- 1Department of Laboratory Science, Binhaiwan Central Hospital of Dongguan, Guangdong, China
- 2Dongguan Key Laboratory of Accurate Etiological Research on the Pathogenesis of Inflammation and Cancer, Guangdong, China
- 3Central Laboratory, Binhaiwan Central Hospital of Dongguan, Guangdong, China
- 4Dongguan Key Laboratory of Precision Medicine, Guangdong, China
Introduction: Biliary Infection in patients is a common and important phenomenon resulting in severe complications and high morbidity, while the distributions and drug resistance profiles of biliary bacteria and related risk factors are dynamic. This study explored the characteristics of and risk factors for biliary infection to promote the rational use of antibiotics in clinically.
Methods: Bacterial identification and drug susceptibility testing were completed using the Vitek 2 Compact analysis system. The distribution and antibiotic-resistant characteristics of 3,490 strains of biliary bacteria in patients at Nankai Hospital from 2019 to 2021 were analyzed using Whonet 5.6 and SPSS 26.0 software. We then retrospectively analyzed the clinical data and risk factors associated with 2,340 strains of Gram-negative bacilli, which were divided into multidrug-resistant bacteria (1,508 cases) and non-multidrug-resistant bacteria (832 cases) by a multivariate Cox regression model.
Results and discussion: A total of 3,490 pathogenic bacterial strains were isolated from bile samples, including 2,340 (67.05%) Gram-negative strains, 1,029 (29.48%) Gram-positive strains, and 109 (4.56%) fungal strains. The top five pathogenic bacteria were Escherichia coli, Klebsiella pneumoniae, Enterococcus faecium, Enterococcus faecalis, and Pseudomonas aeruginosa. The rate of Escherichia coli resistance to ciprofloxacin increased (p < 0.05), while the resistance to amikacin decreased (p < 0.05). The resistance of Klebsiella pneumoniae to cephalosporins, carbapenems, β-lactamase inhibitors, cephalases, aminoglycosides, and quinolones increased (p < 0.05), and the resistance of Pseudomonas aeruginosa to piperacillin, piperacillin/tazobactam, ticacillin/clavulanic acid, and amicacin declined significantly (p < 0.05). The resistance of Enterococcus faecium to tetracycline increased by year (p < 0.05), and the resistance of Enterococcus faecalis to erythromycin and high-concentration gentamicin declined (p < 0.05). Multivariate logistic regression analysis suggested that the administration of third- or fourth-generation cephalosporins was an independent risk factor for biliary infection. In summary, Gram-negative bacilli were the most common pathogenic bacteria isolated from biliary infection patients, especially Escherichia coli, and the rates and patterns of drug resistance were high and in constant flux; therefore, rational antimicrobial drug use should be carried out considering risk factors.
1 Introduction
Biliary infection in patients is common and has been associated with invasive clinical procedures, septicemia, intestinal barrier dysfunction, and gut bacteria translocation (Chi et al., 2023; Ekpanyapong and Reddy, 2023). It continues to be a significant cause of severe complications and morbidity in patients, particularly in elderly patients with underlying comorbidities. The heightened mortality rate among patients with infected biliary secretions underscores the critical need for timely diagnosis and treatment of biliary tract infections. Therefore, for patients with bile infectious symptoms, bile samples are frequently used for clinical microbiological culture tests. However, the bile bacterial spectrum and antibiotic resistance characteristics are constantly changing. Based on previous literature reports, the widespread use of antibiotics globally has increased the likelihood of bacterial species alterations. Data indicates that pathogenic bacteria such as Escherichia coli, Enterococcus faecium, Klebsiella pneumoniae, and Serratia, particularly Enterococcus faecium, have become predominant in recent cases of biliary tract infections. It is noteworthy that the trend of drug resistance is worsening (Ruan et al., 2019).
Gram-negative bacilli, which are some of the most common opportunistic pathogens in hospitals, have shown increasing trends toward drug resistance in recent years. Gram-negative bacilli mainly include those in the Enterobacteriaceae family and nonfermentative bacteria, and easily evolve into multidrug-resistant (Chen S. et al., 2021) organisms (MDROs) that are resistant to three or more types of clinical antibiotics used at the same time. In recent years, with the extensive use of antibiotics for the treatment of bacterial infections, MDROs have become common and important pathogens responsible for clinical infections, resulting in an intractable challenge for clinical diagnosis and treatment (Kawanishi et al., 2017); therefore, it is necessary to clarify the risk factors for MDRO infection in patients.
We conducted this study to understand the characteristics of pathogens causing biliary infections and related risk factors in patients, guide clinical detection, promote the rational use of antibiotics, control multidrug-resistant bacterial infections, and improve the cure rate in biliary tract infectious patients. We analyzed the in-hospital data of patients from January 2019 to December 2021 to retrospectively identify the biliary pathogenic spectrum and drug resistance profiles, Gram-negative multidrug-resistant bacteria distributions and resistance rates, and associations of risk factors in the clinical characteristics of the patients. This analysis aimed to provide valuable insights into the changing trends of biliary tract infections and drug resistance, ultimately informing more effective clinical management, improved detection capability, and prevention strategies in the medical future. The study results are as follows.
2 Materials and methods
2.1 Study subjects
This retrospective cross-sectional study, recruited 1,556 patients, including 962 males and 594 females, who were hospitalized at Tianjin Nankai Hospital, Tianjin Medical University from January 1, 2019, to December 31, 2021. Their data was accessed on January 15, 2022, using the software of the hospital information system and laboratory information system for research purposes within the hospital.
2.2 Sample collection
The present study obtained ethical approval (no. NKYY_YXKT_IRB_2021_155_01) from the Ethical Committee of Tianjin Nankai Hospital, Nankai Clinical College, Tianjin Medical University, following the Helsinki Declaration. Due to its retrospective design, a waiver of participant informed consent was granted by the Ethical Committee of Nankai Clinical College, Tianjin Medical University.
The patients were 18–86 years of age (average age = 62.3 ± 23). Specimens were collected from patients presenting symptoms of potential biliary disease or infection who had not been treated with antibiotics. Bacterial culture identification was conducted by directly extracting 2–10 mL of bile from the gall bladder or through bile duct puncture and drainage. Additionally, newer technologies such as ERCP and other ultrasonographic techniques were utilized to aid in collecting bile samples from the suspected site of infection (Zijlstra, 2022).
2.3 Sample culture and bacterial detection
Bile samples were aseptically injected into aerobic and anaerobic culture bottles, mixed well, placed in a BacT/Alert3D automated blood culture instrument, and continuously monitored for 2 days (Plantamura et al., 2012). Samples with no positive indication were cultured on a blood agar medium and defined as negative if no bacteria were detected and positive if there was evidence of bacterial proliferation and the instrument was able to detect a yellow color on the bottom indicator of the culture bottles at the indicated time. Positive bile samples were transferred to blood agar, Sabouraud fungal medium, McConkey agar, and chocolate agar (Tianjin Jinzhang Biotechnology Development Co., Ltd. Tianjin, China) and incubated in 371 carbon dioxide bacteria incubators at 35°C and 5% CO2 for 18–24 h. Anaerobic culture bags (1.5 L; Qingdao Haibo Biotechnology, Shandong, China) were used for further culture and separation, as needed (Chen S. et al., 2021).
2.4 Bacterial strain identification and drug susceptibility testing
Sole pathogenic colonies were obtained and manually sampled using a VITEK 2 compact automatic microbial analysis system and its supporting identification cards to identify strains and determine the drug susceptibility profiles. The Vitek 2 compact automated microbial identifier is capable of automatically identifying over 400 strains of bacteria, including Gram-negative bacilli, Gram-positive bacteria, fungi, aerobic bacilli, and anaerobic bacteria. It can run thirty cartridges simultaneously and perform susceptibility testing using susceptibility cards. Some antimicrobial drug susceptibility testing that failed to complete in the analyzer was conducted using the paper diffusion method, with interpretation following the American CLSI M100 document (2018–2020). The presence of MDRO, ESBL, VRE, and MRSA was determined according to the National Clinical Laboratory Practice (3rd edition). While complete all microbiological identification still relies on conventional biochemical tests and a combination of methods such as Gram stain observation of morphological colonial characters (identifying hemolytic or non-hemolytic colonies on blood agar), lactose or non-lactose colonies on MacConkey’s agar, oxidase test, catalase test, plasma coagulase test and standard biochemical tests application. Carbapenem resistance was defined when the isolate showed non-susceptibility to any tested carbapenems according to Clinical Standards Institute (CLSI) breakpoints (Carvalhaes et al., 2023). All testing was conducted in strict accordance with international clinical operating procedures.
2.5 Standard quality control
Standard control strains, Staphylococcus aureus ATCC29213, Escherichia coli ATCC25922, and Enterobacter cloacae ATCC700323 (provided by the clinical testing center at the National Health and Family Planning Commission in China), were tested in parallel with the sample testing (Alreshidi et al., 2023).
2.6 Statistical processing methods
The bacterial and drug resistance data were analyzed and statistically processed using the WHONET 5.6 software. Additionally, SPSS 26.0 software (SPSS Inc.) was utilized to conduct the Chi-square test for the drug resistance rates of pathogens among groups isolated from bile specimens during 2019–2021 at a significance level of α = 0.05.
2.7 Independent risk factors for death were identified using a multivariate cox regression model
In the analysis of risk factors for multidrug resistance, 2,340 cases with Gram-negative bacilli isolation during hospitalization from 2019 to 2021 were divided into a multidrug-resistant bacteria group (MDROs, 1,508 cases) and a non-multidrug-resistant bacteria group (Non-MDROs, 832 cases), and their clinical characteristics were retrospectively analyzed to identify potential risk factors. Independent risk factors for mortality were identified using a multivariate Cox regression model. Group comparisons for categorical variables were assessed using χ 2 tests. The findings were reported with a 95% confidence interval (CIs). Statistical analysis was conducted using SPSS Statistics version 26.0 (SPSS Inc.), with statistical significance set at p < 0.05.
3 Results
3.1 General characteristics of the distributions and proportions of bile pathogenic bacteria
During 2019–2021, the positivity rate of the bile culture samples was 72.30% (1,125/1,556), and mixed infections of two or more bacteria accounted for 92.00% (1,035/1125). A total of 3,490 pathogen strains were isolated, of which 2,340 (67.05%) were Gram-negative, 1,029 (29.48%) were Gram-positive, and 109 (3.12%) were fungal, the typical culture images were included in Supplementary Figure S1.
Among the Gram-negative bacteria, Escherichia coli had the highest rate of detection (875 strains/25.07%), followed by Klebsiella pneumoniae (535 strains/15.33%), Pseudomonas aeruginosa (164 strains/4.70%), and Enterobacter cloacae sewer subspecies (145 strains/4.15%). Among the Gram-positive bacteria, Enterococcus faecium (415 strains/11.95%), Enterococcus faecalis (196 strains/5.67%), Enterococcus gallinae (66 strains/1.95%), and Staphylococcus epidermidis (41 strains/1.23%) had the highest rates of detection. Pathogen composition and distribution by year is shown in Figure 1 and Supplementary Figure S2.
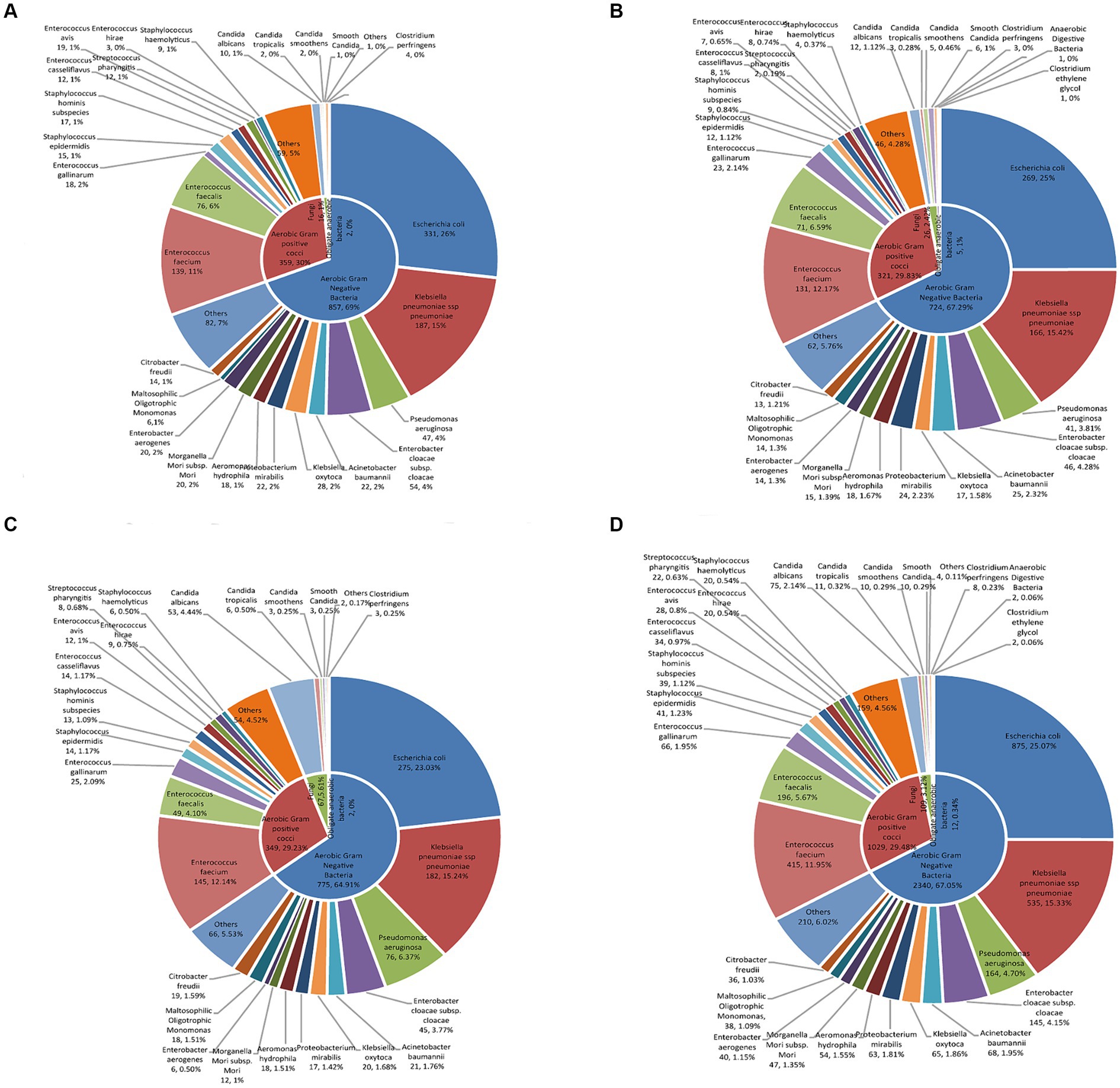
Figure 1. The characteristics of distribution and proportion of pathogenic bacteria in patients with bile tract infection. The distribution and proportion of pathogenic bacteria in patients with bile tract infection in (A) 2019, (B) 2020, (C) 2021, and (D) 2019–2021 using double-layered pie charts. The inner layers in the pie charts were classified based on the proportion of aerobic Gram-positive bacteria, aerobic Gram-negative bacteria, fungi, and anaerobic bacteria. The outer pie charts showed the specific bacterial species under their respective categories.
3.2 Distribution of pathogenic bacteria in the clinical departments
During 2019–2021, the top six specialties with the highest number of pathogen strains detected in bile samples were (from high to low) the clinically major departments in the hospital: hepatobiliary and pancreatic surgery, surgical oncology, minimally invasive surgery, gastrointestinal surgery, intensive care, and oncology (Figure 2).
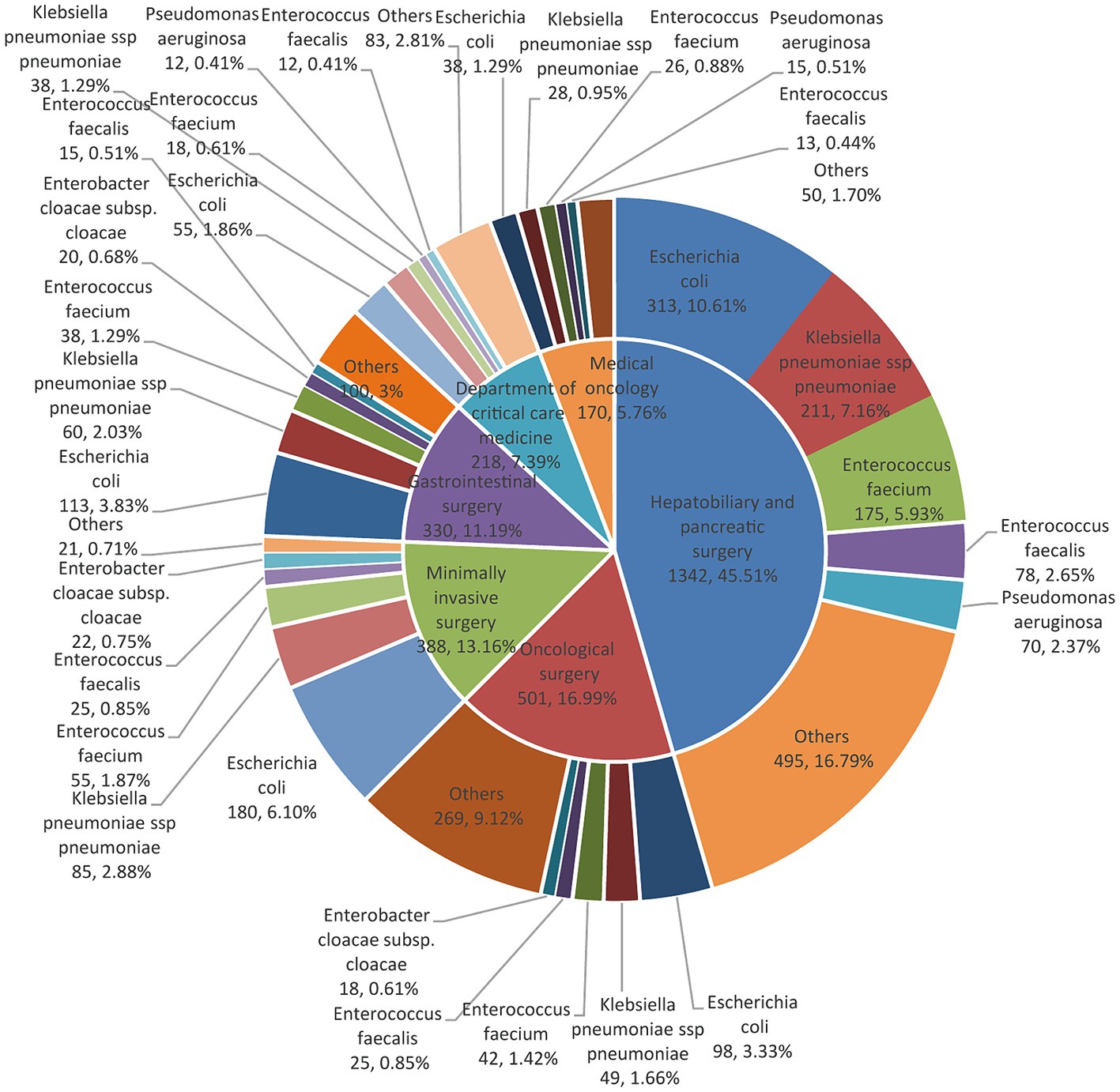
Figure 2. The distribution of major pathogens in patients is seen in each of the top six clinical departments with a pie chart of double layers. The inner layers in the pie charts were classified according to the top six clinical departments. The outer pie charts displayed the top bacterial species under their respective departments.
3.3 Drug-resistant characteristics of the major gram-negative bacteria, Enterobacteriaceae
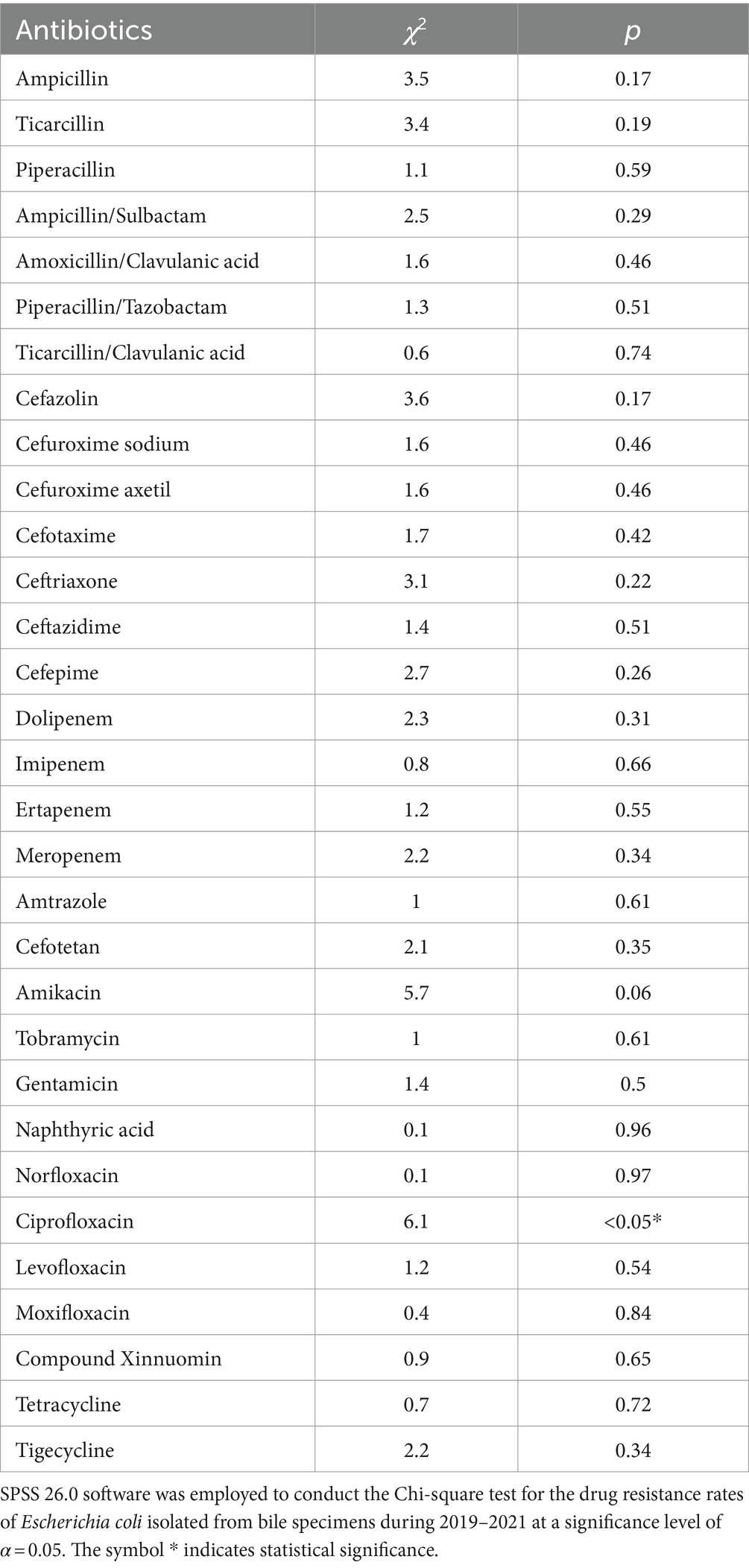
The detection rate of extended-spectrum beta-lactamases (ESBLs) in Escherichia coli was 37.21%. The rate of Escherichia coli resistance to antibiotics remained stable during 2019–2021 (Figure 3A and Table 1), and the average rate of resistance to antibiotics, including ampicillin, ticacillin, piperacillin, ampicillin/sulbactam, cefazolin, cefuroxime sodium, cefuroxime axetil, cefotaxime, ceftriaxone, nalidixic acid, levofloxacin, ciprofloxacin, moxifloxacin, tetracycline, and cotrimoxazole, was >40%. Interestingly, the rate of resistance to ciprofloxacin (42.03–56.20%) increased significantly in 2021 (p < 0.05) (Figure 3A and Table 1). Meanwhile, the rates of Escherichia coli resistance to amoxicillin/clavulanic acid, ticacillin/clavulanic acid, ceftazidime, cefepime, amtronam, and gentamicin, were relatively low. Escherichia coli was more sensitive to piperacillin/tazobactam, dolipenem, imipenem, ertapenem, meropenem, cefotetan, tobramycin, amikacin and tigecycline, with average drug resistant rates of 10.29, 3.29, 1.94, 3.05, 1.92, 5.09, 9.25, 2.05 and 0.23%, respectively.
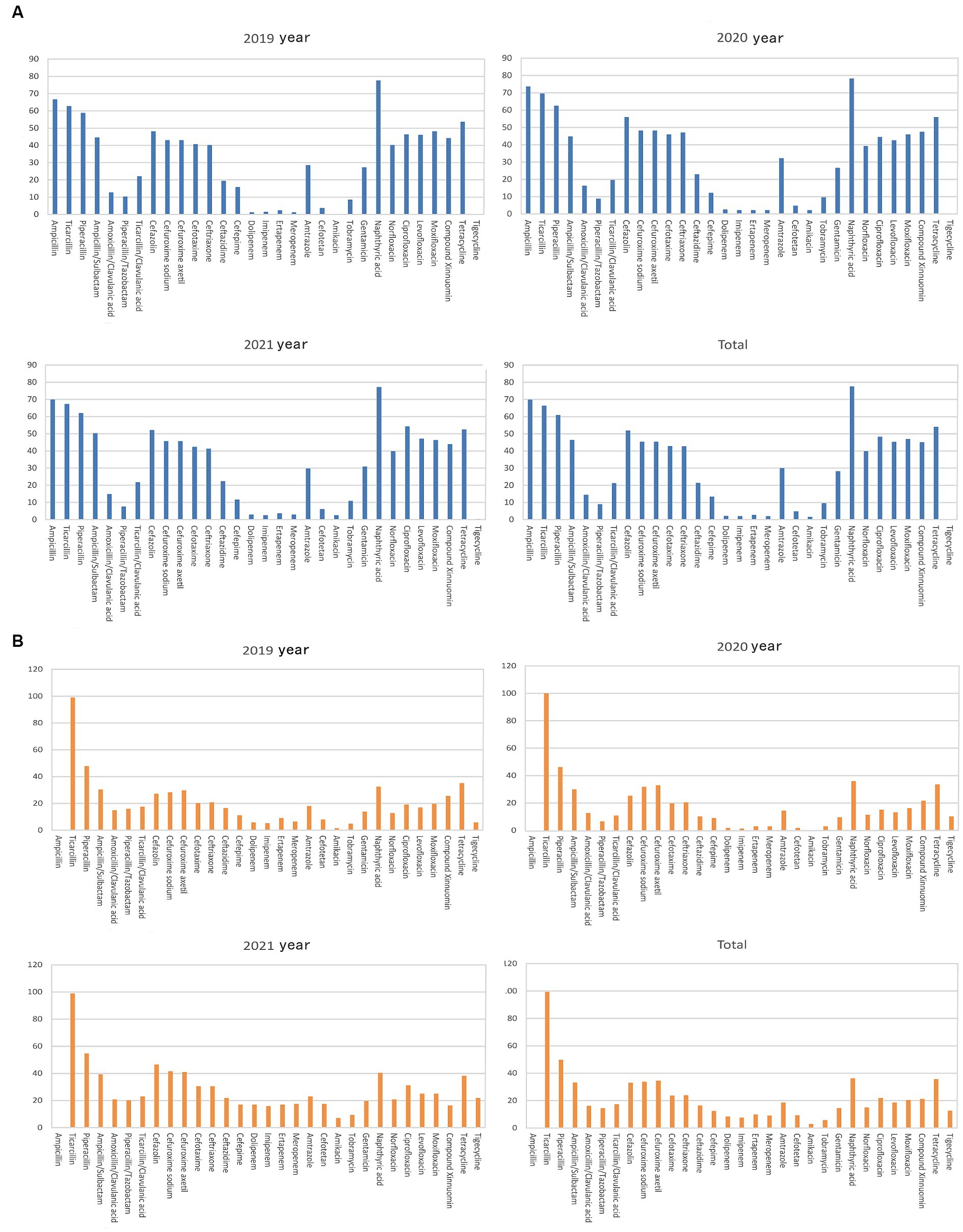
Figure 3. Analysis of the rates of fermenting bacterial drug resistance. On the X-axis of all graphs are antimicrobial agents for Gram-negative bacteria, while the Y-axis displays the percentage of resistant bacterial strains. (A) Analysis of Escherichia coli drug resistance in bile tract infection patients during 2019–2021. Upper left: Escherichia coli drug resistance in 2019. Upper right: Escherichia coli drug resistance in 2020. Left bottom: Escherichia coli drug resistance in 2021. Right bottom: Escherichia coli drug resistance during 2019–2021. (B) Analysis of Klebsiella pneumoniae drug resistance in patients with bile tract infection patients during 2019–2021. Upper left: Klebsiella pneumoniae drug resistance in 2019. Upper right: Klebsiella pneumoniae drug resistance in 2020. Left bottom: Klebsiella pneumoniae drug resistance in 2021. Right bottom: Klebsiella pneumoniae drug resistance during 2019–2021.
Except for tigecycline and carbapenem antibiotics, doripenem, imipenem, ertapenem, and meropenem, Klebsiella pneumoniae had a higher sensitivity than Escherichia coli to most antibiotics (Figure 3B and Table 2). The average rates of Klebsiella pneumoniae resistance to these drugs were 11.34, 8.25, 7.76, 10.02, and 8.94%, respectively, increasing significantly in 2021 (p < 0.01). The rates of Klebsiella pneumoniae resistance to piperacillin/tazobactam, ticacillin/clavulanate, cefazolin, cefuroxime sodium, cefotaxime, ceftriaxone, ceftazidime, cefotetan, amicacin, tobramycin, gentamicin, norfloxacin, and ciprofloxacin showed an increasing trend (p < 0.05). Finally, the rate of ESBL detection in Klebsiella pneumoniae was 17.56%.
3.4 Drug resistance of the non-fermenting bacteria, Pseudomonas aeruginosa
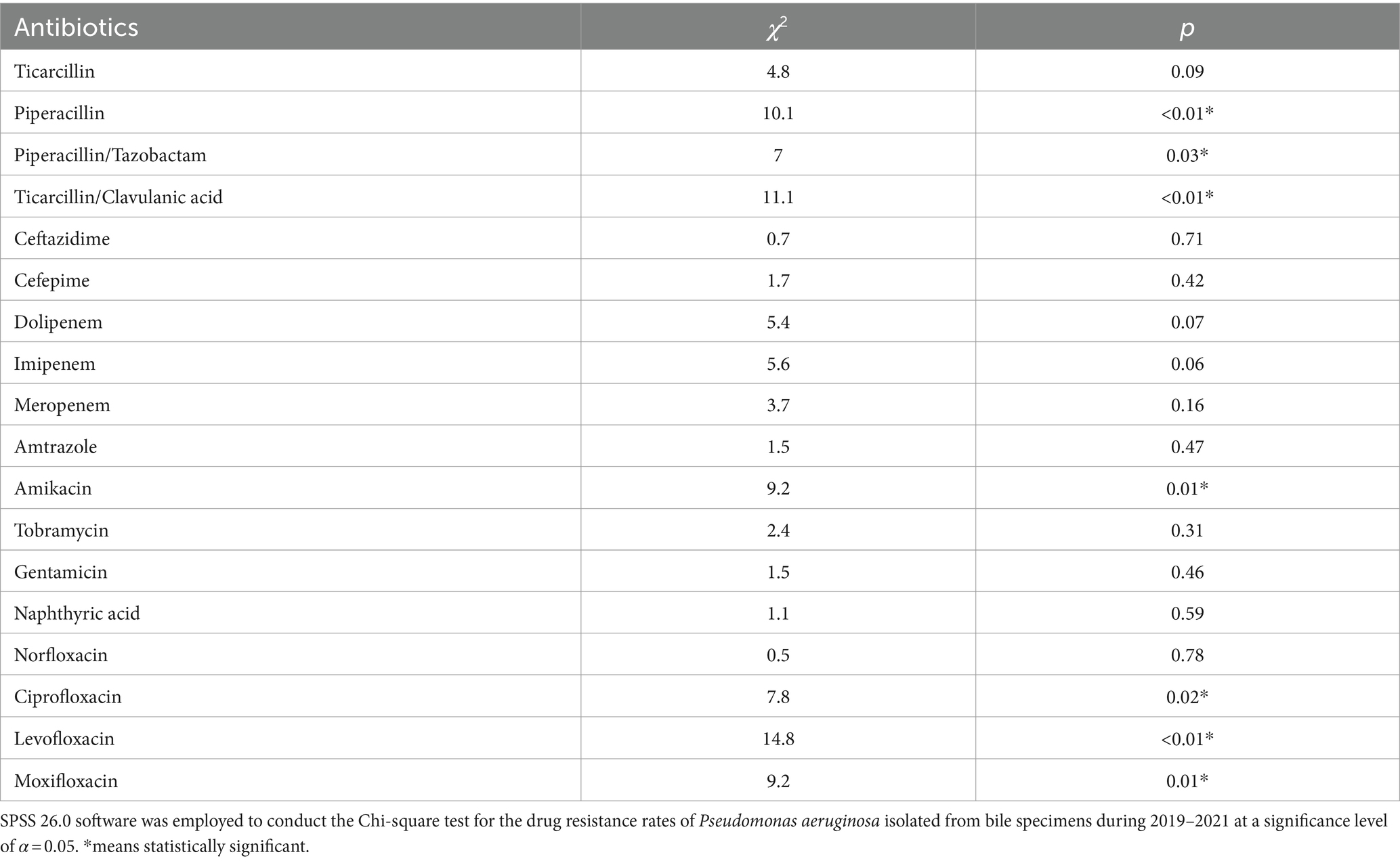
Pseudomonas aeruginosa was the primary non-fermenting bacteria detected in the bile samples, with a higher average 3-year rate of resistance to carbopenicillin antibiotics (>50%) than all the Gram-negative pathogens (Figure 4 and Table 3). Meanwhile, the rates of Pseudomonas aeruginosa resistance to piperacillin, piperacillin/tazobactam, ticacillin/clavulanate, and amikacin decreased significantly over time (p < 0.05). The rates of resistance to quinolone antibiotics, such as ciprofloxacin, levofloxacin, and moxifloxacin, declined in 2020 and rebounded to varying degrees in 2021 (p < 0.05).
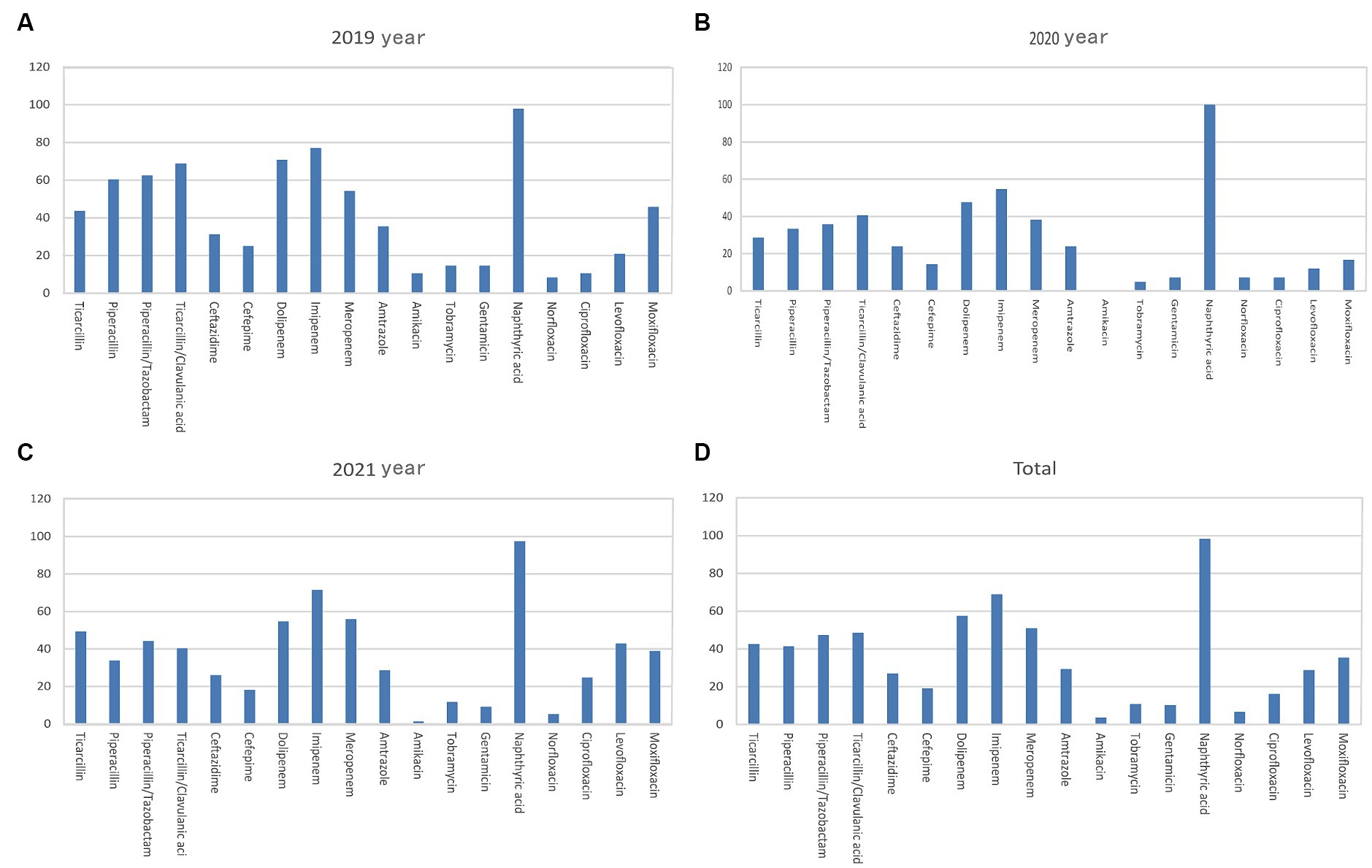
Figure 4. Analysis of the rates of non-fermenting bacterial drug resistance. Rates of Pseudomonas aeruginosa drug resistance in (A) 2019, (B) 2020, (C) 2021, and (D) 2019–2021. The X-axis of all the graphs displays antimicrobial agents, while the Y-axis represents the percentage proportions of resistant bacteria strains.
3.5 Drug-resistant characteristics of the major gram-positive bacteria
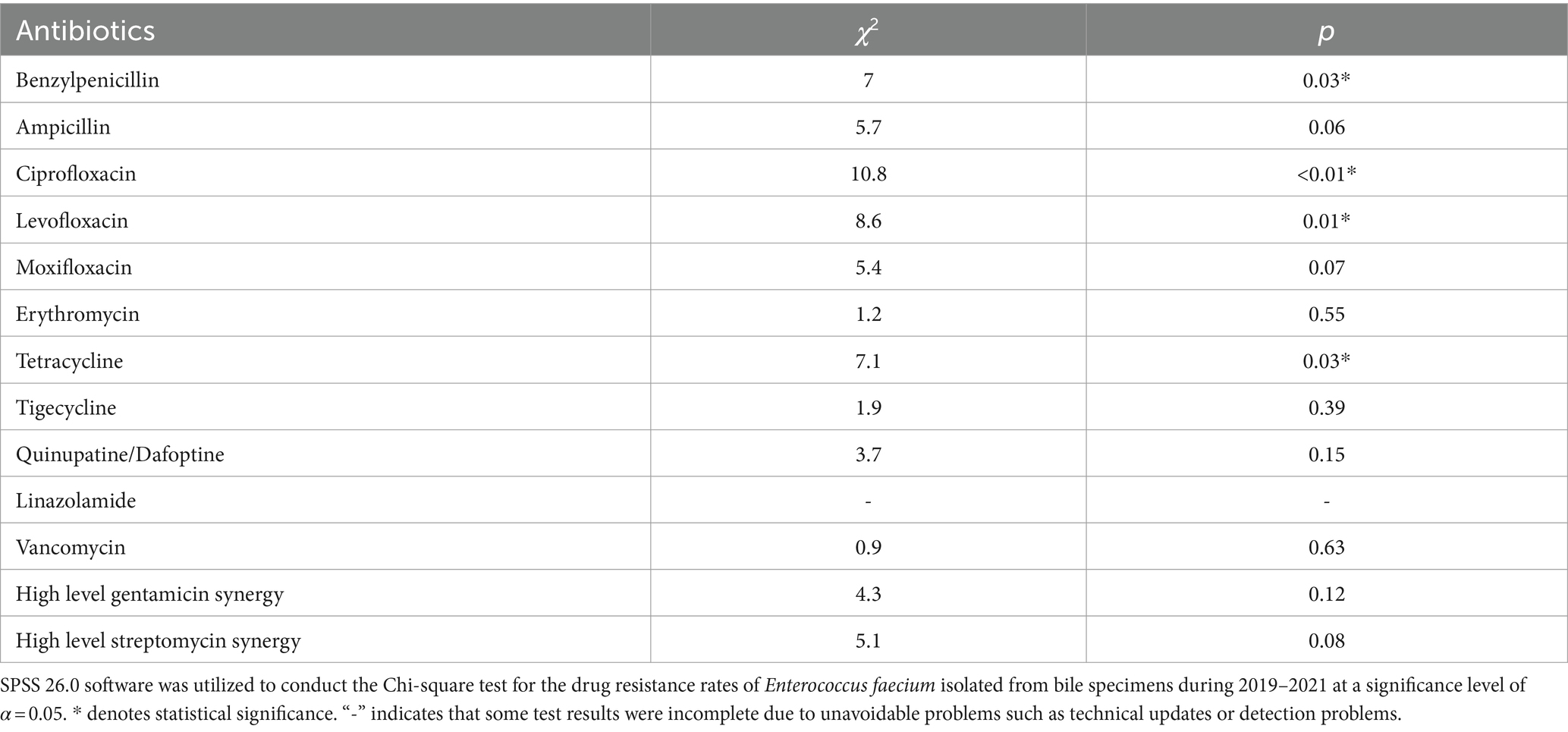
The rate of Enterococcus faecium resistance to a variety of antimicrobials, including benzicillin, ampicillin, levofloxacin, ciprofloxacin, moxifloxacin, and erythromycin, surpassed 50%, and resistance rates to benzicillin, levofloxacin, and ciprofloxacin varied over time (p < 0.05) (Figure 5A and Table 4). There was an increase in the rate of Enterococcus faecium resistance to tetracycline by year (p < 0.05). Enterococcus faecium also showed high sensitivity to tigecycline and vancomycin, with average resistance rates of 0.22 and 0.52%, respectively. Fortunately, no linezolidin-resistant Enterococcus faecium strains were identified.
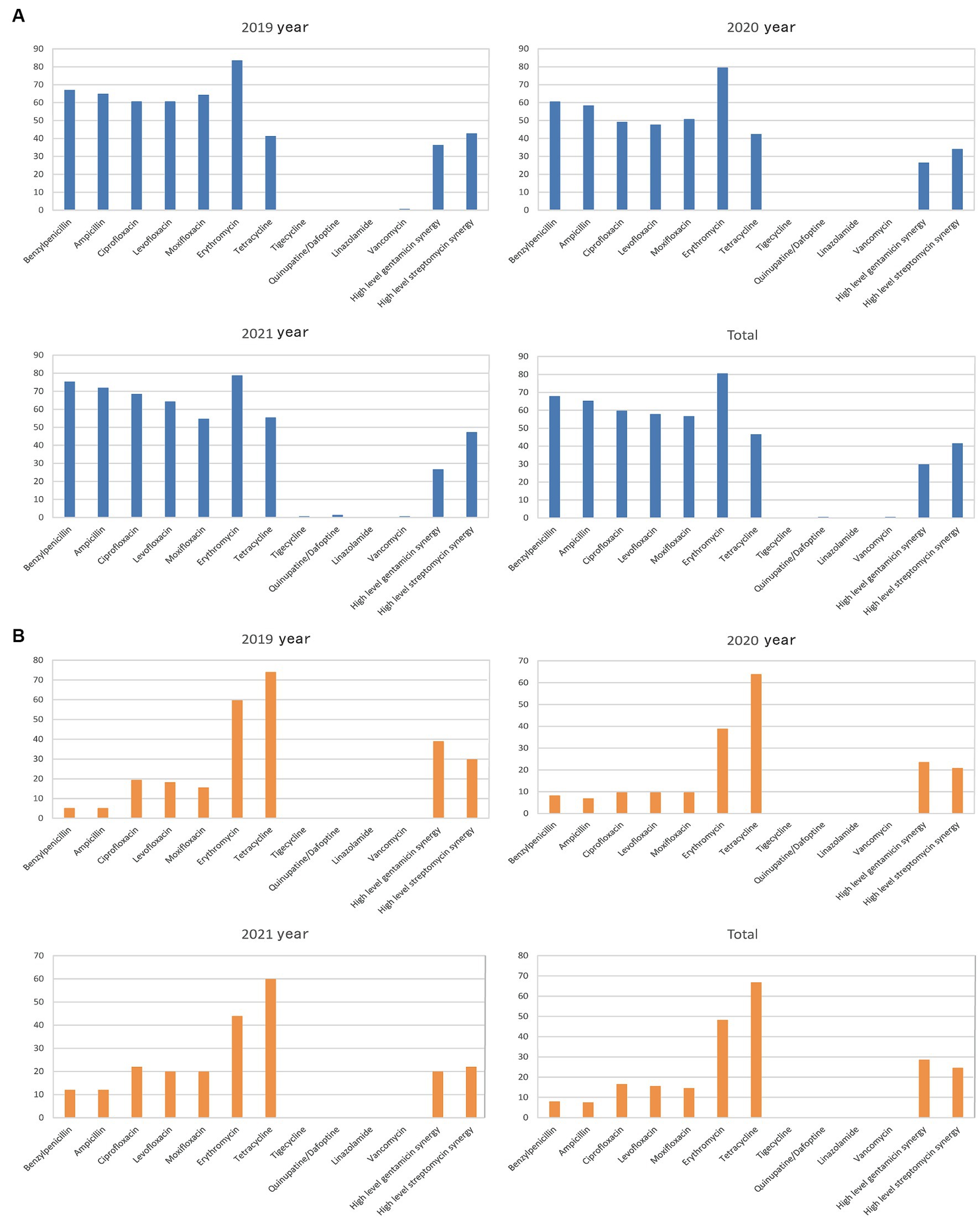
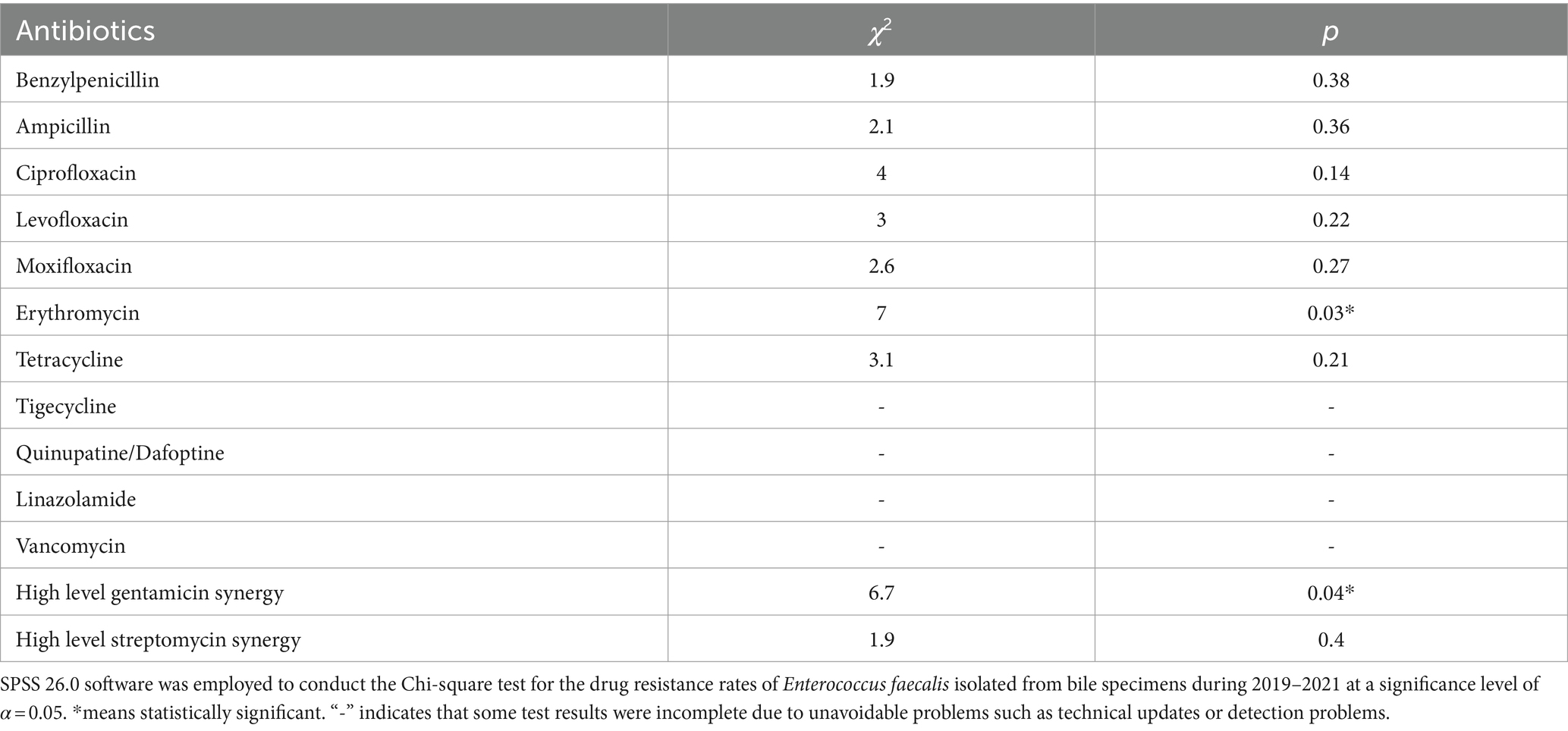
Figure 5. Characteristics of major Gram-positive bacterial drug resistance. The X-axis of all graphs represents antimicrobial agents for Gram-positive bacteria, while the Y-axis depicts the percentages of resistant bacterial strains. (A) Analysis of Enterococcus faecium drug resistance in patients with bile tract infection during 2019–2021. Upper left: Enterococcus faecium drug resistance in 2019. Upper right: Enterococcus faecium drug resistance in 2020. Left bottom: Enterococcus faecium drug resistance in 2021. Right bottom: Enterococcus faecium drug resistance during 2019–2021. (B) Enterococcus faecalis drug resistance in patients with bile tract infection during 2019–2021. Upper left: Enterococcus faecalis drug resistance in 2019. Upper right: Enterococcus faecalis drug resistance in 2020. Left bottom: Enterococcus faecalis drug resistance in 2021. Right bottom: Enterococcus faecalis drug resistance during 2019–2021.
Enterococcus faecalis was more sensitive to most antibiotics than Enterococcus faecium, and showed variability in its resistance to erythromycin (p < 0.05). In contrast, the rate of resistance to high-concentration gentamicin declined by year (p < 0.05) (Figure 5B and Table 5).
3.6 Drug resistance of major fungal pathogens
Candida albicans and Candida tropicalis were the major fungi isolated from the bile samples, and Candida albicans showed 100.00% sensitivity to common antifungal drugs during 2019–2021. Meanwhile, Candida tropicalis showed partial or full resistance to itraconazole, fluconazole, and voriconazole in 2019 and 2021, respectively. Importantly, Candida tropicalis showed no resistance to other antifungal drugs (Table 6).

Table 6. The analysis of the resistance rate of major fungi detected in bile tract infection from 2019 to 2021 year (%).
3.7 Analysis of major risk factors
Analysis of major risk factors was performed according to the literature (Chen S. et al., 2021). The risk factors for common MDRO infections are currently believed to be age, invasive operation, treatment with three or more antimicrobial agents, and previous multiple or long-term hospitalization. In this study, univariate analyses of 14 common potential risk factors were conducted, and the results are shown in Table 7. Multivariate logistic regression analysis was performed for 6 types of risk factors, and the results suggested that the use of third- or fourth-generation cephalosporins was an independent risk factor for Gram-negative MDRO infections in patients with biliary infection (p < 0.05), as shown in Table 8.
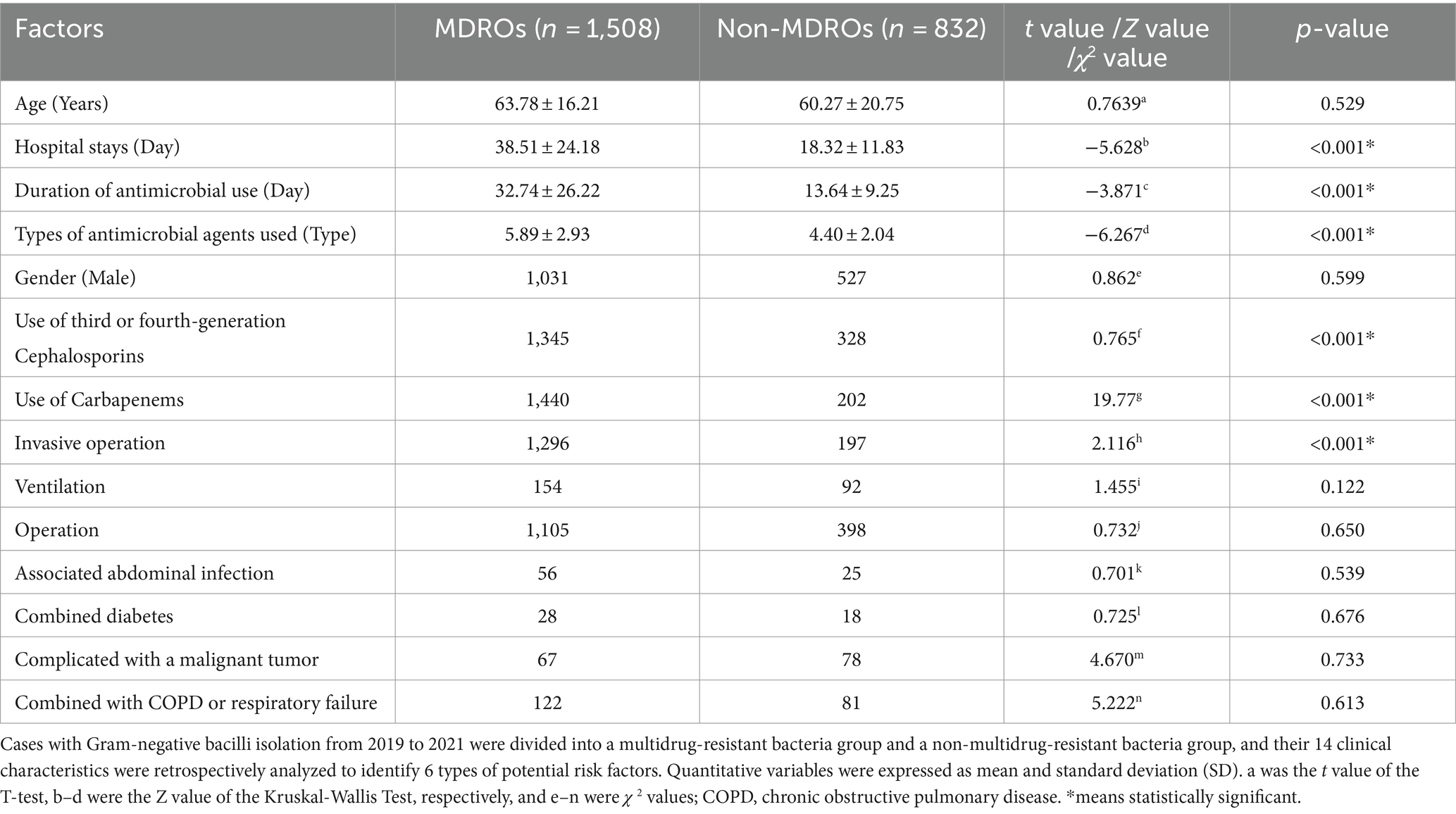
Table 7. Single factor analysis of characteristics associated with bile Gram-negative MDRO infection in patients with biliary tract infection.
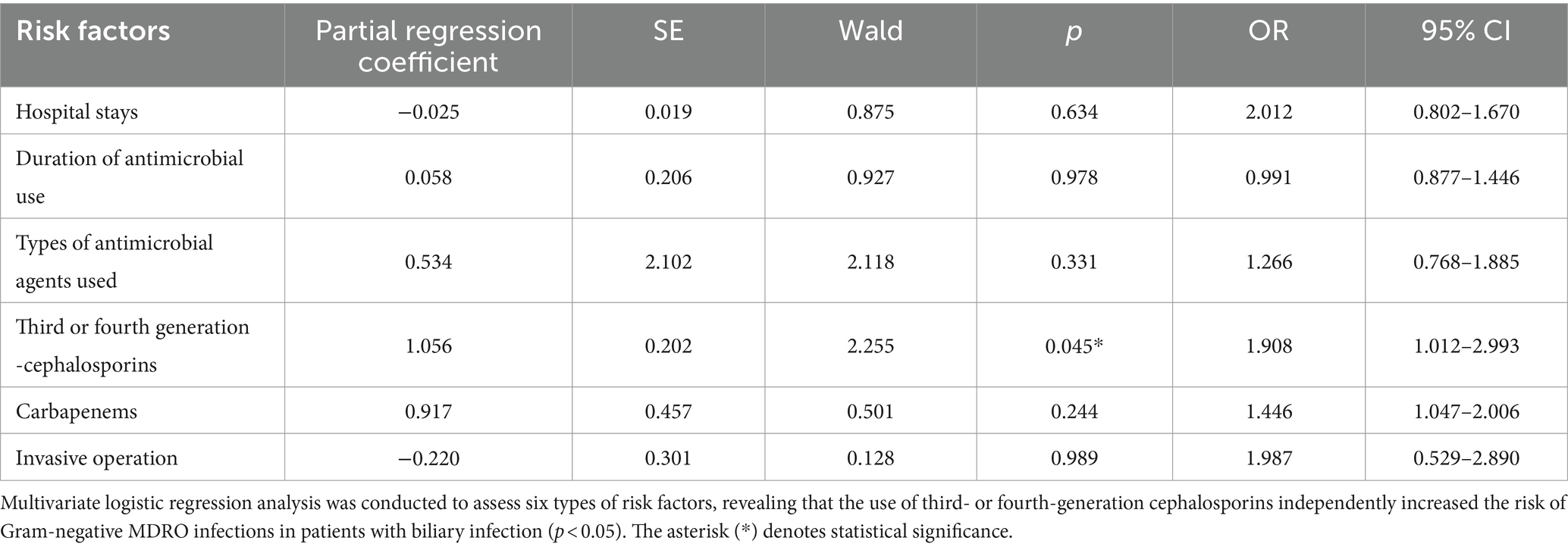
Table 8. Multivariate logistic regression analysis of bile Gram-negative MDRO infection in patients with biliary tract infection.
4 Discussion
Bile is a vital bodily fluid that plays a crucial role in digestion and metabolism within the human body. It typically maintains sterility, and samples are frequently collected during routine in-hospital examinations when infected by pathogens (Jang et al., 2023). Unfortunately, its infection is increasing worldwide. When the normal excretion of bile is blocked by tumors, stones, or worm infections, there is increased pressure in the biliary tract, allowing the invasion of intestinal bacteria through the intestine or portal vein. This can promote the development of acute or chronic cholecystitis, as well as abdominal infection (Zhang et al., 2023).
It is well-known that the rates of resistance to antibiotics vary by bacterial type and time (Shen et al., 2023; Yin et al., 2023). The authors of this paper have extensive experience in clinical microbiological detection within hospital settings. For them, gaining a comprehensive understanding of the distribution and drug resistance patterns of common biliary tract pathogens in China is crucial. This knowledge is essential for enhancing detection capabilities and reducing mortality rates among patients with biliary infections.
To address these issues, the intraoperative collection of bile and bacterial culture samples can greatly improve the accuracy of diagnosis and treatment by providing a more comprehensive understanding of the patient’s condition (Asukai et al., 2023; Papazyan et al., 2023). Moreover, although there are many methods to treat corresponding infectious complications, the effect is not satisfactory (Chi et al., 2023). Therefore, it is necessary to actively prevent pathogen infection in patients, and important to understand the characteristics of pathogen infections and their associated risk factors.
At present, there are scientific studies on clinical biliary infection in humans. The 2000 British study and the 2023 India paper reported ambiguity resulting from limited sample size and a lack of detailed pathogen classification (Kar et al., 2023). In contrast, the 2017 report from Japan, as well as the 2006 report of clinical infections in China, identified Pseudomonas aeruginosa, Enterococcus, Klebsiella pneumoniae, and Escherichia coli as the most prevalent bacteria in biliary tract infection. This aligns with our findings regarding species composition but in an entirely reversed sequence (Chen et al., 2006). Interestingly, the pathogenic dominant strains in our study were similar to those in the Iranian study (Shafagh et al., 2021). There were relatively few controversial studies on pathogen infections in China with the comparison that the dominant strains of acute pancreatic infection were different except for Escherichia coli (Chen H. L. et al., 2021). Hence, it is necessary to conduct in-depth research on infectious pathogens in the bile of biliary infection patients.
In this study, the bile samples were incubated and separated by a BacT/Alert3D automated blood culture instrument by international standard operation procedures for bacterial detection (Chen S. et al., 2021); this method can guarantee the growth and accurate identification of specific organisms, even if the samples contained multiple species.
According to the data obtained from bile specimens from 1,556 patients in hospitals. The bile samples in this study had a positive pathogen detection rate of 72.30% during 2019–2021, supporting prior studies conducted in China (Mavri and Smole Mozina, 2013; Wajima et al., 2022). A total of 3,490 pathogenic bacteria were identified, primarily involving Gram-negative bacilli (67.05%), followed by Gram-positive cocci (29.48%) and fungi (3.12%).
During the 3 years, the predominant Gram-negative bacilli included Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae subspecies seweris, and Pseudomonas aeruginosa. The distribution pattern bears a certain degree of resemblance to the microecological arrangement observed in the gastrointestinal tract in SAP (Chen S. et al., 2021). It is noteworthy that Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae subspecies seweris exhibited a similar pattern of initial decline followed by an increase; however, their data in 2021 did not surpass that of 2019. In contrast, Pseudomonas aeruginosa displayed a distinct pattern of continuous growth. Among the Gram-positive cocci species identified mainly were Enterococcus faecium and Enterococcus faecalis; however, their trends differed. The former showed a slight decline followed by an increase while the latter demonstrated a decreasing trend. Candida albicans was identified as the major fungal species. Regarding obligate anaerobic bacteria, Clostridium perfringens remained the primary species but with a declining trend. The presence of fungi and obligate anaerobes detected, however, is relatively limited in quantity. Therefore, there may be some potential deviation, thus it is not necessary to draw excessive inferences.
These findings are consistent with those described in the National Bacterial Resistance Monitoring Network’s monitoring report (Kawecki et al., 2007; Oldereid et al., 2023). Escherichia coli, Klebsiella pneumoniae, Enterococcus, and other intestinal flora were the primary pathogenic Gram-negative bacilli identified by the current study. Biliary tract infection is often caused by opportunistic intestinal pathogens entering retrograde (Babekir et al., 2023), suggesting that they may be related to the placement of drainage tubes, biliary stents, and other interventional treatments (Quinn et al., 2020; Suh et al., 2021; Behroozian et al., 2022).
Most of the bile pathogens in this study were detected in hepatobiliary and pancreatic surgery and tumor surgery departments by the laboratory clinicians. The majority of patients were experiencing inflammation or tumors of the gallbladder, bile duct, liver, or pancreas, and most underwent surgery or interventional treatments, such as ERCP (Mistry and Zeng, 2022). Older age, a higher number of gallstones, hypoproteinemia, recurrent gallstones, relapsed biliary tract surgery, malignant biliary tract infection, and previous antibiotic exposure are all risk factors for biliary tract infection (Gouveia et al., 2017; Kawanishi et al., 2017; Xing et al., 2022). Thus, more attention should be given to these patient populations, including the implementation of intervention measures to prevent biliary tract infection.
Analysis of the Gram-negative bacteria confirmed that Escherichia coli and Klebsiella pneumoniae Extended-spectrum 𝛽-lactamase (ESBL) detection rates were 37.21 and 17.56%, respectively. While the rate of Escherichia coli antibiotic resistance remained stable during 2019–2021, the rate of resistance to ciprofloxacin increased significantly in 2021 (p < 0.05). This may be due to the high concentration of this drug in bile. Since ciprofloxacin is a common antibiotic used to treat biliary tract infections, more vigilance is required for the use of this drug. While Klebsiella pneumoniae was more susceptible to most antibiotics than Escherichia coli, its resistance to tigecycline and carbapenem increased significantly in 2021 (p < 0.01).
Klebsiella pneumoniae resistance to carbapenem antibiotics was mediated by several different resistance mechanisms, including the production of carbapenemase, increases in effervescent pump activity, and changes in outer membrane protein quantity and function (Yan et al., 2021; Wang et al., 2022; Jing et al., 2023). Tigecycline, one of the newest antibiotic drugs used to treat carbapenem-resistant strains, has been in clinical use in China since 2012 (Fergadaki et al., 2021); however, drug-resistant strains developed quickly and are increasing in number every year. Several mechanisms of resistance, mediated by both chromosomes and plasmids, have been identified (Chen H. L. et al., 2021; Yang J. et al., 2023; Yang Y. et al., 2023). The current study also showed an increase in the resistance of Klebsiella pneumoniae to cephalosporins, carbapenems, lactamase inhibitors, cephalases, sminoglycosides, and quinolones (p < 0.05). The increase in Klebsiella pneumoniae drug resistance indicates a need to intensify the prevention and control of these bacteria in clinical settings.
Pseudomonas aeruginosa was one of the primary causes of infection in bile. The rates of resistance to piperacillin, piperacillin/tazobactam, ticacillin/clavulanate, and amikacin decreased significantly over time (p < 0.05). After the decline in resistance to quinolones in 2020, however, Pseudomonas aeruginosa had varying degrees of resistance in 2021 (p < 0.05). This is an important opportunistic pathogen and a causative agent of clinical infections. Patients who are immunocompromised, have complicated underlying diseases, or have received long-term use of antibiotics or interventional therapy are at higher risk of infection and should be monitored more closely to prevent and effectively treat infection (Araya et al., 2023).
Among Gram-positive bacteria, Enterococcus faecium was the most likely to develop drug resistance, followed by Enterococcus faecalis. The resistance rate of Enterococcus faecium to tetracycline increased over time (p < 0.05), and strains that were resistant to tigecycline and vancomycin also emerged. Low hemoglobin is significantly associated with the production of vancomycin-resistant enterococci, indicating that drug-resistant bacteria may acquire iron to evolve (Brunson et al., 2023). These findings indicate a need to pay more attention to biliary enterococcal infection in patients with anemia.
While Enterococcus faecalis had higher drug sensitivity than Enterococcus faecium, the rate of resistance to erythromycin and high-concentration gentamicin decreased over time (p < 0.05). Meanwhile, Enterococcus faecalis showed no resistance to tigecycline, linezolid, or vancomycin, indicating that these should be the first choice for treatment in severely infected patients. Aminoglycosides can be combined as needed to achieve synergistic effects (Anjum et al., 2022).
Candida albicans and Candida tropicalis were the top fungal strains isolated from bile, and most were sensitive to common antifungal drugs (Jabeen et al., 2023). These findings indicate that the use of conventional antifungal drugs can have strong therapeutic effects. However, clinicians should make sure to use standardized operating and drug application procedures and strive for aseptic infection.
It is important to note that 12 anaerobic bacteria (0.34%) were also isolated from the bile samples. This finding highlights a need to test bile samples for both aerobic and anaerobic bacteria to avoid missing anaerobic infections. In addition, treatment directed at both aerobic and anaerobic bacteria should be considered for patients with a history of bilio-entero-anastomosis (Ligero-Lopez et al., 2023).
With the extensive use of antimicrobial agents at present, MDROs have become important pathogens of clinical infections. MDROs are resistant to ampicillin, first-generation cephalosporins, and second-generation cephalosporins. Moreover, the sensitivity trends of ceftazidime, piperacillin/tazobactam, cefoperazone/sulbactam, gentamicin, and tobramycin were similar. However, resistance to both ceftriaxone and ciprofloxacin was higher than that to ciprofloxacin. In contrast, resistance to aminoglycosides, cefepime, and carbapenems was higher (Xiao et al., 2023). It was concluded that the drug resistance patterns of MDROs were very different, the patterns of MDROs differ slightly between the northern and southern regions in China, even within the same country (Ruan et al., 2019). This variability is less conducive to clinical anti-infection treatment in patients. Merely, analysis of resistance of MDROs and risk factors for Gram-negative MDRO infection can help identify relevant risk factors and prevent infection (Chen S. et al., 2021). Our results showed that six factors, such as the length of hospital stay, time, antibacterial drug use, antimicrobial type, use of third- or fourth-generation cephalosporins, use of penicillium carbon alkene antimicrobial agents, and invasive operations, were risk factors for Gram-negative MDRO infection in biliary infectious patients. Multivariable logistic regression analysis of these factors indicated that the use of third- or fourth-generation cephalosporins in biliary infectious patients with the presence of biliary Gram-negative MDROs was an independent risk factor for infection and high drug resistance. According to a report by Skorochod et al. (2022), independent risk factors for Israeli patients with Staphylococcus aureus hepatobiliary infection included hypertension, bedridden status, and nursing home residence. This suggests that different countries may have their own unique set of independent risk factors for this condition. The rational use of third- or fourth-generation cephalosporins in China was an important measure in preventing MDRO infection in this type of patient.
5 Conclusion
In conclusion, this study has determined that Gram-negative bacteria were the predominant pathogens isolated from patients with biliary infections, particularly Escherichia coli. The rates and patterns of drug resistance were found to be high and constantly changing. In clinical practice, it is crucial to judiciously use third- or fourth-generation cephalosporins as a key measure to prevent multidrug-resistant organism (MDRO) infections in patients with biliary infections. While our study did not investigate resistance mechanisms, the findings have the potential to inform decision-making in the healthcare sector. Furthermore, this information can be utilized to educate both healthcare professionals and the general public about the importance of appropriate antibiotic use. Additionally, the results of this study could lay a foundation for future research efforts, enabling scientists to explore new detection, therapeutic options, and strategies for biliary infections.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethical Committee of Nankai Clinical College, Tianjin Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants' legal guardians/next of kin because Due to its retrospective design, a waiver of participant informed consent was granted by the Ethical Committee of Nankai Clinical College, Tianjin Medical University.
Author contributions
SC: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. WL: Data curation, Writing – original draft, Writing – review & editing. XS: Data curation, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. JLu: Data curation, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. JLi: Data curation, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. HO: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. WZ: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. JC: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. ZY: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. HL: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. YZ: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants 81703846 (SC was the recipient) from the National Science Foundation of China (NSFC), Dongguan Science and Technology of Social Development Program 20231800940062 (SC was the recipient), 20231800937142 (JLi was the recipient), 20231800904242 (XS was the recipient), and 20231800904232 (JLu was the recipient), and grant BS2023001 (SC was its recipient) from the Doctoral Research Foundation of Binhaiwan Central Hospital in Dongguan, Guangdong Province. SC was a recipient of the First Batch of Young Medical Professionals award by the 2018 Tianjin Health Commission.
Acknowledgments
We appreciate the help of technical support from the Tianjin Hospital of Traditional Chinese Medicine. Thank you to all participants involved in the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1404366/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | The culture images of samples culture and sole bacterial colony isolation in detection. (A) The typical aerobic (left) and anaerobic (right) culture bottles with positive indication were shown after aseptically injected by bile samples. (B) The Gram-positive bacteria, such as: Escherichia coli (Left), Klebsiella pneumoniae (middle) and Pseudomonas aeruginosa (right) in the detection were displayed. (C) The major Gram-positive bacterial as Enterococcus faecalis (Left) and Enterococcus faecium (right) were compared. (D) Candida albicans belong with the fungal pathogens were exhibited.
SUPPLEMENTARY FIGURE S2 | The number comparison of pathogenic bacteria in patients with bile tract infection in 2019–2021. The graphs displayed case numbers of biliary bacterial infection on the Y-axis and years for bacterial collection on the X-axis. The different graphs represented the following categories of bacteria: (A) The Gram-negative bacteria, (B) the Gram-positive bacteria, (C) the fungal pathogens, and (D) the obligate anaerobic bacteria.
References
Alreshidi, M., Dunstan, H., Roberts, T., Alreshidi, F., Hossain, A., Bardakci, F., et al. (2023). Cytoplasmic amino acid profiles of clinical and ATCC 29213 strains of Staphylococcus aureus harvested at different growth phases. Biomol. Biomed. 23, 1038–1050. doi: 10.17305/bb.2023.9246
Anjum, J., Zaidi, A., Barrett, K., and Tariq, M. (2022). A potentially probiotic strain of Enterococcus faecalis from human milk that is avirulent, antibiotic sensitive, and nonbreaching of the gut barrier. Arch. Microbiol. 204:158. doi: 10.1007/s00203-022-02754-8
Araya, S., Gebreyohannes, Z., Tadlo, G., Gessew, G. T., and Negesso, A. E. (2023). Epidemiology and multidrug resistance of Pseudomonas aeruginosa and Acinetobacter baumanni isolated from clinical samples in Ethiopia. Infect. Drug Resist. 16, 2765–2773. doi: 10.2147/IDR.S402894
Asukai, K., Akita, H., Mukai, Y., Mikamori, M., Hasegawa, S., Fujii, Y., et al. (2023). The utility of bile juice culture analysis for the management of postoperative infection after pancreaticoduodenectomy. Surgery 173, 1039–1044. doi: 10.1016/j.surg.2022.11.021
Babekir, M. S., Abdelrahim, E. Y., Doush, W. M. A., and Abdelaziz, M. S. (2023). Pattern of bile cultures and antibiotic sensitivity tests in Sudanese patients diagnosed with obstructive jaundice: a single-center prospective study. JGH Open 7, 497–503. doi: 10.1002/jgh3.12937
Behroozian, S., Sampedro, I., Dhodary, B., Her, S., Yu, Q., Stanton, B. A., et al. (2022). Pseudomonas aeruginosa PAO1 is attracted to bovine bile in a novel, cystic fibrosis-derived bronchial epithelial cell model. Microorganisms 10:716. doi: 10.3390/microorganisms10040716
Brunson, D. N., Colomer-Winter, C., Lam, L. N., and Lemos, J. A. (2023). Identification of multiple Iron uptake mechanisms in Enterococcus faecalis and their relationship to virulence. Infect. Immun. 91:e0049622. doi: 10.1128/iai.00496-22
Carvalhaes, C. G., Shortridge, D., Woosley, L. N., Gurung, N., and Castanheira, M. (2023). Performance of the Vitek 2 advanced expert system (AES) as a rapid tool for reporting antimicrobial susceptibility testing (AST) in Enterobacterales from north and Latin America. Microbiol. Spectr. 11:e0467322. doi: 10.1128/spectrum.04673-22
Chen, H. L., Jiang, Y., Li, M. M., Sun, Y., Cao, J. M., Zhou, C., et al. (2021). Acquisition of Tigecycline Resistance by Carbapenem-resistant Klebsiella pneumoniae confers collateral hypersensitivity to aminoglycosides. Front. Microbiol. 12:674502. doi: 10.3389/fmicb.2021.674502
Chen, J., Li, L., Guan, X., Chen, D., Chen, M., Ouyang, B., et al. (2006). The drug resistance of pathogenic bacteria of nosocomial infections in surgical intensive care unit. Zhonghua Wai Ke Za Zhi 44, 1189–1192
Chen, S., Shi, J., Chen, M., Ma, J., Zeng, Z., Wang, R., et al. (2021). Characteristics of and risk factors for biliary pathogen infection in patients with acute pancreatitis. BMC Microbiol. 21:269. doi: 10.1186/s12866-021-02332-w
Chi, J. Y., Ma, L. Y., Zou, J. C., and Ma, Y. F. (2023). Risk factors of pancreatitis after endoscopic retrograde cholangiopancreatography in patients with biliary tract diseases. BMC Surg. 23:62. doi: 10.1186/s12893-023-01953-4
Ekpanyapong, S., and Reddy, K. R. (2023). Liver and biliary tract disease in patients with coronavirus disease-2019 infection. Gastroenterol. Clin. N. Am. 52, 13–36. doi: 10.1016/j.gtc.2022.09.001
Fergadaki, S., Renieris, G., Machairas, N., Sabracos, L., Droggiti, D. I., Misiakos, E., et al. (2021). Efficacy of tigecycline alone or in combination for experimental infections by KPC carbapenemase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents 58:106384. doi: 10.1016/j.ijantimicag.2021.106384
Gouveia, M. J., Pakharukova, M. Y., Laha, T., Sripa, B., Maksimova, G. A., Rinaldi, G., et al. (2017). Infection with Opisthorchis felineus induces intraepithelial neoplasia of the biliary tract in a rodent model. Carcinogenesis 38, 929–937. doi: 10.1093/carcin/bgx042
Jabeen, G., Naz, S. A., Rangel, D. E. N., Jabeen, N., Shafique, M., and Yasmeen, K. (2023). In-vitro evaluation of virulence markers and antifungal resistance of clinical Candida albicans strains isolated from Karachi, Pakistan. Fungal Biol. 127, 1241–1249. doi: 10.1016/j.funbio.2023.04.003
Jang, E., Jung, S., Sohng, W., Choi, D., Hwang, G. S., and Chung, H. (2023). Screening of gall bladder cancer through infrared analysis of bile and examination of varied bile constituent composition by the disease. Spectrochim. Acta A Mol. Biomol. Spectrosc. 286:122030. doi: 10.1016/j.saa.2022.122030
Jing, X. H., Zhao, G. Y., Wang, G. B., Huang, Q. L., Zou, W. S., Huang, L. N., et al. (2023). Naringin alleviates pneumonia caused by Klebsiella pneumoniae infection by suppressing NLRP3 inflammasome. Biomed. Pharmacother. 170:116028. doi: 10.1016/j.biopha.2023.116028
Kar, M., Dubey, A., Patel, S. S., Sahu, C., and Yadav, A. (2023). Multifactorial analysis of biliary infection in patients with Hepato-pancreatico-biliary and associated intraabdominal malignancies admitted to a teaching Hospital in Northern India. Euroasian J. Hepatogastroenterol. 13, 10–17. doi: 10.5005/jp-journals-10018-1389
Kawanishi, K., Kinoshita, J., Abe, H., Kakimoto, T., Yasuda, Y., Hara, T., et al. (2017). Appendectomy as a risk factor for bacteremic biliary tract infection caused by antibiotic-resistant pathogens. Biomed. Res. Int. 2017:3276120. doi: 10.1155/2017/3276120
Kawecki, D., Chmura, A., Pacholczyk, M., Lagiewska, B., Adadynski, L., Wasiak, D., et al. (2007). Bacteria isolated from bile samples of liver recipients in the early period after transplantation: epidemiology and susceptibility of the bacterial strains. Transplant. Proc. 39, 2807–2811. doi: 10.1016/j.transproceed.2007.08.044
Ligero-Lopez, J., Rubio-Mora, E., Ruiz-Bastian, M. D., Quiles-Melero, M. I., Cacho-Calvo, J., and Cendejas-Bueno, E. (2023). Antimicrobial susceptibility testing of anaerobic bacteria causing bacteremia: a 13-year (2010-2022) retrospective study in a tertiary hospital. Anaerobe 84:102803. doi: 10.1016/j.anaerobe.2023.102803
Mavri, A., and Smole Mozina, S. (2013). Resistance to bile salts and sodium deoxycholate in macrolide- and fluoroquinolone-susceptible and resistant campylobacter jejuni and Campylobacter coli strains. Microb. Drug Resist. 19, 168–174. doi: 10.1089/mdr.2012.0217
Mistry, N. P., and Zeng, W. (2022). Intense FDG uptake in the common bile duct post-ERCP mimics acute infectious cholangitis. Tomography 8, 2946–2951. doi: 10.3390/tomography8060248
Oldereid, T. S., Jiang, X., Ogaard, J., Schrumpf, E., Bjornholt, J. V., Rasmussen, H., et al. (2023). Microbial exposure during early life regulates development of bile duct inflammation. Scand. J. Gastroenterol. 59, 192–201. doi: 10.1080/00365521.2023.2278423
Papazyan, R., Ferdyan, N., Srinivasan, K., Gonzalez, C., Shannon, W. D., Blount, K., et al. (2023). Human fecal bile acid analysis after investigational microbiota-based live biotherapeutic delivery for recurrent clostridioides difficile infection. Microorganisms 11:135. doi: 10.3390/microorganisms11010135
Plantamura, E., Huyghe, G., Panterne, B., Delesalle, N., Thepot, A., Reverdy, M. E., et al. (2012). Validation of the BacT/ALERT(R)3D automated culture system for the detection of microbial contamination of epithelial cell culture medium. Cell Tissue Bank. 13, 453–459. doi: 10.1007/s10561-011-9281-1
Quinn, R. A., Melnik, A. V., Vrbanac, A., Fu, T., Patras, K. A., Christy, M. P., et al. (2020). Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129. doi: 10.1038/s41586-020-2047-9
Ruan, H., Liao, G., Peng, P., and Liu, S. (2019). Microbial profiles and risk factors of preexisting biliary infection in patients with therapeutic endoscopy. Gastroenterol. Res. Pract. 2019:1527328. doi: 10.1155/2019/1527328
Shafagh, S., Rohani, S. H., and Hajian, A. (2021). Biliary infection; distribution of species and antibiogram study. Ann. Med. Surg. (Lond). 70:102822. doi: 10.1016/j.amsu.2021.102822
Shen, R., Zhou, Y., Zhang, L., and Yang, S. (2023). The value of bile acid spectrum in the evaluation of hepatic injury in children with infectious mononucleosis caused by Epstein Barr virus infection. Front. Pediatr. 11:1109762. doi: 10.3389/fped.2023.1109762
Skorochod, R., Raveh, D., Wiener-Well, Y., Fteiha, B., Shteingart, S., and Skorochod, Y. (2022). Clinical characteristics of patients with Staphylococcus aureus bile infection. Isr. Med. Assoc. J. 24, 643–648,
Suh, S. W., Choi, Y. S., Choi, S. H., Do, J. H., Oh, H. C., Kim, H. J., et al. (2021). Antibiotic selection based on microbiology and resistance profiles of bile from gallbladder of patients with acute cholecystitis. Sci. Rep. 11:2969. doi: 10.1038/s41598-021-82603-8
Wajima, T., Sugawara, T., Umeda, Y., Hagimoto, A., Tanaka, E., and Nakaminami, H. (2022). Molecular characterisation of carbapenem- and tigecycline-resistant Klebsiella pneumoniae strains isolated from blood and bile samples. J. Infect. Chemother. 28, 187–191. doi: 10.1016/j.jiac.2021.10.005
Wang, Y. P., Chen, Y. H., Hung, I. C., Chu, P. H., Chang, Y. H., Lin, Y. T., et al. (2022). Transporter genes and fosA associated with fosfomycin resistance in carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 13:816806. doi: 10.3389/fmicb.2022.816806
Xiao, S., Fu, Q., Miao, Y., Zhao, M., Lu, S., Xu, J., et al. (2023). Clinical efficacy and drug resistance of ceftazidime-avibactam in the treatment of Carbapenem-resistant gram-negative bacilli infection. Front. Microbiol. 14:1198926. doi: 10.3389/fmicb.2023.1198926
Xing, D., Song, W., Gong, S., Xu, A., and Zhai, B. (2022). Analysis of the bacterial spectrum and key clinical factors of biliary tract infection in patients with malignant obstructive jaundice after PTCD. Dis. Markers 2022:1026254. doi: 10.1155/2022/1026254
Yan, W., Zhang, Q., Zhu, Y., Jing, N., Yuan, Y., Zhang, Y., et al. (2021). Molecular mechanism of polymyxin resistance in multidrug-resistant klebsiella pneumoniae and escherichia coli isolates from Henan Province, China: a multicenter study. Infect. Drug. Resist. 14, 2657–2666. doi: 10.2147/IDR.S314490
Yang, Y., He, R., Wu, Y., Qin, M., Chen, J., Feng, Y., et al. (2023). Characterization of two multidrug-resistant Klebsiella pneumoniae harboring tigecycline-resistant gene tet(X4) in China. Front. Microbiol. 14:1130708. doi: 10.3389/fmicb.2023.1130708
Yang, J., Zhang, K., Ding, C., Wang, S., Wu, W., and Liu, X. (2023). Exploring multidrug-resistant Klebsiella pneumoniae antimicrobial resistance mechanisms through whole genome sequencing analysis. BMC Microbiol. 23:245. doi: 10.1186/s12866-023-02974-y
Yin, G., Guo, Y., Ding, Q., Ma, S., Chen, F., Wang, Q., et al. (2023). Klebsiella quasipneumoniae in intestine damages bile acid metabolism in hematopoietic stem cell transplantation patients with bloodstream infection. J. Transl. Med. 21:230. doi: 10.1186/s12967-023-04068-9
Zhang, X., Han, S., Jiang, X., Duan, S., Gao, Y., Ding, J., et al. (2023). Comparative analysis of bile metabolic profile in patients with biliary obstruction complicated by Clonorchis sinensis infection. Front. Cell. Infect. Microbiol. 13:1254016. doi: 10.3389/fcimb.2023.1254016
Keywords: biliary pathogens, Gram-negative bacteria, distribution of bacteria, multidrug-resistant bacteria, risk factors
Citation: Chen S, Lai W, Song X, Lu J, Liang J, Ouyang H, Zheng W, Chen J, Yin Z, Li H and Zhou Y (2024) The distribution and antibiotic-resistant characteristics and risk factors of pathogens associated with clinical biliary tract infection in humans. Front. Microbiol. 15:1404366. doi: 10.3389/fmicb.2024.1404366
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Dhruv Desai, University of Pennsylvania, United StatesAlexey Porollo, Cincinnati Children's Hospital Medical Center, United States
Paola Di Carlo, University of Palermo, Italy
Juan A. Ayala, Autonomous University of Madrid, Spain
Copyright © 2024 Chen, Lai, Song, Lu, Liang, Ouyang, Zheng, Chen, Yin, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shayan Chen, chenshayan@126.com
 Shayan Chen
Shayan Chen Wenbin Lai1,2
Wenbin Lai1,2