Molecular identification of individual and seasonal variation in incidental ingestion of arthropods by free-ranging goats
- Department of Evolutionary and Environmental Biology, University of Haifa, Haifa, Israel
The extent of direct interactions between large mammalian herbivores (LMH) and plant-dwelling arthropods (PDA), i.e., the ingestion of PDA by LMH, remains largely unexplored. Grazing LMH may ingest a variety of PDA, yet, it is unknown how different foraging strategies (i.e., browsing, grazing, etc.) influence the ingestion of PDA or whether individual variation within herds affects it. Here we examine how individual variation within a herd of browsing LMH impacts PDA ingestion. This was done using a DNA metabarcoding analysis on feces collected monthly from marked individuals within a herd of free-ranging goats. We found that goats frequently ingest PDA while feeding (all samples contains PDA), including a complex food-chain of herbivores, predators and parasites, which differed over the season and among individual goats. In total, 63 families of insects and 9 families of arachnids from 15 orders were ingested by the goats. Most ingested PDA were herbivores with reduced mobility, such as immature or sessile species. Highly mobile and noxious PDA were rarely detected. We show for the first time that ingestion of PDA by LMH is influenced by seasonal and individual variation within the herd and that it is common among LMH, regardless of feeding strategy or habitat.
1. Introduction
Large mammalian herbivores (LMH) and plant-dwelling arthropods (PDA) frequently interact with one another as they both utilize the same plants. Indirect effects of LMH on PDA, occurring through plant-mediated mechanisms, have been studied extensively over the past years (Stewart, 2001; van Klink et al., 2015; Zhu et al., 2019). Yet, direct effects (especially trophic effects), i.e., the ingestion of PDA by LMH, have received much less attention (van Klink et al., 2015; Gish et al., 2017).
A growing body of evidence indicates that direct trophic interactions are prevalent and have considerable consequences for both LMH and PDA (Gish et al., 2017). PDA vulnerable to LMH ingestion have developed specific adaptations to minimize this risk. For example, female butterflies may lay their eggs on lower plant parts which are inaccessible to LMH (Bennett et al., 2015). Aphids can effectively deter LMH by inducing repulsive odors from the galls they induce (Rostás et al., 2013). Certain aphid and ladybug (Coccinellidae) species immediately drop off the plant when they recognize the humid and warm breath typical of LMH (Gish et al., 2010; Ben-Ari and Inbar, 2013). Interestingly, dropping rates of aphids are higher in biotypes that feed exclusively on plant species that are palatable to LMH, implying that PDA who specialize on non-palatable plants may be safe and even benefit from LMH feeding (Ben-Ari et al., 2019). Other PDA, such as ants and bees, may efficiently deter LMH using aggressive behavior (King et al., 2007; Martins, 2010). On the other hand, ingesting noxious or poisonous PDA may harm or even kill LMH (Schmitz, 1989; Nunamaker et al., 2003; Ferrer et al., 2007). Recent studies have demonstrated that LMH possess highly efficient (and innate) behavioral mechanisms that enable them to avoid ingesting such PDA (Berman et al., 2017, 2018, 2019b). Still, many innocuous PDA and species developing within fruits and seeds may be frequently ingested by LMH (Bonal and Muñoz, 2007; Yamazaki and Sugiura, 2008; Gish et al., 2010). Until recently the extent of this direct trophic interaction remained untested.
DNA metabarcoding, which allows the identification of multiple taxa from bulk environmental samples using high-throughput sequencing, has made it possible to efficiently and rapidly resolve a variety of large-scaled trophic interactions (Clare et al., 2014; Pringle and Hutchinson, 2020). Using DNA metabarcoding on feces of free-ranging cattle has recently revealed the extent of PDA ingestion by LMH at the community level (Berman and Inbar, 2022). It was found that PDA ingestion is frequent, as most of the fecal samples of cattle contained sequences belonging to PDA (76%). Cattle (incidentally) ingested a complex multiple trophic level food-chain of PDA, including a large variety of herbivores as well as their predators and parasites. They also ingested aquatic arthropods while drinking. Highly mobile PDA that can escape ingestion (e.g., bees and grasshoppers) were rarely consumed (Berman and Inbar, 2022). While this study revealed important aspects regarding the ingestion of PDA by LMH at the community context, we know little as to whether it is widespread among LMH or how different foraging strategies (i.e., browsing, grazing, etc.) influence PDA ingestion.
Another important feature of DNA metabarcoding of fecal samples is that it provides individual-level data that can be used to understand the individual variance in dietary preferences. Repeatedly collecting fecal samples from known individuals over time offers a rare opportunity to characterize individual variation, while exploiting the non-invasiveness and high taxonomic resolution of this method. Since it is well established that LMH herds are comprised of individuals that vary in resource use and feeding behavior (Searle et al., 2010; Neave et al., 2018), PDA ingestion is also likely to be influenced by individual variance within the herd. Most of the research to date focused on temporal or spatial snapshots of feeding habits (Berman and Inbar, 2022), overlooking the existence of individual differences and their influence on the examined trophic interactions (Pringle and Hutchinson, 2020).
In this study we examine how individual variation of food selection within a herd affects the ingestion of PDA using a DNA metabarcoding analysis (targeting COI gene of arthropods) on fecal samples repeatedly collected from known individuals. We focused on a model system comprised of free-ranging goats. Specifically, we addressed the following questions: (1) which functional feeding groups of PDA (herbivores, predators, parasites, etc.) are ingested by goats? (2) Does the composition of PDA ingested by goats change over the season? (3) Does the pattern of PDA ingestion differ among different individual goats? (4) Do browsers and grazers exhibit a similar pattern of PDA ingestion?
2. Materials and methods
2.1. Study site
We conducted the study at the Ramat Hanadiv Nature Park, located on the southern edge of the Mount Carmel ridge, Israel. The landscape is covered by a Mediterranean woodland (maquis), with steep, rocky slopes and patches of shallow soil. The climate is Mediterranean, characterized by hot, dry summers and mild, rainy winters with a mean annual rainfall of 600 mm (which falls between October and April). The growing season is closely associated with rainfall distribution and green annual herbaceous vegetation covers the soil patches from January to May. The vegetation in the park is dominated by low trees (mainly Phillyrea latifolia L.) and tall shrubs (Pistacia lentiscus L. and Calicotome villosa Poir. Link). Bushes of Ephedra foemina Forskk., Asparagus stipularis Forskk. and Sarcopoterium spinosum L. Spach grow between the trees. Around 500 herbaceous species can be found in the park, mostly Cyclamen persicum, Brachypodium distachyon, Urospermum picroides, Asphodelus ramosus, Anagalis arvensis, and Convolvulus pentapetaloides (Hadar et al., 1999).
2.2. Experimental design and animals
Large areas of the park are grazed by a resident herd of goats containing 150 adult females (Alpine, Damascus and Mamber breeds). The goats forage the park daily, for approximately 4 h, and receive supplementary feed (hay and processed pellets including 16% crude protein, vitamins and minerals) upon their return to the pen.
To trace individual and seasonal variation in ingestion of PDA, we collected fecal samples from a group of 18 marked individual goats, once a month from February to June (end of winter to the beginning of summer) when arthropod abundance is at its highest. We collected the feces (~5 g) directly from the goats’ rectum using sanitary gloves (replaced for each goat to avoid contamination) upon their return from the pasture. The samples were placed in plastic zip-lock bags and kept in a cooler box with ice packs. Upon return to the lab (within 3 h), we pulverized each sample using a mortar and pestle, thoroughly mixed (homogenized) it and transferred it to-80° C until DNA extraction.
2.3. Molecular analyses
DNA extraction. We extracted DNA from the fecal samples using a QIAamp Fast DNA Stool Mini Kit (ID: 51604, Qiagen, Germany) according to the manual of “isolation of DNA from stool for pathogen detection,” with the following modifications: we conducted the lysis with InhibitEX buffer at 95° C and conducted the incubation with proteinase K and AL buffer at 70°C for 20 min. We eluted the DNA twice with 50 μl of AE buffer (total 100 μl) and stored it at-20°C.
PCR amplification. We PCR-amplified DNA using the Zeale et al. (2011) primer set (ZBJ-ArtF1c: 5’-AGATATTGGAACWTTATATTTTATTTTTGG-3′ and ZBJ-ArtR2c: 5’-WACTAATCAATTWCCAAATCCTCC-3′) which targets a 157 bp segment of the mitochondrial cytochrome c oxidase subunit I (COI) gene in arthropods. These COI markers were chosen, despite possible taxonomic biases (Clarke et al., 2014; Deagle et al., 2014; Elbrecht and Leese, 2017) due to their comprehensive reference database (Ratnasingham and Hebert, 2013) and ability to effectively discriminate at the species level (Deagle et al., 2014). Furthermore, they have been successfully applied in many dietary studies involving arthropods (Kaunisto et al., 2017; Alberdi et al., 2018; Vesterinen et al., 2018; Rytkönen et al., 2019).
For PCR amplification we used 12.5 μl EmeraldAmp MAX HS PCR Master Mix (Takara bio Inc., Otsu, Shiga, Japan), 1 μl of forward and reverse primers (0.5 ng/μl), 5 μl of DNA template (10–100 ng), 0.2 μl of BSA (0.1 μg/μl) and 5.3 μl of sterile water for a total reaction volume of 25 μl per sample. The PCR program included an initial denaturation step at 98°C for 2 min, followed by 35 cycles of 10 s at 90°C; 30 s at 52°C; and 30 s at 72°C, and a final elongation step of 1 min at 72° C. Negative controls were carried out for each PCR assay. We verified the amplification by agarose gel electrophoresis and stored the PCR products at-20° C.
2.4. Library construction and sequence analysis
Library construction. COI amplicon sequencing was performed on an Illumina MiniSeq instrument (Illumina, CA, USA) at the Sequencing Core (UICSQC), University of Illinois, Chicago, using a two-stage amplification protocol as described by Naqib et al. (2018). PhiX DNA was used as a spike-in control. A total of 79 fecal samples were sequenced based on gel verification (Supplementary Table S1): 16 samples from February, 15 from March, 17 from April, 18 from May and 13 from June. Sequencing results (pair-ends, 2 × 150 bp) were delivered as FASTQ files.
Sequence Analysis. We processed the Illumina FASTQ files in R using DADA2 (Callahan et al., 2016) to clean them of PCR and Illumina-sequencing errors. We trimmed and filtered the FASTQ reads for quality using the command “filterAndTrim” with the parameters maxN = 0, maxEE = c(5,5), trimLeft = c(30,15). Error rate estimation was carried out using the “learnerror” command with default parameters (randomize parameter was set to “TRUE” to randomly sample nucleotides and reads across all samples). We then implemented the DADA2 algorithm for error correction and merged forward and reverse reads using the “mergePairs” command (minimum overlap of 6 bp). We removed likely chimeras using the command “removeBimeraDenovo” with default parameters. This process generated an amplicon sequence variants (ASV) count table containing 1,313,939 high-quality reads. ASVs with sequences longer or shorter than the expected barcode length (>170 bp or < 150 bp) were eliminated, retaining 409,596 reads binned in 666 ASVs.
Taxonomic identification. We aligned the ASV sequences against the NCBI GenBank nt-database and imported the BLAST output files into “MEGAN community edition” [v.6.21.11 (Huson et al., 2016)] for taxonomic classification. This was done using the Lowest Common Ancestor (LCA) approach with parameters set to: bitscore >100, E value <10^7, min support =1, top percent =10, LCA algorithm = naïve and percent to cover = 100. Any unclassified sequences were examined against the BOLD database (Ratnasingham and Hebert, 2007). We retained only sequences belonging to insects and arachnids. Moreover, we removed any taxa found in a single sample alone (listed in Supplementary Table S2), to provide a more accurate estimate of the arthropod community. This resulted in 76 samples with 342,435 reads and 223 ASVs (Supplementary Table S3). The raw sequence data is available at the NCBI database under BioProject accession number PRJNA579572.
2.5. Data analysis and statistics
To avoid sequencing depth bias, we rarefied the data to 913 reads per sample using the VEGAN R package command “rarefy” (Oksanen et al., 2007), as the curves of most samples reached a plateau at this point (Supplementary Data Sheet 2 Supplementary Figure S1). This process resulted in 57 goat fecal samples (9 samples from February, 10 from March, 13 from April, 13 from May and 12 from June), binned in 215 ASVs (Supplementary Table S4).
Occurrence data (presence/absence) and relative read abundance (RRA) are two approaches currently used to analyze sequence data in DNA metabarcoding dietary studies. Occurrence data is considered to be more cautious, yet RRA may be more accurate at representing population-level diets (Deagle et al., 2018). Indeed, some recent DNA metabarcoding studies of insectivore diets focus on RRA analysis (Vesterinen et al., 2018; Pringle et al., 2019). For these reasons, we chose to use RRA as the main method for analyzing our data (calculated as the proportion of reads of each ASV in each fecal sample). To avoid potential biases of the RRA analysis (which can arise from differential digestibility, amplification biases, etc.), we also performed a supporting analysis of percent of occurrence (POO, calculated as the number of samples containing a given food item, rescaled to 100% across all food items; see Deagle et al., 2018; Supplementary Table S5). In both analysis we removed any sequences with an abundance of <1% (i.e., true presence threshold, see Deagle et al., 2018; Supplementary Table S6). No assumptions were made regarding the actual proportions or biomass of arthropods.
We calculated diversity (Shannon H′) and richness (Fisher’s alpha) indices of the ASVs using PAST (Hammer et al., 2001). Since not all individual goats had a valid sample for each month, we selected goats that had at least three consecutive samples, resulting in six goats with samples from April–May (hereafter “six-goat group”). This group was used to compare the diversity indices among sampling months using a Friedman’s test (post hoc Wilcoxon signed-rank test, Bonferroni corrected for multiple comparisons). The number of arthropod families ingested among sampling months and among individual goats was also compared within the six-goat group using a repeated measures ANOVA (with the individual goats and sampling months as the repeated-measures factors). Both tests were performed using IBM SPSS software v.25.
We examined the similarity of the overall arthropod community (ASVs) among February–June and among all individual goats using a non-metric multidimensional scaling analysis (NMDS) in R (RRA: Bray–Curtis similarity matrix; POO: Jaccard similarity matrix). To test whether the community significantly differed among sampling months and individual goats (and the six-goat group independently) we performed a permutational multivariate analysis of variance (PERMANOVA) with 999 permutations using the R-VEGAN function ADONIS (RRA: Bray-Curtis; POO: Jaccard) for each factor separately. When considering all individual goats, goat ID was set as strata. In the six-goat group, a nested ADONIS function was implemented (goat ID as nested factor).
Finally, we assigned all arthropods to functional feeding groups (Supplementary Table S7, see Supplementary Data Sheet 1 for full details): herbivores, predators, parasites, dung-associated arthropods, aquatic arthropods or unknown. We also determined herbivore feeding niche based on the literature: (1) exophages – feed on the plant surface; (2) endophages – feed within plant parts (such as gall formers and arthropods boring or mining leaves, stems, fruit and seeds); (3) unknown. We compared the mean RRA of the functional feeding groups among sampling months using a Friedman’s test (post hoc Wilcoxon signed-rank test, Bonferroni corrected for multiple comparisons). The number of genera per functional feeding group was compared among sampling months using the Pearson chi-square test. Both tests were performed using IBM SPSS software v.25.
3. Results
Overall, we identified 194 ASVs belonging to arthropods from 57 goat fecal samples (13 ASVs per sample on average), which varied over time and among individual goats. These ASVs represented 103 genera in 63 families and 11 orders of insects (95% RRA; 93% POO; for full occurrence results, see Supplementary Data Sheet 3) and 11 genera in 9 families and 3 orders of arachnids (5% RRA; 7% POO).
3.1. PDA ingested by goats
While grazing, the goats ingested a wide variety of arthropods, mostly plant-dwelling taxa (88% of both RRA and POO, Figure 1A and Supplementary Data Sheet 3: Supplementary Figure S1A) and a few aquatic taxa ingested while drinking (2% of both RRA and POO, Figure 1A and Supplementary Data Sheet 3: Supplementary Figure S1A). A small proportion of the RRA belonged to dung-associated arthropods (2% RRA, Figure 1A; 3% POO, Supplementary Data Sheet 3: Supplementary Figure S1A), which might have contaminated the feces during collection, and to arthropods whos’ functional feeding group could not be determined (8% RRA, Figure 1A; 7% POO, Supplementary Data Sheet 3: Supplementary Figure S1A).
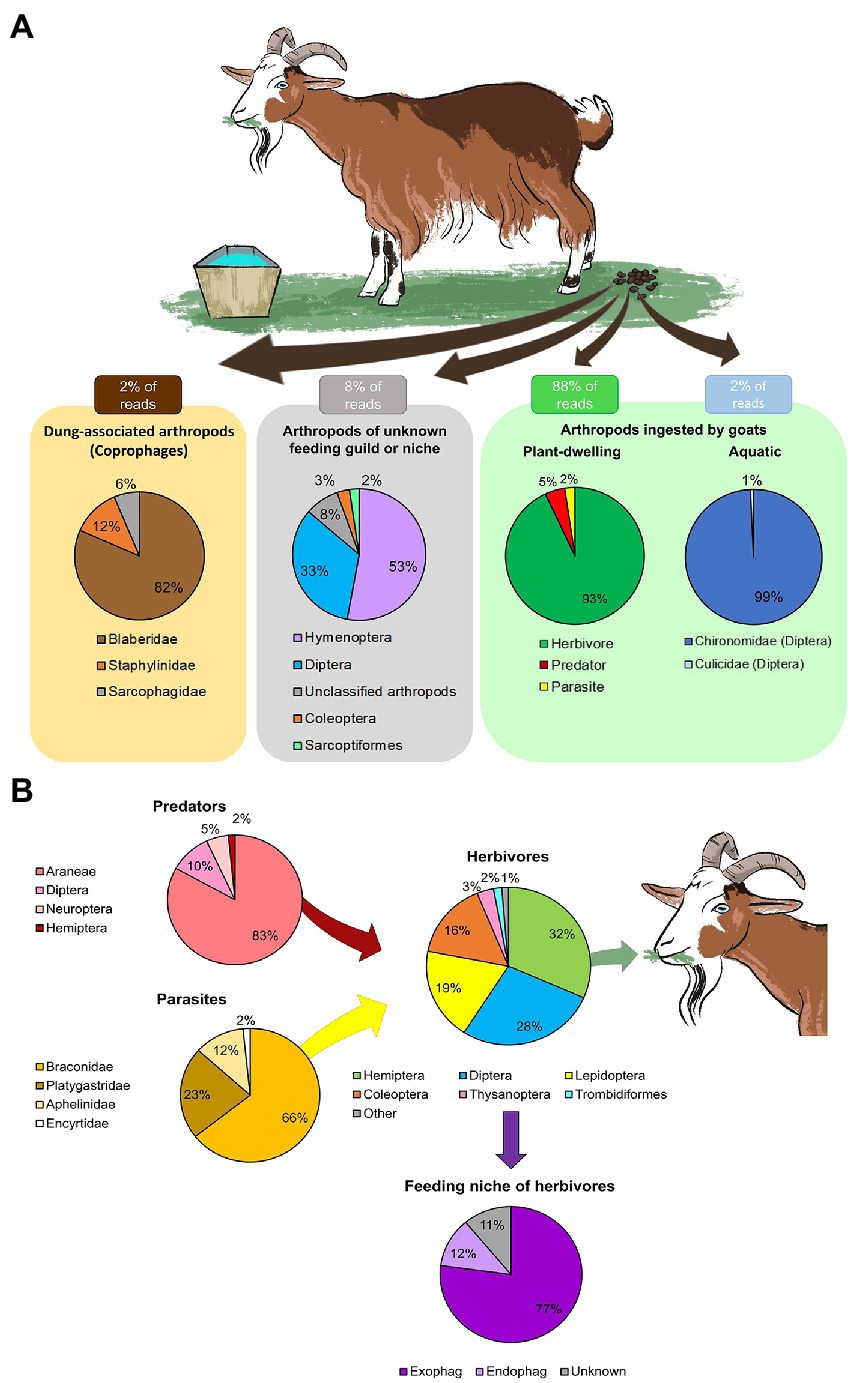
Figure 1. Overall diversity of arthropods ingested by goats. The mean proportion of reads (RRA) detected in the feces was averaged across February–June. Arthropods with few reads were grouped as “other.” (A) The main functional feeding groups of arthropods detected in the goat feces. Most of the RRA belonged to arthropods ingested while grazing (plant-dwelling). A small proportion of the RRA belonged to arthropods of unknown feeding group or niche, dung-associated arthropods (likely a contamination during collection) and aquatic arthropods (ingested while drinking). (B) Assembly of plant-dwelling arthropods detected in the goat feces, including the feeding niche of ingested herbivores (exophages–feed on the plant surface; endophages–feed within plant parts).
The plant-dwelling taxa ingested by goats were comprised of a complex food-chain of herbivores, predators and parasites (Figure 1B). In total, the goats ingested species from 48 families of herbivores (93% RRA of PDA, Figure 1B; 90% POO of PDA, Supplementary Data Sheet 3: Supplementary Figure S1B), mainly of the orders Hemiptera, Diptera, Lepidoptera and Coleoptera (Figure 1B). The majority of these herbivores were exophages (77% RRA, Figure 1B; 74% POO, Supplementary Data Sheet 3: Supplementary Figure S1B). The goats also ingested species from nine families of predators (5% RRA of PDA, Figure 1B; 6% POO of PDA, Supplementary Data Sheet 3: Supplementary Figure S1B), mostly spiders (Araneae), and four families of Hymenopteran parasitoids (2% RRA of PDA, Figure 1B; 4% POO of PDA, Supplementary Data Sheet 3: Supplementary Figure S1B). Larvae of chironomids and mosquitoes (Diptera) were ingested while drinking (Figure 1A). All samples that passed quality control (57/57) contained DNA of PDA, suggesting PDA ingestion is frequent.
3.2. Seasonal variation in PDA ingested by goats
A seasonal shift in the arthropod community, associated with Mediterranean habitats, was strongly evident in the composition of arthropods detected in the goat feces collected over February–June (ADONIS: RRA, R = 0.256, p = 0.001, Figure 2; POO, R = 0.191, p = 0.001; Supplementary Data Sheet 3: Supplementary Figure S2; Supplementary Table S1). This seasonal shift was also visible in the feces of the six-goat group (i.e., goats with at least three consecutive samples from April–June; ADONIS: RRA, R = 0.251, p = 0.001; Figure 2). Interestingly, Diversity and richness estimates (Shannon H′ and Fisher alpha; calculated based on the six-goat group), were similar among sampling months for all individual goats in the six-goat group (Friedman’s test: Shannon H′, χ22 = 1.333, p = 0.513; Fisher’s alpha, χ22 = 0.333, p = 0.846; Supplementary Table S8).
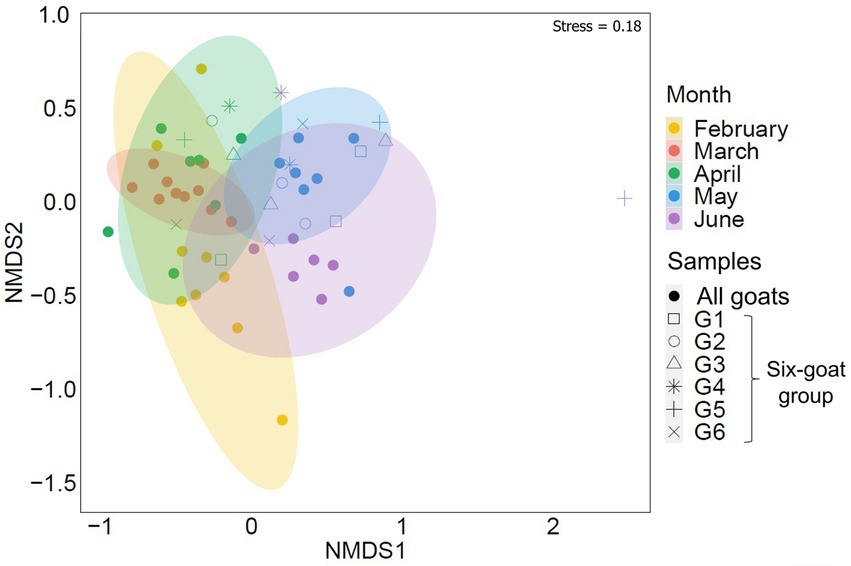
Figure 2. Similarity of the overall arthropod community (ASVs) detected in goat fecal samples by sampling month (February–June). The nonmetric multidimensional scaling analysis (NMDS) is based on Bray–Curtis dissimilarity matrix with 95% confidence ellipses. Each point represents the arthropod composition of a single fecal sample. Samples belonging to the six-goat group (goats with at least three consecutive samples from April–June, G1-G6) are marked by open shapes. The samples clustered together by sampling month.
The majority of PDA ingested by goats in all sampling months were herbivores (Figure 3A). While the overall relative abundance of herbivores remained steady over the season (Friedman’s test: χ2 = 8.898, p = 0.064), endophages were significantly more abundant in May–June (the beginning of summer, Friedman’s test: χ2 = 3.234, p = 0.001, Figure 3C). Parasites were more abundant in April (Friedman’s test: χ2 = 16.288, p = 0.003), whereas predators, like herbivores, remained relatively steady over time (Friedman’s test: χ2 = 8.916, p = 0.063). Although there seemed to be a seasonal trend of ingestion at the genus level for all functional feeding groups (Figures 3B, D), this was non-significant (Chi-square: herbivores, χ21 = 3.64, p = 0.457; exophages, χ21 = 1.683, p = 0. 794; the remainder had less than 5 observations in some cells thus statistics could not be performed).
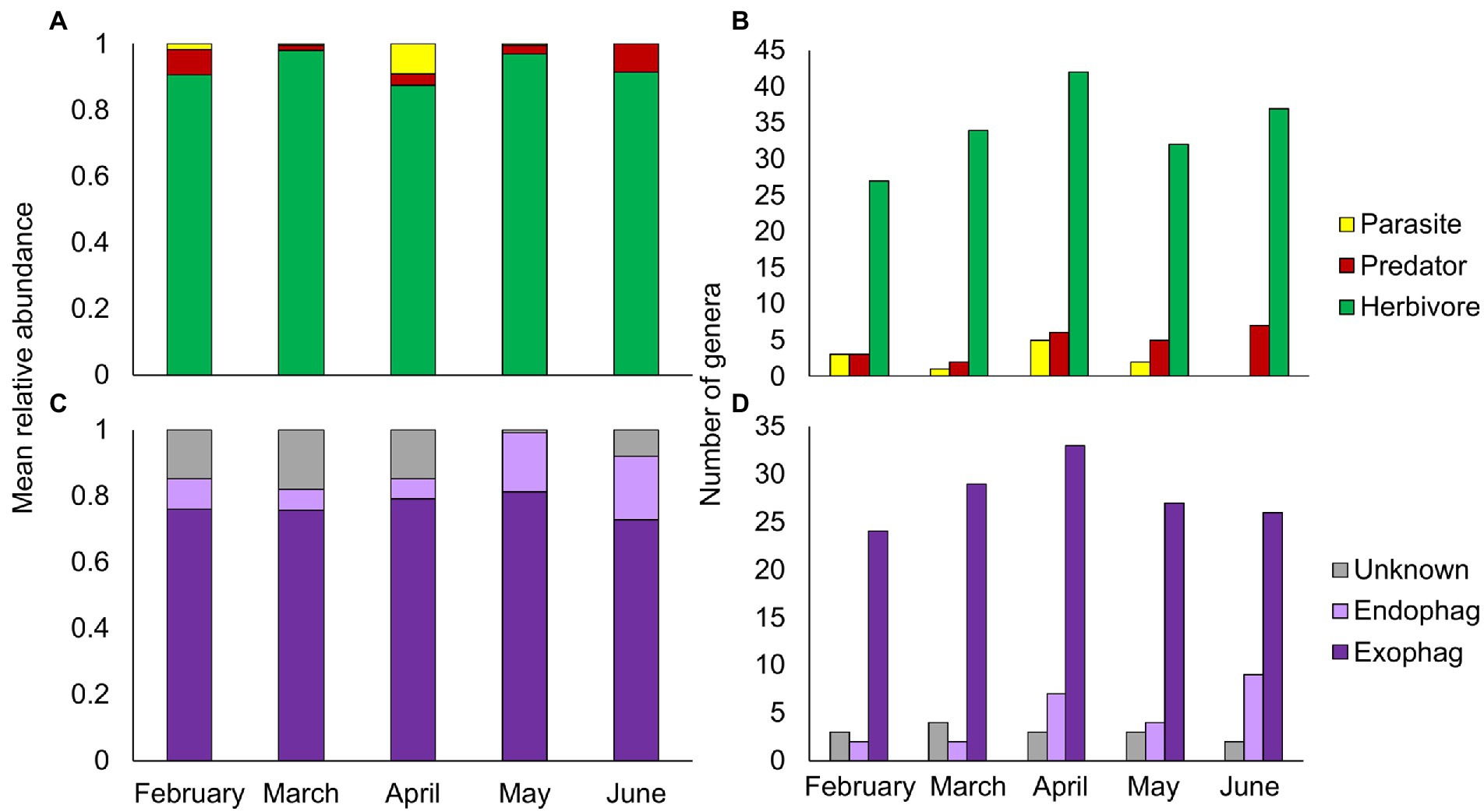
Figure 3. Composition of plant-dwelling arthropods ingested by goats across five sampling months (February–June). (A,B) Mean relative abundance and number of genera of herbivore, predator and parasitic arthropods. (C,D) Mean relative abundance and number of genera of herbivorous arthropods by feeding niche (exophages – feed on the plant surface; endophages – feed within plant parts).
3.3. Individual variation in PDA ingested by goats
Despite feeding together in the same habitat, PDA ingested by individual goats were notably different. The mean number of PDA families ingested by goats varied significantly between individuals in the six-goat group (repeated measures ANOVA: tests of within-subjects’ effects - sampling months, F2,10 = 0.17, p = 0.846; tests of between-subjects’ effects–individual goats: F2,10 = 59.827, p = 0.001; Figure 4A). When observing the actual change in the number of families ingested over April–June, some goats (mostly) remained above the median number of families ingested (8) over time, while others remained below this number (Figure 4B). An individual trend was not apparent when comparing the composition of the total arthropod community (ASVs) among all goats or the six-goat group (ADONIS: all goats - R = 0.019, p = 0.279, Supplementary Data Sheet 2: Supplementary Figure S2; six-goat group - R = 0.058, p = 0.445).
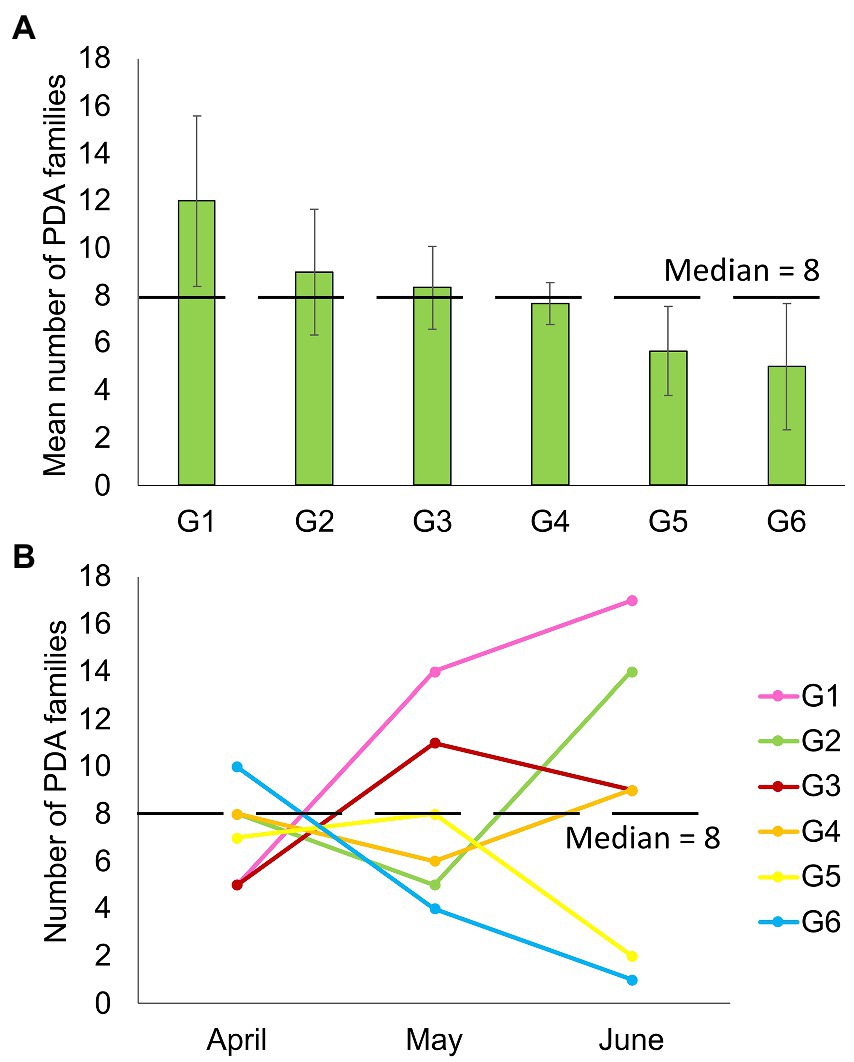
Figure 4. Number of plant-dwelling arthropod families ingested by six different goats (G1–G6). (A) Averaged (+SE) for each goat over three consecutive months (April–June). (B) The variation in the number of families ingested by the goats over three consecutive sampling months. The median number of families detected in all samples (8) is marked by a blacked dashed line in both plots.
Indiviaual (and seasonal) variation especially stood out when examining the ten PDA families with the highest RRA (and POO, Supplementary Data Sheet 3: Supplementary Figure S3; Figure 5; for order abundance for both datasets see Supplementary Data Sheet 3: Supplementary Figure S3). For example, the feces of some goats contained a large proportion (RRA) of Liviidae (plant-parasitic hemipterans), while others contained none at all. Psyllidae (jumping plant lice) and Tephritidae (fruit flies), on the other hand, were present in most of the goats’ feces, while Chrysomelidae (leaf beetles) were present in the feces of only a few goats (Figure 5).
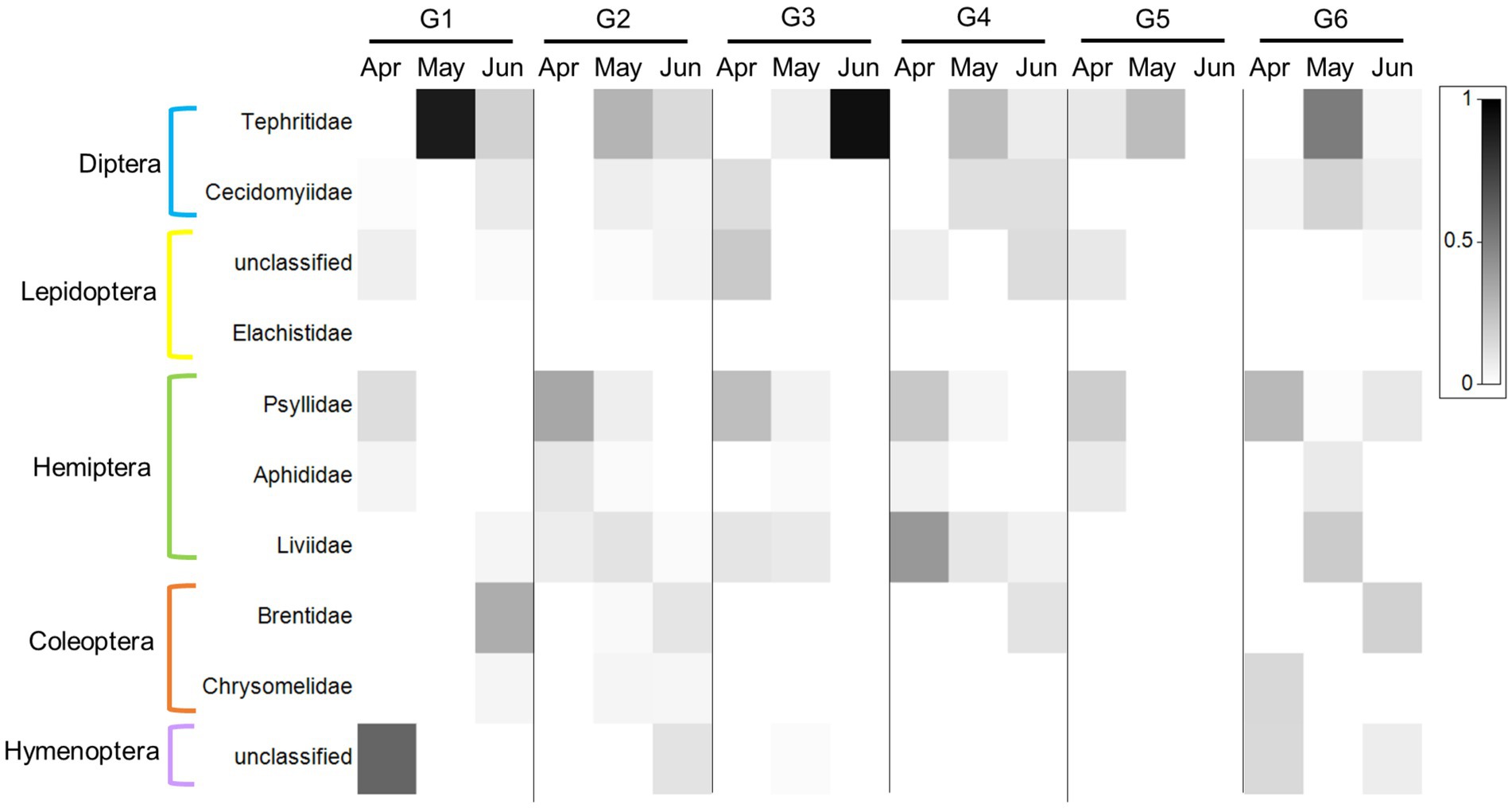
Figure 5. Heat map presentation of the most abundant plant-dwelling arthropod families ingested by the six-goat group (G1–G6) across three consecutive sampling months (columns). The 10 most abundant families based on RRA across all samples are presented in this plot. The six goats are marked by G1–G6. The grey scale intensity represents the mean relative abundance of each family (white squares indicate an absence of reads). The plot was prepared using Primer v7 software (http://www.primer-e.com).
4. Discussion
We show for the first time that browsing LMH frequently ingest PDA while feeding. Free-ranging goats, foraging in the Mediterranean maquis, incidentally ingested a variety of PDA (including herbivores, predators and parasites) which was notably different over the season and among individual goats. All fecal samples (100%) contained DNA belonging to PDA indicating that their ingestion by goats (i.e., direct trophic interaction) is common and thus ecologically important.
4.1. PDA ingested by goats
Goats ingested mostly innocuous PDA characterized by reduced mobility. These included three main groups: (1) Endophagous PDA (12% of PDA, Figure 1B) which reside within plant parts from which they are unable to escape, making them vulnerable to ingestion. This group was comprised mainly of leaf miners (Agromyzidae), plant-parasitic mites (Eriophyidae), gall midges (Cecidomyiidae) and boring caterpillars (Lepidoptera). (2) Immature life stages of PDA with mobile adult stages; such as Thrips (Thripidae), hover flies (Syrphidae), herbivorous moths (Pyralidae, Noctuidae) and parasitic wasps (Hymenoptera, the larval stage was likely ingested together with their arthropod hosts). While adult and young PDA stages (i.e., eggs, pupae, larvae) cannot be distinguished from one another using DNA metabarcoding alone, we can assume that the early and immobile life stages were ingested and not highly mobile adults which can escape danger. (3) Small wingless species, such as aphids (Aphididae) and mites (Acari), which are unable to rapidly escape from the plant, like winged or highly mobile species. Nonetheless, certain aphid species are able to escape feeding LMH (Gish et al., 2010; Ben-Ari et al., 2019). The efficiency of the escaping behavior (within and between PDA species) will determine the outcome of the encounter with LMH. Spiders which are common in the study site (Lubin et al., 2011) were also ingested by the goats; namely crab spiders (Thomisidae) and jumping spiders (Salticidae), that are usually found stalking their prey on plant parts such as flowers and fruit. Goats (and LMH in general) might prefer these nutritious plant parts, making PDA present on them more vulnerable to ingestion.
4.2. PDA that were not ingested by goats
Highly mobile species which are able to rapidly escape the plant when approached by a LMH, such as cicadas, spittlebugs, and grasshoppers, were hardly detected or absent from the fecal samples, despite being common in the study area. Similarly, harmful and noxious PDA [bees and ants, for example, which are also common in the field (Shavit et al., 2009 Finkel et al., 2015)], were also absent from the feces. We recently demonstrated that goats are able to effectively avoid ingesting noxious PDA, such as the spring webworm [Ocnogyna loewii, Lepidoptera (Berman et al., 2017)], while feeding. These caterpillars are covered with long setae which can cause allergic responses in LMH upon contact (Campbell, 2001) and teratogenic diseases if ingested (Webb et al., 2004). While late instar webworms are mobile (moving freely on plants), early instars are not - shortly after hatching the webworms form a common web nest on the vegetation in which they feed and grow. Despite their inability to escape LMH during early instars and their abundance in Mediterranean maquis, especially during the study period (February–June), no webworms (early or late instars) were ingested by the goats. This finding implies that goats are able to sense and avoid caterpillar nests as well as the small webworms within them.
4.3. Seasonal variation in PDA ingested by goats
The population dynamics of arthropods in Mediterranean ecosystems follows the seasonality of the Mediterranean climate (Stamou et al., 2004). Furthermore, Mediterranean ecosystems are characterized by strong oscillations in climatic factors (rainfall, temperature and humidity) and resource availability that are impacted by human activity, including grazing. These changes in plant resources greatly influence herbivore assemblages (Li et al., 2022) and the temporal composition of arthropods in the habitat (Stamou et al., 2004; Lazaro et al., 2016; Zhu et al., 2020). We detected a significant seasonal variation in the composition, but not the diversity, of PDA ingested by goats from the end of winter to the beginning of summer (i.e., February to June; Figure 2 Supplementary Data Sheet 3: Supplementary Figures S2A, B; Supplementary Table S8). Meaning, the assemblage structure of PDA ingested by goats changed over the season, yet the diversity and richness indices of these arthropods remained consistent.
Herbivorous and predatory PDA were consistently ingested by the goats through all sampling months (Figure 3A). However, endophagous herbivores were more prevalent in the feces in the end of spring (May–June, Figure 3C), probably since they develop within plant parts providing protection from the hot Mediterranean summer. Parasites were ingested mostly in spring (April, Figure 3A), probably because they depend on the development of their herbivorous hosts which are plentiful during this period due to warm temperatures and increased vegetation growth. Overall, the detection of PDA in the goat feces during February–June was in accordance with the phenology of PDA in Mediterranean habitats during this time period. Future studies may therefore benefit from using seasonal fecal sampling of LMH as a way to infer seasonal changes in arthropod composition in grazed habitats.
4.4. Individual variation in PDA ingested by goats
By collecting fecal samples directly from marked goats once a month, we were able, for the first time, to observe the extent of individual variation of PDA ingestion in LMH over time. It is important to note that not all goats had a valid sample per month, therefore we focused our analysis on goats with at least three consecutive samples (3 months in a row, i.e., the six-goat group). Despite no significant difference between the six individuals in the overall composition of PDA (ASV’s) detected in the feces (Figure 2), our analysis revealed that the number of PDA families (but not functional feeding groups) ingested by goats (i.e., diversity) varied greatly among individuals (some goats ingested significantly more PDA families than others; Figures 4A, B). In light of our recent studies (Berman et al., 2017, 2019a), it is unlikely that the observed variation is a result of an inability of some goats to detect and avoid PDA ingestion. The fact that the RRA of particular PDA was higher in some goats (such as that of the family Tephritidae which were detected in the feces of all six goats, Figure 5) and that RRA may actually contain some quantitative aspects (Deagle et al., 2018), might suggest, with caution, that different goats ingest different quantities of these PDA. This assumption could be tested when complementing DNA metabarcoding with a quantitative method (Deagle et al., 2005; Guo et al., 2018).
Individual variability in feeding behavior of LMH is influenced by many factors, including availability, distribution and quality of forage or preferred foods, nutritional needs, prior experience with food plants, social interactions (such as competition and learning from conspecifics) and personality (Provenza et al., 2003; Searle et al., 2010; Neave et al., 2018). Since ingestion of PDA by goats is probably incidental, its pattern will likely reflect the dietary choices of each individual, i.e., the plants or plant parts or species consumed by each goat. Individual goats which avoid certain plants based on prior experience, social learning, palatability, or their ability to cope with toxins (Provenza et al., 2003; Neave et al., 2018) will rarely ingest PDA residing on these plants. Personality traits, such as boldness or fearfulness, will influence whether individuals encounter and sample novel food plants (Neave et al., 2018) and the PDA on them. Goats are social animals which feed together under competitive conditions. Thus, intraspecific competition may lead some individuals to exploit resources that are less utilized by conspecifics (Araújo et al., 2011), exposing them to a different variety of PDA. Goat feeding behavior is also influenced by social ranks with dominant individuals having greater access to quality food compared to subordinates (Neave et al., 2018). Such relationships within the herd can affect individual feeding patterns and consequently the variety or amount of PDA ingested.
4.5. Comparing PDA ingestion between different LMH
To date, only one study has examined the extent of PDA ingestion by LMH at the community level using a molecular approach. This study investigated the ingestion of PDA by cattle grazing in Mediterranean grasslands using DNA metabarcoding (Berman and Inbar, 2022). The application of a similar molecular analysis (including the primer set used) to the one used in our current study enabled us to compare between the two.
Goats (i.e., browsers) feed mostly on woody vegetation (fruits, tree and shrub stems and foliage, but also grasses), while cattle (i.e., grazers) feed primarily on grasses and forbs. Beside feed type, these LMH also differ in mouth structure, a fact that may influence bite size and thus feeding rate (Shipley, 1999). Grazers usually have wide muzzles, allowing them to maximize bite size but making it harder for them to select specific plant parts. Browsers, on the other hand, have a narrower muzzle and in some species, such as goats, flexible lips and tongues which allow them to select desired parts but reduce bite size (Lu, 1988; Shipley, 1999). Despite these notable differences, arthropod ingestion was prevalent in both goats and cattle (100 and 76% of feces that passed the quality control contained sequences belonging to PDA, respectively) and their pattern of arthropod ingestion showed high resemblance. Strikingly, goats and cattle ingested similar proportions of herbivores, predators and parasitoids (Figure 6) and a small percentage of aquatic arthropods. The abundance of dung associated arthropods could not be compared between cattle and goats since the time period between defecation and collection was longer in cattle (goat fecal samples were collected at time of defecation), enabling coprophages to arrive and colonize the feces prior to collection. In both studies, PDA characterized by reduced mobility (inability to rapidly escape the plant) were ingested. Harmful or noxious PDA, on the other hand, were largely absent from the fecal samples. This is not surprising as we recently demonstrated that both goats and cattle are able to detect and avoid ingesting noxious PDA (Berman et al., 2017, 2018). Both ingestion patterns also exhibited a similar seasonal shift, with a significant difference in the PDA detected in the feces over time (Berman and Inbar, 2022). Although we did not examine individual variation in cattle, LMH characterized by wide muzzles and large bite size will probably feed in a more homogenous matter than small-muzzled, selective LMH (like goats), reducing the capacity of individual variation in plant consumption (Pansu et al., 2019) and maybe even PDA ingestion. Moreover, goats, which are equipped with flexible lips and tongues, may ingest PDA found in plant parts which are less reachable for other LMH species, including grazers.
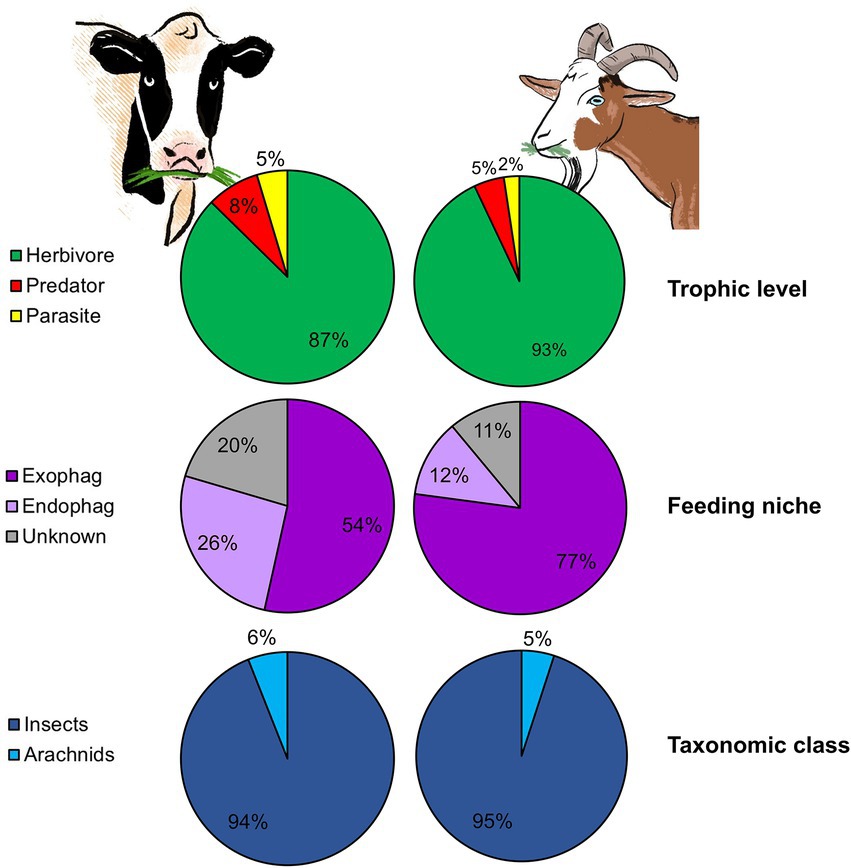
Figure 6. The mean proportion of reads (RRA) of plant-dwelling arthropods (PDA) detected in the feces of two different LMH species according to trophic level, feeding niche, and taxonomic class. The left pie charts show the RRA of PDA detected in cattle feces (reconstructed from Berman and Inbar, 2022). The right pie charts show the RRA of PDA detected in goat feces.
4.6. Strengths and limitations of metabarcoding and the experimental design
The application of DNA metabarcoding on fecal samples collected repeatedly from known individuals enabled us, for the first time, to observe individual variation in the ingestion of PDA by foraging LMH. This finding would be nearly impossible to observe using conventional dietary methods. Despite starting with 18 marked goats, technical issues along the way (notably inability to produce adequate PCR products for part of the samples) prevented us from obtaining a valid sample per month for each individual. Accordingly, we focused our analysis on a sub group of goats with samples from three consecutive months (i.e., the six-goat group). This analysis allowed us to observe individual variation in PDA ingestion at the diversity level (number of different families ingested), but not at the community level (ASV’s). DNA metabarcoding is a strong tool for detecting interactions within food webs (Pringle and Hutchinson, 2020), especially cryptic interactions, such as the presence of endophages and parasitoids which are concealed within their host and would not be detected by trapping or observations. Since the results of DNA metabarcoding studies are affected by recovery biases, contamination of feces by arthropods which were not ingested (i.e., coprophages) may mask actual abundances and interfere with the interpretation of the results. Collecting fecal samples directly from the goats prevented contamination by coprophages, and therefore provides a better picture of the PDA community that was actually ingested.
While DNA metabarcoding enabled us to successfully observe individual variation in the ingestion of PDA by LMH under field conditions, this method has a few disadvantages. For instance, wide range PCR primers may amplify nontarget taxa and may provide less data at the species level (Deagle et al., 2018; Pringle and Hutchinson, 2020; Cuff et al., 2022). We nonetheless chose the Zeale et al. (2011) primers since they enabled us to successfully assign arthropods to functional feeding groups (Berman and Inbar, 2022) and since they are frequently used for arthropod amplification in dietary studies (Alberdi et al., 2018). Future studies should use multiple primer pairs, such as COI and ribosomal markers, to avoid amplification bias (Cuff et al., 2022). Another disadvantage of DNA metabarcoding is that it only provides semi quantitative data (Deagle et al., 2018; Pringle and Hutchinson, 2020), whether using occurrence or RRA data. Both types of analyses used in this study led to a similar outcome in the results and their interpretation (Supplementary Data Sheet 3).
5. Conclusion
Using DNA metabarcoding and a multitrophic approach we demonstrate that the ingestion of PDA by LMH is a widespread phenomenon characterized by seasonal and individual variation. LMH foraging in different habitats incidentally ingest a variety of PDA belonging to different taxonomic and functional groups. These PDA include mostly innocuous herbivorous species characterized by reduced mobility, such as endophages and immature life stages of mobile adults. Noxious PDA were rarely detected in the feces as LMH possess highly efficient behavioral mechanisms which enable them to avoid ingestion of such PDA. The high prevalence of PDA ingestion and the similar pattern of ingestion between LMH, regardless of feeding strategy (i.e., browsing, grazing, etc.) or habitat, suggest that this complex direct trophic interaction is a common and integral part of terrestrial ecosystems.
Data availability statement
All data supporting the results is available at the NCBI database under BioProject accession number PRJNA579572 at: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA579572.
Ethics statement
Ethical review and approval was not required for the animal study because the samples were collected as part of the routine handling of the animals in an experimental goat pen, under the supervision of Ramat Hanadiv employees. The Animal Experimentation Law in Israel does not apply for this case and no ethical review was required.
Author contributions
TB and MI conceived and designed the experiments and wrote the paper. TB performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Israel Science Foundation (grant No. 248/17).
Acknowledgments
We thank the Ramat Hanadiv Nature Park management and staff for their valuable support. We thank Roni Lombrozo Amzallag, Suha Issa, Eva Leman, Daniel Melamed, and Mozy Gish for their technical assistance and fruitful discussions. We thank Maya Lalzar from the University of Haifa Bioinformatics Service Unit for the bioinformatic assistance and Ido Izhaki and Matan Ben-Ari for their statistical assistance. We also thank Lina Gurevich for illustrating Mika the goat (Figure 1). Finally, a special thanks to Rami Reshef (University of Haifa) for the use of his laboratory facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1070088/full#supplementary-material
References
Alberdi, A., Aizpurua, O., Gilbert, M. T. P., and Bohmann, K. (2018). Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 9, 134–147. doi: 10.1111/2041-210X.12849
Araújo, M. S., Bolnick, D. I., and Layman, C. A. (2011). The ecological causes of individual specialisation. Ecol. Lett. 14, 948–958. doi: 10.1111/j.1461-0248.2011.01662.x
Ben-Ari, M., and Inbar, M. (2013). When herbivores eat predators: predatory insects effectively avoid incidental ingestion by mammalian herbivores. PLoS One 8:e56748. doi: 10.1371/journal.pone.0056748
Ben-Ari, M., Outreman, Y., Denis, G., Le Gallic, J.-F., Inbar, M., and Simon, J.-C. (2019). Differences in escape behavior between pea aphid biotypes reflect their host plants’ palatability to mammalian herbivores. Basic Appl. Ecol. 34, 108–117. doi: 10.1016/j.baae.2018.10.005
Bennett, N. L., Severns, P. M., Parmesan, C., and Singer, M. C. (2015). Geographic mosaics of phenology, host preference, adult size and microhabitat choice predict butterfly resilience to climate warming. Oikos 124, 41–53. doi: 10.1111/oik.01490
Berman, T. S., Ben-Ari, M., Glasser, T. A., Gish, M., and Inbar, M. (2017). How goats avoid ingesting noxious insects while feeding. Sci. Rep. 7, 14835–14810. doi: 10.1038/s41598-017-14940-6
Berman, T. S., Ben-Ari, M., Henkin, Z., and Inbar, M. (2018). Immediate and long-term facilitative effects of cattle grazing on a polyphagous caterpillar. Agric. Ecosyst. Environ. 261, 45–53. doi: 10.1016/j.agee.2018.03.019
Berman, T. S., Glasser, T. A., and Inbar, M. (2019a). Goats adjust their feeding behaviour to avoid the ingestion of different insect species. Can. J. Zool. 97, 805–811. doi: 10.1139/cjz-2019-0010
Berman, T. S., and Inbar, M. (2022). Revealing cryptic interactions between large mammalian herbivores and plant-dwelling arthropods via DNA metabarcoding. Ecology 103, e03548–e03549. doi: 10.1002/ecy.3548
Berman, T. S., Messeri, N., Glasser, T. A., and Inbar, M. (2019b). Innate ability of goats to sense and avoid ingestion of noxious insects while feeding. R. Soc. Open Sci. 6:181078. doi: 10.1098/rsos.181078
Bonal, R., and Muñoz, A. (2007). Multi-trophic effects of ungulate intraguild predation on acorn weevils. Oecologia 152, 533–540. doi: 10.1007/s00442-007-0672-8
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Campbell, J. B. (2001). Anti-quality effects of insects feeding on rangeland plants: A review. J. Range Manag. 54, 462–465. doi: 10.2307/4003117
Clare, E. L., Symondson, W. O. C., Broders, H., Fabianek, F., Fraser, E. E., Mackenzie, A., et al. (2014). The diet of Myotis lucifugus across Canada: assessing foraging quality and diet variability. Mol. Ecol. 23, 3618–3632. doi: 10.1111/mec.12542
Clarke, L. J., Soubrier, J., Weyrich, L. S., and Cooper, A. (2014). Environmental metabarcodes for insects: in silico PCR reveals potential for taxonomic bias. Mol. Ecol. Resour. 14, 1160–1170. doi: 10.1111/1755-0998.12265
Cuff, J. P., Kitson, J. J. N., Hemprich-Bennett, D., Tercel, M. P. T. G., Browett, S. S., and Evans, D. M. (2022). The predator problem and PCR primers in molecular dietary analysis: swamped or silenced; depth or breadth? Mol. Ecol. Resour. 23, 41–51. doi: 10.1111/1755-0998.13705
Deagle, B. E., Jarman, S. N., Coissac, E., Pompanon, F., and Taberlet, P. (2014). DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol. Lett. 10:20140562. doi: 10.1098/rsbl.2014.0562
Deagle, B. E., Thomas, A. C., McInnes, J. C., Clarke, L. J., Vesterinen, E. J., Clare, E. L., et al. (2018). Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Mol. Ecol. 28, 391–406. doi: 10.1111/mec.14734
Deagle, B. E., Tollit, D. J., Jarman, S. N., Hindell, M. A., Trites, A. W., and Gales, N. J. (2005). Molecular scatology as a tool to study diet: analysis of prey DNA in scats from captive Steller Sea lions. Mol. Ecol. 14, 1831–1842. doi: 10.1111/j.1365-294X.2005.02531.x
Elbrecht, V., and Leese, F. (2017). Corrigendum: validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Front. Environ. Sci. 5, 1–11. doi: 10.3389/fenvs.2017.00038
Ferrer, L. M., Ortín, A., Loste, A., Fernández, A., Verde, M. T., and Ramos, J. J. (2007). Photosensitisation in sheep grazing alfalfa infested with aphids and ladybirds. Vet. Rec. 161, 312–314. doi: 10.1136/vr.161.9.312
Finkel, M., Ofer, J., Beharav, A., and Nevo, E. (2015). Species interslope divergence of ants caused by sharp microclimatic stresses at ‘evolution canyon’ II, lower Nahal Keziv, western upper Galilee, Israel. Isr. J. Entomol. 44, 63–73.
Gish, M., Ben-Ari, M., and Inbar, M. (2017). Direct consumptive interactions between mammalian herbivores and plant-dwelling invertebrates: prevalence, significance, and prospectus. Oecologia 183, 347–352. doi: 10.1007/s00442-016-3775-2
Gish, M., Dafni, A., and Inbar, M. (2010). Mammalian herbivore breath alerts aphids to flee host plant. Curr. Biol. 20, R628–R629. doi: 10.1016/j.cub.2010.06.065
Guo, Y., Zhang, H., Chen, W., and Zhang, Y. (2018). Herbivore-diet analysis based on illumina MiSeq sequencing: the potential use of an ITS2-barcoding approach to establish qualitative and quantitative predictions of diet composition of mongolian sheep. J. Agric. Food Chem. 66, 9858–9867. doi: 10.1021/acs.jafc.8b02814
Hadar, L., Noy-Meir, I., and Perevolotsky, A. (1999). The effect of shrub clearing and grazing on the composition of a Mediterranean plant community: functional groups versus species. J. Veg. Sci. 10, 673–682. doi: 10.2307/3237082
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). Paleontological statistics software: package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Huson, D. H., Beier, S., Flade, I., Górska, A., El-Hadidi, M., Mitra, S., et al. (2016). MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12:e1004957. doi: 10.1371/journal.pcbi.1004957
Kaunisto, K. M., Roslin, T., Sääksjärvi, I. E., and Vesterinen, E. J. (2017). Pellets of proof: first glimpse of the dietary composition of adult odonates as revealed by metabarcoding of feces. Ecol. Evol. 7, 8588–8598. doi: 10.1002/ece3.3404
King, L. E., Douglas-Hamilton, I., and Vollrath, F. (2007). African elephants run from the sound of disturbed bees. Curr. Biol. 17, R832–R833. doi: 10.1016/j.cub.2007.07.038
Lazaro, A., Tscheulin, T., Devalez, J., Nakas, G., and Petanidou, T. (2016). Effects of grazing intensity on pollinator abundance and diversity, and on pollination services. Ecol. Entomol. 41, 400–412. doi: 10.1111/een.12310
Li, Y., Wang, M. Q., Chesters, D., Anttonen, P., Guo, S.-K., Guo, P.-F., et al. (2022). Differential impacts on herbivore diversity and scale-dependence of tree diversity in subtropical forests. J. Ecol. doi: 10.1111/1365-2745.14054
Lu, C. D. (1988). Grazing behavior and diet selection of goats. Small Rumin. Res. 1, 205–216. doi: 10.1016/0921-4488(88)90049-1
Lubin, Y., Angel, N., and Assaf, N. (2011). Ground spider communities in experimentally disturbed Mediterranean woodland habitats. Arachnol. Mitteilungen 40, 85–93. doi: 10.5431/aramit4010
Martins, D. J. (2010). Not all ants are equal: obligate acacia ants provide different levels of protection against mega-herbivores. Afr. J. Ecol. 48, 1115–1122. doi: 10.1111/j.1365-2028.2010.01226.x
Naqib, A., Poggi, S., Wang, W., Hyde, M., Kunstman, K., and Green, S. J. (2018). “Making and sequencing heavily multiplexed, high-throughput 16S ribosomal RNA gene amplicon libraries using a flexible, two-stage PCR protocol,” in Gene Expression Analysis. eds. N. Raghavachari and N. Garcia-Reyero (New York, NY: Humana Press).
Neave, H. W., Weary, D. M., and Von Keyserlingk, M. A. G. (2018). Review: individual variability in feeding behaviour of domesticated ruminants. Animal 12, S419–S430. doi: 10.1017/S1751731118001325
Nunamaker, R. A., Lockwood, J. A., Stith, C. E., Campbell, C. L., Schell, S. P., Drolet, B. S., et al. (2003). Grasshoppers (Orthoptera: Acrididae) could serve as reservoirs and vectors of vesicular stomatitis virus. J. Med. Entomol. 40, 957–963. doi: 10.1603/0022-2585-40.6.957
Oksanen, J., Legendre, P., O’Hara, B., Stevens, M. H. H., Oksanen, M. J., and Suggests, M. (2007). The vegan package. Commun. Ecol. Packag 10, 631–637.
Pansu, J., Guyton, J. A., Potter, A. B., Atkins, J. L., Daskin, J. H., Wursten, B., et al. (2019). Trophic ecology of large herbivores in a reassembling African ecosystem. J. Ecol. 107, 1355–1376. doi: 10.1111/1365-2745.13113
Pringle, R. M., and Hutchinson, M. C. (2020). Resolving food-web structure. Annu. Rev. Ecol. Evol. Syst. 51, 55–80. doi: 10.1146/annurev-ecolsys-110218-024908
Pringle, R. M., Kartzinel, T. R., Palmer, T. M., Thurman, T. J., Fox-Dobbs, K., Xu, C. C. Y., et al. (2019). Predator-induced collapse of niche structure and species coexistence. Nature 570, 58–64. doi: 10.1038/s41586-019-1264-6
Provenza, F. D., Villalba, J. J., Dziba, L. E., Atwood, S. B., and Banner, R. E. (2003). Linking herbivore experience, varied diets, and plant biochemical diversity. Small Rumin. Res. 49, 257–274. doi: 10.1016/S0921-4488(03)00143-3
Ratnasingham, S., and Hebert, P. (2007). BOLD: the barcode of life data system (www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2006.01678.x
Ratnasingham, S., and Hebert, P. D. N. (2013). A DNA-based registry for all animal species: the barcode index number (BIN) system. PLoS One 8:66213. doi: 10.1371/journal.pone.0066213
Rostás, M., Maag, D., Ikegami, M., and Inbar, M. (2013). Gall volatiles defend aphids against a browsing mammal. BMC Evol. Biol. 13:193. doi: 10.1186/1471-2148-13-193
Rytkönen, S., Vesterinen, E. J., Westerduin, C., Leviäkangas, T., Vatka, E., Mutanen, M., et al. (2019). From feces to data: a metabarcoding method for analyzing consumed and available prey in a bird-insect food web. Ecol. Evol. 9, 631–639. doi: 10.1002/ece3.4787
Schmitz, D. G. (1989). Cantharidin toxicosis in horses. J. Vet. Intern. Med. 3, 208–215. doi: 10.1111/j.1939-1676.1989.tb00859.x
Searle, K. R., Hunt, L. P., and Gordon, I. J. (2010). Individualistic herds: individual variation in herbivore foraging behavior and application to rangeland management. Appl. Anim. Behav. Sci. 122, 1–12. doi: 10.1016/j.applanim.2009.10.005
Shavit, O., Dafni, A., and Ne’Eman, G. (2009). Competition between honeybees (Apis mellifera) and native solitary bees in the Mediterranean region of Israel-implications for conservation. Isr. J. Plant Sci. 57, 171–183. doi: 10.1560/IJPS.57.3.171
Shipley, L. (1999). “Grazers and browsers: how digestive morphology affects diet selection,” in Grazing Behavior of Livestock and Wildlife. eds. K. L. Launchbaugh, J. C. Mosley, and K. O. Sanders (Moscow: University of Idaho).
Stamou, G. P., Stamou, G. V., Papatheodorou, E. M., Argyropoulou, M. D., and Tzafestas, S. G. (2004). Population dynamics and life history tactics of arthropods from Mediterranean-type ecosystems. Oikos 104, 98–108. doi: 10.1111/j.0030-1299.2004.12382.x
Stewart, A. J. A. (2001). The impact of deer on lowland woodland invertebrates: a review of the evidence and priorities for future research. Forestry 74, 259–270. doi: 10.1093/forestry/74.3.259
van Klink, R., van der Plas, F., van Noordwijk, C. G. E., Wallisdevries, M. F., and Olff, H. (2015). Effects of large herbivores on grassland arthropod diversity. Biol. Rev. 90, 347–366. doi: 10.1111/brv.12113
Vesterinen, E. J., Puisto, A. I. E., Blomberg, A. S., and Lilley, T. M. (2018). Table for five, please: dietary partitioning in boreal bats. Ecol. Evol. 8, 10914–10937. doi: 10.1002/ece3.4559
Webb, B. A., Barney, W. E., Dahlman, D. L., DeBorde, S. N., Weer, C., Williams, N. M., et al. (2004). Eastern tent caterpillars (Malacosoma americanum) cause mare reproductive loss syndrome. J. Insect Physiol. 50, 185–193. doi: 10.1016/j.jinsphys.2003.11.008
Yamazaki, K., and Sugiura, S. (2008). Deer predation on leaf miners via leaf abscission. Naturwissenschaften 95, 263–268. doi: 10.1007/s00114-007-0318-z
Zeale, M. R. K., Butlin, R. K., Barker, G. L. A., Lees, D. C., and Jones, G. (2011). Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol. Ecol. Resour. 11, 236–244. doi: 10.1111/j.1755-0998.2010.02920.x
Zhu, Y., Veen, G. F., Wang, D., Wang, L., Zhong, Z., Ma, Q., et al. (2020). Herbivore phenology can predict response to changes in plant quality by livestock grazing. Oikos 129, 811–819. doi: 10.1111/oik.07008
Keywords: DNA metabarcoding, large mammalian herbivores, food webs, grazing, incidental ingestion, trophic interactions
Citation: Berman TS and Inbar M (2023) Molecular identification of individual and seasonal variation in incidental ingestion of arthropods by free-ranging goats. Front. Ecol. Evol. 10:1070088. doi: 10.3389/fevo.2022.1070088
Edited by:
Sebastien Devillard, Université Claude Bernard Lyon 1, FranceCopyright © 2023 Berman and Inbar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tali Sarah Berman, ✉ talisberman@gmail.com
†Present address: Tali Sarah Berman, Department of Plant Pathology and Weed Research, Agricultural Research Organization (ARO), Rishon LeZion, Israel
 Tali Sarah Berman
Tali Sarah Berman Moshe Inbar
Moshe Inbar