cytotoxic T cells, and did not find an association with patient survival (12). Based on these papers it is tempting to suggest that high intra-tumoural infiltration of CD4+ and CD8+ T cells might be associated with a favourable survival, but it should be realised that this was observed in the minority of studies (CD4: 2/6 studies; CD8: 4/8 studies). In the peri-tumoural regions, an association with patient survival has not been observed. The assessment of bias by chemotherapeutical treatment is limited, as not all studies report the use of chemotherapy (Table 1).
FoxP3+ Regulatory T Cells (Tregs)
Tregs are known for their immunosuppressive function in tumour biology. Of the four studies that fulfilled criteria for analysis in this review, three did not show an association between intra-tumoural FoxP3+ T cells and patient survival (16, 18, 21). One other group also studied intra-tumoural FoxP3+ T cells, but did not report on a survival analysis in relation to this marker, strongly suggesting that there is no association with patient survival (15). Contradictory results have been reported for FoxP3+ T cells in peri-tumoural regions. While one group showed an association between high FoxP3+ T cells and a favourable DFS [26], this was not observed in another study (15).
Memory T cells
Memory T cells are T cells that encounter and respond to an antigen, and are therefore primed to react much faster to a second encounter with that antigen (22). The marker CD45RO is used to detect memory T cells. Three independent studies did not show an association between intra-tumoural presence of CD45RO+ T cells and patient survival (11, 15, 20). Two of these studies also analysed the peri-tumoural regions (15, 20). In one study analysing 201 patients by IHC, high infiltration of CD45RO+ T cells near the tumour border was associated with a favourable patient survival (20).
CD20+ B cells
B cells are lymphocytes that can serve as antigen presenting cells, and they are known for secreting antibodies and cytokines. The association of CD20+ B cells with patient survival was analysed in four independent studies. Two studies showed an association between high peri-tumoural CD20+ B cells and a favourable overall survival (15, 19). In addition, one of these studies also showed an association between high intra-tumoural CD20+ B cells and a favourable OS and DFS (15). The two other studies observed no associations with patient survival (12, 17).
Macrophages
The innate immune system is the first line of defence against pathogens, with macrophages as the most-studied immune cell type in oncology, which is mainly stained using CD68 or CD163. In one study, the presence of more intra-tumoural CD68+ cells was associated with a favourable patient DFS (18). Five other studies reported no association between the number of macrophages and patient survival (12, 17, 19, 23, 24). In three out of these five studies, CD68+ (17, 24) and CD163+ (12) macrophages are not mentioned in the survival analysis, strongly suggesting that there is no significant association. Another study specifically analysing the peri-tumoural regions used MAC387 as macrophage marker and found no association with patient survival (23). MAC387 is thought to represent blood-derived macrophages and monocytes recruited to the inflammatory site (25). In conclusion, although there are six studies analysing macrophages in the tumour microenvironment, only one study found an association between favourable survival and high CD68+ macrophages.
Natural Killer (NK) cells
NK cells are a type of cytotoxic lymphocytes that belong to the innate immune system, and they have been linked to antitumour activity in primary CRC (26). One flow cytometry-based study showed no association between patient survival and infiltration of NK cells (CD56+CD3–) (14). In contrast, another study showed that the presence of intra-tumoural NKp46+ cells was associated with a favourable OS after multivariable analysis. They also showed that peri-tumoural NKp46+ cells were not associated with patient survival, suggesting that the NK cells act intra-tumoural (13).
Mast Cells
Although mast cells are mainly known for their IgE response to allergies, they could also be involved in primary CRC biology (27). One study showed that high infiltration of CD117+ mast cells in CRLM tumour tissue is associated with a favourable DFS (17), while another study showed that high infiltration of peri-tumoural tryptase+ mast cells is associated with an unfavourable OS (23). In other cancers, high tumour-infiltrating tryptase+ mast cells generally have an unfavourable association with survival (28). The differences in markers and localisation precludes any conclusion on the role of mast cells in CRLM.
Combining Multiple Immunophenotypical Protein Markers
In primary colorectal cancer a semiquantitative scoring system has been proposed, the Immunoscore, that quantifies CD3+ and CD8+ expression both intra-tumoural and peri-tumoural (29). Moreover, several studies developed their own IHC-based immunological score because it delivered the best explanation for the differences in patient survival they observed in their dataset. Three studies in CRLM showed significant associations between a high immunoscore and favourable patient survival, two in a univariable analysis (17, 30) and one in a multivariable analysis (15). Turcotte et al. showed that the combination of high CD3+ T cells and high MHC class I expression was the best predictor of long term survival (31). Another study showed that a high intra-tumoural expression of both CD3+ T cells and NKp46+ NK cells was associated with a favourable OS (13).
Katz et al. found that the combination of intra-tumoural CD4+ and CD8+ quantification was the best predictor of patient survival (11). In a more recent study, the same authors showed that a high intra-tumoural CD8+/CD3+ ratio was associated with a favourable OS, while a high FoxP3+/CD4+ ratio and a high FoxP3+/CD8+ ratio were independently associated with an unfavourable survival (16). Another study showed an association between a favourable patient survival and a high ratio of CD45RO+ cells concerning localisation (high peri-tumoural CD45RO+ and low CD45RO+ in distant liver parenchyma). In addition, they combine the CD45RO ratio with the histopathological growth pattern to create their best predictor of patient survival (20). Of note, a higher number of peri-tumoural TILs has been reported to be associated with the desmoplastic growth pattern of CRLM, which may explain the relative favourable prognosis compared to the replacement or pushing growth types (32). Finally, Mlecnik et al. show that the combination of high CD8 and CD20 expression, in both the intra-tumoural and peri-tumoural region, was associated with a favourable OS and DFS (15).
Inflammatory Mediators
Cytokines and other inflammatory mediators can attract immune cells to the site of inflammation or have a promoting or repressing effect on the immune system. For example, CTLA-4 and PD-L1 are inhibitory ligands that can de-activate a proper T cell response toward the tumour and are the two primary targets of modern-day immunotherapy. One study in CRLM showed an association between a high PD-L1 expression in the tumour stroma and a favourable DFS (33).
CXC Chemokines and Receptors
CXCR2 is a chemokine receptor that mainly attracts neutrophils and endothelial cells. In addition, CXCL7 is an agonist of CXCR2 and is a cleavage product that is particularly released by platelets. In tumourigenesis however, CXCR2 might facilitate tumour growth and development of metastases by increased angiogenesis (34). Desurmont et al. showed that both a high CXCR2 and CXCL7 expression have an independent association with an unfavourable patient survival (35). Several other chemokines have been studied, but did not show an association with patient survival [e.g., CXCR7/CXCL12 (33), CXCL8 (34)].
CXC Chemokine Receptor 4 (CXCR4)
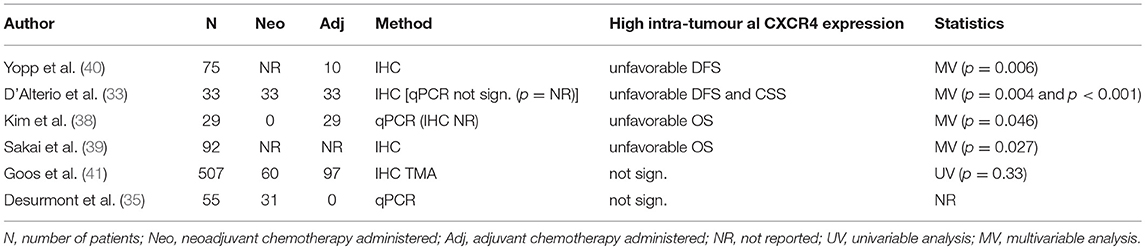
CXCR4 executes its effect by binding to its ligand CXCL12 (SDF-1), allowing downstream signalling that can alter several biological pathways including immune checkpoints (36). It has been shown that an inhibition of CXCR4/CXCL12 can improve the efficacy of immunotherapy, suggesting that high CXCR4 expression inhibits an effective immune response by targeting CTLA-4 (37). Out of the six papers that study CXCR4, four show an association between high CXCR4 expression and unfavourable patient survival (Table 2) (33, 38–40). The two other studies did not find significant associations with patient survival (35, 41). There is considerable variation in the methods used to quantify protein expression by IHC, particularly in the CXCR4 studies. Studies use different cut-off values for assessing positive staining [10% (40) vs. 50% (33)]. Another paper assessed CXCR4 expression using a digital slide computer scanner to score stromal or cancer cells for intensity and frequency (41). Finally, a study classified cytoplasmic CXCR4 expression as low or high relative to the staining intensity of hepatocytes (39). These different scoring methods and cut-off levels make it difficult to draw valid conclusions. However, CXCR4 is associated with an unfavourable prognosis in different scoring systems.
Monocyte Chemoattractant Protein-1 (MCP-1)/CCL2
MCP-1 is a chemokine that mainly influences monocytes and macrophages; it can recruit macrophages via chemotaxis to promote tumour progression (42). One study showed that high MCP-1 expression was associated with unfavourable patient survival and increased hepatic recurrences after surgery. However, a significant correlation between MCP-1 and CD68+ macrophages was not found (24).
COX-2/PTGS2
COX-2 is thought to modulate prostaglandin pathways into promoting tumour immune evasion, and could therefore be a potential target for treatment (43). One group showed that a high intra-tumoural COX-2 expression measured by IHC was associated with an unfavourable OS in multivariable analysis (41, 44).
Molecular RNA Expression Markers
Studies that analyse mRNA or miRNA expression generally only assess intra-tumoural expression of molecular markers. All expression studies that analyse molecular markers initially perform a high-throughput method to scout for interesting genes. In this review, we analysed the overlap between the high-throughput studies to assess if the immune system has a prominent role in identifying differences in patient survival.
Messenger RNA Expression
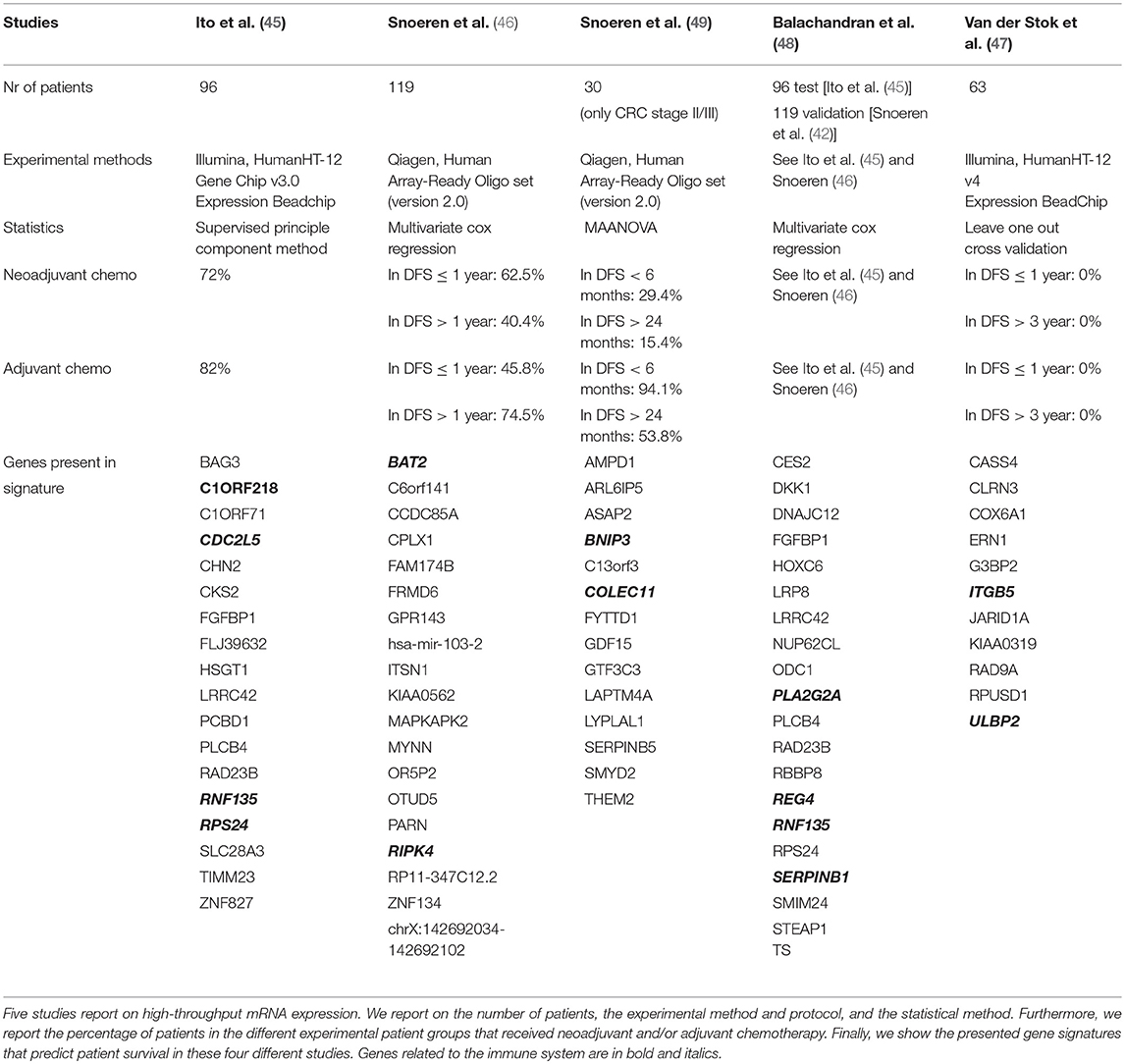
Table 3 shows the results of five papers that describe gene signatures associated with patient survival based on high-throughput expression microarrays (45–49). Balachandran et al. (48) used the test-set from Snoeren et al. (46) and the validation set from Ito et al. (45), thereby creating a validated multi-centre gene signature. Second, Snoeren et al. published two expression studies, with the second zooming in on stage II/III CRC patients (49). Interestingly, the four presented individual centre gene signatures [excluding Balachandran et al. (48)] do not share any gene and do not point toward a specific biological pathway. Of note, because these high-throughput studies do not show the total list of all genes and their association with survival, there could be overlap between the studies. We analysed the enrichment of the immune system in these five gene signatures by selecting the 76 genes and correlated this with the “immune system process” pathway in the Gene Ontology (GO) database (50, 51). Ten signature genes showed overlap with all immune related genes (BNIP3, C1ORF218, CDC2L5, COLEC11, ITGB5, PLA2G2A, RNF135, RPS24, SERPINB1, ULBP2). By specific searches, we found three additional genes related to the immune system (BAT2, REG4, RIPK4). In total, 13 out of 76 (17.1%) genes were present in immune related pathways (marked bold in Table 3), which is comparable to all known immune related genes divided by all human genes (3119/20226; 15.4%) (50, 51). In conclusion, the presented gene signatures based on high-throughput expression profiling generate non-overlapping gene signatures and do not show predominant immunological characteristics that are associated with patient survival.
LIGHT/TNFSF14
Based on expression microarray results (45), it was validated by IHC that both a high LIGHT expression in the tumour cells and in TILs was associated with improved patient survival (52). Tumour necrosis factor superfamily member 14, also known as LIGHT, has been shown to induce T cell proliferation and tumour regression in vivo (53).
MicroRNA Expression
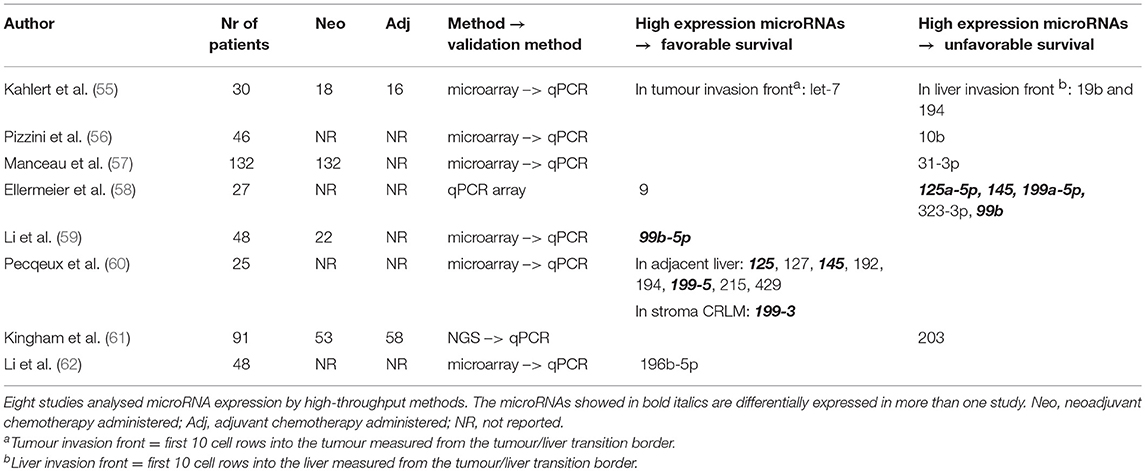
MicroRNAs are small non-coding 18-22nt RNAs that function in the post-transcriptional regulation of gene expression, primarily by silencing mRNA (54). Table 4 shows studies included in this review reporting on microRNAs and patient survival (55–62). There are eight high-throughput studies, of which seven validated their observations by qPCR (55–57, 59–62). In addition, in the earlier described expression study of Snoeren et al. (46), a high microRNA 103-2 expression was associated with an unfavourable DFS (Table 3). There is little overlap between the microRNA studies but, remarkably, the study of Ellermeier et al. shows opposing results compared to other studies (Table 4) (58). For example, Li et al. (59) observed an association between a high intra-tumoural microRNA 99b-5p expression and favourable patient survival, yet Ellermeier et al. show an association with unfavourable patient survival (58). Both studies differ in patient numbers and methodology (Table 4; qPCR array vs. microarrays). In conclusion, there is no overlap in differentially expressed microRNAs between the eight analysed studies.
For many microRNAs, the precise mRNA target is not known. Three papers review microRNAs in the light of immune responses (63), tumour attack and immune escape (64), and PD-1/PD-L1 immune checkpoints (65), giving an overview on the interaction between microRNAs and the immune system. Although there is no overlap in the microRNAs that are differentially expressed in the studies, the downstream effect of the differentially expressed microRNAs might be similar (Supplementary Table 2).
Discussion
We have reviewed the prognostic value of tissue-based immune-related markers after surgery for CRLM. We find that CD3, CD4, and CD8 are the most frequently studied protein markers (Supplementary Table 1). These T cell markers represent the most abundant immune cell types in the tumour microenvironment of CRLM, and are therefore logical markers to study. However, this means that certain immune cell types remain under-analysed in relation to patient survival. The data generated by analysed studies are based on diverse and heterogeneous patient populations as well as various treatment characteristics. This prohibits drawing firm conclusions on the role of the immune system in patients with colorectal liver metastases. Therefore, standardising the quantification of immune infiltration would make it easier to compare studies with each other. In primary colorectal cancer, the consensus Immunoscore is proposed to be implemented in the current TNM staging. This Immunoscore quantifies CD3+ and CD8+ T cells by digital pathology software and has prognostic value and might contribute to compare the results among various study groups (29).
Intra-Tumoural CD3+ T Cells
High numbers of intra-tumoural CD3, CD4, and CD8 were predictive of a favourable patient survival. Out of the eight studies (11, 13, 15–17, 20, 30, 31) that combined immune markers to create the best predictor of patient survival, six include intra-tumoural CD3+ quantification (13, 15–17, 30, 31). These results suggest that intra-tumoural CD3+ T cells have the best prognostic value. However, there are differences in patient and treatment characteristics between the studies. For example, in the studies that observed prognostic value in intra-tumoural CD3+ quantification, neoadjuvant chemotherapy was more often administered compared to studies that did not observe prognostic value (74.9% vs. 31.5%). Previous studies have shown that chemotherapeutic regimens including oxaliplatin or anthracyclins can induce an immune response leading to immunogenic tumour cell death (66–69). This suggests that patients who receive these neoadjuvant treatments potentially have more immune infiltration, e.g., due to an increase in tumour antigen-presentation via MHC (70, 71). However, the majority of the studies included in our review do not explain treatment regimes in detail and do not show associations of neoadjuvant chemotherapy with immune infiltration or patient survival. Thus, the differences in treatment regimes of administering neoadjuvant chemotherapy may have caused bias.
Univariable vs. Multivariable and Clinicopathological Characteristics
Several studies only show univariable analysis of their targeted marker without showing multivariable analysis. This makes it difficult to estimate the importance of the targeted molecular marker in the light of known clinicopathological risk factors. In the studies that do find a significant association between patient survival and an immunological marker, clinicopathological risk factors can also be predictive of patient survival or are strongly related to the immune marker. Out of the 10 studies that analyse CD3/CD4/CD8/FOXP3 by IHC, six studies show multivariable analyses including clinicopathological variables (11, 13, 15, 16, 18, 21). Out of those six studies, four show that known clinical risk factors are also independent predictors of patient survival (11, 16, 18, 21). For example, Katz et al. show by multivariable analysis that high intra-tumoural CD3 expression is associated with a favourable patient survival, but so is the clinical risk score (72). In fact, the clinical risk score is an even better predictor of patient survival compared to high intra-tumoural CD3 expression [p < 0.001; odds ratio 8.8 (3.3–23.5) vs. p = 0.04; odds ratio 4.2 (1.1–16.9)] (11).
Other Molecular Prognostic Markers
Multiple additional molecular markers have shown prognostic value after surgical resection of colorectal liver metastases. It is known that mutations in the KRAS (codon 12 and 13) and BRAF (V600E) genes are associated with unfavourable patient survival (7). In addition, several molecular characteristics are positively associated with an enhanced immune response in (metastatic) colorectal cancer and favourable patient survival (73), like the CpG island methylator phenotype (CIMP) (74) and the microsatellite instability phenotype (75). Patients with microsatellite instable tumours typically have less often metastases, a better survival and a good response to modern immunotherapy (76). Although we primarily studied immune related markers on protein and RNA level, alterations at the DNA level, not discussed in this review, could potentially also be relevant for prognostication (7).
Applicability as Prognostic Biomarker
Modern high-throughput methods like next generation sequencing or microarrays are innovative, but not every hospital has the equipment and knowledge to apply them in a diagnostic setting. Thus, although IHC is a relatively old technology, it is the most used method to predict patient survival. The high-throughput studies of both mRNA and miRNA in CRLM find little overlap in prioritising genes. In contrast, gene signatures designed by high-throughput methods are used to guide treatment decisions in breast cancer: this signature (MammaPrint) is based on the expression of 70 genes and is used to assess whether patients could benefit from adjuvant chemotherapy (77). One could speculate whether high-throughput studies are worthwhile, since genetic markers have the problem that there are potentially many markers. Thus, you need to build up a catalogue of all possible genetic markers, which takes time and cohorts of sufficient size. In contrast, every patient has an immune response (whether effective or not), making immune-related markers as the more obvious immediate target.
Conclusions and Future Perspective
The most important IHC-marker for prognostication in the studies we reviewed is CD3, a general T cell marker. Although intra-tumoural CD3+ quantification is the best immune-related predictor of patient survival, we think that this marker is not accurate enough to guide individual treatment decisions. In addition, while high-throughput expression studies show promise, there is no consensus yet about the important genes for prognostication. A high CXCR4 expression was reported by several studies as an unfavourable prognostic factor, and this might be a future target for improving the efficacy of immunotherapy (37). We know that patients with immunocompetent tumours can potentially benefit from immunotherapy (78, 79). Hopefully, future therapy can enhance the function of the immune system in an effective way to eradicate cancer in all tumour-bearing organs.
Author Contributions
JH performed the analysis and wrote the manuscript. KdJ devised the main conceptual ideas and supervised JH. KK and RS helped supervise the project. All authors discussed the results and contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kate McIntyre for assistance in the editorial process.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00148/full#supplementary-material
References
1. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. (2013) 49:1374–403. doi: 10.1016/j.ejca.2012.12.027
2. Scheele J, Stang R, Altendorf-Hofmann A, Paul M. Resection of colorectal liver metastases. World J Surg. (1995) 19:59–71. doi: 10.1007/BF00316981
3. Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. (2016) 27:1386–422. doi: 10.1093/annonc/mdw235
4. Simmonds PC, Primrose JN, Colquitt JL, Garden OJ, Poston GJ, Rees M. Surgical resection of hepatic metastases from colorectal cancer: a systematic review of published studies. Br J Cancer. (2006) 94:982–99. doi: 10.1038/sj.bjc.6603033
5. Spelt L, Andersson B, Nilsson J, Andersson R. Prognostic models for outcome following liver resection for colorectal cancer metastases: a systematic review. Eur J Surg Oncol. (2012) 38:16–24. doi: 10.1016/j.ejso.2011.10.013
6. Yamashita S, Chun YS, Kopetz SE, Vauthey J-N. Biomarkers in colorectal liver metastases. Br J Surg. (2018) 105:618–27. doi: 10.1002/bjs.10834
7. Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, Moris D, Cloyd J, Spartalis E, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: a systematic review of the current evidence. Surg Oncol. (2018) 27:280–8. doi: 10.1016/j.suronc.2018.05.012
8. Donadon M, Lleo A, Di Tommaso L, Soldani C, Franceschini B, Roncalli M, et al. The shifting paradigm of prognostic factors of colorectal liver metastases: from tumor-centered to host immune-centered factors. Front Oncol. (2018) 8:181. doi: 10.3389/fonc.2018.00181
9. Blok EJ, Bastiaannet E, van den Hout WB, Liefers GJ, Smit VTHBM, Kroep JR, et al. Systematic review of the clinical and economic value of gene expression profiles for invasive early breast cancer available in Europe. Cancer Treat Rev. (2018) 62:74–90. doi: 10.1016/j.ctrv.2017.10.012
10. Neal CP, Garcea G, Doucas H, Manson MM, Sutton CD, Dennison AR, et al. Molecular prognostic markers in resectable colorectal liver metastases: a systematic review. Eur J Cancer. (2006) 42:1728–43. doi: 10.1016/j.ejca.2006.01.056
11. Katz SC, Pillarisetty V, Bamboat ZM, Shia J, Hedvat C, Gonen M, et al. T cell infiltrate predicts long-term survival following resection of colorectal cancer liver metastases. Ann Surg Oncol. (2009) 16:2524–30. doi: 10.1245/s10434-009-0585-3
12. Berthel A, Zoernig I, Valous NA, Kahlert C, Klupp F, Ulrich A, et al. Detailed resolution analysis reveals spatial T cell heterogeneity in the invasive margin of colorectal cancer liver metastases associated with improved survival. Oncoimmunology. (2017) 6:e1286436. doi: 10.1080/2162402X.2017.1286436
13. Donadon M, Hudspeth K, Cimino M, Di Tommaso L, Preti M, Tentorio P, et al. Increased infiltration of natural killer and T cells in colorectal liver metastases improves patient overall survival. J Gastrointest Surg. (2017) 21:1226–36. doi: 10.1007/s11605-017-3446-6
14. Pugh SA, Harrison RJ, Primrose JN, Khakoo SI. T cells but not NK cells are associated with a favourable outcome for resected colorectal liver metastases. BMC Cancer. (2014) 14:180. doi: 10.1186/1471-2407-14-180
15. Mlecnik B, Van den Eynde M, Bindea G, Church SE, Vasaturo A, Fredriksen T, et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst. (2018) 110:97–108. doi: 10.1093/jnci/djx123
16. Katz SC, Bamboat ZM, Maker AV, Shia J, Pillarisetty VG, Yopp AC, et al. Regulatory T cell infiltration predicts outcome following resection of colorectal cancer liver metastases. Ann Surg Oncol. (2013) 20:946–55. doi: 10.1245/s10434-012-2668-9
17. Tanis E, Julié C, Emile J-F, Mauer M, Nordlinger B, Aust D, et al. Prognostic impact of immune response in resectable colorectal liver metastases treated by surgery alone or surgery with perioperative FOLFOX in the randomised EORTC study 40983. Eur J Cancer. (2015) 51:2708–17. doi: 10.1016/j.ejca.2015.08.014
18. Cavnar MJ, Turcotte S, Katz SC, Kuk D, Gönen M, Shia J, et al. Tumor-associated macrophage infiltration in colorectal cancer liver metastases is associated with better outcome. Ann Surg Oncol. (2017) 24:1835–42. doi: 10.1245/s10434-017-5812-8
19. Meshcheryakova A, Tamandl D, Bajna E, Stift J, Mittlboeck M, Svoboda M, et al. B cells and ectopic follicular structures: novel players in anti-tumor programming with prognostic power for patients with metastatic colorectal cancer. PLoS ONE. (2014) 9:e99008. doi: 10.1371/journal.pone.0099008
20. Brunner SM, Kesselring R, Rubner C, Martin M, Jeiter T, Boerner T, et al. Prognosis according to histochemical analysis of liver metastases removed at liver resection. Br J Surg. (2014) 101:1681–91. doi: 10.1002/bjs.9627
21. Nakagawa K, Tanaka K, Homma Y, Nojiri K, Kumamoto T, Takeda K, et al. Low infiltration of peritumoral regulatory T cells predicts worse outcome following resection of colorectal liver metastases. Ann Surg Oncol. (2015) 22:180–6. doi: 10.1245/s10434-014-3974-1
22. Flynn JK, Gorry PR. Stem memory T cells (TSCM)—their role in cancer and HIV immunotherapies. Clin Transl Immunol. (2014) 3:e20. doi: 10.1038/cti.2014.16
23. Suzuki S, Ichikawa Y, Nakagawa K, Kumamoto T, Mori R, Matsuyama R, et al. High infiltration of mast cells positive to tryptase predicts worse outcome following resection of colorectal liver metastases. BMC Cancer. (2015) 15:840. doi: 10.1186/s12885-015-1863-z
24. Yoshidome H, Kohno H, Shida T, Kimura F, Shimizu H, Ohtsuka M, et al. Significance of monocyte chemoattractant protein-1 in angiogenesis and survival in colorectal liver metastases. Int J Oncol. (2009) 34:923–30. doi: 10.3892/ijo_00000218
25. Soulas C, Conerly C, Kim W-K, Burdo TH, Alvarez X, Lackner AA, et al. Recently infiltrating MAC387+ monocytes/macrophages. Am J Pathol. (2011) 178:2121–35. doi: 10.1016/j.ajpath.2011.01.023
26. Coppola A, Arriga R, Lauro D, del Principe MI, Buccisano F, Maurillo L, et al. NK cell inflammation in the clinical outcome of colorectal carcinoma. Front Med. (2015) 2:33. doi: 10.3389/fmed.2015.00033
27. Marech I, Ammendola M, Gadaleta C, Zizzo N, Oakley C, Gadaleta CD, et al. Possible biological and translational significance of mast cells density in colorectal cancer. World J Gastroenterol. (2014) 20:8910–20. doi: 10.3748/wjg.v20.i27.8910
28. Hu G, Wang S, Cheng P. Tumor-infiltrating tryptase + mast cells predict unfavorable clinical outcome in solid tumors. Int J Cancer. (2018) 142:813–21. doi: 10.1002/ijc.31099
29. Pagès F, Mlecnik B, Marliot F, Bindea G, Ou F-S, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. (2018) 391:2128–39. doi: 10.1016/S0140-6736(18)30789-X
30. Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. (2011) 71:5670–7. doi: 10.1158/0008-5472.CAN-11-0268
31. Turcotte S, Katz SC, Shia J, Jarnagin WR, Kingham TP, Allen PJ, et al. Tumor MHC class I expression improves the prognostic value of T-cell density in resected colorectal liver metastases. Cancer Immunol Res. (2014) 2:530–7. doi: 10.1158/2326-6066.CIR-13-0180
32. van Dam P-J, van der Stok EP, Teuwen L-A, Van den Eynden GG, Illemann M, Frentzas S, et al. International consensus guidelines for scoring the histopathological growth patterns of liver metastasis. Br J Cancer. (2017) 117:1427–41. doi: 10.1038/bjc.2017.334
33. D'Alterio C, Nasti G, Polimeno M, Ottaiano A, Conson M, Circelli L, et al. CXCR4–CXCL12–CXCR7, TLR2–TLR4, and PD-1/PD-L1 in colorectal cancer liver metastases from neoadjuvant-treated patients. Oncoimmunology. (2016) 5:e1254313. doi: 10.1080/2162402X.2016.1254313
34. Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. (2008) 267:226–44. doi: 10.1016/j.canlet.2008.04.050
35. Desurmont T, Skrypek N, Duhamel A, Jonckheere N, Millet G, Leteurtre E, et al. Overexpression of chemokine receptor CXCR2 and ligand CXCL7 in liver metastases from colon cancer is correlated to shorter disease-free and overall survival. Cancer Sci. (2015) 106:262–9. doi: 10.1111/cas.12603
36. Walenkamp AME, Lapa C, Herrmann K, Wester H-J. CXCR4 ligands: the next big hit? J Nucl Med. (2017) 58:77S−82S. doi: 10.2967/jnumed.116.186874
37. Lee C-H, Kakinuma T, Wang J, Zhang H, Palmer DC, Restifo NP, et al. Sensitization of B16 tumor cells with a CXCR4 antagonist increases the efficacy of immunotherapy for established lung metastases. Mol Cancer Ther. (2006) 5:2592–9. doi: 10.1158/1535-7163.MCT-06-0310
38. Kim J, Mori T, Chen SL, Amersi FF, Martinez SR, Kuo C, et al. Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg. (2006) 244:113–20. doi: 10.1097/01.sla.0000217690.65909.9c
39. Sakai N, Yoshidome H, Shida T, Kimura F, Shimizu H, Ohtsuka M, et al. CXCR4/CXCL12 expression profile is associated with tumor microenvironment and clinical outcome of liver metastases of colorectal cancer. Clin Exp Metastasis. (2012) 29:101–10. doi: 10.1007/s10585-011-9433-5
40. Yopp AC, Shia J, Butte JM, Allen PJ, Fong Y, Jarnagin WR, et al. CXCR4 expression predicts patient outcome and recurrence patterns after hepatic resection for colorectal liver metastases. Ann Surg Oncol. (2012) 19(Suppl.3):S339–46. doi: 10.1245/s10434-011-1774-4
41. Goos JACM, Coupé VMH, van de Wiel MA, Diosdado B, Delis-Van Diemen PM, Hiemstra AC, et al. A prognostic classifier for patients with colorectal cancer liver metastasis, based on AURKA, PTGS2 and MMP9. Oncotarget. (2016) 7:2123–34. doi: 10.18632/oncotarget.6188
42. Mantovani A, Bottazzi B, Colotta F, Sozzani S, Ruco L. The origin and function of tumor-associated macrophages. Immunol Today. (1992) 13:265–70. doi: 10.1016/0167-5699(92)90008-U
43. Liu B, Qu L, Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. (2015) 15:106. doi: 10.1186/s12935-015-0260-7
44. Goos JACM, Hiemstra AC, Coupé VMH, Diosdado B, Kooijman W, Delis-Van Diemen PM, et al. Epidermal growth factor receptor (EGFR) and prostaglandin-endoperoxide synthase 2 (PTGS2) are prognostic biomarkers for patients with resected colorectal cancer liver metastases. Br J Cancer. (2014) 111:749–55. doi: 10.1038/bjc.2014.354
45. Ito H, Mo Q, Qin L-X, Viale A, Maithel SK, Maker AV, et al. Gene expression profiles accurately predict outcome following liver resection in patients with metastatic colorectal cancer. PLoS ONE. (2013) 8:e81680. doi: 10.1371/journal.pone.0081680
46. Snoeren N, van Hooff SR, Adam R, van Hillegersberg R, Voest EE, Guettier C, et al. Exploring gene expression signatures for predicting disease free survival after resection of colorectal cancer liver metastases. PLoS ONE. (2012) 7:e49442. doi: 10.1371/journal.pone.0049442
47. van der Stok EP, Smid M, Sieuwerts AM, Vermeulen PB, Sleijfer S, Ayez N, et al. mRNA expression profiles of colorectal liver metastases as a novel biomarker for early recurrence after partial hepatectomy. Mol Oncol. (2016) 10:1542–50. doi: 10.1016/j.molonc.2016.09.002
48. Balachandran VP, Arora A, Gönen M, Ito H, Turcotte S, Shia J, et al. A validated prognostic multigene expression assay for overall survival in resected colorectal cancer liver metastases. Clin Cancer Res. (2016) 22:2575–82. doi: 10.1158/1078-0432.CCR-15-1071
49. Snoeren N, Emmink BL, Koerkamp MJG, van Hooff SR, Goos JACM, van Houdt WJ, et al. Maspin is a marker for early recurrence in primary stage III and IV colorectal cancer. Br J Cancer. (2013) 109:1636–47. doi: 10.1038/bjc.2013.489
50. The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. (2017) 45:D331–8. doi: 10.1093/nar/gkw1108
51. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
52. Maker A V, Ito H, Mo Q, Weisenberg E, Qin L-X, Turcotte S, et al. Genetic evidence that intratumoral T-cell proliferation and activation are associated with recurrence and survival in patients with resected colorectal liver metastases. Cancer Immunol Res. (2015) 3:380–8. doi: 10.1158/2326-6066.CIR-14-0212
53. Qiao G, Qin J, Kunda N, Calata JF, Mahmud DL, Gann P, et al. LIGHT elevation enhances immune eradication of colon cancer metastases. Cancer Res. (2017) 77:1880–91. doi: 10.1158/0008-5472.CAN-16-1655
54. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
55. Kahlert C, Klupp F, Brand K, Lasitschka F, Diederichs S, Kirchberg J, et al. Invasion front-specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci. (2011) 102:1799–807. doi: 10.1111/j.1349-7006.2011.02023.x
56. Pizzini S, Bisognin A, Mandruzzato S, Biasiolo M, Facciolli A, Perilli L, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. (2013) 14:589. doi: 10.1186/1471-2164-14-589
57. Manceau G, Imbeaud S, Thiébaut R, Liébaert F, Fontaine K, Rousseau F, et al. Hsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer. Res (2014) 20:3338–47. doi: 10.1158/1078-0432.CCR-13-2750
58. Ellermeier C, Vang S, Cleveland K, Durand W, Resnick MB, Brodsky AS. Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res. (2014) 34:3957–67.
59. Li W, Chang J, Wang S, Liu X, Peng J, Huang D, et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget. (2015) 6:24448–62. doi: 10.18632/oncotarget.4423
60. Pecqueux M, Liebetrau I, Werft W, Dienemann H, Muley T, Pfannschmidt J, et al. A comprehensive MicroRNA expression profile of liver and lung metastases of colorectal cancer with their corresponding host tissue and its prognostic impact on survival. Int J Mol Sci. (2016) 17:E1755. doi: 10.3390/ijms17101755
61. Kingham TP, Nguyen HCB, Zheng J, Konstantinidis IT, Sadot E, Shia J, et al. MicroRNA-203 predicts human survival after resection of colorectal liver metastasis. Oncotarget. (2017) 8:18821–31. doi: 10.18632/oncotarget.13816
62. Li W, Chang J, Tong D, Peng J, Huang D, Guo W, et al. Differential microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Oncotarget. (2017) 8:35783–91. doi: 10.18632/oncotarget.16206
63. Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. (2016) 35:103. doi: 10.1186/s13046-016-0375-2
64. Eichmüller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. (2017) 109:doi: 10.1093/jnci/djx034
65. Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y, et al. The roles of microRNAs in regulating the expression of PD-1/PD-L1 immune checkpoint. Int J Mol Sci. (2017) 18:2540. doi: 10.3390/ijms18122540
66. Riihimäki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. (2016) 6:29765. doi: 10.1038/srep29765
67. Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. (2013) 31:51–72. doi: 10.1146/annurev-immunol-032712-100008
68. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. (2017) 17:97–111. doi: 10.1038/nri.2016.107
69. Garg AD, More S, Rufo N, Mece O, Sassano ML, Agostinis P, et al. Trial watch: immunogenic cell death induction by anticancer chemotherapeutics. Oncoimmunology. (2017) 6:e1386829. doi: 10.1080/2162402X.2017.1386829
70. de la Cruz-Merino L, Henao Carrasco F, Vicente Baz D, Nogales Fernández E, Reina Zoilo JJ, Codes Manuel de Villena M, et al. Immune microenvironment in colorectal cancer: a new hallmark to change old paradigms. Clin Dev Immunol. (2011) 2011:174149. doi: 10.1155/2011/174149
71. Radogna F, Diederich M. Stress-induced cellular responses in immunogenic cell death: implications for cancer immunotherapy. Biochem Pharmacol. (2018) 153:12–23. doi: 10.1016/j.bcp.2018.02.006
72. Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. (1999) 230:309–18. doi: 10.1097/00000658-199909000-00004
73. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. (2015) 21:1350–6. doi: 10.1038/nm.3967
74. Advani SM, Advani P, DeSantis SM, Brown D, VonVille HM, Lam M, et al. Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: a systematic review and meta-analysis. Transl Oncol. (2018) 11:1188–201. doi: 10.1016/j.tranon.2018.07.008
75. George B, Kopetz S. Predictive and prognostic markers in colorectal cancer. Curr Oncol Rep. (2011) 13:206–15. doi: 10.1007/s11912-011-0162-3
76. Lee V, Murphy A, Le DT, Diaz LA Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. (2016) 21:1200–11. doi: 10.1634/theoncologist.2016-0046
77. Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. (2016) 375:717–29. doi: 10.1056/NEJMoa1602253
78. Lynch D, Murphy A. The emerging role of immunotherapy in colorectal cancer. Ann Transl Med. (2016) 4:305. doi: 10.21037/atm.2016.08.29
Keywords: colorectal liver metastases, immune system, prognosis, survival, immunohistochemistry, high-throughput
Citation: Hof J, Kok K, Sijmons RH and de Jong KP (2019) Systematic Review of the Prognostic Role of the Immune System After Surgery of Colorectal Liver Metastases. Front. Oncol. 9:148. doi: 10.3389/fonc.2019.00148
Received: 29 November 2018; Accepted: 21 February 2019;
Published: 19 March 2019.
Edited by:
Khashayarsha Khazaie, Mayo Clinic College of Medicine & Science, United StatesReviewed by:
Niels Halama, German Cancer Research Center (DKFZ), GermanyPashtoon Murtaza Kasi, Mayo Clinic, United States
Copyright © 2019 Hof, Kok, Sijmons and de Jong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joost Hof, j.hof01@umcg.nl
 Joost Hof
Joost Hof Klaas Kok2
Klaas Kok2