- 1Pharmacognosy Department, Faculty of Pharmacy, Cairo University, Giza, Egypt
- 2Department of Phytochemistry and Plant Systematics, Pharmaceutical Division, National Research Centre, Cairo, Egypt
- 3Department of Pharmaceutical Sciences, Faculty of Pharmacy, Almaarefa University, Riyadh, Saudi Arabia
- 4Microbiology and Immunology Department, Faculty of Pharmacy (Girls), Al-Azhar University, Cairo, Egypt
- 5Department of Pharmacy Practice, Faculty of Pharmacy, University of Tabuk, Tabuk, Saudi Arabia
- 6Medicinal Chemistry Department, Theodor Bilharz Research Institute, Giza, Egypt
- 7Microbial Chemistry Department, National Research Centre, Giza, Egypt
Introduction: In recent years, the world’s attention has been drawn to antimicrobial resistance (AMR) because to the frightening prospect of growing death rates. Nanomaterials are being investigated due to their potential in a wide range of technical and biological applications.
Methods: The purpose of this study was to biosynthesis zinc oxide nanoparticles (ZnONPs) using Aspergillus sp. SA17 fungal extract, followed by characterization of the produced nanoparticles (NP) using electron microscopy (TEM and SEM), UV-analysis, X-ray diffraction (XRD), and Fourier-transform infrared spectroscopy (FT-IR).
Results and Discussion: The HR-TEM revealed spherical nanoparticles with an average size of 7.2 nm, and XRD validated the crystalline nature and crystal structure features of the generated ZnONPs, while the zeta potential was 18.16 mV, indicating that the particles’ surfaces are positively charged. The FT-IR was also used to identify the biomolecules involved in the synthesis of ZnONPs. The antibacterial and anticancer properties of both the crude fungal extract and its nano-form against several microbial strains and cancer cell lines were also investigated. Inhibition zone diameters against pathogenic bacteria ranged from 3 to 13 mm, while IC50 values against cancer cell lines ranged from 17.65 to 84.55 M. Additionally, 33 compounds, including flavonoids, phenolic acids, coumarins, organic acids, anthraquinones, and lignans, were discovered through chemical profiling of the extract using UPLC-QTOF-MS/MS. Some molecules, such pomiferin and glabrol, may be useful for antibacterial purposes, according to in silico study, while daidzein 4’-sulfate showed promise as an anti-cancer metabolite.
Introduction
Nanoparticles have gained significant attention due to their unique physiochemical properties which permit them to conjugate other groups due to their charged surface (Chavan et al., 2020; Randive et al., 2020, 2021, 2023). Among these nanoparticles, Zinc nanoparticles (ZnONPs) have gained reasonable interest in recent years due to their unique properties such as their small size and large surface which make them ideal candidates for use in numerous fields like biomedicine, electronics, and agriculture (Gomaa, 2022). Biosynthesis of ZnONPs from endophytic fungi has emerged as a promising alternative way to produce eco-friendly and cost-effective nanometals compared to conventional physical and chemical methods (Yusof et al., 2019).
Several studies have reported the biosynthesis of ZnONPs using endophytic fungi such as Fusarium oxysporum, Aspergillus fumigatus, and Trichoderma viride. The biosynthesis of ZnONPs using endophytic fungi is achieved by reducing the metal ions to their corresponding nanoparticles, which are stabilized by various biomolecules such as proteins, polysaccharides, and enzymes produced by the fungi (Bachheti et al., 2021). ZnONPs have been extensively studied for their potential applications as antimicrobial agents with a broad spectrum toward a large number of pathogens including bacteria, fungi, and viruses (Abdelaziz et al., 2021). ZnONPs have been shown to have antibacterial properties against a variety of harmful microorganisms, including Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli. Furthermore, it has been claimed that the death of cancer cells could possibly be achieved by using ZnONPs through variable pathways, viz. autophagy, necrosis, and apoptosis. ZnONPs have been shown to exhibit anticancer properties against various cancer cell types, including those found in the prostate, lung, and breast tissues (El-Bendary et al., 2021; Abdel-Nasser et al., 2022).
However, no published data currently exist regarding the antimicrobial and anticancer properties of the characterized ZnO nanoparticles with Aspergillus sp. SA17 crude extract. This lack of information has motivated us to explore the phytochemical identity of the crude extract, considering their previously reported data and using molecular docking. Molecular docking is an important computational technique to predict the best interaction between ligand and its functional site.
The objectives of this study were the biosynthesis and characterization of ZnO nanoparticles using Aspergillus sp. SA17 crude extract. Moreover, the chemical constituents of the crude extract were identified using UPLC-QTOF-MS/MS. The antimicrobial and anticancer activities of the crude extract and its nano-form were evaluated to assess the underlying mechanisms by which crude extract inhibited cell proliferation. Also, a molecular docking study was performed.
Materials and methods
Fungal isolation
The separation of endophytic fungi from marine seagrass was performed using the surface sterilization method. The marine seagrass was collected from Hurghada, Egypt, and washed thoroughly with running seawater to remove debris and sand. The samples were then cut into small pieces and surface sterilized via immersion in 70% ethanol for 30 s, followed by washing three times with sterile seawater, then immersion in 5% sodium hypochlorite for 30 s, and then washing three times with sterile seawater. After surface sterilization, the seagrass samples were cut and placed on a potato dextrose agar (PDA) medium supplemented with antibiotics (ampicillin 100 mg/L and streptomycin 50 mg/L) to inhibit bacterial contamination. The plates were incubated at 25°C until fungal colonies appeared (Li and Wang, 2009).
Genetic identification of fungal strain
The fungus strains were isolated and cultivated for 5 days at 25°C in potato dextrose broth media. The suspension was then centrifuged at 10,000 rpm for 10 min at room temperature. DNA extraction was carried out using DNeasy Blood & Tissue Kits manufactured by Qiagen, a leading biotechnology company headquartered in Hilden, Germany. Extraction was performed according to the manufacturer’s instructions. The kit is designed to efficiently purify genomic DNA from various sample sources. Amplification was carried out using two primers, ITS2, GCTGCGTTCTTCATCGATGC, and ITS3, GCATCGATGAAGAACGCAGC (White et al., 1990) and the PCR condition was as follows: Denaturation for 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, then 30 s at 55°C, 90 s at 72°C, and a final 5 min extension step at 72°C. The purified PCR product was sequenced using two primers: ITS1, TCCGTAGGTGAAC-CTGCGG, and ITS4, TCCTCCGCTTATTGATATGC (Op De Beeck et al., 2014; Hamed et al., 2020; Elawady et al., 2023; Khazaal et al., 2023).
Fungal filtrate preparation
The fungus was cultivated in potato dextrose (PD) broth. The PD broth was created by dissolving 24 g of potato dextrose broth (PDB) in 1 L of distilled water. The medium was then poured into sterile flasks. The flasks were sealed with cotton plugs and sterilized by autoclaving at 121°C for 15 min. The endophytic fungi were inoculated into the PD broth by transferring a small piece of fungal mycelium from the PDA plate into the broth using a sterile loop. The flasks were then incubated at 29°C for 7 days with constant shaking at 150 rpm. After 7 days of cultivation, the fungal mycelia were removed from the broth by filtration through filter paper. The filtrate was then gathered and centrifuged at 10,000 rpm for 10 min to eliminate any residual fungal cells or debris. The resulting supernatant was used for further analysis.
The biosynthesis of zinc oxide nanoparticles
Specifically, 1 g of the hydroalcoholic extract was blended with 5 g of a zinc acetate solution, which was dissolvable in 500 mL of bi-distilled water (Attia et al., 2021; Alrabayah et al., 2022). The resulting mixture was heated under stirring for 20 min at 80°C. After that NH4OH drops were added till the formation of yellowish white sediment. The blend was then allowed to sit for 30 min to finalize the interaction. The resulting sediment was centrifuged at 4,000 rpm, cleaned twofold with bi-distilled water, and then cleaned with EtOH to yield a pale yellowish powder of ZnONPs (Ghareeb et al., 2019a; Eskander et al., 2020; Elkhouly et al., 2021a; Hameed et al., 2023).
Characterization of ZnONPs
UV-spectroscopy
The ultraviolet–visible spectral examination was performed using a UV spectrophotometer (Shimadzu Corporation, Japan) to track the transformation of the zinc ion to zinc oxide nanoparticles. The UV spectrum was recorded between 200 and 800 nm.
Fourier-transform infrared spectroscopy
To characterize the functional groups that participated in ZnONPs formation, Fourier transform infrared (FT-IR) analysis was conducted utilizing a FT-IR 6100 spectrometer (Jasco, Japan) operating at a temperature of 25°C, in the domain from 4,000 to 400 cm−1. FT-IR spectroscopy is a potent analytical technique used in nanoparticle characterization because of its ability to identify individual chemical bonds and functional groups in the sample. FT-IR spectroscopy sheds light on molecular structure, composition, and interactions by evaluating the sample’s absorption or emission of infrared radiation. During the analysis, the FT-IR spectrometer fired infrared light at the ZnONPs sample, and the resulting spectrum was recorded. Peaks and patterns in the spectrum corresponded to typical vibrations of chemical bonds within the nanoparticles, such as stretching and bending modes of functional groups such as -OH (hydroxyl), C=O (carbonyl), C-H (alkyl), and others.
Zeta potential measurements
The constancy and charges of the synthesized nanoparticles were assessed using a zeta sizer nano-z’s laser diffractometer (Malvern, United Kingdom). The zeta sizer nano-z’s is equipped with dynamic light scattering (DLS) technology and operates at a temperature of 25°C. Zeta potential measures the electrostatic repulsion forces inside nanostructures and is an important metric for evaluating colloidal stability.
X-ray diffraction
X-ray diffraction (XRD) was carried out using a Bruker D8 Advance Diffractometer (Bruker AXS, Germany) with Cu Ka radiation (k = 1.54) to obtain the XRD pattern of ZnONPs over a 2-theta range of 10–90. XRD detecting and verifying the crystal structure of nanoscale materials, XRD is helpful. The arrangement of atoms in the crystalline lattice can be inferred from the diffraction patterns produced by X-ray diffraction (XRD), which provides important details on the characteristics of the material. The XRD experiment measured a 2-theta range of 10° to 90°. This broad range enabled a complete assessment of the diffraction configurations generated by the ZnONPs, allowing the study of the crystallographic characteristics. X-ray diffraction is operated based on Bragg’s law, with X-rays impacted on a crystalline material diffracted at certain angles given by the spacing of atomic planes inside the crystal lattice. The resulting diffraction pattern contains information on the atoms’ arrangement in the crystal structure, such as lattice parameters, crystal orientation, and phase purity. Using the XRD pattern acquired from the ZnONPs sample, researchers were able to determine the crystal phases present, their relative abundance, and the degree of crystallinity.
Electron microscopy
Finally, the particle dimension and shape of the zinc oxide nanoparticles were examined utilizing transmission electron microscopy (TEM) with a JEOL-JEM-1011 instrument (Japan). Drops of the suspended nanoparticle solution were located on a carbon-coated copper grid, and the solvent was permitted to outlet gradually before the TEM picture was recorded. Additionally, Furthermore, the morphology of the biosynthesized zinc oxide nanoparticles was further studied using scanning electron microscopy (SEM; Quanta FEG-250, Netherlands).
Biological activity
Antimicrobial activity
The antimicrobial activity of the crude extract and ZnONPs was evaluated vs. some pathogenic microbial strains Staphylococcus aureus, Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, Candida albicans, and Aspergillus flavus according to the reported procedures (Madkour et al., 2017; Khalaf et al., 2019). The tested samples were dissolved in methanol and a solution of the concentration 1 mg /ml was prepared separately. Paper discs of Whatman filter paper were prepared in standard size (5 cm) and were cut and sterilized in an autoclave. The paper discs soaked in the desired concentration of the complex solution were placed aseptically in the petri dishes containing nutrient agar media (agar 20 g + beef extract 3 g + peptone 5 g) seeded with the tested microbial strains. The petri dishes were incubated at 36°C and the inhibition zones were recorded after 24 h of incubation. Each treatment was replicated three times. Ampicillin and Clotrimazole were utilized as standard antibiotics. The activity of the standard antibiotics was also recorded using the above-mentioned procedures at the same concentration and solvents. The % efficacy index was estimated using the formula:
Cytotoxicity (MTT assay)
The cytotoxic activity of the tested samples was performed using MTT assay according to the reported procedures (Saad et al., 2017; Ghareeb et al., 2020) using five cell lines namely; human lung fibroblast (WI38), colorectal carcinoma colon cancer (HCT-116), mammary gland breast cancer (MCF-7) and hepatocellular carcinoma (HEPG-2). Doxorubicin was utilized as a standard anticancer drug. The cell lines were obtained from ATCC via Holding company for biological products and vaccines (VACSERA), Cairo, Egypt. This assay is based on the conversion of the yellow tetrazolium bromide (MTT) to a purple formazan derivative by mitochondrial succinate dehydrogenase in viable cells. Cell lines were cultured in RPMI-1640 medium with 10% fetal bovine serum. Antibiotics added were 100 units/mL penicillin and 100 μg/mL streptomycin at 37°C in a 5% CO2 incubator. The cell lines were seeded in a 96-well plate at a density of 1.0 × 104 cells/well. at 37°C for 48 h under 5% CO2. After incubation, the cells were treated with different concentrations of samples and incubated for 24 h. After 24 h of drug treatment, 20 μL of MTT solution at 5 mg/mL was added and incubated for 4 h. Dimethyl sulfoxide (DMSO) in a volume of 100 μL is added into each well to dissolve the purple formazan formed. The colorimetric assay is measured and recorded at the absorbance of 570 nm using a plate reader (EXL 800, United States). The relative cell viability in percentage was calculated as (A570 of treated samples/A570 of untreated sample) × 100.
UPLC–MS/MS analysis
UPLC analysis was performed via using Shimadzu ExionLC system with the following conditions “Mobile phase A: 0.1% formic acid in water and mobile phase B: acetonitrile; Column: GL-Science (100*2.1) mm, 3 μm; Flow rate: 0.35 mL \min; Column oven: 50°C; Time gradient/Mobile phase B %: 0.0/5.0, 5.0/5.0, 25.0/95.0, 30.0/95.0, 32.0/5.0, and 40.0/5.0. The mass instrument specifications are “Instrument Model: X500 QTOF, Source name: TurboIonSpray, Curtain gas (psi): 30, Ion source gas 1 (psi): 50, Ion source gas 2 (psi): 50, Temperature (°C): 500, Scan type: Full scan -SWATH-Screening, Ion spray voltage (V): Negative, −4,000 V, and CAD gas: 7 (Mahmoud et al., 2023).
In silico study
Virtual target identification
The putative target characterization was achieved via Pharmacophore-based Virtual screening using PharmMapper (Wang et al., 2017; El-Sayad et al., 2023). This platform assigns a score to each molecule in the PDB that best fits a pharmacophore model that has been extracted and stored as a library of ligand dataset in mol2 format. After that, when a new molecule is submitted, its fit score is calculated for each pharmacophore, and then each fit score for that pharmacophore is compared to the fit score matrix to determine where it falls on the scale of all the pharmacophore scores. In comparison to chance pharmacophore matching, the pure fit score that results from this procedure carries considerably more weight and assurance. The query structure was submitted to the platform in the PDB format, and the retrieved results were exported as Excel sheet arranging the resulted protein targets according their fit scores.
Docking study
The crystal structures of E. coli GyrB (PDB ID: 6kzv), and both the human PI3K-γ and c-Src (PDB ID: 2v4l and 3en7, respectively) were used for the docking study using AutoDock Vina (Huey et al., 2012). The co-crystallized ligand in each structure was used to determine the binding site and the docking grid-box in each protein structure, respectively. The co-ordinates of the grid-box were set to be: x = −7.86, y = 16.12, z = 2.49; and x = 45.07, y = 13.12, z = 31.49; and x = −5.09, y = 6.34, z = −6.66, respectively. The ligand to binding site shape matching root means square (RMSD) threshold was set to 2.0 Å. The interaction energies were determined using the Charmm force field (v.1.02) with 10.0 Å as a non-bonded cutoff distance and distance-dependent dielectric. Then, 5.0 Å was set as an energy grid extending from the binding site (Huey et al., 2012). The tested compound retinol was energy minimized inside the selected binding pocket. The editing and visualization of the generated binding poses were performed using Pymol software (Yuan et al., 2017).
Molecular dynamics simulation
NAMD 3.0.0. software was used for performing MDS (Yuan et al., 2017; Ribeiro et al., 2018; Soltane et al., 2023). This software applies the Charmm-36 force field. Protein systems were built using the QwikMD toolkit of the VMD software (Humphrey et al., 1996; Ribeiro et al., 2018), where the protein structures were checked for any missing hydrogens, the protonation states of the amino acid residues were set (pH = 7.4), and the co-crystalized water molecules were removed. Thereafter, the whole structures were embedded in an orthorhombic box of TIP3P water together with 0.15 M Na+ and Cl− ions in 20 Å solvent buffer. Afterward, the prepared systems were energy-minimized and equilibrated for 5 ns. The parameters and topologies of the ligands were calculated by using the VMD plugin Force Field Toolkit (ffTK). Afterward, the generated parameters and topology files were loaded to VMD to readily read the protein–ligand complexes without errors and then conduct the simulation steps.
Binding free energy calculations
Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) embedded in the MMPBSA.py module of AMBER18 was utilized to calculate the binding free energy of the docked complex (Humphrey et al., 1996; Ghareeb et al., 2024). 100 frames were processed from the trajectories in total, and the system’s net energy was estimated using the following equation:
Each of the aforementioned terms requires the calculation of multiple energy components, including van der Waals energy, electrostatic energy, internal energy from molecular mechanics, and polar contribution to solvation energy.
Statistical analysis
Results were analyzed statistically using the computerized program SPSS software, version “20” for windows. The one-way analysis of variance “ANOVA” test was done followed by Duncan test. Data were represented as mean ± SE. Values that are less than 0.05 were considered significant, otherwise were non-significant.
Results
Isolation of the fungal isolate from different marine samples
The fungal strain SA17 was isolated and purified from the collected seagrass samples. The fungal colony was selected based on its morphological features. The obtained strain was deposited at 4°C at the Microbial Chemistry Department, National Research Centre, Egypt. Preliminary identification of the isolated fungal isolate SA17 was carried out based on its morphological characteristics. The isolate showed fast-growing colonies with a cottony green appearance. It produces conidiophores that bear numerous conidia (Figure 1).
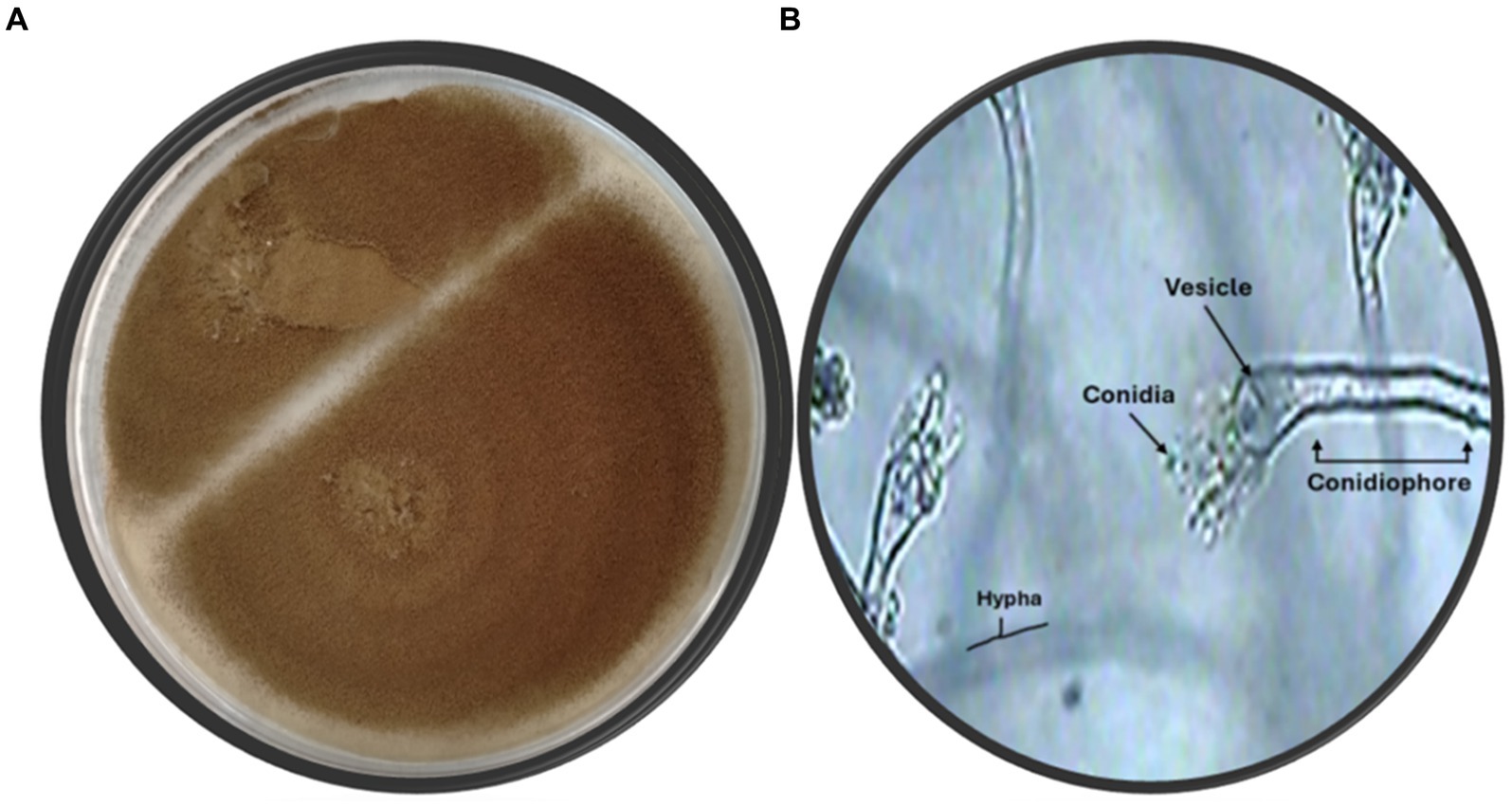
Figure 1. (A) Fungus color on agar plate (B) Microscopic identification of isolate SA17 showing the conidia, vesicle, hypha, and conidiophore of the fungus magnification x500.
The identification of the fungal strain was performed using sequencing of the 18S rRNA gene depending on the initial assessment of the isolated fungi. Subsequently, the DNA of the fungal sample was extracted, amplified, and identified by matching it with other known deposited genes in the GeneBank database using the BLAST approach to define the symmetry record and statistical value of the matches (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 February 2023). The findings disclosed that the 18S rRNA gene arrangements of the isolate were identical, with the Aspergillus sp. having 100% symmetry. The evolution registry (Figure 2) was estimated via the Maximum Likelihood method and the Tamura–Nei model (Humphrey et al., 1996). The proportion of trees in which the correlated taxa are gathered is shown after the branches. The Tamura–Nei model was utilized to produce a matrix of pairwise distances, and the topology with the maximum log-likelihood value was selected as the first tree for the heuristic inspection. MEGA X was utilized to perform the evolutionary exploration (Miller III et al., 2012).
Preparation of fungal crude filtrate and bio-synthesis of zinc oxide nanoparticles
After the fungal culture had grown to the desired phase, the filtrate was obtained by filtering the culture through a sterilized filter paper. The obtained filtrate was mixed with a solution containing zinc ions (zinc acetate). The mixture is then incubated at a specific temperature for a certain period. During this time, the enzymes and biomolecules present in the fungal filtrate act as reducing agents, converting the zinc ions into zinc oxide nanoparticles in solution after heating at 100°C. The dimension and shape of the nanoparticles can be managed by altering the concentration of the precursor solution, the incubation time, and the temperature.
Characterization
UV-analysis
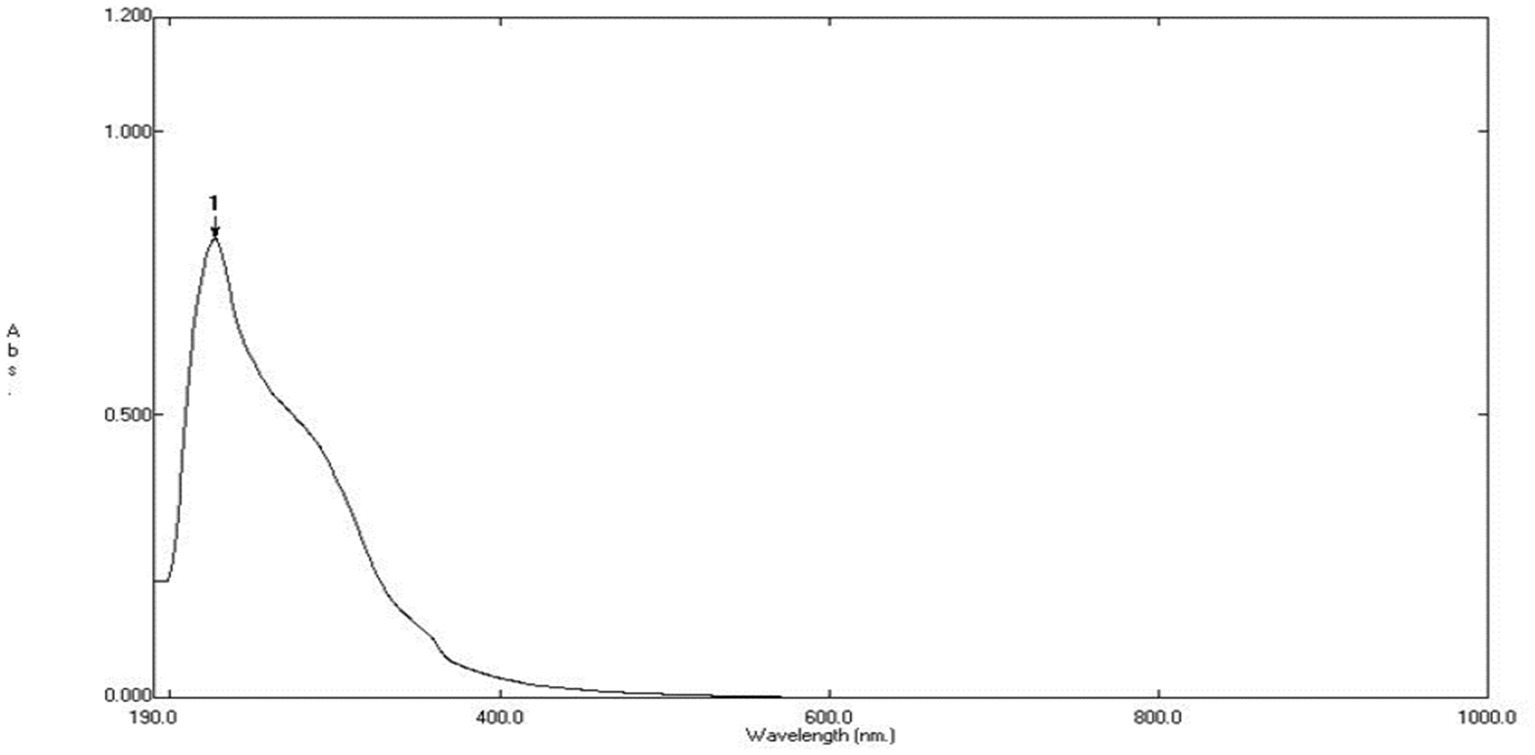
The UV–visible spectroscopy was utilized to monitor the green synthesis of ZnONPs using an aqueous fungal extract. Upon addition of the fungal extract to the filtered zinc acetate solution, the color of the solution changed from brownish red to pale yellow, indicating the successful reduction of Zn(CH3COO)2 to zinc oxide nanoparticles. The synthesized ZnONPs were then characterized by UV–Vis spectroscopy, which revealed a wide absorption band at 280 and 340 nm as shown in Figure 3, confirming the formation of ZnONPs.
The produced ZnONPs were examined using UV–visible spectroscopy, which is the accepted technique for confirming nanoparticle creation and evaluating their optical characteristics. This method works especially well for “green synthesis,” wherein extracts derived from biological processes serve as reducing agents in metal nanoparticles biosynthesis. The study’s conclusions-which are supported by UV–Vis spectroscopy-highlight the usefulness of the fungal extract in the environmentally friendly synthesis of ZnONPs and call attention to its possible uses in the production of sustainable nanomaterials (Harris et al., 2016).
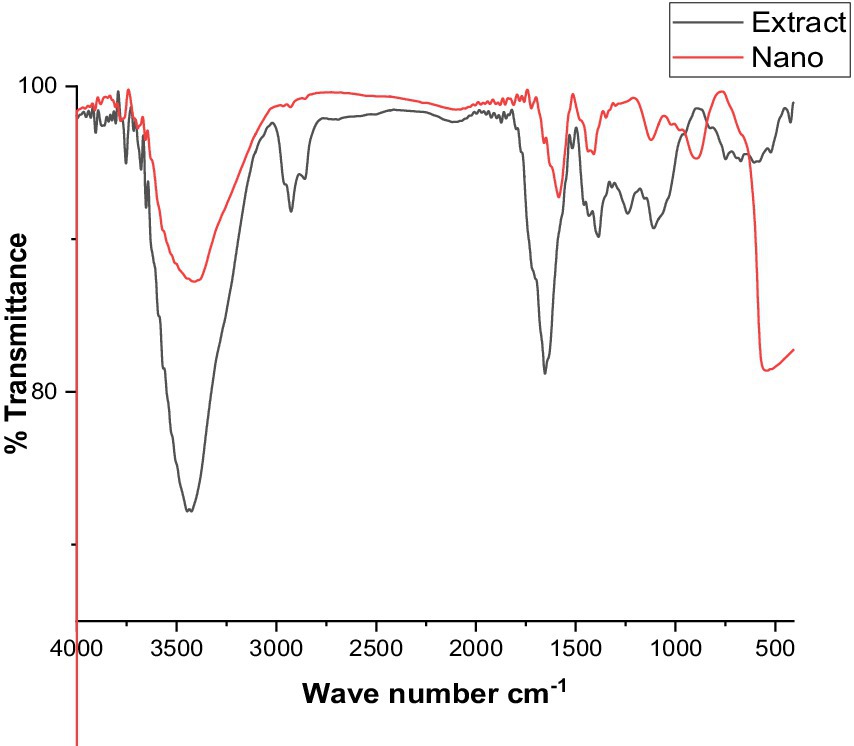
Fourier transforms infrared spectroscopy analysis
The FT-IR analysis of the extract and the produced ZnO nanoparticles is shown in Figure 4. Here, FT-IR analysis has been used to identify the functional groups in charge of ZnONPs production and stability. The FT-IR spectra of the green ZnONPs showed that the various functional groups, CH, C=O, C=C, C-O, C-N, and C-C- in the synthesized ZnONPs were connected to peaks at 3500, 3000, 2,800, 1,700, 1,500, 1335.25, 1218.83, and 1030.65 cm−1. The stretching vibration of the hydroxyl group (O-H) is found at 3320, whereas the stretching vibration of the alkane (C-H) is found at peaks at 2919.92 and 2850.58 cm−1. Similarly, a stretch vibration of carbonyl (C=O) was detected at 1,700 cm−1. At 1730.72 cm−1, the aromatic (C=C) stretch bands were observed. Stretching of the C-N was mentioned in the band at 1,200 cm − 1, while the C-O was detected at 1,305. Furthermore, the C-C is located at 1030.65 cm−1. The presence of flavonoids and phenolic acids in the tested extract can be fundamentally linked to the occurrence of absorption bands related to C=C, C=O, C-O, C-N, OH, and CH (Wu et al., 2022). These functional groups contribute to the stability of the prepared ZnONPs as well as the reduction of Zn metal ions.
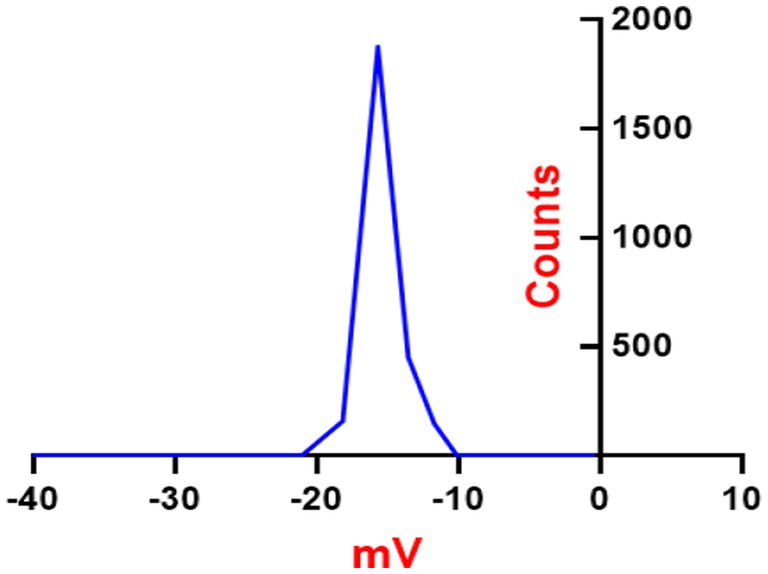
Zeta potential of synthesized ZnONPs
ZnONPs colloidal solutions displayed constancy in the neutral aqueous system (di-distilled water) since it possesses a zeta potential value of −18.16 mV as presented in Figure 5. ZnONPs colloidal stability in a neutral media has important ramifications for possible uses. Stable colloidal dispersions are crucial for drug delivery in the biomedical domains because they inhibit aggregation and preserve the intended particle size distribution, which influences the kinetics of drug release and the effectiveness of targeting (Wilhelm et al., 2016). Furthermore, in environmental applications, the dispersion and interaction of nanoparticles with contaminants, which facilitates their removal and remediation from aqueous systems, are greatly aided by their colloidal stability. It is important to remember that there are additional variables that might affect colloidal stability than the zeta potential, including particle size, surface charge density, and the presence of stabilizing chemicals (Li et al., 2013).
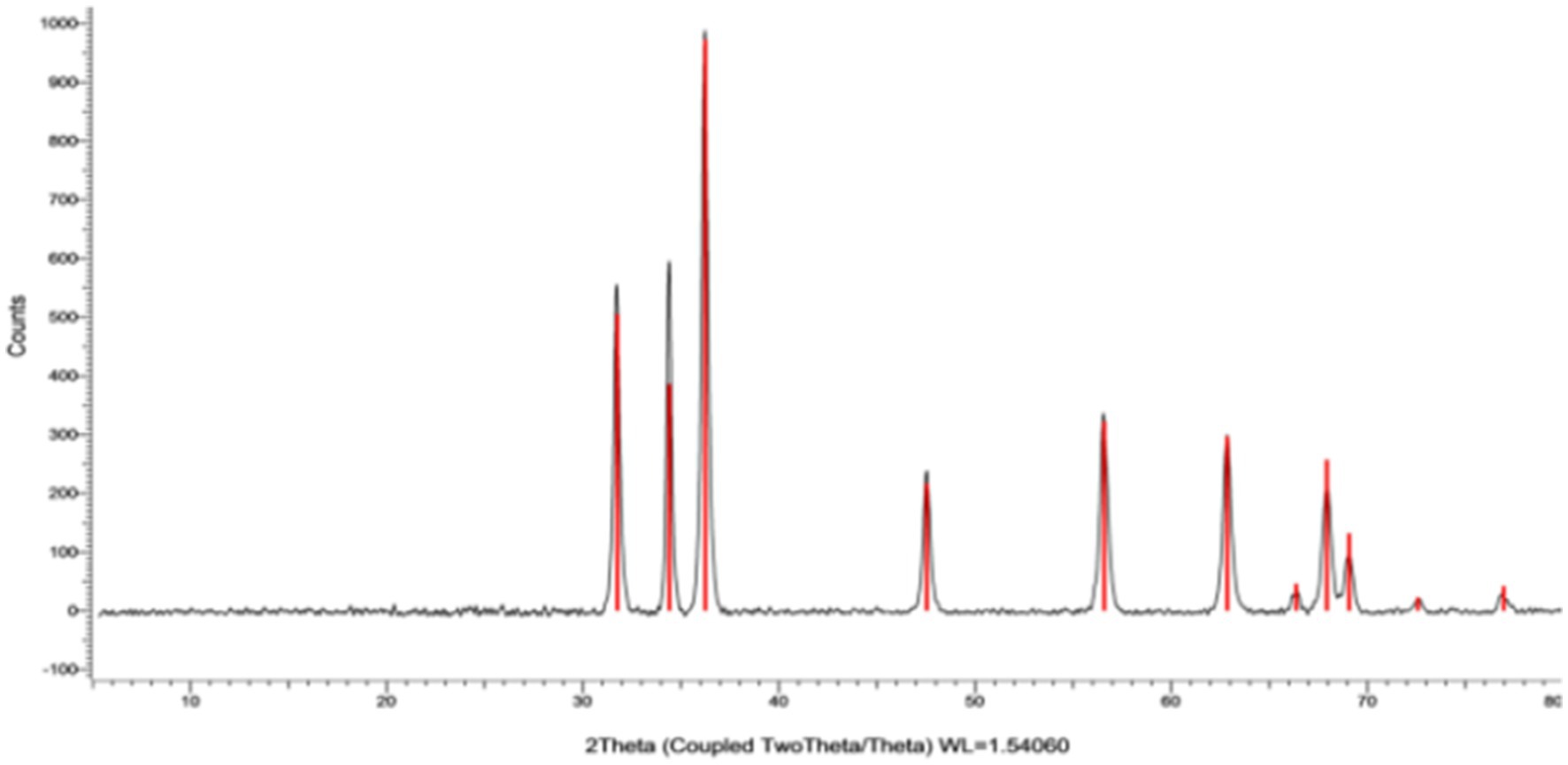
X-ray diffraction pattern examination
The X-ray diffraction (XRD) paradigm was utilized to confirm the crystalline nature of ZnONPs. The XRD pattern displayed distinct peaks at (2θ) angles of 31.77, 34.43, 36.26, 47.55, 56.60, 62.88, 66.39, 67.96, and 69.10°, complemented by indicators (100), (002), (101), (102), (210), and (103), sequentially, as noted in Figure 6. The XRD spectra of ZnONPs revealed that the generated nano-ZnO had a spherical form and matched the profile mentioned by Schreyer et al. (2014).
Transmission and scanning electron microscopy studies
High resolution-TEM was done to investigate the nature of SA-17 ZnONPs particle size distribution and its crystallinity. As shown in Figure 7, the particles consist of a spherical crystal of particle size extending from 3 to 20 nm with an average of 7.2 nm. These spherical crystals contain small particles from 2 to 30 nm with average size of 28.9 nm. The surface morphology of the ZnONPs was explored using a captured SEM image, which showed that the ZnONPs were agglomerated spherical shapes with a diameter ranging from 13 to 55 nm with an average size of 30 nm (Figure 7B). The size detected by SEM was slightly larger than those by TEM as result of surface particles were agglomerated to each other.
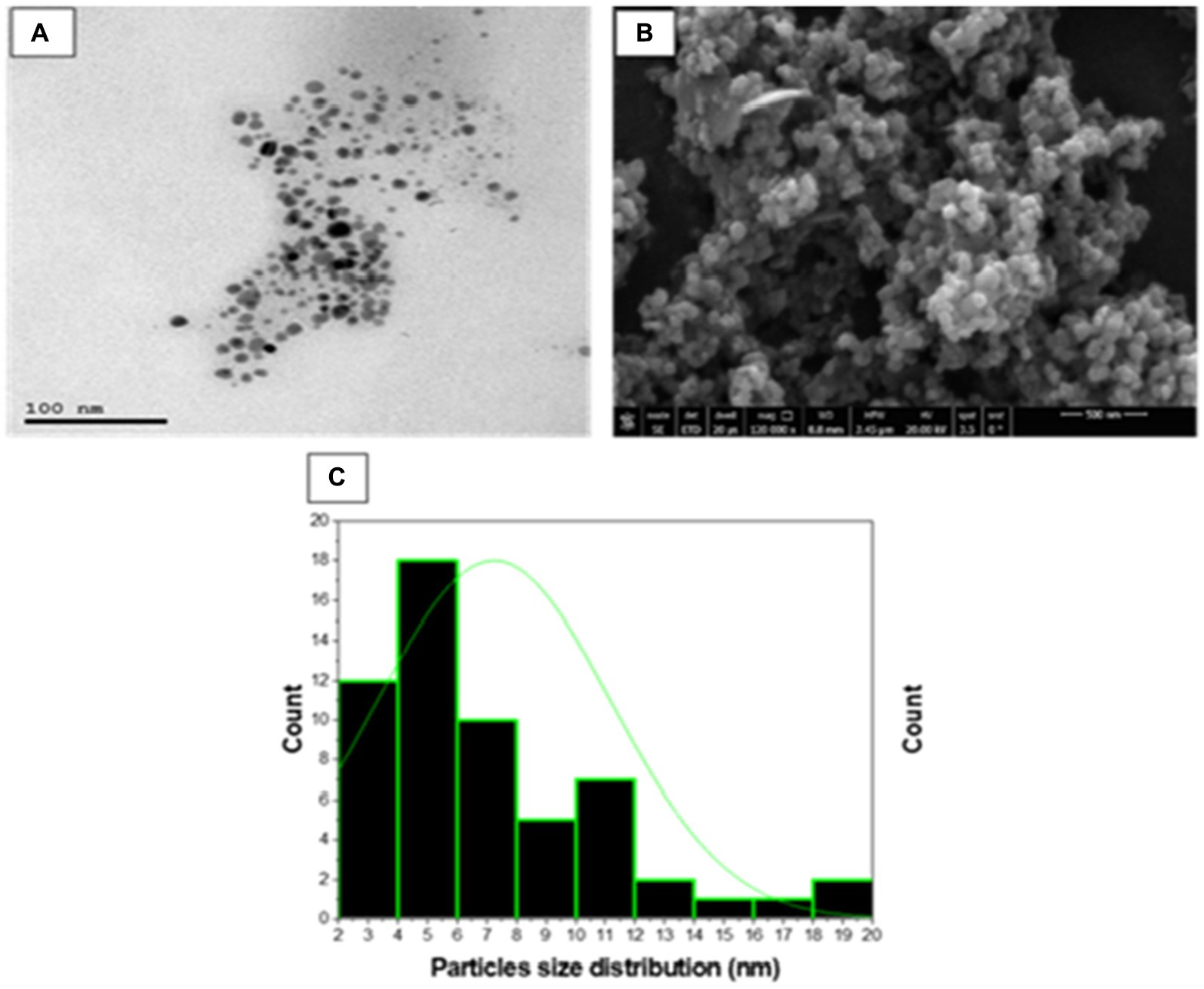
Figure 7. (A) TEM micrograph, (B) SEM micrograph and (C) Particle size distribution of synthesized SA-17 ZnONPs.
Antimicrobial activity
The crude extract and biosynthesized ZnONPs showed a pronounced antimicrobial effect toward E. coli, P. aeruginosa, S. aureus, B. subtilis, C. albicans, and A. flavus, respectively. With inhibition percentages ranging from 96.25% to 99.26%, the data showed that Cipro had the highest antibacterial efficiency against all tested pathogens. By contrast, although to a lesser degree, the extract and ZnONPs likewise showed noteworthy antibacterial activity. ZnONPs showed inhibitory percentages ranging from 12.0% to 39.1%, and the extract showed percentages ranging from 28.0% to 52.2%. Significant variations in the antibacterial activity of the tested medicines against every bacterium were found by statistical analysis (p < 0.05). By contrast, the extract and ZnONPs both showed strong antifungal activity. ZnONPs displayed inhibitory percentages ranging from 37.0% to 32.0%, whereas the extract showed percentages ranging from 48.1% to 48.0%. The antifungal activity of the tested medicines against each microbe varied significantly, according to statistical analysis (p < 0.05). The following letters indicate statistical significance for Aspergillus flavus and Candida albicans: Colitrimazole (a) >Extract (b) >ZnONPs (c). The obtained results were compared with standard antibiotics, including Ciprofloxacin and Colitrimazole. Aspergillus species are known for producing unique and diverse secondary metabolites with potent biological activities (Zhang et al., 2018; Abdel-Razek et al., 2020). Figure 8 displayed the inhibition ratio against all tested microbes.
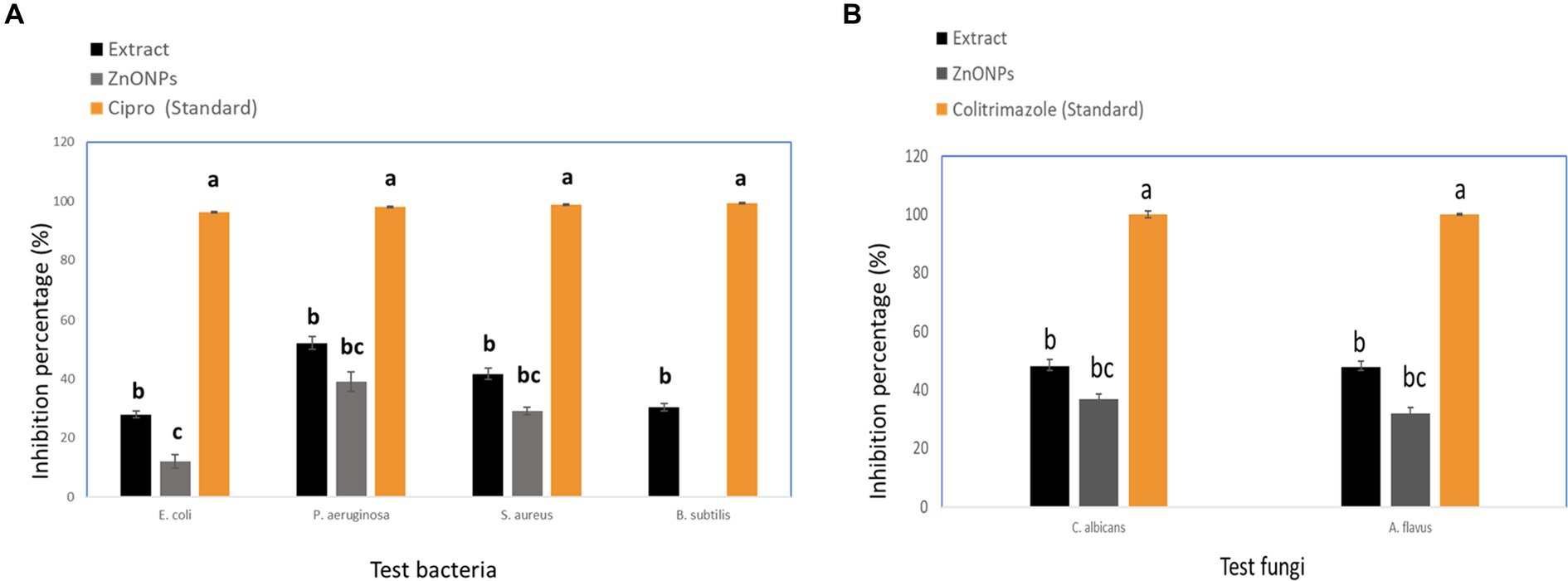
Figure 8. Inhibition percentage against pathogenic microbial strains (A) Tested bacteria, and (B) Tested fungi. While different lowercase letters meant that the antimicrobial effect was significantly different (p < 0.05).
Anticancer activity
The crude extract demonstrated anticancer activity with IC50 values of (84.55 μM), (31.13 μM), (39.06 mm), and (17.65 μM), against WI38, HCT116, HePG2, and MCF7, respectively. On the other hand, the biosynthesized ZnONPs displayed anticancer activity with IC50 values of (59.74 μM), (43.21 μM), (57.03 mm), and (35.66 μM), against WI38, HCT116, HePG2, and MCF7, respectively. The results were compared with Doxorubicin (Figure 9). The cytotoxicity of the tested drugs against each cell line varied significantly, according to statistical analysis (p < 0.05). The letters that indicate statistical significance for the cell lines WI38, HCT116, HePG2, and MCF7 are as follows: ZnONPs (c) > Extract (b) > Doxorubicin (a). Aspergillus sp. has a great ability to produce a wide of secondary metabolites which are considered a potential source of new anticancer compounds (Kusari et al., 2009; El-Sayed et al., 2021; Noman et al., 2021). A study conducted by Almanaa et al. (2021) reported that A. fumigates extract showed a cytotoxic effect against the HepG-2 cell line with IC50 value of 113 μg/mL. Also, the crude extract of Aspergillus tubenginses ASH4 showed anticancer effect against HCT-116, Hep-G2, and MCF-7 with IC50 values of 9.18, 10.41, and 5.89 μg/mL, respectively (Elkhouly et al., 2021a,b).
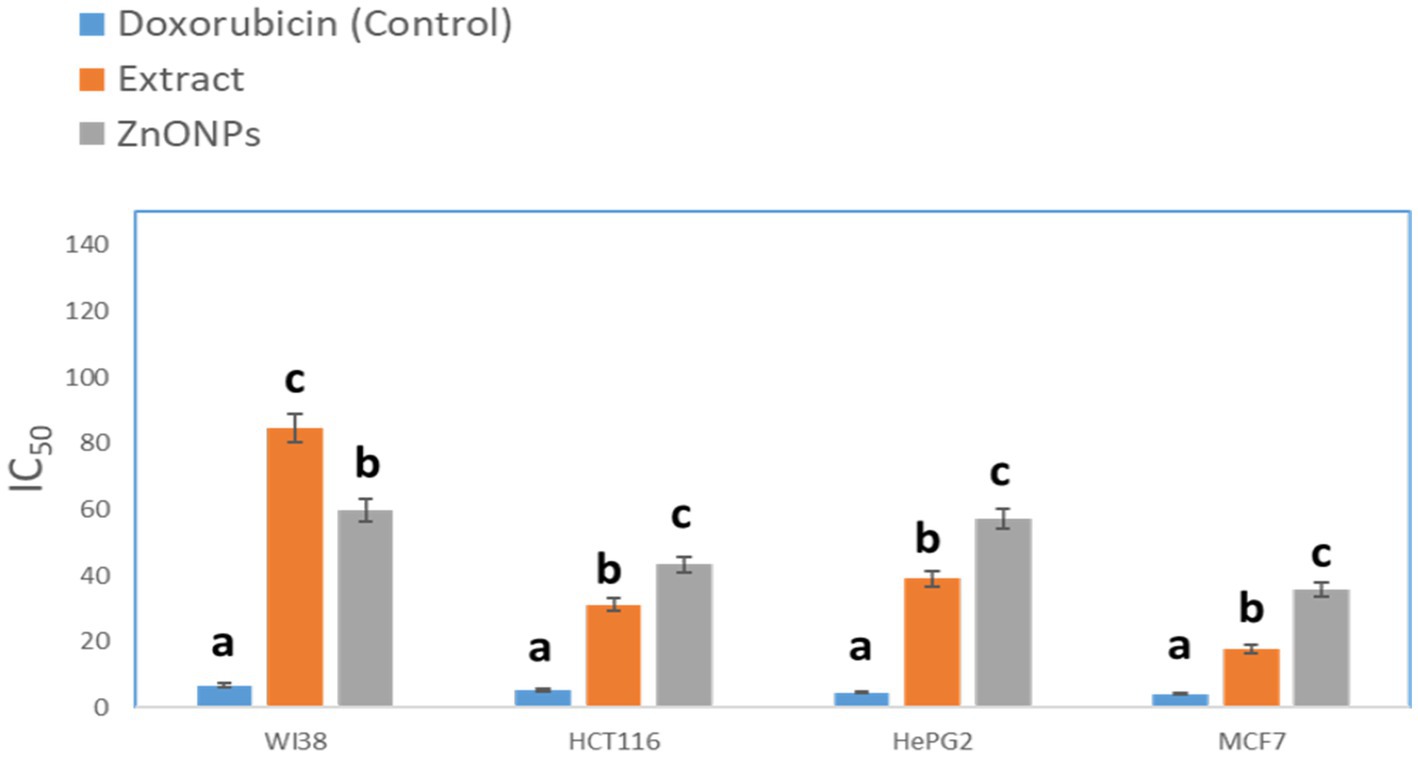
Figure 9. Anticancer activity of the tested extracts against human cell lines. While different lowercase letters meant that the anticancer effect was significantly different (p < 0.05).
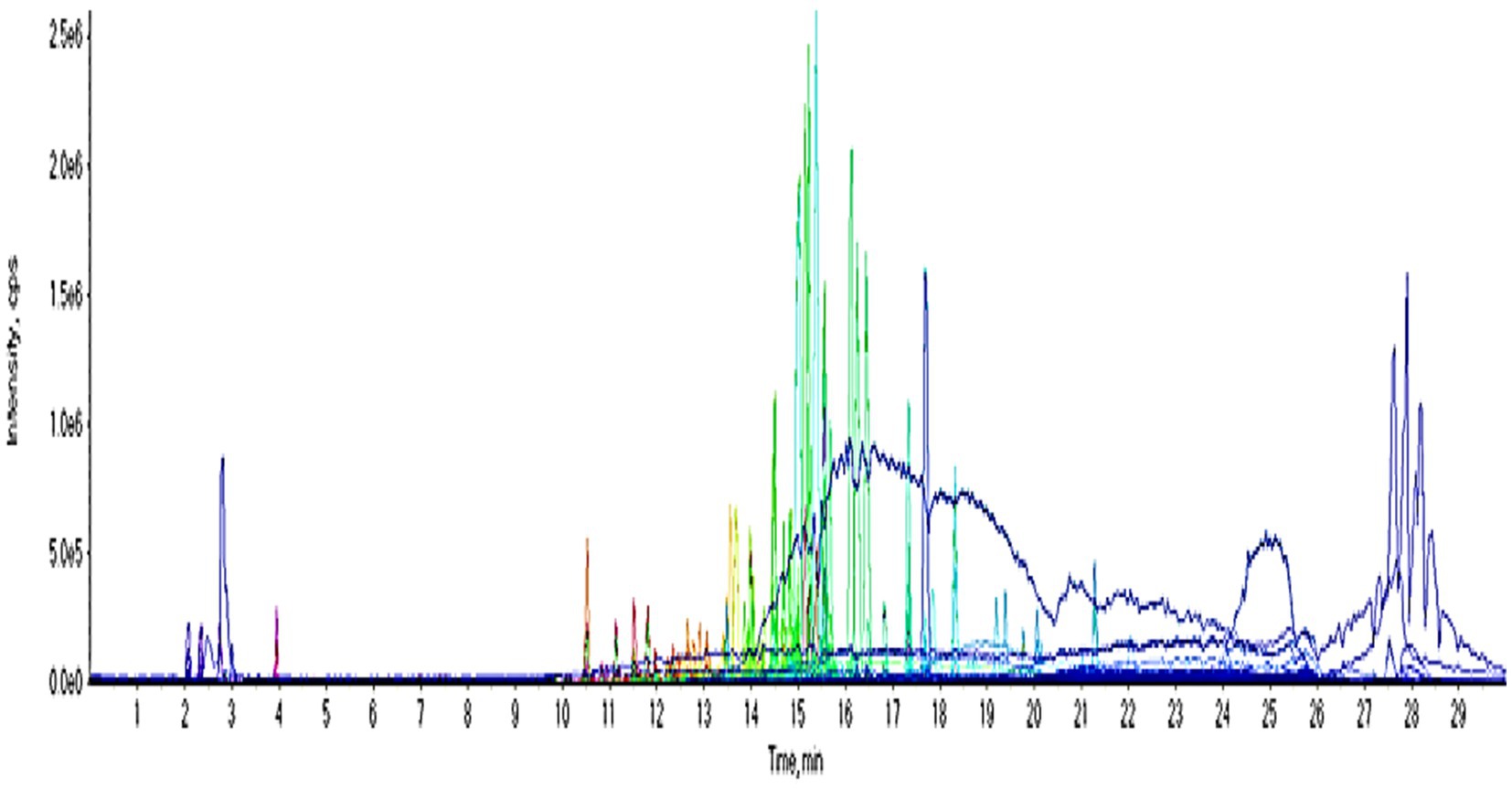
UPLC-QTOF-MS/MS analysis
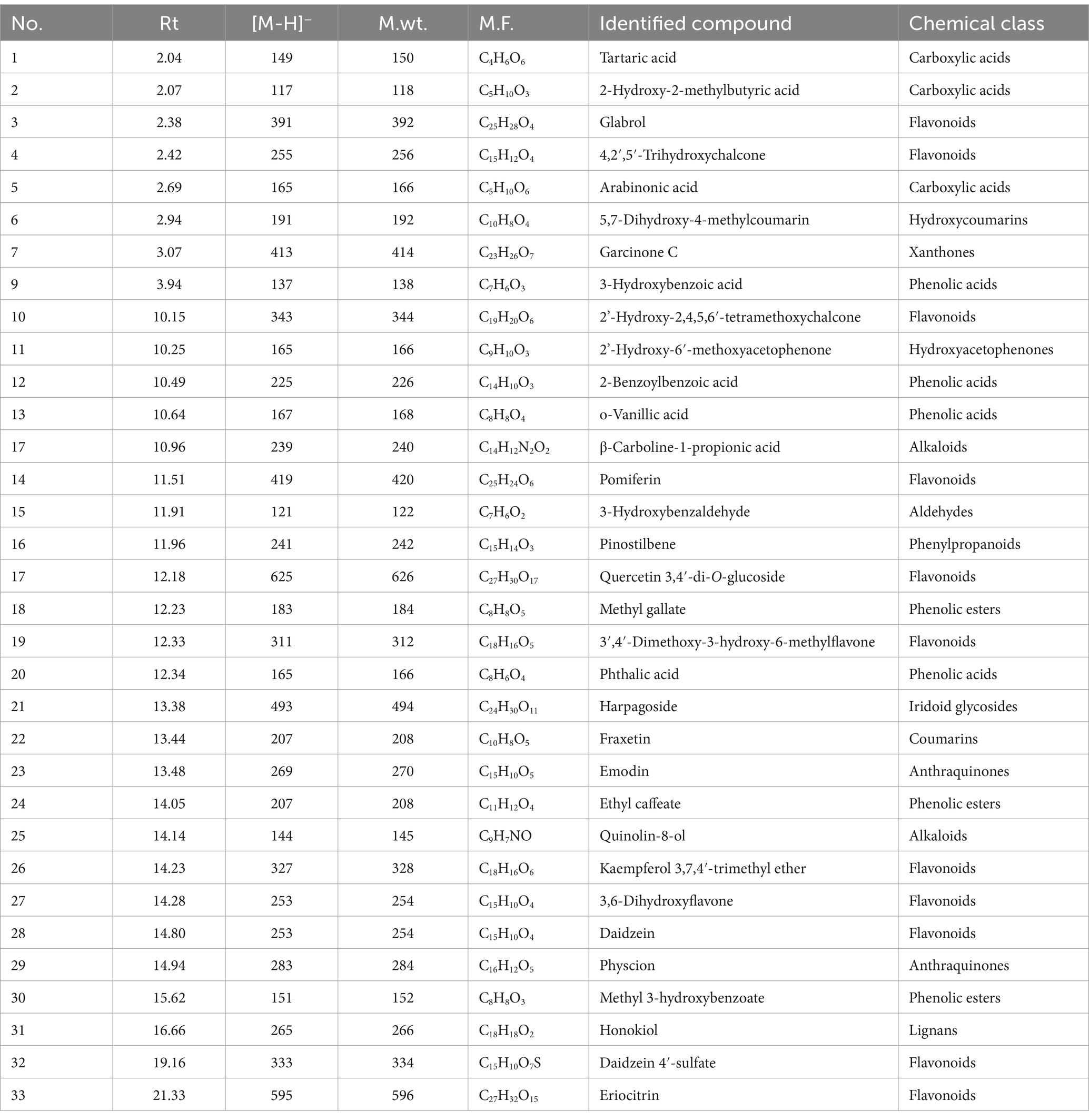
UPLC-QTOF-MS/MS analysis of crude extract in a negative ion mode led to the identification of 33 compounds based on their retention times; fragmentation patters and via comparison with the available reported data. The identified compounds were categorized as carboxylic acids, phenolic acids, flavonoids, benzene derivatives, coumarins, anthraquinones, benzaldehydes, phenols and fatty acids (Figure 10; Table 1). The phenolic acids and flavonoids were the dominant compounds in the extract.
Phenolic, organic and fatty acids and their derivatives
A peak demonstrated an [M-H]- m/z at 149 and daughter ions at m/z 105 [M-H-CO2]-, 87 [M-H-CO2-H2O]-, 71 and 59; it was assigned as tartaric acid (McGovern et al., 2009). A peak demonstrated an [M-H]- m/z at 117 and daughter ions at m/z 73 [M-H-CO2]-, 55 [M-H-CO2-H2O]-, and 61; it was assigned as 2-hydroxy-2-methylbutyric acid (John Wiley & Sons, Inc, 2015). A peak demonstrated an [M-H]- m/z at 317 and daughter ions at m/z 273 [M-H-CO2]-, 165, 113, and 103; it was assigned as 15-Oxo-5Z,8Z,11Z,13E-eicosatetraenoic acid (Sorgi et al., 2018). A peak demonstrated an [M-H]- m/z at 165 and a daughter ion at m/z 119 [M-H-HCOOH]-; it was assigned as arabinonic acid (Moreira et al., 2014). A peak demonstrated an [M-H]- m/z at 115 and a daughter ion at m/z 71 [M-H-CO2]-; it was assigned as 3-oxopentanoic acid (Kallem et al., 2021). A peak demonstrated an [M-H]- m/z at 137 and daughter ions at m/z 109 [M-H-CO]-, 108 [M-2H-CO]-, 93 [M-H-CO2]-, 92 [M-2H-CO2]-, 81, and 65; it was assigned as 3-hydroxybenzoic acid (Ammar et al., 2020; Ghareeb et al., 2023). Hydroxybenzoic acids as a subclass of phenolic acids possess a reliable role in managing neurodegenerative diseases aging, and cancer (e.g., breast cancer; Kalinowska et al., 2021). A peak demonstrated an [M-H]- m/z at 225 and daughter ions at m/z 207 [M-H-H2O]-, 179, 135, and 97; it was assigned as 2-benzoylbenzoic acid (Zengin et al., 2022). A peak demonstrated an [M-H]- m/z at 167 and daughter ions at m/z 152 [M-H-CH3]-, 123 [M-H-CO2]-, 108 [M-H-CH3-CO2]-, and 91 [M-H-COOH-OCH3]-; it was assigned as vanillic acid (Ghareeb et al., 2018a; Sayed et al., 2022). Vanillic acid, an oxidized form of vanillin, is a major active compound isolated from Angelica sinensis and green tea. Vanillin acid is a dietary phenol that can protect biofilms and inhibit lipid peroxidation and eliminates ROS hence act as anti-microbial, anti-inflammatory, anti-cancer factor (Gong et al., 2019). A peak demonstrated an [M-H]- m/z at 183 and daughter ions at m/z 169 [M-H-CH3]-, 168, 125 [M-H-CO2]-, 109, and 93; it was assigned as methyl gallate (Ghareeb et al., 2018b, 2019b). A peak demonstrated an [M-H]- m/z at 165 and daughter ions at m/z 121 [M-H-CO2]-, and 77 [M-H-2CO2]-; it was assigned as phthalic acid (Gillespie et al., 1989). A peak demonstrated an [M-H]- m/z at 207 and daughter ions at m/z 161 [M-H-C2H5OH]-; it was assigned as ethyl caffeate (Barth et al., 2019). A peak demonstrated an [M-H]- m/z at 151 and daughter ions at m/z 136 [M-H-CH3]-, and 92 [M-H-CH3-CO2]-; it was assigned as methyl 3-hydroxybenzoate (Hanley, 2006).
A peak demonstrated an [M-H]- m/z at 191 and daughter ions at m/z 176 [M-H-CH3]-, 147 [M-H- CO2]-, 160 [M-H-OH-CH3]-, 156 [M-H-2OH]-, and 105; it was assigned as 5,7-dihydroxy-4-methylcoumarin (Kerebba et al., 2022). A peak demonstrated an [M-H]- m/z at 207 and daughter ions at m/z 192 [M-H-CH3]-, 164 [M-H-CH3-CO]-, and 102; it was assigned as fraxetin (Wang et al., 2016). Fraxetin is a bioactive molecule present in various natural plants, considered as bioactive molecule that acts as anticancer, antioxidative, anti-inflammatory, antidiabetic and antimicrobial activities (Ha and Son, 2024).
Xanthones
A peak demonstrated an [M-H] m/z at 413 and daughter ions at m/z 323 [M-H-90]-, 285 [M-H-128]-, 257 [M-H-156]-, 229 [M-H-184]-; it was assigned as garcinone C (Khaw et al., 2020). Xanthones as secondary metabolites displayed various biological properties, including cytotoxic, antidiabetic, antioxidant, antimicrobial, antitumor, antihypertensive, and anti-inflammatory (Mohamed and Ibrahim, 2022).
Flavonoids
A peak demonstrated an [M-H]- m/z at 391 and daughter ions at m/z 203 [M-H-188]-, 187 [M-H-204]-, and 159 [M-H-188-44]-; it was assigned as glabrol (Simons et al., 2009). A peak demonstrated an [M-H]- m/z at 255 and daughter ions at m/z 135 [M-H-C7H4O2]-, and 119 [M-H-C7H4O3]-; it was assigned as 4,2′,5′-trihydroxychalcone (Shan et al., 2017). A peak demonstrated an [M-H]- m/z at 343 and daughter ions at m/z 315 [M-H-CO]-, 313 [M-H-2CH3]-, 328 [M-H-CH3]-, 300 [M-H-CH3-CO]-, and 297 [M-H-H2O-CO]-; it was assigned as 2′-hydroxy-2,4,5,6′-tetramethoxychalcone (Liu et al., 2016). A peak demonstrated an [M-H]- m/z at 419; it was assigned as pomiferin (Tian et al., 2006). A peak demonstrated an [M-H]- m/z at 625 and daughter ions at m/z 463 [M-H-Glc]−, and 301 [M-H-2Glc]-, 271, and 255; it was assigned as quercetin 3,4′-di-O-glucoside (Kumar et al., 2017). A peak demonstrated an [M-H]- m/z at 311 and daughter ions at m/z 293, and 209; it was assigned as 3′,4′-dimethoxy-3-hydroxy-6-methylflavone (Reed, 2009). A peak demonstrated an [M-H]− m/z at 327 and daughter ions at m/z 314, 312, 297, 285, 284, 282, 270, and 112; it was assigned as kaempferol 3,7,4′-trimethyl ether (Dehkordi et al., 2020). A peak demonstrated an [M-H]- m/z at 253 and daughter ions at m/z 238, 223, 179, 151, and 123; it was assigned as 3,6-dihydroxyflavone (March and Brodbelt, 2008). A peak demonstrated an [M-H]- m/z at 253 and daughter ions at m/z 225 [M-H-CO]-, 224 [M-H-CHO]-, 209 [M-H-CO2]-, 197 [M-H-2CO]-, 185 [M-H-2CO + C]-, and 135 [M-H-C8H6O]-; it was assigned as daidzein (Zhao et al., 2018). A peak demonstrated an [M-H]- m/z at 333 and daughter ions at m/z 253 [M-H-SO3]-, 225 [M-H-SO3-CO]-, 224 [M-H-SO3-CHO]-, 209 [M-H-SO3-CO2]-, 197 [M-H-SO3-2CO]-, 185 [M-H-SO3-2CO + C]-, and 135 [M-H-SO3-C8H6O]; it was assigned as daidzein 4′-sulfate (Zhao et al., 2018). A peak demonstrated an [M-H]- m/z at 595 and daughter ions at m/z 433 [M-H-Glc]-, 287 [M-H-Glc-Rha]-; it was assigned as eriocitrin (Li et al., 2020).
Hydroxyacetophenones
A peak demonstrated an [M-H]- m/z at 165 and daughter ions at m/z 150 [M-H-CH3]-, 137 [M-H-CO]-, 122 [M-H-CO-CH3]-, and 107 [M-H-CO-2CH3]-; it was assigned as 2′-hydroxy-6′-methoxyacetophenone (Martin, 2013).
Alkaloids
A peak demonstrated an [M-H]- m/z at 239 and daughter ions at m/z 221 [M-H-H2O]-, 193 [M-H-CH2O2]-, 179 [M-H-C2H4O2]-, 165 [M-H-C3H6O2]-, and 138 [M-H-C4H7NO2]-; it was assigned as β-carboline-1-propionic acid (Chua et al., 2011). A peak demonstrated an [M-H]- m/z at 144 and daughter ions at m/z 114, and; it was assigned as quinolin-8-ol (Clugston, 1996).
Hydroxybenzaldehydes
A peak demonstrated an [M-H]- m/z at 121 and daughter ions at m/z 93 [M-H-CO]- and 92 [M-H-CHO]-; it was assigned as 3-hydroxybenzaldehyde (Castillo et al., 2023).
Phenylpropanoids
A peak demonstrated an [M-H]- m/z at 241 and daughter ions at m/z 225, 197, 181, and 169; it was assigned as pinostilbene (Chen et al., 2016).
Iridoids
A peak demonstrated an [M-H]- m/z at 493 and daughter ions at m/z 363 [M-H-cinnamoyl]-, 345 [M-H-cinnamoyl-H2O]-, 201 [M-H-cinnamoyl-Glc]-, 183 [M-H-cinnamoyl-H2O-Glc]-, and 147 [M-H-C15H22O9]-; it was assigned as harpagoside (Xiong et al., 2010).
Anthraquinones
A peak demonstrated an [M-H]- m/z at 269 and daughter ions at m/z 251 [M-H-H2O]-, 241 [M-H-CO]-, 227 [M-H-CO-CH2]-, 225, 223 [M-H-CO-H2O]-, 195 [M-H-CO-H2O-CO]-; it was assigned as emodin (Fu et al., 2015). A peak demonstrated an [M-H]- m/z at 283 and daughter ions at m/z 268 [M-H-CH3]-, 240 [M-H-CH3-CO]-, 212, and 184; it was assigned as physcion (Fu et al., 2015).
Lignans
A peak demonstrated an [M-H]- m/z at 265 and a daughter ion at m/z 224 [M-CH2CH=CH2-H]-; it was assigned as honokiol (Wu et al., 2006).
In silico studies
Virtual screening-based target identification
To find out how crude extract exerts its antibacterial and anticancer activities, all the modeled structures of the UPLC–MS-annotated compounds in these samples were subjected to pharmacophore-based virtual using the PharmMapper platform (Wang et al., 2017).
PharmMapper can screen and recommend the most probable protein targets of a query molecule based on its pharmacophore model by mapping its fundamental pharmacophore characteristics (i.e., the spatial arrangement of structural features).
Accordingly, the compounds that are compatible with these pharmacophore maps have a greater potential for binding to the same protein targets. Therefore, the annotated compounds (Table 2) were run through PharmMapper to find their potential protein targets. The obtained results were arranged according to their degree of conformity to the pre-determined parameters (the Fit score). Only bacterial-relevant and cancer-relevant targets were chosen.

Table 2. Docking scores and ΔGBind (in kcal/mol) of glabrol and pomiferin inside Escherichia coli GyrB’s active sites.
As a result, DNA gyrase subunit-B (GyrB) of E. coli (PDB ID: 6KZV) was found to be the top-scoring bacterial-relevant hit for glabrol and pomiferin (Figure 11). Hence, these metabolites can be considered tentatively as the key antibacterial metabolites in the tested extract.
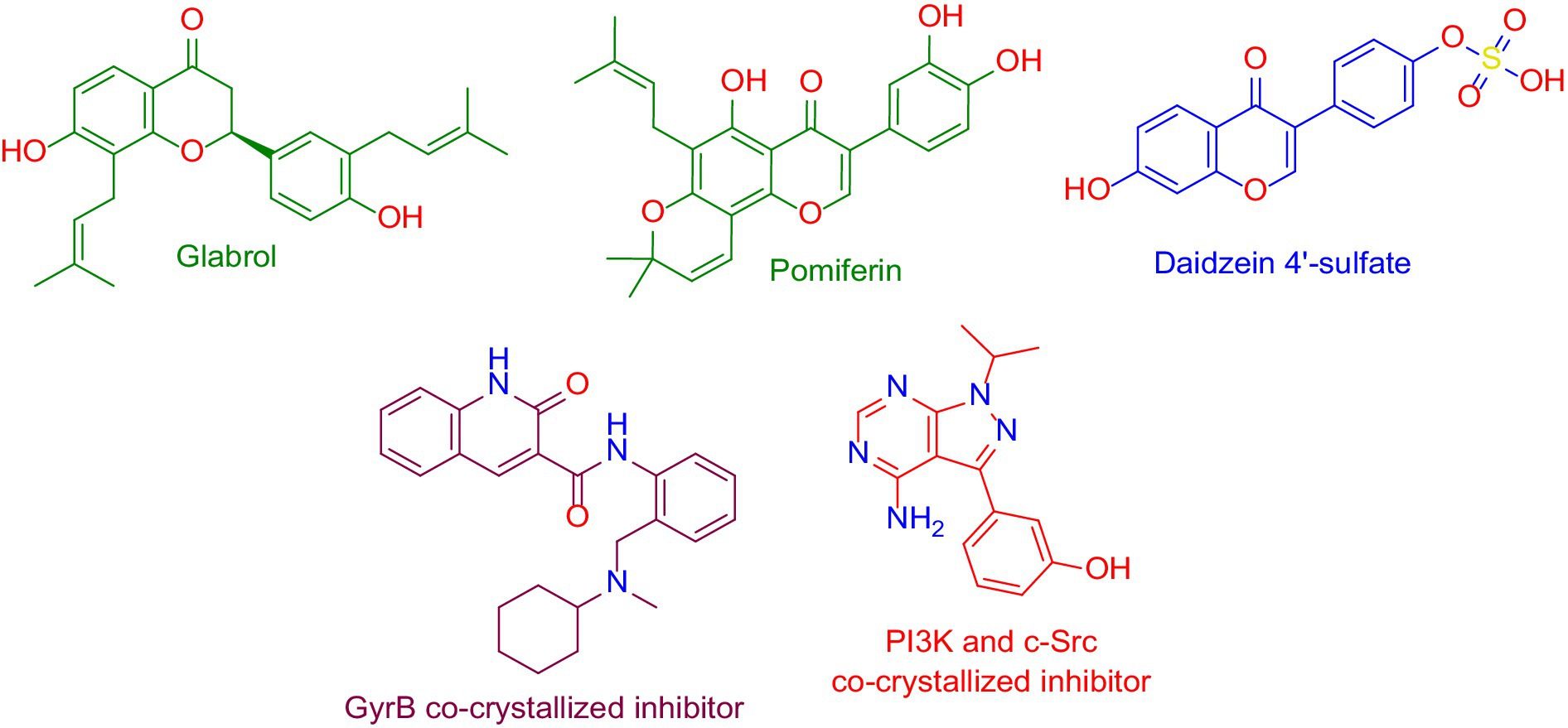
Figure 11. Structures that were found to be probably able to bind to the active sites of Escherichia coli’s GyrB (i.e., glabrol and pomiferin; green-colored structures), and PI3K (i.e., daidzein 4′-sulfate, blue-colored structure), according to the preliminary PharmMapper-based virtual screening alongside the GyrB and PI3K-γ co-crystalized inhibitors (purple-colored and orange-colored structures).
On the other hand, Daidzein 4′-sulfate (Figure 11) was predicted to bind with the active site of human phosphoinositide 3-kinase gamma (PI3K-γ; PDB ID: 2V4L) with a Fit score of 8.28.
It is well-known that GyrB is an essential protein that provides the necessary energy for the GyrA subunit, which in turn unfolds the bacterial DNA, making it available for the DNA replicating enzyme, and thus blocking these enzymes (i.e., GyrA or GyrB) will stop the bacterial DNA replication and eventually bacterial death (Wigley et al., 1991; Ushiyama et al., 2020).
Regarding the cancer-relevant protein, PI3K-γ has been reported in many previous reports to be over-expressed in different types of human tumors, particularly breast cancers (Apsel et al., 2008; Kwak et al., 2019; Miricescu et al., 2020).
Molecular docking and dynamics simulation analysis
To investigate the binding modes of each aforementioned compound (Figure 11) with GyrB and PI3K, their modeled structures were prepared and re-docked into the active sites of each protein. Thereafter, the resulting binding poses were subjected to 50 ns-long MD simulation runs to test the binding affinity and stability of each structure inside the active sites of the suggested protein targets. First, the re-docking of glabrol and pomiferin structures into the GyrB’s active site achieved binding modes and docking scores comparable to those of the co-crystalized inhibitor (Figure 11; Table 2).
As shown in Figures 10A–C, the structures of glabrol and pomiferin were able to achieve binding modes convergent to that of the co-crystalized inhibitor forming comparable hydrophilic and hydrophobic interactions (Table 2). H-bonds with ASP-73 and ARG-76 were the common hydrophilic interactions among the three structures alongside the co-crystalized inhibitor (Figure 12). Second, re-docking of daidzein 4′-sulfate structure into the active sites of PI3K resulted in binding mode and docking score comparable to those of the co-crystalized inhibitor (Figure 13; Table 3). Subsequent experiments using molecular dynamic (MD) simulation (lasting 50 ns) showed that glabrol and pomiferin structures were able to achieve stable binding modes inside the GyrB’s active site, with relative mean square deviations (RMSDs) of 2.523 Å and 1.765 Å, respectively (Figure 14). Hence, the calculated binding free energies (ΔGBinding) these structures in comparison with that of the co-crystallized inhibitor were convergent (ΔGBinding = −7.57, −8.54, and −8.76 kcal/mol, respectively).
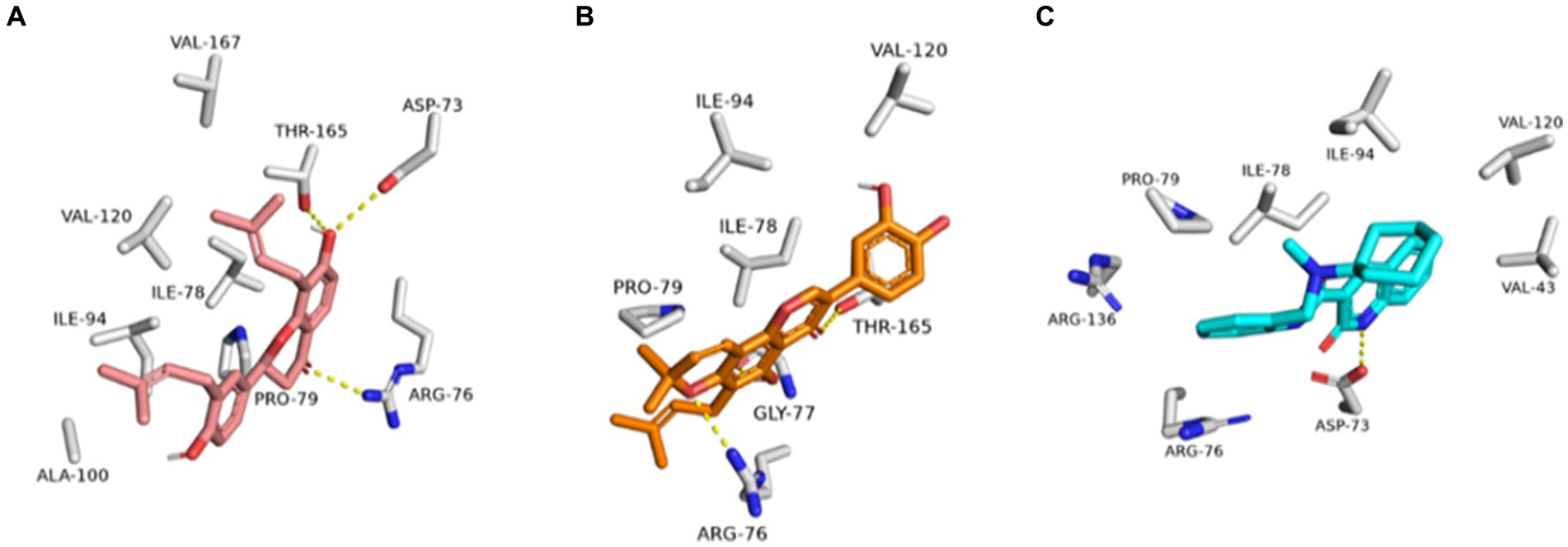
Figure 12. Binding modes of glabrol (brick red-colored structure) (A), and pomiferin (orange-colored structure) (B) along with the co-crystalized inhibitor (cyan-colored structure) (C) inside E. coli GyrB’s active site.
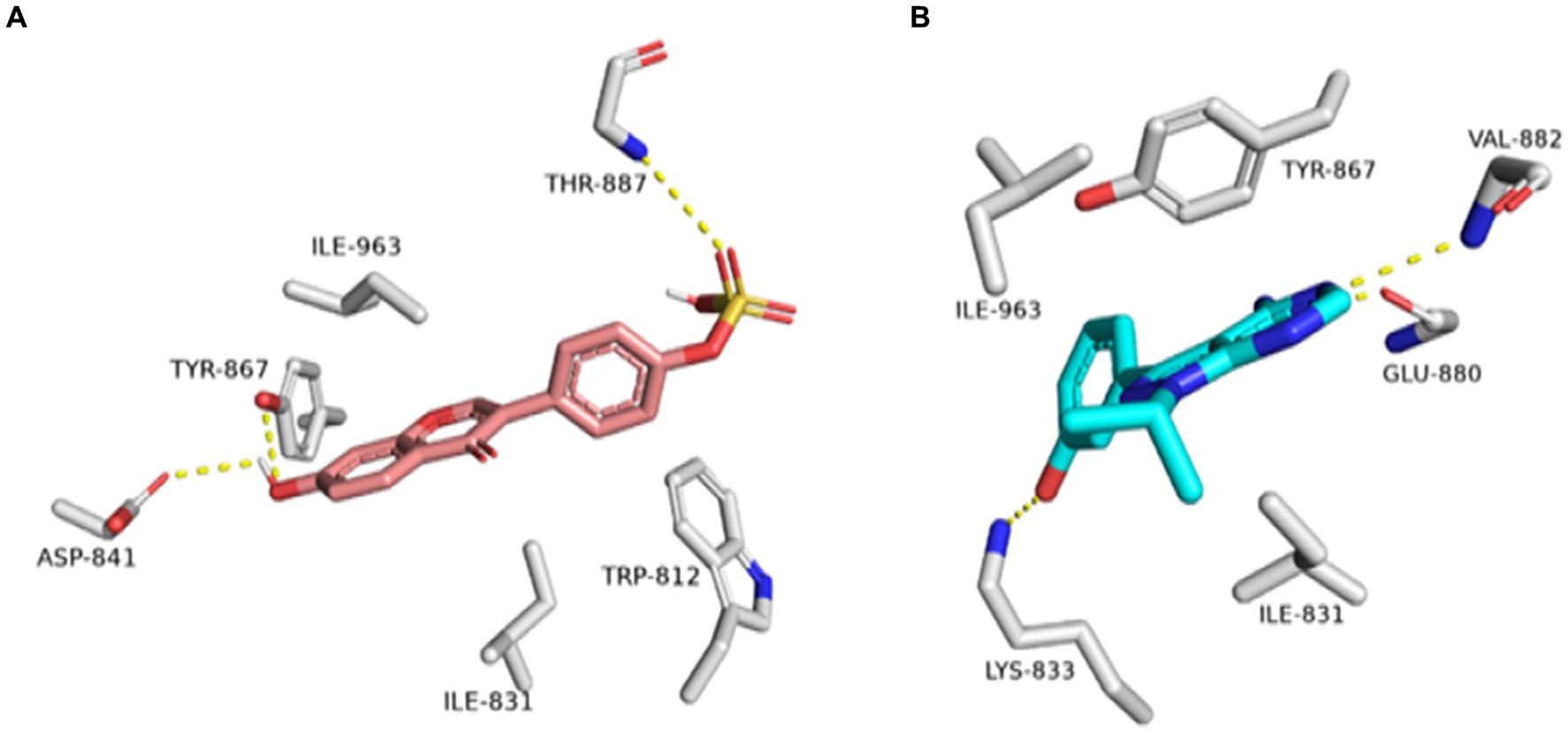
Figure 13. Binding modes of daidzein 4′-sulfate along with the co-crystalized inhibitor (A: brick red-colored structures and B: cyan-colored structure, respectively) inside the active site of PI3K.
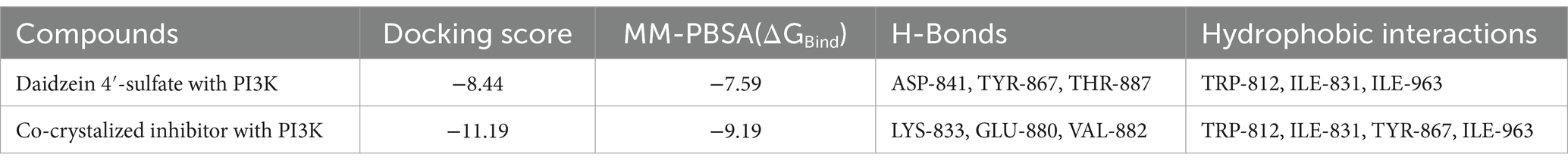
Table 3. Docking scores and ΔGBind (in kcal/mol) of daidzein 4′-sulfate into the active site of PI3K.
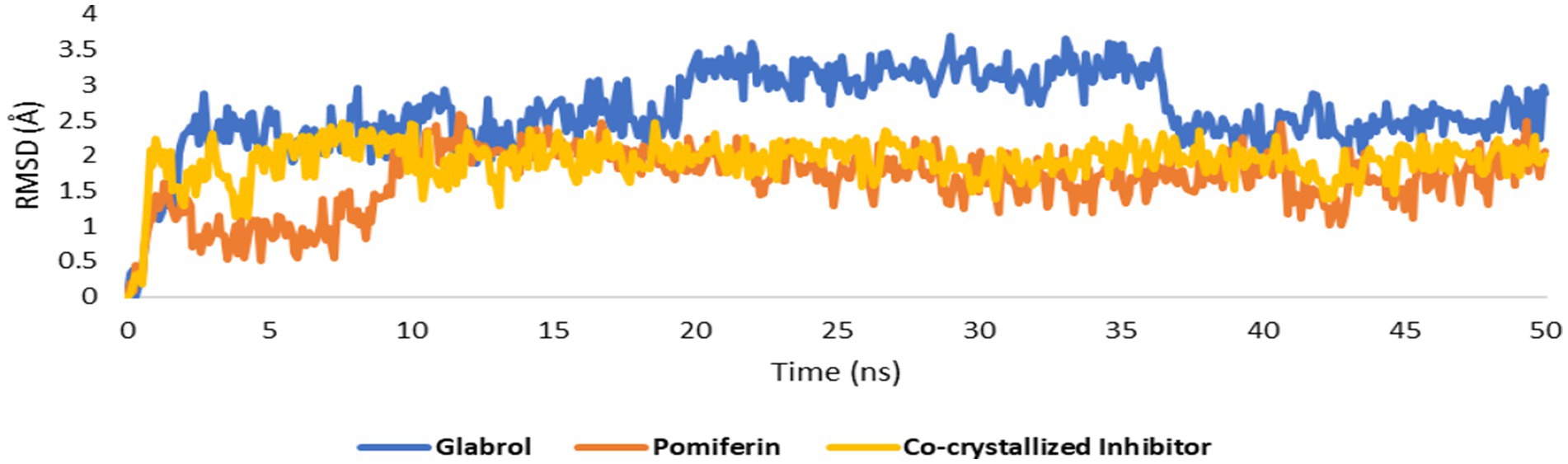
Figure 14. RMSDs of glabrol and pomiferin structures along with that of the co-crystallized inhibitor inside the GyrB’s active site throughout 50 ns-long MD simulation.
Regarding the binding behavior of daidzein 4′-sulfate structure inside the active sites of PI3K throughout 50 ns-long MD simulations in comparison with the co-crystallized inhibitor, it also achieved stable binding concerning the co-crystallized inhibitor with an average RMSD of 2.552 Å. Additionally, their calculated ΔGBinding values were convergent to that of the co-crystalized inhibitor (ΔGBinding = −8.96 and −8.87 kcal/mol for daidzein 4′-sulfate and the co-crystallized inhibitor, respectively; Figure 15).
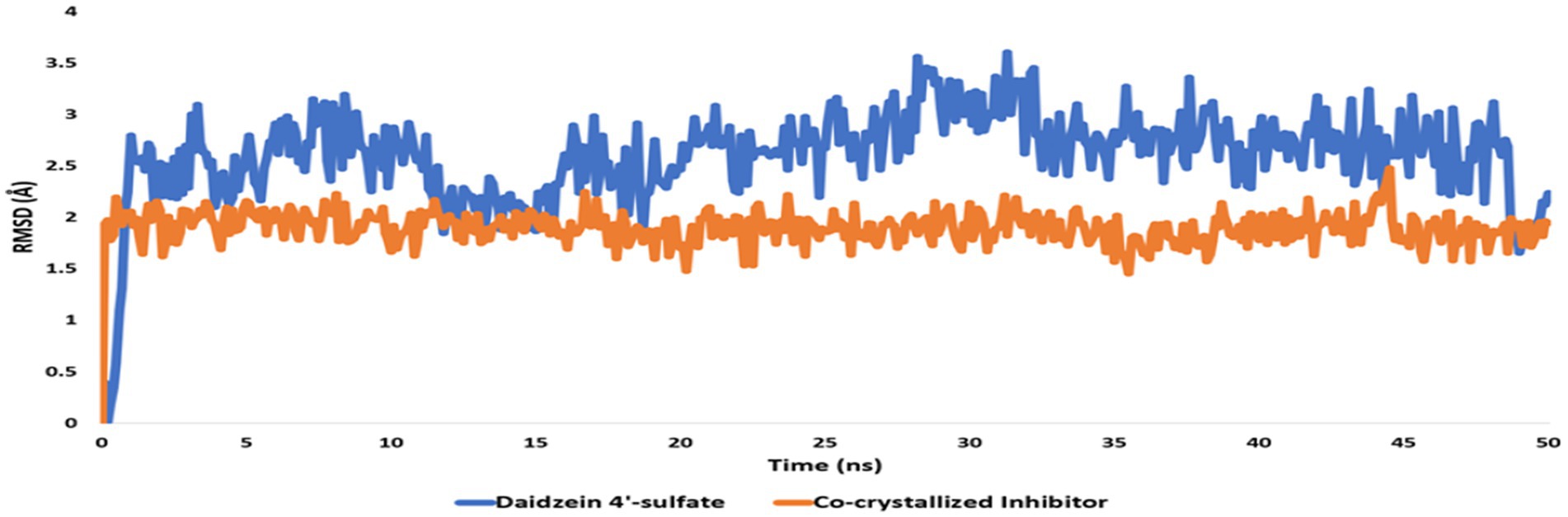
Figure 15. RMSDs of daidzein 4′-sulfate structure alongside that of the co-crystallized inhibitor inside the active site of PI3K throughout 50 ns-long MD.
Moreover, modeling and MD simulation results suggest that glabrol and pomiferin in the tested extract are the antibacterial-relevant metabolites, while daidzein 4′-sulfate is the anticancer-relevant metabolite. In previous reports, glabrol and pomiferin have been shown to exert interesting antibacterial efficacies vs. both Gram-positive and Gram-negative pathogenic bacteria including antibiotics-resistant ones (Gerhäuser, 2005; Stompor and Żarowska, 2016; Wu et al., 2019; Mohamed et al., 2022).
Discussion
Antimicrobial resistance (AMR) has gained global attention in recent years due to the terrifying prospect of rising death rates. Nanomaterials are being studied because of their potential in a variety of technical and biological applications. Zinc nanoparticles (ZnONPs) have attracted reasonable interest as potential candidates for usage in a variety of sectors including medicine, industry, and agriculture. The fungal strain SA17 was isolated and identified based on its morphological characteristics and genetically by sequencing of the 18S rRNA gene as Aspergillus sp. SA17. The fungal crude extract was obtained and used in the green biosynthesis of zinc oxide nanoparticles (ZnONPs). Numerous investigations revealed that after heating the zinc ions to 100°C, the enzymes and biomolecules in the fungal filtrate function as reducing agents to transform the zinc ions into zinc oxide nanoparticles in solution. By adjusting the temperature, incubation period, and precursor solution concentration, one may control the size and form of the nanoparticles. The UV–Vis absorption spectrum provides critical insights into the electronic changes occurring in the produced nanoparticles. The detected absorption bands correspond to specific electronic transitions and can provide information about the nanoparticles’ size, structure, and optical properties (Hameed et al., 2023). The results show that the reduced size of the nanoparticles generates a quantum confinement effect, as indicated by the appearance of an absorption band in the UV region (Qin and Zeng, 2017). In this case, the presence of larger particles or agglomerates is indicated by the absorption band at 340 nm, while the presence of small particles is indicated by the absorption peak at 280 nm (Mitjans et al., 2023). The broad absorption profile that is frequently observed in nanomaterials is caused by the variety of particle sizes in the produced sample. Additionally, the zeta potential value gives important information regarding the surface charge characteristics of ZnONPs, implying their potential for stable dispersion in neutral aqueous environments. Our data was very similar to those reported by Abdelbaky et al. (2022) and Abdelgawad et al. (2023) and more stable than ZnO nanoparticles prepared by Iqbal et al. (2021). The FTIR analysis was also utilized to study the functional groups in charge of ZnONPs production and stability including CH, C=O, C=C, C-O, C-N, and C-C-. while X-ray diffraction (XRD) was used to confirm the crystalline nature of ZnONPs. Moreover, TEM and SEM were done to investigate the nature of SA-17 ZnONPs particle size distribution and its crystallinity.
Biological evaluation of the crude extract and biosynthesized ZnONPs as antimicrobial was investigated toward several pathogens and results showed a pronounced antimicrobial effect toward E. coli, P. aeruginosa, S. aureus, B. subtilis, C. albicans, and A. flavus, respectively. The obtained results were compared with standard antibiotics, including Ciprofloxacin and Colitrimazole. Aspergillus species are known for producing unique and diverse secondary metabolites with potent biological activities (Zhang et al., 2018; Abdel-Razek et al., 2020). Several studies have demonstrated the vital role of some fungal metabolites with antimicrobial activity such as phenolic acids, flavonoids, coumarins, and anthraquinones (Nagia et al., 2012; Beekman and Barrow, 2014; Li et al., 2023). Rodrigues et al. (2022) reported that the ethyl acetate crude extract of Aspergillus sp. has shown antimicrobial activity against E. coli CBAM, S. aureus CBAM, S. aureus ATCC, and S. aureus MRSA with inhibition zone diameter values of 14, 10, 7, and 9 mm, respectively. Moreover, the ethyl acetate extract from Aspergillus unguis SPMD-EGY exhibited a marked antimicrobial effect against S. aureus, P. aeruginosa, and C. albicans (Hamed et al., 2018). In another study, the ethyl acetate extract of Aspergillus fumigatus 3 T-EGY showed an antimicrobial effect against S. aureus, P. aeruginosa, C. albicans, and A. niger with inhibition zone diameter values of 10, 15, 15, and 9 mm, respectively (Abdel-Aziz et al., 2018). Additionally, the dichloromethane extract of Aspergillus tubenginses ASH4 exhibited an antimicrobial effect on P. aeruginosa, S. aureus, E. coli, B. subtilis, and C. albicans with inhibition zone diameter values of 14, 15, 16, 13 and 15 mm, respectively (Elkhouly et al., 2021a). Furthermore, Elkhouly et al. (2021b) stated that the dichloromethane extract of Aspergillus terreus AH1 exhibited antimicrobial efficacy against C. albicans, S. aureus, B. subtilis, P. aeruginosa, and E. coli with inhibition zone diameter values of 17, 16, 14, 15, and 16 mm, respectively. Consequently, the results of the extract under study are to some extent consistent with previous studies. On the other side, several previous studies reported on the antimicrobial efficacy of the biosynthesized ZnONPs against a broad array of microbial strains. For instance, the Camellia japonica leaf extract zinc oxide nanoparticles showed antibacterial effect against S. pneumoniae, B. subtilis, E. coli, and S. typhimurium with inhibition zone values of 21, 13.5, 19, and 14 mm, respectively (Maruthupandy et al., 2018). Also, these ZnONPs showed antifungal effect against A. flavus, A. fumigatus, A. niger, and C. albicans with inhibition zone values of 9.6, 10.5, 13, and 19.1 mm, respectively (Maruthupandy et al., 2018). We can conclude that the tested ZnONPs in our current study showed low antimicrobial efficacy when compared with the findings of Muthuchamy et al. (2020). Also, our results were to some extent with the previous findings (Maruthupandy et al., 2018; Rajivgandhi et al., 2018). On the other hand, The crude extract demonstrated anticancer activity with IC50 values of (84.55 μM), (31.13 μM), (39.06 mm), and (17.65 μM), against WI38, HCT116, HePG2, and MCF7, respectively. On the other hand, the biosynthesized ZnONPs displayed anticancer activity with IC50 values of (59.74 μM), (43.21 μM), (57.03 mm), and (35.66 μM), against WI38, HCT116, HePG2, and MCF7, respectively. The results were compared with Doxorubicin (Figure 9). Aspergillus sp. has a great ability to produce a wide of secondary metabolites which are considered a potential source of new anticancer compounds (Kusari et al., 2009; El-Sayed et al., 2021; Noman et al., 2021). A study conducted by Almanaa et al. (2021) reported that A. fumigates extract showed a cytotoxic effect against the HepG-2 cell line with IC50 value of 113 μg/mL. Also, the crude extract of Aspergillus tubenginses ASH4 showed anticancer effect against HCT-116, Hep-G2, and MCF-7 with IC50 values of 9.18, 10.41, and 5.89 μg/mL, respectively (Elkhouly et al., 2021a,b). Additionally, the crude extract and biosynthesized ZnONPs demonstrated anticancer activity against WI38, HCT116, HePG2, and MCF7. Aspergillus sp. has a great ability to produce a wide range of secondary metabolites which are considered a potential source of new anticancer compounds (Kusari et al., 2009; El-Sayed et al., 2021; Noman et al., 2021). A study conducted by Almanaa et al. (2021) reported that A. fumigates extract showed a cytotoxic effect against the HepG-2 cell line with IC50 value of 113 μg/mL. Also, the crude extract of Aspergillus tubenginses ASH4 showed anticancer effect against HCT-116, Hep-G2, and MCF-7 with IC50 values of 9.18, 10.41, and 5.89 μg/mL, respectively (Elkhouly et al., 2021a,b). Chemical profiling of the extract using UPLC-QTOF-MS/MS also revealed 33 components, including flavonoids, phenolic acids, coumarins, organic acids, anthraquinones, and lignans. Based on previous reports, the different extracts of Aspergillus sp., were screened for their chemical profiles using various chromatographic and spectroscopic tools especially the hyphenated systems like UPLC-QTOF-MS/MS technique (Abdel-Aziz et al., 2018; Abdelgawad et al., 2022). Many compounds have been identified and/ or isolated belonging to several chemical classes such as anthraquinones, phenolic acids, flavonoids, coumarins, alkaloids, lactones, and terpenes (Hussein et al., 2022; Tang et al., 2022; Magot et al., 2023), indicating their unique chemical composition, which is reflected in the biological activities of these extracts. An in silico study suggested that pomiferin and glabrol could have antibacterial properties, and that daidzein 4′-sulfate could be a promising anti-cancer metabolite.
Conclusion
The current study illustrates the manufacture and analysis of zinc nanoparticles utilizing Aspergillus sp. SA17 extract. These nanoparticles demonstrate potential antibacterial and anticancer cell properties, which could have major biomedical uses. Furthermore, the produced zinc nanoparticles’ proven antibacterial and anticancer capabilities open the door for further research into their precise mechanisms of action and potential benefits when combined with current treatment approaches. The chemical examination of the extract indicated chemicals that could be useful in treating bacterial infections and cancer. These discoveries open up new avenues for future research and growth in these fields. Overall, our findings contribute to the growing body of knowledge about tailored nanoparticles and their wide uses in a variety of sectors.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
SA: Methodology, Investigation, Writing – original draft, Writing – review & editing. SE: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Project administration. EM: Conceptualization, Validation, Writing – review & editing. ME: Methodology, Investigation & Writing – original draft. HS: Visualization, Writing – review & editing. AHamd: Visualization, Writing – review & editing. MG: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Supervision, Project administration. AHame: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Supervision, Project administration.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are immensely thankful to their institutions for the unlimited support (1) Faculty of Pharmacy, Cairo University, Egypt; (2) Theodor Bilharz Research Institute, Egypt; and (3) National Research Centre, Egypt.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1366614/full#supplementary-material
Abbreviations
ZnONPs, Zinc oxide nanoparticles; HR-TEM, High-resolution transmission electron microscopy; XRD, X-ray diffraction; FT-IR, Fourier-transform infrared spectroscopy; IC50, Half maximal inhibitory concentration; UPLC-ESI-MS/MS, Ultra performance liquid chromatography-quadrupole time-of-flight-mass-mass; DNA, Deoxyribonucleic acid; PDA, Potato dextrose agar; PCR, Polymerase chain reaction; dNTPs, Deoxynucleotide triphosphates; DMSO, Dimethyl sulfoxide; WI38, Human lung fibroblast; HCT-116, Colorectal carcinoma colon cancer; MCF-7, Mammary gland breast cancer; HEPG-2, Hepatocellular carcinoma; ATCC, American type culture collection; BLAST, Basic local alignment search tool; UV–Vis, Ultraviolet–visible; SEM, Scanning electron microscopy; MD, Molecular dynamic
References
Abdel-Aziz, M. S., Ghareeb, M. A., Saad, A. M., Refahy, L. A., and Hamed, A. A. (2018). Chromatographic isolation and structural elucidation of secondary metabolites from the soil-inhabiting fungus Aspergillus fumigatus 3T-EGY. Acta Chromatogr. 30, 243–249. doi: 10.1556/1326.2017.00329
Abdelaziz, M., Shalabi, A., Radwan, A., Khaled, E., and Hassan, R. (2021). Biosynthesis and bio-sensing applications of silver and gold metal nanoparticles. Egypt. J. Chem. 64, 1057–1063. doi: 10.21608/EJCHEM.2020.43379.2878
Abdelbaky, A. S., Abd El-Mageed, T. A., Babalghith, A. O., Selim, S., and Mohamed, A. M. (2022). Green synthesis and characterization of ZnO nanoparticles using Pelargonium odoratissimum (L.) aqueous leaf extract and their antioxidant, antibacterial and anti-inflammatory activities. Antioxidants 11:1444. doi: 10.3390/antiox11081444
Abdelgawad, F. A. M., El-Hawary, S. S., Abd El-Kader, E. M., Alshehri, S. A., Rabeh, M. A., El-Mosallamy, A. E., et al. (2023). Phytochemical profiling and antiviral activity of green sustainable nanoparticles derived from Maesa indica (Roxb.) sweet against human coronavirus 229E. Plan. Theory 12:2813. doi: 10.3390/plants12152813
Abdelgawad, M. A., Hamed, A. A., Nayl, A. A., Badawy, M. S. E. M., Ghoneim, M. M., Sayed, A. M., et al. (2022). The chemical profiling, docking study, and antimicrobial and antibiofilm activities of the endophytic fungi aspergillus sp. AP5. Molecules 27:1704. doi: 10.3390/molecules27051704
Abdel-Nasser, M., Abdel-Maksoud, G., Abdel-Aziz, M. S., Darwish, S. S., Hamed, A. A., and Youssef, A. M. (2022). Evaluation of the efficiency of nanoparticles for increasing α-amylase enzyme activity for removing starch stain from paper artifacts. J. Cult. Herit. 53, 14–23. doi: 10.1016/j.culher.2021.11.004
Abdel-Razek, A. S., El-Ghonemy, D. H., and Shaaban, M. (2020). Production and purification of bioactive compounds with potent antimicrobial activity from a novel terrestrial fungus aspergillus sp. DHE. Biocatal. Agric. Biotechnol. 28:101726. doi: 10.1016/j.bcab.2020.101726
Almanaa, T. N., Yassin, M. A., El-Mekkawy, R. M., Ahmed, N. S., and Rabie, G. H. (2021). Anticancer and antioxidant activity by secondary metabolites of Aspergillus fumigatus. Adv. Anim. Vet. Sci. 9, 265–273. doi: 10.17582/journal.aavs/2021/9.2.265.273
Alrabayah, I. N., Elhawary, S. S., Kandil, Z. A., El-Kadder, E. M. A., Moemen, Y. S., Saleh, A. M., et al. (2022). Green synthesized zinc oxide nanoparticles based on Cestrum diurnum L. of potential antiviral activity against human Corona 229-E virus. Molecules 28:266. doi: 10.3390/molecules28010266
Ammar, S., Abidi, J., Luca, S. V., Boumendjel, M., Skalicka-Woźniak, K., and Bouaziz, M. (2020). Untargeted metabolite profiling and phytochemical analysis based on RP-HPLC-DAD-QTOF-MS and MS/MS for discovering new bioactive compounds in Rumex algeriensis flowers and stems. Phytochem. Anal. 31, 616–635. doi: 10.1002/pca.2928
Apsel, B., Blair, J. A., Gonzalez, B., Nazif, T. M., Feldman, M. E., Aizenstein, B., et al. (2008). Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 4, 691–699. doi: 10.1038/nchembio.117
Attia, G. H., Moemen, Y. S., Youns, M., Ibrahim, A. M., Abdou, R., and El Raey, M. A. (2021). Antiviral zinc oxide nanoparticles mediated by hesperidin and in silico comparison study between antiviral phenolics as anti-SARS-CoV-2. Colloids Surf. B Biointerfaces 203:111724. doi: 10.1016/j.colsurfb.2021.111724
Bachheti, R. K., Abate, L., Bachheti, A., Madhusudhan, A., and Husen, A. (2021). “Algae-, fungi-, and yeast-mediated biological synthesis of nanoparticles and their various biomedical applications” in Handbook of greener synthesis of nanomaterials and compounds. eds. B. Kharisov and O. Kharissova. (Cambridge: Elsevier Inc.), 701–734.
Barth, C. D. S., Souza, H. G. T. D., Rocha, L. W., Madeira, C. R. D. S., and Bresolin, T. M. B. (2019). RP-HPLC and LC-MS-MS determination of a bioactive artefact from Ipomoea pescaprae extract. Rev. Bras 29, 570–577. doi: 10.1016/j.bjp.2019.05.005
Beekman, A. M., and Barrow, R. A. (2014). Fungal metabolites as pharmaceuticals. Aust. J. Chem. 67, 827–843. doi: 10.1071/CH13639
Castillo, A., Celeiro, M., Lores, M., Grgić, K., Banožić, M., Jerković, I., et al. (2023). Bioprospecting of targeted phenolic compounds of Dictyota dichotoma, Cystoseira barbata, Ericaria amentacea, Sargassum hornschuchii and Ellisolandia elongata from the Adriatic Sea extracted by two green methods. Mar. Drugs 21:97. doi: 10.3390/md21020097
Chavan, R. R., Bhinge, S. D., Bhutkar, M. A., Randive, D. S., Wadkar, G. H., Todkar, S. S., et al. (2020). Characterization, antioxidant, antimicrobial and cytotoxic activities of green synthesized silver and iron nanoparticles using alcoholic Blumea eriantha DC plant extract. Mater. Today Commun. 24:101320. doi: 10.1016/j.mtcomm.2020.101320
Chen, W., Yeo, S. C. M., Chuang, X. F., and Lin, H. (2016). Determination of pinostilbene in rat plasma by LC-MS/MS: application to a pharmacokinetic study. J. Pharm. Biomed. Anal. 120, 316–321. doi: 10.1016/j.jpba.2015.12.051
Chua, L. S., Amin, N. A. M., Neo, J. C. H., Lee, T. H., Lee, C. T., Sarmidi, M. R., et al. (2011). LC-MS/MS-based metabolites of Eurycoma longifolia (Tongkat Ali) in Malaysia (Perak and Pahang). J. Chromatogr. B 879, 3909–3919. doi: 10.1016/j.jchromb.2011.11.002
Clugston, D. M. (1996). Mass spectrometry of organic compounds, part I. Oxygenated quinolines and furoquinoline alkaloids, part II. Lobinaline and its hofmann degradation products. A Thesis Submitted to the Faculty of Graduate Studies in Partial Fulfilment of the Requirements for the Degree Doctor of Philosophy McMaster University.
Dehkordi, F. J., Kharazian, N., and Lorigooini, Z. (2020). Characterization of flavonoid components in Scutellaria L. species (Lamiaceae) using fingerprinting analysis. Acta Biol. Cracov. Bot. 62, 79–96. doi: 10.24425/abcsb.2020.131666
Elawady, M. E., Hamed, A. A., Alsallami, W. M., Gabr, E. Z., Abdel-Monem, M. O., and Hassan, M. G. (2023). Bioactive metabolite from endophytic aspergillus versicolor SB5 with anti-acetylcholinesterase, anti-inflammatory and antioxidant activities: in vitro and in silico studies. Microorganisms. 11:1062. doi: 10.3390/microorganisms11041062
El-Bendary, M. A., Abdelraof, M., Moharam, M. E., Elmahdy, E. M., and Allam, M. A. (2021). Potential of silver nanoparticles synthesized using low active mosquitocidal Lysinibacillus sphaericus as novel antimicrobial agents. Prep. Biochem. Biotechnol. 51, 926–935. doi: 10.1080/10826068.2021.1875236
Elkhouly, H. I., Hamed, A. A., El Hosainy, A. M., Ghareeb, M. A., and Sidkey, N. M. (2021a). Bioactive secondary metabolite from endophytic aspergillus tubenginses ASH4 isolated from Hyoscyamus muticus: antimicrobial, antibiofilm, antioxidant and anticancer activity. Pharmacogenomics J. 13, 434–442. doi: 10.5530/pj.2021.13.55
Elkhouly, H. I., Sidkey, N. M., Ghareeb, M. A., El Hosainy, A. M., and Hamed, A. A. (2021b). Bioactive secondary metabolites from endophytic aspergillus terreus AH1 isolated from Ipomoea carnea growing in Egypt. Egypt. J. Chem. 7511–7520. doi: 10.21608/EJCHEM.2021.85908.4161
El-Sayad, M. H., El-Wakil, E. S., Moharam, Z. H., Abd El-Latif, N. F., Ghareeb, M. A., and Elhadad, H. (2023). Repurposing drugs to treat trichinellosis: in vitro analysis of the anthelmintic activity of nifedipine and Chrysanthemum coronarium extract. BMC Complement. Med. Ther. 23:242. doi: 10.1186/s12906-023-04076-8
El-Sayed, A. S. A., Khalaf, S. A., Azez, H. A., Hussein, H. A., EL-Moslamy, S. H., Sitohy, B., et al. (2021). Production, bioprocess optimization and anticancer activity of Camptothecin from aspergillus terreus and aspergillus flavus, endophytes of Ficus elastica. Process Biochem. 107, 59–73. doi: 10.1016/j.procbio.2021.05.007
Eskander, D. M., Atalla, S. M. M., Hamed, A. A., and ElKhrisy, E. D. A. (2020). Investigation of secondary metabolites and its bioactivity from Sarocladium kiliense SDA20 using shrimp shell wastes. Pharm. J. 12, 636–644. doi: 10.5530/pj.2020.12.95
Fu, Z., Li, Z., Hu, P., Feng, Q., Xue, R., Hu, Y., et al. (2015). A practical method for the rapid detection and structural characterization of major constituents from traditional Chinese medical formulas: analysis multiple constituents in Yinchenhao decoction. Anal. Methods 7, 4678–4690. doi: 10.1039/C5AY00993F
Gerhäuser, C. (2005). Broad spectrum antiinfective potential of xanthohumol from hop (Humulus lupulus L.) in comparison with activities of other hop constituents and xanthohumol metabolites. Mol. Nutr. Food Res. 49, 827–831. doi: 10.1002/mnfr.200500091
Ghareeb, M. A., Hamed, M. M., Saad, A. M., Abdel-Aziz, M. S., Hamed, A. A., and Refahy, L. A. (2019a). Bioactive secondary metabolites from the locally isolated terrestrial fungus Penicillium sp. SAM16-EGY. Pharm. Res. 11, 162–170. doi: 10.4103/pr.pr_102_18
Ghareeb, M. A., Khalaf, O. M., Abdel-Aziz, M. S., Saad, A. M., Madkour, H. M. F., El-Ziaty, A. K., et al. (2020). Chemical profiles and bio-activities of different extracts of Terfezia species and their other associated fungi. Curr. Bioact. Compd. 16, 308–319. doi: 10.2174/1573407214666181009110805
Ghareeb, M. A., Mohamed, T., Saad, A. M., Refahy, L. A., Sobeh, M., and Wink, M. (2018b). HPLC-DAD-ESI-MS/MS analysis of fruits from Firmiana simplex (L.) and evaluation of their antioxidant and antigenotoxic properties. J. Pharm. Pharmacol. 70, 133–142. doi: 10.1111/jphp.12843
Ghareeb, M. A., Mohammed, H. S., Aboushousha, T., Lotfy, D. M., El-Shazly, M. A. M., Sobeh, M., et al. (2024). Ipomoea carnea mitigates ethanol-induced ulcers in irradiated rats via Nrf2/HO-1 pathway: an in vivo and in silico study. Sci. Rep. 14:3469. doi: 10.1038/s41598-024-53336-1
Ghareeb, M. A., Sobeh, M., Aboushousha, T., Esmat, M., Mohammed, H. S., and El-Wakil, E. S. (2023). Polyphenolic profile of Herniaria hemistemon aerial parts extract and assessment of its anti-cryptosporidiosis in a murine model: in silico supported in vivo study. Pharmaceutics. 15:415. doi: 10.3390/pharmaceutics15020415
Ghareeb, M. A., Sobeh, M., El-Maadawy, W. H., Mohammed, H. S., Khalil, H., Botros, S. S., et al. (2019b). Chemical profiling of polyphenolics in Eucalyptus globulus and evaluation of its hepato-renal protective potential against cyclophosphamide induced toxicity in mice. Antioxidants. 8:4153. doi: 10.3390/antiox8090415
Ghareeb, M. A., Sobeh, M., Rezq, S., El-Shazly, A. M., Mahmoud, M. F., and Wink, M. (2018a). HPLC-ESI-MS/MS profiling of polyphenolics of a leaf extract from Alpinia zerumbet (Zingiberaceae) and its anti-inflammatory, anti-nociceptive, and antipyretic activities in vivo. Molecules 23:3238. doi: 10.3390/molecules23123238
Gillespie, T. A., Urogdi, L., Katritzky, A. R., and Yost, R. A. (1989). Studies of phthalic acid and related compounds by positive- and negative-ion chemical ionization tandem mass spectrometry (± CI/MS/MS). Mass Spectrom. 24, 817–822. doi: 10.1002/oms.1210240916
Gomaa, E. Z. (2022). Microbial mediated synthesis of zinc oxide nanoparticles, characterization and multifaceted applications. J. Inorg. Organomet. Polym. Mater. 32, 4114–4132. doi: 10.1007/s10904-022-02406-w
Gong, J., Zhou, S., and Yang, S. (2019). Vanillic acid suppresses HIF-1α expression via inhibition of mTOR/p70S6K/4E-BP1 and Raf/MEK/ERK pathways in human colon cancer HCT116 cells. Int. J. Mol. Sci. 20:465. doi: 10.3390/ijms20030465
Ha, N. M., and Son, N. T. (2024). Health benefits of fraxetin: from chemistry to medicine. Arch. Pharm. e2400092:e2400092. doi: 10.1002/ardp.202400092
Hamed, A. A., Abdel-Aziz, M. S., and Abd El Hady, F. K. (2018). Antimicrobial and antioxidant activities of different extracts from aspergillus unguis SPMD-EGY grown on different media. Bull. Natl. Res. Cent. 42:29. doi: 10.1186/s42269-018-0027-0
Hamed, A. A., Soldatou, S., Qader, M. M., Arjunan, S., Miranda, K. J., Casolari, F., et al. (2020). Screening fungal endophytes derived from under explored Egyptian marine habitats for antimicrobial and antioxidant properties in factionalised textiles. Microorganisms. 8:1617. doi: 10.3390/microorganisms8101617
Hameed, H., Waheed, A., Sharif, M. S., Saleem, M., Afreen, A., Tariq, M., et al. (2023). Green synthesis of zinc oxide (ZnO) nanoparticles from green algae and their assessment in various biological applications. Micromachines. 14:928. doi: 10.3390/mi14050928
Hanley, J. C. J. (2006). Liquid chromatography/mass spectrometric methodologies for metabolomics. A thesis in chemistry submitted in partial fulfillment of the requirements for the degree of doctor of philosophy. The Pennsylvania State University, the Graduate School Eberly College of Science.
Harris, N., Blaber, M. G., and Schatz, G. C. (2016). “Optical properties of metal nanoparticles” in Encyclopedia of nanotechnology. ed. B. Bhushan (Dordrecht: Springer).
Huey, R., Morris, G. M., and Forli, S. (2012). Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial, vol. 10550. United States: The Scripps Research Institute Molecular Graphics Laboratory, 1000.
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Hussein, M. E., Mohamed, O. G., El-Fishawy, A. M., El-Askary, H. I., El-Senousy, A. S., El-Beih, A. A., et al. (2022). Identification of antibacterial metabolites from endophytic fungus Aspergillus fumigatus, isolated from Albizia lucidior leaves (Fabaceae), utilizing metabolomic and molecular docking techniques. Molecules 27:1117. doi: 10.3390/molecules27031117
Iqbal, J., Abbasi, B. A., Yaseen, T., Zahra, S. A., Shahbaz, A., Shah, S. A., et al. (2021). Green synthesis of zinc oxide nanoparticles using Elaeagnus angustifolia L. leaf extracts and their multiple in vitro biological applications. Sci. Rep. 11:20988. doi: 10.1038/s41598-021-99839-z
John Wiley & Sons, Inc. (2015) SpectraBase; SpectraBase Compound ID=H09s6PiaSou SpectraBase Spectrum ID=7FS28zxZ1Qu. Available at: https://spectrabase.com/spectrum/7FS28zxZ1Qu
Kalinowska, M., Gołębiewska, E., Świderski, G., Męczyńska-Wielgosz, S., Lewandowska, H., Pietryczuk, A., et al. (2021). Plant-derived and dietary hydroxybenzoic acids-a comprehensive study of structural, anti−/pro-oxidant, lipophilic, antimicrobial, and cytotoxic activity in MDA-MB-231 and MCF-7 cell lines. Nutrients 13:3107. doi: 10.3390/nu13093107
Kallem, R. R., Pascual, J. M., Primeaux, S., Avila, A., and Putnam, W. C. (2021). Development and validation of a LC-MS/MS method for quantitation of 3-hydroxypentanoic acid and 3-oxopentanoic acid in human plasma and its application to a clinical study of glucose transporter type I deficiency (G1D) syndrome. J. Pharm. Biomed. Anal. 205:114335. doi: 10.1016/j.jpba.2021.114335
Kerebba, N., Oyedeji, A. O., Byamukama, R., Kuria, S. K., and Oyedeji, O. O. (2022). UHPLC-ESI-QTOF-MS/MS characterisation of phenolic compounds from Tithonia diversifolia (Hemsl.) a Gray and antioxidant activity. ChemistrySelect. 7:e202104406. doi: 10.1002/slct.202104406
Khalaf, O. M., Ghareeb, M. A., Saad, A. M., Madkour, H. M. F., El-Ziaty, A. K., and Abdel-Aziz, M. S. (2019). Phenolic constituents, antimicrobial, antioxidant and anticancer activities of ethyl acetate and n-butanol extracts of Senna italica. Acta Chromatogr. 31, 138–145. doi: 10.1556/1326.2018.00412
Khaw, K. Y., Chong, C. W., and Murugaiyah, V. (2020). LC-QTOF-MS analysis of xanthone content in different parts of Garcinia mangostana and its influence on cholinesterase inhibition. J. Enzyme Inhib. Med. Chem. 35, 1433–1441. doi: 10.1080/14756366.2020.1786819
Khazaal, H. T., Khazaal, M. T., Abdel-Razek, A. S., Hamed, A. A., Ebrahim, H. Y., Ibrahim, R. R., et al. (2023). Antimicrobial, antiproliferative activities and molecular docking of metabolites from Alternaria alternata. AMB Express 13:68. doi: 10.1186/s13568-023-01568-1
Kumar, S., Singh, A., and Kumar, B. (2017). Identification and characterization of phenolics and terpenoids from ethanolic extracts of Phyllanthus species by HPLC-ESI-QTOF-MS/MS. J. Pharm. Anal. 7, 214–222. doi: 10.1016/j.jpha.2017.01.005
Kusari, S., Lamshöft, M., and Spiteller, M. (2009). Aspergillus fumigatus Fresenius, an endophytic fungus from Juniperus communis L. Horstmann as a novel source of the anticancer pro-drug deoxypodophyllotoxin. J. Appl. Microbiol. 107, 1019–1030. doi: 10.1111/j.1365-2672.2009.04285.x
Kwak, S. M., Seo, J., Hwang, J. T., Sung, G. J., Song, J. H., Jeong, J. H., et al. (2019). EGFR-c-Src-mediated HDAC3 phosphorylation exacerbates invasion of breast cancer cells. Cells 8:930. doi: 10.3390/cells8080930
Li, L., Fen, R., Fen, X., Chen, Y., Liu, X., Sun, W., et al. (2020). The development and validation of an HPLC-MS/MS method for the determination of eriocitrin in rat plasma and its application to a pharmacokinetic study. RSC Adv. 10, 10552–10558. doi: 10.1039/C9RA10925K
Li, H., Fu, Y., and Song, F. (2023). Marine aspergillus: a treasure trove of antimicrobial compounds. Mar. Drugs 21:277. doi: 10.3390/md21050277
Li, M., Liu, C., Cao, H. B., and Yi, Z. (2013). Surface charge research of graphene oxide, chemically reduced graphene oxide and thermally exfoliated graphene oxide. Adv. Mat. Res. 716, 127–131. doi: 10.4028/www.scientific.net/AMR.716.127
Li, Q., and Wang, G. (2009). Diversity of fungal isolates from three Hawaiian marine sponges. Microbiol. Res. 164, 233–241. doi: 10.1016/j.micres.2007.07.002
Liu, S., Che, Y., Wang, F., Shang, Z., Lu, J., Dai, S., et al. (2016). Identification of metabolites of 6'-Hydroxy-3,4,5,2′,4′-pentamethoxychalcone in rats by a combination of ultra-high-performance liquid chromatography with linear ion trap-orbitrap mass spectrometry based on multiple data processing techniques. Molecules 21:1266. doi: 10.3390/molecules21101266
Madkour, H. M. F., Ghareeb, M. A., Abdel-Aziz, M. S., Khalaf, O. M., Saad, A. M., El-Ziaty, A. K., et al. (2017). Gas chromatography-mass spectrometry analysis, antimicrobial, anticancer and antioxidant activities of n-hexane and methylene chloride extracts from Senna italica. J. Appl. Pharm. Sci. 7, 023–032. doi: 10.7324/JAPS.2017.70604
Magot, F., Van Soen, G., Buedenbender, L., Li, F., Soltwedel, T., Grauso, L., et al. (2023). Bioactivity and metabolome mining of deep-sea sediment-derived microorganisms reveal new hybrid PKS-NRPS macrolactone from aspergillus versicolor PS108-62. Mar. Drugs 21:95. doi: 10.3390/md21020095
Mahmoud, H. N. S., Elshazly, M. A. M., Saad, A. M., Refahy, L. A., Ghareeb, M. A., and Rizk, S. A. (2023). UPLC-QTOF/MS-assisted chemical profiling of Daucus carota leaf extract and evaluation of its antioxidant, antimicrobial and antibiofilm activities: evidence from in vitro and in silico studies. Egypt. J. Chem. 66, 2175–2190. doi: 10.21608/EJCHEM.2023.209102.7935
March, R., and Brodbelt, J. (2008). Analysis of flavonoids: tandem mass spectrometry, computational methods, and NMR. J. Mass Spectrom. 43, 1581–1617. doi: 10.1002/jms.1480
Martin, R. (2013). Handbook of hydroxyacetophenones. Berlin, Germany: Springer Science & Business Media, B.V, 532.
Maruthupandy, M., Rajivgandhi, G., Muneeswaran, T., Song, J. M., and Manoharan, N. (2018). Biologically synthesized zinc oxide nanoparticles as nanoantibiotics against ESBLs producing gram negative bacteria. Microb. Pathog. 121, 224–231. doi: 10.1016/j.micpath.2018.05.041
McGovern, P. E., Mirzoian, A., and Hall, G. R. (2009). Ancient Egyptian herbal wines. Proc. Natl. Acad. Sci. 106, 7361–7366. doi: 10.1073/pnas.0811578106
Miller, B. R. III, McGee, T. D. Jr., Swails, J. M., Homeyer, N., Gohlke, H., and Roitberg, A. E. (2012). MMPBSA. Py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321. doi: 10.1021/ct300418h
Miricescu, D., Totan, A., Stanescu-Spinu, I. I., Badoiu, S. C., Stefani, C., and Greabu, M. (2020). PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int. J. Mol. Sci. 22:173. doi: 10.3390/ijms22010173
Mitjans, M., Marics, L., Bilbao, M., Maddaleno, A. S., Piñero, J. J., and Vinardell, M. P. (2023). Size matters? A comprehensive in vitro study of the impact of particle size on the toxicity of ZnO. Nanomaterials 13:1800. doi: 10.3390/nano13111800
Mohamed, M. S., Abdelkader, K., Gomaa, H. A., Batubara, A. S., Gamal, M., and Sayed, A. M. (2022). Mechanistic study of the antibacterial potential of the prenylated flavonoid auriculasin against Escherichia coli. Arch. Pharm. 355:e2200360. doi: 10.1002/ardp.202200360
Mohamed, G. A., and Ibrahim, S. R. M. (2022). Garcixanthone E and Garcimangophenone C: new metabolites from Garcinia mangostana and their cytotoxic and alpha amylase inhibitory potential. Life. 12:1875. doi: 10.3390/life12111875
Moreira, A. S. P., da Costa, E. V., Evtuguin, D. V., Coimbra, M. A., Nunes, F. M., and Domingues, M. R. M. (2014). Neutral and acidic products derived from hydroxyl radical-induced oxidation of arabinotriose assessed by electrospray ionisation mass spectrometry. J. Mass Spectrom. 49, 280–290. doi: 10.1002/jms.3339
Muthuchamy, M., Muneeswaran, T., Rajivgandhi, G., Franck, Q., Muthusamy, A., and Ji-Ming, S. (2020). Biologically synthesized copper and zinc oxide nanoparticles for important biomolecules detection and antimicrobial applications. Mater. Today Commun. 22:100766. doi: 10.1016/j.mtcomm.2019.100766
Nagia, M. S., El-Metwally, M. M., Shaaban, M., El-Zalabani, S. M., and Hanna, A. G. (2012). Four butyrolactones and diverse bioactive secondary metabolites from terrestrial aspergillus flavipes MM2: isolation and structure determination. Org. Med. Chem. Lett. 2, 1–8. doi: 10.1186/2191-2858-2-9
Noman, E., Al-Shaibani, M. M., Bakhrebah, M. A., Almoheer, R., Al-Sahari, M., Al-Gheethi, A., et al. (2021). Potential of anti-cancer activity of secondary metabolic products from marine fungi. J. Fungi. 7:436. doi: 10.3390/jof7060436
Op De Beeck, M., Lievens, B., Busschaert, P., Declerck, S., Vangronsveld, J., and Colpaert, J. V. (2014). Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:7629. doi: 10.1371/journal.pone.0097629
Qin, J., and Zeng, H. (2017). Photocatalysts fabricated by depositing plasmonic ag nanoparticles on carbon quantum dots/graphitic carbon nitride for broad spectrum photocatalytic hydrogen generation. Appl. Catal. Environ. 209, 161–173. doi: 10.1016/j.apcatb.2017.03.005
Rajivgandhi, G., Vijayan, R., Maruthupandy, M., Vaseeharan, B., and Manoharan, N. (2018). Antibiofilm effect of Nocardiopsis sp. GRG 1 (KT235640) compound against biofilm forming gram negative bacteria on UTIs. Microb. Pathog. 118, 190–198. doi: 10.1016/j.micpath.2018.03.011
Randive, D. S., Gavade, A. S., Shejawal, K. P., Bhutkar, M. A., Bhinge, S. D., and Jadhav, N. R. (2021). Colon targeted dosage form of Capecitabine using folic acid anchored modified carbon nanotube: in vitro cytotoxicity, apoptosis and in vivo roentgen graphic study. Drug Dev. Ind. Pharm. 36, 4894–4907. doi: 10.1557/s43578-021-00431-y
Randive, D. S., Shejawal, K. P., Bhinge, S. D., Bhutkar, M. A., Jadhav, N. R., and Patil, S. B. (2023). Efficient in vitro oxaliplatin delivery with functionalized single-walled carbon nanotube for enhanced colon cancer treatment. Fut J. Pharm. Sci. 9:91. doi: 10.1186/s43094-023-00543-8
Randive, D. S., Shejawal, K. P., Bhinge, S. D., Bhutkar, M. A., Wadkar, G. H., and Jadhav, N. R. (2020). Green synthesis of gold nanoparticles of isolated Citrus bioflavonoid from orange: characterization and in vitro cytotoxicity against colon cancer cell lines COLO 320DM and HT29. Indian Drugs 57, 61–69. doi: 10.53879/id.57.08.12514
Reed, K. A. (2009). Identification of phenolic compounds from Peanut skin using HPLC-MSn. Dissertation submitted to the Faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Food Science and Technology.
Ribeiro, J. V., Bernardi, R. C., Rudack, T., Schulten, K., and Tajkhorshid, E. (2018). QwikMD-gateway for easy simulation with VMD and NAMD. Biophys. J. 114, 673a–674a. doi: 10.1016/j.bpj.2017.11.3632
Rodrigues, J. C., da Silva, W. L., da Silva, D. R., Maia, C. R., Goiabeira, C. V. S., Chagas, H. D. F., et al. (2022). Antimicrobial activity of aspergillus sp. from the Amazon biome: isolation of Kojic acid. Int. J. Microbiol. 2022:4010018. doi: 10.1155/2022/4010018
Saad, A. M., Ghareeb, M. A., Abdel-Aziz, M. S., Madkour, H. M. F., Khalaf, O. M., El-Ziaty, A. K., et al. (2017). Chemical constituents and biological activities of different solvent extracts of Prosopis farcta growing in Egypt. J. Pharmacogn. Phytother. 9, 67–76. doi: 10.5897/JPP2017.0452
Sayed, A. M., El-Hawary, S. S., Abdelmohsen, U. R., and Ghareeb, M. A. (2022). Antiproliferative potential of Physalis peruviana-derived magnolin against pancreatic cancer: a comprehensive in vitro and in silico study. Food Funct. 13, 11733–11743. doi: 10.1039/D2FO01915A
Schreyer, M., Guo, L., Thirunahari, S., Gao, F., and Garland, M. (2014). Simultaneous determination of several crystal structures from powder mixtures: the combination of powder X-ray diffraction, band-target entropy minimization and Rietveld methods. J. Appl. Cryst. 47, 659–667. doi: 10.1107/S1600576714003379
Shan, L., Wu, Y., Yuan, L., Zhang, Y., Xu, Y., and Li, Y. (2017). Rapid screening of chemical constituents in Rhizoma anemarrhenae by UPLC-Q-TOF/MS combined with data post processing techniques. Evid. Based Complement. Alternat. Med. 2017:4032820. doi: 10.1155/2017/4032820
Simons, R., Vincken, J. P., Bakx, E. J., Verbruggen, M. A., and Gruppen, H. (2009). A rapid screening method for prenylated flavonoids with ultra-high-performance liquid chromatography/electrospray ionisation mass spectrometry in licorice root extracts. Rapid Commun. Mass Spectrom. 23, 3083–3093. doi: 10.1002/rcm.4215
Soltane, R., Alasiri, A., Taha, M. N., Abd El-Aleam, R. H., Alghamdi, K. S., Ghareeb, M. A., et al. (2023). Norlobaridone inhibits quorum sensing-dependent biofilm formation and some virulence factors in Pseudomonas aeruginosa by disrupting its transcriptional activator protein LasR dimerization. Biomol. Ther. 13:1573. doi: 10.3390/biom13111573
Sorgi, C. A., Peti, A. P. F., Petta, T., Meirelles, A. F. G., Fontanari, C., de Moraes, L. A. B., et al. (2018). Comprehensive high-resolution multiple-reaction monitoring mass spectrometry for targeted eicosanoid assays. Sci. Data. 5:180167. doi: 10.1038/sdata.2018.167
Stompor, M., and Żarowska, B. (2016). Antimicrobial activity of xanthohumol and its selected structural analogues. Molecules 21:608. doi: 10.3390/molecules21050608
Tang, Z., Wang, Y., Yang, J., Xiao, Y., Cai, Y., Wan, Y., et al. (2022). Isolation and identification of flavonoid-producing endophytic fungi from medicinal plant Conyza blinii H.Lév that exhibit higher antioxidant and antibacterial activities. PeerJ. 8:e8978. doi: 10.7717/peerj.8978
Tian, L., Blount, J. W., and Dixon, R. A. (2006). Phenylpropanoid glycosyltransferases from Osage orange (Maclura pomifera) fruit. FEBS Lett. 580, 6915–6920. doi: 10.1016/j.febslet.2006.11.056
Ushiyama, F., Amada, H., Takeuchi, T., Tanaka-Yamamoto, N., Kanazawa, H., Nakano, K., et al. (2020). Lead identification of 8-(methylamino)-2-oxo-1, 2-dihydroquinoline derivatives as DNA gyrase inhibitors: hit-to-lead generation involving thermodynamic evaluation. ACS Omega. 5, 10145–10159. doi: 10.1021/acsomega.0c00865
Wang, X., Shen, Y., Wang, S., Li, S., Zhang, W., Liu, X., et al. (2017). PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 45, W356–W360. doi: 10.1093/nar/gkx374
Wang, H., Xiao, B., Hao, Z., and Sun, Z. (2016). Simultaneous determination of fraxin and its metabolite, fraxetin, in rat plasma by liquid chromatography-tandem mass spectrometry and its application in a pharmacokinetic study. J. Chromatogr. B 1017-1018, 70–74. doi: 10.1016/j.jchromb.2016.02.030
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: A guide to methods and applications. eds. M. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White. (New York: Academic Press), 315–322.
Wigley, D. B., Davies, G. J., Dodson, E. J., Maxwell, A., and Dodson, G. (1991). Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629. doi: 10.1038/351624a0
Wilhelm, S., Tavares, A. J., Dai, Q., Ohta, S., Audet, J., Dvorak, H. F., et al. (2016). Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1:16014. doi: 10.1038/natrevmats.2016.14
Wu, Y. T., Lin, L. C., and Tsai, T. H. (2006). Simultaneous determination of honokiol and magnolol in Magnolia officinalis by liquid chromatography with tandem mass spectrometric detection. Biomed. Chromatogr. 20, 1076–1081. doi: 10.1002/bmc.644
Wu, S. C., Yang, Z. Q., Liu, F., Peng, W. J., Qu, S. Q., Li, Q., et al. (2019). Antibacterial effect and mode of action of flavonoids from licorice against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 10:2489. doi: 10.3389/fmicb.2019.02489
Wu, X., Zou, K., Liu, X., Fu, S., Zhang, S., Duan, Z., et al. (2022). The novel distribution of intracellular and extracellular flavonoids produced by aspergillus sp. Gbtc 2, an endophytic fungus from Ginkgo biloba root. Front. Microbiol. 13:972294. doi: 10.3389/fmicb.2022.972294
Xiong, Z., Fu, Y., Li, J., Qin, F., and Li, F. (2010). A UPLC-MS-MS method for quantification of harpagoside and cinnamic acid in rat plasma and its application to a pharmacokinetic study after oral administration of Yanyan tablets. Chromatographia 72, 163–169. doi: 10.1365/s10337-010-1600-3
Yuan, S., Chan, H. S., and Hu, Z. (2017). Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 7:e1298. doi: 10.1002/wcms.1298
Yusof, H. M., Mohamad, R., Zaidan, U. H., and Abdul Rahman, N. A. (2019). Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: a review. J. Animal Sci. Biotechnol. 10:57. doi: 10.1186/s40104-019-0368-z
Zengin, G., Fahmy, N. M., Sinan, K. I., Uba, A. I., Bouyahya, A., Lorenzo, J. M., et al. (2022). Differential metabolomic fingerprinting of the crude extracts of three Asteraceae species with assessment of their in vitro antioxidant and enzyme-inhibitory activities supported by in silico investigations. Processes 10:1911. doi: 10.3390/pr10101911
Zhang, X., Li, Z., and Gao, J. (2018). Chemistry and biology of secondary metabolites from aspergillus genus. Nat. Prod. J. 8, 275–304. doi: 10.2174/2210315508666180501154759
Keywords: Aspergillus sp. SA17, ZnONPs, antimicrobial, anticancer, UPLC-QTOF-MS/MS, docking, DNA gyrase, phosphoinositide 3-kinase gamma
Citation: Abdelrahman SESAH, El Hawary S, Mohsen E, El Raey MA, Selim HMRM, Hamdan AME, Ghareeb MA and Hamed AA (2024) Bio-fabricated zinc oxide nanoparticles mediated by endophytic fungus Aspergillus sp. SA17 with antimicrobial and anticancer activities: in vitro supported by in silico studies. Front. Microbiol. 15:1366614. doi: 10.3389/fmicb.2024.1366614
Edited by:
Amr H. Hashem, Al-Azhar University, EgyptReviewed by:
Dalia A. Gaber, University of Applied Sciences Erfurt, GermanyRajivgandhi Govindan, University of Chile, Chile
Asghar Ali, Jamia Hamdard University, India
Copyright © 2024 Abdelrahman, El Hawary, Mohsen, El Raey, Selim, Hamdan, Ghareeb and Hamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Engy Mohsen, engy.mohsen@pharma.cu.edu.eg; Ahmed A. Hamed, ahmedshalbio@gmail.com
 Sally El Said Abo Halawa Abdelrahman
Sally El Said Abo Halawa Abdelrahman Seham El Hawary1
Seham El Hawary1 Mohamed A. El Raey
Mohamed A. El Raey Heba Mohammed Refat M. Selim
Heba Mohammed Refat M. Selim Ahmed M. E. Hamdan
Ahmed M. E. Hamdan Mosad A. Ghareeb
Mosad A. Ghareeb Ahmed A. Hamed
Ahmed A. Hamed