- 1Department of Critical Care Medicine, Jiangnan University Medical Center, Wuxi, China
- 2Department of Critical Care Medicine, Aheqi County People's Hospital, Xinjiang, China
- 3Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 4Institute of Clinical Medicine Research, The Affiliated Suzhou Science and Technology Town Hospital of Nanjing Medical University, Suzhou, China
Background: Acute respiratory distress syndrome (ARDS) is a serious lung ailment marked by significant inflammation and damage in the alveoli and capillaries of the lungs. Recent research suggests a strong correlation between the onset and advancement of ARDS and an imbalance in the gut microbiota (GM).
Methods: In this investigation, Mendelian randomization (MR) analysis was utilized, drawing on data from publicly accessible genome-wide association studies. The primary focus was on examining the interplay between GM, inflammatory factors (IFs) and ARDS. Instrumental variables were established through genetic modifications of GM and IFs. Various statistical analysis methods including the inverse-variance weighted model, MR-Egger method and Wald ratio test were applied for comprehensive data analysis.
Results: Eight bacterial taxa within the GM demonstrated a potential causal link with development of ARDS. Notably, the phylum Actinobacteria and the genus Intestinibacter exhibited a negative association with the risk of ARDS. However, Erysipelotrichales (id. 2,148), Victivallis (id. 2,256), Ruminococcaceae UCG014 (id. 11,371), Eubacterium ruminantium group (id. 11,340), Erysipelotrichaceae (id. 2,149) and Erysipelotrichia (id. 2,147) demonstrated a positive association with ARDS risk. Additionally, the study identified a potential causal relationship between the inflammatory factors interleukin-16 and C-C motif chemokine 3 with the occurrence of ARDS.
Conclusion: This study strongly suggests that the interaction between gut microbiota (GM) and inflammatory factors (IFs) significantly contributes to the pathogenesis of acute respiratory distress syndrome (ARDS). This underscores their crucial involvement in both the initiation and advancement of this severe lung disorder.
1 Introduction
Acute respiratory distress syndrome (ARDS) is a severe pulmonary condition characterized by pronounced lung inflammation and increased permeability of pulmonary blood vessels resulting in respiratory failure, diminished oxygen levels and observable opacities in lung radiographs (Yıldırım et al., 2021; Fujishima, 2023). The etiology of ARDS encompasses various medical conditions such as infections, physical trauma, pancreatitis, pneumonia, aspiration incidents, and extensive blood transfusions, with infections being the predominant cause that is responsible for nearly 40% of all ARDS cases. Globally, ARDS occurs at rates ranging from 7.2 to 34 cases per 100,000 person-years, with a mortality rate between 26 and 35% (Huppert et al., 2019; Yıldırım et al., 2021). In United States ARDS leads to approximately 75,000 deaths annually, affecting around 3 million individuals worldwide. It constitutes 10% of all intensive care unit (ICU) admissions and impacts 23% of ICU patients requiring mechanical ventilation (Huppert et al., 2019).
The pathogenesis of ARDS primarily stems from alveolar injury and increased capillary permeability affecting both the pulmonary endothelium and alveolar epithelial surface. This disruption results in the formation of hyaline membranes and the accumulation of protein-rich fluid within the pulmonary interstitium potentially compromising surfactant molecules, alveolar stability, and gas exchange (Hu et al., 2022). The activation of cytokines and the release of pro-inflammatory mediators exacerbate lung injury and sustain inflammation (Hu et al., 2022). The early phase of ARDS clinically manifests as significant hypoxemia and reduced lung compliance.
A potential association exists between gut microbiota (GM) dysbiosis (GMD) and the initiation and progression of ARDS (Dickson, 2015). The GMD may compromise intestinal barrier function allowing bacteria and toxins to enter the circulatory and lymphatic systems infecting lungs and triggering ARDS. The GM produced metabolites can directly or indirectly induce lung inflammation and GM’s interaction with the immune system influences immune cell activity and inflammatory responses (Sultan et al., 2021). Certain GM bacterial species produce inflammatory factors (IFs) which can influence intestinal epithelial and immune cells (Malesza et al., 2021). The GMD can lead to detrimental changes in intestinal mucosa increasing its permeability and allowing IFs easier access to the bloodstream provoking systemic inflammation (Tilg et al., 2020). Moreover, IFs can impact the GM’s structure and function fostering a self-perpetuating inflammatory cycle (Strati et al., 2022). The GMD negatively affects the immune system heightening inflammatory responses and contributing to ARDS development (Li et al., 2021). The GMD may also influence the lung microbiome contributing to lung inflammation and ARDS. However, the intricate relationship between GM and ARDS remains not entirely understood. Mendelian randomization (MR) employs single nucleotide polymorphisms (SNPs) linked to a specific risk factor as instrumental variables (IVs) to investigate causal relationships between the risk factor and a particular disease (Bowden and Holmes, 2019). Genetic variation occurring from zygote formation engenders considerable variability before disease onset, persisting throughout life and potentially allowing MR studies to bypass biases or confounding factors.
This study aimed to explore the relationship between GM, IFs and ARDS by conducting MR analysis using publicly available summary-level data from genome-wide association studies (GWAS).
2 Materials and methods
2.1 GWAS data
The GM GWAS data was obtained from MiBioGen study1 represent the most extensive multi-ethnic analysis of GM to date. This study, encompassing 16S fecal microbiota data (n = 340) and genotype data (n = 13,263) from 16 cohorts (n = 24,000) aimed to explore the correlation between GM and human health. The findings revealed significant variability in human GM across different regions, ethnicities and age groups. Moreover, GWAS data on 41 IFs were acquired from a cytokine-related GWAS meta-analysis involving three independent cohorts, including the Young Finns Cardiovascular Risk Study (Suhre et al., 2017; Folkersen et al., 2020). To investigate the potential causal relationship between IFs and ARDS, GWAS data for IFs were extracted from meta-analysis datasets from two cytokine-centric GWAS investigations. Initially, 41 cytokine-related factors compiled by Suhre K were gathered, supplemented by additional cytokine factors collated by Folkersen L. Furthermore, the present study utilized ARDS GWAS data from the Finngen database—a Finnish genetic resource integrating genotypic, phenotypic, diagnostic, and prescription information. This platform primarily focuses on identifying gene-disease associations to drive advancements in disease management. It also provides data analysis tools facilitating global collaboration and knowledge sharing. This study analyzed 216,363 samples (inclusive of 16,380,461 SNPs) from Finngen to investigate ARDS. All exposure and outcome data can be obtained online at IEU.2
2.2 Selection of IVs
Bacterial classification and analyses were systematically conducted across five taxonomic levels. The inclusion criteria for IVs in this study were meticulously restricted to ensure the precision and validity of causal relationships between GM and the risk of ARDS development. This involved selecting SNPs with a p-value <1e-05 as IVs for both exposure and outcome in MR studies. Furthermore, the Two Sample MR R package was utilized with parameters r2 = 0.001 and kb = 10,000 to establish IV independence and minimize the potential impact of linkage disequilibrium that could disrupt the random allele assignment process. The SNPs with a minor allele frequency < 0.01 and those exerting a greater influence on outcomes than exposure was subsequently excluded. Furthermore, PhenoScanner V2 was applied to verify that the phenotypes of all SNPs used as IVs were unrelated to the outcomes ensuring adherence to the no-horizontal-pleiotropy assumption in MR analysis (Kamat et al., 2019).
2.3 Statistical analysis
Multiple causality analysis models were applied in this study. The inverse-variance weighted (IVW) model and the MR-Egger method (Birney, 2022) were used for analyzing samples with multiple SNPs whereas, Wald ratio test was applied for samples with only one SNP (Ference et al., 2021).
For sensitivity analyses, heterogeneity was assessed using the Cochran Q method. In case of evident heterogeneity (p < 0.05) MR-Egger regression analysis was applied to evaluate potential pleiotropic effects of the SNPs used as IVs. In MR-Egger regression the intercept term indicated directed horizontal pleiotropy at p < 0.05. The MR-PRESSO methodology was employed to validate MR results. Its use extended to scrutinizing whether the IVs were associated with outliers and assessing the prevalence of horizontal pleiotropy within the global test scope. When pleiotropy was identified, the suggested approach involved excluding outliers, followed by a reiteration of MR and MR-PRESSO analyses. The main objective was to ensure that the p-values obtained with both Egger’s intercept and the global test ascertained through MR-PRESSO exceeded the threshold of 0.05 (Verbanck et al., 2018). Additionally, a leave-one-out analysis was utilized to gauge any potential influence of individual SNPs on the outcomes of each MR study. To visualize MR methodologies’ results clearly and concisely, the TwoSampleMR package was used. This facilitated the generation of scatter plots illustrating the overall correlation between exposure and the resulting outcomes. It also produced funnel plots reflecting the symmetric distribution of IVs, in this case, SNPs. Furthermore, forest plots were generated to depict both individual effects and collective exposure-related outcomes’ benefits. The leave-one-out plots demonstrated the stability of outcomes following the removal of specific SNPs. All statistical analyses in this study were conducted using the R package in the R language application (v4.2.1).
3 Results
3.1 MR analysis of GM and ARDS
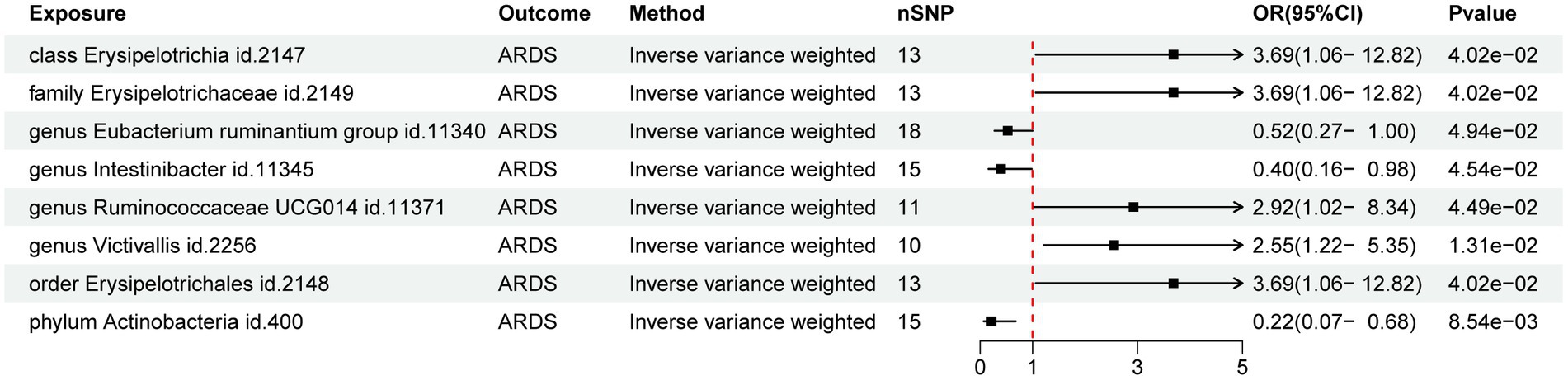
The preliminary findings suggested that 8 of the 211 gut bacterial taxa had a causal association with ARDS (Figure 1). The results of IVW analysis of these 8 bacterial taxa were as follows: phylum Actinobacteria (id.400; p = 8.54E-03; odds ratio [OR] 95% confidence interval [CI] = 0.22 [0.07, 0.68]), order Erysipelotrichales (id. 2,148; p = 4.02E-02; OR [95% CI] = 3.69 [1.06, 12.82]), genus Victivallis (id.2256; p = 1.31E-02; OR [95% CI] = 2.55 [1.22, 5.35]), genus Ruminococcaceae UCG014 (id.11371; p = 4.49E-02; OR [95% CI] = 2.92 [1.02, 8.34]), genus Intestinibacter (id.11345; p = 4.54E-02; OR [95% CI] = 0.40 [0.16, 0.98]), genus Eubacterium ruminantium group (id.11340; p = 4.94E-02; OR [95% CI] = 0.52 [0.27, 0.99]), family Erysipelotrichaceae (id.2149; p = 4.02E-02; OR [95% CI] = 3.69 [1.06, 12.82]), and class Erysipelotrichia (id.2147; p = 4.02E-02; OR [95% CI] = 3.69 [1.06, 12.82]). Among them phylum Actinobacteria (id. 400) and genus Intestinibacter (id. 11,345) were negatively associated with the risk of ARDS development whereas, the other taxa were positively associated with the risk of ARDS development suggesting that their abundance in GM may lead to ARDS development. Detailed information on MR analysis of GM and ARDS is provided in the Supplementary material.
3.2 MR analysis of IFs and ARDS
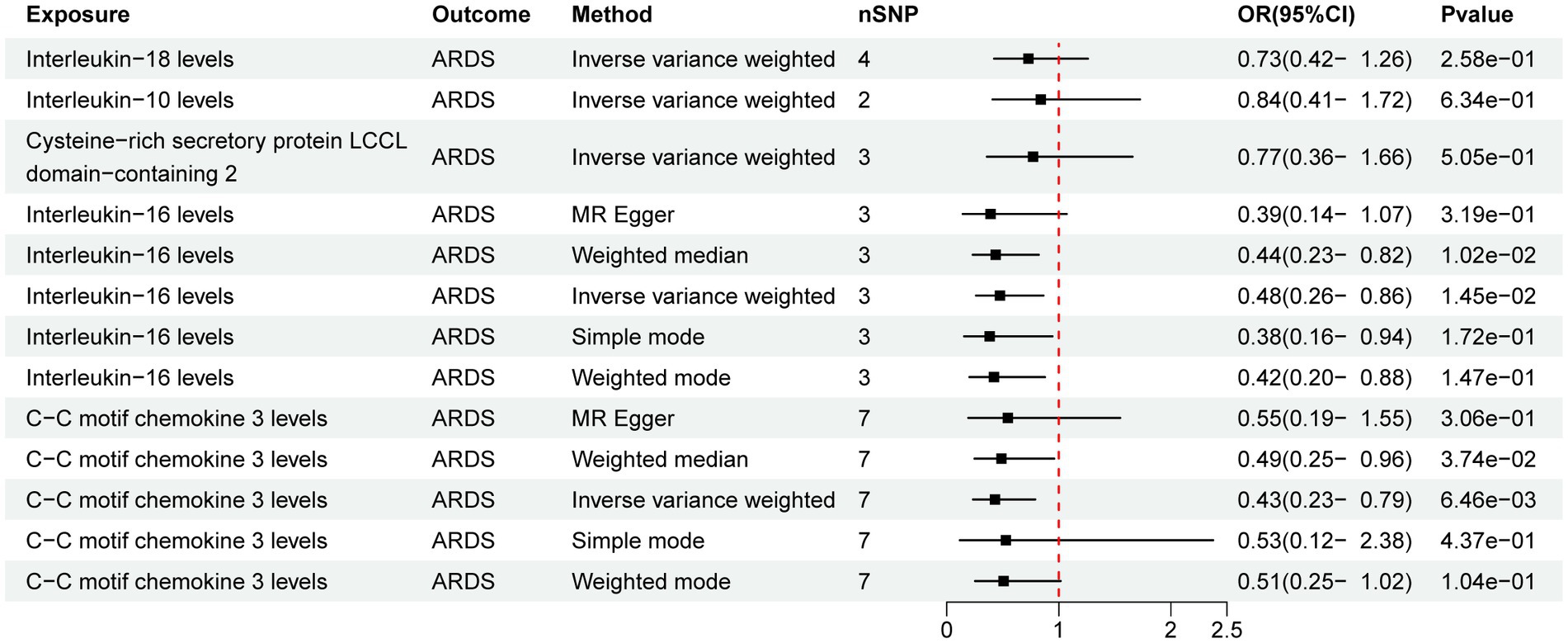
Out of the 41 IFs studied only 2 exhibited a potential causal relationship with ARDS (Figure 2). The results of IVW analysis of the 2 IFs indicated significant associations for IL-16 levels (p = 1.45E-02; OR [95% CI] = 0.48 [0.26, 0.86]) and C-C motif chemokine 3 levels (CCL3, p = 6.46E-03; OR [95% CI] = 0.43 [0.23, 0.79]).
3.3 MR analysis of GM and IFs
The MR analysis of GM and IFs aimed to unveil the role of IFs in the relationship between GM and ARDS. The results of IVW analysis revealed a potential positive correlation between genus Victivallis (id. 2,256) and IL-16 (p = 4.00E-02; OR [95% CI] = 1.16 [1.01, 1.34]) whereas, the other taxa did not exhibit potential relationships with the IFs (Figure 3).

Figure 3. Forest map of the Mendelian randomization analysis of gut microbiota and inflammatory factors.
A sensitivity analysis was conducted to validate our results. Both heterogeneity and multiplicity tests showed a value of p of <0.05, indicating the absence of abnormalities. Furthermore, the findings of the present study remained robust and consistent in subsequent leave-one-out analyses (Figure 4). Detailed results of the sensitivity analysis are presented in the Supplementary material.
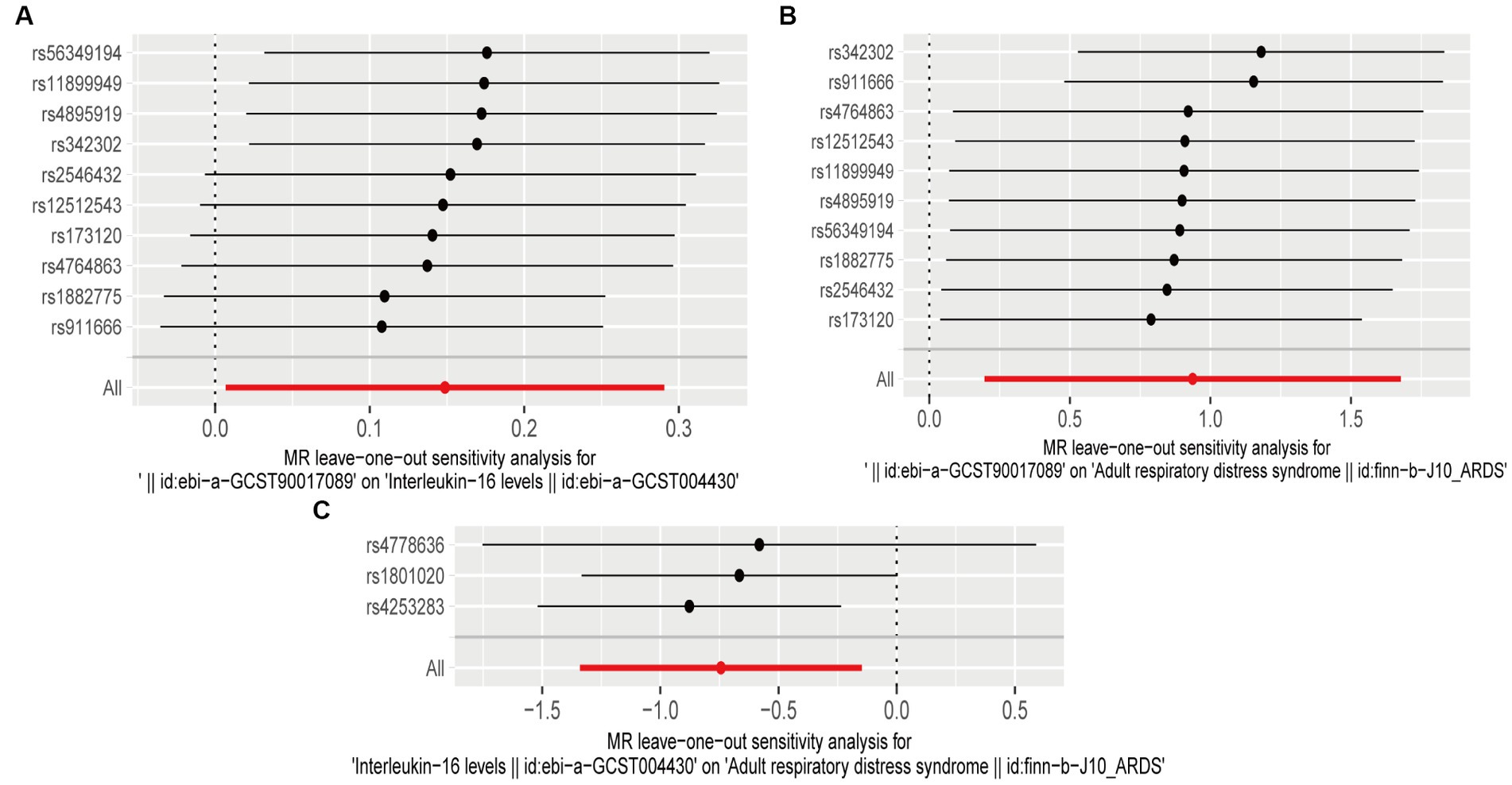
Figure 4. Leave-one-out plots (A) leave-one-out plot of genus Victivallis id.2256 and interleukin (IL)-16; (B) leave-one-out plot of genus Victivallis id.2256 and acute respiratory distress syndrome (ARDS); and (C) leave-one-out plot of IL-16 and ARDS.
4 Discussion
ARDS is a severe lung disease characterized by inflammation and injury to the alveoli and alveolar capillaries (Yıldırım et al., 2021). Infection and inflammation are the major factors for ARDS (Odeyemi et al., 2019; Meyer et al., 2021). Recent studies have unveiled a strong correlation between GMD and the onset of ARDS. The GM is the largest microbial community in body and plays a vital role in maintaining gut health and immune function (Dickson et al., 2016). However, critically ill patients may experience significant alterations in both gut and lung microbiota. The lung microbiota of patients with infection and patients with ARDS has an abundance of gut-related microorganisms which may enter the lungs through bacterial translocation from the gut which is facilitated by increased gut and alveolar-capillary permeability (Dickson et al., 2020). Moreover, the lung microbiota of patients on ventilators also exhibits an enrichment of gut-related microorganisms which are associated with ARDS occurrence. These findings suggest that GMD is a crucial factor in the development and progression of ARDS (Mustansir Dawoodbhoy et al., 2021). The GMD potentially contributes to ARDS development and progression through various mechanisms. First, GMD may lead to the disruption of intestinal barrier function, permitting bacteria and their metabolites to enter the circulatory system, thus affecting immune and inflammatory responses in the lungs (Hu et al., 2023). Second, GMD might prompt the release of intestinal toxins, further activating the inflammatory response and causing inflammation and injury in the lungs. In this study, eight bacterial taxa were identified to have a potential association with ARDS, including Actinobacteria (id. 400), Erysipelotrichales (id. 2,148), Victivallis (id. 2,256), Ruminococcaceae UCG014 (id. 11,371), Intestinibacter (id. 11,345), Eubacterium ruminantium group (id. 11,340), Erysipelotrichaceae (id. 2,149) and Erysipelotrichia (id. 2,147).
Inflammation plays a pivotal role in driving the occurrence and progression of ARDS. The initiation of ARDS is typically linked to pulmonary infections, trauma, and pancreatitis all causing significant damage to the lung tissues and triggering an inflammatory response. This response leads to the accumulation and activation of immune cells such as neutrophils and monocytes which are responsible for releasing inflammatory mediators such as cytokines and chemokines which further exacerbate the inflammatory response (Alghetaa et al., 2021). In the early stages of ARDS, this response disrupts the alveolar and vascular endothelial barriers leading to lung tissue fluid accumulation and pulmonary edema formation (Yin et al., 2017). Furthermore, inflammation contributes to fibrosis within lung tissue compromising pulmonary function. Moreover, it may lead to a cytokine storm which is characterized by extensive activation immune cells as well as excessive release of cytokines. This pathological event frequently causes immune dysregulation, inflammation and pulmonary fibrosis which leads to escalating the severity of inflammation and the resultant lung tissue damage (Martin-Loeches et al., 2020; Siwicka-Gieroba and Czarko-Wicha, 2020). Furthermore, in patients with ARDS peripheral blood mononuclear cells and natural killer cells highly express genes closely associated with the inflammatory response (Zheng et al., 2023; Zou et al., 2023) including genes related to pro-inflammatory cytokines and inflammation-related signaling pathways.
This study suggests that IL-16 and CCL3 play significant roles related to the pathogenesis of ARDS. The IL-16 (a cytokine) and CCL3 (a chemokine, also known as macrophage inflammatory protein-1α) have consistently showed significantly upregulated expression in patients with ARDS (Cruikshank et al., 2000; Houshmandfar et al., 2021; Kono et al., 2023). Increased levels of IL-16 and CCL3 lead to inflammatory cell aggregation and intensifying lung inflammation. Moreover, they can prompt the release of cytokines such as IL-1β, IL-6 and TNF-α resultantly exacerbating lung inflammation (Kono et al., 2023). High IL-16 expression has been linked to increased severity and poor prognosis in ARDS cases (Tsagkaris et al., 2022; Kono et al., 2023). The CCL3 activates inflammatory cells, such as monocytes and lymphocytes and facilitates their entry into the lungs contributing to inflammatory response (Houshmandfar et al., 2021). The CCL3 is also reported to promotes the adhesion and migration of inflammatory cells worsening lung injury (Baier et al., 2004).
This study not only uncovers critical connections between IFs such as IL-16, CCL3, and ARDS but also establishes a link between these factors and the role of GM in ARDS progression. The findings offer a comprehensive understanding related to the pathogenesis of disease. The MR analysis was applied to the publicly available GWAS data for exploring the intricate relationship among GM, IFs, and ARDS. Our findings show that the phylum Actinobacteria (id. 400) and the genus Intestinibacter (id. 11,345) are negatively correlated with the risk of ARDS development whereas, other bacterial groups exhibit positive correlations with the risk of ARDS development. Furthermore, our results indicate a potential causal relationship between IL-16, CCL3 and ARDS. Further analysis between GM and IFs suggests a possible positive correlation between the genus Victivallis and IL-16.
Nevertheless, this initial investigation comes with certain constraints as our analysis is dependent on publicly accessible GWAS data which carries certain limitations and biases. Moreover, the MR technique aids in evaluating causal associations and it may not completely eradicate the influence of confounding variables. Therefore, further research is imperative to corroborate and enhance our comprehension of the association between GM and ARDS. This extended exploration will not only fortify the evidence but also contribute to a more thorough understanding of the mechanisms that interconnect these factors.
5 Conclusion
Mendelian randomization (MR) analysis unveiled the potential significant contributions of gut microbiota (GM) and inflammatory factors (IFs) to both the initiation and advancement of acute respiratory distress syndrome (ARDS). The analysis identified 8 out of 211 gut bacterial taxa demonstrating potential causal associations with ARDS. Actinobacteria (id. 400) and Intestinibacter (id. 11,345) exhibited a negative association with the risk of ARDS development whereas, Erysipelotrichales (id. 2,148), Victivallis (id. 2,256), Ruminococcaceae UCG014 (id. 11,371), Eubacterium ruminantium group (id. 11,340), Erysipelotrichaceae (id. 2,149) and Erysipelotrichia (id. 2,147) showed a positive association with the risk of ARDS development. Furthermore, MR analysis examining IFs and ARDS indicated potential causal links with interleukin-16 (IL-16) and C-C motif chemokine 3 (CCL3). Further exploration uncovered a potential positive correlation between the genus Victivallis (id. 2,256) and IL-16. These findings suggest that GM and IFs may influence immune and inflammatory responses, thereby impacting the development and progression of ARDS.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JM: Writing – original draft. ZZ: Conceptualization, Writing – Original Draft. YY: Formal analysis, Investigation, Writing – original draft. KA: Writing – Review & Editing. LH: Supervision, Writing – review & editing. LL: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received funding and support from the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2023D01F26), Wuxi Medical Innovation Teams (CXTD2021018), Wuxi Key Medical Talents Project (ZDRC007), and scientific research project of Wuxi Municipal Health Commission (Q202221).
Acknowledgments
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript. The authors extend their appreciation to the Researchers supporting project number (RSP2023R185), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1294692/full#supplementary-material
Footnotes
References
Alghetaa, H., Mohammed, A., Zhou, J., Singh, N., Nagarkatti, M., and Nagarkatti, P. (2021). Resveratrol-mediated attenuation of superantigen-driven acute respiratory distress syndrome is mediated by microbiota in the lungs and gut. Pharmacol. Res. 167:105548. doi: 10.1016/j.phrs.2021.105548
Baier, R. J., Majid, A., Parupia, H., Loggins, J., and Kruger, T. E. (2004). CC chemokine concentrations increase in respiratory distress syndrome and correlate with development of bronchopulmonary dysplasia. Pediatr. Pulmonol. 37, 137–148. doi: 10.1002/ppul.10417
Birney, E. (2022). Mendelian randomization. Cold Spring Harb. Perspect. Med. 12:a041302. doi: 10.1101/cshperspect.a041302
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Cruikshank, W. W., Kornfeld, H., and Center, D. M. (2000). Interleukin-16. J. Leukoc. Biol. 67, 757–766. doi: 10.1002/jlb.67.6.757
Dickson, R. P. (2015). The microbiome and critical illness. Lancet Respir. Med. 4, 59–72. doi: 10.1016/S2213-2600(15)00427-0
Dickson, R. P., Schultz, M. J., van der Poll, T., Schouten, L. R., Falkowski, N. R., Luth, J. E., et al. (2020). Biomarker analysis in septic ICU patients (BASIC) consortium. Lung microbiota predict clinical outcomes in critically ill patients. Am. J. Respir. Crit. Care Med. 201, 555–563. doi: 10.1164/rccm.201907-1487OC
Dickson, R. P., Singer, B. H., Newstead, M. W., Falkowski, N. R., Erb-Downward, J. R., Standiford, T. J., et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1:16113. doi: 10.1038/nmicrobiol.2016.113
Ference, B. A., Holmes, M. V., and Smith, G. D. (2021). Using Mendelian randomization to improve the Design of Randomized Trials. Cold Spring Harb. Perspect. Med. 11:a040980. doi: 10.1101/cshperspect.a040980
Folkersen, L., Gustafsson, S., Wang, Q., Hansen, D. H., Hedman, Å. K., Schork, A., et al. (2020). Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat. Metab. 2, 1135–1148. doi: 10.1038/s42255-020-00287-2
Fujishima, S. (2023). Guideline-based management of acute respiratory failure and acute respiratory distress syndrome. J. Intensive Care 11:10. doi: 10.1186/s40560-023-00658-3
Houshmandfar, S., Saeedi-Boroujeni, A., Rashno, M., Khodadadi, A., and Mahmoudian-Sani, M. R. (2021). miRNA-223 as a regulator of inflammation and NLRP3 inflammasome, the main fragments in the puzzle of immunopathogenesis of different inflammatory diseases and COVID-19. Naunyn Schmiedeberg's Arch. Pharmacol. 394, 2187–2195. doi: 10.1007/s00210-021-02163-6
Hu, X., Han, Z., Zhou, R., Su, W., Gong, L., Yang, Z., et al. (2023). Altered gut microbiota in the early stage of acute pancreatitis were related to the occurrence of acute respiratory distress syndrome. Front. Cell. Infect. Microbiol. 13:1127369. doi: 10.3389/fcimb.2023.1127369
Hu, Q., Zhang, S., Yang, Y., Yao, J. Q., Tang, W. F., Lyon, C. J., et al. (2022). Extracellular vesicles in the pathogenesis and treatment of acute lung injury. Mil. Med. Res. 9:61. doi: 10.1186/s40779-022-00417-9
Huppert, L. A., Matthay, M. A., and Ware, L. B. (2019). Pathogenesis of acute respiratory distress syndrome. Semin. Respir. Crit. Care Med. 40, 031–039. doi: 10.1055/s-0039-1683996
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853. doi: 10.1093/bioinformatics/btz469
Kono, H., Asakawa, M., Fujii, H., Maki, A., Amemiya, H., Yamamoto, M., et al. (2023). Edaravone, a novel free radical scavenger, prevents liver injury and mortality in rats administered endotoxin. J. Pharmacol. Exp. Ther. 307, 74–82. doi: 10.1124/jpet.103.053595
Li, S., Yang, S., Zhou, Y., Disoma, C., Dong, Z., Du, A., et al. (2021). Microbiome profiling using shotgun metagenomic sequencing identified unique microorganisms in COVID-19 patients with altered gut microbiota. Front. Microbiol. 12:712081. doi: 10.3389/fmicb.2021.712081
Malesza, I. J., Malesza, M., Walkowiak, J., Mussin, N., Walkowiak, D., Aringazina, R., et al. (2021). High-fat, Western-style diet, systemic inflammation, and gut microbiota: a narrative review. Cells 10:3164. doi: 10.3390/cells10113164
Martin-Loeches, I., Dickson, R., Torres, A., Hanberger, H., Lipman, J., Antonelli, M., et al. (2020). The importance of airway and lung microbiome in the critically ill. Crit. Care 24:537. doi: 10.1186/s13054-020-03219-4
Meyer, N. J., Gattinoni, L., and Calfee, C. S. (2021). Acute respiratory distress syndrome. Lancet 398, 622–637. doi: 10.1016/S0140-6736(21)00439-6
Mustansir Dawoodbhoy, F., Patel, B. K., Patel, K., Bhatia, M., Lee, C. N., and Moochhala, S. M. (2021). Gut microbiota Dysbiosis as a target for improved post-surgical outcomes and improved patient care: a review of current literature. Shock 55, 441–454. doi: 10.1097/SHK.0000000000001654
Odeyemi, Y. E., Herasevich, S., Gong, M. N., and Gajic, O. O. (2019). Clinical strategies to prevent acute respiratory distress syndrome. Semin. Respir. Crit. Care Med. 40, 129–136. doi: 10.1055/s-0039-1683997
Siwicka-Gieroba, D., and Czarko-Wicha, K. (2020). Lung microbiome - a modern knowledge. Cent Eur J Immunol. 45, 342–345. doi: 10.5114/ceji.2020.101266
Strati, F., Lattanzi, G., Amoroso, C., and Facciotti, F. (2022). Microbiota-targeted therapies in inflammation resolution. Semin. Immunol. 59:101599. doi: 10.1016/j.smim.2022.101599
Suhre, K., Arnold, M., Bhagwat, A. M., Cotton, R. J., Engelke, R., Raffler, J., et al. (2017). Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8:14357. doi: 10.1038/ncomms14357
Sultan, M., Wilson, K., Abdulla, O. A., Busbee, P. B., Hall, A., Carter, T., et al. (2021). Endocannabinoid anandamide attenuates acute respiratory distress syndrome through modulation of microbiome in the gut-lung Axis. Cells 10:3305. doi: 10.3390/cells10123305
Tilg, H., Zmora, N., Adolph, T. E., and Elinav, E. (2020). The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 20, 40–54. doi: 10.1038/s41577-019-0198-4
Tsagkaris, C., Bilal, M., Aktar, I., Aboufandi, Y., Tas, A., Aborode, A. T., et al. (2022). Cytokine storm and neuropathological alterations in patients with neurological manifestations of COVID-19. Curr. Alzheimer Res. 2022, 641–657. doi: 10.2174/1567205019666220908084559
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Yıldırım, F., Karaman, İ., and Kaya, A. (2021). Current situation in ARDS in the light of recent studies: classification, epidemiology and pharmacotherapeutics. Tuberk. Toraks 69, 535–546. doi: 10.5578/tt.20219611
Yin, Y., Hountras, P., and Wunderink, R. G. (2017). The microbiome in mechanically ventilated patients. Curr. Opin. Infect. Dis. 30, 208–213. doi: 10.1097/QCO.0000000000000352
Zheng, H., Zhao, Q., Chen, J., Lu, J., Li, Y., and Gao, H. (2023). Gastrointestinal microbiome of ARDS patients induces neuroinflammation and cognitive impairment in mice. J. Neuroinflammation 20:166. doi: 10.1186/s12974-023-02825-7
Keywords: acute respiratory distress syndrome, gut microbiota, inflammatory factors, Mendelian randomization, genome-wide association studies
Citation: Ma J, Zhu Z, Yishajiang Y, Alarjani KM, Hong L and Luo L (2023) Role of gut microbiota and inflammatory factors in acute respiratory distress syndrome: a Mendelian randomization analysis. Front. Microbiol. 14:1294692. doi: 10.3389/fmicb.2023.1294692
Edited by:
Khalid Mehmood, Islamia University of Bahawalpur, PakistanReviewed by:
Xingliang Dai, First Affiliated Hospital of Anhui Medical University, ChinaWen yan Xiao, Second Hospital of Anhui Medical University, China
Copyright © 2023 Ma, Zhu, Yishajiang, Alarjani, Hong and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Hong, becsy123@163.com; Liang Luo, luoliang2117@163.com
†These authors have contributed equally to this work
 Jiawei Ma
Jiawei Ma Zigang Zhu1†
Zigang Zhu1† Lei Hong
Lei Hong Liang Luo
Liang Luo