- 1Key Laboratory of Marine Mineral Resources and Polar Geology, Ministry of Education, China University of Geosciences, Beijing, China
- 2State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Beijing, China
- 3School of Ocean Sciences, China University of Geosciences, Beijing, China
- 4Oil and Gas Survey, China Geological Survey, Beijing, China
Wetlands are an important source of atmospheric methane (CH4) and are sensitive to global climate change. Alpine swamp meadows, accounting for ~50% of the natural wetlands on the Qinghai-Tibet Plateau, were considered one of the most important ecosystems. Methanogens are important functional microbes that perform the methane producing process. However, the response of methanogenic community and the main pathways of CH4 production to temperature rise remains unknown in alpine swamp meadow at different water level in permafrost wetlands. In this study, we investigated the response of soil CH4 production and the shift of methanogenic community to temperature rise in the alpine swamp meadow soil samples with different water levels collected from the Qinghai-Tibet Plateau through anaerobic incubation at 5°C, 15°C and 25°C. The results showed that the CH4 contents increased with increasing incubation temperature, and were 5–10 times higher at the high water level sites (GHM1 and GHM2) than that at the low water level site (GHM3). For the high water level sites (GHM1 and GHM2), the change of incubation temperatures had little effect on the methanogenic community structure. Methanotrichaceae (32.44–65.46%), Methanobacteriaceae (19.30–58.86%) and Methanosarcinaceae (3.22–21.24%) were the dominant methanogen groups, with the abundance of Methanotrichaceae and Methanosarcinaceae having a significant positive correlation with CH4 production (p < 0.01). For the low water level site (GHM3), the methanogenic community structure changed greatly at 25°C. The Methanobacteriaceae (59.65–77.33%) was the dominant methanogen group at 5°C and 15°C; In contrast, the Methanosarcinaceae (69.29%) dominated at 25°C, and its abundance showed a significant positive correlation with CH4 production (p < 0.05). Collectively, these findings enhance the understanding of methanogenic community structures and CH4 production in permafrost wetlands with different water levels during the warming process.
Introduction
Methane (CH4) is a greenhouse gas that has a significant impact on global climate change (Caldwell et al., 2008). Although CH4 only accounts for 1.8 mL/m3 in the atmosphere, its greenhouse effect is about 26 times that of CO2, contributing 15% to global warming (Dlugokencky et al., 2009; Wang et al., 2014). Wetlands are important ecosystems, and have a high potential to affect the climate system (Kadykalo and Findlay, 2016; Li T. T. et al., 2022). Wetland CH4 emissions are the largest natural source in the global CH4 budget, and they may play an increasingly important role in atmospheric CH4 growth in the future (Zhang Z. et al., 2017). Permafrost is quite vulnerable to temperature rise, and is undergoing significant degradation due to climate warming (Dlugokencky et al., 2009; Wang et al., 2014). Therefore, the CH4 emission processes are strongly associated with climate warming and carbon cycling in permafrost wetlands.
Methanogens are the major communities involved in CH4 production. The process of methanogenesis is the terminal step in the decomposition of organic matter by microorganisms in anaerobic environments (Thauer, 1998). Biogenic CH4 accounts for 74% of all global CH4 emissions (Lowe, 2006). CH4 is usually produced by acetate- and H2/CO2-dependent methanogenesis (Cui et al., 2015). Permafrost wetland soils usually contain highly diverse methanogens such as the potentially acetoclastic families Methanosarcinaceae and Methanotrichaceae (formerly Methanosaetaceae) as well as the hydrogenotrophic orders Methanomicrobiales, Methanobacteriales and Methanocellales (formerly Rice cluster I; Cui et al., 2018; Wei et al., 2018; Liu et al., 2019). Methanosarcinales and Methanomicrobiales are the dominant methanogens in the Zoige wetlands on the Tibetan Plateau, where the acetoclastic pathway is the dominant methanogenic pathway of CH4 emissions (Zhang et al., 2008; Cui et al., 2015). Meanwhile, Methanocellales and Methanomassiliicoccales were found in the alpine wetlands of the permafrost region on the Qilian Mountain (Cui et al., 2018; Wei et al., 2018). Moreover, the amount of CH4 generated by H2/CO2 in peat accounts for 50–100% of the total CH4 output (Fey et al., 2001, 2004). Many factors will affect CH4 emissions from wetlands, such as temperature, water conditions and vegetation biomass (Huttunen et al., 2003; Lowe, 2006; Zhang et al., 2019; Li W. et al., 2022; Wu et al., 2022). Previous studies have shown that CH4 emissions from wetlands are more temperature dependent, and that CH4 emissions fluxes are higher at higher temperature (Høj et al., 2008; Cui et al., 2015; Liu et al., 2019). However, soil moisture can affect the availability of oxygen, the diffusion rate of gas, and the activity of microorganisms, so that water-saturated soils tended to release CH4 (Christensen et al., 1997; Li W. et al., 2022). Thus, CH4 production potential varied significantly under different water levels (Kettunen et al., 1999; Huttunen et al., 2003; Wu et al., 2021).
Qinghai-Tibet Plateau is the largest and highest plateau on Earth, with widely distributed permafrost wetlands and alpine swamp meadows, and has been a methane source (Bubier et al., 1995; Chen et al., 2022). Permafrost wetlands play a vital role in terrestrial carbon storage on the Qinghai-Tibet Plateau (Fey et al., 2001; Li X. Q. et al., 2022), and wetlands have the highest CH4 emission rates (0.7 mg CH4-C m−2 h−1) due to the water-logged conditions (Chen et al., 2022). Alpine swamp meadows, with 4.9 × 104 km2 (~50%) of the natural wetlands on the Qinghai-Tibet Plateau, are typically soil-nutrient-rich and water-logged, and were considered one of the most important ecosystems on the Qinghai-Tibet Plateau (Zhao et al., 2005; Bai et al., 2019; Wei et al., 2022). Previous studies investigated the diversity of methanogens in the permafrost wetland soil samples from the Qinghai-Tibet Plateau or their community changes in response to temperature rise based on gene clone libraries (Cui et al., 2018; Wei et al., 2018). However, there are few incubation studies on the influence of warmer temperatures on soil CH4 emissions and microbial communities at the different water conditions in the alpine swamp meadow wetlands on the Qinghai-Tibet Plateau, and the main pathways of CH4 production and the influencing factors remain unclear. Therefore, in this study, continuous anaerobic incubations at 5°C, 15°C and 25°C temperatures were conducted on soil samples with various water levels in this area by means of simulation experiments. The composition and abundance of methanogenic communities in the incubated wetland soils were analyzed by high-throughput gene sequencing and quantitative polymerase chain reaction (qPCR), and the effects of the dominant methanogenic community on CH4 production were further investigated.
Materials and methods
Site description
Our experimental sites are located in the permafrost wetlands of gas hydrate area of Qilian Mountain on the northeastern edge of the Qinghai-Tibet Plateau in southwestern China (Li X. Q. et al., 2022) (Supplementary Figure S1), belonging to the natural ecosystem of alpine swamp meadow (Supplementary Figure S2A). Carbon isotope was applied to show that δ13CH4 in the surface soil of this area ranges from −84.11‰ to −39.81‰, indicating that CH4 is produced by microorganisms (Zhang F. G. et al., 2017). From June to August, surface soil temperatures in this area are between 4.7°C to 21.0°C. Precipitation and glacial melting in this area mainly occurs in summer, forming a natural wetland ecosystem of alpine swamp meadow (Wei et al., 2018). Soil samples from the wetland (collected in June 2017) were provided by the Oil and Gas Survey, China Geological Survey.
Field measurement and sample collection
The atmospheric and the soil surface temperatures were 6.2°C and 5.5°C, respectively, with 4,060 m as the altitude of the sampling points. According to the differences of water level (the depth of the aquifer), three sampling sites were collected and labeled with GHM1 (Supplementary Figure S2B), GHM2 (Supplementary Figure S2C) and GHM3 (Supplementary Figure S2D). Samples were collected by digging into a depth of 0 to 10 cm below the water level. The soil samples were placed into 50 mL centrifuge tubes and sterilized plastic bags. The samples were transported in a box with ice and then stored at 4°C until further use for the experiments (Jiang et al., 2010; Wagner et al., 2013).
Establishment of incubation experiments
The incubation procedure was adopted from previous studies (Keller et al., 2004; Cui et al., 2015). The soil samples were mixed with anoxic sterile water in a 1:1 (v/v) ratio and then homogenized well. Approximately 20 mL of the mixture was placed in 100 mL sterile test bottles. The bottles were sealed with butyl rubber stoppers and aluminum caps, and flushed with N2 to remove the headspace O2. According to the change of surface soil temperatures, three incubation temperatures were set at 5°C, 15°C and 25°C and three parallel slurries were made for each temperature. The bottles were continuously incubated in the dark for 12 weeks, and CH4 concentrations measured at weeks 1, 2, 4, 6, 8, 10, and 12.
Geochemistry analysis
The coordinates of the sampling points, altitude, and the depth of water level were measured during the samples collecting. Total organic carbon (TOC), pH values, and moisture were measured by the method adopted from a previous study (Cui et al., 2018), and soil bulk densities were determined using method adopted from Juottonen et al. (2012). The concentrations of CH4 in the headspace was measured by gas chromatography using the GC 7890A (Agilent Technologies, United States; Cui et al., 2018).
DNA extraction
Total genomic DNA was extracted using the FastDNA SPIN kit (MP Biomedicals, United States) for soil by following the manufacturer’s instructions. The DNA extracted from three parallel samples was mixed in equal amounts to form the DNA of each sample.
QPCR of mcrA gene
The abundance of mcrA gene was determined using an ABI Prism 7,500 qPCR system (Applied Biosystems, United States). The plasmid of methanogen was used as the template DNA to calculate the number of copies; 10 times dilution was carried out to form a standard solution with six concentration gradients of 104 to 109 copies/μL, which served as a standard curve for gene fluorescence quantitative analysis (R2 ≥ 0.99). The qPCR primers were ME3MF and ME2r (Nunoura et al., 2008). The total volume of qPCR reaction was 20 μL containing 10 μL of SYBR Premix Ex Taq (TaKaRa, Japan), 10 ng template DNA, 10 pM each of the forward and reverse primers, 10 pM ROX Reference Dye II and the appropriate volume of ddH2O. The qPCR amplification consisted of 1 cycle of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 34 s (Cui et al., 2018).
PCR amplification and Illumina sequencing
mcrA gene of methanogens were PCR amplified using the primers mcrAF and mcrAR from the DNA incubation samples at week 0 (original soils) and week 12 (Luton et al., 2002). The total volume of PCR reaction was 50 μL, including 5 μL Taq buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 0.25 μM of each primer, 5 U Taq DNA polymerase (Invitrogen, United States) and 20 ng template DNA. PCR amplification conditions: 4 min at 95°C, 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, followed by a final extension step of 10 min at 72°C (Cui et al., 2018; Wei et al., 2018). The mcrA gene amplicons were visualized on 2% agarose gels followed by cutting the gel and purifying it with the AxyPrep DNA Gel Extraction Kit according to the manufacturer’s instructions (Axygen Biosciences, USA). We performed mcrA genes sequencing analysis at week 0 and week 12.
Statistical analysis
The sequencing was carried out on an Illumina Miseq PE300 platform (Shanghai, China). mcrA gene reads were processed using QIIME 2 package (Callahan et al., 2016). OTUs (Operational Taxonomic Units) were defined at the cutoff value of 14.3% for the mcrA gene reads (Barbier et al., 2012; Wei et al., 2018). The analysis process of sequencing data was adopted from Cui et al. (2019). The mcrA OTU data were searched against the Functional Gene Pipeline/Repository database, and sequences were verified in the National Center for Biotechnology Information (NCBI) non-redundant database. Principal Component Analysis (PCA) was used to explain the results by using CANACO for Windows 4.5. The Pearson correlation of methanogenic community compositions and CH4 concentrations was performed using SPSS 22. Program Mothur was used to calculate the Chao 1, Shannon and Simpson diversity indices.
The estimated absolute abundance (EAA) was defined as the product of relative abundance and total number of microbial cells (Dannemiller et al., 2014; Zhang Z. J. et al., 2017). The relative abundance of methanogens was obtained by Illumina sequencing of mcrA gene, while the total number of methanogen cells was quantified by quantitative polymerase chain reaction (qPCR).
Nucleotide sequence accession numbers
The raw reads have been deposited in the NCBI Sequence Read Archive database under accession number of SRP166224.
Results
Soil characteristics of different sampling sites
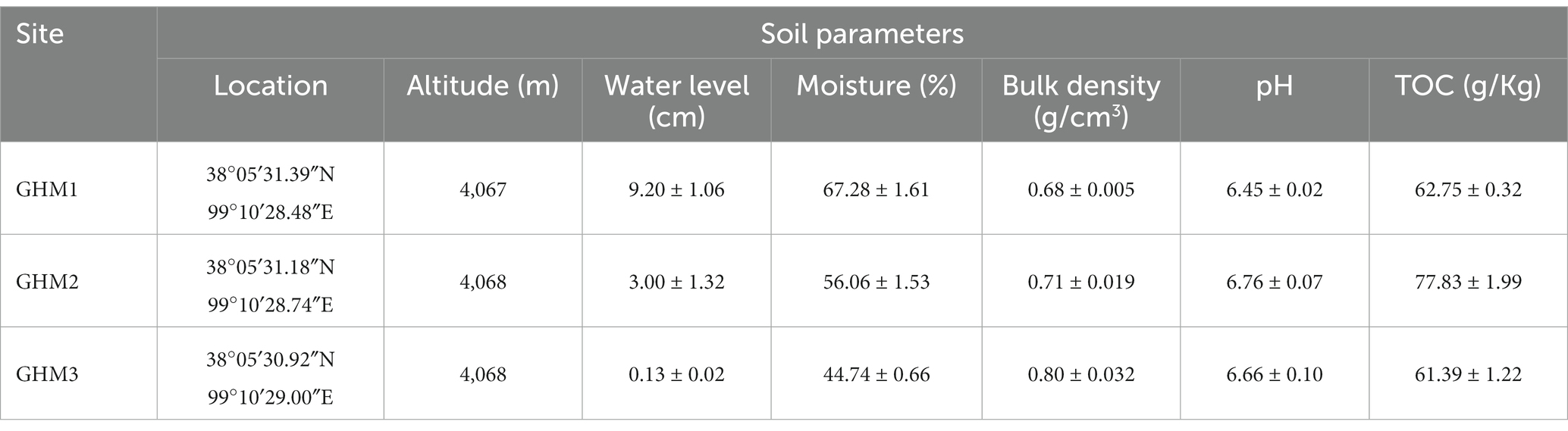
The physical and chemical characteristics of three sampling sites are shown in Table 1. The three sampling sites had the same altitude. Water level and moisture are higher at site GHM1 (9.20 ± 1.06 cm and 67.28 ± 1.61%), and are lower at site GHM3 (0.13 ± 0.02 cm and 44.74 ± 0.66%). The pH of the soil samples ranged from 6.45 ± 0.02 to 6.76 ± 0.07, belonging to slightly acid soil; the pH of GHM1 was the lowest among the three sampling sites. There was no significant difference in the TOC content of the soil from the three sampling sites.
Change regulation of CH4 production
The CH4 accumulation of the three sites increased from 0.01 mmol to 0.31 mmol with the higher incubation temperature (Figure 1). In general, time-course accumulation of CH4 was divided into two phases (the first 6 weeks, and the second 6–12 weeks). CH4 production was higher in the first phase than that in the second phase, and the concentrations of CH4 remained stable. Concentrations of CH4 in the GHM1 treatments at 5°C, 15°C and 25°C peaked at 0.05 mmol, 0.25 mmol and 0.31 mmol, respectively (Figure 1A). Concentrations of CH4 in the GHM2 treatment at 5°C, 15°C, and 25°C peaked at 0.06 mmol, 0.16 mmol and 0.25 mmol, respectively (Figure 1B). Concentrations of CH4 in the GHM3 treatments at 5°C, 15°C, and 25°C peaked at 0.06 mmol, 0.08 mmol and 0.15 mmol, respectively (Figure 1C).
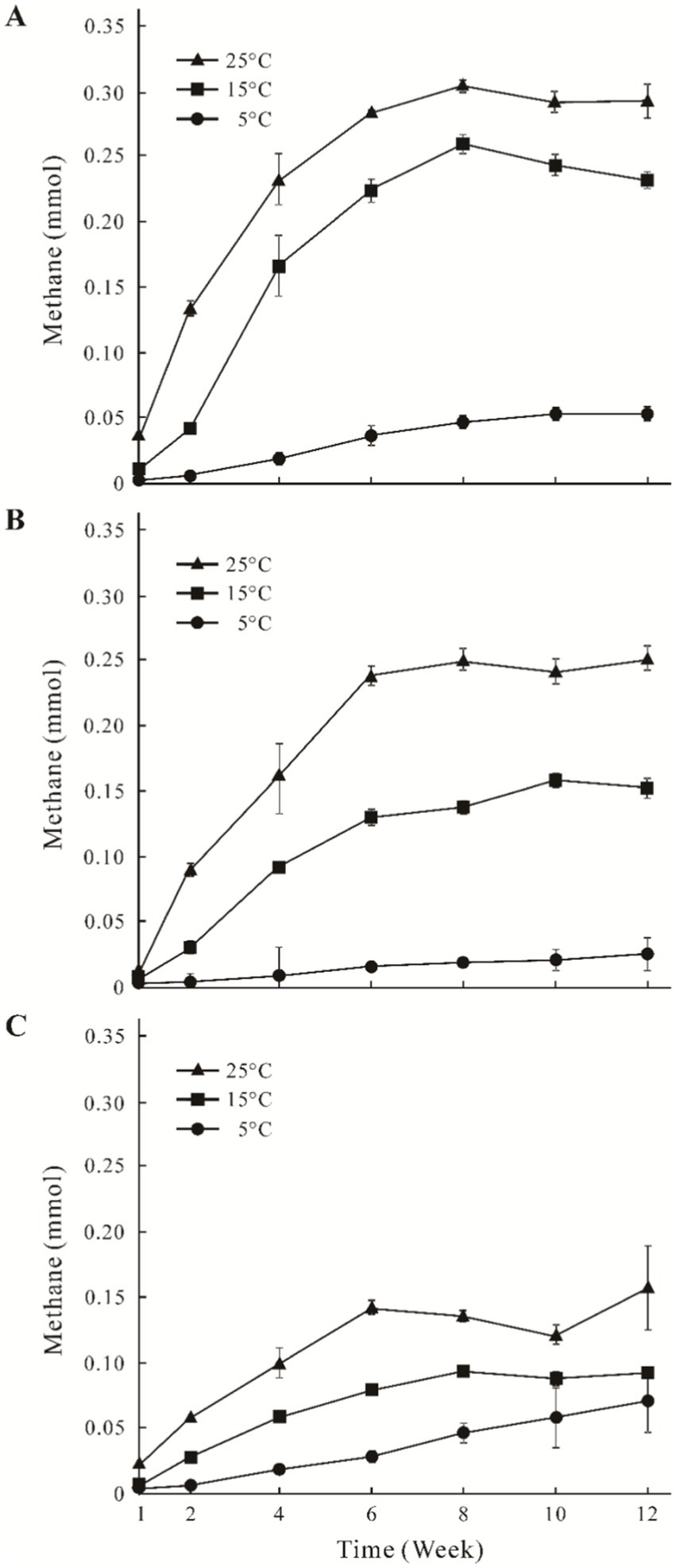
Figure 1. Time course accumulation of CH4 in the incubation microcosms at different temperatures using alpine swamp meadow samples. Panels (A–C) indicate CH4 accumulation of sample GHM1, GHM2, and GHM3, respectively.
Abundance of mcrA gene
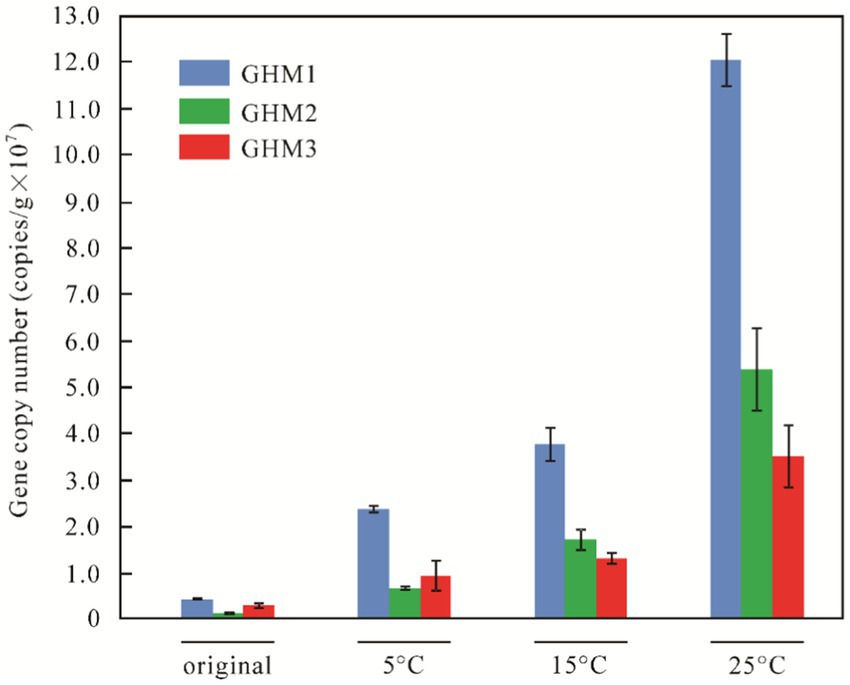
Results of qPCR revealed that the abundance of mcrA genes in three original soil samples was 3.85 ± 0.23 × 106 copies/g soil, 1.06 ± 0.29 × 106 copies/g soil, and 2.53 ± 0.52 × 106 copies/g soil, respectively. The abundance of mcrA genes changed significantly with increasing incubation temperatures (Figure 2). With the production of biogenic CH4 under incubation conditions, the abundance of mcrA gene copies of GHM1 was 6–31 times that of the original sample, while it was 5–48 times for GHM2 and 3–13 times for GHM3. The abundance of mcrA genes in GHM2 increased the most at 25°C, reaching 5.20 ± 0.87 × 107 copies/g soil. The mcrA gene abundance in GHM3 was the least at 5°C, only 3.5 times that of the original sample. The GHM1 sample had the highest abundance of mcrA gene at 25°C, reaching 1.21 ± 0.06 × 108 copies/g soil.
Sequencing and alpha (α) diversity indices
We conducted Illumina sequencing analysis of the mcrA gene of 12 samples from three types of incubations. The number of clean reads was 447,148 with 37,262 average number of reads and an average length of 421 bp. The number of OTUs of the 12 samples ranged between 245 to 377, and the coverage was nearly 0.99. The Chao 1, Simpson, and Shannon diversity indices varied 712.29–1778.98, 0.04–0.13, and 3.27–4.48, respectively (Table 2). The temperature rise had less influence on the diversity of the methanogens of GHM1 and GHM2. However, the diversity of methanogens of GHM3 decreased at 25°C compared to the other two incubation temperatures.
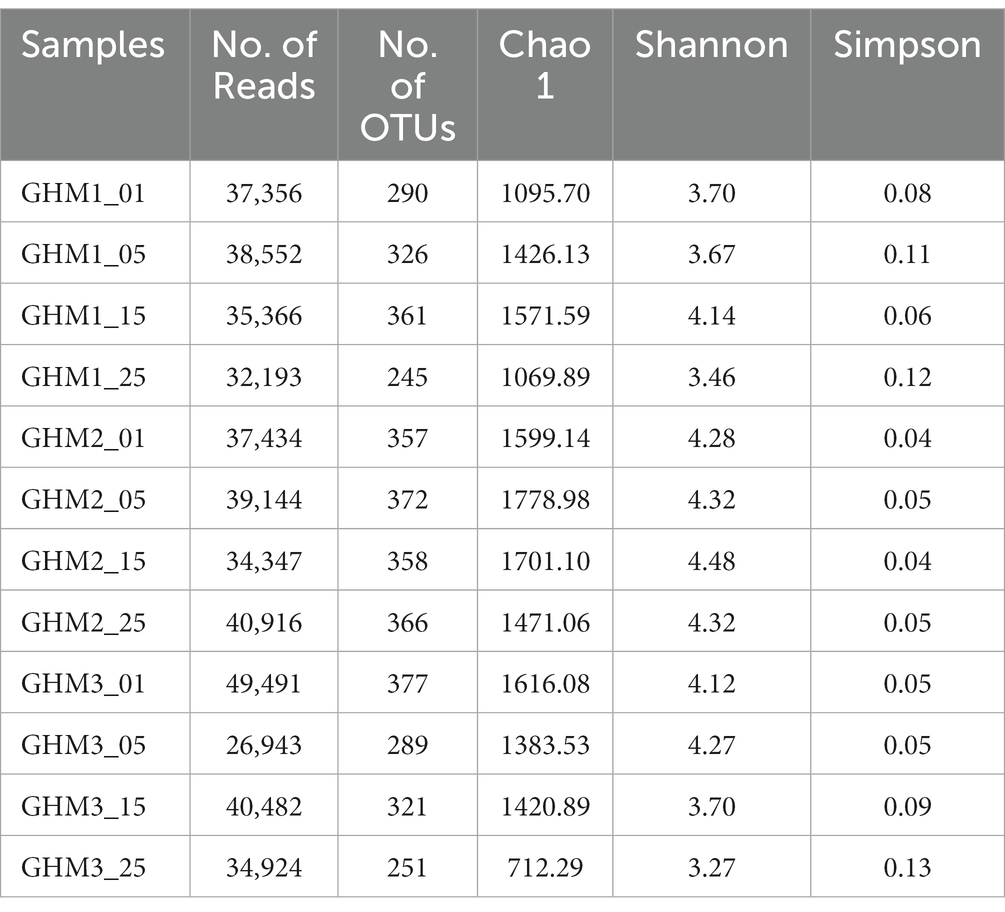
Table 2. Sequencing information and statistical analyses of mcrA gene sequencing of the studied samples.
Methanogen diversity based on mcrA gene
The dominant methanogen group of the GHM1 sample in the original soil was Methanotrichaceae, which accounted for 59.81% of the total sequences, followed by Methanobacteriaceae (20.95%), ZC-I cluster (10.44%), and Methanosarcinaceae (4.99%; Figure 3). The Methanotrichaceae (32.44–65.46%), Methanobacteriaceae (22.05–58.86%) and Methanosarcinaceae (3.22–9.00%) were the dominant groups at the incubation temperatures. With increasing temperature, the relative abundance of Fen cluster increased at 25°C (11.39%). For sample GHM2, Methanotrichaceae (28.64%), Methanobacteriaceae (44.41%), and Methanosarcinaceae (18.58%) were the dominant methanogen groups, followed by Fen cluster (5.94%) in the original soil. The relative abundance of Methanobacteriaceae, Methanosarcinaceae and ZC-I cluster decreased with the increase of incubation temperature, ranging 19.30–23.51%, 8.93–21.24% and 0.93–7.91% in the incubations at 5°C, 15°C and 25°C, respectively. Methanotrichaceae and Methanocellales increased gradually from 33.90 to 45.68%, and from 3.33 to 12.87%, respectively. For sample GHM3, Methanotrichaceae (25.82%), Methanobacteriaceae (40.22%), and Methanosarcinaceae (24.70%) were the dominant methanogen groups, followed by Fen cluster (6.41%) in the original soil. The relative abundance of Methanobacteriaceae increased from 59.65 to 77.33%, while that of Methanotrichaceae and Methanosarcinaceae decreased at 5°C and 15°C from 13.24 to 6.60% and from 12.38 to 9.57%, respectively. Methanosarcinaceae became the dominant methanogen group at 25°C, accounting for 69.29% of the total sequences.
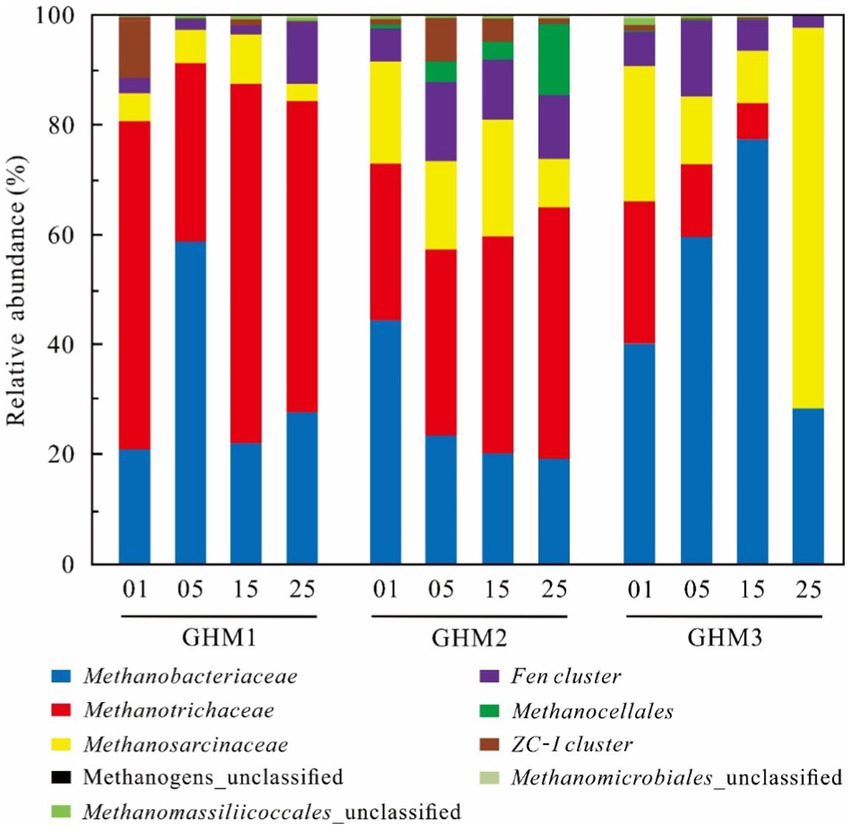
Figure 3. Methanogenic community composition in the studied samples at different incubation temperature. “01” indicates the original soils; “05,” “15,” and “25” indicates the samples incubated at 5°C, 15°C, and 25°C, respectively.
PCA analysis detected differences in the methanogenic community compositional structure in the 12 samples (Figure 4A). After Incubation, the community structure of the methanogens in the GHM1 and GHM2 samples changed slightly, while that of the methanogens in the GHM3 samples changed greatly. Cluster analysis further confirmed that the GHM3 samples incubated at 25°C had lower similarity (40%) than the other samples in terms of community composition of methanogens (Figure 4B).
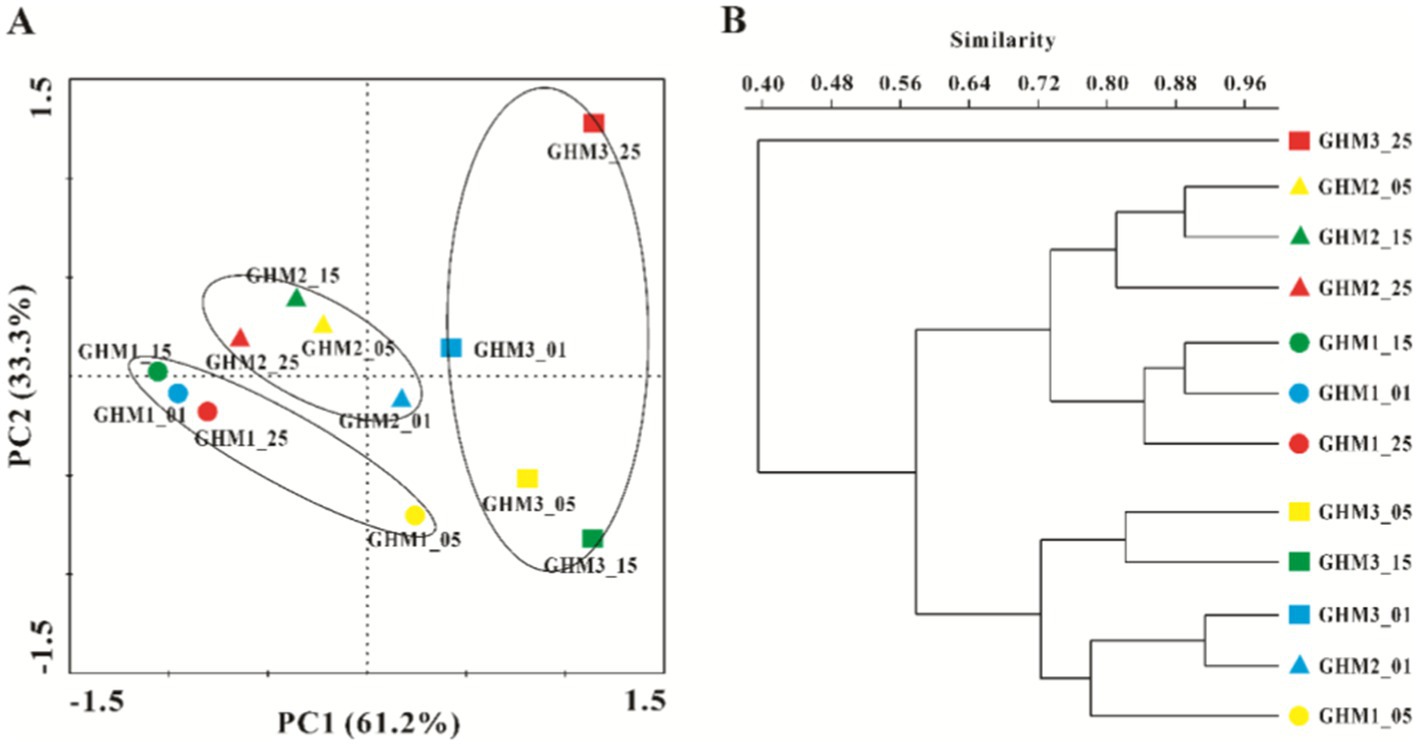
Figure 4. Differences in methanogenic communities structure of all the studied samples. Panel A: PCA analysis of methanogenic communities structure of all the studied samples; Panel B: Cluster analysis of methanogenic communities structure based on Bray–Curtis similarity index (Boot N = 9,999).
Estimated absolute abundance
The estimated absolute abundance (EAA) of the dominant methanogens at different incubation temperatures is displayed in Supplementary Table S1. We further analyzed the correlation between the EAA values of the dominant methanogens and CH4 production (Supplementary Table S2). These results indicated that the EAA values of Methanobacteriaceae, Methanotrichaceae, and Methanosarcinaceae were relatively higher at different incubation temperatures than Fen cluster, Methanocellales, and ZC-I cluster. For the GHM1 and GHM2 samples, the EAA values of four methanogen groups, Methanotrichaceae, Methanosarcinaceae, Fen cluster, and Methanocellales, increased with increasing incubation temperatures, and the EAA values of Methanotrichaceae, Methanosarcinaceae, and Methanocellales were positively correlated with the CH4 production (p < 0.01). For the GHM3 sample, the EAA value of Methanosarcinaceae increased with increasing incubation temperatures, and there was a significant positive correlation between the EAA value and CH4 production (p < 0.05). In addition, the EAA values of Methanotrichaceae and Methanocellales decreased with increasing incubation temperatures, with a significant negative correlation between the EAA value and CH4 production (p < 0.05).
Discussion
Temperature and soil moisture are the major factors affecting the CH4 emissions. Temperature affects the substrate supply of methanogens, thereby affecting their CH4 production rate (Brooks et al., 2003). The CH4 production in the studied soil samples was positively correlated with the incubation temperature. Previous studies showed that the production of CH4 of incubated wetland samples increased with increasing incubation temperature (Jiang et al., 2010; Cui et al., 2018), and that the CH4 production in the alpine wetlands incubated at 15°C was 7 times that of the original sample collected from the Qilian Mountain permafrost on the Qinghai-Tibet Plateau (Wei et al., 2018). In this study, CH4 accumulations in the GHM1, GHM2, and GHM3 samples increased by 5–6, 6–10, and 1–2 times at 5°C, 15°C, and 25°C, respectively (Figure 1). Methanogens are highly sensitive to soil pH and tend to live in weak acidic environments (Metje and Frenzel, 2005, 2007; Xing et al., 2022). Previous studies have revealed that the slightly acidic (pH 6.35–6.75) and high TOC content in permafrost soil can increase CH4 production (Barbier et al., 2012; Wei et al., 2018). In this study, the pH of the three incubation systems ranged 6.45–6.75, belonging slightly acid, and the TOC content was similar between the highest water level site (GHM1) and the lowest site (GHM3). Apparently, the soil moisture might support the CH4 emissions, as the concentrations of CH4 were higher in high water level sites (GHM1 and GHM2) than that in the low water level site (GHM3).
Generally, Methanotrichaceae, Methanobacteriaceae, and Methanosarcinaceae were the dominant methanogenic groups in the high water level sites (GHM1 and GHM2) at high temperature, while the Methanobacteriaceae and Methanosarcinaceae were the major methanogenic groups in the low water level site (GHM3). Acetoclastic and hydrogenotrophic methanogenesis are the two main methanogenic pathways in the wetlands on the Qinghai-Tibet Plateau (Conrad, 1999; Cui et al., 2018). It was found that acetoclastic methanogenesis was widely distributed across various locations, such as Siberian peat bog, flooded Italian rice fields, Zoige wetlands, high Arctic peat, and anaerobic reactors (Fey et al., 2001; Høj et al., 2005; Metje and Frenzel, 2007; Zhang et al., 2008; Qin et al., 2019). In this study, the dominant methanogenic communities were Methanotrichaceae, Methanobacteriaceae, and Methanosarcinaceae. Methanotrichaceae most likely use the acetoclastic pathway, while Methanobacteriaceae the hydrogenotrophic pathway (Thauer et al., 2008; Oren, 2014; Liu et al., 2019). Methanosarcinaceae can use H2, acetate, methanol, and other C1 compounds for methanogenic activity (Liu and Whitman, 2008). After incubation at different temperatures, the composition of methanogenic communities showed little change in the GHM1 and GHM2 samples. The abundance of dominant methanogenic groups Methanotrichaceae and Methanosarcinaceae were positively correlated with CH4 production (p < 0.01) in the GHM1 and GHM2 (Supplementary Table S2). It has been suggested that the occurrence of Methanotrichaceae was related to soil moisture in wetlands (Cui et al., 2018), and they showed the highest relative abundance in the soil where acetate served as the only substrate (Fey and Conrad, 2000). Methanotrichaceae utilized only acetate (Boone and Castenholz, 2001). The relative abundance of Methanotrichaceae was higher at 15°C and 25°C of the GHM1 and GHM2 samples, while another dominant methanogenic group Methanobacteriaceae, which ranged 22.05–58.86% and 19.30–23.51% in the GHM1 and GHM2, respectively, presented an opposite pattern. Methanobacteriaceae can use H2 and CO2 as substrates for methanogenesis, and previous studies have shown that this group can proliferate under certain hydrogen pressure (Conrad and Klose, 2006; Liu and Whitman, 2008). For the GHM3 sample (low water level), the change in incubation temperatures had a great influence on the methanogenic community structure. In general, the abundance of Methanobacteriaceae and Methanosarcinaceae was higher in GHM3 than in GHM1 and GHM2, while the abundance of Methanotrichaceae showed an opposite trend. Methanobacteriaceae (59.65 and 77.33%) was the dominant methanogen group at 5°C and 15°C, while presented a relative low abundance at 25°C. Methanosarcinaceae (69.29%) was the most abundant methanogen group at 25°C. The temperature between 15–25°C might be the threshold for methanogenic community change in the GHM3 sample (low water level). Moreover, the abundance of Methanosarcinaceae had a significant positive correlation with CH4 production (p < 0.05), while the abundance of Methanotrichaceae showed a contrary pattern (Supplementary Table S2). Methanosarcinaceae existed in various wetlands such as peatlands, freshwater marshes, paddy soils and low moisture site, and mainly utilized acetate, various methyl compounds, and hydrogen as methanogenic substrates (Hori et al., 2007; Noll et al., 2010; Liu et al., 2012; Cui et al., 2018).
The minor methanogenic communities, such as Methanocellales, Fen cluster and ZC-I cluster were also detected in the three sites. Moreover, the relative abundance of Fen cluster and Methanocellales of the GHM1 and GHM2 samples increased with the incubation temperature, and had a significant positive correlation with CH4 production (p < 0.05; Supplementary Table S2). Fen cluster belongs to Methanomicrobiales (Conrad et al., 2008), and the relative abundance of Fen Cluster was higher in the GHM1 and GHM2 samples incubated at 25°C. The members of Fen Cluster were relatively tolerant to a changing water table and low pH (Yrjälä et al., 2011). Methanocellales harbored pathways for hydrogenotrophic methanogenesis, and was widely distributed across various locations, such as rice fields, terrestrial as well as marine ecosystems (Krüger et al., 2005; Lu et al., 2005; Sakai et al., 2007; Watanabe et al., 2009). The abundance of Methanocellales was higher in the GHM2 samples, and especially higher at 25°C (Supplementary Table S1). Moreover, the abundance of Methanocellales exhibited a negative correlation with methane production in the GHM3 samples (p < 0.05; Supplementary Table S2). ZC-I cluster belonged to Methanosarcinales (Conrad et al., 2008), and ZC-I cluster was mainly found in the Zoige wetlands with its main metabolic substrate including methanol, trimethylamine, acetate, and H2/CO2, excluding formate (Zhang et al., 2008). In this study, the abundance of ZC-I cluster had no significant positive correlation with CH4 production of the three sites.
Conclusion
Soil moisture and temperature are the main factors affecting CH4 production in alpine swamp meadow of the permafrost wetland on the Qinghai-Tibet Plateau. Enhanced soil moisture facilitates the production of methane. Temperature rise has an inhibitory effect on the growth and methanogenic activity of Methanotrichaceae and Methanocellales. These findings may help us better understand the methane cycle in alpine swamp meadow of the permafrost wetland and its response to climate warming.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XS and HJ conceived the work. SP, YZ, and SZ performed on-site measurements and collected the samples. HC, YW, CM, and WH analyzed geochemistry and microbiology of the samples. HC, SW, and YW analyzed the sequencing data. HC, SW, XS, and HJ drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Funds of Oil and Gas Survey, China Geological Survey (Grant nos. GZH201400308, DD20160222, and DD20160226), the 111 project (no. B20011), and the Fundamental Research Funds for the Central Universities (no. 2-9-2020-030).
Acknowledgments
The authors are grateful to the Muli Field Scientific Observation and Research Station for Gas Hydrate.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1181658/full#supplementary-material
References
Bai, W., Xi, J. Y., and Wang, G. X. (2019). Effects of short-term warming and nitrogen addition on CO2 emission during growing season in an alpine swamp meadow ecosystem of Qinghai-Tibetan Plateau. Chin. J. Ecol. 38, 927–936. doi: 10.13292/j.1000-4890.201904.001
Barbier, B. A., Dziduch, I., Liebner, S., Ganzert, L., Lantuit, H., Pollard, W., et al. (2012). Methane-cycling communities in a permafrost-affected soil on Herschel Island, Western Canadian Arctic: active layer profiling of mcrA and pmoA genes. FEMS Microbiol. Ecol. 82, 287–302. doi: 10.1111/j.1574-6941.2012.01332.x
Boone, D., and Castenholz, R. W. (2001). The Archaea and the Deep Branching and Phototrophic Bacteria. New York, NY: Springer.
Brooks, A. G., Shannon, R. D., White, J. R., Martens, C. S., and Alperin, M. J. (2003). Controls on methane production in a tidal freshwater estuary and a peatland: methane production via acetate fermentation and CO2 reduction. Biogeochemistry 62, 19–37. doi: 10.1023/A:1021128400602
Bubier, J. L., Moore, T. R., Bellisario, L., Comer, N. T., and Crill, P. M. (1995). Ecological controls on methane emissions from a northern peatland complex in the zone of discontinuous permafrost, Manitoba, Canada. Global. Biogeochem. Cycles 9, 455–470. doi: 10.1029/95GB02379
Caldwell, S. L., Laidler, J. R., Brewer, E. A., Eberly, J. O., Sandborgh, S. C., and Colwell, F. S. (2008). Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ. Sci. Technol. 42, 6791–6799. doi: 10.1021/es800120b
Callahan, B., McMurdie, P., Rosen, M., Han, A., Johnson, A. J., and Holmes, S. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, H., Ju, P. J., Zhu, Q., Xu, X. L., Wu, N., Gao, Y. H., et al. (2022). Carbon and nitrogen cycling on the Qinghai–Tibetan Plateau. Nat. Rev. Earth. Env. 3, 701–716. doi: 10.1038/s43017-022-00344-2
Christensen, T. R., Michelsen, A., Jonasson, S., and Schmidt, I. K. (1997). Carbon dioxide and methane exchange of a subarctic heath in response to climate change related environmental manipulations. Oikos 79, 34–44. doi: 10.2307/3546087
Conrad, R. (1999). Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28, 193–202. doi: 10.1111/j.1574-6941.1999.tb00575.x
Conrad, R., and Klose, M. (2006). Dynamics of the methanogenic archaeal community in anoxic rice soil upon addition of straw. Eur. J. Soil Sci. 57, 476–484. doi: 10.1111/j.1365-2389.2006.00791.x
Conrad, R., Klose, M., Noll, M., Kemnitz, D., and Bodelier, P. L. E. (2008). Soil type links microbial colonization of rice roots to methane emission. Glob. Change Biol. 14, 657–669. doi: 10.1111/j.1365-2486.2007.01516.x
Cui, M. M., Ma, A. Z., Qi, H. Y., Zhuang, X. L., Zhuang, G. Q., and Zhao, G. H. (2015). Warmer temperature accelerates methane emissions from the Zoige wetland on the Tibetan plateau without changing methanogenic community composition. Sci. Rep. 5:11616. doi: 10.1038/srep11616
Cui, H. P., Su, X., Chen, F., Holland, M., Yang, S. X., Liang, J. Q., et al. (2019). Microbial diversity of two cold seep systems in gas hydrate-bearing sediments in the South China Sea. Mar. Environ. Res. 144, 230–239. doi: 10.1016/j.marenvres.2019.01.009
Cui, H. P., Su, X., Wei, S. P., Zhu, Y. H., Lu, Z. Q., Wang, Y. F., et al. (2018). Comparative analyses of methanogenic and methanotrophic communities between two different water regimes in controlled wetlands on the Qinghai-Tibetan Plateau, China. Curr. Microbiol. 75, 484–491. doi: 10.1007/s00284-017-1407-7
Dannemiller, K. C., Lang-Yona, N., Yamamoto, N., Rudich, Y., and Peccia, J. (2014). Combining real-time PCR and next-generation DNA sequencing to provide quantitative comparisons of fungal aerosol populations. Atmos. Environ. 84, 113–121. doi: 10.1016/j.atmosenv.2013.11.036
Dlugokencky, E. J., Bruhwiler, L., White, J. W. C., Emmons, L. K., Novelli, P. C., Montzka, S. A., et al. (2009). Observational constraints on recent increases in the atmospheric CH4 burden. Geophys. Res. Lett. 36, 252–260. doi: 10.1029/2009GL039780
Fey, A., Chin, K. J., and Conrad, R. (2001). Thermophilic methanogens in rice field soil. Environ. Microbiol. 3, 295–303. doi: 10.1046/j.1462-2920.2001.00195.x
Fey, A., Claus, P., and Conrad, R. (2004). Temporal change of 13C-isotope signatures and methanogenic pathways in rice field soil incubated anoxically at different temperatures. Geochim. Cosmochim. Acta 68, 293–306. doi: 10.1016/S0016-7037(03)00426-5
Fey, A., and Conrad, R. (2000). Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl. Environ. Microbiol. 66, 4790–4797. doi: 10.1128/AEM.66.11.4790-4797.2000
Høj, L., Olsen, R. A., and Torsvik, V. L. (2005). Archaeal communities in high Arctic wetlands at Spitsbergen, Norway (78°N) as characterized by 16S rRNA gene fingerprinting. FEMS Microbiol. Ecol. 53, 89–101. doi: 10.1016/j.femsec.2005.01.004
Høj, L., Olsen, R. A., and Torsvik, V. L. (2008). Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2, 37–48. doi: 10.1038/ismej.2007.84
Hori, T., Noll, M., Igarashi, Y., Friedrich, M. W., and Conrad, R. (2007). Identification of acetate-assimilating microorganisms under methanogenic conditions in anoxic rice field soil by comparative stable isotope probing of RNA. Appl. Environ. Microbiol. 73, 101–109. doi: 10.1128/AEM.01676-06
Huttunen, J. T., Nykänen, H., Turunen, J., and Martikainen, P. J. (2003). Methane emissions from natural peatlands in the northern boreal zone in Finland, Fennoscandia. Atmos. Environ. 37, 147–151. doi: 10.1016/S1352-2310(02)00771-9
Jiang, N., Wang, Y. F., and Dong, X. Z. (2010). Methanol as the primary methanogenic and acetogenic precursor in the cold Zoige wetland at Tibetan plateau. Microb. Ecol. 60, 206–213. doi: 10.1007/s00248-009-9602-0
Juottonen, H., Hynninen, A., Nieminen, M., Tuomivirta, T. T., Tuittila, E. S., Nousiainen, H., et al. (2012). Methane-cycling microbial communities and methane emission in natural and restored peatlands. Appl. Environ. Microbiol. 78, 6386–6389. doi: 10.1128/AEM.00261-12
Kadykalo, A. N., and Findlay, C. S. (2016). The flow regulation services of wetlands. Ecosyst. Serv. 20, 91–103. doi: 10.1016/j.ecoser.2016.06.005
Keller, J. K., White, J. R., Bridgham, S. D., and Pastor, J. (2004). Climate change effects on carbon and nitrogen mineralization in peatlands through changes in soil quality. Glob. Change Biol. 10, 1053–1064. doi: 10.1111/j.1529-8817.2003.00785.x
Kettunen, A., Kaitala, V., Lehtinen, A., Lohila, A., Alm, J., Silvola, J., et al. (1999). Methane production and oxidation potentials in relation to water table fluctuations in two boreal mires. Soil Biol. Biochem. 31, 1741–1749. doi: 10.1016/S0038-0717(99)00093-0
Krüger, M., Frenzel, P., Kemnitz, D., and Conrad, R. (2005). Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51, 323–331. doi: 10.1111/j.1462-2920.2004.00634.x
Li, T. T., Canadell, J. G., Yang, X. Q., Zhai, P. M., Chao, Q. C., Lu, Y. Y., et al. (2022). Methane emissions from wetlands in China and their climate feedbacks in the 21st century. Environ. Sci. Technol. 56, 12024–12035. doi: 10.1021/acs.est.2c01575
Li, W., Shi, R., Yuan, L. C., Lan, X. L., Feng, D. F., and Chen, H. (2022). Effect of short-term warming and drought on the methanogenic communities in degraded peatlands in Zoige plateau[J]. Front. Microbiol. 13:880300. doi: 10.3389/fmicb.2022.880300
Li, X. Q., Xing, J. W., Pang, S. J., Zhu, Y. H., Zhang, S., Xiao, R., et al. (2022). Carbon isotopic evidence for gas hydrate release and its significance on seasonal wetland methane emission in the Muli permafrost of the Qinghai-Tibet Plateau. Int. J. Environ. Res. Public Health 19:2437. doi: 10.3390/ijerph19042437
Liu, D. Y., Ding, W. X., Jia, Z. J., and Cai, Z. C. (2012). The impact of dissolved organic carbon on the spatial variability of methanogenic archaea communities in natural wetland ecosystems across China. Appl. Microbiol. Biotechnol. 96, 253–263. doi: 10.1007/s00253-011-3842-x
Liu, P., Klose, M., and Conrad, R. (2019). Temperature-dependent network modules of soil methanogenic bacterial and archaeal communities. Front. Microbiol. 10:496. doi: 10.3389/fmicb.2019.00496
Liu, Y. C., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Lowe, D. C. (2006). Global change: a green source of surprise. Nature 439, 148–149. doi: 10.1038/439148a
Lu, Y. H., Lueders, T., Friedrich, M. W., and Conrad, R. (2005). Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7, 326–336. doi: 10.1111/j.1462-2920.2005.00697.x
Luton, P. E., Wayne, J. M., Sharp, R. J., and Riley, P. W. (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148, 3521–3530. doi: 10.1099/00221287-148-11-3521
Metje, M., and Frenzel, P. (2005). Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl. Environ. Microbiol. 71, 8191–8200. doi: 10.1128/AEM.71.12.8191-8200.2005
Metje, M., and Frenzel, P. (2007). Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ. Microbiol. 9, 954–964. doi: 10.1111/j.1462-2920.2006.01217.x
Noll, M., Klose, M., and Conrad, R. (2010). Effect of temperature change on the composition of the bacterial and archaeal community potentially involved in the turnover of acetate and propionate in methanogenic rice field soil. FEMS Microbiol. Ecol. 73, 215–225. doi: 10.1111/j.1574-6941.2010.00883.x
Nunoura, T., Oida, H., Miyazaki, J., Miyashita, A., Imachi, H., and Takai, K. (2008). Quantification of mcrA by fluorescent PCR in methanogenic and methanotrophic microbial communities. FEMS Microbiol. Ecol. 64, 240–247. doi: 10.1111/j.1574-6941.2008.00451.x
Oren, A. (2014). “The family Methanotrichaceae” in The Prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 298–306.
Qin, X. C., Li, C. J., Gao, Y. S., Zhang, Z. J., and Zhang, X. J. (2019). Divergence of granular sludges and microbial communities in two types of anaerobic reactors treating different wastewaters. J. Microbiol. Biotechnol. 29, 633–644. doi: 10.4014/jmb.1807.07007
Sakai, S., Imachi, H., Sekiguchi, Y., Ohashi, A., Harada, H., and Kamagata, Y. (2007). Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster Rice cluster I. Appl. Environ. Microbiol. 73, 4326–4331. doi: 10.1128/AEM.03008-06
Thauer, R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144, 2377–2406. doi: 10.1099/00221287-144-9-2377
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Wagner, D., Schirmack, J., Ganzert, L., Morozova, D., and Mangelsdorf, K. (2013). Methanosarcina soligelidi sp. nov., a desiccation and freeze-thaw-resistant methanogenic archaeon from a Siberian permafrost-affected soil. Int. J. Syst. Evol. Microbiol. 63, 2986–2991. doi: 10.1099/ijs.0.046565-0
Wang, C. J., Han, G. D., Wang, S. P., Zhai, X. J., Brown, J., Havstad, K. M., et al. (2014). Sound management may sequester methane in grazed rangeland ecosystems. Sci. Rep. 4:4444. doi: 10.1038/srep04444
Watanabe, T., Kimura, M., and Asakawa, S. (2009). Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol. Biochem. 41, 276–285. doi: 10.1016/j.soilbio.2008.10.025
Wei, S. P., Cui, H. P., Zhu, Y. H., Lu, Z. Q., Pang, S. J., Zhang, S., et al. (2018). Shifts of methanogenic communities in response to permafrost thaw results in rising methane emissions and soil property changes. Extremophiles 22, 447–459. doi: 10.1007/s00792-018-1007-x
Wei, J. Q., Li, X. Y., Liu, L., Christensen, T. R., Jiang, Z. Y., Ma, Y. J., et al. (2022). Radiation, soil water content, and temperature effects on carbon cycling in an alpine swamp meadow of the northeastern Qinghai–Tibetan plateau. Biogeosciences 19, 861–875. doi: 10.5194/bg-19-861-2022
Wu, F. T., Cao, S. K., Cao, G. C., Chen, K. L., and Peng, C. H. (2021). The characteristics and seasonal variation of methane fluxes from an alpine wetland in the Qinghai Lake watershed, China. Wetlands 41, 1–11. doi: 10.1007/s13157-021-01415-8
Wu, M. H., Xue, K., Wei, P. J., Jia, Y. L., Zhang, Y., and Chen, S. Y. (2022). Soil microbial distribution and assembly are related to vegetation biomass in the alpine permafrost regions of the Qinghai-Tibet Plateau. Sci. Total Environ. 834:155259. doi: 10.1016/j.scitotenv.2022.155259
Xing, T. T., Liu, P. F., Ji, M. K., Deng, Y. C., Liu, K. S., Wang, W. Q., et al. (2022). Sink or source: alternative roles of glacier foreland meadow soils in methane emission is regulated by glacier melting on the Tibetan plateau. Front. Microbiol. 13:862242. doi: 10.3389/fmicb.2022.862242
Yrjälä, K., Tuomivirta, T., Juottonen, H., Putkinen, A., Lappi, K., Tuittila, E. S., et al. (2011). CH4 production and oxidation processes in a boreal fen ecosystem after long-term water table drawdown. Glob. Change Biol. 17, 1311–1320. doi: 10.1111/j.1365-2486.2010.02290.x
Zhang, L. Y., Adams, J. M., Dumont, M. G., Li, Y. T., Shi, Y., He, D., et al. (2019). Distinct methanotrophic communities exist in habitats with different soil water contents. Soil Boil. Biochem. 132, 143–152. doi: 10.1016/j.soilbio.2019.02.007
Zhang, Z. J., Qu, Y. Y., Li, S. Z., Feng, K., Wang, S., Cai, W. W., et al. (2017). Soil bacterial quantification approaches coupling with relative abundances reflecting the changes of taxa. Sci. Rep. 7:4837. doi: 10.1038/s41598-017-05260-w
Zhang, G. S., Tian, J. Q., Jiang, N., Guo, X. P., Wang, Y. F., and Dong, X. Z. (2008). Methanogen community in Zoige wetland of Tibetan plateau and phenotypic characterization of a dominant uncultured methanogen cluster ZC-I. Environ. Microbiol. 10, 1850–1860. doi: 10.1111/j.1462-2920.2008.01606.x
Zhang, F. G., Zhang, S. Y., Tang, R. L., Wang, H. Y., Yang, Z. B., Zhou, Y. L., et al. (2017). Methane emission characteristics of active layer in wetland permafrost area of the Tibetan plateau. Geophys. Geochem. Explor. 41, 1027–1036. doi: 10.11720/wtyht.2017.6.06
Zhang, Z., Zimmermann, N. E., Stenke, A., and Poulter, B. (2017). Emerging role of wetland methane emissions in driving 21st century climate change. Proc. Natl. Acad. Sci. U. S. A. 114, 9647–9652. doi: 10.1073/pnas.1618765114
Keywords: Qinghai-Tibet Plateau, permafrost wetland, alpine swamp meadow, methane production, methanogen
Citation: Cui H, Wang Y, Su X, Wei S, Pang S, Zhu Y, Zhang S, Ma C, Hou W and Jiang H (2023) Response of methanogenic community and their activity to temperature rise in alpine swamp meadow at different water level of the permafrost wetland on Qinghai-Tibet Plateau. Front. Microbiol. 14:1181658. doi: 10.3389/fmicb.2023.1181658
Edited by:
Yiguo Hong, Guangzhou University, ChinaReviewed by:
Jie Lv, Beijing University of Chemical Technology, ChinaPengfei Liu, Lanzhou University, China
Copyright © 2023 Cui, Wang, Su, Wei, Pang, Zhu, Zhang, Ma, Hou and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongchen Jiang, hongchen.jiang@gmail.com; Xin Su, xsu@cugb.edu.cn
†These authors share first authorship
 Hongpeng Cui
Hongpeng Cui Yanfa Wang1,2,3†
Yanfa Wang1,2,3† Xin Su
Xin Su Shiping Wei
Shiping Wei Youhai Zhu
Youhai Zhu Weiguo Hou
Weiguo Hou Hongchen Jiang
Hongchen Jiang