- 1Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2College of Veterinary Medicine, Yunnan Agricultural University, Kunming, Yunnan, China
- 3Xishuangbanna Vocational and Technical College, Yunnan, China
Enterococcus spp., as an opportunistic pathogen, are widely distributed in the environment and the gastrointestinal tracts of both humans and animals. Captive Asian elephants, popular animals at tourist attractions, have frequent contact with humans. However, there is limited information on whether captive Asian elephants can serve as a reservoir of antimicrobial resistance (AMR). The aim of this study was to characterize AMR, antibiotic resistance genes (ARGs), virulence-associated genes (VAGs), gelatinase activity, hemolysis activity, and biofilm formation of Enterococcus spp. isolated from captive Asian elephants, and to analyze the potential correlations among these factors. A total of 62 Enterococcus spp. strains were isolated from fecal samples of captive Asian elephants, comprising 17 Enterococcus hirae (27.4%), 12 Enterococcus faecalis (19.4%), 8 Enterococcus faecium (12.9%), 7 Enterococcus avium (11.3%), 7 Enterococcus mundtii (11.3%), and 11 other Enterococcus spp. (17.7%). Isolates exhibited high resistance to rifampin (51.6%) and streptomycin (37.1%). 50% of Enterococcus spp. isolates exhibited multidrug resistance (MDR), with all E. faecium strains demonstrating MDR. Additionally, nine ARGs were identified, with tet(M) (51.6%), erm(B) (24.2%), and cfr (21.0%) showing relatively higher detection rates. Biofilm formation, gelatinase activity, and α-hemolysin activity were observed in 79.0, 24.2, and 14.5% of the isolates, respectively. A total of 18 VAGs were detected, with gelE being the most prevalent (69.4%). Correlation analysis revealed 229 significant positive correlations and 12 significant negative correlations. The strongest intra-group correlations were observed among VAGs. Notably, we found that vancomycin resistance showed a significant positive correlation with ciprofloxacin resistance, cfr, and gelatinase activity, respectively. In conclusion, captive Asian elephants could serve as significant reservoirs for the dissemination of AMR to humans.
1. Introduction
Enterococcus spp., a genus of Gram-positive, spherical or elliptical bacteria, are commonly found in the digestive tracts of humans and animals, and are considered an opportunistic pathogen. Although many strains of Enterococcus spp. are harmless commensals, some strains can cause nosocomial and community-acquired human infections, including bacteremia, peritoneal and intra-abdominal infections, and urinary tract infections, among others (Arias and Murray, 2012). The first global mortality estimates for 33 bacterial pathogens and 11 infection types indicate that 539,000 deaths in 2019 were associated with enterococcal infections, with the predominant pathogens being Enterococcus faecalis and Enterococcus faecium, which account for approximately 80% of enterococcal infections (Ikuta et al., 2022). Enterococcus spp. are also used as probiotics, starters in food fermentation, bio-preservatives and indicators of fecal contamination of food or water (Foulquié Moreno et al., 2006; Ben Braïek and Smaoui, 2019). Additionally, Enterococcus spp. are important key indicator bacterium in some human and veterinary antimicrobial resistance (AMR) monitoring systems.
Enterococcus spp. possess inherent resistance to a range of antimicrobial agents, spanning from β-lactam, cephalosporin, aminoglycoside, to lincosamide, exhibiting varying degrees of susceptibility at different levels (Murray, 1990). They can also acquire resistance to penicillin, chloramphenicol, tetracyclines, and vancomycin through mobile genetic elements carrying antimicrobial resistance genes (ARGs). Additionally, Enterococcus spp. play a crucial role in acquiring, storing, and disseminating resistance determinants (Werner et al., 2013; Malik et al., 2022; Sagor et al., 2022). The pathogenesis and biofilm formation of Enterococcus spp. are attributed to a multitude of virulence determinants, including aggregation substance (asa1), collagen-binding protein (ace), enterococcal surface protein (esp), gelatinase (gelE), hyaluronidase (hyl), E. faecalis and E. faecium endocarditis antigen A (efaAfs and efaAfm), pili (ebpABC locus, srt, and pil), the quorum sensing (fsrA, fsrB and fsrC), and sex pheromones (cpd, cob, and ccf), serine protease (sprE), cytolysin (cylA, cylB, cylM and cylLL; Stępień-Pyśniak et al., 2019b). Biofilms endow Enterococcus spp. with enhanced AMR and survival capabilities, while also facilitating their more effective invasion of host and the initiation of infections. Infections related to biofilms of Enterococcus spp. are not only difficult to eradicate but also act as centers for bacterial transmission and reservoirs of antibiotic resistance genes (Ch'ng et al., 2019). The process of enterococcal biofilm formation involves the participation of multiple genes, such as esp., gelE, fsrABC, ace, ebpABC (Ch'ng et al., 2019). Furthermore, gelatinase and hemolysin enhance its survival and dissemination capabilities within the host, while also inflicting damage on host tissues and the immune system.
The correlation between AMR and virulence traits in Enterococcus spp. has been investigated in previous studies (Baylan et al., 2011; Arabestani et al., 2017; Say Coskun, 2019; Alzahrani et al., 2022). In E. faecium, there is a certain correlation between ampicillin and vancomycin resistance, where ampicillin-resistant E. faecium is often detected prior to vancomycin resistance (Mundy et al., 2000). This suggests that the use of cephalosporins may facilitate the emergence of vancomycin-resistant E. faecium (Loeb et al., 1999). Previous studies also have shown that the vanA and erm(B) genes are often located on the same transferable plasmid (Aarestrup et al., 2000). In E. faecalis, most of cytolytic strains also express aggregation substance (Chow et al., 1993). In enterococcal clinical isolates, it was found that the agg and fsrB genes are positively correlated with biofilm formation and gelatinase activity, respectively (Hashem et al., 2017). A positive correlation between gentamicin resistance and hemolysis was demonstrated in E. faecalis blood isolates (Huycke et al., 1991). It has been found that a certain correlation between antibiotic resistance and biofilm formation (Fallah et al., 2017). Overall, there may be a potential synergistic interaction between this virulence traits and AMR in pathogenesis or survival. Moreover, there are certain variations in AMR and virulence characteristics among different species of Enterococcus spp. Enterococcus faecalis exhibits a higher occurrence of virulence factors, including cytolysin, aggregation substance, gelatinase, extracellular superoxide, and extracellular surface protein, compared to E. faecium. Conversely, E. faecium shows a higher AMR than E. faecalis (Jett et al., 1994; Mundy et al., 2000).
In recent years, the monitoring of AMR in wild animals has been performed (Smoglica et al., 2022). Research suggests that wild animals are one of the potential reservoirs for transmitting AMR pathogens to humans, such as wild birds (Stępień-Pyśniak et al., 2019a), non-human primates (Zhu et al., 2021), wild boar (Dias et al., 2022), rodents (Gwenzi et al., 2021). Meanwhile, close contact between humans and animals has been shown to lead to the mutual transmission of antimicrobial resistant microorganisms (van den Bogaard et al., 2001; Kim et al., 2016; Fagre et al., 2022). The recent studies have shown that captive Asian elephants harbor potential pathogenic species as well as a wide range of ARGs (Li et al., 2022; Cao et al., 2023). Being a popular wildlife species, captive Asian elephants have close interactions with humans at tourist attractions, which may lead to the spread of AMR pathogens between humans and animals. To the best of our knowledge, there is currently no research available on AMR and Virulence Characteristics of Enterococcus spp. in captive Asian elephants. Therefore, our objective is to determine AMR, ARGs, virulence-associated genes (VAGs), gelatinase activity, hemolysis activity, and biofilm formation, as well as the correlation between them, in Enterococcus isolates obtained from captive Asian elephants.
2. Materials and methods
2.1. Sample collection and processing
From June 2022 to December 2022, a total of 69 fecal samples from elephants were collected from eight sites located in southwestern China, including Kunming Zoo, Yunnan Nationalities Village Ji Xiang Garden, Menglia Shelter, Fengqing Park, Manting Park, Bifengxia Wildlife Park, Jiuding Mountain Wildlife Zoo, and Chengdu Zoo. All captive Asian elephants involved in this study were in a healthy state and did not exhibit any abnormal symptoms. Each elephant was only sampled once for fecal collection. After defecation, fecal samples were collected within 24 h by keepers wearing sterile gloves. The samples were immediately transported with ice bags to the Clinical Veterinary Laboratory at Sichuan Agricultural University, where isolation of Enterococcus spp. was performed.
2.2. Isolation and identification of Enterococcus spp.
Enterococcus spp. was obtained from the collected samples utilizing a medium specifically designed for the isolation of Enterococcus, as described previously (Stępień-Pyśniak et al., 2019a). The species identification of hypothetical Enterococcus spp. isolates was performed by multiplex PCR amplification of the groES-EL intergenic spacer region and the E. hirae-specific muramidase gene (mur-2), as described by Holman et al. (2021). Strains were identified as E. hirae when both groES-EL and mur-2 exhibited positive amplification. In cases where only groES-EL showed positive amplification, the PCR product was subjected to Sanger sequencing at Shanghai Shenggong Biotechnology Co., Ltd. The sequencing results will be analyzed using BLAST on the National Center for Biological Information1 to determine the species of Enterococcus spp.
2.3. Antibiotic susceptibility testing
According to the recommended guidelines by the Clinical and Laboratory Standards Institute (CLSI) in 2020 (M100), antimicrobial susceptibility testing of the isolated Enterococcus spp. was performed using the disk diffusion method for 10 antibiotic classes, comprising tetracycline (30 μg; TE), vancomycin (30 μg; VA), ampicillin (10 μg; AMP), rifampin (5 μg; RD), linezolid (30 μg; LZD), erythromycin (15 μg; E), teicoplanin (30 μg; TEC), nitrofurantoin (300 μg; F), chloramphenicol (30 μg; C), streptomycin (300 μg; S), ciprofloxacin (5 μg; CIP), and gentamicin (120 μg; CN). Specifically, the overnight bacterial culture was diluted to a concentration of 0.5 McFarland, and then 100 μL of the diluted suspension was evenly spread on Muller-Hinton agar, followed by incubation at 35°C for 16–18 h. AMR of Enterococcus spp. was determined by measuring the size of inhibition zones. Staphylococcus aureus ATCC 25923 was used as the quality control strain. Enterococcus spp. isolates that exhibit resistance to three or more classes of antibiotics will be defined as multiple drug resistance (MDR). Enterococcus spp. isolates that exhibit resistance to at least one class of antimicrobials was defined as antimicrobial resistance (AR).
2.4. Identification of ARGs and VAGs
PCR was performed to detect ARGs, including oxazolidinones (optrA and cfr), tetracyclines [tet(M) and tet(L)], chloramphenicol (cat), erythromycin (ermA and ermB), ampicillin (pbp5), aminoglycosides (aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, and str), and vancomycin (vanA and vanB). In accordance with Stępień-Pyśniak et al. (2019b), a total of 23 VAGs in Enterococcus spp. were detected by PCR, including ebpA, ebpB, ebpC, pil, srt, sprE, efaAfm, efaAfs, asa1, ace, esp., gelE, hyl, cylA, cylB, cylM, cylLL, fsrA, fsrB, fsrC, cpd, cob, and ccf. PCR tests were performed using a 25 μL reaction mixture composed of 12.5 μL of Premix Taq™ (TaKaRa Bio, Otsu, Japan), 8.5 μL of sterile ddH2O, 1 μL of each primer, and 2 μL of genomic DNA. The primers, cycling conditions, and amplicon sizes for ARGs are summarized in Supplementary Table 1. Negative controls were used in each PCR run. PCR-positive products were sent to Shanghai Shenggong Biotechnology Co., Ltd. for Sanger sequencing, and the resulting sequences were aligned with reference sequences on NCBI using BLAST.
2.5. Phenotypic detection of enterococcal virulence factors
2.5.1. Hemolysin and gelatinase activities
Hemolysin production was assessed by inoculating Enterococcus strains onto Columbia agar containing 5% defibrinated horse blood and incubating at 37°C for 24 h. Bacterial colonies displaying transparent hemolytic zones (β-hemolysis) and grass-green hemolytic zones (γ-hemolysis) on agar plates are, respectively, regarded as positive and negative colonies for hemolysin production. Staphylococcus aureus ATCC 25923 was utilized as the positive control.
The isolates of Enterococcus spp. were inoculated into tubes containing the Nutrient Gelatin (Thermo Scientific™, China). The tubes were incubated at 37°C for 24 to 72 h, followed by a 30-min refrigeration at 4°C. Gelatinase-producing Enterococcus spp. still cause liquefaction of the nutrient gelatin medium, even after refrigeration. Conversely, a semisolid consistency of the nutrient gelatin medium after refrigeration indicated a negative result of gelatinase production. Proteus mirabilis ATCC®29,906 and Escherichia coli ATCC®25,922 were employed as positive and negative controls, respectively.
2.5.2. Biofilm formation assay
The ability of Enterococcus spp. to form biofilms was assessed based on a method described by Aladarose et al. (2019). Briefly, after overnight incubation at 37°C on Columbia blood agar plates, a single colony was transferred into tryptic soy broth (TSB) with 0.25% glucose, and incubated overnight at 37°C. The bacterial suspension was adjusted to an optical density (OD600) of 0.2–0.257 using TSB. This was then inoculated into a 96-well plate (200 μL per well) with three replicate wells per strain. An additional 200 μL of TSB served as the negative control. After 24 h at 37°C, the wells were washed with PBS, fixed with methanol, and stained with 1% crystal violet dye. Following destaining, the optical density (OD570) was measured, and the final OD value for each strain was calculated as the average of three wells. This experiment was conducted in triplicate on separate days. According to previous studies (Stępień-Pyśniak et al., 2019b), the biofilm formation ability of Enterococcus spp. can be classified into four levels: negative, weak, moderate, and strong.
2.6. Statistical analysis
Statistical analysis was performed using the Fisher’s exact test or Chi-square test in SPSS version 22.0 (IBM Armonk Corp., Armonk, NY, United States), with significance defined as p < 0.05. The correlation analysis was performed using the corrplot package in RStudio.
3. Results
3.1. Isolation and identification of Enterococcus spp.
1–2 strains of Enterococcus spp. could be isolated from each elephant fecal sample, with no variation in isolation rates across the different regions. A total of 62 strains of Enterococcus spp. were isolated from 44 elephant fecal samples, and were then molecularly identified as belonging to 11 different species. The predominant species was E. hirae (17/62, 27.4%), followed by E. faecalis (12/62, 19.4%), E. faecium (8/62, 12.9%), E. avium (7/62, 11.3%), E. mundtii (7/62, 11.3%), E. gallinarum (4/62, 6.5%), E. casseliflavus (3/62, 4.8%), E. asini (1/62, 1.6%), E. flavescens (1/62, 1.6%), E. malodoratus (1/62, 1.6%), and E. raffinosus (1/62, 1.6%).
3.2. Antibiotic susceptibility test
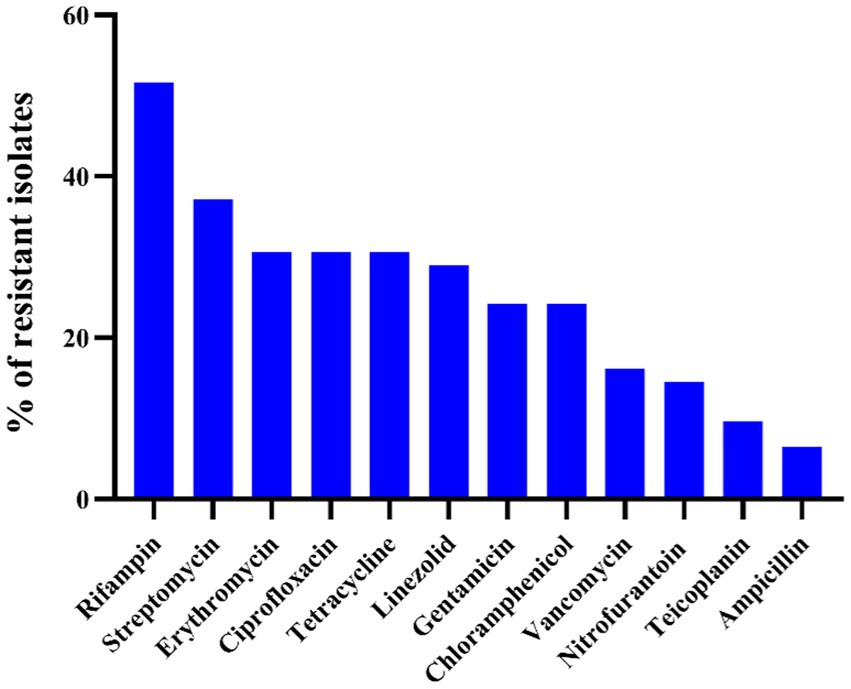
The results of the antibiotic susceptibility test of Enterococcus spp. isolates are presented in Table 1 and Figure 1. Among the 62 isolates of Enterococcus spp., the highest resistance was observed against RD (32/62, 51.6%), followed by S (23/62, 37.1%), TE (19/62, 30.6%), E (19/62, 30.6%), CIP (19/62, 30.6%), LZD (18/62, 29.0%), CN (15/62, 24.2%), C (15/62, 24.2%), VA (10/62, 16.1%), F (9/62, 14.5%), TEC (6/62, 9.7%), and AMP (4/62, 6.5%). Enterococcus faecalis was more resistant to RD (91.7%), VA (58.3%), and CIP (50.0%), whereas E. faecium was more resistant to CIP (87.5%), RD (75%), and E (62.5%). In 62 strains of Enterococcus spp., 53 strains (85.5%) exhibited resistance to at least one antibiotic, 20 strains (32.3%) were resistant to two antibiotics, and the remaining 33 strains (53.2%) were classified as MDR. Notably, the rate of MDR was significantly higher in E. faecium (8/8, 100%) when compared other Enterococcus species (n ≥ 7; p < 0.05). There were no significant differences in the prevalence of AR among different species of Enterococcus spp. (p > 0.5). For E. hirae, E. faecalis, E. faecium, E. avium, and E. mundtii, the mean multiple antibiotic resistance (MAR) indices were 0.20 (range: 0–0.92), 0.31 (range: 0.08–0.5), 0.39 (range: 0.25–0.58), 0.29 (range: 0–0.83), and 0.15 (range: 0–0.5), respectively.
3.3. Characterization of enterococcal virulence factors
No isolates exhibited β-hemolytic activity, while 9 isolates demonstrated α-hemolytic activity. For gelatinase activities, 15 isolates of Enterococcus spp. were found to produce gelatinase, of which 11 strains were identified as E. faecalis (73.3%). The gelatinase activity of E. faecalis was significantly higher that of other species of Enterococcus spp. (n ≥ 7; p < 0.01).
Overall, among the 62 strains of Enterococcus spp., 49 strains (79.0%) were found to be capable of producing biofilms, including 14 strains of E. hirae, 12 strains of E. faecalis, 5 strains of E. faecium, 6 strains of E. avium, 4 strains of E. mundtii, and 8 strains of other Enterococcus species. Among the 49 biofilm-producing Enterococcus spp., 36 strains (73.5%) were identified as weak biofilm producers, 10 strains (20.4%) as moderate biofilm producers, and 3 strains (6.1%) as strong biofilm producers. The strong biofilm producers include one strain of E. mundtii and two strains of E. faecalis. The biofilm-forming ability of E. faecalis was significantly higher than other species of Enterococcus spp. (p < 0.05, Wilcoxon rank-sum test), as all 12 isolates of E. faecalis were capable of biofilm formation, with 2 strains identified as strong biofilm producers.
3.4. Detection of ARGs and VAGs
Figure 2 and Table 2 show the detection rates of 23 VAGs and 14 ARGs across all isolates. Overall, 9 out of 14 ARGs were detected in Enterococcus spp. isolates. The detection rates of tet(M) were the highest at 51.6%, followed by erm(B) (24.2%), cfr (21.0%), str (9.7%), tet(L) (9.7%), pbp5 (8.1%), aph(3′)-IIIa (6.5%), ant(6)-Ia (6.5%), and aac(6′)-Ie-aph(2″)-Ia (4.8%). No erm(A), VanA, VanB, cat, optrA were detected. Interestingly, the detection rates of cfr were significantly higher in E. faecalis than in E. faecium (p < 0.01). Conversely, the detection rates of str and pbp5 were significantly higher in E. faecium than in E. faecalis (p < 0.05). The detection rates of ant(6)-Ia and pbp5 were significantly higher in MDR Enterococcus spp. than in non-MDR Enterococcus spp. (p < 0.05).
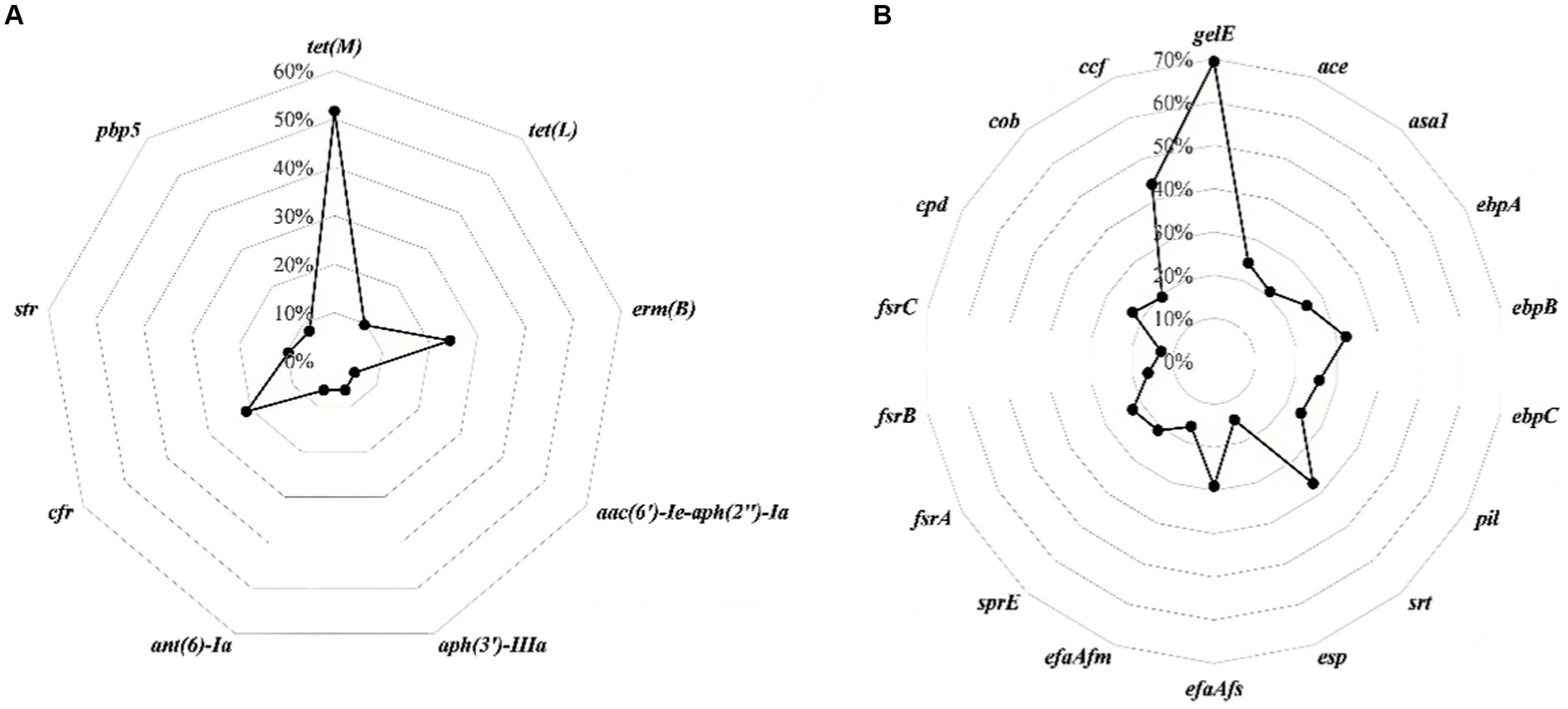
Figure 2. The detection rates of antimicrobial resistance genes (A) and virulence-associated genes (B) in all isolates of Enterococcus spp.
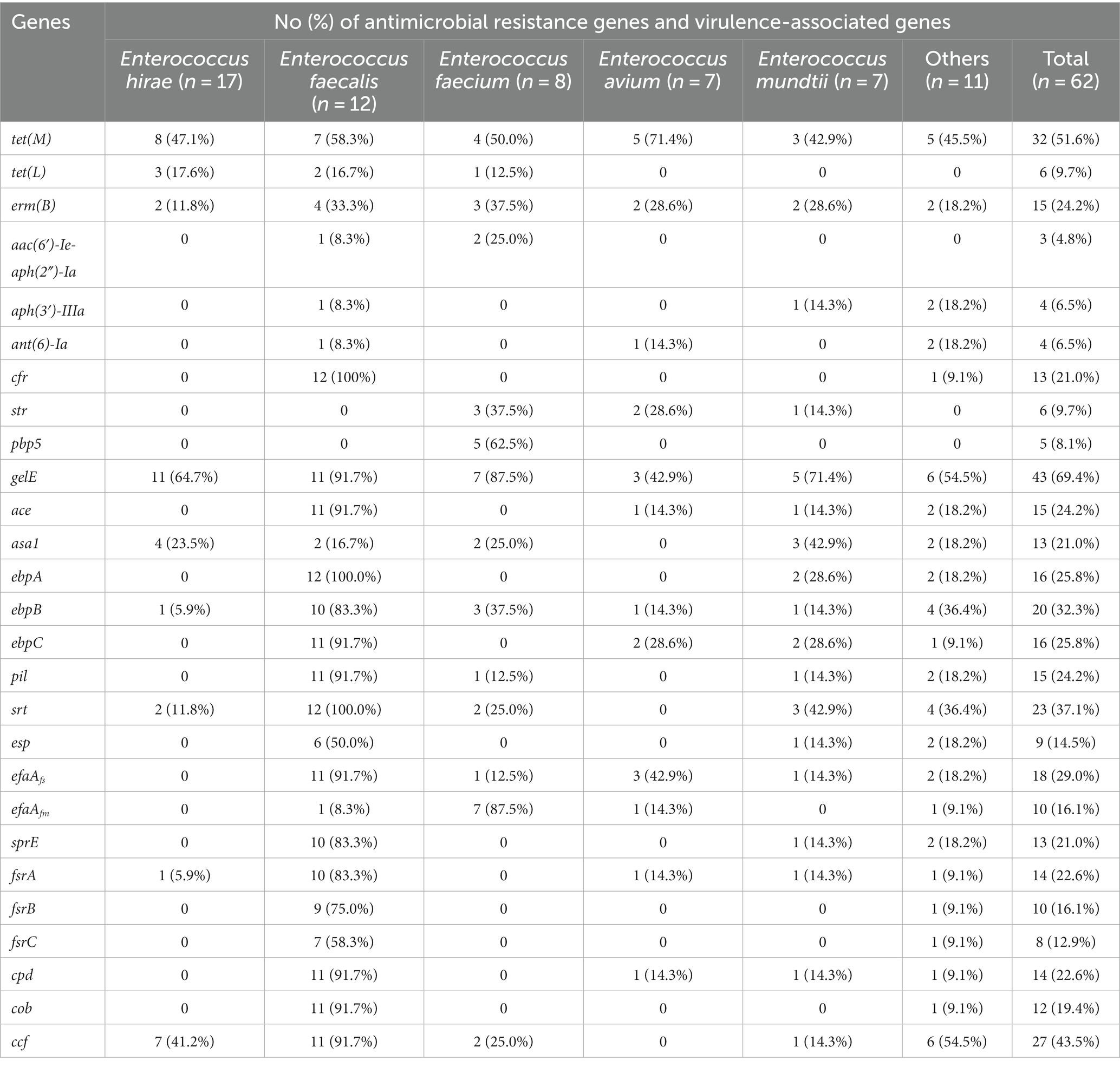
Table 2. Distribution of antimicrobial resistance genes and virulence-associated genes among different Enterococcus spp.
For the 23 VAGs, the detection rate of gelE was the highest at 69.4%, followed by ccf (43.5%), srt (37.1%), ebpB (32.3%), efaAfs (29.0%), ebpA (25.8%), ebpC (25.8%), ace (24.2%), pil (24.2%), cpd (22.6%), fsrA (22.6%), sprE (21.0%), asa1 (21.0%), cob (19.4%), efaAfm (16.1%), fsrB (16.1%), esp (14.5%), and fsrC (12.9%). No hyl, cylA, cylB, cylM, and cylL genes were detected in any of the tested isolates. The detection rates of VAGs (ccf, ace, pil, srt, cpd, fsrA, fsrB, sprE, fsrC, cob, ebpA, ebpB, ebpC, efaAfs, and esp) in E. faecalis were significantly higher than in other species of Enterococcus spp. (n ≥ 7; p < 0.01). There was no significant difference in the detection rates of VAGs between MDR Enterococcus spp. and non-MDR Enterococcus spp. (p > 0.05).
3.5. Correlation analysis
Figure 3 shows the results of the intragroup and intergroup correlation analysis among AMR, ARGs, VAGs, gelatinase activity, hemolysis activity, and biofilm formation. Correlation analysis within the group of 12 antibiotics revealed 17 pairs of antibiotics showing significant positive correlation and no significant negative correlation. The most significant positive correlation was observed between CIP and VA (r = 0.47, p < 0.001). A significant correlation was found between gelatinase activity and biofilm formation (r = 0.29, p < 0.05). There were six pairs of significant positive correlations among ARGs, without any significant negative correlations. The most significant positive correlation was observed between aph(3′)-IIIa and ant(6)-Ia, both of which determine aminoglycoside resistance (r = 0.73, p < 0.001). A total of 91 pairs of VAGs exhibited significant positive correlations, while no significant negative correlations were observed. The most significant correlation was found between ebpC and cpd (r = 0.92, p < 0.001). Interestingly, the number of significant correlations within VAGs was significantly higher than that within AMR, ARGs, and virulence factors (p < 0.01).
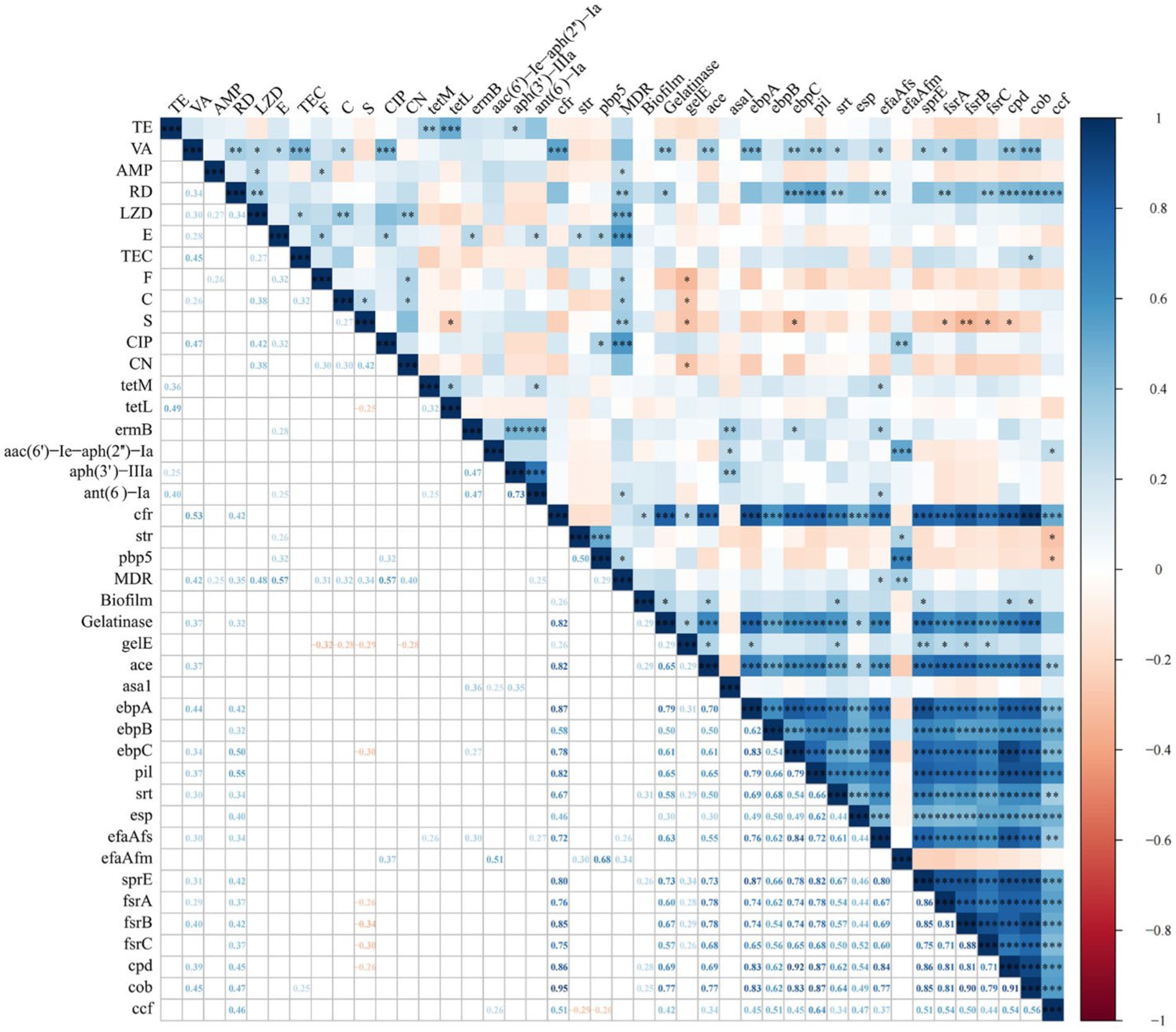
Figure 3. Correlation between antimicrobial resistance, antimicrobial resistance genes, virulence factors, and virulence-associated genes in Enterococcus spp. isolates (*, p < 0.05; **, p < 0.01; ***, p < 0.001). The numbers in the heatmap represent the correlation coefficients (r) between two objects. Blue color represents positive correlation (r > 0), while red color represents negative correlation (r < 0).
We further analyzed the intergroup correlations. All antibiotics showed a positive correlation with MDR, with significant positive correlations observed between MDR and all antibiotics except for VA, TE, CN and TEC. Only two antibiotics showed a significant correlation with gelatinase activity and biofilm formation, namely VA with gelatinase activity (r = 0.37, p < 0.01), RD with gelatinase activity (r = 0.32, p < 0.05). For AMR and ARGs, a total of 9 significantly correlated pairs were identified, with the strongest correlation observed between VA and cfr (r = 0.53, p < 0.001). Further analysis of the correlation between AMR and VAGs revealed 21 significant positive correlations and 9 significant negative correlations. The most significant positive and negative correlations were observed between RD and pil (r = 0.55, p < 0.001), and S and frsB (r = −0.34, p < 0.01), respectively. We only identified two significant correlations between ARGs and MDR (ant(6)-Ia and pbp5; p < 0.05). ARGs and virulence factors exhibit only two significant correlations: between cfr and biofilm formation (r = 0.26, p < 0.05), and between cfr and gelatinase activity (r = 0.82, p < 0.001). Between ARGs and VAGs, a total of 27 significantly positive correlations and 2 negative correlations were identified. Notably, cfr showed significant positive correlations with 16 VAGs, with the strongest correlation observed between cfr and cob (r = 0.95, p < 0.001). MDR showed significant positive correlations with efaAfs (r = 0.26, p < 0.05) and efaAfm (r = 0.34, p < 0.01), respectively. For biofilm formation and VAGs, we observed significant positive correlations between five VAGs (ace, srt, sprE, cpd, and cob) and biofilm formation (all p < 0.05). For gelatinase activity and VAGs, we identified significant positive correlations between 15 ARGs and gelatinase activity, with ebpA showing the strongest correlation with gelatinase activity (r = 0.79, p < 0.001).
4. Discussion
AMR poses a significant and urgent threat to global public health, ranking among the top 10 global public health threats identified by the World Health Organization. Recent research revealed that bacterial AMR was responsible for 1.27 million direct fatalities in 2019 (Murray et al., 2022). Enterococcus spp., as indicators of bacterial AMR, play a crucial role in monitoring the spread of AMR. The interaction between tourists and wild animals may lead to the mutual transmission of AMR. A study demonstrating higher AMR in tourists or local resident relative to that of primates suggests that this high level of AMR may be spreading to primates (Chong et al., 2020). Captive elephants at tourist attractions, being popular animals, frequently come into contact with visitors, which may lead to the transmission of AMR from humans to captive elephants. Therefore, captive Asian elephants may exhibit high levels of AMR. In this study, we characterized AMR and virulence profiles of Enterococcus spp. isolated from captive Asian elephants in zoos, while also exploring the correlations between AMR and virulence profiles.
Enterococcus hirae was the most common Enterococcus species found in the gastrointestinal tract of captive Asian elephants in this study, rather than the commonly observed E. faecalis and E. faecium in human or other mammalian feces (Lebreton et al., 2014). Enterococcus hirae is also the major Enterococcus species in the feces of cats (Jackson et al., 2009), and cattle (Jackson et al., 2011), and white-backed stilt (Medeiros et al., 2017). The species differences between elephants and human clinical isolates indicate that elephant-associated Enterococcus spp. may not be a significant source or origin of human-associated Enterococcus species. In addition to E. faecalis and E. faecium, other Enterococcus species, such as E. hirae, E. avium, and E. mundtii, are deemed infrequent agents of human clinical infections (Ruoff et al., 1990). More E. faecalis than E. faecium were isolated in this study, possibly due to the higher natural abundance of E. faecalis in the gastrointestinal tract (Goh et al., 2017). The majority of clinical enterococcal infections are caused by E. faecalis and E. faecium, indicating that the transmission of these pathogens from captive Asian elephants to humans could pose a potential risk to human health. However, this study lacks environmental and human samples, making it impossible to determine whether there is a risk of the outward transmission of elephant-associated Enterococcus spp.
The intrinsic antibiotic resistance and the capacity to acquire additional antibiotic resistance in Enterococcus spp. make infections challenging to manage. This study revealed that Enterococcus spp. showed high levels of resistance to RD, which is consistent with the resistance patterns observed in Enterococcus spp. previously isolated from pets (Tumpa et al., 2022), ducks (Kissinga et al., 2018), food (Russo et al., 2018), and water environments (Saingam et al., 2021). However, RD in combination with other antibiotics is commonly used to treat E. faecium resistant to both VA and LZD (Pankey et al., 2005). This study found VA resistance in 10 strains of Enterococcus spp., with 7 of them being specifically identified as E. faecalis. However, no VA resistance genes were detected, suggesting that the enhanced VA resistance may be attributed to the biofilm formation of E. faecalis. This is consistent with the results of the biofilm formation assay, where 100% E. faecalis isolates were identified as biofilm producers. In this study, E. flavescens, E. gallinarum, and E. casseliflavus exhibited intermediate susceptibility to VA, which could be their intrinsic resistance to VA (Vincent et al., 1992). LZD plays a critical role in combating infections caused by vancomycin resistant Enterococcus (VRE). Among oxazolidinones, linezolid is notably influenced by the cfr gene. Notably, all 12 E. faecalis isolates in this study carried the cfr gene, with 10 strains exhibiting resistance or intermediate resistance to LZD. It may pose a potential threat to public health due to cfr being a transferable resistant gene. We also found that Enterococcus spp. isolates capable of forming strong/moderate biofilms were either intermediate or resistant to LZD, which was also found in the report of Aladarose et al. (2019). Different species of Enterococcus spp. also exhibit varying susceptibilities to distinct antibiotics. As demonstrated in this study, E. faecium, E. faecalis, E. hirae, and E. avium exhibited the highest resistance to CIP, RD, S, and TE, respectively. Enterococcus faecium exhibits higher resistance to AMP compared to E. faecalis, and AMP-resistant E. faecalis strains have been scarcely reported in animals (Miller et al., 2014; Torres et al., 2018). As observed in this study, E. faecium (25.0%) isolates were resistant to AMP, whereas no E. faecalis isolates showed resistance to AMP. Conversely, E. faecium is more prevalent than E. faecalis for intrinsic aminoglycosides resistance (Abat et al., 2016), as demonstrated in this study. Interestingly, all the E. faecium isolates in this study were found to be MDR, whereas in E. mundtii, E. hirae, E. faecalis, and E. avium, MDR was detected at a lower rate. The inherent tenacity, genomic flexibility, and the widespread use of antibiotics have propelled E. faecium to become the predominant MDR species among Enterococcus spp. (Zhou et al., 2020). A higher prevalence of MDR isolates in E. faecium has also been found in other animals (Chotinantakul et al., 2018; Cui et al., 2020). This demonstrates that E. faecium is a major source of AMR in the gastrointestinal tract of captive Asian elephants.
Biofilm serves as a significant virulence factor in Enterococcus spp., particularly in the pathogenicity of Enterococcus spp. Biofilms can enhance bacterial resistance to antibiotics and anti-phagocytosis, posing significant challenges for infection treatment (Lewis, 2001). In the present study, we found that the proportion of biofilm-producing isolates was significantly higher in E. faecalis (100%) compared to E. faecium (62.5%). Consistent with most studies, E. faecalis exhibited a higher propensity to form biofilms than E. faecium (Mohamed and Huang, 2007). The formation of biofilm has been correlated with various environmental and genetic factors. Among them, VAGs (ace, gelE, esp., fsrABC, ebpABC, sprE, and asa1) are associated with the formation of biofilms in Enterococcus spp. Indeed, VAGs (ace and sprE) were found to be associated with biofilm formation in this study. Interestingly, we found a highly significant association (p < 0.001) between VAGs (ace, sprE, fsrA, fsrB, and cob) and the formation of strong/moderate biofilms (data not shown). Gelatinase represents a crucial virulence factor in E. faecalis, tightly linked to the formation of biofilms. According to our study, all E. faecalis strains with gelatinase activity carried gelE, which is consistent with the results reported by other authors (Kim et al., 2016; Stępień-Pyśniak et al., 2019b). The encoding of gelE and sprE occurs within the same operon, and their expression is regulated by the density-sensing system encoded by the fsr locus. Thus, we observed that all gelatinase-positive E. faecalis almost carries gelE, sprE and fsrABC genes. Previous studies have shown that gelE alone is insufficient to predict enterococcal gelatinase activity unless Enterococcus spp. carries fsrAB or fsrB (Hashem et al., 2017). As discovered in this study, 60.5% of Enterococcus spp. that were simultaneously positive for gelE and negative for fsrAB did not exhibit gelatinase activity. In line with the report of Barbosa et al. (2010), the efaAfs and efaAfm genes were detected not only in E. faecalis and E. faecium but also in a few other Enterococcus species. The virulence factors of Enterococcus spp., such as cytolysin and adhesive substances, can be transferred through the process of gene exchange. The virulence characteristics of E. faecalis in this study were significantly stronger than those of other enterococcal species, indicating that the E. faecalis isolated from captive Asian elephants should be considered a potential source of some virulence determinants.
Currently, no research has indicated the reasons for the correlation between virulence characteristics and AMR in Enterococcus spp. The most synergistic combinations of various virulence traits and AMR may enhance colonization and tissue invasion in the process of enterococcal infection or facilitate adaptation to environmental changes, thereby promoting survival. In general, statistically significant positive correlations were more prevalent in this study, which differs from the findings observed in Escherichia coli (Zhang et al., 2021). We observed the strongest positive associations within the following categories: AMR (CIP and VA), ARGs (aph(3′)-IIIa and ant(6)-Ia), VAGs (ebpC and cpd), AMR and MDR (E and CIP), AMR and ARGs (VA and cfr), AMR and virulence factors (VA and gelatinase), AMR and VAGs (RD and pil), MDR and ARGs (MDR and pbp5), virulence factors and ARGs (gelatinase and cfr), virulence factors and VAGs (gelatinase and ebpA), as well as ARGs and VAGs (cfr and cob). CIP or nitrofurantoin are viable options for managing uncomplicated urinary tract infections caused by VRE. The strong positive correlation between CIP and VA may be the result of CIP use leading to an increase in strains resistant to CIP and VA. The gene ant(6) is frequently identified within a gene cluster ant(6)-sat4-aph(3′)-III that is part of Tn5405 and other related transposons (Ramirez and Tolmasky, 2010). Furthermore, a prior investigation revealed that the predominant Aminoglycoside modifying enzyme gene profile detected among clinical isolates of E. faecalis, E. faecium, and E. avium was ant(6)-Ia + aph(3′)-IIIa (Kobayashi et al., 2001). This could be attributed to the strong correlation between aph(3′)-IIIa and ant(6)-Ia. E, CIP, and pbp5 exhibit a highly significant correlation with MDR, indicating that they represent the predominant antibiotic-resistant phenotypes and genotype within MDR Enterococcus spp. isolates. Some studies have indicated that VA inhibits gelatinase activity (Hashem et al., 2017). Therefore, VA resistance might potentially lead to an elevation in gelatinase activity. This study observed a significant correlation between cfr and cob, which differs from the correlation between cfr and asa1 observed within LZD-resistant E. faecium strains (Krawczyk et al., 2020). The potential statistical association between ARGs and VAGs may indicate a physical connection of genes on the same genetic element (Diarra et al., 2010). Nevertheless, the correlation presented in this study is based solely on statistical findings, and further research is required to elucidate the underlying mechanisms of this correlation, especially concerning the relationship between biofilm formation and AMR.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Sichuan Agricultural University Animal Ethical and Welfare Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JY: Writing – original draft, Formal analysis, Investigation, Methodology. YC: Investigation, Writing – original draft. ZD: Writing – original draft, Software. WZ: Writing – original draft, Data curation. LL: Funding acquisition, Resources, Supervision, Writing – review & editing. WM: Funding acquisition, Resources, Supervision, Writing – review & editing. QL: Funding acquisition, Resources, Supervision, Writing – review & editing. KF: Funding acquisition, Resources, Supervision, Writing – review & editing. ZiZ: Project administration, Validation, Writing – review & editing. HL: Project administration, Validation, Writing – review & editing. ZhZ: Project administration, Validation, Writing – review & editing. XX: Investigation, Writing – review & editing. JZ: Investigation, Writing – review & editing. GP: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Science and Technology Department’s “13th Five-Year” Special Subproject of China (no. 2016YFD0501009).
Acknowledgments
Many thanks to Yunjiang Liu and Jialiang Xin for their assistance in this research. We also need to thank the elephant keepers for their contribution to the collection of samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1277221/full#supplementary-material
Footnotes
References
Aarestrup, F. M., Kruse, H., Tast, E., Hammerum, A. M., and Jensen, L. B. (2000). Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6, 63–70. doi: 10.1089/mdr.2000.6.63
Abat, C., Raoult, D., and Rolain, J. M. (2016). Low level of resistance in enterococci isolated in four hospitals, Marseille, France. Microb. Drug Resist. 22, 218–222. doi: 10.1089/mdr.2015.0121
Aladarose, B. E., Said, H. S., and Abdelmegeed, E. S. (2019). Incidence of virulence determinants among Enterococcal clinical isolates in Egypt and its association with biofilm formation. Microb. Drug Resist. 25, 880–889. doi: 10.1089/mdr.2018.0320
Alzahrani, O. M., Fayez, M., Alswat, A. S., Alkafafy, M., Mahmoud, S. F., al-Marri, T., et al. (2022). Antimicrobial resistance, biofilm formation, and virulence genes in Enterococcus species from small backyard chicken flocks. Antibiotics 11:380. doi: 10.3390/antibiotics11030380
Arabestani, M. R., Nasaj, M., and Mousavi, S. M. (2017). Correlation between infective factors and antibiotic resistance in enterococci clinical isolates in west of Iran. Chonnam Med. J. 53, 56–63. doi: 10.4068/cmj.2017.53.1.56
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Barbosa, J., Gibbs, P. A., and Teixeira, P. (2010). Virulence factors among enterococci isolated from traditional fermented meat products produced in the north of Portugal. Food Control 21, 651–656. doi: 10.1016/j.foodcont.2009.10.002
Baylan, O., Nazik, H., Bektöre, B., Citil, B. E., Turan, D., Ongen, B., et al. (2011). The relationship between antibiotic resistance and virulence factors in urinary Enterococcus isolates. Mikrobiyol. Bul. 45, 430–445.
Ben Braïek, O., and Smaoui, S. (2019). Enterococci: between emerging pathogens and potential probiotics. Biomed. Res. Int. 2019, 5938210–5938213. doi: 10.1155/2019/5938210
Cao, K., Wang, Y., Bai, X., Wang, J., Zhang, L., Tang, Y., et al. (2023). Comparison of fecal antimicrobial resistance genes in captive and wild Asian elephants. Antibiotics 12:859. doi: 10.3390/antibiotics12050859
Ch'ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., and Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. doi: 10.1038/s41579-018-0107-z
Chong, C. W., Alkatheeri, A. H. S., Ali, N., Tay, Z. H., Lee, Y. L., Paramasivam, S. J., et al. (2020). Association of antimicrobial resistance and gut microbiota composition in human and non-human primates at an urban ecotourism site. Gut Pathog. 12:14. doi: 10.1186/s13099-020-00352-x
Chotinantakul, K., Chansiw, N., and Okada, S. (2018). Antimicrobial resistance of Enterococcus spp. isolated from Thai fermented pork in Chiang Rai Province, Thailand. J Glob Antimicrob Resist. 12, 143–148. doi: 10.1016/j.jgar.2017.09.021
Chow, J. W., Thal, L. A., Perri, M. B., Vazquez, J. A., Donabedian, S. M., Clewell, D. B., et al. (1993). Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37, 2474–2477. doi: 10.1128/aac.37.11.2474
Cui, P., Feng, L., Zhang, L., He, J., An, T., Fu, X., et al. (2020). Antimicrobial resistance, virulence genes, and biofilm formation capacity among Enterococcus species from yaks in aba Tibetan autonomous prefecture, China. Front. Microbiol. 11:1250. doi: 10.3389/fmicb.2020.01250
Diarra, M. S., Rempel, H., Champagne, J., Masson, L., Pritchard, J., and Topp, E. (2010). Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Appl. Environ. Microbiol. 76, 8033–8043. doi: 10.1128/aem.01545-10
Dias, D., Fonseca, C., Mendo, S., and Caetano, T. (2022). A closer look on the variety and abundance of the faecal resistome of wild boar. Environ. Pollut. 292:118406. doi: 10.1016/j.envpol.2021.118406
Fagre, A. C., Cohen, L. E., Eskew, E. A., Farrell, M., Glennon, E., Joseph, M. B., et al. (2022). Assessing the risk of human-to-wildlife pathogen transmission for conservation and public health. Ecol. Lett. 25, 1534–1549. doi: 10.1111/ele.14003
Fallah, F., Yousefi, M., Pourmand, M. R., Hashemi, A., Nazari Alam, A., and Afshar, D. (2017). Phenotypic and genotypic study of biofilm formation in enterococci isolated from urinary tract infections. Microb. Pathog. 108, 85–90. doi: 10.1016/j.micpath.2017.05.014
Foulquié Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Goh, H. M. S., Yong, M. H. A., Chong, K. K. L., and Kline, K. A. (2017). Model systems for the study of Enterococcal colonization and infection. Virulence 8, 1525–1562. doi: 10.1080/21505594.2017.1279766
Gwenzi, W., Chaukura, N., Muisa-Zikali, N., Teta, C., Musvuugwa, T., Rzymski, P., et al. (2021). Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics 10:68. doi: 10.3390/antibiotics10010068
Hashem, Y. A., Amin, H. M., Essam, T. M., Yassin, A. S., and Aziz, R. K. (2017). Biofilm formation in enterococci: genotype-phenotype correlations and inhibition by vancomycin. Sci. Rep. 7:5733. doi: 10.1038/s41598-017-05901-0
Holman, D. B., Klima, C. L., Gzyl, K. E., Zaheer, R., Service, C., Jones, T. H., et al. (2021). Antimicrobial resistance in Enterococcus Spp. isolated from a beef processing plant and retail ground beef. Microbiol Spectr. 9:e0198021. doi: 10.1128/Spectrum.01980-21
Huycke, M. M., Spiegel, C. A., and Gilmore, M. S. (1991). Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 35, 1626–1634. doi: 10.1128/aac.35.8.1626
Ikuta, K. S., Swetschinski, L. R., Robles Aguilar, G., Sharara, F., Mestrovic, T., Gray, A. P., et al. (2022). Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400, 2221–2248. doi: 10.1016/S0140-6736(22)02185-7
Jackson, C. R., Fedorka-Cray, P. J., Davis, J. A., Barrett, J. B., and Frye, J. G. (2009). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 107, 1269–1278. doi: 10.1111/j.1365-2672.2009.04310.x
Jackson, C. R., Lombard, J. E., Dargatz, D. A., and Fedorka-Cray, P. J. (2011). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 52, 41–48. doi: 10.1111/j.1472-765X.2010.02964.x
Jett, B. D., Huycke, M. M., and Gilmore, M. S. (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7, 462–478. doi: 10.1128/cmr.7.4.462
Kim, D. H., Chung, Y. S., Park, Y. K., Yang, S. J., Lim, S. K., Park, Y. H., et al. (2016). Antimicrobial resistance and virulence profiles of Enterococcus spp. isolated from horses in Korea. Comp. Immunol. Microbiol. Infect. Dis. 48, 6–13. doi: 10.1016/j.cimid.2016.07.001
Kissinga, H. D., Mwombeki, F., Said, K., Katakweba, A. A. S., Nonga, H. E., and Muhairwa, A. P. (2018). Antibiotic susceptibilities of indicator bacteria Escherichia coli and enterococci spp. isolated from ducks in Morogoro municipality, Tanzania. BMC. Res. Notes 11:87. doi: 10.1186/s13104-018-3201-4
Kobayashi, N., Alam, M., Nishimoto, Y., Urasawa, S., Uehara, N., and Watanabe, N. (2001). Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126, 197–204. doi: 10.1017/s0950268801005271
Krawczyk, B., Wysocka, M., Kotłowski, R., Bronk, M., Michalik, M., and Samet, A. (2020). Linezolid-resistant Enterococcus faecium strains isolated from one hospital in Poland -commensals or hospital-adapted pathogens? PloS One 15:e0233504. doi: 10.1371/journal.pone.0233504
Lebreton, F., Willems, R. J. L., and Gilmore, M. S. (2014). “Enterococcus diversity, origins in nature, and gut colonization” in Enterococci: From commensals to leading causes of drug resistant infection. eds. M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston: Massachusetts Eye and Ear Infirmary)
Lewis, K. (2001). Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45, 999–1007. doi: 10.1128/aac.45.4.999-1007.2001
Li, G., Jiang, Y., Li, Q., An, D., Bao, M., Lang, L., et al. (2022). Comparative and functional analyses of fecal microbiome in Asian elephants. Antonie Van Leeuwenhoek 115, 1187–1202. doi: 10.1007/s10482-022-01757-1
Loeb, M., Salama, S., Armstrong-Evans, M., Capretta, G., and Olde, J. (1999). A case-control study to detect modifiable risk factors for colonization with vancomycin-resistant enterococci. Infect. Control Hosp. Epidemiol. 20, 760–763. doi: 10.1086/501580
Malik, F., Nawaz, M., Anjum, A. A., Firyal, S., Shahid, M. A., Irfan, S., et al. (2022). Molecular characterization of antibiotic resistance in poultry gut origin enterococci and horizontal gene transfer of antibiotic resistance to Staphylococcus aureus. Pakistan Vet. J. 42, 383–389. doi: 10.29261/pakvetj/2022.035
Medeiros, A. W., Blaese Amorim, D., Tavares, M., de Moura, T. M., Franco, A. C., d’Azevedo, P. A., et al. (2017). Enterococcus species diversity in fecal samples of wild marine species as determined by real-time PCR. Can. J. Microbiol. 63, 129–136. doi: 10.1139/cjm-2016-0427
Miller, W. R., Munita, J. M., and Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Mohamed, J. A., and Huang, D. B. (2007). Biofilm formation by enterococci. J. Med. Microbiol. 56, 1581–1588. doi: 10.1099/jmm.0.47331-0
Mundy, L. M., Sahm, D. F., and Gilmore, M. (2000). Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13, 513–522. doi: 10.1128/cmr.13.4.513
Murray, B. E. (1990). The life and times of the Enterococcus. Clin. Microbiol. Rev. 3, 46–65. doi: 10.1128/cmr.3.1.46
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Pankey, G., Ashcraft, D., and Patel, N. (2005). In vitro synergy of daptomycin plus rifampin against Enterococcus faecium resistant to both linezolid and vancomycin. Antimicrob. Agents Chemother. 49, 5166–5168. doi: 10.1128/aac.49.12.5166-5168.2005
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ruoff, K. L., de la Maza, L., Murtagh, M. J., Spargo, J. D., and Ferraro, M. J. (1990). Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28, 435–437. doi: 10.1128/jcm.28.3.435-437.1990
Russo, N., Caggia, C., Pino, A., Coque, T. M., Arioli, S., and Randazzo, C. L. (2018). Enterococcus spp. in Ragusano PDO and pecorino Siciliano cheese types: A snapshot of their antibiotic resistance distribution. Food Chem. Toxicol. 120, 277–286. doi: 10.1016/j.fct.2018.07.023
Sagor, S., Hossain, M., Islam, T., Mahmud, M., Miah, M., Karim, M., et al. (2022). Phenotypic and genotypic antibiotic resistance and virulence profiling of Enterococcus faecalis isolated from poultry at two major districts in Bangladesh. Pakistan Vet. J. 42, 2074–7764. doi: 10.29261/pakvetj/2022.019
Saingam, P., Di, D. Y. W., and Yan, T. (2021). Diversity and health risk potentials of the Enterococcus population in tropical coastal water impacted by hurricane lane. J. Water Health 19, 990–1001. doi: 10.2166/wh.2021.209
Say Coskun, U. S. (2019). Investigation of the relationship between virulence factors and antibiotic resistance of enterococci isolates. Cell. Mol. Biol. 65, 14–17. doi: 10.14715/cmb/2019.65.2.3
Smoglica, C., Vergara, A., Angelucci, S., Festino, A. R., Antonucci, A., Marsilio, F., et al. (2022). Evidence of linezolid resistance and virulence factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics 11:223. doi: 10.3390/antibiotics11020223
Stępień-Pyśniak, D., Hauschild, T., Dec, M., Marek, A., and Urban-Chmiel, R. (2019a). Clonal structure and antibiotic resistance of Enterococcus spp. from wild birds in Poland. Microb. Drug Resist. 25, 1227–1237. doi: 10.1089/mdr.2018.0461
Stępień-Pyśniak, D., Hauschild, T., Kosikowska, U., Dec, M., and Urban-Chmiel, R. (2019b). Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci. Rep. 9:11204. doi: 10.1038/s41598-019-47602-w
Torres, C., Alonso, C. A., Ruiz-Ripa, L., León-Sampedro, R., Del Campo, R., and Coque, T. M. (2018). Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol Spectr. 6. doi: 10.1128/microbiolspec.ARBA-0032-2018
Tumpa, A., Štritof, Z., and Pintarić, S. (2022). Prevalence and antimicrobial susceptibility of Enterococcus spp. from urine of dogs and cats in northwestern Croatia. Res. Vet. Sci. 151, 42–46. doi: 10.1016/j.rvsc.2022.04.015
van den Bogaard, A. E., London, N., Driessen, C., and Stobberingh, E. E. (2001). Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47, 763–771. doi: 10.1093/jac/47.6.763
Vincent, S., Minkler, P., Bincziewski, B., Etter, L., and Shlaes, D. M. (1992). Vancomycin resistance in Enterococcus gallinarum. Antimicrob. Agents Chemother. 36, 1392–1399. doi: 10.1128/aac.36.7.1392
Werner, G., Coque, T. M., Franz, C. M., Grohmann, E., Hegstad, K., Jensen, L., et al. (2013). Antibiotic resistant enterococci-tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 303, 360–379. doi: 10.1016/j.ijmm.2013.03.001
Zhang, S., Chen, S., Rehman, M. U., Yang, H., Yang, Z., Wang, M., et al. (2021). Distribution and association of antimicrobial resistance and virulence traits in Escherichia coli isolates from healthy waterfowls in Hainan, China. Ecotoxicol. Environ. Saf. 220:112317. doi: 10.1016/j.ecoenv.2021.112317
Zhou, X., Willems, R. J. L., Friedrich, A. W., Rossen, J. W. A., and Bathoorn, E. (2020). Enterococcus faecium: from microbiological insights to practical recommendations for infection control and diagnostics. Antimicrob. Resist. Infect. Control 9:130. doi: 10.1186/s13756-020-00770-1
Keywords: Enterococcus spp., Asian elephants, antimicrobial resistance, virulence-associated genes, Enterococcus faecalis, Enterococcus faecium
Citation: Yang J, Chen Y, Dong Z, Zhang W, Liu L, Meng W, Li Q, Fu K, Zhou Z, Liu H, Zhong Z, Xiao X, Zhu J and Peng G (2023) Distribution and association of antimicrobial resistance and virulence characteristics in Enterococcus spp. isolates from captive Asian elephants in China. Front. Microbiol. 14:1277221. doi: 10.3389/fmicb.2023.1277221
Edited by:
Manuela Oliveira, University of Lisbon, PortugalReviewed by:
Mashkoor Mohsin, University of Agriculture, Faisalabad, PakistanSilpak Biswas, Calcutta School of Tropical Medicine, India
Copyright © 2023 Yang, Chen, Dong, Zhang, Liu, Meng, Li, Fu, Zhou, Liu, Zhong, Xiao, Zhu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangneng Peng, pgn.sicau@163.com
†These authors have contributed equally to this work
 Jinpeng Yang
Jinpeng Yang Yanshan Chen1†
Yanshan Chen1† Wenqing Zhang
Wenqing Zhang Qianlan Li
Qianlan Li Ziyao Zhou
Ziyao Zhou Haifeng Liu
Haifeng Liu Zhijun Zhong
Zhijun Zhong Guangneng Peng
Guangneng Peng