Establishment of methods for rapid detection of Prymnesium parvum by recombinase polymerase amplification combined with a lateral flow dipstick
- 1Key Laboratory of Marine Biotechnology of Zhejiang Province, School of Marine Science, Ningbo University, Ningbo, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
Prymnesium parvum is a toxic algal bloom (HAB)-forming species. The toxicity of this alga is a result of a collection of compounds known as prymnesins. Prymnesins exert harmful effects upon fish, shellfish, and mollusks, causing huge economic losses. In the present study, a new method was developed for the detection of P. parvum. The novel method utilizes isothermal amplification, known as recombinase polymerase amplification (RPA), in combination with lateral-flow dipstick (LFD). Herein, a set of primers and probes were designed for internal transcribed spacer (ITS) sequences, and a specific and sensitive RPA-LFD rapid detection method was established for P. parvum. Meanwhile, we verified its feasibility for the detection of environmental samples. It was demonstrated that the optimal amplification temperature and time for RPA were 39°C and 15 min. RPA/RPA-LFD was experimentally verified to be specific, demonstrating no cross-reaction with distinct control microalgae, and furthermore, the total time required for the RPA-LFD experiment was 20 min. Meanwhile, the detection limit for the genomic DNA of P. parvum was 1.5×10-1 pg/μL, and the detection limit for plasmids was 2.35 pg/μL. In addition, the results herein revealed that the RPA-LFD assay was 100 times more sensitive than PCR for detection of P. parvum. In conclusion, we developed an RPA-LFD that does not require precision instruments, and can be utilized for rapid on-site detection of P. parvum. In the future, the RPA-LFD can be considered for practical application for environmental detection of the toxic algal species.
Introduction
The haptophyte Prymnesium parvum is a freshwater marine species that usually exists in brackish and inland high-mineral waters. It first reported in a Brackish water pond in 1937 (Carter, 1937). As a toxic and harmful alga, the toxin produced by P. parvum, prymnesins, exhibits potent neurotoxic, ichthyotoxic, cytotoxic and hemolytic effects (Manning and Claire, 2010b; Seoane et al., 2016). Prymnesins exert a potent toxic effect on fish, regularly resulting in the deaths of a large number of fish and subsequently causing great losses to farmers. Moreover, in humans toxins can be accumulated in by the consumption of seafood leading to the dangerous biomagnification of reactive molecules (Sharifan and Ma, 2017). For phytoplankton, mixed blooms of the killer alga P. parvum and known harmful cyanobacteria were one of the major causes of catastrophic changes in Europe’s protected wetlands. The alga and cyanobacteria combination caused the phytoplankton to collapse (Maria et al., 2022). Over the previous two decades, the harmful algal blooms of P. parvum have been reported in the North America regions, such as Hawaii, Nebraska, etc., causing tens of millions of dollars in economic damage (Southard et al., 2010; Roelke et al., 2016). In China, algal blooms resulting from P. parvum occur frequently in Ningxia, Tianjin, Zhejiang, Shandong, Shanxi and other provinces. P. parvum has been known to cause the mass mortality of fish in China since 1963 (Guo et al., 1996). In a comparison of the toxicity of three kinds of harmful algae frequently occurring in Sanmen Bay and Xiangshan Bay, P. parvum exhibited the most toxic effect, causing acute death within five hours (Yang et al., 2018). Furthermore, P. parvum was isolated from the Pearl River Estuary in China (Qin et al., 2020). Therefore, it is necessary to establish a detection method of P. parvum that is fast and sensitive enough to detect the algae in advance prior to the outbreak of blooms; the assay must also meet the requirements of field detection, so as to prevent loss as much as possible.
At present, the methods used for the detection of microbiota are mainly divided into two categories; the first are based on the detection of algae morphology, primarily by microscopic examination (Töbe et al., 2006); the second are molecular detection methods based on nucleic acid amplification, including the polymerase chain reaction (PCR) (Manning and Claire, 2010a), quantitative PCR (qPCR) (Manning and Claire, 2013; Beyer et al., 2019), molecular probe techniques (Eckford-Soper and Daugbjerg, 2015) and isothermal amplification techniques (IAT). Upon the basis of these techniques, the detection of P. parvum has been broadly established. However, when considering IAT, loop-mediated isothermal amplification (LAMP) is the only method used for P. parvum detection (Zhu et al., 2019). Furthermore, all the existing detection methods have some limitations. For example, P. parvum is relatively small and fragile and can become distorted during preservation, making its identification and detection difficult (Galluzzi et al., 2008). Examination by microscope is highly uncertain and subjective (Zamor et al., 2012). Only experts skilled in algal identification are qualified for this task. In addition to microscopy methods, PCR, qPCR and other non-isothermal nucleic acid amplification methods possess the advantages of being fast, sensitive, and specific. Although the above molecular methods are useful additions to traditional identification tools, they still need to be performed under laboratory conditions by professional technicians. Moreover, it is difficult to achieve on-site detection of environmental samples. Although a loop-mediated isothermal amplification and lateral flow dipstick (LAMP-LFD) method may possess promise in solving the problems associated with detection in the field, primer design for LAMP technology is complicated (4-6 pairs of primers), and the reaction requires incubation at 65–67°C for 30–60 min. These shortcomings have limited the application of the aforementioned molecular methods for the rapid detection of P. parvum. Fortunately, in addition to LAMP, several other isothermal amplification techniques (IAT) have been widely employed for the detection of harmful algal bloom species, including quadruplex priming amplification (QPA) (Lomidze et al., 2018), helicase-dependent amplification (HDA) (Liu et al., 2020), nucleic acid sequence-based amplification (NASBA) (Nai et al., 2022), rolling circle amplification (RCA) (Yang et al., 2022), and recombinase polymerase amplification (RPA) (Cao et al., 2022).
The RPA method was first developed by Piepenburg in 2006 as an isothermal nucleic acid amplification technique (Piepenburg et al., 2006). The RPA reaction process can be completed within 20 minutes at a constant temperature of 37°C without the requirement for high-temperature denaturation. Previous studies have demonstrated that RPA has the same or greater sensitivity and specificity as PCR for the detection of algae, and it only requires a single pair of primers (Fu et al., 2019; Zhai et al., 2021; Toldrà et al., 2018). Finally, RPA can be combined with a variety of endpoint detection techniques for the rapid and simultaneous detection of multiple samples, these include agarose gel electrophoresis (Fu et al., 2020), real-time fluorescence quantitative assays (Wang X.F. et al., 2020), dye assays (Kang et al., 2020), and lateral flow dipsticks (LFD) (Ayfan Abdulrahman et al., 2021). LFDs are simple biosensors that involve building a ternary complex consisting of a biotinylated RPA product hybridized with a fluorescein isothiocyanate (FITC) probe and further complexed with a gold-labeled anti-FITC antibody in non-laboratory environments (Lin et al., 2022). When the control line and the test line simultaneously color, a positive result is indicated; a result than can be detected by the human eye. Indeed, LFD is a convenient assay for immediate visualization after the rapid amplification of RPA (Xia et al., 2022). Compared with traditional gel electrophoresis detection, LFDs main advantages are that a result can be observed within 1-2 minutes, without the need for precision instruments such as imagers (Wang L. et al., 2020).
At present, due to the technical advantages exhibited by RPA-LFD, the technique has been widely used in various testing fields (Daher et al., 2014; Karakkat et al., 2018; Li et al., 2019; Yang et al., 2020; Jiang T.T. et al., 2022). However, the use of RPA-LFD in detecting harmful algae is still in its infancy. In the present study, we establish a specific and sensitive RPA-LFD assay for the detection of P. parvum and investigated its performance in application in the field. The ultimate purpose of this study was to explore novel methods for the rapid, low cost, and convenient field detection of harmful algae blooms species.
Materials and methods
Algae culture
A total of 9 algae species were selected for RPA/RPA-LFD specificity analysis in the study. Of these, Prymnesium parvum (NMBjih029), Thalassiosira pseudonana (NMBguh006), Chaetoceros curvisetus (NMBguh003-10), Skeletonema costatum (NMBguh0042), Pseudo-nitzschia multiseries (NMBguh002-1-1), and Isochrysis galbana (NMBjih021-2) were supplied by the Microalgae Collection of Ningbo University. Meanwhile, Chaetoceros debilis (MMDL50116), and Thalassiosira rotula (MMDL50319) were supplied by Xiamen University, Trichodesmium erythraeum (IMS101) was supplied by the University of Southern California and Phaeodactylum tricornutum (CCMP2561) was supplied by Westlake University. The culture medium used was f/2 medium. All algal strains were cultured in a 100 ml flask, the light intensity was 15~20 μmol/(m2·s), the light-dark period was 12 h:12 h, and the temperature was 16°C, with shaking 1–2 times daily, and stationary culture.
DNA extraction, PCR, cloning and sequencing of target gene
Genomic DNA was obtained from P. parvum cells prepared by harvesting algal culture during their stationary phase, by centrifugation at 12000 × g for 2 min, with the Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China). DNA concentrations were measured using a Nano-300 micro spectrophotometer (Allsheng, Hangzhou, China), the concentration was 1.5×104 pg/μL. The genomic DNA was stored at -20°C.
The ITS of the target sequence was investigated and two PCR primers were designed; these were named Pr-PCR-F (5’-TCTCGCAACGATGAAGAACG-3’) and Pr-PCR-R (5’-TCGATGGCACAACGACTTG-3’). The PCR reaction procedure was as follows: an initial denaturation at 95°C for 3 min, 30 cycles of 95°C for 15 s, 56°C for 15 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR products were purified by Hipure PCR Pure Kits (Magen, Guangzhou, China). The purified PCR products were linked using a pMD 18-T Vector (TaKaRa, Dalian, China) to transform the competent Escherichia coli (DH5α). The positive clones identified by colony PCR were sent to Zhejiang Youkang Biotechnology Co., Ltd. for sequencing.
Design of RPA primers and probes
According to the principles of primer probe design in TwistAmp® DNA Amplification Kits (TwistDX, UK), 5 pairs of primers were designed for ITS sequencing of P. parvum. The designed primers underwent further screening experiments to determine the final optimal primers. To obtain specific RPA primers, the Clustalw tool in the Biodeit software package (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was utilized in addition to BLAST in the NCBI. The sequences of similar species involved in multiple alignment analysis included Isochrysis galbana (JX393298.1), Chrysochromulina ephippum (KT390082.1) and Pleurochrysis scherffelii (AM936922.1). Primer Premier 5 (www.PremierBiosoft.com) was used to design the primers and probes. For the RPA-LFD reaction, biotin was tagged to the 5’-end of the optimum anti-sense primer. The 5’-end of the probe was next labeled with a carboxy fluorescein (FAM), the middle of the probe was labeled with a tetrahydrofuran (THF) site, and the 3’-end was labeled with a C3-spacer blocking group. Synthesis of the primers and probe were performed by Zhejiang Youkang Biotechnology Co., Ltd. The primer design results are shown in Table 1.
The RPA reaction and system optimization
The RPA reactions were conducted using TwistAmp® Basic kits (TwistDX,UK). RPA reactions were performed in a total volume of 50 µL, containing 29.5 μL of rehydration buffer, 11.2 μL of nuclease-free water, 2.4 μL of sense primer (10 μM), 2.4 μL of anti-sense primer (10 μM), and 2 μL of genomic DNA extracted from P. parvum. The reaction mixture was briefly vortexed and centrifuged in a 1.5 ml centrifuge tube. Next, the reaction mixture was transferred to a tube containing lyophilized enzyme pellets that were supplied in the TwistAmp® Basic kits, 2.5 μL of MgOAc (280 mM) was rapidly added to the pellets to resuspend. After vortexing and spinning, the solution was incubated in a metal bath (Allsheng, Hangzhou, China) at 39°C for 20 min. According to the recommended process (Lillis et al., 2016), the reaction tubes were removed after 4 min and the reaction was continued after vortexing. At the end of the RPA reaction, the product was purified using Hipure PCR Pure Kits (Magen, Guangzhou, China) and detected using 2% agarose gel electrophoresis.
System optimization was performed in two parts: reaction temperature optimization and reaction time optimization. Optimization of amplification temperature was performed using the optimum primers, and a total of 3 temperature gradients of 35°C, 37°C, and 39°C. Next, the optimal times were further stratified by setting different amplification times (10 min, 15 min, and 20 min). Subsequent experiments were all the optimum primers and reaction set-up.
The RPA-LFD reaction
To confirm the RPA results, the LFD (Milenia biotec, Germany) was employed to detect the RPA products. RPA reactions were conducted using the DNA thermostatic rapid amplification kit (Amplification Future, China). Moreover, RPA reactions were performed with a total volume of 50 µL, containing 29.4 μL of buffer A, 11.5 μL of nuclease-free water, 2 μL of sense primer (10 μM), 2 μL of anti-sense primer labeled with biotin (10 μM), 0.6 μL of LFD probe (10 μM) and 2 μL of genomic DNA extracted from P. parvum. The subsequent procedure refers to the RPA reaction, but the RPA products were not purified using kits: the 8 μL RPA reaction product was added directly to 92 μL of assay buffer and mixed well in a well of a microtiter plate. The dipsticks were place in sample application assay buffer wells and incubated for 1 minutes in an upright position and the results could be read by eye after the control line was fully colored. The result was positive if both the control line and the detection line were observed. On the contrary, the result was negative if only the control line is observed.
Specificity and sensitivity analysis
The specificity study was performed using the 10 aforementioned algal species. ddH2O was selected as the template for the negative control. After extracting the algal genomic DNA using the method mentioned in the “DNA extraction, PCR, cloning and sequencing of target genes” section, RPA, RPA-LFD and PCR amplification were performed. Simultaneously, to ensure the validity of genomic DNA, PCR amplification was first performed with universal primers TW81 (5’-GGGATCCGTTTCCGTAGGTGAACCTGC-3’) and AB28 (5’-GGGATCCATATGCTTAAGTTCAGCGGGT-3’) (White et al., 1990) before performing the specificity study.
Both the genomic DNA of P. parvum and the recombinant plasmids consisting of the ITS sequence of P. parvum were used to determine sensitivity. Genomic DNA and a recombinant plasmid containing the P. parvum ITS sequence were diluted in a ten-fold serial dilution in ddH2O for 8 gradients. The result was genomic DNA of concentration ranging from 1.5×104 pg/μL to 1.5×10-3 pg/μL, and recombinant plasmid DNA of concentration ranging from 2.35×105 pg/μL to 2.35×10-2 pg/μL. The plasmids were extracted with a PurePlasmid Mini Kit (CWBIO, Suzhou, China). Sensitivity of PCR, RPA, and RPA-LFD was detected using the serially diluted genomic DNA, while the plasmid was used as a template with ddH2O as a blank control. The primers used were the optimal RPA primers identified by earlier screening. The sensitivity of RPA-LFD was evaluated by comparing the lower limits of detection for PCR, RPA, and RPA-LFD.
Practicality study for the RPA-LFD assay
The established RPA-LFD method was applied to both the spiked samples and the real sample to verify the feasibility of this study for field application. The natural seawater used for the simulated water sample experiment was obtained from the South China Sea on August – September in 2021, and after microscopic examination it was confirmed that the sample contained no target algal cells. A number of P. parvum cells were harvested by centrifugation at 12000 × g for 2 min before the obtained cells were used to prepare the spiked samples using natural seawater as diluents. After a ten-fold gradient dilution of the water sample was generated (1.12×105 cells/ml-1.12×10-2 cells/ml), genomic DNA was extracted and sensitive detection was performed. Finally, the real environmental water samples from Xiangshan and Sanmen Bay were applied to the practical study. Among them, Xiangshan water samples represented the red tide period. The accuracy of the RPA-LFD results was verified using PCR results.
Results
Primer screening and system optimization
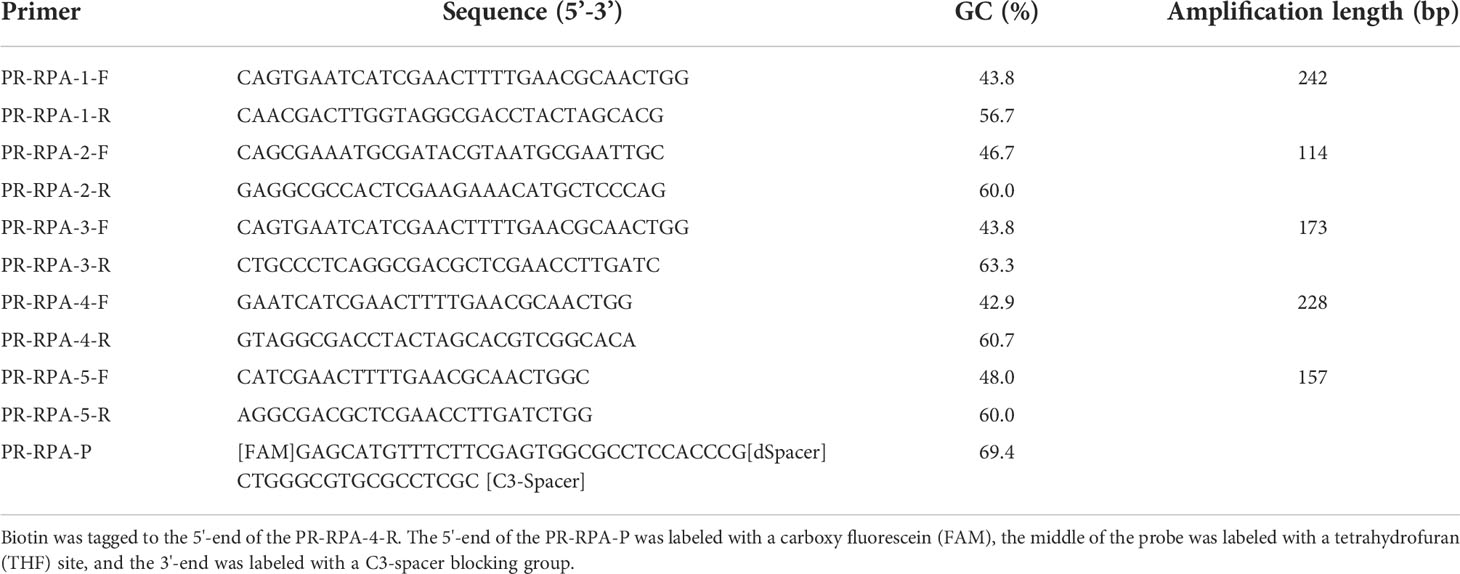
As revealed by Figure 1, of the five pairs of primers, only PR-RPA-4-F/R could amplify a clear and bright band. Thus, PR-RPA-4-F/R was selected as the optimum primer for subsequent RPA reactions. For the RPA-LFD reactions, biotin was tagged to the 5’-end of the PR-RPA-4-R, named PR-RPA-4-biotion.
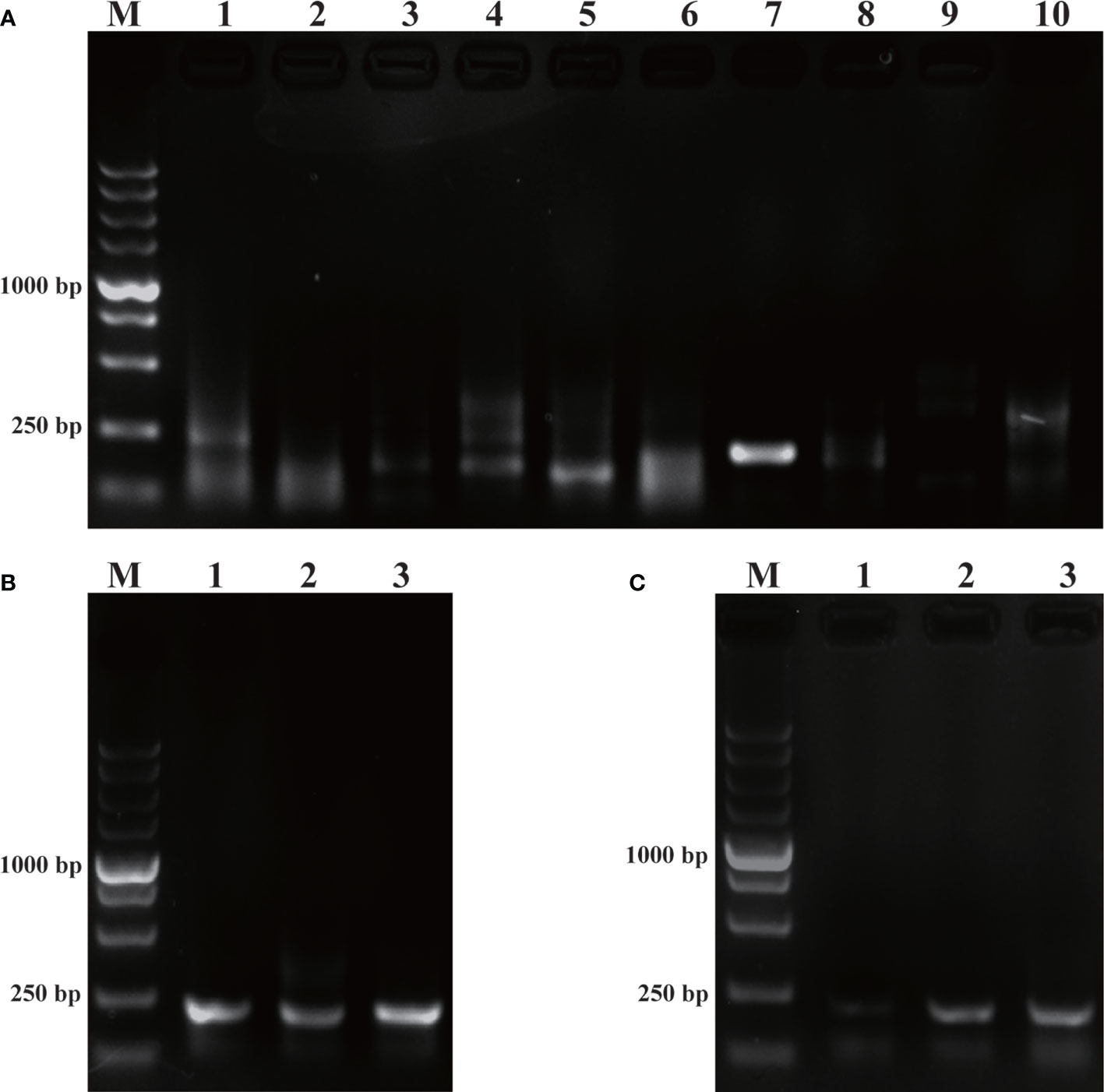
Figure 1 Primer screening and system optimization. (A): Primer screening results for P. parvum. M: DL5000 DNA marker; 1, 3, 5, 7, 9: positive control of primers 1-5; 2, 4, 6, 8, 10: negative control of primers 1-5. (B): Optimization of amplification temperature. 1: 35°C; 2: 37°C; 3: 39°C. (C): Optimization of amplification time. 1: 10 min; 2: 15 min; 3: 20 min.
Using the screened optimal primers, the RPA amplification temperature was initially optimized at a 20 min time-point. Meanwhile, the temperature was set to a range of 35°C, 37°C, 39°C (Figure 1B). Among the three amplification temperatures, the amplification band representing the 39°C setting was the brightest, so 39°C was selected as the optimal temperature for the RPA amplification system. Time optimization experiments were next performed at the optimal 39°C temperature. The time gradient tested was 10 min, 15 min, 20 min (Figure 1C). When the amplification time was 15 min or 20 min, the LFD bands were brighter than for 10 min, thus 15 min was selected as the optimal amplification time from an economical perspective. Subsequent RPA/RPA-LFD were amplified using the described optimal temperature and time.
Specificity study
The purpose of this study was to ensure accurate detection of P. parvum in complex environments. Thus, the primers and probes designed were required to be highly specific. As shown in Figure 2, the genomic DNA of the 10 species of algae used for specificity testing was successfully amplified with the universal primers (TW28 and AB28) for their ITS sequences; these data rule out possible interference due to degradation of genomic DNA of algae. Meanwhile, through interpretation of the results of PCR and RPA, it was identified that apart from for P. parvum, the results of the other algae amplification of all other algae species resulted in negative results in the RPA-LFD are all negative (Figures 3A, B). In addition, during the RPA-LFD detection of P. parvum, the control line and the test line were colored at the same time, while analysis of the other algae species resulted in coloring of the control line only (Figure 3C). Therefore, it was concluded that the primers and probes designed in this test are highly specific for P. parvum and could be used for further experimentation.
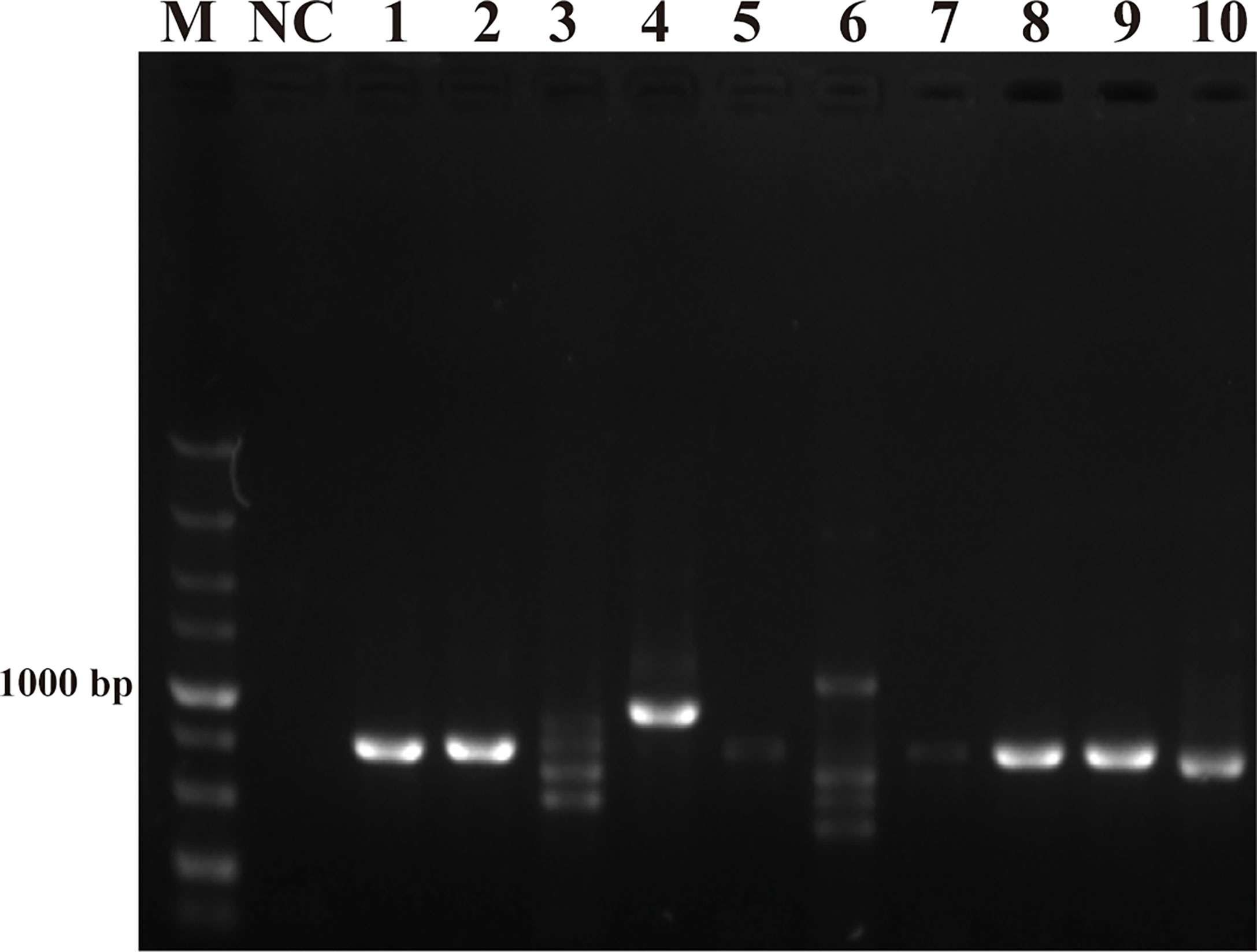
Figure 2 Universal primers for amplification of ITS sequences. M: DL5000 DNA marker; NC: negative control; 1: Prymnesium parvum; 2: Thalassiosira pseudonana; 3: Chaetoceros debilis; 4: Phaeodactylum tricornutum; 5: Pseudo-nitzschia multiseries; 6: Trichodesmium; 7: Skeletonema costatum; 8: Thalassiosira rotula; 9: Chaetoceros curvisetus; 10: Isochrysis galbana.
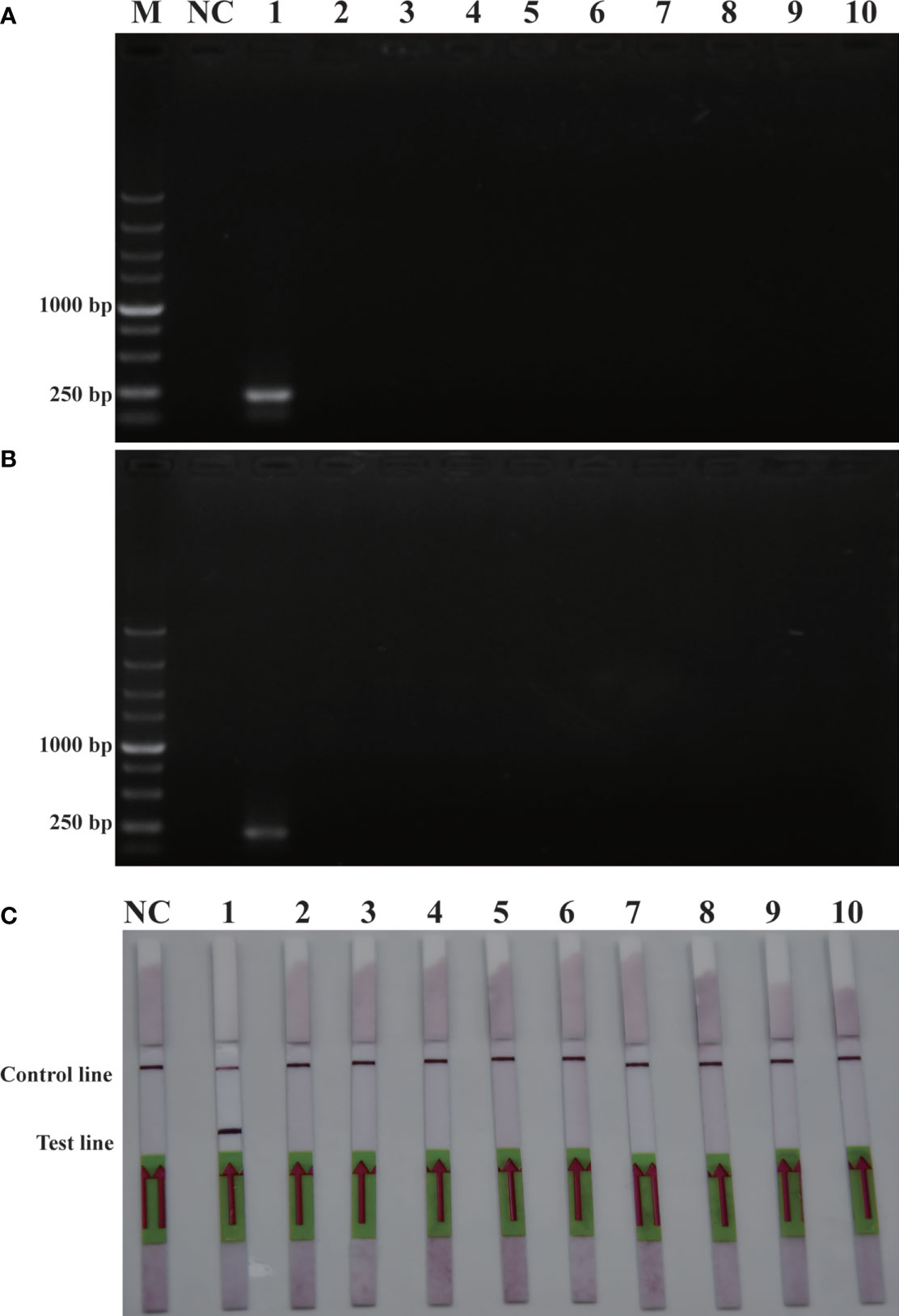
Figure 3 Specificity validation. (A): PCR; (B): RPA; (C): RPA-LFD. M: DL5000 DNA marker; NC: negative control; 1: P. parvum; 2: T. pseudonana; 3: C. debilis; 4: P. tricornutum; 5: P. multiseries; 6: Trichodesmium; 7: S. costatum; 8: T. rotula; 9: C. curvisetus; 10: Isochrysis galbana.
Sensitivity study
Sensitivity is an important parameter for on-site detection of harmful algae. The methodological sensitivity and applicability determine whether the method could be utilized to predict the occurrence of harmful algal blooms. The detection limit of the RPA-LFD was first quantified using the genomic DNA of P. parvum (1.5×104 pg/μL-1.5×10-3 pg/μL) in comparison to PCR and RPA (Figure 4). The results indicated that the detection limit of RPA-LFD (1.5×10-1 pg/μL) with genomic DNA was 1000 times higher than that of RPA alone (1.5×102 pg/μL) and 100 times that of PCR (1.5×101 pg/μL).
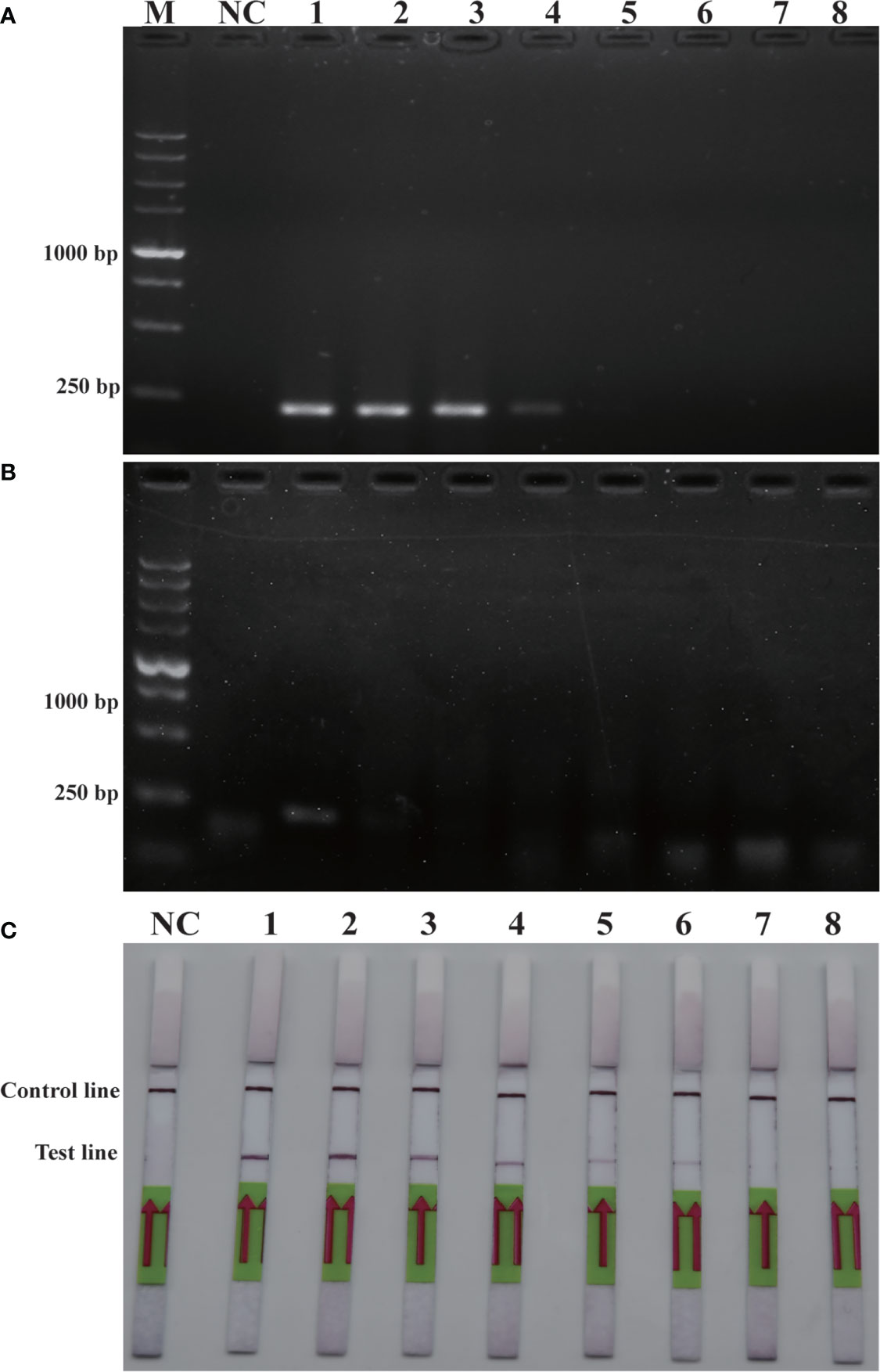
Figure 4 Comparison of sensitivities of PCR (A), RPA (B) and RPA-LFD (C) with gDNA. M: DL5000 DNA marker; NC: negative control; 1-8: gDNA concentration ranges from 1.5×104 pg/μL-1.5×10-3 pg/μL.
Similarly, sensitivity tests were performed using recombinant plasmid (2.35×105 pg/μL-2.35×10-2 pg/μL). After performing conventional PCR, RPA or RPA-LFD amplification, the results obtained were illustrated in Figure 5. The detection limit for PCR was 2.35×100 pg/μL, while the detection limit for RPA was 2.35×101 pg/μL, and the detection limit for RPA-LFD was 2.35×100 pg/μL. The plasmid detection sensitivity of RPA-LFD was therefore same as that of PCR, with both techniques being 10 times more sensitive than RPA. Compared to the sensitivity for genomic DNA detection, the sensitivity of RPA for plasmid DNA was significantly improved. Overall, the results were suggested that the sensitivity of RPA-LFD is higher than that of conventional PCR.
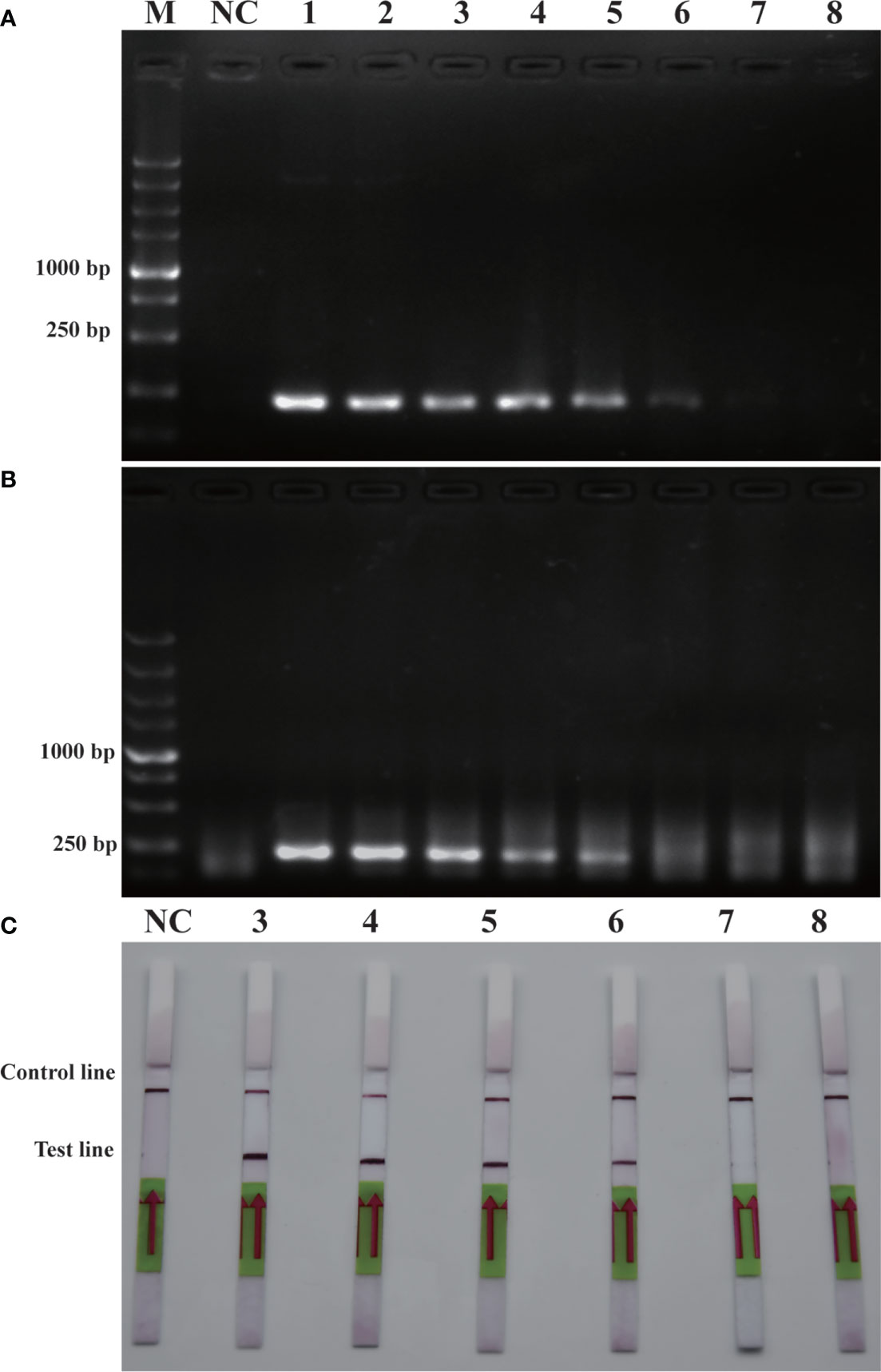
Figure 5 Comparison of sensitivities of PCR (A), RPA (B) and RPA-LFD (C) with plasmid. M: DL5000 DNA marker; NC: negative control; 1-8: plasmid concentration ranges from 2.35×105 pg/μL-2.35×10-2 pg/μL.
Practicality experiments on RPA-LFD
To verify the feasibility of the RPA-LFD assay for on-site environmental detection of P. parvum, practical experiments were conducted using spiked samples (Figure 6) and real samples (Figure 7). The results revealed that using the primers and probes optimized in this study, PCR, RPA and RPA-LFD successfully detected P. parvum in spiked samples effectively applied during on-site detection. In addition, RPA-LFD was more sensitive than PCR and RPA, with a detection limit of 1.12×10-1 cells/ml. However, in the experiment with real environment water samples, both PCR and RPA-LFD test results were negative, which demonstrated that the environmental water sample currently did contain of P. parvum cells.
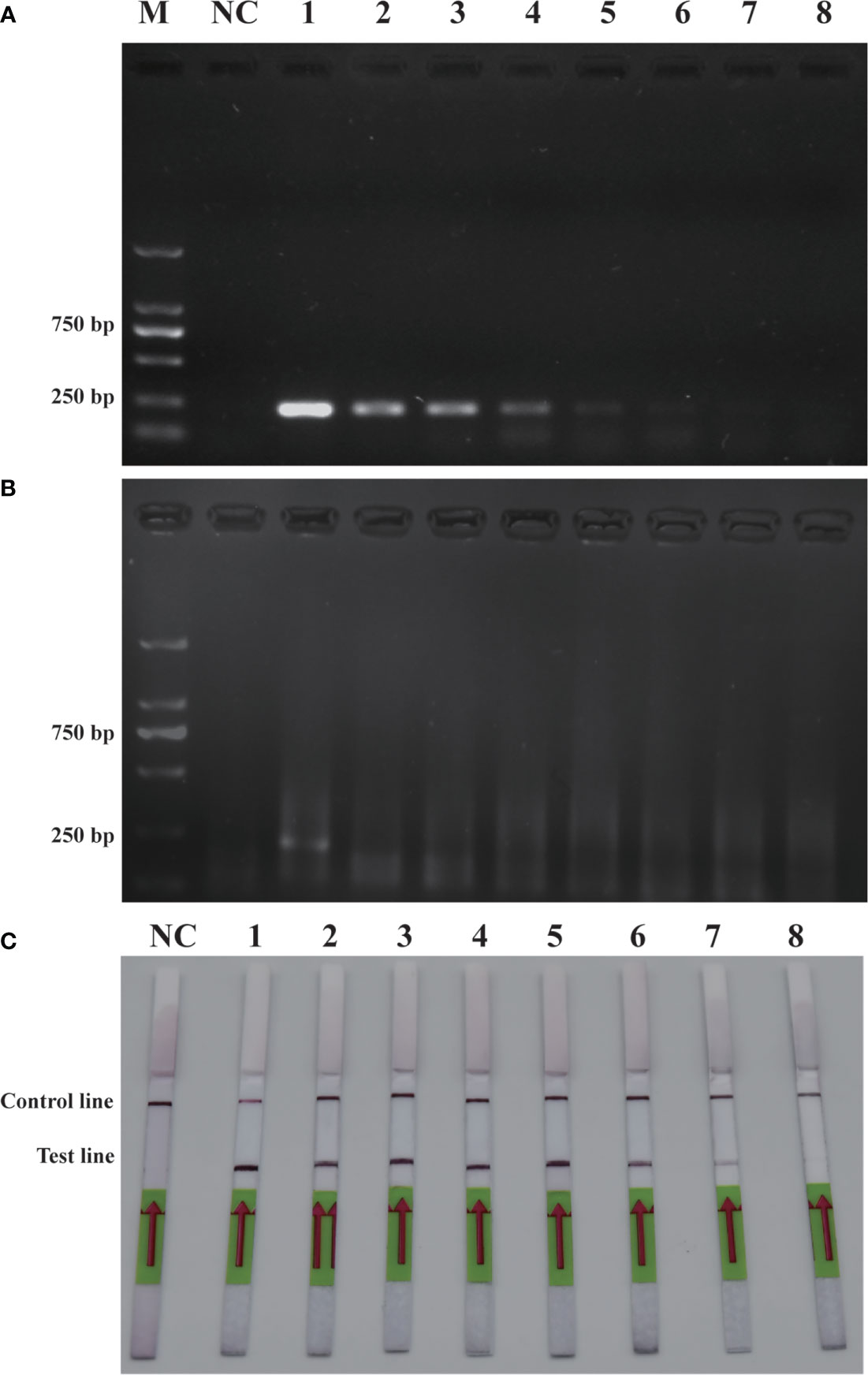
Figure 6 Comparison of sensitivities of PCR (A), RPA (B) and RPA-LFD (C) with gDNA of spiked sample. M: DL2000 DNA marker; NC: negative control; 1-8: P. parvum cell concentration ranges from 1.12×105 cells/ml-1.12×10-2 cells/ml.
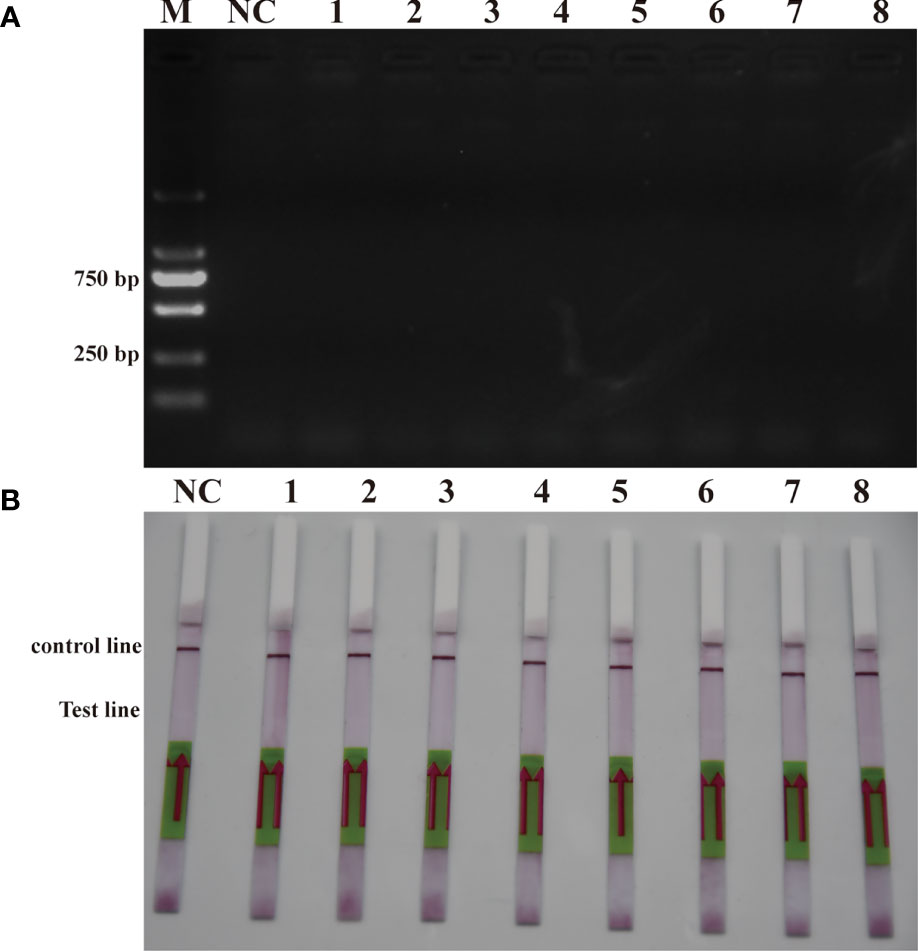
Figure 7 Real sample detection (A): PCR; (B): RPA-LFD. M: DL2000 DNA marker; NC: negative; 1: Xiangshan 1; 2: Xiangshan 2; 3: Sanmen Bay 1; 4: Sanmen Bay 2; 5: Sanmen Bay 3; 6: Sanmen Bay 4; 7: Sanmen Bay 5; 8: Sanmen Bay 6.
Discussion
As a toxic haptophyte alga, Prymnesium parvum has caused cases of algal bloom outbreaks globally, causing huge economic losses. Therefore, it has attracted increasing levels of attention from researchers. However, most of the current research on P. parvum is focused on toxins and growth factors (Larsen and Bryant, 1998; McCoy Gary et al., 2014; Liu et al., 2022). To date, many tools have been developed for the detection of P. parvum; microscopic detection and PCR are still widely utilized, however, these methods are generally time-consuming and laborious. To the best of our knowledge, RPA-LFD-based approaches have not yet been established for monitoring aquatic environments for P. parvum.
In the present study, an RPA-LFD detection method targeted to the ITS sequence of P. parvum was established. The detection limit of the RPA-LFD for the genomic DNA of the algae was 1.5×10-1 pg/μL, and the detection limit for plasmids was 2.35×100 pg/μL, not inferior to conventional PCR. In addition, the RPA-LFD assay took approximately 20 min in its entirety, substantially faster than the 1.5 hours taken for conventional PCR. Moreover, in comparison with other isothermal amplification methods for detecting P. parvum, RPA-LFD method possesses the significant advantage of high sensitivity. For example, Zhu et al. (2019) demonstrated that the limit of detection was 3×101 pg/μL when LAMP-LFD was used to detect P. parvum. The detection limit of the RPA-LFD utilized in the present study (1.5×10-1 pg/μL) was 200 times higher than that of the that previously recorded for LAMP-LFD. Therefore, the RPA-LFD may be useful in rapid and sensitive detection of P. parvum in the laboratory or in the field while maintaining high specificity. As a caveat, in our study, the results of sensitivity of RPA-AGE were much lower than that of RPA-LFD and PCR, which was inconsistent with findings from other studies. The reason for this phenomenon may be related to the method of product purification used in RPA. In this study, we used a PCR product purification kit, while others have used phenol-chloroform mixtures for purification; the latter may have resulted in a loss of RPA product. This result in fact reflected the advantages of RPA-LFD, because the technique does not require purification of RPA products.
Unfortunately, RPA reactions produce more non-specific amplifications relative to PCR, a difficult phenomenon to avoid (Kim et al., 2017). Therefore, it is necessary to screen primers to minimize the interference of non-specific amplification for subsequent experiments. RPA is an emerging method, and many researchers have conducted studies regarding the optimization of this method, including buffered conditions, specificity enhancement, stirred conditions, and additional methods of detection such as RPA combined with LFD (Moody et al., 2016; Luo et al., 2019; Tomar et al., 2022). It is worth noting that combination of RPA with the latest CRISPR technology has been applied to various bioassay fields (Patchsung et al., 2020; Meng et al., 2021; Jiang H.J. et al., 2022; Zhao et al., 2022). For example, an RPA-CRISPR/Cas13a detection system named Sherlock has been developed (Gootenberg et al., 2017). The combination of CRISPR and RPA resulted in high specificity and sensitivity of detection of RPA assays. At present, however, there are still few applications employed for the detection of harmful algal blooms species, and there is a huge potential for development (Durán-Vinet et al., 2021). In the future, it would be of great benefit to design RPA-CRISPR-LFD method for the rapid on-site detection of harmful algal bloom species.
To verify that the application of the RPA-LFD is reliable and stable for the analysis of field samples, mock environment sample groups were established by spiking P. parvum into natural seawater in the absence of target algal cells. In the present study, we initially spiked samples to verify the feasibility of the method in the detection of field samples, and then tested using a real sample. However, the final experimental results demonstrated no presence of P. parvum in the real samples used in this study. The most likely reason for this result was that the environmental water sample contained no P. parvum. Therefore, additional field samples isolated during different seasons should be used for testing, perhaps during periods of algal bloom.
Conclusion
This study established a rapid, highly specific and sensitive RPA-LFD method for detection of Prymnesium parvum. The method exhibits advantageous simple operation, short detection time and visual detection results. RPA-LFD requires just 20 min for the experimental process, substantially less than for PCR. Furthermore, this method can be employed in field for detection of P. parvum, and compared with traditional PCR, its sensitivity is substantially improved. These advantages mean this novel method can be used for early detection of P. parvum and prevention of algal bloom. Meanwhile, the RPA-LFD established in this study may also be utilized in largescale field screening for P. parvum.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
NL: Conceptualization, Methodology, Investigation, Data Curation, Writing-Original Draft; HH: Conceptualization, Methodology, Writing-Review and Editing, Supervision, Funding acquisition; HJ: Conceptualization, Methodology, Writing-Review and Editing, Supervision, Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (Grant No. 91951111, No. 32170108, and No. 42188102) and the Independent Research Projects of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai, Grant No. SML2021SP204).
Acknowledgments
We are grateful to Prof. H.J Zhang from Ningbo University for help in field sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1032847/full#supplementary-material
References
Ayfan Abdulrahman K., Macdonald J., Harris P. N. A., Heney C., Paterson D. L., Trembizki E., et al. (2021). Rapid detection of NDM and VIM carbapenemase encoding genes by recombinase polymerase amplification and lateral flow-based detection. Eur. J. Clin. Microbiol. Infect. Dis.: Off. Publ. Eur. Soc. Clin. Microbiol. 40 (11), 2447–2453. doi: 10.1007/s10096-021-04267-6
Beyer J. E., Zamor R. M., Hambright K. D. (2019). Detection limits affect the predictability of the presence of an invasive harmful alga across geographic space. Biol. Invasions 21 (7), 2301–2311. doi: 10.1007/s10530-019-01977-z
Cao G. H., Huo D. Q., Chen X. L., Wang X. F., Zhou S. Y., Zhao S. X, et al. (2022). Automated, portable, and high-throughput fluorescence analyzer (APHF-analyzer) and lateral flow strip based on CRISPR/Cas13a for sensitive and visual detection of SARS-CoV-2. Talanta 248, 123594–123594. doi: 10.1016/j.talanta.2022.123594
Daher R. K., et al, Stewart G., Boissinot M., Bergeron M. G. (2014). Isothermal recombinase polymerase amplification assay applied to the detection of group b streptococci in vaginal/anal samples. Clin. Chem. 60 (4), 660–666. doi: 10.1373/clinchem.2013.213504
Durán-Vinet B., Araya-Castro K., Chao T. C., Wood S. A., Gallardo V., Godoy K, et al. (2021). Potential applications of CRISPR/Cas for next-generation biomonitoring of harmful algae blooms: A review. Harmful Algae 103, 102027–102027. doi: 10.1016/j.hal.2021.102027
Eckford-Soper L. K., Daugbjerg N. (2015). Examination of six commonly used laboratory fixatives in HAB monitoring programs for their use in quantitative PCR based on taqman probe technology. Harmful Algae 42, 52–59. doi: 10.1016/j.hal.2014.12.007
Fu M. Q., Chen G. F., Zhang C. Y., Wang Y. Y., Sun R., Zhou J. (2019). Rapid and sensitive detection method for Karlodinium veneficum by recombinase polymerase amplification coupled with lateral flow dipstick. Harmful Algae 84, 1–9. doi: 10.1016/j.hal.2019.01.011
Fu M. Q., Yang Y. Y., Zhang C. Y., Chen G. F., Wang Y. Y., et al. (2020). Recombinase polymerase amplification combined with lateral-flow dipstick for rapid detection of Prorocentrum minimum. J. Appl. Phycol. 32, 1–14. doi: 10.1007/s10811-020-02079-3
Galluzzi L., Bertozzini E., Penna A., Perini F., Pigalarga A., Graneli E, et al. (2008). Detection and quantification of Prymnesium parvum (Haptophyceae) by real-time PCR. Lett. Appl. Microbiol. 46 (2), 261–266. doi: 10.1111/j.1472-765X.2007.02294.x
Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356 (6336), 438–442. doi: 10.1126/science.aam9321
Guo M., Harrison P. J., Taylor F. J. R. (1996). Fish kills related to Prymnesium parvum n. carter (Haptophyta) in the people's republic of China. J. Appl. Phycol. 8 (2), 111–117. doi: 10.1007/BF02186313
Jiang T. T., Wang Y. C., Jiao W. W., Song Y. Q., Zhao Q., Wang T. Y., et al. (2022). Recombinase polymerase amplification combined with real-time fluorescent probe for Mycoplasma pneumoniae detection. J. Clin. Med. 11 (7), 1780–1780. doi: 10.3390/jcm11071780
Jiang H. J., Tan R., Jin M., Yin J., Gao Z. X., Li H. B, et al. (2022). Visual detection of Vibrio parahaemolyticus using combined CRISPR/Cas12a and recombinase polymerase amplification. Biomed. Environ. Sci. 35 (6), 518–527. doi: 10.3967/bes2022.069
Kang J. Y., Jang H., Yeom G., Kim M. G. (2020). Ultrasensitive detection platform of disease biomarkers based on recombinase polymerase amplification with h-sandwich aptamers. Analytical Chem. 93 (2), 992–1000. doi: 10.1021/acs.analchem.0c03822
Karakkat B., Hockemeyer K., Franchett M., Olson M., Mullenberg C., Koch P., et al. (2018). Data for designing two isothermal amplification assays for the detection of root-infecting fungi on cool-season turfgrasses. Data Brief 20, 471–479. doi: 10.1016/j.dib.2018.08.021
Kim J. M., truongatai Yoon B. (2017). Comparison between specific DNA-amplifications using recombinase polymerase amplification (RPA) and using polymerase chain reaction (PCR). J. Apicult. 32 (1), 41–50. doi: 10.17519/apiculture.2017.04.32.1.41
Larsen A., Bryant S. (1998). Growth rate and toxicity of Prymnesium parvum and Prymnesium patelliferum(haptophyta) in response to changes in salinity, light and temperature. Sarsia 83 (5), 409–418. doi: 10.1080/00364827.1998.10413700
Li J. J., Wang C. M., Yu X., Lin H. R., Hui C., Shuai L, et al. (2019). Rapid detection of Cyanobacteria by recombinase polymerase amplification combined with lateral flow strips. Water Supply 19 (4), 1181–1186. doi: 10.2166/ws.2018.174
Lillis L., Siverson J., Lee A., Cantera J., Parker M., Piepenburg O., et al. (2016). Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol. Cell. Probes 30, 74–78. doi: 10.1016/j.mcp.2016.01.009
Lin H., Zhao S ., Liu Y. H., Shao L., Ye Y. Y., Jiang N. Z., et al. (2022). Rapid visual detection of Plasmodium using recombinase-aided amplification with lateral flow dipstick assay. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.922146
Liu M., Li C. C., Luo X. L., Ma F., Zhang C. Y. (2020). 5-hydroxymethylcytosine glucosylation-triggered helicase-dependent amplification-based fluorescent biosensor for sensitive detection of β-glucosyltransferase with zero background signal. Analytical Chem. 92 (24), 16307–16313. doi: 10.1021/acs.analchem.0c04382
Liu S. Y., Zhao R. Z., Qiu X. C., Guo Q. (2022). Optimization analysis to evaluate the relationships between different ion concentrations and Prymnesium parvum growth rate. Water 14 (6), 928–928. doi: 10.3390/w14060928
Lomidze L., Williford T. H., Musier-Forsyth K., Kankia B. (2018). Isothermal amplification of long DNA segments by quadruplex priming amplification. Analytical Methods: Advancing Methods Appl. 10 (25), 2972–2979. doi: 10.1039/c8ay00843d
Luo G. C., Yi T. T., Jiang B., Guo X. L., Zhang G. Y. (2019). Betaine-assisted recombinase polymerase assay with enhanced specificity. Analytical Biochem. 575, 36–39. doi: 10.1016/j.ab.2019.03.018
Manning S. R., Claire J. W. (2010a). Multiplex PCR methods for the species-specific detection and quantification of Prymnesium parvum (Haptophyta). J. Appl. Phycol. 22 (5), 587–597. doi: 10.1007/s10811-009-9498-6
Manning S. R., La Claire J. W. (2010b). Prymnesins: Toxic metabolites of the golden alga, Prymnesium parvum carter (Haptophyta). Mar. Drugs 8 (3), 678–704. doi: 10.3390/md8030678
Manning S. R., La Claire J. W.I. I. (2013). Isolation of polyketides from Prymnesium parvum (Haptophyta) and their detection by liquid chromatography/mass spectrometry metabolic fingerprint analysis. Analytical Biochem. 442 (2), 189–195. doi: 10.1016/j.ab.2013.07.034
Maria D., Genitsaris S., Mazaris A. D., Kyparissis A., Voutsa D., Kozari A., et al. (2022). A catastrophic change in a european protected wetland: From harmful phytoplankton blooms to fish and bird kill. Environ. Pollution 312, 120038. doi: 10.1016/j.envpol.2022.120038
McCoy Gary R., Kegel J. U., Touzet N., Fleming G. T. A., Medlin L K., Raine R., et al. (2014). An assessment of RNA content in Prymnesium parvum, prymnesium polylepis, cf. chattonella sp. and Karlodinium veneficum under varying environmental conditions for calibrating an RNA microarray for species detection. FEMS Microbiol. Ecol. 88 (1), 140–159. doi: 10.1111/1574-6941.12277
Meng Q. Z., Yang H. M., Zhang G. Q., Sun W. J., Ma P. X., Liu X. Y., et al. (2021). CRISPR/Cas12a-assisted rapid identification of key beer spoilage bacteria. Innovative Food Sci. Emerg. Technol. 74, 1–9. doi: 10.1016/j.ifset.2021.102854
Moody C., Newell H., Viljoen H., et al (2016). A mathematical model of recombinase polymerase amplification under continuously stirred conditions. Biochem. Eng. J. 112, 193–201. doi: 10.1016/j.bej.2016.04.017
Nai Y. H., Doeven E. H., Guijt R. M. (2022). An improved nucleic acid sequence-based amplification method mediated by T4 gene 32 protein. PloS One 17 (3), e0265391–e0265391. doi: 10.1371/journal.pone.0265391
Patchsung M., Jantarug K., Pattama A., Aphicho K ., Suraritdechachai S., Meesawat P., et al. (2020). Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 4 (12), 1140–1149. doi: 10.1038/s41551-020-00603-x
Piepenburg O., Williams C. H., Stemple D. L., Armes N. A. (2006). DNA Detection using recombination proteins. PloS Biol. 4 (7), e204. doi: 10.1371/journal.pbio.0040204
Qin J. L., Hu Z. X., Zhang Q., Xu N., Yang Y. F. (2020). Toxic effects and mechanisms of prymnesium parvum (Haptophyta) isolated from the pearl river estuary, China. Harmful Algae 96 (C), 101844. doi: 10.1016/j.hal.2020.101844
Roelke D. L., Barkoh A., Brooks B. W., Grover J. P., Hambright K. D., Laclaire J. W. II, et al. (2016). A chronicle of a killer alga in the west: ecology, assessment, and management of Prymnesium parvum blooms. Hydrobiologia 764 (1), 29–50. doi: 10.1007/s10750-015-2273-6
Seoane S., Riobó P., Franco J. (2016). Haemolytic activity in different species of the genus prymnesium (Haptophyta). J. Mar. Biol. Assoc. United Kingdom 97 (3), 491–496. doi: 10.1017/S0025315416001077
Sharifan H., Ma X. M. (2017). Potential photochemical interactions of UV filter molecules with multichlorinated structure of prymnesins in harmful algal bloom events. Mini-Rev. Org. Chem. 14 (5), 391–399. doi: 10.2174/1570193X14666170518124658
Southard G. M., Fries L. T., Barkoh A. (2010). Prymnesium parvum: the Texas experience. J. Am. Water Resour. Assoc. 46 (1), 14–23. doi: 10.1111/j.1752-1688.2009.00387.x
Töbe K., Eller G., Medlin L. K. (2006). Automated detection and enumeration for toxic algae by solid-phase cytometry and the introduction of a new probe for Prymnesium parvum (Haptophyta: Prymnesiophyceae). J. Plankton Res. 28 (7), 643–657. doi: 10.1093/plankt/fbi147
Toldrà A., Jauset-Rubio M., Andree K. B., Fernandez-Tejedor M., Diogene J., Katakis I., et al. (2018). Detection and quantification of the toxic marine microalgae Karlodinium veneficum and Karlodinium armiger using recombinase polymerase amplification and enzyme-lined oligonucleotide assay. Analytica Chim. Acta 1039, 140–148. doi: 10.1016/j.aca.2018.07.057
Tomar S., Lavickova B., Guiducci C. (2022). Recombinase polymerase amplification in minimally buffered conditions. Biosensors Bioelectronics 198, 113802–113802. doi: 10.1016/j.bios.2021.113802
Wang L., Chen G. F., Zhang C. Y., Wang Y. Y., Sun R., et al. (2020). Application of loop-mediated isothermal amplification combined with lateral flow dipstick to rapid and sensitive detection of Alexandrium catenella. Environ. Sci. pollut. Res. Int. 27 (4), 4246–4257. doi: 10.1007/s11356-019-06889-y
Wang X. F., Xie S. L., Chen X. Y., Peng C., Xu X. L., Wei W., et al. (2020). A rapid and convenient method for on-site detection of MON863 maize through real-time fluorescence recombinase polymerase amplification. Food Chem. 324, 126821. doi: 10.1016/j.foodchem.2020.126821
White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications. Eds. Innis M., Gel-fand J., Sainsky J. (San Diego: Academic Press), 315–322.
Xia W. L., Chen K., Liu W. S., Yin Y., Yao Q., Ban Y., et al. (2022). Rapid and visual detection of Mycoplasma synoviae by recombinase-aided amplification assay combined with a lateral flow dipstick. Poultry Sci. 101 (7), 101860–101860. doi: 10.1016/j.psj.2022.101860
Yang X. Q., Wen X., Zhou C. X., Zhu X. J., Meng R., Luo Q.J, et al. (2018). Comparative study of brine shrimp bioassay-based toxic activities of three harmful microalgal species that frequently blooming in aquaculture ponds. J. Oceanol. Limnol. 36 (5), 1697–1706. doi: 10.1007/s00343-018-7140-7
Yang X. H., Zhang X., Wang Y., Shen H., Jiang G., Dong J. Q, et al. (2020). A real-time recombinase polymerase amplification method for rapid detection of Vibrio vulnificus in seafood. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.586981
Yang Q., Yang H. Y., Yuan N., Zuo S. A., Zhang Y. Z., Zhang W. (2022). Closed-tube saltatory rolling circle amplification with hydroxynaphthol blue for visual on-site detection of peanut as an allergenic food. Food Chem. 393, 133408–133408. doi: 10.1016/j.foodchem.2022.133408
Zamor R. M., Glenn K. L., Hambright K.D. (2012). Incorporating molecular tools into routine HAB monitoring programs: Using qPCR to track invasive prymnesium. Harmful Algae 15, 1–7. doi: 10.1016/j.hal.2011.10.028
Zhai Y. D., Li R. Q., Liu F. G., Zhang C. Y., Wang Y. Y., Chen G. F. (2021). Recombinase polymerase amplification combined with lateral flow dipstick for the rapid detection of Prorocentrum donghaiense. Mar. Biol. Res. 17, 7–8. doi: 10.1080/17451000.2021.2010279
Zhao G., Wang J., Yao C. Y., Xie P. C., Li X. M., Xu Z. L, et al. (2022). Alkaline lysis-recombinase polymerase amplification combined with CRISPR/Cas12a assay for the ultrafast visual identification of pork in meat products. Food Chem. 383, 132318–132318. doi: 10.1016/j.foodchem.2022.132318
Keywords: Prymnesium parvum, rapid detection, recombinase polymerase amplification, lateral flow dipstick, internal transcribed spacer (ITS)
Citation: Luo N, Huang H and Jiang H (2022) Establishment of methods for rapid detection of Prymnesium parvum by recombinase polymerase amplification combined with a lateral flow dipstick. Front. Mar. Sci. 9:1032847. doi: 10.3389/fmars.2022.1032847
Received: 07 September 2022; Accepted: 10 October 2022;
Published: 24 October 2022.
Edited by:
Tony Gutierrez, Heriot-Watt University, United KingdomCopyright © 2022 Luo, Huang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailong Huang, huanghailong@nbu.edu.cn; Haibo Jiang, jianghaibo@nbu.edu.cn
 Ningjian Luo
Ningjian Luo Hailong Huang
Hailong Huang Haibo Jiang
Haibo Jiang