CO1 barcodes resolve an asymmetric biphyletic clade for Diabrotica undecimpunctata subspecies and provide nucleotide variants for differentiation from related lineages using real-time PCR
- 1Department of Agricultural Biology, Colorado State University, Fort Collins, CO, United States
- 2Pest Identification Technology Laboratory, USDA-APHIS-PPQ-Science and Technology, Fort Collins, CO, United States
- 3Systematic Entomology Laboratory, USDA-ARS, Washington, DC, United States
- 4Plant Pest Diagnostics Branch, California Department of Food and Agriculture, Sacramento, CA, United States
Diabrotica undecimpunctata is a multivoltine polyphagous beetle species that has long been documented as a significant agricultural pest throughout its native range in North America. This beetle can vector bacterial and viral plant pathogens that result in major losses to crops such as cucumber and soybean. Many countries outside the Americas treat D. undecimpunctata as a species of quarantine importance, while in the USA only the subspecies D. u. duodecimnotata is subject to quarantine, to prevent introduction from Mexico. Identification of D. undecimpunctata on the basis of morphology alone can be complicated given the use of conflicting characters in the description of some subspecific taxa. To better understand relationships among D. undecimpunctata subspecies and other related species, we sequenced mitochondrial cytochrome oxidase 1 (CO1) and nuclear internal transcribed spacer 2 (ITS2) DNA from individuals in different subspecific taxa and across different parts of the species range using museum samples and interceptions. When our data were combined with publicly available Diabrotica data, no pattern of divergence consistent with the currently recognized subspecific designations was found. In addition, we compared phylogenetic patterns in CO1 data from the congener D. virgifera to demonstrate the utility of mitochondrial data in resolving subspecies. From the CO1 data, a diagnostic real-time PCR assay was developed that could successfully identify all haplotypes within the large D. undecimpunctata clade for use in surveys and identification at ports of entry. These findings underscore the need to resolve molecular and morphological datasets into cogent, lineage-based groupings. Such efforts will provide an evolutionary context for the study of agriculturally important attributes of Diabrotica such as host preferences, xenobiotic metabolism, and natural and anthropogenic patterns of dispersal.
1 Introduction
The leaf beetle species Diabrotica undecimpunctata Mannerheim (Coleoptera, Chrysomelidae) has been documented as a serious pest of agricultural importance in North America for well over 100 years (1–3). Adult D. undecimpunctata feed on more than 50 plant species, including food, feed, and fiber crops such as alfalfa, apple, beans, beets, cotton, cucurbits, hemp, maize, peach, peas, potato, sunflower, and tomato, as well as causing damage to ornamentals such as canna, carnation, dahlia, peony, rose, and wisteria (4–7). The larvae are root feeders and are often found on maize roots but have also been frequently noted on other species, including several grass species, alfalfa, and peas (5, 7, 8). While the economic impact of D. undecimpunctata is less than the US$1 billion annual estimate associated with D. virgifera (9), D. undecimpunctata does account for serious economic losses during large infestations (6, 10, 11). One of the most problematic aspects of D. undecimpunctata feeding is the transmission of the virulent bacterial pathogen Erwinia tracheiphila (12, 13), which can cause rapid crop failure among cucurbitaceous species (14). In addition to transmission of E. tracheiphila, D. undecimpunctata is a competent vector of the bean pod mottle virus, which can cause losses in soybean (15). On account of these concerns and the potential for establishment outside the historic species range (16), D. undecimpunctata was recently categorized as an A1 quarantine pest under Annex IIA by the Panel on Plant Health of the European Food Safety Authority (17).
Given the importance of D. undecimpunctata to agriculture, it has been the subject of several taxonomic revisions [summarized in (18)]. The currently accepted taxonomic arrangement of D. undecimpunctata subdivides the species into four subspecies based primarily on morphological differences (19): D. u. undecimpunctata Mannerheim 1843, D. u. duodecimnotata Harold 1875, D. u. howardi Barber 1947, and D. u. tenella LeConte 1858. Difficulty in differentiating between D. undecimpunctata subspecies, such as D. u. howardi and D. u. tenella, on the basis of morphology has been previously noted [e.g. (20)]. Some molecular phylogenetic work has been conducted in Diabrotica and related lineages, but much of this has been at the genus level and above, with very little at the subspecies level (21–23). Generating DNA sequence data sampled at the subspecific level for D. undecimpunctata will provide a genetic genealogical history that pertains to subspecific designations for this group and will ultimately improve taxonomic, agronomic, and phylogeographic studies.
Historically, Diabrotica has been split into the three species groups fucata, signifera, and virgifera (24). While less well studied than the spread of species in the virgifera species group, it is thought that the expansion and establishment of D. undecimpunctata in North American agroecosystems has also been the result of human activities, such as the large-scale cultivation of maize, cucurbits, and other host plants, as well as trade in agricultural commodities (25–27). To this point, D. u. duodecimnotata is frequently intercepted on agricultural commodities imported from Mexico into the USA during border inspections. The subspecies D. u. duodecimnotata is considered a quarantine pest by the United States Department of Agriculture (USDA) based on its potential to cause damage to agricultural crops. Diabrotica beetles have also been a concern outside the Americas since the introduction of D. virgifera virgifera to Europe in 1992, with its subsequent impact on agricultural production (28, 29). Given the limited molecular data sampled at the subspecific level for D. undecimpunctata and the concerns regarding introduction and establishment outside its native range, we generated mitochondrial cytochrome oxidase 1 (CO1) and nuclear 45S internal transcribed spacer 2 (ITS2) sequence data from intercepted and museum specimens to address the following questions: (1) can current or novel subspecies of D. undecimpunctata be recognized via CO1 and/or ITS2 sequence data? and (2) do CO1 data provide fixed nucleotide variants for the development of a reliable real-time PCR assay to separate all subspecies of D. undecimpunctata from closely related species?
2 Materials and methods
2.1 Sample acquisition
From 2018 to 2019, 254 Diabrotica beetles were identified at ports of entry coming from Mexico into the USA. Of these, 115 samples were preserved in absolute ethanol for later study. In addition to these beetles acquired from interceptions, loans of Diabrotica beetles were made from the Smithsonian National Museum of Natural History, the American Museum of Natural History, the California Department of Food and Agriculture, and the University of California at Berkeley and Davis. The 18 most recently collected museum specimens, mainly from the Smithsonian, were sampled for non-destructive DNA extraction and sequencing. Specimens used are summarized in Table 1 and sample details for sequences generated in this study can be found using GenBank accessions OQ649623–OQ649739 for CO1 and OQ641612–OQ641622 for ITS2.
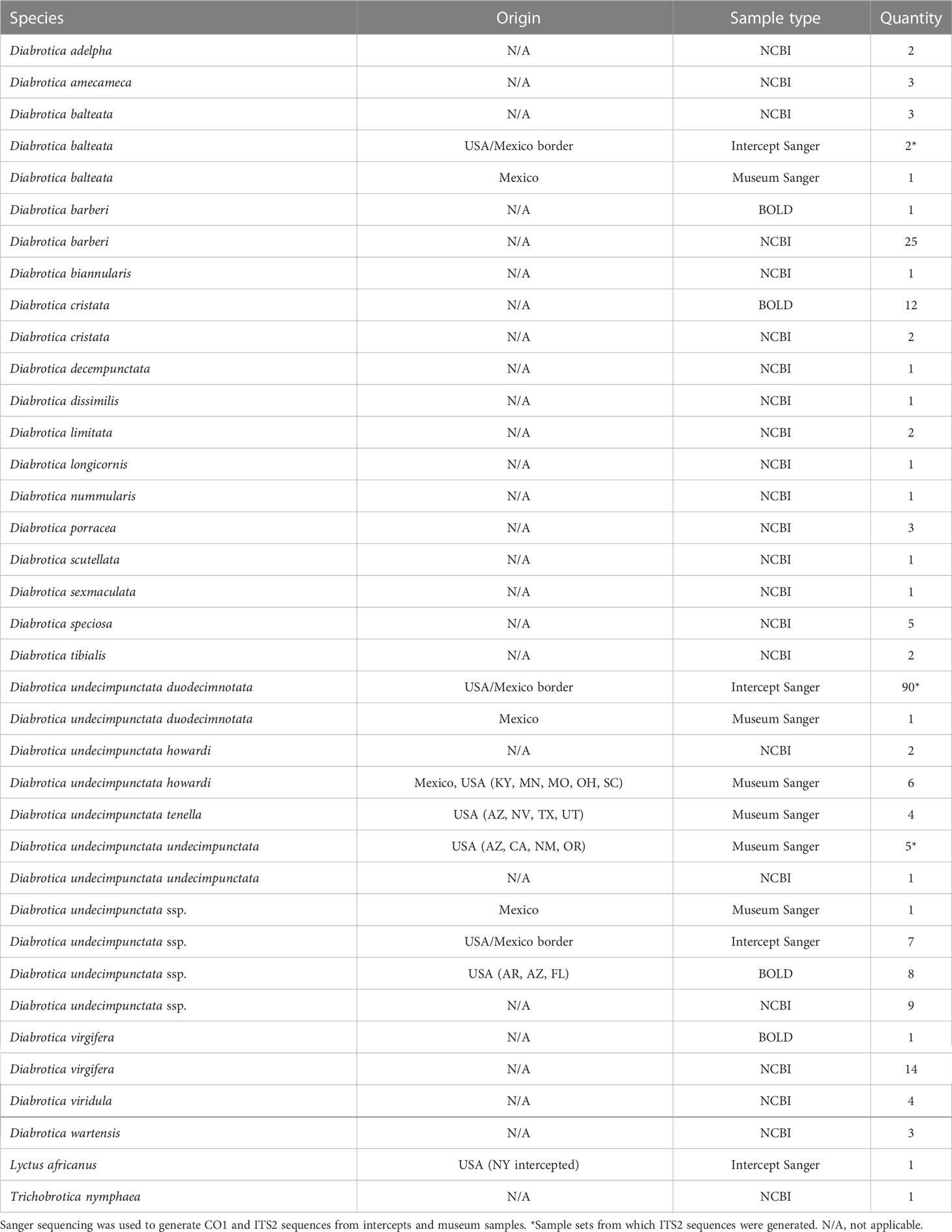
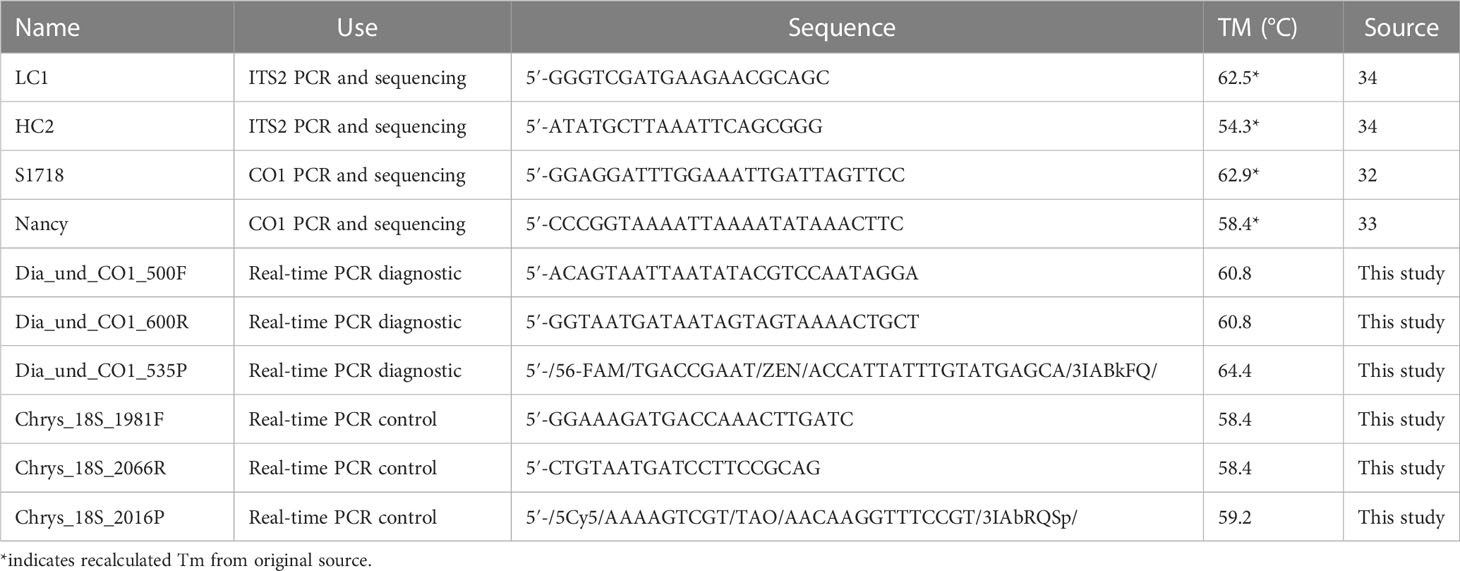
Table 1 Samples used to resolve maternal relationships among Diabrotica species and for development and/or testing of a real-time PCR assay.
2.2 DNA extraction
DNA was extracted from individual adult specimens of Diabrotica preserved in ethanol or from pinned museum specimens using a Lucigen MasterPure DNA extraction kit (Lucigen Corp., Middleton, WI, USA). Isolation of DNA from intercepted specimens preserved in ethanol was preceded by drying the specimens in 1.5mL microcentrifuge tubes on a digital dry bath set to 55°C for 20 to 30 minutes. Museum specimens were removed from their pins. All individual, whole dried specimens were placed in 1.5mL microcentrifuge tubes, immersed in 300 μL of Tissue and Cell Lysis Solution and 1 μL of Proteinase K, and heated to 65°C overnight on an Eppendorf ThermoMixer FP (Eppendorf SE, Hamburg, Germany) at 500 rpm. After overnight incubation, specimens were removed from the lysis buffer, rinsed with absolute ethanol, and returned to pins (museum specimens) or ethanol (intercepted specimens). The remaining extraction was carried out following the manufacturer’s instructions with modifications as described by Zink et al. (30). After elution, DNA concentration and purity were measured for a 2μL sample using a NanoDrop 2000 v 1.6 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Two readings were taken to ensure machine consistency. Throughout these and the following steps, all equipment and materials were sanitized between steps and filter tips were used to handle any liquids containing DNA to prevent contamination. Negative controls were employed during all DNA extraction and PCR steps to test for contamination.
2.3 CO1 and ITS2 PCR amplification and sequencing
All PCRs were performed on a Bio-Rad C1000 Touch thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). In order to generate sequences that could be aligned to previous Diabrotica CO1 datasets (22, 31) we employed the universal primers S1718 (32) and Nancy (33; Table 2). An optimized PCR protocol was used due to the degraded nature of many of the samples. The 50 μL reactions contained 32.75 μL molecular grade H2O, 5.00 μL 10× Ex Taq buffer, 4.00 μL deoxynucleoside triphosphate (dNTP) mixture at 2.5 mM, 200 nM S1718 forward primer, 200 nM Nancy reverse primer, 1.25 mM MgCl2, 0.05 mg bovine serum albumin, 1 unit of TaKaRa Ex Taq HS polymerase (Takara Bio Inc., Shiga, Japan), and 1 μL of DNA template of varying concentration or autoclaved ddH2O for no-tissue controls. The thermocycler protocol included an initial denaturation step of 94°C for 3 min, followed by 40 cycles of 94°C for 20 s, 52°C for 20 s, 72°C for 30 s, and a final extension at 72°C for 5 min. A lid temperature of 105°C was maintained throughout all cycles. A subset of samples amplified for CO1 showing haplotypic diversity were also amplified for ITS2 so that mitochondrial relationships could be compared against nuclear data. The general primers (located in 5.8S and 28S) and thermocycler protocol described in Navajas et al. (34) were used to generate complete ITS2-spanning amplicons for comparison with previously generated data (21); the reaction mixture employed was otherwise identical to that employed for the CO1 PCRs described above. Success of PCRs was confirmed on 1% agarose gels containing ethidium bromide and imaged with a UV light source (Analytik Jena, LLC, Jena, Germany). Reactions with visible bands were purified using a Qiagen QIAquick PCR purification kit following the manufacturer’s instructions (Qiagen Inc., Hilden, Germany). After purification, CO1 PCR products were sequenced at the University of Chicago Comprehensive Cancer Center DNA Sequencing Facility on an Applied Biosystems 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA) using the same primers as for amplification. The ITS2 amplicons were sequenced with a 3730XL by Genewiz (Azenta Life Sciences Inc., Chelmsford, MA, USA). Sequences were manually trimmed to remove poor-quality base calls from the 5′ and 3′ ends (including primer sequences), assembled into contigs, and converted into consensus sequences in Geneious Prime 2021.0.31 for use in subsequent analyses.
2.4 Phylogenetic and network analyses of DNA sequence data
The 117 consensus CO1 sequences generated in this study were combined with 85 Diabrotica sequences from GenBank and 22 from the Barcode of Life Data System (35). The combined dataset was aligned using MAFFT v 7.450 (36, 37) with default settings. Once aligned, the matrix was trimmed to exclude non-overlapping sequences to a size of 227 samples by 420 nucleotides and realigned using the same method as above. With the symmetrical matrix, a neighbor joining (NJ) distance tree was resolved using the Tamura-Nei distance model (38) with another chrysomelid in the same subtribe Trichobrotica nymphaea (AY242440) set as the outgroup and 1,000 jackknife replicates to assess branch support. Using MrBayes 3.2.6 (39), Bayesian inference (BI) was run with a general time reversible + invariable sites + gamma rate variation among sites (GTR+I+G) substitution and rate variation model, four gamma categories, four heated chains, a chain length of 1,100,000, a subsampling frequency of 200, a burn-in of 100,000, and an unconstrained branch length prior. A second MrBayes run was conducted with the JC69 nucleotide substitution model (all other settings the same as above) to assess for any differences in tree topology and branch support when using a less complex substitution model. A subset of 10 D. undecimpunctata samples were selected for amplification of ITS2 based on CO1 haplotype diversity. The same steps as for analysis of CO1 sequences were followed for ITS2 but with a trimmed matrix of 26 samples by 530 loci (including gaps) and the use of Acalymma vittatum (AF278557) as an outgroup. A dataset downloaded from GenBank (318 samples by 617 nucleotides) of CO1 sequences from the two subspecies of D. virgifera was analyzed (along with three outgroup taxa) using the above techniques and was employed as a basis for comparison with D. undecimpunctata given the phylogenetic and taxonomic similarities between these lineages. In order to visually resolve informative single nucleotide variants (SNVs) between CO1 haplotypes most closely related to D. undecimpunctata (based on the BI tree), the original trimmed matrix was reduced to a 156 sample by 420 nucleotide matrix and analyzed with the TCS statistical parsimony algorithm (40) as implemented in PopART 1.7 (41).
2.5 Diagnostic real-time PCR primer and probe design
The 227 sample by 420 nucleotide alignment of CO1 sequences (Supplementary Data Sheet 1) was used for primer and probe design to target D. undecimpunctata and exclude all other Diabrotica lineages. The alignment was manually scanned for regions of high SNV density with respect to differences outside D. undecimpunctata and low SNV density within D. undecimpunctata using consensus base calls as a guide. From the regions meeting these criteria, separate ~30-bp windows for forward primers, reverse primers, and a probe were selected within 100- to 150-bp segments of the alignment for automated primer and probe design. Five primers and probes were designed using Primer 3 v 2.3.7 (42) within each window with the following parameter settings for primers: monovalent salt 50 mM (divalent 1.5 mM); primer DNA 50 nM; dNTP 0.6 mM; Tm °C [calculated using the SantaLucia method (43)] min. 50, opt. 55.5, max. 60; GC% min. 20, opt. 50, max. 80; primer length nt min. 14, opt. 22, max. 35; max. dimer Tm 47; max. poly-X 5; and max. 3′ stability 9. All settings were the same for probe design except for: Tm °C min. 57, opt. 60, max. 63; GC % min. 20, opt. 50, max. 80; and probe length nt min. 18, opt. 20, max. 36. From the five primers and probes designed from each window, the best primers and probe were chosen based on the highest number of nucleotide variants with respect to other closely related species, location of the nucleotide variants (primer/probe sequence ends preferred), highest Tm differential between primers and probe, and lowest level of self-dimerization, hairpin formation, self-annealing, and hetero-dimerization to the other oligos. These structural and thermodynamic tests were conducted using the IDT OligoAnalyzer2 and/or OligoCalc (44). BLASTn searches of the entire nt database were also employed to ensure specificity of the selected primer and probe sequences in combination. Control primers and probe were also designed using the same parameters but with an 18S rDNA dataset retrieved from a BLASTn alignment constrained by ‘Coleoptera’. Manual optimization employed similarity instead of difference in window selection and lower Tm with respect to the diagnostic primers and probe as final selection criteria. Primer and probe designs were submitted to IDT (Integrated DNA Technologies, Coralville, IA, USA) for synthesis and purification. Final primer and probe sequences can be found in Table 2.
2.6 Duplex real-time PCR optimization and testing
The Minimum Information for Publication of Quantitative Real-time PCR Experiments (MIQE) guidelines (45) were followed wherever applicable. All optimization and testing processes were carried out on a Bio-Rad CFX96 Touch Real-time PCR Detection System (Bio-Rad Laboratories, Inc.) in 96-well, thin-walled, white-well, hard-shell PCR plates (Bio-Rad Laboratories, Inc.) sealed with optically clear Microseal ‘B’ seals (Bio-Rad Laboratories Inc.). The assay was optimized for annealing temperature and primer and probe concentration for both loci. The reaction mix used was as follows: 10.00 μL of 2× iTaq Universal Probe Supermix (Bio-Rad Laboratories, Inc.); control primers and probe of 500nM Chrys_18S_1981F, 500nM Chrys_18S_2066R, 250nM Chrys_18S_2016P (labeled with 56-FAM reporter); diagnostic primers and probe of 500nM Dia_und_500Fa, 500nM Dia_und_600R, 13.75nM Dia_und_CO1_535P (labeled with 5Cy5 reporter); 1.00 μL DNA template of varying concentration or ddH2O for no-tissue controls; and ddH2O to complete the dilution of the supermix to a final volume of 20.00 μL. The optimized thermocycler conditions were as follows: an initial denaturation step for 5 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 15 s, followed by data capture. A lid temperature of 105°C was maintained throughout all cycles. The assay was tested on 128 individuals for two replicates conducted by different laboratory technicians on different days.
The sensitivity of the assay was tested with serial dilutions of D. undecimpunctata DNA at concentrations from 100 ng/µL to 0.0001 ng/µL using both the diagnostic and control probes in duplexed reactions. Results were averaged from four independent runs and the Cq values were compared to DNA concentration on a logarithmic scale to determine the slope, y-intercept, and correlation of DNA concentration to assay sensitivity (30, 46).
3 Results
From a combination of intercepted and museum chrysomelid specimens, we were able to generate 118 CO1 DNA barcodes for use in analyses and testing (Table 1). During the course of creation of sequence contigs and quality control, 10 specimens (not included in Table 1) were found to contain Centistes sp. (Braconidae) parasitoid wasp larvae [see (47)]. These specimens were excluded from phylogenetic analyses but were later used in testing the real-time PCR assay. From the CO1 alignment (Supplementary Data Sheet 1), the BI and NJ analyses (Figure 1 and Supplementary Figures 1–3) resolved the Diabrotica species into two clades (fucata and virgifera species groups) and the D. undecimpunctata lineage (in the fucata group) into a well-supported monophyletic clade (posterior probability (PP) 1 GTR, PP 1 JC69, and NJ jackknife (JK) 99.8). Within the D. undecimpunctata clade, an early-diverging lineage containing two samples from western California was supported as distinct (PP 0.99 GTR, PP 1 JC69, and NJ JK 100). Network analysis of the subclade containing D. undecimpunctata and the most closely related species further confirmed a nearly homogeneous D. undecimpunctata and a next-nearest haplogroup containing the two western California specimens of D. undecimpunctata separated by 20 informative nucleotide variants along the shortest branch from the larger group (Figure 2). In comparison, for the large D. undecimpunctata haplogroup, excluding the western California samples, the longest path separating haplotypes was three nucleotide variants. Across this entire haplogroup, only 14 informative nucleotide variants of either one or two steps described all the haplotypic diversity. None of the D. undecimpunctata subspecies designations were isolated to a single haplogroup in the network analysis (Figure 2) or to any of the subclades in any of the BI trees (Figure 1 and Supplementary Figures 1, 2) or NJ trees (Supplementary Figure 3). From the CO1 data, a subset of samples were selected to be sequenced for nuclear ITS2 to confirm (1) that the western California lineage was an outlier to other D. undecimpunctata samples, and (2) that the ingroup made up of different D. undecimpunctata subspecies was homogeneous. Samples for this test were selected from different haplotypic lineages (including the western California lineage) to maximize the possibility of finding ITS2 nucleotide variants. The results from ITS2 confirmed that the western California lineage was an outlier to all other D. undecimpunctata (Supplementary Figures 4–6) through the retention of an ancestral thymine at position 326 shared with all other Diabrotica species (Supplementary Data Sheet 2). The remaining D. undecimpunctata samples in the dataset were homogeneous in sequence except for an autapomorphic cytosine at 171 in D. u. undecimpunctata (AF278571) and an adenine at 430 in the same sample, which was a heterozygotic ‘R’ (adenine or guanine) peak in one intercepted D. u. duodecimnotata (DIA-007).
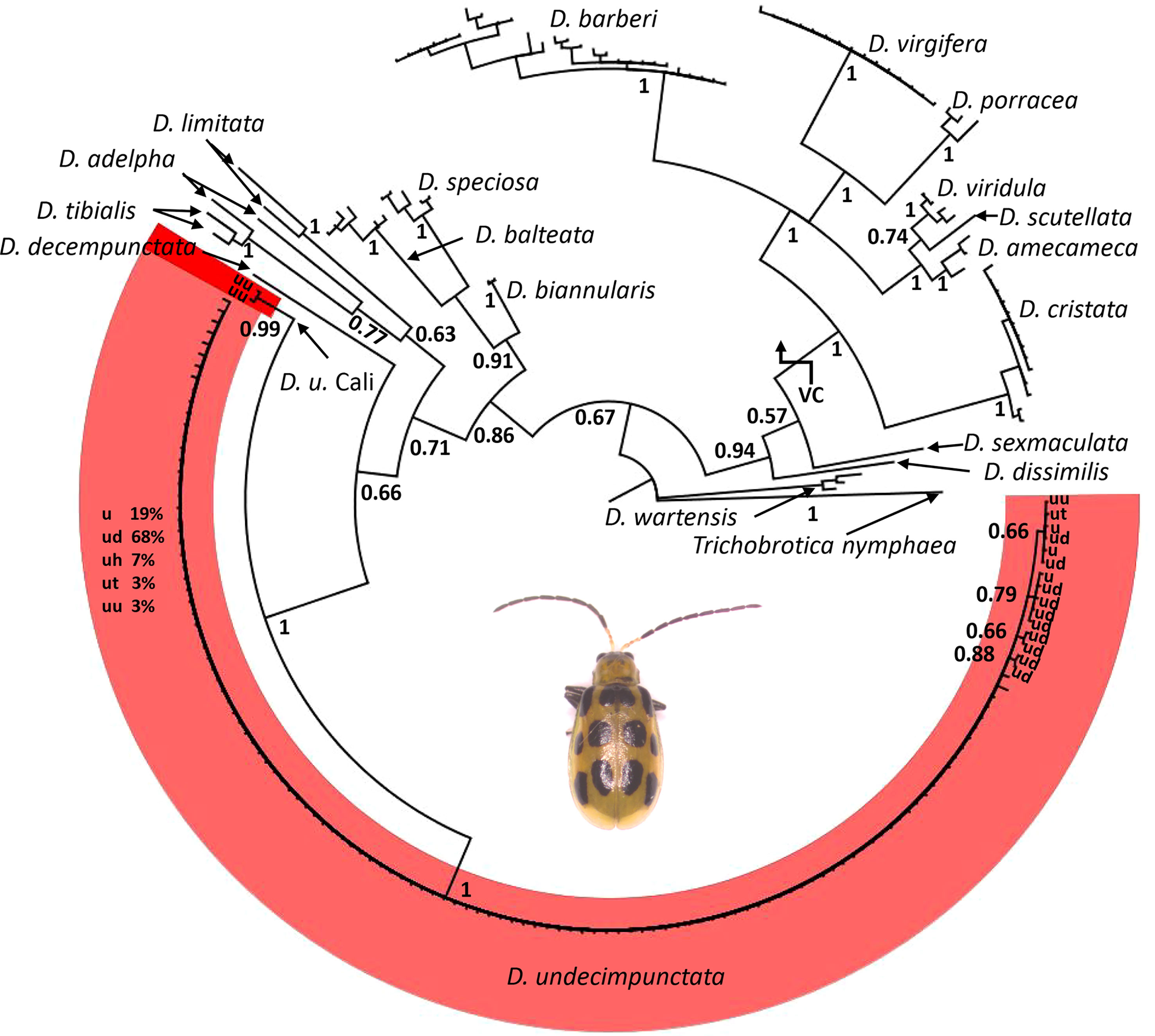
Figure 1 A phylogenetic tree for Diabrotica generated via Bayesian inference with a general time reversible (GTR) substitution model using an alignment of 227 CO1 DNA barcodes. Values at nodes are posterior probabilities; Trichobrotica nymphaea was set as an outgroup. Within D. undecimpunctata, subclade terminals are labeled by subspecies: D. undecimpunctata (u), D. u. duodecimnotata (ud), D. u. howardi (uh), D. u. tenella (ut), and D. u. undecimpunctata (uu). The large unstructured grade is labeled by percentage of terminals with a given subspecific designation. The virgifera species group is denoted VC in the tree, with all other Diabrotica terminals belonging to the fucata species group. Inset image is an adult D. undecimpunctata.
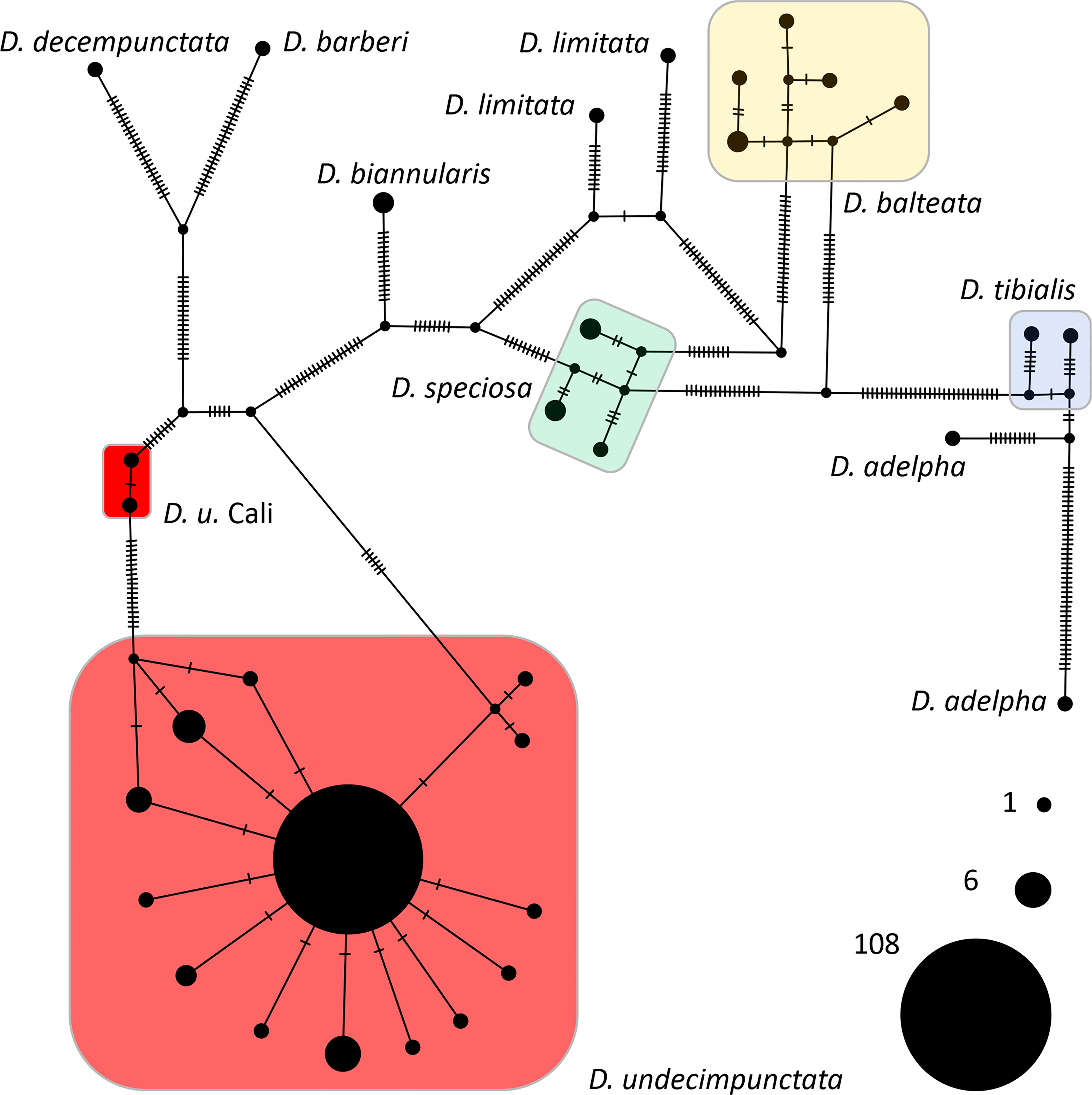
Figure 2 A TCS network from an alignment of 156 Diabrotica CO1 sequences. Each hatch mark along a branch indicates an informative nucleotide variant between haplotypes. Network terminals are scaled by the number of individuals sharing an identical haplotype; a key to the size of these terminals is provided in the lower right. The D. undecimpunctata haplogroups are color coded as in Figure 1.
The phylogenetic tree of D. virgifera subspecies, generated to assess the correlation of CO1 data to subspecific designations for use as a comparator to the congeneric D. undecimpunctata subspecies, resolved three well-supported clades from a CO1 alignment (Supplementary Data Sheet 3) across all tree-building methods (Figure 3 and Supplementary Figures 7–9). The first lineage was early-diverging and consisted of four individuals from Guatemala, all identified as D. v. zeae. The second lineage contained individuals from Mexico (23%), the USA (35%), and Croatia (42%, which is inferred as the primary lineage from which the introduced populations were sourced), with individuals identified as both D. v. virgifera (63%) and D. v. zeae (37%). The third lineage consisted almost exclusively of individuals from Mexico (97%), along with a small number from Croatia (3%), with all Mexican samples identified as D. v. zeae and all Croatian samples as D. v. virgifera. Thus, the CO1 data from D. virgifera did not entirely correspond to subspecific designation but rather appeared to be associated with attributes such as unidirectional reproductive incompatibility and geographic isolation (both of which can be involved with speciation), which are discussed in detail below.
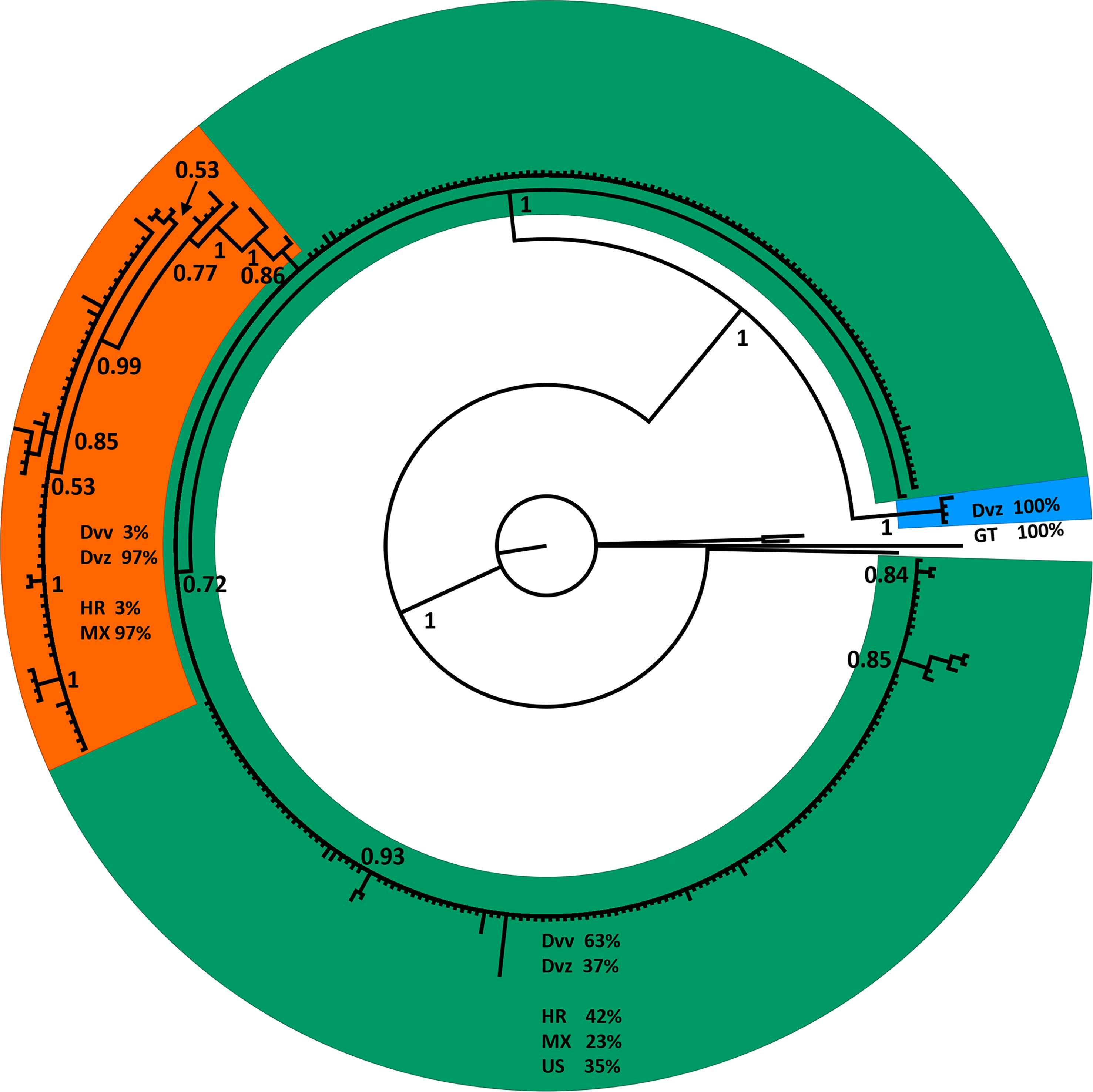
Figure 3 A phylogenetic tree generated via Bayesian inference with a general time reversible (GTR) substitution model using an alignment of CO1 barcodes for 314 Diabrotica virgifera identified to subspecies downloaded from GenBank. Values at nodes are posterior probabilities; one D. undecimpunctata, two D. amecameca, and one D. porracea were used as outgroup taxa. Clades are color coded as follows: orange, a mainly D. v. zeae clade from Mexico; green, a mixed D. v. virgifera and D. v. zeae clade from Mexico and the US; and blue, a D. v. zeae-only clade from Guatemala. For each colored clade, the percentage of terminals identified as D. v. virgifera (Dvv) and D. v. zeae (Dvz) is indicated, as well as the percentage of terminals from Guatemala (GT), Croatia (HR), Mexico (MX), and the United States (US).
Of the 110 target D. undecimpunctata samples tested with the real-time assay described here, four produced anomalous results. For these anomalous samples, very low (1.6–5.6) and very high (32.9–37.1) Cq values were produced for the diagnostic and control probes, respectively. Such an outcome is most likely the result of abnormal differences between CO1 and 18S copy number. Fortunately, in these rare cases the probes can be rerun in separate reactions to confirm the results. Of the 10 D. undecimpunctata samples (not included in the 110 target samples) that had been parasitized by Centistes, only one produced an anomalous result, which was nearly identical to that of the four samples described above. The non-target D. balteata (2), Lyctus africanus (1), and early-diverging D. undecimpunctata (2) from western California all produced positive results for the control probe and negative results for the diagnostic probe. A third D. balteata sample failed for both probes, likely due to deteriorated DNA. All samples that showed amplification for each probe were used to set the relative fluorescence unit (RFU) threshold above which a sample is considered positive. This value was set to 500 for the diagnostic probe and to 250 for the control probe. For each, a Cq between 5 and 30 was considered positive. Background values originated from either NTC samples or non-targets in the case of the diagnostic probe and were used to inform threshold setting. No non-target samples produced amplification of the diagnostic assay. Using these thresholds, the Cq values across all targets ranged from 6.18 to 28.93 (mean 15.71 ± SD 4.38) for the diagnostic probe (Figure 4A) and from 10.72 to 29.41 (mean 17.18 ± SD 3.99) for the control probe (Figure 4B). The end RFU values across all replicates ranged from 813.94 to 1934.33 (mean 1359.33 ± SD 209.07) for the diagnostic probe (Figure 4A) and 315.21 to 3169.02 (mean 1533.68 ± SD 546.79) for the control probe (Figure 4B). In addition, ΔCq (|control probe Cq – diagnostic probe Cq|) was calculated for each reaction containing target DNA using the thresholds described above for which values ranged from 0.01 to 6.56 (mean 1.50 ± SD 0.65). From this, the ΔCq cutoff to confirm a sample as positive was set to 7.
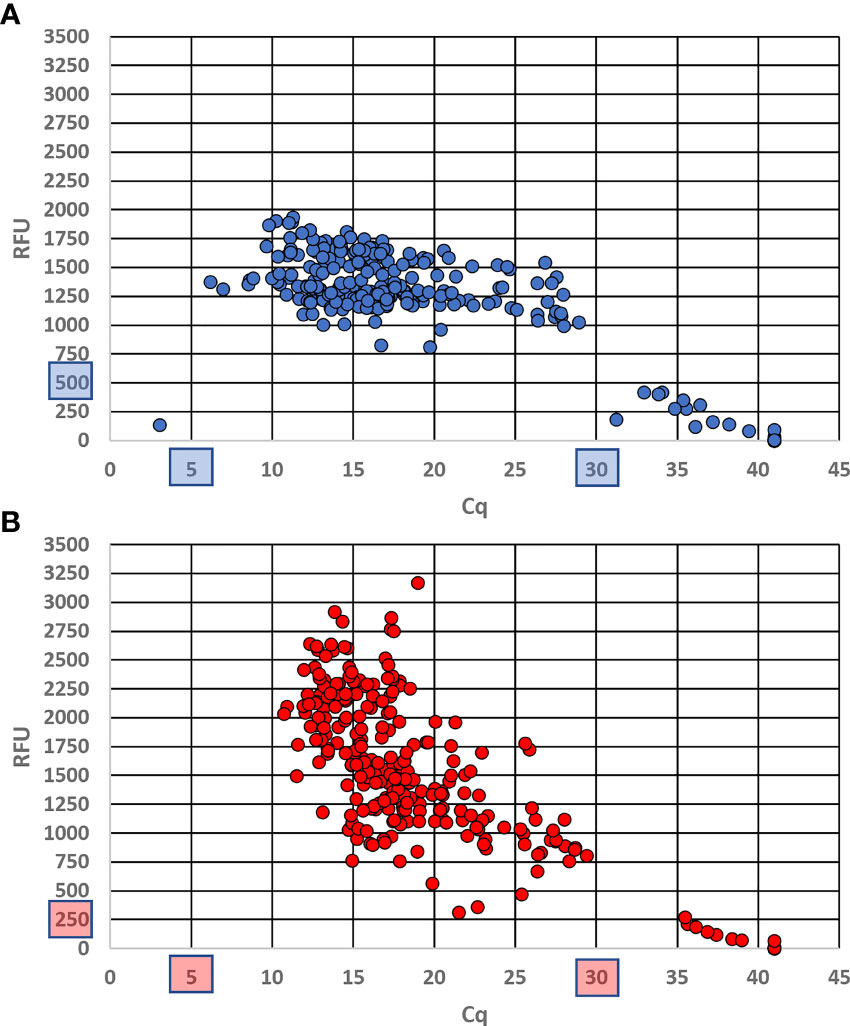
Figure 4 End relative fluorescence unit (RFU) values plotted against Cq values for each probe run in duplex across all replicates using DNA from museum specimens and intercepted Diabrotica. (A) Results from the CO1 diagnostic probe labeled with a 56-FAM fluorophore; (B) results from the 18S control probe labeled with a 5Cy5 fluorophore. Cutoff values are shown in blue (A) and red (B) boxes.
The real-time PCR assay, when run in duplex, showed a linear dose response in Cq for both probes across a DNA dilution series (Figure 5). From the standard curve, the assay developed here should provide reliable target detection at DNA concentrations ≥ 0.1 ng/µL.
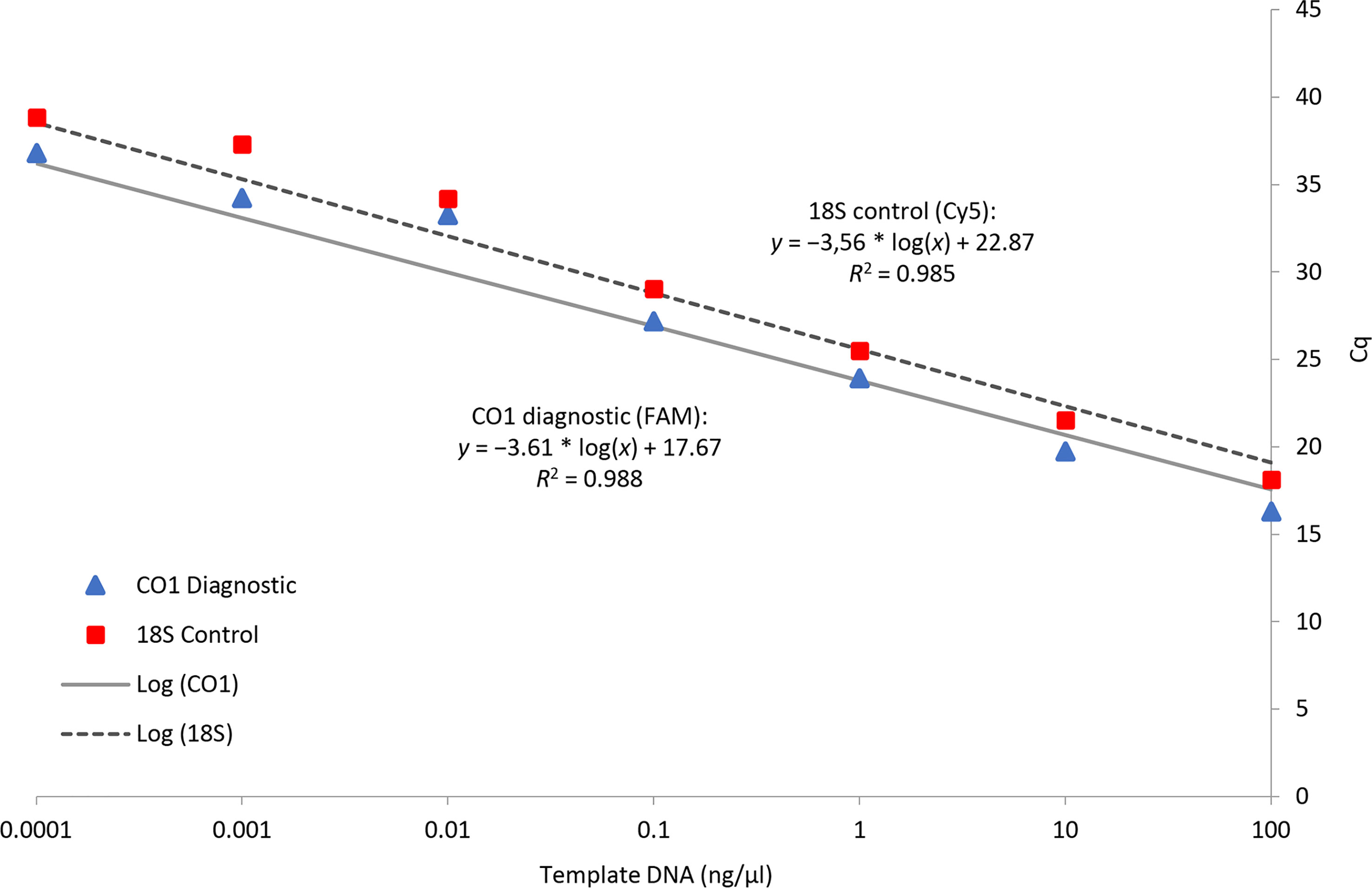
Figure 5 Serial dilutions of template DNA for three D. undecimpunctata individuals with a standard curve based on response in Cq for the CO1 diagnostic and 18S control probes run in duplex.
4 Discussion
A set of Diabrotica CO1 sequences originating from multiple different public data repositories and studies, as well as sequences newly generated as part of this study from museum specimens and port-of-entry interceptions, showed no clear association with current subspecies designations and genetic divergence when multiple phylogenetic approaches were applied (Figure 1). The nuclear ITS2 sequence data did not provide any additional resolution among these subspecies; however, like the CO1 data, ITS2 separated all D. undecimpunctata samples from other Diabrotica species and resolved an outlier lineage from western California (Supplementary Figures 4–6). While complete genome data may provide additional resolution (48) and further insight into the relationships among D. undecimpunctata lineages, such data cannot be rapidly or inexpensively generated, nor will the level of sampling be equivalent to the currently available CO1 databases for some time. Additionally, CO1 data have proven to be highly effective in the identification of insect species that were originally described based on morphology when applying a monophyly criteria (49, 50). While CO1 is often congruent with morphology-based species descriptions, such data have also been useful in detecting cryptic insect lineages (51–53). Thus, our CO1 and ITS2 results are not intended to immediately refute the validity of the current morphology-based D. undecimpunctata subspecific categories, but rather (1) to provide a framework for the re-examination of morphological and biological attributes in these taxonomic categories (54, 55) and (2) to supply data for the development of a molecular assay for use in inspection of transshipped commodities or high-throughput field surveys.
We compared our CO1 data for D. undecimpunctata sampled at the subspecific level to D. virgifera, the only other Diabrotica species with extensive mtDNA sampling at the subspecific level, to examine how genetic and taxonomic patterns correlated within and between these lineages (Figures 1, 3). In both species, subspecific designation did correspond to well-supported CO1 lineages, although more so in D. virgifera. The subspecific split between D. v. virgifera and D. v. zeae is thought to be largely driven by the infection of D. v. virgifera males by Wolbachia bacteria, resulting in cytoplasmic incompatibility when they mate with uninfected D. v. zeae females and thus the perpetuation of a unidirectional reproductive barrier (56). The divergence resulting from this prolonged reproductive incompatibility is evident in the matrilineal CO1 data (Figure 3), with the resolution of a D. v. zeae lineage restricted to Mexico (save two D. v. virgifera from Croatia, which might be the result of misidentification or post-invasion mitochondrial introgression from the loss of Wolbachia infection) and a polyphyletic lineage of D. v. virgifera and D. v. zeae found in both Mexico and the USA. All D. v. zeae in the polyphyletic lineage are from either Texas or Mexico, an area which is thought to be a hybrid zone given the intermediacy of traits found among individuals collected in this region (57). As such, the polyphyly in this lineage may be the result of reciprocal crosses between D. v. virgifera females and D. v. zeae males in which rates of viability are known to be much higher (56). A third early-diverging lineage of D. v. zeae from Guatemala was resolved in the dataset and may represent an ancestral haplotype, as it shares several nucleotide variants with outgroup taxa not present in the other two lineages (Supplementary Data Sheet 3). This third, evidently relictual, lineage may in part be the result of mitochondrial displacement induced via Wolbachia-driven cytoplasmic incompatibility (58, 59), geographic isolation (60), and/or positive selection (61). By extension, the early-diverging lineage resolved in D. undecimpunctata may have evolved via similar processes given similarities in genetic and geographic patterns between these congeneric lineages, but more work is needed to assess what factors are driving these patterns of asymmetrical mitochondrial haplotype counts in these Diabrotica lineages. Furthermore, the question of whether these early-diverging monophyletic lineages should be treated as distinct species needs additional research. Interestingly, the species range of D. u. undecimpunctata has been considered to be restricted to the west coast of the USA and it has also been considered the most morphologically divergent and easily identified subspecies (20). Beyond the taxonomic need to study these early-diverging Diabrotica lineages, phylogeographic and ecological studies may provide additional insights into the relictual status and biology of these lineages. For instance, if Guatemala and western California were the locations in which D. virgifera and D. undecimpunctata first evolved, then the dispersal of D. undecimpunctata may not have initially followed maize cultivation from Mexico as was the case for D. virgifera (26). Study of Diabrotica populations in their centers of origin/diversity may also yield insights into natural enemies or other aspects of their biology applicable to controlling their populations in agroecosystems [e.g. (62)].
Like the early-diverging CO1 lineages found in D. virgifera and D. undecimpunctata, several similar patterns of divergence were noted in our analyses of CO1 from other Diabrotica taxa; these may have been the result of operational error and/or biological processes. For example, within the species D. adelpha and D. limitata, haplotypes were separated by 53 and 28 SNVs, respectively (Figure 2). The publicly available CO1 sequences for D. biannularis, D. balteata, D. speciosa, and D. tibialis were, by contrast, similar to what is generally observed for intraspecific variability in beetles and other insects (49, 63), although increased sampling is needed to more fully describe the nucleotide variability in these Diabrotica lineages. Given the discrepancies observed between taxonomic designation and CO1 haplotypes, both here and in other phylogenetic appraisals of Diabrotica and allied species (22), a concerted effort should be made to better understand how the sorting and evolution of mitochondrial and other genetic and genomic data corresponds to morphological, behavioral, and adaptive divergence. Such efforts will provide an evolutionary context to the study of agriculturally important attributes of Diabrotica such as host preferences, xenobiotic metabolism, and natural and anthropogenic patterns of dispersal and migration.
The real-time PCR assay described here was effective at distinguishing D. undecimpunctata from related species. Given the known challenges of separating some D. undecimpunctata subspecies on the basis of morphology and the frequency with which these beetles are intercepted on agricultural commodities, the real-time PCR assay presented here represents a useful tool for rapid identification at ports of entry. Similarly, given concerns about the introduction of D. undecimpunctata outside North America, this assay could also be used for prescreening of agricultural commodities before export. The assay was designed to exclude the early-diverging western California lineage at present, until follow-up work can clarify the species status of these individuals; however, given that these samples are rare and geographically isolated, this should not affect routine screening efforts. If this haplotype needs to be included in future assays, design of a probe should be straightforward given the number of SNVs separating this haplotype from the next-nearest specimens.
When other Diabrotica species were tested with our real-time PCR assay there was no signal for the diagnostic probe, indicating that these samples did not produce off-target amplification. In a small number of target samples (five), large differences between diagnostic and control probe Cq might have been related to early stages of Centistes parasitism, as one of these samples was known to have been parasitized (47), or to other factors known to affect CO1 copy number (64). In the future, using only legs in DNA extraction might limit such imbalances in CO1 and 18S copy number to the extent that these were associated with the whole-body extractions employed throughout this study. Testing of more Diabrotica species and of D. undecimpunctata at different life stages should be carried out to further validate the findings reported here. A broader diversity of sample types should also be tested with the primers and probes developed for this study so that this assay can be adapted for other applications, such as testing of soil samples for the presence of D. undecimpunctata larvae [e.g. (65)] or the processing of bulk samples using ddPCR [e.g. (66)]. Soil sampling of this type could be an important planning tool for producers, helping them to decide to plant resistant crops when evidence of D. undecimpunctata is found. The assay described here could be expanded to identify not only D. undecimpunctata but also associated plant pathogens such as E. tracheiphila in multiplex PCRs.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers are as follows: NCBI; OQ641612–OQ641622 and OQ649623–OQ649739.
Author contributions
FZ, LT, and TG conceived and designed the study. CW, FZ, and LT prepared and processed the samples used in the study. CW, FZ, and LT performed analyses. LT and TG acquired funding and administered the project. AK, FZ, CW, LT, and TG wrote the original draft of the manuscript and LT, CW, FZ, AET, TG, AK, and AKT helped to write and edit all subsequent versions. All authors contributed to the article and approved the submitted version.
Funding
This work was made possible by a Cooperative Agreement (AP19PPQS&T00C136) from the United States Department of Agriculture’s Animal and Plant Health Inspection Service (APHIS) to LT at Colorado State University.
Acknowledgments
We thank Casey Hubble at the Essig Museum of Entomology, Lynn Kimsey at the Bohart Museum of Entomology, Lee Herman and Corey Smith at the American Museum of Natural History, Floyd Shockley at the Smithsonian, and Marc Epstein at the California Department of Food and Agriculture for assistance with museum specimens. We also thank Anthony Boughton, Tyler Hedlund, and Garrett Hughes for their ongoing work to collect, identify, and preserve Diabrotica beetles for use in this and similar studies. Special thanks to Charlotte Aldebron and Allan H. Smith-Pardo for their help with this manuscript. Thanks to the Joe Tembrock family for providing logistical support while work was being conducted in California.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
This work may not necessarily express the views of APHIS. Mentions of trade names or commercial products in this publication are solely for the purpose of providing specific information and do not imply recommendation or endorsement by the USDA; the USDA is an equal opportunity provider and employer.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2023.1168586/full#supplementary-material
Footnotes
References
2. Garcia F. Injurious insects. D. 12-punctata [press release]. New Mex Agr Expt St Bul (1908) 68:17.
3. Britton W. Insects attacking squash, cucumbers, and allied plants in Connecticut. D. 12-punctata: Conn Agr Expt Sta Bul. (1919), 37. doi: 10.5962/bhl.title.50906
4. Sell RA. Some notes on the Western twelve-spotted and the Western striped cucumber beetles. J Econ Ent. (1915) 8(6):515–20. doi: 10.1093/jee/8.6.515
5. Clark SM, LeDoux DG, Seeno TN, Riley EG, Gilbert AJ, Sullivan JM. Host plants of leaf beetle species occurring in United States and Canada (Coleoptera: Orsodacnidae, Megalopodidae, Chrysomelidae exclusive of Bruchinae). Sp Pub Col Soc (2004) 2:476.
6. Luna JM, Xue L. Aggregation behavior of western spotted cucumber beetle (Coleoptera: Chrysomelidae) in vegetable cropping systems. Environ Entomol (2009) 38(3):809–14. doi: 10.1603/022.038.0334
7. Pedersen AB, Godfrey LD. Field and vegetable crops as hosts of larval western spotted cucumber beetle (Coleoptera: Chrysomelidae). Environ Entomol (2011) 40(3):633–8. doi: 10.1603/EN10143
9. Burchett A. Operation rootworm: Can biotechnology beat the one-billion dollar bug? Farm J (2001) 125:16–8.
10. Metcalf CL, Flint WP, Metcalf RL. Destructive and useful insects. 4th ed. New York, NY: McGraw-Hill (1962).
11. Porter P, Cronholm GB, Parker RD, Troxclair N, Patrick CD, Biles SP, et al. Managing insect and mite pests of Texas corn (2010). Available at: http://hdl.handle.net/1969.1/87672.
12. Yao C, Zehnder G, Bauske E, Kloepper J. Relationship between cucumber beetle (Coleoptera: Chrysomelidae) density and incidence of bacterial wilt of cucurbits. J Econ Entomol (1996) 89(2):510–4. doi: 10.1093/jee/89.2.510
13. Shapiro LR, Paulson JN, Arnold BJ, Scully ED, Zhaxybayeva O, Pierce NE, et al. An introduced crop plant is driving diversification of the virulent bacterial pathogen Erwinia tracheiphila. mBio (2018) 9(5):e01307–18. doi: 10.1128/mBio.01307-18
14. Shapiro LR, Scully ED, Straub TJ, Park J, Stephenson AG, Beattie GA, et al. Horizontal gene acquisitions, mobile element proliferation, and genome decay in the host-restricted plant pathogen Erwinia tracheiphila. Genome Biol Evol (2016) 8(3):649–64. doi: 10.1093/gbe/evw016
15. Milbrath GM, Mclaughlin MR, Goodman RM. Identification of bean pod mottle virus from naturally infected soybeans in Illinois. Plant Dis Rep (1975) 59:982–3.
16. Marchioro CA, Krechemer FS. Potential global distribution of Diabrotica species and the risks for agricultural production. Pest Manag Sci (2018) 74:2100–9. doi: 10.1002/ps.4906
17. EFSA PLH Panel (EFSA Pannel on Plant Health), Bragard C, Dehnen-Schmutz K, Di Serio F, Gonthier P, Jacques M-A, et al. Scientific opinion on the pest categorization of Diabrotica undecimpunctata undecimpunctata. EFSA J (2020) 18(10):26. doi: 10.2903/j.efsa.2020.6291
18. Tubbs PK. Diabrotica undecimpunctata Mannerheim 1843 (Insects, Coleoptera): proposed conservation of the specific name and of the subspecific name howardi Barber 1947. Bul Zoo nom. (1991) 48:219–21. doi: 10.5962/bhl.part.729
19. Derunkov A, Konstantinov A, Tishechkin A, Hartje L, Redford AJ. Diabrotica ID: Identification of Diabrotica species (Coleoptera: Chrysomelidae) from North and Central America (2013). USDA APHIS PPQ Center for Plant Health Science and Technology, USDA Agricultural Research Service, University of Maryland, and Louisiana State University. Available at: http://idtools.org/id/beetles/diabrotica/ (Accessed 09/02/2022).
20. Krysan JL. Introduction: Biology, distribution, and identification of pest Diabrotica. In: Krysan JL, Miller TA, editors. Methods for the study of pest Diabrotica; Springer. springer series in experimental entomology . New York, NY: Springer-Verlag (1986). p. 1–23.
21. Clark TL, Meinke LJ, Foster JE. Molecular phylogeny of Diabrotica beetles (Coleoptera: Chrysomelidae) inferred from analysis of combined mitochondrial and nuclear DNA sequences. Insect Mol Biol (2001) 10(4):303–14. doi: 10.1046/j.0962-1075.2001.00269.x
22. Eben A, Espinosa de Los Monteros A. Tempo and mode of evolutionary radiation in diabroticina beetles (genera Acalymma, Cerotoma, and Diabrotica). Zookeys (2013) 19(332):207–321. doi: 10.3897/zookeys.332.5220
23. Nie RE, Breeschoten T, Timmermans MJTN, Nadein K, Xue HJ, Bai M, et al. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics (2018) 34(2):113–30. doi: 10.1111/cla.12196
24. Krysan JL, Smith RF. Systematica of the virgifera species group of Diabrotica (Coleoptera: Chrysomelidae: Galerucinae). Entomography (1987) 5:375–484.
25. Smith RF. Distributional patterns of selected western North American insects: the distribution of diabroticites in western North America. Bul Entomol Soc Am (1966) 12(2):108–10. doi: 10.1093/besa/12.2.108
26. Branson TF, Krysan JL. Feeding and oviposition behavior and life cycle strategies of Diabrotica: an evolutionary view with implications for pest management. Env Entomol (1981) 10(6):826–31. doi: 10.1093/ee/10.6.826
27. Lombaert E, Ciosi M, Miller NJ, Sappington TW, Blin A, Guillemaud T. Colonization history of the western corn rootworm (Diabrotica virgifera virgifera) in North America: insights from random forest ABC using microsatellite data. Biol Invasions (2017) 20(3):665–77. doi: 10.1101/117424
28. Kiss J, Komáromi J, Bayar K, Edwards CR, Hatala-Zsellér I. Western Corn rootworm (Diabrotica virgifera virgifera) and the crop rotation systems in Europe. In: Vidal S, Kuhlmann U, Edwards CR, editors. Western Corn rootworm ecology and management. Oxfordshire, UK: CABI Publishing (2005).
29. Bažok R, Lemić D, Chiarini F, Furlan L. Western Corn rootworm (Diabrotica virgifera virgifera LeConte) in Europe: current status and sustainable pest management. Insects (2021) 12(3):195.
30. Zink FA, Tembrock LR, Timm AE, Gilligan TM. A real-time PCR assay for rapid identification of Tuta absoluta (Lepidoptera: Gelechiidae). J Econ Entomol (2020) 113(3):1479–85. doi: 10.1093/jee/toaa040
31. Swigoňová Z, Kjer KM. Phylogeny and host-plant association in the leaf beetle genus Trirhabda LeConte (Coleoptera: Chrysomelidae). Mol Phylogenet Evol (2004) 32(1):358–74. doi: 10.1016/j.ympev.2004.02.010
32. Jordal BH, Hewitt GM. The origin and radiation of macaronesian beetles breeding in Euphorbia: the relative importance of multiple data partitions and population sampling. Syst Biol (2004) 53(5):711–34. doi: 10.1080/10635150490468710
33. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am (1994) 87(6):651–701. doi: 10.1093/aesa/87.6.651
34. Navajas M, Cotton D, Kreiter S, Gutierrez J. Molecular approach in spider mites (Acari: Tetranychidae): preliminary data on ribosomal DNA sequences. Exp Appl Acarol. (1992) 15(4):211–8. doi: 10.1007/BF01246563
35. Ratnasingham S, Hebert PDN. BOLD: The barcode of life data system. Mol Ecol Notes (2007) 7(3):355–64. doi: 10.1111/j.1471-8286.2007.01678.x
36. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res (2002) 30(14):3059–66. doi: 10.1093/nar/gkf436
37. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol (2013) 30(4):772–80. doi: 10.1093/molbev/mst010
38. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol (1993) 10(3):512–26.
39. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (2001) 17(8):754–5. doi: 10.1093/bioinformatics/17.8.754
40. Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol (2000) 9(10):1657–60. doi: 10.1046/j.1365-294x.2000.01020.x
41. Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Meth Ecol Evol (2015) 6(9):1110–6. doi: 10.1111/2041-210X.12410
42. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res (2012) 40(15):e115. doi: 10.1093/nar/gks596
43. SantaLucia J Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA (1998) 95(4):1460–5. doi: 10.1073/pnas.95.4.1460
44. Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res (2007) 35(Web Server issue):W43–6. doi: 10.1093/nar/gkm234
45. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem (2009) 55(4):611–22. doi: 10.1373/clinchem.2008.112797
46. Barr NB, Ledezma LA, Farris RE, Epstein ME, Gilligan TM. A multiplex real-time polymerase chain reaction assay to diagnose Epiphyas postvittana (Lepidoptera: Tortricidae). J Econ Entomol (2011) 104(5):1706–19. doi: 10.1603/EC11093
47. Wilson CR, Cooke-McEwen C, Gilligan TM, Tembrock LR. Detection of Centistes sp. (Hymenoptera, Braconidae) from intercepted Diabrotica undecimpunctata (Coleoptera, Chrysomelidae) using CO1 DNA barcodes and larval morphology. J Hymenopt Res (2022) 91:69–81. doi: 10.3897/jhr.91.84139
48. Yang H, You CJ, Tsui CKM, Tembrock LR, Wu ZQ, Yang P. Phylogeny and biogeography of the Japanese rhinoceros beetle, Trypoxylus dichotomus (Coleoptera: Scarabaeidae) based on SNP markers. Ecol Evol (2020) 11(1):153–73.
49. Hendrich L, Morinière J, Haszprunar G, Hebert PD, Hausmann A, Kohler F, et al. A comprehensive DNA barcode database for central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD. Mol Ecol Resour (2015) 15(4):795–818. doi: 10.1111/1755-0998.12354
50. Lopez-Vaamonde C, Kirichenko N, Cama A, Doorenweerd C, Godfray HCJ, Guiguet A, et al. Evaluating DNA barcoding for species identification and discovery in European gracillariid moths. Front Ecol Evol (2021) 9:626752. doi: 10.3389/fevo.2021.626752
51. Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PD. DNA Barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci U S A. (2006) 103(10):3657–62. doi: 10.1073/pnas.0511318103
52. Smith MA, Fernandez-Triana J, Roughley R, Hebert PDN. DNA Barcode accumulation curves for understudied taxa and areas. Mol Ecol Resour (2009) 9:208–16. doi: 10.1111/j.1755-0998.2009.02646.x
53. Bisconti R, Canestrelli D, Tenchini R, Belfiore C, Buffagni A, Nascetti G. Cryptic diversity and multiple origins of the widespread mayfly species group Baetis rhodani (Ephemeroptera: Baetidae) on northwestern Mediterranean islands. Ecol Evol (2016) 6(21):7901–10. doi: 10.1002/ece3.2465
54. Hennig W. Phylogenetic systematics. Davis DD, Zangerl R, editors. Urbana, Illinois: Urbana University of Illinois Press (1966).
55. DePinna M. Concepts and tests of homology in the cladistic paradigm. Cladistics (1991) 7(4):367–94.
56. Giordano R, Jackson JJ, Robertson HM. The role of Wolbachia bacteria in reproductive incompatibilities and hybrid zones of Diabrotica beetles and Gryllus crickets. Proc Natl Acad Sci USA (1997) 94(21):11439–44. doi: 10.1073/pnas.94.21.11439
57. Krysan JL, Smith RF, Branson TF, Guss PL. A new subspecies of Diabrotica virgifera (Coleoptera: Chrysomelidae): description, distribution, and sexual compatibility. Ann Entomol Soc Am (1980) 73(2):123–30. doi: 10.1093/aesa/73.2.123
58. Kodandaramaiah U, Simonsen TJ, Bromilow S, Wahlberg N, Sperling F. Deceptive single-locus taxonomy and phylogeography: Wolbachia-associated divergence in mitochondrial DNA is not reflected in morphology and nuclear markers in a butterfly species. Ecol Evol (2013) 3(16):5167–76. doi: 10.1002/ece3.886
59. Deng J, Assandri G, Chauhan P, Futahashi R, Galimberti A, Hansson B, et al. Wolbachia-driven selective sweep in a range expanding insect species. BMC Ecol Evol (2021) 21(1):181. doi: 10.1186/s12862-021-01906-6
60. Yamaguchi R, Iwasa Y. First passage time to allopatric speciation. Interface Focus (2013) 3(6):20130026. doi: 10.1098/rsfs.2013.0026
61. Li XD, Jiang GF, Yan LY, Li R, Mu Y, Deng WA. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front Genet (2018) 9:605. doi: 10.3389/fgene.2018.00605
62. Vavilov N. Origin and geography of cultivated plants. Löve D, editor. Cambridge, England: Cambridge University Press (1992).
63. Blekhman A, Goryacheva I, Schepetov D, Zakharov I. Variability of the mitochondrial CO1 gene in native and invasive populations of Harmonia axyridis Pall. Comparative analysis. PloS One (2020) 15(4):e0231009. doi: 10.1371/journal.pone.0231009
64. Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet (2001) 17(4):199–205. doi: 10.1016/S0168-9525(01)02238-7
65. Baidoo R, Yan G, Nagachandrabose S, Skantar AM. Developing a real-time PCR assay for direct identification and quantification of Pratylenchus penetrans. Soil. Plant Dis (2017) 101(8):1432–41. doi: 10.1094/PDIS-01-17-0117-RE
Keywords: spotted cucumber beetle, maternal inheritance, species concept, plant pathogen vector, ITS2, Diabrotica virgifera, monophyly
Citation: Tembrock LR, Wilson CR, Zink FA, Timm AE, Gilligan TM, Konstantinov AS and Tishechkin AK (2023) CO1 barcodes resolve an asymmetric biphyletic clade for Diabrotica undecimpunctata subspecies and provide nucleotide variants for differentiation from related lineages using real-time PCR. Front. Insect Sci. 3:1168586. doi: 10.3389/finsc.2023.1168586
Received: 17 February 2023; Accepted: 28 March 2023;
Published: 20 April 2023.
Edited by:
Satnam Singh, Punjab Agricultural University, IndiaReviewed by:
Karen Frances Armstrong, Lincoln University, New ZealandGagandeep Brar, North Dakota State University, United States
Copyright © 2023 Tembrock, Wilson, Zink, Timm, Gilligan, Konstantinov and Tishechkin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luke R. Tembrock, tembrock@colostate.edu
 Luke R. Tembrock
Luke R. Tembrock Christina R. Wilson1
Christina R. Wilson1  Alicia E. Timm
Alicia E. Timm Todd M. Gilligan
Todd M. Gilligan