Cell-type expression and activation by light of neuropsins in the developing and mature Xenopus retina
- Department of Cell Biology and Anatomy, Hotchkiss Brain Institute, Alberta Children’s Hospital Research Institute, University of Calgary, Calgary, AB, Canada
Photosensitive opsins detect light and perform image- or nonimage-forming tasks. Opsins such as the “classical” visual opsins and melanopsin are well studied. However, the retinal expression and functions of a novel family of neuropsins are poorly understood. We explored the developmental time-course and cell-type specificity of neuropsin (opn5, 6a, 6b, and 8) expression in Xenopus laevis by in situ hybridization and immunohistochemistry. We compared the Xenopus results with publicly available single cell RNA sequencing (scRNA-seq) data from zebrafish, chicken, and mouse. Additionally, we analyzed light-activation of neuropsin-expressing cells through induction of c-fos mRNA. opn5 and opn8 expression begins at stage 37/38 when the retinal circuits begin to be activated. Once retinal circuits connect to the brain, opn5 mRNA is distributed across multiple retinal cell types, including bipolar (~70%–75%), amacrine (~10%), and retinal ganglion (~20%) cells, with opn8 present in amacrine (~70%) and retinal ganglion (~30%) cells. opn6a and opn6b mRNAs emerge in newborn-photoreceptors (stage 35), and are colocalized in rods and cones by stage 37/38. Interestingly, in the mature larval retina (stage 43/44), opn6a and opn6b mRNAs become preferentially localized to rods and cones, respectively, while newborn photoreceptors bordering the proliferative ciliary marginal zone express both genes. In zebrafish, opn6a and opn6b are also expressed in photoreceptors, while Müller glia and amacrine cells express opn8c. Most neuropsin-expressing retinal ganglion cells display c-fos expression in response to light, as do over half of the neuropsin-expressing interneurons. This study gave a better understanding of retinal neuropsin-expressing cells, their developmental onset, and light activation.
Introduction
Opsins are photosensitive proteins that capture and decode light information for visual and non-visual tasks. In the retina, the function and physiology of classical opsins [rhodopsin (RHO) and cone-opsins] and melanopsin (opn4) are well known (Thoreson and Dacey, 2019). Less understood, however, is the physiological role of the novel neuropsin family (opn5, 6, 7, 8, and 9). opn5 is well studied as the only neuropsin conserved in the mammalian lineage, while opn6, 7 and 8 are present in non-mammalian tetrapods and teleost fish, and opn9 is conserved only in teleost (Davies et al., 2015; Beaudry et al., 2017). The pressure selection associated with the evolutionary conservation of neuropsins is unknown. Additionally, in the retina when neuropsin expression initiates developmentally the cell types that express the different neuropsins, and when these cells become integrated into light-activated circuits is poorly understood. This knowledge is important to our understanding of neuropsin physiology and the evolutionary processes which favoured particular neuropsins over others in the vertebrate retina.
The vertebrate retina detects light and converts it to neural signals that are sent to the brain to allow the organism to visualize its environment and regulate biological rhythms (Díaz et al., 2016; Thoreson and Dacey, 2019). Common to the retina of all vertebrates are five specialized types of neurons organized in three layers: (i) photoreceptors (rods and cones) located in the outer nuclear layer (ONL); (ii) horizontal (HC), bipolar (BC) and amacrine (AC) cells in the inner nuclear layer (INL); and (iii) retinal ganglion cells (RGCs) in the ganglion cell layer (GCL) (Lamb, 2013). Light entering the eye stimulates photoreceptors that then transmit electrical signals to RGCs through BCs. RGC axons exit the eye as the optic nerve and terminate at downstream brain targets. HC and AC interneurons modulate the neural signals within the retina. Muller glia (MG) are the predominant macroglial cells of the vertebrate neural retina and serve mainly in retinal support (Morera et al., 2016; Hoang et al., 2020).
Previous expression analyses localize neuropsins to particular retinal laminae (Davies et al., 2015), with only a few studies identifying the specific cell types that express the different neuropsins (Yamashita et al., 2010; Rios et al., 2019; D’Souza et al., 2022). In mouse, a cre recombinant system shows that opn5 is expressed exclusively in RGCs (Calligaro et al., 2022; D’Souza et al., 2022), where it regulates retinal photoentrainment (Buhr et al., 2015). In birds, however, opn5m (“m” for mammalian-like) mRNA is additionally present in INL cells during development and after hatching (Rios et al., 2019; Calligaro et al., 2022). In fish and amphibians, opn5 is expressed mainly in the inner portion of the INL, with a small subset of opn5+ cells located in the GCL (Davies et al., 2015; Sato et al., 2016; Bertolesi et al., 2021). Less is known about the retinal expression of opn6, 7, 8 and 9 that do not have mammalian counterparts. In Xenopus laevis, opn6, a paralogue of opn5, underwent gene duplication to produce opn6a and opn6b. In the mature larval retina, we find both genes expressed in a handful of ONL cells that border the ciliary marginal zone (CMZ) (Bertolesi et al., 2021). These cells are likely newborn photoreceptors that arise from the CMZ, a proliferative neuroepithelium characteristic of fish and amphibians that provides new neurons for the retina over the life span of the organism (Perron and Harris, 1999). opn6 mRNA is expressed by photoreceptors of the adult zebrafish retina (Davies et al., 2015). The amphibian opn8 gene and its homologues in bony fish (opn8a, opn8b and opn8c) are detected in cells of the INL and GCL (Davies et al., 2015; Bertolesi et al., 2021), while opn9 is expressed by cells of the outer INL of the mature teleost (Davies et al., 2015). These findings suggest that the spatial localization of neuropsin expression may differ between vertebrate lineages and species.
X. laevis is an excellent model organism to study how the function of neuropsins within the retina may compare to those of other opsins. First, the neuropsins and other opsin genes are identified (Chang and Harris, 1998; Bertolesi et al., 2020). After accounting for the duplication of genes resulting from the allotetraploid nature of X. laevis, there exists 22 distinct type-II opsin genes (Session et al., 2016; Bertolesi et al., 2020). Of the type-II opsins, the neuropsin family includes opn5, opn6a, opn6b, opn7a, opn7b and opn8 (Bertolesi et al., 2020). The opn7 genes, however, exhibit little or no expression in the retina at stage 43/44 (Bertolesi et al., 2021). Second, retinal neurogenesis and lamination, and the onset of expression of the classical opsins (rhodopsin and cone opsins) and melanopsins (opn4 and opn4a; a.k.a. as opn4m and opn4x for mammalian- and Xenopus-like, respectively) are characterized in the developing Xenopus retina (Table 1). The expression of the classical opsins in the mature larval Xenopus retina (stage 43/44 tadpoles, mature, functional retina) and in adults is also known. For example, reports have defined both the distribution of the classical opsins in cones and rods in the ONL, and of opn4/a in HCs and RGCs (Table 1) (Chang and Harris, 1998; Gábriel and Wilhelm, 2001; Bertolesi et al., 2014).
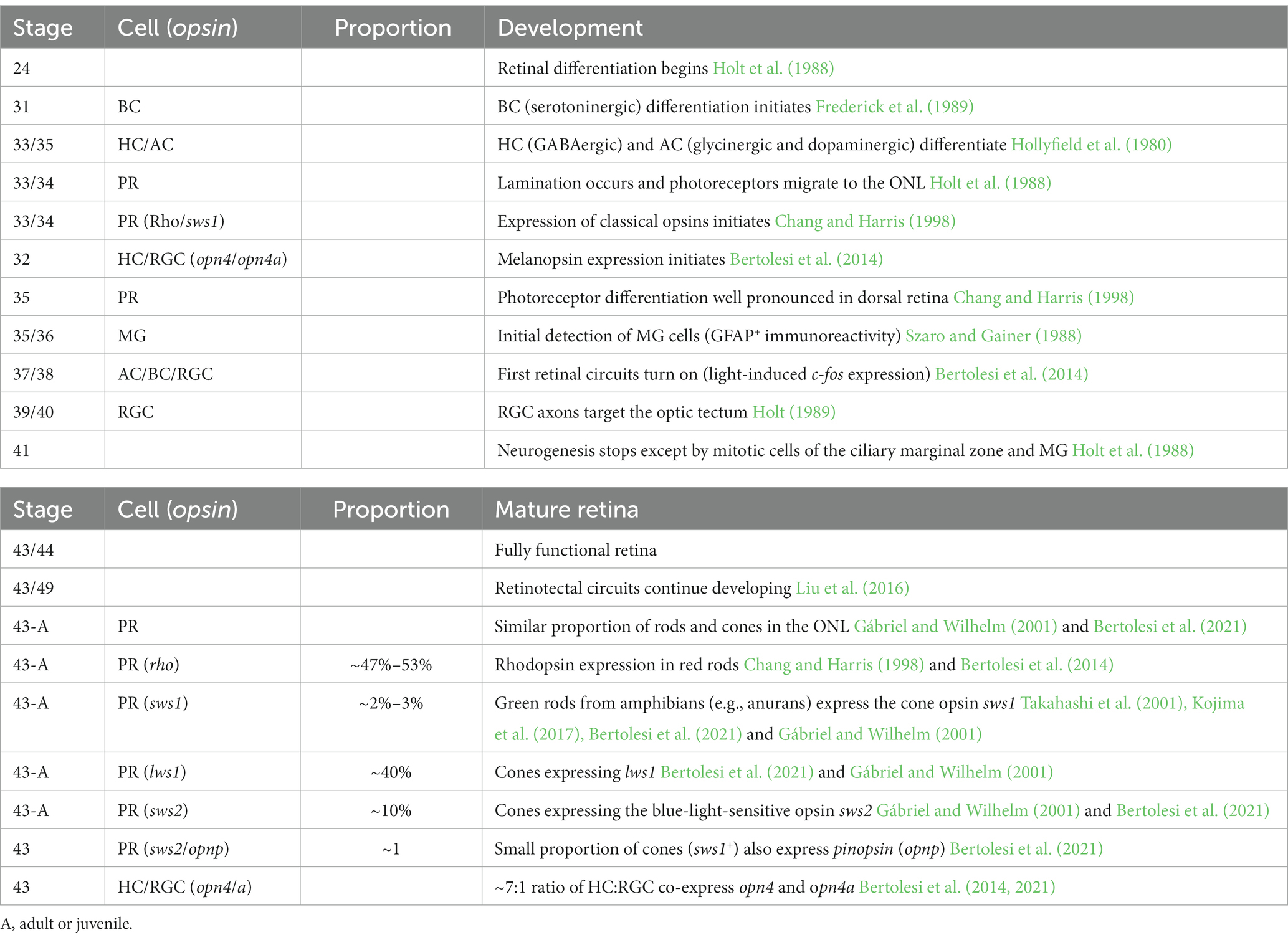
Table 1. Timeline of eye development, onset of “classical” opsin and melanopsins and opsin expression/distribution in the Xenopus laevis “mature” retina.
Here, we determined when neuropsin expression begins in the embryonic X. laevis retina and identified the cell types involved. Additionally, we investigated co-expression between neuropsins and opn4, and the activation of neuropsin-expressing cells in light-stimulated circuitry by looking at expression of the immediate early gene marker c-fos (Yu et al., 1994; Huerta et al., 1997). Finally, we compared our findings in X. laevis to other species with publicly available whole-eye single-cell RNA-sequencing (scRNA-seq) data. For Xenopus, this powerful technique was used to study gene expression and cell type diversity during embryogenesis (Briggs et al., 2018; Sonam et al., 2022), though retinal data is not yet publicly available. Gene expression, however, was recently analyzed for adult zebrafish, chicken and mouse retinas (Hoang et al., 2020). We compared our data from larval X. laevis with that of a scRNA-seq dataset from the adult zebrafish retina (Hoang et al., 2020) to assess differences in neuropsin expression between vertebrate lineages. Our results contribute to the understanding of retinal neuropsins in two novel aspects: (1) by comparing with the known expression of opsins in the Xenopus retina, we provide insight into neuropsin physiology. Specifically, we describe new photoreceptor cell types and neuropsin-expressing cells in the INL and GCL, the developmental time-course of expression, and whether expressing cells are integrated into light-activated circuits; (2) by comparing neuropsin expression between different species we provide insight into evolutionary differences between neuropsins.
Materials and methods
Embryos
All procedures involving frogs and tadpole embryos were approved by the Animal Care and Use Committee at the University of Calgary. X. laevis egg production was induced by chorionic gonadotrophin injection (Intervet Canada Ltd.) and in vitro fertilization in accordance with standard procedures (see protocols at www.xenbase.org/ RRID:SCR_003280). Embryos were stored at 16°C or 22°C and were kept on a 12:12 h cycle of light and dark. For light exposure experiments, embryos were kept in the dark from stage 24 until they reached stage 43 and were exposed to 30 min of white light (approximately 800 lux) immediately before fixing. Embryo stages were determined using Nieuwkoop and Faber’s developmental data for X. laevis stages on Xenbase.1
Single RNA in-situ hybridization
Embryos at desired stages were collected and fixed in MEMFA [(0.1 M 3-N-morpholino propanesulfonic acid (MOPS), 2 mM ethylene glycol tetra-acetic acid (EGTA), 1 mM MgSO4, and 4% formaldehyde in diethylpyrocarbonate (DEPC)-treated H2O)] at 4°C. After 24 h, embryos were placed in 30% sucrose in DEPC water until sunken and then frozen in optimal cutting temperature (OCT) compound (Tissue Tek, Sakura Finetek Inc., United States) and stored at −80°C. OCT-embedded embryos underwent cryostat sectioning (12 μm thick sections) for slide in situ hybridization (ISH). Sense and antisense riboprobes were synthesized from linearized plasmids containing the partial sequences of X. laevis opn4a, opn5, opn6a, opn6b, opn8, and c-fos (GenBank access numbers: MN820837, MN820850, MN820855, MN820856, MN820855 and BC079689.1, respectively). SP6 and T7 RNA polymerases (Roche, Quebec, Canada) with digoxigenin (DIG)-labeled nucleotides (Roche) were used to generate the riboprobes. Riboprobes were hybridized at 65°C and detected with anti-DIG antibodies (1:2500, Roche) conjugated to alkaline phosphatase, and the signal developed with BCIP (Roche; 5-bromo-4-chloro-3-indolyl phosphate) and NBT (nitro blue tetrazolium chloride; Roche) in a NTMT solution (100 mM NaCl, 100 mM Tris-Cl pH 9.5, 50 mM MgCl2, and 1% Tween 20).
Double fluorescent in-situ hybridization (FISH)
DIG- and fluorescein isothiocyanate (FITC)-labeled probes were synthesized and detected by using anti-DIG (1:500 dilution) and anti-FITC (1:500 dilution) specific antibodies coupled to horse radish peroxidase. The signal was developed with tyramide signal amplification (TSA) cyanine 3 or TSA fluorescein system kits, respectively (both Perkin Elmer, United States), following the manufacturer’s protocol.
Immunohistochemistry post fluorescent in-situ hybridization
DIG-labeled probes developed with TSA cyanine 3 were used for single FISH of retina sections. Following single FISH, immunohistochemistry was performed by incubating the slides with antibodies for Otx2 (rabbit polyclonal 1:200; ab21990, Abcam), Pax6 (rabbit polyclonal 1:100, Cedarlane), Rhodopsin (clone 4D2; mouse 1:200; Millipore) or Calbindin (rabbit polyclonal 1:200; Swant) for 1 hour. Alexa Fluor-tagged secondary antibodies (1:1000, green, 488 nm excitation, anti-mouse or anti-rabbit, Molecular Probes) were used to identify the primary antibody. DAPI staining after ISH and immunohistochemistry labeled cell nuclei and guided cell counting.
Microscopy
For single RNA ISH, bright field optics on a Zeiss compound microscope were used to examine slides through a 20× or 40× objective. Digital images were obtained with Axiovision software. A Zeiss LSM 900 confocal laser scanning microscope was used to image both FISH followed by immunohistochemistry, and double FISH. Z-stacks of digital images (1 μm thick) were generated by using Zen Blue software (Zeiss) on the confocal microscope. A minimum of 8 embryos (N) were analyzed for single ISH, FISH, double FISH, or FISH followed by immunohistochemistry.
Single cell RNA sequencing analysis
Publicly available single cell RNA sequencing (scRNA-seq) whole eye datasets for the adult zebrafish (Danio rerio), chicken (P10) (Gallus gallus) and mouse (P60) (Mus musculus) were downloaded from GitHub at https://github.com/jiewwwang/Single-cell-retinal-regeneration (Hoang et al., 2020). For each species, RStudio (R version 4.3.0) and Seurat V3 were used to generate a Seurat object. All metadata, including cell annotations and sample identification, were preserved from the original study. The Seurat objects were subset to include only control (uninjured) retinae. Quality control was performed to include only cells with between 500 and 2,500 gene features (standard values) and less than 15% mitochondrial genes. A standard Seurat integration pipeline with the “vst” selection method and 2000 variable features was performed for each species. Integrated data were scaled, and an unbiased principal component analysis was performed. RunUMAP was performed with 19 dimensions determined by JackStraw statistics. Original annotation was re-evaluated and confirmed by examining the expression of common cell type markers (Supplementary Table S1) in UMAP clusters indicated by the “FindAllMarkers” function in Seurat.
Neuropsin expression in distinct clusters was evaluated using a custom R script which parses through a gene count matrix to find the number of cells of a particular cell type that express a gene at or above an expression level of one UMI. A list of genes was provided to the script which were then iteratively analyzed, and a data-frame was generated for all results. Github repository including all information pertaining to the downloading and instructions for how to use the function of the “blackshaw” dataset is linked to the following website https://github.com/s-storey/opn_expression/tree/main.
Cell counting and analysis
Digital images from confocal microscopy that captured expression in single transverse central retinal sections containing the optic nerve were collected for analysis of neuropsin-positive cells in the retina. If the optic nerve section was not preserved, an adjacent anterior or posterior section was used to count cells The number of retinal slices analyzed (n) and the mean with 95% confidence interval are indicated in the corresponding figures. Immunohistochemical analysis were performed at less two times (N ≥ 2) containing a minimum of 4 independent tadpoles in each experiment. The number of central retina sections analyzed (n) is indicated in figure legends. GraphPad Prism software (v.9.1.2, GraphPad Software, San Diego, CA, United States) was used for statistical analysis.
Results
Developmental expression of neuropsins in the Xenopus retina
The onset of neuropsin expression in the retina is unknown. We analyzed the developmental expression of retinal neuropsins found in X. laevis: opn5, opn6a, opn6b and opn8 (Bertolesi et al., 2020, 2021). Neuronal differentiation initiates at stage 24 (1 day of development), classical opsins are detected a day later at stage 35, and the RGC axons target the optic tectum by stage 39/40 (3 days of development) (Table 1). Two opn5 genes exist in X. laevis. These genes likely originated from the X. laevis specific tetraploid in that both localize to homologous chromosome 5; opn5.L (gene number 100500792) and opn5.S (gene number 100500792) located on homologous chromosomes “long” and “short,” respectively, which correspond to the sub-genomes (2n each) provided by the distinct progenitors that gave rise to the tetraploid. ISH on 12 μm retinal sections using an opn5.L riboprobe (1,222 bp length; GenBank number: MN820850) revealed a specific, yet sparse, expression pattern first detected at stage 37/38 (Figure 1A), when the retinal circuits initially become light-activated (Bertolesi et al., 2014). At stage 41, when the retinal-brain circuits are functional, opn5 expression was strongest in cells located in the outer region of the INL (Figure 1A). By stage 43, opn5 was expressed sparsely in the INL, and several cells (~20% of total opn5+ cells) were present in the GCL (Figure 2). Of note, the opn5.L probe possessed 91% identity to opn5.S (GenBank number XM_018264817.1), therefore expression of both opn5 genes was likely detected. opn5 expression in the INL and GCL in X. laevis resembles the pattern reported previously in post-hatching chick retina (Yamashita et al., 2010), but differs from mammals, where opn5 expression appears restricted to the GCL (Calligaro et al., 2022; D’Souza et al., 2022).
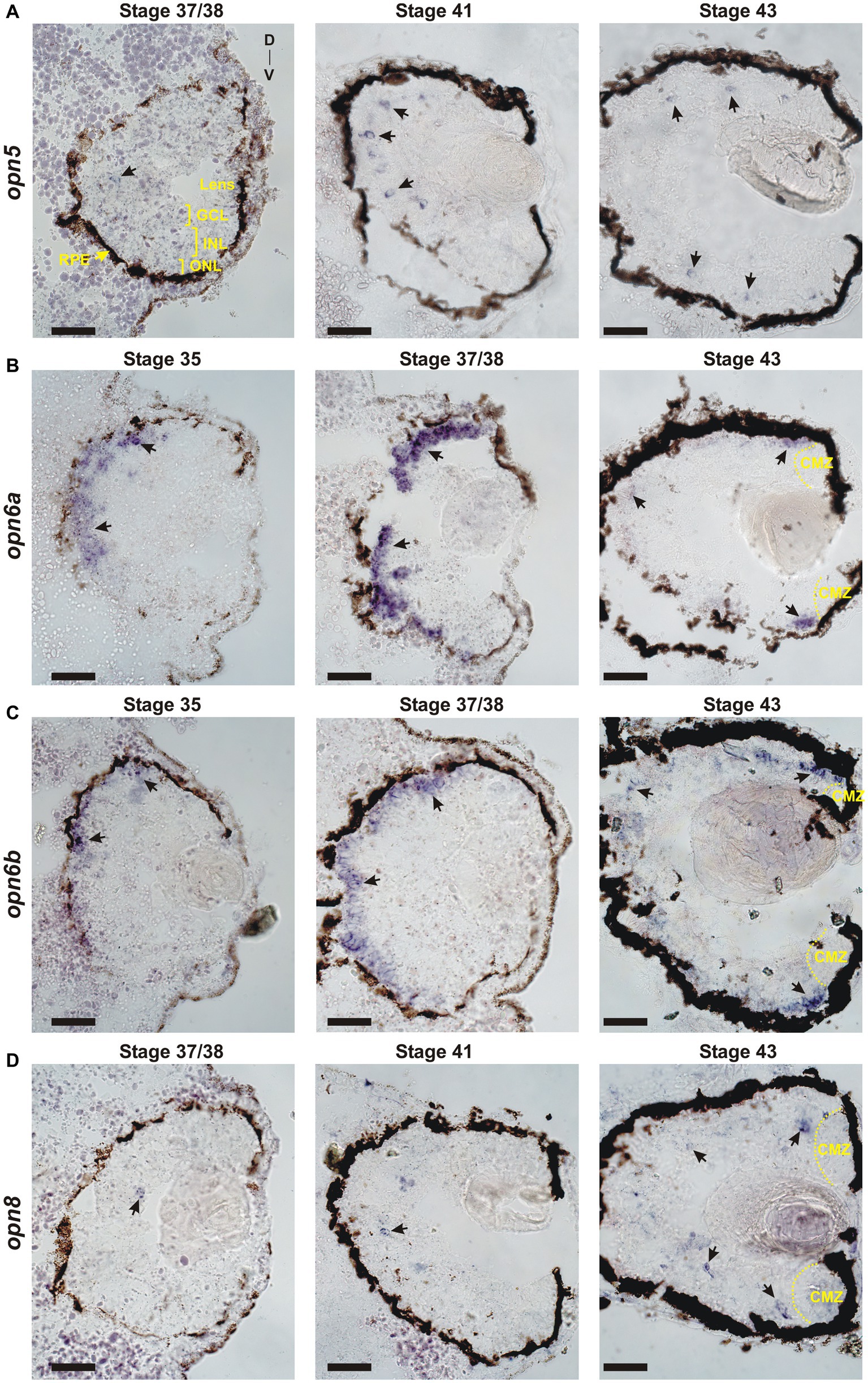
Figure 1. Developmental expression of neuropsins. ISH with specific antisense riboprobes against opn5 (A), opn6a (B), opn6b (C) and opn8 (D) on transverse retinal sections at stages 35, 37/38, 41 and 43. Black arrows point to neuropsin-expressing cells. CMZ, ciliary marginal zone; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium; D, dorsal; V, ventral. Scale bar = 100 μm.
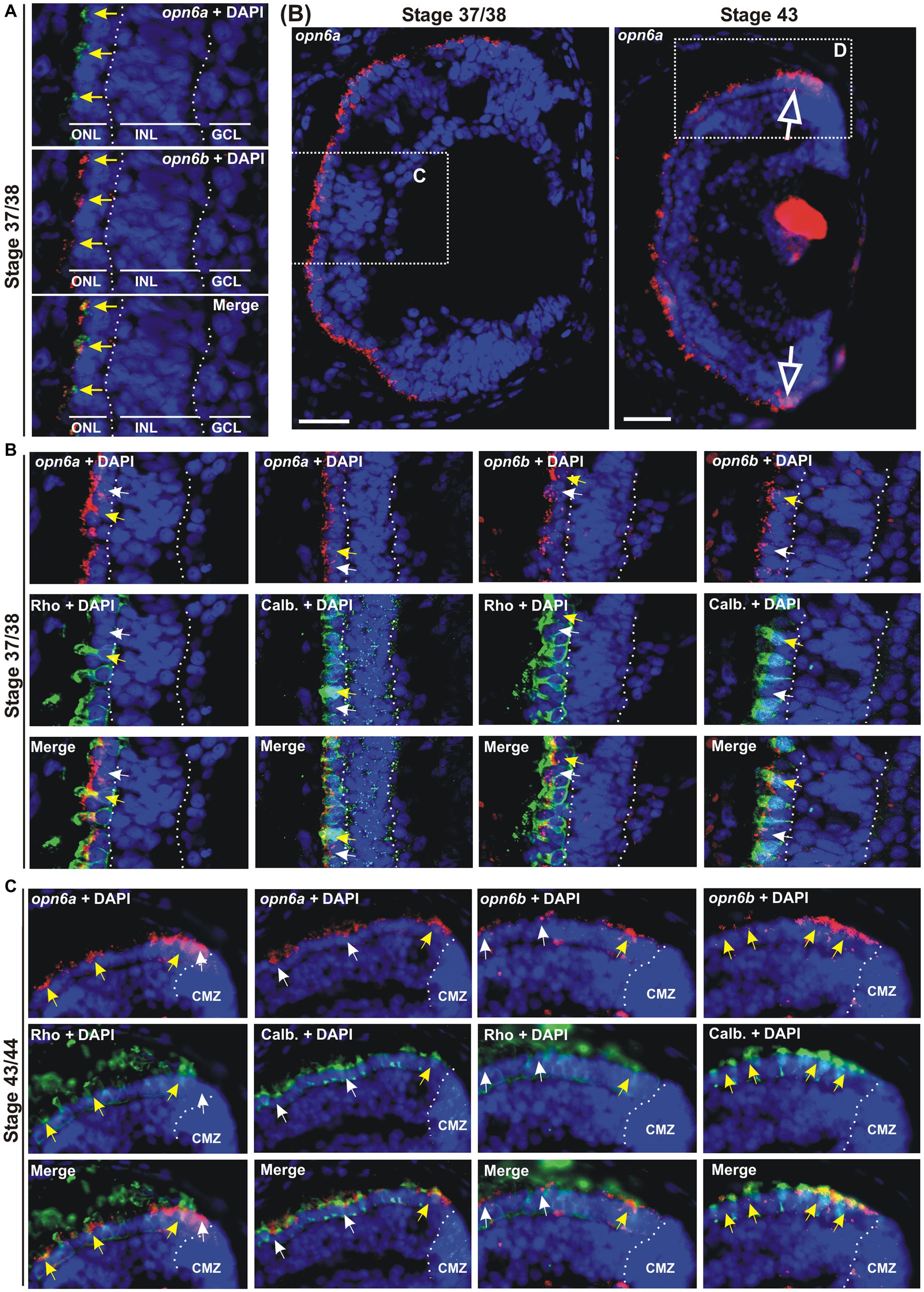
Figure 2. opn6a and opn6b colocalize in newly born photoreceptors at stage 37/38 but are differentially expressed in differentiated rods and cones at stage 43/44. (A–D) Fluorescent ISH with specific riboprobes against opn6a or opn6b on stage 37/38 (A,C) and 43/44 Xenopus laevis transverse retinal sections (B,D), followed by immunohistochemistry against a rod marker (rhodopsin; green) or a cone marker (calbindin; green) (C,D). DAPI (blue) stains nuclei. (A) Yellow arrows point to ONL cells that express both opn6a and opn6b. (B) Higher magnification of the dotted-line boxes are shown in bottom panels (C,D). The open white arrows point to opn6a+ cells that are adjacent to the ciliary marginal zone (CMZ). (C,D) White small arrows point to opn6a or opn6b expressing cells that are either not rhodopsin+ or not calbindin+, while yellow arrows indicate opn6a+ or opn6b+ cells that are also positive for rhodopsin or calbindin. GLC, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar in (B): 100 μm.
We previously identified three opn6 genes in X. laevis that are phylogenetically related, and that we named opn6a.L, opn6a.S, and opn6b.L (Bertolesi et al., 2020). Two homologous genes exist for opn6a, located on the homologous chromosomes 5 L and 5 S (opn6a.L, gene number 108716317 and opn6a.S, gene number 108717820), while opn6b.L (gene number 108698982) is located on chromosome 8 L as a single copy. opn6a and opn6b are not homologous genes resulting from X. laevis tetraploidy. Instead opn6b.L was potentially generated by inter-chromosomal transposition. The probe generated to detect opn6a (972 bp) was produced from the opn6a.L template (GenBank number MN820855) and contained 94% identity with the predicted opn6a.S transcript sequence. Therefore, the opn6a probe likely recognized both opn6a.L and opn6a.S homologous transcripts. In contrast, no significant similarity was found between the recognition sequence of the opn6a.L probe and that of opn6b.L (gene number 108698982). Furthermore, the sequence recognized by the opn6b probe (1,047 bp length) showed no significant similarity to opn6a.L or opn6a.S. Of note, at the time of this publication, opn6b.L is classified as an opn5 gene in the NCBI database and should be renamed.
opn6a expression was first detected at stage 35 as diffuse staining, mainly in cells of the dorsal-most outer layer of the retina, with a few positive cells located in the inner portion of the retina (Figure 1B). By stage 37/38, opn6a was expressed strongly by cells of the ONL (Figure 1B). opn6a was present in the ONL by stage 43, but apparently in a smaller number of cells than at stage 37/38, with the strongest expression observed in cells that border the CMZ (Figures 1B, 2B). The CMZ-associated expression at stage 43/44 is consistent with our previous data suggesting that opn6a is robustly expressed in a transient manner in newborn photoreceptors (Bertolesi et al., 2021). Interestingly, the opn6b expression was similar to opn6a, re-enforcing the idea that opn6b is related to opn6a and not to opn5. opn6b expression was first detected at stage 35, and most strongly expressed at stage 37/38 in the ONL. Fewer cells expressed opn6b mRNA in the ONL by stage 43, with the strongest label located at the CMZ border (Figure 1C). Interestingly, the developmental time-course of opn6a/b expression at stage 35/36 and 37/38 matches the characterized processes of specification, migration, and differentiation of X. laevis cone and rod photoreceptors (Table 1) (Chang and Harris, 1998). The decrease in expression seen in maturing photoreceptors integrated into functional retinal circuits (stages 41 and 43/44), however, is different from what is observed for the classical opsins that continue to be highly expressed with ongoing larval development (Chang and Harris, 1998).
We detected three related opn7 genes in Xenopus laevis: opn7a.L and opn7b.L and opn7b.S (gene numbers 108718575, 108700890 and 108702595, respectively). opn7b.L and opn7b.S are homologous genes located on chromosomes 9–10 L and S, respectively, while opn7a.L is located on chromosome 6 L (Bertolesi et al., 2020). mRNA for these genes is almost undetectable in the eye at stage 43/44 (Bertolesi et al., 2021), and therefore we did not analyze them here.
Two opn8 genes (opn8.L; gene number 108717224 and opn8.S; gene number 108718281) are located on chromosome 5 L and 5 S, respectively. Expression of the opn8 genes was analyzed by ISH using a probe generated against opn8.L (904 bp length; GenBank number MN820854; with 91% identity to opn8.S). opn8 expression was first detected at stage 37/38 in a few cells located in the INL. opn8 remained sparsely expressed in the INL, GCL and in cells bordering the CMZ with ongoing development (stage 41 and 43) (Figure 1D).
Together, our results show a specific pattern of neuropsin expression in the retina. opn6a and opn6b are expressed by photoreceptors in the ONL, with expression beginning about the same time as the classical opsins at ~stage 35. With further development, however, opn6a and opn6b expression diminishes in fully differentiated photoreceptors, but is present likely in newborn photoreceptors that arise from the CMZ. The expression of opn5 and opn8 turns on slightly later at stage 37/38. In the mature larval retina (stage 43), opn5 and opn8 are both expressed mainly in cells located in the INL, with the remainder of cells in the GCL.
opn6a and opn6b mRNAs initially colocalize in rods and cones, but in the maturing larval retina become differentially expressed by rods and cones, respectively
Next, we identified the specific cell types that express the different neuropsins. We started with the analysis of which photoreceptors in the ONL express opn6. Our ISH data show strong opn6a and opn6b expression at stage 37/38. Thus, we chose this stage to determine whether these two opsins are co-expressed and in which photoreceptor classes. Co-expression was determined by double fluorescent ISH (dFISH). At stage 37/38, most photoreceptors appeared to express both opn6a and opn6b (Figure 2A). Cell types were assigned through FISH, followed by immunohistochemistry either against the known rod marker, rhodopsin, or calbindin, a calcium binding protein expressed by cones (Chang and Harris, 1998). Initially, opn6a and opn6b were expressed by both rhodopsin+ and calbindin+ cells, with the fluorescent signal commonly localizing to the inner segments of photoreceptors as a cluster of fluorescent dots (Figures 2B,C). However, by stage 43/44, once retinal circuits are functional and connected to the brain, opn6a mRNA was found preferentially in differentiated rods (rhodopsin+), and in only a few differentiated cones (calbindin+) (Figures 2B,D). In the opposite manner, opn6b mRNA was found preferentially in differentiated calbindin+ cones and only a few differentiated rhodopsin+ rods (Figure 2D). Interestingly, for the newly born photoreceptors located proximal to the CMZ opn6 opsins were expressed in both rhodopsin+ and calbindin+ cells, similar to what was observed in the central retina at a younger developmental stage (stage 37/38) (Figure 2D).
We find opn6a and opn6b are co-expressed by all newly generated photoreceptors shortly after their birth, either in the central embryonic retina (stage 37/38) or those generated subsequently by the CMZ (stage 43). With differentiation in the central retina, however, opn6a and opn6b become selectively expressed in rod and cone photoreceptors, respectively.
opn5 is expressed by bipolar and amacrine cells, opn8 is expressed solely by amacrine cells, and Pax6+ RGCs express both opsins
Next, we identified the cell types that express opn5 and opn8 in the INL and GCL. The ISH data indicated that by stage 41, when retinal differentiation and circuity are defined (Table 1), opn5 and opn8 are mainly expressed in the INL and a subset of cells in the GCL. To determine the cell types located in the INL, we performed FISH followed by immunohistochemistry against the transcription factors Otx2 and Pax6 that in the INL label BCs (Viczian et al., 2003) and ACs (Hirsch and Harris, 1997), respectively.
opn5 mRNA was seen by FISH as clusters of subcellular red fluorescent dots that were either nuclear or associated with the nuclear membrane (Figure 3A). Most of the opn5+cells were in the outer portion of the INL (Figures 1, 3A). Approximately 70%–75% of the opn5+ cells were BCs, as they colocalized with Otx2 (75.25%, 149 opn5+ Otx2+ cells/198 opn5+ cells; n = 6 retinal sections) (Figures 3A,B). The remaining populations (24.74%, 49 opn5+ Otx2− cells/198 opn5+ cells) were distributed either in the inner part of the INL or the GCL (Figures 3A,B). We found that these cells were Pax6+ (Figure 3C); the opn5+ Pax6+ cells in the INL were ACs (10.8%, 21 cells opn5+ Pax6+ in the INL/194 opn5+cells; Figure 3D), and the remaining were located in the GCL (21.13%, 41 cells opn5+ Pax6+ in the GCL/194 opn5+cells, Figure 3E).
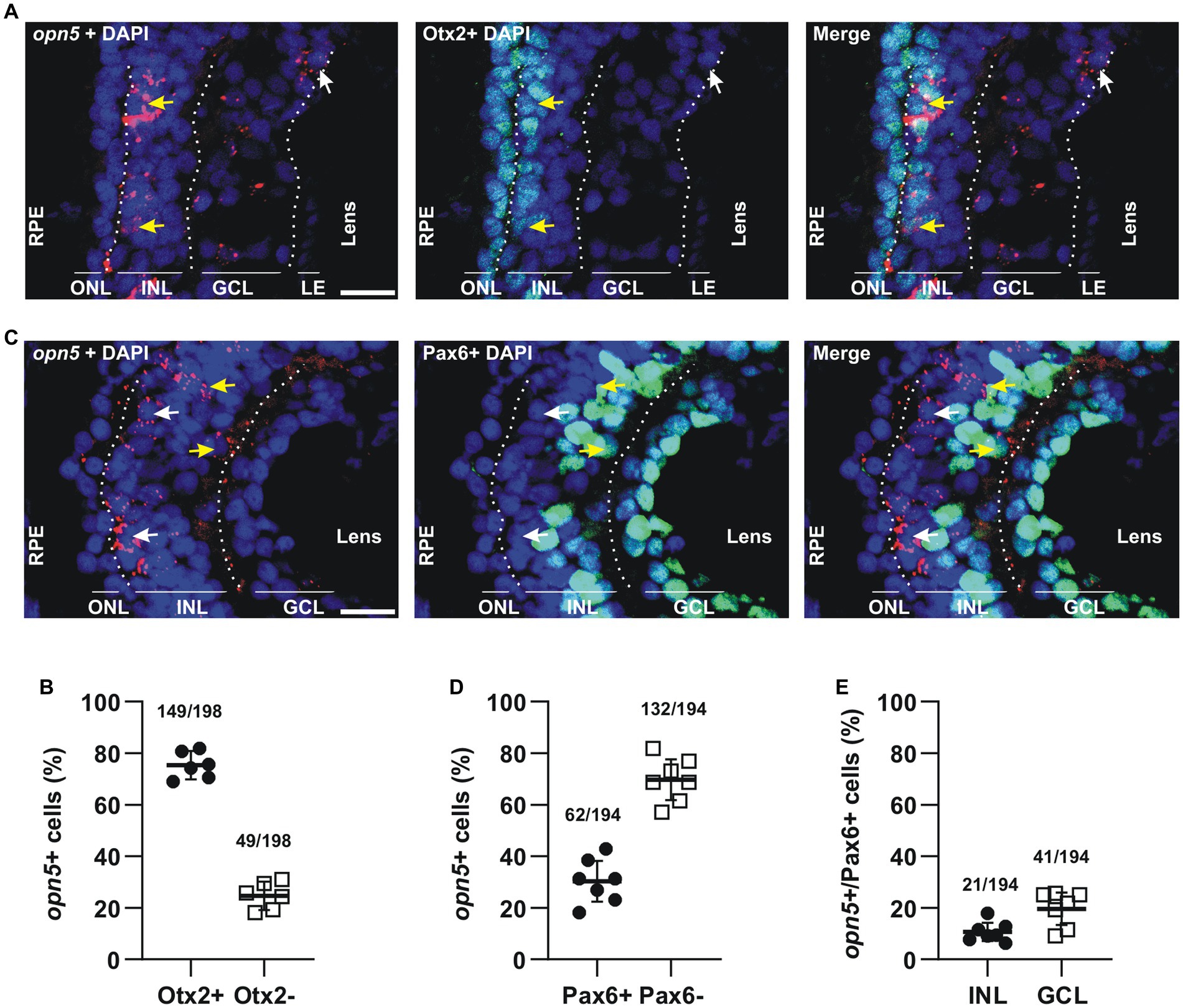
Figure 3. opn5 is expressed in bipolar and amacrine cells in the INL and in RGCs. (A,C) FISH against opn5 (red) followed by immunohistochemistry against a bipolar cell marker (Otx2; green) (A) or an amacrine and retinal ganglion cell marker (Pax6; green) (C), on stage 41 Xenopus laevis transverse retinal sections. DAPI (blue) stains the nucleus. ONL (outer nuclear layer), INL (inner nuclear layer), and GCL (ganglion cell layer) are indicated between dotted lines. White arrows point to opn5-expressing cells that are either Otx2− or Pax6−, while yellow arrows indicate opn5+ cells that are also Otx2+ or Pax6+. RPE, retinal pigment epithelium. Scale bar = 50 μm. (B,D,E) Graphs showing percentage of opn5+ cells that are Otx2+ (B) or Pax6+ (D), and the distribution of Pax6+/opn5+cells between the INL and GCL of stage 41 retina (E). Each dot represents the percentage of cells in one central retina section [(B), n = 6 sections; (D,E), n = 7 sections]. Lines are the mean with 95% confidence interval.
For opn8-expressing cells, we used immunohistochemistry against Pax6 after FISH at stage 41, when opn8 is expressed in both the INL and the GCL. Like opn5, opn8 expression was seen by FISH as subcellular red fluorescent dots that were either nuclear localized or associated with a nuclear membrane (Figure 4A). opn8 was expressed by only a few retinal cells per section. These cells were all Pax6+ and were found in either the AC-populated inner portion of the INL or the GCL (Figures 4B,C). Thus, for the opn8+ cells, 70% were INL-located cells, likely ACs, and 30% were GCL-located cells (Figure 4B).
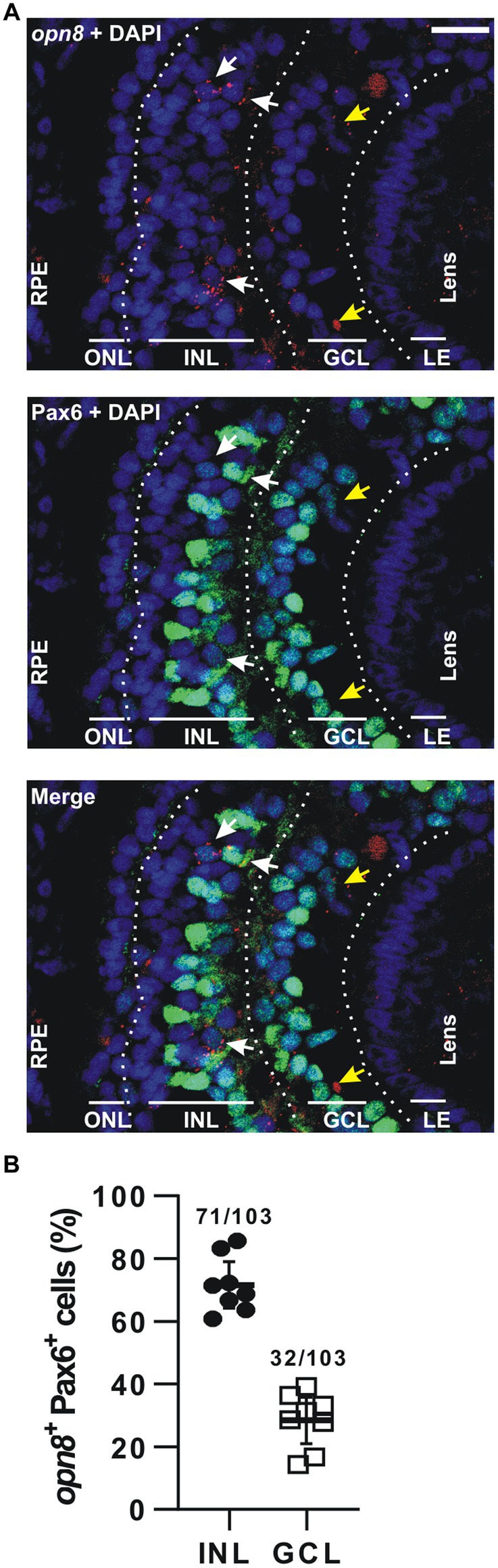
Figure 4. opn8 is expressed in Pax6+ cells in the INL and GCL. (A) Fluorescent ISH against opn8 (red) followed by immunohistochemistry against an amacrine and RGC marker (Pax6; green) on stage 41 Xenopus laevis transverse retinal section. DAPI (blue) stained the nucleus. ONL, INL, and GCL indicated with bars. White arrows point to opn8-expressing cells that are Pax6+ INL cells, while yellow arrows indicate Pax6+ GCL cells. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. Scale bar = 50 μm. (B) Distribution of opn8+/Pax6+ cells between the INL and the GCL of stage 41 retinas. Each dot represents the percentage of cells in one central retina section (n = 8 sections); Lines are the mean with 95% confidence interval.
Neuropsins and melanopsin show distinct and partially overlapping expression in retinal cells
The expression of opn5 and opn8 in Pax6+cells in the INL and the GCL led us investigate possible neuropsin co-expression by performing double ISH against opn5 and opn8 on stage 43/44 retinal sections. Interestingly, we found that opn5 and opn8 mRNAs did not colocalize in the INL (2% co-localization; 5 cells opn5+ opn8+/248 opn5+ cells; 52 opn8+ cells counted) and only a few cells expressed both genes in the GCL (17% co-localization; 8 cells opn5+ opn8+/47 opn5+ cells; n = 8 retinal sections) (Figures 5A,B).
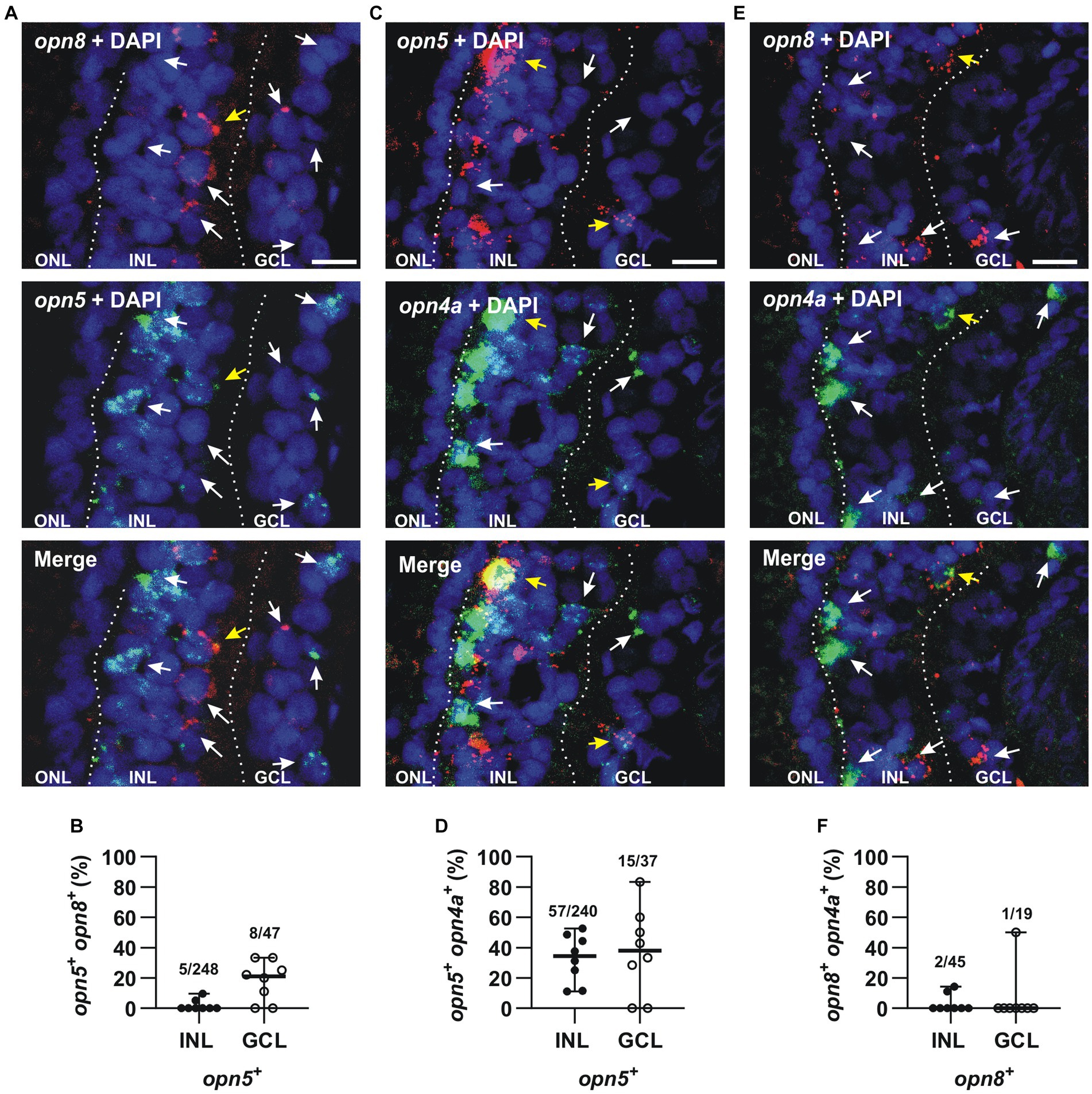
Figure 5. opn5 and opn8 sparsely co-express in the GCL but not INL, while opn4 and opn5 are partially co-expressed in both GCL and INL, and opn4 and opn8 do not co-express. (A,C,E) Double FISH against opn5 (green) and opn8 (red) (A), opn5 (red) and opn4a (green) (C), and opn8 (red) and opn4a (green) (E) on stage 43/44 X. laevis transverse retinal sections. DAPI (blue) stained the nucleus and is shown with the merged image. White arrows point to cells that express a single opsin, while yellow arrows point to cells that express two opsins. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar = 20 μm. (B,D,F) Graphs showing distribution between the INL and GCL of cells that co-express opn5 and opn8 (B), opn5 and opn4a (D) and opn8 and opn4a (F). Each dot represents the percentage of cells in one central retina section [n = 8 sections, (B,D,F)]. Lines are the mean with 95% confidence interval.
We also analyzed if neuropsins were co-expressed with melanopsin. In the Xenopus retina, opn4 and opn4a are co-expressed (91% colocalization) in a subset of HCs and RGCs (Bertolesi et al., 2014). Because of the high level of co-expression, we could use the opn4a antisense probe alone in order to determine both opn4 and opn4a colocalization with opn5 and opn8. Double FISH at stage 43/44 revealed that only 24% of opn5+ cells also expressed opn4a in the INL (57 opn5+ opn4a+cells/240 opn5+ cells), while 41% of cells expressed opn4a and opn5 in the GCL (15 opn5+ opn4a+ cells/37 opn5+cells; n = 8 retina central sections) (Figures 5C,D). In contrast, opn8 was not expressed by melanopsin+ cells of the INL or GCL (4% in the INL; 2 cells opn8+ opn4a+/45 opn8+ cells; n = 148 opn4a+cells counted; n = 8 retinal sections) (5% in the GCL; 1 cell opn8+ opn4a+/19 opn8+ cells; n = 19 opn4a+ cells counted in eight retinal sections) (Figures 5E,F).
Together, these results suggest that neuropsins are differentially expressed in distinct cells, with opn5 in BCs, ACs, and RGCs, and opn8 in ACs and in a different subset of RGCs. Additionally, mRGCs do not express opn8 and only a few express opn5. Thus, our results suggest that the neuropsins opn5 and opn8 allow for a greater total number of potentially photosensitive RGCs.
Light induces c-fos expression in opn5- and opn8-expressing cells
Next, we investigated whether the neuropsin-expressing cells participate in light-stimulated retinal circuits. To identify light-activated cells, we used c-fos, an immediate early gene marker expressed in neurons activated in response to synaptic stimuli (Yoshida et al., 1996; Huerta et al., 1997). We showed previously that several cells express c-fos after 30 min of white light illumination, particularly in the retina of dark-adapted X. laevis embryos (Bertolesi et al., 2014). Specifically, light-induced activation occurs in at least two cells of the INL (ACs and BCs) for each activated RGC. No c-fos expression is induced in photoreceptors (Bertolesi et al., 2014). Thus, we used double FISH to analyze at stage 43/44 the co-expression of c-fos with opn5 and opn8 upon light exposure.
We found that the majority of opn5-expressing cells in the INL (64%) expressed c-fos (n = 363 cells counted) after 30 min of light stimulation, as did almost all cells (87%) in the GCL (n = 71 cells counted) (Figures 6A,B). Similarly, 70% and 77% of opn8-expressing cells also expressed c-fos in the INL (n = 77 cells counted) and the GCL (n = 35 cells counted), respectively (Figures 6C,D). It is worth noting that without light stimulation, almost no c-fos expression was detected in the retina of dark-adapted embryos (Supplementary Figure S1 and Bertolesi et al., 2014).
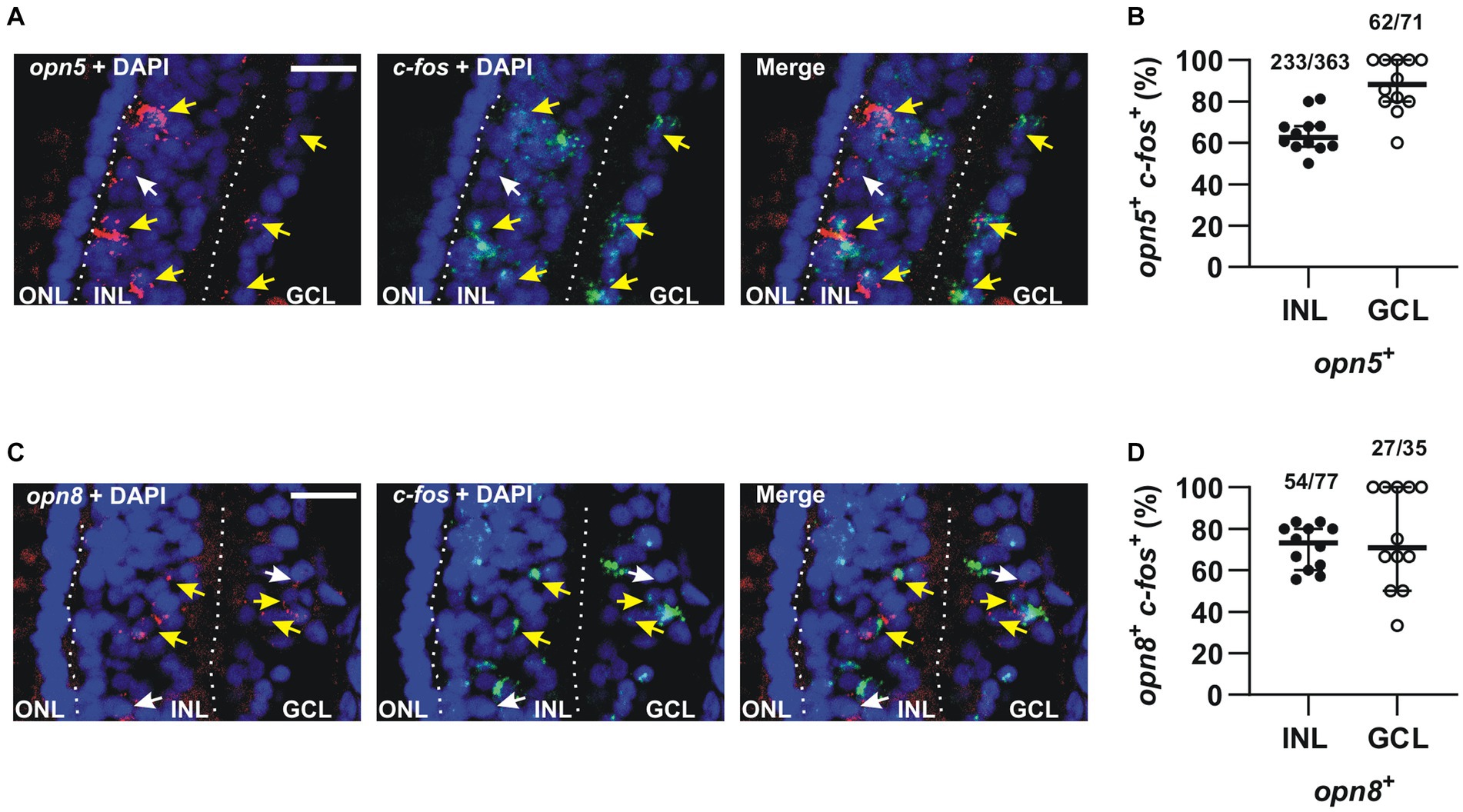
Figure 6. The majority of opn5+ and opn8+ cells were activated in response to light. (A,C) Double FISH against opn5 (red) and c-fos (green) (A) and opn8 (red) and c-fos (green) (C) on stage 43/44 Xenopus laevis transverse retinal sections after 30 min of light stimulation. DAPI (blue) stains the nucleus. White arrows point to opn5+ or opn8+ cells that do not express c-fos, while yellow arrows point to opn5+ or opn8+ cells that are also c-fos+. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar = 50 μm. (B,D) Quantification and distribution between the INL and GCL of cells co-expressing opn5 and c-fos (B) and opn8 and c-fos (D). Each dot represents the percentage of cells in one central retina section n = 12 sections; Lines are the mean with 95% confidence interval.
Together, our data show that the majority of opn5+ and opn8+ cells in the INL and GCL express c-fos after light stimulation. We also define a second group of opsin-expressing RGCs, in addition to the larger number of melanopsin-expressing cells in the GCL (Bertolesi et al., 2014), which are in light-activated circuits.
Expression of neuropsins in the retina of adult zebrafish
We next compared our data obtained in X. laevis to publicly available scRNA-seq datasets from the adult zebrafish, mouse, and chicken retina (Hoang et al., 2020). The datasets were generated recently to analyze gene expression of normal and injured retinas (Hoang et al., 2020). We isolated the scRNA-seq data of the control (uninjured) retina and analyzed the expression of neuropsins in different retinal cell-type clusters. opn5 was the only neuropsin gene detected in the chicken and mouse dataset, however, almost no cells showed expression (5 opn5+ AC and 1 opn5+ RGC of 8,895 chicken retinal cells; no opn5+ neuropsin gene of 14,891 mouse retinal cells). These results can be explained by a lack of evolutionary conservation of neuropsins, the small number of retinal cells in the chicken and mouse datasets, and low opn5 mRNA levels. In contrast, the zebrafish dataset contained more retinal cells (18,057 cells), and several neuropsins were detected (Figures 7A,B). Of note, some of the retinal cluster annotations present in the original dataset, such as GABAergic and glycinergic ACs as well as activated and resting MG cells, were grouped as ACs and MG cells, respectively, in our analysis.
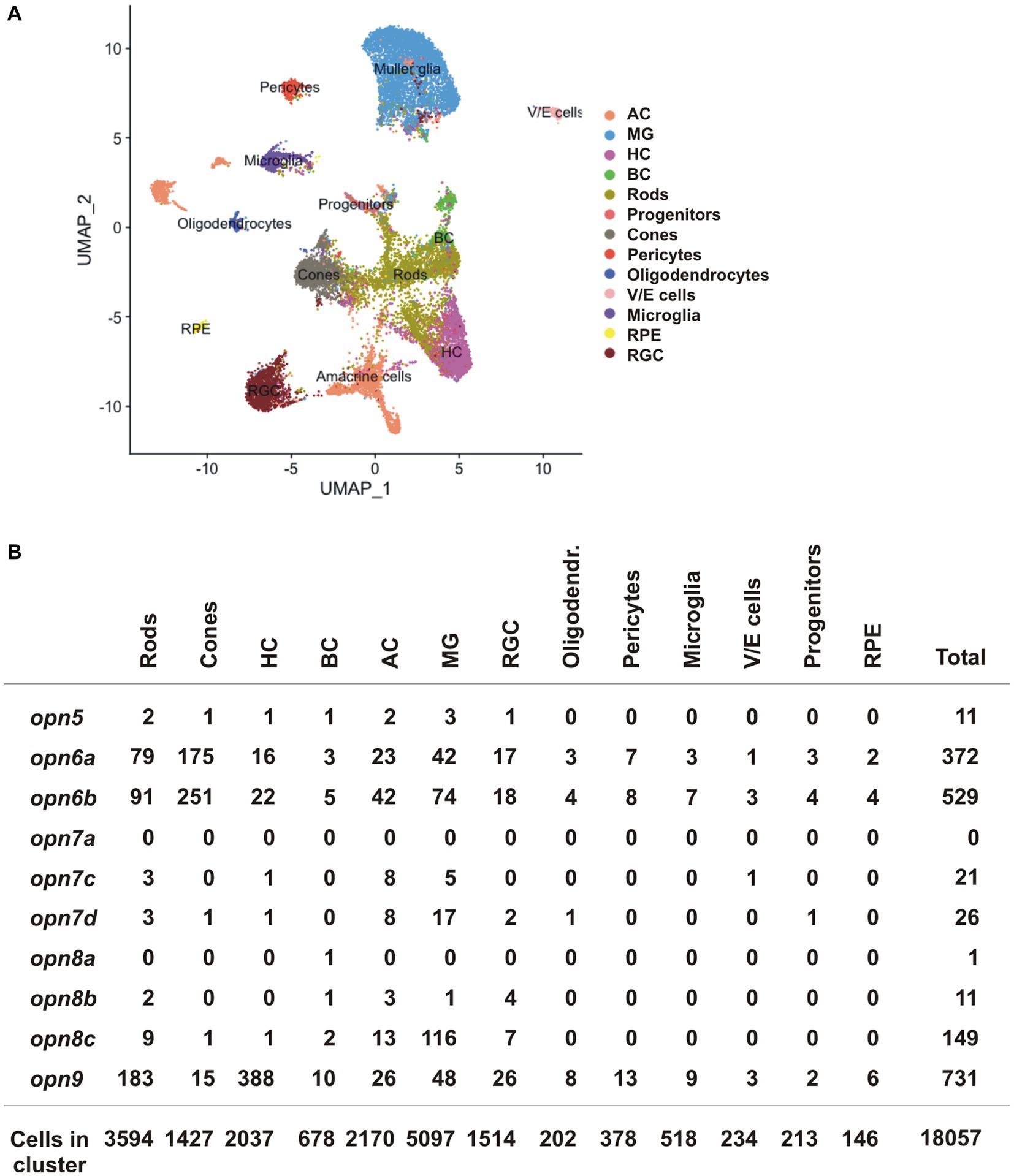
Figure 7. scRNA-seq data from the adult zebrafish (Hoang et al., 2020). (A) UMAP plot featuring annotated retinal cell types (Supplementary Table S1). (B) Quantification of cells which express neuropsin mRNA organized by cell type detected by scRNA-seq.
opn5 was detected in only a handful of cells in the adult zebrafish retina (11 opn5+ cells/18,057 total cells), spread between different cell types (rods, cones, BC, HC, AC and MG cells) (Figure 7B). In contrast, other neuropsins were expressed in more cells and the data was supportive of our results in X. laevis: (i) opn6a and opn6b were detected mainly in cones and rods [opn6a: 175/1427 cones (12.26%), 79/3,594 rods (2.2%); opn6b: 251/1,427 cones (17.6%); 91/3,594 rods (2.5%)]. Of note, zebrafish opn6b is homologous to opn6a generated during the teleost whole genome duplication and differs from the opn6b in X. laevis; (ii) the three opn7 genes were barely expressed in the retina; and (iii) opn8c was expressed mainly in MG cells (116 cells/5,097 MG, 2.27%) and ACs (13/2,170 cells, 0.6%). Additionally, opn9 that is present in teleosts but lost in tetrapods, was expressed in HCs (388/2037 cells, 19%), which agrees with findings from ISH performed in adult zebrafish (Davies et al., 2015).
Discussion
Here, we studied the expression of the neuropsin family members in the X. laevis retina and compared our results with scRNA-seq data from zebrafish, chicken, and mouse. We demonstrated that: (1) opn6a and opn6b expression initiates at stage 35, at the same time as the classical opsins and melanopsin (Figure 1 and Table 1). opn5 and opn8 mRNA induction occurs several hours later (stage 37), (2) opn6a and opn6b are the neuropsins of newly born photoreceptors (Figure 2). The number of cells expressing opn6a/b diminished as eye circuity and lamination progresses, such that, by stage 43/44 (mature retina), the strongest expression is within newly born photoreceptors that border the CMZ (Figure 2). The opn6 genes are also expressed in adult zebrafish in photoreceptors, but mRNA is not detected in chicken and mouse scRNA-seq data (Figure 7), (3) opn5 mRNA is present both in the INL in a subpopulation of BCs and ACs, and in a small proportion of cells in the GCL (Figure 3). Similarly, the chicken scRNA-seq data point to opn5 mRNA in ACs and RGCs (Figure 7), (4) opn8 is expressed only by Pax6+ cells of the INL and the GCL (Figure 4), while in zebrafish the two clusters with highest expression are MG and ACs (Figure 7), (5) opn8 shows little or no co-localization with either opn5 or opn4a, while opn5 shows moderate expression (~40%) in opn4a + RGCs (Figure 5), and (6) opn5- and opn8- expressing cells (>60%) induce c-fos expression and are part of retinal circuits that activate with a light stimulus (Figure 6).
Particularly interesting is the expression of opn6a and opn6b in what appears to be newly generated photoreceptors that emerge in the central retina near the end of embryogenesis, and subsequently are generated at the periphery of the mature retina by the progenitors of the CMZ. These data suggest that opn6 functions in early photoreceptor differentiation, at the same time that expression of rod and cone opsins become pronounced (stage 35/36) (Chang and Harris, 1998). Expression subsequently refines to specific in mature and differentiated photoreceptors of the central retina (Figure 2), while less differentiated photoreceptors express both opn6a (rods; Rho+) and opn6b (cones; Calbindin+) mRNAs.
In combination with known photoreceptor opsins, opn6a and opn6b add complexity to the classical photoreceptor subtypes located in the Xenopus ONL, with seven different types now possible (Figure 8): (1) red rods, Rhodopsin (Rho)-expressing cells at ~47%–50% of the total photoreceptors, (2) green rods, atypical photoreceptors (2%–3%) found in amphibians that express the short-wavelength-sensitive 2 opsin (sws2), a blue-sensitive opsin located in cells with a rod-shaped structure (Gábriel and Wilhelm, 2001; Kojima et al., 2017), (3) cones that express the long wavelength cone opsin (lws; ~40% of photoreceptors), (4) cones that express the short wavelength-sensitive 1 opsin (sws1; ~10% of photoreceptors) (Starace and Knox, 1998; Bertolesi et al., 2021), (5) a small proportion of cones (<1%) that express pinopsin (opnp) alongside sws1 (Bertolesi et al., 2021), (6) rods that co-express rho and opn6a, and (7) cones likely expressing lws, the most abundant Calbindin+ cell, and opn6b.
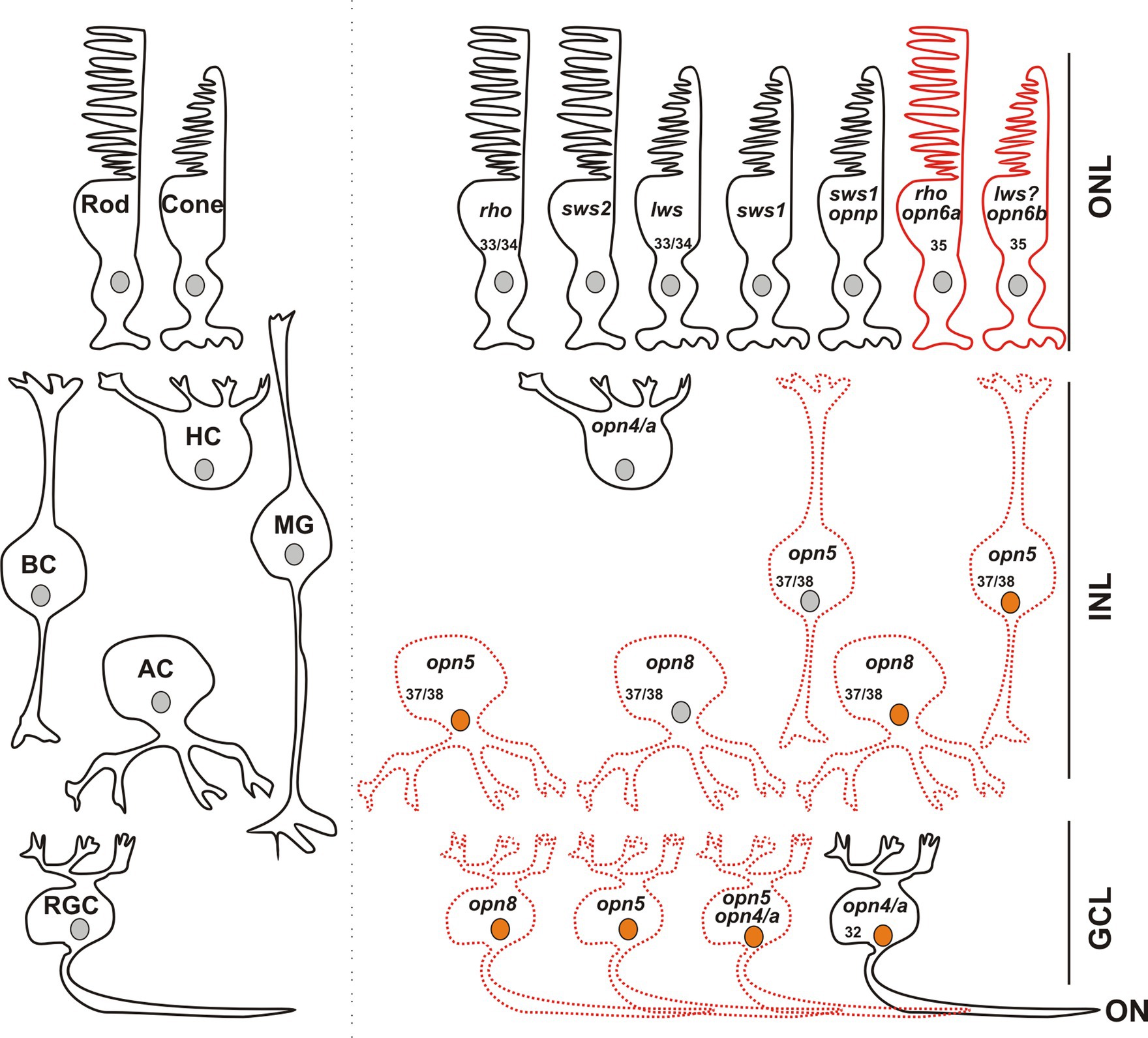
Figure 8. Opsin-expressing cells in the retina of a stage 43/44 Xenopus tadpole. Photoreceptors located in the outer nuclear layer (ONL) express the classical opsins in rods (rho or sws2; Kojima et al., 2017) and cones (lws and sws1). Pinopsin (opnp) is co-expressed in a few sws1+ cells (Bertolesi et al., 2021). Labeled with red are cells expressing opn6a and opn6b in rods (rho+) and cones, likely lws+, respectively. In the inner nuclear layer (INL), horizontal cells (HC) co-express the two melanopsin genes (opn4 and opn4a), bipolar cells (BC) express opn5, while amacrine cells (AC) express opn5 or opn8. Different cells in the ganglion cell layer (GCL) express opsins, including opn5, opn8, opn4/opn4a, and a few cells that contain opn4/a and opn5. Cells that express c-fos in response to exposure to white light are shown with an orange nucleus. Although classical photoreceptors are c-fos negative, they are highly likely activated by the white light exposure. We propose that opsin-expressing cells in the INL or GCL that exhibit light-induced c-fos expression serve as second- or third-order neurons, which receive synaptic inputs from photoreceptors or other retinal cells. We cannot rule out, however, that the opsin-expressing cells of the INL and GCL are intrinsically photosensitive and induce c-fos mRNA upon light stimulation, acting as first-order neurons. opn4/a+ HCs do not express c-fos while opn4/a+ RGCs do (Bertolesi et al., 2014). Opsin-expressing cells that we describe in this manuscript are either labeled with red solid lines (PR; photoreceptors), or red dotted lines as having the potential to be photosensitive in Xenopus, since direct light activation has not yet been demonstrated. Excluded are all the photoreceptors, and HCs and RGCs expressing melanopsins, for which photosensitivity was demonstrated in fish (Cheng et al., 2009) and mammals (Thoreson and Dacey, 2019). Numbers in the cells indicate the development stage of Xenopus when opsin expression is first detected (See also Table 1).
During evolution, opn6 was maintained from teleosts to birds. opn6 disappeared in the mammalian lineage (Davies et al., 2015; Beaudry et al., 2017), a feature that correlates with the loss of an adult retinal stem cell niche, the CMZ. The CMZ functions to add retinal neurons to the periphery of the neural retina in larval and adult fish and frogs (Fischer et al., 2013). Reptiles maintain a CMZ that differs among species and between embryonic and postnatal retina, and with a small proliferative capacity in the adult (Eymann et al., 2019). Proliferating progenitor-like cells are also present in the retinas of some birds, although located in a smaller structure than the CMZ detected in amphibians and fish, while mammals do not have a CMZ (Fischer et al., 2013). The presence of the neuroepithelium of the CMZ and the evolutionary conservation of opn6 (expressed by photoreceptors abutting the CMZ) may be explained by the requirement for continued expression of opn6 with ongoing production of new photoreceptors by the CMZ of the frog and fish retina over the lifetime.
The presence of opn5 in cells located in the INL appears to decrease during evolution, though expression in RGCs is maintained. We find opn5 mRNA is first detected at stage 37/38, is pronounced at stage 41, and sparse by stage 43, which agrees with our previous data showing minimal expression of opn5 in the INL and RGC layer in a mature X. laevis retina (Bertolesi et al., 2021). At the peak of its expression at stage 41, opn5 mRNA is distributed between BCs (~70%–75%), ACs (~10%) and RGCs (~20%). Similarly, in teleost fish, opn5m mRNA is present in the inner region of the INL and opn5m2 mRNA in the outer edge of the INL (Sato et al., 2016). In mammals, however, opn5 expression is restricted to RGCs (Calligaro et al., 2022; D’Souza et al., 2022), where opn5 is used for photoentrainment (Buhr et al., 2015) and eye growth during development (Nguyen et al., 2019). opn5m mRNA is detected in RGCs, ACs, and MG within the avian retina during development and after hatching (Rios et al., 2019; Guido et al., 2020; Calligaro et al., 2022). Thus, opn5 expression is present in INL cells of teleosts and amphibians, but is reduced in birds and absent in mammals.
We first detected opn8 in the retina at stage 37/38 in a handful of ACs and RGCs, in a ratio of 7:3. The expression of the paralogues in zebrafish (opn8a, 8b, and 8c) differs somewhat. For example, the scRNA-seq data indicates that opn8c is most highly expressed in MGs, followed by ACs and photoreceptors, while opn8a and opn8b exhibit minimal expression in retinal cells. The laminar localization of opn8 transcripts in the adult zebrafish retina support this cellular localization; opn8a and opn8c mRNAs are in the outer INL where BCs and HCs reside, opn8b and opn8c mRNAs are in the inner INL where the somas of ACs and MG are present, and opn8b mRNA is in the GCL (Davies et al., 2015). Thus, the opn8 transcript in X. laevis appears to show a more restricted expression (ACs and RGCs) than its counterparts in zebrafish (potentially BCs, ACs, HCs, MG and RGCs). Evolutionarily, opn8 genes are found in vertebrates from Holostei to birds, but are gone from mammals (Davies et al., 2015; Beaudry et al., 2017). The three opn8 genes found in zebrafish also exist in the spotted gar (Beaudry et al., 2017), the common ancestor of Holostei and Teleostei. The evolutionary process of opn8 in vertebrates is unknown, and the question of which fish opn8 gene corresponds to the one conserved in tetrapods remains unanswered. What we do know is that the evolutionary rate of opn8c is higher than the other homologues (opn8a and opn8b) in the basal branch of teleost (Beaudry et al., 2017), which suggests an adaptative advantage. When zebrafish and Xenopus opn8 are compared at the amino acid level, however, opn8c shows the least conservation (37% identity for opn8c, 42% for opn8b, and 56% for opn8a). Genetic and biochemical characterizations of opn8 in these two model organisms may help us understand its physiology and evolution.
Our data suggests a lack of co-expression of neuropsins, which increases considerably the repertoire of potential photosensitive cells in the INL and GCL. We find that in the INL and the GCL, most cells express only one opsin. Indeed, opn8+ ACs and RGCs do not express either opn5 or opn4/a and only a modest proportion (<40%) of opn5+ cells co-express melanopsin (Figures 5, 8). Thus, in the GCL, potential photosensitive RGCs are opn4+, opn5+ and opn8+, while in the INL the repertoire of photosensitive cells includes opn5+ or opn8+ACs, opn5+ BCs, and opn4/a+ HCs (Figure 8). opn5+ MG and opn4+ HCs in birds and zebrafish, respectively, are known to be photosensitive (Cheng et al., 2009; Guido et al., 2020). This is also true of melanopsin-expressing RGCs in mammals (Thoreson and Dacey, 2019). Of note, the two Xenopus melanopsin genes, opn4 and opn4a, are co-expressed in HCs and RGCs (Bertolesi et al., 2014). Interestingly, the opn4a gene was lost in mammals, but was positively selected for, through unknown evolutionary selection pressures, in other tetrapods and teleosts (Borges et al., 2012).
Co-expression of opsins in photoreceptors is more common, and would increase their range of wavelength sensitivity. For example, co-expression of a few cone opsins contributes to the UV-sensitivity of anemonefish (Mitchell et al., 2021), and tropical fish that live in clear waters with broad light environments express up to five opsin genes, thus increasing the gradient of visual sensitivity (Torres-Dowdall et al., 2021). In Xenopus, the four classical photoreceptors classified by their expression of rho, sws2, sws1, and lws are now increased to seven by the co-expression of pinopsin with sws1 (Bertolesi et al., 2021), and opn6a and opn6b with rho and likely with lws, respectively. Understanding the photosensitive properties of these photoreceptors will require electrophysiological and/or biochemical studies to define the unknown spectral characteristics of opn6a and opn6b.
Our previous work suggested that with light stimulation, c-fos is not induced in first-order neurons of the retina, such as the photoreceptors that express opn6a and opn6b, but is in second-order and third-order neurons of the INL and GCL (Bertolesi et al., 2014). For the INL, some but not all opsin-expressing cells respond to white light activation with c-fos induction. For instance, c-fos is not induced in opn4/a+-expressing HCs (Bertolesi et al., 2014) that are found in amphibians (Provencio et al., 1998; Bertolesi et al., 2014), birds (Verra et al., 2011), and teleosts (Cheng et al., 2009), but not mammals. These cells exhibit intrinsic photosensitivity in fish (Cheng et al., 2009), act as photoreceptors in birds (Morera et al., 2016), and in amphibians we postulated that opn4/a+ HCs recognize light reflected from the substrate during background adaptation (Bertolesi and McFarlane, 2018). Interestingly, several c-fos+ cells located in the outer region of the INL, which are likely the classical HCs (Prox1+) that turn on c-fos when the eye receives light stimulation via synaptic inputs. Additionally, several ACs and BCs that express neuropsins are c-fos+ after white light illumination. This is true of 60% of the opn5+ Otx2+ BCs. The observation that 40% of the cells do not turn on c-fos suggests that the opn5+ Otx2+ BCs consist of both ON (Otx2+ Isl1+) and OFF (Otx2+) BCs (Bertolesi et al., 2014); the latter population participate in circuits that shut down with light stimulation. Similarly, only a subpopulation of the opn8+ ACs show c-fos induction, suggesting that opn8+ ACs participate in both ON and OFF circuits (Figure 6). In contrast, almost all neuropsin- and melanopsin- expressing cells in the GCL are activated upon light stimulation (Bertolesi et al., 2014) (Figures 6, 8).
This study gave a better understanding of retinal neuropsin-expressing cells, their developmental onset and activation by light. By using specific markers, we defined the cell types that express neuropsins. The expression of neuropsins in the ONL (opn6a and opn6b) increase the number of photoreceptors present in Xenopus, while expression in cells in the INL and GCL increase dramatically the number of potentially photosensitive cells. The onset of expression of opn6a and opn6b (stage 35) coincides with that of ‘classical’ opsins, and suggests a role in the differentiation and/or function of newborn photoreceptors. The onset of expression of opn5 (BC, AC and RGC) and opn8 (AC and RGC) occurs slightly later (stage 37) when retinal circuits become “wired” (stage 37/38). The fact that neuropsins are present as the retinal circuits are established argues for the importance of intrinsic photosensitivity within interneurons in modulating activity in the circuits, an interesting idea that needs exploring. Finally, by comparing neuropsin expression between different species we provide insight into evolutionary differences. Our data suggest that the numbers of different types of neuropsin-expressing cells in the INL and GCL likely diminished during evolution (teleost > amphibians > birds > mammals). In mammals, opsin-expressing cells are in photoreceptors and RGCs (opn4 and opn5) while the modulatory interneurons do not express opsins. Understanding the evolution of retinal circuits in vertebrates will be helped by a better grasp of neuropsin physiology.
Data availability statement
The names of the repository/repositories and accession number(s) are mentioned in materials and methods section or can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Animal Care and Use Committee at the University of Calgary. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review & editing. SS: Software, Data curation, Investigation, Methodology, Writing–original draft, Writing–review & editing. GB: Validation, Data curation, Investigation, Methodology, Writing–original draft, Writing–review & editing, Conceptualization, Formal Analysis, Supervision. SM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–original draft, Writing–review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to SM (PIN03909-2018). LM was a recipient of an NSERC summer studentship award.
Acknowledgments
The authors would like to thank Carrie L. Hehr for her technical contributions to the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2023.1266945/full#supplementary-material
Abbreviations
BCIP, 5-Bromo-4-chloro-3-indolyl phosphate; CMZ, Ciliary marginal zone; DEPC, Diethylpyrocarbonate; DIG, Digoxigenin; D-FISH, Double fluorescent in-situ hybridization; FITC, Fluorescein isothiocyanate; GCL, Ganglion cell layer; INL, Inner nuclear layer; LWS, Long-wavelength sensitive; MOPS, 3-N-morpholino propanesulfonic acid; NBT, Nitro blue tetrazolium chloride; OCT, Optimal cutting temperature; ONL, Outer nuclear layer; RGC, Retinal ganglion cell; RT-PCR, Reverse-transcription polymerase chain reaction; SWS, Short-wavelength sensitive; TSA, Tyramide signal amplification; UV, Ultra-violet.
Footnotes
References
Beaudry, F., Iwanicki, T. W., Mariluz, B., Darnet, S., Brinkmann, H., Schneider, P., et al. (2017). The non-visual opsins: eighteen in the ancestor of vertebrates, astonishing increase in ray-finned fish, and loss in amniotes. J. Exp. Zool. B 328, 685–696. doi: 10.1002/jez.b.22773
Bertolesi, G. E., Atkinson-Leadbeater, K., Mackey, E. M., Song, Y. N., Heyne, B., and McFarlane, S. (2020). The regulation of skin pigmentation in response to environmental light by pineal type II opsins and skin melanophore melatonin receptors. J. Photochem. Photobiol. B 212:112024. doi: 10.1016/j.jphotobiol.2020.112024
Bertolesi, G. E., Debnath, N., Atkinson-Leadbeater, K., Niedzwiecka, A., and McFarlane, S. (2021). Distinct type II opsins in the eye decode light properties for background adaptation and behavioural background preference. Mol. Ecol. 30, 6659–6676. doi: 10.1111/mec.16203
Bertolesi, G. E., Hehr, C. L., and McFarlane, S. (2014). Wiring the retinal circuits activated by light during early development. Neural Dev. 9:3. doi: 10.1186/1749-8104-9-3
Bertolesi, G. E., and McFarlane, S. (2018). Seeing the light to change colour: an evolutionary perspective on the role of melanopsin in neuroendocrine circuits regulating light-mediated skin pigmentation. Pigment Cell Melanoma Res. 31, 354–373. doi: 10.1111/pcmr.12678
Borges, R., Johnson, W. E., O’Brien, S. J., Vasconcelos, V., and Antunes, A. (2012). The role of gene duplication and unconstrained selective pressures in the melanopsin gene family evolution and vertebrate circadian rhythm regulation. PLoS One 7:e52413. doi: 10.1371/jour-nal.pone.0052413
Briggs, J. A., Weinreb, C., Wagner, D. E., Megason, S., Peshkin, L., Kirschner, M. W., et al. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, 1–23. doi: 10.1126/science.aar5780
Buhr, E. D., Yue, W. W. S., Ren, X., Jiang, Z., Liao, H.-W. R., Mei, X., et al. (2015). Neuropsin (opn5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. Proc. Natl. Acad. Sci. U. S. A. 112, 13093–13098. doi: 10.1073/pnas.1516259112
Calligaro, H., Dkhissi-Benyahya, O., and Panda, S. (2022). Ocular and extraocular roles of neuropsin in vertebrates. Trends Neurosci. 45, 200–211. doi: 10.1016/J.TINS.2021.11
Chang, W. S., and Harris, W. A. (1998). Sequential genesis and determination of cone and rod photoreceptors in Xenopus. J. Neurobiol. 35, 227–244.
Cheng, N., Tsunenari, T., and King-Wai, Y. (2009). Intrinsic light response of retinal horizontal cells of teleosts. Nature 460, 899–903. doi: 10.1038/nature08175
D’Souza, S. P., Swygart, D. I., Wienbar, S. R., Upton, B. A., Zhang, K. X., Mackin, R. D., et al. (2022). Retinal patterns and the cellular repertoire of neuropsin (opn5) retinal ganglion cells. J. Comp. Neurol. 530, 1247–1262. doi: 10.1002/CNE.25272
Davies, W. I. L., Tamai, T. K., Zheng, L., Fu, J. K., Rihel, J., Foster, R. G., et al. (2015). An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 25, 1666–1679. doi: 10.1101/gr.189886.115
Díaz, N. M., Morera, L. P., and Guido, M. E. (2016). Melanopsin and the non-visual photochemistry in the inner retina of vertebrates. Photochem. Photobiol. 92, 29–44. doi: 10.1111/php.12545
Eymann, J., Salomies, L., and Macrì, S. (2019). Variations in the proliferative activity of the peripheral retina correlate with postnatal ocular growth in squamate reptiles. J. Comp. Neurol. 527, 2356–2370. doi: 10.1002/cne.24677
Fischer, A. J., Bosse, J. L., and El-hodiri, H. M. (2013). The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp. Eye Res. 116, 199–204. doi: 10.1016/j.exer.2013.08.018
Frederick, J. M., Rayborn, M. E., and Hollyfield, J. G. (1989). Serotoninergic neurons in the retina of Xenopus laevis: selective staining, identification, development, and content. J. Comp. Neurol. 281, 516–531. doi: 10.1002/cne.902810403
Gábriel, R., and Wilhelm, M. (2001). Structure and function of photoreceptor and second-order cell mosaics in the retina of Xenopus. Int. Rev. Cytol. 210, 77–120. doi: 10.1016/s0074-7696(01)10004-5
Guido, M. E., Marchese, N. A., Rios, M. N., Morera, L. P., Diaz, N. M., Garbarino-Pico, E., et al. (2020). Non-visual opsins and novel photo-detectors in the vertebrate inner retina mediate light responses within the blue spectrum region. Cell. Mol. Neurobiol. 42, 59–83. doi: 10.1007/s10571-020-00997-x
Hirsch, N., and Harris, W. A. (1997). Xenopus Pax-6 and retinal development. J. Neurobiol. 32, 45–61.
Hoang, T., Wang, J., Boyd, P., Wang, F., Santiago, C., Yoo, S., et al. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science 370, 1–34. doi: 10.1126/science.abb8598.Gene
Hollyfield, J. G., Rayborn, M. E., Sarthy, P. V., and Lam, D. M. K. (1980). Retinal development: time and order of appearance of specific neuronal properties. Neurochem. Int. 1, 93–101. doi: 10.1016/0197-0186(80)90053-4
Holt, C. E. (1989). A single-cell analysis of early retinal ganglion cell differentiation Xenopus: from soma to axon tip. J. Neurosci. 9, 3123–3145. doi: 10.1523/JNEUROSCI.09-09-03123.1989
Holt, C. E., Bertsch, T. W., Ellis, H. M., and Harris, W. A. (1988). Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1, 15–26. doi: 10.1016/0896-6273(88)90205-x
Huerta, J. J., Llamosas, M. M., Cernuda-Cernuda, R., and García-Fernández, J. M. (1997). Fos expression in the retina of rd/rd mice during the light/dark cycle. Neurosci. Lett. 232, 143–146. doi: 10.1016/s0304-3940(97)00595-8
Kojima, K., Matsutani, Y., Yamashita, T., Yanagawa, M., Imamoto, Y., Yamano, Y., et al. (2017). Adaptation of cone pigments found in green rods for scotopic vision through a single amino acid mutation. Proc. Natl. Acad. Sci. U.S.A. 114, 5437–5442. doi: 10.1073/pnas.1620010114
Lamb, T. D. (2013). Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119. doi: 10.1016/j.preteyeres.2013.06.001
Liu, Z., Hamodi, A. S., and Pratt, K. G. (2016). Early development and function of the Xenopus tadpole retinotectal circuit. Curr. Opin. Neurobiol. 41, 17–23. doi: 10.1016/J.CONB.2016.07.002
Mitchell, L. J., Cheney, K. L., Cortesi, F., Lu, M., Marshall, J., and Michie, K. (2021). Molecular evolution of ultraviolet visual opsins and spectral tuning of photoreceptors in anemonefishes (Amphiprioninae). Genome Biol. Evol. 13, 1–14. doi: 10.1093/gbe/evab184
Morera, L. P., Díaz, N. M., and Guido, M. E. (2016). Horizontal cells expressing melanopsin x are novel photoreceptors in the avian inner retina. Proc. Natl. Acad. Sci. U.S.A. 113, 13215–13220. doi: 10.1073/pnas.1608901113
Nguyen, M. T., Vemaraju, S., Nayak, G., Odaka, Y., Buhr, E. D., Alonzo, N., et al. (2019). An opsin 5-dopamine pathway mediates light-dependent vascular development in the eye. Nat. Cell Biol. 21, 420–429. doi: 10.1038/s41556-019-0301-x
Perron, M., and Harris, W. A. (1999). “Cellular determination in amphibian retina” in Cell lineage and fate determination. ed. S. A. Moody . 1st ed (New York: Academic Press), 353–368.
Provencio, I., Jiang, G., De Grip, W. J., Hayes, W. P., and Rollag, M. D. (1998). Melanopsin: an opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. U. S. A. 95, 340–345. doi: 10.1073/pnas.95.1.340
Rios, M. N., Marchese, N. A., and Guido, M. E. (2019). Expression of non-visual opsins opn3 and opn5 in the developing inner retinal cells of birds. Light-responses in Müller glial cells. Front. Cell. Neurosci. 13:376. doi: 10.3389/fncel.2019.00376
Sato, K., Yamashita, T., Haruki, Y., Ohuchi, H., and Kinoshita, M. (2016). Two UV-sensitive photoreceptor proteins, opn5m and opn5m2 in ray-finned fish with distinct molecular properties and broad distribution in the retina and brain. PLoS One 11:e0155339. doi: 10.1371/journal.pone.0155339
Session, A. M., Uno, Y., Kwon, T., Chapman, J. A., Toyoda, A., Takahashi, S., et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. doi: 10.1038/nature19840
Sonam, S., Bangru, S., Perry, K. J., Chembazhi, U. V., Kalsotra, A., and Henry, J. J. (2022). Cellular and molecular profiles of larval and adult Xenopus corneal epithelia resolved at the single-cell level. Dev. Biol. 491, 13–30. doi: 10.1016/j.ydbio.2022.08.007
Starace, D. M., and Knox, B. E. (1998). Cloning and expression of a Xenopus short wavelength cone pigment. Exp. Eye Res. 67, 209–220. doi: 10.1006/exer.1998.0507
Szaro, B. G., and Gainer, H. (1988). Immunocytochemical identification of non-neuronal intermediate filament proteins in the developing Xenopus laevis nervous system. Dev. Brain Res. 43, 207–224. doi: 10.1016/0165-3806(88)90100-9
Takahashi, Y., Hisatomi, O., Sakakibara, S., Tokunaga, F., and Tsukahara, Y. (2001). Distribution of blue-sensitive photoreceptors in amphibian retinas. FEBS Lett. 501, 151–155. doi: 10.1016/S0014-5793(01)02632-1
Thoreson, W. B., and Dacey, D. M. (2019). Diverse cell types, circuits, and mechanisms for color vision in the vertebrate retina. Physiol. Rev. 99, 1527–1573. doi: 10.1152/physrev.00027.2018
Torres-Dowdall, J., Karagic, N., Härer, A., and Meyer, A. (2021). Diversity in visual sensitivity across Neotropical cichlid fishes via differential expression and intraretinal variation of opsin genes. Mol. Ecol. 30, 1880–1891. doi: 10.1111/mec.15855
Verra, D. M., Contin, M. A., Hicks, D., and Guido, M. E. (2011). Early onset and differential temporospatial expression of melanopsin isoforms in the developing chicken retina. Invest. Ophthalmol. Vis. Sci. 52, 5111–5120. doi: 10.1167/iovs.11-75301
Viczian, A. S., Vignali, R., Zuber, M. E., Barsacchi, G., and Harris, W. A. (2003). XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development 130, 1281–1294. doi: 10.1242/dev.00343
Yamashita, T., Ohuchi, H., Tomonari, S., Ikeda, K., Sakai, K., and Shichida, Y. (2010). opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc. Natl. Acad. Sci. U. S. A. 107, 22084–22089. doi: 10.1073/pnas.1012498107
Yoshida, K., Imaki, J., Hitoshi, F., Harada, T., Ohki, K., Matsuda, H., et al. (1996). Differential distribution of CaM kinases and induction of c-fos expression by flashing and sustained light in rat retinal cells. Invest. Ophthalmol. Vis. Sci. 37, 174–179.
Keywords: retina, opsin, eye, neuronal circuit, frog, evolution, development
Citation: Man LLH, Storey SS, Bertolesi GE and McFarlane S (2023) Cell-type expression and activation by light of neuropsins in the developing and mature Xenopus retina. Front. Cell. Neurosci. 17:1266945. doi: 10.3389/fncel.2023.1266945
Edited by:
Matthew Van Hook, University of Nebraska Medical Center, United StatesReviewed by:
Shijun Weng, Fudan University, ChinaGladys Y.-P. Ko, Texas A and M University, United States
Copyright © 2023 Man, Storey, Bertolesi and McFarlane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel E. Bertolesi, gbertole@ucalgary.ca; Sarah McFarlane, smcfarla@ucalgary.ca
†Present address: Lawrence L. H. Man, Herbert Wertheim School of Optometry and Vision Science, University of California, Berkeley, CA, United States
 Lawrence L. H. Man
Lawrence L. H. Man Samuel S. Storey
Samuel S. Storey Gabriel E. Bertolesi
Gabriel E. Bertolesi Sarah McFarlane
Sarah McFarlane