- 1Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 2Arthritis and Clinical Immunology, Oklahoma Medical Research Foundation, Oklahoma City, OK, United States
- 3Department of Biochemistry and Molecular Biology and Geriatric Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 4Division of Rheumatology, Immunology and Allergy, Department of Cell Biology and Internal Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 5Department of Medicine and Pathology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
Memory B cells that are generated during an infection or following vaccination act as sentinels to guard against future infections. Upon repeat antigen exposure memory B cells differentiate into new antibody-secreting plasma cells to provide rapid and sustained protection. Some pathogens evade or suppress the humoral immune system, or induce memory B cells with a diminished ability to differentiate into new plasma cells. This leaves the host vulnerable to chronic or recurrent infections. Single cell approaches coupled with next generation antibody gene sequencing facilitate a detailed analysis of the pathogen-specific memory B cell repertoire. Monoclonal antibodies that are generated from antibody gene sequences allow a functional analysis of the repertoire. This review discusses what has been learned thus far from analysis of diverse pathogen-specific memory B cell compartments and describes major differences in their repertoires. Such information may illuminate ways to advance the goal of improving vaccine and therapeutic antibody design.
Introduction
The long-term efficacy of vaccines is determined in large part by the generation of B and T cell memory (1, 2). Memory B cells (Bmem) defend against previously experienced pathogens by differentiation into antibody (Ab)-secreting plasma cells (PCs) (3, 4). However, certain pathogens drive functional changes in the Bmem compartment that may be age-dependent and contribute to chronic or recurrent infections (5, 6). Understanding the characteristics and the diversity of protective, ineffective, and pathogenic Bmem responses is likely to aid in the development of efficacious vaccines and therapeutic Abs. In this review we present an overview of Bmem cellular subsets in humans. We highlight recent methods that have allowed us to explore the Bmem Ab gene repertoire. We then examine what is known about the human Bmem Ab repertoires that have been observed following vaccination or infection.
Memory B Cell Generation
B cell receptor (BCR) diversity within the naïve B cell compartment results from the recombination of the variable (V), diversity (D), and joining (J) genes in the heavy (H) chain and VJ genes in the light (L) chain (kappa and lambda) during B cell maturation. During an infection, naïve B cells are exposed to antigen (Ag) in the secondary lymphoid organs and undergo activation and differentiation including somatic hypermutation (SHM) and immunoglobulin (Ig) class switching to produce high affinity Abs. Ag-activated B cells may undergo differentiation into Bmem or short and long-lived PCs. The differentiation into Bmem may occur with or without T cell help and in a germinal center (GC)-dependent or independent manner (7) (Figure 1). This results in Bmem subsets that differ in their effector function and overall capacity for protection (3, 8).
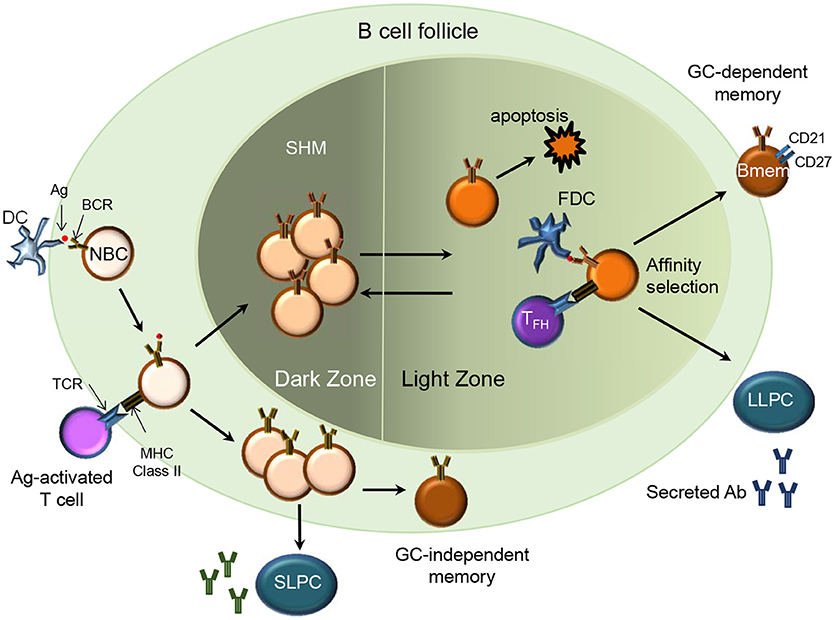
Figure 1. Memory B cell generation (T cell-dependent). In a secondary lymphoid organ, Ag-activated naïve B cells (NBC) via their MHC class II receptors and T cells via their T cell receptors (TCR) form stable interactions upon migrating to the borders of the B cell follicle and T cell zone respectively. B cells then proliferate and form the germinal center (GC)-dependent Bmem, GC-independent Bmem or differentiate into Ab-secreting short-lived plasma cells (SLPC) in an extra-follicular foci. The Ag-specific B cells undergo clonal expansion and SHM in the dark zone. B cells with diversified BCR may relocate to the light zone where they encounter immune complex-coated follicular dendritic cells (FDC) and Ag-specific follicular helper T cells (TFH). Through the process of affinity selection, the B cells with high affinity for the Ag survive while those with low affinity undergo apoptosis. After affinity maturation, the B cells can either re-enter the GC or exit the GC as Bmem or Ab-secreting long-lived plasma cells (LLPC) that home to their survival niche in the bone marrow.
T-Dependent Bmem
Ag-activated B cells receiving T cell help may differentiate into extrafollicular short-lived PCs, GC-dependent Bmem or GC-independent Bmem (7) (Figure 1). Imaging studies in mice indicate that durable interactions of B cells with cognate follicular helper T cells (Tfh cells) at the B cell- T cell border in spleen and lymph nodes provides T cell help and promotes entry of the B cells in to the GC (9, 10). GC B cells undergo affinity maturation and SHM and upon cognate interaction with GC Th cells differentiate into Bmem or long-lived PCs (11). Mechanisms regulating the differentiation of GC B cells to Bmem are not yet well understood. While most Bmem result from a GC reaction, mouse studies indicate that CD40 signaling via T cells can drive differentiation of GC-independent Bmem (12) and that these cells express Abs with fewer mutations and low affinity (13). However, no clear evidence from human studies supports a GC-independent path for the generation of human Bmem.
T-Independent Bmem
T-independent Ags such as polysaccharides and other molecules displaying repeating epitopes have long been considered incapable of generating a “memory” response. However, based on murine studies, B-1b cells found predominantly in the peritoneal cavity and marginal zone B cells are the primary precursors for T-independent memory (14). In humans, IgM+/IgD+/CD27+ B cells observed in human peripheral blood and spleen are the only known B cell subset associated with an Ab response to a T-independent Ag (15). Several studies including one involving immunization with a polysaccharide vaccine, demonstrated that the IgM+ Bmem in the periphery were derived from splenic marginal zone B cells with a pre-diversified Ig repertoire (15–19).
Memory B Cell Subsets
While early studies on Bmem in humans and mice predominantly involved IgG+ cells, later studies revealed the existence of IgM+ Bmem. The use of CD27 as a Bmem surface marker in humans further distinguished the memory from naïve IgM+ B cells. These surface markers help identify the potential origin and function of each Bmem subset. A detailed discussion of human Bmem generation, subsets and function has been provided in other reviews (1, 8). Here, we highlight the human Bmem subsets based on their Ig isotypes and their importance during repertoire analysis.
IgM+
The human Bmem compartment was believed to be exclusively or mostly composed of class-switched B cells, however studies have demonstrated the presence of somatically mutated IgM+ B cells (20). B cells expressing CD27 and either IgM alone or IgM and IgD represent the IgM+ Bmem subset. While the GC-dependence of IgM only Bmem is accepted, the origin of the IgM+/IgD+ subset is contentious. In their review of human Bmem, Seifert and Küppers (8) discuss evidence for a GC origin of this IgD+ subset including: (a) their ability to reenter the GC upon secondary challenge, (b) their tendency to differentiate to PCs, (c) their longevity, and (d) their transcriptome profile. Thus, the GC-dependent origin of IgM+/IgD+ cells supports their characterization as Bmem.
The peripheral blood IgM+/IgD+/CD27+ Bmem correspond to circulating splenic marginal zone B cells and contribute to protection against blood-borne T-independent pathogens (15, 21). Pneumococcal polysaccharide-specific IgM+/IgD+/CD27+ Bmem displayed somatic mutations when isolated post immunization with a T-independent vaccine (Pneumovax) (22). A vesicular stomatitis virus-Ebola vaccine generated a neutralizing IgM response that persisted despite administration of a booster vaccine (23). These studies point to a previously under-appreciated contribution of hyper-mutated IgM+ Bmem to protection against pathogens.
IgG+
IgG-expressing Bmem constitute 15–20% of the peripheral blood B lineage cells in adults. The structure and composition of the Ag as well as regulatory factors determine how the IgG+ Bmem cells class switch to express mainly IgG1, IgG2, or IgG3 and to a lesser extent IgG4. Some examples include: the importance of the polysaccharide-specific IgG2 response in protection against Streptococcus pneumoniae; the predominance of IgG1 and IgG3 subclasses generated against HIV, Ebola virus, and Plasmodium falciparum; and IgG4 responses to Schistosoma mansoni (24). However, not all the subclasses expressed are equally efficacious. For example, IgG3 was found to be more effective at neutralizing HIV than IgG1 (25). While the majority of the IgG expressing Bmem are CD27+, 20–25% lack CD27 expression (26). IgG+/CD27− Bmem cells have fewer mutations in their V regions and predominantly express the IgG3 subclass (26, 27). This subpopulation is increased in the elderly and is hypothesized to represent an “exhausted” Bmem pool (28). IgG+ Bmem upon reactivation typically differentiate into PCs rather than re-enter the GC. Therefore, the IgG subclass is also an important aspect of the Ab repertoire that should be considered in analyses of data sets.
IgA+
IgA-expressing Bmem are associated with mucosal immune responses and tend to arise from and localize in the intestine and mucosa-associated lymphoid tissue. They make up ~10% of the B cells in the periphery. While most IgA+ Bmem are CD27+, there is evidence of less mutated IgA+ CD27− cells undergoing low levels of proliferation and expressing poly-reactive Abs (29, 30). This phenotype is indicative of cells generated independent of the GC. Alternatively, an early exit from the GC allows for a broader and less mutated IgA+ Bmem which could cross-protect against related pathogens such as enterotoxigenic E. coli and Vibrio cholera (31). A recent study demonstrated that IgM+ Bmem shared gut-specific gene signatures with IgA+ Bmem, were related to some IgA+ clonotypes and could switch to IgA upon T-dependent or independent signals (32). Sustained Ag presence could drive a protective IgA response and could be utilized to improve oral vaccines.
IgE+
Although the presence of IgE antibodies and their causal relationship with atopic diseases such as allergy and asthma is well established, their generation is not well understood and they are detected at very low levels in human peripheral blood. Studies in mouse models have demonstrated the potential for “sequential switching” wherein IgG1 cells switch to IgE Ab-secreting cells (33–35). Another study examined the repertoire of human parental Bmem and their progenies. In that study, it was demonstrated that high affinity IgE-secreting PC clones were derived from the selection and expansion of rare high affinity IgG1 Bmem clones without undergoing further mutation (36). Antibody repertoire analysis of IgE+ B cells in patients with seasonal rhinitis demonstrated that the V gene usage was limited and similar across multiple patients (37). Furthermore, people with parasitic infections and patients with atopic dermatitis had less clonal diversity and lower frequency of SHM in their IgE repertoires than those with asthma (38). These differences reiterate the importance of examining the pathogen-directed IgE repertoire in the context of specific pathological events.
Atypical, Tissue-Like, or Exhausted Memory B Cells
HIV, Mycobacterium tuberculosis, Plasmodium falciparum, and Hepatitis C virus cause chronic infections and account for more than five million deaths a year. The chronic presence of Ag, prematurely aborted GC, extra-follicular differentiation or loss of survival niche may drive the expansion of a phenotypically and functionally altered Bmem subset referred to as “exhausted,” “tissue-like,” or “atypical” Bmem (Figure 2) (39–42). Distinct from typical CD27+ Bmem, these atypical Bmem do not express CD27 and cannot be stimulated via their BCR to subsequently produce Ab. HIV-associated CD21lo/CD27− cells expressed high levels of CD20 and their expression of CD11c, T-bet and inhibitory receptors of the Fc receptor like (FcRL) family distinguished them from other B cell subsets (40). Their resemblance to the FcRL4-expressing cells resident in the tonsils defined them as “tissue-like” Bmem. The tonsillar CD20hi/CD21lo/CD27−/FcRL4+ B cells had undergone isotype switching and SHM similar to CD27+ Bmem but were non-responsive to stimulation through BCR cross-linking (43). Atypical FcRL4-expressing Bmem were also observed to be increased in frequency in individuals with chronic HCV infection (44) and in those with active and latent TB infection (45). A similarly expanded subset of atypical Bmem was observed in those repeatedly infected with P. falciparum (46, 47). The atypical Bmem in patients with malaria express FcRL5 rather than the FcRL4 expression observed on tissue-like Bmem in HIV (48). In keeping with the “exhausted” phenotype, these FcRL5+ atypical cells were more refractive to BCR crosslinking and CpG stimulation as compared to FcRL5− Bmem. While T-bet-expressing, CD21low/− B cells have been observed in individuals with autoimmune conditions such as rheumatoid arthritis (49) and systemic lupus erythematosus (50), they likely differ phenotypically and functionally from cells arising during chronic infections (6). Chronic immune stimulation and inflammation (Figure 2) are thought to contribute to the expansion of atypical Bmem that are unable to secrete Ab which could explain lack of acquisition of immunity against chronic infections.
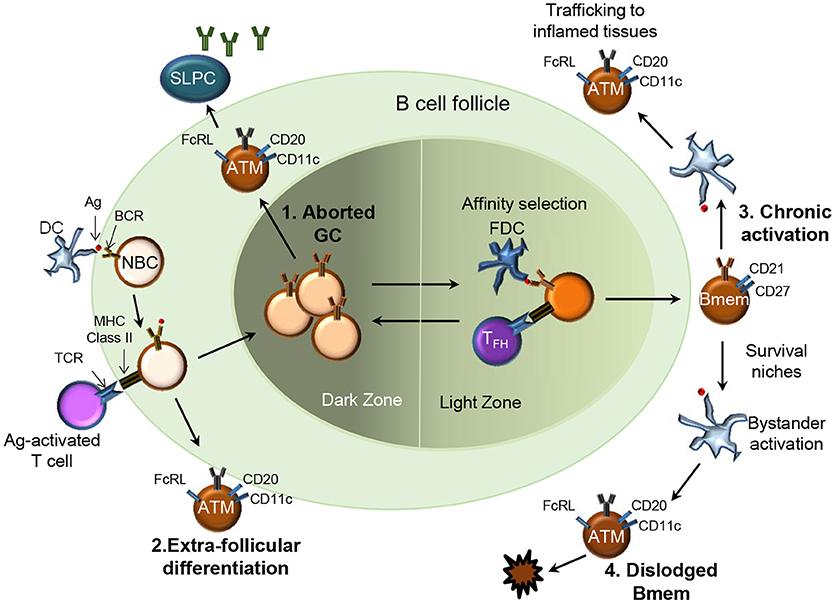
Figure 2. Four potential pathways for generation of atypical memory B cells in chronic infections. Several pathways for the origin of atypical Bmem (ATM) have been proposed: 1.They may be Bmem derived from prematurely aborted GCs (top left), 2. Bmem derived from an extra-follicular differentiation pathway (bottom left); 3. Chronic Ag-mediated activation of previously functional Bmem may drive the expansion of ATM (top right), or 4. Represent the end stage for Bmem dislodged from survival niches due to repeated bystander activation (bottom right).
Human Monoclonal Antibody Production Methods to Sample the Memory B Cell Repertoire
Methods used to produce human monoclonal Abs (mAbs) include combinatorial display libraries, B cell immortalization, single-cell expression cloning and hybridoma generation (Figure 3) (51–53). Each of these methods has been used to improve our understanding of the Bmem repertoire.
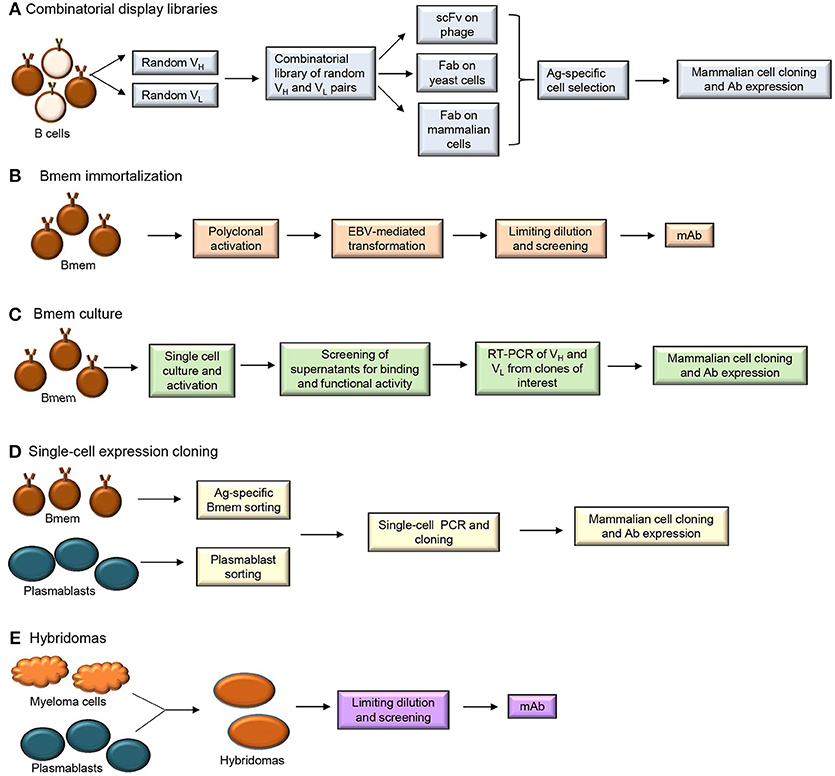
Figure 3. Human monoclonal antibody isolation methods (A) Combinatorial display libraries (B) Bmem immortalization (C) Bmem cell culture (D) Single-cell expression cloning and (E) Hybridoma approaches.
Generation of combinatorial display libraries of single-chain variable fragments (scFvs) and Ag-binding fragments (Fabs) (Figure 3A) is a high-throughput screening method that has allowed discovery of several pathogen-neutralizing Abs (54–57). Libraries of Ab V genes are generated from B cells isolated from an individual post-immunization or post-infection. These random pairs of H and L chain genes are then expressed on phage, yeast or mammalian cells. The Ag-specific Ab-expressing cells or particles are selected and cloned into mammalian cells for mAb expression. Since the H and L chain pairing occurs randomly during this process, the resulting Ab does not represent a pairing that may occur naturally in the immune system. This technique can allow generation of higher affinity Abs but does not allow characterization of the properly-paired Ag-specific B cell repertoire.
B cell immortalization (Figure 3B) involves transformation of Bmem using Epstein Barr Virus (EBV) in the presence of TLR (Toll-like receptor) 9 agonist CpG DNA (58). Total blood lymphocytes or isolated IgG+ Bmem are typically cultured with EBV and CpG and/or additional co-stimulants for 1–2 weeks to allow proliferation of B cells and secretion of Abs. Supernatants from the cultures are then harvested to test for Ag-specific Ab after which the individual Ig H and L chain pairs are cloned and sequenced. EBV-transformed B cells allow Bmem analysis but require screening of significant numbers of cells (~1,000) to identify the very few Bmem (~10) of interest. Bmem can also be immortalized by forced expression of BCL6 and BCL-XL and when cultured in the presence of CD40L/IL-21 these plasmablast-like transduced cells secrete Ab in the supernatant (59). While plasmablasts are not amenable to immortalization, recent studies have demonstrated that both Bmem and plasmablasts can be cultured in vitro (Figure 3C), without immortalization, for Ab production (60–62).
Single-cell expression cloning involves application of reverse transcription-polymerase chain reaction (RT-PCR) at the single-cell level for the amplification of Ig H and L chain genes that can be cloned and expressed in mammalian cell lines (Figure 3D). Plasmablasts and Bmem have been sorted from individuals post-vaccination or post-infection to generate human mAbs by single-cell expression cloning. This technique involves specific PCR amplification of Ab transcripts from individual cells and is technically challenging and dependent upon high quality primers (63). Application of single-cell expression cloning for repertoire analyses can be difficult in peripheral B cell subsets that have fewer Ig transcripts per cell than plasmablasts (64). Plasmablasts that secrete larger quantities of Ig are more amenable to these types of protocols. Although number of circulating plasmablasts are typically low in healthy individuals, 5–8 days after vaccination (or infection) there is a transient but large population of Ag-specific Ig-secreting cells that can be easily isolated (63, 65). This population can be distinguished from the IgG+ Bmem population which peaks at 2–3 weeks post-immunization. Plasmablasts sorted from human PBMC 1 week after vaccination against influenza, Bacillus anthracis and Streptococcus pneumoniae have successfully yielded Ag-specific and high affinity mAbs (52).
Ag-specific plasmablasts and Bmem have been isolated by fluorescence-activated cell sorting (FACS) using reagents that bind Ag-specific cells (Ag baiting) (66–71). IgG+ PCs and plasmablasts are difficult to isolate in an Ag-specific manner due to their limited surface expression of Ig (72). However, a flow cytometry-based Ig capture assay was established to sort IgG+ Ag-specific plasmablasts for three vaccine Ags, namely HIV gp140, tetanus toxin and Hepatitis B surface Ag (73). This method captured the secreted Ab from the plasmablast enabling the fluorescent Ag probe to identify Ag-specific cells. Isolating Ag-specific human Bmem is also challenging since they circulate at very low frequencies in the periphery (74) but they can be expanded in vitro and immortalized using EBV-transformation techniques mentioned earlier. A recent study isolated human CD27+ Bmem and demonstrated that “an in vitro booster vaccination” consisting of streptavidin-coated nanoparticles conjugated with biotinylated Ag and CpG DNA generated Ag-specific mAbs in a period of 6 days (75).
Traditional mAb generation methods such as hybridoma technology (Figure 3E) involve the production of Ab from a hybridoma clone formed by the fusion of a myeloma cell with B cell from a donor or an immunized animal (53, 76). Although this method can generate high affinity mAb, it has to be combined with transgenic humanized mouse strains to obtain fully human mAb (77).
Next Generation Sequencing to Study Memory B Cell Repertoire
The ability to accurately perform deep sequencing of Ab VDJ regions has provided valuable insights into the regulation and evolution of the Bmem repertoire. Rapidly developing high-throughput next-generation sequencing technologies have achieved high resolution in the number of single chains that can be sequenced in one experiment (78). However, information about the endogenous pairing of IgH/IgL genes cannot be obtained by single chain sequencing which then hinders an accurate representation of the B cell repertoire. This suggests that improved methods for template preparation, sequencing and imaging, and data analysis are required for more accurate and efficient large scale analysis of functional Ab repertoires (79).
A recently developed “ultra high-throughput” method amplifies IgH/IgL genes from a B cell captured within a droplet to create a single DNA amplicon allowing sequencing of paired chains (80). In addition, simultaneous sequencing of barcoded Ig genes along with co-expressed functional genes will better represent the Ab repertoire (79). Combining large datasets from several reads (deep sequencing) of B cell Ab repertoire sequences (BCR/Rep-seq) with data from mass spectrometry of serum Ab (Ig-seq) will allow comparison of the Ab repertoires in the two compartments (81, 82).
Single-cell RNA-seq technologies typically generate large datasets that require bioinformatics tools and expertise for deconvolution and interpretation. Several “pipelines” have been developed for BCR sequence analysis and will likely continue to evolve to accommodate data complexity (83, 84). As the cost per kilobase of high-throughput sequencing (HTS) has dropped, the past 3 years alone has seen a rapid increase in the number of published resources for analysis of B-cell sequence data (Table 1).
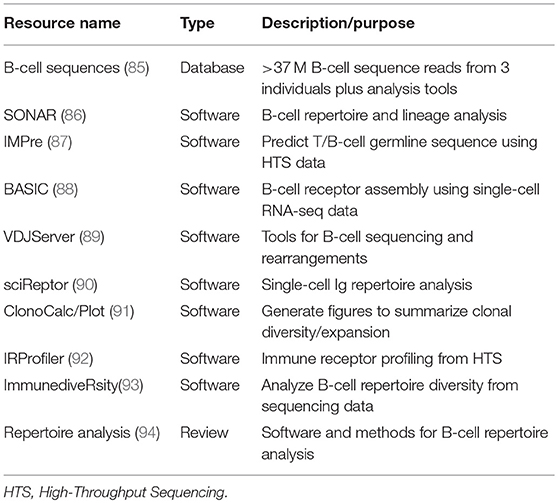
Table 1. Bioinformatics resources for analysis and comparison of B-cell antibody sequence diversity.
As we will discuss, the techniques outlined above have been applied to Bmem subsets, allowing an improved understanding of their development and differentiation during specific diseases and post-vaccination.
Infection-Induced Memory B Cell Antibody Repertoires
While there has been significant research on mAbs as therapeutics for multi-drug resistant Gram negative bacteria [reviewed in (95)], our knowledge on how they shape human Bmem repertoires is lacking and deserves further attention. The reader is also referred to three studies on the Bmem repertoire against Vibrio cholerae, Klebsiella pneumoniae, and Hemophilus influenzae (96–98). In this article, we have focused on Gram positive infections for which a larger number of published studies was available for analysis.
Gram Positive Bacterial Infections
Bacillus anthracis
Bacillus anthracis is a gram positive, rod-shaped bacteria that causes a serious, often fatal, infection in humans. Alarmingly, B. anthracis has been used as a biological weapon highlighting the seriousness of anthrax disease. Antibiotics are used for anthrax treatment and as post-exposure prophylaxis. The use of FDA approved Anthrax Vaccine adsorbed (AVA) is restricted to military personnel and those who might face occupational anthrax exposure. AVA is predominantly composed of protective Ag (PA), which is a component of the tripartite anthrax exotoxin. AVA elicits toxin neutralizing Abs that are protective, but generating the protective response requires multiple vaccinations and ongoing annual boosters (99). The vaccine generates a protective short-lived anti-PA IgG response in humans, but both human and animal studies using AVA demonstrated a long-lived anti-PA Bmem response (100, 101). Using mAbs from PA-specific B cells isolated from seven AVA-vaccinated individuals, Reason and coworkers showed that although a majority of the mAbs bound a specific 20 kDa region on the PA monomer, their VH region sequence analysis revealed 64 unique gene rearrangements (102). Ab-secreting cells were also isolated from individuals 7 days following AVA or recombinant PA vaccination to generate and characterize human PA-specific mAbs (103, 104). Several of those mAbs demonstrated in vitro toxin neutralization and were protective in vivo following challenge of mice with bio-active anthrax toxin. While several PA-specific mAbs are in development, the recombinant human mAb raxibacumab is the only one approved for use in treatment of inhalational anthrax (105).
Clostridium tetani
Clostridium tetani secretes tetanus toxin, which is responsible for the symptoms associated with tetanus disease. Since the introduction of tetanus toxoid (TT)-containing vaccines in the mid-1940s, the incidence of reported tetanus cases in the United States declined by over 98% from 0.39 per 100,000 population in 1947 to 0.01 per 100,000 population in 2016 (106). The tetanus-specific Ab repertoire has been characterized by several groups using different methods. Analysis of Fab libraries created from plasmablasts isolated day 6 post-TT vaccination indicated that 100 B cell clones and their hyper-mutated variants comprised the human polyclonal Ab repertoire (107). The follow-up study examined the plasmablast V gene region after 3 TT vaccinations and suggested that the majority of the B cell clones developed in response to a single vaccination event (108). Libraries created using a single human VL region paired with a collection of VH regions resulted in diverse and high affinity Abs where the TT-specificity was encoded solely by VH (109). Lavinder and coworkers used high-resolution liquid chromatography tandem MS proteomic analyses of serum Abs coupled with next-generation sequencing of the V gene repertoire in peripheral B cells to understand the serum IgG and B cell repertoire following TT booster vaccination (110). This study confirmed the previous finding that TT+ serum IgG comprised ~100 clonotypes, but they found that only 3 clonotypes accounted for >40% of the response and that <5% of plasmablast clonotypes account for the Abs detected in the serum 9 months post-vaccination. As TT vaccines are very effective in preventing tetanus, characterizing the Bmem repertoire generated in response to this vaccine will provide information that is valuable in the improvement of vaccines to other pathogens.
Streptococcus pneumoniae
Streptococcus pneumoniae is a causative agent for community-acquired pneumonia, a health care burden worsened by antibiotic resistance. Despite effective vaccines, the lack of adequate coverage especially in adults results in a large at-risk population (111). This problem is compounded by some serotypes not being represented in current vaccines (112).
The Ab repertoire in response to vaccination with the 23-valent polysaccharide vaccine and the conjugate vaccine has been examined (113–116). Full-length fully human serotype-specific mAbs and anti-cell wall polysaccharide mAbs generated from individuals 7 days post-vaccination were examined for their V gene usage and clonal families (113). Zhou and colleagues generated human Fab fragments specific to the capsular polysaccharide of S. pneumoniae strains 23F and 6B to determine their H and L chain variable gene usage (115, 116). Bmem-derived human hybridomas were generated from recipients of the conjugate vaccine to study the structure-function correlation of mAbs and how that may affect the B cell repertoire during invasive pneumococcal infection (114). While different methods were utilized to examine the Ab repertoire in the studies cited above, they all demonstrated an increased frequency of SHM in response to primary vaccination. This suggested that the recipients were either asymptomatic carriers or previously exposed to a wild-type strain of S. pneumoniae which allowed the vaccination to elicit a memory response. Furthermore, the studies also revealed that the Ab repertoire across individuals was oligoclonal based on their limited usage of VH and VL families and similar H and L pairing for specific serotypes. Two studies identified IgG2 and IgA H chains matching the Ab isotype and response found in the donor sera indicating that the Fabs and Abs derived were from a response to S. pneumoniae infection (114, 115). Further characterization of the full-length fully human serotype-specific mAbs revealed a preferential use of lambda over kappa light chains in response to certain serotypes (117).
The emergence of S. pneumoniae serotypes not covered in the vaccine has driven the efforts to develop a “universal” vaccine that is not serotype-specific. Pneumococcal surface protein A (PspA) is found in all S. pneumoniae isolates and studies examining the Ab response against its proline-rich region have indicated that PspA may be a good vaccine candidate (118–120). A detailed examination of the human anti-PspA Ab repertoire would allow a better understanding of the efficacy of PspA-based vaccines.
Parasitic Infection
Plasmodium falciparum
Plasmodium falciparum is the mosquito-borne parasite that causes malaria. According to the CDC, malaria caused an estimated 216 million cases of malaria and 445,000 deaths worldwide in 2016. With repeated exposure, older children and adults slowly develop resistance to severe illness and death but never achieve complete resistance to infection (121). In humans, P. falciparum infection generates a long-lived atypical Bmem response which develops slowly, after many years of malaria exposure, and is limited in magnitude (122). The neutralizing IgG+ Abs produced by atypical Bmem seen during malaria infection in adults differ in their V region repertoire from Abs produced by classical Bmem (123). A similar study that examined the V gene of Abs in children, found no differences between the classical and atypical Bmem repertoire (121). Infant immune repertoires are poorly characterized and thought to be limited in their ability to respond to Ag challenge (124). A recent study isolated Bmem from infants and toddlers pre-infection and during acute malaria infection and examined the Ab repertoire using a method known as molecular identifiers clustering-based immune repertoire sequencing (125). This study found that infant Bmem could acquire over 20 mutations per H chain V region in response to malaria infection and the breadth of repertoire achieved was comparable to that in young adults exposed to malaria. It was also observed that upon malaria re-challenge, Bmem from toddlers previously exposed to malaria could undergo further mutations while retaining IgM expression. The ability to sequence and analyze the immune repertoire even from small samples has allowed a better understanding of the infant Bmem compartment which in turn can inform vaccine design and strategy to prevent malaria in this age group.
Viral Infections
The role of serum Ab as an immunotherapy in viral infections has been studied for over 50 years and its importance in prophylaxis is well recognized (126). Improved investigative tools have enabled us to identify neutralizing viral epitopes that can influence vaccine design and Ab engineering. Herein, we focus on the B cell and Bmem repertoires of some more commonly studied viruses.
Influenza Virus
Influenza virus causes annual epidemics and poses a significant risk for mortality in immunocompromised individuals, the elderly (>65 years) and young children (<5 years). Due to its ability to constantly evolve and adapt, a “universal Influenza vaccine” has eluded investigators. The neutralizing Ab response to viral infection or vaccination mainly targets the glycoprotein hemagglutinin (HA) which prevents entry into target cells, while Abs directed against the second glycoprotein neuraminidase (NA) prevent virus exit and spread (52, 127). The Abs targeting the HA globular head are limited in breadth. In an effort to generate broadly neutralizing Abs, the focus has been on targeting the HA stem region which is conserved across strains. Abs neutralizing most strains in H1, H5, and H9 clades have been discovered by several groups using phage display and immortalization of Bmem cells (128–131). High affinity human Abs have been cloned from IgG+ Ab-secreting cells 1 week after vaccination. In that study it was observed that the repertoire had limited clonality but an accumulation of somatic mutations within each clone, resulting in extensive intraclonal diversity (63). A recent study examined the HA-specific Bmem repertoire from three unrelated individuals that were known to have BCRs cross-reactive to HA from group 1 and 2 influenza viruses (132). They characterized the clonal lineage and determined that germline receptors had the capacity to recognize epitopes on the head of the HA trimer of divergent Influenza subtypes (H1 and H3 subtype). Furthermore, they showed that the clonally related Abs bound both H1 and H3 HA via their long HCDR3 loop which had the same length and related amino acid sequence. The cross-reactive Abs bound H1 and H3 with differing affinities, suggesting that sequential exposure to each of the Influenza subtypes may have driven affinity maturation of those Abs. Other Ab repertoire analyses have revealed the presence of signature sequences and key somatic mutations in HCDR3 and the preferential VH gene usage for the generation of broadly neutralizing anti-influenza Abs (127, 133).
Human Immunodeficiency Virus
Human Immunodeficiency Virus infected an estimated 1.1 million people in the United States at the end of 2015 (134). With an error-prone reverse transcriptase and its ability to replicate rapidly, multiple variants of HIV co-exist in the host and allow successful evasion of the innate and adaptive immune response. The exposed HIV envelope glycoproteins (Env) are the most accessible to Ab and HIV has evolved mechanisms to avoid recognition by Ab. Evasion mechanisms employed by HIV include generating amino acid changes in the variable regions of the Env and close spacing of the N-glycans to prevent Ab recognition of Env (135). Chronically HIV-1-infected individuals develop anti-Env broadly neutralizing Abs. These Abs have increased SHM, activation-induced cytidine aminase- mediated insertions, deletions in the H or L chain sequences and long HCDR3s or self-reactivity (135). Env-specific single Bmem were sorted from macaques 14 days after immunization and VDJ sequences were examined to reveal 502 unique clonotypes and more than 600 functional sequences (136). Another study examining the Bmem repertoire generated from a chronically infected individual reported that the response was oligoclonal with about 50 clonotypes observed and that the high levels of SHM observed were not restricted to broadly-neutralizing Abs (137). Atypical or exhausted Bmem identified in chronically HIV-infected individuals were demonstrated to have fewer SHM and lower HIV neutralization capacity than resting Bmem-derived mAbs (138). Bmem immortalization, in vitro Bmem differentiation and next-generation sequencing techniques have also been applied to samples obtained from infected donors to generate and examine the Ab repertoire with HIV-neutralizing capacity (52, 139). Detailed information regarding differences in the Ab repertoire from “vaccinated” models and chronically infected individuals has been reviewed elsewhere (135).
Dengue Virus
Dengue Virus (DENV) is a mosquito-borne virus that infects an estimated 390 million people yearly (140). The traditional understanding of Bmem function is that these cells provide the means to rapidly respond to a pathogen upon repeat exposure and provide protection against disease. In DENV infections, however, where sequential infection with more than one serotype is common, the Abs from a memory response to a primary infection can worsen a secondary infection caused by a heterologous dengue virus (141). Halstead proposed Ab-dependent enhancement (ADE) to explain this severity during secondary infection (142). Cross-reactive non-neutralizing Ab generated from a primary response forms Ag-Ab complexes upon secondary infection with a heterologous DENV serotype. The cross-reactive Ab binds Fc receptors allowing more efficient viral entry and replication in Fc-expressing cells resulting in an exacerbated disease. The role of ADE was underscored by the discovery of non-neutralizing cross-reactive Abs generated by sorting IgG+ Bmem from DENV-infected individuals (143).
Acute DENV infection results in a large and rapid virus-specific plasmablast response (144) and mAbs derived from plasmablasts of patients with secondary DENV infection revealed a repertoire that was highly affinity matured and clonally expanded (145). These mAbs also demonstrated cross-reactivity to DENV serotypes in binding and neutralization. Appanna and colleagues sorted plasmablasts and Ag-specific Bmem from patients re-infected with DENV and examined their Ab repertoires to conclude that the plasmablast response represented only a fraction of the Bmem repertoire (146). That study emphasized the need to examine each B lineage compartment for a complete understanding of the humoral response. Recently, a DENV-exposed volunteer was vaccinated with a live attenuated tetravalent vaccine (Butantan-DV) and 15 days later plasmablasts were isolated to generate neutralizing Abs (147). The immunization resulted in neutralizing Abs to the previous exposure with DENV type 3 as well as six other mAbs that neutralized three or more serotypes. Additionally, the DENV surface glycoprotein E, the main target for neutralizing Abs, contains regions that are highly conserved between DENV and the re-emerging Zika virus (ZIKV) which results in immunological cross-reactivity (148). The mAbs generated from plasmablasts of DENV-infected individuals cross-reacted with ZIKV (149, 150) and ZIKV-specific mAbs from Bmem of primary ZIKV-infected individuals with no history of DENV infection cross-reacted with DENV E proteins (151).
Future Directions: Sequencing Antibody Repertoires to Target Urgent and Emerging Threats
Urgent Threats
In 2013, CDC designated Clostridioides difficile (C. difficile; previously Clostridium difficile), Carbapenem-resistant enterobacteriaceae and antibiotic-resistant Neisseria gonorrhea as “urgent threats.” Due to the challenge of antibiotic resistance in these pathogens, there has been an emphasis on generating novel therapeutics (95, 152, 153). However, to the best of our knowledge, the Ab repertoire in response to these infections have yet to be examined.
Clostridioides difficile, a spore-forming gram positive bacteria, is the leading cause of nosocomial infections in industrialized nations. With at least 500,000 new cases estimated in the US every year, C. difficile infection (CDI) is no longer restricted to health care facilities (154). CDI is a potentially debilitating illness ranging from mild diarrhea to a life-threatening pseudomembranous enterocolitis caused by the two C. difficile exotoxins Toxin A (TcdA) and Toxin B (TcdB) (154). The existence of recurrent CDI, suggesting mis-directed memory responses and emergence of new hyper-virulent ribotypes, poses a significant challenge to developing appropriate vaccines and therapeutics (155). Several vaccines and mAbs targeting TcdA and TcdB have been introduced in clinical trials and several more are being developed to address treatment and prevention of infection (152). Bezlotoxumab is a humanized mAb that binds TcdB and has been approved by the US FDA for treating recurrent CDI (156). Targeting the toxins for therapy seems appropriate considering studies have demonstrated anti-toxin circulating Ab and Bmem responses in certain individuals with C. difficile-associated diarrhea (157) and Bmem response to C-terminal domain (CTD) fragment of TcdB in those with history of CDI (158).
Results from murine studies indicate that while immunization with CTD from the historical strains such as VPI10463 generates a robust recall response with neutralizing Abs that protect against lethal challenge, the CTD from hyper-virulent strains such as BI/NAP1/027 generates a slow Bmem response with toxin-neutralizing Ab that only delays death post-lethal challenge (158). Interestingly, although the TcdB from the two strains share 92% homology, the anti-CTD Abs generated in immunized animal models do not cross neutralize toxin (159). Furthermore a low serum concentration of anti-TcdA and –TcdB Abs have been linked to recurrence (160) underscoring the need to examine in depth the Ab responses in individuals with history of CDI and compare responses in those that suffer recurrences to those that do not. Specifically understanding the Bmem compartment will provide insights into disease recurrence and support better treatment strategies.
Emerging Threats
Recent outbreaks of Ebola and Zika virus have accelerated the research and development of treatments to counter these threats (161). These renewed research efforts have led to several therapeutic and vaccine candidates against Ebola (162–164) and flaviviruses such as Zika and West Nile virus (165–167) and provided important information about the immune repertoire of individuals vaccinated against or infected with these viruses (23, 168–170). Chikungunya Virus (CHIKV), a mosquito-borne alpha virus first isolated in Tanzania in 1952, is now emerging as a world-wide threat. There are no licensed vaccines or therapies available currently to prevent or treat CHIKV. While the immune response to CHIKV infection is not fully understood, multiple studies have demonstrated the role of passive immunotherapy in controlling CHIKV infection (171–176).
Although great strides have been made to identify and characterize neutralizing mAbs as therapeutics, these Abs are typically derived from very few individuals. Expanding Ab repertoire studies by including more samples and accounting for variables such as age, gender, immune-competency, co-morbidities and time post-infection would be invaluable in designing appropriate prevention and treatment strategies for these and other emerging infections.
Concluding Remarks
Recent advances in cellular, molecular and computational techniques have provided detailed insights into the specificity and breadth of Ab responses. This in-depth knowledge has revealed the nuances of our immune system and its capacity to generate a pathogen-specific response, as described in this review.
Several considerations need to be taken into account when undertaking the arduous task of examining specific B cells at a single cell level. In addition, a successful transition toward clinical applications requires an understanding of the infecting pathogen or vaccine, the cell subset to be examined, the technology available and ultimately careful interpretation of the data.
Pathogens such as HIV and malaria lead to atypical memory, HIV and Influenza constantly adapt to evade the immune system, S. pneumoniae has several strains, Dengue virus worsens clinical disease upon reinfection and a primary infection with C. difficile renders the host prone to recurrent infections. Therefore, understanding the pathogen and its mechanism of action will allow better selection of antigenic targets to generate broadly neutralizing Ab.
The selection of the cell subset to be examined is crucial; specific Bmem are rare, but available months after the acute response has subsided and their Ab repertoire is reflective of a wider response providing a more “historical” perspective of the specific response. Plasmablasts peak shortly after vaccination or infection, do not require Ag-specific baiting for isolation and their repertoire is typically representative of the current response only. PCs are rare in the periphery but can be generated in vitro by Bmem differentiation and while they provide information on the existing memory, this methodology does not take into account the influence of the tissue microenvironment which cannot be fully recreated ex vivo.
While we have acquired considerable information of the Ag-specific B cell repertoire, we have just now begun to explore the possible existence of BCR “public lineages/clonotypes” in response to infection (177). Public clonotypes are described as the presence of dominant or nearly identical VDJ amino acid sequences across multiple individuals and have been better studied in the context of T cells rather than B cells. Analyzing Ig sequences shared across multiple individuals will be invaluable in understanding why these clonotypes emerge and the overall adaptive responses to specific pathogens.
In addition to infectious diseases, the analysis of B cell repertoires has improved our ability to detect immune system disorders and to elucidate the possible mechanisms causing them. For example, rheumatoid arthritis (RA) is a systemic autoimmune disease that often involves the production of anti-citrullinated protein Abs. Ab sequencing in RA patients has defined autoantibody specificities (178) and provided insights into clinical disease and the mechanisms leading to breaks in tolerance (179) and increased pro-inflammatory activity (180). In common variable immune deficiency, analysis of Ig H chain gene rearrangements demonstrated abnormal VDJ rearrangement and CDR3 formation, suggesting early differences in B cell selection and development in these patients (181). Finally, Ab studies have been pivotal in demonstrating the clonal origin of multiple myeloma (182) and more recently, sequencing technologies to define a patient's Ab repertoire have been applied clinically for disease detection and assessment of therapeutic response in myelomas (183) as well as leukemias (184).
Considering the knowledge and tools available to us, the goal of developing precise and personalized therapeutics is coming closer to fruition.
Author Contributions
HS, KS, JW, CW, JB, RB, JJ, and ML contributed to the writing and editing of this review article.
Funding
The ideas discussed herein were developed as part of a Team Science research award from the Presbyterian Health Foundation of Oklahoma City.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Weisel F, Shlomchik M. Memory B cells of mice and humans. Annu Rev Immunol. (2017) 35:255–84. doi: 10.1146/annurev-immunol-041015-055531
2. Jandus C, Usatorre AM, Vigano S, Zhang L, Romero P. The Vast Universe of T cell diversity, subsets of memory cells and their differentiation. Methods Mol Biol. (2017) 1514:1–17. doi: 10.1007/978-1-4939-6548-9_1
3. Kurosaki T, Kometani K, Ise W. Memory B cells. Nat Rev Immunol. (2015) 15:149–59. doi: 10.1038/nri3802
4. Crotty S, Ahmed R. Immunological memory in humans. Semin Immunol. (2004) 16:197–203. doi: 10.1016/j.smim.2004.02.008
5. Nothelfer K, Sansonetti PJ, Phalipon A. Pathogen manipulation of B cells: the best defence is a good offence. Nat Rev Microbiol. (2015) 13:173–84. doi: 10.1038/nrmicro3415
6. Portugal S, Obeng-Adjei N, Moir S, Crompton PD, Pierce SK. Atypical memory B cells in human chronic infectious diseases: an interim report. Cell Immunol. (2017) 321:18–25. doi: 10.1016/j.cellimm.2017.07.003
7. Takemori T, Kaji T, Takahashi Y, Shimoda M, Rajewsky K. Generation of memory B cells inside and outside germinal centers. Eur J Immunol. (2014) 44:1258–64. doi: 10.1002/eji.201343716
8. Seifert M, Kuppers R. Human memory B cells. Leukemia (2016) 30:2283–92. doi: 10.1038/leu.2016.226
9. Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science (2007) 315:528–31. doi: 10.1126/science.1136736
10. Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. (2011) 208:1243–52. doi: 10.1084/jem.20102477
11. Victora GD, Nussenzweig MC, Germinal centers. Annu Rev Immunol. (2012) 30:429–57. doi: 10.1146/annurev-immunol-020711-075032
12. Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. (2012) 209:597–606. doi: 10.1084/jem.20111696
13. Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. (2012) 209:2079–97. doi: 10.1084/jem.20120127
14. Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity (2001) 14:617–29. doi: 10.1016/S1074-7613(01)00129-7
15. Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood (2004) 104:3647–54. doi: 10.1182/blood-2004-01-0346
16. Defrance T, Taillardet M, Genestier L. T cell-independent B cell memory. Curr Opin Immunol. (2011) 23:330–6. doi: 10.1016/j.coi.2011.03.004
17. Alugupalli KR, Leong JM, Woodland RT, Muramatsu M, Honjo T, Gerstein RM. B1b lymphocytes confer T cell-independent long-lasting immunity. Immunity (2004) 21:379–90. doi: 10.1016/j.immuni.2004.06.019
18. Obukhanych TV, Nussenzweig MC. T-independent type II immune responses generate memory B cells. J Exp Med. (2006) 203:305–10. doi: 10.1084/jem.20052036
19. Yang Y, Ghosn EE, Cole LE, Obukhanych TV, Sadate-Ngatchou P, Vogel SN, et al. Antigen-specific memory in B-1a and its relationship to natural immunity. Proc Natl Acad Sci USA. (2012) 109:5388–93. doi: 10.1073/pnas.1121627109
20. Weill JC, Weller S, Reynaud CA. Human marginal zone B cells. Annu Rev Immunol. (2009) 27:267–85. doi: 10.1146/annurev.immunol.021908.132607
21. Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. (2005) 23, 161–96. doi: 10.1146/annurev.immunol.23.021704.115728
22. Lucas AH, Moulton KD, Tang VR, Reason DC. Combinatorial library cloning of human antibodies to Streptococcus pneumoniae capsular polysaccharides, variable region primary structures and evidence for somatic mutation of Fab fragments specific for capsular serotypes 6B, 14, and 23F. Infect Immun. (2001) 69:853–64. doi: 10.1128/IAI.69.2.853-864.2001
23. Khurana S, Fuentes S, Coyle EM, Ravichandran S, Davey RT Jr Beigel JH. Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat Med. (2016) 22:1439–47. doi: 10.1038/nm.4201
24. Irani V, Guy AJ, Andrew D, Beeson JG, Ramsland PA, Richards JS. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol Immunol. (2015) 67(2 Pt A):171–82. doi: 10.1016/j.molimm.2015.03.255
25. Cavacini LA, Kuhrt D, Duval M, Mayer K, Posner MR. Binding and neutralization activity of human IgG1 and IgG3 from serum of HIV-infected individuals. AIDS Res Hum Retroviruses. (2003) 19:785–92. doi: 10.1089/088922203769232584
26. Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol. (2006) 177:3728–36. doi: 10.4049/jimmunol.177.6.3728
27. Wu YC, Kipling D, Dunn-Walters DK. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol. (2011) 2:81. doi: 10.3389/fimmu.2011.00081
28. Colonna-Romano G, Bulati M, Aquino A, Pellicano M, Vitello S, Lio D, et al. A double-negative (IgD-CD27-) B cell population is increased in the peripheral blood of elderly people. Mech Ageing Dev. (2009) 130:681–90. doi: 10.1016/j.mad.2009.08.003
29. Berkowska MA, Driessen GJ, Bikos V, Grosserichter-Wagener C, Stamatopoulos K, Cerutti A, He B, et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood (2011) 118:2150–8. doi: 10.1182/blood-2011-04-345579
30. Berkowska MA, Schickel JN, Grosserichter-Wagener C, de Ridder D, Ng YS, van Dongen JJ, et al. Circulating human CD27-IgA+ memory B cells recognize bacteria with polyreactive Igs. J Immunol. (2015) 195:1417–26. doi: 10.4049/jimmunol.1402708
31. Clemens JD, Sack DA, Harris JR, Chakraborty J, Neogy PK, Stanton B, et al. Cross-protection by B subunit-whole cell cholera vaccine against diarrhea associated with heat-labile toxin-producing enterotoxigenic Escherichia coli: results of a large-scale field trial. J Infect Dis. (1988) 158:372–7.
32. Magri G, Comerma L, Pybus M, Sintes J, Llige D, Segura-Garzon D, et al. Human secretory IgM emerges from plasma cells clonally related to Gut memory B cells and targets highly diverse commensals. Immunity (2017) 47:118–34 e8. doi: 10.1016/j.immuni.2017.06.013
33. Xiong H, Dolpady J, Wabl MM, Curotto de Lafaille A, Lafaille JJ. Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med. (2012) 209:353–64. doi: 10.1084/jem.20111941
34. Erazo A, Kutchukhidze N, Leung M, Christ AP, Urban JF Jr de Lafaille MA, et al. Unique maturation program of the IgE response in vivo. Immunity (2007) 26:191–203. doi: 10.1016/j.immuni.2006.12.006
35. Wesemann DR, Portuguese AJ, Magee JM, Gallagher MP, Zhou X, Panchakshari RA, et al. Reprogramming IgH isotype-switched B cells to functional-grade induced pluripotent stem cells. Proc Natl Acad Sci USA. (2012) 109:13745–50. doi: 10.1073/pnas.1210286109
36. He JS, Subramaniam S, Narang V, Srinivasan K, Saunders SP, Carbajo D, et al. Curotto de Lafaille, IgG1 memory B cells keep the memory of IgE responses. Nat Commun. (2017) 8:641. doi: 10.1038/s41467-017-00723-0
37. Davies JM, O'Hehir RE. VH gene usage in immunoglobulin E responses of seasonal rhinitis patients allergic to grass pollen is oligoclonal and antigen driven. Clin Exp Allergy (2004) 34:429–36. doi: 10.1111/j.1365-2222.2004.01900.x
38. Davies JM, Platts-Mills TA, Aalberse RC. The enigma of IgE+ B-cell memory in human subjects. J Allergy Clin Immunol. (2013) 131:972–6. doi: 10.1016/j.jaci.2012.12.1569
39. Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, et al. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J Immunol. (2007) 178:6624–33. doi: 10.4049/jimmunol.178.10.6624
40. Moir S, Malaspina A, Ho J, Wang W, Dipoto AC, O'Shea MA, et al. Normalization of B cell counts and subpopulations after antiretroviral therapy in chronic HIV disease. J Infect Dis. (2008) 197:572–9. doi: 10.1086/526789
41. Sohn HW, Krueger PD, Davis RS, Pierce SK. FcRL4 acts as an adaptive to innate molecular switch dampening BCR signaling and enhancing TLR signaling. Blood (2011) 118:6332–41. doi: 10.1182/blood-2011-05-353102
42. Nogaro SI, Hafalla JC, Walther B, Remarque EJ, Tetteh KK, Conway DJ, et al. The breadth, but not the magnitude, of circulating memory B cell responses to P. falciparum increases with age/exposure in an area of low transmission. PLoS ONE (2011) 6:e25582. doi: 10.1371/journal.pone.0025582
43. Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med. (2005) 202:783–91. doi: 10.1084/jem.20050879
44. Oliviero B, Mantovani S, Ludovisi S, Varchetta S, Mele D, Paolucci S, et al. Skewed B cells in chronic hepatitis C virus infection maintain their ability to respond to virus-induced activation. J Viral Hepat. (2015) 22:391–8. doi: 10.1111/jvh.12336
45. Joosten SA, van Meijgaarden KE, Del Nonno F, Baiocchini A, Petrone L, Vanini V, et al. Patients with tuberculosis have a dysfunctional circulating B-Cell compartment, which normalizes following successful treatment. PLoS Pathog. (2016) 12:e1005687. doi: 10.1371/journal.ppat.1005687
46. Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, et al. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J Immunol. (2009) 183:2176–82. doi: 10.4049/jimmunol.0901297
47. Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. elife (2015) 4:e07218. doi: 10.7554/eLife.07218
48. Sullivan RT, Kim CC, Fontana MF, Feeney ME, Jagannathan P, Boyle MJ, et al. FCRL5 Delineates functionally impaired memory B cells associated with Plasmodium falciparum exposure. PLoS Pathog. (2015) 11:e1004894. doi: 10.1371/journal.ppat.1004894
49. Isnardi I, Ng YS, Menard L, Meyers G, Saadoun D, Srdanovic I, et al. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood (2010) 115:5026–36. doi: 10.1182/blood-2009-09-243071
50. Becker AM, Dao KH, Han BK, Kornu R, Lakhanpal S, Mobley AB, et al. SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature. PLoS ONE (2013) 8:e67003. doi: 10.1371/journal.pone.0067003
51. Corti D, Lanzavecchia A. Efficient methods to isolate human monoclonal antibodies from memory B cells and plasma cells. Microbiol Spectr. (2014) 2. doi: 10.1128/microbiolspec.AID-0018-2014
52. Wilson PC, Andrews SF. Tools to therapeutically harness the human antibody response. Nat Rev Immunol. (2012) 12:709–19. doi: 10.1038/nri3285
53. Zhang C. Hybridoma technology for the generation of monoclonal antibodies. Methods Mol Biol. (2012) 901:117–35. doi: 10.1007/978-1-61779-931-0_7
54. Burton DR, Barbas CF III. Human antibodies from combinatorial libraries. Adv Immunol. (1994) 57:191–280. doi: 10.1016/S0065-2776(08)60674-4
55. Marasco WA, Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat Biotechnol. (2007) 25:1421–34. doi: 10.1038/nbt1363
56. Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol Biol. (2002) 178, 1–37. doi: 10.1385/1-59259-240-6:001
57. Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. (1997) 15:553–7. doi: 10.1038/nbt0697-553
58. Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. (2004) 10:871–5. doi: 10.1038/nm1080
59. Kwakkenbos MJ, van Helden PM, Beaumont T, Spits H. Stable long-term cultures of self-renewing B cells and their applications. Immunol Rev. (2016) 270:65–77. doi: 10.1111/imr.12395
60. Jin A, Ozawa T, Tajiri K, Obata T, Kondo S, Kinoshita K, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. (2009) 15:1088–92. doi: 10.1038/nm.1966
61. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science (2011) 333:850–6. doi: 10.1126/science.1205669
62. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science (2009) 326:285–9. doi: 10.1126/science.1178746
63. Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature (2008) 453:667–71. doi: 10.1038/nature06890
64. Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. (2015) 16:663–73. doi: 10.1038/ni.3154
65. Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. (2009) 4:372–84. doi: 10.1038/nprot.2009.3
66. Casali P, Inghirami G, Nakamura M, Davies TF, Notkins AL. Human monoclonals from antigen-specific selection of B lymphocytes and transformation by EBV. Science (1986) 234:476–9.
67. Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, et al. Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods (2003) 275:223–37. doi: 10.1016/S0022-1759(03)00013-9
68. Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods (2008) 329:112–24. doi: 10.1016/j.jim.2007.09.017
69. Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods (2009) 343:65–7. doi: 10.1016/j.jim.2008.11.012
70. Di Niro R, Mesin L, Raki M, Zheng NY, Lund-Johansen F, Lundin KE, et al. Rapid generation of rotavirus-specific human monoclonal antibodies from small-intestinal mucosa. J Immunol. (2010) 185:5377–83. doi: 10.4049/jimmunol.1001587
71. Di Niro R, Mesin L, Zheng NY, Stamnaes J, Morrissey M, Lee JH, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med. (2012) 18:441–5. doi: 10.1038/nm.2656
72. Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. (2011) 121:1946–55. doi: 10.1172/JCI44447
73. Pinder CL, Kratochvil S, Cizmeci D, Muir L, Guo Y, Shattock RJ, et al. Isolation and characterization of antigen-specific plasmablasts using a novel flow cytometry-based Ig capture assay. J Immunol. (2017) 199:4180–8. doi: 10.4049/jimmunol.1701253
74. Lanzavecchia A, Sallusto F. Human B cell memory. Curr Opin Immunol. (2009) 21:298–304. doi: 10.1016/j.coi.2009.05.019
75. Nandin IS, Fong C, Deantonio C, Torreno-Pina JA, Pecetta S, Maldonado P, et al. Novel in vitro booster vaccination to rapidly generate antigen-specific human monoclonal antibodies (vol 214, 2017). J Exp Med. (2017) 214:2811. doi: 10.1084/jem.2017063307262017c
76. Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature (1975) 256:495–7. doi: 10.1038/256495a0
77. Jakobovits A, Amado RG, Yang X, Roskos L, Schwab G. From XenoMouse technology to panitumumab, the first fully human antibody product from transgenic mice. Nat Biotechnol. (2007) 25:1134–43. doi: 10.1038/nbt1337
78. Turchaninova MA, Davydov A, Britanova OV, Shugay M, Bikos V, Egorov ES, et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc. (2016) 11:1599–616. doi: 10.1038/nprot.2016.093
79. Robinson WH, Mao R. Decade in review-technology: technological advances transforming rheumatology. Nat Rev Rheumatol. (2015) 11:626–8. doi: 10.1038/nrrheum.2015.137
80. DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. (2015) 21:86–91. doi: 10.1038/nm.3743
81. Lavinder JJ, Horton AP, Georgiou G, Ippolito GC. Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Curr Opin Chem Biol. (2015) 24:112–20. doi: 10.1016/j.cbpa.2014.11.007
82. Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. (2012) 30:447–52. doi: 10.1038/nbt.2167
83. Chaudhary N, Wesemann DR. Analyzing Immunoglobulin Repertoires. Front Immunol. (2018) 9:462. doi: 10.3389/fimmu.2018.00462
84. Upadhyay AA, Kauffman RC, Wolabaugh AN, Cho A, Patel NB, Reiss SM, et al. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med. (2018) 10:20. doi: 10.1186/s13073-018-0528-3
85. DeWitt WS, Lindau P, Snyder TM, Sherwood AM, Vignali M, Carlson CS, et al. A Public database of memory and naive B-Cell receptor sequences. PLoS ONE (2016) 11:e0160853. doi: 10.1371/journal.pone.0160853
86. Schramm CA, Sheng Z, Zhang Z, Mascola JR, Kwong PD, Shapiro L. SONAR:A high-throughput pipeline for inferring antibody ontogenies from longitudinal sequencing of B cell transcripts. Front Immunol. (2016) 7:372. doi: 10.3389/fimmu.2016.00372
87. Zhang W, Wang IM, Wang C, Lin L, Chai X, Wu J, et al. IMPre: an accurate and efficient software for prediction of T- and B-cell receptor germline genes and alleles from rearranged repertoire data. Front Immunol. (2016) 7:457. doi: 10.3389/fimmu.2016.00457
88. Canzar S, Neu KE, Tang Q, Wilson PC, Khan AA. BASIC: BCR assembly from single cells. Bioinformatics (2017) 33:425–7. doi: 10.1093/bioinformatics/btw631
89. Christley S, Scarborough W, Salinas E, Rounds WH, Toby IT, Fonner JM, et al. VDJServer: a cloud-based analysis portal and data commons for immune repertoire sequences and rearrangements. Front Immunol. (2018) 9:976. doi: 10.3389/fimmu.2018.00976
90. Imkeller K, Arndt PF, Wardemann H, Busse CE. sciReptor: analysis of single-cell level immunoglobulin repertoires. BMC Bioinformatics (2016) 17:67. doi: 10.1186/s12859-016-0920-1
91. Fahnrich A, Krebbel M, Decker N, Leucker M, Lange FD, Kalies K, et al. ClonoCalc and ClonoPlot: immune repertoire analysis from raw files to publication figures with graphical user interface. BMC Bioinformatics (2017) 18:164. doi: 10.1186/s12859-017-1575-2
92. Maramis C, Gkoufas A, Vardi A, Stalika E, Stamatopoulos K, Hatzidimitriou A, et al. IRProfiler-a software toolbox for high throughput immune receptor profiling. BMC Bioinformatics (2018) 19:144. doi: 10.1186/s12859-018-2144-z
93. Cortina-Ceballos B, Godoy-Lozano EE, Samano-Sanchez H, Aguilar-Salgado A, Velasco-Herrera Mdel C, Vargas-Chavez C, et al. Reconstructing and mining the B cell repertoire with ImmunediveRsity. MAbs (2015) 7:516–24. doi: 10.1080/19420862.2015.1026502
94. Yaari G, Kleinstein SH. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. (2015) 7:121. doi: 10.1186/s13073-015-0243-2
95. Motley MP, Fries BC. A new take on an old remedy: generating antibodies against multidrug-resistant gram-negative bacteria in a postantibiotic world. mSphere (2017) 2:e00397-17. doi: 10.1128/mSphere.00397-17
96. Kauffman RC, Bhuiyan TR, Nakajima R, Mayo-Smith LM, Rashu R, Hoq MR, et al. Single-cell analysis of the plasmablast response to vibrio cholerae demonstrates expansion of cross-reactive memory B cells. MBio (2016) 7:e02021-16. doi: 10.1128/mBio.02021-16
97. Rollenske T, Szijarto V, Lukasiewicz J, Guachalla LM, Stojkovic K, Hartl K, et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat Immunol. (2018) 19:617–24. doi: 10.1038/s41590-018-0106-2
98. Lucas AH, Reason DC. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol Rev. (1999) 171:89–104. doi: 10.1111/j.1600-065X.1999.tb01343.x
99. Wright JG, Quinn CP, Shadomy S, Messonnier N. C. Centers for disease and prevention: use of anthrax vaccine in the united states, recommendations of the advisory committee on immunization practices (ACIP) 2009. MMWR Recomm Rep. (2010) 59:1–30.
100. Garman L, Smith K, Farris AD, Nelson MR, Engler RJ, James JA. Protective antigen-specific memory B cells persist years after anthrax vaccination and correlate with humoral immunity. Toxins (Basel). (2014) 6:2424–31. doi: 10.3390/toxins6082424
101. Quinn CP, Sabourin CL, Niemuth NA, Li H, Semenova VA, Rudge TL, et al. A three-dose intramuscular injection schedule of anthrax vaccine adsorbed generates sustained humoral and cellular immune responses to protective antigen and provides long-term protection against inhalation anthrax in rhesus macaques. Clin Vaccine Immunol. (2012) 19:1730–45. doi: 10.1128/CVI.00324-12
102. Reason DC, Ullal A, Liberato J, Sun J, Keitel W, Zhou J. Domain specificity of the human antibody response to Bacillus anthracis protective antigen. Vaccine (2008) 26:4041–7. doi: 10.1016/j.vaccine.2008.05.023
103. Smith K, Crowe SR, Garman L, Guthridge CJ, Muther JJ, McKee E, et al. Human monoclonal antibodies generated following vaccination with AVA provide neutralization by blocking furin cleavage but not by preventing oligomerization. Vaccine (2012) 30:4276–83. doi: 10.1016/j.vaccine.2012.03.002
104. Chi X, Li J, Liu W, Wang X, Yin K, Liu J, et al. Generation and characterization of human monoclonal antibodies targeting anthrax protective antigen following vaccination with a recombinant protective antigen vaccine. Clin Vaccine Immunol. (2015) 22:553–60. doi: 10.1128/CVI.00792-14
105. Kummerfeldt CE. Raxibacumab: potential role in the treatment of inhalational anthrax. Infect Drug Resist. (2014) 7:101–9. doi: 10.2147/IDR.S47305
106. Liang JL, Tiwari T, Moro P, Messonnier NE, Reingold A, Sawyer M, et al. Prevention of Pertussis, tetanus, and diphtheria with vaccines in the united states: recommendations of the advisory committee on immunization practices (ACIP). MMWR Recomm Rep. (2018) 67:1–44. doi: 10.15585/mmwr.rr6702a1
107. Poulsen TR, Meijer PJ, Jensen A, Nielsen LS, Andersen PS. Kinetic, affinity, and diversity limits of human polyclonal antibody responses against tetanus toxoid. J Immunol. (2007) 179:3841–50. doi: 10.4049/jimmunol.179.6.3841
108. Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. (2011) 187:4229–35. doi: 10.4049/jimmunol.1000928
109. de Kruif J, Kramer A, Visser T, Clements C, Nijhuis R, Cox F, van der Zande V, et al. Human immunoglobulin repertoires against tetanus toxoid contain a large and diverse fraction of high-affinity promiscuous V(H) genes. J Mol Biol. (2009) 387:548–58. doi: 10.1016/j.jmb.2009.02.009
110. Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. (2014) 111:2259–64. doi: 10.1073/pnas.1317793111
111. Williams WW, Lu PJ, O'Halloran A, Kim DK, Grohskopf LA, Pilishvili T, et al. Surveillance of vaccination coverage among adult populations-United States, 2015. MMWR Surveill Summ. (2017) 66:1–28. doi: 10.15585/mmwr.ss6611a1
112. Eletu SD, Sheppard CL, Thomas E, Smith K, Daniel P, Litt DJ, et al. Development of an extended-specificity multiplex immunoassay for detection of streptococcus pneumoniae serotype-specific antigen in urine by use of human monoclonal antibodies. Clin Vaccine Immunol. (2017) 24:e00262-17. doi: 10.1128/CVI.00262-17
113. Smith K, Muther JJ, Duke AL, McKee E, Zheng NY, Wilson PC, et al. Fully human monoclonal antibodies from antibody secreting cells after vaccination with Pneumovax(R)23 are serotype specific and facilitate opsonophagocytosis. Immunobiology (2013) 218:745–54. doi: 10.1016/j.imbio.2012.08.278
114. Baxendale HE, Goldblatt D. Correlation of molecular characteristics, isotype, and in vitro functional activity of human antipneumococcal monoclonal antibodies. Infect Immun. (2006) 74:1025–31. doi: 10.1128/IAI.74.2.1025-1031.2006
115. Zhou J, Lottenbach KR, Barenkamp SJ, Lucas AH, Reason DC. Recurrent variable region gene usage and somatic mutation in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae type 23F. Infect Immun. (2002) 70:4083–91. doi: 10.1128/IAI.70.8.4083-4091.2002
116. Zhou J, Lottenbach KR, Barenkamp SJ, Reason DC. Somatic hypermutation and diverse immunoglobulin gene usage in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae Type 6B. Infect Immun. (2004) 72:3505–14. doi: 10.1128/IAI.72.6.3505-3514.2004
117. Smith K, Shah H, Muther JJ, Duke AL, Haley K, James JA. Antigen nature and complexity influence human antibody light chain usage and specificity. Vaccine (2016) 34:2813–20. doi: 10.1016/j.vaccine.2016.04.040
118. Melin M, Coan P, Hollingshead S. Development of cross-reactive antibodies to the proline-rich region of pneumococcal surface protein A in children. Vaccine (2012) 30:7157–60. doi: 10.1016/j.vaccine.2012.10.004
119. Daniels CC, Coan P, King J, Hale J, Benton KA, Briles DE, et al. The proline-rich region of pneumococcal surface proteins A and C contains surface-accessible epitopes common to all pneumococci and elicits antibody-mediated protection against sepsis. Infect Immun. (2010) 78:2163–72. doi: 10.1128/IAI.01199-09
120. Mukerji R, Hendrickson C, Genschmer KR, Park SS, Bouchet V, Goldstein R, et al. The diversity of the proline-rich domain of pneumococcal surface protein A (PspA): potential relevance to a broad-spectrum vaccine. Vaccine (2018) 36:6834–43. doi: 10.1016/j.vaccine.2018.08.045
121. Zinocker S, Schindler CE, Skinner J, Rogosch T, Waisberg M, Schickel JN, et al. The V gene repertoires of classical and atypical memory B cells in malaria-susceptible West African children. J Immunol. (2015) 194:929–39. doi: 10.4049/jimmunol.1402168
122. Wipasa J, Suphavilai C, Okell LC, Cook J, Corran PH, Thaikla K, et al. Long-lived antibody and B Cell memory responses to the human malaria parasites, Plasmodium falciparum and Plasmodium vivax. PLoS Pathog. (2010) 6:e1000770. doi: 10.1371/journal.ppat.1000770
123. Muellenbeck MF, Ueberheide B, Amulic B, Epp A, Fenyo D, Busse CE, et al. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J Exp Med. (2013) 210:389–99. doi: 10.1084/jem.20121970
124. PrabhuDas M, Adkins B, Gans H, King C, Levy O, Ramilo O, et al. Challenges in infant immunity: implications for responses to infection and vaccines. Nat Immunol. (2011) 12:189–94. doi: 10.1038/ni0311-189
125. Wendel BS, He C, Qu M, Wu D, Hernandez SM, Ma KY, et al. Accurate immune repertoire sequencing reveals malaria infection driven antibody lineage diversification in young children. Nat Commun. (2017) 8:531. doi: 10.1038/s41467-017-00645-x
126. Walker LM, Burton DR. Passive immunotherapy of viral infections: 'super-antibodies' enter the fray. Nat Rev Immunol. (2018) 18:297–308. doi: 10.1038/nri.2017.148
127. Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol. (2017) 24:60–69. doi: 10.1016/j.coviro.2017.03.002
128. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science (2009) 324:246–51. doi: 10.1126/science.1171491
129. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. (2009) 16:265–73. doi: 10.1038/nsmb.1566
130. Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE (2008) 3:e3942. doi: 10.1371/journal.pone.0003942
131. Corti D, Suguitan AL Jr Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. (2010) 120:1663–73. doi: 10.1172/JCI41902
132. McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, et al. Memory B cells that cross-react with Group 1 and Group 2 influenza a viruses are abundant in adult human repertoires. Immunity (2018) 48:174–184 e9. doi: 10.1016/j.immuni.2017.12.009
133. Sano K, Ainai A, Suzuki T, Hasegawa H. The road to a more effective influenza vaccine: up to date studies and future prospects. Vaccine (2017) 35:5388–95. doi: 10.1016/j.vaccine.2017.08.034
134. Dailey AF, Hoots BE, Hall HI, Song R, Hayes D, Fulton P Jr . Vital signs: human immunodeficiency virus testing and diagnosis delays-United States. MMWR Morb Mortal Wkly Rep. (2017) 66:1300–6. doi: 10.15585/mmwr.mm6647e1
135. Karlsson Hedestam GB, Guenaga J, Corcoran M, Wyatt RT. Evolution of B cell analysis and Env trimer redesign. Immunol Rev. (2017) 275:183–202. doi: 10.1111/imr.12515
136. Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, et al. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol. (2014) 192:3637–44. doi: 10.4049/jimmunol.1303334
137. Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature (2009) 458:636–40. doi: 10.1038/nature07930
138. Meffre E, Louie A, Bannock J, Kim LJ, Ho J, Frear CC, et al. Maturational characteristics of HIV-specific antibodies in viremic individuals. JCI Insight. (2016) 1:e84610. doi: 10.1172/jci.insight.84610
139. Hammond PW. Accessing the human repertoire for broadly neutralizing HIV antibodies. MAbs (2010) 2:157–64.
140. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature (2013) 496:504–7. doi: 10.1038/nature12060
141. Yam-Puc JC, Cedillo-Barron L, Aguilar-Medina EM, Ramos-Payan R, Escobar-Gutierrez A, Flores-Romo L. The cellular bases of antibody responses during dengue virus infection. Front Immunol. (2016) 7:218. doi: 10.3389/fimmu.2016.00218
142. Halstead SB, O'Rourke EJ. Antibody-enhanced dengue virus infection in primate leukocytes. Nature (1977) 265:739–41. doi: 10.1038/265739a0
143. Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science (2010) 328:745–8. doi: 10.1126/science.1185181
144. Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol. (2012) 86:2911–8. doi: 10.1128/JVI.06075-11
145. Priyamvada L, Cho A, Onlamoon N, Zheng NY, Huang M, Kovalenkov Y, et al. B Cell Responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived plasmablasts. J Virol. (2016) 90:5574–85. doi: 10.1128/JVI.03203-15
146. Appanna R, Kg S, Xu MH, Toh YX, Velumani S, Carbajo D, et al. Plasmablasts During Acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine (2016) 12:178–88. doi: 10.1016/j.ebiom.2016.09.003
147. Magnani DM, Silveira CGT, Ricciardi MJ, Gonzalez-Nieto L, Pedreno-Lopez N, Bailey VK, et al. Potent plasmablast-derived antibodies elicited by the National Institutes of health dengue vaccine. J Virol. (2017) 91:e00867-17. doi: 10.1128/JVI.00867-17
148. Priyamvada L, Hudson W, Ahmed R, Wrammert J. Humoral cross-reactivity between Zika and dengue viruses: implications for protection and pathology. Emerg Microbes Infect. (2017) 6:e33. doi: 10.1038/emi.2017.42
149. Priyamvada L, Suthar MS, Ahmed R, Wrammert J. Humoral immune responses against zika virus infection and the importance of preexisting flavivirus immunity. J Infect Dis. (2017) 216(Suppl. 10):S906–S911. doi: 10.1093/infdis/jix513
150. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. (2016) 17:1102–8. doi: 10.1038/ni.3515
151. Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science (2016) 353:823–6. doi: 10.1126/science.aaf8505
152. Martin J, Wilcox M. New and emerging therapies for Clostridium difficile infection. Curr Opin Infect Dis. (2016) 29:546–554. doi: 10.1097/QCO.0000000000000320
153. Gala RP, Zaman RU, D'Souza MJ, Zughaier SM. Novel whole-cell inactivated neisseria gonorrhoeae microparticles as vaccine formulation in microneedle-based transdermal immunization. Vaccines (Basel). (2018) 6:E60. doi: 10.3390/vaccines6030060
154. Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. (2016) 2:16020. doi: 10.1038/nrdp.2016.20
155. Lang ML, Shrestha B. Adaptive immune constraints on C. difficile vaccination. Expert Rev Vaccines (2017) 16:1053–5. doi: 10.1080/14760584.2017.1379397
156. Navalkele BD, Chopra T. Bezlotoxumab: an emerging monoclonal antibody therapy for prevention of recurrent Clostridium difficile infection. Biologics (2018) 12:11–21. doi: 10.2147/BTT.S127099
157. Monaghan TM, Robins A, Knox A, Sewell HF, Mahida YR. Circulating antibody and memory B-Cell responses to C. difficile toxins A and B in patients with C. difficile-associated diarrhoea, inflammatory bowel disease and cystic fibrosis. PLoS ONE (2013) 8:e74452. doi: 10.1371/journal.pone.0074452
158. Devera TS, Lang GA, Lanis JM, Rampuria P, Gilmore CL, James JA, et al. Memory B cells encode neutralizing antibody specific for Toxin B from the Clostridium difficile strains VPI 10463 and NAP1/BI/027 but with superior neutralization of VPI 10463 Toxin B. Infect Immun. (2015) 84:194–204. doi: 10.1128/IAI.00011-15
159. Lanis JM, Barua S, Ballard JD. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. (2010) 6:e1001061. doi: 10.1371/journal.ppat.1001061
160. Bauer MP, Nibbering PH, Poxton IR, Kuijper EJ, van Dissel JT. Humoral immune response as predictor of recurrence in Clostridium difficile infection. Clin Microbiol Infect. (2014) 20:1323–8. doi: 10.1111/1469-0691.12769
161. Mehand MS, Millett P, Al-Shorbaji F, Roth C, Kieny MP, Murgue B. World health organization methodology to prioritize emerging infectious diseases in need of research and development. Emerg Infect Dis. (2018) 24. doi: 10.3201/eid2409.171427
162. Dhama K, Karthik K, Khandia R, Chakraborty S, Munjal A, Latheef SK, et al. Advances in designing and developing vaccines, drugs, and therapies to counter ebola virus. Front Immunol. (2018) 9:1803. doi: 10.3389/fimmu.2018.01803
163. Pascal KE, Dudgeon D, Trefry JC, Anantpadma M, Sakurai Y, Murin CD, et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in non-human primates. J Infect Dis (2018) 218(Suppl. 5):S612–26. doi: 10.1093/infdis/jiy285
164. Sivapalasingam S, Kamal M, Slim R, Hosain R, Shao W, Stoltz R, et al. Safety, pharmacokinetics, and immunogenicity of a co-formulated cocktail of three human monoclonal antibodies targeting Ebola virus glycoprotein in healthy adults, a randomised, first-in-human phase 1 study. Lancet Infect Dis. (2018) 18:884–93. doi: 10.1016/S1473-3099(18)30397-9
165. Sun H, Chen Q, Lai H. Development of antibody therapeutics against flaviviruses. Int J Mol Sci. (2017) 19:E54. doi: 10.3390/ijms19010054
166. das Neves Almeida R, Racine T, Magalhaes KG, Kobinger GP. Zika virus vaccines, and challenges and perspectives. Vaccines (Basel). (2018) 6:E62. doi: 10.3390/vaccines6030062
167. Martinez AA, Espinosa BA, Adamek RN, Thomas BA, Chau J, Gonzalez E, et al. Breathing new life into West Nile virus therapeutics; discovery and study of zafirlukast as an NS2B-NS3 protease inhibitor. Eur J Med Chem. (2018) 157:1202–13. doi: 10.1016/j.ejmech.2018.08.077
168. Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med. (2016) 8:369ra179. doi: 10.1126/scitranslmed.aai8336
169. Tsioris K, Gupta NT, Ogunniyi AO, Zimnisky RM, Qian F, Yao Y, et al. Neutralizing antibodies against West Nile virus identified directly from human B cells by single-cell analysis and next generation sequencing. Integr Biol (Camb). (2015) 7:1587–97. doi: 10.1039/C5IB00169B
170. Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, et al. Recurrent potent human neutralizing antibodies to Zika Virus in Brazil and Mexico. Cell (2017) 169:597–609 e11. doi: 10.1016/j.cell.2017.04.024
171. Akahata W, Yang ZY, Andersen H, Sun SY, Holdaway HA, Kong WP, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. (2010) 16:334–134. doi: 10.1038/nm.2105
172. Couderc T, Khandoudi N, Grandadam M, Visse C, Gangneux N, Bagot S, et al. Prophylaxis and therapy for Chikungunya virus infection. J Infect Dis. (2009) 200:516–23. doi: 10.1086/600381
173. Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia KA, Gardner J, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. (2013) 7:e2423. doi: 10.1371/journal.pntd.0002423
174. Fric J, Bertin-Maghit S, Wang CI, Nardin A, Warter L. Use of human monoclonal antibodies to treat Chikungunya virus infection. J Infect Dis. (2013) 207:319–22. doi: 10.1093/infdis/jis674
175. Warter L, Lee CY, Thiagarajan R, Grandadam M, Lebecque S, Lin RT, et al. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol. (2011) 186:3258–64. doi: 10.4049/jimmunol.1003139
176. Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, et al. Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against Chikungunya Virus. Cell Host Microbe. (2015) 18:86–95. doi: 10.1016/j.chom.2015.06.009
177. Jackson KJ, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol. (2013) 4:263. doi: 10.3389/fimmu.2013.00263
178. Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, Lu DR, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. (2014) 66:2706–15. doi: 10.1002/art.38754
179. Lu DR, McDavid AN, Kongpachith S, Lingampalli N, Glanville J, Ju CH, et al. T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol (2018) 70:1732–44. doi: 10.1002/art.40578
180. Elliott SE, Kongpachith S, Lingampalli N, Adamska JZ, Cannon BJ, Mao R, et al. Affinity maturation drives epitope spreading and generation of pro-inflammatory anti-citrullinated protein antibodies in rheumatoid arthritis. Arthritis Rheumatol (2018) 70:1946–58. doi: 10.1002/art.40587
181. Roskin KM, Simchoni N, Liu Y, Lee JY, Seo K, Hoh RA, et al. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med. (2015) 7:302ra135. doi: 10.1126/scitranslmed.aab1216
182. Kubagawa H, Vogler LB, Capra JD, Conrad ME, Lawton AR, Cooper MD. Studies on the clonal origin of multiple myeloma. Use of individually specific (idiotype) antibodies to trace the oncogenic event to its earliest point of expression in B-cell differentiation. J Exp Med. (1979) 150:792–807. doi: 10.1084/jem.150.4.792
183. Nishihori T, Song J, Shain KH. Minimal residual disease assessment in the context of multiple myeloma treatment. Curr Hematol Malig Rep. (2016) 11:118–26. doi: 10.1007/s11899-016-0308-3
Keywords: memory B cells, vaccination, monoclonal antibody, antibody repertoires, next generation sequencing
Citation: Shah HB, Smith K, Wren JD, Webb CF, Ballard JD, Bourn RL, James JA and Lang ML (2019) Insights From Analysis of Human Antigen-Specific Memory B Cell Repertoires. Front. Immunol. 9:3064. doi: 10.3389/fimmu.2018.03064
Received: 01 November 2018; Accepted: 11 December 2018;
Published: 15 January 2019.
Edited by:
Kim Good-Jacobson, Monash University, AustraliaReviewed by:
Kevin M. Nickerson, University of Pittsburgh, United StatesKai-Michael Toellner, University of Birmingham, United Kingdom
Copyright © 2019 Shah, Smith, Wren, Webb, Ballard, Bourn, James and Lang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hemangi B. Shah, hemangi-shah@ouhsc.edu
Mark L. Lang, mark-lang@ouhsc.edu
 Hemangi B. Shah
Hemangi B. Shah Kenneth Smith
Kenneth Smith Jonathan D. Wren
Jonathan D. Wren Carol F. Webb
Carol F. Webb Jimmy D. Ballard
Jimmy D. Ballard Rebecka L. Bourn
Rebecka L. Bourn Judith A. James
Judith A. James Mark L. Lang
Mark L. Lang