- 1Qingdao Institute of Marine Geology, China Geologic Survey, Qingdao, China
- 2Laboratory for Marine Geology, Qingdao Marine Science and Technology Center, Qingdao, China
- 3School of Water Resources and Environment, China University of Geosciences (Beijing), Beijing, China
- 4Department of Environmental Sciences, College of the Coast and Environment, Louisiana State University, Baton Rouge, LA, United States
The impact of climate warming on soil microbial communities can significantly influence the global carbon cycle. Coastal wetlands, in particular, are susceptible to changes in soil microbial community structure due to climate warming and the presence of invasive plant species. However, there is limited knowledge about how native and invasive plant wetland soil microbes differ in their response to warming. In this study, we investigated the temporal dynamics of soil microbes (prokaryotes and fungi) under experimental warming in two coastal wetlands dominated by native Phragmites australis (P. australis) and invasive Spartina alterniflora (S. alterniflora). Our research indicated that short-term warming had minimal effects on microbial abundance, diversity, and composition. However, it did accelerate the succession of soil microbial communities, with potentially greater impacts on fungi than prokaryotes. Furthermore, in the S. alterniflora wetland, experimental warming notably increased the complexity and connectivity of the microbial networks. While in the P. australis wetland, it decreased these factors. Analysis of robustness showed that experimental warming stabilized the co-occurrence network of the microbial community in the P. australis wetland, but destabilized it in the S. alterniflora wetland. Additionally, the functional prediction analysis using the Faprotax and FunGuild databases revealed that the S. alterniflora wetland had a higher proportion of saprotrophic fungi and prokaryotic OTUs involved in carbon degradation (p < 0.05). With warming treatments, there was an increasing trend in the proportion of prokaryotic OTUs involved in carbon degradation, particularly in the S. alterniflora wetland. Therefore, it is crucial to protect native P. australis wetlands from S. alterniflora invasion to mitigate carbon emissions and preserve the health of coastal wetland ecosystems under future climate warming in China.
1 Introduction
Coastal wetlands are important blue carbon ecosystems that regulate carbon sequestration, support biodiversity, and enhance ecosystem productivity globally (McLeod et al., 2011). However, they are highly vulnerable to climate change and human activities because of their location in intertidal zones (Osland et al., 2016; Schuerch et al., 2018). Among these threats, plant invasion and warming are particularly concerning threats. Spartina alterniflora (S. alterniflora), a globally invasive species, has spread from its native habitat along the Atlantic coast of North America to coastal areas in Asia, Europe, Oceania, southern Africa, and the Pacific coast of the United States over the past two centuries (Wang et al., 2006). Researches have shown that the invasion of S. alterniflora has changed the community structure of soil microorganisms, which in turn affects the carbon and nitrogen cycling processes in ecosystems (Yuan et al., 2016; Gao et al., 2022). For example, a study in Yancheng, Jiangsu Province, China, found that the invasion of S. alterniflora increased the abundance of soil ammonia-oxidizing archaea, methanogens, and sulfate-reducing bacteria, leading to accelerated soil carbon and nitrogen mineralization compared to native soil habitats (Yuan et al., 2016). Gao et al. (2022) discovered that invasion of S. alterniflora resulted in important reductions in the complexity and stability of microbial networks, decoupling the associations between microbes and carbon pools. While the influences of plant invasions on soil microbial communities and functions are well-documented (Lin et al., 2021; Gao et al., 2022; Hussain et al., 2023), there is limited knowledge about the influence of climate warming on soil microbes in coastal wetlands (Song et al., 2020). Specifically, it is unclear whether native and invasive plant wetlands respond differently to climate warming. Soil microbes are essential in the carbon cycle by influencing carbon mineralization and stabilization (Bardgett et al., 2008; Zhou et al., 2012). Changes in soil microbial communities can significantly impact greenhouse gas emissions from the soil (Davidson and Janssens, 2006). Therefore, studying the differential response of soil microbes in invasive and native plant wetlands to global warming can provide a more accurate assessment of the adaptability and stability of coastal wetland ecosystems under future climate change. This research can also serve as a scientific basis for projecting future carbon sequestration in coastal wetlands and developing effective management strategies.
Most studies on the impact of climate warming on ecosystems have focused on changes in microbial communities after several years warming (Xue et al., 2016; Song et al., 2020; Zhou S.-Y.-D. et al., 2023). Yet, little attention has been given to the impact of climate warming on the temporal succession of microbial communities. The succession can depend on factors such as spatial and temporal scales, ecosystem characteristics, types of disturbances, and functional traits (Zhou et al., 2014; Li et al., 2016). Previous research has shown that climate warming accelerates the turnover rates of soil microbial communities in grassland ecosystem, leading to a divergent succession of microbial communities (Guo et al., 2018). Understanding the dynamics of microbial communities over time is crucial for predicting how ecosystem stability, functionality, and services will be affected by climate warming (Prach and Walker, 2011). Therefore, to determine whether there are differences in the response of soil microbial communities in wetlands invaded by plants versus native plant wetlands to global warming, it is necessary to consider the temporal succession of microbial communities in addition to the conventional comparison of community structures.
Within soil ecosystems, microbial species interact with one another through mechanisms such as commensalism, cooperation, mutualism, competition, and predation (Faust and Raes, 2012; Banerjee et al., 2016). Such complex ecological relationships can be illustrated as networks that are able to explain the co-variation of species abundances (Faust and Raes, 2012; Pržulj and Malod-Dognin, 2016). Recent studies have demonstrated that changes in the structure of microbial networks in forest, grassland, and agricultural soils can have a significant impact on ecosystem functionality and stability (Yuan et al., 2021; Guo et al., 2022; Tian et al., 2022; Zhou S.-Y.-D. et al., 2023). For example, competing interactions among key taxa can lead to increased diversity of bacterial and fungal communities while minimizing soil organic carbon loss (Gao et al., 2020). Additionally, studies have shown that ecological networks properties can impact how soil microbial communities respond to environmental changes (de Vries et al., 2018; Yuan et al., 2021; Zhou et al., 2021). For example, Yonatan et al. (2022) have suggested that natural microbiomes with weaker interactions tend to be more stable compared to those with stronger interactions. Given the pivotal role of microbial networks for determining the ecosystem functionality and responses of wetland communities to disturbances, a comprehensive understanding of how climate warming influence microbial networks among native plant wetlands and invasive plant wetlands can shed light on the mechanisms and the direction of response of the different coastal wetland ecosystems to climate change in the future.
To investigate whether there are differences in the response of soil microbes (prokaryotes and fungi) to climate warming between native plant wetlands and invasive plant wetlands, we studied the temporal dynamics and networks of soil microbes under experimental warming in two coastal wetlands dominated by native Phragmites australis (P. australis) and invasive S. alterniflora, respectively. Our objectives were to determine: (1) How experimental warming affects soil microbes in coastal wetlands; (2) whether there are variations in the response of soil microbes to warming between native P. australis and invasive S. alterniflora; (3) how warming-induced changes shape ecosystem stability and functioning.
2 Materials and methods
2.1 Study area and experimental design
The field experiment is a part of Coastal-wetland Research On Warming Network (CROWN), located in the coastal wetland of Yancheng, Jiangsu Province, China, with an average annual tidal ranged 3–5 m (Figure 1). As described previously, the experiment was carried out in a temperate-subtropical climate transition zone, experiencing four distinct seasons (Zhou P. et al., 2023). The average air temperature is 15.5°C, and the average annual precipitation is 1,015 mm. The experimental site, CROWN S (33°22′N, 120°43′E) and CROWN P (33°36′N, 120°32′E) was dominated by C3 grasses (S. alterniflora) and C4 grasses (P. australis), respectively. Starting from December 2017, the experimental plots were subjected to temperature manipulation to simulate climate warming. Each site consisted of six blocks, with each block containing one control plot and one warming plot. The warming plots were designed using open-top chambers (OTCs), which were octagonal structures measuring 2.7 m in height and covering an area of 5.54 m2. As delineated in our prior studies (Yu et al., 2023), these structures can be analogized to diminutive greenhouses. Each of the octagonal facets spans 1.07 meters in width, unified by the interconnection of aluminum alloy frames. The facets are constructed from 4 mm thick stalinite glass, which is clear and transparent, boasting a transmittance rate of 92%. This design facilitates the absorption and retention of incoming solar radiation, thereby enhancing heating without inhibiting photosynthetic processes. Furthermore, by blocking turbulent exchanges and reflecting a portion of the long-wave radiation emitted from the ground back to plants and surface soil, the internal temperature of the chambers is naturally elevated. In the control plots, an equivalent-sized woody structure was installed to minimize environmental disturbances caused by the OTC installation. Monitoring data from 2018 to 2020 showed that the OTCs led to a significant average increase in air temperature of 0.54°C at CROWN P and 0.69°C at CROWN S (Zhou P. et al., 2023). Notably, Due to the passive warming nature of OTCs, the degree of temperature increase cannot be precisely controlled, resulting in variations across different seasons. In this study, the highest temperature increase was observed in the summer, while the lowest was in the winter (Zhou P. et al., 2023).
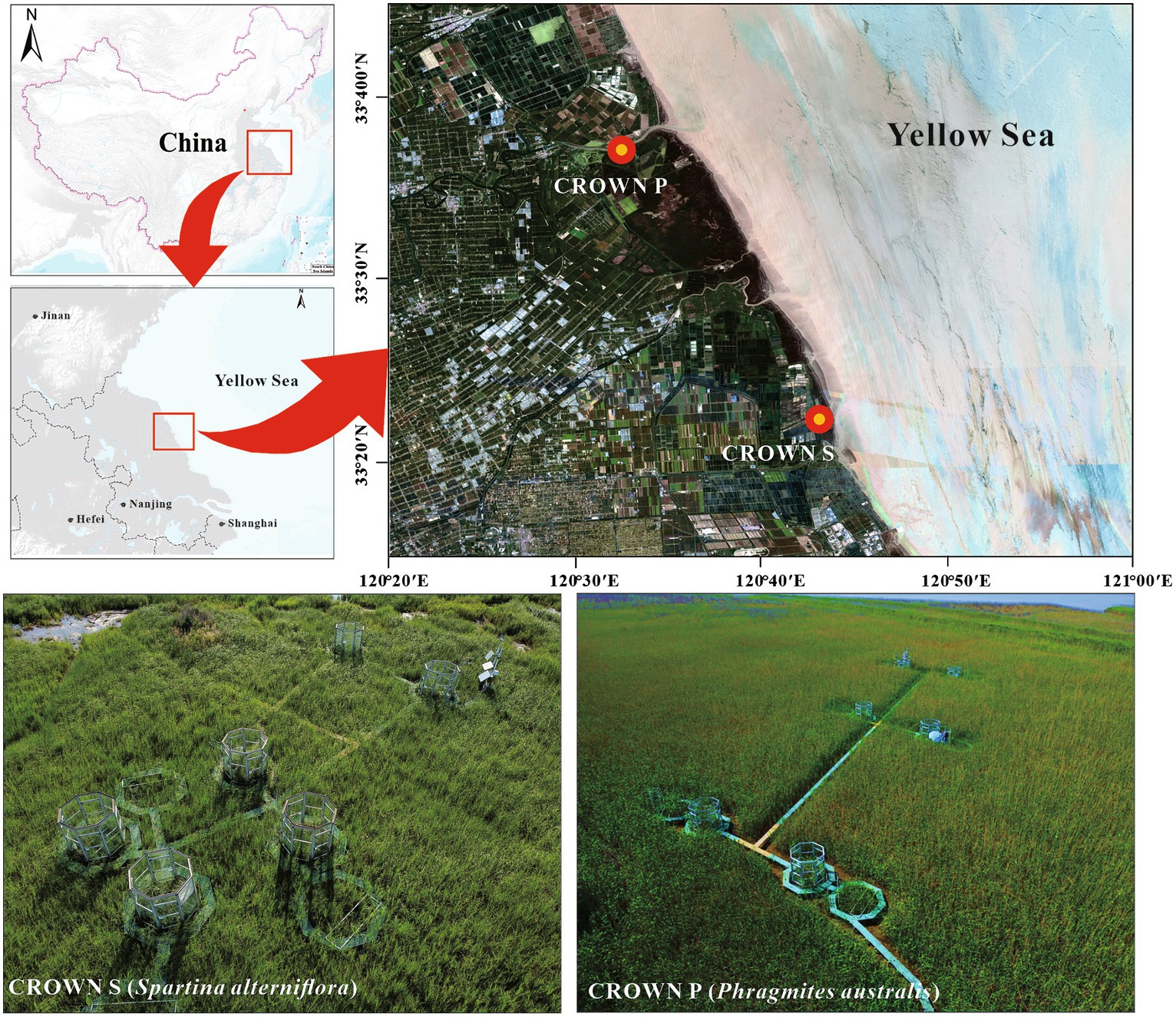
Figure 1. Experimental locations and design of CROWN S site (Spartina alterniflora) and CROWN P site (Phragmites australis) in the coastal wetland of Yancheng, Jiangsu Province, China.
2.2 Field measurements, sampling, and physicochemical analysis
Temperature sensors (HMP155A, Vaisala, Finland) placed at the center of each plot were used to record air temperature every 10 min in the warming/control plots. The pen salinity meter (SA287, HAZFULL, China) was used to measure the salinity of wetland water in the field. Ecosystem carbon exchanges, including net ecosystem exchange (NEE), ecosystem respiration (Reco), and gross primary productivity (GPP), were seasonally measured by an Ultra-Portable Greenhouse Gas Analyzer (Los Gatos Research, Quebec, Canada) along with a static chamber.
From May 2018 to January 2020, soil samples were collected seasonally (May 2018, July 2018, September 2018, November 2018, April 2019, July 2019, September 2019, November 2019, and January 2020) from the surface layer (0–10 cm) in three control and three warming plots at each CROWN site. Each sample was a composite of five soil cores (Diameter × Depth = 2.5 cm × 10 cm) obtained using a soil drill. Finally, 108 soil surface samples (9 batches) were collected and stored in a freezer at −80°C until processing. Before microbial and physicochemical analysis, visible roots and stones were manually removed from the composited soil in the lab.
Soil moisture content was determined by calculating the weight loss after freeze drying the samples. The bulk density (BD) of the soil was determined by the ring knife method, where the dry weight of the soil sample in the ring knife was divided by the volume of the ring knife. Soil pH was measured with a pH meter (Mettler-Toledo model GmbH 8,603) in a 1:2.5 sediment-to-water suspension. Total organic carbon (TOC) and total nitrogen (TN) were determined by combusting the dried samples in a CHNS analyzer (Perkin-Elmer model 2,400). Major components SiO2, Al2O3, Fe, K2O, CaO, Na2O, MgO, and P2O5 in the soils were measured using X-ray fluorescence spectroscopy (ZSX Primus II, Rigaku, Japan).
2.3 DNA extraction, PCR, Illumina sequencing, and quantitative PCR
We extracted microbial DNA from 250 mg of freeze-dried, well-mixed soil for each sample (top 10 cm, bulk soil) using the DNeasy Powersoil Pro Kit (QIAGEN, Germany). The quality and purity of the extracted DNA were assessed using an Ultramicro spectrophotometer (DS-11, DeNovix, USA).
To determine the microbial composition of soil, 16S ribosomal RNA (rRNA) gene and ITS fragment amplicons were prepared and sequenced with the Illumina Miseq platform. Following a previously established protocol, 515FmodF (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806RmodR (5′-GGACTACNVGGGTWTCTAAT-3′) targeting the V4 hypervariable region of prokaryotic (bacterial and archaeal) 16S rRNA genes (Walters et al., 2015), and ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) for fungal ITS fragment (Adams et al., 2013) were used in this study. The amplicons from each sample underwent gel purification and were utilized for the preparation of the sequencing library. Subsequently, the libraries were sequenced using the Illumina Miseq platform with 2 × 250 bp reads. The amplicon sequencing data were processed using the online platform of the Majorbio Cloud Platform.1 This involved demultiplexing, trimming, and quality score-based cleaning. The forward and reverse sequences were merged and clustered into operational taxonomic units (OTUs) at 97% similarity for both the 16S rRNA gene and the ITS. Singleton OTUs was eliminated, and to normalize the samples, 28,500 sequences of the 16S rRNA gene and 33,000 sequences of the ITS fragment were randomly resampled for each sample.
Total prokaryotic 16S rRNA gene and fungal ITS fragment abundances were measured for samples from 2019 by quantitative PCR (Q-PCR) using 515FmodF and 806RmodR primers for prokaryotes and ITS1F and ITS2F primers for fungi. All sample were run as technical triplicates on ABI7300 Q-PCR, using the 2X ChamQ SYBR color qPCR Master Mix with 2 uL DNA per 20-ul reaction and primers at a final concentration of 0.4 uM. The PCR program was 3 min at 95°C initially, followed by 40 × (5 s at 95°C, 30 s at 60°C, 60 s at 72°C), and a melt curve (60°C–95°C). Efficiencies were 97.39% for prokaryotes, with an R2 of 0.9919, and 101.47% for fungal, with an R2 of 0.9997.
2.4 Microbial ecological network analysis
We analyzed prokaryotic and fungal co-occurrence networks for each site and treatment separately. Each network consisted of 27 samples from various sampling times. To ensure reliable data associations, we included only OTUs that were present in 21 or more (>75%) samples for analysis. All networks were constructed using the Molecular Ecological Network Analysis pipeline (MENAP2) and were based on Spearman correlations (Deng et al., 2012). A threshold of r ≥ 0.820 and p < 0.001 were determined for network construction using an approach based on random matrix theory (Zhou et al., 2010; Yuan et al., 2021). Visualization of the networks was performed using Gephi software (0.10.1). The roles of nodes in the network were identified by calculating within-module connectivity (Zi) and among-module connectivity (Pi) (Guimerà and Nunes Amaral, 2005). The criteria used for categorizing nodes into module hubs, connectors, network hubs, and peripherals were based on previous studies (Zhou et al., 2011; Shi et al., 2016; Yuan et al., 2021). To assess the robustness of the microbial network, we calculated the proportion of the remaining species in the network after the removal of random or targeted nodes. In this study, we calculated the robustness when 50% of random nodes or keystone nodes were removed, as previously described (Yuan et al., 2021; Guo et al., 2022).
2.5 Statistical analysis
The impact of warming and plant invasion on soil variables and ecosystem carbon fluxes were assessed through one-way analysis of variance (ANOVA) in SPSS Statistics 27.0. The combined impacts of research site, sampling time, and experimental warming on gene abundance and alpha diversity indices were analyzed using linear mixed-effects models (Guo et al., 2022; Wu et al., 2022). The effects on the microbial community were tested using Adonis based on the Bray-Curtis distance. Additionally, we utilized Principal coordinate analysis (PCoA) to visually represent the patterns of the microbial community across different sampling times, coupled with ANOSIM for inter-group difference testing. Time-decay relationships (TDRs) were calculated as the slopes of ordinary least-squares regression lines for the relationships between the time interval and community similarities (Bray–Curtis metric). We conducted Adonis/ANOSIM to test for differences in the entire microbial community composition (Anderson, 2001). This analysis considered the factors of experimental warming, sampling time. To explore the associations between microbial community composition and soil variables, as well as ecosystem carbon fluxes, we employed Mantel-tests. All statistical analyses were performed using R version 4.0.5, along with specific packages such as vegan, picante, nlme, and lmerTest, unless otherwise stated. Finally, we utilized the FAPROTAX and Funguild databases for functional annotations of prokaryotes and fungal taxa, respectively.
3 Results
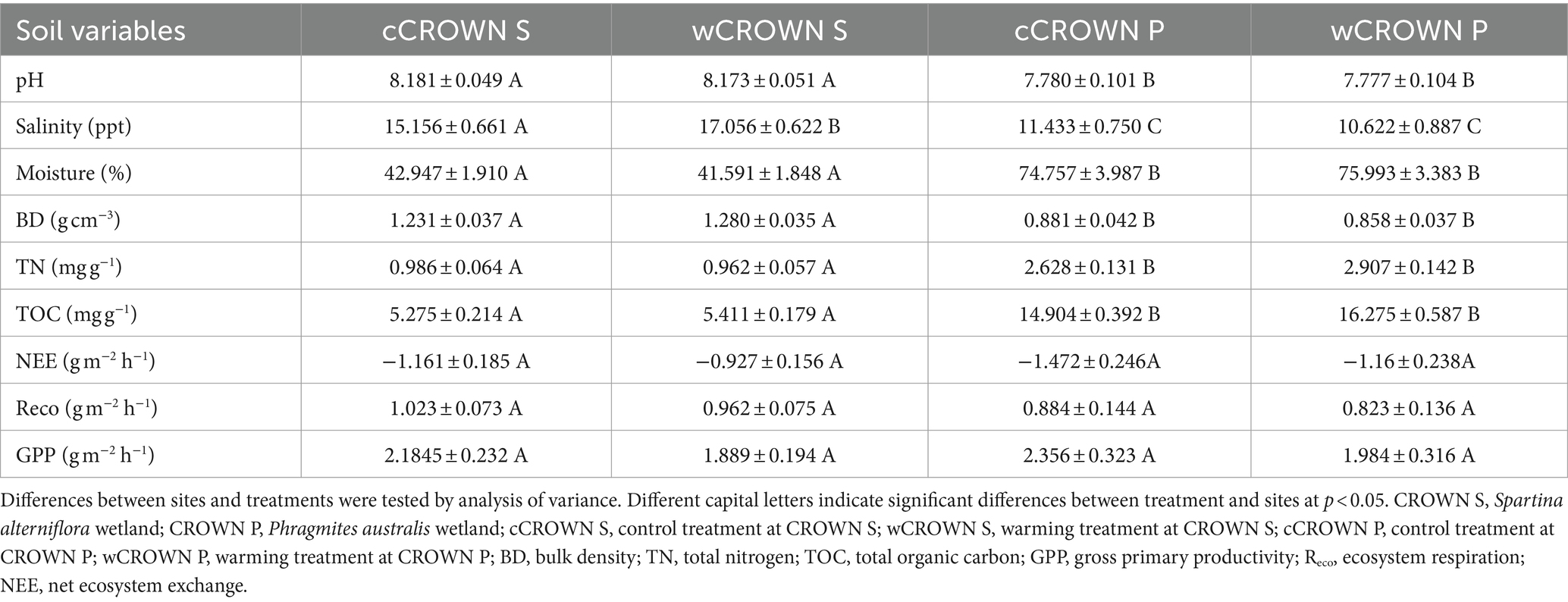
3.1 Soil variables and ecosystem carbon flux (NEE, Reco, and GPP)
In coastal wetlands, the majority of soil variables remain relatively constant under both experimental warming and varying time conditions (Table 1; Supplementary Table S1; Supplementary Figure S1). There were no significant differences in soil variables between the warming and control treatments, except for a significant increase in salinity from 15.2 ± 0.7 to 17.1 ± 0.6 ppt in the warming treatment at CROWN S (p = 0.041, Table 1). Despite the levels of NEE, Reco, and GPP appeared to decrease in the warming versus control sites, the warming effects of all those metrics were not statistically supported (p > 0.05). In Table 1, no matter in the warming or control treatments, CROWN P exhibited lower NEE and Reco values compared to CROWN S. Combined with significantly higher TOC at CROWN P, the carbon uptake capacity of CROWN P was greater than that of CROWN S. Over the period 2018–2020, aside from the pronounced seasonal fluctuations of ecosystem carbon fluxes (NEE, Reco, and GPP), TOC, TN, and Moisture (p = 0.000–0.0350), metrics including soil pH, salinity, SiO2, Al2O3, K2O, Fe2O3, and P2O5 remained constant in the coastal wetlands, with marginally significant or insignificant seasonality (p = 0.082–0.804, Supplementary Figure S1; Supplementary Table S1). However, at the CROWN P site, there was an abrupt accumulation of sediment that started in April 2019, and the soil pH, TOC, and TN decreased significantly compared to 2018, while the soil moisture increased substantially (Supplementary Figure S1). The levels of TOC, TN, and water content were higher and the pH, salinity, and BD were lower at the CROWN P site compared to the CROWN S site (Table 1).
3.2 Microbial abundance, diversity, and composition,
We investigated the influence of warming and wetland type (native Phragmites australis wetland vs. invasive Spartina alterniflora wetland) on soil prokaryotic and fungal communities at CROWN sites over nine consecutive seasons from 2018 to 2020. Our findings showed that the wetland type had the most significant influence on the abundance, diversity, and composition of soil microorganisms, followed by sampling time (Table 2). The effect of warming on the abundance, diversity, and composition of soil microorganisms was found to be insignificant (p > 0.083, Table 2; Supplementary Figure S2).
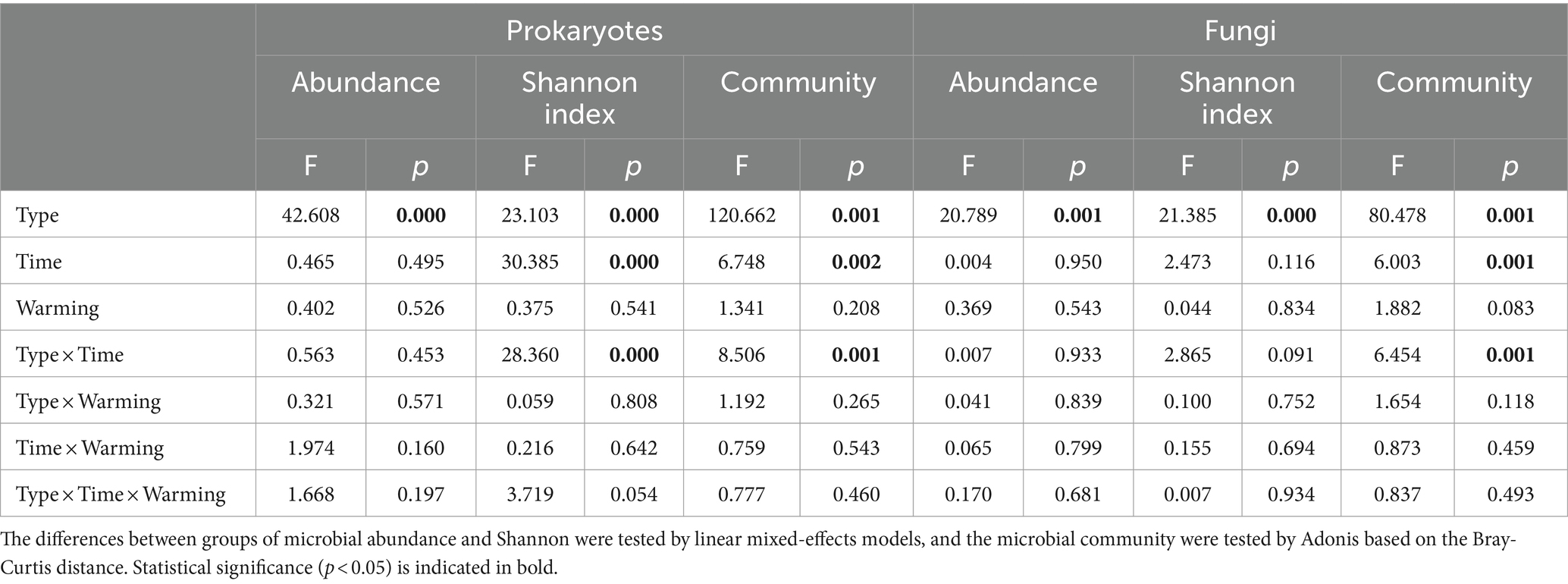
Table 2. The combined impacts of wetland type (native Phragmites australis wetland vs. invasive Spartina alterniflora wetland), sampling time, and experimental warming on soil microbes.
The absolute abundances of the 16S rRNA gene and ITS fragment in the wetland soil remained relatively constant over time and under the warming treatment (Table 2; Supplementary Figure S3). However, a markedly higher abundance of the 16S rRNA gene and ITS fragment was observed at CROWN P compared to CROWN S (Figures 2A,D). Furthermore, the results of Pearson analysis showed that the absolute abundance of these genes were strongly correlated with the levels of TOC, P2O5 and TN (r = 0.659–0.853, p < 0.001, Supplementary Table S2). For alpha diversity, the richness and diversity of prokaryotic microorganisms were significantly higher at CROWN S, while the opposite trend was observed for fungi (Supplementary Table S3). Experimental warming did not have a significant impact on the alpha diversity of both prokaryotes and fungi (Table 2; Supplementary Table S3), indicating their resilience to warming. Moreover, the diversity (e.g., Shannon index) of prokaryotes exhibited temporal shifts (Table 2), with a significant increasing trend over time at the CROWN P site (Figure 2B). This trend was strongly correlated with salinity (r = 0.685, p < 0.001). Conversely, diversity at the CROWN S site remained relatively constant throughout the study period (Figures 2B,E; Supplementary Table S4).
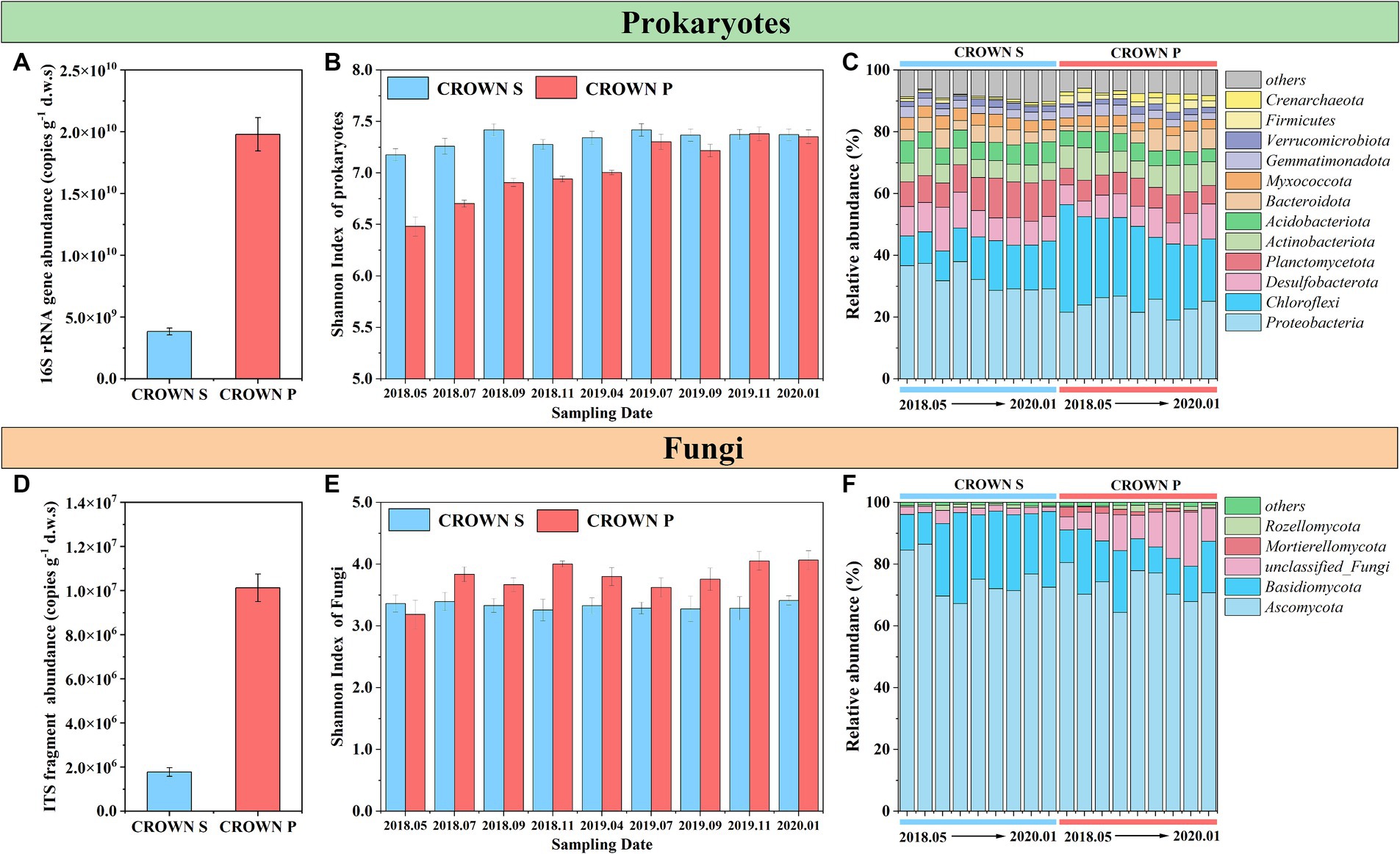
Figure 2. Microbial abundance, diversity, and composition at CROWN S and CROWN P site. (A) The absolute abundance of the 16S rRNA gene. (B) The Shannon index of prokaryote diversity at the OTU level across all sampling dates. (C) Prokaryotic taxonomic composition at the phylum level across all sampling dates. (D) The absolute abundance of the ITS fragment. (E) The Shannon index of fungal diversity at the OTU level across all sampling dates. (F) Taxonomic composition of fungi at the phylum level across all sampling dates. Error bars represent standard errors. To increase the statistical power, this analysis no longer differentiates between the group subjected to warming and the control treatment. CROWN S, Spartina alterniflora wetland; CROWN P, Phragmites australis wetland.
From Figures 2C,F, the dominant phyla in the prokaryotic microbial community were Proteobacteria, Chloroflexi, Desulfobacterota, Planctomycetota, and Actinobacteriota, while the fungal community was mainly composed of Ascomycota and Basidiomycota. At the class level, AlphaProteobacteria and GammaProteobacteria contributed to nearly all phylum Proteobacteria, while the most abundant class in the phylum Chloroflexi was Anaerolineae. The microbial community composition of both prokaryotes and fungi showed marked differences between CROWN S and CROWN P (Table 2). Specifically, for prokaryotes, Proteobacteria, Desulfobacterota, and Planctomycetota, Acidobacteriota were more abundant in the soil at CROWN S compared to CROWN P (p < 0.007), whereas Chloroflexi, Actinobacteriota, and Firmicutes were more abundant at CROWN P than at CROWN S (p < 0.001). As for fungi, Basidiomycota was notably more abundant at CROWN S than at CROWN P (p < 0.001). However, Mortierellomycota were significantly more abundant at CROWN P than at CROWN S (p < 0.05). The correlation analysis indicated that the phyla, which differ significantly between CROWN S and CROWN P, were highly related with the variation of soil properties (Supplementary Table S5). The relative abundances of Proteobacteria, Planctomycetota, and Acidobacteriota were negatively correlated with the levels of TOC, TN, P2O5, and other nutrient elements. On the other hand, Chloroflexi, Actinobacteriota and Firmicutes showed an opposite trend, with their relative abundances positively correlated with the levels of TOC, TN, P2O5, and other nutrient elements. Desulfobacterota only showed a significant positive correlation with salinity and Na2O. As for fungi, Mortierellomycota showed significant correlations with various soil properties, while Basidiomycota showed relatively weaker correlations with soil parameters.
Statistical tests (ANOSIM and Adonis) using Bray-Curtis, weighted Unifrac, and unweighted Unifrac distance metrics (Supplementary Table S6) revealed that the community composition of both prokaryotes and fungi remained constant in the experimental warming treatments. However, there was an exception to this constant. The fungal community at CROWN P was significantly different in the warming treatment when the analysis was performed with Bray-Curtis distance metrics. That difference accounted for 4.3% of the total variance (Supplementary Table S6).
Experimental warming had no significant effect on the structure of the soil microbial communities. However, the timing of sampling did have a significant impact (Table 2). Sampling time explained 20–36% of the variance in the prokaryotic microbial community and 20–33% of the variance in the fungal community (Supplementary Table S7). It is important to note that the microbial community composition in the study area has undergone significant changes over time, but it has not exhibited seasonal cyclic patterns (Figures 3A–D; Supplementary Figures S4, S5). To better understand the temporal changes in microbial community composition, we analyzed the time-decay relationships (TDRs) of the soil prokaryotes and fungi using linear regressions between community similarity and temporal distance. The slope of the linear regression, represented by the TDRs value (v), indicates the turnover rate of soil microbes over time. Our results showed that the v value of the microbial community was significantly higher at CROWN P site compared to the CROWN S site, and the v value was significantly higher for the fungi compared to the prokaryotes (Figures 3E–H). Furthermore, the v values for prokaryotes and fungi at the CROWN S and CROWN P sites were considerably higher in the warming treatments (Figures 3E–H).
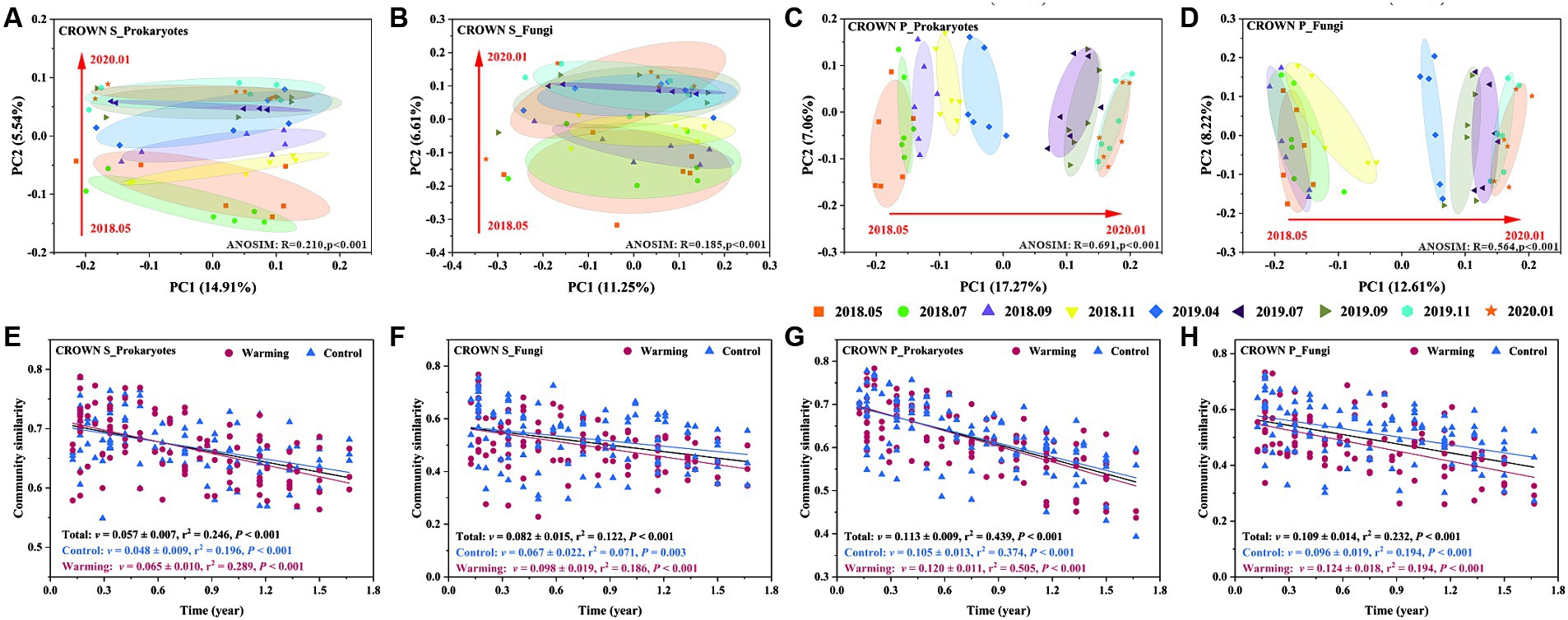
Figure 3. The temporal evolution of soil microbial communities at the CROWN S and CROWN P sites. (A–D) Principal coordinate analysis of the temporal changes in microbial communities at CROWN S and CROWN P. The analysis was performed based on unweighted UniFrac distance metrics. The information for other distance metrics (Bray-curtis and weighted UniFrac) is shown in Supplementary Figures S4, S5. To increase the statistical power, this analysis no longer differentiates between the group subjected to warming and the control treatment. (E–H) The time-decay relationships (TDRs) of soil prokaryotic and fungal communities under warming and control conditions. Community similarity was calculated as Bray-Curtis distance metrics. The TDR value (v) represents the absolute slope of the linear regression and indicates the rate of temporal turnover of the soil microbes. CROWN S, Spartina alterniflora wetland; CROWN P, Phragmites australis wetland.
3.3 Microbial community co-occurrence network
Using the abundance of prokaryotic and fungal OTUs from each site, we built twenty-two co-occurrence networks to explore the associations of microorganisms in different sites, treatments, and sampling time (Figure 4A; Supplementary Figure S6). The co-occurrence networks exhibited scale-free (R2, 0.927–0.993) and small-word characteristics (the average path distance, 1–7.180), indicating that they were not formed by random chance. As illustrated in Figure 4A and Supplementary Figure S6, bacterial dominance was observed in the microbial network of the coastal wetland soil, with fungi and archaea having lower representation. In addition, the CROWN S site had a larger and more complex network compared to the CROWN P site (Figure 4). Moreover, the effects of warming treatments on the topological properties of soil microbial co-occurrence networks differed significantly between CROWN S and CROWN P sites (Figures 4A–E; Supplementary Table S8), with warming enhanced complexity and connectivity in CROWN S and diminished them in CROWN P. On the temporal scale, the complexity of microbial network showed a significantly increasing trend over time (p < 0.026), while other topological properties (e.g., Average connectivity and Modularity) remained constant (Supplementary Figure S6).
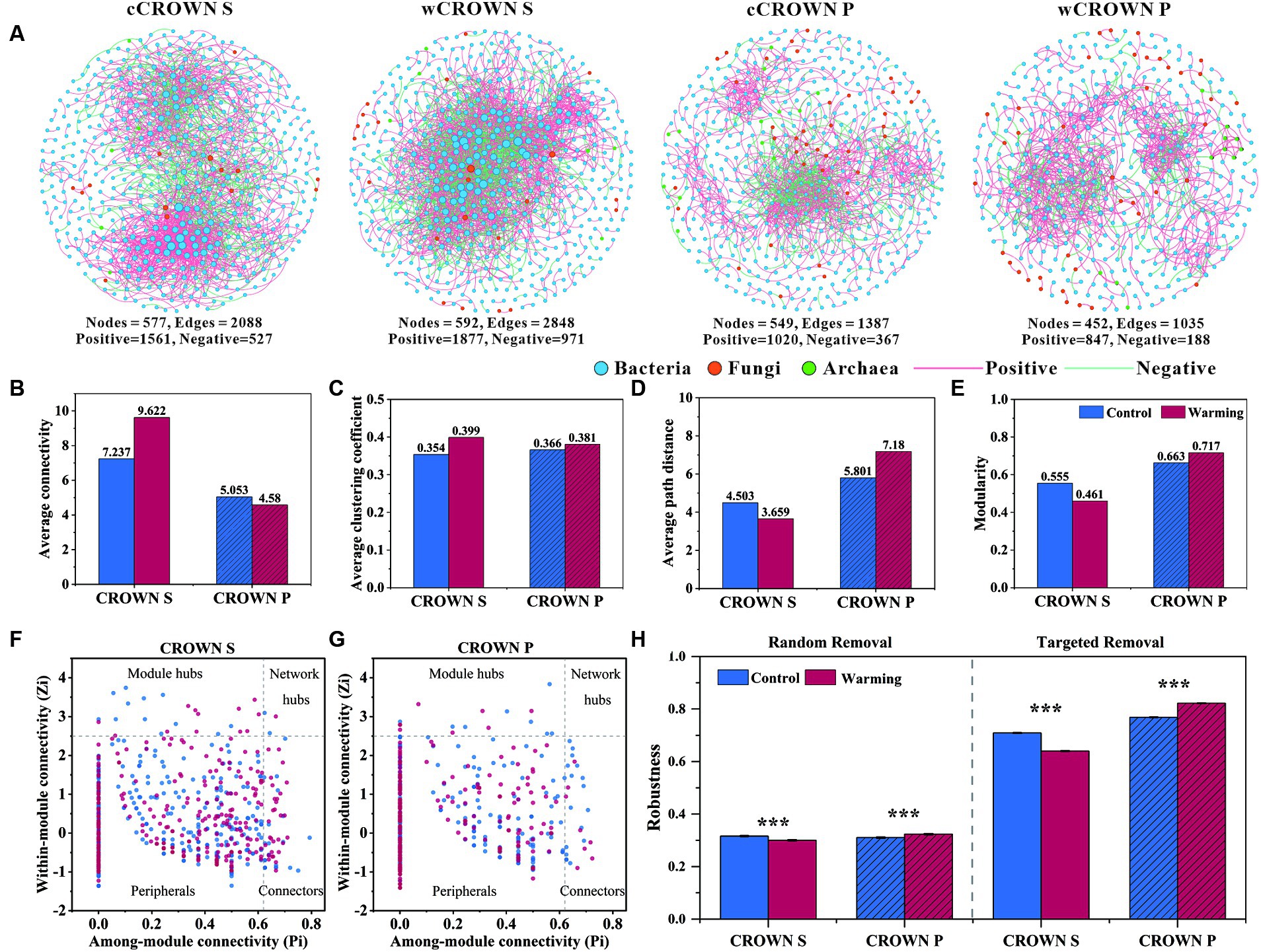
Figure 4. The co-occurrence networks of microbial communities in warming and control treatments at CROWN S and CROWN P sites. (A) Overall dynamics of microbial co-occurrence networks under warming and control. The nodes represent unique OTUs in the datasets. The size of each node is proportional to the connectivity. (B–E) Topological properties of soil microbial co-occurrence networks in the warming and control treatments. (F,G) Zi-Pi plots to identify putative keystone taxa under warming (red) and control (blue) treatments. (H) Stability of microbial networks measured by robustness. In the case of random removal, robustness was equated to the proportion of taxa that remained when 50% of the taxa were randomly removed from each of the microbial networks. In the case of targeted removal, robustness was equated to the proportion of taxa that remained when 50% of the keystone nodes (module hubs, connectors, and network hubs) were removed from each of the microbial networks. CROWN S, Spartina alterniflora wetland; CROWN P, Phragmites australis wetland; cCROWN S, control treatment at CROWN S; wCROWN S, warming treatment at CROWN S; cCROWN P, control treatment at CROWN P; wCROWN P, warming treatment at CROWN P.
The altered complexity of a network could lead to changes in the role of individual members within the network. In this study, we identified a total of 24 (16) module hubs, 53 (25) connectors, and 3 (0) network hubs in the CROWN S (CROWN P) microbial network. These nodes can be identified as keystone nodes that was essential in shaping network structure (Figures 4F–G; Supplementary Table S9). The CROWN S site had the highest number of module hubs, connectors, and network hubs. In addition, warming caused the number of keystone nodes to increase (decrease) at the CROWN S (CROWN P) site (Figures 4F–G; Supplementary Table S9). Most of the key taxa in both the CROWN S and CROWN P networks belonged to abundant phyla: 25.4% were Proteobacteria; 21.1% were Chloroflexi; and 17.5% were Planctomycetota (Supplementary Table S9). Some OTUs were found in multiple networks, serving as module hubs, connectors, or network hubs (Supplementary Table S9). For instance, OTU720, which belonged to the genus Halioglobus in the Proteobacteria, was a module hub in both the cCROWN S and wCROWN S networks. However, it is noteworthy that only 3.4% of all the keystone nodes were common between the warming and control treatments (Supplementary Table S9), indicating that warming induced alterations in the network structure at the level of keystone node.
All the microbial community co-occurrence networks that we detected were scale-free, modular, and small-world. We hypothesized that this fact might have significant implications for microbially mediated ecosystem stability. To test this hypothesis, we simulated species extinctions and calculated the robustness of the microbial community co-occurrence networks. When species were randomly lost or keystone nodes (module hubs, connectors, and network hubs) were targeted removed, the networks of CROWN P showed significantly greater robustness in the warming treatment compared to the control treatment (Figure 4H). Conversely, the networks of CROWN S showed significantly lower robustness in the warming treatment compared to the control treatment (Figure 4H). These results suggested that under experimental warming, the microbial community co-occurrence network become more stable in CROWN P but less stable in CROWN S.
3.4 Discerning drivers and functions of the soil microbial community
Differences in soil microbial communities were observed between CROWN S and CROWN P sites (Table 2). Our analysis, using Mantel test, revealed that the composition of the soil microbial communities was mainly correlated with TOC, followed by Na2O, K2O, P2O5, and TN (Supplementary Table S10). Additionally, we found that the soil microbial communities at CROWN S showed generally stronger correlations with various environmental parameters compared to CROWN P (Figures 5A,B). Specifically, at CROWN S, the microbial community composition was significantly influenced by the major components of the soil (K2O, CaO, MgO, Mn, Fe2O3), moisture, and TOC (Figure 5A). At CROWN P, the microbial community composition was significantly shaped by CaO, salinity, and pH (Figure 5B). Notably, these correlations were generally robust in both the warming and the control treatment, although the correlations in some cases were relatively weak (Supplementary Figure S7). Furthermore, at both the CROWN S and CROWN P sites, there was a close association between the composition of the soil microbial community and Reco (Figures 5A,B).
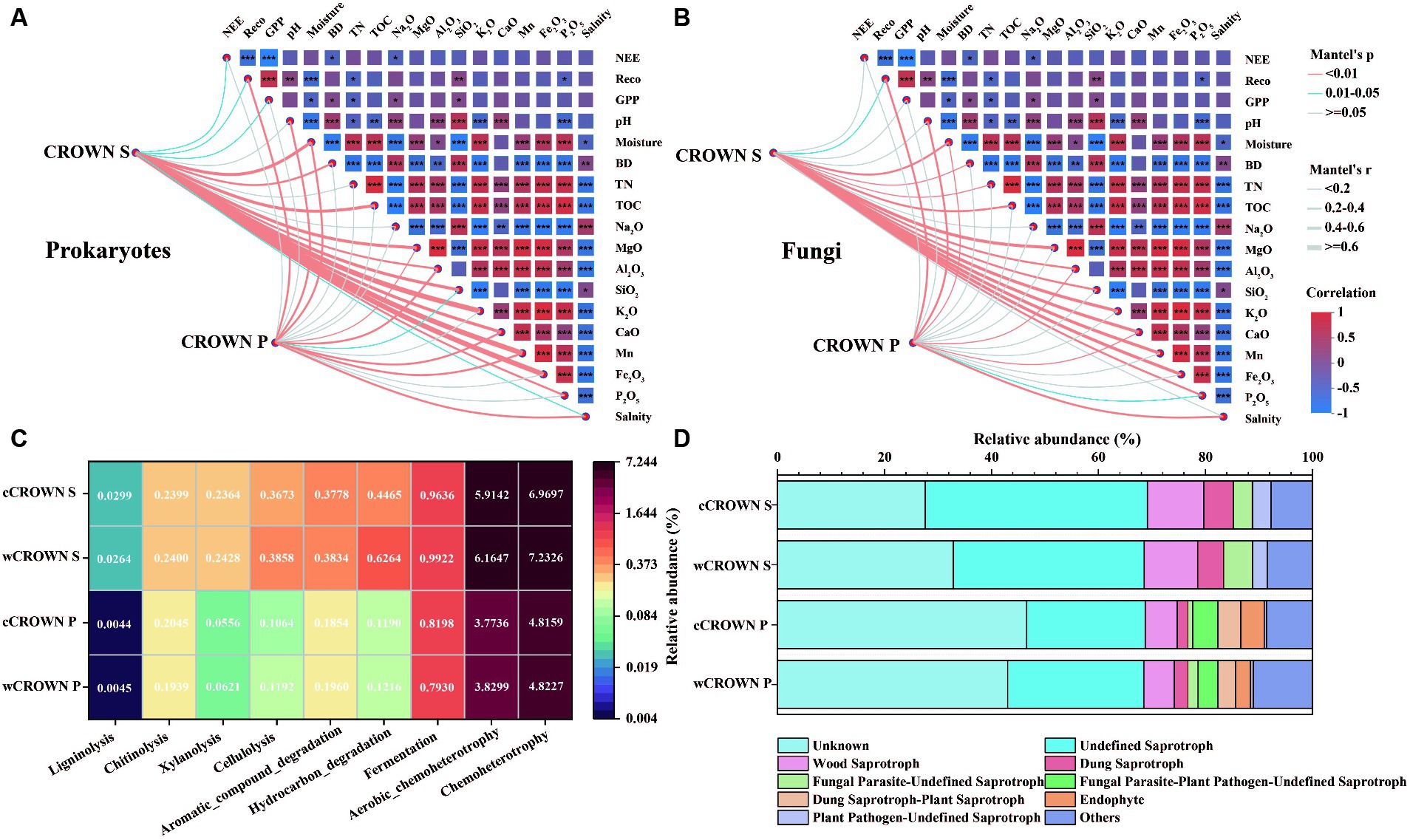
Figure 5. Relationships between the microbial community, soil variables, ecosystem processes, and community functional traits. (A) Correlations of the prokaryotic community composition (Bray-Curtis distance) with soil variables and ecosystem carbon fluxes. (B) Correlations of the fungal community composition (Bray-Curtis distance) with soil variables and ecosystem carbon fluxes. (C) Functional prediction for prokaryotes with the Faprotax database. (D) Functional prediction for fungi with the FUNGuild database. CROWN S, Spartina alterniflora wetland; CROWN P, Phragmites australis wetland; cCROWN S, control treatment at CROWN S; wCROWN S, warming treatment at CROWN S; cCROWN P, control treatment at CROWN P; wCROWN P, warming treatment at CROWN P. NEE, net ecosystem exchange; Reco, ecosystem respiration; GPP, gross primary productivity; BD, bulk density; TN, total nitrogen; TOC, total organic carbon.
To evaluate the differences in carbon degradation potential between sites and treatments, we used the Faprotax and FunGuild databases to predict the functions of the prokaryotes and fungi, respectively, with OTUs. The results showed that the proportion of prokaryotic OTUs involved in carbon degradation were significantly higher at CROWN S than at CROWN P (p < 0.05, Figure 5C; Supplementary Table S11), and the warming treatments generally resulted in an increased proportion of OTUs involved in carbon degradation (Figure 5C). Based on their nutritional modes, eight guilds of fungi (abundances >2%) were identified from the three major fungal categories: saprotrophs, symbiotrophs, and pathotrophs (Figure 5D). Among these categories, the relative abundances of undefined saprotrophs (22.2–41.5%) and wood saprotrophs (5.7–10.6%) were the highest. A comparison of the two sites revealed that the relative abundance of saprotrophs was significantly higher at the CROWN S site (p < 0.05, Supplementary Table S12), whereas the proportion of symbiotrophs was significantly higher at the CROWN P site (p < 0.01, Supplementary Table S12). There was no discernible evidence of significant differences in the relative abundances of the various guilds between the control and warming treatments at either site (Supplementary Table S12).
4 Discussion
4.1 Distinct soil microorganisms across invasive Spartina alterniflora and native Phragmites australis
In the present study, prokaryotic and fungal communities were studied in quantitative and qualitative terms over the coastal wetland. Compared to the CROWN P site, the CROWN S site exhibited higher microbial abundance (Figures 2A,D). This can be attributed to the higher levels of nutrients (such as TOC, TP, TN, etc.) in the soil at the CROWN P site (Supplementary Table S2). Organic carbon acts as the primary carbon source for microbial growth and reproduction, while phosphorus and total nitrogen are essential nutrients for microbial growth (Stotzky and Norman, 1961; Li et al., 2015). The favorable conditions of high TOC, P2O5, TN create a conducive environment for microbial growth and reproduction (Yang et al., 2019a,b). It is worth emphasizing that in this study, we found minimal differences between seasons in the abundances of soil microorganisms in coastal wetlands (Supplementary Figure S3). Whether in the hot summer (July) or cold winter (November), the fact that they were able to maintain a relatively constant abundance indicated that these microbial communities were able to adapt to the environmental changes associated with the different seasons while retaining their ecological functions. This relatively constant abundance may also be attributed to the homogeneity of soil properties in coastal wetlands, which may in part reflect the influence of robust hydrological fluctuations like tidal cycles, which mitigate the effects of soil heterogeneity.
The diversity of prokaryotic microbes at CROWN P gradually increased over time and was closely associated with changes in salinity (R = 0.685, p < 0.001) (Figure 2B). Interestingly, when the soil salinity at CROWN P approached that of CROWN S, the diversity index at CROWN P also approached that of CROWN S. These findings indicated that salinity in the study area played a crucial role in shaping the prokaryotic microbial diversity. In the soil, salinity can alter the distribution of roots, enhance soil heterogeneity, and consequently lead to higher diversity and richness of soil microorganisms (Yang et al., 2016). In Figure 2E and Supplementary Table S3, the richness and diversity of fungi were significantly higher at CROWN P than that at CROWN S. The difference in fungal richness and diversity could be due to the stronger symbiotic relationship between P. australis and soil fungi, whereas in comparison, the symbiotic relationship between S. alterniflora and soil fungi is less common or harmful to fungal survival (Figure 5D; Supplementary Table S12), leading to reduced fungal richness and diversity. This discovery was consistent with the results of earlier research, which have suggested that S. alterniflora exhibits lower rates of fungal colonization than P. australis (Liang et al., 2016).
Based on microbial taxonomic information, most of the prokaryotic and fungal groups observed in other coastal areas (An et al., 2019; Yang et al., 2019b; Lin et al., 2022; Hussain et al., 2023) were also identified in this study. Nevertheless, noticeable differences were evident in the composition of the prokaryotic and fungal communities between CROWN S and CROWN P soils. Among them, AlphaProteobacteria, Planctomycetota, and Acidobacteriota are typical oligotrophic bacteria, capable of adapting well to low-nutrient environments (Cui et al., 2019; Zhang et al., 2019; Wang et al., 2021a), whereas Actinobacteriota and Firmicutes are classified as copiotrophic bacteria, growing rapidly in nutrient-rich environments (Li et al., 2021; Yang et al., 2023). This corresponds well with our Pearson analysis results (Supplementary Table S5), and effectively explaining the differences in their relative abundance between CROWN S (low-nutrient environments) and CROWN P (high-nutrient environments). Surprisingly, Chloroflexi, typically oligotrophic bacteria (Yang et al., 2023), were more abundant in the nutrient-rich environment of CROWN P compared to CROWN S, and their abundance was positively correlated with levels of TOC, TN, and other nutrients (Supplementary Table S5). This may be due to hydrological factors, as CROWN P soil has high soil moisture content (Table 1), and subsequence anaerobic conditions favor the growth of Anaerolineae (the most abundant class in Chloroflexi) (Yamada and Sekiguchi, 2009). Pearson correlation analysis also showed a significant positive correlation between the relative abundance of Chloroflexi and soil moisture content (Supplementary Table S5). Desulfobacterota, dominant among sulfate-reducing bacteria communities in salt marshes and known to increase in abundance after invasion by S. alterniflora (Zeleke et al., 2013; Zhang et al., 2020), was consistent with our study. Pearson correlation analysis indicated that its relative abundance was significantly and positively correlated only with salinity and Na2O (Supplementary Table S5), suggesting that salinity primarily controls its distribution. Regarding fungi, Supplementary Table S5 shows that Mortierellomycota is highly correlated with various soil parameters, while Basidiomycota shows weaker correlations. Therefore, the greater relative abundance of Basidiomycota at CROWN S compared to CROWN P may be mainly attributed to plant factors, as previous research has indicated that S. alterniflora is rich in recalcitrant carbon compounds (such as lignin and cellulose) (Schneider et al., 2012), and Basidiomycota plays a pivotal role in breaking down these compounds (Yang et al., 2009).
4.2 Changes in soil microbial communities over time
As shown in Table 2 and Supplementary Table S7, the soil microbial communities in the coastal wetlands changed significantly over time. However, unlike grass and forest ecosystems (Waldrop and Firestone, 2006; Fu et al., 2020; Luo et al., 2020; Zhou S.-Y.-D. et al., 2023), the composition of soil microbial communities in the coastal wetlands did not exhibit a seasonal cyclic pattern over time (Figures 3A–D; Supplementary Figures S4, S5). Generally, the seasonal periodicity of soil microbial communities is attributed mainly to environmental factors that undergo seasonal changes, such as soil moisture and type of vegetation (Brockett et al., 2012; Thoms and Gleixner, 2013; Zhou S.-Y.-D. et al., 2023). Within coastal wetland ecosystems, however, vegetation is uniform, and soil homogenization due to semi-diurnal tides is prevalent. We thus inferred that the relatively stable environment of the coastal wetland soils may have been one of the main factors contributing to the absence of seasonal periodicity in the microbial communities. Additionally, based on TDR analysis (Figures 3E–H) and three microbial communities time-dissimilarity patterns which proposed by Gao et al. (2024), we found the microbial communities time-dissimilarity dynamics in study area was linear pattern. This implied in coastal wetlands, the succession of community composition is driven by factors such as continuous selection, constant dispersal rate, and fixed speciation probability, etc. (Sprockett et al., 2020; Custer et al., 2022). Moreover, the TDR results revealed that replacement of native P. australis by invasive S. alterniflora slowed the rate of microbial succession (Figures 3E–H). This phenomenon may be attributed to the uniform soil properties observed in the CROWN S (Supplementary Figure S1), which do not possess the necessary characteristics to drive microbial succession. On the other hand, the decrease in spatial heterogeneity within soil microbial communities caused by plant invasion led to a lower rate of succession (Li et al., 2022; Zhang et al., 2023). Lastly, our co-occurrence network analysis of soil microbes showed a linear increase in network complexity over time, while connectivity and modularity remained consistent (Supplementary Figure S6). This outcome suggests that the interrelationships within the microbial community in coastal wetland soils grow more intricate as time advances, yet the overall network structure maintains relative stability. Specifically, the linear escalation in network complexity over time signifies the gradual broadening and intensification of interrelationships within the microbial community. This could be attributed to the accumulation and enhancement of ecological processes and interactions within the microbial community at varying time points. Despite the rise in complexity, the network’s connectivity and modularity remain unvarying, denoting the relative stability of the microbial community’s internal structure. This could indicate that while microbial interactions continue to augment, the fundamental patterns and organizational methods of the overall network structure remain largely unaltered.
4.3 Warming shapes coastal wetland soil microbial communities
Our analyses found that short-term warming did not have a significant impact on the abundance, diversity, and composition of soil microbial communities in coastal wetlands (Table 2). However, it did accelerate the turnover rates of soil prokaryotes and fungal communities (Figure 3). This result was consistent with a recent study conducted in a grassland ecosystem (Guo et al., 2018). According to the metabolic theory of ecology, temperature plays a key role in determining metabolic rates, which in turn affect various biological processes, including species turnover (Brown et al., 2004; Ren et al., 2017). Specifically, higher temperatures enhance enzyme activity, leading to faster metabolic rates, cell division, nutrient uptake, and metabolite production. This enables microorganisms to reproduce and grow more rapidly in high-temperature environments, thereby promoting faster succession of microbial communities. It is important to note that warming appeared to increase the rates of species turnover, potentially leading to greater spatiotemporal variations in the soil microbial community (Guo et al., 2018, 2019, 2022). As reported in a montane community and a coastal ecosystem coastal, warming resulted in divergent community succession and accelerated temporal scaling (Hillebrand et al., 2010; Gibson-Reinemer et al., 2015). Therefore, the effects of warming on microbial communities may be progressive (DeAngelis et al., 2015), and as warming persists, its influence may become more pronounced over time. The implication is that future climate change scenarios may lead to microbial communities deviate significantly from their current states. There is hence a possibility that these communities will diverge toward multiple alternative states, as noted by Guo et al. (2018). In addition, the significantly larger increased slope of the TDR for fungi than prokaryotes in the warming treatments suggested that fungi were more sensitive to temperature than prokaryotes, which is consistent with previous studies (Guo et al., 2018; Feng et al., 2022; Zhou S.-Y.-D. et al., 2023). This heightened sensitivity of fungi may be attributed to their greater susceptibility to alterations in surface vegetation (Dassen et al., 2017), as supported by our previous research demonstrating the substantial impact of short-term warming on surface vegetation phenological traits (Zhou et al., 2023a).
The analysis of co-occurrence networks of soil prokaryotic and fungal communities revealed that with warming, the topologic parameters of the networks became more (less) complex and connective at CROWN S (CROWN P) (Figures 4A–E). Furthermore, the microbial community co-occurrence network appeared more (less) stable in CROWN P (CROWN S) under experimental warming (Figure 4H). Collectively, these findings showed that as network complexity and connectivity increase, the stability of microbial ecosystems decreases. This phenomenon can be explained by May’s stability theory (May, 2019). In other words, higher complexity and connectivity in highly diverse ecosystems destabilize ecological systems (May, 2019; Yonatan et al., 2022). Similarly, research by de Vries et al. (2018) has revealed that microbial co-occurrence networks characterized by high connectivity and low modularity are associated with low stability. The implication is that warming may increase the resilience of P. australis wetland soil to environmental perturbations and lead to greater stability of microbial communities and their functionality (Thébault and Fontaine, 2010; Coyte et al., 2015). Conversely, under future climate change scenarios, S. alterniflora wetlands may become more vulnerable to perturbations. Based on robustness analysis (Figure 4H) and May’s stability theory (May, 2019; Yonatan et al., 2022), we inferred that the soil microbial communities in the P. australis wetland were more stable compared to those in the S. alterniflora wetland. Compared with the microbial network at ambient temperature, experimental warming increased (decreased) the percentage of negative links between species at CROWN S (CROWN P) (Supplementary Table S8). If these associations indicate biological interactions, the negative associations could be due to competition for limited resources (Fuhrman, 2009; Berry and Widder, 2014). This conclusion aligned precisely with the relatively nutrient-poor environment in the CROWN S soil, where experimental warming accelerated the metabolism and succession of soil microbes and thereby intensified interspecies competition for resources (Faust and Raes, 2012; Berry and Widder, 2014). It should be noted that predicting the status of key species in microbial networks is challenging due to their dynamic nature. Nonetheless, our findings indicated that the keystone nodes within these networks differed between the warming and control conditions. These results suggested that experimental warming had the potential to induce structural changes at the level of the keystone node, as documented by Yuan et al. (2021). Of the 121 keystone nodes we identified (Supplementary Table S9), 82 had a relative abundance of less than 0.1% and were classified as rare. This observation underscores the potential significance of species rarity within ecological networks, as previously highlighted (Fahey et al., 2020; Zhou S.-Y.-D. et al., 2023).
4.4 Connections between microbial communities, soil properties, and ecosystem function
The composition of the soil microbial community showed a stronger correlation with environmental factors in CROWN S compared to CROWN P (Figure 5). This difference was mainly attributable to the lower levels of soil nutrients in CROWN S. In low-nutrient environments, microorganisms need to intricately regulate their metabolism and resource acquisition to adapt to the constraints of limited nutrient availability (Dai et al., 2022). Consequently, the composition and activity of microbial communities become more vulnerable to environmental factors, including soil moisture and concentrations of nitrogen, phosphorus, and organic carbon (Philippot et al., 2023). Microorganisms in such environments depend heavily on efficient resource utilization and compete with one another for resources (Faust and Raes, 2012; Berry and Widder, 2014; Lima-Mendez et al., 2015). This scenario corresponds to the intricate microbial networks observed in CROWN S. Conversely, in high-nutrient soil environments (CROWN P), the greater ease with which microorganisms can access requisite nutrients results in a diminished impact of environmental factors on their community composition. This conclusion, however, does not imply an absence of a relationship between microorganisms and environmental factors in high-nutrient environments. Microorganisms under these conditions may display a higher degree of adaptability to resource availability that leads to a less pronounced response to environmental factors. Collectively, this analysis provided further evidence of the greater stability of the soil microorganisms in the P. australis wetland compared to the S. alterniflora wetland in this study.
The high correlation between soil microbial communities and ecosystem respiration (Reco) indicated a close association between changes in community composition and the potential for carbon degradation. As anticipated, predictive functions based on taxonomic analysis using the Faprotax and FunGuild databases revealed that the soil microorganisms in CROWN S had a significantly higher carbon decomposition potential (Supplementary Tables S11, S12) that aligned with the relatively high Reco values in CROWN S (Table 1). Similarly, previous research has indicated that the invasion of S. alterniflora enhanced microbial carbon metabolism (Zhang et al., 2023). As illustrated in Figure 5C, experimental warming had a positive effect on the carbon degradation potential at both CROWN S and CROWN P site, suggesting that climate warming may exacerbate the decomposition of organic carbon in coastal wetland soils, as observed in other ecosystems (Giardina et al., 2014; Xue et al., 2016; Wang et al., 2021b), and consequently might reduce the carbon sequestration potential of coastal wetlands. Notably, the invasive S. alterniflora wetlands were more sensitive to warming than the native P. australis wetlands, with the response of carbon degradation potential was more pronounced in CROWN S (Figure 5C). As reported in our previous study, the carbon sequestration capacity of S. alterniflora wetlands decreases and the wetland may even become a carbon source under a warming scenario (Zhou P. et al., 2023). The implication is that S. alterniflora wetlands may release more CO2 into the atmosphere as the climate warms and exacerbate rather than mitigate the negative effects of climate change. This possibility underscores the importance of wetland management and conservation and particularly emphasizes the need to avoid indiscriminate use of S. alterniflora for coastal protection.
5 Conclusion
This study revealed that climate warming has a significant impact on the turnover rates of soil prokaryotic and fungal communities. The response of soil microbial network patterns to experimental warming differed between native P. australis wetlands and invasive S. alterniflora wetlands. Specifically, the network patterns in the P. australis wetland were found to be more stable under warming, while in the S. alterniflora wetland, they were less stable. The greater increase of the relative abundance of prokaryotic OTUs involved in carbon degradation in response to warming in the invasive S. alterniflora wetlands could lead to positive feedback and a release rather than uptake of CO2. We thus suggest that in the context of climate warming, P. australis wetlands exhibit greater adaptability and stability compared to S. alterniflora wetlands. The former are best suited to maintaining carbon sequestration functions in coastal wetlands. Overall, preserving native P. australis wetlands from S. alterniflora invasion could be crucial for mitigating the detrimental effects of warming-induced changes on ecosystem functions in China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1042794 and PRJNA1042799.
Author contributions
LP: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. SiY: Conceptualization, Funding acquisition, Project administration, Resources, Writing – review & editing. LX: Investigation, Supervision, Writing – review & editing. PZ: Formal analysis, Investigation, Writing – review & editing. LH: Investigation, Writing – review & editing. ShY: Investigation, Writing – review & editing. XD: Investigation, Writing – review & editing. HY: Investigation, Writing – review & editing. TD: Software, Writing – review & editing. EL: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was jointly funded by National Forestry and Grassland Administration Emergency Leading the Charge with Open Competition Project (202302), National Natural Science Foundation of China (U22A20558), Laoshan Laboratory (LSKJ202204003), Shandong Provincial Natural Science Foundation (ZR2023MD060 and ZR2022MD024), National Key R&D Program of China (2016YFE0109600), and the China Geological Survey Program (DD20221775 and DD20189503).
Acknowledgments
We thank Yunying Duan, Xinghao Jiang, Mingwen Liao, Shuyu Yuan, Saisai Li, Zhiqiang Zhu, and other staff of our working group for the hard working in the field and in the laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1347821/full#supplementary-material
Footnotes
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273. doi: 10.1038/ismej.2013.28
An, J., Liu, C., Wang, Q., Yao, M., Rui, J., Zhang, S., et al. (2019). Soil bacterial community structure in Chinese wetlands. Geoderma 337, 290–299. doi: 10.1016/j.geoderma.2018.09.035
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Bardgett, R. D., Freeman, C., and Ostle, N. J. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814. doi: 10.1038/ismej.2008.58
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Brockett, B. F. T., Prescott, C. E., and Grayston, S. J. (2012). Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 44, 9–20. doi: 10.1016/j.soilbio.2011.09.003
Brown, J. H., Gillooly, J. F., Allen, A. P., Savage, V. M., and West, G. B. (2004). Toward a metabolic theory ecology. Ecology 85, 1771–1789. doi: 10.1890/03-9000
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Cui, Y., Fang, L., Guo, X., Han, F., Ju, W., Ye, L., et al. (2019). Natural grassland as the optimal pattern of vegetation restoration in arid and semi-arid regions: evidence from nutrient limitation of soil microorganisms. Sci. Total Environ. 648, 388–397. doi: 10.1016/j.scitotenv.2018.08.17
Custer, G. F., Bresciani, L., and Dini-Andreote, F. (2022). Ecological and evolutionary implications of microbial dispersal. Front. Microbiol. 13:855859. doi: 10.3389/fmicb.2022.855859
Dai, T., Wen, D., Bates, C. T., Wu, L., Guo, X., Liu, S., et al. (2022). Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 13:175. doi: 10.1038/s41467-021-27857-6
Dassen, S., Cortois, R., Martens, H., de Hollander, M., Kowalchuk, G. A., van der Putten, W. H., et al. (2017). Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol. Ecol. 26, 4085–4098. doi: 10.1111/mec.14175
Davidson, E. A., and Janssens, I. A. (2006). Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173. doi: 10.1038/nature04514
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. doi: 10.1038/s41467-018-05516-7
DeAngelis, K. M., Pold, G., Topçuoğlu, B. D., van Diepen, L. T. A., Varney, R. M., Blanchard, J. L., et al. (2015). Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 6:104. doi: 10.3389/fmicb.2015.00104
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Fahey, C., Koyama, A., Antunes, P. M., Dunfield, K., and Flory, S. L. (2020). Plant communities mediate the interactive effects of invasion and drought on soil microbial communities. ISME J. 14, 1396–1409. doi: 10.1038/s41396-020-0614-6
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Feng, Y., Zhang, J., Berdugo, M., Guirado, E., Guerra, C. A., Egidi, E., et al. (2022). Temperature thresholds drive the global distribution of soil fungal decomposers. Glob. Chang. Biol. 28, 2779–2789. doi: 10.1111/gcb.16096
Fu, D., Wu, X., Qiu, Q., Duan, C., and Jones, D. L. (2020). Seasonal variations in soil microbial communities under different land restoration types in a subtropical mountains region, Southwest China. Appl. Soil Eco. 153:103634. doi: 10.1016/j.apsoil.2020.103634
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199. doi: 10.1038/nature08058
Gao, Y., Li, Y., Shang, J., and Zhang, W. (2024). Temporal profiling of sediment microbial communities in the three gorges reservoir area discovered time-dissimilarity patterns and multiple stable states. Water Res. 252:121225. doi: 10.1016/j.watres.2024.121225
Gao, G. F., Li, H., Shi, Y., Yang, T., Gao, C.-H., Fan, K., et al. (2022). Continental-scale plant invasions reshuffle the soil microbiome of blue carbon ecosystems. Glob. Chang. Biol. 28, 4423–4438. doi: 10.1111/gcb.16211
Gao, G. F., Peng, D., Tripathi, B. M., Zhang, Y., and Chu, H. (2020). Distinct community assembly processes of abundant and rare soil bacteria in coastal wetlands along an inundation gradient. mSystems 5, e01150–20. doi: 10.1128/mSystems.01150-20
Giardina, C. P., Litton, C. M., Crow, S. E., and Asner, G. P. (2014). Warming-related increases in soil CO2 efflux are explained by increased below-ground carbon flux. Nat. Clim. Chang. 4, 822–827. doi: 10.1038/nclimate2322
Gibson-Reinemer, D. K., Sheldon, K. S., and Rahel, F. J. (2015). Climate change creates rapid species turnover in montane communities. Ecol. Evol. 5, 2340–2347. doi: 10.1002/ece3.1518
Guimerà, R., and Nunes Amaral, L. A. (2005). Functional cartography of complex metabolic networks. Nature 433, 895–900. doi: 10.1038/nature03288
Guo, X., Feng, J., Shi, Z., Zhou, X., Yuan, M., Tao, X., et al. (2018). Climate warming leads to divergent succession of grassland microbial communities. Nat. Clim. Chang. 8, 813–818. doi: 10.1038/s41558-018-0254-2
Guo, X., Yuan, M., Lei, J., Shi, Z., Zhou, X., Li, J., et al. (2022). Climate warming restructures seasonal dynamics of grassland soil microbial communities. mLife 1, 245–256. doi: 10.1002/mlf2.12035
Guo, X., Zhou, X., Hale, L., Yuan, M., Ning, D., Feng, J., et al. (2019). Climate warming accelerates temporal scaling of grassland soil microbial biodiversity. Nat. Ecol. Evol. 3, 612–619. doi: 10.1038/s41559-019-0848-8
Hillebrand, H., Soininen, J., and Snoeijs, P. (2010). Warming leads to higher species turnover in a coastal ecosystem. Glob. Chang. Biol. 16, 1181–1193. doi: 10.1111/j.1365-2486.2009.02045.x
Hussain, S., Chen, M., Liu, Y., Mustafa, G., Wang, X., Liu, J., et al. (2023). Composition and assembly mechanisms of prokaryotic communities in wetlands, and their relationships with different vegetation and reclamation methods. Sci. Total Environ. 897:166190. doi: 10.1016/j.scitotenv.2023
Li, C., Bo, H., Song, B., Chen, X., Cao, Q., Yang, R., et al. (2022). Reshaping of the soil microbiome by the expansion of invasive plants: shifts in structure, diversity, co-occurrence, niche breadth, and assembly processes. Plant Soil 477, 629–646. doi: 10.1007/s11104-022-05445-x
Li, P., Cadotte, M., Meiners, S., Pu, Z., Fukami, T., and Jiang, L. (2016). Convergence and divergence in a long-term old-field succession: the importance of spatial scale and species abundance. Ecol. Lett. 19, 1101–1109. doi: 10.1111/ele.12647
Li, J., Li, Z., Wang, F., Zou, B., Chen, Y., Zhao, J., et al. (2015). Effects of nitrogen and phosphorus addition on soil microbial community in a secondary tropical forest of China. Biol. Fertil. Soils 51, 207–215. doi: 10.1007/s00374-014-0964-1
Li, H., Yang, S., Semenov, M. V., Yao, F., Ye, J., Bu, R., et al. (2021). Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob Change Biol. 27, 2763–2779. doi: 10.1111/gcb.15593
Liang, X., He, C., Zhu, X., Chen, X., Lei, Y., Zhang, H., et al. (2016). Effect of exotic Spartina alterniflora on fungal symbiosis with native plants Phragmites australis and Scirpus mariqueter, and model plants Lolium perenne L. and Trifolium repens. Aquat. Bot. 130, 50–58. doi: 10.1016/j.aquabot.2015.10.003
Lima-Mendez, G., Faust, K., Henry, N., Decelle, J., Colin, S., Carcillo, F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348:1262073. doi: 10.1126/science.1262073
Lin, G., He, Y., Lu, J., Chen, H., and Feng, J. (2021). Seasonal variations in soil physicochemical properties and microbial community structure influenced by Spartina alterniflora invasion and Kandelia obovata restoration. Sci. Total Environ. 797:149213. doi: 10.1016/j.scitotenv.2021.149213
Lin, Y., Hu, H.-W., Yang, P., and Ye, G. (2022). Spartina alterniflora invasion has a greater impact than non-native species, Phragmites australis and Kandelia obovata, on the bacterial community assemblages in an estuarine wetland. Sci. Total Environ. 822:153517. doi: 10.1016/j.scitotenv.2022.153517
Luo, S.-P., He, B.-H., Zeng, Q.-P., Li, N.-J., and Yang, L. (2020). Effects of seasonal variation on soil microbial community structure and enzyme activity in a Masson pine forest in Southwest China. J. Mt. Sci-ngl. 17, 1398–1409. doi: 10.1007/s11629-019-5825-9
May, R. M. (2019). Stability and complexity in model ecosystems. New Jersey: Princeton University Press.
McLeod, E., Chmura, G. L., Bouillon, S., Salm, R., Björk, M., Duarte, C. M., et al. (2011). A blueprint for blue carbon: toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Front. Ecol. Environ. 9, 552–560. doi: 10.1890/110004
Osland, M. J., Enwright, N. M., Day, R. H., Gabler, C. A., Stagg, C. L., and Grace, J. B. (2016). Beyond just sea-level rise: considering macroclimatic drivers within coastal wetland vulnerability assessments to climate change. Glob. Chang. Biol. 22, 1–11. doi: 10.1111/gcb.13084
Philippot, L., Chenu, C., Kappler, A., Rillig, M. C., and Fierer, N. (2023). The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 22, 226–239. doi: 10.1038/s41579-023-00980-5
Prach, K., and Walker, L. R. (2011). Four opportunities for studies of ecological succession. Trends Ecol. Evol. 26, 119–123. doi: 10.1016/j.tree.2010.12.007
Pržulj, N., and Malod-Dognin, N. (2016). Network analytics in the age of big data. Science 353, 123–124. doi: 10.1126/science.aah3449
Ren, L., He, D., Chen, Z., Jeppesen, E., Lauridsen, T. L., Søndergaard, M., et al. (2017). Warming and nutrient enrichment in combination increase stochasticity and beta diversity of bacterioplankton assemblages across freshwater mesocosms. ISME J. 11, 613–625. doi: 10.1038/ismej.2016.159
Schneider, T., Keiblinger, K. M., Schmid, E., Sterflinger-Gleixner, K., Ellersdorfer, G., Roschitzki, B., et al. (2012). Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 6, 1749–1762. doi: 10.1038/ismej.2012.11
Schuerch, M., Spencer, T., Temmerman, S., Kirwan, M. L., Wolff, C., Lincke, D., et al. (2018). Future response of global coastal wetlands to sea-level rise. Nature 561, 231–234. doi: 10.1038/s41586-018-0476-5
Shi, S., Nuccio, E. E., Shi, Z. J., He, Z., Zhou, J., and Firestone, M. K. (2016). The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936. doi: 10.1111/ele.12630
Song, S., Zhang, C., Gao, Y., Zhu, X., Wang, R., Wang, M., et al. (2020). Responses of wetland soil bacterial community and edaphic factors to two-year experimental warming and Spartina alterniflora invasion in Chongming Island. J. Clean. Prod. 250:119502. doi: 10.1016/j.jclepro.2019.119502
Sprockett, D. D., Martin, M., Costello, E. K., Burns, A. R., Holmes, S. P., Gurven, M. D., et al. (2020). Microbiota assembly, structure, and dynamics among Tsimane horticulturalists of the Bolivian Amazon. Nat. Commun. 11:3772. doi: 10.1038/s41467-020-17541-6
Stotzky, G., and Norman, A. G. (1961). Factors limiting microbial activities in soil. Archiv. Microbial. 40, 341–369. doi: 10.1007/BF00422050
Thébault, E., and Fontaine, C. (2010). Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856. doi: 10.1126/science.1188321
Thoms, C., and Gleixner, G. (2013). Seasonal differences in tree species' influence on soil microbial communities. Soil Biol. Biochem. 66, 239–248. doi: 10.1016/j.soilbio.2013.05.018
Tian, B., Zhu, M., Pei, Y., Ran, G., Shi, Y., and Ding, J. (2022). Climate warming alters the soil microbial association network and role of keystone taxa in determining wheat quality in the field. Agric. Ecosyst. Environ. 326:107817. doi: 10.1016/j.agee.2021.107817
Waldrop, M. P., and Firestone, M. K. (2006). Seasonal dynamics of microbial community composition and function in oak canopy and open grassland soils. Microb. Ecol. 52, 470–479. doi: 10.1007/s00248-006-9100-6
Walters, W., Hyde Embriette, R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2015). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 1, e00009-15. doi: 10.1128/msystems.00009-15
Wang, Q., An, S., Ma, Z., Zhao, B., Chen, J., and Li, B. (2006). Invasive Spartina alterniflora: biology, ecology and management. Acta Phytotaxonomica Sinica. 44, 559–588. doi: 10.1360/aps06044
Wang, C., Morrissey, E. M., Mau, R. L., Hayer, M., Piñeiro, J., Mack, M. C., et al. (2021a). The temperature sensitivity of soil: microbial biodiversity, growth, and carbon mineralization. ISME J. 15, 2738–2747. doi: 10.1038/s41396-021-00959-1
Wang, C., Wang, S., Wu, B., Wei, M., Rong, X., Li, Y., et al. (2021b). Ecological restoration treatments enhanced plant and soil microbial diversity in the degraded alpine steppe in northern Tibet. Land Degrad. Dev. 32, 723–737. doi: 10.1002/ldr.3754
Wu, L., Zhang, Y., Guo, X., Ning, D., Zhou, X., Feng, J., et al. (2022). Reduction of microbial diversity in grassland soil is driven by long-term climate warming. Nat. Microbiol. 7, 1054–1062. doi: 10.1038/s41564-022-01147-3
Xue, K., Yuan, M. M., Shi, Z. J., Qin, Y., Deng, Y., Cheng, L., et al. (2016). Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat. Clim. Chang. 6, 595–600. doi: 10.1038/nclimate2940
Yamada, T., and Sekiguchi, Y. (2009). Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi ‘subphylum I’ with natural and biotechnological relevance. Microbes Environ. 24, 205–216. doi: 10.1264/jsme2.ME09151S
Yang, Y., Dou, Y., Wang, B., Xue, X., Wang, Y., An, S., et al. (2023). Deciphering factors driving soil microbial life-history strategies in restored grasslands. iMeta 2:e66. doi: 10.1002/imt2.66
Yang, H., Hu, J., Long, X., Liu, Z., and Rengel, Z. (2016). Salinity altered root distribution and increased diversity of bacterial communities in the rhizosphere soil of Jerusalem artichoke. Sci. Rep-uk. 6:20687. doi: 10.1038/srep20687
Yang, W., Jeelani, N., Zhu, Z., Luo, Y., Cheng, X., and An, S. (2019a). Alterations in soil bacterial community in relation to Spartina alterniflora Loisel. Invasion chronosequence in the eastern Chinese coastal wetlands. Appl. Soil Eco. 135, 38–43. doi: 10.1016/j.apsoil.2018.11.009
Yang, S., Li, J., Zheng, Z., and Meng, Z. (2009). Characterization of Spartina alterniflora as feedstock for anaerobic digestion. Biomass Bioenergy 33, 597–602. doi: 10.1016/j.biombioe.2008.09.007
Yang, W., Zhang, D., Cai, X., Xia, L., Luo, Y., Cheng, X., et al. (2019b). Significant alterations in soil fungal communities along a chronosequence of Spartina alterniflora invasion in a Chinese Yellow Sea coastal wetland. Sci. Total Environ. 693:133548. doi: 10.1016/j.scitotenv.2019.07.354
Yonatan, Y., Amit, G., Friedman, J., and Bashan, A. (2022). Complexity–stability trade-off in empirical microbial ecosystems. Nat. Ecol. Evol. 6, 693–700. doi: 10.1038/s41559-022-01745-8
Yu, X., Ye, S., Pei, L., Xie, L., Krauss, K., Chapman, S., et al. (2023). Biophysical warming patterns of an open-top chamber and its short-term influence on a Phragmites wetland ecosystem in China. China Geol. 610. doi: 10.31035/cg2022064
Yuan, J., Ding, W., Liu, D., Kang, H., Xiang, J., and Lin, Y. (2016). Shifts in methanogen community structure and function across a coastal marsh transect: effects of exotic Spartina alterniflora invasion. Sci. Rep. 6:18777. doi: 10.1038/srep18777
Yuan, M. M., Guo, X., Wu, L., Zhang, Y., Xiao, N., Ning, D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 11, 343–348. doi: 10.1038/s41558-021-00989-9
Zeleke, J., Sheng, Q., Wang, J.-G., Huang, M.-Y., Xia, F., Wu, J.-H., et al. (2013). Effects of Spartina alterniflora invasion on the communities of methanogens and sulfate-reducing bacteria in estuarine marsh sediments. Front. Microbiol. 4:243. doi: 10.3389/fmicb.2013.00243
Zhang, G., Bai, J., Zhao, Q., Jia, J., Wang, W., and Wang, X. (2020). Bacterial succession in salt marsh soils along a short-term invasion chronosequence of Spartina alterniflora in the Yellow River estuary, China. Microb. Ecol. 79, 644–661. doi: 10.1007/s00248-019-01430-7
Zhang, G., Jia, J., Zhao, Q., Wang, W., Wang, D., and Bai, J. (2023). Seasonality and assembly of soil microbial communities in coastal salt marshes invaded by a perennial grass. J. Environ. Manag. 331:117247. doi: 10.1016/j.jenvman.2023.117247
Zhang, W., Liu, W., Xu, M., Deng, J., Han, X., Yang, G., et al. (2019). Response of forest growth to C:N:P stoichiometry in plants and soils during Robinia pseudoacacia afforestation on the Loess Plateau, China. Geoderma 337, 280–289. doi: 10.1016/j.geoderma.2018.09.042
Zhou, J., Deng, Y., Luo, F., He, Z., Tu, Q., and Zhi, X. (2010). Functional molecular ecological networks. mBio 1, e00169–e00110. doi: 10.1128/mbio.00169-10
Zhou, J., Deng, Y., Luo, F., He, Z., and Yang, Y. (2011). Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio 2, e00122–e00111. doi: 10.1128/mbio.00122-11
Zhou, J., Deng, Y., Zhang, P., Xue, K., Liang, Y., Nostrand, J., et al. (2014). Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. USA 111, E836–E845. doi: 10.1073/pnas.1324044111
Zhou, S.-Y.-D., Lie, Z., Liu, X., Zhu, Y.-G., Peñuelas, J., Neilson, R., et al. (2023). Distinct patterns of soil bacterial and fungal community assemblages in subtropical forest ecosystems under warming. Glob. Chang. Biol. 29, 1501–1513. doi: 10.1111/gcb.16541
Zhou, Y., Sun, B., Xie, B., Feng, K., Zhang, Z., Zhang, Z., et al. (2021). Warming reshaped the microbial hierarchical interactions. Glob. Chang. Biol. 27, 6331–6347. doi: 10.1111/gcb.15891
Zhou, J., Xue, K., Xie, J., Deng, Y., Wu, L., Cheng, X., et al. (2012). Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Chang. 2, 106–110. doi: 10.1038/nclimate1331
Keywords: coastal wetland, climate change, microbial network, microorganism, carbon sequestration
Citation: Pei L, Ye S, Xie L, Zhou P, He L, Yang S, Ding X, Yuan H, Dai T and Laws EA (2024) Differential effects of warming on the complexity and stability of the microbial network in Phragmites australis and Spartina alterniflora wetlands in Yancheng, Jiangsu Province, China. Front. Microbiol. 15:1347821. doi: 10.3389/fmicb.2024.1347821
Edited by:
Zhanfei He, Zhejiang University of Technology, ChinaCopyright © 2024 Pei, Ye, Xie, Zhou, He, Yang, Ding, Yuan, Dai and Laws. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siyuan Ye, siyuanye@hotmail.com
 Lixin Pei
Lixin Pei Siyuan Ye
Siyuan Ye Liujuan Xie1,2
Liujuan Xie1,2 Shixiong Yang
Shixiong Yang Tianjiao Dai
Tianjiao Dai Edward A. Laws
Edward A. Laws