- Department of Neurology, Shengjing Hospital of China Medical University, Shenyang, China
Multiple system atrophy (MSA) is an adult onset, fatal disease, characterized by an accumulation of alpha-synuclein (α-syn) in oligodendroglial cells. MicroRNAs (miRNAs) are small non-coding RNAs involved in post-translational regulation and several biological processes. Disruption of miRNA-related pathways in the central nervous system (CNS) plays an important role in the pathogenesis of neurodegenerative diseases, including MSA. While the exact mechanisms underlying miRNAs in the pathogenesis of MSA remain unclear, it is known that miRNAs can repress the translation of messenger RNAs (mRNAs) that regulate the following pathogenesis associated with MSA: autophagy, neuroinflammation, α-syn accumulation, synaptic transmission, oxidative stress, and apoptosis. In this review, the metabolism of miRNAs and their functional roles in the pathogenesis of MSA are discussed, thereby highlighting miRNAs as potential new biomarkers for the diagnosis of MSA and in increasing our understanding of the disease process.
Introduction
Multiple system atrophy (MSA) is a progressive, fatal neurodegenerative disease. Two types of MSA are clinically distinguished: the parkinsonian variant (MSA-P), associated with striatonigral degeneration, and the cerebellar variant (MSA-C), related to olivopontocerebellar atrophy (Gilman et al., 2008; Stefanova et al., 2009; Jellinger, 2014). In Western countries, MSA-P is the most common variant of MSA (Gilman et al., 2008). In contrast, a recent study in Chinese patients found no significant difference between the number of MSA-P and MSA-C patients (Zhang et al., 2018). MSA is primarily a sporadic disease; familial MSA has also been reported (Multiple-System Atrophy Research Collaboration, 2013). Currently, there are no effective therapies for MSA treatment, only symptomatic therapy (Fanciulli and Wenning, 2015).
Multiple system atrophy is characterized by the accumulation of misfolded alpha-synuclein (α-syn) in oligodendroglial cells. Abnormal α-syn is also the pathological feature of other neurodegenerative diseases including Parkinson’s disease (PD) (Nuber et al., 2018; Palma and Kaufmann, 2018). While abnormal α-syn is evident in both MSA and PD cases, when brain extracts of α-syn from MSA and PD cases were injected into transgenic mice, only the α-syn from MSA cases induced neurodegeneration. These results indicate that MSA strains of α-syn are more toxic than the PD strains of α-syn in terms of neurodegeneration (Prusiner et al., 2015).
MicroRNAs (miRNAs) are small non-coding RNAs (19–24 nucleotides in length) that regulate messenger RNA (mRNA) expression and control post-translational regulation (Treiber et al., 2019). Interestingly, it is possible that miRNAs may modulate MSA-related gene expression, for example, SNCA encoding α-syn (Asi et al., 2014; Tagliafierro and Chiba-Falek, 2016). However, MSA-related genes, such as small nuclear ribonucleoprotein polypeptide N (SNRPN), may, in turn, regulate miRNA processing (Xiong et al., 2015; Hama et al., 2017). It is well known that miRNAs in serum, plasma, and cerebrospinal fluid (CSF) are tissue-specific, highly stable, and quantifiable, indicating that these miRNAs may be used to provide early and a more accurate diagnosis of several diseases, including neurodegenerative diseases (Ramaswamy et al., 2018). Targeting miRNAs by anti-miRs have shown positive results in several preclinical studies in cancer and various other diseases (Rupaimoole and Slack, 2017). The role of antisense oligonucleotides (ASO), structurally similar to anti-miRs, is also being investigated in preclinical and clinical trials in several neurodegenerative diseases including PD, amyotrophic lateral sclerosis (ALS), Huntington disease (HD), and Alzheimer disease (AD) (Kordasiewicz et al., 2012; Hinrich et al., 2016; Becker et al., 2017; Zhao et al., 2017). The therapeutic potential of anti-miRs and miR mimics in neurodegenerative diseases has been demonstrated (Zhou et al., 2016; Valera et al., 2017).
The pathophysiological mechanisms of MSA remain unknown; however, studies have demonstrated that α-syn toxicity contributes to the disruption of multiple organelles, including mitochondria, synaptic vesicles, lysosomes, and autophagosomes, and the nucleus, all of which are involved in the pathogenesis of MSA (Abati et al., 2018). Furthermore, α-syn toxicity has been shown to contribute to neuroinflammation, loss of neurotrophic support and neuronal dysfunction, resulting ultimately in neuronal death (Wong and Krainc, 2017). It is possible that miRNAs may regulate targeted genes and play a role in the pathogenesis of MSA. In this review, the role of MSA-related genes and the regulatory network between miRNAs and mRNAs will be discussed in the hope of providing new insight into the early diagnosis and therapeutic treatment of MSA.
Biology of miRNAs
It is known that miRNAs control the expression of more than 50% of protein-coding genes by acting as post-translational regulators (Krol et al., 2010). Disruption of miRNAs can cause mitochondrial dysfunction, oxidative stress, and cell death (Arshad et al., 2017; Treiber et al., 2019). More specifically, miRNAs also play a role in the proliferation of neural stem cells, the maturation of neurons, and the formation of synapses (Bian and Sun, 2011).
Gene-encoding miRNAs are transcribed by RNA polymerase II into primary miRNAs (pri-miRNAs). The pri-miRNAs then undergo cleavage by ribonuclease (RNase) III Drosha and cofactor protein DiGeorge Critical Region 8 (DGCR8) to form pre-miRNAs that are released from the nucleus to the cytoplasm. The pre-miRNAs in the cytoplasm are then sliced by RNase III protein Dicer and trans-activation-responsive RNA binding protein (TRBP) to form miRNA duplexes. The miRNA duplexes are incorporated into the RNA-induced silencing complex (RISC) mediated by the Argonaute (AGO) family. After unwinding and strand selection, the mature one-strand miRNA is capable of target recognition. The mature miRNAs then guide RISC to the complementary sequences for the 3′-untranslated region of the target mRNAs, resulting in the repression of mRNA-induced genes (Bartel, 2009; Huntzinger and Izaurralde, 2011; Figure 1).
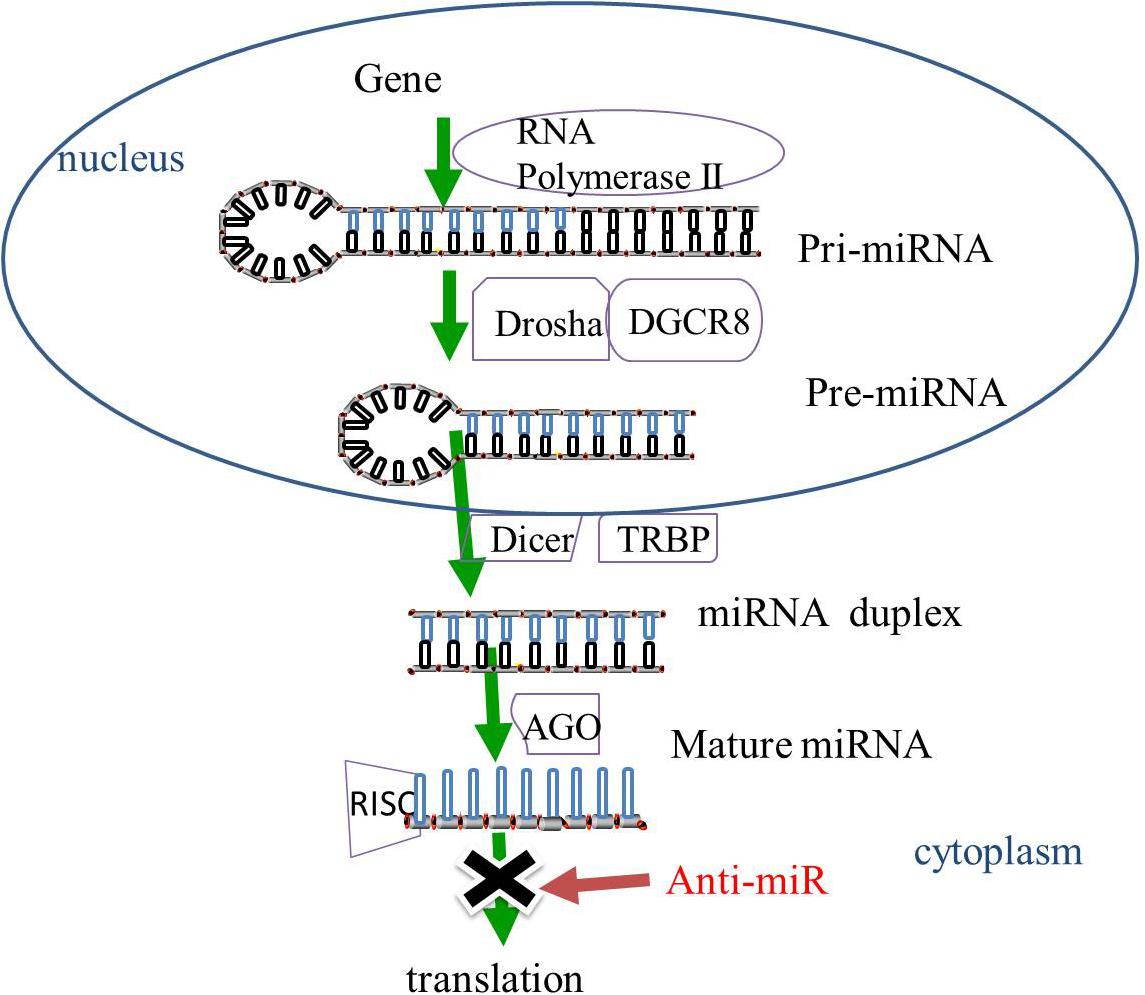
Figure 1. The biogenesis of miRNAs. In the nucleus, miRNAs are transcribed by RNA polymerase II into pri-miRNAs, and cleaved by RNase III Drosha and DGCR8 to pre-miRNAs. The pre-miRNAs are released from the nucleus to the cytoplasm and sliced by RNase III protein Dicer and TRBP to form miRNA duplexes. The miRNA duplexes are incorporated into the RISC, which is mediated by the AGO family, leading to the inhibition of gene expression. MiRNA-based treatment, including anti-miR can rescue the repression of miRNA-targeted genes. Pri-miRNA, primary miRNA; Pre-miRNA RNase, ribonuclease; DGCR8, cofactor DiGeorge Critical Region 8; TRBP, trans-activation-responsive RNA binding protein; RISC, RNA-induced silencing complex, AGO, Argonaute; anti-miR, anti-microRNA.
Mutation of the genes encoding the miRNAs biogenesis enzymes, Drosha, Dicer, and AGO has been reported to be linked to several types of cancer and neurodegenerative diseases (Tan et al., 2015). The absence of Dicer has been shown to cause a reduction in the expression of dopamine neurons and a stimulation of miRNAs, thereby promoting neuronal survival, resulting in implications for the treatment of PD (Chmielarz et al., 2017).
The trans-activating response region (TAR) DNA-binding protein-43 (TDP-43) facilitates the production of pre-miRNAs via interaction with the Drosha complex and has been shown to be important in the pathogenesis of ALS (Kawahara and Mieda-Sato, 2012). The colocalization of TDP-43 and α-syn in glial cytoplasmic inclusions (GCIs) has been reported in a small number of MSA patients (Geser et al., 2011; Koga et al., 2018).
Induction of mRNA degradation and inhibition of mRNA translation by miRNAs highlight the important roles of miRNAs in several biological processes. In addition, neurodegenerative disease-related protein has also been reported to affect miRNA expression (Kawahara and Mieda-Sato, 2012; Rinchetti et al., 2018).
The Role of miRNAs in MSA Pathogenesis
Several genes may be related to the pathogenesis of MSA; however, the exact mechanisms and the roles of these genes remain unknown (Mitsui et al., 2015; Sailer et al., 2016; Gu et al., 2018; Table 1). Altered expression of transcripts related to myelination and neuroinflammation has been observed in MSA striatum (Kim et al., 2019). In addition, a progressive decay of genes related to glutamate transport was reported (Kim et al., 2019). Therefore, miRNAs may regulate translation in neuronal processes and activation in synaptic transmission, thus taking part in various biological processes in neurodegenerative diseases, including MSA. The potential mechanisms underlying miRNA dysregulation will now be discussed, and the different roles of miRNAs in MSA pathogenesis will be further described in the hope of highlighting the development of potential biomarkers and therapeutic approaches (Figure 2 and Table 2).
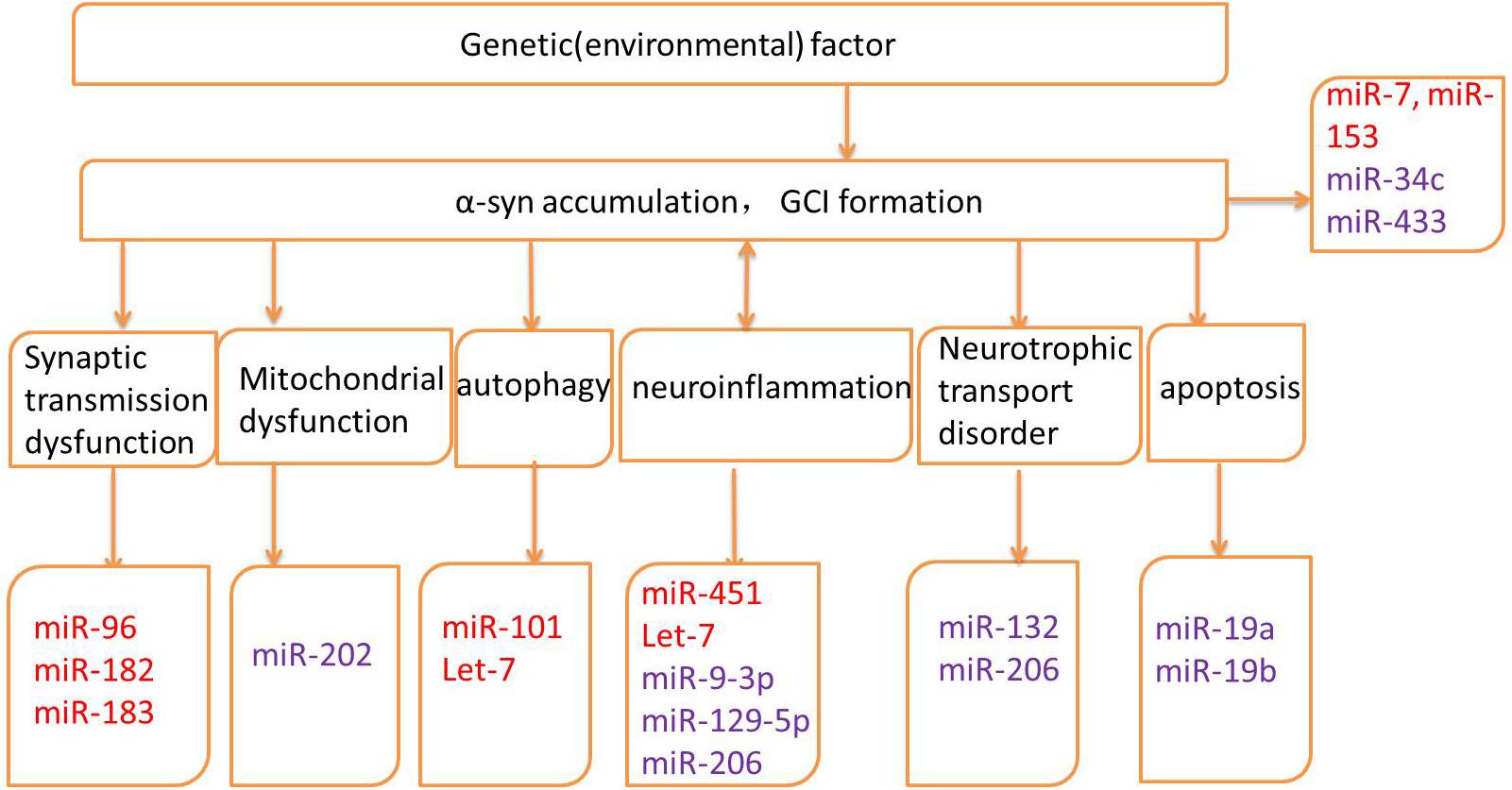
Figure 2. Pathogenic mechanisms and related microRNAs (miRNAs) of MSA. Red words indicate the up-regulated miRNAs and purple words indicate the down-regulated miRNAs.
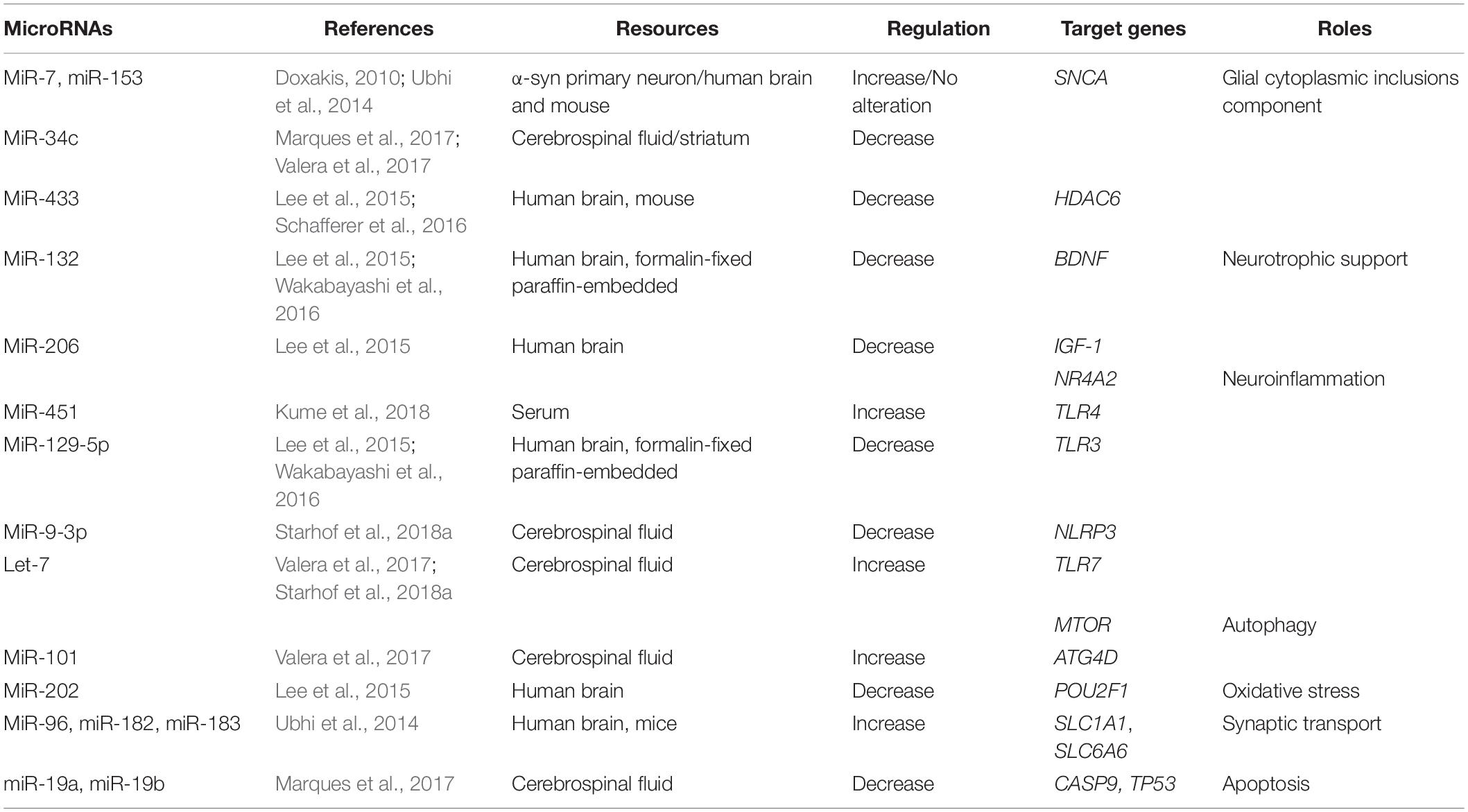
Table 2. Summary of altered microRNAs (miRNAs) and their targeted genes in Multiple system atrophy (MSA).
miRNAs and GCI Aggregation
The aggregation of α-syn promotes the relocalization of tubulin polymerization-promoting protein (TPPP/p25α) from myelin to oligodendroglia, resulting in oligodendrocyte swelling and abnormal uptake of α-syn by oligodendrocytes (Asi et al., 2014; Reyes et al., 2014). The interaction between α-syn and p25α also promotes the phosphorylation of α-syn and the formation of insoluble GCIs (Jellinger and Wenning, 2016). The formation of GCIs disrupts neuronal support, neuroinflammation, and neurotrophic support (Fanciulli and Wenning, 2015). It has been shown that miRNAs play important roles in the pathogenesis of MSA by regulating the expression of GCI components, α-syn, heat-shock protein (HSP), and p25α (Wong and Krainc, 2017). Both miR-7 and miR-153 have a similar expression and distribution pattern with α-syn in both neural and non-neural tissues. In addition, it has been shown that overexpression of miR-7 and miR-153 significantly reduces the level of α-syn in primary neurons (Doxakis, 2010). A study in MSA patients was unable to replicate these findings (Ubhi et al., 2014). Interestingly, it has been reported that miR-34c is reduced in the striatum of MSA-P patients (Valera et al., 2017) and in the CSF of MSA patients (Marques et al., 2017). Indeed, inhibition of miR-34c has been shown to result in the aggregation of α-syn in a cellular model (Kabaria et al., 2015).
Along with hyperphosphorylated α-syn, GCIs also contain ubiquitin, HSP, and p25α (Jellinger and Lantos, 2010). It has been suggested that down-regulation of miR-433 in the striatum observed in an MSA transgenic mouse model may be associated with the regulation of histone deacetylase 6 (HDAC6), a microtubule-associated deacetylase (Schafferer et al., 2016). These findings were further supported by a study showing colocalization of GCIs immunolabeled with anti-HDAC6 antibody in the striatum from MSA patients (Chiba et al., 2012). To conclude, miRNAs are possibly involved in the pathogenesis of MSA via the regulation of GCI components. It is therefore possible that miR-7, miR-153, and miR-34c may act as neuroprotective agents by regulating the SNCA; however, the precise roles of these miRs in MSA require further investigation.
miRNAs and Neuroinflammation
Neuroinflammation is a dynamic response including activation of microglia and astroglia, the expression of proinflammatory cytokines and chemokines (O’Callaghan et al., 2008). The role of neuroinflammation in the pathogenesis of MSA has been demonstrated in several studies (Vieira et al., 2015). Recent studies have suggested that microglial activation is essential to α-syn accumulation and promotes cell degeneration in neurodegenerative diseases (Sanchez-Guajardo et al., 2015). Indeed pro-inflammatory cytokines are elevated in serum, CSF, and brain tissue of MSA patients (Kaufman et al., 2013; Rydbirk et al., 2017; Starhof et al., 2018b).
Toll-like receptor (TLR) mediates the activation of innate immunity (Takeda and Akira, 2005). Furthermore, TLR4 mediates α-syn-induced microglial activation and pro-inflammatory cytokines. The mRNA expression of several TLRs is elevated in MSA, including TLR-3 and TLR-4 (Fellner et al., 2013).
Up-regulation of miRNAs can repress neuroinflammation and act as a positive feedback, reducing cell death in the pathogenesis of MSA. Overexpression of miR-451 has been shown to inhibit the release of cytokines through microglial activation, via the targeting of TLR4 (Sun and Zhang, 2018). Levels of miR-451 are elevated in brains of MSA patients (Kume et al., 2018).
Other miRNAs have also been shown to promote neuroinflammation and induce neurodegeneration. Up-regulation of Let-7 in MSA is associated with the activation of microglia and the induction of neurodegeneration by activating TLR7 (Lehmann et al., 2012). Elevated miR-129-5p can decrease cytokine activation and ameliorate inflammation-induced neuronal damage via TLR3 (Li et al., 2017). In addition, expression of miR-129-5p is decreased in MSA patients (Lee et al., 2015; Wakabayashi et al., 2016). In human astrocytes, overexpression of miR-206 increases pro-inflammatory cytokine expression (Duan et al., 2015), and miR-206 is down-regulated in the brains of MSA patients (Lee et al., 2015).
The nucleotide-binding domain leucine-rich repeats protein family (NLRP3) inflammasome is a multiprotein cytosolic complex that induces the release of cytokines (Duewell et al., 2010). The number of NLRP3 inflammasome-related proteins is increased in the brains of MSA patients postmortem. In a study by Li et al., NLRP3 inflammasome-related proteins played a role in astroglial activation and the release of interleukin-1 beta (IL-1β) (Li et al., 2018). It has also been shown that miR-9 can decrease NLRP3 expression, resulting in the suppression of pro-inflammatory cytokines (Wang et al., 2017). Levels of miR-9-3p are down-regulated in the CSF of MSA patients compared to PD patients, indicating that this miR may be used as a biomarker to discriminate between MSA and PD (Starhof et al., 2018a).
Release of α-syn by degenerating neurons may induce neuroinflammation, and simultaneously, neuroinflammation may trigger cytokine release, thus producing a pro-inflammatory environment, leading to the formation of intracellular α-syn aggregates (Vieira et al., 2015). Furthermore, the miRNA-based therapeutic strategies have been reported to be successful via regulating neuroinflammation in several neurodegenerative diseases, further highlighting a role of miRNAs in neuroinflammation in the treatment of MSA (Gaudet et al., 2018).
miRNAs and Autophagy
Disruption in the clearance of aggregated proteins is well known in the pathogenesis of neurodegenerative diseases (Scrivo et al., 2018). Selective autophagy may therefore act as a potential target for some neurodegenerative diseases. The inhibition of autophagy has been related to elevated secretion and transmission of α-syn (Lee et al., 2013). Autophagy is primarily controlled by autophagy-related proteins (ATG) and the mammalian/mechanistic target of rapamycin (mTOR) family (Sarkar, 2013). It has been shown that the mTOR pathway is involved in the pathogenesis of MSA and that mRNA levels of mTOR are decreased in the striatum of MSA patients (Valera et al., 2017). The conjugation of ATGs with microtubule-associated protein 1 light chain 3 (LC3) forms autophagosome, a double-membrane vesicle. Immunoreactivity of LC3 has been associated with α-syn-positive GCIs in neuropathological examination of MSA brains (Schwarz et al., 2012). The phosphorylation regulated by kinase complex beclin-1 has also been shown to be important in the formation of autophagosomes (Scrivo et al., 2018). Furthermore, expression of beclin-1 is decreased in MSA patients (Kaji et al., 2018; Miki et al., 2018).
It has been shown that miRNAs are associated with the pathogenesis of MSA by specifically targeting autophagy, including miR-101 and Let-7. In association with the increase of miR-101, levels of the autophagy markers beclin-1 and LC3 were decreased in the striatum of MSA-P patients. In an MSA mouse model, treatment with anti-miR-101 increased levels of LC3 and beclin-1, resulting in improved autophagy (Valera et al., 2017). Furthermore, treatment with the Let-7 family has been shown to inhibit the mTOR signaling pathway, in conjunction with a significant increase in the levels of Let-7b in the brains of MSA patients (Dubinsky et al., 2014; Valera et al., 2017).
Autophagy is involved in the process of neuroinflammation in neurodegenerative diseases (Su et al., 2016). Enhancing autophagy, as indicated by the up-regulation of beclin-1 and autophagy-related 5 (ATG5), facilitates the shift from deleterious microglial response M1 to neuroprotective microglial response M2 (Ji et al., 2018). Inhibition of the mTOR pathway has been shown to reduce neuronal death and microglial activation (Srivastava et al., 2016).
To conclude, levels of miR-101 and Let-7b are up-regulated through the ATG and the mTOR family in the pathogenesis of MSA. Results of a cellular model demonstrated the suppression of autophagy through regulation of miRNAs, indicating a role for anti-miR-101 in the treatment of MSA (Valera et al., 2017). The miRNAs, such as Let-7, show potential in the regulation of the pathogenesis of MSA via different biological processes.
miRNAs and Deregulation of Neurotrophic Factors
Neurotrophic factors, including glial-derived neurotrophic factor (GDNF), brain-derived neurotrophic factor (BDNF), and insulin-like growth factor (IGF) can mediate the development, survival, and maintenance of the peripheral system and the CNS (Bianchi et al., 2017; Sampaio et al., 2017). Neurotrophic factors are produced by oligodendrocytes (Ubhi et al., 2010) and the deregulation of these factors play important roles in the pathogenesis of MSA. Serum IGF-1 is significantly higher in MSA patients and is associated with disease progression (Numao et al., 2014). It has been reported that BDNF and other neurotrophic factors are elevated in MSA patients, while a specific reduction in the expression of GDNF was observed in α-syn transgenic mice and in MSA patients (Ubhi et al., 2010; Rydbirk et al., 2017). In conjunction, these findings suggest that increased expression of IGF-1 and BDNF may act as a compensatory response in MSA.
The expression of miR-132 was down-regulated in the brain of postmortem MSA patients (Lee et al., 2015; Wakabayashi et al., 2016). Up-regulation of miR-132 decreased the expression of BDNF, thereby inhibiting neuronal survival (Lungu et al., 2013). The relationship between the down-regulation of miR-206 in MSA patients and neuroinflammation has been previously reported (Lee et al., 2015). An additional study has shown that miR-206 is negatively associated with the IGF-1 signaling pathway, suggesting that IGF-1 may be another target gene for miR-206 in the pathogenesis of MSA (Xing et al., 2016; Liu et al., 2018).
miRNAs and Oxidative Stress
Oxidative stress can promote α-syn accumulation in oligodendrocytes (Pukass et al., 2015). Progressive microglial activation can cause chronic oxidative stress and ultimately lead to neuronal cell death in an MSA transgenic mouse model (Stefanova et al., 2007). The organic cation transporter proteins (OCT) are transcription factors that regulate gene levels by regulating reactive oxygen species (ROS). OCT is widely expressed by Purkinje cells in the cerebellum. Low levels of OCT1 in the MSA cerebellum has been shown to result in reduced resistance of neurons to oxidative stress (Lee et al., 2015). Up-regulation of miR-202 has been shown to down-regulate the expression of OCT1, thus contributing to cerebellar degeneration in the cerebellum of MSA patients (Lee et al., 2015).
miRNAs and Synaptic Transmission Dysfunction
Elevated α-syn expression has been shown to be increased in the presynaptic terminal, thus disrupting dopamine and γ-aminobutyric acid (GABA) release (Guatteo et al., 2017). Neurotransmitters at synapses are transported to presynaptic terminals through several transporters, including SLC1 and SLC6 (Lin et al., 2015). The SLC1A1 and SLC6A6 genes, which encode for the neuronal/epithelial high-affinity glutamate transporter (EAAT3/EAAC1) and taurine transporter, respectively, are down-regulated by the miR-96 cluster (miR-96, miR-182, miR-183) in MSA brain tissues and MSA mouse models, indicating a role for miRNAs in neurotransmitter release (Ubhi et al., 2014).
miRNAs and Apoptosis
The accumulation of apoptosome-related protein can occur in the neuronal and oligodendroglial elements of the MSA brain. Activated caspase-9 and -8, and increased expression of p53, have been observed in MSA brain tissue (Kawamoto et al., 2016). A reduction of miR-19a and miR-19b has been reported to result in a significant increase in apoptosis in SH-SY5Y cells and increase the level of p53 protein in MSA brain tissue (Zhu et al., 2016). In addition, levels of both miR-19a and miR-19b have been found to be lower in the CSF of MSA patients in the early stage of the disease (Marques et al., 2017). A recent study also found that miR-19b was decreased in the plasma of MSA patients compared to controls and PD patients. Interestingly, levels of miR-24 were also decreased in the plasma of MSA patients, with a strong correlation between miR-24 and miR-19b levels, suggesting that both miR-24 and miR-19b play a role in the regulation of apoptosis and the pathogenesis of MSA (Uwatoko et al., 2019).
miRNAs as Biomarkers of MSA
Several parameters, including α-syn, total tau, phosphorylated tau, the 42-amino-acid form of Aβ, neurofilament light chain protein, fms-like tyrosine kinase ligand, homocysteine, uric acid, and coenzyme Q10, have previously been identified as potential biomarkers for the diagnosis of patients with MSA (Laurens et al., 2015). However, these results are inconsistent and several biomarkers are unable to differentiate among neurodegenerative diseases characterized by the abnormal accumulation of α-syn.
Recently, miRNAs have been recognized as potential biomarkers for MSA due to their small size, stability, accuracy, and tissue-specific nature (Mitchell et al., 2008). Several studies have shown that miR-9-3p, miR-19a, miR-19b, and miR-24 are potential biomarkers that can be used to distinguish patients with MSA from those with PD and healthy people (Marques et al., 2017; Starhof et al., 2018a; Uwatoko et al., 2019). However, the expression of miRNAs is differentially expressed in the CSF and peripheral blood of patients with MSA (Vallelunga et al., 2014; Marques et al., 2017). The inconsistency of results may be due to the small sample sizes of the studies. Further clinical investigations with a large sample size, along with functional studies, are needed to confirm the role of miRNAs in the pathogenesis of MSA.
Interestingly, miRNAs can also be used as prognostic factors for nervous system diseases. For example, similar to the results reported using samples from the CSF of patients with MSA, miR-19a and miR-19b were also down-regulated in the prodromal stage of patients with synucleinopathies (Fernández-Santiago et al., 2015). In contrast, the up-regulation of both miR-19a and miR-19b has been reported in MSA patients at postmortem (Ubhi et al., 2014). Together, these findings show that the expression of miRNAs is not consistent throughout the course of the disease, indicating that miRNAs may be useful in determining disease stage.
miRNA-Based Therapeutic Approaches for MSA
Based on current studies, blocking α-syn aggregation (rifampicin), enhancing neuroprotection (riluzole), and reducing neuroinflammation (minocycline) were expected to have positive effects on the treatment of MSA (Bensimon et al., 2009; Dodel et al., 2010; Low et al., 2014). Unfortunately, most of these treatments failed in clinical trials and more effective therapies are therefore urgently needed.
RNA interference-based clinical trials by miRNA mimics for various diseases are progressing well (Bobbin and Rossi, 2016). Reducing gene expression by miRNA has also been shown to be effective in neurodegenerative diseases (Keiser et al., 2016). For example, anti–miR-101 improved autophagy and reduced α-syn accumulation in an MSA mouse model (Valera et al., 2017). MiR-7 reduced α-syn accumulation and had a neuroprotective effect in both cellular and mouse models, which demonstrate that miR-7 may be helpful in the treatment of diseases characterized by α-syn accumulation (Junn et al., 2009; Titze-de-Almeida and Titze-de-Almeida, 2018). However, an additional challenge exists for miRNA mimics in the treatment of neurodegenerative diseases due to the blood–brain barrier. Virus vector-mediated delivery and specifically designed polymeric nanoparticles may be useful; however, further studies are needed (Saraiva et al., 2016; Nakamori et al., 2019).
Conclusion
Several studies have shown that miRNAs have the potential to be considered as biomarkers and prognostic factors in MSA. MiR-9-3p, miR-19a, miR-19b, and miR-24 may be regarded as potential biomarkers to distinguish between MSA and PD patients, and healthy people, while the expression of miR-19a and miR-19b may be related to the development of the disease. Furthermore, miRNA mimics and anti-miR could reduce α-syn expression and play a beneficial role in both cellular and mouse models. Thus, the development of miRNA-based treatment may provide a new therapeutic model for disease modification in MSA. In addition, the role of miRNAs in MSA remains to be further elucidated by large-scale comparative studies.
Author Contributions
CX and SC contributed to the drafting and revising the manuscript. SH contributed to drafting and modifying the table. JN contributed to drafting and modifying the figure. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81371271), and was also sponsored by the “Liaoning BaiQianWan Talents Program.”
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abati, E., Di Fonzo, A., and Corti, S. (2018). In vitro models of multiple system atrophy from primary cells to induced pluripotent stem cells. J. Cell. Mol. Med. 22, 2536–2546. doi: 10.1111/jcmm.13563
Al-Chalabi, A., Dürr, A., Wood, N. W., Parkinson, M. H., Camuzat, A., Hulot, J. S., et al. (2009). Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS One 4:e7114. doi: 10.1371/journal.pone.0007114
Arshad, A. R., Sulaiman, S. A., Saperi, A. A., Jamal, R., Mohamed Ibrahimm, N., and Abdul Murad, N. A. (2017). MicroRNAs and target genes as biomarkers for the diagnosis of early onset of Parkinson disease. Front. Mol. Neurosci. 10:352. doi: 10.3389/fnmol.2017.00352
Asi, Y. T., Simpson, J. E., Heath, P. R., Wharton, S. B., Lees, A. J., Revesz, T., et al. (2014). Alpha-synuclein mRNA expression in oligodendrocytes in MSA. Glia 62, 964–970. doi: 10.1002/glia.22653
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Becker, L. A., Huang, B., Bieri, G., Ma, R., Knowles, D. A., Jafar-Nejad, P., et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. doi: 10.1038/nature22038
Bensimon, G., Ludolph, A., Agid, Y., Vidailhet, M., Payan, C., Leigh, P. N., et al. (2009). Riluzole treatment, survival and diagnostic criteria in Parkinson plus disorders: the NNIPPS study. Brain 132, 156–171. doi: 10.1093/brain/awn291
Bian, S., and Sun, T. (2011). Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 44, 359–373. doi: 10.1007/s12035-011-8211-3
Bianchi, V. E., Locatelli, V., and Rizzi, L. (2017). Neurotrophic and neuroregenerative effects of GH/IGF1. Int. J. Mol. Sci. 11:E2441. doi: 10.3390/ijms18112441
Bobbin, M. L., and Rossi, J. J. (2016). RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu. Rev. Pharmacol. Toxicol. 56, 103–122. doi: 10.1146/annurev-pharmtox-010715-103633
Chen, Y., Cao, B., Ou, R., Wei, Q., Chen, X., Zhao, B., et al. (2018). Determining the effect of the HNMT, STK39, and NMD3 polymorphisms on the incidence of Parkinson’s disease, amyotrophic lateral sclerosis, and multiple system atrophy in Chinese populations. J. Mol. Neurosci. 4, 574–580. doi: 10.1007/s12031-018-1048-8
Chiba, Y., Takei, S., Kawamura, N., Kawaguchi, Y., Sasaki, K., Hasegawa-Ishii, S., et al. (2012). Immunohistochemical localization of aggresomal proteins in glial cytoplasmic inclusions in multiple system atrophy. Neuropathol. Appl. Neurobiol. 38, 559–571. doi: 10.1111/j.1365-2990.2011.01229.x
Chmielarz, P., Konovalova, J., Najam, S. S., Alter, H., Piepponen, T. P., Erfle, H., et al. (2017). Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 8:e2813. doi: 10.1038/cddis.2017.214
Dodel, R., Spottke, A., Gerhard, A., Reuss, A., Reinecker, S., Schimke, N., et al. (2010). Minocycline 1-year therapy in multiple system atrophy: effect on clinical symptoms and [(11)C] (R)-PK11195 PET (MEMSA-trial). Mov. Disord. 1, 97–107. doi: 10.1002/mds.22732
Doxakis, E. (2010). Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 285, 12726–12734. doi: 10.1074/jbc.M109.086827
Duan, X., Zohaib, A., Li, Y., Zhu, B., Ye, J., Wan, S., et al. (2015). miR-206 modulates lipopolysaccharide-mediated inflammatory cytokine production in human astrocytes. Cell. Signal. 27, 61–68. doi: 10.1016/j.cellsig.2014.10.006
Dubinsky, A. N., Dastidar, S. G., Hsu, C. L., Zahra, R., Djakovic, S. N., Duarte, S., et al. (2014). Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell. Metab. 20, 626–638. doi: 10.1016/j.cmet.2014.09.001
Duewell, P., Kono, H., Rayner, K. J., Sirois, C. M., Vladimer, G., Bauernfeind, F. G., et al. (2010). NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361. doi: 10.1038/nature08938
Fellner, L., Irschick, R., Schanda, K., Reindl, M., Klimaschewski, L., Poewe, W., et al. (2013). Toll-like receptor 4 is required for alpha-synuclein dependent activation of microglia and astroglia. Glia 61, 349–360. doi: 10.1002/glia.22437
Ferguson, M. C., Garland, E. M., Hedges, L., Womack-Nunley, B., Hamid, R., and Phillips, J. A. III. (2014). SHC2 gene copy number in multiple system atrophy (MSA). Clin. Auton. Res. 24, 25–30. doi: 10.1007/s10286-013-0216-8
Fernández-Santiago, R., Iranzo, A., Gaig, C., Serradell, M., Fernández, M., Tolosa, E., et al. (2015). MicroRNA association with synucleinopathy conversion in rapid eye movement behavior disorder. Ann. Neurol. 5, 895–901. doi: 10.1002/ana.24384
Gaudet, A. D., Fonken, L. K., Watkins, L. R., Nelson, R. J., and Popovich, P. G. (2018). MicroRNAs: roles in regulating neuroinflammation. Neuroscientist 3, 221–245. doi: 10.1177/1073858417721150
Geser, F., Malunda, J. A., Hurtig, H. I., Duda, J. E., Wenning, G. K., Gilman, S., et al. (2011). TDP-43 pathology occurs infrequently in multiple system atrophy. Neuropathol. Appl. Neurobiol. 37, 358–365. doi: 10.1111/j.1365-2990.2010.01136.x
Gilman, S., Wenning, G. K., Low, P. A., Brooks, D. J., Mathias, C. J., Trojanowski, J. Q., et al. (2008). Second consensus statement on the diagnosis of multiple system atrophy. Neurology 71, 670–676. doi: 10.1212/01.wnl.0000324625.00404.15
Gu, X., Chen, Y., Zhou, Q., Lu, Y. C., Cao, B., Zhang, L., et al. (2018). Analysis of GWAS-linked variants in multiple system atrophy. Neurobiol. Aging 67, 201.e1–201.e4. doi: 10.1016/j.neurobiolaging.2018.03.018
Guatteo, E., Rizzo, F. R., Federici, M., Cordella, A., Ledonne, A., Latini, L., et al. (2017). Functional alterations of the dopaminergic and glutamatergic systems in spontaneous alpha-synuclein overexpressing rats. Exp. Neurol. 287, 21–33. doi: 10.1016/j.expneurol.2016.10.009
Guo, X. Y., Chen, Y. P., Song, W., Zhao, B., Cao, B., Wei, Q. Q., et al. (2014). SNCA variants rs2736990 and rs356220 as risk factors for Parkinson’s disease but not for amyotrophic lateral sclerosis and multiple system atrophy in a Chinese population. Neurobiol. Aging 35, 2882.e1–2882.e6. doi: 10.1016/j.neurobiolaging.2014.07.014
Hama, Y., Katsu, M., Takigawa, I., Yabe, I., Matsushima, M., Takahashi, I., et al. (2017). Genomic copy number variation analysis in multiple system atrophy. Mol. Brain 1:54. doi: 10.1186/s13041-017-0335-6
Hinrich, A. J., Jodelka, F. M., Chang, J. L., Brutman, D., Bruno, A. M., Briggs, C. A., et al. (2016). Therapeutic correction of ApoER2 splicing in Alzheimer’s disease mice using antisense oligonucleotides. EMBO Mol. Med. 8, 328–345. doi: 10.15252/emmm.201505846
Huntzinger, E., and Izaurralde, E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. doi: 10.1038/nrg2936
Jellinger, K. A. (2014). Neuropathology of multiple system atrophy: new thoughts about pathogenesis. Mov. Disord. 14, 1720–1741. doi: 10.1002/mds.26052
Jellinger, K. A., and Lantos, P. L. (2010). Papp-Lantos inclusions and the pathogenesis of multiple system atrophy: an update. Acta Neuropathol. 119, 657–667. doi: 10.1007/s00401-010-0672-3
Jellinger, K. A., and Wenning, G. K. (2016). Multiple system atrophy: pathogenic mechanisms and biomarkers. J. Neural. Transm. 123, 555–572. doi: 10.1007/s00702-016-1545-2
Ji, J., Xue, T. F., Guo, X. D., Yang, J., Guo, R. B., Wang, J., et al. (2018). Antagonizing peroxisome proliferator-activated receptor gamma facilitates M1-to-M2 shift of microglia by enhancing autophagy via the LKB1-AMPK signaling pathway. Aging Cell. 17:e12774. doi: 10.1111/acel.12774
Junn, E., Lee, K. W., Jeong, B. S., Chan, T. W., Im, J. Y., and Mouradian, M. M. (2009). Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. U.S.A. 31, 13052–13057. doi: 10.1073/pnas.0906277106
Kabaria, S., Choi, D. C., Chaudhuri, A. D., Mouradian, M. M., and Junn, E. (2015). Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 589, 319–325. doi: 10.1016/j.febslet.2014.12.014
Kaji, S., Maki, T., Kinoshita, H., Uemura, N., Ayaki, T., Kawamoto, Y., et al. (2018). Pathological endogenous alpha-synuclein accumulation in oligodendrocyte precursor cells potentially induces inclusions in multiple system atrophy. Stem Cell Rep. 10, 356–365. doi: 10.1016/j.stemcr.2017.12.001
Kaufman, E., Hall, S., Surova, Y., Widner, H., Hansson, O., and Lindqvist, D. (2013). Proinflammatory cytokines are elevated in serum of patients with multiple system atrophy. PLoS One 8:e62354. doi: 10.1371/journal.pone.0062354
Kawahara, Y., and Mieda-Sato, A. (2012). TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 3347–3352. doi: 10.1073/pnas.1112427109
Kawamoto, Y., Ayaki, T., Urushitani, M., Ito, H., and Takahashi, R. (2016). Activated caspase-9 immunoreactivity in glial and neuronal cytoplasmic inclusions in multiple system atrophy. Neurosci. Lett. 628, 207–212. doi: 10.1016/j.neulet.2016.06.036
Keiser, M. S., Kordasiewicz, H. B., and McBride, J. L. (2016). Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia. Hum. Mol. Genet. R1, R53–R64. doi: 10.1093/hmg/ddv442
Kim, T., Valera, E., and Desplats, P. (2019). Alterations in striatal microRNA-mRNA networks contribute to neuroinflammation in multiple system atrophy. Mol. Neurobiol. 56, 7003–7021. doi: 10.1007/s12035-019-1577-3
Koga, S., Lin, W. L., Walton, R. L., Ross, O. A., and Dickson, D. W. (2018). TDP-43 pathology in multiple system atrophy: colocalization of TDP-43 and alpha-synuclein in glial cytoplasmic inclusions. Neuropathol. Appl. Neurobiol. 44, 707–721. doi: 10.1111/nan.12485
Kordasiewicz, H. B., Stanek, L. M., Wancewicz, E. V., Mazur, C., McAlonis, M. M., Pytel, K. A., et al. (2012). Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044. doi: 10.1016/j.neuron.2012.05.009
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 9, 597–610. doi: 10.1038/nrg2843
Kume, K., Iwama, H., Deguch, K., Ikeda, K., Takata, T., Kokudo, Y., et al. (2018). Serum microRNA expression profiling in patients with multiple system atrophy. Mol. Med. Rep. 17, 852–860. doi: 10.3892/mmr.2017.7995
Labbe, C., Heckman, M. G., Lorenzo-Betancor, O., Murray, M. E., Ogaki, K., Soto-Ortolaza, A. I., et al. (2016). MAPT haplotype diversity in multiple system atrophy. Parkinsonism Relat. Disord. 30, 40–45. doi: 10.1016/j.parkreldis.2016.06.010
Laurens, B., Constantinescu, R., Freeman, R., Gerhard, A., Jellinger, K., Jeromin, A., et al. (2015). Fluid biomarkers in multiple system atrophy: a review of the MSA Biomarker Initiative. Neurobiol. Dis. 80, 29–41. doi: 10.1016/j.nbd.2015.05.004
Lee, H. J., Cho, E. D., Lee, K. W., Kim, J. H., Cho, S. G., and Lee, S. J. (2013). Autophagic failure promotes the exocytosis and intercellular transfer of alpha-synuclein. Exp. Mol. Med. 45:e22. doi: 10.1038/emm.2013.45
Lee, S. T., Chu, K., Jung, K. H., Ban, J. J., Im, W. S., Jo, H. Y., et al. (2015). Altered expression of miR-202 in cerebellum of multiple-system atrophy. Mol. Neurobiol. 51, 180–186. doi: 10.1007/s12035-014-8788-4
Lehmann, S. M., Krüger, C., Park, B., Derkow, K., Rosenberger, K., Baumgart, J., et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835. doi: 10.1038/nn.3113
Li, F., Ayaki, T., Maki, T., Sawamot, N., and Takahashi, R. (2018). NLRP3 inflammasome-related proteins are upregulated in the putamen of patients with multiple system atrophy. J. Neuropathol. Exp. Neurol. 77, 1055–1065. doi: 10.1093/jnen/nly090
Li, X. Q., Chen, F. S., Tan, W. F., Fang, B., Zhang, Z. L., and Ma, H. (2017). Elevated microRNA-129-5p level ameliorates neuroinflammation and blood-spinal cord barrier damage after ischemia-reperfusion by inhibiting HMGB1 and the TLR3-cytokine pathway. J. Neuroinflammation 14:205. doi: 10.1186/s12974-017-0977-4
Lin, L., Yee, S. W., Kim, R. B., and Giacomini, K. M. (2015). SLC transporters as therapeutic targets: emerging opportunities. Nat. Rev. Drug Discov. 14, 543–560. doi: 10.1038/nrd4626
Liu, T. J., Wang, B., Li, Q. X., Dong, X. L., Han, X. L., and Zhang, S. B. (2018). Effects of microRNA-206 and its target gene IGF-1 on sevoflurane-induced activation of hippocampal astrocytes in aged rats through the PI3K/AKT/CREB signaling pathway. J. Cell Physiol. 233, 4294–4306. doi: 10.1002/jcp.26248
Low, P. A., Robertson, D., Gilman, S., Kaufmann, H., Singer, W., Biaggioni, I., et al. (2014). Efficacy and safety of rifampicin for multiple system atrophy: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 3, 268–275. doi: 10.1016/S1474-4422(13)70301-6
Lungu, G., Stoica, G., and Ambrus, A. (2013). MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci. Lett. 553, 153–158. doi: 10.1016/j.neulet.2013.08.001
Marques, T. M., Kuiperij, H. B., Bruinsma, I. B., van Rumund, A., Aerts, M. B., Esselink, R. A. J., et al. (2017). MicroRNAs in cerebrospinal fluid as potential biomarkers for Parkinson’s disease and multiple system atrophy. Mol. Neurobiol. 54, 7736–7745. doi: 10.1007/s12035-016-0253-0
Miki, Y., Tanji, K., Mori, F., Tatara, Y., Utsumi, J., Sasaki, H., et al. (2018). AMBRA1, a novel alpha-synuclein-binding protein, is implicated in the pathogenesis of multiple system atrophy. Brain Pathol. 28, 28–42. doi: 10.1111/bpa.12461
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 30, 10513–10518. doi: 10.1073/pnas.0804549105
Mitsui, J., Matsukawa, T., Sasaki, H., Yabe, I., Matsushima, M., Dürr, A., et al. (2015). Variants associated with Gaucher disease in multiple system atrophy. Ann. Clin. Transl. Neurol. 2, 417–426. doi: 10.1002/acn3.185
Multiple-System Atrophy Research Collaboration, (2013). Mutations in COQ2 in familial and sporadic multiple-system atrophy. N. Engl. J. Med. 369, 233–244. doi: 10.1056/NEJMoa1212115
Nakamori, M., Junn, E., Mochizuki, H., and Mouradian, M. M. (2019). Nucleic acid-based therapeutics for Parkinson’s disease. Neurotherapeutics 2, 287–298. doi: 10.1007/s13311-019-00714-7
Nuber, S., Rajsombath, M., Minakaki, G., Winkler, J., Müller, C. P., Ericsson, M., et al. (2018). Abrogating native alpha-synuclein tetramers in mice causes a L-DOPA-Responsive motor syndrome closely resembling Parkinson’s disease. Neuron 100, 75.e5–90.e5. doi: 10.1016/j.neuron.2018.09.014
Numao, A., Suzuki, K., Miyamoto, M., Miyamoto, T., and Hirata, K. (2014). Clinical correlates of serum insulin-like growth factor-1 in patients with Parkinson’s disease, multiple system atrophy and progressive supranuclear palsy. Parkinsonism Relat. Disord. 20, 212–216. doi: 10.1016/j.parkreldis.2013.11.005
O’Callaghan, J. P., Sriram, K., and Miller, D. B. (2008). Defining “neuroinflammation”. Ann. N. Y. Acad. Sci. 1139, 318–330. doi: 10.1196/annals.1432.032
Palma, J. A., and Kaufmann, H. (2018). Treatment of autonomic dysfunction in Parkinson disease and other synucleinopathies. Mov. Disord. 33, 372–390. doi: 10.1002/mds.27344
Prusiner, S. B., Woerman, A. L., Mordes, D. A., Watts, J. C., Rampersaud, R., Berry, D. B., et al. (2015). Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 38, E5308–E5317. doi: 10.1073/pnas.1514475112
Pukass, K., Goldbaum, O., and Richter-Landsberg, C. (2015). Mitochondrial impairment and oxidative stress compromise autophagosomal degradation of alpha-synuclein in oligodendroglial cells. J. Neurochem. 135, 194–205. doi: 10.1111/jnc.13256
Ramaswamy, P., Christopher, R., Pal, P. K., and Yadav, R. (2018). MicroRNAs to differentiate Parkinsonian disorders: advances in biomarkers and therapeutics. J. Neurol. Sci. 394, 26–37. doi: 10.1016/j.jns.2018.08.032
Reyes, J. F., Rey, N. L., Bousset, L., Melki, R., Brundin, P., and Angot, E. (2014). Alpha-synuclein transfers from neurons to oligodendrocytes. Glia 3, 387–398. doi: 10.1002/glia.22611
Rinchetti, P., Rizzuti, M., Faravelli, I., and Corti, S. (2018). MicroRNA metabolism and dysregulation in amyotrophic lateral sclerosis. Mol. Neurobiol. 3, 2617–2630. doi: 10.1007/s12035-017-0537-z
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222. doi: 10.1038/nrd.2016.246
Rydbirk, R., Elfving, B., Andersen, M. D., Langbøl, M. A., Folke, J., Winge, K., et al. (2017). Cytokine profiling in the prefrontal cortex of Parkinson’s disease and multiple system atrophy patients. Neurobiol. Dis. 106, 269–278. doi: 10.1016/j.nbd.2017.07.014
Sailer, A., Scholz, S. W., Nalls, M. A., Schulte, C., Federoff, M., Price, T. R., et al. (2016). A genome-wide association study in multiple system atrophy. Neurology 87, 1591–1598. doi: 10.1212/WNL.0000000000003221
Sampaio, T. B., Savall, A. S., Gutierrez, M. E. Z., and Pinton, S. (2017). Neurotrophic factors in Alzheimer’s and Parkinson’s diseases: implications for pathogenesis and therapy. Neural Regen. Res. 4, 549–557. doi: 10.4103/1673-5374.205084
Sanchez-Guajardo, V., Tentillier, N., and Romero-Ramos, M. (2015). The relation between alpha-synuclein and microglia in Parkinson’s disease: recent developments. Neuroscience 302, 47–58. doi: 10.1016/j.neuroscience.2015.02.008
Saraiva, C., Paiva, J., Santos, T., Ferreira, L., and Bernardino, L. (2016). MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 235, 291–305. doi: 10.1016/j.jconrel.2016.06.005
Sarkar, S. (2013). Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 41, 1103–1130. doi: 10.1042/BST20130134
Sasaki, H., Emi, M., Iijima, H., Ito, N., Sato, H., Yabe, I., et al. (2011). Copy number loss of (src homology 2 domain containing)-transforming protein 2 (SHC2) gene: discordant loss in monozygotic twins and frequent loss in patients with multiple system atrophy. Mol. Brain 10, 4–24. doi: 10.1186/1756-6606-4-24
Schafferer, S., Khurana, R., Refolo, V., Venezia, S., Sturm, E., Piatti, P., et al. (2016). Changes in the miRNA-mRNA regulatory network precede motor symptoms in a mouse model of multiple system atrophy: clinical implications. PLoS One 11:e0150705. doi: 10.1371/journal.pone.0150705
Scholz, S. W., Houlden, H., Schulte, C., Sharma, M., Li, A., Berg, D., et al. (2009). SNCA variants are associated with increased risk for multiple system atrophy. Ann. Neurol. 65, 610–614. doi: 10.1002/ana.21685
Schwarz, L., Goldbaum, O., Bergmann, M., Probst-Cousin, S., and Richter-Landsberg, C. (2012). Involvement of macroautophagy in multiple system atrophy and protein aggregate formation in oligodendrocytes. J. Mol. Neurosci. 47, 256–266. doi: 10.1007/s12031-012-9733-5
Scrivo, A., Bourdenx, M., Pampliega, O., and Cuervo, A. M. (2018). Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 17, 802–815. doi: 10.1016/S1474-4422(18)30238-2
Soma, H., Yabe, I., Takei, A., Fujiki, N., Yanagihara, T., and Sasaki, H. (2008). Associations between multiple system atrophy and polymorphisms ofSLC1A4,SQSTM1, andEIF4EBP1Genes. Mov. Disord. 23, 1161–1167. doi: 10.1002/mds.22046
Srivastava, I. N., Shperdheja, J., Baybis, M., Ferguson, T., and Crino, P. B. (2016). mTOR pathway inhibition prevents neuroinflammation and neuronal death in a mouse model of cerebral palsy. Neurobiol. Dis. 85, 144–154. doi: 10.1016/j.nbd.2015.10.001
Starhof, C., Hejl, A. M., Heegaard, N. H. H., Carlsen, A. L., Burton, M., Lilje, B., et al. (2018a). The biomarker potential of cell-free microRNA from cerebrospinal fluid in Parkinsonian syndromes. Mov. Disord. 2, 246–254. doi: 10.1002/mds.27542
Starhof, C., Winge, K., Heegaard, N. H. H., Skogstrand, K., Friis, S., and Hejl, A. (2018b). Cerebrospinal fluid pro-inflammatory cytokines differentiate parkinsonian syndromes. J. Neuroinflammation 15:305. doi: 10.1186/s12974-018-1339-6
Stefanova, N., Bücke, P., Duerr, S., and Wenning, G. K. (2009). Multiple system atrophy: an update. Lancet Neurol. 8, 1172–1178. doi: 10.1016/S1474-4422(09)70288-1
Stefanova, N., Reindl, M., Neumann, M., Kahle, P. J., Poewe, W., and Wenning, G. K. (2007). Microglial activation mediates neurodegeneration related to oligodendroglial alpha-synucleinopathy: implications for multiple system atrophy. Mov. Disord. 15, 2196–2203. doi: 10.1002/mds.21671
Su, P., Zhang, J., Wang, D., Zhao, F., Cao, Z., Aschner, M., et al. (2016). The role of autophagy in modulation of neuroinflammation in microglia. Neuroscience 319, 155–167. doi: 10.1016/j.neuroscience.2016.01.035
Sun, X., and Zhang, H. (2018). miR-451 elevation relieves inflammatory pain by suppressing microglial activation-evoked inflammatory response via targeting TLR4. Cell Tissue Res. 3, 487–495. doi: 10.1007/s00441-018-2898-7
Tagliafierro, L., and Chiba-Falek, O. (2016). Up-regulation of SNCA gene expression: implications to synucleinopathies. Neurogenetics 17, 145–157. doi: 10.1007/s10048-016-0478-0
Takeda, K., and Akira, S. (2005). Toll-like receptors in innate immunity. Int. Immunol. 17, 1–14. doi: 10.1093/intimm/dxh186
Tan, L., Yu, J. T., and Tan, L. (2015). Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol. Neurobiol. 51, 1249–1262. doi: 10.1007/s12035-014-8803-9
Titze-de-Almeida, R., and Titze-de-Almeida, S. S. (2018). MiR-7 replacement therapy in Parkinson’s disease. Curr. Gene Ther. 3, 143–153. doi: 10.2174/1566523218666180430121323
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 1, 5–20. doi: 10.1038/s41580-018-0059-1
Ubhi, K., Rockenstein, E., Kragh, C., Inglis, C., Spencer, B., Michael, S., et al. (2014). Widespread microRNA dysregulation in multiple system atrophy - disease-related alteration in miR-96. Eur. J. Neurosci. 39, 1026–1041. doi: 10.1111/ejn.12444
Ubhi, K., Rockenstein, E., Mante, M., Inglis, C., Adame, A., Patrick, C., et al. (2010). Neurodegeneration in a transgenic mouse model of multiple system atrophy is associated with altered expression of oligodendroglial-derived neurotrophic factors. J. Neurosci. 30, 6236–6246. doi: 10.1523/JNEUROSCI.0567-10.2010
Uwatoko, H., Hama, Y., Iwata, I. T., Shirai, S., Matsushima, M., Yabe, I., et al. (2019). Identification of plasma microRNA expression changes in multiple system atrophy and Parkinson’s disease. Mol. Brain. 1:49. doi: 10.1186/s13041-019-0471-2
Valera, E., Spencer, B., Mott, J., Trejo, M., Adame, A., Mante, M., et al. (2017). MicroRNA-101 modulates autophagy and oligodendroglial alpha-synuclein accumulation in multiple system atrophy. Front. Mol. Neurosci. 10:329. doi: 10.3389/fnmol.2017.00329
Vallelunga, A., Ragusa, M., Di Mauro, S., Iannitti, T., Pilleri, M., Biundo, R., et al. (2014). Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front. Cell Neurosci. 8:156. doi: 10.3389/fncel.2014.00156
Vieira, B. D., Radford, R. A., Chung, R. S., Guillemin, G. J., and Pountney, D. L. (2015). Neuroinflammation in multiple system atrophy: response to and cause of alpha-synuclein aggregation. Front. Cell Neurosci. 9:437. doi: 10.3389/fncel.2015.00437
Vilarino-Guell, C., Soto-Ortolaza, A. I., Rajput, A., Mash, D. C., Papapetropoulos, S., Pahwa, R., et al. (2011). MAPT H1 haplotype is a risk factor for essential tremor and multiple system atrophy. Neurology 76, 670–672. doi: 10.1212/WNL.0b013e31820c30c1
Wakabayashi, K., Mori, F., Kakita, A., Takahashi, H., Tanaka, S., Utsumi, J., et al. (2016). MicroRNA expression profiles of multiple system atrophy from formalin-fixed paraffin-embedded samples. Neurosci. Lett. 635, 117–122. doi: 10.1016/j.neulet.2016.10.034
Wang, Y., Han, Z., Fan, Y., Zhang, J., Chen, K., Gao, L., et al. (2017). MicroRNA-9 Inhibits NLRP3 inflammasome activation in human atherosclerosis inflammation cell models through the JAK1/STAT signaling pathway. Cell. Physiol. Biochem. 41, 1555–1571. doi: 10.1159/000470822
Wong, Y. C., and Krainc, D. (2017). alpha-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat. Med. 23, 1–13. doi: 10.1038/nm.4269
Xing, H., Guo, S., Zhang, Y., Zheng, Z., and Wang, H. (2016). Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-beta by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 14, 1357–1364. doi: 10.3892/mmr.2016.5369
Xiong, X. P., Vogler, G., Kurthkoti, K., Samsonova, A., and Zhou, R. (2015). SmD1 modulates the miRNA pathway independently of its pre-mRNA splicing function. PLoS Genet. 8:e1005475. doi: 10.1371/journal.pgen.1005475
Zhang, L., Cao, B., Zou, Y., Wei, Q. Q., Ou, R., Liu, W., et al. (2018). Causes of death in Chinese patients with multiple system atrophy. Aging Dis. 9, 102–108. doi: 10.14336/AD.2017.0711
Zhao, H. T., John, N., Delic, V., Ikeda-Lee, K., Kim, A., Weihofen, A., et al. (2017). LRRK2 antisense oligonucleotides ameliorate alpha-synuclein inclusion formation in a Parkinson’s disease mouse model. Mol. Ther. Nucleic Acids 8, 508–519. doi: 10.1016/j.omtn.2017.08.002
Zhou, X., Wang, C., Chen, Z., Peng, Y., Peng, H., Hou, X., et al. (2018). Association of TNF-alpha rs1799964 and IL-1beta rs16944 polymorphisms with multiple system atrophy in Chinese Han population. Int. J. Neurosci. 128, 761–764. doi: 10.1080/00207454.2017.1418346
Zhou, Y., Lu, M., Du, R. H., Qiao, C., Jiang, C. Y., and Zhang, K. Z. (2016). MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 11:28. doi: 10.1186/s13024-016-0094-3
Keywords: microRNA, multiple system atrophy, alpha-synuclein, autophagy, neuroinflammation
Citation: Xiang C, Han S, Nao J and Cong S (2019) MicroRNAs Dysregulation and Metabolism in Multiple System Atrophy. Front. Neurosci. 13:1103. doi: 10.3389/fnins.2019.01103
Received: 07 April 2019; Accepted: 30 September 2019;
Published: 17 October 2019.
Edited by:
Vincenzo La Bella, University of Palermo, ItalyReviewed by:
Srijit Das, National University of Malaysia, MalaysiaClaudio Toma, Neuroscience Research Australia, Australia
Nicola Ticozzi, University of Milan, Italy
Copyright © 2019 Xiang, Han, Nao and Cong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyan Cong, congshuyan@hotmail.com
 Chunchen Xiang
Chunchen Xiang Shunchang Han
Shunchang Han Jianfei Nao
Jianfei Nao Shuyan Cong
Shuyan Cong