- 1Department of Oncology and Pathology, Karolinska Institutet, SciLifeLab, Solna, Sweden
- 2Laboratory of Transplantation Immunology, National Research Centre for Hematology, Moscow, Russia
- 3Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia
- 4Institute of Fundamental Medicine and Biology, Kazan Federal University, Kazan, Russia
FOXP1 is ubiquitously expressed in the human body and is implicated in both physiological and pathological processes including cancer. However, despite its importance the role of FOXP1 in T-cells has not been extensively studied. Although relatively few phenotypic and mechanistic details are available, FOXP1 role in T-cell quiescence and differentiation of CD4+ subsets has recently been established. FOXP1 prevents spontaneous T-cell activation, preserves memory potential, and regulates the development of follicular helper and regulatory T-cells. Moreover, there is growing evidence that FOXP1 also regulates T-cell exhaustion. Altogether this makes FOXP1 a crucial and highly undervalued regulator of T-cell homeostasis. In this review, we discuss the biology of FOXP1 with a focus on discoveries made in T-cells in recent years.
FOXP1 biology overview
The gene encoding forkhead box protein P1 (FOXP1) was originally discovered and cloned in 2001 (1, 2). The full-length FOXP1 protein is 677 amino acids long, and its gene is located in 3p14.1 chromosomal region. FOXP1 is ubiquitously expressed in normal tissues and belongs to the family of forkhead box (FOX) proteins, which all share a conserved DNA-binding domain (winged helix) (3). In addition, FOXP1 is a member of a smaller FOXP subgroup, which includes FOXP1-4 and has leucine zipper and zinc finger domains (4). FOXP1 can form homo- and heterodimers with itself and other members of the FOXP family through the leucine zipper motif, and the dimerization is necessary for the transcriptional activity (5–7). Further, it can interact with CtBP-1:NuRD complex and generally acts as a transcriptional repressor (8, 9).
Alternative splicing of FOXP1 can generate 7 isoforms (total number of predicted isoforms is 23) in human and 23 isoforms in murine cells (10, 11). FOXP1 DNA-binding site is highly conserved (90% homology) between humans and mice (10). Besides, human FOXP1 isoforms 3 and 4 share 50% homology to murine isoforms 2, 3, 4 and 5.
FOXP1 analysis in neurological disorders identified 7 mutations in the protein (A339Sfs*4, V423Hfs*37, Y439*, R465G, R514C, R525*, W534R) (12, 13). All of them disrupted the transcriptional activity of FOXP1, whereas three also disrupted FOXP1 ability to homo- and heterodimerize (A339Sfs*4, R525*, W534R). V423Hfs*37, R465G, and R514C variants, on the other hand, bound and sequestered wild type FOXP1 and FOXP2, hence, exerting a dominant-negative effect. ChIP-seq analysis of murine CD4+ T-cells revealed 3071 FOXP1-bound sites in the Treg subset and 1088 sites in the naïve subset (14). Interestingly, FOXP1 de novo motif discovery performed by the same group showed that only 17% of binding sites contained canonical forkhead motif, (G/A)T(A/C)AA(C/T)A. Whereas, over 26% of binding sites contained ETS motif, GGA(A/T). Relatively low occurrence of forkhead motif in FOXP1 binding sites suggests the important role of other binding partners.
FOXP1 is implicated in various physiological settings including B-cell development, monocyte differentiation, and lung epithelia regeneration (15–17). In cancer it can act as both an oncogene (B-cell lymphoma, ovarian cancer, hepatocellular carcinoma) and a tumor suppressor (T-cell lymphoma, NSCLC, colorectal cancer) (18–22). Moreover, there is evidence that in breast cancer cells, FOXP1 expression dampens the synthesis of T-cell attracting cytokines and, as a result, impairs T-cell infiltration (23).
Further in the first section we review the molecular biology of FOXP1 in human CD4+ T-cells and then in the second section we discuss FOXP1 role in T-cell quiescence and exhaustion. In the third section we have a closer look at FOXP1 in the context of CD4+ T-cell differentiation. Finally, in the fourth section we briefly cover discrepancies among various isoforms.
FOXP1 biology in human CD4+ T-cells
Durek et al. (2016) was the first to look at FOXP1 role in human CD4+ T-cells and found 4 isoforms of FOXP1 in their RNA-seq dataset (24). 3 of those isoforms contained complete protein coding sequences, whereas the 4th isoform contained only 3 exons. Expression levels of all detected FOXP1 isoforms were the highest in the naïve (Tn) subset, substantially decreased in the central memory (Tcm) subset and the lowest in the effector memory (Tem) subset. This expression downregulation was accompanied by the increased DNA methylation in FOXP1 promoter suggesting epigenetic shut-down of FOXP1 in more differentiated CD4+ T-cell subsets.
Garaud et al. (2017) performed another comprehensive investigation of FOXP1 biology in human CD4+ T-cells (25). They identified that 6 out of 23 predicted FOXP1 splice isoforms are expressed in human PBMCs and CD4+ T-cells and concluded that the full-length isoform was the most abundant one. In agreement with Durek et al. (2016), FOXP1 expression in CD45RA+CD4+ T-cells (mostly naïve) was greater than in CD45RA-CD4+ subpopulation (memory). Also, in line with Durek et al. (2016), memory T-cells showed increased DNA methylation in FOXP1 promoter. FOXP1 RNA and protein levels were shown to decrease in naïve and memory CD4+ T-cells upon T-cell receptor (TCR) stimulation. Bypassing TCR stimulation and activation of PKC-mediated Ca2+ influx directly did not achieve the same level of FOXP1 repression, suggesting that other TCR pathways play a role in FOXP1 downregulation. Interestingly, the downregulation in naïve T-cells was more pronounced than in memory T-cells. Another interesting observation by the authors is that the expression of a shorter isoform (65 kDa) increased 24 hours after activation but then decreased back to initial levels. FOXP1 repression by shRNA increased the number of cycling and activated cells but did not affect apoptosis. The authors performed FOXP1 ChIP-qPCR analysis and found that FOXP1 bound and repressed ID2, STAT6, IL-13, and IL-17A in naïve T-cells, but only ID2 in memory T-cells. The products of these genes play role in T-cell differentiation and effector functions, suggesting that FOXP1 can preserve stemness in T-cells (26). Importantly, FOXP1 did not bind FOXP3 promoter but FOXP3 expression was increased in FOXP1-knockdown memory T-cells but not naïve CD4+ T-cells, indicating that FOXP1 can repress FOXP3 expression.
Although the aforementioned studies elucidated many aspects of FOXP1 in human CD4+ T-cells, additional studies are needed to investigate FOXP1 in the context of CD4+ T-cell memory formation, antitumor function, and autoimmunity.
FOXP1 role in T-cell quiescence
Early evidence of FOXP1 role in T-cells came from a study of lymphocytic variant of hypereosinophilic syndrome (L-HES) patients. L-HES is a benign lymphoproliferative disease that is characterized by clonally expanded CD3-CD4+ T-cells and, in some cases, can progress to T-cell lymphoma (27). Expansion of these abnormal Th2 cells and their IL-5 secretion increase eosinophil counts. Gene expression profile of abnormal T-cells was compared to that of normal CD3+CD4+ T-cells (28). This analysis showed significant downregulation of FOXP1 in abnormal T-cells suggesting a potential role in T-cell quiescence. Indeed, in patients with peripheral T-cell lymphoma FOXP1 expression inversely correlated with proliferation marker Ki-67 (19). FOXP1 overexpressing tumors were associated with better survival. Surprisingly, this was not the case for B-cell lymphomas, which suggests different role of FOXP1 in these cell types (29, 30). Another explanation of this discrepancy is preferential expression of N-terminal truncated FOXP1 isoforms (60 and 65 kDa) by malignant B-cells, which seem to have an oncogenic role (31, 32). However, another report showed that, although short FOXP1 isoform is preferentially expressed in B-cell lymphomas, overexpressed full-length FOXP1 has a similar oncogenic activity (33). Feng et al. (2010) used a mouse model with conditional deletion of FOXP1 at CD4+CD8+ stage of thymocyte development and showed that both mature FOXP1-/- CD4+ and CD8+ T-cells lost their quiescence (34). These cells had larger size, downregulated CD62L, upregulated CD44, and were more prone to IFN-γ production and apoptosis. Heterozygous FOXP1 deletion led to more activated phenotype only in CD8+ T-cells, suggesting that CD4+ T-cells are less sensitive to the changes in FOXP1 levels. The authors did not observe any defects in Tregs formation and function. Similar activated CD62L-CD44+ phenotype in CD4+ cells was observed by an independent group in a mouse model with conditional FOXP1 deletion (24). In their subsequent study, Feng et al. (2011) used tamoxifen-mediated deletion of FOXP1 in mature T-cells (35). They showed that FOXP1 directly repressed IL-7Ra expression and, thus, prevented IL-7-driven antigen-independent proliferation of naïve T-cells in vitro and in vivo. Upon FOXP1 deletion, both naïve CD8+ and CD4+ subpopulations demonstrated activated phenotype but only CD8+ proliferated in response to IL-7. FOXP1 was shown to inhibit MEK/Erk pathway activation independently from IL-7Ra expression. They also confirmed that complete (homozygous) deletion of FOXP1 in CD8+ T-cells was necessary for the change in phenotype. In summary, it was shown that in CD8+ T-cells FOXP1 induces quiescence through downregulation of MAPK pathway and responsiveness to IL-7. Interestingly, FOXP1 protein bound the same enhancer as FOXO1, antagonizing IL-7Ra expression activation by FOXO1. FOXO1 deletion can also lead to spontaneous T-cell activation, but FOXO1 is necessary for IL-7Ra expression (36). Given FOXO1 role in the restriction of T-cell effector differentiation and in memory formation, it seems that FOXP1 is more important for the naïve T-cell quiescence (37). On the other hand, memory T-cells have reduced FOXP1 and increased FOXO1 expression, which potentially contributes to homeostatic proliferation. In another study, this group elucidated the mechanistic details of FOXP1-induced T-cell quiescence (38). They used the same conditional FOXP1 deletion in mature CD8+ T-cells to demonstrate that FOXP1 deficient cells activated PI3K/Akt/mTOR pathway in response to IL-7 and that FOXP1 is necessary for the expression of PI3K inhibitor, Pik3ip1. They also showed that FOXP1 deficient cells had higher expression of cell-cycle E2F factors and their target genes (MCM5, CDK1, PCNA).
Although FOXP1 biology in T-cells is only beginning to reveal its secrets, there is evidence to claim FOXP1 role in maintaining T-cell quiescence in mice (Figure 1). T-cell quiescence is essential not only for preventing spontaneous activation and autoimmunity but also for maintaining a pool of naïve and memory T-cells that can mount immune response. The constantly proliferating T-cells are expected eventually reach Hayflick limit and lose their ability to expand further, whereas FOXP1 counteracts this process and preserves T-cell expansion capacity.
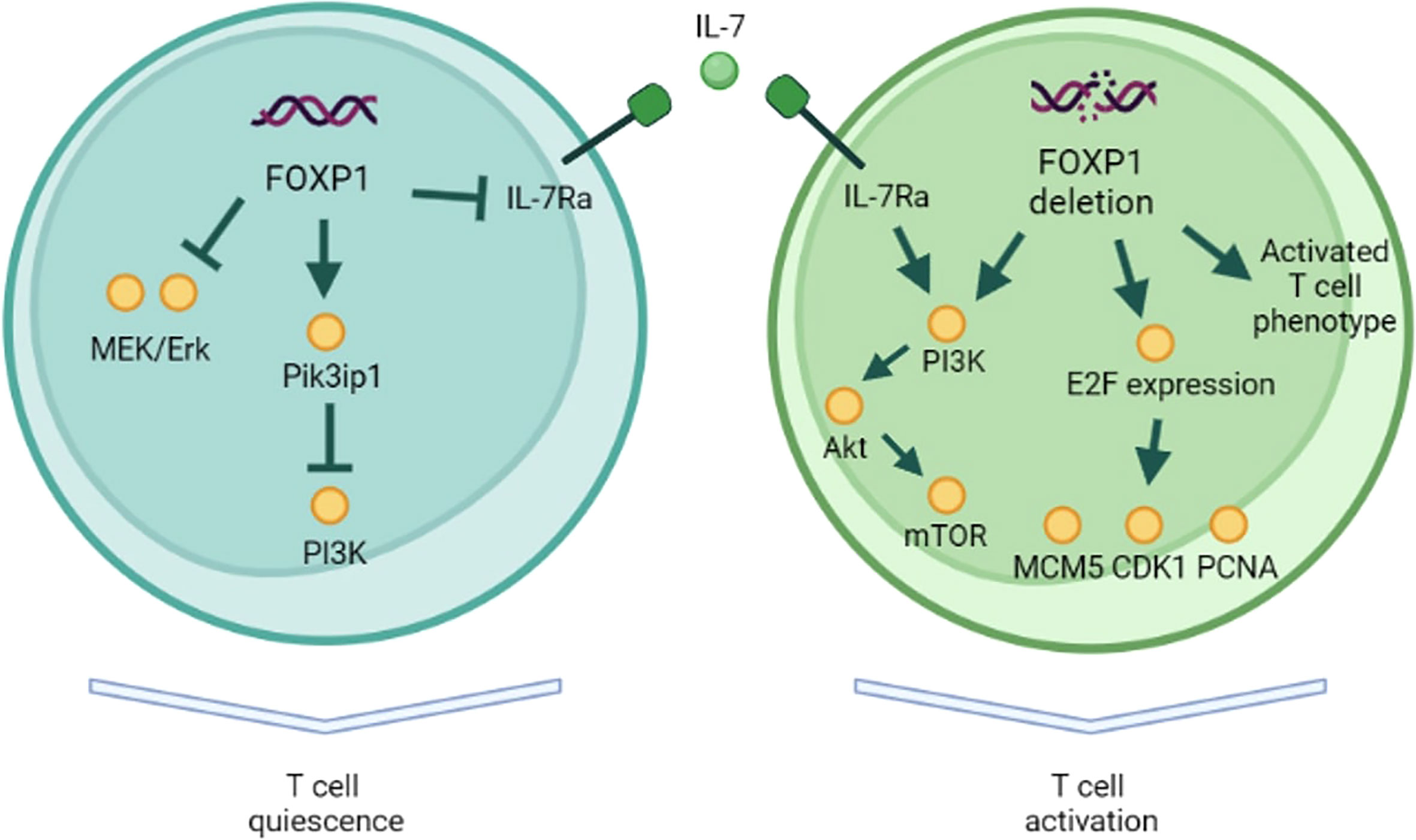
Figure 1 FOXP1 role in T-cell quiescence. In naïve T-cells FOXP1 inhibits MAPK and PI3K pathways. FOXP1 also represses IL-7Ra expression, preventing IL-7-driven antigen-independent proliferation. FOXP1-deficient naïve T-cells demonstrate activated surface phenotype, PI3K pathway activation in response to IL-7, and the higher expression of E2F factors and their target genes (MCM5, CDK1, PCNA).
FOXP1 role in T-cell exhaustion and antitumor immunity
The FOXP1-induced quiescence in T-cells was shown to have implications for the antitumor immunity. Stephen et al. (2014) analyzed FOXP1 in the context of TGF-β signaling in the tumor microenvironment (39). They showed that human CD8+ T-cells expressed both FOXP1A (full-length) and FOXP1D. FOXP1A was downregulated upon activation, whereas both isoforms were upregulated in tumor-infiltrating lymphocytes. IL-2, IL-6, IL-7, IL-15, IL-17, IL-23 and VEGF-α did not affect FOXP1 expression, while ICAM1, CXCL12, and TGF-β signaling induced FOXP1 upregulation. Moreover, FOXP1 interacted with SMAD2/3, and this interaction was required for TGF-β repressive activity. Accordingly, FOXP1 deficiency improved antitumor immunity in ovarian cancer and sarcoma models. In agreement with this work, another group showed that the TGF-β pathway activity as well as FOXP1 expression in human CD8+ T-cells were elevated after co-culture with M2 inhibitory macrophages (40). As TGF-β signaling is implicated in T-cell exhaustion, it is reasonable to assume that FOXP1 (possibly in cooperation with SMAD2/3) also regulates this dysfunction state in T-cells. Similarly to human TILs, FOXP1 expression was higher in CD8+ T-cells from a tumor-bearing mouse supporting its role in T-cell exhaustion (41). Overexpression of miR-149-3p which inhibits PD-1, TIM-3, BTLA, and FOXP1 translation, counteracted CD8+ T-cell exhaustion. Although additional work is needed to expand our knowledge, there is already growing evidence that FOXP1 may play an important role in T-cell exhaustion, affecting T-cell ability to fight tumor.
FOXP1 role in CD4+ T-cell differentiation
In addition to the abovementioned functions FOXP1 also regulates differentiation of CD4+ T-cells. Wang et al. (2014) showed that in the murine system FOXP1 suppresses differentiation of CD4+ T follicular helper cells (Tfh) (42). Tfh cells facilitate B-cell maturation and germinal centers formation, and also express high levels of Bcl-6, CXCR5, ICOS and IL-21 (43–46). Contrary to what has been shown by Garaud et al. (2017) in human CD4+ T-cells, the expression of full-length FOXP1 isoform (FOXP1A) in mouse CD4+ T-cells did not change upon TCR stimulation, whereas the levels of another isoform (FOXP1D) increased both in vitro and in vivo. Because both FOXP1A and FOXP1D can repress Tfh, this increase in total FOXP1 levels led to indirectly reduced expression of ICOS, which is necessary for Tfh differentiation. The authors hypothesized that lower ICOS expression was due to FOXP1 inhibitory effect on MEK/Erk signaling that was shown to induce ICOS expression (47). FOXP1 also directly repressed the expression of IL-21, which is important for both Tfh differentiation and B-cell maintenance. Later this group demonstrated that FOXP1 directly regulates CTLA4, which is important for Tfh differentiation and function (48). They also showed that FOXP1-deficient CD4+ T-cells downregulated CCR7 receptor and, hence, migrated to B-cell follicles earlier during the infection. Similarly, FOXP1 was also demonstrated to negatively regulate Th9 differentiation (49). Th9 CD4+ T-cell subset is characterized by IL-9 and IL-21 secretion and plays a role in antitumor immunity (50–52). The authors showed that in mouse CD4+ T-cells FOXP1 directly repressed IL-9 expression by binding to its promoter. FOXO1, on the other hand, induced IL-9 expression and antagonized FOXP1 at IL-9 promoter. It is noteworthy that IL-7 signaling was responsible for FOXP1 nuclear exclusion and FOXO1 nuclear localization. Interestingly, the expression of full-length FOXP1 isoform (FOXP1A) increased after murine CD4+ T-cell activation, which contradicts the abovementioned results of Wang et al. (2014).
FOXP1 plays an important role in homeostasis of regulatory T-cells. In inducible Tregs (iTregs) FOXP1 was essential for both the proper differentiation and the stability of mature Treg lineage (53). Mechanistically, upon TGF-β treatment, FOXP1 directly bound FOXP3 promoter and enhancer, and this interaction was associated with an increase in permissive chromatin modifications (H3K4me3, H3K9/K14Ac), sustaining FOXP3 expression. On the other hand, in thymus Tregs (tTregs) FOXP3 expression was not affected by FOXP1 deletion, and this cell population was stable in the absence of FOXP1. Dependence of iTregs on FOXP1-driven FOXP3 expression was confirmed by another group (54). They demonstrated that, as opposed to naïve murine CD4+ cells, activated Tregs downregulated FOXP1A expression similar to human CD4+ T-cells. They also showed that FOXP1 deletion led to increased number of activated Tregs with impaired suppressive function and that genes downregulated in FOXP1 deficient cells were significantly enriched in TGF-β pathway. FOXP1 was shown to increase FOXP3 recruitment to target genes and regulate CTLA4 expression through FOXP1-FOXP3 axis (14). FOXP1 and FOXP3 shared the majority of promoter binding sites and regulated target genes expression largely in a cooperative manner. FOXP1 deficiency reduced FOXP3 DNA binding ability and led to decreased CD25 expression. Interestingly, some features of Treg differentiation in FOXP1-deficient cells were rescued by stronger IL-2 signaling, which suggests an existence of compensatory mechanisms.
Summarizing all the above, FOXP1 plays an important role in Tfh and Treg differentiation in mouse CD4+ T-cells (Figure 2). Its role in other CD4+ T-cell subsets has not been studied extensively so far. Moreover, further studies are needed to confirm whether FOXP1 also regulates differentiation of human CD4+ T-cells.
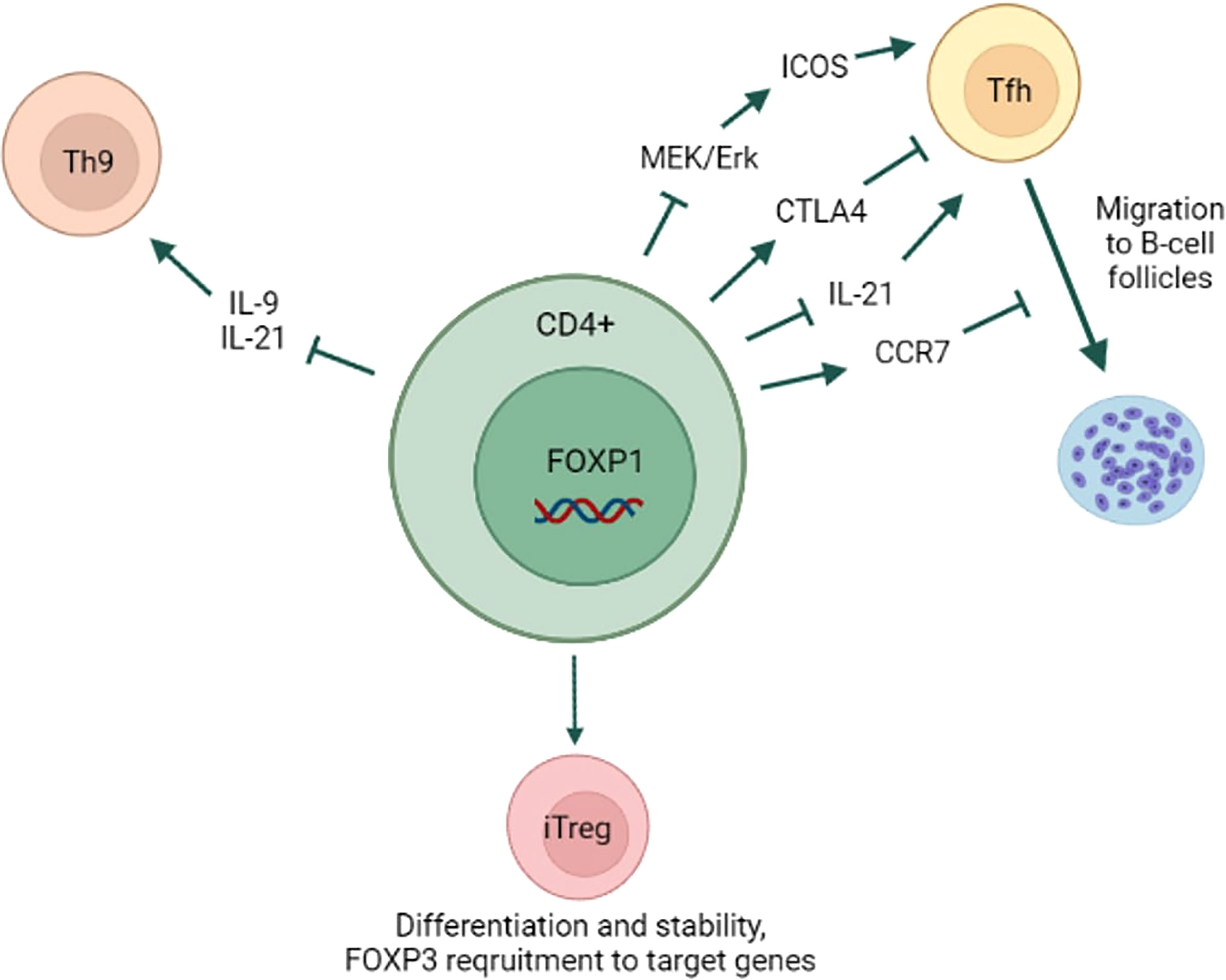
Figure 2 FOXP1 role in CD4+ T-cell differentiation. FOXP1 prevents naïve CD4+ T-cell differentiation into Tfh subset by inhibiting MAPK/ICOS axis and IL-21 expression. FOXP1 promotes CTLA4 and CCR7 expression, which inhibit Tfh formation and the migration to B-cell follicles, respectively. By repressing IL-9 and IL-21 expression FOXP1 prevents CD4+ T-cell differentiation into Th9 subset. FOXP1 is essential for iTregs differentiation and lineage stability likely through FOXP3 recruitment to its target genes. Yet, FOXP1 is dispensable for thymus Tregs differentiation and lineage stability.
Confusion with FOXP1 isoforms
It is noteworthy that a discrepancy exists in FOXP1 expression kinetics after T-cell activation (Table 1). Some reports demonstrated FOXP1 downregulation, while others showed the opposite effect (25, 39, 42). TCR activation presumably induces expression of short FOXP1 isoform that can act as a dominant-negative regulator with respect to full-length FOXP1 (25). The full-length FOXP1 preserves the quiescent state, therefore it should be inhibited during antigen-driven activation and expansion (34–38). However, both isoforms (full-length FOXP1 and truncated FOXP1D) share certain functionality given that either can repress Tfh differentiation (42). Similar to T-cells, B-cells also upregulate short FOXP1 isoforms upon activation that can act as oncogenes in B-cell malignancies (31, 32).
It is essential to clarify which FOXP1 isoforms are expressed in human and mouse T-cells and how conserved they are between the species. This will remove an ambiguity that is currently present around FOXP1 isoforms.
Conclusion
In the light of recent evidence, it would be safe to say that FOXP1 is one of the essential intrinsic factors that keep mouse CD8+ T-cells in quiescent state. In human CD8+ T-cells, FOXP1 modulates TGF-β signaling transduction but its role in quiescence is yet to be validated. In mouse CD4+ T-cells FOXP1 plays a role in Tfh and Treg differentiation. However, FOXP1 may not be as critical in murine thymus Tregs as in Tregs of a different origin. Given the extensive cooperativity between FOXP1 and lineage defining FOXP3, this may point to fundamental differences between Treg subsets. Intriguingly, unlike in mice, human CD4+ T-cells FOXP1 may repress FOXP3 expression, and this discrepancy should be carefully addressed by the future studies.
Although FOXP1 is not a therapeutic target as of now, it can potentially be manipulated for the therapeutic benefit especially if its connection to T-cell exhaustion is further validated. For example, FOXP1 knockout in CAR-T-cells may improve their expansion and persistence. Alternatively, through the inhibition of Tfh differentiation, the reduction of FOXP1 levels may help alleviate autoimmune diseases driven by abnormal B-cell responses. Nevertheless, any therapeutic manipulation of FOXP1 mostly remains contingent on the future studies of its biology.
Author contributions
YK and VK conceived the idea and wrote the manuscript. AK assisted with finalizing the manuscript. EZ prepared the figures. EB proofread the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation (grant agreement no. 075-15-2020-795, state contract no. 13.1902.21.0027 of 29.09.2020 unique project ID: RF-190220X0027).
Acknowledgments
The work was performed in frames of Kazan Federal University Strategic Academic Leadership Program (PRIORITY-2030).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ChIP-seq, Chromatin immunoprecipitation (ChIP); with DNA sequencing. FOXO1, Forkhead box protein O1; FOXP1, Forkhead box protein P1; FOXP3, Forkhead box protein P3; ICOS, Inducible T-cell costimulatory; IFN-γ, Interferon gamma; NSCLC, Non-Small Cell Lung Cancer; PKC, Protein kinase C; TGF-β, Transforming growth factor beta; Tfh, T follicular helper cells; Tn, naïve T-cells; Tcm, central memory T-cells; Tem, effector memory T-cells; Tregs, Regulatory T-cells.
References
1. Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res (2001) 61(24):8820–9.
2. Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem (2001) 276(29):27488–97. doi: 10.1074/jbc.M100636200
3. Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol (2002) 250(1):1–23. doi: 10.1006/dbio.2002.0780
4. Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol (2004) 24(2):809–22. doi: 10.1128/MCB.24.2.809-822.2004
5. Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol (2007) 19(7):825–35. doi: 10.1093/intimm/dxm043
6. Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol (2012) 13(10):1010–9. doi: 10.1038/ni.2402
7. Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. Cell (2003) 278(27):24259–68. doi: 10.1074/jbc.M207174200
8. Chokas AL, Trivedi CM, Lu MM, Tucker PW, Li S, Epstein JA, et al. Foxp1/2/4-NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of interleukin-6. J Biol Chem (2010) 285(17):13304–13. doi: 10.1074/jbc.M109.088468
9. Kim J-H, Hwang J, Jung JH, Lee H-J, Lee DY, Kim S-H. Molecular networks of FOXP family: Dual biologic functions, interplay with other molecules and clinical implications in cancer progression. Mol Cancer (2019) 18(1):1–19. doi: 10.1186/s12943-019-1110-3
10. The UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res (2021) 49(D1):D480–D9. doi: 10.1093/nar/gkaa1100
11. Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet (2005) 37(6):588–9. doi: 10.1038/ng1575
12. Sollis E, Graham SA, Vino A, Froehlich H, Vreeburg M, Dimitropoulou D, et al. Identification and functional characterization of de novo FOXP1 variants provides novel insights into the etiology of neurodevelopmental disorder. Hum Mol Genet (2015) 25(3):546–57. doi: 10.1093/hmg/ddv495
13. Lozano R, Vino A, Lozano C, Fisher SE, Deriziotis P. A de novo FOXP1 variant in a patient with autism, intellectual disability and severe speech and language impairment. Euro J Hum Genet (2015) 23(12):1702–7. doi: 10.1038/ejhg.2015.66
14. Konopacki C, Pritykin Y, Rubtsov Y, Leslie CS, Rudensky AY. Transcription factor Foxp1 regulates Foxp3 chromatin binding and coordinates regulatory T-cell function. Nat Immunol (2019) 20(2):232–42. doi: 10.1038/s41590-018-0291-z
15. Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, et al. Foxp1 is an essential transcriptional regulator of b-cell development. Nat Immunol (2006) 7(8):819–26. doi: 10.1038/ni1358
16. Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. (2008) 112(12):4699–711. doi: 10.1182/blood-2008-01-137018
17. Fujino N, Ota C, Suzuki T, Suzuki S, Hegab AE, Yamada M, et al. Analysis of gene expression profiles in alveolar epithelial type II-like cells differentiated from human alveolar epithelial progenitor cells. Respir Investig (2012) 50(3):110–6. doi: 10.1016/j.resinv.2012.07.002
18. Brown PJ, Wong KK, Felce SL, Lyne L, Spearman H, Soilleux EJ, et al. FOXP1 suppresses immune response signatures and MHC class II expression in activated b-cell-like diffuse large b-cell lymphomas. Leukemia (2015) 30(3):605–16. doi: 10.1038/leu.2015.299
19. Yamada S, Sato F, Xia H, Takino H, Kominato S, Ri M, et al. Forkhead box P1 overexpression and its clinicopathologic significance in peripheral T-cell lymphoma, not otherwise specified. Hum Pathol (2012) 43(8):1322–7. doi: 10.1016/j.humpath.2011.10.013
20. Choi EJ, Seo EJ, Kim DK, Lee SI, Kwon YW, Jang IH, et al. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget. (2016) 7(3):3506–19. doi: 10.18632/oncotarget.6510
21. Wang X, Sun J, Cui M, Zhao F, Ge C, Chen T, et al. Downregulation of FOXP1 inhibits cell proliferation in hepatocellular carcinoma by inducing G1/S phase cell cycle arrest. Int J Mol Sci (2016) 17(9):1534. doi: 10.3390/ijms17091501
22. Xiao J, He B, Zou Y, Chen X, Lu X, Xie M, et al. Prognostic value of decreased FOXP1 protein expression in various tumors: a systematic review and meta-analysis. Sci Rep (2016) 6:30437. doi: 10.1038/srep30437
23. De Silva P, Garaud S, Solinas C, de Wind A, Van den Eyden G, Jose V, et al. FOXP1 negatively regulates tumor infiltrating lymphocyte migration in human breast cancer. EBioMedicine. (2019) 39:226–38. doi: 10.1016/j.ebiom.2018.11.066
24. Durek P, Nordström K, Gasparoni G, Salhab A, Kressler C, de Almeida M, et al. Epigenomic profiling of human CD4(+) T-cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity (2016) 45(5):1148–61. doi: 10.1016/j.immuni.2016.10.022
25. Garaud S, Roufosse F, De Silva P, Gu-Trantien C, Lodewyckx J-N, Duvillier H, et al. FOXP1 is a regulator of quiescence in healthy human CD4+ T-cells and is constitutively repressed in T-cells from patients with lymphoproliferative disorders. Eur J Immunol (2017) 47(1):168–79. doi: 10.1002/eji.201646373
26. Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8(+) T-cell subsets. Nat Immunol (2011) 12(12):1221–U117. doi: 10.1038/ni.2158
27. Lefèvre G, Copin M-C, Staumont-Sallé D, Avenel-Audran M, Aubert H, Taieb A, et al. The lymphoid variant of hypereosinophilic syndrome: study of 21 patients with CD3-CD4+ aberrant T-cell phenotype. Med (Baltimore). (2014) 93(17):255–66. doi: 10.1097/MD.0000000000000088
28. Ravoet M, Sibille C, Gu C, Libin M, Haibe-Kains B, Sotiriou C, et al. Molecular profiling of CD3–CD4+ T-cells from patients with the lymphocytic variant of hypereosinophilic syndrome reveals targeting of growth control pathways. Blood (2009) 114(14):2969–83. doi: 10.1182/blood-2008-08-175091
29. Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large b-cell lymphoma (DLBCL) patients with poor outcome. Blood. (2004) 104(9):2933–5. doi: 10.1182/blood-2004-03-1209
30. Han SL, Wu XL, Wan L, Zeng QQ, Li JL, Liu Z. FOXP1 expression predicts polymorphic histology and poor prognosis in gastric mucosa-associated lymphoid tissue lymphomas. Dig Surg (2009) 26(2):156–62. doi: 10.1159/000212058
31. Brown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, et al. Potentially oncogenic b-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated b-cell-like subtype of DLBCL. Blood, J Am Soc Hematol (2008) 111(5):2816–24. doi: 10.1182/blood-2007-09-115113
32. Green MR, Gandhi MK, Courtney MJ, Marlton P, Griffiths L. Relative abundance of full-length and truncated FOXP1 isoforms is associated with differential NFkappaB activity in follicular lymphoma. Leuk Res (2009) 33(12):1699–702. doi: 10.1016/j.leukres.2009.05.004
33. van Keimpema M, Grüneberg LJ, Schilder-Tol EJM, Oud MECM, Beuling EA, Hensbergen PJ, et al. The small FOXP1 isoform predominantly expressed in activated b-cell-like diffuse large b-cell lymphoma and full-length FOXP1 exert similar oncogenic and transcriptional activity in human b-cells. Haematologica. (2017) 102(3):573–83. doi: 10.3324/haematol.2016.156455
34. Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T-cells during thymocyte development. Blood. (2010) 115(3):510–8. doi: 10.1182/blood-2009-07-232694
35. Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T-cells. Nat Immunol (2011) 12(6):544–50. doi: 10.1038/ni.2034
36. Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the forkhead-box transcription factor Foxo1 in control of T-cell homeostasis and tolerance. Immunity. (2009) 30(3):358–71. doi: 10.1016/j.immuni.2009.02.003
37. Rajesh RR, Li Q, Bupp MRG, Shrikant PA. Transcription factor Foxo1 represses T-bet-Mediated effector functions and promotes memory CD8+ T-cell differentiation. Immunity (2012) 36(3):374–87. doi: 10.1016/j.immuni.2012.01.015
38. Wei H, Geng J, Shi B, Liu Z, Wang YH, Stevens AC, et al. Cutting edge: Foxp1 controls naive CD8+ T-cell quiescence by simultaneously repressing key pathways in cellular metabolism and cell cycle progression. J Immunol (2016) 196(9):3537–41. doi: 10.4049/jimmunol.1501896
39. Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, et al. Transforming growth factor beta-mediated suppression of antitumor T-cells requires FoxP1 transcription factor expression. Immunity. (2014) 41(3):427–39. doi: 10.1016/j.immuni.2014.08.012
40. Xun X, Zhang C, Wang S, Hu S, Xiang X, Cheng Q, et al. Cyclooxygenase-2 expressed hepatocellular carcinoma induces cytotoxic T lymphocytes exhaustion through M2 macrophage polarization. Am J Transl Res (2021) 13(5):4360–75.
41. Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu Q, et al. miR-149-3p reverses CD8(+) T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol (2019) 9(10):190061. doi: 10.1098/rsob.190061
42. Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T-cells. Nat Immunol (2014) 15(7):667–75. doi: 10.1038/ni.2890
43. Liu XD, Yan XW, Zhong B, Nurieva RI, Wang AB, Wang XH, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med (2012) 209(10):1841–52. doi: 10.1084/jem.20120219
44. Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. (2008) 29(1):138–49. doi: 10.1016/j.immuni.2008.05.009
45. Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, et al. Follicular b helper T-cells express CXC chemokine receptor 5, localize to b-cell follicles, and support immunoglobulin production. J Exp Med (2000) 192(11):1545–52. doi: 10.1084/jem.192.11.1545
46. Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, et al. T Follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for b-cells. J Immunol (2004) 173(1):68–78. doi: 10.4049/jimmunol.173.1.68
47. Tan AH, Wong SC, Lam KP. Regulation of mouse inducible costimulator (ICOS) expression by fyn-NFATc2 and ERK signaling in T-cells. J Biol Chem (2006) 281(39):28666–78. doi: 10.1074/jbc.M604081200
48. Shi B, Geng J, Wang Y-H, Wei H, Walters B, Li W, et al. Foxp1 negatively regulates T follicular helper cell differentiation and germinal center responses by controlling cell migration and CTLA-4. J Immunol (Baltimore Md 1950). (2018) 200(2):586–94. doi: 10.4049/jimmunol.1701000
49. Bi E, Ma X, Lu Y, Yang M, Wang Q, Xue G, et al. Foxo1 and Foxp1 play opposing roles in regulating the differentiation and antitumor activity of TH9 cells programmed by IL-7. Sci Signaling (2017) 10(500):eaak9741. doi: 10.1126/scisignal.aak9741
50. Lu Y, Hong S, Li H, Park J, Hong B, Wang L, et al. Th9 cells promote antitumor immune responses in vivo. J Clin Invest (2012) 122(11):4160–71. doi: 10.1172/JCI65459
51. Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T-cells. Nat Med (2012) 18(8):1248–53. doi: 10.1038/nm.2856
52. Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T-cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T-cells. Nat Immunol (2008) 9(12):1347–55. doi: 10.1038/ni.1677
53. Ghosh S, Roy-Chowdhuri S, Kang K, Im SH, Rudra D. The transcription factor Foxp1 preserves integrity of an active Foxp3 locus in extrathymic treg cells. Nat Commun (2018) 9(1):4473. doi: 10.1038/s41467-018-07018-y
Keywords: FoxP1, T cell, Treg, quiescence, T cell differentiation, isoforms, antitumor, exhaustion
Citation: Kaminskiy Y, Kuznetsova V, Kudriaeva A, Zmievskaya E and Bulatov E (2022) Neglected, yet significant role of FOXP1 in T-cell quiescence, differentiation and exhaustion. Front. Immunol. 13:971045. doi: 10.3389/fimmu.2022.971045
Received: 08 July 2022; Accepted: 20 September 2022;
Published: 04 October 2022.
Edited by:
Andrew D. Wells, Children’s Hospital of Philadelphia, United StatesReviewed by:
Pushpamali De Silva, Dana–Farber Cancer Institute, United StatesAgeliki Tsagaratou, University of North Carolina at Chapel Hill, United States
Copyright © 2022 Kaminskiy, Kuznetsova, Kudriaeva, Zmievskaya and Bulatov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emil Bulatov, chembio.kazan@gmail.com
 Yaroslav Kaminskiy
Yaroslav Kaminskiy Varvara Kuznetsova2
Varvara Kuznetsova2 Emil Bulatov
Emil Bulatov