- 1Department of Genetics and Genome Sciences and Case Comprehesive Cancer Center, Case Western Reserve University, Cleveland, OH, United States
- 2Centre for Digital Transformation, Imperial College, London, United Kingdom
Immune checkpoint inhibitors (ICIs) have revolutionized cancer immunotherapy by reinvigorating antitumor immune responses, but their efficacy remains limited in most patients. To address this challenge and optimize Immune check inhibitor treatment, understanding the underlying molecular intricacies involved is crucial. The emergence of CRISPR-Cas9 technology has empowered researchers to precisely investigate gene function and has introduced transformative shifts in identifying key genes for various physiological and pathological processes. CRISPR screenings, particularly in vivo CRISPR screenings, have become invaluable tools in deciphering molecular networks and signaling pathways governing suppressive immune checkpoint molecules. In this review, we provide a comprehensive overview of in vivo CRISPR screenings in cancer immunotherapy, exploring how this cutting-edge technology has unraveled potential novel therapeutic targets and combination strategies. We delve into the latest findings and advancements, shedding light on immune checkpoint regulation and offering exciting prospects for the development of innovative and effective treatments for cancer patients.
1 Introduction
Immune checkpoints play a crucial role in maintaining immune system homeostasis by preventing prolonged immune cell activation and safeguarding normal tissue integrity (Emens et al., 2017). However, cancer cells exploit these checkpoints to evade immune surveillance and establish immune tolerance. To counteract this evasion strategy, immune checkpoint inhibitors (ICIs) have emerged as a transformative approach to reinvigorate antitumor immune responses (Finck et al., 2020; Sharma et al., 2023). The FDA approval of Ipilimumab (Yervoy), the first ICI targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4), marked a significant milestone in cancer immunotherapy (Hodi et al., 2010). Subsequently, the approval of several anti-programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1) therapies, including Nivolumab, Pembrolizumab, Atezolizumab, Avelumab, Durvalumab, and Cemiplimab, further revolutionized the field (Hodi et al., 2010; Topalian et al., 2012; Herbst et al., 2014) Despite the remarkable clinical successes achieved with anti-CTLA-4 and anti-PD-1/PD-L1 therapies, their efficacy remains limited for most patients (Emens et al., 2017),and some individuals experience acquired resistance or immune-related adverse events (irAEs) (Schoenfeld and Hellmann, 2020). The quest to understand and overcome resistance mechanisms while optimizing the clinical efficacy of ICIs has become increasingly urgent in recent years. Addressing this challenge requires innovative approaches to elucidate the underlying molecular intricacies involved. The advent of the Clustered Regularly Interspaced Short Palindromic-associated endonuclease 9 (CRISPR-Cas9) mediated genome editing technology has revolutionized the field of genetics and emerged as a powerful tool for precisely investigating gene function. CRISPR screenings, represented by the landmark development of the genome-scale CRISPR-Cas9 knockout (GeCKO) library by Shalem et al., have introduced a transformative shift in our ability to identify key genes responsible for various physiological and pathological processes (Shalem et al., 2014). Leveraging the power of CRISPR technology, researchers have harnessed its potential to investigate novel therapeutic targets and combination strategies that can augment the effectiveness of ICIs. CRISPR screenings have proven invaluable in dissecting molecular networks and signaling pathways underlying immune checkpoint regulation, paving the way for the development of innovative cancer therapeutics (Manguso et al., 2017).
In this review, we summarize the broad methodologies and approaches involved in CRISPR screening libraries, with a particular emphasis on creating and utilizing in vivo CRISPR screenings within the realm of cancer immunotherapy. Through the lens of these innovative tools, we will delve into the latest findings and advancements that contribute to our understanding of cancer immunotherapy and offer exciting prospects for the development of novel and effective treatments for cancer patients.
2 The CRISPR-Cas9 screening strategy
While next-generation sequencing (NGS) technologies have greatly improved the understanding of genetic variants, many identified genes turned out to be “passenger gene alterations” that do not harbor any functional significance (McDaniel et al., 2013; Vogelstein et al., 2013), but can often elicit immunoediting when presented as neoepitopes. The breakthrough discovery of the CRISPR-Cas9 system by Charpentier and Doudna in 2012 fundamentally reshaped genome editing technology (Jinek et al., 2012), The CRISPR-Cas9 system enables precise modification of genomes, including gene removal, introduction of mutations, as well as gene silencing or activation (Doudna and Charpentier, 2014). This technology has been continuously modified to enhance efficiency, reduce off-target effects, and simplify delivery methods (Zhang, 2019). Large-scale genomic screening strategies have utilized the CRISPR/Cas9 system’s capability to edit multiple sites simultaneously (Shalem et al., 2014). The development of the first genome-scale CRISPR-Cas9 knockout library in 2014 enabled comprehensive screening of 18,080 human genes, facilitating negative and positive selection screening in human cells ((Shalem et al., 2014). Since then, CRISPR technology has been widely applied in various fields, including development and evolutionary biology, immunology, tissue regeneration, oncogenesis, metastasis, and drug discovery (Xu and Li, 2020).
In the landscape of genetic functional testing, CRISPR/Cas9-based screens present significant advantages over conventional RNAi or cDNA library screens, primarily owing to their superior genetic editing capability with reduced off-target effects (Gilbert et al., 2014; Doench et al., 2016). This revolutionary technology enables precise targeting of both coding and non-coding regions throughout the genome, offering unprecedented versatility (Gilbert et al., 2014; Doench et al., 2016). For loss-of-function screenings involving protein-coding genes, researchers can opt for two distinct approaches: CRISPR knockout (CRISPRko) or CRISPR interference (CRISPRi), each tailored to specific research goals (Koike-Yusa et al., 2014; Shalem et al., 2014). CRISPRi proves valuable for gene dosage studies, exploring reversible gene expression, or when working with cell lines exhibiting highly rearranged genomes. In contrast, CRISPRko stands as the preferred choice for investigating gene essentiality and achieving stable gene knockout (Koike-Yusa et al., 2014; Shalem et al., 2014).
2.1 Methodological insights: In vitro and in vivo CRISPR/Cas9 screenings
In tissue culture experiments, the use of pooled screenings employing single guide (sgRNA) libraries targeting multiple genes simultaneously has emerged as a powerful tool to attribute functional phenotypes to various gene perturbations (Doench et al., 2016). Conducting in vitro screening experiments is relatively straightforward, involving cell transduction with the packaged library of interest, followed by antibiotic selection. To ensure individual sgRNA expression, it is crucial to maintain a low viral load during transduction, typically ranging between 30% and 50% (Doench et al., 2016)Subsequently, NGS is employed to determine the relative sgRNA frequency across the transduced cells, effectively establishing stable cellular barcodes (Figure 1) (Doench et al., 2016). In vitro screenings offer the advantage of using broader libraries, facilitating comprehensive exploration of gene functions. In contrast, in vivo screens often necessitate more focused libraries due to increased noise and delivery challenges when targeting multiple tissues (Kuhn et al., 2021). However, in vivo screening offers a deeper understanding of the functional outcomes of gene perturbation, considering the influence of cell-extrinsic stimuli that can significantly impact cellular phenotypes (Doench et al., 2016).
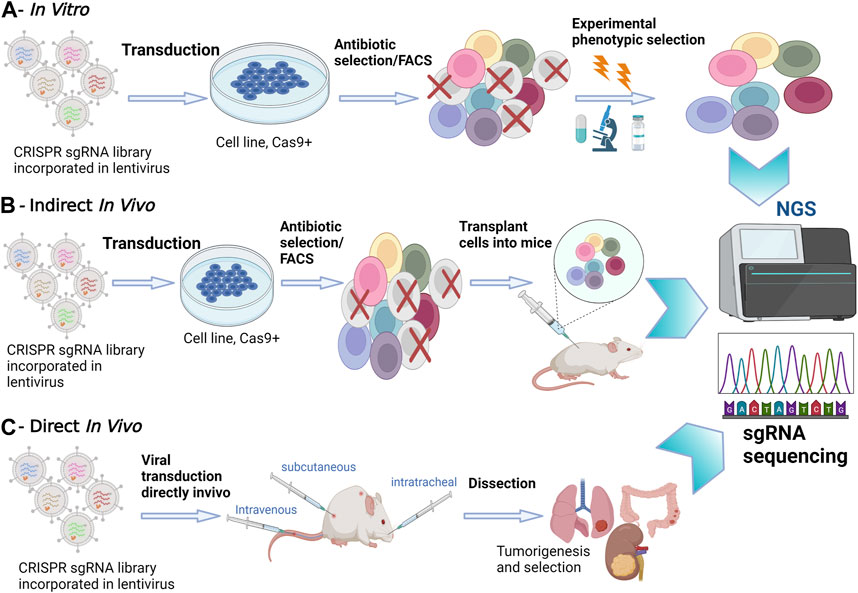
FIGURE 1. In Vitro versus In Vivo CRISPR/Cas9 Screenings. (A) In Vitro Screening: In vitro experiments take place in a controlled laboratory environment. CRISPR-Cas9 technology is applied to manipulate genes of interest in cell cultures or isolated genetic material. NGS is utilized to analyze the outcomes, providing a comprehensive view of genetic changes. (B) Indirect In Vivo Screening: In this method, CRISPR-edited cells are first cultured in vitro. Subsequently, these edited cells are transplanted into a living organism, often a model organism such as a mouse model. NGS is employed to monitor the changes in gene expression and genetic modifications in the host organism. This approach allows for assessing the in vivo impact of edited genes. (C) Direct In Vivo Screening: CRISPR-Cas9 editing is performed directly within the living organism. NGS is applied to analyze genetic changes within the host organism itself, eliminating the need for prior in vitro culturing. This approach provides real-time insights into gene editing effects within the organism’s natural context.
Options for in vivo screening involve either transplanting CRISPRko-modified cells or directly delivering CRISPR compounds into living tissues. Transplantation entails introducing transduced cancer cells into mice and analyzing sgRNA abundance in surviving tumors through deep sequencing. However, this method has limitations in recapitulating the gradual and spatially determined interplay of local and cell-extrinsic niche factors leading to oncogenesis. Additionally, injecting a large number of cells can damage the animal’s tissues and compromise the validity of transplantation-based metastasis screenings (Chow and Chen, 2018). To address these challenges, direct in vivo screenings have been developed to provide a more precise recapitulation of human body conditions. Most direct in vivo screenings involve delivering CRISPR libraries to Cas9 transgenic mice, offering advantages in discovering genes involved in oncogenesis, cancer evolution, tumor initiation and progression (Figure 1) (Platt et al., 2014; Adamson et al., 2016). The delivery method often involves the use of viral vectors, such as lentiviruses, to introduce CRISPR components into the target cells. These screenings ensure the retention of the tumor’s native microenvironment and the natural response of the immune system to tumorigenesis. However, a limitation of direct in vivo screens is the relatively poor delivery efficacy of viral vectors into solid tumors (Doench et al., 2016). Despite this challenge, direct in vivo screens remain instrumental in unraveling the intricacies of gene function within the physiological complexities of living organisms, providing crucial insights for therapeutic developments and precision medicine.
2.2 In vivo precision: CRISPR screening studies in tumor immunotherapy advancement
Despite the significant progress of ICIs in cancer treatment, a substantial proportion of patients still do not show modification of tumor progression (Gandhi et al., 2018). The high heterogeneity and corresponding evolvability of tumors, along with the presence of multiple inhibitory immune checkpoint molecules that collectively promote immune escape, has led to the exploration of combination therapies as a future direction for tumor immunotherapy (Pauken and Wherry, 2015). Performing unbiased CRISPR screenings for potential targets to enhance the antitumor activity of ICIs can be achieved by utilizing sgRNA libraries targeting the whole genome or at least genes implicated in multiple functional classes (Li et al., 2014). However, tumor evolution is a complex and dynamic process that cannot be accurately replicated by in vitro CRISPR systems alone (Liu et al., 2020). In vivo CRISPR screenings offer several advantages for exploring candidate targets by manipulating the direct and indirect interactions between immune cells and tumor cells, providing a better representation of the immune microenvironment(Schumann et al., 2015; Manguso et al., 2017). Thus, this review exclusively focuses on the promising realm of all in vivo screenings that have been done in the field of tumor immunotherapy.
2.2.1 CRISPR-Cas9 screenings using CRISPRko-modified cancer cells
As solid tumors can avoid detection by the various arms of the immune system or limit the extent of immunological killing, evading immune responses is recognized as a hallmark of cancers (Hanahan and Weinberg, 2011). Cancer cells escape immune surveillance by several mechanisms that fall under three major principles: a) lack of tumor antigen recognition, b) induction of immunological tolerance particularly through immunosuppressive factors, c) resistance to cell death (Fouad and Aanei, 2017; Jhunjhunwala et al., 2021). To discover potential targets that can sensitize cancer cells to immunotherapies, multiple CRISPR/Cas9-based in vivo screenings were conducted in cancer cells with genome-scale or focused sgRNA libraries (Table 1).
2.2.1.1 CRISPR/Cas9 screenings with genome-scale sgRNA libraries
Genome-scale CRISPRko screening offers a powerful tool for investigating the connection between genotype and a specific phenotype, such as response to ICIs, in a highly efficient and parallel manner. Mouse GeCKO v2 genome-wide library and Brie mouse genome-wide library are two widely used sgRNA libraries. Dubrot et al. performed eight genome-scale screenings in mouse-transplantable tumors using the Brie mouse genome-wide library. Eight cell lines from five cancer types (melanoma, pancreatic, lung, renal, and colon cancers) were infected with the lentiviral Brie mouse library and implanted into untreated and ICI-treated immunocompetent mice, with NOD SCID mice as controls. Although the comparison between the ICI-treated and wild-type (WT) groups did not reveal a substantial number of hits for six of eight models, ICI treatment produced more significantly depleted or enriched sgRNAs than the untreated condition compared to the NOD SCID group. Comparing ICI treatment and NOD SCID groups, two pathways stood out with shared regulators among different cancer models: the Interferon (IFN) sensing and signaling pathway [Janus kinase 1 (Jak1), Jak2, Signal transducer and activator of transcription 1 (Stat1) and Interferon gamma receptor 1 (Ifngr1)] and antigen processing and presentation pathway [Transporter associated with antigen processing 1 (Tap1), Tap2, TAP-associated glycoprotein (Tapbp), Calreticulin (Calr), Protein disulfide-isomerase A3 (Pdia3), Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator (Erap1), Beta-2-Microglobulin (B2m) and histocompatibility 2, T region locus 23 (H2-T23)] (Dubrot et al., 2022). On the other hand, Li et al. infected ovalbumin-expressing B16 cells (B16-OVA) with the lentiviral mouse GeCKO v2 library. The infected B16-OVA cells were then subcutaneously injected into the right flank of mice mixed with an equal number of SIINFEKL-specific CD8 + T cells (OT-I T cells). SgRNA abundance was quantified by NGS 1 month after transplanting. Ubiquitin-specific peptidase 22 (Usp22) ranked 1 and 7 from two independent repeated in vivo screenings, suggesting that Usp22 deficiency mediated resistance to T cell killing in B16-OVA cells (M. Li et al., 2021b). Usp22 directly interacted with Stat1, deubiquitinated it and improved its stability in melanoma cells. JAK-STAT-IFN signaling pathway highlighted in both genome-wide screenings indicated that targeting the IFN signaling pathway might be a promising strategy to sensitize cancer cells to ICIs.
2.2.1.2 CRISPR/Cas9 screenings with immune-related sgRNA libraries
Immune evasion is a key characteristic of cancer (Hanahan and Weinberg, 2011). While the innate and adaptive immune system can normally recognize and eliminate tumors, certain cancer cells possess mechanisms to escape immune detection or limit immunological killing (Mittal et al., 2014). To identify the genes and signaling pathways involved in immune evasion, Kearney et al. first conducted a genome-wide CRISPR screening in vitro using MC38-OVA cells incubated with activated OT-I T cells and MC38 cells incubated with natural killer (NK) cells in the presence of IgG or anti-PD-1 (Kearney et al., 2018). The top-ranked enriched genes in both groups included tumor necrosis factor receptor superfamily member 1A (Tnfrsf1a) and Caspase 8 (Casp8) in the TNF signaling pathway, Jak1 and Stat1 in the IFN-γ signaling pathway, as well as Tap1 and B2m related to antigen presentation (Kearney et al., 2018)In a subsequent experiment, the top 2000 sgRNAs from both screens were cloned into a custom library, named the immune evasion library, and introduced into MC38-OVA cells expressing Cas9. These cells were then implanted into recipient NSG mice, followed by injection of OT-I T cells after tumor formation (Kearney et al., 2018). Sequencing of harvested tumors revealed enrichment of sgRNAs targeting genes involved in IFN-γ signaling (Stat1), antigen presentation (Tap1), and TNF signaling [Casp8, tnfrsf1a, and 2-aminoethanethiol dioxygenase (Ado)] (Kearney et al., 2018). Deletion of key genes within the TNF signaling, IFN-γ signaling, and antigen presentation pathways provided protection for tumor cells against CD8+ T cell-mediated killing and impaired antitumor immune responses in vivo (Kearney et al., 2018). To uncover the immune-related genes in the antitumor or protumor process that might indicate potential therapeutic strategies, Ji et al. designed and generated a mouse sgRNA library corresponding to 2,796 human disease-related immune genes, which termed the disease-related immune gene library (DrIM library). DrIM-transduced 4T1-Cas9 cells were subcutaneously transplanted into immunocompetent BALB/c mice and immune-deficient NOD-NGP mice. After 14 days, the tumors were harvested for high-throughput sgRNA library sequencing. 227 candidate genes were dichotomized into immune escape genes and immune surveillance genes and constructed into a mini-DrIM sgRNA library. A second round in vivo screening was performed in BALB/c mice, BALB/c-Nude mice only lacking T cells, CB-17 scid mice lacking T cells and B cells, and NPG mice lacking T cells, B cells, and NK cells. By comparing different groups, the screening provided chances to reveal candidates responsible for responsiveness to specific immune cells. Five genes were identified as potential regulators of immune surveillance in all screening settings, among which galectin 2 (Lgals2) induced the increased number of tumor-associated macrophages and resulted in the immunosuppressive microenvironment (Ji et al., 2022). The result provided a theoretical basis for LGALS2 as a potential immunotherapy target.
2.2.1.3 CRISPR/Cas9 screenings with epigenetic sgRNA libraries
Epigenetic modulation genes play crucial roles in cancer biology, and accumulating evidence indicates their involvement in modulating the tumor immune microenvironment (TIME) and regulating the antitumor immune response (Zhu et al., 2015). To comprehensively assess cell-intrinsic epigenetic regulators of tumor immunity, Li et al. conducted an in vivo CRISPR screen using an epigenetic-focused sgRNA library targeting 524 epigenetic regulators (Li et al., 2020). By comparing sgRNAs recovered from subcutaneous xenografts derived from KrasG12D/Trp53−/− mouse lung cancer cells in mice treated with control IgG and anti-PD-1 antibody, several hits were identified, including Tap2, Jak2, Stat1, Catenin Beta 1 (Ctnnb1), Anti-Silencing Function 1A Histone Chaperone (Asf1a), Mitogen-Activated Protein Kinase 3 (Mapk3), and TSC Complex Subunit 1 (Tsc1), that influenced the sensitivity to anti-PD-1 treatment. Loss of Asf1a induced immunogenic macrophage differentiation in the TIME and enhanced T cell activation in combination with anti-PD-1(Li et al., 2020). Another gene hit, Tsc1, was further investigated by Huang et al., who found that TSC1/2 deficiency upregulated PD-L1 expression, making TSC1/2-deficient lung cancer cells more responsive to anti-PD-1 therapy (Huang et al., 2022). Moreover, Griffin et al. employed a sgRNA library targeting chromatin genes to identify ICI sensitizers. They transduced the library into B16 melanoma and Lewis lung carcinoma (LLC) cells, which were then transplanted into mice for tumor-cell vaccination and PD-1 blockade or combination PD-1/CTLA-4 blockade (Griffin et al., 2021). The H3K9-methyltransferase SET Domain Bifurcated Histone Lysine Methyltransferase 1 (Setdb1) emerged as the top-ranked sensitizer in both B16 and LLC models, as its loss led to the de-repression of transposable elements (TEs) capable of generating major histocompatibility complex class I (MHC-I) peptides and triggering T cell responses (Seoane et al., 2019). Another epigenetic library targeting 850 epigenetic factors and RNA-binding factors was applied by Li et al. in the subcutaneous pancreatic ductal adenocarcinoma (PDAC) model. By comparing sgRNA abundance in GAFCP (gemcitabine, G; nab-paclitaxel, A; anti-CD40 agonist, F; anti-CTLA-4, C; and anti-PD1–1, p) treatment group and control group, they identified Lysine Demethylase 3A (Kdm3a) as a potent epigenetic regulator of immunotherapy response in PDAC (J. Li et al., 2021a). These discoveries emphasize the promising opportunity of directing attention towards epigenetic factors to boost the effectiveness of immune checkpoint inhibitors. This can be achieved not only by regulating PD-L1 expression but also by reshaping the immune microenvironment.
2.2.1.4 CRISPR/Cas9 screenings with kinome sgRNA libraries
As kinases are a major drug target and a major control point in cell behavior, the kinase has also been the target of large-scale functional genomics with CRISPRko screenings and drug discovery efforts, especially in cancer therapeutics (Workman, 2005). To investigate the contribution of glioma cell-intrinsic kinases in T cell recognition, GL261 cells were intracranially implanted into WT and CD8 deficient C57BL/6 mice after they had been transfected with a CRISPRko library for all 713 known kinases (Dmello et al., 2023). Among the kinase KO clones depleted in WT mice relative to the CD8 deficient mice, which contributed towards resistance to CD8 T-cell-mediated killing, checkpoint kinase 2 (Chek2) had the most depleted sgRNA. Mechanistically, loss of Chek2 enhances antigen presentation, STING pathway activation, and PD-L1 expression in mouse gliomas, supporting Chek2 as a promising target for enhancement of response to immune checkpoint blockade therapy in glioblastoma (GBM) (Dmello et al., 2023).
2.2.1.5 CRISPR/Cas9 screenings with other custom sgRNA libraries
A commonly employed approach to overcome the high cost and achieve comprehensive coverage of sgRNAs in whole-genome in vivo CRISPR screens is the utilization of custom sgRNA libraries (Li et al., 2014). Wang et al. utilized a custom murine CRISPR-Cas9 knockout (MusCK) library containing 5 sgRNAs for each of the over 4,500 genes implicated in tumor initiation, progression, and immune modulation. They performed in vivo CRISPR screens in 4T1 cells implanted in syngeneic BALB/c mice. A subsequent library (MusCK 2.0) focused on 79 candidate genes identified in the primary screen, with 8 sgRNAs per gene Significant depletion of immune evasion mediators (Cd274/Pd-l1), components of the IFN-γ signaling pathway [Jak1, Jak2, Stat1, and Interferon regulatory factor 1 (Irf1)], an E3 ubiquitin ligase (Cop1), and an oncogenic transcriptional activator [Tripartite Motif Containing 24 (Trim24)] was observed (Wang et al., 2021). Further investigation of Cop1 inhibition revealed reduced macrophage-associated chemokine secretion, decreased tumor macrophage infiltration, and synergy with ICI through polyubiquitination and proteasomal degradation of the CCAAT/enhancer-binding protein (Cebpδ) (Wang et al., 2021). In another study by Manguso et al., lentiviral vectors encoding 9,872 sgRNAs targeting 2,368 genes were used to engineer B16 melanoma cells expressing Cas9. These cells were transplanted into T cell receptor (Tcra)−/− mice or WT mice treated with GVAX and PD-1 blockade. The enrichment of five genes involved in sensing and signaling through the IFNγ pathway (Stat1, Jak1, Ifngr2, Ifngr1, and Jak2) in immunotherapy-treated mice suggested their role in MHC-I presentation upregulation. Further investigation confirmed that Protein Tyrosine Phosphatase Non-Receptor Type 2 (Ptpn2) loss activated IFN-γ signaling and increased antigen presentation (Manguso et al., 2017). Following that, Ishizuka et al. observed a significant depletion of Adenosine deaminase acting on RNA 1 (Adar1)-targeting sgRNA in tumors of immunocompetent mice treated with GVAX and PD-1 blockade. Loss of ADAR1 overcame resistance to PD-1 checkpoint blockade by restoring antigen presentation. The absence of ADAR1 reduced A-to-I editing of interferon-inducible RNA species, leading to melanoma differentiation-associated protein 5 (MDA5)-mediated immune microenvironment inflammation through the secretion of Interferon β (Ishizuka et al., 2019).
2.2.2 CRISPR-Cas9 screenings in CRISPRko-modified T cells
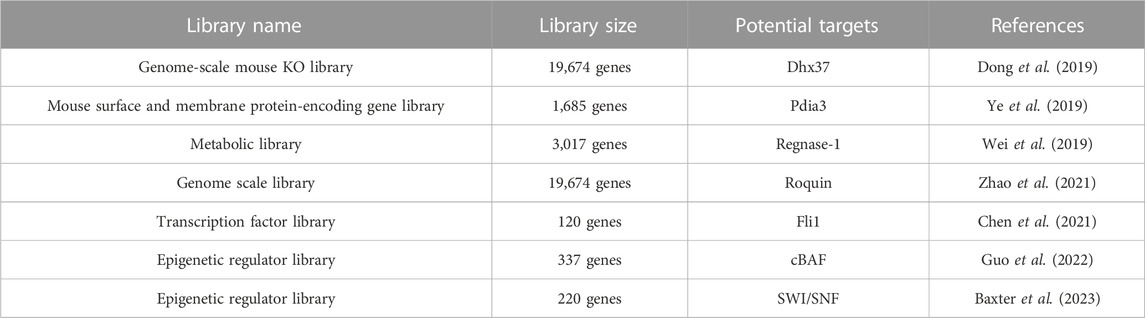
T cells play a central role in the adaptive immune response and immunotherapy. Cytotoxic CD8+ T cells are responsible for destroying virus-infected cells and tumor cells. Matthew et al. designed and generated a lentiviral CRISPR vector, which contains a sgRNA expression cassette and Thy1.1, a surface antigen marker for thymocytes, resulting in a more robust efficiency in determining transduction efficiency by fluorescence-activated cell sorting (FACS) (Table 2) (Dong et al., 2019). They isolated native CD8+ T cells from OT-I, Cas9 mice, infected the T cells with genome-scale mouse KO library, and then transferred these T cells into Recombination activating gene 1 (Rag1)-deficient mice bearing E0771-OVA transplanted tumors (Dong et al., 2019). By harvesting tumors with tumor-infiltrating lymphocytes and analyzing the sgRNA frequency, they re-identified canonical immunotherapy targets such as PD-1 and T cell immunoglobulin and mucin-domain containing-3 (Tim-3), along with genes that have not been characterized in T cells like DEAH-Box Helicase 37 (Dhx37). Dhx37 suppressed effector functions, cytokine production, and T cell activation by modulating nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) pathway (Dong et al., 2019). Ye et al., in the same group, applied the Sleeping Beauty transposon system to enable efficient randomized gene and regulatory-element knockout in primary murine T cells and genomic integration of the sgRNA cassette for screen readout (Ye et al., 2019). They focused on a mouse surface and membrane protein-encoding gene library targeting 1,685 genes and demonstrated that adoptive transfer of CD8+ T cells with Pdia3, Mannoside Acetylglucosaminyltransferase 5 (Mgat5), Epithelial Membrane Protein 1 (Emp1) or Lymphocyte-activation gene 3 (Lag3) gene editing enhanced the survival of GBM-bearing mice in both syngeneic and T cell receptor transgenic models (Ye et al., 2019). Similar strategies were applied by other researchers to conduct in vivo CRISPR screens in CD8+ T cells with different libraries. Targeting Zinc finger CCCH-type containing 12A (ZC3H12A), which encodes the ribonuclease REGNASE-1 was revealed to program long-lived effector T cells for cancer therapy by screening with a metabolic library targeting 3,017 genes(Wei et al., 2019). Rc3h1 encoding the E3 ubiquitin ligase Roquin-1 was found to be the strongest repressor of CD8+ T cell expansion by a genome scale CRISPR screen(Zhao et al., 2021). Surprisingly, the Fli1 (Friend leukemia integration 1) protooncogene was identified to improve effector T cells differentiation and protective immunity in cancer by screen with a library targeting 120 transcription factors (Chen et al., 2021). Multiple components of the mammalian canonical BRG1/BRM-associated factors were discovered to be essential for the differentiation of activated CD8+ T cells into effector T cells by screening with a library targeting 337 epigenetic regulators (Guo et al., 2022). Another chromatin-remodeling complex SWI/SNF (SWItch/Sucrose Non-Fermentable) was identified by screen with a library targeting 220 epigenetic regulators, indicating that chromatin-remodeling complex is a potential target to regulate differentiation of T cells(Baxter et al., 2023).
2.2.3 Direct CRISPR-Cas9 screenings
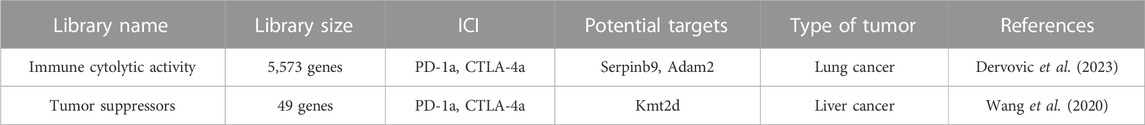
Because the immune microenvironment within a subcutaneously implanted tumor model diverges significantly from that found in a tumor in its autochthonous (natural) location, the advantage lies with in situ CRISPR-Cas9 screens. The primary essence of in situ CRISPR-Cas9 screens revolves around prompting the transformation of ordinary cells into tumorous ones within their native context and subsequently employing CRISPR-Cas9 technology to modify the genomes of these cells. Wang et al. developed a CRISPR-mediated genetically engineered mouse model (CRISPR-GEMM) of liver cancer by utilizing an adeno-associated virus (AAV)-CRISPR vector to drive autochthonous liver tumorigenesis in fully immunocompetent mice through pooled mutagenesis (Wang et al., 2020).(Table 3). The vector contained a liver-specific thyroxine-binding globulin (TBG) promoter driving Cre recombinase expression, along with two sgRNA expression cassettes. The Cre recombinase thus can induce Cas9 and firefly luciferase expression in the liver of the Lox-stop-Lox (LSL)-Cas9, LSL-Fluc mouse. As for the two sgRNAs, one targeted Trp53 to induce tumorigenesis, while the other served as a backbone for cloning and expressing specific sgRNAs targeting the top 49 most frequently mutated tumor suppressor genes in pan-cancer TCGA dataset (Wang et al., 2020). Comparing the anti-CTLA4-treated mice with the PBS-treated mice, the mutation frequencies of B2m, Glucocorticoid receptor DNA-binding factor 1 (Grlf1), BCL6 Corepressor (Bcor), and Kdm5c were significantly increased, while the mutation frequency of AT-Rich Interaction Domain 1A (Arid1a) was significantly decreased. Similarly, comparing anti-PD1-treated mice with PBS-treated mice, the knockout of B2m, Grif1, Von Hippel-Lindau Tumor Suppressor (Vhl), Cyclin-Dependent Kinase Inhibitor 1B (Cdkn1b), and Bcor was associated with anti-PD1 resistance, whereas knockout of Kmt2d, Arid1a, Ring Finger Protein 43 (Rnf43), and ATRX Chromatin Remodeler (Atrx) was linked to anti-PD1 responsiveness(Wang et al., 2020). Loss-of-function mutations in Lysine Methyltransferase 2D (KMT2D) were found to potentiate anti-PD1 checkpoint immunotherapy by enhancing immune infiltration in the tumor microenvironment. This was achieved through multiple mechanisms, including increased DNA damage and mutation burden, activation of TEs, elevation of IFNγ-stimulated antigen presentation, and upregulation of myeloid-recruiting cytokines (Wang et al., 2020). Dervovic et al. designed a similar vector, LV-sgRNA-Cre-OVA, to induce tumorigenesis in mouse lung tissue and conduct a CRISPR-Cas9 screen. They administered the lentiviral sgRNA library generated by LV-sgRNA-Cre-OVA vector at postnatal day 2 to the lungs of LSL-KrasG12D or LSL-BrafV600E mice(Dervovic et al., 2023). The Cre recombinase activated the expression of KrasG12D or LSL-BrafV600E in the lung tissue and induced lung cancer. OVA was applied to increase efficient antigen presentation and induce immune responses. OT-I T cells were injected in the mice via the tail-vein and anti-PD-1 treatment, anti-CTLA4 treatment or IgG control was administered 4 weeks after lung tumor induction by LV-Cre-OVA inhalation. These mice were then immunized with OVA-peptide emulsified in Complete Freund’s adjuvant on day 1 and primed with OVA/Incomplete Freund’s adjuvant emulsion on day 7 post adoptive cell transfer to stimulate specific OT-I T cell responses (Dervovic et al., 2023). sgRNAs were quantified at 6.5 weeks after tumor induction. They recovered the known immune evasion factors Stat1 and Serpin Family B Member 9 (Serpinb9) and identified the cancer testis antigen ADAM Metallopeptidase Domain 2 (Adam2) as an immune modulator, whose expression is induced by KrasG12D and further elevated by immunotherapy (Dervovic et al., 2023).
3 Concluding remarks and future perspectives
In conclusion, the application of CRISPR/Cas9 technology within the intricate landscape of immunology offers a tantalizing pathway towards advancing the realm of immunotherapy and presents a promising avenue for enhancing ICIs efficacy, as it enables the identification of novel targets for cancer treatment.
Furthermore, the continuous advancements and application of CRISPR/Cas9 technologies such as those that pairs CRISPR-ko with lineage tracing via random barcodes hold significant potential if combined with studies that tackle the mechanisms of immunomodulatory drugs (Rogers et al., 2017). This is a promising field to revolutionize cancer treatment, offering more precise and effective therapies for patients. Apart from the CRISPRko screens covered in this review, numerous in vitro screenings utilizing CRISPRa and CRISPRi have unveiled potential targets capable of enhancing CAR-T efficacy (Ye et al., 2022), mediating T cell stimulation (Schmidt et al., 2022), or acting as potential drivers of cancer resistance to NK and T cell-mediated cytotoxicity (Joung et al., 2022; Lee et al., 2023). However, it is noteworthy that the in vivo screening results for CRISPRa and CRISPRi libraries in this field are yet to be identified.
Despite the immense potential of CRISPR/Cas9 high-throughput screening in improving immunotherapy efficacy, certain safety and efficacy concerns remain, impeding its full translation into clinical settings. The main challenges facing the use of CRISPR/Cas9-based treatments include the ethical concerns, the identification of a proper delivery technique, the occurrence of off-targeting modifications, and the possibility of causing autoimmune disease (Rasul et al., 2022). Therefore, it is crucial to leverage extensive screening, molecular, and clinical data to deepen our understanding of tumor immunity and expedite progress in the field of cancer therapy. Through diligent research and collaboration, we can usher in a new era of personalized and targeted cancer treatments.
Author contributions
YW: Conceptualization, Writing–original draft, Writing–review and editing. AKh: Conceptualization, Writing–original draft, Writing–review and editing. AKa: Conceptualization, Writing–original draft, Writing–review and editing. MD: Writing–original draft, Writing–review and editing. TD: Writing–review and editing. CM: Writing–review and editing, Conceptualization. ZW: Writing–review and editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grants R01CA271540 and R00CA226506 to CM and R01CA196643, R01CA264320, R01CA260629, P50CA150964, and P30 CA043703 to ZW.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamson, B., Norman, T. M., Jost, M., Cho, M. Y., Nuñez, J. K., Chen, Y., et al. (2016). A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882. doi:10.1016/j.cell.2016.11.048
Baxter, A. E., Huang, H., Giles, J. R., Chen, Z., Wu, J. E., Drury, S., et al. (2023). The SWI/SNF chromatin remodeling complexes BAF and PBAF differentially regulate epigenetic transitions in exhausted CD8+ T cells. Immunity 56 (6), 1320–1340.e10. doi:10.1016/j.immuni.2023.05.008
Chen, Z., Arai, E., Khan, O., Zhang, Z., Ngiow, S. F., He, Y., et al. (2021). In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184 (5), 1262–1280.e22. doi:10.1016/j.cell.2021.02.019
Chow, R. D., and Chen, S. (2018). Cancer CRISPR screens in vivo. Trends Cancer 4 (5), 349–358. doi:10.1016/j.trecan.2018.03.002
Dervovic, D., Malik, A. A., Chen, E. L. Y., Narimatsu, M., Adler, N., Afiuni-Zadeh, S., et al. (2023). In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer. Nat. Commun. 14 (1), 3150. doi:10.1038/s41467-023-38841-7
Dmello, C., Zhao, J., Chen, L., Gould, A., Castro, B., Arrieta, V. A., et al. (2023). Checkpoint kinase 1/2 inhibition potentiates anti-tumoral immune response and sensitizes gliomas to immune checkpoint blockade. Springer U. S. 14 (1), 1566. doi:10.1038/s41467-023-36878-2
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology 34 (2), 184–191. doi:10.1038/nbt.3437
Dong, M. B., Wang, G., Chow, R. D., Ye, L., Zhu, L., Dai, X., et al. (2019). Systematic immunotherapy target discovery using genome-scale In Vivo CRISPR screens in CD8 T cells. Cells 178 (5), 1189–1204. doi:10.1016/j.cell.2019.07.044
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213), 1258096. doi:10.1126/science.1258096
Dubrot, J., Du, P. P., Lane-Reticker, S. K., Kessler, E. A., Muscato, A. J., Mehta, A., et al. (2022). In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat. Immunol. 23 (10), 1495–1506. doi:10.1038/s41590-022-01315-x
Emens, L. A., Ascierto, P. A., Darcy, P. K., Demaria, S., Eggermont, A. M. M., Redmond, W. L., et al. (2017). Cancer immunotherapy: opportunities and challenges in the rapidly evolving clinical landscape. Eur. J. Cancer 81, 116–129. doi:10.1016/j.ejca.2017.01.035
Finck, A., Gill, S. I., and June, C. H. (2020). Cancer immunotherapy comes of age and looks for maturity’, Nature Communications. Springer U. S. 11 (1), 1–4. doi:10.1038/s41467-020-17140-5
Fouad, Y. A., and Aanei, C. (2017). Revisiting the hallmarks of cancer. Am. J. cancer Res. 7 (5), 1016–1036.
Gandhi, L., Rodríguez-Abreu, D., Gadgeel, S., Esteban, E., Felip, E., De Angelis, F., et al. (2018). Pembrolizumab plus chemotherapy in metastatic non–small-cell lung cancer. N. Engl. J. Med. 378 (22), 2078–2092. doi:10.1056/nejmoa1801005
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159 (3), 647–661. doi:10.1016/j.cell.2014.09.029
Griffin, G. K., Wu, J., Iracheta-Vellve, A., Patti, J. C., Hsu, J., Davis, T., et al. (2021). Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature 595 (7866), 309–314. doi:10.1038/s41586-021-03520-4
Guo, A., Huang, H., Zhu, Z., Chen, M. J., Shi, H., Yuan, S., et al. (2022). cBAF complex components and MYC cooperate early in CD8+ T cell fate. Nature 607 (7917), 135–141. doi:10.1038/s41586-022-04849-0
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Herbst, R. S., Soria, J. C., Kowanetz, M., Fine, G. D., Hamid, O., Gordon, M. S., et al. (2014). Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515 (7528), 563–567. doi:10.1038/nature14011
Hodi, F. S., O’Day, S. J., McDermott, D. F., Weber, R. W., Sosman, J. A., Haanen, J. B., et al. (2010). Improved survival with Ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723. doi:10.1056/nejmoa1003466
Huang, Q., Li, F., Hu, H., Fang, Z., Gao, Z., Xia, G., et al. (2022). Loss of TSC1/TSC2 sensitizes immune checkpoint blockade in non–small cell lung cancer. Sci. Adv. 8 (5), eabi9533–11. doi:10.1126/sciadv.abi9533
Ishizuka, J. J., Manguso, R. T., Cheruiyot, C. K., Bi, K., Panda, A., Iracheta-Vellve, A., et al. (2019). Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565(7737), pp. 43–48. doi:10.1038/s41586-018-0768-9
Jhunjhunwala, S., Hammer, C., and Delamarre, L. (2021). Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 21 (5), 298–312. doi:10.1038/s41568-021-00339-z
Ji, P., Gong, Y., Jin, M. L., Wu, H. L., Guo, L. W., Pei, Y. C., et al. (2022). In vivo multidimensional CRISPR screens identify Lgals2 as an immunotherapy target in triple-negative breast cancer. Sci. Adv. 8 (26), eabl8247. doi:10.1126/sciadv.abl8247
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E., et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Joung, J., Kirchgatterer, P. C., Singh, A., Cho, J. H., Nety, S. P., Larson, R. C., et al. (2022). CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity. Nat. Commun. 13 (1), 1606–1614. doi:10.1038/s41467-022-29205-8
Kearney, C. J., Vervoort, S. J., Hogg, S. J., Ramsbottom, K. M., Freeman, A. J., Lalaoui, N., et al. (2018). Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 3 (23), 1–15. doi:10.1126/sciimmunol.aar3451
Koike-Yusa, H., Li, Y., Tan, E. P., Velasco-Herrera, M. D. C., and Yusa, K. (2014). Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32 (3), 267–273. doi:10.1038/nbt.2800
Kuhn, M., Santinha, A. J., and Platt, R. J. (2021). Moving from in vitro to in vivo CRISPR screens. Gene and Genome Editing 2 (May), 100008. doi:10.1016/j.ggedit.2021.100008
Lee, D-H., Ahn, H., Sim, H. I., Choi, E., Choi, S., Jo, Y., et al. (2023). A CRISPR activation screen identifies MUC-21 as critical for resistance to NK and T cell-mediated cytotoxicity. J. Exp. Clin. Cancer Res. 42 (1), 272. doi:10.1186/s13046-023-02840-9
Li, F., Huang, Q., Luster, T. A., Hu, H., Zhang, H., Ng, W. L., et al. (2020). In vivo epigenetic crispr screen identifies asf1a as an immunotherapeutic target in kras-mutant lung adenocarcinoma. Cancer Discov. 10 (2), 270–287. doi:10.1158/2159-8290.CD-19-0780
Li, J., Yuan, S., Norgard, R. J., Yan, F., Sun, Y. H., Kim, I. K., et al. (2021a). Epigenetic and transcriptional control of the epidermal growth factor receptor regulates the tumor immune microenvironment in pancreatic cancer. Cancer Discov. 11 (3), 736–753. doi:10.1158/2159-8290.CD-20-0519
Li, M., Xu, Y., Liang, J., Lin, H., Qi, X., Li, F., et al. (2021b). USP22 deficiency in melanoma mediates resistance to T cells through IFNγ-JAK1-STAT1 signal axis. Mol. Ther. 29 (6), 2108–2120. doi:10.1016/j.ymthe.2021.02.018
Li, W., Xu, H., Xiao, T., Cong, L., Love, M. I., Zhang, F., et al. (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15 (12), 554. doi:10.1186/s13059-014-0554-4
Liu, D., Zhao, X., Tang, A., Xu, X., Liu, S., Zha, L., et al. (2020). CRISPR screen in mechanism and target discovery for cancer immunotherapy. BBA - Rev. Cancer 1874 (1), 188378. doi:10.1016/j.bbcan.2020.188378
Manguso, R. T., Pope, H. W., Zimmer, M. D., Brown, F. D., Yates, K. B., Miller, B. C., et al. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547 (7664), 413–418. doi:10.1038/nature23270
McDaniel, A. L., Alger, H. M., Sawyer, J. K., Kelley, K. L., Kock, N. D., Brown, J. M., et al. (2013). Phytosterol feeding causes toxicity in ABCG5/G8 knockout mice. Am. J. Pathology 182 (4), 1131–1138. doi:10.1016/j.ajpath.2012.12.014
Mittal, D., Gubin, M. M., Schreiber, R. D., and Smyth, M. J. (2014). New insights into cancer immunoediting and its three component phases — elimination, equilibrium and escape. Physiology Behav. 27, 16–25. doi:10.1016/j.coi.2014.01.004
Pauken, K. E., and Wherry, E. J. (2015). Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 36 (4), 265–276. doi:10.1016/j.it.2015.02.008
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159 (2), 440–455. doi:10.1016/j.cell.2014.09.014
Rasul, M. F., Hussen, B. M., Salihi, A., Ismael, B. S., Jalal, P. J., Zanichelli, A., et al. (2022). Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Mol. Cancer 21 (1), 64–30. doi:10.1186/s12943-021-01487-4
Rogers, Z. N., McFarland, C. D., Winters, I. P., Naranjo, S., Chuang, C. H., Petrov, D., et al. (2017). A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat. Methods 14 (7), 737–742. doi:10.1038/nmeth.4297
Schmidt, R., Steinhart, Z., Layeghi, M., Freimer, J. W., Bueno, R., Nguyen, V. Q., et al. (2022). CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 375 (6580), eabj4008–30. doi:10.1126/science.abj4008
Schoenfeld, A. J., and Hellmann, M. D. (2020). Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37 (4), 443–455. doi:10.1016/j.ccell.2020.03.017
Schumann, K., Lin, S., Boyer, E., Simeonov, D. R., Subramaniam, M., Gate, R. E., et al. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U. S. A. 112 (33), 10437–10442. doi:10.1073/pnas.1512503112
Seoane, J. A., Kirkland, J. G., Caswell-Jin, J. L., Crabtree, G. R., and Curtis, C. (2019). Chromatin regulators mediate anthracycline sensitivity in breast cancer. Nat. Med. 25 (11), 1721–1727. doi:10.1038/s41591-019-0638-5
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. doi:10.1126/science.1247005
Sharma, P., Goswami, S., Raychaudhuri, D., Siddiqui, B. A., Singh, P., Nagarajan, A., et al. (2023). Immune checkpoint therapy—current perspectives and future directions. Cell 186 (8), 1652–1669. doi:10.1016/j.cell.2023.03.006
Topalian, S., et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 365, 687–696.
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339 (6127), 1546–1558. doi:10.1126/science.1235122
Wang, G., Chow, R. D., Zhu, L., Bai, Z., Ye, L., Zhang, F., et al. (2020). Crispr-gemm pooled mutagenic screening identifies kmt2d as a major modulator of immune checkpoint blockade. Cancer Discov. 10 (12), 1912–1933. doi:10.1158/2159-8290.CD-19-1448
Wang, X., Tokheim, C., Gu, S. S., Wang, B., Tang, Q., Li, Y., et al. (2021). In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target. Cell 184 (21), 5357–5374.e22. doi:10.1016/j.cell.2021.09.006
Wei, J., Long, L., Zheng, W., Dhungana, Y., Lim, S. A., Guy, C., et al. (2019). Targeting Regnase-1 programs long-lived effector T cells for cancer therapy. Nature 576 (7787), 471–476. doi:10.1038/s41586-019-1821-z
Workman, P. (2005). Drugging the cancer kinome: progress and challenges in developing personalized molecular cancer therapeutics. Cold Spring Harb. Symposia Quantitative Biol. 70, 499–515. doi:10.1101/sqb.2005.70.020
Xu, Y., and Li, Z. (2020). CRISPR-Cas systems: overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 18, 2401–2415. doi:10.1016/j.csbj.2020.08.031
Ye, L., Park, J. J., Dong, M. B., Yang, Q., Chow, R. D., Peng, L., et al. (2019). In vivo CRISPR screening in CD8 T cells with AAV–Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 37 (11), 1302–1313. doi:10.1038/s41587-019-0246-4
Ye, L., Park, J. J., Peng, L., Yang, Q., Chow, R. D., Dong, M. B., et al. (2022). A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab. 34 (4), 595–614.e14. doi:10.1016/j.cmet.2022.02.009
Zhang, F. (2019). Development of CRISPR-Cas systems for genome editing and beyond. Q. Rev. Biophysics 52, e6. doi:10.1017/s0033583519000052
Zhao, H., Liu, Y., Wang, L., Jin, G., Zhao, X., Xu, J., et al. (2021). Genome-wide fitness gene identification reveals Roquin as a potent suppressor of CD8 T cell expansion and anti-tumor immunity’, Cell Reports. ElsevierCompany 37 (10), 110083. doi:10.1016/j.celrep.2021.110083
Keywords: CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (CRISPR associated protein 9)-mediated genome editing, PD-1, immune check inhibitor (ICI), CTLA-4, in vivo CRISPR screen, invitro CRISPR screen, tumor immunotherapy
Citation: Wang Y, Khalil A, Kamar A, Du M, Dinh T, McFarland C and Wang Z (2023) Unveiling immune checkpoint regulation: exploring the power of in vivo CRISPR screenings in cancer immunotherapy. Front. Genet. 14:1304425. doi: 10.3389/fgene.2023.1304425
Received: 29 September 2023; Accepted: 27 November 2023;
Published: 14 December 2023.
Edited by:
Rodrigo Alexandre Panepucci, University of Sao Paulo, BrazilReviewed by:
Sunil K Malonia, University of Massachusetts Medical School, United StatesKatiuchia Sales, University of São Paulo, Brazil
Copyright © 2023 Wang, Khalil, Kamar, Du, Dinh, McFarland and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher McFarland, christopher.mcfarland@case.edu; Zhenghe Wang, zhenghe.wang@case.edu
†These authors have contributed equally to this work
 Yuxiang Wang
Yuxiang Wang Athar Khalil
Athar Khalil Amina Kamar
Amina Kamar Mengyan Du
Mengyan Du Trang Dinh
Trang Dinh Christopher McFarland1*
Christopher McFarland1* Zhenghe Wang
Zhenghe Wang