- 1College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2College of Veterinary Medicine Sichuan Key Laboratory of Animal Epidemic Disease and Human Health, Sichuan Agricultural University, Chengdu, China
- 3Animal Breeding and Genetics Key Laboratory of Sichuan Province, Sichuan Animal Science Academy, Chengdu, China
- 4Livestock and Poultry Biological Products Key Laboratory of Sichuan Province, Sichuan Animal Science Academy, Chengdu, China
Porcine circovirus 4 (PCV4) was identified in 2019 as a novel circovirus species and then proved to be pathogenic to piglets. However, there is a lack of its prevalence in the Southwest of China. To investigate whether PCV4 DNA existed in the Southwest of China, 374 samples were collected from diseased pigs during 2021–2022 and detected by a real-time PCR assay. The results showed that the positive rate of PCV4 was 1.34% (5/374) at sample level, and PCV4 was detected in two of 12 cities, demonstrating that PCV4 could be detected in pig farms in the Southwest of China, but its prevalence was low. Furthermore, one PCV4 strain (SC-GA2022ABTC) was sequenced in this study and shared a high identity (98.1–99.7%) with reference strains at the genome level. Combining genetic evolution analysis with amino acid sequence analysis, three genotypes PCV4a, PCV4b, and PCV4c were temporarily identified, and the SC-GA2022ABTC strain belonged to PCV4c with a specific amino acid pattern (239V for Rep protein, 27N, 28R, and 212M for Cap protein). Phylogenetic tree and amino acid alignment showed that PCV4 had an ancient ancestor with mink circovirus. In conclusion, the present study was the first to report the discovery and the evolutionary analysis of the PCV4 genome in pig herds of the Southwest of China and provide insight into the molecular epidemiology of PCV4.
Introduction
Porcine circoviruses (PCVs) are small, circular and single-strand DNA viruses belonging to the genus Circovirus of the family Circoviridae (Hamel et al., 1998; Meng, 2013; Opriessnig et al., 2020). At present, four species (PCV1, porcine circovirus 1; PCV2, porcine circovirus 2; PCV3, porcine circovirus 3; and PCV4, porcine circovirus 4) were recognized with similar structure. The genome contains two major open reading frames (ORFs): ORF1 and ORF2. The ORF1 gene encodes the replication-associated protein (Rep), and the ORF2 gene, in the opposite orientation from Rep, encodes the capsid protein (Cap) which was associated with the antigenic characteristics of circoviruses (Lekcharoensuk et al., 2004; Cheung, 2012).
Porcine circovirus 1 was first reported in 1974 and subsequently recognized non-pathogenic to pigs (Tischer et al., 1974, 1986; Allan et al., 1995), whereas, PCV2 has been deemed as one of the main agents of PCV-associated disease (PCVAD) (Nayar et al., 1997; Allan et al., 1998; Ellis et al., 1998; Kiupel et al., 1998; Morozov et al., 1998). PCVAD included post-weaning multisystem wasting syndrome (PMWS), porcine dermatitis and nephrotic syndrome (PDNS), and other syndromes (Nayar et al., 1997; Allan et al., 1998; Ellis et al., 1998; Kiupel et al., 1998; Morozov et al., 1998). In 2015, PCV3 was identified by next-generation sequencing analysis, and Jiang et al. (2019) have recently reported that PDNS-like disease can be reproduced in pigs infected with a cloned PCV3 virus (Phan et al., 2016; Palinski et al., 2017). In 2019, a novel circovirus species was identified in farmed pigs designated as PCV4, Hunan Province, China (Zhang D. et al., 2020), and later was also reported in several provinces and cities in China and South Korea (Chen et al., 2021; Sun et al., 2021a; Tian et al., 2021; Kim et al., 2022; Xu et al., 2022b). Recently, PCV4 was successfully rescued by Niu et al. (2022) from an infectious clone and demonstrated to be pathogenic to piglets. Nevertheless, genetic diversity and prevalence of PCV4 strains circulating in the Southwest of China have not been studied.
Therefore, in this study, a total of 374 samples were collected from diseased pigs with clinical signs of gastroenteritis (diarrhea) in pig farms in the Southwest of China and screened for the presence of PCV4 using a real-time PCR assay. Meanwhile, a complete genome sequence of PCV4 strain was generated and compared with published sequences in GenBank to study the genetic diversity of the PCV4 currently circulating in the Southwest of China during 2021–2022.
Materials and methods
Clinical samples collection and screening for porcine circovirus 4
A total of 374 clinical samples from pigs with clinical signs (PDNS, respiratory disease, and diarrhea) were collected from 37 farms in 12 cities (Mianyang, Suining, Chengdu, Deyang, Luzhou, Dazhou, Guangan, Guangyuan, Yibin, Nanchong, Neijiang, and Leshan) in the southwest of China from 2021 to 2022. The sample types in different farm comprised heart, liver, spleen, lung, kidney, brain, intestine, serum, nasal swab, and throat swab.
The viral genome was extracted using the FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme Biotech Co. Ltd., Nanjing, China) according to the manufacturer’s instructions. The RNA viral genome was extracted using Total RNA Kit I R6834 (Omega, Guangzhou, China) and then reverse-transcribed to cDNA using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). DNA was detected for the presence of PCV4 using a SYBR Green I-based qPCR assay as described previously (Xu et al., 2022b). In addition, cDNA or DNA was also detected for eight viruses in pigs including porcine epidemic diarrhea virus (PEDV), porcine reproductive and respiratory syndrome virus (PRRSV), Porcine deltacoronavirus (PDCoV), swine acute diarrhea syndrome-coronavirus (SADS-CoV) pseudorabies virus (PRV), porcine circovirus 2 (PCV2), and porcine circovirus 3 (PCV3) using previously described PCR or qPCR assays (Kim et al., 2000; Wang et al., 2018; Zhao Y. et al., 2019; Zhou et al., 2019; Tian et al., 2020; Zheng et al., 2020).
Complete genome sequencing of porcine circovirus 4
The complete genome of PCV4 was amplified as described previously (Xu et al., 2022a). The primers for amplifying the whole genome were summarized in Supplementary Table 1. The PCR reaction mixture consisted of 10 μl of PrimeSTAR® Max DNA Polymerase (Takara, Beijing, China), 0.5 μl (25 μM) of forward and reverse primers, 1 μl of template DNA, and 8 μl of ddH2O. The thermistor parameters were as follows: initial incubation at 98°C for 3 min; 35 cycles of 20 s at 98°C, 60°C for 20 s, and 72°C for 45 s, and a final extension for 5 min at 72°C. The PCR products were purified using Gel Extraction Kit D2500 (Omega Bio-Tek) and then cloned into the pMD18-T Vector (Takara, Dalian, China) in accordance with the manufacturer’s instructions. Finally, three positive clones were sequenced directly by Sangon Biotech Shanghai Co. Ltd., China.
Sequence alignment and phylogenetic analysis
The PCV4 whole genome was assembled using DNASTAR Lasergene. All unique PCV4 strains (Supplementary Table 2), available in GenBank database (accessed April 2, 2022) were analyzed with PCV4 strains in this study. To infer relationship of PCV4 and eight families of CRESS DNA viruses, Rep gene of 59 reference strains (Supplementary Table 2) that belong to eight families were downloaded from GenBank database. Molecular Evolutionary Genetics Analysis (MEGA) software (version 7.0) was used to the assemble, align, and analyze of the sequences. A neighbor-joining (NJ) phylogenetic tree was constructed with a p-distance model, and a bootstrap of 1,000 replicates using MEGA7.0.
Results and discussion
In 2019, the first case of PCV4 infection was reported in Hunan Province, China. Since then, the genome of PCV4 has been identified in several provinces of China, including Jiangsu, Shanxi, Henan, Anhui, Guangxi, and Inner Mongolia (Zhang D. et al., 2020; Chen et al., 2021; Ha et al., 2021; Tian et al., 2021). Moreover, PCV4 DNA was also detected in Korea but not in Spain, Italy, and Colombia (Franzo et al., 2020; Nguyen et al., 2022; Vargas-Bermudez et al., 2022). Niu et al. (2022) have further identified that PCV4 was pathogenic to piglets. However, the prevalence and genetic evolution of PCV4 in the Southwest of China remains unclear. Therefore, extensive epidemiological studies of PCV4 in the Southwest of China should be conducted to better address the potential threat of this novel virus.
In this study, 374 samples from 37 pig farms located in 12 cities in the Southwest of China during 2021–2022 were screened to verify the presence of PCV4. The results showed that the positive rate of PCV4 was 1.34% (5/374), which was far lower than that of PCV4 (25.40%, 16/63) in pigs of Henan and Shanxi Provinces described by Tian et al. (2021). The prevalence of PCV4 at sample level was 1.32% (2/151) in 2021 and 1.34% (3/223) in 2022. These results suggested that PCV4 existed in the Southwest of China, but it is not widespread. In my opinion, it is necessary to take some measures to prevent and control the potential threat of PCV4. Two positive samples in 2021 were collected from newborn piglets from a farm suffered from severe watery and projectile diarrhea, and PEDV was positive in this farm (Table 1). Three positive samples in 2022 were collected from different growth stages from a pig farm suffered from PDNS, and PCV2 was also positive in this farm (Table 1). The pathogenicity of PCV4 infection alone warrants further study.
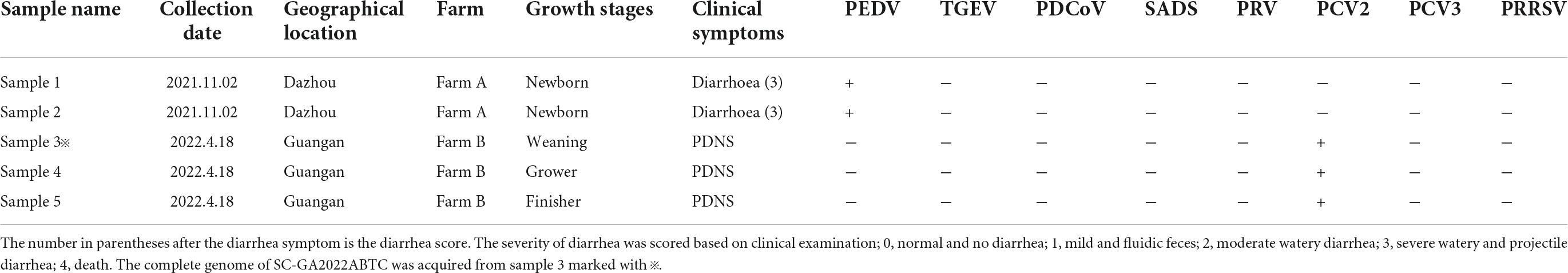
Table 1. Origin, clinical manifestation information and detection results of porcine circovirus 4 (PCV4) in pig clinical samples from the Southwest of China during 2021–2022.
The complete genome of one PCV4 strain (SC-GA2022ABTC) was amplified from a pig suffering from PDNS in Guangan, the Southwest of China, in 2022, and submitted to the GenBank database with the accession number OP497960. The complete genome of SC-GA2022ABTC was 1,770 nt in length without deletions and insertions of nucleotides. Compared with all 41 unique PCV4 strains (Supplementary Table 2) available in the GenBank database (accessed April 2, 2022), the SC-GA2022ABTC strain showed a high identity (98.1–99.8%) with PCV4 reference strains at the complete genome level. Among 41 PCV4 reference strains, SC-GA2022ABTC strain displayed the highest complete genome homology (99.8%) with HNU-AHG1-2019 derived from Hunan Province. As is depicted in Figure 1, among the provinces where the PCV4 whole genome has been reported, Hunan is the closest to Sichuan. Besides, 26 other representative circovirus strains (Supplementary Table 2) were selected for further analysis (Table 2). The genome of SC-GA2022ABTC strain showed the highest homology (68%) to the Mink circovirus, followed by 62.6% with Bat associated circovirus (accession no. NC 038385) and then to 35.8–51.6% with other circovirus species (Table 2), which was consistent with a previous report (Zhang H. H. et al., 2020). Additionally, SC-GA2022ABTC exhibited amino acid homology with these circovirus strains ranging from 16.2 to 80.1% for Rep proteins and 12.7 to 70.2% for Cap Proteins.
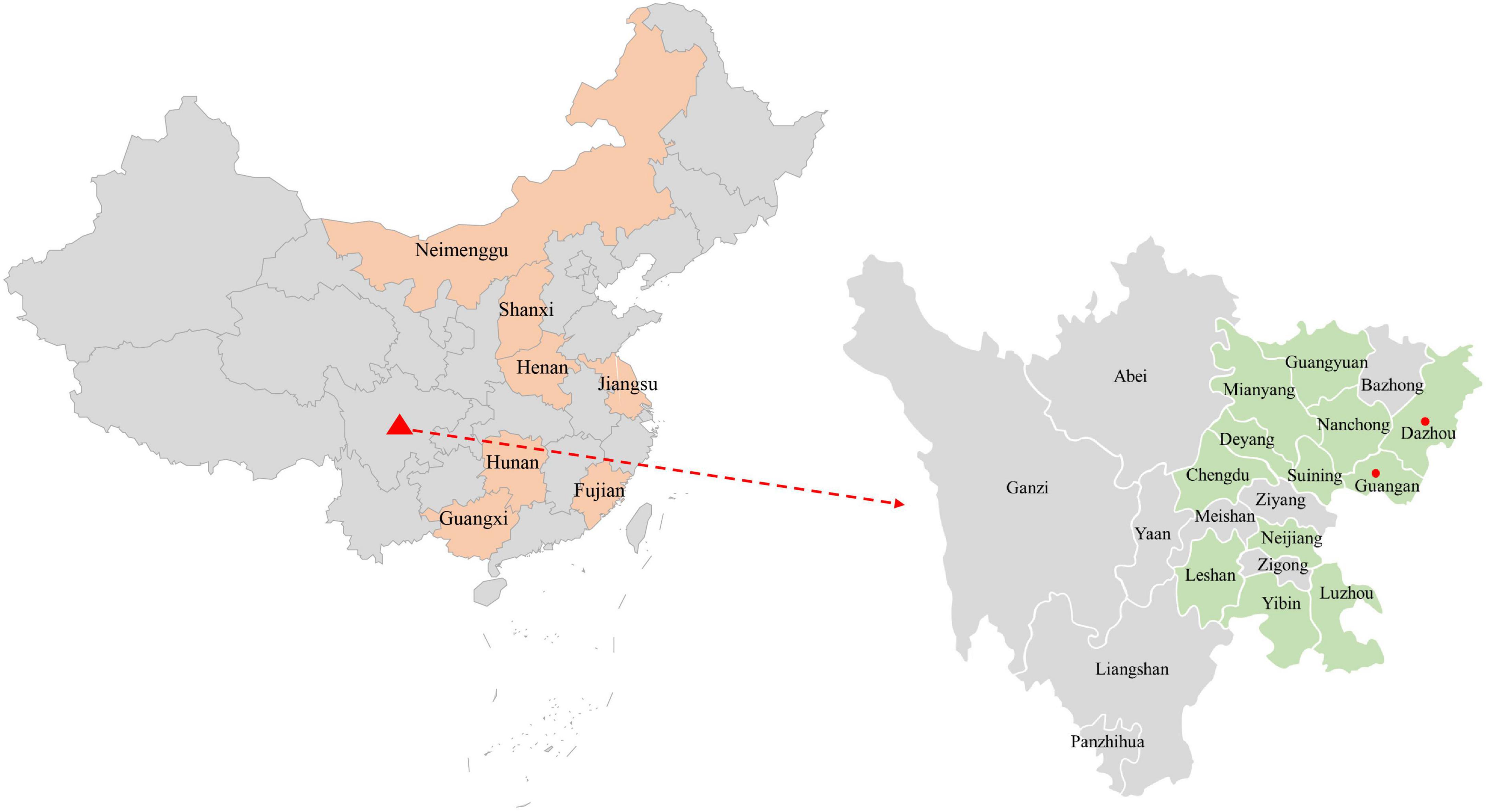
Figure 1. The geographical distribution of the 374 samples in the Southwest of China. In China, provinces with PCV4 complete genome amplified are filled with orange. In the Southwest of China, cities with sample collections are filled with light green, and cities with positive sample detection are marked with red solid circles.
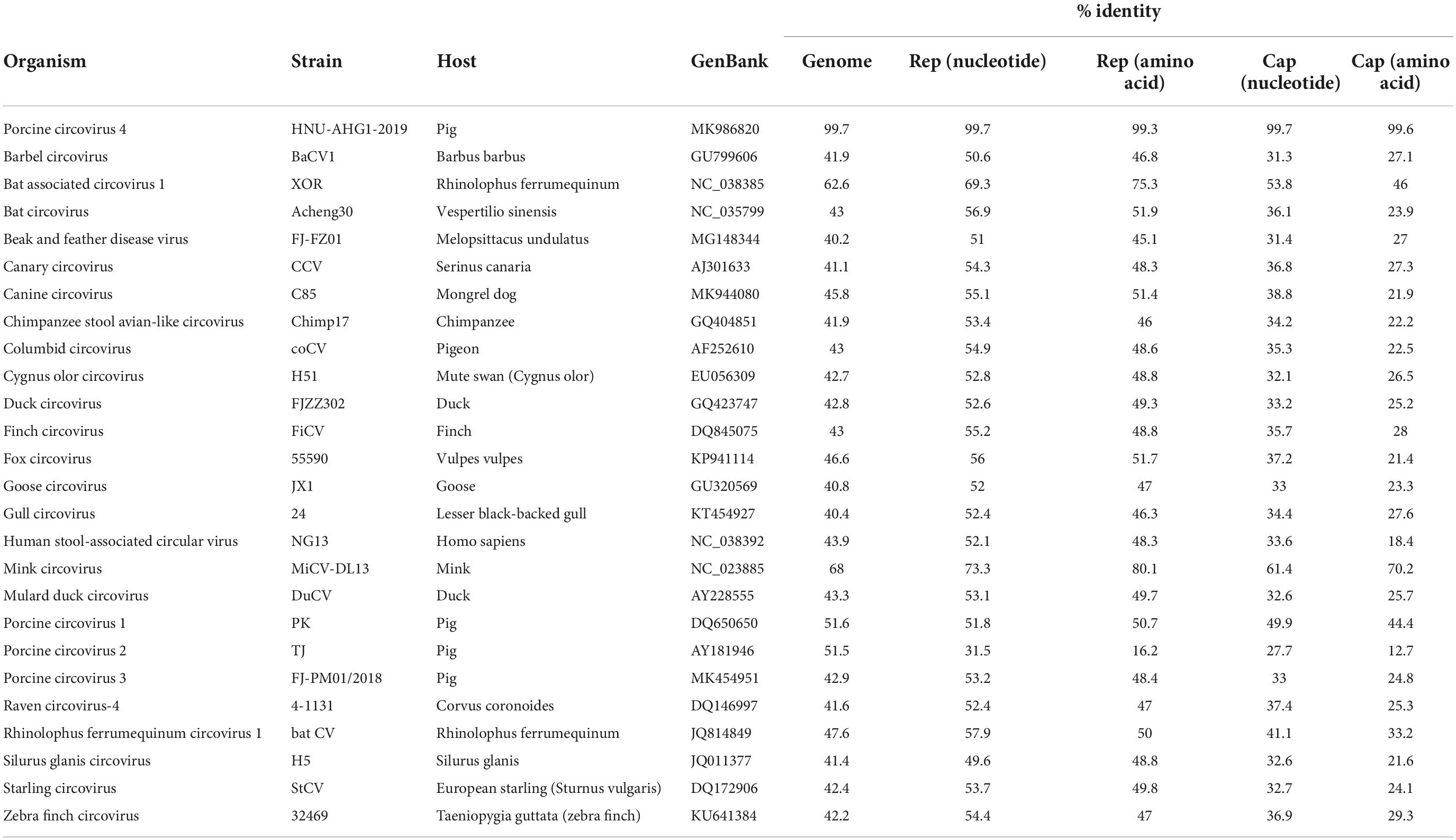
Table 2. Percent nucleotide and amino acid identity (% Identity) between porcine circovirus 4 (PCV4) strain in this study and reference strains.
To address the evolutionary relationship between the SC-GA2022ABTC strain and PCV4 strains in other regions, a phylogenetic tree was constructed based on the complete genome sequences of the SC-GA2022ABTC strain and 41 reference strains, and PCV4 was temporarily divided into three genotypes (PCV4a, PCV4b, and PCV4c) (Figure 2; Xu et al., 2022a). A total of 22 PCV4 strains from four provinces (Henan, Hebei, Guangxi, and Jiangsu) of China belonged to PCV4a; 13 reference strains derived from two provinces (Henan and Hebei) belonged to PCV4b. Nevertheless, the SC-GA2022ABTC strain and six PCV4 strains from three provinces (Fujian, Hunan, and Inner Mongolia) of China were clustered into PCV4c together with one South Korea strain.
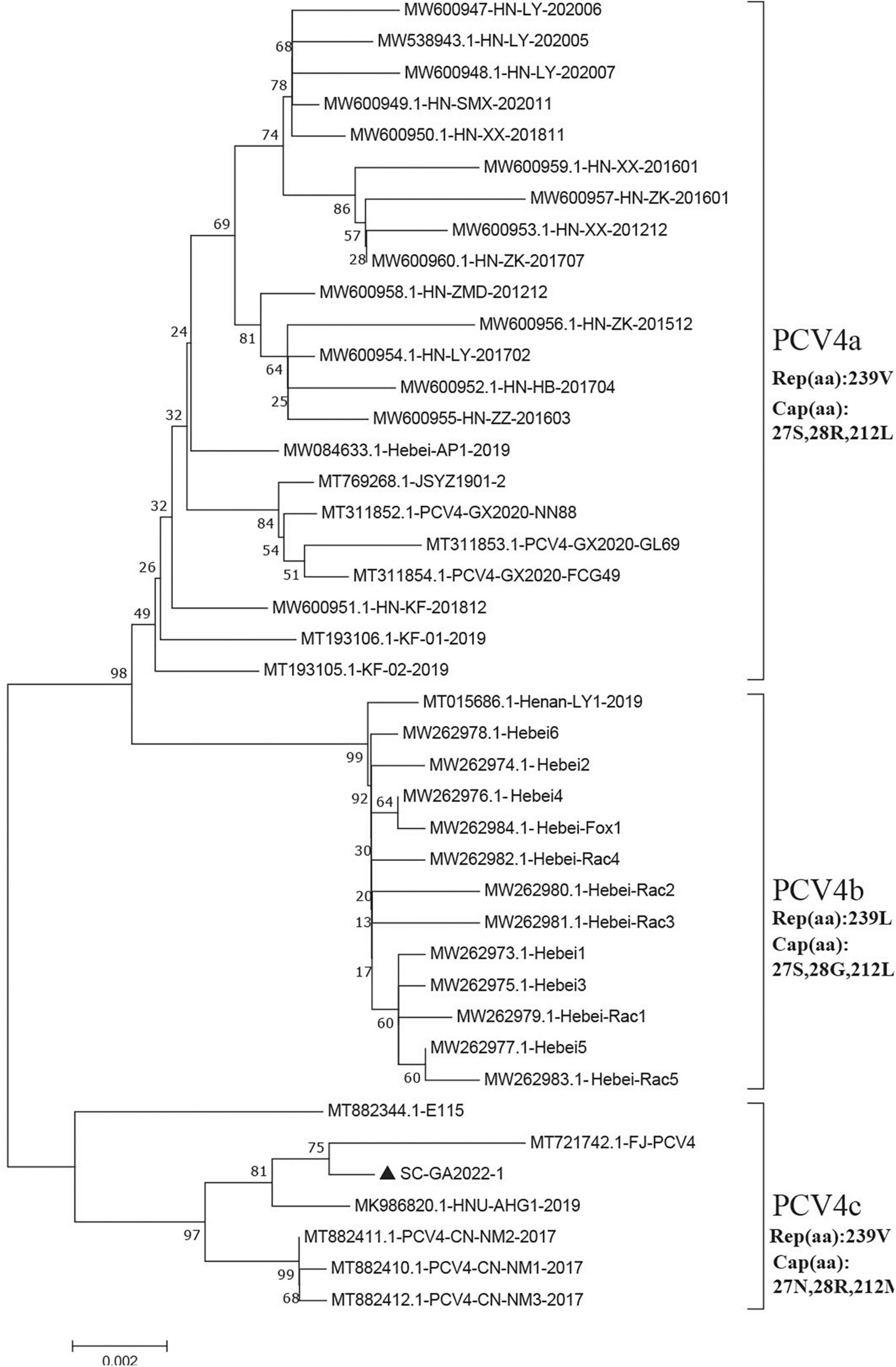
Figure 2. Neighbor-joining trees were constructed with a p-distance model and bootstrapping at 1,000 replicates. Phylogenetic tree was constructed based on the complete genome of 42 PCV4 strains. SC-GA2022ABTC strain was marked with the black solid triangle (▲). Scale bar indicates nucleotide substitutions per site.
Moreover, four amino acid mutations (V239L in Rep, S27N, R28G, and L212M in Cap) could serve as a molecular marker for PCV4 clade divisions (Xu et al., 2022a), which was also observed in this study (Figure 2). Summarizing the marker codons in Rep and Cap resulted in a specific amino acid pattern for PCV4a (239V in Rep, 27S, 28R, and 212L in Cap), for PCV4b (239L in Rep, 27S, 28G, and 212L in Cap) and for PCV4c (239V for Rep protein, 27N, 28R, and 212M for Cap protein). Amino acid substitutions as markers for clade divisions of other viruses were also reported as described previously, such as PCV3 and CPV (Parrish et al., 1988, 1991; Zhao et al., 2016; Fu et al., 2018; Kwan et al., 2021). Compared with previous studies (Hou et al., 2021; Xu et al., 2022b), more whole-genome sequences were available in the Genbank database, and the genetic tree was also richer, which could aid an easier interpretation of PCV-4 molecular epidemiology. Therefore, greater efforts must be made to provide more representative and structured sampling campaigns and to increase the sharing of correctly annotated sequences in free databases, in order to establish more accurate and scientific typing schemes.
The circular Rep-encoding single stranded (CRESS) DNA virus emergence in diverse host has been associated with severe diseases (Zhao L. et al., 2019). Seven family members of the CRESS DNA viruses have been reported, including Circovidae, Nanoviridae, Smacoviridae, Genomoviridae, Bacilladnaviridae, Geminiviridae, and Redondoviridae, established by the International Committee on the Taxonomy of Viruses (ICTV). Recently, a proposed family Kirkoviridae emerged and was reported in several recent studies (Liu et al., 2021; Sun et al., 2021b). Moreover, porcine circovirus-like virus P1 is an unclassified circovirus and might be one of the agents of PCVAD, while its replication-related proteins remained unknown (Wen et al., 2018). To better understand the evolutionary origin of PCV4, a data set including the PCV4 strain in our study, three PCV4 reference strain and 59 other representative CRESS DNA virus strains was analyzed. Then, a phylogenetic tree was constructed based on the amino acids of Rep to explore the origin of PCV 4 (Figure 3A). As is depicted in Figure 3A, four PCV4 strains (including SC-GA2022ABTC) were clustered into Circovidae. The genus Redondoviridae includes the species Brisavirus and Vientovirus (Figure 3A), and Circovidae two genera (Cyclovirus and Circovirus) (Figure 3B). The phylogenetic tree based on the amino acids of Rep of Circovidae strains showed that PCV4 was distantly related to other PCVs but closely related to mink circovirus (Figure 3B), which was corroborated by complete genome homology of PCV4 strain in this study with reference strains (Table 1). These results suggested that PCV4 shares an ancient ancestor with mink circovirus, which was consistent with a previous study (Li et al., 2022).
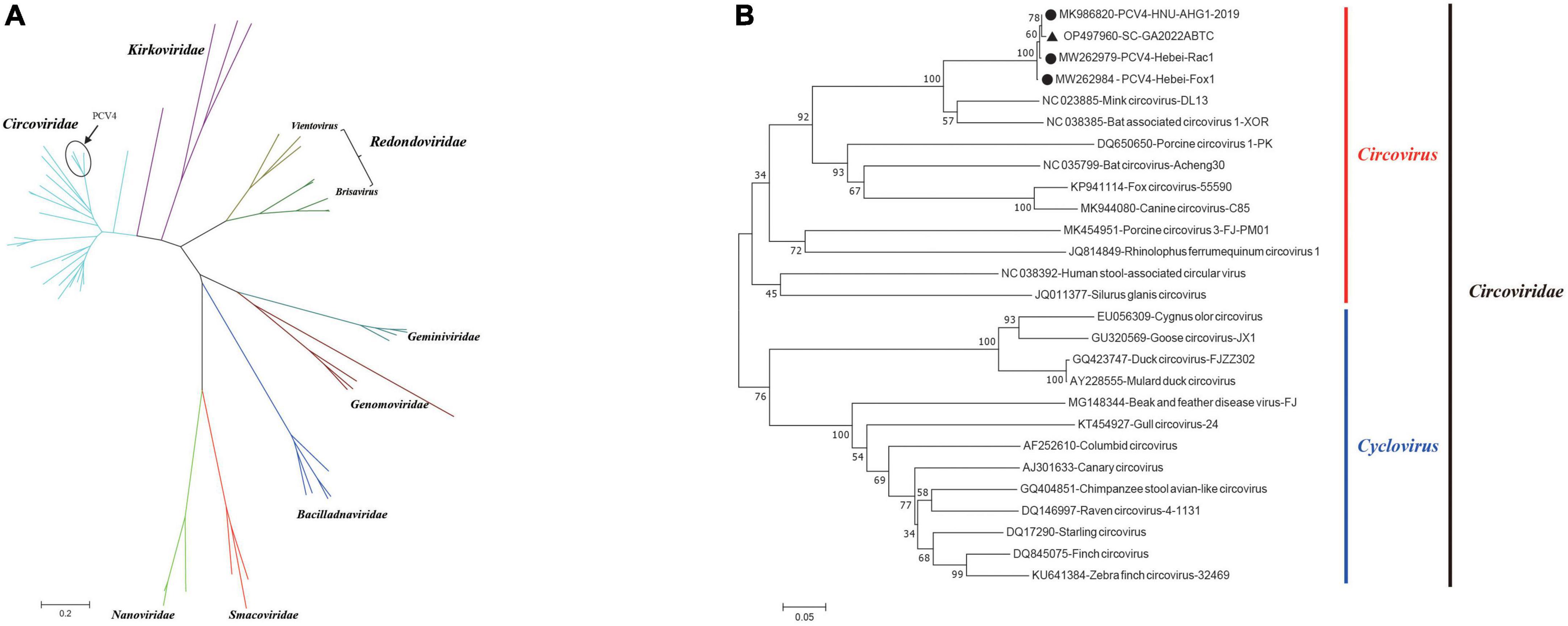
Figure 3. Neighbor-joining trees were constructed with a p-distance model and bootstrapping at 1,000 replicates. (A) Phylogenetic tree was constructed based on amino acids of Rep of 63 CRESS DNA virus strains. (B) Phylogenetic tree was constructed based on amino acids of Rep of 27 Circoviridae strains. SC-GA2022ABTC strain was marked with the black solid triangle (▲). The other three Porcine circovirus 4 (PCV4) reference strains were marked with black filled circles (⬤). Scale bar indicates nucleotide substitutions per site.
Among 42 PCV4 strains, 32 amino acid mutations were observed in the Rep (Figure 4). For Rep, the N-terminal endonuclease domain comprised three conserved motifs (motifI-13FTLNN17, motifII-50PHLQG54, and motifIII-90YCSK93) and the helicase domain of superfamily 3 (SF3) containing three Walker motifs (Walker A-168GxxxxGKS175, Walker B-207DDY209, and Walker C-245ITSN248) were observed in SC-GA2022ABTC strain, which was consistent with a previous study (Nguyen et al., 2022).
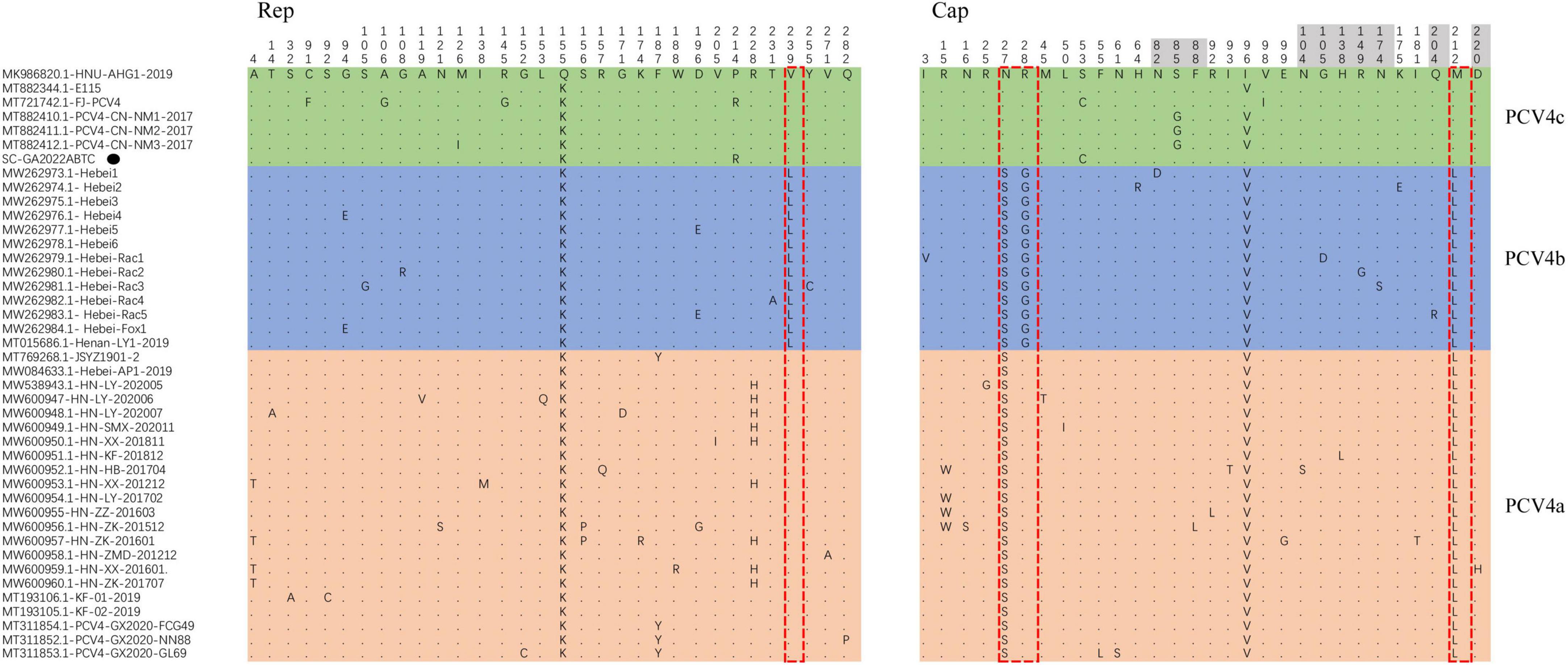
Figure 4. All amino acid mutation sites of Rep protein and Cap of 42 Porcine circovirus 4 (PCV4) strains. PCV4 strains were divided into three genotypes, comprising PCV4a (light orange), PCV4b (light green), PCV4c (light blue). The specific amino acid patterns of genotype were displayed in the red dotted box. Amino acid residues contained in potential linear B-cell epitopes are highlighted in light grey. SC-GA2022ABTC strain investigated in this study. SC-GA2022ABTC strain sequenced in this study was marked with the black solid triangle (▲).
For the Caps of PCVs (PCV1–PCV3), the nuclear localization signal (NLS) region, an arginine-rich region within the circovirus genus, mediate nuclear targeting of viral genomes and was experimentally confirmed (Liu et al., 2001; Shuai et al., 2008; Mou et al., 2019), which was also confirmed in the N-terminus of PCV4 Cap that ranged from 1 to 20 amino acid (Zhou et al., 2021). Compared to other PCV4 reference strains, no amino acid mutations occurred in the NLS region of the SC-GA2022ABTC strain. However, the amino acid mutations (I3V for Hebei-Rac1 strain, R15W for HN-HB-201704 strain, HN-LY-201702 strain, HN-ZZ-201603 strain, and HN-ZK-201512 strain, and N16S for HN-ZK-201512) were different from other PCV4 strains and occurred in the NLS region, indicating these PCV4 strains might differ in cell tropism, manner and speed of cell entry. Five potential linear B-cell epitopes with high antigenicity were predicted in a recent report, including Epitope A: 72F-88F; Epitope B: 104N-112Y; Epitope C: 122D-177N; Epitope D: 199N-205V; and Epitope E: 219F-225P (Wang et al., 2021). As shown in Figure 4, there were 30 amino acid mutations in Cap of 42 PCV4 strains, 11 of which were located in the predicted epitope region, which may alter the antigenicity of the PCV4 Cap. However, potential immunogenic changes due to amino acid mutations in epitope regions of Cap remained to be determined.
Conclusion
Overall, this study was the first to report the presence of PCV4 in the Southwest of China. The first complete genome sequence in the Southwest of China was successfully sequenced and named SC-GA2022ABTC strain. The SC-GA2022ABTC strain shared a high homology (98.1–99.7%) with other PCV4 strains. SC-GA2022ABTC strain belonged to PCV4c with a specific amino acid pattern (239V for Rep protein, 27N, 28R, and 212M for Cap protein) by combining genetic evolution with amino acid sequence. These findings help us to understand the prevalence and genomic characteristics of PCV4 in pig farms in the Southwest of China.
Data availability statement
The data presented in this study are deposited in the GenBank repository, accession number: OP497960.
Ethics statement
All experimental procedures were reviewed and approved by the Sichuan Agricultural University Animal Care and Use Committee [license number SCXK (Sichuan) 2013-0001].
Author contributions
X-GS, S-YL, and LZ contributed to the conceptualization. Y-CZ and Y-RA contributed to the methodology. TX contributed to the software and writing—original draft preparation. FW and DY contributed to the validation and supervision. Z-WX contributed to the project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chongqing Municipal Technology Innovation and Application Development Project (No. cstc2021jscx-dxwt BX0007), the Key K&D Program of Sichuan Science and Technology Plan (No. 2022YFN0007), the Porcine Major Science and Technology Project of Sichuan Science and Technology Plan (No. 2021ZDZX0010-3), the Sichuan Provincial Department of Science and Technology Rural Area Key R&D Program (No. 2020YFN0147), and the Agricultural Industry Technology System of Sichuan Provincial Department of Agriculture (Project No. CARS-SVDIP).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1052533/full#supplementary-material
References
Allan, G. M., Mcneilly, F., Cassidy, J. P., Reilly, G. A., Adair, B., Ellis, W. A., et al. (1995). Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet. Microbiol. 44, 49–64. doi: 10.1016/0378-1135(94)00136-k
Allan, G. M., Mcneilly, F., Kennedy, S., Daft, B., Clarke, E. G., Ellis, J. A., et al. (1998). Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagnost. Investig. 10, 3–10. doi: 10.1177/104063879801000102
Chen, N., Xiao, Y., Li, X., Li, S., Xie, N., Yan, X., et al. (2021). Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 68, 1615–1624. doi: 10.1111/tbed.13833
Cheung, A. K. (2012). Porcine circovirus: Transcription and DNA replication. Virus Res. 164, 46–53. doi: 10.1016/j.virusres.2011.10.012
Ellis, J., Hassard, L., Clark, E., Harding, J., Allan, G., Willson, P., et al. (1998). Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39, 44–51.
Franzo, G., Ruiz, A., Grassi, L., Sibila, M., Drigo, M., and Segalés, J. (2020). Lack of Porcine circovirus 4 Genome Detection in Pig Samples from Italy and Spain. Pathogens 9:433. doi: 10.3390/pathogens9060433
Fu, X., Fang, B., Ma, J., Liu, Y., Bu, D., Zhou, P., et al. (2018). Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound. Emerg. Dis. 65, e296–e303. doi: 10.1111/tbed.12752
Ha, Z., Yu, C., Xie, C., Wang, G., Zhang, Y., Hao, P., et al. (2021). Retrospective surveillance of porcine circovirus 4 in pigs in Inner Mongolia, China, from 2016 to 2018. Arch. Virol. 166, 1951–1959. doi: 10.1007/s00705-021-05088-w
Hamel, A. L., Lin, L. L., and Nayar, G. P. (1998). Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72, 5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998
Hou, C. Y., Zhang, L. H., Zhang, Y. H., Cui, J. T., Zhao, L., Zheng, L. L., et al. (2021). Phylogenetic analysis of porcine circovirus 4 in Henan Province of China: A retrospective study from 2011 to 2021. Transbound. Emerg. Dis. 69, 1890–1901. doi: 10.1111/tbed.14172
Jiang, H., Wang, D., Wang, J., Zhu, S., She, R., Ren, X., et al. (2019). Induction of Porcine Dermatitis and Nephropathy Syndrome in Piglets by Infection with Porcine Circovirus Type 3. J. Virol. 93, e02045–18. doi: 10.1128/jvi.02045-18
Kim, D. Y., Kim, H. R., Park, J. H., Kwon, N. Y., Kim, J. M., Kim, J. K., et al. (2022). Detection of a novel porcine circovirus 4 in Korean pig herds using a loop-mediated isothermal amplification assay. J. Virol. Methods 299:114350. doi: 10.1016/j.jviromet.2021.114350
Kim, O., Choi, C., Kim, B., and Chae, C. (2000). Detection and differentiation of porcine epidemic diarrhoea virus and transmissible gastroenteritis virus in clinical samples by multiplex RT-PCR. Vet. Rec. 146, 637–640. doi: 10.1136/vr.146.22.637
Kiupel, M., Stevenson, G. W., Mittal, S. K., Clark, E. G., and Haines, D. M. (1998). Circovirus-like viral associated disease in weaned pigs in Indiana. Vet. Pathol. 35, 303–307. doi: 10.1177/030098589803500411
Kwan, E., Carrai, M., Lanave, G., Hill, J., Parry, K., Kelman, M., et al. (2021). Analysis of canine parvoviruses circulating in Australia reveals predominance of variant 2b and identifies feline parvovirus-like mutations in the capsid proteins. Transbound. Emerg. Dis. 68, 656–666. doi: 10.1111/tbed.13727
Lekcharoensuk, P., Morozov, I., Paul, P. S., Thangthumniyom, N., Wajjawalku, W., and Meng, X. J. (2004). Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 78, 8135–8145. doi: 10.1128/jvi.78.15.8135-8145.2004
Li, X., Chen, S., Niu, G., Zhang, X., Ji, W., Ren, Y., et al. (2022). Porcine Circovirus Type 4 Strains Circulating in China Are Relatively Stable and Have Higher Homology with Mink Circovirus than Other Porcine Circovirus Types. Int. J. Mol. Sci. 23:3288. doi: 10.3390/ijms23063288
Liu, Q., Tikoo, S. K., and Babiuk, L. A. (2001). Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 285, 91–99. doi: 10.1006/viro.2001.0922
Liu, X., Zhang, X., Xu, G., Wang, Z., Shen, H., Lian, K., et al. (2021). Emergence of porcine circovirus-like viruses associated with porcine diarrheal disease in China. Transbound. Emerg. Dis. 68, 3167–3173. doi: 10.1111/tbed.14223
Meng, X. J. (2013). Porcine circovirus type 2 (PCV2): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 1, 43–64. doi: 10.1146/annurev-animal-031412-103720
Morozov, I., Sirinarumitr, T., Sorden, S. D., Halbur, P. G., Morgan, M. K., Yoon, K. J., et al. (1998). Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36, 2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998
Mou, C., Wang, M., Pan, S., and Chen, Z. (2019). Identification of Nuclear Localization Signals in the ORF2 Protein of Porcine Circovirus Type 3. Viruses 11:1086. doi: 10.3390/v11121086
Nayar, G. P., Hamel, A., and Lin, L. (1997). Detection and characterization of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Can. Vet. J. 38, 385–386.
Nguyen, V. G., Do, H. Q., Huynh, T. M., Park, Y. H., Park, B. K., and Chung, H. C. (2022). Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound. Emerg. Dis. 69, 538–548. doi: 10.1111/tbed.14017
Niu, G., Zhang, X., Ji, W., Chen, S., Li, X., Yang, L., et al. (2022). Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo. Transbound. Emerg. Dis. 69, e1632–e1641. doi: 10.1111/tbed.14498
Opriessnig, T., Karuppannan, A. K., Castro, A., and Xiao, C. T. (2020). Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 286:198044. doi: 10.1016/j.virusres.2020.198044
Palinski, R., Piñeyro, P., Shang, P., Yuan, F., Guo, R., Fang, Y., et al. (2017). A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 91, e01879–16. doi: 10.1128/jvi.01879-16
Parrish, C. R., Aquadro, C. F., Strassheim, M. L., Evermann, J. F., Sgro, J. Y., and Mohammed, H. O. (1991). Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65, 6544–6552. doi: 10.1128/JVI.65.12.6544-6552.1991
Parrish, C. R., Have, P., Foreyt, W. J., Evermann, J. F., Senda, M., and Carmichael, L. E. (1988). The global spread and replacement of canine parvovirus strains. J. Gen. Virol. 69, 1111–1116. doi: 10.1099/0022-1317-69-5-1111
Phan, T. G., Giannitti, F., Rossow, S., Marthaler, D., Knutson, T. P., Li, L., et al. (2016). Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 13:184. doi: 10.1186/s12985-016-0642-z
Shuai, J., Wei, W., Jiang, L., Li, X., Chen, N., and Fang, W. (2008). Mapping of the nuclear localization signals in open reading frame 2 protein from porcine circovirus type 1. Acta Biochim. Biophys. Sin. 40, 71–77. doi: 10.1111/j.1745-7270.2008.00377.x
Sun, W., Du, Q., Han, Z., Bi, J., Lan, T., Wang, W., et al. (2021a). Detection and genetic characterization of porcine circovirus 4 (PCV4) in Guangxi. China. Gene 773:145384. doi: 10.1016/j.gene.2020.145384
Sun, W., Wang, W., Cao, L., Zheng, M., Zhuang, X., Zhang, H., et al. (2021b). Genetic characterization of three porcine circovirus-like viruses in pigs with diarrhoea in China. Transbound. Emerg. Dis. 68, 289–295. doi: 10.1111/tbed.13731
Tian, R. B., Jin, Y., Xu, T., Zhao, Y., Wang, Z. Y., and Chen, H. Y. (2020). Development of a SYBR green I-based duplex real-time PCR assay for detection of pseudorabies virus and porcine circovirus 3. Mol. Cell Probes 53:101593. doi: 10.1016/j.mcp.2020.101593
Tian, R. B., Zhao, Y., Cui, J. T., Zheng, H. H., Xu, T., Hou, C. Y., et al. (2021). Molecular detection and phylogenetic analysis of Porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 68, 276–282. doi: 10.1111/tbed.13714
Tischer, I., Mields, W., Wolff, D., Vagt, M., and Griem, W. (1986). Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91, 271–276. doi: 10.1007/bf01314286
Tischer, I., Rasch, R., and Tochtermann, G. (1974). Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol.Orig. A 226, 153–167.
Vargas-Bermudez, D. S., Mogollón, J. D., and Jaime, J. (2022). The Prevalence and Genetic Diversity of PCV3 and PCV2 in Colombia and PCV4 Survey during 2015-2016 and 2018-2019. Pathogens 11:633. doi: 10.3390/pathogens11060633
Wang, D., Mai, J., Lei, B., Zhang, Y., Yang, Y., and Wang, N. (2021). Structure, Antigenic Properties, and Highly Efficient Assembly of PCV4 Capsid Protein. Front. Vet. Sci. 8:695466. doi: 10.3389/fvets.2021.695466
Wang, X. Y., Ji, C. J., Zhang, X., Xu, D. P., and Zhang, D. L. (2018). Infection, genetic and virulence characteristics of porcine epidemic diarrhea virus in northwest China. Infect. Genet. Evol. 62, 34–39. doi: 10.1016/j.meegid.2018.04.001
Wen, L., Mao, A., Jiao, F., Zhang, D., Xie, J., and He, K. (2018). Detection of porcine circovirus-like virus P1 in Hebei. China. Transbound. Emerg. Dis. 65, 1133–1136. doi: 10.1111/tbed.12896
Xu, T., Hou, C. Y., Zhang, Y. H., Li, H. X., Chen, X. M., Pan, J. J., et al. (2022b). Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 808:145991. doi: 10.1016/j.gene.2021.145991
Xu, T., Chen, X. M., Fu, Y., Ai, Y., Wang, D. M., Wei, Z. Y., et al. (2022a). Cross-species transmission of an emerging porcine circovirus (PCV4): First molecular detection and retrospective investigation in dairy cows. Vet. Microbiol. 273:109528. doi: 10.1016/j.vetmic.2022.109528
Zhang, D., Bai, C., Ge, K., Li, Y., Gao, W., Jiang, S., et al. (2020). Establishment of an SYBR Green-based real-time PCR assay for porcine circovirus type 4 detection. J. Virol. Methods 285:113963. doi: 10.1016/j.jviromet.2020.113963
Zhang, H. H., Hu, W. Q., Li, J. Y., Liu, T. N., Zhou, J. Y., Opriessnig, T., et al. (2020). Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 67, 1057–1061. doi: 10.1111/tbed.13446
Zhao, L., Rosario, K., Breitbart, M., and Duffy, S. (2019). Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses With Small Genomes and a Diverse Host Range. Adv. Virus Res. 103, 71–133. doi: 10.1016/bs.aivir.2018.10.001
Zhao, Y., Han, H. Y., Fan, L., Tian, R. B., Cui, J. T., Li, J. Y., et al. (2019). Development of a TB green II-based duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of porcine circovirus 2 and 3. Mol. Cell Probes 45, 31–36. doi: 10.1016/j.mcp.2019.04.001
Zhao, Z., Liu, H., Ding, K., Peng, C., Xue, Q., Yu, Z., et al. (2016). Occurrence of canine parvovirus in dogs from Henan province of China in 2009-2014. BMC Vet. Res. 12:138. doi: 10.1186/s12917-016-0753-1
Zheng, L. L., Chai, L. Y., Tian, R. B., Zhao, Y., Chen, H. Y., and Wang, Z. Y. (2020). Simultaneous detection of porcine reproductive and respiratory syndrome virus and porcine circovirus 3 by SYBR Green I-based duplex real-time PCR. Mol. Cell Probes 49:101474. doi: 10.1016/j.mcp.2019.101474
Zhou, J., Qiu, Y., Zhu, N., Zhou, L., Dai, B., Feng, X., et al. (2021). The Nucleolar Localization Signal of Porcine Circovirus Type 4 Capsid Protein Is Essential for Interaction With Serine-48 Residue of Nucleolar Phosphoprotein Nucleophosmin-1. Front. Microbiol. 12:751382. doi: 10.3389/fmicb.2021.751382
Keywords: porcine circovirus 4, the Southwest of China, molecular detection, genetic analysis, genotype
Citation: Xu T, You D, Wu F, Zhu L, Sun X-G, Lai S-Y, Ai Y-R, Zhou Y-C and Xu Z-W (2022) First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021–2022. Front. Microbiol. 13:1052533. doi: 10.3389/fmicb.2022.1052533
Received: 24 September 2022; Accepted: 10 October 2022;
Published: 03 November 2022.
Edited by:
Jue Liu, Yangzhou University, ChinaReviewed by:
Naidong Wang, Hunan Agricultural University, ChinaChangxu Song, South China Agricultural University, China
Copyright © 2022 Xu, You, Wu, Zhu, Sun, Lai, Ai, Zhou and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Wen Xu, abtcxzw@126.com
†These authors have contributed equally to this work
 Tong Xu1†
Tong Xu1† Ling Zhu
Ling Zhu Yuan-Cheng Zhou
Yuan-Cheng Zhou Zhi-Wen Xu
Zhi-Wen Xu