Corrigendum: Comparative genomics revealed fluoroquinolone resistance determinants and OmpF deletion in carbapenem-resistant Escherichia coli
- 1Division of Infection, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan
- 2Department of Computer Science and Information Engineering, National Chung Cheng University, Chia-Yi, Taiwan
- 3Ph.D. Program in Translational Medicine, National Chung Hsing University, Taichung, Taiwan
- 4Rong Hsing Research Center for Translational Medicine, National Chung Hsing University, Taichung, Taiwan
Escherichia coli (E. coli) is a major causative organism of complicated urinary tract infections, bloodstream infections, and pneumonia. With the widespread use of antimicrobial agents, the prevalence of carbapenem resistance in E. coli has been increasing with limited therapeutic options. Fluoroquinolone remains a choice in carbapenem-resistant E. coli (CREc) that were once susceptible to the drug. Despite robust studies on the fluoroquinolone-resistant mechanisms of E. coli, few studies focused specifically on the group of CREc. In this study, we used comparative genomics to identify the fluoroquinolone-resistant mechanisms of CREc and detected gyrA D87N mutation in all the fluoroquinolone-resistant and CREc. Moreover, to investigate the mechanism underlying non-carbapenemase-producing carbapenem-resistant E. coli, we targeted the complete genome sequences for in-depth analysis and found a deletion in OmpF (DEL264-269) that might contribute to carbapenem resistance, which has not been reported before. Further studies focusing on the impact of these mutations on the expression levels are warranted. We further investigate the MLST, serotype, fimH type, phylogroup, and clinical characteristics of the CREc. Combination analysis of clinical and genomic characteristics suggests the polyclonal and highly diverse nature of the CREc in Taiwan. This study provides an insight into the molecular epidemiology of CREc in Taiwan.
Introduction
Escherichia coli (E. coli) is a major causative organism of complicated urinary tract infections, bloodstream infections, and pneumonia. Due to the advent of antimicrobial resistance, therapeutic options have become limited. The carbapenems are sometimes the only effective regimens against the severe infection caused by drug-resistant E. coli. However, with the widespread use of antimicrobial agents, the prevalence of carbapenem resistance in E. coli has increased. The reported occurrence of carbapenem-non-susceptible or -resistant E. coli varies by region. The prevalence rate of carbapenem-non-susceptible E. coli was 0–3% among the 12 Asia-Pacific nations (Mendes et al., 2013), 4.3% among hospitals in the US (McCann et al., 2018; Castanheira et al., 2020), and 0–7% in Europe (Kostyanev et al., 2019). According to surveillance conducted in Taiwan in 2009, the rate of carbapenem non-susceptibility in E. coli was 1.6–7.1% (Jean et al., 2013). The proportion of carbapenem-non-susceptible and -resistant E. coli has been increasing (Martirosov and Lodise, 2016). Moreover, patients infected with carbapenem-resistant pathogens, when compared to those infected with susceptible pathogens, showed increased morbidity and mortality (Nordmann and Poirel, 2019). The crude mortality of patients with carbapenem-resistant Enterobacterales infections is estimated to be 70% (Friedman et al., 2017). Carbapenem resistance increases the medical cost and causes an enormous economic burden (Bartsch et al., 2017).
Fluoroquinolone remains a choice in carbapenem-resistant E. coli (CREc) if the bacteria is found to be susceptible to the drug. Despite robust studies on fluoroquinolone-resistant mechanisms of E. coli, few studies specifically addressed the CREc group.
Two important mechanisms of resistance are observed in CREc. One is mediated by the carbapenemase genes, and the isolates are termed as carbapenemase-producing carbapenem-resistant E. coli (CP-CREc). They are classified according to the Ambler classification into three groups: class A, class B, and class D enzymes (Ambler, 1980). The mechanism of resistance in non-carbapenemase-producing carbapenem-resistant E. coli (non-CP-CREc) is mediated by ESBL or AmpC beta-lactamase associated with an overexpression of efflux pumps (such as AcrAB) or a loss of expression of porin (such as OprD and OprM) (Chetri et al., 2019; Theuretzbacher et al., 2020).
The prevalence of CP-CREc has increased in recent years in Taiwan (Jean et al., 2013), and the reported types of carbapenemases in CP-CREc is also increasing (Dortet et al., 2017). However, some mechanisms that are responsible for carbapenem resistance in E. coli are not well understood. In addition, the phylogenetic relationship, clinical phenotypes, and resistome characteristics still warrant further investigation. Whole-genome sequencing (WGS) provides a comprehensive method to identify the mechanisms underlying carbapenem resistance in E. coli, in particular non-CP-CREc, such as porin-encoding gene alternation along with ESBL or AmpC (Larkin et al., 2020). In this study, we aim to use WGS to characterize the molecular resistance mechanisms, virulence determinants, and phylogenetic relatedness of CREc.
Methods
Bacterial Isolates and Antibiotic Susceptibility Testing
This study was conducted at a tertiary hospital in Taichung, Taiwan. The CREc samples were collected from patients with bacteremia admitted during the period from January 2015 to May 2019, and we randomly selected five quinolone-resistant E. coli and five quinolone-susceptible E. coli. The E. coli isolates resistant to any of the ertapenem (minimum inhibitory concentrations [MICs] of ertapenem ≤ 2 μg/ml), imipenem, or meropenem (MIC of imipenem and meropenem ≤ 4 μg/ml) derivatives were enrolled in the study. We used MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) mass spectrometry analysis for the identification of bacteria. The MICs of gentamicin, amikacin, ceftriaxone, ceftazidime, cefepime, piperacillin-tazobactam, ertapenem, imipenem, ciprofloxacin, and trimethoprim-sulfamethoxazole were determined by using VITEK 2 Automated System (BioMe rieux, Marcy l'Etoile, France). We used Clinical and Laboratory Standards Institute-established criteria (CLSI M100-S29) to determine the susceptibility of bacteria to each antibiotic. Susceptibility to ceftazidime-avibactam was determined by broth microdilution method, as suggested by the Clinical and Laboratory Standards Institute (CLSI).
Epidemiological and Clinical Characteristics
We retrospectively reviewed the medical files of the patients with positive isolates of CREc. We collected demographic data, including age, gender, underlying conditions, and the use of antibiotics. In addition, antimicrobial susceptibility results, clinical parameters, and outcomes were also collected. The Pitt bacteremia score (PBS) is a severity of acute illness index and is broadly used in studies related to bloodstream infections. It scores from 0 to 14 points, and score ≥4 denotes critical condition and increased mortality (Chow and Yu, 1999). The sources of bacteremia were determined by a combination of clinical, radiological, and laboratory investigations. Intra-abdominal infection (IAI) encompasses a number of infectious processes, including peritonitis, cholecystitis, liver abscess, and bacterial translocation. Community-acquired CRE isolates were collected within 48 h of the patient's admission to the hospital. Hospital-acquired CRE isolates were collected after 48 h of admission (Garner et al., 1988; Weinstein et al., 1997).
DNA Extraction, Whole-Genome Sequencing, and Assembly of Genomes
Ten isolates of CREc were subjected to WGS using Nanopore GridION. The DNA library was prepared using the rapid barcoding kit (SQK-RBK004) by following the manufacturer's instructions, and the sequencing was performed in a R9.4.1 flowcell for 48 h. The sequenced reads were basecalled using Guppy 4.4.2. The adaptor sequences left in the reads were trimmed using Porechop (v0.2.4). The clean reads were assembled into contigs via Flye (v2.7) (Kolmogorov et al., 2019), and the remaining sequencing errors were sequentially polished by Racon, Medaka, and Homopolish (Vaser et al., 2017; Huang et al., 2021b). The assembled contigs were classified into chromosomes and plasmids using both NCBI BLAST and PlasmidFinder. Protein-coding genes, coding and non-coding RNAs in the chromosomes, and plasmids were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). Antibiotic-resistant genes (ARGs) were predicted by aligning protein-coding genes against the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2019). Only ARGs with alignment coverage greater than 90 % were retained. Efflux pumps were excluded from the ARG analysis. Beta-lactamases were separately predicted by using the curated hidden Markov models in NCBI AMRFinderPlus (Feldgarden et al., 2021).
The phylogeny of two outer membrane proteins, OmpC and OmpF, was reconstructed by MEGA (v11) and visualized by the Interactive Tree of Life (ITOL) (Tamura et al., 2021). Point mutations, insertions, and deletions in these porins were manually identified from the multiple sequence alignment in MEGA. Mutations within the quinolone resistance-determining region in GyrA, GyrB, ParC, and ParE of each CREc isolate were found by BLAST alignment against each other. The protein effects of each mutation (i.e., neutral or deleterious) were assessed by PROVEN (Choi et al., 2012).
Phylogenetic Grouping, MLST, Serotyping, and Virulence Determinant Analyses
We uploaded the assembled genomes into the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/), and the plasmid replicon type, MLST, serotype, and fimH type were determined using the following programs: PlasmidFinder (version 2.0.1), MLST (version 2.0.4), SerotypeFinder (version 2.0.1), VirulenceFinder 2.0 (version: 2.0.3), and FimTyper (version 1.0), respectively (Larsen et al., 2012; Carattoli et al., 2014; Joensen et al., 2014; Roer et al., 2017; Malberg Tetzschner et al., 2020). We uploaded the assembled genomes to the ClermonTyping v1.4.0 web tool to assess the phylogroups (Alikhan et al., 2018; Beghain et al., 2018).
Results
Demographic and Genetic Data
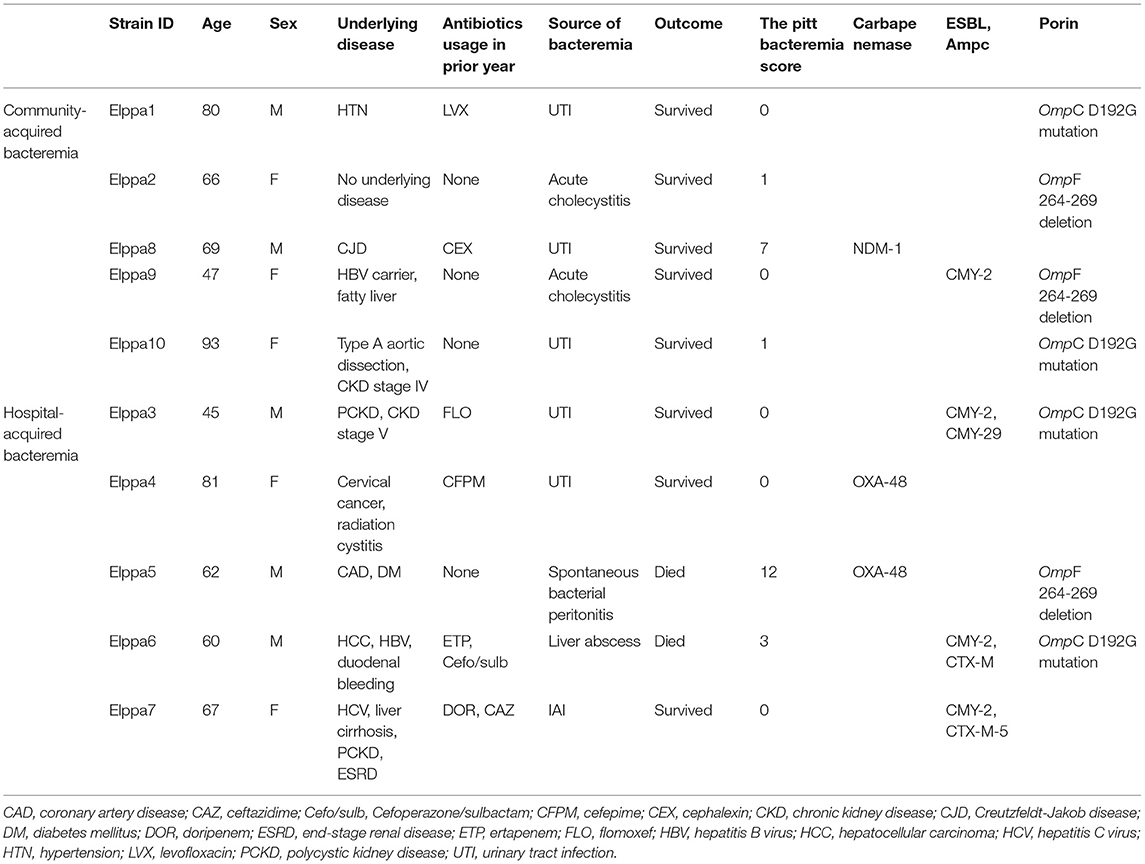
The clinical characteristics are summarized in Table 1. An equal number of strains were isolated from men and women. The median age of the patient was 66.5 years (IQR = 60–80). Half of the isolates suffered from community-acquired bacteremia, while the other half contracted hospital-acquired bacteremia. The sources of bacteremia in five isolates were urinary tract, while the intra-abdominal infection was the source of bacteremia in the remaining five isolates. Two patients expired, one of them was diagnosed with spontaneous bacterial peritonitis and the other had a liver abscess. Six out of the 10 patients were exposed to a carbapenem and broad-spectrum cephalosporin in the previous year. The median of the PBS was 0.5 (IQR = 0–3). Elppa 4, which harbored OXA-48 carbapenemase, had the highest PBS 12, and the second-highest PBS 7 was found in Elppa 8, which harbored NDM-1 mutation. Three out of 10 CREc isolates were carbapenemase-producing bacteria (OXA-48, n = 2; NDM-1, n = 1). Among non-CP-CREc, four out of seven isolates have ESBL or AmpC beta-lactamase. Comparative genomics revealed OmpC D192G mutation in four isolates and OmpF 264-269 deletion in another three isolates.
Mechanism of Carbapenem Resistance in E. coli
Among the 10 CREc isolates, three were carbapenemase-producing E. coli (CP-CREc, n = 3/10), with two harboring OXA-48 gene and one harboring NDM-1 gene, as presented in Table 2. None of the CP-CREc isolates had ESBL or AmpC enzymes. The OXA-48 genes are located on plasmids, and the NDM-1 gene is located on the chromosome. Seven isolates had no known carbapenemase genes. Among these non-CP-CREc, two isolates harbored AmpC enzymes (Elppa 3 and 9), and two isolates held the combination of both ESBL and AmpC (Elppa6 and 7). In comparison, three isolates neither had AmpC nor ESBL but had only broad-spectrum beta-lactamase (Elppa 1, 2, and 10).
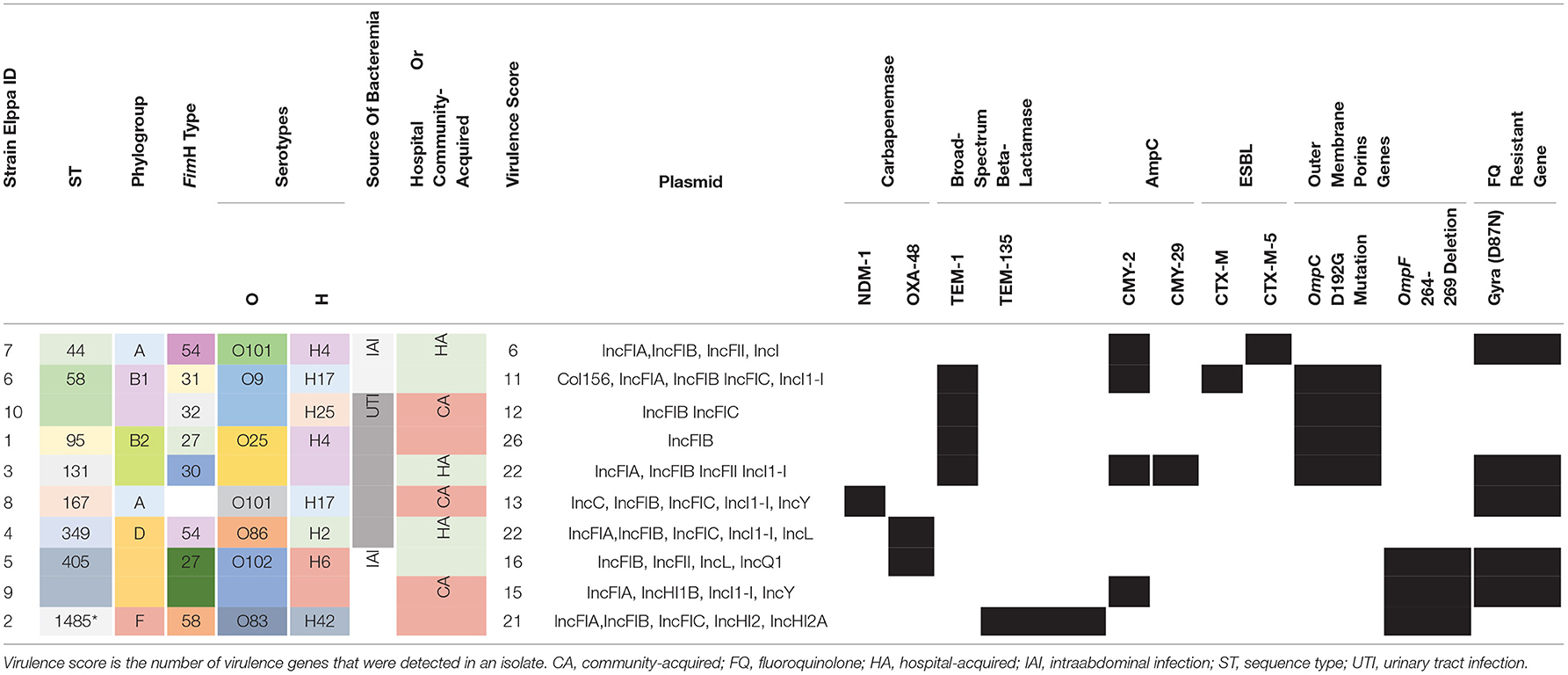
Table 2. Genetic diversity, source of bacteremia, and antibiotic-resistant mechanisms identified in the E. coli strains.
Loss of porin can also contribute to carbapenem resistance. In addition to the known OmpC D192G mutation (e.g., Elppa 1, 3, 6, and 10), we identified a novel deletion in OmpF (DEL264-269) in three isolates (Elppa 2, 5, and 9) (Figure 1A), where two of them lack carbapenemase. These carbapenemase-negative isolates may contribute to carbapenem resistance via loss of OmpF in conjunction with other beta-lactamases (e.g., TEM in Elppa 2 and CTX-M in Elppa 9). In particular, we observed that Elppa 2, 5, and 9 are phylogenetically clustered in OmpF (Figure 1B), implying that a subgroup resistant to carbapenems using a new mechanism is emerging. Similarly, the three isolates carrying the D192G mutation in OmpC are also grouped together (Supplementary Figure 1). These results suggested that multiple pathways leading to carbapenem resistance are emerging in these isolates.
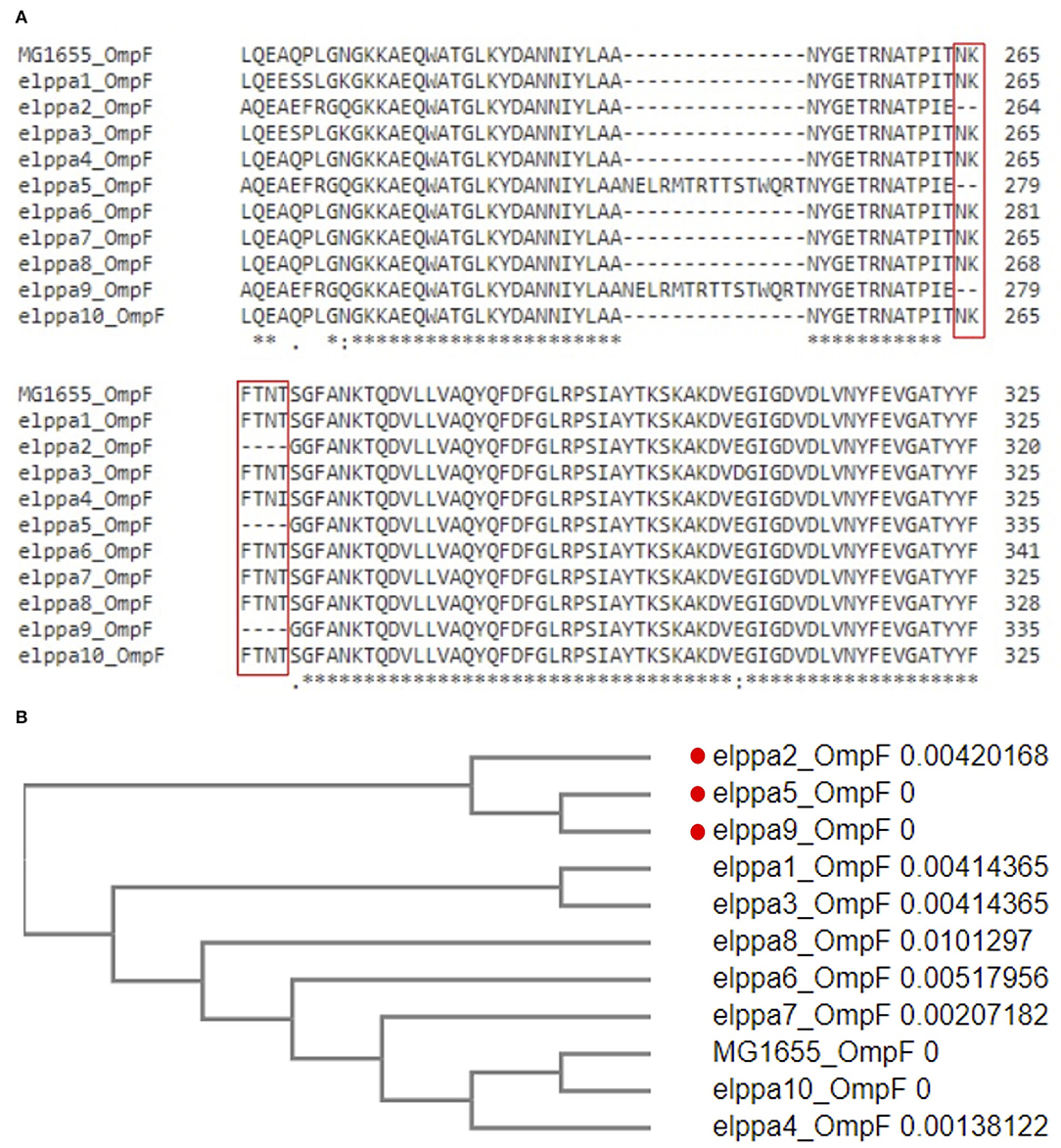
Figure 1. (A) Multiple sequence alignment of OmpF revealed deletion in 264-269. (B) Phylogenetic clusters in deleted OmpF.
Antimicrobial Resistance Profile and Resistance Mechanism Among CREc Isolates in This Study
Supplementary Table 1 presents the susceptibility testing results of CREC to antimicrobial agents. All the CREc isolates were resistant to ceftriaxone, ceftazidime, cefepime, piperacillin-tazobactam, ertapenem, and imipenem. The susceptibility rate for ciprofloxacin was 50%, in concordance with the isolation of gyrA (D87N) mutation (Table 2). The only isolate resistant to ceftazidime-avibactam was Elppa8, which had NDM-1 mutation.
We further compared the susceptibility patterns to different antimicrobial agents between non-carbapenemase-producing and carbapenemase-producing carbapenem-resistant E. coli isolates, and the results are presented in Table 3. The susceptible rate of ciprofloxacin (CP-CREc vs. Non-CP-CREc, 33.3 vs. 57.1%), amikacin (100 vs. 66.7%), and ceftazidime-avibactam (100 vs. 66.7%) was lower in CP-CREc when compared to Non-CP-CREc isolates.
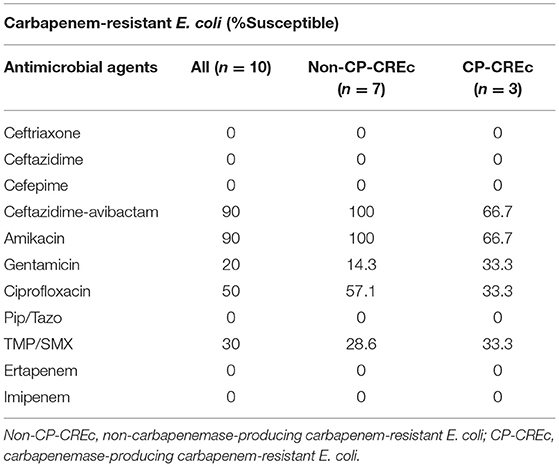
Table 3. The susceptibility patterns to different antimicrobial agents between non-carbapenemase-producing and carbapenemase-producing carbapenem-resistant E. coli isolates.
Phylogenetic Grouping, MLST, and Serotyping
Different typing approaches were used to unravel population structure and genetic diversity, including MLST, phylogenetic grouping, and serotyping, presented in Table 2. The CREc strains in this study showed a great variation in the sequence type, and eight ST types were identified. ST 58 and ST405 were detected in two isolates, while the remaining were singletons. The three carbapenemase-producing strains belong to different ST types. The E. coli isolate carrying NDM-1 gene was categorized as ST167, and the two OXA-48-positive E. coli strains were classified as ST 349 and ST 405, respectively. The ST types of non-carbapenemase-producing E. coli were also highly diverse. This finding implicates the polyclonal pathogenic carbapenem-resistant E. coli population.
In addition, we detected five phylogenic groups. Three isolates belonged to phylogroup D and two belonged to phylogroups B1 and B2. Phylogroup B2, which is in line with phylogroup D, was reported to include virulent extra-intestinal strains (Chakraborty et al., 2015). Phylogroups B2, F, and D had the highest number of virulence factors, with an average of 24, 21, and 17, respectively.
Virulence Genes
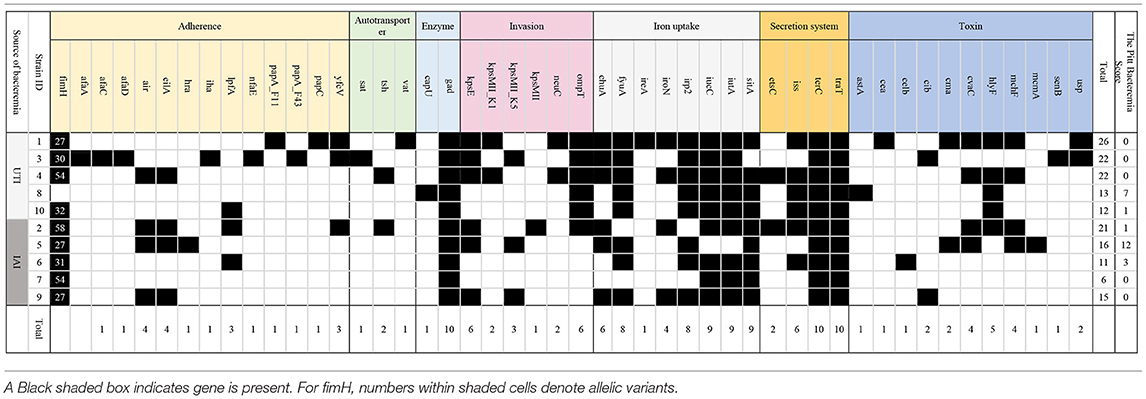
In total, we identified 47 virulence genes, and these genes impact different aspects of functions in E. coli, including adherence, autotransporter, enzyme, invasion, iron acquisition, secretion system, and toxin generation, which is presented in Table 4. The maximum number of detected virulence genes was 26 in Elppa1, which belonged to phylogroup B2. Elppa 7 had the smallest number of virulence genes (n = 6), and it belonged to phylogroup A.
Among the adhesion proteins of pathogenic E. coli, type 1 fimbrin D-mannose specific adhesin protein plays an important role and is encoded by the fimH gene. FimH typing provides a method for epidemiological typing of pathogenic E. coli. In this study, six types of fimH gene were identified in nine isolates, with fimH27 being the commonest (n = 3).
The most common genes that are sequenced are genes linked to adherence and toxin. All E. coli isolates in the present studies harbor gad, terC, and traT (Figure 2), which are all protection factors, including genes that mediate resistance to substances harmful to bacterial growth and are associated with the capsule. Glutamate decarboxylase (gad) is one of the most effective acid resistance systems in E.coli.
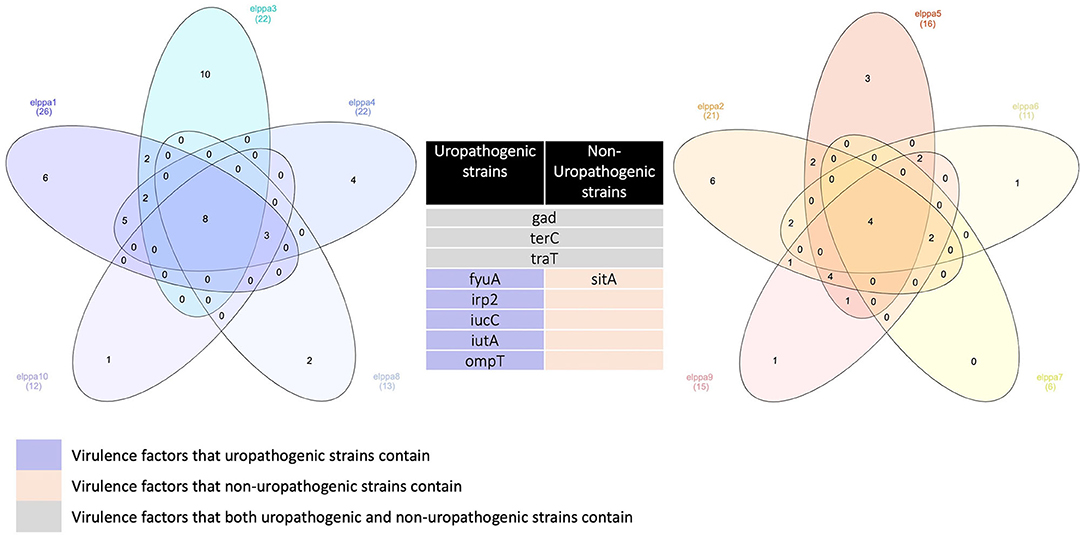
Figure 2. Venn diagrams comparing virulence factors of bloodstream infections from abdominal origin or urologic origin. Five E. coli isolates from the urinary tract shared eight virulence factors, including gad, terC, traT, fyuA, irp2, iucC, iutA, and ompT. Five E. coli isolates from abdominal origin shared four virulence factors, including gad, terC, traT, and sitA. Bloodstream infection from both sources contained virulence factors of gad, terC, and traT. Venn diagrams were created by InteractiVenn (Heberle et al., 2015).
The expression of glutamate decarboxylase is induced to maintain the physiological pH under acidic conditions (Capitani et al., 2003). Gad is particularly relevant for enteropathic E. coli, but it has been identified in patients with UTI, IAI, and bacteremia (Chen et al., 2021). Tellurite is deleterious to most bacteria. Tellurium ion resistance (TerC) has been implicated in tellurium resistance. It is hypothesized to promote the efflux of tellurium ions (Nguyen et al., 2021). TraT is an outer membrane protein that hinders the formation of membrane attack complex in the later stages (Miajlovic and Smith, 2014).
We further compared the virulence factors between uropathogenic and non-uropathogenic E. coli strains. Uropathogenic E. coli strains were the strains that recovered from the urinary sources of bacteremia, while non-uropathogenic E. coli strains were those that recovered from the intra-abdominal source. Uropathogenic E. coli strains (n = 5, totally contain 95 virulence factors) when compared to non-uropathogenic E. coli strains (n = 5, totally contain 69 virulence factors) had higher frequencies of virulence genes, as presented in Table 4 and Figure 2. All the uropathogenic strains had fyuA, irp2, iucC, iutA, ompT. FyuA, irp2, iucC, and iutA genes, and all were related to iron uptake.
To increase virulence, all the extra-intestinal pathogenic E. coli strains harbor siderophores, such as enterobactin, salmochelin, yersiniabactin, and aerobactin (Sarowska et al., 2019). Irp2 and fyuA were associated with yersiniabactin (Schubert et al., 1998), while iucC and iutA were associated with aerobactin (Ling et al., 2013). In addition, all the non-uropathogenic strains discovered from intra-abdominal infection harbor SitA. SitA is a subunit of an ABC-type ferric iron transport system (Reigstad et al., 2007), which mediates iron uptake from the periplasm into the cytoplasm.
Discussion
Carbapenems are the last resort to treat multidrug-resistant Gram-negative E. coli. With the increasing use of carbapenems, the prevalence of carbapenem-resistant E. coli is growing and poses a threat to public health and challenges to physicians. Only a few antibiotics are left for physicians to treat severe infectious diseases (Huang et al., 2021a). An understanding of the resistance mechanisms is essential to therapy and improved control measures. In this study, we demonstrated a high level of genetic diversity in CREc in both community and hospital settings. A previous study found that 0.04–29.5% of the CRE isolates were associated with the community (Kelly et al., 2017). Another report from Taiwan identified that 29.5% of CRE cases were community-acquired infections (Tang et al., 2016). In line with this study, CRE can be acquired from the community and hence poses an urgent public health threat.
In our study, the majority of CREc are not non-carbapenemase-producing strains. Two main molecular mechanisms contribute to carbapenem resistance in Enterobacteriaceae. The resistance is mediated either by carbapenemase production or by ESBL or AmpC in combination with increased efflux pump activity or porin alternation, as observed in the cases of non-carbapenemase-producing CRE. Among carbapenem-resistant and non-susceptible E. coli, the proportion of carbapenemase-producing strains varies across different regions of the world, as shown in Figure 3. Despite the fact that the definition of carbapenem non-susceptibility or resistance slightly varied between the studies, a trend toward an increasing number of CP-CREc cases is observed. The predominant carbapenemase varies in different geographic areas, as shown in Figure 3. Among the E. coli isolates that were non-susceptible/resistant to carbapenem in Taiwan, the rate of CP-CREc occurrence increased from 1.4% during 2010–2012 (Wang et al., 2015), to 7.59% during 2012–2015 (Chang et al., 2019), and to 29.5% during 2016–2018 (Huang et al., 2021a). According to previous studies in Taiwan, NDM-1 is the most common carbapenemase and accounts for 39.1% of CP-CREc (Chang et al., 2019; Huang et al., 2021a). In this study, 30% of the CREc are CP-CREc. Among the CP-CREc strains, OXA-48 and NDM-1 were presented. The data provide insight into the molecular characteristic of CREc in Central Taiwan.
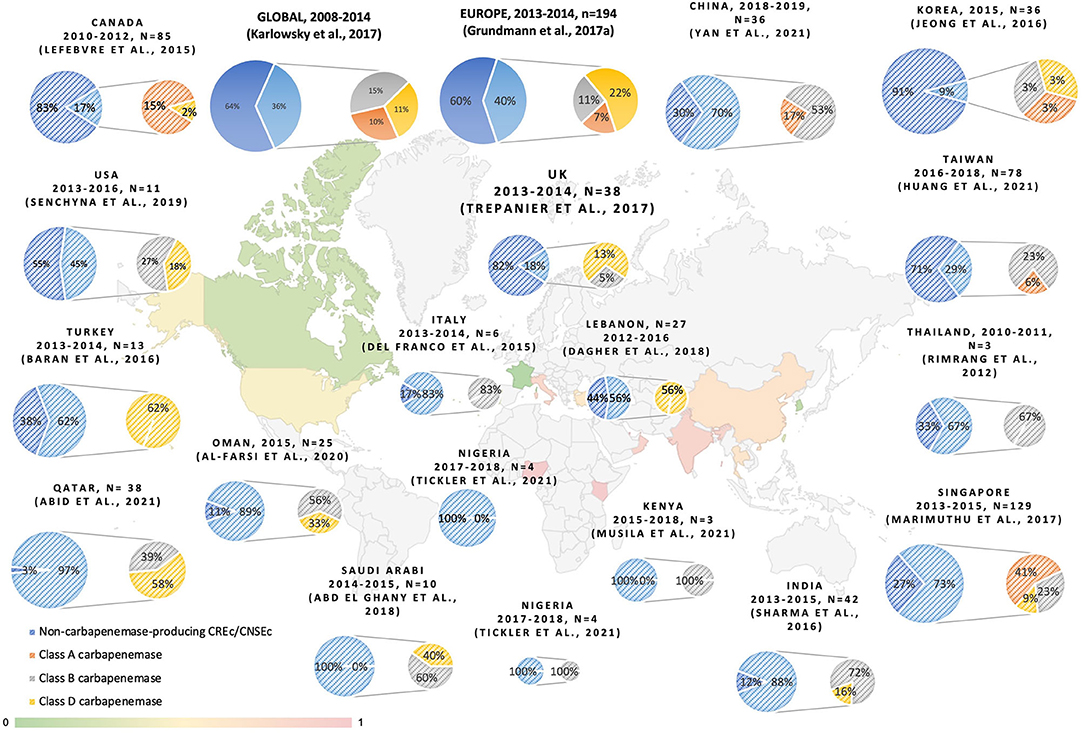
Figure 3. The reporting proportion of non-CP-CRE and carbapenemase among CREc/CNSEc (Rimrang et al., 2012; Del Franco et al., 2015; Lefebvre et al., 2015; Baran and Aksu, 2016; Dupont et al., 2016; Jeong et al., 2016; Sharma et al., 2016; Grundmann et al., 2017; Karlowsky et al., 2017; Marimuthu et al., 2017; Trepanier et al., 2017; Abd El Ghany et al., 2018; Dagher et al., 2018; Senchyna et al., 2019; Al-Farsi et al., 2020; Abid et al., 2021; Huang et al., 2021a; Musila et al., 2021; Tickler et al., 2021; Yan et al., 2021).
In this study, we detected OmpC D192G and OmpF 264-269 deletions using comparative genomics in non-CP-CREc. Porins, which are outer membrane proteins, block antibiotic permeation into bacteria and contribute to antibiotic resistance. Mutation in porins mediates antibiotic resistance by three different mechanisms: an alteration in the electrostatics of the channel, a reduction in the size of the porin channel, or a decrease in the expression level of porins or loss of porins (Fernández and Hancock Robert, 2012). In E. coli, two modifications of porin were observed: loss of OmpC and OmpF and mutation in OmpC (Vergalli et al., 2020). Lou et al. (2011) and Bajaj et al. (2016) proved that a single mutation in OmpC proteins could decrease the permeability of an antibiotic, by using liposome permeation assays and single-channel electrophysiology. OmpC D192G has been identified in a previous study and verified by PROVEAN (Al-Farsi et al., 2020; Tian et al., 2020). To our best knowledge, OmpF 264-269 deletion has not been reported before. OmpF 264-269 deletion might contribute to carbapenem resistance. Further studies regarding the impact of these mutations on the expression level are warranted.
In the present study, 10 isolates belonged to eight different ST types, indicating polyclonal spreading. Previous studies reported that 55.6% of NDM-positive E. coli isolates were classified as ST410 type (Huang et al., 2021a), and among carbapenem non-susceptible E. coli, ST131 is the predominant ST type in Taiwan (Ma et al., 2013; Mathers et al., 2015). The NDM-1-producing E. coli in the present study belonged to ST167, which differed from the results of previous studies. These findings suggest that pathogenic E. coli in Taiwan is polyclonal and highly genetically diverse.
The NDM-1 gene identified in this study is located within the chromosome, and the isolate belongs to ST167. Usually, blaNDM genes are located in various types of plasmids, such as IncA/C, IncF, IncNIncL/M, and untypable/IncR (Poirel et al., 2011; Khan et al., 2017). Shen et al. reported the first case of a chromosomally integrated blaNDM−1 E. coli which originated in China. The blaNDM−1 gene was inserted between two tandem copies of an insertion sequence common region 1 element, which originated from the plasmid of Proteus mirabilis. The chromosomally encoded blaNDM−1 E. coli strain belonged to ST167, which is identical to the blaNDM−1 E. coli strain of this study (Shen et al., 2017). In this study, we demonstrated that blaNDM−1 in E. coli can be chromosomally integrated and transmitted vertically. Close surveillance and monitoring are warranted to prevent the spread of the blaNDM−1 chromosomally integrated E. coli strains.
We also searched for the mechanism of quinolone resistance among the five quinolone-resistant CREc, and no qnr-like, horizontally transferred resistant genes were detected by BLAST in all the plasmids. We found GyrA D87N mutation within the chromosome in all the quinolone-resistant CREc strains in this study.
Whole-genome sequencing is an emerging technology to investigate molecular resistance mechanisms. While most previous studies used PCR to identify ESBL and AmpC beta-lactamase (Chia et al., 2009; Dupont et al., 2016; Ho et al., 2016; Tian et al., 2020), some studies employed WGS to identify molecular resistant mechanisms in non-carbapenemase-producing CREC (non-CP-CREc)/carbapenem non-susceptible E.coli (non-CP-CNEc) (Dagher et al., 2018; Senchyna et al., 2019; Al-Farsi et al., 2020). The major mechanisms of the non-CP-CRE/CNEC are AmpC or ESBL β-lactamase in combination with alternation or loss of an outer membrane porin (OmpC and OmpF), as shown in Figure 4. In Taiwan, plasmid-encoded AmpC β-lactamase CMY-2 and DHA-1 combined with OmpC/F loss was the primary mechanism (Chia et al., 2009; Ma et al., 2013). Despite intensive surveys, an average of 16% of the non-CP-CREc/CNEc isolates in the literature cannot be fully explained by the above-mentioned mechanism. In the present study, we found OmpF 264-269 deletion and verified via the online PROVEAN platform that this deletion was deleterious. There are still knowledge gaps regarding the mechanism underlying non-CP-CREc. Further studies on the efflux pump and regulatory protein, which regulate the expression of outer membrane porin, may improve our understanding of these issues.
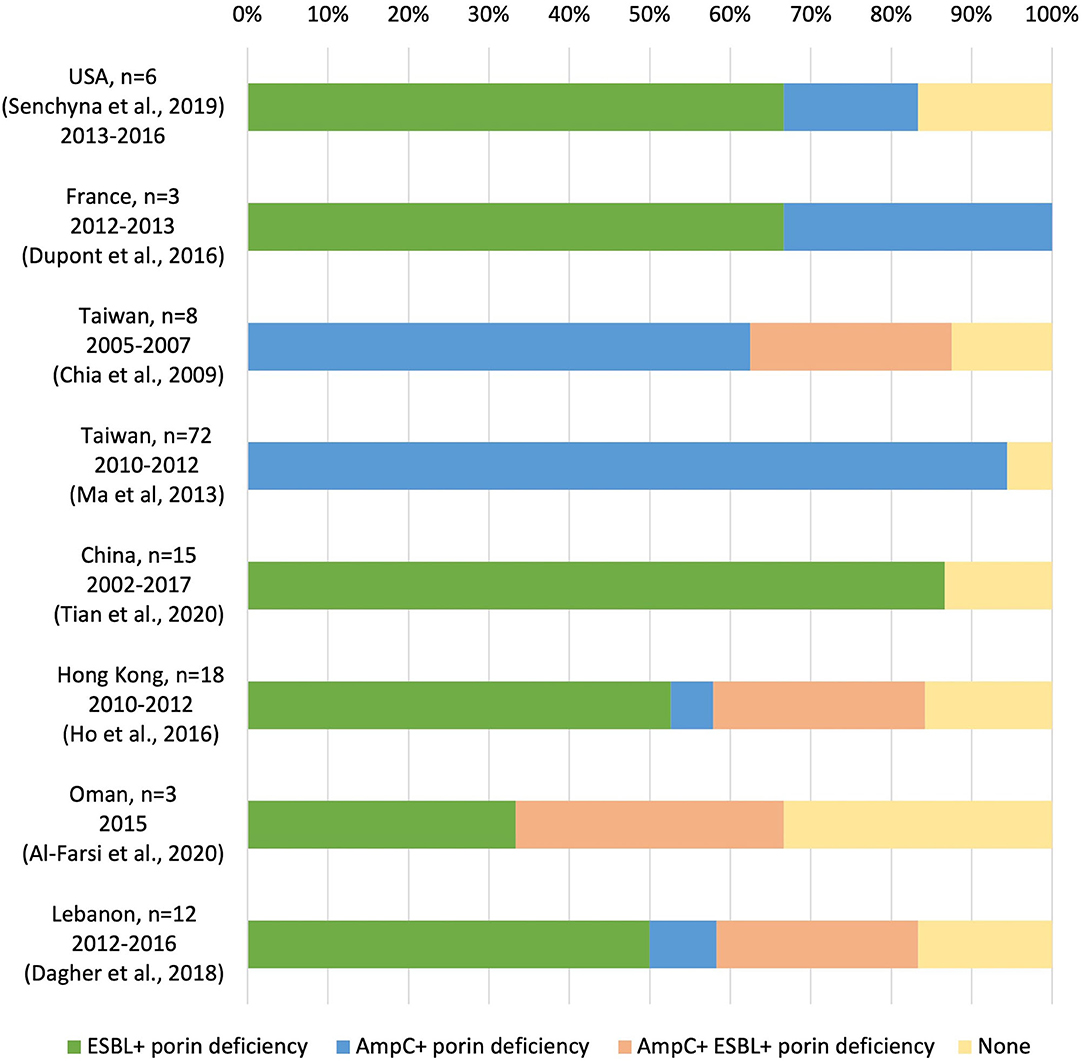
Figure 4. Molecular resistance mechanisms in non-carbapenemase-producing carbapenem-resistant E. coli/carbapenem non-susceptible E.coli (non-CP-CREc /non-CP-CNEc) (Chia et al., 2009; Ma et al., 2013; Dupont et al., 2016; Ho et al., 2016; Dagher et al., 2018; Senchyna et al., 2019; Al-Farsi et al., 2020; Tian et al., 2020).
A previous study demonstrated that CP-CRE strains had greater potency to hydrolyze carbapenems than non-CP-CRE strains and were more likely to resist other non-β-lactam antibiotics (Tamma et al., 2017). However, in our study, we could not identify differences in the MIC distribution of carbapenems. We did find that the susceptible rate of ciprofloxacin was lower in CP-CREc (33.3%) than in Non-CP-CREc (57.1%). Previous studies have proved that the mode of CREc acquisition differs between CP-CREc and non-CP-CREc isolates. The mode of acquisition of non-CP-CREc was through an endogenous pathway, which involves selective pressure of antibiotics on the intestinal microbiota and is primarily associated with recent exposure to antibiotics. While for CP-CREc, the mode of acquisition was primarily exogenous through “patient-to-patient” horizontal spread (Goodman et al., 2016; van Duin and Doi, 2017; Bouganim et al., 2020). By investigating the molecular mechanisms of CREc, we can better predict the risk factors among patients who are at risk of being infected with CREc and prevent CREc.
All E. coli strains in the present study harbor gad, terC, and traT. In this study, we found that uropathogenic E. coli strains (n = 5, totally contain 95 virulence factors), compared with non-uropathogenic E. coli strains (n = 5, totally contain 69 virulence factors), had higher frequencies of virulence genes. Extra-intestinal pathogenic E. coli (ExPEC) are multifaceted pathogens that can cause infections at extra-intestinal sites. The ExPEC group includes avian pathogenic E. coli, neonatal meningitis E. coli, uropathogenic E. coli (UPEC), and sepsis-associated E. coli (SEPEC). ExPEC are pathogenic to humans due to the presence of a constellation of virulence genes. A previous study discovered that E. coli that caused IAIs had fewer virulence genes than those that caused UTIs or primary bacteremia (Chen et al., 2021). It was postulated that during IAI, enteric E. coli may inadvertently translocate from the intestine to the bloodstream. A previous study had suggested that CP-CRE may be more virulent than non-CP-CRE and are associated with poorer outcomes (Tamma et al., 2017). Analysis has indicated zoonotic origin for ExPEC, as evident by the fact that avian and human E. coli isolates encompass similar virulence genes and that they belong to the same phylogenetic groups. This finding reminds us that to control the spread of ExPEC, poultry product surveillance and monitoring are important steps (Sarowska et al., 2019).
This study has several limitations. First, a genotype is not equal to a phenotype. Isolates harboring carbapenemase-producing genes are not identical to the isolates that express carbapenemase. Second, we did not conduct a drug susceptibility test for tigecycline and colistin. Third, the efflux pump was not analyzed in this study. Fourth, no function assessment of porin mutation was done in this study. Fifth, we defined community- and hospital-acquired CRE according to the previous studies (Garner et al., 1988; Weinstein et al., 1997). Thus, a patient with multiple exposures to the healthcare system can be classified into community-acquired CRE. Sixth, we could not explain the carbapenem-resistant mechanisms underlying three CREc isolates.
Conclusion
Complete genome sequencing detected that gyrA D87N mutation mediated fluoroquinolone resistance in all the CREc isolates in this study, suggesting a common mechanism. We also identified that OmpF 264-269 deletions might contribute to carbapenem resistance in CREc isolates. This study further provides insight into the clonal diversity, resistant mechanisms, and virulence factors of CREc strains isolated from patients with bacteremia in Taiwan. In E. coli, BlaNDM−1 mutation can be observed in patients with community-acquired bacteremia and in patients without risk factors for CRE infection. Combined clinical and genomic analyses indicate the highly diverse nature of the CREc in Taiwan.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP084378.
Author Contributions
W-TY, P-YL, and Y-TH contributed to the conception and design of the study and wrote the first draft of the manuscript. W-TY and P-YL contributed to the collection of clinical isolates. P-YL and Y-TH contributed to the experiments. Y-TH performed the bioinformatic analysis. All authors contributed to revising the manuscript and have approved the submitted version.
Funding
Y-TH was supported in part by the Ministry of Science and Technology (109-2221-E-194-038-MY3). P-YL was supported in part by the Ministry of Science and Technology (110-2314-B-075A-011) and the Taichung Veterans General Hospital (TCVGH-1113901C, TCVGH-1113901D, TCVGH-NK1109004, TCVGH-NK1099002, and TCVGH-1093902D).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.886428/full#supplementary-material
Abbreviations
CA, community-acquired; CAD, coronary artery disease; CAZ, ceftazidime; Cefo/sulb, Cefoperazone/sulbactam; CEX, cephalexin; CFPM, cefepime; CJD, Creutzfeldt-Jakob disease; CKD, chronic kidney disease; CLSI, Clinical and Laboratory Standards Institute; CNSE, carbapenem non-susceptible Enterobacteriaceae; CP-CRE, carbapenemase-producing carbapenem-resistant Enterobacteriaceae; CP-CREc, carbapenemase-producing carbapenem-resistant E. coli; DM, diabetes mellitus; DOR, doripenem; ESRD, end-stage renal disease; ETP, ertapenem; ExPEC, extra-intestinal pathogenic E. coli; FLO, flomoxef; HA, hospital-acquired; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HTN, hypertension; IAI, intra-abdominal infection; LVX, levofloxacin; MIC, minimum inhibitory concentration; Non-CP-CRE, non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae; Non-CP-CREc, non-carbapenemase-producing carbapenem-resistant E. coli; PCKD, polycystic kidney disease; UPEC, uropathogenic E. coli; UTI, urinary tract infection.
References
Abd El Ghany, M., Sharaf, H., Al-Agamy, M. H., Shibl, A., Hill-Cawthorne, G. A., and Hong, P. Y. (2018). Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS ONE. 13, e0201613. doi: 10.1371/journal.pone.0201613
Abid, F. B., Tsui, C. K. M., Doi, Y., Deshmukh, A., McElheny, C. L., Bachman, W. C., et al. (2021). Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. Eur. J. Clin. Microbiol. Infect. Dis. 40, 1779–1785. doi: 10.1007/s10096-021-04185-7
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2019). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucl. Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Al-Farsi, H. M., Camporeale, A., Ininbergs, K., Al-Azri, S., Al-Muharrmi, Z., Al-Jardani, A., et al. (2020). Clinical and molecular characteristics of carbapenem non-susceptible Escherichia coli: a nationwide survey from Oman. PLoS ONE. 15, e0239924. doi: 10.1371/journal.pone.0239924
Alikhan, N.-F., Zhou, Z., Sergeant, M. J., and Achtman, M. J. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14, e1007261. doi: 10.1371/journal.pgen.1007261
Ambler, R. P. (1980). The structure of beta-lactamases. Philos. Trans. R Soc. Lond. B Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Bajaj, H., Scorciapino, M. A., Moyni,é, L., Page, M. G., Naismith, J. H., Ceccarelli, M., et al. (2016). Molecular Basis of Filtering Carbapenems by Porins from β-Lactam-resistant Clinical Strains of Escherichia coli. J. Biol. Chem. 291, 2837–2847. doi: 10.1074/jbc.M115.690156
Baran, I., and Aksu, N. (2016). Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann. Clin. Microbiol. Antimicr. 15, 1–11. doi: 10.1186/s12941-016-0136-2
Bartsch, S. M., McKinnell, J. A., Mueller, L. E., Miller, L. G., Gohil, S. K., Huang, S. S., et al. (2017). Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin. Microbiol. Infect. 23, 48.e49–48.e16. doi: 10.1016/j.cmi.2016.09.003
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. J. M.g. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Micro. Genom. 4. doi: 10.1099/mgen.0.000192
Bouganim, R., Dykman, L., Fakeh, O., Motro, Y., Oren, R., Daniel, C., et al. (2020). The clinical and molecular epidemiology of noncarbapenemase-producing carbapenem-resistant enterobacteriaceae: a case-case-control matched analysis. In: Open Forum Infectious Diseases. Oxford University Press US. ofaa299. doi: 10.1093/ofid/ofaa299
Capitani, G., De Biase, D., Aurizi, C., Gut, H., Bossa, F., and Grütter, M. G. (2003). Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 22, 4027–4037. doi: 10.1093/emboj/cdg403
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicro. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Castanheira, M., Doyle Timothy, B., Kantro, V., Mendes Rodrigo, E., and Shortridge, D. (2020). Meropenem-Vaborbactam Activity against Carbapenem-Resistant Enterobacterales Isolates Collected in U.S. Hospitals during 2016 to 2018. Antimicro. Agents Chemother. 64, e01951–19. doi: 10.1128/AAC.01951-19
Chakraborty, A., Saralaya, V., Adhikari, P., Shenoy, S., Baliga, S., and Hegde, A. (2015). Characterization of escherichia coli phylogenetic groups associated with extraintestinal infections in south indian population. Ann. Med. Health Sci. Res. 5, 241–246. doi: 10.4103/2141-9248.160192
Chang, Y. T., Siu, L. K., Wang, J. T., Wu, T. L., Chen, Y. H., Chuang, Y. C., et al. (2019). Resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible Escherichia coli in Taiwan, 2012-2015. Infect. Drug. Resist. 12, 2113–2123. doi: 10.2147/IDR.S208231
Chen, F., Lv, T., Xiao, Y., Chen, A., Xiao, Y., and Chen, Y. (2021). Clinical characteristics of patients and whole genome sequencing-based surveillance of escherichia coli community-onset bloodstream infections at a non-tertiary hospital in CHINA. Front. Microbiol. 12, 748471. doi: 10.3389/fmicb.2021.748471
Chetri, S., Bhowmik, D., Paul, D., Pandey, P., Chanda, D. D., Chakravarty, A., et al. (2019). AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol. 19, 210. doi: 10.1186/s12866-019-1589-1
Chia, J. H., Siu, L. K., Su, L. H., Lin, H. S., Kuo, A. J., Lee, M. H., et al. (2009). Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J. Chemother. 21, 621–626. doi: 10.1179/joc.2009.21.6.621
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., and Chan, A. P. (2012). Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE. 7, e46688. doi: 10.1371/journal.pone.0046688
Chow, J. W., and Yu, V. L. (1999). Combination antibiotic therapy versus monotherapy for gram-negative bacteraemia: a commentary. Int. J. Antimicrob. Agents. 11, 7–12. doi: 10.1016/S0924-8579(98)00060-0
Dagher, C., Salloum, T., Alousi, S., Arabaghian, H., Araj, G. F., and Tokajian, S. (2018). Molecular characterization of Carbapenem resistant Escherichia coli recovered from a tertiary hospital in Lebanon. PLoS ONE. 13, e0203323. doi: 10.1371/journal.pone.0203323
Del Franco, M., Paone, L., Novati, R., Giacomazzi, C. G., Bagattini, M., Galotto, C., et al. (2015). Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol. 15, 1–9. doi: 10.1186/s12866-015-0597-z
Dortet, L., Cuzon, G., Ponties, V., and Nordmann, P. (2017). Trends in carbapenemase-producing Enterobacteriaceae, France, 2012 to 2014. Eurosurveillance. 22, 30461. doi: 10.2807/1560-7917.ES.2017.22.6.30461
Dupont, H., Gaillot, O., Goetgheluck, A. S., Plassart, C., Emond, J. P., Lecuru, M., et al. (2016). Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in france and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob. Agents Chemother. 60, 215–221. doi: 10.1128/AAC.01559-15
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11, 12728. doi: 10.1038/s41598-021-91456-0
Fernández, L., and Hancock Robert, E. W. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681. doi: 10.1128/CMR.00043-12
Friedman, N. D., Carmeli, Y., Walton, A. L., and Schwaber, M. J. (2017). Carbapenem-resistant enterobacteriaceae: a strategic roadmap for infection control. Infect. Control Hosp. Epidemiol. 38, 580–594. doi: 10.1017/ice.2017.42
Garner, J. S., Jarvis, W. R., Emori, T. G., Horan, T. C., and Hughes, J. M. (1988). CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control. 16, 128–140. doi: 10.1016/0196-6553(88)90053-3
Goodman, K., Simner, P., Tamma, P., and Milstone, A. (2016). Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev. Anti-Infect. Ther. 14, 95–108. doi: 10.1586/14787210.2016.1106940
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasevi,ć, A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. doi: 10.1016/S1473-3099(16)30257-2
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P., and Minghim, R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 16, 169. doi: 10.1186/s12859-015-0611-3
Ho, P., Cheung, Y., Wang, Y., Lo, W., Lai, E., Chow, K., et al. (2016). Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 35, 379–385. doi: 10.1007/s10096-015-2550-3
Huang, Y.-S., Tsai, W.-C., Li, J.-J., Chen, P.-Y., Wang, J.-T., Chen, Y.-T., et al. (2021a). Increasing New Delhi metallo-β-lactamase-positive Escherichia coli among carbapenem non-susceptible Enterobacteriaceae in Taiwan during 2016 to 2018. Sci. Rep. 11, 2609. doi: 10.1038/s41598-021-82166-8
Huang, Y.-T., Liu, P.-Y., and Shih, P.-W. (2021b). Homopolish: a method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 22, 1–17. doi: 10.1186/s13059-021-02282-6
Jean, S. S., Hsueh, P. R., Lee, W. S., Yu, K. W., Liao, C. H., Chang, F. Y., et al. (2013). Carbapenem susceptibilities and non-susceptibility concordance to different carbapenems amongst clinically important Gram-negative bacteria isolated from intensive care units in Taiwan: results from the Surveillance of Multicentre Antimicrobial Resistance in Taiwan (SMART) in 2009. Int. J. Antimicrob. Agents. 41, 457–462. doi: 10.1016/j.ijantimicag.2013.02.001
Jeong, S. H., Kim, H. S., Kim, J. S., Shin, D. H., Kim, H. S., Park, M. J., et al. (2016). Prevalence and molecular characteristics of carbapenemase-producing enterobacteriaceae from five hospitals in Korea. Ann. Lab. Med. 36, 529–535. doi: 10.3343/alm.2016.36.6.529
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Karlowsky, J. A., Lob, S. H., Kazmierczak, K. M., Badal, R. E., Young, K., Motyl, M. R., et al. (2017). In vitro activity of imipenem against carbapenemase-positive enterobacteriaceae isolates collected by the SMART global surveillance program from 2008 to 2014. J. Clin. Microbiol. 55, 1638–1649. doi: 10.1128/JCM.02316-16
Kelly, A. M., Mathema, B., and Larson, E. L. (2017). Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int. J. Antimicrob. Agents. 50, 127–134. doi: 10.1016/j.ijantimicag.2017.03.012
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 17, 101. doi: 10.1186/s12866-017-1012-8
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kostyanev, T., Vilken, T., Lammens, C., Timbermont, L., Van't Veen, A., and Goossens, H. (2019). Detection and prevalence of carbapenem-resistant Gram-negative bacteria among European laboratories in the COMBACTE network: a COMBACTE LAB-Net survey. Int. J. Antimicrob. Agents. 53, 268–274. doi: 10.1016/j.ijantimicag.2018.10.013
Larkin, P. M. K., Realegeno, S., Ward, K. W., Garner, O. B., and Yang, S. (2020). An unusual carbapenem resistant escherichia coli carrying plasmid-mediated ampc and mutated ompc in a patient with recurrent urinary tract infections. IDCases. 20, e00781–e00781. doi: 10.1016/j.idcr.2020.e00781
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lefebvre, B., Lévesque, S., Bourgault, A.-M., Mulvey, M. R., Mataseje, L., Boyd, D., et al. (2015). Carbapenem non-susceptible enterobacteriaceae in Quebec, Canada: results of a laboratory surveillance program (2010–2012). PLoS ONE. 10, e0125076. doi: 10.1371/journal.pone.0125076
Ling, J., Pan, H., Gao, Q., Xiong, L., Zhou, Y., Zhang, D., et al. (2013). Aerobactin synthesis genes iucA and iucC contribute to the pathogenicity of avian pathogenic Escherichia coli O2 strain E058. PLoS ONE. 8, e57794. doi: 10.1371/journal.pone.0057794
Lou, H., Chen, M., Black, S. S., Bushell, S. R., Ceccarelli, M., Mach, T., et al. (2011). Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coli. PLoS ONE. 6, e25825. doi: 10.1371/journal.pone.0025825
Ma, L., Siu, L. K., Lin, J. C., Wu, T. L., Fung, C. P., Wang, J. T., et al. (2013). Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect. Dis. 13, 599. doi: 10.1186/1471-2334-13-599
Malberg Tetzschner, A. M., Johnson, J. R., Johnston, B. D., Lund, O., and Scheutz, F. (2020). In Silico Genotyping of escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58. doi: 10.1128/JCM.01269-20
Marimuthu, K., Venkatachalam, I., Khong, W. X., Koh, T. H., Cherng, B. P. Z., Van La, M., et al. (2017). Clinical and molecular epidemiology of carbapenem-resistant enterobacteriaceae among adult inpatients in Singapore. Clin. Infect. Dis. 64, S68–S75. doi: 10.1093/cid/cix113
Martirosov, D. M., and Lodise, T. P. (2016). Emerging trends in epidemiology and management of infections caused by carbapenem-resistant Enterobacteriaceae. Diagnost. Microbiol. Infect. Dis. 85, 266–275. doi: 10.1016/j.diagmicrobio.2015.10.008
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
McCann, E., Srinivasan, A., DeRyke, C. A., Ye, G., DePestel, D. D., Murray, J., et al. (2018). Carbapenem-nonsusceptible gram-negative pathogens in ICU and Non-ICU settings in US hospitals in 2017: a multicenter study. Open Forum Infect. Dis. 5, ofy241–ofy241. doi: 10.1093/ofid/ofy241
Mendes, R. E., Mendoza, M., Banga Singh, K. K., Castanheira, M., Bell, J. M., Turnidge, J. D., et al. (2013). Regional resistance surveillance program results for 12 Asia-Pacific nations. Antimicrob. Agents Chemother. 57, 5721–5726. doi: 10.1128/AAC.01121-13
Miajlovic, H., and Smith, S. G. (2014). Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol. Lett. 354, 1–9. doi: 10.1111/1574-6968.12419
Musila, L., Kyany'a, C., Maybank, R., Stam, J., Oundo, V., and Sang, W. (2021). Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS ONE. 16, e0246937. doi: 10.1371/journal.pone.0246937
Nguyen, T. T. H., Kikuchi, T., Tokunaga, T., Iyoda, S., and Iguchi, A. (2021). Diversity of the tellurite resistance gene operon in Escherichia coli. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.681175
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011). Genetic features of bla NDM-1-positive Enterobacteriaceae. Antimicr. Agents Chemother. 55, 5403–5407. doi: 10.1128/AAC.00585-11
Reigstad, C. S., Hultgren, S. J., and Gordon, J. I. (2007). Functional genomic studies of uropathogenic < em>escherichia coli < /em> and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282, 21259–21267. doi: 10.1074/jbc.M611502200
Rimrang, B., Chanawong, A., Lulitanond, A., Wilailuckana, C., Charoensri, N., Sribenjalux, P., et al. (2012). Emergence of NDM-1- and IMP-14a-producing Enterobacteriaceae in Thailand. J. Antimicrob. Chemother. 67, 2626–2630. doi: 10.1093/jac/dks267
Roer, L., Tchesnokova, V., Allesøe, R., Muradova, M., Chattopadhyay, S., Ahrenfeldt, J., et al. (2017). Development of a web tool for Escherichia coli subtyping based on fimH alleles. J. Clin. Microbiol. 55, 2538–2543. doi: 10.1128/JCM.00737-17
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathogens. 11, 1–16. doi: 10.1186/s13099-019-0290-0
Schubert, S., Rakin, A., Karch, H., Carniel, E., and Heesemann, J. (1998). Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66, 480–485. doi: 10.1128/IAI.66.2.480-485.1998
Senchyna, F., Gaur, R. L., Sandlund, J., Truong, C., Tremintin, G., Kültz, D., et al. (2019). Diversity of resistance mechanisms in carbapenem-resistant Enterobacteriaceae at a health care system in Northern California, from 2013 to 2016. Diagnost. Microbiol. Infect. Dis. 93, 250–257. doi: 10.1016/j.diagmicrobio.2018.10.004
Sharma, A., Bakthavatchalam, Y. D., Gopi, R., An, S., Verghese, V. P., Veeraraghavan, B., et al. (2016). Mechanisms of carbapenem resistance in K. pneumoniae and E. coli from bloodstream infections in India. J. Infect. Dis. Ther. 4, 293 doi: 10.4172/2332-0877.1000293
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2017). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of bla NDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/JCM.01581-16
Tamma, P. D., Goodman, K. E., Harris, A. D., Tekle, T., Roberts, A., Taiwo, A., et al. (2017). Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant enterobacteriaceae bacteremia. Clin. Infect. Dis. 64, 257–264. doi: 10.1093/cid/ciw741
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Molec. Biol. Evolut. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, H. J., Hsieh, C. F., Chang, P. C., Chen, J. J., Lin, Y. H., Lai, C. C., et al. (2016). Clinical significance of community- and healthcare-acquired carbapenem-resistant enterobacteriaceae isolates. PLoS ONE. 11, e0151897. doi: 10.1371/journal.pone.0151897
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucl. Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Theuretzbacher, U., Bush, K., Harbarth, S., Paul, M., Rex, J. H., Tacconelli, E., et al. (2020). Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 18, 286–298. doi: 10.1038/s41579-020-0340-0
Tian, X., Zheng, X., Sun, Y., Fang, R., Zhang, S., Zhang, X., et al. (2020). Molecular mechanisms and epidemiology of carbapenem-resistant escherichia coli isolated from chinese patients during 2002-2017. Infect. Drug Resist. 13, 501–512. doi: 10.2147/IDR.S232010
Tickler, I. A., Shettima, S. A., dela Cruz, C. M., Le, V. M., Dewell, S., Sumner, J., et al. (2021). Characterization of carbapenem-resistant gram-negative bacterial isolates from nigeria by whole genome sequencing. Diagnost. Microbiol. Infect. Dis. 101, 115422. doi: 10.1016/j.diagmicrobio.2021.115422
Trepanier, P., Mallard, K., Meunier, D., Pike, R., Brown, D., Ashby, J. P., et al. (2017). Carbapenemase-producing Enterobacteriaceae in the UK: a national study (EuSCAPE-UK) on prevalence, incidence, laboratory detection methods and infection control measures. J. Antimicr. Chemother. 72, 596–603. doi: 10.1093/jac/dkw414
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 8, 460–469. doi: 10.1080/21505594.2016.1222343
Vaser, R., Sovi,ć, I., Nagarajan, N., and Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Vergalli, J., Bodrenko, I. V., Masi, M., Moyni,é, L., Acosta-Gutiérrez, S., Naismith, J. H., et al. (2020). Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176. doi: 10.1038/s41579-019-0294-2
Wang, J.-T., Wu, U.-I., Lauderdale, T.-L. Y., Chen, M.-C., Li, S.-Y., Hsu, L.-Y., et al. (2015). Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS ONE. 10, e0121668. doi: 10.1371/journal.pone.0121668
Weinstein, M. P., Towns, M. L., Quartey, S. M., Mirrett, S., Reimer, L. G., Parmigiani, G., et al. (1997). The Clinical Significance of Positive Blood Cultures in the 1990s: A Prospective Comprehensive Evaluation of the Microbiology, Epidemiology, and Outcome of Bacteremia and Fungemia in Adults. Clin. Infect. Dis. 24, 584–602. doi: 10.1093/clind/24.4.584
Yan, W. J., Jing, N., Wang, S. M., Xu, J. H., Yuan, Y. H., Zhang, Q., et al. (2021). Molecular characterization of carbapenem-resistant Enterobacteriaceae and emergence of tigecycline non-susceptible strains in the Henan province in China: a multicentrer study. J. Med. Microbiol. 70. doi: 10.1099/jmm.0.001325
Keywords: carbapenem-resistant, whole-genome sequencing, Escherichia coli, carbapenemase, virulence, epidemiology
Citation: Yang W-T, Chiu I-J, Huang Y-T and Liu P-Y (2022) Comparative Genomics Revealed Fluoroquinolone Resistance Determinants and OmpF Deletion in Carbapenem-Resistant Escherichia coli. Front. Microbiol. 13:886428. doi: 10.3389/fmicb.2022.886428
Received: 28 February 2022; Accepted: 22 March 2022;
Published: 18 April 2022.
Edited by:
Jiun-Ling Wang, National Cheng Kung University, TaiwanReviewed by:
Theodoros Karampatakis, Papanikolaou General Hospital of Thessaloniki, GreeceSandip Kumar Mukherjee, University of California, Los Angeles, United States
Bela Kocsis, Semmelweis University, Hungary
Copyright © 2022 Yang, Chiu, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Po-Yu Liu, pyliu@vghtc.gov.tw; Yao-Ting Huang, ythuang@cs.ccu.edu.tw
 Wan-Ting Yang
Wan-Ting Yang I-Ju Chiu2
I-Ju Chiu2 Yao-Ting Huang
Yao-Ting Huang Po-Yu Liu
Po-Yu Liu