- School of Marine Science and Policy, College of Earth, Ocean, and Environment, University of Delaware, Lewes, DE, United States
Nitrate reductase (NR) catalyzes the rate-limiting step in nitrate assimilation. Plant and algal NRs have a highly conserved domain architecture but differ in regulation. In plants, NR activity is regulated by reversible phosphorylation and subsequent binding of 14-3-3 proteins at a conserved serine residue. Algal NRs typically lack 14-3-3 binding motifs, which have only recently been identified in a few algal species. Previous research indicates that the alga, Chattonella subsalsa, possesses a novel NR, NR2-2/2HbN (NR2), which incorporates a 2/2 hemoglobin domain. A second NR (NR3) in C. subsalsa lacks the cytochrome b5 (heme-Fe) domain but includes a putative binding motif for 14-3-3 proteins. The expression of NR2 and NR3 genes indicates that NR2 transcript abundance was regulated by light, nitrogen source, and temperature, while NR3 transcript levels were only regulated by light. Here, we measured total NR activity in C. subsalsa and the potential for regulation of NR activity by putative 14-3-3 binding proteins. Results indicate that NR activity in C. subsalsa was regulated by light, nitrogen source, and temperature at the translational level. NR activity was also regulated by endogenous rhythm and temperature at the post-translational level, supporting the hypothesis that NR3 is regulated by 14-3-3 binding proteins. Together with a previous report describing the regulation of NR gene expression in C. subsalsa, results suggest that C. subsalsa responds to environmental conditions by differential regulation of NRs at transcriptional, translational, and post-translational levels. This flexibility may provide a competitive advantage for this species in the environment. To date, this is the first report which provides evidence for the potential post-translational regulation of NR by 14-3-3 proteins in algal species and suggests that regulatory mechanisms for NR activity may be shared between plants and some algal species.
1. Introduction
Nitrate and ammonium are major nitrogen sources for phytoplankton in the environment (reviewed by Shen et al., 2015; Rezvani et al., 2017; Salbitani and Carfagna, 2021). It has been estimated that 40% of global primary production resulted from the assimilation of these two nitrogen sources by phytoplankton (Malerba et al., 2015 and references therein). These essential nitrogen types also play a role in the stimulation and succession of harmful algal blooms (HABs) and are key factors affecting HAB dynamics (reviewed by Davidson et al., 2012; Wang, 2016; Wurtsbaugh et al., 2019; Coyne et al., 2022). The toxic microalga Chattonella subsalsa is among the HAB species implicated in mass fish kills in coastal waters worldwide (reviewed by Imai and Yamaguchi, 2012; Lum et al., 2021). Recent research indicated nitrogen as a major factor affecting C. subsalsa growth (Kok et al., 2019), pigmentation (Kok et al., 2019), and competition in natural phytoplankton assemblies (Wang, 2016; Coyne et al., 2022).
For phytoplankton using nitrate as a nitrogen source, nitrate is reduced to nitrite by nitrate reductase (NR), and nitrite is then reduced to ammonium by nitrite reductase (NiR), followed by amino acid assimilation (reviewed by Kumar and Bera, 2020). Nitrate assimilation represents one of the most important redox processes that support 20% of the algal growth in the ocean (reviewed by Bellido-Pedraza et al., 2020). In this process, NR catalyzes the first enzymatic and also rate-limiting step; regulation of this step is essential for the coordination of nitrogen and carbon assimilation in higher plants, algae, and fungi (reviewed by Campbell, 1999; Parker and Armbrust, 2005; Wang et al., 2018). Recent research also indicated NR to be a moonlighting protein that also participates in the production of nitric oxide (reviewed by Sanz-Luque et al., 2015b; Tejada-Jimenez et al., 2019; Bellido-Pedraza et al., 2020, 2022), as well as the conversion of nitric oxide back to nitrate (e.g., Stewart and Coyne, 2011; reviewed by Tejada-Jimenez et al., 2019; Bellido-Pedraza et al., 2020).
It has been well established that the expression and activity of NR are tightly regulated by extra- and intracellular cues (reviewed by Young and Berges, 2016; Wang et al., 2018; Coyne et al., 2022; Yu et al., 2022). The tight regulation of NR is essential for organisms to optimize nitrogen assimilation and prevent the accumulation of toxic compounds, such as nitrite, as well as reactive oxygen and nitrogen species (reviewed by Wang et al., 2018). Nitrogen sources are among the key factors that regulate NR. For instance, nitrate and ammonium have been reported to act oppositely on NR, where NR gene expression and activity can be enhanced by nitrate, while repressed or inhibited by ammonium (reviewed by Sanz-Luque et al., 2015a). When both nitrate and ammonium exist, NR is regulated by the balance between these nitrogen sources (reviewed by Sanz-Luque et al., 2015a; Glibert et al., 2016). The light-to-dark cycle also regulates NR; some algal NRs respond to the light during diurnal changes (e.g., Chow et al., 2004; Wang et al., 2015, 2018), while others change due to an endogenous circadian rhythm (Falcão et al., 2010). Temperature is another factor that may affect NR gene expression and activities in algae (Wang et al., 2018; Zhong et al., 2020). These factors may affect NR independently or synergistically (Parker and Armbrust, 2005). For instance, Parker and Armbrust (2005) demonstrated higher NR gene transcript abundance in diatoms under high light and low temperature conditions, suggesting NR catalyzed nitrogen assimilation may play a role in dissipating excess energy from the photosynthetic light reactions.
In higher plants, NRs are regulated at the post-translational level by the binding of a 14-3-3 protein following the reversible phosphorylation of a serine residue located at the Hinge-1 region (Chi et al., 2015). NR activity can be inhibited by the 14-3-3 binding by obstructing the electron flow between domains through conformational change (Chi et al., 2015). The 14-3-3-mediated NR regulation is crucial for the rapid response of plants to environmental conditions, such as excess nitrate stress (Xu et al., 2016), C/N balance changes (Huarancca Reyes et al., 2018), and light to dark cycles (reviewed by Gao et al., 2014). Although highly conserved in higher plants, 14-3-3 binding motifs were not identified in algal species until recent research (Wang et al., 2018).
NRs are multidomain proteins that are only active as dimers; a 5-domain-and-3-region structure is conserved among NRs of algae, plants, and fungi (reviewed by Campbell, 1999; Chamizo-Ampudia et al., 2017; Wang et al., 2018; Figure 1). These domains include a molybdenum-molybdopterin cofactor domain (Mo-MPT), a dimer interface (DI), a cytochrome b5-binding domain containing Heme-Fe (the Heme-Fe domain), a flavin adenine dinucleotide domain (FAD), and an NAD(P)H domain (reviewed by Campbell, 1999; Chamizo-Ampudia et al., 2017). The three regions include an N-terminal preceding the Mo-MPT domain, a Hinge-1 region between the DI and Heme domains, and a Hinge-2 region between the Heme and FAD domains (reviewed by Campbell, 1999; Chamizo-Ampudia et al., 2017).
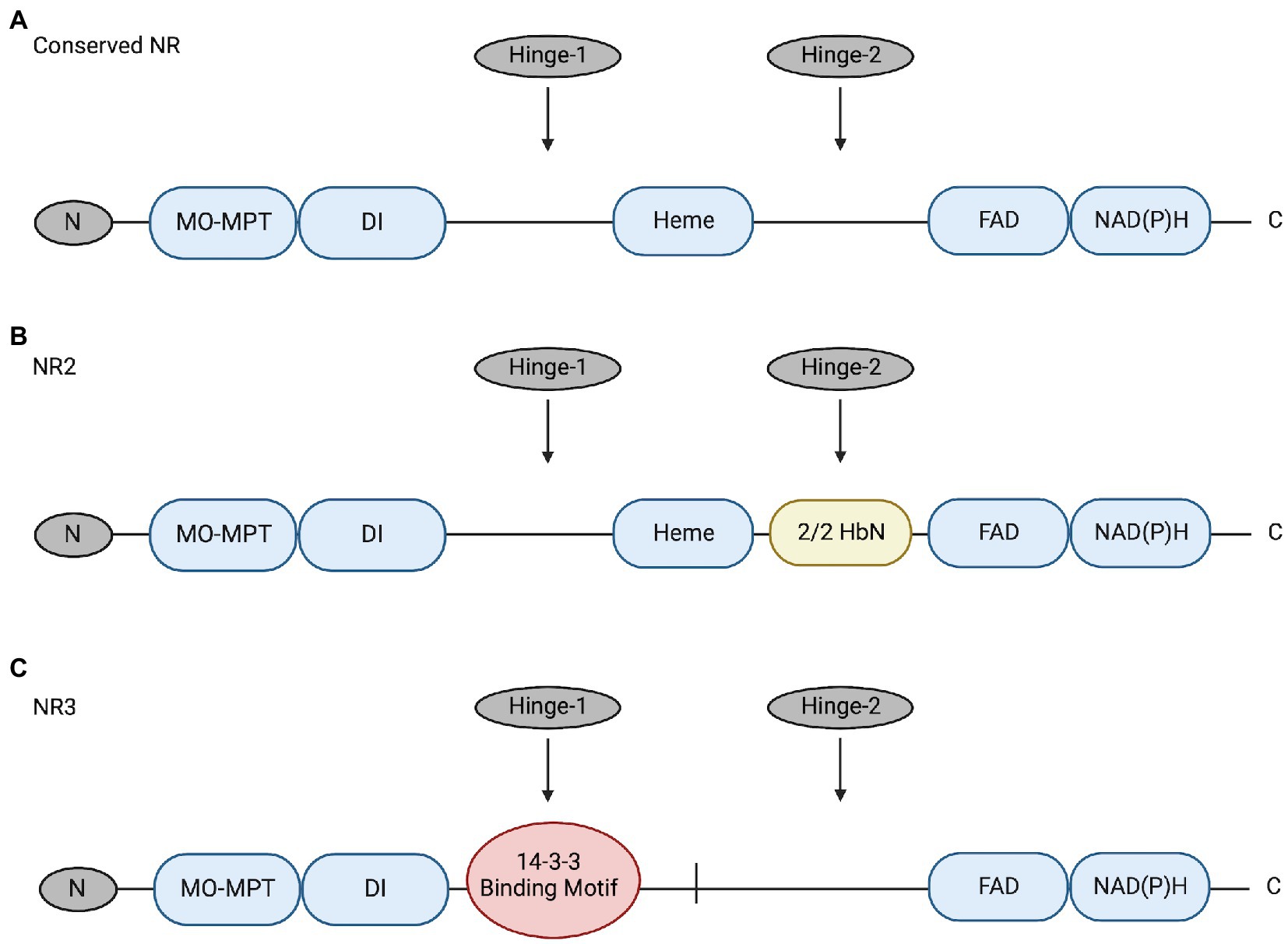
Figure 1. (A) Conserved NR structure includes 5 domains (blue; Mo-MPT, DI, Heme, FAD, and NAD[P]H) and 3 regions (grey; N, Hinge-1, and Hinge-2) (Campbell, 1999). (B) Structure of NR2 has a 2/2HbN domain (yellow) inserted in the Hinge-2 region. This enzyme was identified in raphidophytes Heterosigma akashiwo and Chattonella subsalsa (Stewart and Coyne, 2011). (C) Structure of the novel NR3 enzyme in C. subsalsa that lacks the conserved Heme domain and has a 14-3-3 binding motif (red) in the Hinge-1 region (Wang et al., 2018). The figure was created with Biorender.com and modified from Wang et al. (2018).
Recent research identified novel NRs with alternative structures in C. subsalsa and Heterosigma akashiwo (Raphidophyceae) (Stewart and Coyne, 2011; Wang et al., 2018; Figure 1). NR2-2/2HbN (referred to hereafter as NR2) identified in C. subsalsa and H. akashiwo has a 2/2 hemoglobin (2/2Hb) domain inserted in the Hinge-2 region that has been proposed to confer nitric oxide dioxygenase activity to this enzyme (Stewart and Coyne, 2011; Healey et al., 2023). Following research identified another novel NR in C. subsalsa (NR3) (Wang et al., 2018). The deduced amino acid sequence of NR3 lacked the 2/2HbN domain found in NR2 as well as the conserved Heme-Fe domain present in all other eukaryotic NRs (Wang et al., 2018). As mentioned earlier, dimerization is necessary for NR to function (reviewed by Chamizo-Ampudia et al., 2017), with a tendency to form tetramers (reviewed by Campbell, 2001). Here, without the conserved Heme-Fe domain, NR3 may form tetramers with NR2 to perform necessary functions as discussed in Wang et al. (2018). Additional sequence analysis also revealed that NR3 included a potential phosphorylation site and a canonical 14-3-3 binding protein motif in the Hinge-1 region (Wang et al., 2018), suggesting the potential for post-translational regulation of this enzyme in a manner similar to plant NRs. Expression of NR2 and NR3 in C. subsalsa indicated that NR2 transcript abundance changed in response to light, temperature, and nitrogen source, while NR3 transcript levels only responded to light (Wang et al., 2018). No research has been conducted to investigate the response of NR enzyme activity in C. subsalsa to environmental cues or the potential regulation of putative 14-3-3 protein binding in any algal NRs.
In this study, the post-transcriptional regulation of NR enzyme activity and the potential role of the presumed 14-3-3 proteins in post-translational regulation of NR activity in C. subsalsa were examined. NR regulation by 14-3-3 proteins in plants is a two-step process (Figure 2), where NR is first phosphorylated by kinases to allow the subsequent recognition and binding of the 14-3-3 proteins (Paul et al., 2012). Additionally, the binding of 14-3-3 proteins to phosphorylated NR requires divalent cations, such as Mg2+, at millimolar concentrations (Athwal and Huber, 2002). The requirement for divalent cations provides a means to evaluate the differential regulation of NR activity by putative 14-3-3 protein binding in cellular homogenates (Mackintosh et al., 1995). In other words, the addition of excess Mg2+ would allow the formation of the NR-Ser-P/14-3-3 protein complex in the cellular homogenate. This complex inhibits the activity of the phosphorylated NR, so that the decrease in NR activity is a measure of the remaining non-phosphorylated NR (the actual NR activity) (Mackintosh et al., 1995). On the other hand, adding EDTA to the cellular homogenates would sequester Mg2+ through chelation to yield the activity of total NR comprised of both phosphorylated and non-phosphorylated NR (Mackintosh et al., 1995). The difference in activity in the presence and absence of Mg2+ provides a measure of NR activity that is inhibited by 14-3-3 protein binding to phosphorylated NR (Mackintosh et al., 1995). Here, changes in NR activity in C. subsalsa due to putative 14-3-3 protein binding were investigated in response to light, nitrogen source, and temperature. Results of this study will contribute to our understanding of nitrogen utilization in algal species, and its response to vital environmental cues.
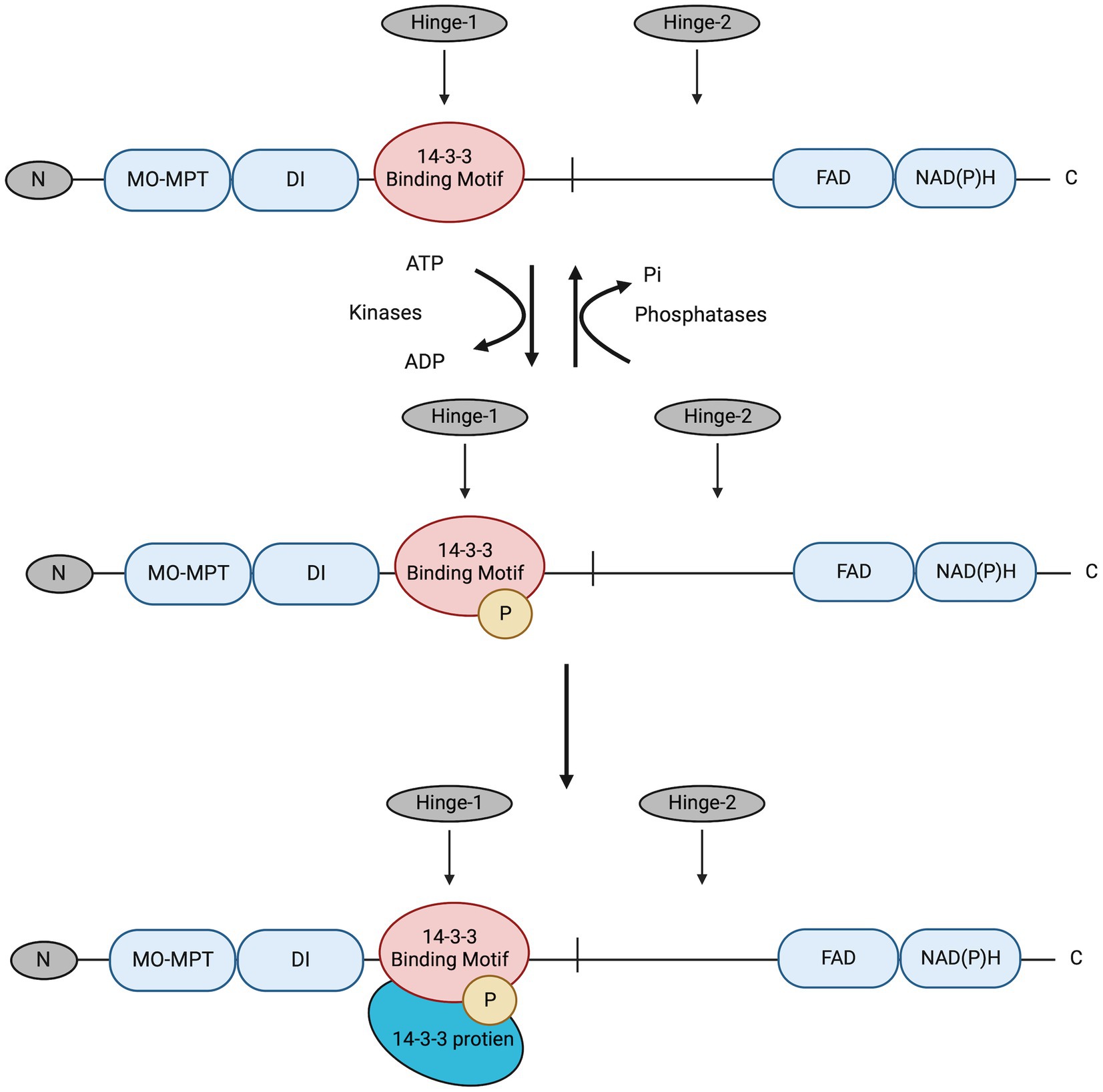
Figure 2. The deduced process for NR3 regulation by reversible phosphorylation and subsequent binding of putative 14-3-3 proteins. The 14-3-3 protein binding motif in the Hinge-1 region is first phosphorylated by kinases; then putative 14-3-3 proteins recognize and bind to the motif for NR3 enzyme activity regulation. This was deduced from the conserved mechanism for NR regulation by 14-3-3 proteins in higher plants (Paul et al., 2012). Pi, phosphate. The figure was created with Biorender.com.
2. Materials and methods
2.1. Culture conditions
Stock culture of Chattonella subsalsa (National Center for Marine Algae and Microbiota, CCMP21911 [accessed on 8/8/2022]) was maintained semi-continuously in exponential growth phase in f/2 medium (without silica) (Guillard and Ryther, 1962), with either f/2 levels of nitrate (882 μM), or 100 μM nitrate or ammonium (only for the nitrogen source experiment below). The salinity of the medium was 20, and the temperature for incubation was 25°C. Stock cultures were maintained without agitation at approximately 100 μE m−2 s−1 irradiance with a 12:12 h light: dark cycle.
2.2. NR activity under different light conditions
2.2.1. Long-term light experiment
C. subsalsa was cultured as above with f/2 levels of nitrate as the nitrogen source. To test for changes in activity after acclimation to the light regime, replicate cultures (N = 3; culture volume = 500 ml) were exposed to either a 12:12 h light: dark cycle or to constant light at 100 μmol photons m−2 s−1 for 24 h before the initial sampling. Cultures in constant light remained so for the entire sampling period. Samples were collected at five time points to cover a wide range of timing before and after the light changes for cultures under the light: dark cycle: 1 h before lights on (1:00), 1 h after lights on (3:00), 6 h after lights on (8:00), 1 h after dark (15:00), and 6 h after dark (20:00). Samples from cultures under the constant light were also collected at the same time-points to compare with those under the light to dark cycle. At each time point, samples of 50 ml were collected and centrifuged at 6000 RPM for 5 min. The supernatant was discarded, the cell pellet was immediately frozen in liquid nitrogen and stored at −80°C until analysis (see below).
2.2.2. Short-term light experiment
To test for short-term regulation of NR activity, C. subsalsa was cultured as above in f/2 medium with a 12:12 light: dark cycle. At 6 h after lights on, replicate cultures (N = 3; culture volume = 50 ml) were transferred to the dark. Controls (N = 3) remained in the light. Samples were collected at 15 min after transferring to the dark and processed as above for the NR activity assay described below.
2.3. NR activity with different nitrogen sources
C. subsalsa was cultured without agitation in f/2 medium with either 100 μM NO3− or 100 μM NH4+ as the nitrogen source (N = 3; culture volume > 100 ml). The temperature and light conditions for these cultures were described above as for the stock cultures. The semi-continuous cultures were acclimated to growth with each nitrogen type and maintained in exponential growth before harvesting. Cultures were harvested at 6 h after the start of the light cycle and processed as described above for the NR activity assay.
2.4. NR activity at different temperatures
2.4.1. Long-term temperature acclimation experiment
To test for changes in activity after acclimation to temperature, C. subsalsa was cultured without agitation in f/2 medium with nitrate (f/2 level) at 28°C, 25°C, and 18°C (N = 3; culture volume > 100 ml). The light intensity was the same as the stock cultures. Semi-continuous cultures were maintained in exponential growth at each temperature condition for 2 months before harvesting. Samples were collected at 6 h after the start of the light cycle as described above for NR activity analysis.
2.4.2. Short-term temperature change experiment
To test for short-term regulation of NR activity, C. subsalsa was cultured as above in f/2 medium at 25°C. At 6 h after lights on, replicate cultures (N = 3; culture volume = 50 ml) were transferred to a water bath at 8°C. Controls (N = 3) remained at 25°C. To be noted, 8°C is below C. subsalsa’s growth temperature (10 to 30°C; Zhang et al., 2006) and was used here to assess its NR activity under cold stress. Samples were collected at 15 min after transferring to the cold and processed for the NR activity assay as described above. Only total NR activity was assayed for the controls.
2.5. NR activity assay
The NR activity assay was optimized for C. subsalsa according to the methods published by Berges and Harrison (1995). The assay was optimized for homogenization methods (grinding vs. sonication), extraction buffers (KPi [potassium phosphate] buffer, BTP [Bis-tris propane] buffer, TAPS {[tris(hydroxymethyl)methylamino] propanesulfonic acid}) buffer, EPPS [HEPPS] buffer, Tris [Tris(hydroxyl-methyl)aminomethane] buffer, and MOPS [3-morpholinopropane-1-sulfonic acid] buffer), stabilizing additives (BSA [Bovine Serum Albumin], DTT [dithiothreitol], and BSA plus DTT), specificity for NADH versus NADPH, as well as assay temperatures and pH. Ca2+ was also tested as an alternative divalent cation for Mg2+ to measure the activity of non-phosphorylated NR but did not improve the assay. The optimized protocol included lysing cells by sonication, followed by extraction in 200 mM KPi buffer [200 mM KH2PO4, 2.5% (w/v) KOH], pH 7.9, using NADH as a reductant, and with no stabilizing additives. Mg2+ was used as the divalent cation when required in the assay below. The assay was conducted at 25°C.
The frozen cell pellet was resuspended in 1 ml 200 mM KPi extraction buffer (pH 7.9) on ice, homogenized by sonication, and centrifuged to clarify at 4°C. The supernatants were assayed immediately. The homogenate was divided to evaluate total NR activity (with EDTA to chelate endogenous Mg2+) or NR activity in the presence of excess Mg2+ to measure non-phosphorylated NR (Berges and Harrison, 1995; Nemie-Feyissa et al., 2013). The optimized assay consisted of 100 μl extract, an equal volume of assay buffer (500 mM KPi, pH 7.9, amended with 2 mM EDTA or 6 mM MgCl2), 0.2 mM NADH (Sigma Chem. Co., St. Louis, MO) and 10 mM KNO3 as substrate. Sterile water was added to the negative control reaction in place of KNO3. The reaction was incubated for 30 min at room temperature in the dark. Zinc acetate (12.5 μl of 1 M stock) was added to stop the reaction. The samples were centrifuged, and the supernatants were used below.
The concentration of nitrite product in the supernatants was measured colorimetrically. Ten microliters of 750 μM N-Methylphenazonium methyl sulfate (Sigma–Aldrich Fluka, Buchs SG, Switzerland) were added to 200 μl supernatants. The reaction was incubated in the dark for 20 min, followed by the addition of 12.5 μl 5 M HCl and 125 μl 58 mM Sulfanilamide (Sigma Chem. Co.). After an additional 5 min of incubation in the dark, 125 μl 4 mM N-(1-Naphthyl) ethylenediamine dihydrochloride (Sigma Chem. Co.) was added to the reaction. The samples were incubated for 10 min and then were aliquoted into 96 well plates. The absorbance was measured at 540 nm on a microplate reader (BMG Labtech, Ortenberg, Germany). NR activity was normalized to the protein content of the cellular homogenate as measured using the Pierce BCA Protein Assay Kit (Pierce, Rockford, IL, United States).
2.6. Statistical analysis
Repeated measures ANOVA was used to test if the activity of total NR and non-phosphorylated NR changed significantly over time in the long-term light experiment. If the change was significant, then a paired t-test was conducted to analyze the significance of the differences between the values at each time point. Two sample t-test was used to test the significance of the difference between total or non-phosphorylated NR activity in the light to dark cycle compared to constant light at each time point in the long-term light experiment. Two sample t-test was also used to assess the significance of difference between total or non-phosphorylated NR activity comparing the light conditions (light vs. dark) in the short-term experiment.
One-way ANOVA was used to test if different temperatures had a significant effect on the total or non-phosphorylated NR activity in the long-term temperature experiment. If the effect was significant, then Tukey’s HSD test was used to analyze the difference between values in all possible pairs of groups. Two sample t-test was used to assess the significance of difference between the total NR activity comparing different temperatures in the short-term experiment.
Additionally, a paired t-test was used to test if there was a significant difference between total and non-phosphorylated NR in all experiments under each of the light, nitrogen, and temperature conditions. A significance level of 0.05 was used in all tests. All statistical analyses were performed in R (V.3.2.4 Revised, R Core Team 2016).
3. Results
3.1. Diel activity of NR under different light conditions
3.1.1. Long-term light experiment
There was a significant difference in activity between total NR activity (assay conducted with EDTA) and non-phosphorylated NR activity (assay conducted with excess Mg2+) in samples under constant light at 20:00 (p < 0.05; Figure 3). No significant difference between total NR and non-phosphorylated NR activity was observed at any other time points under constant light or at any time point in the light:dark treatment (p > 0.05).
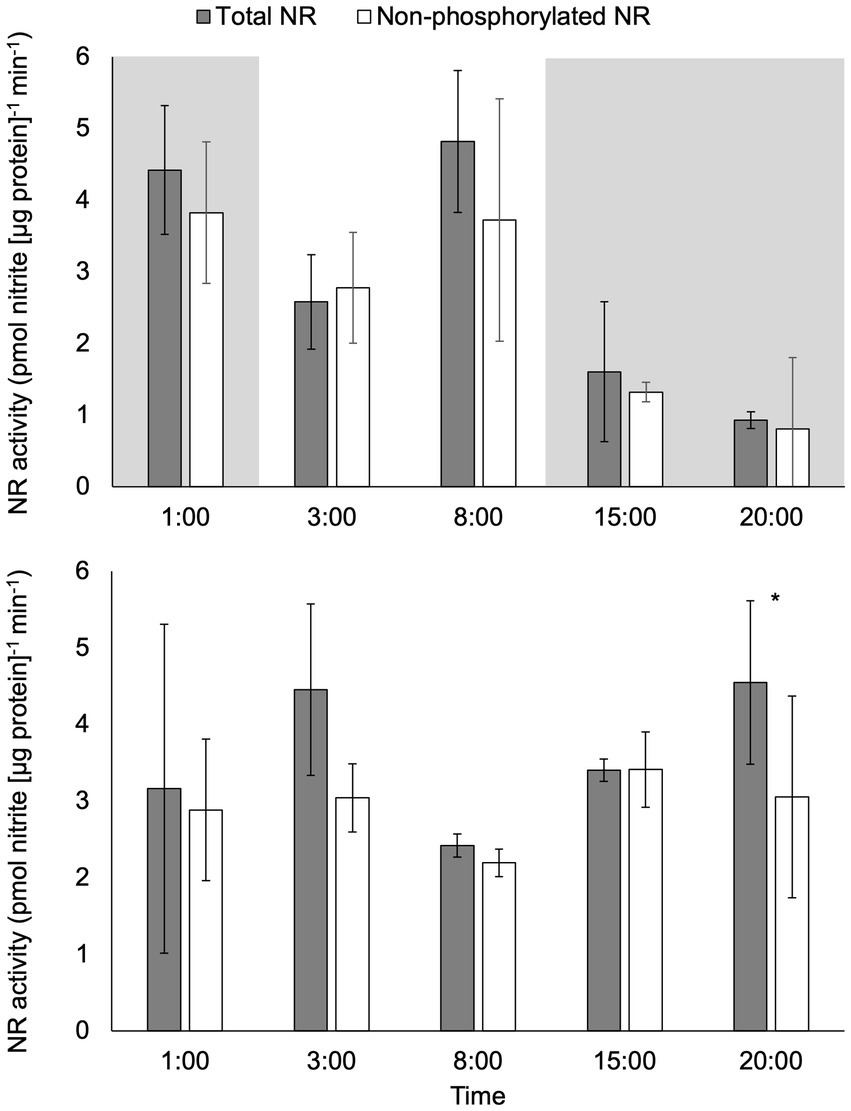
Figure 3. Activity (pmol nitrite [μg protein]−1 min−1) of total NR and non-phosphorylated NR in C. subsalsa cultured in a 12:12 h light: dark cycle (upper panel) or constant light (lower panel) in the long-term light experiment. The portion with shade indicates the time that was in dark for algae cultured with the light: dark cycle. Samples were taken at 1 h before lights on (1:00), 1 h after lights on (3:00), 6 h after lights on (8:00), 1 h after dark (15:00), and 6 h after dark (20:00). Asterisk (*) reveals the significant difference in NR activity between total and non-phosphorylated NR, indicating the regulation by reversible phosphorylation and the subsequent binding of putative 14-3-3 proteins, at the indicated time point (p < 0.05). Error bars are standard deviation of three biological replicates.
Total NR activity decreased significantly over time in the light:dark treatment (p < 0.05) from mid-day (8:00) to 6 h after dark (20:00, p < 0.05; Figure 3). There were no significant changes over time in total NR activity under constant light (p > 0.05). Comparison of total NR activity in light:dark treatment to constant light revealed significantly higher activity in the light:dark treatment at 8:00 (6 h after lights on) and significantly lower activity at 20:00 (6 h after dark, p < 0.05).
The activity of non-phosphorylated NR for the light:dark treatment was significantly higher at 1 h (1:00) before lights on than at 1 h after dark (15:00 p < 0.05; Figure 3). There were no significant changes over time in the activity of non-phosphorylated NR under constant light (p > 0.05). Non-phosphorylated NR activity was significantly lower at 1 h after dark (15:00) in the light:dark treatment than non-phosphorylated NR activity from the same time point in constant light treatment (p < 0.05).
3.1.2. Short-term light experiment
In the short-term experiment, there was no significant difference in total or non-phosphorylated NR activity between samples in light vs. those transferred to dark for 15 min (p > 0.05; Supplementary Figure S1). Additionally, there was no significant difference in total and non-phosphorylated NR activity within samples for either light condition (p > 0.05; Supplementary Figure S1).
3.2. NR activity during growth with different nitrogen sources
Only the treatments cultured with NO3− exhibited NR activity (Supplementary Figure S2), while activity in treatments with NH4+ was below detection. For the nitrate treatments, there was no significant difference between the total and non-phosphorylated NR activity (p > 0.05).
3.3. NR activity when growing at different temperatures
3.3.1. Long-term temperature acclimation experiment
After acclimation to temperatures ranging from 18 ° to 28°C, total NR activity was significantly higher in cultures at 18°C compared to those at 28°C (p < 0.05; Figure 4), but no significant difference was observed in total NR activities in treatments cultured at 25°C compared to those at 18°C or 28°C (p > 0.05; Figure 4). Furthermore, non-phosphorylated NR activity was significantly lower than total NR activity in cultures acclimated to 18°C (p < 0.05), but not in the other two temperature treatments (p > 0.05).
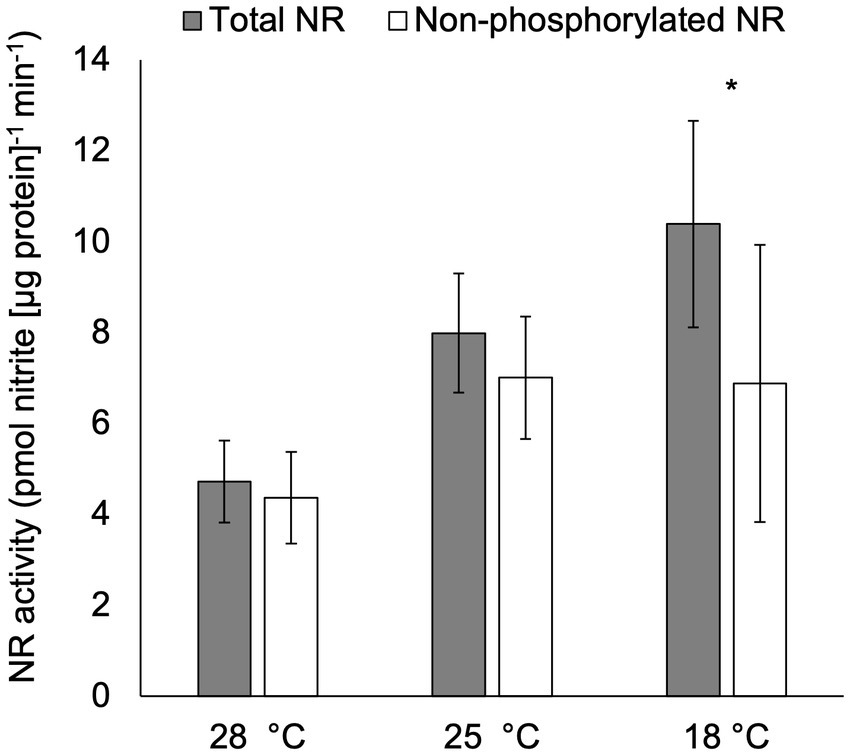
Figure 4. Activity (pmol nitrite [μg protein]−1 min−1) of total NR and non-phosphorylated NR in C. subsalsa acclimated to different temperatures (28°C, 25°C, and 18°C) in the long-term temperature experiment. Samples for NR activity assay were collected at 6 h after lights on. Asterisk (*) reveals the significant difference in NR activity between total and non-phosphorylated NR, indicating the regulation by reversible phosphorylation and the subsequent binding of putative 14-3-3 proteins, at the indicated temperature (p < 0.05). Error bars are standard deviation of three biological replicates.
3.3.2. Short-term temperature change experiment
There were no significant differences in total NR activity for cultures moved from 25°C to 8°C in the short-term temperature manipulation experiment (p > 0.05; Figure 5). However, non-phosphorylated NR activity was significantly lower than total NR activity for cultures moved from 25°C to 8°C (p < 0.05).
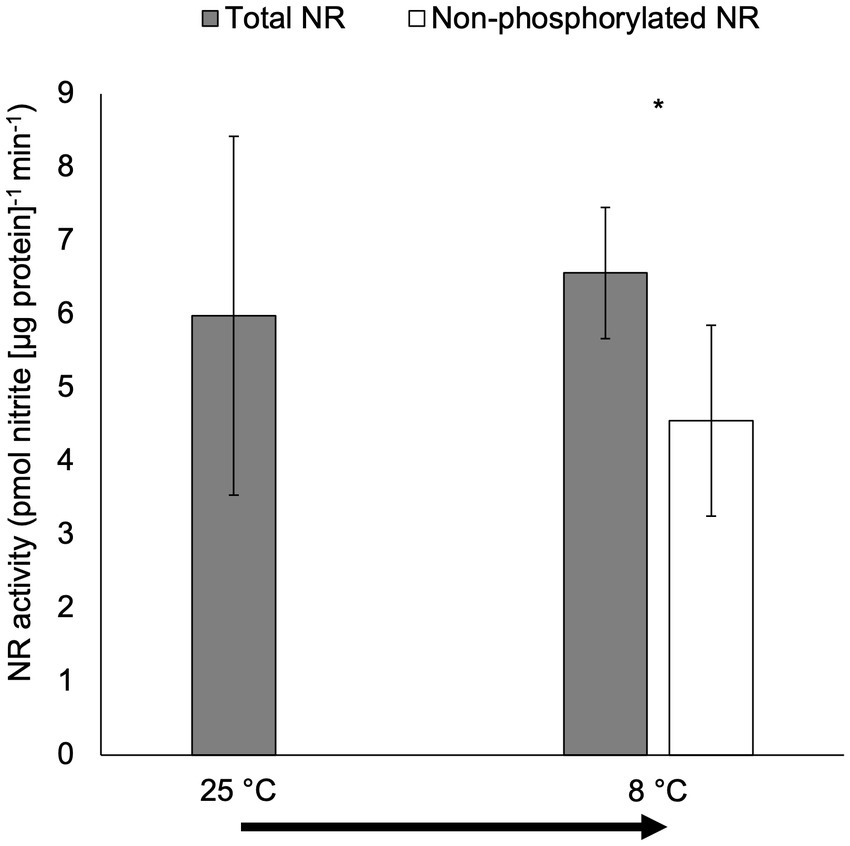
Figure 5. Activity (pmol nitrite [μg protein]−1 min−1) of total NR and non-phosphorylated NR in the short-term temperature experiment when the culture was transferred from 25°C to 8°C (15-min incubation after the transfer; only total NR activity was measured for the culture under 25°C). The arrow indicates the short-term temperature change. Samples for NR activity assay were collected at 6 h after lights on. Asterisk (*) reveals the significant difference in NR activity between total and non-phosphorylated NR, indicating the regulation by reversible phosphorylation and the subsequent binding of putative 14-3-3 proteins, at the indicated temperature (p < 0.05). Error bars are standard deviation of three biological replicates.
4. Discussion
This study aimed to investigate post-transcriptional (translational and/or post-translational) regulation of nitrate reductase enzymes (NR) in the harmful alga C. subsalsa. In contrast to NR1 and NR2 in H. akashiwo, which are identical except for the insertion of a 2/2HbN domain in NR2 (Coyne, 2010; Stewart and Coyne, 2011), analysis of NR amino acid sequences in C. subsalsa shows that NR3 was distinct from NR2 (Wang et al., 2018; Figure 1). C. subsalsa NR3 contained a potential phosphorylation site and a canonical 14-3-3 protein binding motif in the Hinge-1 region similar to those found in plant NRs (Wang et al., 2018), suggesting that this enzyme may be regulated by 14-3-3 protein binding (Figure 2). Here, the maximum, or total, NR enzyme activity (phosphorylated and non-phosphorylated) was compared to non-phosphorylated NR as a measure of “actual” or uninhibited activity within the cell. Differences between total and non-phosphorylated NR activity indicate a response to environmental conditions or nutrient status regulated by reversible phosphorylation and putative binding of 14-3-3 proteins. In this investigation, responses to light regime, nutrient source, and temperature were evaluated in order to identify factors involved in post-translational regulation of NR activity by putative 14-3-3 protein binding.
The previous study on the gene expression profile of NR2 and NR3 in C. subsalsa under light: dark cycle vs. constant light demonstrated that the gene expression of both enzymes was regulated by light rather than the circadian clock (Wang et al., 2018). A similar pattern was observed for the activity of total and non-phosphorylated NR in C. subsalsa, with activity responding to the light regime and not regulated by an endogenous diel rhythm (Figure 3). However, NR activity was decoupled from gene expression in C. subsalsa, with the highest activity near mid-day (Figure 3 upper panel), following the gene expression maximum in the early morning (Wang et al., 2018). Decoupling of expression and activity has been observed in other plants and algae (Chow et al., 2004; Lea et al., 2006; Falcão et al., 2010), in which the highest levels of NR activity matched the high photosynthetic rates accompanying the light phase (e.g., Granbom et al., 2004; Lea et al., 2006; Young et al., 2007). The similar rhythm of photosynthesis and nitrate assimilation is likely due to a combination of the requirement for ATP and reduced ferredoxin generated by photosynthesis for nitrite reduction, while NR serves as a crucial sink for excess reductant produced by the light reactions of photosynthesis (reviewed by Chow et al., 2004; Sanz-Luque et al., 2015a; Glibert et al., 2016).
There were no significant differences between the activity of total and non-phosphorylated NR in response to a 12:12 light: dark cycle (Figure 3), or in the short-term light shift experiment (Supplementary Figure S1), indicating that light-driven changes in NR activity were not regulated post-translationally by putative 14-3-3 protein binding at time points investigated here. These results differed from the reports on plant NRs, where these enzymes are transiently inactivated by binding of 14-3-3 proteins responding to a change in light intensity (Lea et al., 2006; Nemie-Feyissa et al., 2013). It is likely that other mechanisms played a role in regulating NR activity in C. subsalsa in response to changes in the light regime. For instance, research in plants demonstrated NR phosphorylation by mitogen-activated protein kinases (MAPKs) play a role in the regulation of its activity (Wang et al., 2010), and recent research suggested MAPK kinase kinases (MAPKKKs) in the green alga Chlamydomonas reinhardtii may also be involved in nitrogen assimilation (Gomez-Osuna et al., 2020). The role of alternative mechanisms involved in the regulation of C. subsalsa NR activity is unknown and requires future research.
In higher plants, 14-3-3 proteins modulate the circadian activation of hydraulic conductivity, which is essential for plants to synchronize water supply to optimize growth with diurnal cycles (Prado et al., 2019). This regulatory mechanism is also evident in studies demonstrating a post-translational oscillator and protein phosphorylation involved in the circadian-clock-mediated cellular processes in cyanobacteria (Chang et al., 2015; Tseng et al., 2017; Brenna and Albrecht, 2020). In the research conducted here, the significant difference in total vs. non-phosphorylated NR activity at 6 h after dark (20:00) in cultures maintained at constant light suggests that putative 14-3-3 binding of NR3 is influenced by an endogenous rhythm that is not affected by light regime (Figure 3).
NR activity in C. subsalsa was responsive to nitrogen sources, with measurable NR activity in treatments cultured with nitrate (Supplementary Figure S2) but no detectable activity in ammonium treatments, demonstrating the translational regulation of NR by nitrogen sources in this species. This is in contrast to previous research showing constitutive expression of NR2 and NR3 transcripts in cultures of C. subsalsa supplied only with ammonium (Wang et al., 2018), and suggests feedback inhibition by the downstream products of nitrogen assimilation at the level of translation (Ali et al., 2007). These results are consistent with reports on other algal species, where NR activity is strongly suppressed by high concentrations of ammonium (Young et al., 2007; Chow and de Oliveira, 2008). Additionally, there was no indication for putative 14-3-3 binding involved in post-translational regulation in C. subsalsa cultured with nitrate here. To be noted, the nitrate concentration used in this study (100 μM) was much higher than concentrations typically measured in the environment. It has been well established that in higher plants, NRs are regulated at the post-translational level with reversible phosphorylation by the relative proportions of nitrate to ammonium, as well as the carbon to nitrate or ammonium balances (Engelsberger and Schulze, 2012; Huarancca Reyes et al., 2018). For instance, in Arabidopsis, phosphorylation levels of NR were higher in cultures resupplied with ammonium compared to nitrate after nitrogen starvation (Engelsberger and Schulze, 2012), and NR was inhibited by reversible phosphorylation and subsequent binding of 14-3-3 proteins when treated with elevated glucose and nitrate or ammonium as the nitrogen source (Huarancca Reyes et al., 2018). Additional research will further our understanding of NR regulation in C. subsalsa exposed to more environmentally relevant nitrate and ammonium concentrations.
The role of protein phosphorylation and 14-3-3 protein binding in regulating NR activities in response to temperature has also been reported in higher plants. For example, NR was activated by low temperatures through protein dephosphorylation and the subsequent dissociation of 14-3-3 binding proteins in leaves from winter wheat (Triticum aestivum L. cv. Sadovo-1) (Yaneva et al., 2002). Reports indicate that 14-3-3 proteins also play other roles in temperature-mediated regulation; these proteins regulate freezing tolerance and cold acclimation in higher plants by controlling cold-induced gene expression (Catalá et al., 2014) and protect animal cells from heat stress by dissolving thermal-aggregated proteins (Yano et al., 2006). Little is known, however, about the temperature-mediated regulatory mechanisms involved in enzyme activity in unicellular algae. In C. subsalsa, a significant difference between total and non-phosphorylated NR was observed at lower temperatures in both the long- (18°C) and short-term (8°C) experiments in this study (Figures 4, 5), providing the strongest evidence for post-translational regulation of NR activity involving reversible phosphorylation and putative 14-3-3 protein binding.
Elevated total NR activity at lower temperatures observed in the long-term temperature acclimation experiment (Figure 4) could be partially driven by regulation at the transcriptional level, where expression of NR2 in C. subsalsa was highest in cultures acclimated to lower temperatures (Wang et al., 2018). Higher NR activity at lower temperatures was also observed in other studies and suggested that increased NR activity may act to dissipate excess energy generated under high-light and low-temperature conditions (Parker and Armbrust, 2005). Light conditions here, however, were not excessive for C. subsalsa (100 μE m−2 s−1) (Zhang et al., 2006). This may have contributed to the fine-tuning of post-translational regulation of NR activity at 18°C by putative 14-3-3 proteins. Evidence presented here and in Wang et al. (2018) suggests that the up-regulation of transcription (Wang et al., 2018) and the higher total activity of NR at lower temperatures (Figure 4) after long-term acclimation were modulated by post-translational mechanisms that occurred over a short period of time. That is, the observed significant difference in total vs. non-phosphorylated NR within 15 min after the shift from 25°C to 8°C (Figure 5) persisted after long-term acclimation to low temperature (Figure 4). Under low light, low temperature conditions, results presented here suggest post-translational mechanisms involving reversible phosphorylation and putative 14-3-3 protein binding to inhibit NR activity. This inhibition in activity occurred in spite of higher transcript abundance (Wang et al., 2018) and total NR activity, and resulted in no significant differences in non-phosphorylated (uninhibited) NR enzyme activity over the temperature range tested here (Figure 4).
In contrast to the long-term temperature acclimation experiment, there was no increase in the activity of total NR when cultures were shifted to a lower temperature during the short-term experiment (Figure 5), demonstrating that mechanisms involved in the translational regulation of NRs in C. subsalsa responding to temperatures require a longer period of cold stress. Together, these results indicate that the (relatively) slower translational response results in elevated total NR activity at lower temperatures after long-term acclimation, but that putative reversible phosphorylation and 14-3-3 protein binding can rapidly modulate NR activity in response to other environmental factors.
In conclusion, the results of this study demonstrated translational and post-translational regulation of NR enzymes in C. subsalsa by different environmental factors. Total NR activity was regulated at the level of translation by light, nitrogen source, and temperature. At the post-translational level, evidence was presented supporting the regulation of NR activity in C. subsalsa through the reversible phosphorylation and subsequent binding of 14-3-3 proteins under constant light and low-temperature conditions. The ability of C. subsalsa to differentially regulate NRs at multiple levels (transcriptional, translational, and post-translational levels) may allow this species to fine tune nitrogen assimilation with photosynthetic activities in response to environmental cues. To date, this is the first report of NR regulation at the post-translational level involving reversible phosphorylation and subsequent binding of putative 14-3-3 proteins in algal species and suggests that regulatory mechanisms for NR activity may be shared between plants and some algal species.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YW conducted the research, analyzed the data, and wrote the manuscript. GJ conducted the research and contributed to data analysis. AP processed the samples. KC conceived of and supervised the project and contributed to manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the Gordon and Betty Moore Foundation through grant #2637 to the National Center for Genome Resources. This research was also funded by Delaware Sea Grant College Program (grant #R/SCD-2 to KC). Additional funding was provided through the National Oceanic and Atmospheric Administration National Center for Coastal Ocean Science (NCCOS) Ecology and Oceanography of Harmful Algal Blooms (ECOHAB) program (grant # NA18NOS4780165 to KJC); publication #ECO1042.
Acknowledgments
We thank Julie Rose (US NOAA) for her assistance with statistical analysis. The content of this manuscript previously appeared online as a Master’s thesis by YW from the University of Delaware (Title: Expression and activity of novel nitrate reductase enzymes in Chattonella subsalsa and implications for competitive dynamics in marine environments).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1059074/full#supplementary-material
Footnotes
References
Ali, A., Sivakami, S., and Raghuram, N. (2007). Effect of nitrate, nitrite, ammonium, glutamate, glutamine and 2-oxoglutarate on the RNA levels and enzyme activities of nitrate reductase and nitrite reductase in rice. Physioll. Mol. Biol. Plants 13, 17–25.
Athwal, G. S., and Huber, S. C. (2002). Divalent cations and polyamines bind to loop 8 of 14-3-3 proteins, modulating their interaction with phosphorylated nitrate reductase. Plant J. 29, 119–129. doi: 10.1046/j.0960-7412.2001.01200.x
Bellido-Pedraza, C. M., Calatrava, V., Llamas, A., Fernandez, E., Sanz-Luque, E., and Galvan, A. (2022). Nitrous oxide emissions from nitrite are highly dependent on nitrate reductase in the microalga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 23:9412. doi: 10.3390/ijms23169412
Bellido-Pedraza, C. M., Calatrava, V., Sanz-Luque, E., Tejada-Jiménez, M., Llamas, Á., Plouviez, M., et al. (2020). Chlamydomonas reinhardtii, an algal model in the nitrogen cycle. Plan. Theory 9, 1–14. doi: 10.3390/plants9070903
Berges, J., and Harrison, P. J. (1995). Nitrate reductase activity quantitatively predicts the rate of nitrate incorporation under steady state light limitation: a revised assay and characterization of the enzyme in three species of marine phytoplankton. Limnol. Oceanogr. 40, 82–93. doi: 10.4319/lo.1995.40.1.0082
Brenna, A., and Albrecht, U. (2020). Phosphorylation and circadian molecular timing. Front. Physiol. 11:1539. doi: 10.3389/FPHYS.2020.612510/BIBTEX
Campbell, W. H. (1999). Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 277–303. doi: 10.1146/annurev.arplant.50.1.277
Campbell, W. H. (2001). Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 58, 194–204. doi: 10.1007/PL00000847
Catalá, R., López-Cobollo, R., Mar Castellano, M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. doi: 10.1105/TPC.114.127605
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Chang, Y. G., Cohen, S. E., Phong, C., Myers, W. K., Kim, Y. I., Tseng, R., et al. (2015). A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science. 349, 324–328. doi: 10.1126/science.1260031
Chi, J. C., Roeper, J., Schwarz, G., and Fischer-Schrader, K. (2015). Dual binding of 14-3-3 protein regulates Arabidopsis nitrate reductase activity. J. Biol. Inorg. Chem. 20, 277–286. doi: 10.1007/s00775-014-1232-4
Chow, F., and de Oliveira, M. C. (2008). Rapid and slow modulation of nitrate reductase activity in the red macroalga Gracilaria chilensis (Gracilariales, Rhodophyta): influence of different nitrogen sources. J. Appl. Phycol. 20, 775–782. doi: 10.1007/s10811-008-9310-z
Chow, F., De Oliveira, M. C., and Pedersén, M. (2004). In vitro assay and light regulation of nitrate reductase in red alga Gracilaria chilensis. J. Plant Physiol. 161, 769–776. doi: 10.1016/j.jplph.2004.01.002
Coyne, K. J. (2010). Nitrate reductase (NR1) sequence and expression in the harmful alga Heterosigma akashiwo (Raphidophyceae). J. Phycol. 46, 135–142. doi: 10.1111/j.1529-8817.2009.00781.x
Coyne, K. J., Wang, Y., Wood, S. A., Countway, P. D., and Greenlee, S. M. (2022). “Current applications and technological advances in quantitative real-time PCR (qPCR): a versatile tool for the study of phytoplankton ecology,” in Advances in phytoplankton ecology: Applications of emerging technologies. eds. L. Clementson, R. Eriksen, and A. Willis (Elsevier), 303–351.
Davidson, K., Gowen, R. J., Tett, P., Bresnan, E., Harrison, P. J., McKinney, A., et al. (2012). Harmful algal blooms: how strong is the evidence that nutrient ratios and forms influence their occurrence? Estuar. Coast. Shelf Sci. 115, 399–413. doi: 10.1016/j.ecss.2012.09.019
Engelsberger, W. R., and Schulze, W. X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen- starved Arabidopsis seedlings. Plant J. 69, 978–995. doi: 10.1111/j.1365-313X.2011.04848.x
Falcão, V. R., Oliveira, M. C., and Colepicolo, P. (2010). Molecular characterization of nitrate reductase gene and its expression in the marine red alga Gracilaria tenuistipitata (Rhodophyta). J. Appl. Phycol. 22, 613–622. doi: 10.1007/s10811-010-9501-2
Gao, J., van Kleeff, P. J. M., Oecking, C., Li, K. W., Erban, A., Kopka, J., et al. (2014). Light modulated activity of root alkaline/neutral invertase involves the interaction with 14-3-3 proteins. Plant J. 80, 785–796. doi: 10.1111/TPJ.12677
Glibert, P. M., Wilkerson, F. P., Dugdale, R. C., Raven, J. A., Dupont, C. L., Leavitt, P. R., et al. (2016). Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol. Oceanogr. 61, 165–197. doi: 10.1002/lno.10203
Gomez-Osuna, A., Calatrava, V., Galvan, A., Fernandez, E., and Llamas, A. (2020). Identification of the MAPK cascade and its relationship with nitrogen metabolism in the green alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 21:3417. doi: 10.3390/ijms21103417
Granbom, M., Chow, F., Lopes, P. F., De Oliveira, M. C., Colepicolo, P., De Paula, E. J., et al. (2004). Characterisation of nitrate reductase in the marine macroalga Kappaphycus alvarezii (Rhodophyta). Aquat. Bot. 78, 295–305. doi: 10.1016/j.aquabot.2003.11.001
Guillard, R. R. L., and Ryther, J. H. (1962). Studies of marine planktonic diatoms I. Cyclotella nana Hustedt, and Detonula confervasea (Cleve). Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Healey, E. M., Flood, S., Bock, P. K., Fulweiler, R. W., York, J. K., and Coyne, K. J. (2023). Effects of nitrate and ammonium on assimilation of nitric oxide by Heterosigma akashiwo. Sci. Rep. 13:621. doi: 10.1038/s41598-023-27692-3
Huarancca Reyes, T., Scartazza, A., Pompeiano, A., Ciurli, A., Lu, Y., Guglielminetti, L., et al. (2018). Nitrate reductase modulation in response to changes in C/N balance and nitrogen source in Arabidopsis. Plant Cell Physiol. 59, 1248–1254. doi: 10.1093/pcp/pcy065
Imai, I., and Yamaguchi, M. (2012). Life cycle, physiology, ecology and red tide occurrences of the fish-killing raphidophyte Chattonella. Harmful Algae 14, 46–70. doi: 10.1016/j.hal.2011.10.014
Kok, J. W. K., Yeo, D. C. J., and Leong, S. C. Y. (2019). Growth, pigment, and chromophoric dissolved organic matter responses of tropical Chattonella subsalsa (Raphidophyceae) to nitrogen enrichment. Phycol. Res. 67, 134–144. doi: 10.1111/pre.12360
Kumar, A., and Bera, S. (2020). Revisiting nitrogen utilization in algae: a review on the process of regulation and assimilation. Bioresour Technol Rep 12:100584. doi: 10.1016/j.biteb.2020.100584
Lea, U. S., Leydecker, M.-T., Quilleré, I., Meyer, C., and Lillo, C. (2006). Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol. 140, 1085–1094. doi: 10.1104/pp.105.074633
Lum, W. M., Benico, G., Doan-Nhu, H., Furio, E., Leaw, C. P., Leong, S. C. Y., et al. (2021). The harmful raphidophyte Chattonella (Raphidophyceae) in Western Pacific: its red tides and associated fisheries damage over the past 50 years (1969–2019). Harmful Algae 107:102070. doi: 10.1016/j.hal.2021.102070
Mackintosh, C., Douglas, P., and Lillo, C. (1995). Identification of a protein that inhibits the phosphorylated form of nitrate reductase from spinach (Spinacia oleracea) leaves. Plant Physiol. 107, 451–457. doi: 10.1104/pp.107.2.451
Malerba, M. E., Connolly, S. R., and Heimann, K. (2015). An experimentally validated nitrate-ammonium-phytoplankton model including effects of starvation length and ammonium inhibition on nitrate uptake. Ecol. Model. 317, 30–40. doi: 10.1016/j.ecolmodel.2015.08.024
Nemie-Feyissa, D., Królicka, A., Førland, N., Hansen, M., Heidari, B., and Lillo, C. (2013). Post-translational control of nitrate reductase activity responding to light and photosynthesis evolved already in the early vascular plants. J. Plant Physiol. 170, 662–667. doi: 10.1016/j.jplph.2012.12.010
Parker, M. S., and Armbrust, E. V. (2005). Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae)1. J. Phycol. 41, 1142–1153. doi: 10.1111/j.1529-8817.2005.00139.x
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., and Ferl, R. J. (2012). 14-3-3 phosphoprotein interaction networks–does isoform diversity present functional interaction specification? Front. Plant Sci. 3:190. doi: 10.3389/fpls.2012.00190
Prado, K., Cotelle, V., Li, G., Bellati, J., Tang, N., Tournaire-Roux, C., et al. (2019). Oscillating aquaporin phosphorylation and 14-3-3 proteins mediate the circadian regulation of leaf hydraulics. Plant Cell 31, 417–429. doi: 10.1105/TPC.18.00804
Rezvani, F., Sarrafzadeh, M. H., Seo, S. H., and Oh, H. M. (2017). Phosphorus optimization for simultaneous nitrate-contaminated groundwater treatment and algae biomass production using Ettlia sp. Bioresour. Technol. 1, 785–792. doi: 10.1016/j.biortech.2017.08.053
Salbitani, G., and Carfagna, S. (2021). Ammonium utilization in microalgae: a sustainable method for wastewater treatment. Sustainability (Switzerland) 13, 1–17. doi: 10.3390/su13020956
Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A., and Fernandez, E. (2015a). Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 6:899. doi: 10.3389/fpls.2015.00899
Sanz-Luque, E., Ocaña-Calahorro, F., de Montaigu, A., Chamizo-Ampudia, A., Llamas, Á., Galván, A., et al. (2015b). THB 1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity. Plant J. 81, 467–479. doi: 10.1111/tpj.12744
Shen, Q. H., Gong, Y. P., Fang, W. Z., Bi, Z. C., Cheng, L. H., Xu, X. H., et al. (2015). Saline wastewater treatment by Chlorella vulgaris with simultaneous algal lipid accumulation triggered by nitrate deficiency. Bioresour. Technol. 1, 68–75.doi: 10.1016/j.biortech.2015.06.050
Stewart, J. J., and Coyne, K. J. (2011). Analysis of raphidophyte assimilatory nitrate reductase reveals unique domain architecture incorporating a 2/2 hemoglobin. Plant Mol. Biol. 77, 565–575. doi: 10.1007/s11103-011-9831-8
Tejada-Jimenez, M., Llamas, A., Galván, A., and Fernández, E. (2019). Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plan. Theory 8:56. doi: 10.3390/plants8030056
Tseng, R., Goularte, N. F., Chavan, A., Luu, J., Cohen, S. E., Chang, Y. G., et al. (2017). Structural basis of the day-night transition in a bacterial circadian clock. Science. 355, 1174–1180. doi: 10.1126/science.aag2
Wang, Y. (2016). Expression and Activity of Novel nitrate Reductase Enzymes in Chattonella subsalsa and Implications for Competitive Dynamics in Marine Environments. Master’s Thesis University of Delaware.
Wang, Y., Bouchard, J. N., and Coyne, K. J. (2018). Expression of novel nitrate reductase genes in the harmful alga, Chattonella subsalsa. Sci. Rep. 8, 13417–13412. doi: 10.1038/s41598-018-31735-5
Wang, P., Du, Y., Li, Y., Ren, D., and Song, C. P. (2010). Hydrogen peroxide–mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22, 2981–2998. doi: 10.1105/tpc.109.072959
Wang, D. S., Xu, D., Wang, Y. T., Fan, X., Ye, N. H., Wang, W. Q., et al. (2015). Adaptation involved in nitrogen metabolism in sea ICE alga Chlamydomonas sp. ICE-L to Antarctic extreme environments. J. Appl. Phycol. 27, 787–796. doi: 10.1007/s10811-014-0372-9
Wurtsbaugh, W. A., Paerl, H. W., and Dodds, W. K. (2019). Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum. WIREs Water 6, 1–27. doi: 10.1002/wat2.1373
Xu, H., Zhao, X., Guo, C., Chen, L., and Li, K. (2016). Spinach 14-3-3 protein interacts with the plasma membrane H+-ATPase and nitrate reductase in response to excess nitrate stress. Plant Physiol. Biochem. 106, 187–197. doi: 10.1016/j.plaphy.2016.04.043
Yaneva, I. A., Wolf Hoffmann, G., and Tischner, R. (2002). Nitrate reductase from winter wheat leaves is activated at low temperature via protein dephosphorylation. Physiol. Plant. 114, 65–72. doi: 10.1034/J.1399-3054.2002.1140110.X
Yano, M., Nakamuta, S., Wu, X., Okumura, Y., and Kido, H. (2006). A novel function of 14-3-3 protein: 14-3-3ζ is a heat-shock–related molecular chaperone that dissolves thermal-aggregated proteins. Mol. Biol. Cell 17, 4769–4779. doi: 10.1091/MBC.E06-03-0229
Young, E. B., and Berges, J. A. (2016). “Nitrate reductase: a nexus of disciplines, organisms, and metabolism” in Aquatic Microbial Ecology and Biogeochemistry: A Dual Perspective, eds. P. Glibert and T. Kana (Cham: Springer) 105–116. doi: doi: 10.1007/978-3-319-30259-1_9
Young, E. B., Dring, M. J., and Berges, J. A. (2007). Distinct patterns of nitrate reductase activity in brown algae: light and ammonium sensitivity in Laminaria digitata is absent in Fucus species. J. Phycol. 43, 1200–1208. doi: 10.1111/j.1529-8817.2007.00403.x
Yu, D., Yin, J., Wang, Y., Lu, A., He, Y., and Shen, S. (2022). Nitrogen assimilation-associated enzymes and nitrogen use efficiency of Pyropia yezoensis (Rhodophyta) in nitrate-sufficient conditions. Algal Res. 64:102682. doi: 10.1016/j.algal.2022.102682
Zhang, Y., Fu, F. X., Whereat, E., Coyne, K. J., and Hutchins, D. A. (2006). Bottom-up controls on a mixed-species HAB assemblage: a comparison of sympatric Chattonella subsalsa and Heterosigma akashiwo (Raphidophyceae) isolates from the Delaware inland bays, USA. Harmful Algae 5, 310–320. doi: 10.1016/j.hal.2005.09.001
Keywords: Chattonella subsalsa, nitrate reductase, 14-3-3 binding, harmful algal blooms, Heterosigma akashiwo, truncated 2/2 hemoglobin, NR3, phosphorylation
Citation: Wang Y, Johnson GI, Postles A and Coyne KJ (2023) Nitrate reductase enzymes in alga Chattonella subsalsa are regulated by environmental cues at the translational and post-translational levels. Front. Microbiol. 14:1059074. doi: 10.3389/fmicb.2023.1059074
Edited by:
Adriane Clark Jones, Mount St. Mary’s University, United StatesReviewed by:
Angel Llamas, University of Cordoba, SpainMolly Miller, University of South Alabama, United States
Copyright © 2023 Wang, Johnson, Postles and Coyne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathryn J. Coyne, kcoyne@udel.edu
†Present address: Anna Postles, LabVantage Solutions, Inc., Somerset, NJ, United States
 Yanfei Wang
Yanfei Wang Gretchen I. Johnson
Gretchen I. Johnson Anna Postles†
Anna Postles† Kathryn J. Coyne
Kathryn J. Coyne