- 1Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education China, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 2Nursing Department, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 3Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 4Department of Clinical Laboratory, Zhoukou Maternal and Child Health Hospital, Zhoukou, China
- 5School of Nursing, Wenzhou Medical University, Wenzhou, China
- 6Institute of Translational Medicine, Baotou Central Hospital, Baotou, China
Florfenicol is widely used for the treatment of bacterial infections in domestic animals. The aim of this study was to analyze the molecular mechanisms of florfenicol and oxazolidinone resistance in Enterococcus isolates from anal feces of domestic animals. The minimum inhibitory concentration (MIC) levels were determined by the agar dilution method. Polymerase chain reaction (PCR) was performed to analyze the distribution of the resistance genes. Whole-genome sequencing and comparative plasmid analysis was conducted to analyze the resistance gene environment. A total of 351 non-duplicated enteric strains were obtained. Among these isolates, 22 Enterococcus isolates, including 19 Enterococcus. faecium and 3 Enterococcus. faecalis, were further studied. 31 florfenicol resistance genes (13 fexA, 3 fexB, 12 optrA, and 3 poxtA genes) were identified in 15 of the 19 E. faecium isolates, and no florfenicol or oxazolidinone resistance genes were identified in 3 E. faecalis isolates. Whole-genome sequencing of E. faecium P47, which had all four florfenicol and oxazolidinone resistance genes and high MIC levels for both florfenicol (256 mg/L) and linezolid (8 mg/L), revealed that it contained a chromosome and 3 plasmids (pP47-27, pP47-61, and pP47-180). The four florfenicol and oxazolidinone resistance genes were all related to the insertion sequences IS1216 and located on two smaller plasmids. The genes fexB and poxtA encoded in pP47-27, while fexA and optrA encoded in the conjugative plasmid pP47-61. Comparative analysis of homologous plasmids revealed that the sequences with high identities were plasmid sequences from various Enterococcus species except for the Tn6349 sequence from a Staphylococcus aureus chromosome (MH746818.1). The current study revealed that florfenicol and oxazolidinone resistance genes (fexA, fexB, poxtA, and optrA) were widely distributed in Enterococcus isolates from animal in China. The mobile genetic elements, including the insertion sequences and conjugative plasmid, played an important role in the horizontal transfer of florfenicol and oxazolidinone resistance.
Introduction
Enterococci are Gram-positive facultative anaerobes that typically colonize the gastrointestinal tracts of most animals and humans. However, along with staphylococci, enterococci are among the leading causes of nosocomial infections, such as bacteremia, peritonitis, endocarditis, urinary tract, wound, and device-related infections (Sava et al., 2010). Multidrug-resistant (MDR) enterococci are significant nosocomial pathogens that can develop resistance to many antimicrobials currently used in the clinic (Arias and Murray, 2012).
Florfenicol is a fluorinated derivative of chloramphenicol and has a broad-spectrum bacteriostatic activity for a wide range of microorganisms by binding to the peptidyl transferase center at the 50S ribosomal subunit of the 70S complex, which inhibits bacterial protein biosynthesis (Syriopoulou et al., 1981). In order to control the respiratory tract microbial infection of cattle and pigs, florfenicol has been licensed in Europe in 1995 and 2000, respectively (Schwarz and Chaslus-Dancla, 2001; Schwarz et al., 2004). China also approved its use in 1999 for the treatment of bacterial infection in pigs, cattle, chickens, and fish. Since then, florfenicol has been excessively used in veterinary medicine and as a feed additive, and the prevalence and abundance of florfenicol resistance genes has increased in bacteria from different sources (Zhao et al., 2016).
Oxazolidinones are antimicrobial agents that are effective in treating various Gram-positive bacterial infections as those caused by methicillin-resistant Staphylococcus (MRSA) and vancomycin-resistant enterococci (VRE; Fung et al., 2001). The most common mechanism contributing to linezolid resistance involves point mutations in domain V of the 23S rRNA gene, and it has been found that the most frequent mutation is on G2576T. The oxazolidinone-resistant phenotype is also related to spontaneous mutations in the ribosomal subunit proteins L3, L4, and L22 encoded by the rplC, rplD, and rplV genes, respectively (Long et al., 2006; Mendes et al., 2010). It was reported that the cfr, cfr(B), cfr(C), cfr(D), cfr(E), optrA, and poxtA genes are related to oxazolidinone resistance. The cfr gene is the first transferable oxazolidinone resistance gene, which encodes a rRNA methyltransferase (Schwarz et al., 2000; Deshpande et al., 2015; Wang et al., 2015; Tamang et al., 2017; Tang et al., 2017; Antonelli et al., 2018; Stojkovic et al., 2019). Homologs of cfr have been identified in nonpathogenic Bacillales, and three additional cfr-like genes that share less than 80% protein sequence identity to Cfr have been described in Clostridium and Enterococcus. These genes are known as cfr(B), cfr(C), and cfr(D). A new determinant termed cfr(E) was recovered in Clostridium difficile strains across Latin America (Stojkovic et al., 2019). However, a novel ABC transporter gene optrA was first reported to be encoded in a plasmid from Enterococcus faecalis in 2015 in China (Wang et al., 2015). Since it was first described in 2015, optrA has been found in the human and animal enterococci, and it has environmental origins from Ireland, Italy, Korea, Malaysia, Denmark, and different regions of China, and it has been detected in pet food in Europe and in different sources from Tunisia, India, Czech Republic, and Sweden (Elghaieb et al., 2020; Bakthavatchalam et al., 2021; Fang et al., 2021; Freitas et al., 2021; Malisova et al., 2021). Furthermore, many optrA variants have been collected from enterococci and staphylococci (Cai et al., 2015; Brenciani et al., 2016; Cui et al., 2016; Li et al., 2016; Mendes et al., 2016; Cavaco et al., 2017; Lazaris et al., 2017; Lee et al., 2017). Recently, a novel oxazolidinone resistance gene named poxtA, encoding an ATP-binding cassette (ABC) protein, was identified in a MRSA that originated from the clinical (Antonelli et al., 2018).
Domestic animals are widely recognized as a source of antimicrobial resistance. Worldwide reports of optrA-mediated oxazolidinone resistance among bacterial isolates of animal origin have largely focused on animals from farms (Fan et al., 2016; Torres et al., 2018; Elghaieb et al., 2020). The wide distribution of oxazolidinone resistance optrA and cfr genes in domestic animals has been reported in rural areas of China (Sun et al., 2018). The occurrence of the linezolid resistance gene optrA in linezolid-resistant E. faecalis and E. faecium isolates from food animals and animal carcasses was first reported in Korea (Tamang et al., 2017).
In this study, we used multiple approaches, including molecular cloning, whole-genome sequencing, and comparative plasmid analysis, to investigate the molecular resistance mechanism against amphenicol and oxazolidinone from Enterococcus isolated from domestic animal samples in China.
Materials and methods
Bacterial isolates
Samples were collected from the anal feces of cattle, chickens, ducks, and pigs from five animal farms in Zhejiang (Hangzhou), Henan (Zhoukou), Shanxi (Linfen), Shandong (Liaocheng), and Sichuan (Bazhong) provinces in 2016, respectively. Only one farm was chosen in each sampling site. Then, the feces samples were streaked onto LB agar plates. A total of 351 non-duplicated enteric strains were obtained. Then, the 351 strains were cultured on LB agar plates supplemented with 8 mg/L florfenicol to screen the florfenicol-resistant strains. The strains were first identified by biochemical methods and then verified by homology comparisons of the 16S rRNA gene with the nucleotide sequence database by using BLASTN program1 (Altschul et al., 1990).
Detection of resistance genes
The presence of resistance genes (fexA, fexB, cfr, and optrA) was investigated using PCR, as previously described (Kehrenberg and Schwarz, 2006; Liu et al., 2012; Wang et al., 2015). The primers (Table 1) were designed by using Primer 5.0 and synthesized by Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China), which included a pair of flanking restriction endonuclease adapters. PCR amplification was carried out under the following conditions: an initial 10 min denaturation at 94°C followed by 35 cycles of denaturation (94°C for 45 s), annealing (58°C for 45 s), and extension (72°C for 90 s), and a final extension step at 72°C for 10 min. The PCR products were further confirmed by Sanger sequencing with an ABI 3730 automated sequencer (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China). Both strands of the PCR products were sequenced with the forward and reverse primers (Table 1). The sequence data were compared to the NCBI nucleotide sequence database using the BLAST program (Altschul et al., 1990).
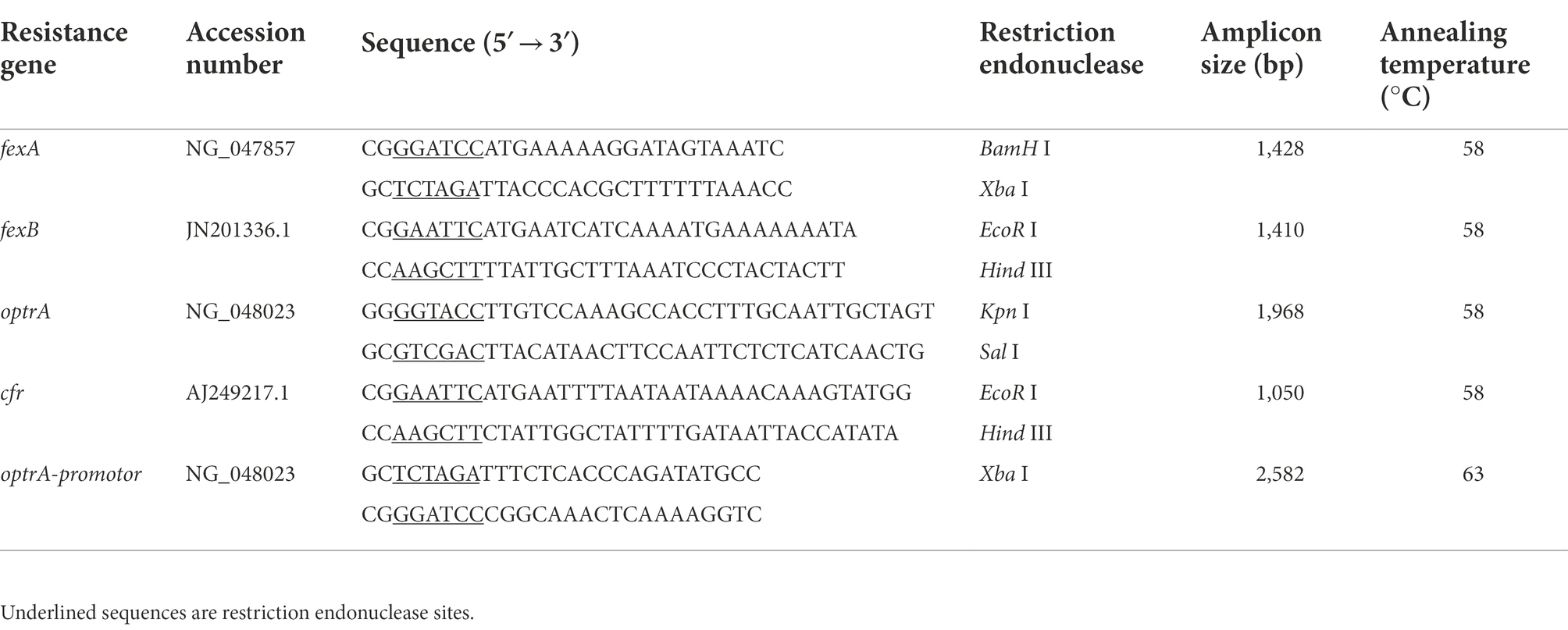
Table 1. Primer sequences and PCR product sizes of the florfenicol and oxazolidinones resistance genes.
Pulsed field gel electrophoresis
Pulsed field gel electrophoresis (PFGE) was carried out on Enterococcus isolates according to the method described previously with minor modifications (Liu et al., 2013). Enterococcal cells were treated with lysozyme at a final concentration of 10 mg/ml for 1 h at 37°C and then incubated with the restriction enzyme SmaI (Takara, Japan) for 4 h with shaking. Briefly, genomic DNA fragments were resolved on a 1% agarose (SeaKem Gold Agarose, Lonza) gel at 14°C, and electrophoresis was conducted using a CHEF-Mapper XA PFGE system (Bio-Rad, United States) at a pulse time gradient of 3.5–23.5 s and a total running time of 18 h within a molecular weight of 10–300 kb. The DNA band patterns were visualized by a Gel Doc XR gel imaging system (Bio-Rad, United States) and further analyzed by QualityOne software (Bio-Rad Laboratories, United States). Phylogenetic trees then allow the comparison of genetic relatedness and clonal assignment (Neoh et al., 2019). A similarity coefficient of 80% was selected to define the pulsed-field type (PFT) clusters (McDougal et al., 2003).
Whole-genome sequencing and multi-locus sequence typing
Due to E. faecium P47 showing high resistance to florfenicol and linezolid and carrying the most resistance genes, it was selected for whole-genome sequencing. The genomic DNA of E. faecium P47 was extracted by an AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, CA, United States) according to the manufacturer’s instructions. A 20-kb library was generated using the SMRTbell Template Prep Kit (Pacific Biosciences, CA, United States) according to the PacBio standard protocol and was sequenced on a PacBio RS II instrument (Pacific Biosciences). In addition, a paired-end library with 300-bp insert sizes was constructed and sequenced from both ends using the HiSeq 2,500 Illumina sequencing platform (Illumina, CA, United States). The PacBio long reads were initially assembled using Canu software (Koren et al., 2017). The Illumina sequencing reads were then mapped onto the assembled contigs to correct the primary assembly by using Bwa and the Genome Analysis Toolkit (McKenna et al., 2010). The potential ORFs were predicted using Glimmer software and annotated against a non-redundant protein database using the BLASTX program (Altschul et al., 1990).
Genotyping by the multi-locus sequence typing (MLST) method for E. faecium P47 was determined using 7 housekeeping genes (atpA, ddl, gdh, purK, gyd, pstS, and adk). Alleles and sequence types (STs) were assigned using the MLST database2 (Homan et al., 2002).
Plasmid transfer and cloning experiments
The transferability of linezolid and florfenicol resistance was assessed by filter mating using E. faecium P47 as a donor and E. faecalis JH2-2 as a recipient, following the method described previously (Huys et al., 2004). The transconjugants were selected on brain heart infusion (BHI) plates containing 16 mg/L florfenicol, 25 mg/L rifampicin, and 50 mg/L fusidic acid. The size of the transferred plasmid was estimated by S1 nuclease-PFGE as previously reported with some modifications (Liu et al., 2013). Whole-cell DNA was incubated with 32 U S1-nuclease (TaKaRa, Japan) for 25 min at 30°C. The resulting restriction fragments were separated using a CHEF-DRIII system (Bio-Rad) with a clamped homogeneous electric field of 6 V/cm, using a 120° switch angle for 17 h at 14°C, with the pulse time linearly ramped from 3.5 to 23.5 s. PCR amplifications of the fexA, fexB, poxtA and optrA genes were performed to confirm its presence in the transconjugants.
To confirm the function of the optrA gene in E. faecium P47, open reading frames with upstream promoter regions were amplified using the optrA promoter primers (Table 1). The purified PCR products were digested and ligated into the shuttle vector pAM401, and further transformed into E. faecalis JH2-2 by electrotransformation and grown on BHI plates supplemented with 16 mg/L chloramphenicol. The recombinant plasmids were verified by XbaI and BamHI (TaKaRa, Japan) digestion and then sequencing.
Antimicrobial susceptibility testing
The MICs for 22 Enterococcus isolates, transconjugants, and transformants were determined by the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2021: M100).3 A total of 16 antimicrobial agents, including chloramphenicol, florfenicol, linezolid, ampicillin, gentamicin, erythromycin, tetracycline, vancomycin, fosfomycin, rifampicin, fusidic acid, spectinomycin, norfloxacin, levofloxacin, clarithromycin, and tigecycline, were used. Enterococcus faecalis ATCC 29212 was used as a quality control strain.
Comparative plasmid analysis
The sequences used for the comparative plasmid analysis of pP47-27 or pP47-61 were retrieved from the NCBI nucleotide database using the complete nucleotide sequence of pP47-27 or pP47-61 as a query based on a cutoff value of 40% coverage and 97% identity for pP47-61 and 90% coverage and 99% identity for pP47-27. Plasmid incompatibility groups were predicted by PlasmidFinder4 (Carattoli et al., 2014). Nucleotide sequence comparisons were carried out using BLASTN. Gview5 was used to construct basic genomic features that were then employed in comparative plasmid analysis (Petkau et al., 2010). For the comparative plasmid analysis of the optrA-encoding fragment, sequences containing the optrA gene were also obtained from the NCBI nucleotide database using “optrA gene” as a keyword. The resulting sequences were filtered, and only sequences that contained a complete optrA gene and harbored at last one complete or truncated IS1216 gene were retained. The plasmid pE349 which was first reported to carry optrA gene was also retained. Multiple sequence alignments were performed by MAFFT using the 16 kb optrA gene-related fragment (approximately 8 kb upstream and downstream of the optrA gene) of pP47-61 as a reference (Katoh and Standley, 2013). The 16 kb fragment was flanked by two IS1216 insertion sequences. The sequence with the highest sequence similarity to the other sequences in each cluster was chosen as the candidate for ortholog analysis. Orthologous groups of genes from the candidate sequences were identified using BLASTP and InParanoid (Altschul et al., 1990; Remm et al., 2001). The sequence retrieval, statistical analysis, and other bioinformatic tools used in this study were applied with Python and Biopython scripts (Cock et al., 2009).
Nucleotide sequence accession numbers
The genome sequence of P47 has been deposited in GenBank with the following accession numbers CP091100 (P47 chromosome), CP091101 (pP47-27), CP091102 (pP47-61), and CP091103 (pP47-180).
Results
Antibiotic resistance and resistance gene detection of the isolates
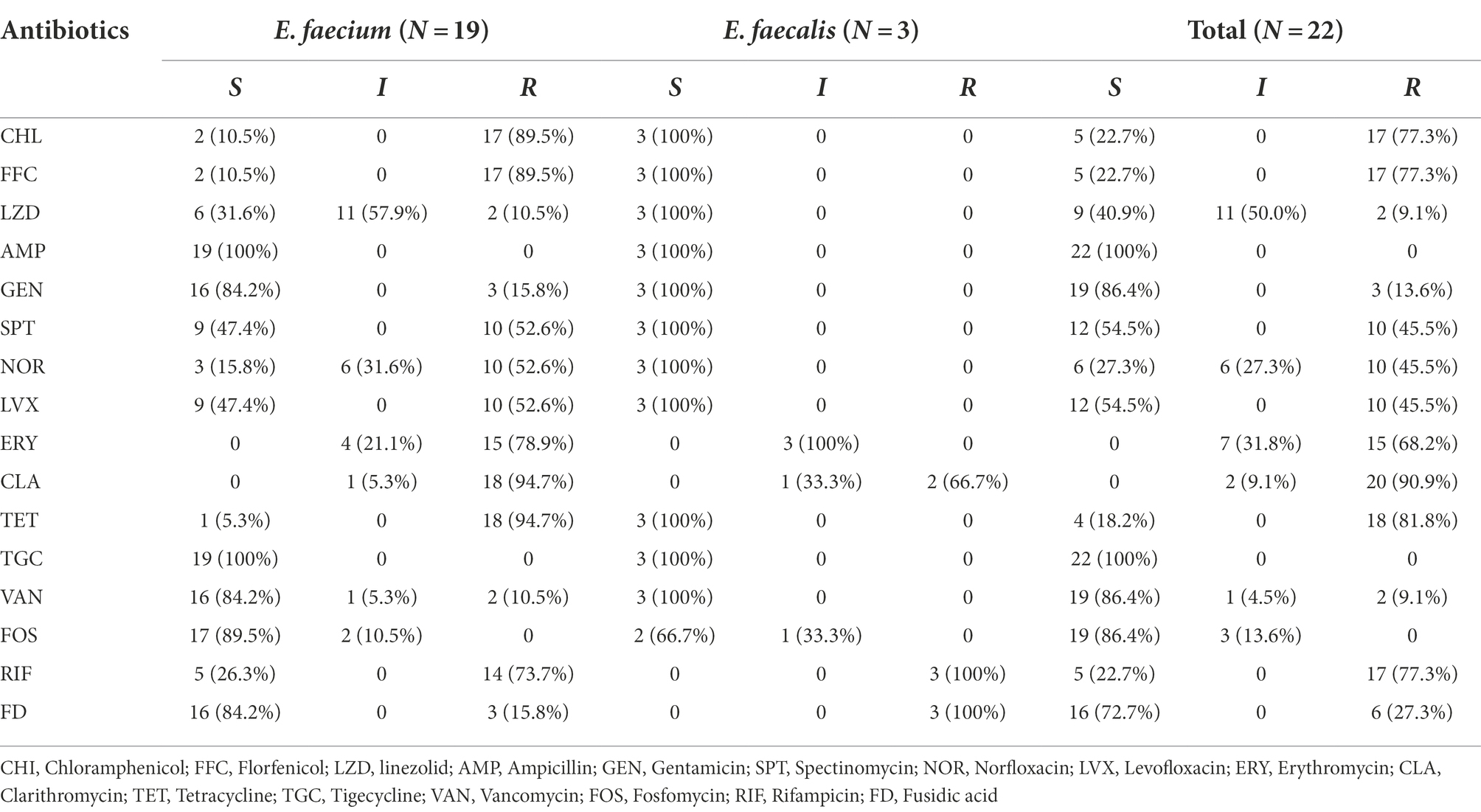
We collected a total of 351 non-duplicated enteric bacterial strains in five farms from five provinces of China, including 237 Escherichia coli, 45 Bacillus, 38 Staphylococcus, 22 Enterococcus (3 Enterococcus faecalis and 19 Enterococcus faecium), 4 Klebsiella pneumoniae, 4 Shigella, 4 Planctomycetes, 3 Proteus, and 4 unclassified strains. The source and regional distribution of 351 strains are shown in Supplementary Table S1. We have investigated the resistance profile of animal-derived Gram-negative strains in previous studies (Wu et al., 2019; Li et al., 2020; Liu et al., 2020; Zhu et al., 2020). In this study, we mainly focused on the resistance characteristics of Enterococcus in the animals. Therefore, 22 Enterococcus strains, including 19 E. faecium and 3 E. faecalis, were subjected to further antimicrobial resistance investigation. Among 22 Enterococcus isolates, most (81.8%, 18/22) were isolated from chickens, and the remaining 4 strains were pig-origin. More than 68.2% (15/22) of the strains showed resistance to 5 antibiotics, including chloramphenicol (77.3%, 17/22), florfenicol (77.3%, 17/22), erythromycin (68.2%, 15/22), tetracycline (81.8%, 18/22), and clarithromycin (90.9%, 20/22), while only 2 strains were resistant to linezolid (9.1%, 2/22), as shown in Table 2.
A total of 31 florfenicol and oxazolidinone resistance genes (including 13 fexA, 3 fexB, 12 optrA, and 3 poxtA genes) were identified in 15 of the 19 E. faecium isolates, and no florfenicol or oxazolidinone resistance genes were identified in any of the E. faecalis isolates. In the resistance gene-positive isolates, more than two-thirds (72.3%, 11/15) carried two genes, most of which (10/11, 90.1%) carried fexA and optrA, and one strain (E. faecium FH67) harbored fexB and poxtA simultaneously. Among the other four isolates, two strains (E. faecium FH56 and E. faecium FH58) carried fexA each, one strain (E. faecium FH59) carried three genes (fexB, optrA, and poxtA), and one strain (E. faecium P47) carried four genes (fexA, fexB, optrA, and poxtA). All the florfenicol-resistant isolates (77.3%, 17/22), which exhibited MIC levels ranging from 64–2048 mg/L, were E. faecium, while 5 isolates including 2 E. faecium and 3 E. faecalis isolates were susceptible to florfenicol and exhibited MIC levels (≤ 4 mg/L). None of the known florfenicol resistance genes were identified in the 5 florfenicol-susceptible isolates. However, two florfenicol-resistant isolates which did not harbor any known florfenicol resistance gene had a MIC of 256 mg/L (H38) and 64 mg/L (FH72), respectively (Supplementary Table S2).
The florfenicol resistance gene-positive strains also showed higher MIC levels to linezolid than those strains which did not contain florfenicol resistance genes. A total of 86.7% (13/15) of florfenicol resistance gene-positive strains (excluding FH75 and H32 which had MIC levels of 2 mg/l with linezolid) exhibited MIC levels of ≥4 mg/L against linezolid; however, in the florfenicol resistant gene-negative strains, none of the strains exhibited this MIC level against linezolid.
Molecular typing by pulsed field gel electrophoresis
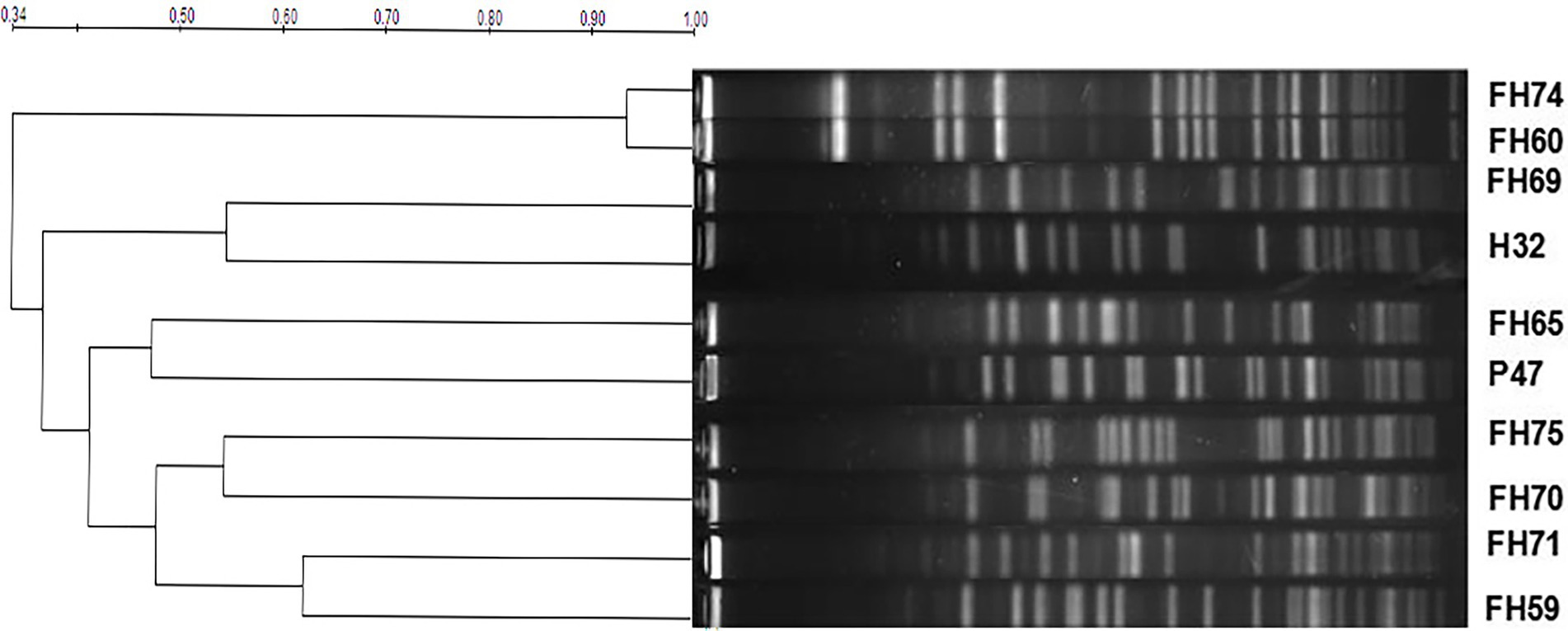
Pulsed field gel electrophoresis (PFGE) typing for 10 optrA-positive E. faecium strains was analyzed. The band sizes were mainly between 20–700 kb, and the number of bands was approximately 15–22. Based on a similarity coefficient of 0.80, the 10 E. faecium isolates showed 9 pulsed-field type (PFT) clusters (Figure 1). There were two isolates (FH74 and FH60) harboring the same florfenicol resistance genes (optrA and fexA) from the same chicken farm in Hangzhou, China, sharing over 90% similarity. They could be considered the same PFT, which indicated that there may be a clone spread of the strains harboring the optrA and fexA genes in the farm.
General features of the Enterococcus faecium P47 genome
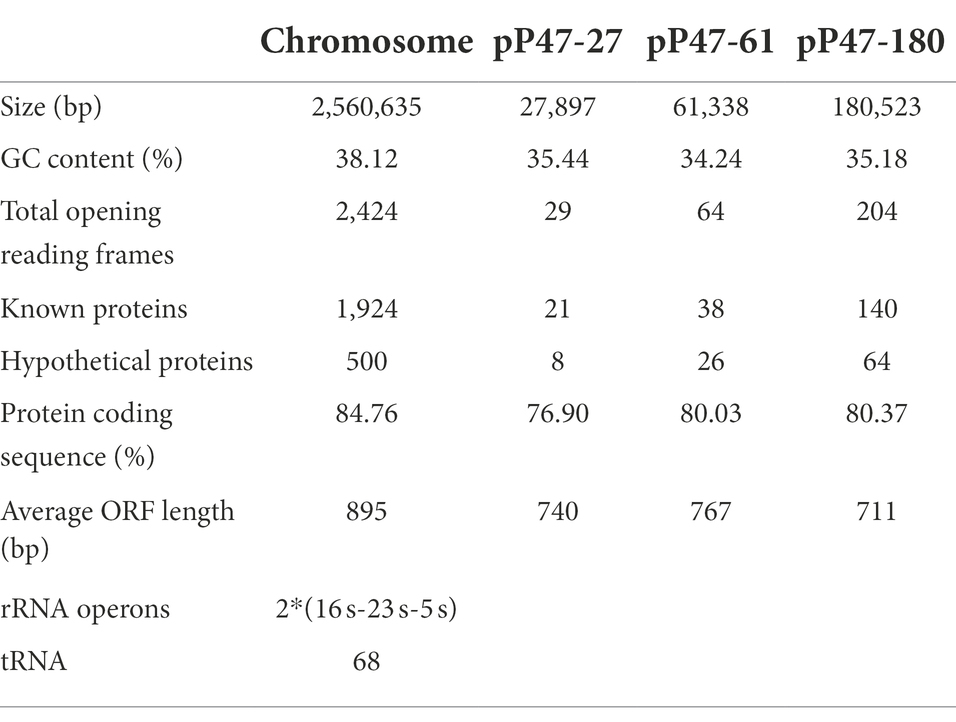
To elucidate the molecular resistance mechanism of the Enterococcus strains to antimicrobials, especially florfenicol and oxazolidinones, the whole-genome sequence of E. faecium P47 was chosen for whole-genome sequencing (WGS) analysis. E. faecium P47 co-carries four florfenicol and oxazolidinone resistance genes, fexA, fexB, optrA, and poxtA, and has a wide resistance spectrum and high MIC values to the antibiotics tested (especially linezolid with an MIC level of 8 mg/L). The general features of the P47 genome are shown in Table 3. The complete E. faecium P47 genome consisted of one chromosome and three circular plasmids (pP47-27, pP47-61, and pP47-180). The chromosome had an average G + C content of 38.12% and was 2,560,635 bp in length, encoding 2,424 ORFs. Of the three plasmids, pP47-27 was 27,897 bp in length, encoding 29 ORFs; pP47-61 was 61,338 bp in length, encoding 64 ORFs; and pP47-180 was 180,523 bp in length, encoding 204 ORFs. Functional annotation revealed that the whole genome encoded 14 resistance genes with 2 (aac(6′)-Ii and dfrG) in the chromosome and 12 in the plasmids, including 4 (fexB, poxtA, tetL and tetM) encoded on the plasmid pP47-27 (Figures 2, 3), 5 (ermB, fosB, ermA, optrA and fexA) on the plasmid pP47-61 (Figure 4) and 3 [aac(6′)-aph(2″), ant(6)-Ia and ermB] on the plasmid pP47-180 (Supplementary Table S2). MLST analysis revealed that E. faecium P47 contained the adk-1, atpA-4, ddi-5, gdh-1, gyd-1, pstS-1, and purK-3 alleles and belonged to the sequence type ST29.
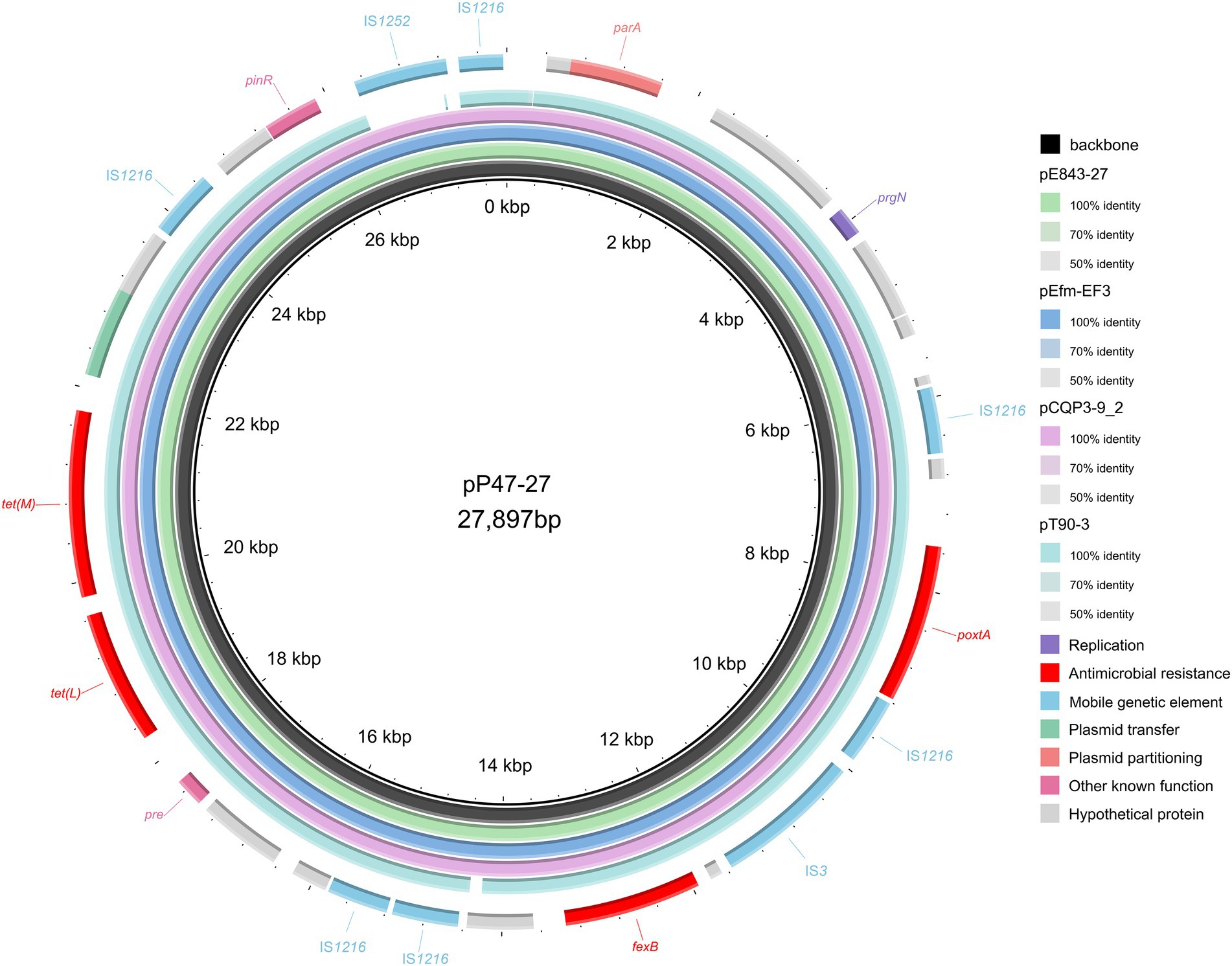
Figure 2. Comparative plasmid analysis of pP47-27 and other similar plasmids. The inside to the outside is as follows: circle 1, the backbone of pP47-27; circle 2, pE843-27 (the plasmid of Enterococcus lactis E843 isolated from swine, CP082268.1); circle 3, pEfm-EF3 (the plasmid of Enterococcus faecium EF3 isolated from marine sediment, MT683615.1); circle 4, pCQP3-9_2 (the plasmid of Enterococcus hirae pCQP3-9 isolated from fecal sample, CP037957.1); circle 5, pT90-3 (the plasmid of Enterococcus faecalis strain T90-3 isolated from swine, CP069131.1); and circle 6, the genes encoded on pP47-27.
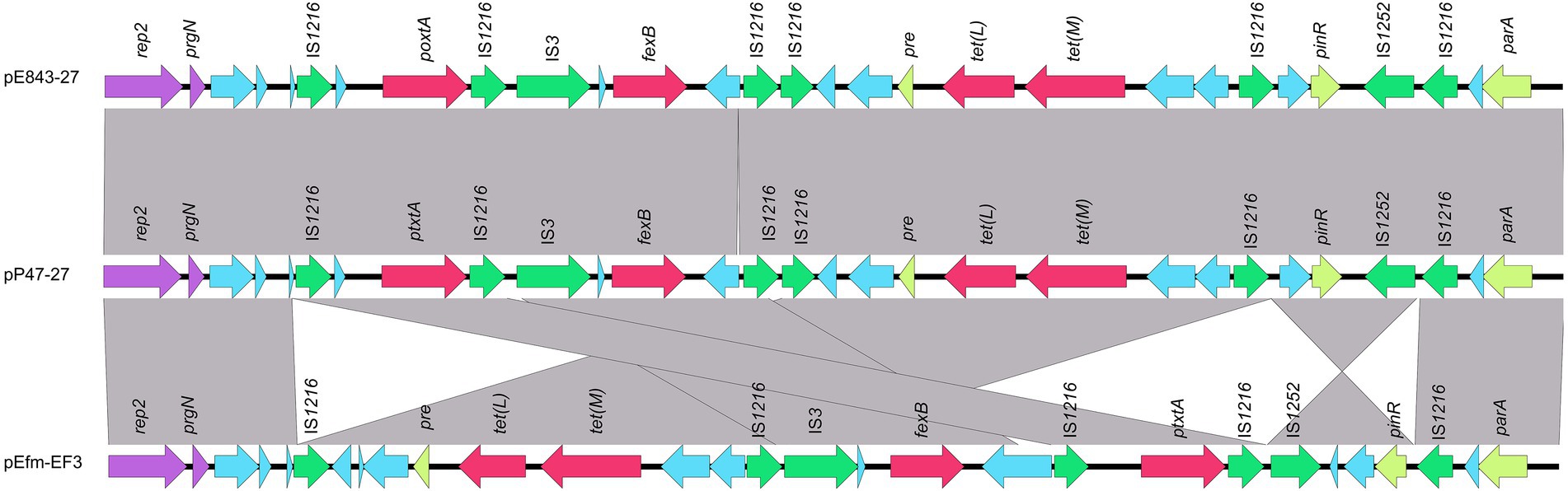
Figure 3. Genetic environments of the poxtA and fexB genes encoded in different plasmids. The ORFs are shown as arrows, and the arrowheads indicate the direction of transcription. The same color represents the same elements and genes, with antimicrobial resistance genes in red, mobile genetic elements in green, replications in purple, and other genes in blue. Gray-shaded areas represent regions with >95% nucleotide sequence identities. The sequences and their origins are Enterococcus lactis E843 (the plasmid of Enterococcus lactis E843 isolated from swine, CP082268.1) and Enterococcus faecium EF3 (the plasmid of Enterococcus faecium EF3 isolated from marine sediment, MT683615.1).
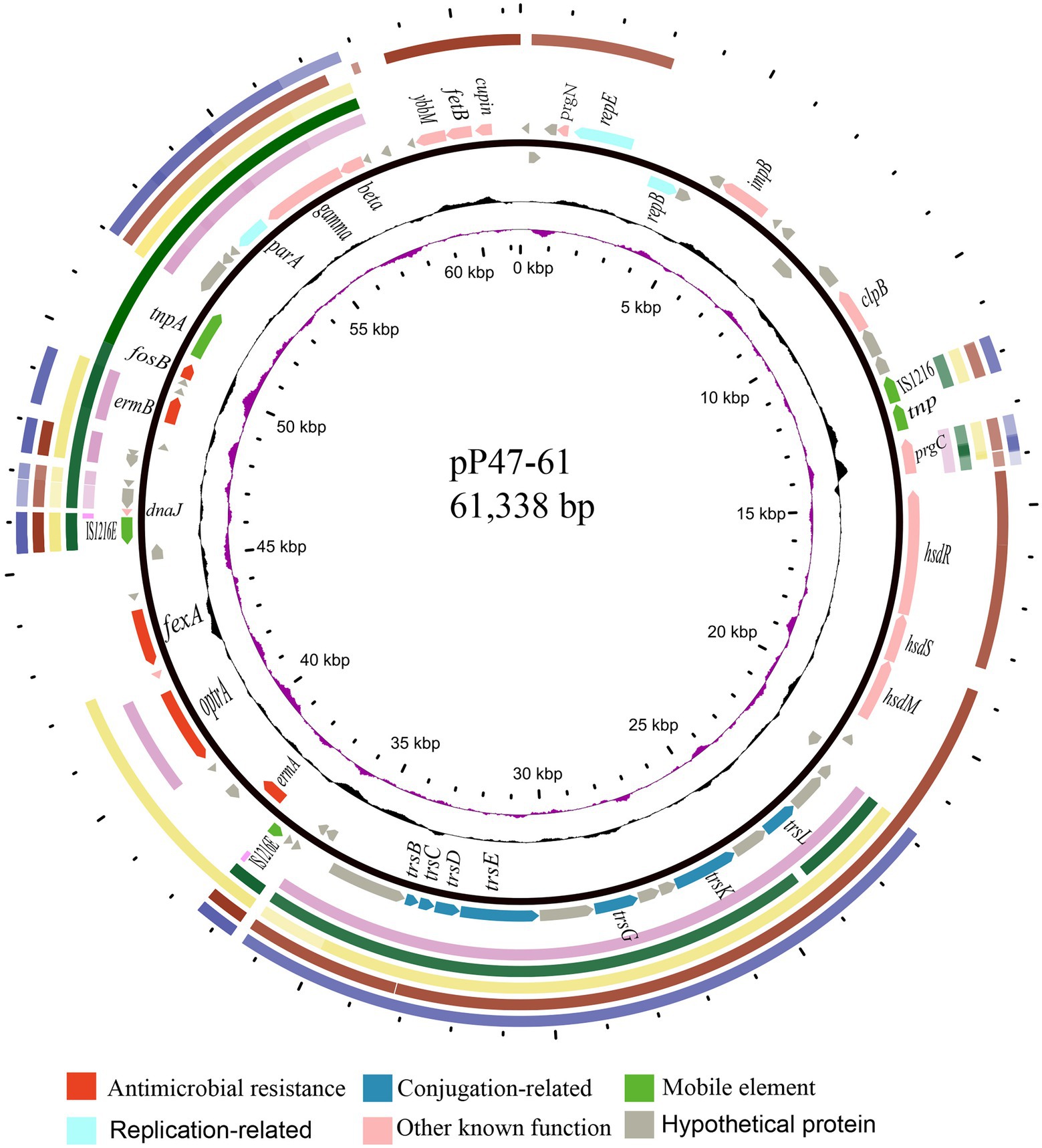
Figure 4. Comparative plasmid analysis of pP47-61 and other similar sequences. Circles from outside to inside indicate (1) the regions of the four plasmids (all from Enterococcus, including pCR1B, CP030934.1; pF120805, KY579372.1; pKUB3007-3, AP018546.1; and pE35048-oc, MF580438.1) and a transposon sequence of the Staphylococcus aureus AOUC-0915 chromosome (Tn6349, MH746818.1), which share high homology with pP47-61. (2) Predicted coding sequences encoded on the forward and reverse strands of pP47-61. (3) G + C content (with an average of 50%, in which a G + C content of more than 50% is shown toward the outside and less is shown inward) and G + C skew (with a positive GC skew toward the outside and a negative GC skew toward the inside) of pP47-61. (4) The innermost circle shows the position in kbp.
Transferability of the resistance plasmid and resistance activity of the transconjugant
Through conjugation experiments, a transconjugant carrying transferable pP47-61 was obtained and further determined by S1 nuclease-PFGE. The MIC results revealed that the transconjugant (pP47-61/JH2-2) exhibited at least a 4-fold increase in MIC levels for florfenicol, chloramphenicol, and linezolid and a > 512-fold increase in MIC levels for macrolides (erythromycin and clarithromycin) compared to that of the recipient strain JH2-2, which was in accordance with the resistance genes (fexA, optrA, ermA, and ermB) encoded on the plasmid (Supplementary Table S2).
The WGS sequencing results demonstrated that the deduced OptrA protein from the plasmid pP47-61 included three amino acid substitutions at position 3 (K3E), 176(Y176D), and 481(T481P; EDP variant) compared with the first reported optrA gene in E. faecalis E349 of human origin (Wang et al., 2015). To identify the resistance activity of the optrA gene (EDP variant) of E. faecium P47, the ORF with its promoter region was cloned. The MIC levels of the recombinant strain expressing the cloned resistance gene against 16 antimicrobial agents are shown in Supplementary Table S2. The recombinant strain harboring the cloned optrA (optrA-pAM401/JH2-2) exhibited 8-and 2-fold increases in MIC levels with florfenicol and linezolid compared to the control strain JH2-2/pAM401, respectively, while no MIC changes were found for the other antibiotics tested compared with the control strain JH2-2/pAM401 (E. faecalis JH2-2 harboring the vector pAM401).
Comparative plasmid analysis and the genetic environment of the resistance genes on the plasmids
Four plasmids and a transposon sharing the highest nucleotide sequence similarity (≥ 40% coverage and ≥ 97% identity) with pP47-61 were retrieved from the NCBI nucleotide database. The plasmids belonged to the Inc18 incompatibility group, including pCR1B of E. gilvus CR1 (CP030934.1, 80 kb) isolated from Japan, pF120805 of E. faecium F120805 (KY579372.1, 72 kb) from Ireland, pKUB3007-3 of E. faecalis KUB3007 (AP018546.1, 59 kb) from Japan, pE35048-oc of E. faecium E35048 (MF580438.1, 41 kb) from Italy, and Tn6349 from Staphylococcus aureus AOUC-0915 chromosome (MH746818.1, 48 kb) from Italy (Morroni et al., 2018; D’Andrea et al., 2019). The plasmids displayed highly global genomic synteny and shared a conserved backbone sequence of a typical Inc18 plasmid, which included two homologous regions (regions A and B, Figure 4; Supplementary Table S3). Region A, approximately 10 kb in size (from 23 to 37 kb), contained a cluster of genes related to conjugation (trsBCDEGKL) and a series of hypothetical proteins. Region B, approximately 10 kb in size (from 45 to 57 kb), encoded genes of dnaJ (a molecular chaperone), ermB, and parA (for partition), and this region was interrupted by the insertion of tnpA (transposase) and fosB (fosfomycin resistance enzyme) into plasmids pP47-61 and pKUB3007-3. The segment (from 37 to 45 kb) of an approximately 8.0 kb sequence encoding a complex transposon with a gene array of IS1216E-hp-fexA-hp-optrA-hp-hp-ermA-IS1216E was unique in pP47-61, although part of the sequence was found in the plasmids pF120805 and pE35048-oc. Comparative genomic analysis of the complex transposon encoding resistance genes (fexA, optrA, and ermA) of pP47-61 with similar sequences from the NCBI nucleotide database revealed that the core gene array of the complex transposon (hp-fexA-hp-optrA-hp-hp-ermA) was virtually identical to the sequences in the plasmids pE419 (E. faecalisE419), pSF35 (E. faecalisSF35), p10-2-2 (E. faecalis 10-2-2), and pE121 (E. faecalis E121), while in the other plasmid sequences (pFX13 of E. faecalis, pE121 of E. faecalis and pE349 of E. faecalis), they shared similar sequence segments (Figure 5; Supplementary Table S3).
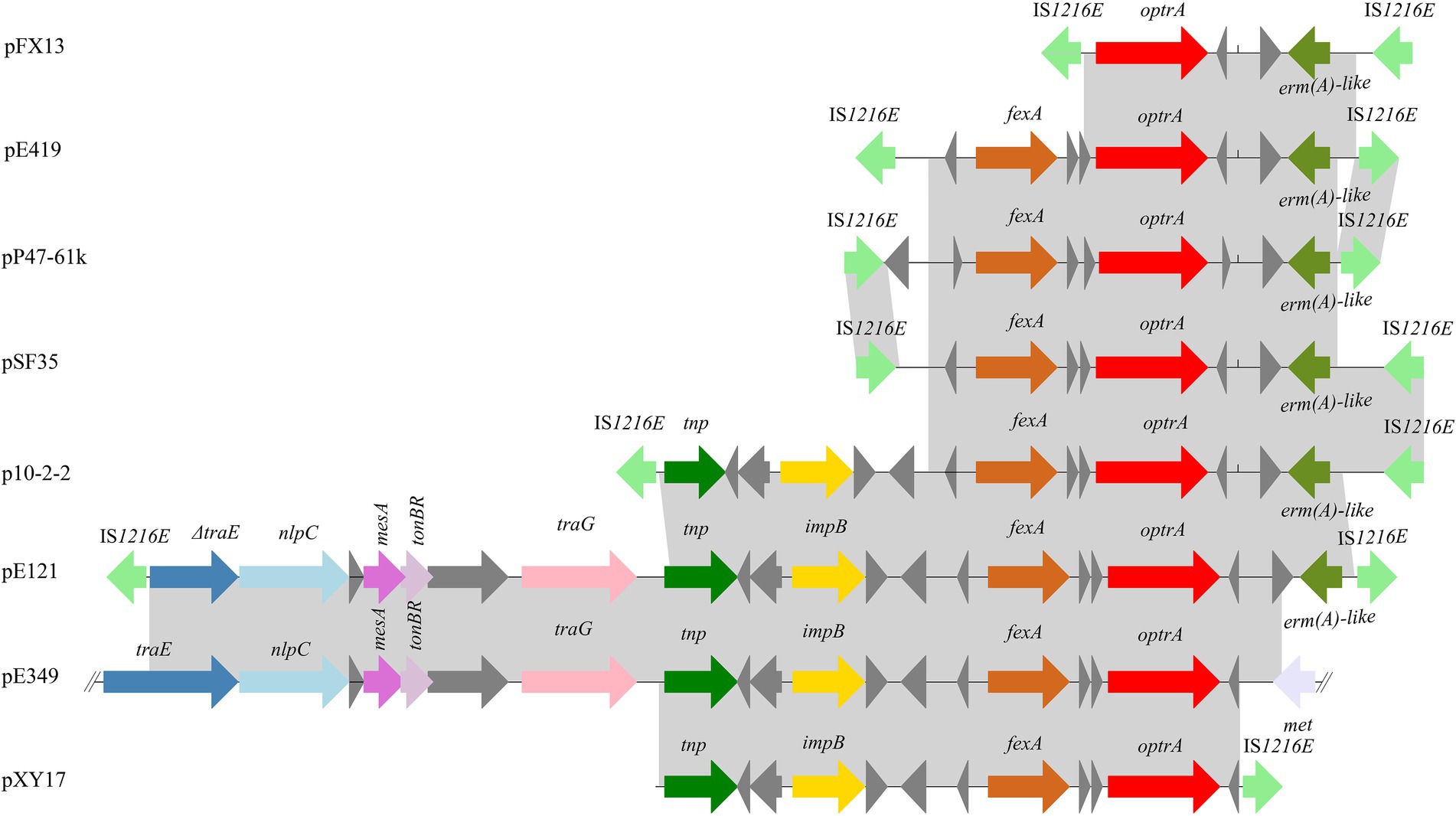
Figure 5. Schematic representation of the genetic environments surrounding the optrA gene in different sequences. The ORFs are shown as arrows, with the arrowhead indicating the direction of transcription. The same color represents the same elements and genes. Regions are drawn to scale. The sequences were retrieved from GenBank with accession numbers KT862775.1 (p10-2-2), T862776.1 (pE121), KP399637.1 (pE394), KT862777.1 (pE419), KT862778.1 (pFX13), KT862779.1 (pSF35), and KT862780.1 (pXY17).
When searching for sequences similar to plasmid pP47-27, we retrieved 25 sequences with identities >99.0% and coverage >90.0% from the NCBI nucleotide database (Supplementary Table S3). They were all plasmid sequences from the genus Enterococcus, including species of Enterococcus faecium (n = 17), Enterococcus faecalis (n = 5), Enterococcus hirae (n = 2), and Enterococcus lactis (n = 1). Of the 25 sequences, only two (pE843-27 from E. lactis and pEfm-EF3 of E. faecium) had a similar length of approximately 27 kb, while all the other plasmids were longer than 32 kb. Comparative genomic analysis revealed that all sequences encoded the florfenicol resistance genes fexB and poxtA, although mostly with different gene contexts or gene orders (Figures 2, 3; Supplementary Table S3).
Discussion
In this work, 77.3% (17/22) and 9.1% (2/22) of the Enterococcus isolates tested were resistant to florfenicol and linezolid, respectively. This result is similar to the results that showed resistance rates of 78.3% (for florfenicol) and 11.3% (for linezolid) against 106 enterococcal isolates from pigeon and duck feces in Egypt (Osman et al., 2019). Most of the florfenicol-resistant isolates also showed resistance to the other four antimicrobials, chloramphenicol, erythromycin, clarithromycin, and tetracycline. These antimicrobials are commonly consumed in the poultry industry as animal growth additives or therapeutic drugs (Cheng et al., 2014). It was significant to discover that two isolates (FH69 and P47) showed resistances to linezolid (≥8 mg/L). Linezolid is a very important antimicrobial used in the treatment of human enterococcal infections and is constantly being monitored by surveillance programs such as the global Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS; Mendes et al., 2016). The lower level of linezolid resistance among enterococci could be due to resistance genes (optrA and poxtA) and commonly found spontaneous mutations, such as the G2576T mutation in 23S rRNA (Biedenbach et al., 2010; Patel et al., 2013). Enterococci contain 4–6 copies of the 23S rRNA gene, and therefore, mutations in multiple copies are necessary to confer resistance (Biedenbach et al., 2010).
None of the enterococcal isolates in this work carried a cfr gene, while more than half (54.5%, 12/22) contained optrA genes. The optrA-positive rate was much higher than that reported by Kang et al. (2019), who found that only 3.7% (6/158) of enterococcal strains of swine origin harbored optrA. All of the optrA-positive isolates also co-carried the amphenicol exporter gene fexA. The presence of both optrA and fexA in the same isolate has been described in Enterococcus from humans and animals from China (Cai et al., 2015; Wang et al., 2015). To the best of our knowledge, this is the first report of one E. faecium isolate, P47, carrying four different amphenicol and oxazolidinone resistance genes (fexA, optrA, fexB, and poxtA). The most recently described linezolid resistance gene, poxtA, in S. aureus encodes a protein that is 32% identical to OptrA. Expression of poxtA in E. faecalis was able to decrease susceptibility to phenicols and oxazolidinones (Antonelli et al., 2018). As reported previously, Enterococcus isolates in the present study simultaneously carried several resistance genes that conferred the host bacterium resistance phenotypes, such as the tet(M) and tet(L) genes for tetracycline resistance and the erm(A) and erm(B) genes for macrolide-lincosamide-streptogramin B resistance (Almeida et al., 2020). Oxazolidinones are not approved for veterinary use in China, whereas tetracyclines, macrolides, and florfenicol are consistently being used in swine and poultry production in China (Kang et al., 2019). As mentioned above, florfenicol has been widely used for the treatment of gastrointestinal tract and respiratory infections in food-producing animals. The use of florfenicol has caused selective pressure and has likely been utilized for resistance carriage by some common constituent of the swine intestinal flora (Almeida et al., 2020). Therefore, the prevalence of optrA and poxtA in Enterococcus originating from household animals in the current study may be explained by co-selection. In particular, the regions encoding optrA and poxtA harbor multiple antimicrobial resistance genes. Selection for any of these resistance genes could also result in co-selection for the genes (Sun et al., 2018).
The resistance genes encoded in both pP47-27 [poxtA, fexB, tet(L), and tet(M)] and pP47-61 (ermA, optrA, fexA, ermB, and fosB) were all related to mobile genetic elements (MGEs). Comparative genomic analysis demonstrated that the similar genetic context of the resistance genes has been found in many plasmids (or chromosomes) of different bacterial species with animal and human origins (He et al., 2016; Kang et al., 2019), which indicated that mobile genetic elements participated in spreading the resistance genes, including amphenicol and oxazolidinone resistance genes, among the DNA molecules of different bacteria.
Conclusion
In conclusion, the results of this study demonstrated that the occurrence of oxazolidinone and amphenicol resistance is still frequent among Enterococcus isolates from animals. Mobile genetic elements, such as conjugative plasmids, insertion sequences, and transposons, may facilitate the horizontal transmission of oxazolidinone and amphenicol resistance genes. The presence of other resistance genes, including tet(L), tet(M), ermA, and ermB encoded on the plasmids, may lead to their co-selection. Therefore, the use of amphenicol antibiotics in food-producing animals should be closely monitored. Livestock husbandry might be recognized as a source of transferable oxazolidinone and amphenicol resistance determinants that could be spread to humans through the food chain.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: GenBank, accession numbers CP091100-CP091103.
Ethics statement
The animal study was reviewed and approved by Animal Welfare and Ethics Committee of Wenzhou Medical University, Zhejiang Province, China (Animal protocol number: wydw2021-0323).
Author contributions
PL, MG, WS, TY, SL, and XL collected the strains and performed the experiments. CF, LZ, JL, and TX analyzed the experimental results. CF and AL performed the bioinformatics analysis. PL, TX, KL, JL, and QB co-led the writing of the manuscript. QB and CS designed the work. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science and Technology Project of Wenzhou City, China (N20210001), Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001), and the National Natural Science Foundation of China (81960381).
Acknowledgments
The authors would like to acknowledge all the study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.811692/full#supplementary-material
Footnotes
1. ^https://blast.ncbi.nlm.nih.gov/blast.cgi
2. ^https://pubmlst.org/efaecium/
3. ^https://clsi.org/standards/
References
Almeida, L. M., Lebreton, F., Gaca, A., Bispo, P. M., Saavedra, J. T., Calumby, R. N., et al. (2020). Transferable resistance gene optrA in Enterococcus faecalis from swine in Brazil. Antimicrob. Agents Chemother. 64:20. doi: 10.1128/AAC.00142-20
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antonelli, A., D’Andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Bakthavatchalam, Y. D., Vasudevan, K., Babu, P., Neeravi, A. R., Narasiman, V., and Veeraraghavan, B. (2021). Genomic insights of optrA-carrying linezolid-resistant Enterococcus faecium using hybrid assembly: first report from India. J. Glob. Antimicrob. Resist. 25, 331–336. doi: 10.1016/j.jgar.2021.04.005
Biedenbach, D. J., Farrell, D. J., Mendes, R. E., Ross, J. E., and Jones, R. N. (2010). Stability of linezolid activity in an era of mobile oxazolidinone resistance determinants: results from the 2009 Zyvox(R) annual appraisal of potency and Spectrum program. Diagn. Microbiol. Infect. Dis. 68, 459–467. doi: 10.1016/j.diagmicrobio.2010.09.018
Brenciani, A., Morroni, G., Vincenzi, C., Manso, E., Mingoia, M., Giovanetti, E., et al. (2016). Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 71, 1118–1119. doi: 10.1093/jac/dkv438
Cai, J., Wang, Y., Schwarz, S., Lv, H., Li, Y., Liao, K., et al. (2015). Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010–2014. Clin. Microbiol. Infect. 21, 1095.e1091–1094.e1091. doi: 10.1016/j.cmi.2015.08.007
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cavaco, L. M., Korsgaard, H., Kaas, R. S., Seyfarth, A. M., Leekitcharoenphon, P., and Hendriksen, R. S. (2017). First detection of linezolid resistance due to the optrA gene in enterococci isolated from food products in Denmark. J. Glob. Antimicrob. Resist. 9, 128–129. doi: 10.1016/j.jgar.2017.04.001
Cheng, G., Hao, H., Xie, S., Wang, X., Dai, M., Huang, L., et al. (2014). Antibiotic alternatives: the substitution of antibiotics in animal husbandry? Front. Microbiol. 5:217. doi: 10.3389/fmicb.2014.00217
CLSI, (2021). Performance Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100. 31th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Cui, L., Wang, Y., Lv, Y., Wang, S., Song, Y., Li, Y., et al. (2016). Nationwide surveillance of novel Oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 60, 7490–7493. doi: 10.1128/AAC.01256-16
D’Andrea, M. M., Antonelli, A., Brenciani, A., Di Pilato, V., Morroni, G., Pollini, S., et al. (2019). Characterization of Tn6349, a novel mosaic transposon carrying poxtA, cfr and other resistance determinants, inserted in the chromosome of an ST5-MRSA-II strain of clinical origin. J. Antimicrob. Chemother. 74, 2870–2875. doi: 10.1093/jac/dkz278
Deshpande, L. M., Ashcraft, D. S., Kahn, H. P., Pankey, G., Jones, R. N., Farrell, D. J., et al. (2015). Detection of a new cfr-Like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 59, 6256–6261. doi: 10.1128/AAC.01473-15
Elghaieb, H., Tedim, A. P., Abbassi, M. S., Novais, C., Duarte, B., Hassen, A., et al. (2020). From farm to fork: identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals and retail meat. J. Antimicrob. Chemother. 75, 30–35. doi: 10.1093/jac/dkz419
Fan, R., Li, D., Wang, Y., He, T., Fessler, A. T., Schwarz, S., et al. (2016). Presence of the optrA gene in methicillin-resistant Staphylococcus sciuri of porcine origin. Antimicrob. Agents Chemother. 60, 7200–7205. doi: 10.1128/AAC.01591-16
Fang, H., Froding, I., Ullberg, M., and Giske, C. G. (2021). Genomic analysis revealed distinct transmission clusters of vancomycin-resistant Enterococcus faecium ST80 in Stockholm, Sweden. J. Hosp. Infect. 107, 12–15. doi: 10.1016/j.jhin.2020.10.019
Freitas, A. R., Finisterra, L., Tedim, A. P., Duarte, B., Novais, C., Peixe, L., et al. (2021). Linezolid-and multidrug-resistant enterococci in raw commercial dog food, Europe, 2019-2020. Emerg. Infect. Dis. 27, 2221–2224. doi: 10.3201/eid2708.204933
Fung, H. B., Kirschenbaum, H. L., and Ojofeitimi, B. O. (2001). Linezolid: an oxazolidinone antimicrobial agent. Clin. Ther. 23, 356–391. doi: 10.1016/s0149-2918(01)80043-6
He, T., Shen, Y., Schwarz, S., Cai, J., Lv, Y., Li, J., et al. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Homan, W. L., Tribe, D., Poznanski, S., Li, M., Hogg, G., Spalburg, E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40, 1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002
Huys, G., D’Haene, K., Collard, J. M., and Swings, J. (2004). Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70, 1555–1562. doi: 10.1128/AEM.70.3.1555-1562.2004
Kang, Z. Z., Lei, C. W., Kong, L. H., Wang, Y. L., Ye, X. L., Ma, B. H., et al. (2019). Detection of transferable oxazolidinone resistance determinants in Enterococcus faecalis and Enterococcus faecium of swine origin in Sichuan Province, China. J. Glob. Antimicrob. Resist. 19, 333–337. doi: 10.1016/j.jgar.2019.05.021
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fex A and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Lazaris, A., Coleman, D. C., Kearns, A. M., Pichon, B., Kinnevey, P. M., Earls, M. R., et al. (2017). Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J. Antimicrob. Chemother. 72, 3252–3257. doi: 10.1093/jac/dkx292
Lee, S. M., Huh, H. J., Song, D. J., Shim, H. J., Park, K. S., Kang, C. I., et al. (2017). Resistance mechanisms of linezolid-nonsusceptible enterococci in Korea: low rate of 23S rRNA mutations in Enterococcus faecium. J. Med. Microbiol. 66, 1730–1735. doi: 10.1099/jmm.0.000637
Li, D., Wang, Y., Schwarz, S., Cai, J., Fan, R., Li, J., et al. (2016). Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 71, 1474–1478. doi: 10.1093/jac/dkw040
Li, P., Zhu, T., Zhou, D., Lu, W., Liu, H., Sun, Z., et al. (2020). Analysis of resistance to Florfenicol and the related mechanism of dissemination in different animal-derived Bacteria. Front. Cell. Infect. Microbiol. 10:369. doi: 10.3389/fcimb.2020.00369
Liu, Y., Wang, Y., Schwarz, S., Li, Y., Shen, Z., Zhang, Q., et al. (2013). Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob. Agents Chemother. 57, 42–48. doi: 10.1128/AAC.01605-12
Liu, H., Wang, Y., Wu, C., Schwarz, S., Shen, Z., Jeon, B., et al. (2012). A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J. Antimicrob. Chemother. 67, 322–325. doi: 10.1093/jac/dkr481
Liu, Y., Wu, F., Chen, Q., Ying, Y., Jiang, Y., Lu, J., et al. (2020). Comparative genomics analysis of Raoultella planticola S25 isolated from duck in China, with florfenicol resistance. Comp. Immunol. Microbiol. Infect. Dis. 68:101398. doi: 10.1016/j.cimid.2019.101398
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Malisova, L., Jakubu, V., Pomorska, K., Musilek, M., and Zemlickova, H. (2021). Spread of linezolid-resistant Enterococcus spp. in human clinical isolates in the Czech Republic. Antibiotics 10:219. doi: 10.3390/antibiotics10020219
McDougal, L. K., Steward, C. D., Killgore, G. E., Chaitram, J. M., McAllister, S. K., and Tenover, F. C. (2003). Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41, 5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mendes, R. E., Deshpande, L. M., Farrell, D. J., Spanu, T., Fadda, G., and Jones, R. N. (2010). Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome Italy. J. Antimicrob. Chemother. 65, 2329–2335. doi: 10.1093/jac/dkq331
Mendes, R. E., Hogan, P. A., Jones, R. N., Sader, H. S., and Flamm, R. K. (2016). Surveillance for linezolid resistance via the Zyvox(R) annual appraisal of potency and Spectrum (ZAAPS) programme (2014): evolving resistance mechanisms with stable susceptibility rates. J. Antimicrob. Chemother. 71, 1860–1865. doi: 10.1093/jac/dkw052
Morroni, G., Brenciani, A., Antonelli, A., D’Andrea, M. M., Di Pilato, V., Fioriti, S., et al. (2018). Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes From an Enterococcus faecium clinical isolate. Front. Microbiol. 9:2189. doi: 10.3389/fmicb.2018.02189
Neoh, H. M., Tan, X. E., Sapri, H. F., and Tan, T. L. (2019). Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 74:103935. doi: 10.1016/j.meegid.2019.103935
Osman, K. M., Badr, J., Orabi, A., Elbehiry, A., Saad, A., Ibrahim, M. D. S., et al. (2019). Poultry as a vector for emerging multidrug resistant Enterococcus spp.: first report of vancomycin (van) and the chloramphenicol-florfenicol (cat-fex-cfr) resistance genes from pigeon and duck faeces. Microb. Pathog. 128, 195–205. doi: 10.1016/j.micpath.2019.01.006
Patel, S. N., Memari, N., Shahinas, D., Toye, B., Jamieson, F. B., and Farrell, D. J. (2013). Linezolid resistance in Enterococcus faecium isolated in Ontario Canada. Diagn. Microbiol. Infect. Dis. 77, 350–353. doi: 10.1016/j.diagmicrobio.2013.08.012
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Remm, M., Storm, C. E., and Sonnhammer, E. L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052. doi: 10.1006/jmbi.2000.5197
Sava, I. G., Heikens, E., and Huebner, J. (2010). Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 16, 533–540. doi: 10.1111/j.1469-0691.2010.03213.x
Schwarz, S., and Chaslus-Dancla, E. (2001). Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32, 201–225. doi: 10.1051/vetres:2001120
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Schwarz, S., Werckenthin, C., and Kehrenberg, C. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000
Stojkovic, V., Ulate, M. F., Hidalgo-Villeda, F., Aguilar, E., Monge-Cascante, C., Pizarro-Guajardo, M., et al. (2019). Cfr(B), cfr(C), and a new cfr-Like gene, cfr(E), in Clostridium difficile strains recovered across Latin America. Antimicrob. Agents Chemother. 64:19. doi: 10.1128/AAC.01074-19
Sun, C., Zhang, P., Ji, X., Fan, R., Chen, B., Wang, Y., et al. (2018). Presence and molecular characteristics of oxazolidinone resistance in staphylococci from household animals in rural China. J. Antimicrob. Chemother. 73, 1194–1200. doi: 10.1093/jac/dky009
Syriopoulou, V. P., Harding, A. L., Goldmann, D. A., and Smith, A. L. (1981). In vitro antibacterial activity of fluorinated analogs of chloramphenicol and thiamphenicol. Antimicrob. Agents Chemother. 19, 294–297. doi: 10.1128/AAC.19.2.294
Tamang, M. D., Moon, D. C., Kim, S. R., Kang, H. Y., Lee, K., Nam, H. M., et al. (2017). Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet. Microbiol. 201, 252–256. doi: 10.1016/j.vetmic.2017.01.035
Tang, Y., Fang, L., Xu, C., and Zhang, Q. (2017). Antibiotic resistance trends and mechanisms in the foodborne pathogen Campylobacter. Anim. Health Res. Rev. 18, 87–98. doi: 10.1017/S1466252317000135
Torres, C., Alonso, C. A., Ruiz-Ripa, L., Leon-Sampedro, R., Del Campo, R., and Coque, T. M. (2018). Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 6:2018. doi: 10.1128/microbiolspec.ARBA-0032-2018
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wu, F., Ying, Y., Yin, M., Jiang, Y., Wu, C., Qian, C., et al. (2019). Molecular characterization of a multidrug-resistant Klebsiella pneumoniae strain R46 isolated from a rabbit. Int. J. Genom. 2019, 5459190. doi: 10.1155/2019/5459190
Zhao, Q., Wang, Y., Wang, S., Wang, Z., Du, X. D., Jiang, H., et al. (2016). Prevalence and abundance of Florfenicol and linezolid resistance genes in soils adjacent to swine feedlots. Sci. Rep. 6, 32192. doi: 10.1038/srep32192
Zhu, T., Liu, S., Ying, Y., Xu, L., Liu, Y., Jin, J., et al. (2020). Genomic and functional characterization of fecal sample strains of Proteus cibarius carrying two flo R antibiotic resistance genes and a multiresistance plasmid-encoded cfr gene. Comp. Immunol. Microbiol. Infect. Dis. 69:101427. doi: 10.1016/j.cimid.2020.101427
Keywords: florfenicol, oxazolidinone, fexA, fexB, optrA, poxtA, Enterococcus, IS1216
Citation: Li P, Gao M, Feng C, Yan T, Sheng Z, Shi W, Liu S, Zhang L, Li A, Lu J, Lin X, Li K, Xu T, Bao Q and Sun C (2022) Molecular characterization of florfenicol and oxazolidinone resistance in Enterococcus isolates from animals in China. Front. Microbiol. 13:811692. doi: 10.3389/fmicb.2022.811692
Edited by:
Kristina Kadlec, Independent researcher, Wunstorf, GermanyReviewed by:
Patricia Alba, Experimental Zooprophylactic Institute of the Lazio and Tuscany Regions (IZSLT), ItalyIlana L. B. C. Camargo, University of São Paulo, Brazil
Copyright © 2022 Li, Gao, Feng, Yan, Sheng, Shi, Liu, Zhang, Li, Lu, Lin, Li, Xu, Bao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caixia Sun, 512177824@qq.com; Teng Xu, xuteng@wmu.edu.cn; Qiyu Bao, baoqy@genomics.cn
†These authors have contributed equally to this work
 Pingping Li
Pingping Li Mengdi Gao
Mengdi Gao Chunlin Feng1,3
Chunlin Feng1,3 Zhiqiong Sheng
Zhiqiong Sheng Anqi Li
Anqi Li Xi Lin
Xi Lin Kewei Li
Kewei Li Teng Xu
Teng Xu Qiyu Bao
Qiyu Bao