- 1School of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu, China
- 2School of Environment and Safety Engineering, Jiangsu University, Zhenjiang, Jiangsu, China
- 3Ningbo Women and Children's Hospital, Ningbo, China
Cyanobacteria are an excellent microbial photosynthetic platform for sustainable carbon dioxide fixation. One bottleneck to limit its application is that the natural carbon flow pathway almost transfers CO2 to glycogen/biomass other than designed biofuels such as ethanol. Here, we used engineered Synechocystis sp. PCC 6803 to explore CO2-to-ethanol potential under atmospheric environment. First, we investigated the effects of two heterologous genes (pyruvate decarboxylase and alcohol dehydrogenase) on ethanol biosynthesis and optimized their promoter. Furthermore, the main carbon flow of the ethanol pathway was strengthened by blocking glycogen storage and pyruvate-to-phosphoenolpyruvate backflow. To recycle carbon atoms that escaped from the tricarboxylic acid cycle, malate was artificially guided back into pyruvate, which also created NADPH balance and promoted acetaldehyde conversion into ethanol. Impressively, we achieved high-rate ethanol production (248 mg/L/day at early 4 days) by fixing atmospheric CO2. Thus, this study exhibits the proof-of-concept that rewiring carbon flow strategies could provide an efficient cyanobacterial platform for sustainable biofuel production from atmospheric CO2.

Graphical Abstract. Carbon flux rewiring enables high rate of CO2-to-ethanol in photosynthetic microorganism.
1. Introduction
The increased level of atmospheric greenhouse gas arises the concern of seeking environmentally friendly technologies to fix and even reuse CO2 as an energy chemical (Fang et al., 2021). Microbial CO2 fixation has received much attention because of its highly renewable reaction under mild conductions (Gassler et al., 2020; Satanowski and Bar-Even, 2020; Chen et al., 2023a,b). Among those biotechnologies, photo-driven CO2 bioconversion represents one of the sustainable strategies to generate carbon-neutral biofuels, such as ethanol and butanol (Liu et al., 2019; Velmurugan and Incharoensakdi, 2020; Fang et al., 2022). Thus, it is urgent to develop a photo-driven biosynthesis platform for CO2-to-biofuel production.
Cyanobacterium owns high photosynthesis efficiency (theoretical maximum is 8–10%) and has the potential to convert CO2 into biofuels through the Calvin–Benson–Bassham (CBB) cycle (Santos-Merino et al., 2021). Notably, it has successfully engineered cyanobacteria to assimilate CO2 and produce value-added chemicals, such as ethylene (Li et al., 2021), isoprene (Lindberg et al., 2010), ethanol (Gao et al., 2012), isobutanol (Miao et al., 2017), acetone (Lee et al., 2020), and p-coumaric acid (Gao et al., 2021). Ethanol as a simple but major renewable biofuel can be easily produced by introducing two heterologous enzymes (pyruvate decarboxylase and alcohol dehydrogenase) in cyanobacteria (Gao et al., 2012). The model cyanobacterium of Synechocystis sp. PCC 6803 (hereafter Synechocystis) shows double ethanol yield compared to other cyanobacteria such as Synechococcus elongatus PCC 7942 (Dexter and Fu, 2009). Furthermore, Synechocystis owns clear genetic background to assemble and engineer heterologous pathways, indicating the promising future of CO2-to-ethanol production (Zhang and Bryant, 2011).
Recently, many efforts have been explored to promote ethanol production in Synechocystis. Optimization of abiotic and biotic factors showed positive effects on cell growth and ethanol synthesis (Heidorn et al., 2011; Gao et al., 2012). Overexpressing the ethanol-producing steps or blocking the production of storage polymers (glycogen and polyhydroxybutyrate) was able to increase ethanol production (Namakoshi et al., 2016; Velmurugan and Incharoensakdi, 2020). The enhancement of carbon fixation in the CBB cycle also significantly improved the ethanol yield as well as cell growth (Liang et al., 2018; Roussou et al., 2021). In addition, co-culture engineering and modular engineering were systematic strategies to achieve high-level ethanol production in photosynthetic microorganisms (Liu et al., 2019; Velmurugan and Incharoensakdi, 2020). However, it is difficult to channel the fixed carbon atoms into the target product because of the imbalance of cell growth rate and ethanol byproduct accumulation (Luan et al., 2020). The above strategies are still challenging to adjust the ethanol pathway in one system and are rarely explored in systematic investigations on promoter optimization, byproduct blocking, and cofactor regeneration on ethanol accumulation. In addition, efficiently fixing atmospheric CO2 into ethanol via Synechocystis is still due to a lack of study.
Herein, to investigate the ethanol-producing potential of optimizing metabolic pathways, the engineered Synechocystis cells were genetically modified in a stepwise approach via inhibiting the phosphoenolpyruvate pathway from pyruvate, removing glycogen storage, and shunting carbon metabolic flux of the tricarboxylic acid cycle. This approach leads to proof-of-concept with high-efficient ethanol production directly from solar energy and atmospheric CO2 and significantly contributes to the sustainability of CO2-to-biofuel conversion.
2. Materials and methods
2.1. Strains and growth conditions
Escherichia coli DH5α carrying various plasmids were grown in LB medium, which contained special antibiotics such as 50 μg/ml spectinomycin (SpR), 50 μg/ml kanamycin (KmR), or 25 μg/ml chloramphenicol (CmR). Synechocystis cells were grown in the BG11 medium and cultured at light conductions (50 μmol photons m−2 s−1 and 30°C). Unless otherwise noted, appropriate antibiotics were added to the BG11 medium.
2.2. Plasmid construction for gene knockout
The pMD18-T vector (TaKaRa, Dalian) is used as a backbone to construct cyanobacterial plasmids, which are presented in Supplementary Table S1. Using PCR to amplify the fragments, the fragment and the vector were double-digested by recombinase (NEW ENGLAND BioLabs Beijing, China). Corresponding primers (SupplementaryTable S2) were used to clone up/down-fragments of the Synechocystis genome, and T4 ligase (NEB, Beijing) was used for ligation. The recombinant pBE406 plasmid (containing 600 bp upstream/downstream slr0168 and spectinomycin resistance gene) for gene knockout is shown in Supplementary Figure S2. Similarly, pMD-slr0301-Ω and pMD-slr1176-Ω were constructed. Otherwise, to construct a recombinant ethanol pathway, the synthesized pdc and yqhD genes (Sangon Biotech Co Ltd., Shanghai) coupled with promoters (PpetE or PpsbA2s) and TrbcL terminator were designed in pBE02/pBE03 (see the target genes including other resistance genes in Supplementary Table S1). Plasmid pBE09 was constructed by inserting the PpsbA2s-maeB expression cassette (maeB gene cloned from E. coli) and TrbcL terminator into the pMD-slr1176-Ω vector.
2.3. Engineered cyanobacteria construction
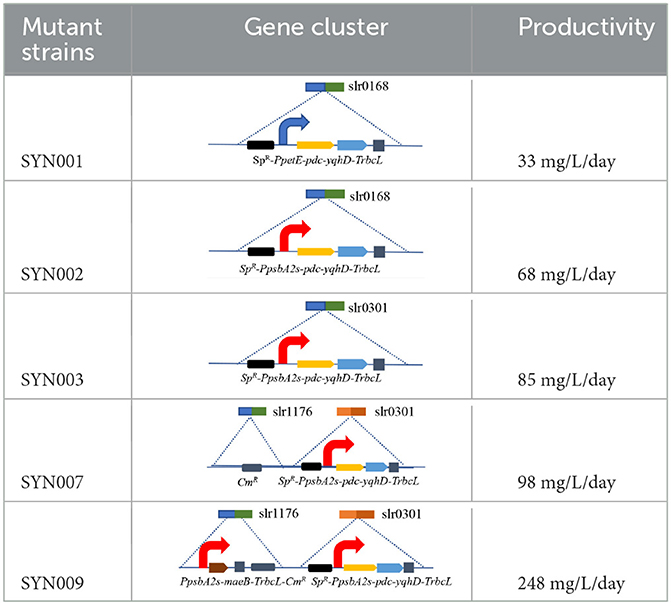
Synechocystis cells collected at the exponential phase (~1 OD730) were washed with a fresh BG11 medium three times, and then mixed with plasmids (100 ng DNA to 100 μl cyanobacteria) for 5 h and illuminated incubation at 30°C. The above mixture was streaked on a sterile filter membrane for another 24 h of illuminated incubation on the BG11 solid medium. To select the corrected mutant, the filter membrane was further transferred to a solid BG11 medium with corresponding antibiotics. After 2 weeks, single clones sub-cultured on solid plates were isolated in a liquid BG11 medium for analysis. All the strains referred to in this study are presented in Table 1.
The slr0301 gene is a gene encoding phosphoenolpyruvate synthase (PpsA) in the genome of Synechocystis sp. PCC6803, which catalyzes the conversion of pyruvate to phosphoenolpyruvate. The absence of this gene increases the accumulation of the intermediate pyruvate. The slr1176 gene is a gene encoding glucose-1-phosphate adenylate transferase in the genome of PCC6803, which catalyzes the conversion of glucose-1-phosphate (G1P) to ADP-glucose. It is a major rate-limiting enzyme in glycogen synthesis, and its absence can lead to complete inhibition of glycogen synthesis. The slr0168 is a neutral gene in PCC6803 algae cells, and knocking out this gene has no effect on the growth of algae cells, making it a commonly used expression platform.
2.4. Ethanol production and analytical methods
For ethanol production, all the mutants were cultured in a fresh BG11 medium with an initial 0.1 OD730 and cultivated photoautotrophically in a flask (50 μmol photons m−2 s−1 without additional CO2 injection). Notably, the BG11 medium of SYN001 contains 500 nM copper ions to induce the expression of ethanol-producing genes (Ghassemian et al., 1994; Choi and Park, 2016). After centrifugation and filtration, supernatant with ethanol was submitted for high-performance liquid chromatography (HPLC) analysis using an Aminex HPX-87H column (Bio-Rad, United States) (Seo et al., 2017).
2.5. Transcription level analysis
Synechocystis wild-type culture and mutants at 0.6 OD730 were collected after centrifugation (3,500 × g, 15 min, 4°C). RNA extraction and quantitative reverse transcription PCR (RT-qPCR) analysis were performed according to the previous methods (Gao et al., 2012). The relative transcription levels of targeted genes were estimated using the calculation method of 2−ΔΔCT, in which a higher ΔCT value means low transcription (Livak and Schmittgen, 2001). The endogenous 16S rRNA was set as a reference gene. All experimental groups were carried out with three biological replicates.
2.6. Statistical analysis
All statistical analyses were performed using GraphPad Prism (version 8.01, United States). The difference in this study was compared by unpaired t-test and statistical significance was set at p < 0.05. ** represents p < 0.01 and *** represents p < 0.001.
3. Results and discussion
3.1. Synthetic ethanol pathway optimization
Generally, there are two precursors (pyruvate and acetyl-CoA) that are involved in ethanol synthesis (Gao et al., 2012). To determine the optimal ethanol pathway in engineered Synechocystis, the concentrations of those two metabolites were investigated (Supplementary Figure S1). Interestingly, pyruvate linking to the CBB cycle and tricarboxylic acid (TCA) cycle exhibited higher concentration (1.05 μmol/gDW, approximately three times that of acetyl-CoA), indicating that pyruvate was more suitable to serve as the ethanol precursor. Therefore, we attempted to optimize the carbon flow network by selecting a strong pyruvate-acetaldehyde-ethanol pathway.
The synthetic ethanol pathway contained pyruvate decarboxylase (PDC) from Zymomonas mobilis and alcohol dehydrogenase (YqhD) from Escherichia coli (Supplementary Figure S2). Those two enzymes exhibited high activities to convert pyruvate into acetaldehyde and subsequently reduce acetaldehyde into ethanol in other microorganisms (Atsumi et al., 2009; Gao et al., 2012). The slr0168 gene not affecting cell growth or photosynthesis was primarily chosen as an exchange site according to previous reports (Dexter and Fu, 2009; Gao et al., 2012). Vectors containing upstream/downstream slr0168 gene, antibiotic resistance gene (spectinomycin, SpR), various promoters, target genes (pdc-yqhD), and terminator TrbcL were constructed and integrated into the Synechocystis genome to obtain stable ethanol-producing recombinants (Supplementary Figure S3). Those two strains (SYN001 and SYN002) showed similar growth rates to wild-type Synechocystis (Figure 1A).

Figure 1. Construction of synthetic ethanol pathway in cyanobacteria. (A) Cell growth curves of wild-type and recombinant cyanobacteria. (B) Transcription levels of pbd-yqhd in various Synechocystis recombinants. (C) Ethanol yields under the control of various promoters. An unpaired t-test (**p < 0.01, ***p < 0.001) was used.
Subsequently, promoter optimization to adjust the transcription level of pdc and yqhD was conducted. The RT-qPCR results showed that strong promoter PpsbA2s in recombinant SYN002 obviously improved pdc-yqhD transcription levels, ~2-fold compared to medium-level promoter PpetE in SYN001 (Figure 1B and Supplementary Figure S4). As expected, both recombinants achieved obvious accumulation of ethanol (rarely detected in wild-type Synechocystis), and SYN002 yielded the highest titer (474 mg/L) at 7 days (Figure 1C). Thus, a basic cyanobacterium with photosynthetic CO2-to-ethanol ability was obtained.
3.2. Effect of phosphoenolpyruvate synthase and glycogen synthesis knockout
Blocking carbon loss (phosphoenolpyruvate backflow and glycogen synthesis) was conducted to learn their effects on CO2-to-ethanol production (Figure 2A). From metabolic network analysis (Supplementary Figures S1, S2) and literature investigation (Angermayr et al., 2014; Dienst et al., 2014), we learned that native Synechocystis could remarkably turn pyruvate back into the upstream module via highly active phosphoenolpyruvate synthase (PpsA, referred to slr0301 gene). To abolish competitive consumption of pyruvate, we constructed a new cassette (pMD-PpsbA2s-pdc-yqhD) to exchange the slr0301 gene on the genome (Figure 2B). The newly obtained recombinant SYN003 (Δslr0301) exhibited 600 mg/L ethanol yield after 7 days of photosynthetic CO2 conversion (Figure 2C) and approximately 1.3-fold improvement compared to PpsA-existed SYN002. Furthermore, to enhance photosynthetic carbon flux toward the CO2-to-ethanol pathway, a key gene slr1176 related to glycogen synthesis was knocked out via gene exchange cassette (pMD-slr1176-Ω) (Figure 2B). Impressively, the ethanol yield was further improved via double-knockout recombinant SYN007 (Δslr0301 Δslr1176) and obtained more than 700 mg/L titer (Figure 2C). Pyruvate was deduced as a carbon sink in the Embden–Meyerhof–Parnas pathway according to the previous study of glycogen synthesis abolishment (Van Der Woude et al., 2014). The increased carbon flux of CO2-to-pyruvate probably supported the pyruvate-utilizing reaction of ethanol accumulation. Interestingly, it slightly inhibited cell growth when blocking glycogen synthesis at the slr1176 site (Figure 2C). We deduced that the shift of excessive carbon from the glycolytic pathway and pentose phosphate pathway to ethanol pathway resulted in carbon deficiency of biomass synthesis (Young et al., 2011).

Figure 2. Effects of blocking carbon loss on CO2-to-ethanol production. (A) Knockout sites on the pathway map. The gray font words of GlgC and PpsA are glycogen synthase and phosphoenolpyruvate synthase, respectively. (B) Heterologous genes exchange and cyanobacteria gene knockout schematics. (C) Cell density and ethanol yield of various Synechocystis recombinants under photosynthetic process. An unpaired t-test (**p < 0.01, ***p < 0.001) was used.
3.3. Effect of malic enzyme overexpression on ethanol production
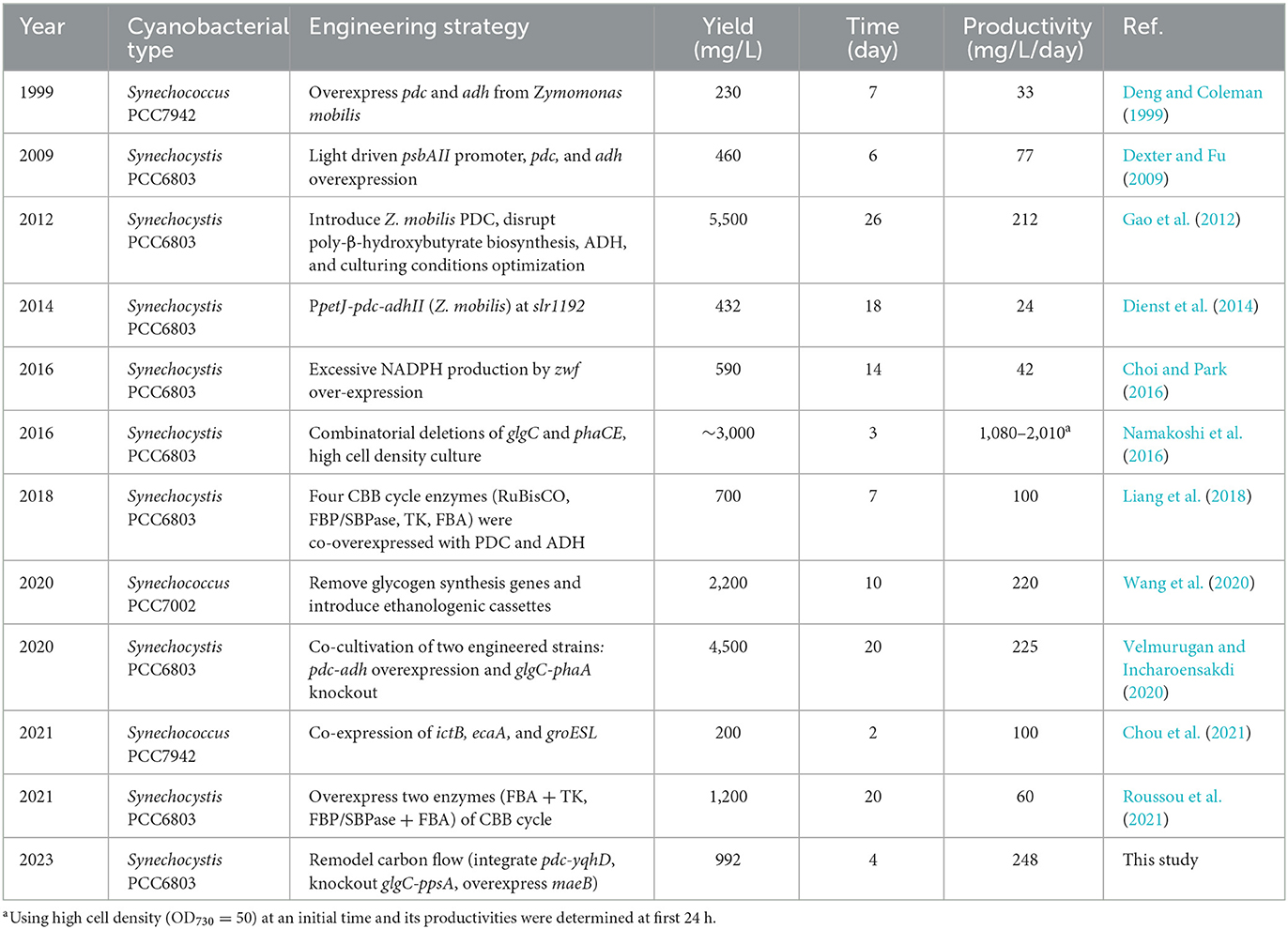
The engineered strains in cell proliferation stage should use the TCA cycle to support biomass synthesis, however, resulting in carbon atom loss (Zhang and Bryant, 2011). Thus, we designed a simple pathway to modify the TCA cycle by improving glyoxylate flux to reduce the carbon loss between isocitrate and succinate (Supplementary Figure S2). Malate close to the end of the TCA cycle was selected as the key metabolite for carbon recycling through an NADP+-dependent malic enzyme from E. coli (Yoshikawa et al., 2015), which not only converted malate into pyruvate but also increased pyruvate and NADPH pool (Figure 3A). The maeB gene was introduced into the slr1176 site of SYN003, establishing the recombinant SYN009 (Figure 3B). Intriguingly, the cell growth of Synechocystis was rescued, and the SYN009 showed a little fast proliferation after 3 days of photoautotrophic growth (Supplementary Figure S6). Under simulated sunlight source, SYN009 used CO2 as a sole carbon source to produce 1.09 g/L ethanol (Figure 3C and Supplementary Figure S5). Impressively, the time curve showed that before 4 days, SYN009 achieved 248 mg/L/day productivity, the fastest accumulation rate compared to other literature studies (Table 2).

Figure 3. Effects of malic enzyme overexpression on ethanol production. (A) Pathway construction schematics. (B) The gene exchange of cassette of new recombinant. (C) The cell growth and photosynthetic ethanol production time curve using atmospheric CO2.
The metabolic flux imbalance between metabolism and synthesis is a big challenge limiting target product yield in microbial cell factories (Oliver et al., 2013). Choosing suitable promoters to overexpress PDC/YqhD has been usually considered to enhance the carbon flux toward ethanol (Dexter and Fu, 2009; Gao et al., 2012), which also exhibited positive results of ethanol production in this study (Supplementary Figure S5). In addition, the overexpression of key enzymes in the CBB cycle was another important strategy to supply sufficient carbon flux in the form of 3-phosphoglycerate (Liang et al., 2018; Roussou et al., 2021). Compared to previous reports (Table 2), the carbon flow optimization strategy in our study consumed the minimum number of days to achieve the highest ethanol productivity of ~248 mg/L/day (Table 1). It indicated that a comprehensive and precise adjustment of carbon flow is promising to improve CO2-to-ethanol production in cyanobacteria. Notably, this photosynthetic cell factory still faces the challenge of cell density (Supplementary Figure S6), such as only ~0.6 OD730 increase after 7 days of cultivation under light and atmospheric CO2. New strategies, such as co-cultivation and batch culture with high density, can probably yield outstanding ethanol production (Namakoshi et al., 2016; Velmurugan and Incharoensakdi, 2020).
4. Conclusion
We developed a cyanobacterial platform that was entitled to convert atmospheric CO2 into ethanol at high efficiency via stepwise optimization of carbon flow. It showed that carbon flow rewiring strategies, such as integrating strong pyruvate-acetaldehyde-ethanol pathway, blocking carbon loss via inhibition of PEP synthase activity and glycogen synthesis, and recycling carbon atoms via overexpression of exogenous malic enzyme, were beneficial to ethanol synthesis. This study provides a proof-of-concept to create a photosynthetic cell factory that could be further remodeled and optimized for higher CO2-to-biofuel production.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
E-BG conceived the original idea, carried out the experiment, and wrote the manuscript with input from all authors. ZF and HQ interpreted the results contributed to the final version of the manuscript. JW and HC aided in interpreting the results and worked on the manuscript. ZF and PY contributed to the analysis of the results, the writing of the manuscript, and supervised the project. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31200019), Ningbo Clinical Research Center for Children's Health and Diseases (2019A21002), Ningbo Top Medical and Health Research Program (No. 2022020405), Project of Faculty of Agricultural Equipment of Jiangsu University (NZXB20210203), and Young Talents Cultivation Program of Jiangsu University.
Acknowledgments
We thank Degang Ning (Institute of Hydrobiology, Chinese Academy of Sciences) for the Synechocystis sp. PCC6803 wild-type strain and his technical expertise.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1211004/full#supplementary-material
References
Angermayr, S. A., Van Der Woude, A. D., Correddu, D., Vreugdenhil, A., Verrone, V., Hellingwerf, K. J., et al. (2014). Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp PCC6803. Biotechnol. Biofuels 7, 99. doi: 10.1186/1754-6834-7-99
Atsumi, S., Higashide, W., and Liao, J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180. doi: 10.1038/nbt.1586
Chen, H., Huang, Y., Sha, C., Moradian, J. M., Yong, Y. C., Fang, Z., et al. (2023a). Enzymatic carbon dioxide to formate: mechanisms, challenges and opportunities. Ren. Sustain. Energy Rev. 178, 113271. doi: 10.1016/j.rser.2023.113271
Chen, H., Li, J. W., Fan, Q. C., Zheng, T., Zhang, Y. F., Yong, Y. C., et al. (2023b). A feasible strategy for microbial electrocatalytic CO2 reduction via whole-cell-packed and exogenous-mediator-free rGO/Shewanella biohydrogel. Chem. Eng. J. 460, 141863. doi: 10.1016/j.cej.2023.141863
Choi, Y. N., and Park, J. M. (2016). Enhancing biomass and ethanol production by increasing NADPH production in Synechocystis sp. PCC 6803. Bioresour. Technol. 213, 54–57. doi: 10.1016/j.biortech.2016.02.056
Chou, H. H., Su, H. Y., Chow, T. J., Lee, T. M., Cheng, W. H., Chang, J. S., et al. (2021). Engineering cyanobacteria with enhanced growth in simulated flue gases for high-yield bioethanol production. Biochem. Eng. J. 165, 107823. doi: 10.1016/j.bej.2020.107823
Deng, M. D., and Coleman, J. R. (1999). Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65, 523–528. doi: 10.1128/AEM.65.2.523-528.1999
Dexter, J., and Fu, P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2, 857–864. doi: 10.1039/b811937f
Dienst, D., Georg, J., Abts, T., Jakorew, L., Kuchmina, E., Borner, T., et al. (2014). Transcriptomic response to prolonged ethanol production in the cyanobacterium Synechocystis sp PCC6803. Biotechnol. Biofuels 7, 21. doi: 10.1186/1754-6834-7-21
Fang, Z., Tang, Y. J. J., and Koffas, M. A. G. (2022). Harnessing electrical-to-biochemical conversion for microbial synthesis. Curr. Opin. Biotechnol. 75, 102687. doi: 10.1016/j.copbio.2022.102687
Fang, Z., Zhou, J., Zhou, X., and Koffas, M. a. G. (2021). Abiotic-biotic hybrid for CO2 biomethanation: from electrochemical to photochemical process. Sci. Total Environ. 791, 148288. doi: 10.1016/j.scitotenv.2021.148288
Gao, E. B., Kyere-Yeboah, K., Wu, J. H., and Qiu, H. Y. (2021). Photoautotrophic production of p-Coumaric acid using genetically engineered Synechocystis sp. Pasteur Culture Collection 6803. Algal Res. 54, 102180. doi: 10.1016/j.algal.2020.102180
Gao, Z., Zhao, H., Li, Z., Tan, X., and Lu, X. (2012). Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 5, 9857–9865. doi: 10.1039/C2EE22675H
Gassler, T., Sauer, M., Gasser, B., Egermeier, M., Troyer, C., Causon, T., et al. (2020). The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38, 210–216. doi: 10.1038/s41587-019-0363-0
Ghassemian, M., Wong, B., Ferreira, F., Markley, J. L., and Straus, N. A. (1994). Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120. Microbiology 140, 1151–1159. doi: 10.1099/13500872-140-5-1151
Heidorn, T., Camsund, D., Huang, H. H., Lindberg, P., Oliveira, P., Stensjo, K., et al. (2011). Synthetic biology in cyanobacteria engineering and analyzing novel functions. Meth. Enzymol. 497, 539–579. doi: 10.1016/B978-0-12-385075-1.00024-X
Lee, H. J., Son, J., Sim, S. J., and Woo, H. M. (2020). Metabolic rewiring of synthetic pyruvate dehydrogenase bypasses for acetone production in cyanobacteria. Plant Biotechnol. J. 18, 1860–1868. doi: 10.1111/pbi.13342
Li, Z., Wu, C., Gao, X., Addison, B., Shinde, S., Wang, X., et al. (2021). Exogenous electricity flowing through cyanobacterial photosystem I drives CO2 valorization with high energy efficiency. Energy Environ. Sci. 14, 5480–5490. doi: 10.1039/D1EE01526E
Liang, F., Englund, E., Lindberg, P., and Lindblad, P. (2018). Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio. Metab. Eng. 46, 51–59. doi: 10.1016/j.ymben.2018.02.006
Lindberg, P., Park, S., and Melis, A. (2010). Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79. doi: 10.1016/j.ymben.2009.10.001
Liu, X., Miao, R., Lindberg, P., and Lindblad, P. (2019). Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO2 in cyanobacteria. Energy Environ. Sci. 12, 2765–2777. doi: 10.1039/C9EE01214A
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, G., Zhang, S., and Lu, X. (2020). Engineering cyanobacteria chassis cells toward more efficient photosynthesis. Curr. Opin. Biotechnol. 62, 1–6. doi: 10.1016/j.copbio.2019.07.004
Miao, R., Liu, X., Englund, E., Lindberg, P., and Lindblad, P. (2017). Isobutanol production in Synechocystis PCC 6803 using heterologous and endogenous alcohol dehydrogenases. Metab. Engin. Commun,. 5, 45–53. doi: 10.1016/j.meteno.2017.07.003
Namakoshi, K., Nakajima, T., Yoshikawa, K., Toya, Y., and Shimizu, H. (2016). Combinatorial deletions of glgC and phaCE enhance ethanol production in Synechocystis sp. PCC 6803. J. Biotechnol. 239, 13–19. doi: 10.1016/j.jbiotec.2016.09.016
Oliver, J. W., Machado, I. M., Yoneda, H., and Atsumi, S. (2013). Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. USA. 110, 1249–1254. doi: 10.1073/pnas.1213024110
Roussou, S., Albergati, A., Liang, F., and Lindblad, P. (2021). Engineered cyanobacteria with additional overexpression of selected Calvin-Benson-Bassham enzymes show further increased ethanol production. Metabolic engineering communications, 12, e00161–e00161. doi: 10.1016/j.mec.2021.e00161
Santos-Merino, M., Torrado, A., Davis, G. A., Rottig, A., Bibby, T. S., Kramer, D. M., et al. (2021). Improved photosynthetic capacity and photosystem I oxidation via heterologous metabolism engineering in cyanobacteria. Proc. Natl. Acad. Sci. USA. 118, e2021523118. doi: 10.1073/pnas.2021523118
Satanowski, A., and Bar-Even, A. (2020). A one-carbon path for fixing CO2. EMBO Rep. 21, e50273. doi: 10.15252/embr.202050273
Seo, S. O., Wang, Y., Lu, T., Jin, Y. S., and Blaschek, H. P. (2017). Characterization of a Clostridium beijerinckii spo0A mutant and its application for butyl butyrate production. Biotechnol. Bioeng. 114, 106–112. doi: 10.1002/bit.26057
Van Der Woude, A. D., Angermayr, S. A., Puthan Veetil, V., Osnato, A., and Hellingwerf, K. J. (2014). Carbon sink removal: increased photosynthetic production of lactic acid by Synechocystis sp. PCC6803 in a glycogen storage mutant. J. Biotechnol. 184, 100–102. doi: 10.1016/j.jbiotec.2014.04.029
Velmurugan, R., and Incharoensakdi, A. (2020). Co-cultivation of two engineered strains of Synechocystis sp. PCC 6803 results in improved bioethanol production. Ren. Energy 46, 1124–1133. doi: 10.1016/j.renene.2019.07.025
Wang, M., Luan, G., and Lu, X. (2020). Engineering ethanol production in a marine cyanobacterium Synechococcus sp. PCC7002 through simultaneously removing glycogen synthesis genes and introducing ethanolgenic cassettes. J. Biotechnol. 317, 1–4. doi: 10.1016/j.jbiotec.2020.04.002
Yoshikawa, K., Hirasawa, T., and Shimizu, H. (2015). Effect of malic enzyme on ethanol production by Synechocystis sp. PCC 6803. J. Biosci. Bioeng. 119, 82–84. doi: 10.1016/j.jbiosc.2014.06.001
Young, J. D., Shastri, A. A., Stephanopoulos, G., and Morgan, J. A. (2011). Mapping photoautotrophic metabolism with isotopically non-stationary 13C flux analysis. Metab. Eng. 13, 656–665. doi: 10.1016/j.ymben.2011.08.002
Keywords: cyanobacteria, metabolic engineering, cofactor regeneration, CO2 fixation, photosynthetic cell factory
Citation: Gao E-B, Wu J, Ye P, Qiu H, Chen H and Fang Z (2023) Rewiring carbon flow in Synechocystis PCC 6803 for a high rate of CO2-to-ethanol under an atmospheric environment. Front. Microbiol. 14:1211004. doi: 10.3389/fmicb.2023.1211004
Received: 24 April 2023; Accepted: 09 May 2023;
Published: 31 May 2023.
Edited by:
Yong Jiang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Shrameeta Shinde, Miami University, United StatesYue Yi, Beijing Institute of Technology, China
Copyright © 2023 Gao, Wu, Ye, Qiu, Chen and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhua Wu, wudata@163.com; Zhen Fang, zhenfang@ujs.edu.cn
 E-Bin Gao1,2
E-Bin Gao1,2 Junhua Wu
Junhua Wu Huayou Chen
Huayou Chen Zhen Fang
Zhen Fang