CircHIPK3: Key Player in Pathophysiology and Potential Diagnostic and Therapeutic Tool
- Hunan Key Laboratory of Oral Health Research & Hunan 3D Printing Engineering Research Center of Oral Care & Hunan Clinical Research Center of Oral Major Diseases and Oral Health & Xiangya Stomatological Hospital & Xiangya School of Stomatology, Central South University, Changsha, China
A large number of studies in China and other countries have confirmed that circularHIPK3 (circHIPK3) plays an important role in the pathophysiological processes of various diseases. Through the action of sponge miRNA (miR), circHIPK3 regulates cell proliferation, differentiation, and migration, and plays a key role in disease processes. By referring to a large number of research reports, this article explores the specific functional role of circHIPK3 in fibrotic diseases, cancer, and other diseases. This review aims to clarify the role of circHIPK3 in disease processes in order to aid further studies into the specific pathogenesis and clinical diagnosis of various diseases and provide new ideas for treatments.
Introduction
In 1976, Sanger et al. proposed the viroid to be a single-stranded covalently closed circRNA molecule and was the first human discovery of circRNA, a new type of non-coding endogenous RNA molecule. CircRNA, which exists in all eukaryotic cells, is formed by exon or intron cyclization, which connects the 3′ and 5′ ends to complete the circular structure. In the early stages of its discovery, circRNA was not generally recognized as being important because of the limitations of the traditional research methods (1–3). With the development of bioinformatics, research on circRNA is becoming increasingly in-depth. Numerous studies have shown that circRNA plays an important role in the pathophysiological processes of various diseases, such as cancer and cardiovascular disease, and in the development of neurons. Furthermore, studies have shown that circRNA also affects the occurrence and development of fibrotic diseases of organs such as the heart, lungs, and kidneys, playing a key role in organ fibrosis (1, 4–9). It has been shown that a member of the circRNA family, circHIPK3, is abnormally upregulated in organ fibrosis, cancer, and other diseases, suggesting that circHIPK3 is involved in these diseases. This review will primarily discuss the function of circHIPK3 in these conditions and the current relevant research progress and future prospects to provide new ideas for exploring the circHIPK3-related pathogenesis, clinical diagnosis, and treatment of cancer, fibrosis, and other diseases.
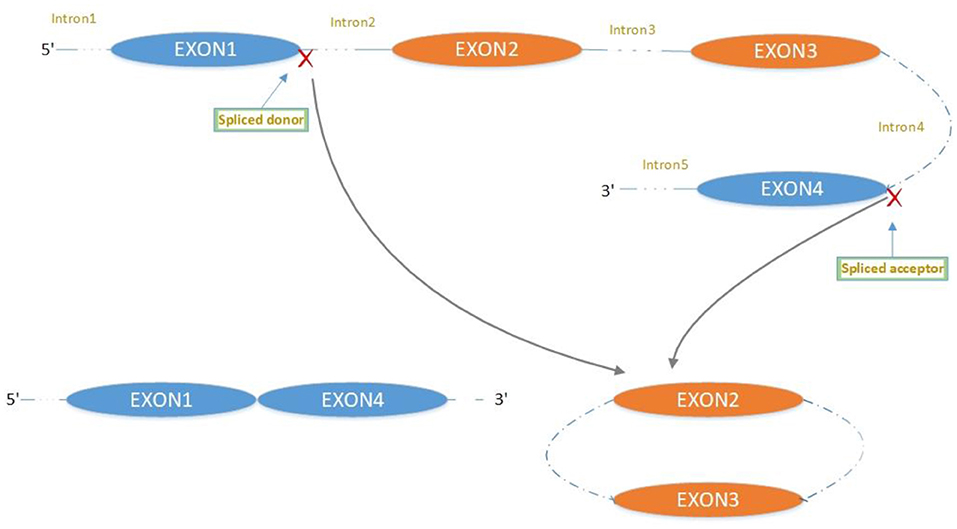
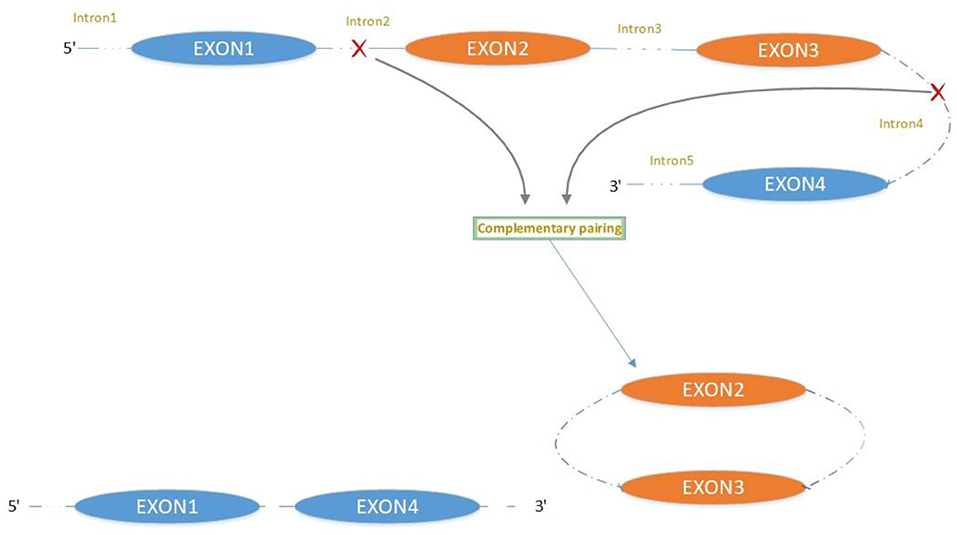
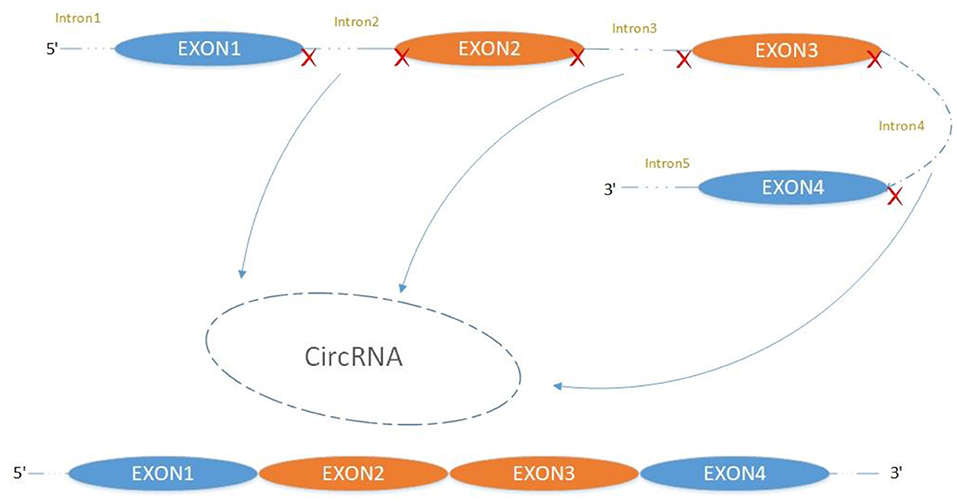
CircRNA Biogenesis
The majority of circRNAs are produced by complete protein-coding gene exons. CircRNA undergoes circularization in two ways: lasso-protein-driven or intron-pairing-driven circularization. The former method of circularization involves exon skipping, i.e., the 3′ splice donor of one exon is covalently bound to the 5′ splice acceptor of the other exon—the lasso (10, 11). Intron-pairing-driven circularization starts with the codons of a splice donor and a splice acceptor, which are exonerated (Figure 1). After the circular structure is formed, the introns are removed to form circRNA (Figure 2). Studies have also confirmed that circRNA can form from intron circularization (Figure 3) (3–6). Because of the lack of a free 3′ or 5′ end, circRNA has a long half-life and is more stable than linear RNA. In terms of gene regulation, circRNAs have multiple functions: they (1) act as microRNA sponges that can competitively bind to miRNA; (2) regulate splicing or transcription; (3) interact with RNA-binding proteins; (4) are involved in translation; (5) regulate epigenetic alterations; and (6) transport substances and information (12, 13). Studies have shown that, because of the high production and accumulation rate, nervous tissue is rich in circRNA. In addition, circRNA is highly enriched in synapses, and its levels are regulated by neuronal activity, suggesting that circRNA has a specific role in the brain (2, 14, 15), and the latest research has shown this in the mammalian brain. Some circRNAs control gene expression by regulating mRNA production from host genes. Other circRNAs have trans-functionality, for example, CDR1 and Sry bind to specific miRNA as well as acting as miRNA sponges and/or transporters, and circ-muscleblind (circMbl) may bind the multifunctional protein muscleblind (MBL) (16, 17). Furthermore, other circRNAs are thought to bind to transcription factors, participate in muscle development or viral transcription, or have other functions.
CircHIPK3 Biogenesis
The circularization of the second exon of the homology domain-interacting protein kinase 3 (HIPK3) gene produces circHIPK3, which is abundantly expressed in the cytoplasm of several human tissues including the lungs, heart, stomach, colon, brain, and other organs. The content of HIPK3 in the human body is particularly abundant, and the reverse splicing rate is high. HIPK3 is located on chromosome 11p13 and consists of 7,551 base pairs. Long flanking introns with complementary Alu repeat sequences are necessary for the formation of circHIPK3. Using luciferase experiments, Zheng et al. found that circHIPK3 has nine direct miRNA binding sites and 18 potential binding sites, and they concluded that circHIPK3 participates in mediating cell growth, proliferation, and migration through the sponging mechanism and plays an important role in the occurrence and development of various diseases (18–20). A large number of reports show that circHIPK3 plays a key role in the occurrence and development of fibrosis and tumors.
The Role of circHIPK3 in Fibrosis Diseases
Cardiac Fibrosis
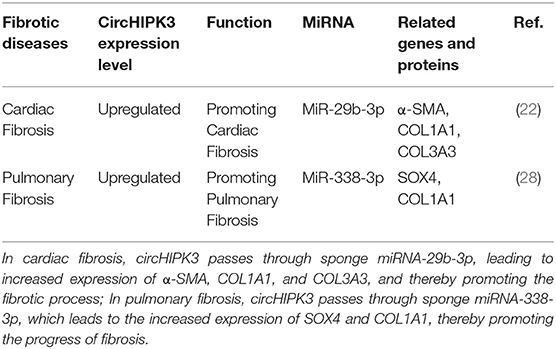
Cardiac fibrosis, which is a key feature of the pathological processes of cardiovascular disease, is due to the deposition of a large amount of extracellular matrix in the heart interstitium. This leads to the contraction and relaxation of the heart, ultimately leading to a series of heart conditions, including heart failure (21). Ni et al. found that, compared with normal heart tissue, circHIPK3 is expressed at a high level in cardiac fibrotic tissue and is induced by angiotensin II. Silencing circHIPK3 can inhibit the proliferation and migration of cardiac fibroblasts, but overexpression of circHIPK3 can promote the proliferation and migration of cardiac fibroblasts; circHIPK3 regulates the proliferation and migration of cardiac fibroblasts through the circHIPK3-miR-29b-3p pathway. A combination of circHIPK3 silencing and miR-29b-3p overexpression had a better inhibitory effect on the proliferation and migration of cardiac fibroblasts (22). Si et al. found that circHIPK3 is upregulated in the hearts of fetuses and newborns, and it is necessary for neonatal cardiomyocyte proliferation. In in vitro experiments, the overexpression of circHIPK3 promoted the proliferation and migration of cardiomyocytes. Furthermore, the overexpression of circHIPK3 promoted heart regeneration in adult mice. The circHipk3-miR-133a (the circHipk3-miR-133a-connective tissue growth factor axis) pathway in the myocardium participates in mediating the function and angiogenesis of human coronary artery endothelial cells (HCAECs) during infarction, while circHIPK3 induces cardiomyocyte proliferation by increasing the stability of notch1 intracellular domain (N1ICD) (23). Bai et al. found that circHIPK3 promotes myocardial ischemia/reperfusion injury by sponge miR-124-3p (24). In experiments using mice with myocardial infarction, Deng et al. found that circHIPK3 downregulation can reduce cardiac fibrosis and maintain cardiac function (25). Whether studying cardiac fibrosis or other cardiovascular diseases, scholars have found that circHIPK3 binds miRNA through the sponge mechanism to change the expression of the corresponding protein, thereby affecting the progression of the disease. These findings suggest that circHIPK3 plays an essential role in the process of cardiac fibrosis. However, these experiments were in vitro or animal experiments, so further studies are needed to verify this conclusion.
Pulmonary Fibrosis
Pulmonary fibrosis is a chronic, progressive, and fibrotic interstitial lung disease of unknown etiology that can lead to an irreversible decline in lung function, progressive respiratory failure, and even death. The specific pathogenesis of pulmonary fibrosis is still unclear. Recent studies have found that smoking and other factors lead to continuous alveolar epithelial cell damage and abnormal repair, the proliferation of fibroblasts, and the accumulation of extracellular matrix, leading to structural disorders of the lungs and, finally, fibrosis (26, 27). A study by Zhang et al. found that, in the bleomycin-induced pulmonary fibrosis mouse model, the expression level of circHIPK3 was upregulated in lung fibroblast-derived myofibroblasts.circHIPK3 participated in regulating the differentiation of fibroblasts into myofibroblasts and the proliferation of fibroblasts through the circHIPK3-miR338-3p/SOX4/COL1A1 pathway (28). Similarly to cardiac fibrosis, circHIPK3 increases the expression levels of SOX4 and COL1A1 by binding miRNA-338-3p in pulmonary fibrosis and thus promoting the progress of pulmonary fibrosis. The functional characteristics of circHIPK3 in fibrosis diseases are detailed in Table 1.
The Role of circHIPK3 in Cancer/Tumoral Diseases
Numerous studies have shown that circHIPK3 is involved in the pathogenesis and prognosis of various cancers. Zeng et al. found that circHIPK3 promotes the growth and metastasis of colorectal cancer through sponge miR-7 (29), and Zhang et al. found that colorectal cancer patients resistant to oxaliplatin and those with colorectal cancer recurrence showed high levels of circHIPK3 expression and that circHIPK3, colorectal tumor size, regional lymph node metastasis, distant metastasis, and prognosis were closely related (30). In patients with chronic myeloid leukemia, circHIPK3 was found at high expression levels, suggesting that circHIPK3 is closely related to the prognosis of the disease (31). When Fang Teng et al. silenced circHIPK3, the proliferation, migration, and invasion of ovarian cancer cells and normal ovarian epithelial cells were promoted, and apoptosis was inhibited. Gao Wenzhe et al. found that the abnormal expression of circHIPK3 may be closely related to the prognosis of patients with malignant tumors (32).
Compared with adjacent tissues, the expression level of circHIPK3 was higher in gastric cancer cells compared with gastric mucosal cells, and the expression level of circHIPK3 was negatively correlated with the overall prognosis of gastric cancer patients. Silencing circHIPK3 weakened the proliferation and migration ability of gastric cancer cells. In addition, knocking out circHIPK3 significantly downregulated the levels of WNT1, TCF4, and β-catenin (33). Moreover, circHIPK3 was found to participate in the regulation of brain-derived neurotrophic factors by sponge miR-107, and it plays a further key role in the development of gastric cancer (34).
In another study, circHIPK3 promoted glioma progression by regulating the miR-654/IGF2BP3 signaling pathway and could be used as a prognostic indicator of glioma (35). Additionally, circHIPK3 was found to promote the proliferation and metastasis of gliomas through sponge miR-124-3p (36). Yawei Li et al. showed that sponge miR-558 and circHIPK3 inhibited the expression of heparinase in bladder cancer cells (37). The overexpression of circHIPK3 was found to promote the proliferation and metastasis of prostate cancer cells through sponge miR-338-3p (38); however, Dong Chen et al. demonstrated that the circHIPK3-miR-193a-3p-MCL1 signaling axis mediates the occurrence and development of prostate cancer (39).
Bioinformatics analysis has revealed the circHIPK3-miRNA-mRNA axis to be a potential signal pathway involved in the effects of circHIPK3 (40). In one study, circHIPK3 promoted the proliferation of human oral squamous cell carcinoma cells through sponge miR-124 (41). Whereas, in another study, circHIPK3 inhibited the progression of lung cancer via sponge miR-124 (42). In addition, circHIPK3 was shown to regulate the expression level of IGF1 through miR-379 to promote the proliferation of non-small cell lung cancer cell lines, NCI-H1299 and NCI-H2170 (43). According to Jiale Wang et al., circHIPK3 promoted acinar cell carcinoma apoptosis by regulating the miR-193a-5p/GSDMD pathway (41, 44). Jinjin Lai et al. showed that circHIPK3 promotes the proliferation and metastasis of renal cancer cells by downregulating miR-485-3p (45). The metastasis of osteosarcoma cells is mediated through the circHIPK3-miR-637/STAT3 signaling pathway (46, 47). In animal experiments, circHIPK3 promoted the proliferation and metastasis of esophageal squamous cell carcinoma cells by mediating the circHIPK3-miR-599/c-MYC signaling pathway (48).
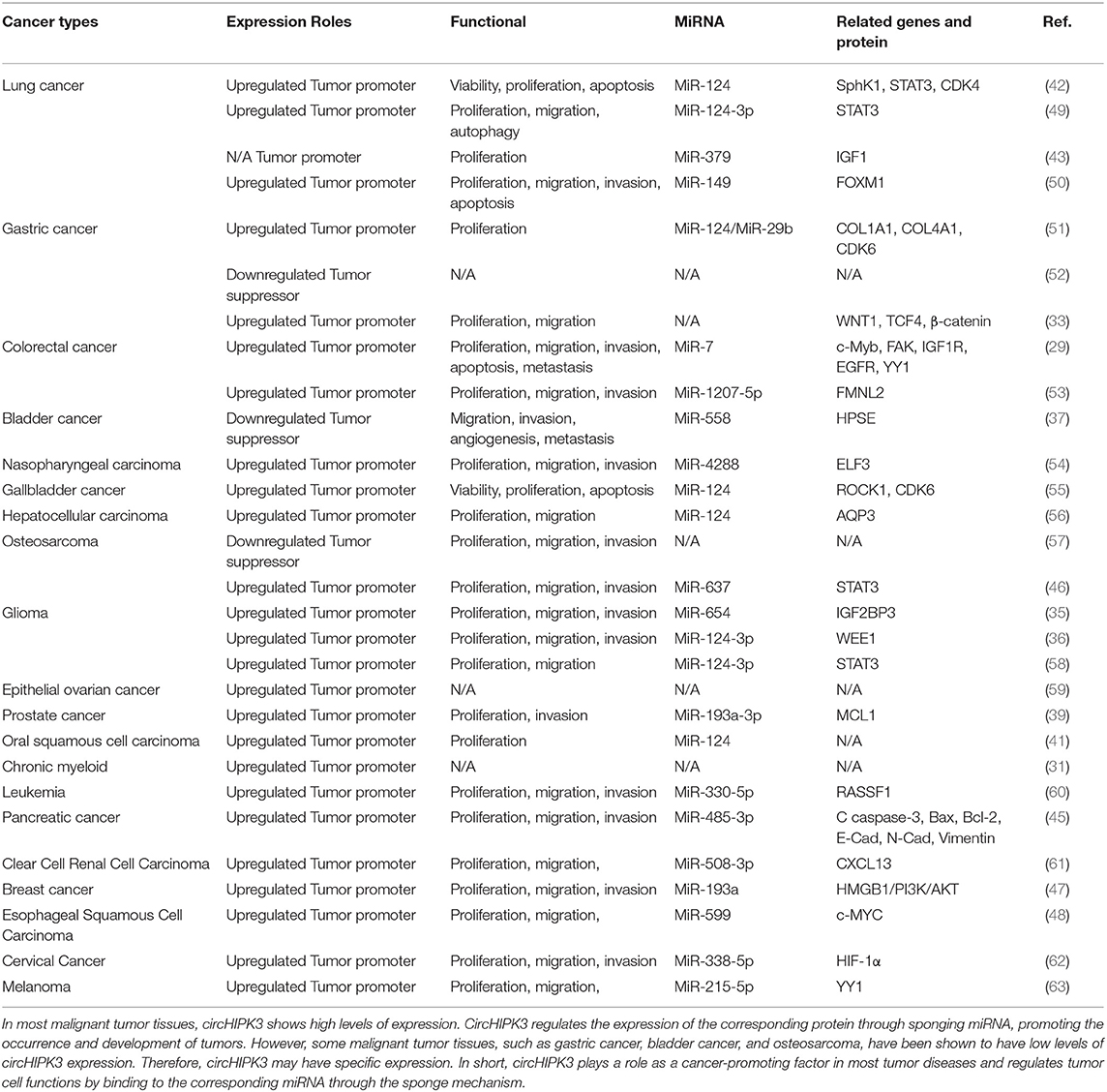
High expression levels of circHIPK3 in tumor patients indicate that the tumor is at a relatively advanced stage. In addition, in the prognostic follow-up of patients with high levels of circHIPK3 expression, the probability of distant tumor metastasis and lymph node metastasis was higher than for other patients. Thus, the expression level of circHIPK3 plays an important role in the progression and prognosis of tumor diseases, suggesting that circHIPK3 may be a target gene for the diagnosis and treatment of related tumors. The functional characteristics of circHIPK3 in cancer are detailed in Table 2 (refer to Wen J, et al. Circular RNA HIPK3: A Key Circular RNA in a Variety of Human Cancers. Front Oncol. 2020 May 15;10:773, and supplement) (64).
In short, these lines of evidence indicate that circHIPK3 is closely related to the progression and prognosis of various malignant tumors. In in vitro and in vivo experiments, the silencing and overexpression of circHIPK3 had significant effects on the proliferation, migration, and invasion of tumor cells, indicating that expression levels of circHIPK3 are closely related to the biological behavior of tumor cells. However, the recent experiments have had many limitations. To explore and further demonstrate the specific mechanisms of circHIPK3 in human diseases, additional experiments are needed.
The Role of circHIPK3 in Other Diseases
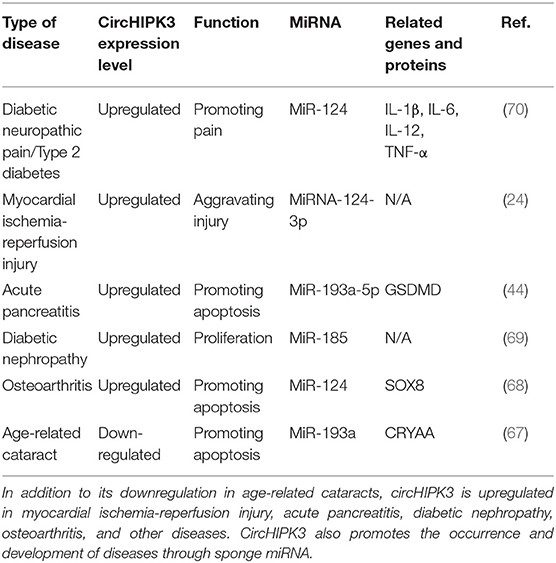
CircHIPK3 is involved in the pathophysiological process of other diseases, including fibrotic diseases, cancer, and other diseases. Research found circHIPK3 promoted the expression of vascular endothelial growth factor C, FZD4, and WNT2 by sponge miR-30a-3p and, consequently, mediated diabetic retinal vascular dysfunction (65). High blood glucose levels led to downregulated circHIPK3 expression, promoting damage to vascular endothelial cells, while the overexpression of circHIPK3 inhibited vascular endothelial cell damage (66). Other studies have found that circHIPK3 is involved in the pathophysiological processes of diseases such as diabetic neuropathic pain, type 2 diabetes, diabetic nephropathy, acute pancreatitis, osteoarthritis, myocardial ischemia reperfusion injury, and age-related cataract, and plays an important role in these conditions (24, 44, 67–70). Although circHIPK3 is downregulated in age-related cataracts, and it is upregulated in all other fibrotic diseases. It also regulates cell proliferation, differentiation, and metastasis through the sponging mechanism, which affects the progression of diseases. The functional characteristics of circHIPK3 in other diseases are detailed in Table 3.
Conclusions and Future Prospects
CircRNA, a type of non-coding RNA, can regulate gene expression through a variety of mechanisms and may also have roles in regulating protein expression. The function of circRNA is complex, and its regulatory mechanisms are not fully understood. Current research has focused on changes in circRNA expression during physiological and pathological processes, and circRNA has become a new tool for understanding different diseases and their progression. Many scholars have been exploring the potential uses of the molecules in detecting disease states and for gene therapy. Functional research into circRNA in fibrotic diseases, cancer, and other diseases is a rapidly developing field, but these research avenues are still in the early stages and require follow-up efforts. CircHIPK3 is a member of the circRNA family. Numerous studies have shown that circHIPK3 is abnormally expressed in fibrotic diseases, cancer, and other diseases, which indicates that circHIPK3 may play an important role in pathogenesis. This review focused on the role of circHIPK3 in the pathophysiological processes of fibrotic diseases, cancer, and other diseases. As we have discussed, there is substantial information on the role of circHIPK3 in various diseases, which provide ideas for studying the pathogenesis of various diseases and clinical diagnosis and treatment of various diseases. Except for its downregulation in age-related cataracts, circHIPK3 is upregulated in all other diseases studied and can control cells through the sponging mechanism, affecting the cell cycle, cell proliferation, cell apoptosis, and invasion and migration. Furthermore, circHIPK3 can be used as a prognostic indicator of disease. Considering circHIPK3's unique stability, miRNA sponging function, and regulation ability, it may be a promising therapeutic drug carrier and a potential diagnosis and treatment target. We have discussed the role of circHIPK3 in heart and pulmonary fibrosis, but there are no reports on its role in oral submucosal fibrosis, and further studies are needed. Although there has been a plethora of research efforts to clarify the mechanisms of circHIPK3 action in various diseases, because of technological limitations, research has focused on the role of circHIPK3 as a microRNA sponge, and its other functions require further investigation. Based on the large number of studies that have shown correlation between circHIPK3 and various diseases, we have deepened our understanding of the role of circHIPK3 in pathogenesis. At the same time, it also provides a new way to explore the pathogenesis of different diseases. In addition, circHIPK3 has great clinical value in the diagnosis, treatment, and prognosis of fibrotic diseases, cancer, and other diseases. Although the specific mechanism through which circHIPK3 participates in various diseases is still unknown, we believe that, with the development of bioinformatics technology, the link between circHIPK3 and diseases will eventually be revealed.
Author Contributions
JZ designed the manuscript, a major contributor in writing the manuscript, and drew the figures and tables. JZ, BW, XB, CX, and BL mainly revised the manuscript. JZ, OL, and ZT participated in the design of the review and made some revisions of the review. All the authors read and approved the final version of the review.
Funding
This work was supported by National Natural Science Fund (81671003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. (2018) 25:1–7. doi: 10.1007/s12282-017-0793-9
2. Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. (2018) 9:e1478. doi: 10.1002/wrna.1478
3. Rajappa A, Banerjee S, Sharma V, Khandelia P. Circular RNAs: emerging role in cancer diagnostics and therapeutics. Front Mol Biosci. (2020) 7:577938. doi: 10.3389/fmolb.2020.577938
4. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. (2018) 71:428–42. doi: 10.1016/j.molcel.2018.06.034
5. Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. (2019) 13:669–80. doi: 10.1002/1878-0261.12468
6. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
7. Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
8. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. (2020) 77:1661–80. doi: 10.1007/s00018-019-03345-5
9. Zhang C, Hu J, Yu Y. CircRNA Is a rising star in researches of ocular diseases. Front Cell Dev Biol. (2020) 8:850. doi: 10.3389/fcell.2020.00850
10. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. (2014) 32:453–61. doi: 10.1038/nbt.2890
11. Cortés-López M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. (2016) 89:527–37.
12. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. (2017) 66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017
13. Xie Y, Yuan X, Zhou W, Kosiba AA, Shi H, Gu J, et al. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: review. Life Sci. (2020) 254:117252. doi: 10.1016/j.lfs.2019.117252
14. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
15. Rybak-Wolf A, Stottmeister C, GlaŽar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. (2015) 58:870–85. doi: 10.1016/j.molcel.2015.03.027
16. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. (2017) 7:4183–91. doi: 10.7150/thno.21299
17. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
18. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. doi: 10.1038/ncomms11215
19. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
20. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. (2014) 28:2233–47. doi: 10.1101/gad.251926.114
21. Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci. (2014) 71:549–74. doi: 10.1007/s00018-013-1349-6
22. Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. (2019) 292:188–96. doi: 10.1016/j.ijcard.2019.04.006
23. Si X, Zheng H, Wei G, Li M, Li W, Wang H, et al. circRNA Hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to Notch1 and miR-133a. Mol Ther Nucleic Acids. (2020) 21:636–55. doi: 10.1016/j.omtn.2020.06.024
24. Bai M, Pan CL, Jiang GX, Zhang YM, Zhang Z. CircHIPK3 aggravates myocardial ischemia-reperfusion injury by binding to miRNA-124-3p. Eur Rev Med Pharmacol Sci. (2019) 23:10107–14. doi: 10.26355/eurrev_201911_19580
25. Deng Y, Wang J, Xie G, Zeng X, Li H. Circ-HIPK3 strengthens the effects of adrenaline in heart failure by MiR-17-3p - ADCY6 Axis. Int J Biol Sci. (2019) 15:2484–96. doi: 10.7150/ijbs.36149
26. Meyer KC. Pulmonary fibrosis, part I: epidemiology, pathogenesis, and diagnosis. Expert Rev Respir Med. (2017) 11:343–59. doi: 10.1080/17476348.2017.1312346
27. Thannickal VJ, Toews GB, White ES, Lynch JP III, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med. (2004) 55:395–417. doi: 10.1146/annurev.med.55.091902.103810
28. Zhang JX, Lu J, Xie H, Wang DP, Ni HE, Zhu Y, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. (2019) 10:182. doi: 10.1038/s41419-019-1430-7
29. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. (2018) 9:417. doi: 10.1038/s41419-018-0454-8
30. Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. (2019) 48:277–88. doi: 10.1016/j.ebiom.2019.09.051
31. Feng XQ, Nie SM, Huang JX, Li TL, Zhou JJ, Wang W, et al. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. (2020) 67:171–7. doi: 10.4149/neo_2018_181129N908
32. Wenzhe G, Jiahao X, Cheng P, Hongwei Z, Xiao Y. Circular RNA HIPK3 is a prognostic and clinicopathological predictor in malignant tumor patients. J Cancer. (2020) 11:4230–9. doi: 10.7150/jca.40001
33. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. (2019) 23:7905–12. doi: 10.26355/eurrev_201909_19004
34. Wei J, Xu H, Wei W, Wang Z, Zhang Q, De W, et al. circHIPK3 promotes cell proliferation and migration of gastric cancer by sponging miR-107 and regulating BDNF expression. Onco Targets Ther. (2020) 13:1613–24. doi: 10.2147/OTT.S226300
35. Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. (2018) 503:1570–4. doi: 10.1016/j.bbrc.2018.07.081
36. Xia L, Yi F, Zhai X, Zhang M. Circular RNA homeodomain-interacting protein kinase 3 (circHIPK3) promotes growth and metastasis of glioma cells by sponging miR-124-3p. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. (2020) 36:609–15.
37. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. (2017) 18:1646–59. doi: 10.15252/embr.201643581
38. Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. (2019) 12:3363–72. doi: 10.2147/OTT.S196931
39. Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. (2019) 11:1415–23. doi: 10.2147/CMAR.S190669
40. Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. (2019) 112:8–17. doi: 10.1016/j.biocel.2019.04.011
41. Wang J, Zhao SY, Ouyang SS, Huang ZK, Luo Q, Liao L. Circular RNA circHIPK3 acts as the sponge of microRNA-124 to promote human oral squamous cell carcinoma cells proliferation. Zhonghua Kou Qiang Yi Xue Za Zhi. (2018) 53:546–51. doi: 10.3760/cma.j.issn.1002-0098.2018.08.009
42. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. (2018) 506:455–62. doi: 10.1016/j.bbrc.2018.10.087
43. Tian F, Wang Y, Xiao Z, Zhu X. Circular RNA CircHIPK3 Promotes NCI-H1299 and NCI-H2170 Cell Proliferation through miR-379 and its Target IGF1. Zhongguo Fei Ai Za Zhi. (2017) 20:459–67. doi: 10.3779/j.issn.1009-3419.2017.07.04
44. Wang J, Li X, Liu Y, Peng C, Zhu H, Tu G, et al. CircHIPK3 promotes pyroptosis in acinar cells through regulation of the miR-193a-5p/GSDMD axis. Front Med (Lausanne). (2020) 7:88. doi: 10.3389/fmed.2020.00088
45. Lai J, Xin J, Fu C, Zhang W. CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int. (2020) 20:248. doi: 10.1186/s12935-020-01319-3
46. Huang Z, Yuan C, Gu H, Cheng X, Zhou K, Xu J, et al. Circular RNA circHIPK3 promotes cell metastasis through miR-637/STAT3 axis in osteosarcoma. Biomed Res Int. (2020) 2020:2727060. doi: 10.1155/2020/2727060
47. Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, Wang GS. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. (2020) 11:2660–71. doi: 10.1111/1759-7714.13603
48. Ba Y, Liu Y, Li C, Zhu Y, Xing W. HIPK3 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-599/c-MYC axis. Onco Targets Ther. (2020) 13:1967–78. doi: 10.2147/OTT.S217087
49. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. (2020) 16:659–71. doi: 10.1080/15548627.2019.1634945
50. Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. (2020) 21:113–21. doi: 10.1080/15384047.2019.1669995
51. Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. (2018) 16:216. doi: 10.1186/s12967-018-1582-8
52. Ghasemi S, Emadi-Baygi M, Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. (2019) 49:687–95. doi: 10.3906/sag-1806-50
53. Yan Y, Su M, Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun. (2020) 524:839–46. doi: 10.1016/j.bbrc.2020.01.055
54. Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. (2019) 234:1699–706. doi: 10.1002/jcp.27041
55. Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. (2018) 503:863–9. doi: 10.1016/j.bbrc.2018.06.088
56. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. (2018) 9:175. doi: 10.1038/s41419-017-0204-3
57. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. (2018) 9:1856–62. doi: 10.7150/jca.24619
58. Hu D, Zhang Y. Circular RNA HIPK3 promotes glioma progression by binding to miR-124-3p. Gene. (2019) 690:81–9. doi: 10.1016/j.gene.2018.11.073
59. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. (2018) 22:3713–8. doi: 10.26355/eurrev_201806_15250
60. Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X, et al. CircHIPK3 promotes Gemcitabine (GEM) resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag Res. (2020) 12:921–9. doi: 10.2147/CMAR.S239326
61. Han B, Shaolong E, Luan L, Li N, Liu X. CircHIPK3 promotes clear cell Renal Cell Carcinoma (ccRCC) cells proliferation and metastasis via altering of miR-508-3p/CXCL13 signal. Onco Targets Ther. (2020) 13:6051–62. doi: 10.2147/OTT.S251436
62. Qian W, Huang T, Feng W. Circular RNA HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p to up-regulate HIF-1α. Cancer Manag Res. (2020) 12:177–87. doi: 10.2147/CMAR.S232235
63. Zhu X, Sun J. CircHIPK3 regulates melanoma cell behaviors by binding with miR-215-5p to upregulate YY1. Mol Cell Probes. (2020) 53:101644. doi: 10.1016/j.mcp.2020.101644
64. Wen J, Liao J, Liang J, Chen XP, Zhang B, Chu L. Circular RNA HIPK3: a key circular RNA in a variety of human cancers. Front Oncol. (2020) 10:773. doi: 10.3389/fonc.2020.00773
65. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. (2017) 136:1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004
66. Cao Y, Yuan G, Zhang Y, Lu R. High glucose-induced circHIPK3 downregulation mediates endothelial cell injury. Biochem Biophys Res Commun. (2018) 507:362–8. doi: 10.1016/j.bbrc.2018.11.041
67. Liu X, Liu B, Zhou M, Fan F, Yu M, Gao C, et al. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/CRYAA axis. Biochem Biophys Res Commun. (2018) 503:2277–85. doi: 10.1016/j.bbrc.2018.06.149
68. Wu Q, Yuan ZH, Ma XB, Tang XH. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur Rev Med Pharmacol Sci. (2020) 24:7937–45. doi: 10.26355/eurrev_202008_22476
69. Liu R, Zhang M, Ge Y. Circular RNA HIPK3 exacerbates diabetic nephropathy and promotes proliferation by sponging miR-185. Gene. (2020) 765:145065. doi: 10.1016/j.gene.2020.145065
Keywords: CircHIPK3, miRNA, fibrosis, cancer, pathophysiological, treatment
Citation: Zhou J, Wang B, Bin X, Xie C, Li B, Liu O and Tang Z (2021) CircHIPK3: Key Player in Pathophysiology and Potential Diagnostic and Therapeutic Tool. Front. Med. 8:615417. doi: 10.3389/fmed.2021.615417
Received: 09 October 2020; Accepted: 02 February 2021;
Published: 22 February 2021.
Edited by:
Luigi M. Terracciano, University of Basel, SwitzerlandReviewed by:
Axel zur Hausen, Maastricht University Medical Centre, NetherlandsMaria Contaldo, University of Campania Luigi Vanvitelli, Italy
Copyright © 2021 Zhou, Wang, Bin, Xie, Li, Liu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ousheng Liu, liuousheng@163.com; Zhangui Tang, tangzhangui@aliyun.com
 Jiang Zhou
Jiang Zhou Baisheng Wang
Baisheng Wang