- 1State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2Joint International Research Laboratory of Metabolic and Developmental Sciences, Shanghai Jiao Tong University, Shanghai, China
- 3Department of Civil, Chemical, Environmental and Materials Engineering – DICAM, University of Bologna, Bologna, Italy
- 4SUS Environment Co., Ltd., Shanghai, China
The extensive use of acids in a variety of manufacturing industries results in the increase of discharged acidic waste stream into the environment. Such co-pollution of acids and other organic pollutants limits the biodegradation capability of neutrophilic degraders. With high-throughput genetic techniques, we aim to improve the acid tolerance of a pollutant-degrading bacterium, Pseudomonas putida S16 by genetically engineering it with the glutamate decarboxylase (GAD)-dependent system and the global regulator (IrrE) of extreme radiation resistance. The engineered strains holding either GAD system or irrE regulator could grow under pH 4.5, compared to the wild type. They could also degrade over 90% of a selected pollutant (benzoate or nicotine) under pH 5.0 in 48 h, while no biodegradation was detected with the wild type under the same conditions. We conclude that acid stress tolerance by the possession of the GAD system or IrrE regulator in pollutant-degrading bacteria would be a promising approach to enhance their viability and biodegrading activities in bioremediation of acidic wastes.
Introduction
Co-pollution between acids and organic wastes often occurs in the effluents discharged from agriculture and manufacturing industries, and persistence of such pollutants are harmful to any living organisms (Kjeldsen et al., 2002; Foo and Hameed, 2009). In food industries, benzoate is a typical aromatic pollutant found in the waste stream at typical pH 4.0 (Oie et al., 2007; Xie et al., 2009). Another example is nicotine, which is a predominant compound contaminated in the effluent from tobacco factories. These pollutants usually exist in the acidic waste stream at shallow pH values (pH < 5) (Dixon et al., 2000; Zhong et al., 2010; Li et al., 2019). These acidic conditions limit the survival and biodegradation activities of neutrophilic pollutant-degrading bacteria like pseudomonads that generally dominate the polluted sites. Genetics improvement of these bacteria by enhancing their acid stress resistance would be a promising strategy to optimize biological waste treatments.
Some bacteria have evolved various acid-resistance mechanisms, such as the glutamate decarboxylase (GAD)-dependent system, F1-F0-ATPase proton pump, and protection or repair of macromolecules (Su et al., 2011; De Biase and Pennacchietti, 2012). The GAD-dependent system exists in various bacteria, and it involves one or two glutamate decarboxylases (GadA/B) and one antiporter, GadC (Ma et al., 2012; Kanjee and Houry, 2013). GadA/B catalyzes the conversion of protonated glutamate to γ-aminobutyric acid (GABA), and GadC exports GABA in exchange for a new extracellular glutamate molecule and consuming intracellular protons. Deinococcus radiodurans has a global regulator, IrrE, which plays a vital role in the protection of this bacterium against radiation (Lin et al., 2013). This IrrE regulator also involves diverse resistance mechanisms of Escherichia coli toward osmotic pressure, oxidative stress, ethanol, butanol, acetate, and inorganic acid (Gao et al., 2003; Pan et al., 2009; Chen et al., 2011; Ma et al., 2011).
Previous studies revealed that Pseudomonas putida S16 is a non-pathogenic member of the genus Pseudomonas, and utilizes a variety of organic wastes as a sole source of carbon and/or nitrogen (Wang et al., 2004, 2007). For instance, P. putida S16 effectively degraded nicotine through the pyrrolidine pathway (Tang et al., 2013; Hu et al., 2019). However, this bacterium cannot grow well under acidic conditions (optimal pH7.5), which depletes its value in bioremediation of acidic wastes. The lack of acid stress tolerance of P. putida S16 was subsequently confirmed with the absence of genes that play crucial roles in the GAD system (gadBC) and the IrrE regulator (irrE) in its genome (Yu et al., 2011).
In this study, we employed synthetic biology strategies to improve the acid stress tolerance of a pollutant-degrading bacterium, P. putida S16 by cloning with the expressing plasmid of the GAD system or the IrrE regulator. Viability, growth, and pollutant-degrading activities of the engineered strains under acidic conditions were tested in comparison with the wild type. A typical set of organic pollutants found co-polluting in the acidic waste stream produced by industries in China, i.e., nicotine and benzoate were used in the biodegradation experiments. The possibility and benefits of using genetically modified microbes to optimize bioremediation technology were addressed and discussed in this work.
Materials and Methods
Materials
L-Nicotine (purity, ≥99%) was purchased from Fluka Chemie GmbH (Buchs Corp., Switzerland). Restriction enzymes, NcoI and XhoI were purchased from TaKaRa (Dalian, China). Sodium benzoate and glycerin were purchased from Sinopharm Chemical Reagent Co., Ltd. Nucleotide sequencing service was provided by BioSune Company (Shanghai, China).
Bacterial Strains and Growth Conditions
P. putida S16 was grown in Luria-Bertani (LB) broth (composition per L: 5 g Yeast extract, 10 g Typtone, 10 g NaCl) at 30°C or in mineral salt medium (MSM, composition per L: 13.3 g K2HPO4⋅3H2O, 4 g KH2PO4, 0.2 g MgSO4⋅7H2O, and 0.5 mL trace elements solution) supplemented with 0.1% (w/v) L-nicotine as for the nicotine medium pH5.5 or 7 (Tang et al., 2013) or 1% (w/v) glycerin and 0.1% (w/v) ammonium sulfate as for glycerin medium pH4, 4.5, and 5. The supplemented compounds served as the sole source of carbon, nitrogen, and energy. The trace elements solution contained 0.1 g ZnSO4, 0.1 g Na2MoO4⋅2H2O, 0.05 g CuCl2⋅2H2O, 0.05 g CaCl2⋅2H2O, 0.004 g FeSO4⋅7H2O, 0.008 g MnSO4⋅H2O, and 0.05 g Na2WO4⋅2H2O per 1 L of 0.1 M HCl.
E. coli BL21(DE3) was the source of the GAD system, which was grown in LB broth or agar (solidified with 1.5% (w/v) agar powder to the liquid medium) at 37°C. D. radiodurans R1 (ATCC 13939) served as the source of the IrrE regulator, which was cultivated in tryptone-glucose-yeast extract (TGY) broth or agar, containing 0.5% tryptone, 0.3% yeast extract, and 0.1% glucose, and 1.5% agar powder for the agar medium. The liquid culture of any bacteria was incubated with shaking at 200 rpm (30 mg/mL tetracycline might be added to the media if necessary).
Construction of Acid Stress Tolerance Bacteria
The genomic DNA templates of E. coli BL21(DE3) and D. radiodurans R1 were extracted by using Wizard Genomic DNA Purification Kit (PROMEGA, A1125). Following, the target gene expressing the GAD system was the gadBC gene, which was amplified by PCR using a template from the genomic DNA of E. coli BL21 with primers: 5′-CCGCCATGGGATAATTCAGGAGGCACAGAA-3′ and 5′ -GTGCTCGAGTTAGTGTTTCTTGTCATTCAT-3′. For the IrrE regulator, irrE gene and the GroESL promoter from the genomic DNA of D. radiodurans R1 was extracted and used as the template. The PCR primers were 5′-AGGCGACCGCGATGTGCCCAGTGCCAA-3′ and 5′-GTGCT CGAGTCCAGTTCACTGTGCAGC-3′ for the irrE gene; and 5′-CCGCCATGGGGATACCCCCATTCCCCG-3′ and 5′-AC TGGGCACATCGCGGTCGCCTAAAGG-3′ for the GroESL promoter. To generate the DNA sequence of GroESL-irrE, the recombinant PCR was then performed using EasyGeno Assembly Cloning Kit (TIANGEN, VI1201) with primers 5′-GTGCTCGAGTCCAGTTCACTGTGCAGC-3′ and 5′- CCG CCATGGGGATACCCCCATTCCCCG-3′.
The PCR products were firstly double digested with NcoI and XhoI, and then ligated into the NcoI and XhoI sites of plasmid pME6032 (Tang et al., 2013). The recombinant plasmid pME-Gad or pME-GirrE was introduced into the competent cells of P. putida S16 (Tang et al., 2013) by electroporation with a field strength of 125,000 V/cm, the electric resistance of 200 Ω, and a time constant of ∼5.0 ms. The original plasmid pME6032 served as the control. All products were cultured on LB selective plates with tetracycline after electroporation. The positive clones picked from the plates and sent for sequencing.
Growth Assays at Different pH Values
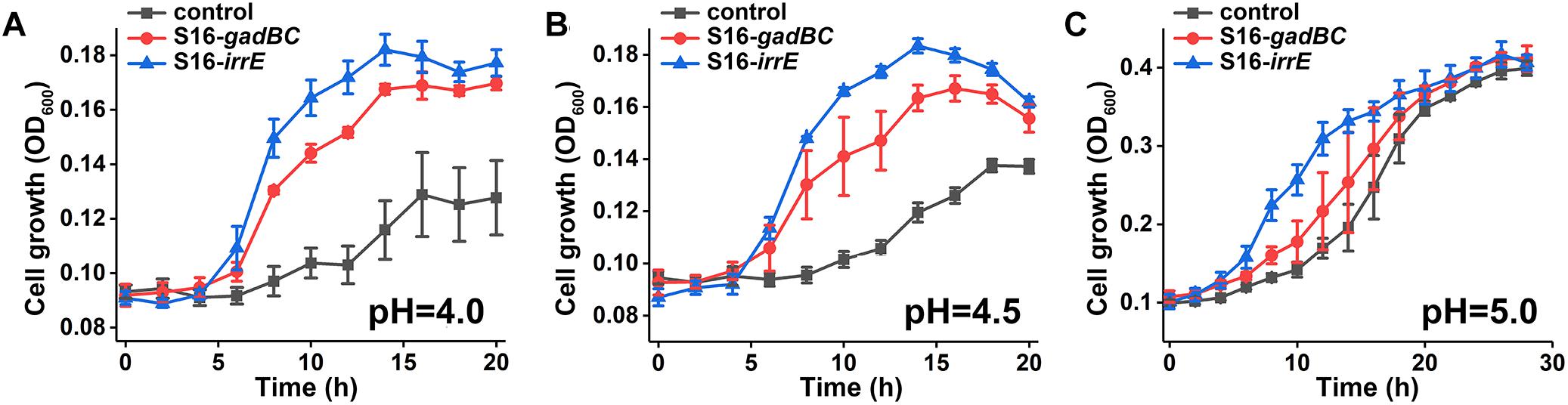
Viability and growth of the engineered bacteria, P. putida S16 harboring the recombinant plasmid pME-Gad or pME-GirrE were tested under acidic conditions compared with the wildtype holding the plasmid pME6032 as a control. The assays were carried out by growing each bacterial strain in MSM supplemented with 10 g/L glycerin (as a carbon source) and 1 g/L (NH4)2SO4 (as a nitrogen source) at different pH values of 4.0, 4.5, and 5.0. The pH was adjusted with 88 mM H3PO4. Growth conditions were monitored in the Bioscreen C MBR (Finland) at 30°C, with absorbance at 600 nm, and shaking continuously at a medium speed. The experimental set was independently prepared and measured in triplicate.
Biodegradation Experiments
The biodegradation assay was carried out under acidic conditions at pH 5.5 in comparison with the neutral conditions (pH 7). Either engineered or wildtype strain of P. putida S16 was grown in sodium benzoate medium (MSM plus 1 g/L sodium benzoate and 1 g/L ammonium sulfate) at 30°C with shaking at 200 rpm incubator. Equal portions of the bacterial cultures were sampled, which were centrifuged at 12,000 × g to remove precipitates. The supernatant was filtered with 0.22 μm membrane filter (PALL) before measuring the UV spectra and the concentration of sodium benzoate by high-performance liquid chromatography (HPLC). The HPLC system was an Eclipse XDB-C18 column (column size: 4.6 mm × 250 mm; particle size, 5 μm; Agilent), and the detection wavelength of the UV detector was 259 nm. A mixture of acetonitrile and 1 mM H2SO4 (15:85, v/v) served as the mobile phase, with a flow rate of 0.5 mL/min at 30°C. Qualitative and quantitative analysis of sodium benzoate relied on the retention time and peak areas of the samples in comparison with the known concentrations of standards (Supplementary Figure S2).
To test the efficiency of nicotine degradation, either engineered or wild-type strains of P. putida S16 were cultivated separately in the nicotine medium pH 7.0 and pH 5.5 at 30°C. The culture broth was sampled during cultivation, in which bacterial cells were removed by centrifugation at 12,000 × g and 4°C for 5 min. The supernatant was used for measuring ultraviolet (UV) absorption and HPLC analysis. The supernatant was diluted with 0.1 M HCl and scanned with UV2550 (SHIMADZU) spectrophotometer to record the UV spectrogram. Qualitative and quantitative analysis of nicotine relied on the retention time and peak areas of the samples in comparison with the known concentrations of standards (Supplementary Figure S3).
Evaluation of Gene Expression by Reverse Transcription-Quantitative PCR (RT-qPCR)
Experiments in this section were performed routinely in triplicate with control and nicotine induction cultures of P. putida S16 harboring pME6032, pME-Gad, or pME-GirrE. A single colony of recombinants or control was picked randomly from MSM plate, and a 1:100 dilution of a fresh overnight culture was inoculated in three 250-mL flasks containing 50 mL MSM (control) or MSM plus 1 g/L nicotine (induction).
Batch cultures were incubated at 30°C with shaking (200 rpm) to early-exponential phase (OD600 ∼0.5). The early-exponential cells were harvested by centrifugation at 14,000 × g for 2 min. Total RNA was extracted from ∼1 × 108 cells using an RNAprep pure cell/bacteria kit (Tiangen), and quantified by NanoDrop (Thermo Fisher Scientific). Then, total RNA was treated by 0.8 mg of DNase (Fermentas), and reverse transcribed to cDNA using FastKing cDNA Kit (Tiangen). The cDNA was diluted 1:10 and served as the template for qPCR analysis using the CFX96 Real-Time PCR Detection system (Bio-Rad) with SYBR Green RealMasterMix (Tiangen) and qPCR primers (Tang et al., 2013). The threshold cycle (CT) values for each target gene were normalized with the reference 16S rRNA gene. The 2ΔΔCT method was used to calculate the relative expression level, where ΔΔCT = (CTtarget − CT16S)induction − (CTtarget − CT16S)control (Livak and Schmittgen, 2001).
Results and Discussion
Pseudomonas putida is a widely used strain for degrading persistent organic compounds. Up to now, no report has been published for enhancing the acid stress resistance for P. putida. Applications of a P. putida strain to curb environmental pollution under acid stress has therefore been restricted.
Engineered Strains of P. putida S16 and Their Acid Resistance
We constructed two different potential acid-resistance devices, gadBC and irrE, to evaluate the degradation ability of P. putida S16 under acidic conditions. The recombinant plasmid pME-Gad constructed by insertion of a 3.3-kb fragment containing the gene gadBC and 230 bp upstream from the initiation site of gadB is shown in Supplementary Figure S1A, while the recombinant plasmid pME-GirrE holding the gene irrE and the promoter GroESL is shown in Supplementary Figure S1C. The possession of the gene expressing gadBC or irrE was confirmed with PCR (Supplementary Figures S1B,D) and DNA sequencing. The overview of the verification strategy if two different acid resistance systems is shown in Figure 1. Over more than 20 h of cultivation, P. putida S16 harboring pME-Gad or pME-GirrE displayed better acid tolerance than the control. At pH 4.5 and 4.0, the control bacterium grew much slower compared to the transformant strains with gadBC or irrE that grew faster and reached to higher cell densities (Figure 2). These results revealed that gadBC and irrE increased the acid resistance ability in P. putida S16, and irrE displayed a stronger effect on acid tolerance than gadBC in strain S16.
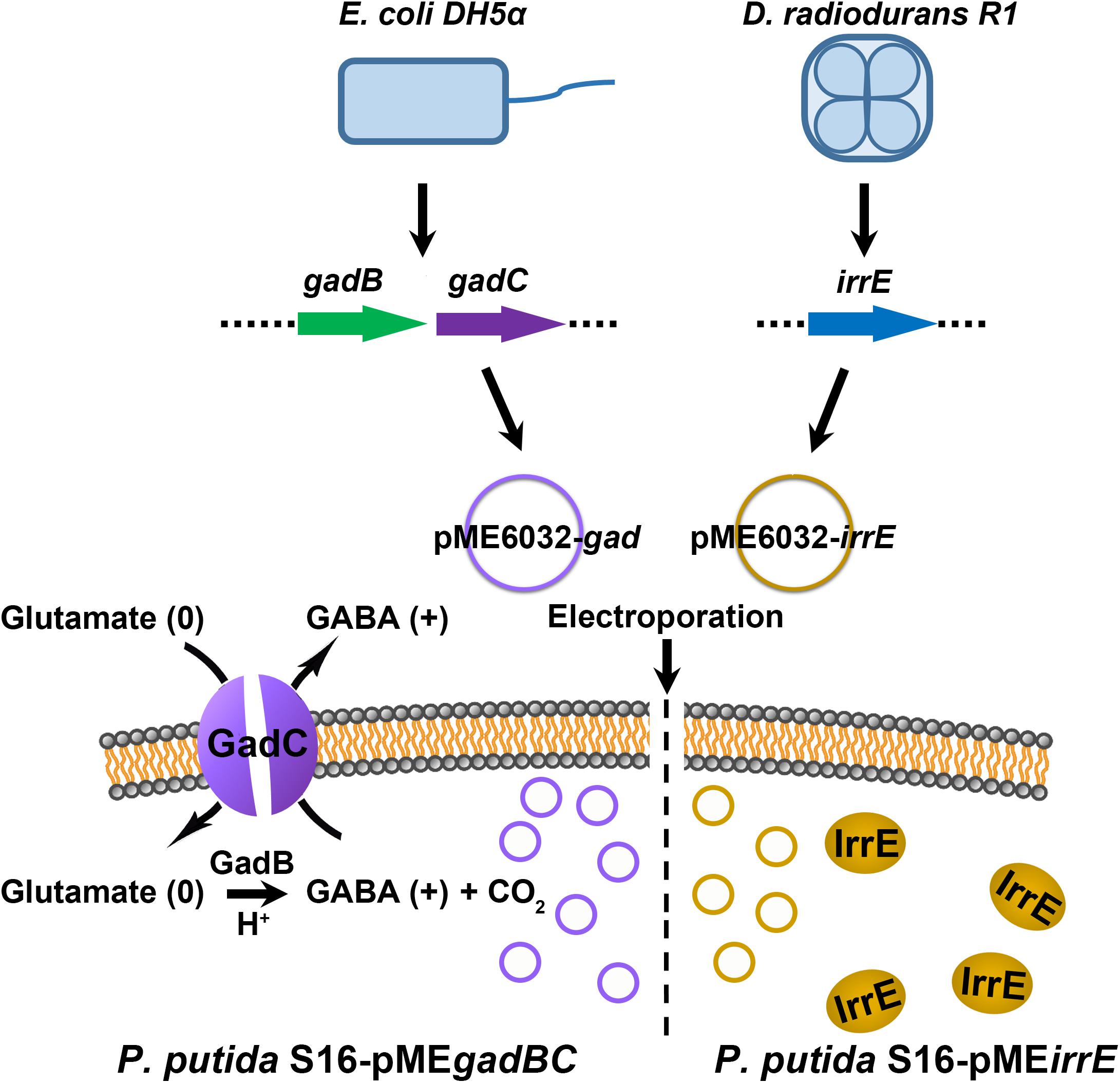
Figure 1. The verification strategy for two different acid resistance systems. The Gad system was obtained from E. coli BL21(DE3), and molecular chaperone system was amplified from D. radiodurans R1. The plasmid pME6032 was used as empty control or transporter to construct recombinants.
Pollutant-Degrading Potential of Engineered P. putida S16
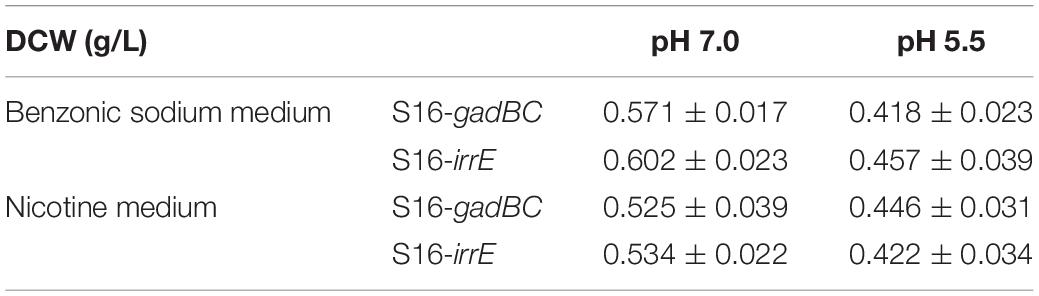
Either control or engineered P. putida S16 strains displayed similar growth rates and benzoate-degrading efficiencies under pH 7 (Figures 3A,B), which demonstrated that the acid-resistant components did not influence these bacterial strains under neutral conditions. At pH 5.5, either gadBC or irrE holding strains showed better growth characteristics in comparison to the control. For example, strains with the gadBC gene had a maximum growth of 1.0 within 30 h, and strains with the irrE gene reached a maximum growth of 0.9 within the same incubation time, whereas no growth was observed for the control. Sodium benzoate degradation rates for gadBC and irrE were 0.09 and 0.087 g/L/h, respectively (Figures 3C,D). Strains harboring the irrE gene have an advantage at early degradation phase, while the ones with the gadBC gene had a longer lag phase.
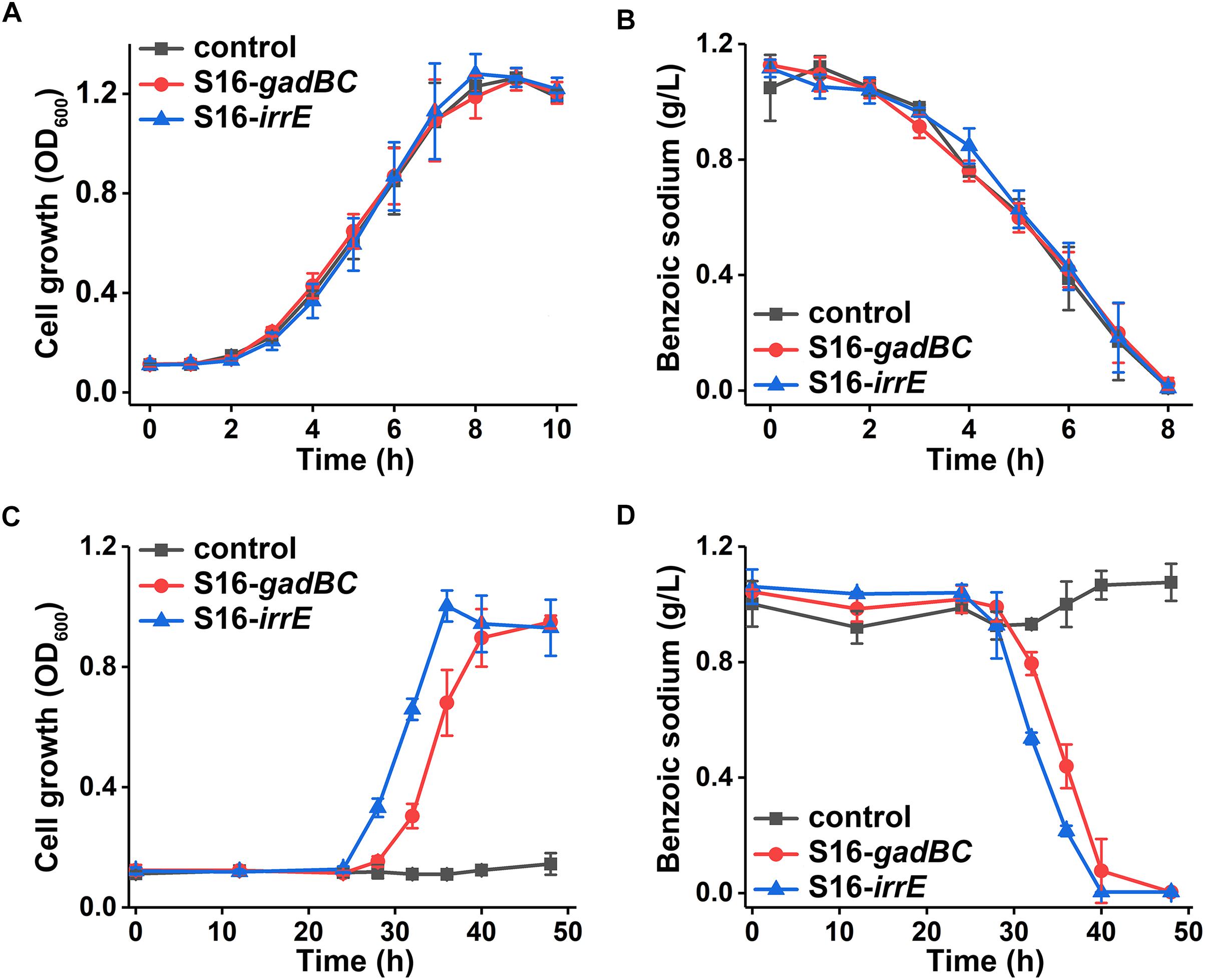
Figure 3. The growth curves in sodium benzoate medium (A) and (C) and sodium benzoate degradation curves (B) and (D) in pH 7.0 and pH 5.5, respectively.
Every test strain displayed no difference in either growth rate or nicotine degradation under a neutral environment (Figure 4). However, their growth characteristics differed under an acidic environment, pH 5.5 (Figure 4A). After 56 h of growth, cells harboring pME-Gad or pME-GirrE plasmid reached a maximum growth of 1.0 and grew better than the control. Furthermore, nicotine was entirely degraded by the engineered strains with the gadBC or irrE gene at pH 5.5 within 60 h, showing the significant acid tolerance compared to the control (Figure 4C). The gadBC and irrE genes significantly enhanced both the growth rate and degradation efficiency of P. putida S16 under acidic conditions.
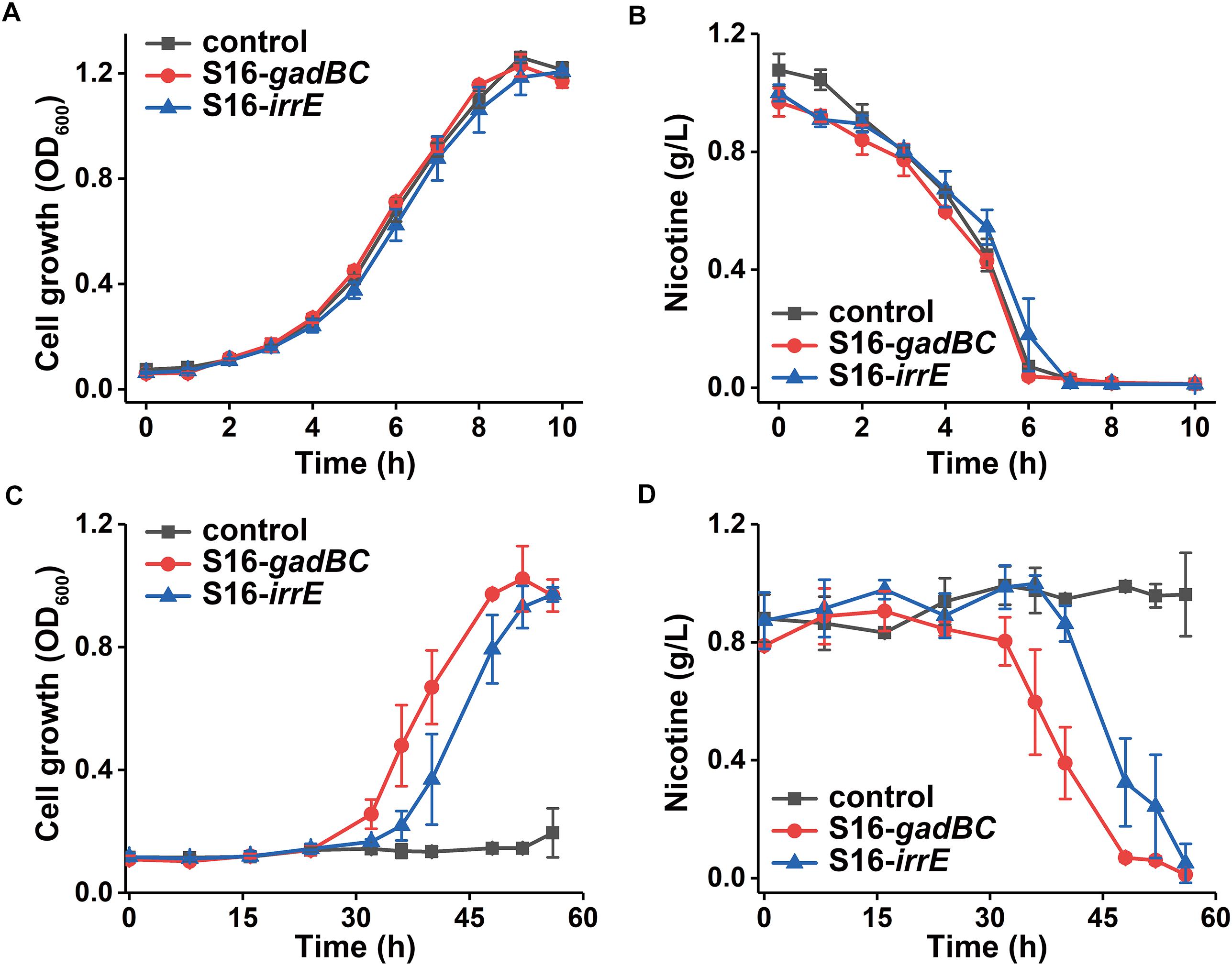
Figure 4. The growth curve in nicotine medium (A) and (C) and nicotine degradation curve (B) and (D) under pH 7.0 and pH 5.5, respectively.
Based on the results of cell growth (Table 1) and nicotine degradation, both gadBC and irrE could improve acid-resistance in P. putida S16. Under acidic conditions (pH 5.5), both growth and degradation rate of P. putida S16 harboring pME-Gad were better than the strain harboring pME-GirrE, and showed a shorter lag phase in nicotine medium. The results were similar to a previous study showing that the GAD system not only contributes to pH homeostasis but, by transiently accumulating GABA in the cell, it counteracts illicit entry of protons by inversion of the membrane potential, a strategy similar to that adopted by extreme acidophiles (Foster, 2004). Past studies of the molecular mechanisms of gadBC or irrE inducing stress resistance in bacterial cells focused on the enhancement rate of stress resistance ability. In this study, we combined theoretical research with practical environmental degradation to better understand the real affects.
Transcriptional Expression of Nicotine-Degrading Genes
To validate the nicotine degradation and assess relative transcription levels of genes related to nicotine degradation in P. putida S16 expressing gadBC or irrE, we utilized RT-qPCR to compare mRNA levels of genes nicA2, pnao, sapd, spmA, and spmC at pH7 and pH5.5. The RT-qPCR analysis revealed that all target genes were up-regulated in cells harboring pME-GirrE. The spmC mRNA expression was 3.2-fold up-regulated which is the highest one in those genes, following were nicA2 2.4-fold, spmA 1.9-fold, pnao 1.7-fold and sapd 1.2-fold (Figure 5). On the contrary, in gadBC holding strain, those nicotine-degrading genes were down-regulated. These results indicated that the GAD system promoted a different role in nicotine degradation of strain S16 (Figure 5). Combined with the result of growth curves, the strain harboring irrE at pH 5.5 had worse growth behavior than the condition of pH 7.0. Interestingly, the entire nicotine degrading genes were upregulated in the strain harboring irrE at pH 5.5 at the mRNA level, with the same degradation rate at pH 7.0. We suppose that acidic conditions decreased the optimal enzyme activities, so that the global regulator IrrE has to upregulate expression of nicotine-degrading genes to make up for deficiency of the enzyme activities under acidic conditions.
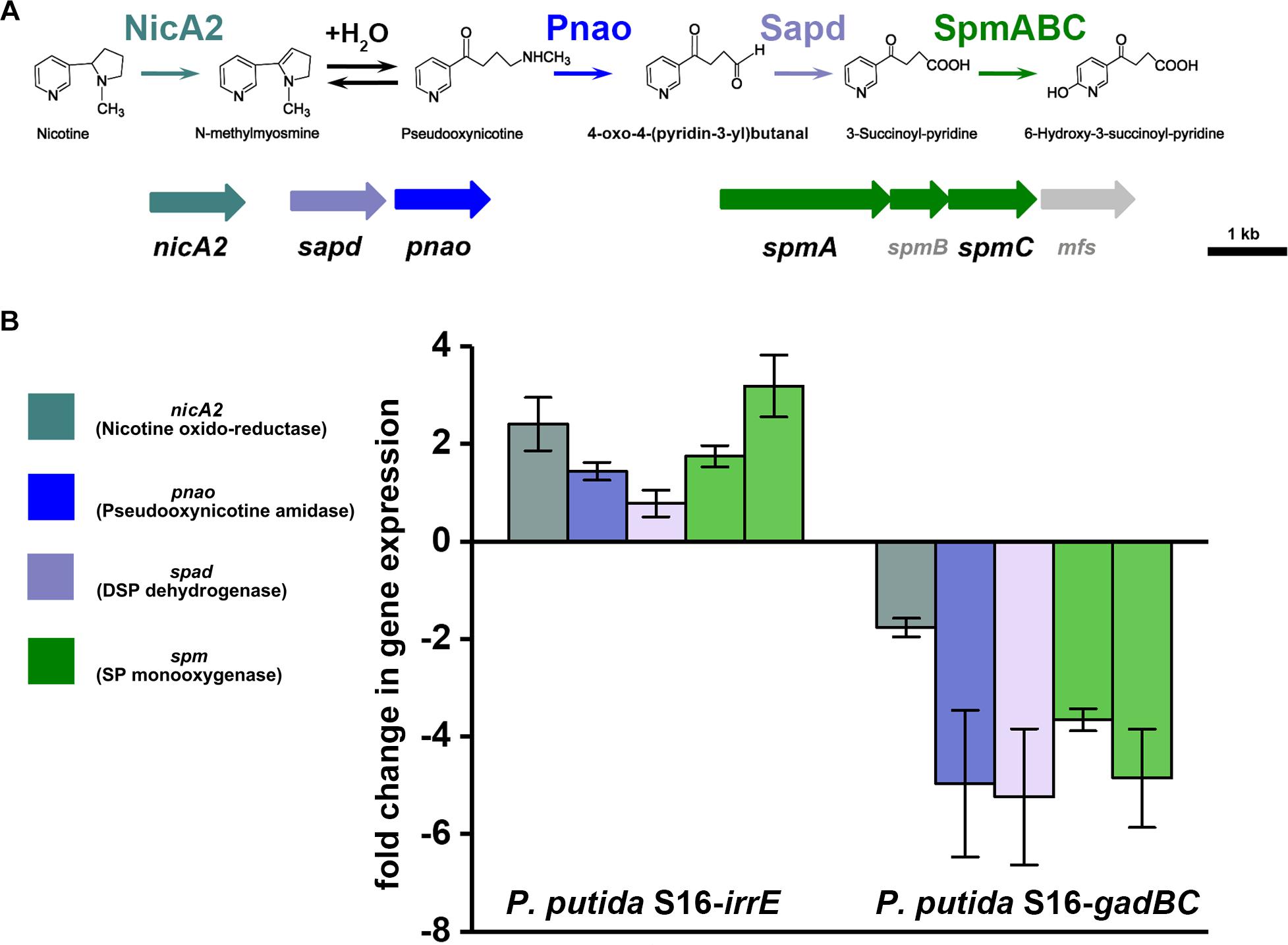
Figure 5. Validation of transcriptional regulation of differentially expressed proteins. RT-qPCR analysis of target gene transcripts produced in P. putida S16 harboring irrE or GAD systems grown at pH 7 and pH 5.5. (A) Nicotine degradation pathway (partial) (B) mRNA expression levels of 5 target genes involved in nicotine degradation of P. putida S16 were estimated using RT-qPCR and the 2Δ Δ CT method. The 16S rRNA gene was used as the reference gene. Results presented in these histograms are the means of three independent experiments, and error bars indicate the standard deviations.
Conclusion
Optimization of the GAD system and IrrE expression as tools to promote bacterial growth in acidic environments will be important for further studies. Synthetic biology has shown enormous potential to generate biological modules with unprecedented applications by combining the basic elements of biology (Khalil and Collins, 2010). A more thorough understanding of the acid tolerance mechanisms of microorganisms is needed and, in combination with synthetic biology, may contribute greatly to the industrial processes involving acid-resistant microbes. What’s more, it could not be ignored that horizontal gene transfer would bring unexpected influences for natural microorganisms. Introducing engineered bacteria with mobile genetic elements of strong resistance traits into nature may reach to the increase of resistant pathogens coexisted out there, which is a global threat of antimicrobial resistance problem. For avoiding this risk, we are trying to manipulate engineered bacteria to death using smart sensor modules and suicide modules when the concentrations of pollutants are reduced to normal environmental levels.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
ZZ, PX, and HT conceived the project and wrote the manuscript. ZZ, HT, and YL designed and performed all the experiments. ZZ and YL analyzed the results. ZW and GZ provided samples and materials. GZ revised the manuscript.
Funding
This work was supported by grants from National Key Research and Development Project (SQ2018YFA090024), the Science and Technology Commission of Shanghai Municipality (17JC1403300), by the “Shuguang Program” (17SG09) supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, and by grants from the Chinese National Science Foundation for Excellent Young Scholars (31422004).
Conflict of Interest Statement
ZW was employed by company SUS Environment Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02033/full#supplementary-material
References
Chen, T., Wang, J., Yang, R., Li, J., Lin, M., and Lin, Z. (2011). Laboratory-evolved mutants of an exogenous global regulator, IrrE from Deinococcus radiodurans, enhance stress tolerances of Escherichia coli. PLoS One 6:e16228. doi: 10.1371/journal.pone.0016228
De Biase, D., and Pennacchietti, E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86, 770–786. doi: 10.1111/mmi.12020
Dixon, M., Lambin, K., and Seeman, J. (2000). Mini-review: on the transfer of nicotine from tobacco to the smoke: a brief review of ammonia and “pH” factors. Beitr. Tabak 19, 103–113. doi: 10.2478/cttr-2013-0700
Foo, K. Y., and Hameed, B. H. (2009). An overview of landfill leachate treatment via activated carbon adsorption process. J. Hazard. Mater. 171, 54–60. doi: 10.1016/j.jhazmat.2009.06.038
Foster, J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. doi: 10.1038/nrmicro1021
Gao, G., Tian, B., Liu, L., Sheng, D., Shen, B., and Hua, Y. (2003). Expression of Deinococcus radiodurans PprI enhances the radioresistance of Escherichia coli. DNA Repair 2, 1419–1427. doi: 10.1016/j.dnarep.2003.08.012
Hu, H., Wang, L., Wang, W., Wu, G., Tao, F., Xu, P., et al. (2019). Regulatory mechanism of nicotine degradation in Pseudomonas putida. mBio 10, e602–e619. doi: 10.1128/mBio.00602-19
Kanjee, U., and Houry, W. A. (2013). Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81. doi: 10.1146/annurev-micro-092412-155708
Khalil, A. S., and Collins, J. J. (2010). Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379. doi: 10.1038/nrg2775
Kjeldsen, P., Barlaz, M. A., Rooker, A. P., Baun, A., Ledin, A., and Christensen, T. H. (2002). Present and long-term composition of MSW landfill leachate: a review. Crit. Rev. Environ. Sci. Technol. 32, 297–336. doi: 10.1080/10643380290813462
Li, J., Wang, J., Li, S., Yi, F., Xu, J., Shu, M., et al. (2019). Co-occurrence of functional modules derived from nicotine-degrading gene clusters confers additive effects in Pseudomonas sp. JY-Q. Appl. Microbiol. Biotechnol. 103, 4499–4510. doi: 10.1007/s00253-019-09800-4
Lin, Z., Zhang, Y., and Wang, J. (2013). Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol. Adv. 31, 986–991. doi: 10.1016/j.biotechadv.2013.02.010
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, D., Lu, P., Yan, C., Fan, C., Yin, P., Wang, J., et al. (2012). Structure and mechanism of a glutamate-GABA antiporter. Nature 483, 632–636. doi: 10.1038/nature10917
Ma, R., Zhang, Y., Hong, H., Lu, W., Lin, M., Chen, M., et al. (2011). Improved osmotic tolerance and ethanol production of ethanologenic Escherichia coli by IrrE, a global regulator of radiation-resistance of Deinococcus radiodurans. Curr. Microbiol. 62, 659–664. doi: 10.1007/s00284-010-9759-2
Oie, C. S., Albaugh, C. E., and Peyton, B. M. (2007). Benzoate and salicylate degradation by Halomonas campisalis, an alkaliphilic and moderately halophilic microorganism. Water Res. 41, 1235–1242. doi: 10.1016/j.watres.2006.12.029
Pan, J., Wang, J., Zhou, Z., Yan, Y., Zhang, W., Lu, W., et al. (2009). IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One 4:e4422. doi: 10.1371/journal.pone.0004422
Su, M. S., Schlicht, S., and Ganzle, M. G. (2011). Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 10:S8. doi: 10.1186/1475-2859-10-S1-S8
Tang, H., Wang, L., Wang, W., Yu, H., Zhang, K., Yao, Y., et al. (2013). Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genetics 9:e1003923. doi: 10.1371/journal.pgen.1003923
Wang, S., Liu, Z., Tang, H., Meng, J., and Xu, P. (2007). Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology SGM 153, 1556–1565. doi: 10.1099/mic.0.2006/005223-0
Wang, S., Xu, P., Tang, H., Meng, J., Liu, X., Huang, J., et al. (2004). Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 26, 1493–1496. doi: 10.1023/b:bile.0000044450.16235.65
Xie, N., Tang, H., Feng, J., Tao, F., Ma, C., and Xu, P. (2009). Characterization of benzoate degradation by newly isolated bacterium Pseudomonas sp. XP-M2. Biochem. Eng. J. 46, 79–82.
Yu, H., Tang, H., Wang, L., Yao, Y., Wu, G., Xu, P., et al. (2011). Complete genome sequence of the nicotine-degrading Pseudomonas putida strain S16. J. Bacteriol. 193, 5541–5542. doi: 10.1128/JB.05663-11
Keywords: acid resistance, bioremediation, glutamate decarboxylase (GAD) system, gadBC, global regulator, IrrE
Citation: Zhou Z, Liu Y, Zanaroli G, Wang Z, Xu P and Tang H (2019) Enhancing Bioremediation Potential of Pseudomonas putida by Developing Its Acid Stress Tolerance With Glutamate Decarboxylase Dependent System and Global Regulator of Extreme Radiation Resistance. Front. Microbiol. 10:2033. doi: 10.3389/fmicb.2019.02033
Received: 28 May 2019; Accepted: 19 August 2019;
Published: 04 September 2019.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Weihong Zhong, Zhejiang University of Technology, ChinaAsifullah Khan, Abdul Wali Khan University Mardan, Pakistan
Jiangxin Wang, Shenzhen University, China
Rungroch Sungthong, University of Glasgow, United Kingdom
Copyright © 2019 Zhou, Liu, Zanaroli, Wang, Xu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongzhi Tang, tanghongzhi@sjtu.edu.cn
 Zikang Zhou1,2
Zikang Zhou1,2 Ping Xu
Ping Xu Hongzhi Tang
Hongzhi Tang