Approaches and considerations of studying neuronal ensembles: a brief review
- 1William Beaumont School of Medicine, Oakland University, Rochester, MI, United States
- 2Department of Psychiatry and Behavioral Neurosciences, Wayne State University School of Medicine, Detroit, MI, United States
- 3Neuronal Ensembles in Addiction Section, Behavioral Neuroscience Research Branch, Intramural Research Program/National Institute on Drug Abuse/National Institutes of Health, Bethesda, MD, United States
- 4Department of Biomedical Sciences, Mercer University, Macon, GA, United States
First theorized by Hebb, neuronal ensembles have provided a framework for understanding how the mammalian brain operates, especially regarding learning and memory. Neuronal ensembles are discrete, sparsely distributed groups of neurons that become activated in response to a specific stimulus and are thought to provide an internal representation of the world. Beyond the study of region-wide or projection-wide activation, the study of ensembles offers increased specificity and resolution to identify and target specific memories or associations. Neuroscientists interested in the neurobiology of learning, memory, and motivated behavior have used electrophysiological-, calcium-, and protein-based proxies of neuronal activity in preclinical models to better understand the neurobiology of learned and motivated behaviors. Although these three approaches may be used to pursue the same general goal of studying neuronal ensembles, technical differences lead to inconsistencies in the output and interpretation of data. This mini-review highlights some of the methodologies used in electrophysiological-, calcium-, and protein-based studies of neuronal ensembles and discusses their strengths and weaknesses.
Introduction
The idea of “cell assemblies” or “neuronal ensembles” was postulated three-quarters of a century ago in theoretical form (Hebb, 1949) and refined in the 90’s after the discovery of Fos and other immediate early genes induced by neuronal activation (Murphy et al., 1991). The study of “neuronal ensembles” has recently gained momentum in the fields of learning, memory, and motivated behavior thanks mainly to new IEGs-based, electrophysiological, and calcium imaging techniques. Neuronal ensembles represent the distributed, sparse, and distinct groups of individual neurons that are activated in response to a specific stimulus in the outside world.1
Our aim in this mini-review is to highlight some recent advancements in techniques for the detection and manipulation of neuronal ensembles, discuss important factors inherent to each (positive and negative), and briefly compare them. Due to constraints of the mini-review and the rapid development of this quickly growing field, some techniques or specific studies may not be included.
Early detection methods for identifying neuronal ensembles involved recording activated neurons in a physical space in the rodent hippocampus using electrophysiological recordings (Wilson and McNaughton, 1993). Wilson and McNaughton demonstrated that external space activated only particular neurons within the hippocampus, considerably influencing how we understand physiologically encoded memory. Electrophysiological recordings have demonstrated that action potentials generate calcium influx into neurons (Smetters et al., 1999). In parallel, advances in calcium sensors (Cornell-Bell et al., 1990) with sophisticated microscopy (Denk et al., 1990) allowed access to neuronal ensembles using non-electrophysiological means. This led to increased use of other methods for localizing second messengers and gene products as molecular markers correlated with neuronal electrical activity (Worley et al., 1991). In these correlational approaches, the expression of genes regulated by the cell’s electrical activity was measured in response to ongoing behavior. However, causal role experiments were not possible until neuronal ensemble-specific ablation was accomplished through genetic approaches (e.g., Daun02 chemoablation), demonstrating for the first time the co-dependence between neuronal ensembles and learned behavior (Koya et al., 2009). Fortunately, recent developments in microscopy and optogenetics have enabled researchers to study cause-and-effect relationships with non-chemogenic means, although, not all of these approaches are feasible in freely moving animal models (Adesnik and Abdeladim, 2021).
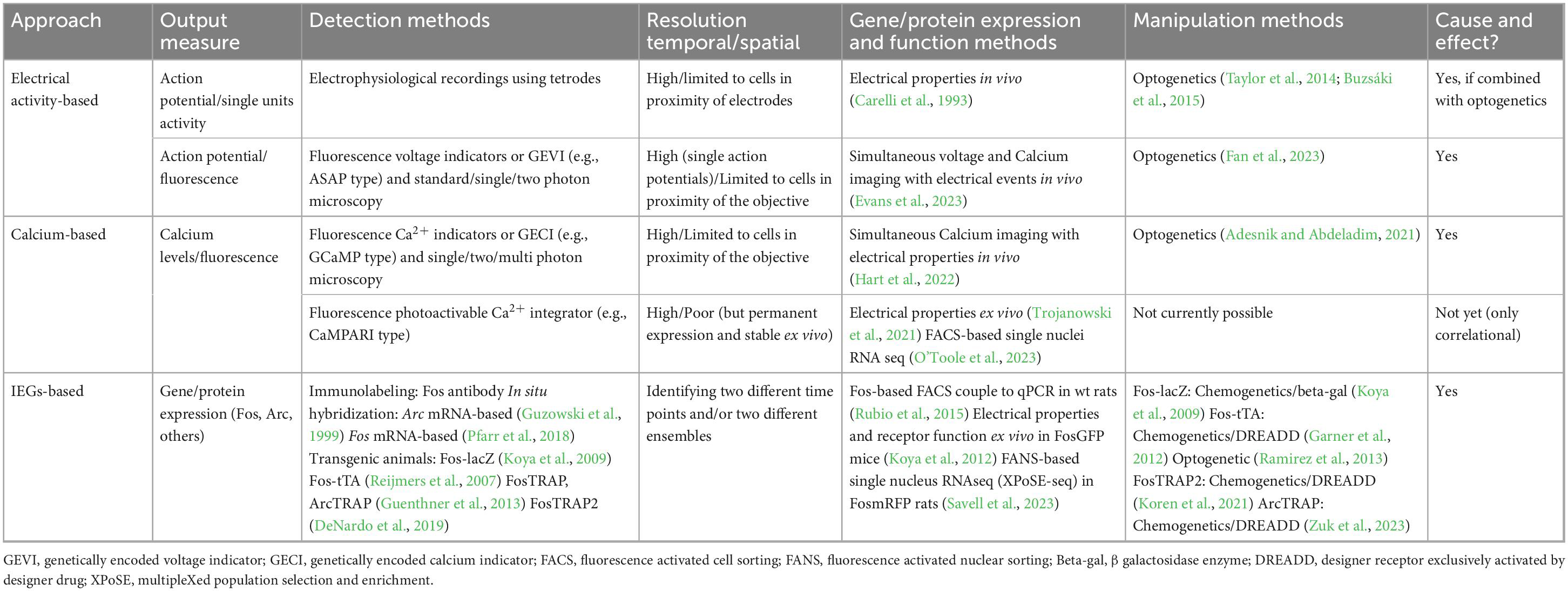
Recent experimental findings have shown that disruption of neuronal ensembles involved in behaviors related to drug abuse (Bossert et al., 2011; Fanous et al., 2012; Cruz et al., 2014; Pfarr et al., 2015; de Guglielmo et al., 2016; Caprioli et al., 2017; Xue et al., 2017; Warren et al., 2019; Gobin et al., 2022), natural reward, e.g., food (Suto et al., 2016; Quintana-Feliciano et al., 2021), social reward (Sakurai et al., 2016), and fear cues (Giannotti et al., 2019), all result in reduced corresponding related behaviors, such as reward-seeking or conditioned fear response. Electrophysiological-, calcium-, and Immediate Early Gene (IEG) -based approaches to study neuronal ensembles all present an exciting opportunity to investigate the neuronal mechanisms and connections that underlie these learned and motivated behaviors. This brief review details the strengths and weaknesses (Table 1) of the three related but different approaches to identify and manipulate neuronal ensembles.
Electrophysiological in vivo recordings in freely moving rodents
Electrophysiological approaches, such as tetrode recordings, provided early evidence of neuronal ensembles and insight into relevant neurobiology by demonstrating that neuronal ensemble activation directly correlates with learned and motivated behaviors across multiple associated brain regions (Cruz et al., 2013; Kamarajan and Porjesz, 2015). Electrophysiological techniques serve to measure cellular or neuronal function in a quantitative and versatile manner (Kamarajan and Porjesz, 2015). Through electrode implantation in brain regions of interest, changes in electrical activity in the form of action potentials can be mapped to measure patterns of neuronal activity over time. Signals from multiple neurons (both excitatory and inhibitory) close in proximity to the probe comprise recordings from active brain regions in animals (Schoenbaum et al., 1999; Takahashi et al., 2003; Wikenheiser et al., 2021). Electrophysiological recordings of distinct neuronal ensembles allow for high temporal resolution in detecting ensemble activity, mapping at several depths within the brain over an extended duration of time (Buzsaki, 2004; Wenzel and Hamm, 2021). Despite the high temporal resolution, the accuracy of electrode implantation and limited recording sites restrict spatial resolution, such as when ensembles from several different brain regions are to be assessed.
Genetically encoded voltage indicators (GEVIs)
Another promising avenue is genetically encoded voltage indicators (GEVIs). Unlike the cytosolic calcium indicators that transduce changes in intracellular calcium levels in transient fluorescence variation, the transmembrane GEVIs can sense voltage changes across the cell membrane and therefore depolarization as consequence of neuronal activity with higher temporal resolution to detect single action potentials (Liu et al., 2022; Evans et al., 2023; Fan et al., 2023). The latest improvements in GEVIs allow 2-photon deep tissue in vivo imaging in multiple neurons for longer times. An example of this is a jellyfish-derived electricity-reporting designer indicator for 2-photon or JEDI-2P (Liu et al., 2022). GEVIs can detect action potentials, hyperpolarization, subthreshold depolarization, and sustained depolarization (Kannan et al., 2019). Fortunately, much like calcium indicators GEVIs can be used alongside optogenetics (Fan et al., 2023).
In vivo calcium imaging
Due to the direct and quantifiable association between electrical spiking and calcium influx into the neuron, calcium imaging presents an additional approach to assess cellular activation and therefore, activation of neuronal ensembles (Grienberger et al., 2022). Therefore, assessing cellular activation with real time changes in intracellular calcium levels provides an opportunity to identify activation of neuronal ensembles. This second approach to track neuronal ensembles is aided by the genetic over-expression of organic fluorescent Ca2+ indicators, (e.g., GCaMP; Nakai et al., 2001), and engineering advances in microscopy technology (Barbera et al., 2019; Yang et al., 2019).
Recent advancements in two-photon imaging coupled with optogenetics have seen impressive advancement in the field (Russell, 2011; Carrillo-Reid et al., 2016; Adesnik and Abdeladim, 2021; Pettit et al., 2022; Bollmann et al., 2023). Two-photon imaging can reduce image scatter endemic to traditional light and has even contributed to the development of holographic optogenetics, where sparse and distributed groups of ensembles at different brain depths are both imaged and manipulated in vivo (Adesnik and Abdeladim, 2021). This methodology requires a large setting, high-cost equipment, well-trained personnel, and head-fixed animals. As such, a large number of these studies have been done using sensory tasks and recording of primary sensory areas of the mouse. Thus, to perform these experiments in freely moving animals who are engaged in complex behaviors would require improvement to current designs. Fortunately, other researchers have worked on changing behavioral paradigms to fit multi-photon microscopy (Vollmer et al., 2021). This behavioral paradigm now allows in vivo calcium imaging using drugs as a reinforcement. A few other points of consideration are related to spatial resolution. In some of these imaging types, more superficial layers of the brain have been examined as the optical probe has penetration limits, only ensembles confined to the field of view (usually a few hundred μm can be imaged and manipulated). As the field of optical physics and microscopy advances, we are likely to see better spatial resolution to accompany the high temporal resolution that calcium imaging provides, with the ultimate goal of being conducted in freely moving animals.
More recently, the development of a calcium-modulated photoactivatable radiometric integrator (CaMPARI) has allowed identifying neuronal ensembles in free moving animals (Fosque et al., 2015; Moeyaert et al., 2018). This calcium and light dependent approach requires the use of the same tools used for optogenetic approaches but with the added advantage of being able to see the outcome ex vivo, in contrast to the classical GCaMP-based calcium imaging. Therefore, the CaMPARI approach allows neuronal ensembles to be detected ex vivo using specific antibodies to detect both states of fluorescence (active and inactive) following fluorescence photoconversion in the presence of calcium in vivo. Additionally, harvested tissue could be subjected to further analytical techniques such as electrophysiology, cell sorting (FACS) and RNA-seq. One advantage of this calcium-dependent photoactivation approach is the temporal specificity; the experimenter can tag the photoactive calcium sensor neuronal ensembles as soon as a few seconds after a stimulus. Additionally, CaMPARI has the potential to detect different calcium-dependent activation states by quantifying the red-to-green ratio (only red: active state, only green; inactive state) between the two fluorescent protein forms demonstrating a graduation of firing as opposed to simple binary firing indication from GCaMP sensors. A newer generation of CaMPARI, called CaMPARI2 has seen enhancements in photoconversion increasing the specificity of these graduations and allowing for a more nuanced picture of ensemble construction and activation (Trojanowski et al., 2021; Berndt et al., 2023; O’Toole et al., 2023). Researchers performing CaMPARI2 focused on activity in the cortex of head fixed mice navigating a virtual reality. Single neurons photoconverted functionally in vivo were sorted by fluorescence using FACS for RNA-seq analysis (O’Toole et al., 2023) demonstrating an enrichment in specific neuronal cell types within the CaMPARI2-tagged neuronal ensemble. While these calcium-based approaches are related to firing activity and offer many benefits to studying neuronal ensembles in vivo and ex vivo, they may overestimate the number of neuronal ensembles by tagging cells that, while active, are not explicitly responding to the specific stimuli (Liang et al., 2018; Tsuda, 2020; Zhou et al., 2021).
Immediate early genes (IEGs) based approaches
As with calcium imaging and calcium-based approaches, IEGs (e.g., Fos, Arc, Zif-268) can be used as a proxy measurement for neuronal, and thus ensemble activity (Morgan et al., 1987; Guzowski et al., 1999; Guenthner et al., 2013; Beverley et al., 2014; Cruz et al., 2014; Rubio et al., 2015). IEGs have specific synaptic (Jones et al., 2001; Shepherd et al., 2006) and cellular functions (Christy et al., 1988) through their actions as transcription factors and/or effector proteins. IEG expression is rapidly induced after neuronal activation (Morgan and Curran, 1991a,b) and presents opportunities not only for identification and manipulation of ensembles, but also to study molecular alterations exclusively in neuronal ensembles (Liu et al., 2014; Li et al., 2015; Rubio et al., 2015). Traditional IEG-based techniques for neuronal ensemble detection have used IEG protein products or their nascent mRNA as indicators to identify active cells that participate in a neuronal ensemble (Kubik et al., 2007; Sauvage et al., 2019).
Additionally, given that IEGs are highly expressed in strongly activated cells, IEG-based techniques may overcome the inclusion of non-ensemble activated neurons common to both electrophysiological and calcium-based techniques. Approaches that combine the use of techniques to quantify IEG-expressing neurons and functional calcium imaging in vivo have shown a good correlation between IEG protein expression and neuronal (calcium) activity (Mahringer et al., 2019, 2022; Wang et al., 2021; Pettit et al., 2022). Recently, this has been conducted using 2-photon imaging to capture in vivo Fos ensembles (Lee et al., 2021). Though most of the brain can be visualized with this approach, the activity history of ensembles prior to the test day cannot be determined. IEG based techniques such as catFISH (Cellular compartment Analysis of Temporal activity by Fluorescent in situ Hybridization) based on IEGs Arc (Guzowski et al., 1999) or Fos (Pfarr et al., 2018), and Fos-based TRAP (Targeted Recombination of Activated Populations) mitigate this limitation and offer the opportunity to tag a specific ensemble at distinct time points or be able to differentiate two different ensembles in response to two distinct stimuli (Guenthner et al., 2013; Vassilev et al., 2020). The improved version, TRAP2, improves upon the first iteration of the TRAP system, including better spatial resolution as well as preservation of endogenous Fos (DeNardo et al., 2019).
Perturbation of neuronal ensembles via IEG methodology has been achieved both optogenetically (Ramirez et al., 2013) and chemogenetically (Koya et al., 2009). The Daun02 chemoablation technique allows for a timed and selective ablation of Fos-expressing ensembles, providing a powerful and causal approach to studying ensembles (Koya et al., 2009; Cruz et al., 2013). The IEG approach to identify and manipulate neuronal ensembles has been selected by different labs to demonstrate functionality and causality. The Fos-based TRAP system can be combined with the use of the Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) technique. For example, a TRAP-DREADD approach has recently been applied to ameliorate neurological symptoms in a rodent model of Rett syndrome (Achilly et al., 2021). The TRAP-DREADD approach demonstrates the dual possibilities of identifying and manipulating these highly activated neurons in order to demonstrate the function of neuronal ensembles in encoding memories (Giannotti et al., 2019). Although using IEGs-based approaches to study ensembles offers optimal spatial resolution making them better suited to study ensembles across several brain regions or within one region at one or a few selected timepoints, the temporal resolution is not as good as can be achieved by using the calcium and electrophysiological approaches. Combining IEG-based approaches with calcium or electrophysiology techniques can solve the dissociation between spatial and temporal resolution.
Discussion
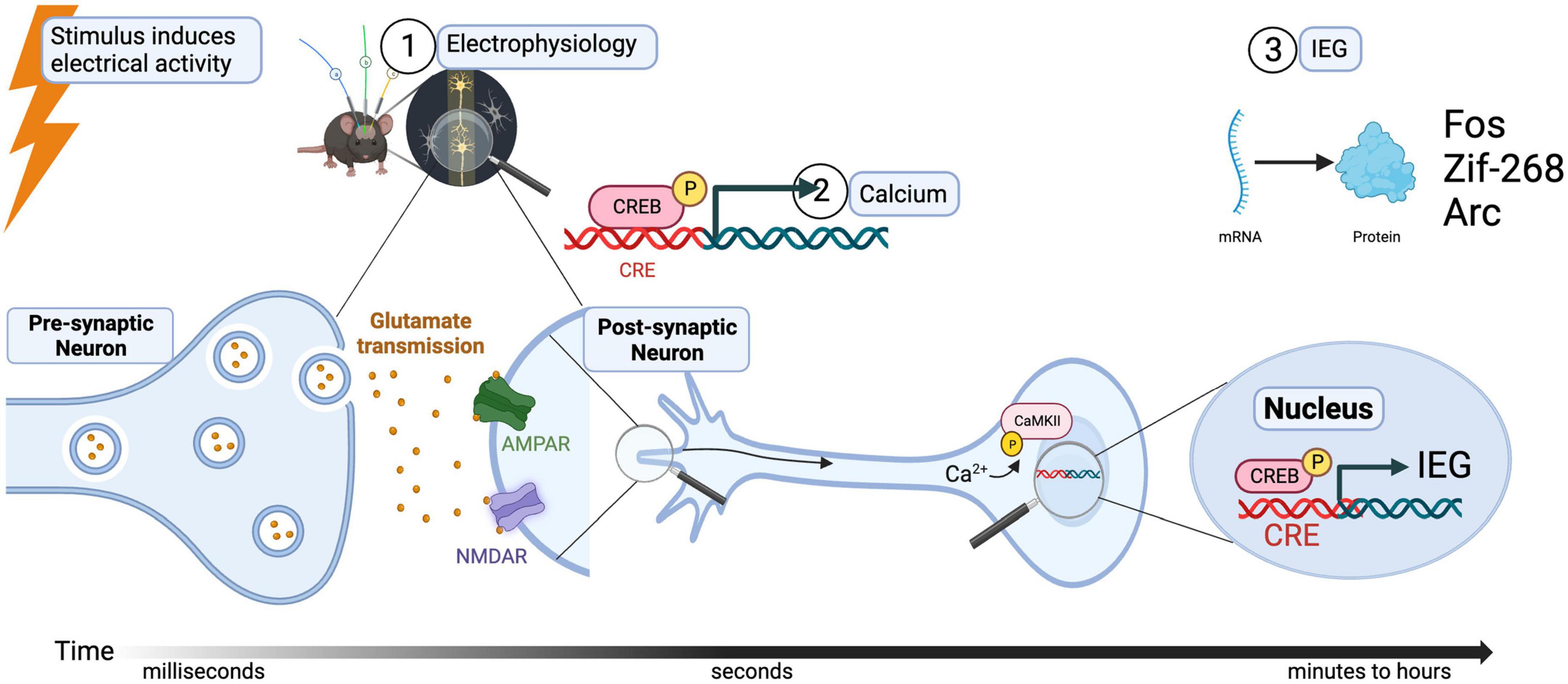
When an action potential occurs near the cell body of a neuron ①, calcium-activated signaling can follow, either through ligand- or voltage-gated channels (Buzsaki, 2004). This influx of calcium, also known as a master switch, will ② phosphorylate Calmodulin (CaM) and in turn, Calcium/calmodulin-dependent protein kinase II (or CamKII). The destination of this second messenger cascade (starting from, for example occupation of NMDA or calcium receptors) is the cellular transcription factor cAMP response element-binding protein (or CREB) which, along with other transcription factors, ③ induces the expression of early immediate genes Arc, Fos, and BDNF (see Figure 1). It is important to notice that before this cascade of events occurs in the cell body of a postsynaptic neuron, information in the form of neurotransmitters released through the axon terminals of a presynaptic neuron is necessary. Therefore, signals from synapses to nuclei are also ultimately activating the soma with the consequent increase in expression of IEGs (Colgan et al., 2023). The design of new techniques and approaches to identify and modulate neuronal activation at the synaptic level is a recent area of development (Perez-Alvarez et al., 2020; Rubio et al., 2023), and although it is not the aim of this review, it is necessary to be stated as one exciting advance in the field of activated synapses.
The choice of approach or combination of techniques to study neuronal ensembles relies on the “type of activity marker” selected to identify an activated neuron (electrical activity, increase in calcium levels or IEG expression). Each activation marker represents not an undoubtedly activation state but a proxy that reveals one of the events within the activation-dependent cascade. Thus, the accuracy in identifying a neuronal ensemble is related not only to methodological aspects, feasibility, and behavioral basis of signaling to assess that question (Kim and Schnitzer, 2022), but also to the time gap between the stimulus that induces the neuronal activation and the cellular or molecular event (consequence of the activation) that the marker is revealing. The choice of methodology partly depends on whether ex vivo analysis will be done from the ensembles recorded in vivo. We may want to use electrophysiology recordings to identify functionally activated neurons in real time immediately after a stimulus, but this might overestimate the actual neuronal ensembles that are encoding a specific memory. Similarly, calcium imaging techniques detect the increase in calcium in the neuron but can’t indicate whether all neurons undergoing calcium influx are part of a specific neuronal ensemble. As the gene expression and protein translation in the nucleus is one of the ultimate events that happens after the action potential and calcium influx, and therefore needs minutes (mRNA) or hours (protein) to be detected, this methodology might reflect better the extension of a neuronal ensemble (1–5% of neurons in a region).
To overcome the limitation of individual techniques, a combination of two different approaches, such as CaMPARI as a calcium-dependent photoactivatable indicator with IEG expression can be the best way to accurately label a specific neuronal ensemble. For instance, the photoconversion of CaMPARI in vivo a few seconds after a specific stimulus generates a permanent change in fluorescence can be assessed ex vivo a few hours later in combination with Fos immunohistochemistry. Analyzing the neurons that express both the active CaMPARI form and Fos protein might give us better resolution to identify specific neuronal ensembles. This method will control for neuronal ensembles that are actually composed of mixed populations of neurons activated by specific stimuli and those triggered by cues, contexts which are also picked up by classical electrophysiology or calcium imaging methods. Some of these combined approaches have recently started, the combination of 2-photon imaging the contribution of GABAergic vs. Glutamatergic neurons in shaping ensembles in the barrel cortex (Bollmann et al., 2023; Kourdougli et al., 2023), two-photon imaging has also established a cause -and-effect link between Fos and place cell activity (Pettit et al., 2022), Fos positive neurons are a more reliable index of place places than non-positive Fos cells. A surprising twist to the story has also come from the combined use of in vivo 2-photon and Fos imaging (Lee et al., 2021). The authors show that whisker-dependent sensory association learning in the primary somatosensory cortex does not alter Fos, and in fact synaptic changes are predominant in non-Fos cells. As referenced earlier in this paper, the approaches are not necessarily competing; but rather, each may be used to answer unique questions about how neuronal ensembles contribute to the neurobiology of motivated behavior. Future investigations, especially those that are purporting to study “neuronal ensembles” need to agree upon a common, operational definition of what this entails and distinction from engrams as we have included. Integration and exploration of these approaches, guided by their prior use in other domains, can provide great insights into future research on “neuronal ensembles/cell assemblies.”
Author contributions
CD: Conceptualization, Writing – original draft, Writing – review and editing. AM: Conceptualization, Software, Writing – original draft, Writing – review and editing. MY: Writing – original draft, Writing – review and editing. FJR: Formal analysis, Supervision, Writing – original draft, Writing – review and editing. AG: Funding acquisition, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by the National Institute on Drug Abuse of the National Institutes of Health under award number K01 DA055068 to AG. This research was also supported in part by the Intramural Research Program of the NIH, NIDA to FJR.
Acknowledgments
We would like to thank Shane A. Perrine, Ph.D. for his feedback on this manuscript and BioRender.com was used to construct the figure.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ We would like to make a clear distinction between Neuronal Ensembles and Engrams. We are defining ensembles using the theoretical definition from Hebb (1949) as a sparse distributed pattern of cells/synapses that are selectively activated by cues or stimuli to mediate specific information, while engrams are defined as long-lasting molecular and/or cellular adaptations that can physically encode learned associations within these ensembles.
References
Achilly, N. P., Wang, W., and Zoghbi, H. Y. (2021). Presymptomatic training mitigates functional deficits in a mouse model of Rett syndrome. Nature 592, 596–600. doi: 10.1038/s41586-021-03369-7
Adesnik, H., and Abdeladim, L. (2021). Probing neural codes with two-photon holographic optogenetics. Nat. Neurosci. 24, 1356–1366. doi: 10.1038/s41593-021-00902-9
Barbera, G., Liang, B., Zhang, L., Li, Y., and Lin, D. T. (2019). A wireless miniScope for deep brain imaging in freely moving mice. J. Neurosci. Methods 323, 56–60. doi: 10.1016/j.jneumeth.2019.05.008
Berndt, M., Trusel, M., Roberts, T. F., Pfeiffer, B. E., and Volk, L. J. (2023). Bidirectional synaptic changes in deep and superficial hippocampal neurons following in vivo activity. Neuron 111, 2984.e4–2994.e4. doi: 10.1016/j.neuron.2023.08.014
Beverley, J. A., Piekarski, C., Van Waes, V., and Steiner, H. (2014). Potentiated gene regulation by methylphenidate plus fluoxetine treatment: Long-term gene blunting (Zif268, Homer1a) and behavioral correlates. Basal Ganglia 4, 109–116. doi: 10.1016/j.baga.2014.10.001
Bollmann, Y., Modol, L., Tressard, T., Vorobyev, A., Dard, R., Brustlein, S., et al. (2023). Prominent in vivo influence of single interneurons in the developing barrel cortex. Nat. Neurosci. 26, 1555–1565. doi: 10.1038/s41593-023-01405-5
Bossert, J. M., Stern, A. L., Theberge, F. R., Cifani, C., Koya, E., Hope, B. T., et al. (2011). Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nat. Neurosci. 14, 420–422. doi: 10.1038/nn.2758
Buzsaki, G. (2004). Large-scale recording of neuronal ensembles. Nat. Neurosci. 7, 446–451. doi: 10.1038/nn1233
Buzsáki, G., Stark, E., Berényi, A., Khodagholy, D., Kipke, D. R., Yoon, E., et al. (2015). Tools for probing local circuits: High-density silicon probes combined with optogenetics. Neuron 86, 92–105. doi: 10.1016/j.neuron.2015.01.028
Caprioli, D., Venniro, M., Zhang, M., Bossert, J. M., Warren, B. L., Hope, B. T., et al. (2017). Role of Dorsomedial striatum neuronal ensembles in incubation of methamphetamine craving after voluntary abstinence. J. Neurosci. 37, 1014–1027. doi: 10.1523/jneurosci.3091-16.2016
Carelli, R. M., King, V. C., Hampson, R. E., and Deadwyler, S. A. (1993). Firing patterns of nucleus accumbens neurons during cocaine self-administration in rats. Brain Res. 626, 14–22.
Carrillo-Reid, L., Yang, W., Bando, Y., Peterka, D. S., and Yuste, R. (2016). Imprinting and recalling cortical ensembles. Science 353, 691–694. doi: 10.1126/science.aaf7560
Christy, B. A., Lau, L. F., and Nathans, D. (1988). A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc. Natl. Acad. Sci. U.S.A. 85, 7857–7861. doi: 10.1073/pnas.85.21.7857
Colgan, L. A., Parra-Bueno, P., Holman, H. L., Tu, X., Jain, A., Calubag, M. F., et al. (2023). Dual regulation of spine-specific and synapse-to-nucleus signaling by PKCδ during plasticity. J. Neurosci. 43, 5432–5447. doi: 10.1523/jneurosci.0208-22.2023
Cornell-Bell, A. H., Finkbeiner, S. M., Cooper, M. S., and Smith, S. J. (1990). Glutamate induces calcium waves in cultured astrocytes: Long-range glial signaling. Science 247, 470–473. doi: 10.1126/science.1967852
Cruz, F. C., Babin, K. R., Leao, R. M., Goldart, E. M., Bossert, J. M., Shaham, Y., et al. (2014). Role of nucleus accumbens shell neuronal ensembles in context-induced reinstatement of cocaine-seeking. J. Neurosci. 34, 7437–7446. doi: 10.1523/jneurosci.0238-14.2014
Cruz, F. C., Koya, E., Guez-Barber, D. H., Bossert, J. M., Lupica, C. R., Shaham, Y., et al. (2013). New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nat. Rev. Neurosci. 14, 743–754. doi: 10.1038/nrn3597
de Guglielmo, G., Crawford, E., Kim, S., Vendruscolo, L. F., Hope, B. T., Brennan, M., et al. (2016). Recruitment of a neuronal ensemble in the central nucleus of the amygdala is required for alcohol dependence. J. Neurosci. 36, 9446–9453. doi: 10.1523/jneurosci.1395-16.2016
DeNardo, L. A., Liu, C. D., Allen, W. E., Adams, E. L., Friedmann, D., Fu, L., et al. (2019). Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat. Neurosci. 22, 460–469. doi: 10.1038/s41593-018-0318-7
Denk, W., Strickler, J. H., and Webb, W. W. (1990). Two-photon laser scanning fluorescence microscopy. Science 248, 73–76. doi: 10.1126/science.2321027
Evans, S. W., Shi, D. Q., Chavarha, M., Plitt, M. H., Taxidis, J., Madruga, B., et al. (2023). A positively tuned voltage indicator for extended electrical recordings in the brain. Nat. Methods 20, 1104–1113. doi: 10.1038/s41592-023-01913-z
Fan, L. Z., Kim, D. K., Jennings, J. H., Tian, H., Wang, P. Y., Ramakrishnan, C., et al. (2023). All-optical physiology resolves a synaptic basis for behavioral timescale plasticity. Cell 186, 543.e19–559.e19. doi: 10.1016/j.cell.2022.12.035
Fanous, S., Goldart, E. M., Theberge, F. R., Bossert, J. M., Shaham, Y., and Hope, B. T. (2012). Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. J. Neurosci. 32, 11600–11609. doi: 10.1523/jneurosci.1914-12.2012
Fosque, B. F., Sun, Y., Dana, H., Yang, C. T., Ohyama, T., Tadross, M. R., et al. (2015). Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science 347, 755–760. doi: 10.1126/science.1260922
Garner, A. R., Rowland, D. C., Hwang, S. Y., Baumgaertel, K., Roth, B. L., Kentros, C., et al. (2012). Generation of a synthetic memory trace. Science 335, 1513–1516. doi: 10.1126/science.1214985
Giannotti, G., Heinsbroek, J. A., Yue, A. J., Deisseroth, K., and Peters, J. (2019). Prefrontal cortex neuronal ensembles encoding fear drive fear expression during long-term memory retrieval. Sci. Rep. 9:10709. doi: 10.1038/s41598-019-47095-7
Gobin, C., Sortman, B., Rakela, S., Quintana-Feliciano, R., and Warren, B. L. (2022). Fos-expressing neuronal ensembles in rat infralimbic cortex encode initial and maintained oxycodone seeking in rats. Addict. Biol. 27:e13148. doi: 10.1111/adb.13148
Grienberger, C., Giovannucci, A., Zeiger, W., and Portera-Cailliau, C. (2022). Two-photon calcium imaging of neuronal activity. Nat. Rev. Methods Primers 2:67. doi: 10.1038/s43586-022-00147-1
Guenthner, C. J., Miyamichi, K., Yang, H. H., Heller, H. C., and Luo, L. (2013). Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78, 773–784. doi: 10.1016/j.neuron.2013.03.025
Guzowski, J. F., McNaughton, B. L., Barnes, C. A., and Worley, P. F. (1999). Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 2, 1120–1124. doi: 10.1038/16046
Hart, E. E., Gardner, M. P. H., Panayi, M. C., Kahnt, T., and Schoenbaum, G. (2022). Calcium activity is a degraded estimate of spikes. Curr. Biol. 32, 5364.e4–5373.e4. doi: 10.1016/j.cub.2022.10.037
Jones, M. W., Errington, M. L., French, P. J., Fine, A., Bliss, T. V., Garel, S., et al. (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4, 289–296. doi: 10.1038/85138
Kamarajan, C., and Porjesz, B. (2015). Advances in electrophysiological research. Alcohol. Res. Curr. Rev. 37, 53–87.
Kannan, M., Vasan, G., and Pieribone, V. A. (2019). Optimizing strategies for developing genetically encoded voltage indicators. Front. Cell Neurosci. 13:53. doi: 10.3389/fncel.2019.00053
Kim, T. H., and Schnitzer, M. J. (2022). Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185, 9–41. doi: 10.1016/j.cell.2021.12.007
Koren, T., Yifa, R., Amer, M., Krot, M., Boshnak, N., Ben-Shaanan, T. L., et al. (2021). Insular cortex neurons encode and retrieve specific immune responses. Cell 184, 5902.e17–5915.e17. doi: 10.1016/j.cell.2021.10.013
Kourdougli, N., Suresh, A., Liu, B., Juarez, P., Lin, A., Chung, D. T., et al. (2023). Improvement of sensory deficits in fragile X mice by increasing cortical interneuron activity after the critical period. Neuron 111, 2863.e6–2880.e6. doi: 10.1016/j.neuron.2023.06.009
Koya, E., Cruz, F. C., Ator, R., Golden, S. A., Hoffman, A. F., Lupica, C. R., et al. (2012). Silent synapses in selectively activated nucleus accumbens neurons following cocaine sensitization. Nat. Neurosci. 15, 1556–1562. doi: 10.1038/nn.3232
Koya, E., Golden, S. A., Harvey, B. K., Guez-Barber, D. H., Berkow, A., Simmons, D. E., et al. (2009). Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat. Neurosci. 12, 1069–1073. doi: 10.1038/nn.2364
Kubik, S., Miyashita, T., and Guzowski, J. F. (2007). Using immediate-early genes to map hippocampal subregional functions. Learn. Mem. 14, 758–770. doi: 10.1101/lm.698107
Lee, J., Urban-Ciecko, J., Park, E., Zhu, M., Myal, S. E., Margolis, D. J., et al. (2021). FosGFP expression does not capture a sensory learning-related engram in superficial layers of mouse barrel cortex. Proc. Natl. Acad. Sci. U.S.A. 118:e2112212118. doi: 10.1073/pnas.2112212118
Li, X., Rubio, F. J., Zeric, T., Bossert, J. M., Kambhampati, S., Cates, H. M., et al. (2015). Incubation of methamphetamine craving is associated with selective increases in expression of Bdnf and trkb, glutamate receptors, and epigenetic enzymes in cue-activated fos-expressing dorsal striatal neurons. J. Neurosci. 35, 8232–8244. doi: 10.1523/jneurosci.1022-15.2015
Liang, B., Zhang, L., Barbera, G., Fang, W., Zhang, J., Chen, X., et al. (2018). Distinct and dynamic ON and OFF neural ensembles in the prefrontal cortex code social exploration. Neuron 100, 700.e9–714.e9. doi: 10.1016/j.neuron.2018.08.043
Liu, Q. R., Rubio, F. J., Bossert, J. M., Marchant, N. J., Fanous, S., Hou, X., et al. (2014). Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS). J. Neurochem. 128, 173–185. doi: 10.1111/jnc.12381
Liu, Z., Lu, X., Villette, V., Gou, Y., Colbert, K. L., Lai, S., et al. (2022). Sustained deep-tissue voltage recording using a fast indicator evolved for two-photon microscopy. Cell 185, 3408.e29–3425.e29. doi: 10.1016/j.cell.2022.07.013
Mahringer, D., Petersen, A. V., Fiser, A., Okuno, H., Bito, H., Perrier, J.-F., et al. (2019). Expression of c-Fos and Arc in hippocampal region CA1 marks neurons that exhibit learning-related activity changes. bioRxiv [Preprint]. doi: 10.1101/644526
Mahringer, D., Zmarz, P., Okuno, H., Bito, H., and Keller, G. B. (2022). Functional correlates of immediate early gene expression in mouse visual cortex. Peer Community J. 2:e45. doi: 10.24072/pcjournal.156
Moeyaert, B., Holt, G., Madangopal, R., Perez-Alvarez, A., Fearey, B. C., Trojanowski, N. F., et al. (2018). Improved methods for marking active neuron populations. Nat. Commun. 9:4440. doi: 10.1038/s41467-018-06935-2
Morgan, J. I., Cohen, D. R., Hempstead, J. L., and Curran, T. (1987). Mapping patterns of c-fos expression in the central nervous system after seizure. Science 237, 192–197. doi: 10.1126/science.3037702
Morgan, J. I., and Curran, T. (1991a). Proto-oncogene transcription factors and epilepsy. Trends Pharmacol. Sci, 1z, 343–349. doi: 10.1016/0165-6147(91)90594-i
Morgan, J. I., and Curran, T. (1991b). Stimulus-transcription coupling in the nervous system: Involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 14, 421–451. doi: 10.1146/annurev.ne.14.030191.002225
Murphy, T. H., Worley, P. F., Nakabeppu, Y., Christy, B., Gastel, J., and Baraban, J. M. (1991). Synaptic regulation of immediate early gene expression in primary cultures of cortical neurons. J. Neurochem. 57, 1862–1872. doi: 10.1111/j.1471-4159.1991.tb06396.x
Nakai, J., Ohkura, M., and Imoto, K. (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141. doi: 10.1038/84397
O’Toole, S. M., Oyibo, H. K., and Keller, G. B. (2023). Molecularly targetable cell types in mouse visual cortex have distinguishable prediction error responses. Neuron 111, 2918.e8–2928.e8. doi: 10.1016/j.neuron.2023.08.015
Perez-Alvarez, A., Fearey, B. C., O’Toole, R. J., Yang, W., Arganda-Carreras, I., Lamothe-Molina, P. J., et al. (2020). Freeze-frame imaging of synaptic activity using SynTagMA. Nat. Commun. 11:2464. doi: 10.1038/s41467-020-16315-4
Pettit, N. L., Yap, E. L., Greenberg, M. E., and Harvey, C. D. (2022). Fos ensembles encode and shape stable spatial maps in the hippocampus. Nature 609, 327–334. doi: 10.1038/s41586-022-05113-1
Pfarr, S., Meinhardt, M. W., Klee, M. L., Hansson, A. C., Vengeliene, V., Schönig, K., et al. (2015). Losing control: Excessive alcohol seeking after selective inactivation of cue-responsive neurons in the Infralimbic cortex. J. Neurosci. 35, 10750–10761. doi: 10.1523/jneurosci.0684-15.2015
Pfarr, S., Schaaf, L., Reinert, J. K., Paul, E., Herrmannsdorfer, F., Rossmanith, M., et al. (2018). Choice for drug or natural reward engages largely overlapping neuronal ensembles in the Infralimbic prefrontal cortex. J. Neurosci. 38, 3507–3519. doi: 10.1523/JNEUROSCI.0026-18.2018
Quintana-Feliciano, R., Gobin, C., Kane, L., Sortman, B., Rakela, S., Genovese, A., et al. (2021). Food-seeking behavior is mediated by fos-expressing neuronal ensembles formed at first learning in rats. eNeuro 8:ENEURO.0373-20.2021. doi: 10.1523/eneuro.0373-20.2021
Ramirez, S., Liu, X., Lin, P. A., Suh, J., Pignatelli, M., Redondo, R. L., et al. (2013). Creating a false memory in the hippocampus. Science 341, 387–391. doi: 10.1126/science.1239073
Reijmers, L. G., Perkins, B. L., Matsuo, N., and Mayford, M. (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. doi: 10.1126/science.1143839
Rubio, F. J., Liu, Q. R., Li, X., Cruz, F. C., Leão, R. M., Warren, B. L., et al. (2015). Context-induced reinstatement of methamphetamine seeking is associated with unique molecular alterations in Fos-expressing dorsolateral striatum neurons. J. Neurosci. 35, 5625–5639. doi: 10.1523/jneurosci.4997-14.2015
Rubio, F. J., Olivares, D. E., Dunn, C., Zhang, S., Hilaire, E. M., Henry, A., et al. (2023). Flow Cytometry of Synaptoneurosomes (FCS) reveals increased ribosomal S6 and Calcineurin proteins in activated medial prefrontal cortex to nucleus Accumbens synapses. J. Neurosci. 43, 4217–4233. doi: 10.1523/jneurosci.0927-22.2023
Russell, J. T. (2011). Imaging calcium signals in vivo: A powerful tool in physiology and pharmacology.. Br. J. Pharmacol. 163, 1605–1625. doi: 10.1111/j.1476-5381.2010.00988.x
Sakurai, K., Zhao, S., Takatoh, J., Rodriguez, E., Lu, J., Leavitt, A. D., et al. (2016). Capturing and manipulating activated neuronal ensembles with CANE delineates a hypothalamic social-fear circuit. Neuron 92, 739–753. doi: 10.1016/j.neuron.2016.10.015
Sauvage, M., Kitsukawa, T., and Atucha, E. (2019). Single-cell memory trace imaging with immediate-early genes. J. Neurosci. Methods 326:108368. doi: 10.1016/j.jneumeth.2019.108368
Savell, K. E., Madangopal, R., Saravanan, P., Palaganas, R. G., Woods, K. D., Thompson, D. J., et al. (2023). MultipleXed population selection and enrichment single nucleus RNA sequencing (XPoSE-seq) enables sample identity retention during transcriptional profiling of rare populations. bioRxiv [Preprint]. doi: 10.1101/2023.09.27.559834
Schoenbaum, G., Chiba, A. A., and Gallagher, M. (1999). Neural encoding in orbitofrontal cortex and basolateral amygdala during olfactory discrimination learning. J. Neurosci., 19, 1876–1884. doi: 10.1523/jneurosci.19-05-01876.1999
Shepherd, J. D., Rumbaugh, G., Wu, J., Chowdhury, S., Plath, N., Kuhl, D., et al. (2006). Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484. doi: 10.1016/j.neuron.2006.08.034
Smetters, D., Majewska, A., and Yuste, R. (1999). Detecting action potentials in neuronal populations with calcium imaging. Methods 18, 215–221. doi: 10.1006/meth.1999.0774
Suto, N., Laque, A., De Ness, G. L., Wagner, G. E., Watry, D., Kerr, T., et al. (2016). Distinct memory engrams in the infralimbic cortex of rats control opposing environmental actions on a learned behavior. eLife 5:e21920. doi: 10.7554/eLife.21920
Takahashi, S., Anzai, Y., and Sakurai, Y. (2003). A new approach to spike sorting for multi-neuronal activities recorded with a tetrode–how ICA can be practical. Neurosci. Res., 46, 265–272. doi: 10.1016/s0168-0102(03)00103-2
Taylor, H., Schmiedt, J. T., Carcak, N., Onat, F., Di Giovanni, G., Lambert, R., et al. (2014). Investigating local and long-range neuronal network dynamics by simultaneous optogenetics, reverse microdialysis and silicon probe recordings in vivo. J. Neurosci. Methods 235, 83–91. doi: 10.1016/j.jneumeth.2014.06.031
Trojanowski, N. F., Bottorff, J., and Turrigiano, G. G. (2021). Activity labeling in vivo using CaMPARI2 reveals intrinsic and synaptic differences between neurons with high and low firing rate set points. Neuron 109, 663.e5–676.e5. doi: 10.1016/j.neuron.2020.11.027
Tsuda, S. (2020). Chapter 17 - Optogenetics. Cambridge, MA: Academic Press, doi: 10.1016/b978-0-12-817528-6.00017-6
Vassilev, P., Avvisati, R., Koya, E., and Badiani, A. (2020). Distinct populations of neurons activated by heroin and cocaine in the striatum as assessed by catFISH. eNeuro 7:ENEURO.0394-19.2019. doi: 10.1523/ENEURO.0394-19.2019
Vollmer, K. M., Doncheck, E. M., Grant, R. I., Winston, K. T., Romanova, E. V., Bowen, C. W., et al. (2021). A novel assay allowing drug self-administration, extinction, and reinstatement testing in head-restrained mice. Front. Behav. Neurosci. 15:744715. doi: 10.3389/fnbeh.2021.744715
Wang, G., Xie, H., Hu, Y., Chen, Q., Liu, C., Liu, K., et al. (2021). Egr1-EGFP transgenic mouse allows in vivo recording of Egr1 expression and neural activity. J. Neurosci. Methods 363:109350. doi: 10.1016/j.jneumeth.2021.109350
Warren, B. L., Kane, L., Venniro, M., Selvam, P., Quintana-Feliciano, R., Mendoza, M. P., et al. (2019). Separate vmPFC ensembles control cocaine self-administration versus extinction in rats. J. Neurosci. 39, 7394–7407. doi: 10.1523/jneurosci.0918-19.2019
Wenzel, M., and Hamm, J. P. (2021). Identification and quantification of neuronal ensembles in optical imaging experiments. J. Neurosci. Methods 351:109046. doi: 10.1016/j.jneumeth.2020.109046
Wikenheiser, A. M., Gardner, M. P. H., Mueller, L. E., and Schoenbaum, G. (2021). Spatial representations in rat orbitofrontal cortex. J. Neurosci. 41, 6933–6945. doi: 10.1523/jneurosci.0830-21.2021
Wilson, M. A., and McNaughton, B. L. (1993). Dynamics of the hippocampal ensemble code for space. Science 261, 1055–1058.
Worley, P. F., Christy, B. A., Nakabeppu, Y., Bhat, R. V., Cole, A. J., and Baraban, J. M. (1991). Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc. Natl. Acad. Sci. U.S.A. 88, 5106–5110.
Xue, Y. X., Chen, Y. Y., Zhang, L. B., Zhang, L. Q., Huang, G. D., Sun, S. C., et al. (2017). Selective inhibition of amygdala neuronal ensembles encoding nicotine-associated memories inhibits nicotine preference and relapse. Biol. Psychiatry 82, 781–793. doi: 10.1016/j.biopsych.2017.04.017
Yang, Y., Zhang, L., Wang, Z., Liang, B., Barbera, G., Moffitt, C., et al. (2019). A two-step GRIN lens coating for in vivo brain imaging. Neurosci. Bull. 35, 419–424. doi: 10.1007/s12264-019-00356-x
Zhou, J., Zong, W., Jia, C., Gardner, M. P. H., and Schoenbaum, G. (2021). Prospective representations in rat orbitofrontal ensembles. Behav. Neurosci. 135, 518–527. doi: 10.1037/bne0000451
Keywords: cell assemblies, immediate early gene, Fos, calcium imaging, electrophysiology
Citation: Davidson CJ, Mascarin AT, Yahya MA, Rubio FJ and Gheidi A (2023) Approaches and considerations of studying neuronal ensembles: a brief review. Front. Cell. Neurosci. 17:1310724. doi: 10.3389/fncel.2023.1310724
Received: 09 October 2023; Accepted: 27 November 2023;
Published: 14 December 2023.
Edited by:
Kasia M. Bieszczad, Rutgers, The State University of New Jersey, United StatesReviewed by:
Nazim Kourdougli, University of California, Los Angeles, United StatesCopyright © 2023 Davidson, Mascarin, Yahya, Rubio and Gheidi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Gheidi, Gheidi_a@mercer.edu
 Cameron J. Davidson1
Cameron J. Davidson1  F. Javier Rubio
F. Javier Rubio Ali Gheidi
Ali Gheidi