- 1Department of Optometry and Vision Science, University of Alabama at Birmingham, Birmingham, AL, United States
- 2Department of Anesthesiology and Perioperative Medicine, University of Alabama at Birmingham, Birmingham, AL, United States
Introduction: As a metabolic disease, diabetes often leads to health complications such as heart failure, nephropathy, neurological disorders, and vision loss. Diabetic retinopathy (DR) affects as many as 100 million people worldwide. The mechanism of DR is complex and known to impact both neural and vascular components in the retina. While recent advances in the field have identified major cellular signaling contributing to DR pathogenesis, little has been reported on the protein post-translational modifications (PTM) - known to define protein localization, function, and activity - in the diabetic retina overall. Protein glycosylation is the enzymatic addition of carbohydrates to proteins, which can influence many protein attributes including folding, stability, function, and subcellular localization. O-linked glycosylation is the addition of sugars to an oxygen atom in amino acids with a free oxygen atom in their side chain (i.e., threonine, serine). To date, more than 100 congenital disorders of glycosylation have been described. However, no studies have identified the retinal O-linked glycoproteome in health or disease. With a critical need to expedite the discovery of PTMomics in diabetic retinas, we identified both global changes in protein levels and the retinal O-glycoproteome of control and diabetic mice.
Methods: We used liquid chromatography/mass spectrometry-based proteomics and high throughput screening to identify proteins differentially expressed and proteins differentially O-glycosylated in the retinas of wildtype and diabetic mice.
Results: Changes in both global expression levels of proteins and proteins differentially glycosylated in the retinas of wild-type and diabetic mice have been identified. We provide evidence that diabetes shifts both global expression levels and O-glycosylation of metabolic and synaptic proteins in the retina.
Discussion: Here we report changes in the retinal proteome of diabetic mice. We highlight alterations in global proteins involved in metabolic processes, maintaining cellular structure, trafficking, and neuronal processes. We then showed changes in O-linked glycosylation of individual proteins in the diabetic retina.
Introduction
Diabetes is a metabolic disorder associated with the development of hyperglycemia due to a high blood glucose level. The global diabetes prevalence in 2019 was estimated to be 463 million people, rising to 578 million by 2030 and 700 million by 2045 (1). Type-1 (T1D) and Type-2 diabetes (T2D) differ in their pathogenesis. T1D, or juvenile diabetes, is autoimmune in nature and results in the pancreas producing very little insulin. T2D patients produce insulin but have tissues that are resistant to the effects of insulin. Diabetic retinopathy (DR) is a life-limiting complication of diabetes, a disease estimated to affect 100 million people worldwide and is associated with neuromicrovascular dysfunction (2). DR pathogenesis is characterized as nonproliferative (NPDR) -or early stage, and advanced proliferative (PDR) (reviewed in (3)]. Retinas of patients with NPDR may show clear clinical signs such as microaneurysms, hemorrhages and intraretinal microvascular abnormalities. Patients with PDR develop pathological preretinal neovascularization. Neurodegeneration and retinal vascular dysfunction are hallmarks of DR, with neovascularization being the primary clinical focus. However, recent research projects have identified various neuronal abnormalities present in models of DR. These include cone dysfunction and thinning of the retinal ganglion cell (RGC) layer (4, 5). Diabetes is a systemic disease with multiple features having the ability to impact the retina including hyperglycemia, advanced glycation end-products (AGE) adduct accumulation, dyslipidemia, and hypertension. Hyperglycemia alone accounts for 10% of the risk factor for developing DR (6). Abnormal glucose flux and hyperglycemia are believed to activate protein kinase C signaling, hexose monophosphate shunt, and AGE, which demonstrates the incredibly complex molecular mechanisms of DR pathogenesis (7). Unfortunately, the identification of relevant treatments for DR has been delayed by the inaccessibility of suitable animal models (8–10).
Recent breakthroughs in protein purification strategies and current proteomic technologies (11) make it possible to identify the proteomics of healthy and diseased retinas. Despite these advances, the research field is significantly lagging when it comes to identifying proteins modified in diseased retinas, despite knowledge of the changes in protein regulation in the retina being critical for drug development (12).
Protein modifications that occur either co-translationally or posttranslationally (post-translational modifications or PTMs) constitute a key step in protein biosynthesis and in the regulation of protein function, activity, and localization. Phosphorylation can, for example, dynamically alter enzymatic activity. In addition, methylation and acetylation, specifically of histones, can regulate gene expression. Protein ubiquitination can vastly impact protein function or even signal for its degradation through the ubiquitin proteosome system.
Protein glycosylation is the enzymatic addition of carbohydrate chains to proteins, which greatly influences protein folding, stability, function, and subcellular localization [reviewed in (13–16)]. Glycosylation is dependent on the cytosolic synthesis of nucleotide-sugars, with the exception of CMP-NeuAc being synthesized in the nucleus, though N-linked glycosylation is initiated in the ER, co-translationally while O-glycosylation is initiated in the Golgi. (17). Glycans are assembled and added to proteins for further modifications. N-linked glycosylation involves the enzymatic linkage of glycans to proteins, begins in the ER, continues in the Golgi, and greatly influences protein stability and function. As compared to N-linked glycosylation, O-linked glycosylation occurs on the side chain of serine or threonine instead of asparagine residues. O-linked glycans are incredibly diverse as they are initiated by a plethora of different monosaccharides that yields even higher order diversity when forming O-glycans (18). For example, the most common O-linked glycosylation, O-GalNAc, is initiated in the Golgi and has significant influence on the cellular localization, binding partners, and function of proteins. O-Linked β-N-acetylglucosamine (O-GlcNAcylation) occurs in the cytoplasm and nucleus. O-glycosylation is one of the most abundant PTMs; however, detailed analyses of this PTM has been historically limited due to the shortage of appropriate methods.
Studies of glycosylation disorders have led to the discovery of tissue specific glycosylation pathways [reviewed in (15)]. In fact, more than 100 disorders of glycosylation have been described to date. Even with clear evidence that altered glycosylation can lead to disease, the functional glycoproteome remains poorly studied in both healthy and diseased tissue. Furthermore, no extensive studies have identified the retinal O-linked glycoproteome in health or disease, although a critical need of discovery glycoproteomics exists in the field. A few exceptions have been presented by published work of Gurel and Sheibani, (19) Xing et al., (20), and Lei et al. (21) which have highlighted the importance of O-glycosylation in diabetic retinas in general and have not identified O-glycosylation of individual proteins. In the present study, we identify O-glycosylation protein pattern in healthy and diseased retinas with nano high-performance liquid chromatography (HPLC)/mass spectrometry (MS) and detected differential retinal global protein expression in the streptozotocin (STZ)-induced diabetes mouse model. Though, we report evidence of robust changes in multiple cellular pathways, in this study, we primarily focus on the changes in metabolic and synaptic proteins at both the protein and O-glycosylation levels.
Results
Discovery proteomics in the retinas of diabetic mice
To facilitate discovery proteomics of diabetic retinopathy, we utilized a well characterized mouse model of diabetes, STZ-induced diabetes (3, 22–25) using a non-biased proteomics approach. Proteins were extracted from retinal tissue of diabetic mice, trypsin digested and analyzed using nano-HPLC/MS at 12 weeks post injection. We first identified differences in global proteomics between retinas from control and STZ mice (Figure 1, Tables 1, 2, and Supplementary Table S1). In this dataset, we identified significant changes in peptides corresponding to 145 proteins. Figure 1 shows a heatmap of global protein changes in diabetic retinas. Table 1 highlights global top hits, 30 proteins of elevated abundance and 30 proteins with reduced abundance— and indicates their fold changes and p-values.

Figure 1 A heatmap of major proteins modified in the diabetic retina. Increased and decreased proteins are indicated in red and green.
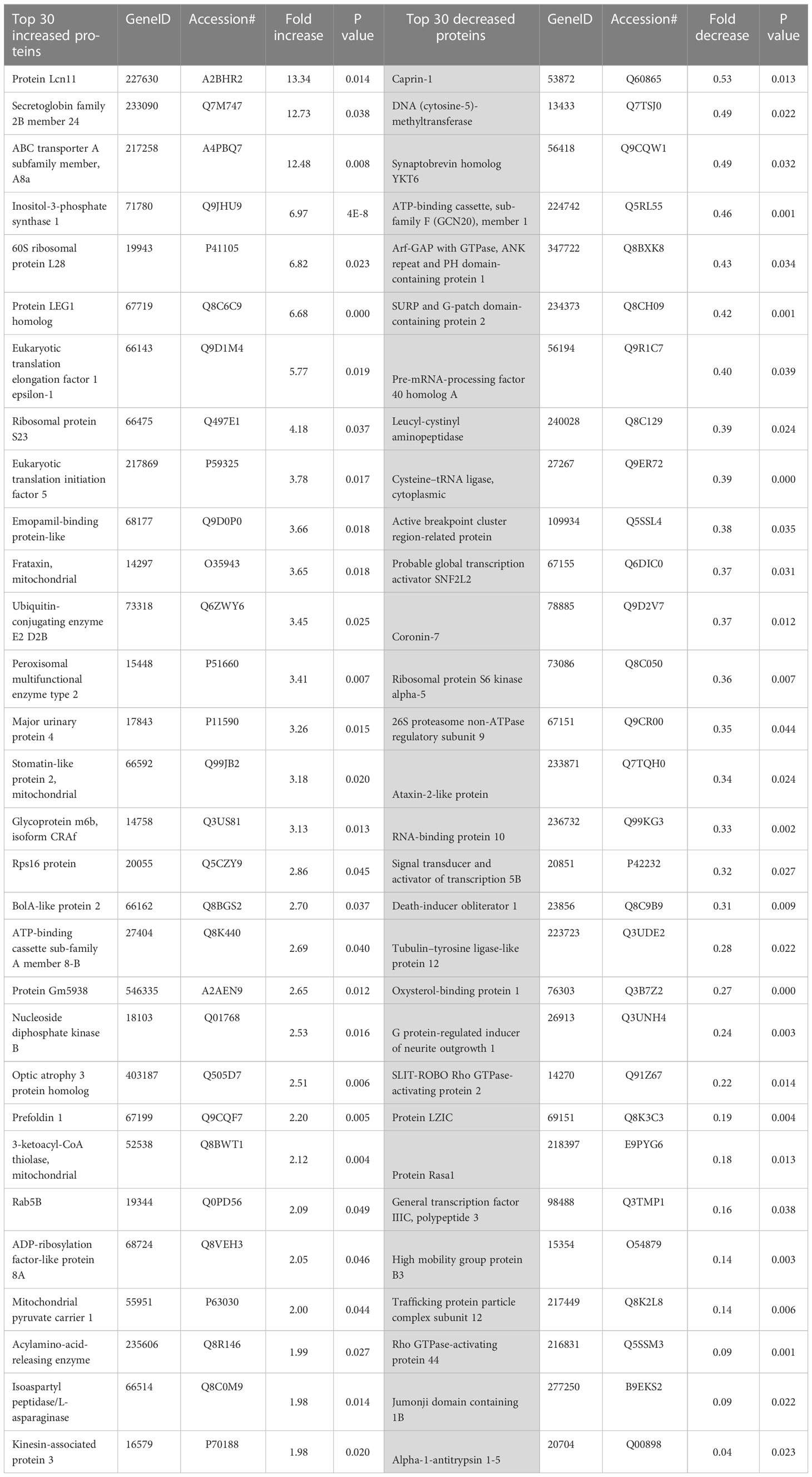
Table 1 The top 30 increased and decreased differentially expressed proteins in the diabetic retinas.
Because diabetes is a metabolic disorder that alters metabolic pathway in the retina leading to neurodegeneration, in this manuscript, we primarily focus on the results of global protein changes in retinal metabolism and neuronal processes.
Global changes in metabolic pathways in diabetic retinas
Upon analyzing the datasets, it became clear that many of the top hits are involved in one or more metabolic processes. For instance, diabetic mice had elevated levels of inositol-3-phosphate synthase 1 (Isyna1), Nucleoside diphosphate kinase A (Nme1), 3-ketoacyl-CoA-thiolase- mitochondrial (Hadhb), Aldehyde dehydrogenase, Isoaspartyl peptidase/L-asparaginase (Asrgl1), Adenylosuccinate synthetease isozyme 1 (AdSS1), Inositol-3-phosphate synthase 1(Isyna1), long-chain fatty acid transport protein 1(Fatp1), and Acylamino-acid-releasing enzyme (Acph) suggesting that multiple metabolic pathways could be altered in diabetic retinas. Protein Red, high mobility group protein HMG-I/HMG-Y and RNA binding protein 3 — proteins involved in RNA processing, were also elevated in diabetic retinas. Anti-oxidant superoxide dismutase Sod1 was also elevated in diabetic retina (Figure 1). In addition, levels of proteins involved in protein processing and folding, Prefoldin 1 and Prefoldin subunit 2, were also increased. Taken together, these elevated proteins indicate an increase of components involved in a significant number of metabolic processes in diabetic retinas.
Certain cellular processes appeared to be globally reduced in the retinas of diabetic mice. Of note, a number of proteins involved in metabolic processes were reduced in diabetic retinas. Some of the top hits reduced in diabetic retinas include Vigilin, Splicing factor 3A subunit 1 (Sf3a1), ATP-binding cassette sub-family F member 1 (Abcf1), SURP and G-patch domain-containing protein 2 (Sugp2), Eukaryotic translation initiation factor 4 gamma 2 (Eif4g2), Ribosomal protein S6 kinase alpha-5 (Rps6ka5), RNA-binding protein 10 (Rbm10), Serine/threonine-protein kinase mTOR (Figures 1, 2A and Table 1).
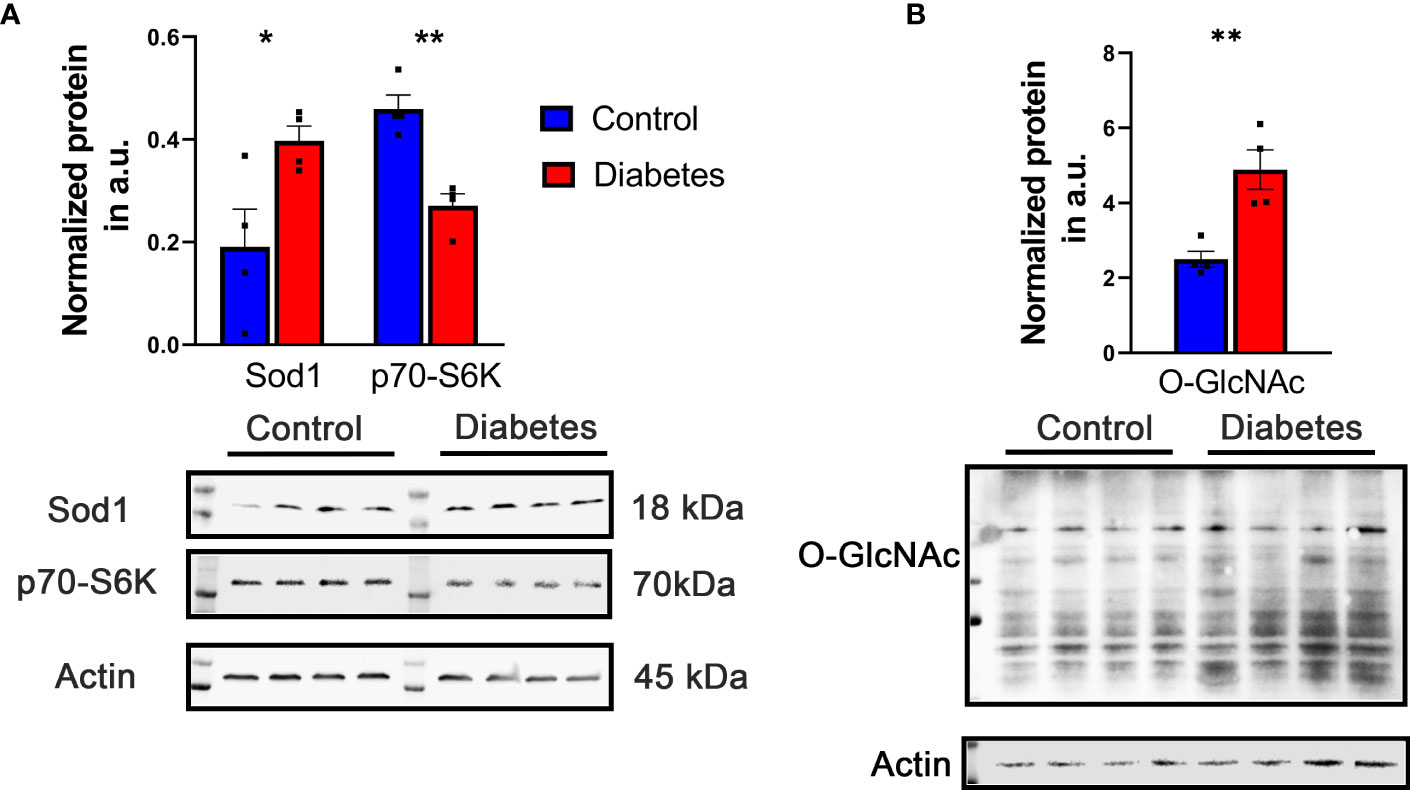
Figure 2 Representative increased and decreased proteins were detected by western blot. (A) Increase in Sod1 and decrease in S6K level were detected in diabetic retina validating results obtained with LC-MS -based approach. (B) The level of O-GlcNAc proteins detected with CDT110.6 antibody was enhanced in diabetic retina. *p<0.05 , **p<0.01.
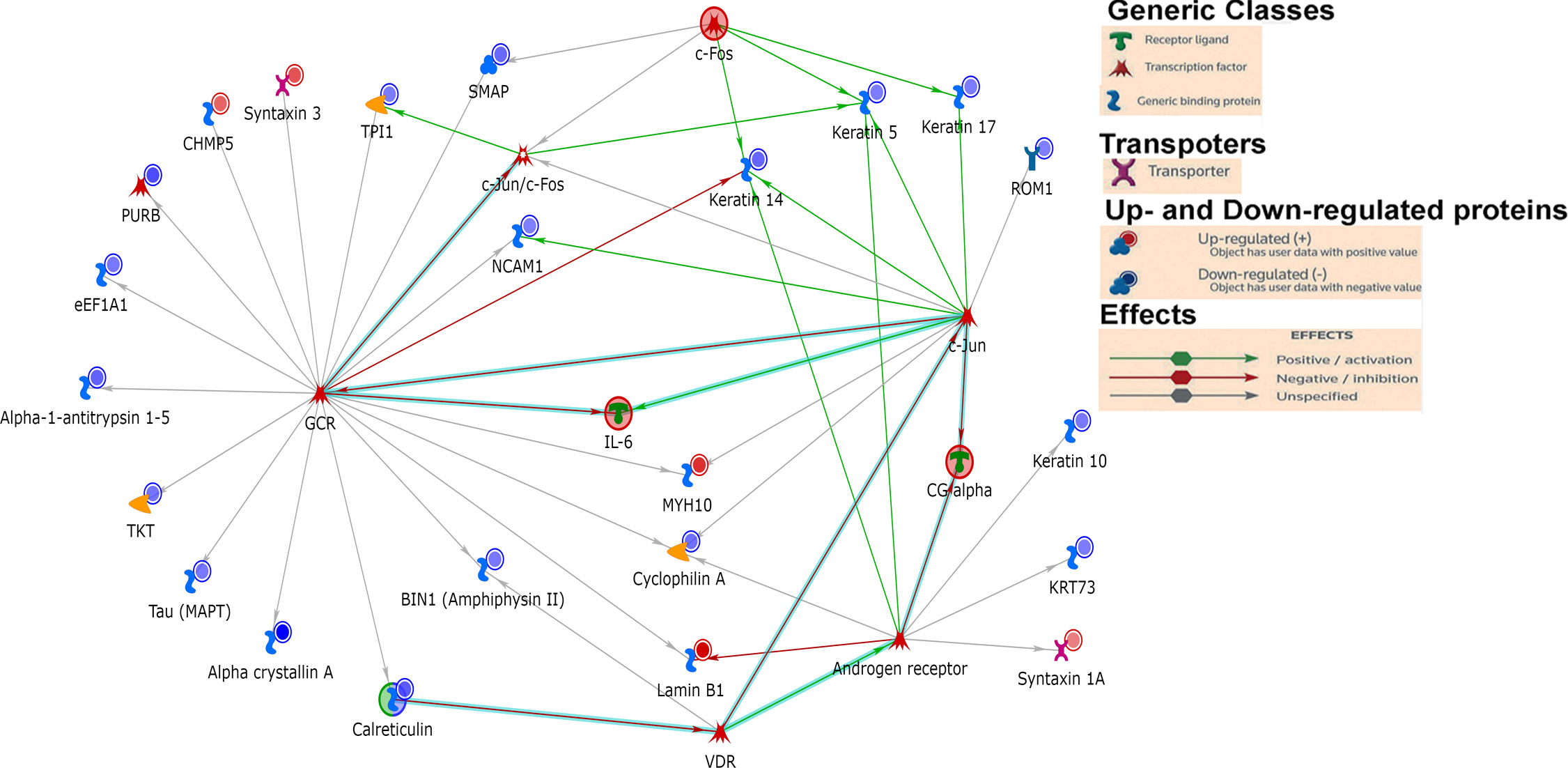
Statistically significant increased and decreased proteins in diabetic retinas were then plotted into ShinyGo program to group them by Gene Ontology (GO) biological processes using a 1.5 threshold for increased and 30% for decreased proteins (Figure 3). Figure 3 depicts the most abundant biological process occurring in diabetic retinas using the fold enrichment criterion. The negative regulation of p38K cascade was one of the drastically decreased biological processes in the diabetic retinas.
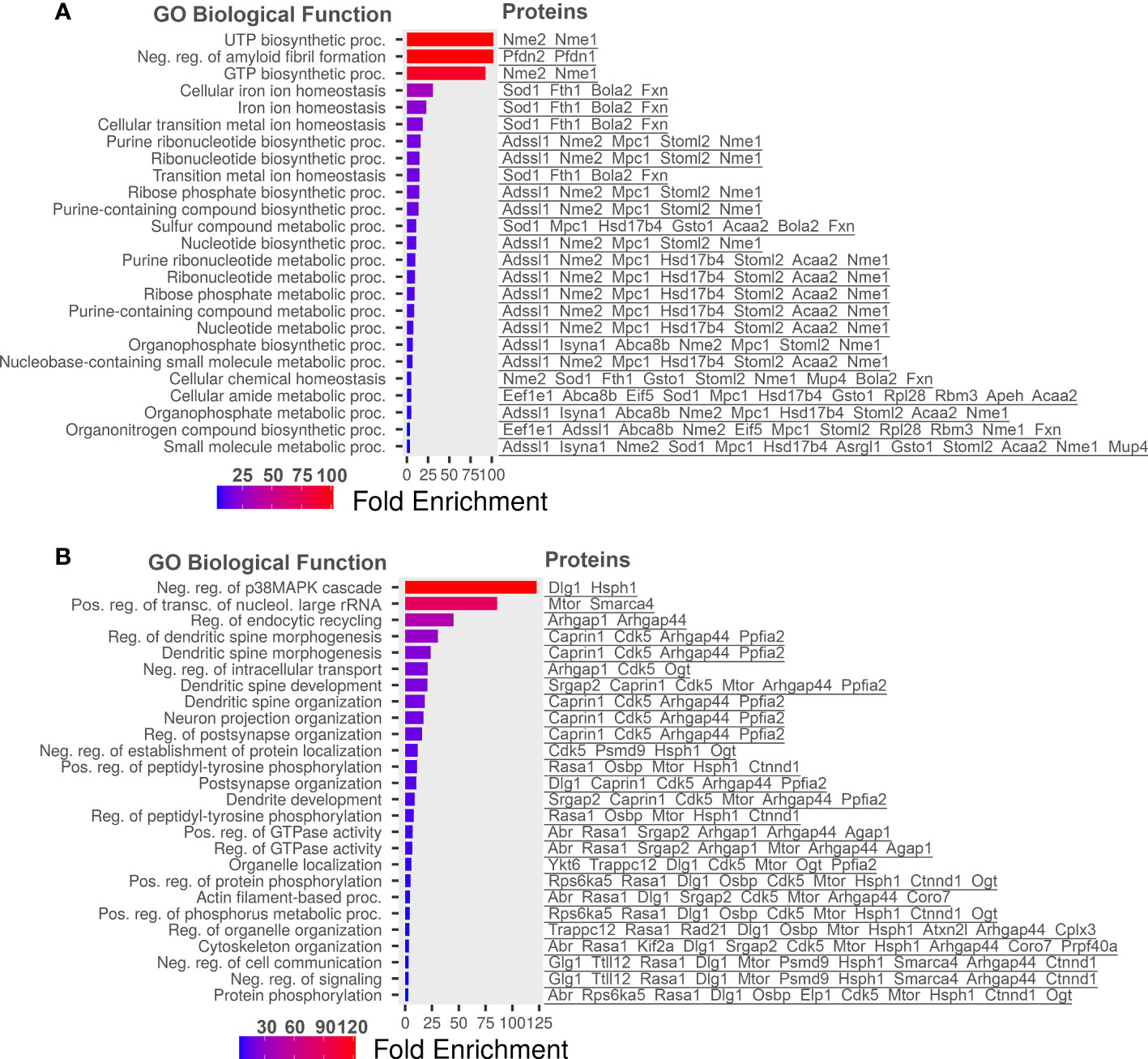
Figure 3 Go Biological processes for proteins significantly changed in our dataset. (A) Biological processes of proteins increased in diabetic retinas. (B) Go Biological processes of proteins reduced in diabetic retinas. Left is list of GO Biological Processes and right is list of proteins related to the biological process. Graphs and lists generated with ShinyGO software.
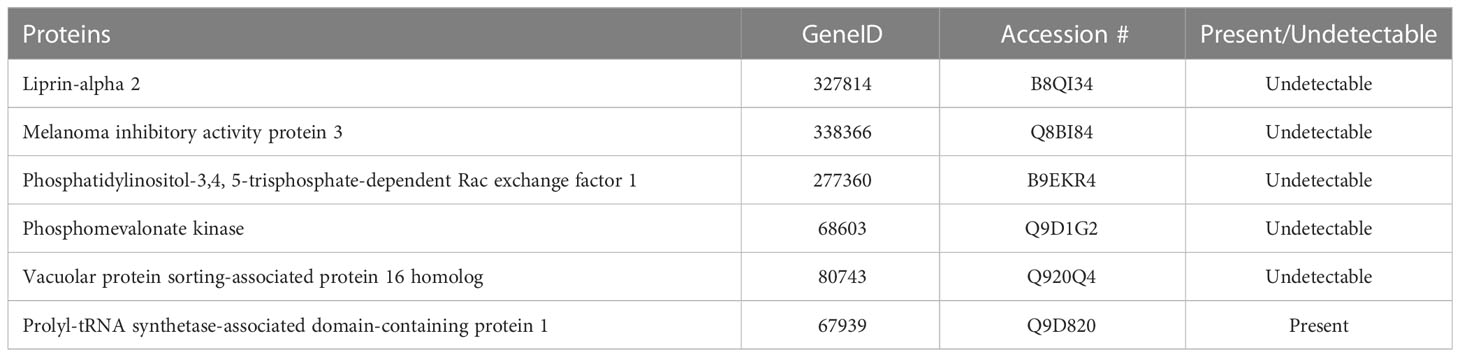
Figure 4 highlights global changes in metabolic processes in more depth. All significant changes in global proteomics can be found in Supplementary Table S1. Table 2 depicts proteins either exclusively identified or undetectable in the diabetic retinas. For example, we learned that VPS16 that plays an important role in segregation of intracellular molecules into distinct organelles was undetectable in diabetic retinas. The VPS16 is known to be predominantly associated with late endosomes/lysosomes, and therefore, may mediate vesicle trafficking steps in the endosome/lysosome pathway in diabetic retinas. Moreover, loss of function of VPS16 gene causes early onset dystonia associated with lysosomal abnormalities (26). Another example is PRXD1 present exclusively in diabetic retinas. We also noted significant changes in levels of many proteins involved in metabolic processes (Figure 4). The major changes occurred in the expression of proteins responsible for lipid metabolism: fatty acid β-oxidation (3-fold), phosphatidylcholine pathway (2.7-fold), triacylglycerol pathway (2.6-fold), ceramide pathway (2.3-fold), and carbohydrate metabolism: propionate pathway and transport (2-fold). Figures 1, 2, 4 depict just a fraction of the proteins and cellular processes altered in our dataset, for a complete list of all proteins, cellular processes, functions, and more, see dataset 1 and Supplementary File 1.

Figure 4 The major cellular pathways altered in diabetic retina are shown. Carbohydrate and amino acid metabolism presented almost 50% of all endogenous metabolic pathways modified in the diabetic mouse retina. The alteration in vitamin metabolism, carnitine & ceramide pathways, and phosphatidic acid pathways accounted about 35%. The remaining 15% of metabolism altered in the diabetic retinas was associated with lipid metabolism and alkyl glycerol phosphate and ethanolamine homeostasis.
Global shift in levels of proteins involved in cellular structure, trafficking and neuronal processes
Various proteins involved in structural integrity, cellular trafficking and/or synaptic function were also reduced in diabetic mice. For example, CLIP-associating protein 2 (Clasp2), Complexin-3, G protein-regulated inducer of neurite outgrowth 1 (Grin1), Cytoskeleton-associated protein 5 (Ckap5), Caprin-1, Synaptobrevin homolog YKT6, Kinesin-like protein KIF2A, Coronin-7, Catenin delta-1, and TOM1-like protein 2 (Tom1l2) were significantly diminished. Considering the highlighted above changes in levels of proteins responsible for metabolic signaling and of those involved in structural, neuronal processes and trafficking, we theorize that the retinal neurons in diabetic mice are altering their metabolism and neuronal processes to shift the energy expenditure towards molecular mechanisms responsible for the life support of the somas.
Identifying the retinal O-glycoproteome in diabetic mice
Protein glycosylation is the enzymatic addition of carbohydrates to proteins. Glycosylation can influence protein folding, stability, function, and localization. There are diseases associated with faulty glycosylation machinery as well as changes in glycosylation of key proteins. For example, defect in O-linked glycosylation causes familial tumoral calcinosis, severe autosomal recessive metabolic disorders showing massive calcium deposits in the skin and subcutaneous tissues (27). The disorder is due to mutations in GALNT3, one of the O-GalNAc transferases in mucin O-glycosylation. Another example is Tn-syndrome, which is caused by somatic mutations in the X-linked gene COSMC encoding a highly specific chaperone required for the proper folding and normal activity of β1-3 galactosyltransferase needed for the synthesis of core 1 O-glycans (28). Though studies on retinal diseases have highlighted proteins whose glycosylation is vital for their functions, to date, no large-scale datasets pertaining to O-glycosylation in the retina have been published. Therefore, we next sought to elucidate how the retinal O-glycoproteome was altered in diabetic retinas.
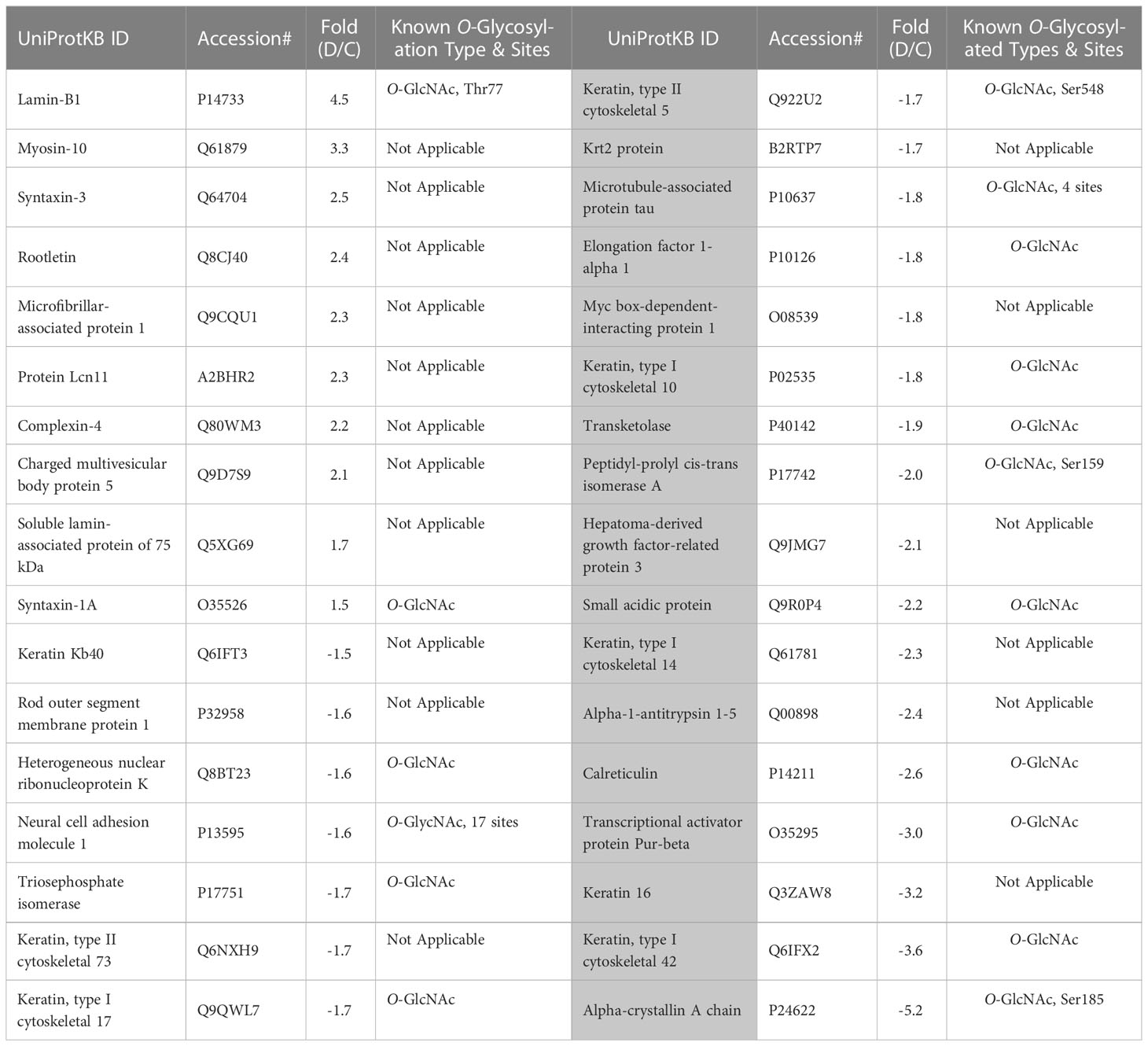
First, we analyzed the level of O-GlcNAc-modified proteins by western blot analysis and learned that O-GlcNAcylation is increased in diabetic retina (Figure 2B). Because our LC-MS technique does not distinguish the different types of O-glycosylation, we next refer to proteins identified by this technique as O-linked glycosylated proteins. We again used an unbiased proteomics approach utilizing LC/MS to identity glycosylated peptides in both groups and compared the results (Figures 4, 5, Table 3). We identified significant changes in O-glycosylation of peptides corresponding to 37 proteins. As expected, this is much fewer than detected in our global dataset. We identified differentially O-glycosylated proteins involved in metabolism, neuronal structure, and presynaptic vesicle docking. For example, diabetic retinas displayed reduced O-glycosylation of Triosephosphate isomerase (Tpi), Elongation factor 1-alpha 1(Eef1a1), and Transketolase demonstrating a reduction of O-glycosylation of proteins representing an array of metabolic processes ranging from glycolysis to protein synthesis. On the contrary, we found that O-glycosylation of Microfibrillar-associated protein 1 (Mfap1), a protein involved in Pre-mRNA splicing, and Charged multivesicular body protein 5 (Chmp5), a protein seemingly involved in a plethora of cellular processes including endosomal sorting, was elevated (29).
To our surprise, we found elevated O-glycosylation of Syntaxin-1A, Syntaxin-3, and Complexin-4, proteins involved in synaptic vesicle docking and fusion (30, 31), in diabetic retinas. Syntaxin-1A is involved in neurotransmitter dependent endocytosis and exocytosis and part of the Soluble NSF Attachment Receptor (SNARE) complex (32). Syntaxin-3 is reportedly involved in the docking of synaptic vesicles (33, 34). Syntaxin-3 is essential for photoreceptor survival (35, 36), so elevated O-glycosylation of this protein in DR is certainly intriguing. O-glycosylation of Rootletin was also increased in the retinas of diabetic mice. Rootletin is an essential component of the ciliary rootlet (37, 38), which is present at the base of the primary cilium including those of photoreceptor outer segments (OS) (39).
We also detected reduced O-glycosylation of Neural cell adhesion molecule 1(Ncam1), Rod outer segment membrane protein 1 (Rom1) and Microtubule-associated protein tau. As its name suggests, Ncam1 is involved in neuron-neuron adhesion but also participates in the outgrowth of neurites (40). Rom1 is a protein vital to a range of processes in the rod photoreceptor OS including organization and disk maintenance. Of note, ROM1 mutations have been associated with retinitis pigmentosa (41). Another example is Tau, while best known for its association with neurodegenerative diseases (42), is also involved in various neuronal processes such as establishing neuronal polarity, structurally connecting components to the plasma membrane, and cytoskeletal stability (42). Reduced O-glycosylated pattern of Tau has been reported in postmortem brain of Alzheimer patients (43). It has been proposed that the glycosylation is responsible for its hyperphosphorylation and aggregation (44). Though it is unclear what O-glycosylation of these proteins regulates in the retina, it is interesting to see a reduction of these proteins as global levels of proteins with similar neuronal involvement was also observed.
Discussion
Here we reported changes in the retinal proteome of mice with diabetes. We highlighted alterations in global proteins involved in metabolic processes, maintaining cellular structure, trafficking, and neuronal processes. We then showed alterations in the O-linked glycosylation of individual proteins in the diabetic retina.
We learn that biological processes such as UTP/GTP biosynthesis and cellular iron homeostasis were markedly enriched in diabetic retinas while negative regulation of p38/MAPK cascade was the most downregulated pathway. The alterations of metabolic pathways in the diabetic retina consist of major changes in the carbohydrate metabolism including TCA and glycolysis. These changes are usually associated with T1D and therefore, there is no wonder that STZ-induced diabetic mice manifest these alterations. Although presented by a smaller portion, the alterations in signaling pathways such as lipid metabolism detected in the diabetic retina of mice with T1D are primarily associated with T2D overall indicating that changes in fatty acid β-oxidation and TAG pathways could also be identified in the STZ-induce diabetes.
Diabetes is a complex systemic metabolic disorder with an interplay between several metabolic pathways in different organs of the human body. The diabetic retina is not an exclusion and together with neuropathy, nephropathy, and cardiopathy presents a complication of diabetes. Dyslipidemia involving both central as well as the local retinal specific mechanisms plays a critical role in the development of DR. Proteins responsible for fatty acid β-oxidation, phosphatidylcholine singling, triacylglycerol metabolism, and ceramide biogenesis are top-modified proteins in the retina of mice with 3 months of diabetes. The ABC transporter subunit A8 is one of them. Over 12-fold increase of these proteins is found in diabetic retinas (Table 1). ABCA8 as well as ABCA1 facilitate the efflux of cholesterol to lipid-free ApoA-I (45) and is proposed to participate in sphingomyelin (SM) production (46). SM is predominantly stored in membrane lipid rafts and significant increase in its level could impact membrane rigidity and permeability. Excess SM could also be converted into ceramide by the catalytic action of sphingomyelinase. This metabolic pathway is altered in diabetic retinas (Figure 2). Both altered sphingolipid and ceramide metabolism leading to inflammation and apoptotic cell death in diabetic retinas have been reported earlier (47, 48).
Another example is oxysterol binding protein 1 (OSBP1) which participates in lipid metabolism, regulation of secretory vesicle generation and signal transduction, and acts as a sterol sensor controlling a variety of sterol-dependent cellular processes. The level of this protein drops over 3-fold in the diabetic retina. Altogether these results suggest that lipid metabolism is dysregulated in the diabetic retina.
Recent studies on O-glycosylation revealed an abnormal protein PTM in varied human and mouse diabetic tissues(49–51). Moreover, in mice with T1D and T2D, the enhancement of protein O-glycosylation has been also reported (52–54) which is in agreement with our findings demonstrating increase in O-GlcNac-modified proteins (Figure 2). Despite the importance of the mentioned above studies and the present information on individual retinal cell types manifesting enhanced O-glycosylation, these studies, unfortunately, did not provide a list of individual proteins subjected to aberrant O-glycosylation. Therefore, to the best our knowledge, this is the first comprehensive study on the O-glycosylation of individual retinal proteins in early diabetes.
Knowing to be associated with the access of glucose, the O-GlcNAc modification is a dynamic process occurring in response to environmental stimuli. Two enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) participate in dynamic O-glycosylation. OGT catalyzes the addition of the O-GlcNAc moiety while the OGA is responsible for its removal. Surprisingly, while the OGA level was not changed in diabetic retinas, the production of nuclear OGT1 (110 kD) was reduced by 30% as compared to the control (Supplementary Table S1). Although this reduction is in agreement with a reduction in mTOR, S6K, and eIF4G proteins known to be responsible for mRNA translation (Figure 1 and Table 1) and coincide with the data on selected and overall reduced mRNA translation reported in a hyperglycemic environment (55), our results and the reported studies indicated an increase in O-GlcNAc proteins overall (Figure 2 and (52–54). One of the potential explanations for this phenomenon is that STZ could inhibit OGA (56) and the reduction in OGT is a compensatory feedback to this inhibition. It is possible that the OGT/OGA ratio is highly dynamic, and we caught the changes in OGT and did not detect ones in OGA. Nevertheless, like the study with Akita mice, future experiments should identify an OGT1/OGA ratio in the retina of STZ-induced mice at different stages of DR progression, since the abnormal cycling has been reported to induce early cell death of retinal pericytes, one of the earliest signs of DR (52, 57).
The O-glycosylation could occur either in the cytoplasm or the nucleus. In our study, nuclear Lamin B1 demonstrates the highest hit for the modified O-glycosylated proteins in the diabetic retina. It has been reported that Lamins (A, B, and C) have the highest O-glycosylation levels among all nuclear proteins in the cell (58). Therefore, it is possible that the threshold for O-Glycosylation detection used in the study was high and this led us to limited numbers of over-glycosylated proteins overall (Table 3). Despite this potential limitation, we observed an increase in the O-glycosylation pattern of rootletin. Rootletin is known to localize at the basal body of photoreceptors and extend to its synaptic terminal. Little is known about PTMs of this protein in healthy and in diseased retinas. Besides the knowledge of 2 O-glycosylated sites (Ser1065 and Ser1892) located in rootletin (GlyGen, Q5TZA2-1), no study has been conducted to elucidate the role of O-Glycosylation in this protein. Therefore, future studies should reveal the role of rootletin O-glycosylation in photoreceptor homeostasis.
Another discovery of our study was the enhanced O-glycosylation of Syntaxins (1A and 3) in the diabetic retina. While the role of O-glycosylation in the syntaxin activity has not been explored, the study by Delacour et al. has demonstrated that treatment of polarized HT-29 cells with 1-benzyl-2-acetamido-2-deoxy-D-galactopyranoside (GalNAc-O-bn), an inhibitor of glycosyltransferase incorporating glucosamine into O-glycans, induce a shift into intracellular distribution of Syntaxin 3 vs apical membrane localization (59). The study proposed that the aberrant O-glycosylation may affect Syntaxins cellular localization. Of note, the expression of Syntaxin 1A is reduced in the T1D (STZ-induced) Wistar rat and T2D db/db mouse retinas (60, 61).
Calreticulin demonstrates reduced O-glycosylated pattern in diabetic retinas as compared to control. Although glycosylation of calreticulin has been shown in the liver and brain, it has been also reported to be lacking in other tissues including human lymphocytes (62). Here we reported that the retinal tissue also manifests calreticulin O-glycosylation similar to the brain. However, the certain type of O-Glycosylation and the potential role this PTM could play in varied tissues and cells have not been revealed. Recently, a study conducted with tunicamycin-induced inhibition of glycosylation has proposed that aberrant glycosylation enhances pro-inflammatory activity and immunogenicity without affecting the monomeric status of calreticulin (63). For example, treatment of mouse peritoneal macrophages with recombinant calreticulin manifesting reduced O-glycosylation results in elevated secretion of pro-inflammatory cytokines TNFa and Il-6. Of note, inflammation is known to be a hallmark of diabetes that promotes the development of diabetic retinopathy in patients. Therefore, it is possible, that the observed 2.6-fold reduction in calreticulin glycosylation contributes to the immune response in the diabetic retina.
Another example of the reduced O-glycosylate pattern in our study was alpha-1-antitrypsin (A1AT). Although this protein is mainly produced by hepatocytes, it could also derive from macrophages, monocytes, and other cells. Thus, the retinas of C57BL6 mice showed A1AT expression mainly distributed in the inner nuclear layer and is mostly co-localized with CD68 and CD11b positive staining, markers of microglia in the retina (64). Knowing to be a therapeutic target for the treatment of chronic obstructive pulmonary disease, recently A1AT has been proposed as a candidate target to treat diabetic retinopathy due to its involvement in anti-inflammatory processes, anti-apoptotic activity, extracellular matrix remodeling, and protection of vessel walls and capillaries via NOS-mediated NO suppression (65, 66) The study by Ortiz and colleagues demonstrated that treatment of diabetic mice with A1AT results in reduced inflammation and retinal degeneration (67). Consequently, more information on the mechanism of A1AT action in diabetic retinas should be gained including glycosylation as a PTM.
The A1AT molecule is composed of 394 amino acids and is posttranslationally modified by N-glycosidically linked oligosaccharides at three asparagine residues at positions 70, 107, and 271 (67). Several glycoforms of A1AT exist which are assigned different properties regarding half-life, stability, and the immune modulatory properties of A1AT (68). Because it is well studied, N-glycosylation is a primary focus of A1AT PTM research. It is known that recombinant A1AT with an extended half-life and protection from agglomeration is differently glycosylated than human-purified A1AT which is prone to polymerization. Therefore, glycosylation is needed for A1AT secretion. In addition, A1AT plays a multifaceted role in inflammation by affecting the expression of TNFα, IL-6, IL-8, nFN-γ, and IL-1-β. The study demonstrated that non-glycosylated recombinant A1AT does not possess the same anti-inflammatory effect as glycosylated human-purified A1AT. Interestingly, both A1AT expression and O-glycosylation were significantly reduced in our study. While low A1AT expression (25-fold decrease) is in agreement with the observed diminishing A1AT in rd1 mice with severe retinopathy (64), the role of reduced O-glycosylation (over 2-fold decrease) needs to be explored further in detail.
In the diabetic retina, alpha Crystallin A (Cryaa) manifests the lowest level of O-glycosylation (over 5-fold as compared to control). Cryaa is a small heat shock protein acting as a molecular chaperone holding misfolded proteins in large soluble aggregates. It has been originally proposed that Cryaa expression is mostly restricted to the lens in the human body. Later, Deretic and colleagues reported that alpha A- and alpha B-Crystallins bound specifically to the photoreceptor post-Golgi membranes to mediate the transport of newly synthesized rhodopsin in frog retinal photoreceptors (69). The studies reported that Cryaa serves as a protective factor in retinal pathological processes and is strongly elevated after injury and light damage (70, 71).
While the mechanism of Cryaa-mediated protection is still under investigation, the Cryaa chaperoning activity could be associated with its PTM. For example, nonenzymatic glycation of Cryaa is 2-fold higher in the diabetic lens than in the normal lenses (72). It has been proposed that increased glycation correlates with an increase in the size of aggregates formed by Cryaa leading to a decrease in chaperoning activity (73). Altogether, these data suggest that Cryaa loses its cytoprotective during diabetes. Oppositely, metabolic product methylglyoxal binds and post-translationally enhances the Cryaa activity (74). These data suggest that the functional role of Cryaa PMTs should be carefully examined. For example, the functional role of O-GlcNac in Cryaa is unknown. Attachment of O-GlcNac occurring at serine 162 has been proposed to increase the resistant of Cryaa to PNGase F cleavage (75). Therefore, the observed reduction in O-Glycosylation in Cryaa in the diabetic retina could be associated with an increase in Cryaa agglomeration and/or solubility leading to the loss of its protective effect. Additionally, rapid phosphorylation prolonging its half-life could also be a reason as well. Notably, cone opsins, main retinal proteins, did not manifest changes in the O-glycosylation pattern in diabetes; both O- and N-glycosylation occur on these proteins at the N-terminal (76).
Conclusion
We have identified top-rated proteins with altered O-glycosylation in the diabetic retina. One of the limitations of the current study is that it doesn’t not distinguish between different types of O-Glycosylation. A search of the literature demonstrated that little is known about the posttranslational O-glycosylation of these proteins in the diabetic retina, for example O-GlcNAcylation or O-GalNAcylation. In addition to the discovery of new sites within proteins for example, O-GalNAcylation and O-GlcNAcylation, future studies should provide information on the role of individual protein O-Glycosylation to understand the consequences of altered PTM in diabetic retinas. Not only these studies are critical to enhancing our knowledge of protein structure, function, and localization but also they ensure the future development of therapeutic strategies targeting individual protein PTMomes in the diabetic retina.
Materials and methods
Animals
All animal procedures were approved by The University of Alabama at Birmingham institutional animal use and care (IACUC) committee and in accordance with the statement for the Use of Animals in Ophthalmic and Vision Research by The Association for Research in Vision and Ophthalmology (ARVO). C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME). Diabetes was induced in mice as described previously. Briefly, 2 month-old-C57BL6 males were subjected to daily injections of 50 mg/kg STZ (streptozotocin) or vehicle (0.1mol/L citrate buffer,pH4.5) during 5 consecutive days. Hyperglycemia in mice was recorded by measuring blood glycose levels (Hb1Ac) as previously described (5).
Proteomics analysis
Sample preparation
Proteomics analysis was carried out as previously referenced with minor changes [(77), within section 2.5 nLC-ESI-MS2 under Protein IDs for GeLC]. Mice were euthanized by CO2 asphyxiation, then retinas were harvested and proteins were extracted using T-PER™ Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Cat.# 78510) supplemented with HALT protease inhibitor cocktail (Thermo Fisher Scientific, Cat.# 78425), and benzonase nuclease (Sigma, E1014) following manufacturers instructions. Lysates were quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Cat.# 23227). Protein extracts were divided to carry out both “global” and “O-glycan enriched” proteomics analysis, whereby the later preparation included overnight digestion of 300ug of protein with PNGase-F (Sigma, P7367) as per manufacturers recommendation. To detect O-glycosylation, the PNGase-F treated lysates were then processed to enrich O-linked glycosylated protein using a ConA glycoprotein isolation kit (Thermo Fisher Scientific, Cat.# 89804). ConA is known to be useful for the separating O-Glycosylated from N-Glycosylation proteins removed by PNGase-F treatment in the first place. Therefore, the ConA bound fraction was released and analyzed further as per the global proteome workflow. For global analysis 20ug of protein was separated half way on the gel, and for the O-glycan enriched proteins 1ug was run as a short stack into gel. In both cases, protein per sample was diluted to 25µL using NuPAGE LDS sample buffer (1x final conc., Invitrogen, Cat.# NP0007). Proteins were then reduced with DTT and denatured at 70°C for 10min prior to loading everything onto Novex NuPAGE 10% Bis-Tris Protein gels (Invitrogen, Cat.# NP0315BOX) and separated appropriately (@ 200 constant V). The gels were stained overnight with Novex Colloidal Blue Staining kit (Invitrogen, Cat.# LC6025). Following de-staining, each entire lane was cut into multiple MW fractions (1-fraction for the O-glycan enriched, and 3 fractions for the global), and equilibrated in 100 mM ammonium bicarbonate (AmBc), each gel plug was then digested overnight with Trypsin Gold, Mass Spectrometry Grade (Promega, Cat.# V5280) following manufacturer’s instruction. Peptide extracts were reconstituted in 0.1% Formic Acid/ddH2O at 0.1µg/µL.
Mass spectrometry
Peptide digests (8µL each) were injected onto a 1260 Infinity nHPLC stack (Agilent Technologies), and separated using a 75 micron I.D. x 15 cm pulled tip C-18 column (Jupiter C-18 300 Å, 5 micron, Phenomenex). This system runs in-line with a Thermo Q Exactive HFx mass spectrometer, equipped with a Nanospray Flex™ ion source (Thermo Fisher Scientific), and all data were collected in CID mode. The nHPLC is configured with binary mobile phases that includes solvent A (0.1%FA in ddH2O), and solvent B (0.1%FA in 15% ddH2O/85% ACN), programmed as follows; 10min @ 5%B (2µL/min, load), 30min @ 5%-40%B (linear: 0.5nL/min, analyze), 5min @ 70%B (2µL/min, wash), 10min @ 0%B (2µL/min, equilibrate). Following each parent ion scan (300-1200m/z @ 60k resolution), fragmentation data (MS2) were collected on the top most intense 18 ions @7.5K resolution. For data dependent scans, charge state screening and dynamic exclusion were enabled with a repeat count of 2, repeat duration of 30s, and exclusion duration of 90s.
MS data conversion and searches
The XCalibur RAW files were collected in profile mode, centroided and converted to MzXML using ReAdW v. 3.5.1. The mgf files were created using MzXML2Search (included in TPP v. 3.5) for all scans. The data was searched using SEQUEST (Thermo Fisher Scientific), which is set for three maximum missed cleavages, a precursor mass window of 20ppm, trypsin digestion, variable modification C @ 57.0293, and M @ 15.9949 as a base setting. Searches were performed with the mus musculus species specific subset of the UniProtKB database.
Peptide filtering, grouping, and quantification
The list of peptide IDs generated based on SEQUEST search results were filtered using Scaffold (Protein Sciences, Portland Oregon). Scaffold filters and groups all peptides to generate and retain only high confidence IDs while also generating normalized spectral counts (N-SC’s) across all samples for the purpose of relative quantification. The filter cut-off values were set with minimum peptide length of >5 AA’s, with no MH+1 charge states, with peptide probabilities of >80% C.I., and with the number of peptides per protein ≥2. The protein probabilities will be set to a >99.0% C.I., and an FDR<1.0. Scaffold incorporates the two most common methods for statistical validation of large proteome datasets, the false discovery rate (FDR) and protein probability (Keller, Nesvizhskii, Weatherly). Relative quantification across experiments were then performed via spectral counting (Old, Liu), and when relevant, spectral count abundances will then be normalized between samples (Hyde).
Statistical analysis
For the proteomic data generated, two separate non-parametric-like statistical analyses were performed between each pair-wise comparison. These analyses included; 1) the calculation of weight values by significance analysis of microarray (SAM; cut off >|0.8| combined with, 2) T-Test (single tail, unequal variance, cut off of p < 0.05), which are then sorted according to the highest statistical relevance in each comparison. For SAM (Golub and Xu), whereby the weight value (W) is a statistically derived function that approaches significance as the distance between the means (μ1-μ2) for each group increases, and the SD (δ1-δ2) decreases using the formula, W=(μ1-μ2)/(δ1-δ2). For protein abundance ratios determined with N-SC’s, we set a 1.5-fold change as the threshold for significance, determined empirically by analyzing the inner-quartile data from the control experiments using ln-ln plots, where the Pierson’s correlation coefficient (R) is 0.98, and >99% of the normalized intensities fell between the set fold change. In each case, all three tests (SAM, Ttest, and fold change) have to pass in order to be considered significant.
Systems Analysis: Gene ontology assignments and pathway analysis will be carried out using MetaCore (GeneGO Inc., St. Joseph, MI) and ShinyGo (78). Interactions identified within MetaCore are manually correlated using full text articles. Detailed algorithms have been described previously [Bhatia and Ekins]. doi: 10.1093/bioinformatics/btz931.
Immunoblotting
Retinas from control and STZ induced diabetic animals were dissected and lysed with RIPA buffer (Cell signaling, Cat#9806) supplemented with Halt protease and phosphatase inhibitor cocktail following the manufacturer’s instructions. Homogenized retina extracts rotated for 30 min at 4 °C, centrifuged 12000g for 10min at 4 °C and the supernatant was collected for protein estimation (BioRad Cat#5000001). 70 μg of protein was separated by SDS-PAGE and transferred to a PVDF membrane for immunoblotting. Primary antibodies were obtained from Cell Signaling Technology (p70-S6K;Cat#2708T, O-GlcNac (CDT110,6;Cat#9875) or from Millipore-Sigma (beta actin; Cat#2066). HRP- conjugated secondary antibodies were obtained from LI-COR (HRP Goat anti-Mouse IgG;Cat#926-80010, HRP Goat ant-Rabbit;Cat#926-80011).
Data availability statement
The global mass spectrometry proteomics data presented in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, accession number PXD044269. The mass spectrometry proteomics data pertaining to the glycoproteomics presented in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, accession number PXD044270.
Ethics statement
The animal study was reviewed and approved by UAB IACUC committee.
Author contributions
CS, JM, and MG analyzed data, wrote, edited, and approved the manuscript, MG designed experiments. CS, JM, AZ, and MG prepared figures. CS and AZ conducted experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Eye Institute, grants RO1 EY027763.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1229089/full#supplementary-material
Supplementary Table 1 | Global proteins modified in diabetic retinas.
References
1. Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract (2019) 157:107843. doi: 10.1016/j.diabres.2019.107843
2. Duh EJ, Sun JK, Stitt AW. Diabetic retinopathy: current understanding, mechanisms, and treatment strategies. JCI Insight (2017) 2(14). doi: 10.1172/jci.insight.93751
3. Wang W, Lo ACY. Diabetic retinopathy: pathophysiology and treatments. Int J Mol Sci (2018) 19(6). doi: 10.3390/ijms19061816
4. Gorbatyuk OS, Pitale PM, Saltykova IV, Dorofeeva IB, Zhylkibayev AA, Athar M, et al. A novel tree shrew model of diabetic retinopathy. Front Endocrinol (Lausanne) (2021) 12:799711. doi: 10.3389/fendo.2021.799711
5. Pitale PM, Saltykova IV, Adu-Agyeiwaah Y, Li Calzi S, Satoh T, Akira S, et al. Tribbles homolog 3 mediates the development and progression of diabetic retinopathy. Diabetes (2021) 70(8):1738–53. doi: 10.2337/db20-1268
6. Hirsch IB, Brownlee M. Beyond hemoglobin A1c–need for additional markers of risk for diabetic microvascular complications. JAMA (2010) 303(22):2291–2. doi: 10.1001/jama.2010.785
7. Pitale PM, Gorbatyuk MS. Diabetic retinopathy: from animal models to cellular signaling. Int J Mol Sci (2022) 23(3). doi: 10.3390/ijms23031487
8. Verma A, Rani PK, Raman R, Pal SS, Laxmi G, Gupta M, et al. Is neuronal dysfunction an early sign of diabetic retinopathy? Microperimetry and spectral domain optical coherence tomography (SD-OCT) study in individuals with diabetes, but no diabetic retinopathy. Eye (Lond) (2009) 23(9):1824–30. doi: 10.1038/eye.2009.184
9. Vujosevic S, Midena E. Retinal layers changes in human preclinical and early clinical diabetic retinopathy support early retinal neuronal and Muller cells alterations. J Diabetes Res (2013) 2013:905058. doi: 10.1155/2013/905058
10. Verbraak FD. Neuroretinal degeneration in relation to vasculopathy in diabetes. Diabetes (2014) 63(11):3590–2. doi: 10.2337/db14-0888
11. Bradberry MM, Peters-Clarke TM, Shishkova E, Chapman ER, Coon JJ. N-glycoproteomics of brain synapses and synaptic vesicles. Cell Rep (2023) 42(4):112368. doi: 10.1016/j.celrep.2023.112368
12. Murray AR, Vuong L, Brobst D, Fliesler SJ, Peachey NS, Gorbatyuk MS, et al. Glycosylation of rhodopsin is necessary for its stability and incorporation into photoreceptor outer segment discs. Hum Mol Genet (2015) 24(10):2709–23. doi: 10.1093/hmg/ddv031
13. Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta (2013) 1833(11):2430–7. doi: 10.1016/j.bbamcr.2013.04.001
14. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol (2019) 15(6):346–66. doi: 10.1038/s41581-019-0129-4
15. van Tol W, Wessels H, Lefeber DJ. O-glycosylation disorders pave the road for understanding the complex human O-glycosylation machinery. Curr Opin Struct Biol (2019) 56:107–18. doi: 10.1016/j.sbi.2018.12.006
16. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol (2020) 21(12):729–49. doi: 10.1038/s41580-020-00294-x
17. Abeijon C, Hirschberg CB. Topography of initiation of N-glycosylation reactions. J Biol Chem (1990) 265(24):14691–5. doi: 10.1016/S0021-9258(18)77357-2
18. Starr CR, Gorbatyuk MS. Posttranslational modifications of proteins in diseased retina. Front Cell Neurosci (2023) 17:1150220. doi: 10.3389/fncel.2023.1150220
19. Gurel Z, Sheibani N. O-Linked β-N-acetylglucosamine (O-GlcNAc) modification: a new pathway to decode pathogenesis of diabetic retinopathy. Clin Sci (Lond) (2018) 132(2):185–98. doi: 10.1042/cs20171454
20. Xing X, Wang H, Zhang Y, Niu T, Jiang Y, Shi X, et al. O- glycosylation can regulate the proliferation and migration of human retinal microvascular endothelial cells through ZFR in high glucose condition. Biochem Biophys Res Commun (2019) 512(3):552–7. doi: 10.1016/j.bbrc.2019.03.135
21. Liu C, Dong W, Li J, Kong Y, Ren X. O-glcNAc modification and its role in diabetic retinopathy. Metabolites (2022) 12(8). doi: 10.3390/metabo12080725
22. Blumenthal SA. Observations on sodium retention related to insulin treatment of experimental diabetes. Diabetes (1975) 24(7):645–9. doi: 10.2337/diab.24.7.645
23. Orth JM, Murray FT, Bardin CW. Ultrastructural changes in Leydig cells of streptozotocin-induced diabetic rats. Anat Rec (1979) 195(3):415–30. doi: 10.1002/ar.1091950302
24. Ko CY, Wu CH, Huang WJ, Lo YM, Lin SX, Wu JS, et al. Alleviative effects of α-lipoic acid on muscle atrophy via the modulation of TNF-α/JNK and PI3K/AKT pathways in high-fat diet and streptozotocin-induced type 2 diabetic rats. Food Sci Nutr (2023) 11(4):1931–9. doi: 10.1002/fsn3.3227
25. Xiang W, Li L, Hong F, Zeng Y, Zhang J, Xie J, et al. N-cadherin cleavage: A critical function that induces diabetic retinopathy fibrosis via regulation of β-catenin translocation. FASEB J (2023) 37(4):e22878. doi: 10.1096/fj.202201664RR
26. Steel D, Zech M, Zhao C, Barwick KES, Burke D, Demailly D, et al. Loss-of-function variants in HOPS complex genes VPS16 and VPS41 cause early onset dystonia associated with lysosomal abnorMalities. Ann Neurol (2020) 88(5):867–77. doi: 10.1002/ana.25879
27. Chefetz I, Sprecher E. Familial tumoral calcinosis and the role of O-glycosylation in the maintenance of phosphate homeostasis. Biochim Biophys Acta (2009) 1792(9):847–52. doi: 10.1016/j.bbadis.2008.10.008
28. Stotter BR, Talbot BE, Capen DE, Artelt N, Zeng J, Matsumoto Y, et al. Cosmc-dependent mucin-type O-linked glycosylation is essential for podocyte function. Am J Physiol Renal Physiol (2020) 318(2):F518–f530. doi: 10.1152/ajprenal.00399.2019
29. Vild CJ, Li Y, Guo EZ, Liu Y, Xu Z. A novel mechanism of regulating the ATPase VPS4 by its cofactor LIP5 and the endosomal sorting complex required for transport (ESCRT)-III protein CHMP5. J Biol Chem (2015) 290(11):7291–303. doi: 10.1074/jbc.M114.616730
30. Babai N, Sendelbeck A, Regus-Leidig H, Fuchs M, Mertins J, Reim K, et al. Functional roles of complexin 3 and complexin 4 at mouse photoreceptor ribbon synapses. J Neurosci (2016) 36(25):6651–67. doi: 10.1523/jneurosci.4335-15.2016
31. Yang X, Tu W, Gao X, Zhang Q, Guan J, Zhang J. Functional regulation of syntaxin-1: An underlying mechanism mediating exocytosis in neuroendocrine cells. Front Endocrinol (Lausanne) (2023) 14:1096365. doi: 10.3389/fendo.2023.1096365
32. Vardar G, Salazar-Lázaro A, Zobel S, Trimbuch T, Rosenmund C. Syntaxin-1A modulates vesicle fusion in mamMalian neurons via juxtamembrane domain dependent palmitoylation of its transmembrane domain. Elife (2022) 11. doi: 10.7554/eLife.78182
33. Yu H, Zhou J, Takahashi H, Yao W, Suzuki Y, Yuan X, et al. Spatial control of proton pump H,K-ATPase docking at the apical membrane by phosphorylation-coupled ezrin-syntaxin 3 interaction. J Biol Chem (2014) 289(48):33333–42. doi: 10.1074/jbc.M114.581280
34. Soo Hoo L, Banna CD, Radeke CM, Sharma N, Albertolle ME, Low SH, et al. The SNARE protein syntaxin 3 confers specificity for polarized axonal trafficking in neurons. PloS One (2016) 11(9):e0163671. doi: 10.1371/journal.pone.0163671
35. Kakakhel M, Tebbe L, Makia MS, Conley SM, Sherry DM, Al-Ubaidi MR, et al. Syntaxin 3 is essential for photoreceptor outer segment protein trafficking and survival. Proc Natl Acad Sci U.S.A. (2020) 117(34):20615–24. doi: 10.1073/pnas.2010751117
36. Janecke AR, Liu X, Adam R, Punuru S, Viestenz A, Strauß V, et al. Pathogenic STX3 variants affecting the retinal and intestinal transcripts cause an early-onset severe retinal dystrophy in microvillus inclusion disease subjects. Hum Genet (2021) 140(8):1143–56. doi: 10.1007/s00439-021-02284-1
37. Ko D, Kim J, Rhee K, Choi HJ. Identification of a structurally dynamic domain for oligomer formation in rootletin. J Mol Biol (2020) 432(13):3915–32. doi: 10.1016/j.jmb.2020.04.012
38. Turn RE, Linnert J, Gigante ED, Wolfrum U, Caspary T, Kahn RA. Roles for ELMOD2 and rootletin in ciliogenesis. Mol Biol Cell (2021) 32(8):800–22. doi: 10.1091/mbc.E20-10-0635
39. Akiyama T, Inoko A, Kaji Y, Yonemura S, Kakiguchi K, Segawa H, et al. SHG-specificity of cellular Rootletin filaments enables naïve imaging with universal conservation. Sci Rep (2017) 7:39967. doi: 10.1038/srep39967
40. Perez-Rando M, Guirado R, Tellez-Merlo G, Carceller H, Nacher J. Estradiol regulates polysialylated form of the neural cell adhesion molecule expression and connectivity of O-LM interneurons in the hippocampus of adult female mice. Neuroendocrinology (2022) 112(1):51–67. doi: 10.1159/000515052
41. Bascom RA, Liu L, Heckenlively JR, Stone EM, McInnes RR. Mutation analysis of the ROM1 gene in retinitis pigmentosa. Hum Mol Genet (1995) 4(10):1895–902. doi: 10.1093/hmg/4.10.1895
42. Sohn PD, Tracy TE, Son HI, Zhou Y, Leite RE, Miller BL, et al. Acetylated tau destabilizes the cytoskeleton in the axon initial segment and is mislocalized to the somatodendritic compartment. Mol Neurodegener (2016) 11(1):47. doi: 10.1186/s13024-016-0109-0
43. Robertson LA, Moya KL, Breen KC. The potential role of tau protein O-glycosylation in Alzheimer’s disease. J Alzheimers Dis (2004) 6(5):489–95. doi: 10.3233/jad-2004-6505
44. Cantrelle FX, Loyens A, Trivelli X, Reimann O, Despres C, Gandhi NS, et al. Phosphorylation and O-GlcNAcylation of the PHF-1 epitope of tau protein induce local conformational changes of the C-terminus and modulate tau self-assembly into fibrillar aggregates. Front Mol Neurosci (2021) 14:661368. doi: 10.3389/fnmol.2021.661368
45. Trigueros-Motos L, van Capelleveen JC, Torta F, Castaño D, Zhang LH, Chai EC, et al. ABCA8 regulates cholesterol efflux and high-density lipoprotein cholesterol levels. Arterioscler Thromb Vasc Biol (2017) 37(11):2147–55. doi: 10.1161/atvbaha.117.309574
46. Kim WS, Hsiao JH, Bhatia S, Glaros EN, Don AS, Tsuruoka S, et al. ABCA8 stimulates sphingomyelin production in oligodendrocytes. Biochem J (2013) 452(3):401–10. doi: 10.1042/bj20121764
47. Fox TE, Han X, Kelly S, Merrill AH 2nd, Martin RE, Anderson RE, et al. Diabetes alters sphingolipid metabolism in the retina: a potential mechanism of cell death in diabetic retinopathy. Diabetes (2006) 55(12):3573–80. doi: 10.2337/db06-0539
48. Mandal N, Grambergs R, Mondal K, Basu SK, Tahia F, Dagogo-Jack S. Role of ceramides in the pathogenesis of diabetes mellitus and its complications. J Diabetes Complications (2021) 35(2):107734. doi: 10.1016/j.jdiacomp.2020.107734
49. Konrad RJ, Kudlow JE. The role of O-linked protein glycosylation in beta-cell dysfunction. Int J Mol Med (2002) 10(5):535–9.
50. Clark RJ, McDonough PM, Swanson E, Trost SU, Suzuki M, Fukuda M, et al. Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J Biol Chem (2003) 278(45):44230–7. doi: 10.1074/jbc.M303810200
51. Walgren JL, Vincent TS, Schey KL, Buse MG. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am J Physiol Endocrinol Metab (2003) 284(2):E424–434. doi: 10.1152/ajpendo.00382.2002
52. Gurel Z, Sieg KM, Shallow KD, Sorenson CM, Sheibani N. Retinal O-linked N-acetylglucosamine protein modifications: implications for postnatal retinal vascularization and the pathogenesis of diabetic retinopathy. Mol Vis (2013) 19:1047–59.
53. Xu C, Liu G, Liu X, Wang F. O-GlcNAcylation under hypoxic conditions and its effects on the blood-retinal barrier in diabetic retinopathy. Int J Mol Med (2014) 33(3):624–32. doi: 10.3892/ijmm.2013.1597
54. Kim SJ, Yoo WS, Choi M, Chung I, Yoo JM, Choi WS. Increased O-GlcNAcylation of NF-κB enhances retinal ganglion cell death in streptozotocin-induced diabetic retinopathy. Curr Eye Res (2016) 41(2):249–57. doi: 10.3109/02713683.2015.1006372
55. Dierschke SK, Miller WP, Favate JS, Shah P, Imamura Kawasawa Y, Salzberg AC, et al. O-GlcNAcylation alters the selection of mRNAs for translation and promotes 4E-BP1-dependent mitochondrial dysfunction in the retina. J Biol Chem (2019) 294(14):5508–20. doi: 10.1074/jbc.RA119.007494
56. Mohan R, Jo S, Da Sol Chung E, Oribamise E, Lockridge A, Abrahante-Llorens JE, et al. Pancreatic beta-cell O-glcNAc transferase overexpression increases susceptibility to metabolic stressors in female mice. Cells (2021) 10(10). doi: 10.3390/cells10102801
57. Gurel Z, Zaro BW, Pratt MR, Sheibani N. Identification of O-GlcNAc modification targets in mouse retinal pericytes: implication of p53 in pathogenesis of diabetic retinopathy. PloS One (2014) 9(5):e95561. doi: 10.1371/journal.pone.0095561
58. Cejas RB, Lorenz V, Garay YC, Irazoqui FJ. Biosynthesis of O-N-acetylgalactosamine glycans in the human cell nucleus. J Biol Chem (2019) 294(9):2997–3011. doi: 10.1074/jbc.RA118.005524
59. Delacour D, Gouyer V, Leteurtre E, Ait-Slimane T, Drobecq H, Lenoir C, et al. 1-benzyl-2-acetamido-2-deoxy-alpha-D-galactopyranoside blocks the apical biosynthetic pathway in polarized HT-29 cells. J Biol Chem (2003) 278(39):37799–809. doi: 10.1074/jbc.M305755200
60. Gaspar JM, Baptista FI, Galvão J, Castilho AF, Cunha RA, Ambrósio AF. Diabetes differentially affects the content of exocytotic proteins in hippocampal and retinal nerve terminals. Neuroscience (2010) 169(4):1589–600. doi: 10.1016/j.neuroscience.2010.06.021
61. Ramos H, Bogdanov P, Sabater D, Huerta J, Valeri M, Hernández C, et al. Neuromodulation induced by sitagliptin: A new strategy for treating diabetic retinopathy. Biomedicines (2021) 9(12). doi: 10.3390/biomedicines9121772
62. Denning GM, Leidal KG, Holst VA, Iyer SS, Pearson DW, Clark JR, et al. Calreticulin biosynthesis and processing in human myeloid cells: demonstration of signal peptide cleavage and N-glycosylation. Blood (1997) 90(1):372–81. doi: 10.1182/blood.V90.1.372.372_372_381
63. Gong FY, Gong Z, Duo CC, Wang J, Hong C, Gao XM. Aberrant glycosylation augments the immuno-stimulatory activities of soluble calreticulin. Molecules (2018) 23(3). doi: 10.3390/molecules23030523
64. Zhou T, Huang Z, Zhu X, Sun X, Liu Y, Cheng B, et al. Alpha-1 antitrypsin attenuates M1 microglia-mediated neuroinflammation in retinal degeneration. Front Immunol (2018) 9:1202. doi: 10.3389/fimmu.2018.01202
65. Zuddas A, Corsini GU, Schinelli S, Johannessen JN, di Porzio U, Kopin IJ. MPTP treatment combined with ethanol or acetaldehyde selectively destroys dopaminergic neurons in mouse substantia nigra. Brain Res (1989) 501(1):1–10. doi: 10.1016/0006-8993(89)91020-2
66. Potilinski MC, Ortíz GA, Salica JP, López ES, Fernández Acquier M, Chuluyan E, et al. Elucidating the mechanism of action of alpha-1-antitrypsin using retinal pigment epithelium cells exposed to high glucose. Potential use in diabetic retinopathy. PloS One (2020) 15(2):e0228895. doi: 10.1371/journal.pone.0228895
67. Ortiz G, Lopez ES, Salica JP, Potilinski C, Fernández Acquier M, Chuluyan E, et al. Alpha-1-antitrypsin ameliorates inflammation and neurodegeneration in the diabetic mouse retina. Exp Eye Res (2018) 174:29–39. doi: 10.1016/j.exer.2018.05.013
68. McCarthy C, Saldova R, Wormald MR, Rudd PM, McElvaney NG, Reeves EP. The role and importance of glycosylation of acute phase proteins with focus on alpha-1 antitrypsin in acute and chronic inflammatory conditions. J Proteome Res (2014) 13(7):3131–43. doi: 10.1021/pr500146y
69. Deretic D, Aebersold RH, Morrison HD, Papermaster DS. Alpha A- and alpha B-crystallin in the retina. Association with the post-Golgi compartment of frog retinal photoreceptors. J Biol Chem (1994) 269(24):16853–61. doi: 10.1016/S0021-9258(19)89469-3
70. Sakaguchi H, Miyagi M, Darrow RM, Crabb JS, Hollyfield JG, Organisciak DT, et al. Intense light exposure changes the crystallin content in retina. Exp Eye Res (2003) 76(1):131–3. doi: 10.1016/s0014-4835(02)00249-x
71. Vázquez-Chona F, Song BK, Geisert EE Jr. Temporal changes in gene expression after injury in the rat retina. Invest Ophthalmol Vis Sci (2004) 45(8):2737–46. doi: 10.1167/iovs.03-1047
72. Garlick RL, Mazer JS, Chylack LT Jr., Tung WH, Bunn HF. Nonenzymatic glycation of human lens crystallin. Effect of aging and diabetes mellitus. J Clin Invest (1984) 74(5):1742–9. doi: 10.1172/jci111592
73. Swamy MS, Tsai C, Abraham A, Abraham EC. Glycation mediated lens crystallin aggregation and cross-linking by various sugars and sugar phosphates in vitro. Exp Eye Res (1993) 56(2):177–85. doi: 10.1006/exer.1993.1025
74. Nagaraj RH, Oya-Ito T, Padayatti PS, Kumar R, Mehta S, West K, et al. Enhancement of chaperone function of alpha-crystallin by methylglyoxal modification. Biochemistry (2003) 42(36):10746–55. doi: 10.1021/bi034541n
75. Roquemore EP, Dell A, Morris HR, Panico M, Reason AJ, Savoy LA, et al. Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. J Biol Chem (1992) 267(1):555–63. doi: 10.1016/S0021-9258(18)48530-4
76. Salom D, Jin H, Gerken TA, Yu C, Huang L, Palczewski K. Human red and green cone opsins are O-glycosylated at an N-terminal Ser/Thr-rich domain conserved in vertebrates. J Biol Chem (2019) 294(20):8123–33. doi: 10.1074/jbc.RA118.006835
77. Ludwig MR, Kojima K, Bowersock GJ, Chen D, Jhala NC, Buchsbaum DJ, et al. Surveying the serologic proteome in a tissue-specific kras(G12D) knockin mouse model of pancreatic cancer. Proteomics (2016) 16(3):516–31. doi: 10.1002/pmic.201500133
Keywords: diabetes, retina, proteomics, diabetic retinopathy, glycosylation
Citation: Starr CR, Zhylkibayev A, Mobley JA and Gorbatyuk MS (2023) Proteomic analysis of diabetic retinas. Front. Endocrinol. 14:1229089. doi: 10.3389/fendo.2023.1229089
Received: 25 May 2023; Accepted: 28 July 2023;
Published: 25 August 2023.
Edited by:
Rajesh H. Amin, Auburn University, United StatesReviewed by:
Gladys Y.-P. Ko, Texas A and M University, United StatesRajakrishnan Veluthakal, City of Hope National Medical Center, United States
Copyright © 2023 Starr, Zhylkibayev, Mobley and Gorbatyuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina S. Gorbatyuk, mgortk@uab.edu
 Christopher R. Starr
Christopher R. Starr Assylbek Zhylkibayev
Assylbek Zhylkibayev James A. Mobley
James A. Mobley Marina S. Gorbatyuk
Marina S. Gorbatyuk