- 1Beijing Neurosurgical Institute, Capital Medical University, Beijing, China
- 2Department of Neurosurgery, Beijing Tongren Hospital Affiliated to Capital Medical University, Beijing, China
- 3Department of Neurosurgery, Beijing Tiantan Hospital Affiliated to Capital Medical University, Beijing, China
- 4Center of Brain Tumor, Beijing Institute for Brain Disorders, Beijing, China
- 5China National Clinical Research Center for Neurological Diseases, Beijing, China
Introduction: The invasive behavior of nonfunctioning pituitary neuroendocrine tumors (NF-PitNEts) affects complete resection and indicates a poor prognosis. Cancer immunotherapy has been experimentally used for the treatment of many tumors, including pituitary tumors. The current study aimed to screen the key immune-related genes in NF-PitNEts with invasion.
Methods: We used two cohorts to explore novel biomarkers in NF-PitNEts. The immune infiltration-associated differentially expressed genes (DEGs) were obtained based on high/low immune scores, which were calculated through the ESTIMATE algorithm. The abundance of immune cells was predicted using the ImmuCellAI database. WGCNA was used to construct a coexpression network of immune cell-related genes. Random forest analysis was used to select the candidate genes associated with invasion. The expression of key genes was verified in external validation set using quantitative real-time polymerase chain reaction (qRT‒PCR).
Results: The immune and invasion related DEGs was obtained based on the first dataset of NF-PitNEts (n=112). The immune cell-associated modules in NF-PitNEts were calculate by WGCNA. Random forest analysis was performed on 81 common genes intersected by immune-related genes, invasion-related genes, and module genes. Then, 20 of these genes with the highest RF score were selected to construct the invasion and immune-associated classification model. We found that this model had high prediction accuracy for tumor invasion, which had the largest area under the receiver operating characteristic curve (AUC) value in the training dataset from the first dataset (n=78), the self-test dataset from the first dataset (n=34), and the independent test dataset (n=73) (AUC=0.732/0.653/0.619). Functional enrichment analysis revealed that 8 out of the 20 genes were enriched in multiple signaling pathways. Subsequently, the 8-gene (BMP6, CIB2, FABP5, HOMER2, MAML3, NIN, PRKG2 and SIDT2) classification model was constructed and showed good efficiency in the first dataset (AUC=0.671). In addition, the expression levels of these 8 genes were verified by qRT‒PCR.
Conclusion: We identified eight key genes associated with invasion and immunity in NF-PitNEts that may play a fundamental role in invasive progression and may provide novel potential immunotherapy targets for NF-PitNEts.
Introduction
Pituitary neuroendocrine tumors (PitNEts) account for approximately 10-20% of intracranial tumors and are the second most common neoplasms of the central nervous system (1, 2). The prevalence of PitNEts ranges from 76-116 cases per 100,000 population, and the incidence is between 3.9 and 7.4 cases per 100,000 per year (3). These tumors are classified into functional and nonfunctioning pituitary tumor subtypes according to endocrine status (4). Nonfunctional pituitary neuroendocrine tumors (NF-PitNEts) account for 36%-54% of PitNEts and are usually detected based on signs and symptoms (headache, visual disturbance, and/or hypopituitarism) related to the effects of tumor mass because of the lack of excessive hormone secretion (2, 4–6). In this context, surgery is the treatment of choice because it can rapidly achieve decompression and symptomatic improvement (7–9). Because most macro-NF-PitNEts have the potential to invade the surrounding structures, such as the cavernous sinus or the sphenoid sinus, complete resection is often challenging and is achieved in up to 60-73% of patients (10, 11). Moreover, invasive tumors have an increased recurrence rate due to tumor residues, which require additional surgery or radiation therapy and thus pose a further risk of complications (12–14). As a result, it is necessary to explore the pathogenesis of invasive NF-PitNEts to optimize the treatment of this tumor.
The tumor immune microenvironment (TIME) plays a crucial role in tumor development, progression, and immunotherapy (15, 16). The TIME is composed of immune cells (lymphocytes and macrophages), immune-related pathways and cytokines secreted by tumor cells or immune cells (17). Pituitary tumor cells have been shown to recruit a variety of tumor-infiltrating immune cells, such as macrophages, T lymphocytes, B lymphocytes, FOXP3+ cells, neutrophils, and NK cells, into the tumor microenvironment (18–20). Moreover, the TIME has many effector functions and may promote the proliferation, migration and invasion of pituitary tumors (18, 21, 22). Therefore, it is essential to comprehensively analyze immunological genes affecting the abundance of immune cells in the invasive NF-PitNEts microenvironment.
In the current study, differentially expressed genes (DEGs) were identified at the tumor invasive and immune levels. Weighted correlation network analysis (WGCNA) was used to screen immune cell-related genes. The key invasive and immunological genes were further investigated by constructing a classification model and enrichment analysis. Our study screened out critical invasive-immune associated genes, which could provide new ideas for exploring immunological studies and some potential treatment strategies for NF-PitNEts patients.
Materials and methods
Human tissue samples and clinical data
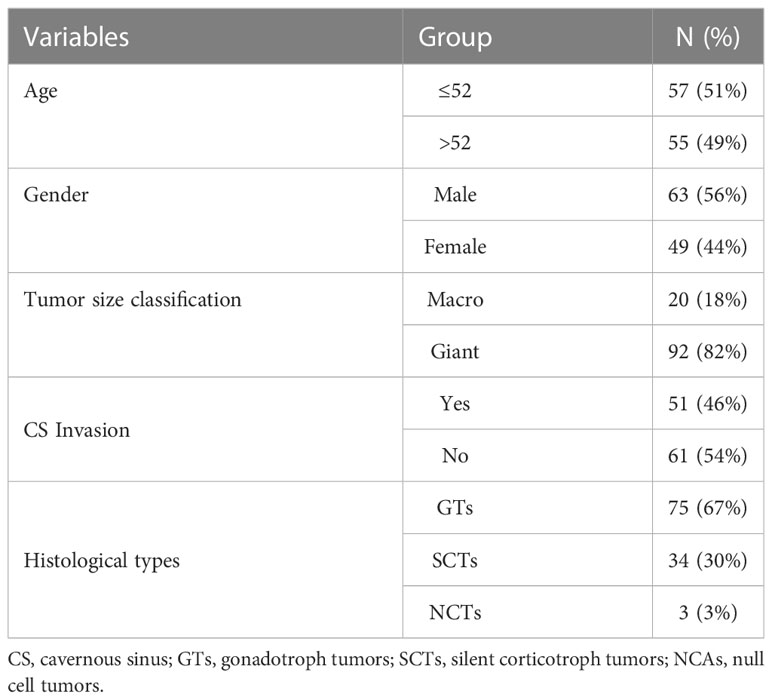
In this study, we used two cohorts to explore novel biomarkers in pituitary tumors. The first dataset included 112 patients, and another independent test dataset contained 73 patients. Tumor specimens were obtained from patients with NF-PitNEts (n=112) who underwent transsphenoidal surgical resection at Beijing Tiantan Hospital between June 2018 and July 2019. The diagnosis of NF-PitNEts is defined as the absence of clinical and biochemical evidence of overproduction of adenohypophysis hormone. The mean age of these 112 patients was 52 years (range, 21-75), and there were 63 males and 49 females. The demographics and clinicopathological features of the patients are summarized in Table 1. Tumor cavernous sinus (CS) invasion was defined as Knosp grade 3 and 4 or intraoperative evidence (23). The expression profiles and matching clinical information of independent test datasets (n=73) were described previously (24). In addition, 16 NF-PitNEts specimens (8 invasive and 8 noninvasive) were collected from the same hospital as an independent validation cohort (Table S3), and their expression levels were verified by quantitative real-time polymerase chain reaction (qRT-PCR). This study recruitment process and protocol were approved by the Medical Ethics Committee of Beijing Tiantan Hospital, and informed consent was obtained from all individual participants.
Total RNA extraction and RNA sequencing
A total of 1-3 μg RNA per sample was extracted and purified from the collected specimens of NF-PitNEts. According to the instructions provided, sequencing libraries were constructed using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (#E7530L, NEB, USA). After the library was successfully generated (effective concentration >10 nM), the index-coded samples were clustered on the cBot cluster generation system using HiSeq PE Cluster Kit v4-cBot-HS (Illumina). The library was then sequenced on an Illumina platform, and 150-bp paired-end reads were generated. Raw data were filtered with FAST-QC, and the clean reads were then mapped to the human genome hg19 sequence (GRCh37) using HISAT2 (25). HTseq was used to generate gene counts, and the RPKM method was used to determine gene expression (26).
Differential expression analysis
The Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) algorithm (27) was used to obtain the immune levels of 112 pituitary tumor patients. Based on the median immune score, patients were divided into high and low groups. These 112 patients were also divided into invasion and noninvasion groups based on their clinical invasion information.
The “limma” R package was used to obtain the DEGs from the high vs. low immune score groups and invasion vs. noninvasion groups. Genes with an adjusted P value < 0.05 and |log2-fold change| ≥ 0.585 were filtered as DEGs.
Weighted gene coexpression network analysis
First, the ImmuCellAI database was applied to estimate the abundance of 24 immune cell types in the 112 patients with NF-PitNEts (28). The abundance of these immune cells in the invasion and noninvasion groups was then used for WGCNA.
Second, the “WGCNA” R package was applied to build a coexpression network of immune cell-related genes. Next, several gene modules were detected based on their similar expression patterns. Finally, the abundance of immune cells was associated with these gene expression modules, and genes in the modules were selected for further analysis.
Random forest analysis
The “randomForest” R package was used to select the candidate genes associated with invasion. We first divided the 112 patients into a training dataset (n=78) and a self-test (n=34) dataset based on the “sample” function of the R program. Then, random forest analysis (RF) was performed through the “randomForest” function in the training dataset, the error rate curve was drawn and changes in the error rate of different numbers of genes selected were observed. Finally, the “ggplot” R package was used to show the MeanDecreaseAccuracy and the best RF model of these genes.
Genes with the top 20 MeanDecreaseAccuracy were used to build the SVM model through the “e1071” R package, and the “pROC” package was used to perform the classification efficiency of the model in the training dataset, self-test dataset and independent test dataset.
Consensus clustering
The R package “ConsensusClusterPlus” was applied to explore the classification efficiency of 8 crucial genes. Subsequently, the “pROC” package was used to show the classification efficiency of these genes.
Enrichment analysis
Functional enrichment analysis was carried out by the “clusterProfiler” R package, and only functions with a p value<0.05 were selected.
qRT-PCR assay
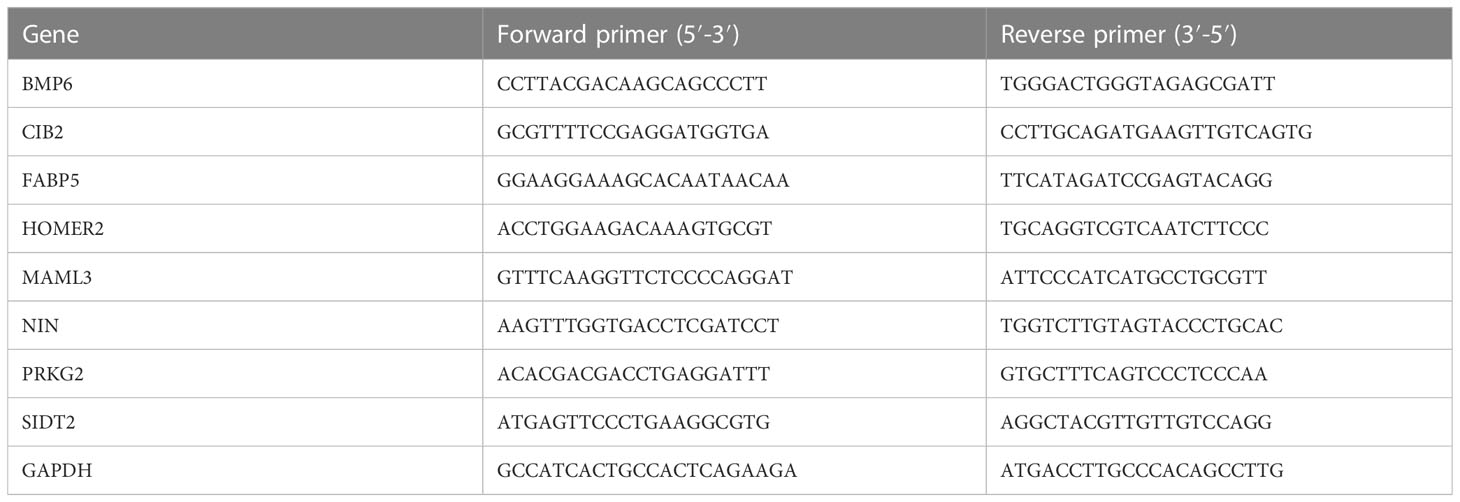
The QIAGEN RNeasy Kit and High Capacity cDNA Reverse Transcription Kit were used to extract total RNA from the verified samples and conduct reverse transcription reactions. qRT-PCR was performed in a volume of 20 µl with Power SYBR™ Green PCR Master Mix on a QuantStudio 3 and 5 System (Applied Biosystems). GAPDH was used as a housekeeping control. The sequences of the primers are shown in Table 2.
Statistical analysis
All statistical analyses were performed by R software (version 3.6.3). The bar plot between two different groups was drawn by the “ggplot” package, and a T test was used to compare the differences. A P value <0.05 was considered significant.
Results
Identification of immune infiltration-associated and invasion-associated DEGs in pituitary tumor patients
ESTIMATE analysis was performed in the 112 NF-PitNEts expression data, and the samples were divided into high- and low-immune score groups. Next, 3152 DEGs were obtained from the high- vs. low-immune score groups, including 662 upregulated and 2490 downregulated genes (Figure 1A and Table S1). The enrichment analysis indicated that these DEGs were involved in Th1 and Th2 cell differentiation, the relaxin signaling pathway, and the FoxO signaling pathway (Figure 1B). Then, we obtained 525 DEGs (352 upregulated and 173 downregulated) associated with invasion (Figure 1C and Table S2). These genes are related to the FoxO signaling pathway and Th17 cell differentiation (Figure 1D). This finding reveals that invasion-associated DEGs are involved in the immune-related functions of pituitary tumors.
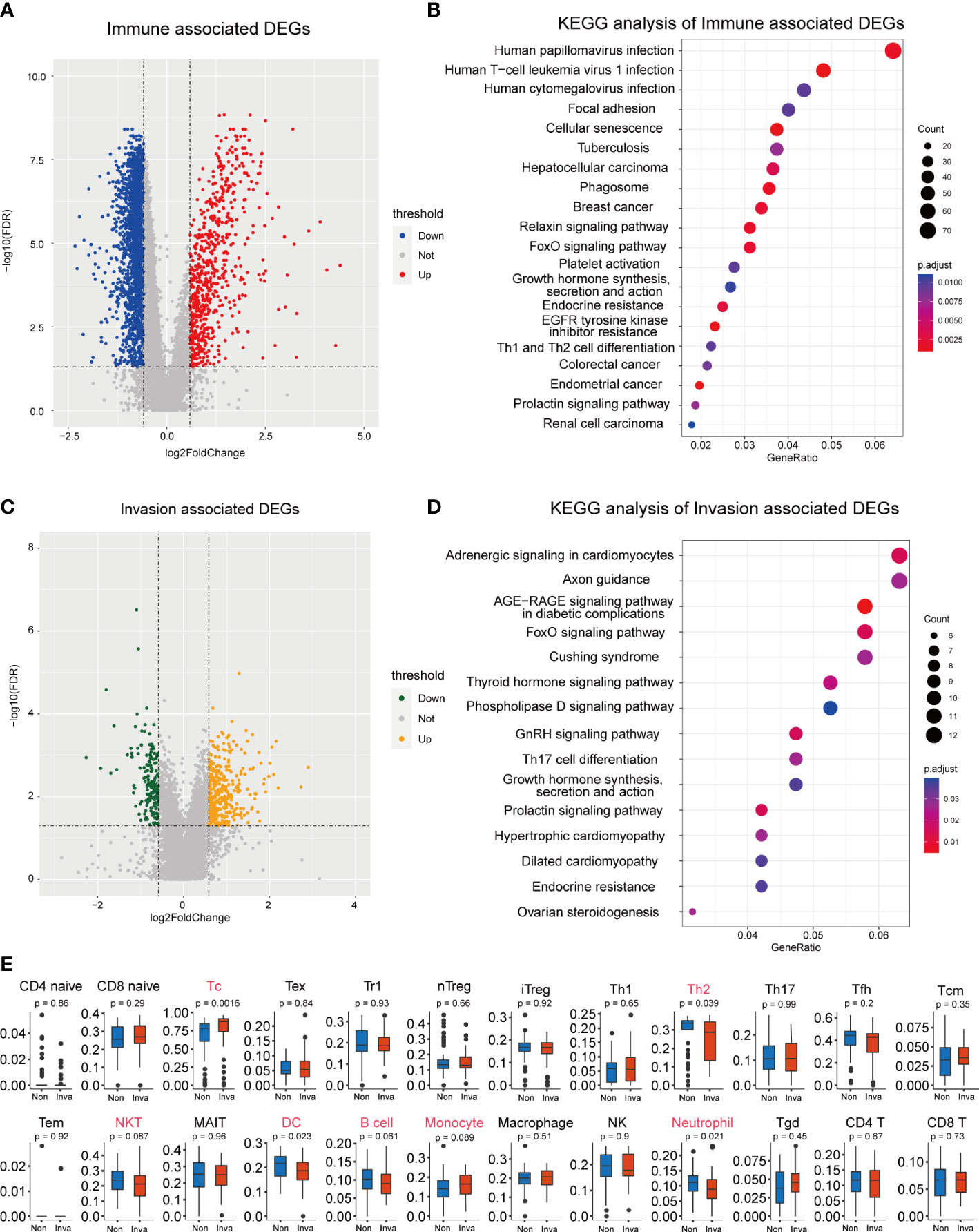
Figure 1 Screening for immune and invasion-related genes in pituitary tumors. (A) Volcano plot showing the differentially expressed genes (DEGs) between high Immune-score samples and low Immune-score samples. (B) The functional enrichment analysis of the Immune-related DEGs. (C) Volcano plot showing the differentially expressed genes (DEGs) between patients with and without invasion. (D) The functional enrichment analysis of the Invasion-related DEGs. (E) Multiple immune cell differences between patients with and without invasion.
Subsequently, we predicted the immune cell abundance of these patients. Cytotoxic T cells (Tc), Th2 cells, natural killer T cells (NKT), dendritic cells (DC), B cells, monocytes and neutrophils differed between the invasion and noninvasion groups (Figure 1E).
Immune cell-associated modules in pituitary tumors
We used the “WGCNA” package to calculate the immune cell-associated modules in pituitary tumors. The soft power of the coexpression network was calculated through the “pickSoftThreshold” function and was set to 20 (Figure 2A). At this power, the R2 of the scale-free topology model under the soft threshold was 0.92, which indicates that the network conformed to the scale-free feature (Figure 2A). Then, the network was constructed, and seven coexpression modules (green, turquoise, blue, yellow, black, brown, and red) were built (Figure 2B). Subsequently, the correlation between these seven modules and immune cell abundance was calculated. The green module was significantly correlated with neutrophils; the turquoise module was significantly correlated with monocytes; the blue module was significantly correlated with cytotoxic cells; and the yellow, black, brown, and red modules were significantly associated with NKT cells (Figure 2C). These results suggest that these genes are immune-related in the tumor microenvironment of pituitary tumors.
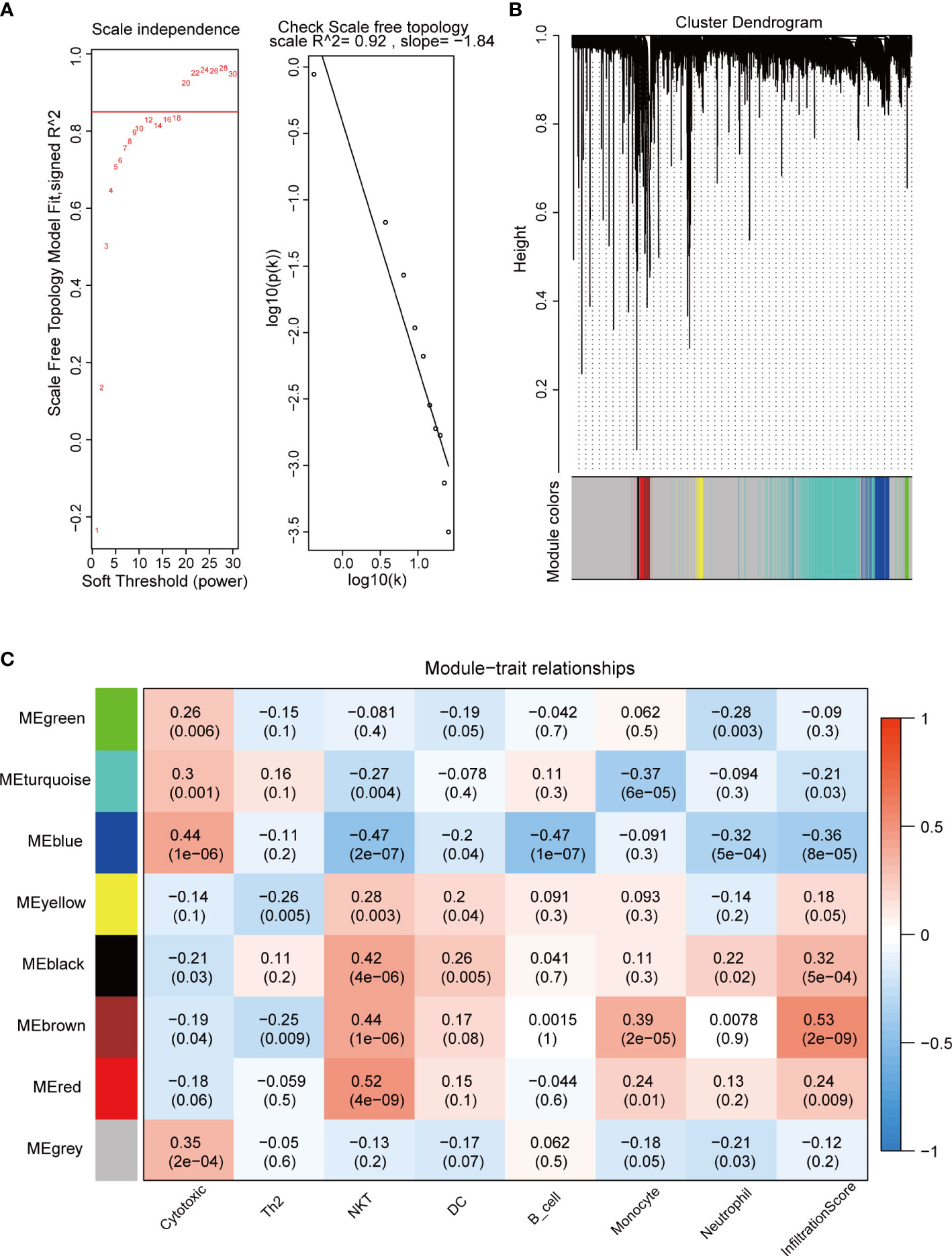
Figure 2 WGCNA of crucial immune cells in pituitary tumors. (A) Soft power selection of the WGCNA network. Here, we selected 20 as the power. (B) Clustering dendrogram of genes with dissimilarity based on topological overlap and assigned module colors. (C) The relationships between gene modules and immune cells. The P value is shown in parentheses.
Construction of the invasion and immune-associated classification model (IICM)
To select the candidate crucial genes in pituitary tumors, we first set the intersection of immune-related DEGs, invasion-related DEGs and module genes and found 81 common genes (Figure 3A). Next, we used the 81 common genes to perform RF analysis. We started the RF analysis by generating 1000 decision trees, which showed a lower error rate (Figure 3B). Then, the random forest results indicated that, when the number of candidate genes was 20, the classification efficiency error rate of the model was the lowest. We showed the top 50 accuracies of these genes and selected the top 20 for further analysis (Figures 3C, D).
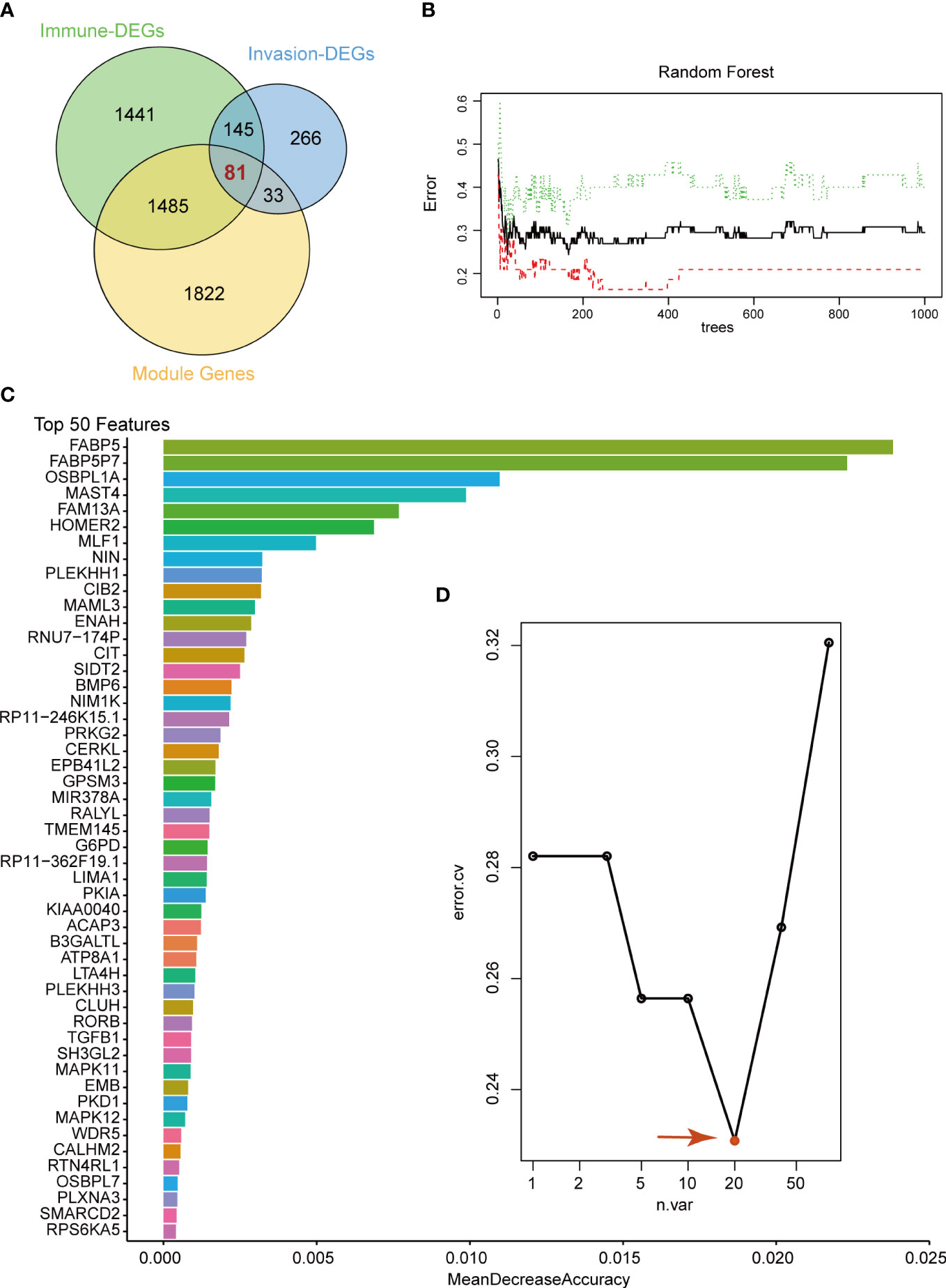
Figure 3 Random Forest analysis. (A) Venn diagram showing the candidate genes between immune-related DEGs, invasion-related DEGs and WGCNA modules. We selected 81 common genes for random forest analysis. (B) Random Forest analysis of the 81 genes. (C) Top 50 genes with the lowest MeanDecreaseAccuracy in random forest analysis. (D) The lowest error rate model contains 20 candidate genes based on random forest analysis.
Subsequently, SVM analysis was used to construct the IICM (Figure 4A). We found that this model could distinguish the invasive and noninvasive patients in the training dataset (AUC=0.732, Figure 4A). It also exhibited good efficacy in the self-test dataset (AUC=0.653, Figure 4B) and independent test dataset (AUC=0.619, Figure 4C). The results showed that this model could be used to predict the invasion state of pituitary tumors.
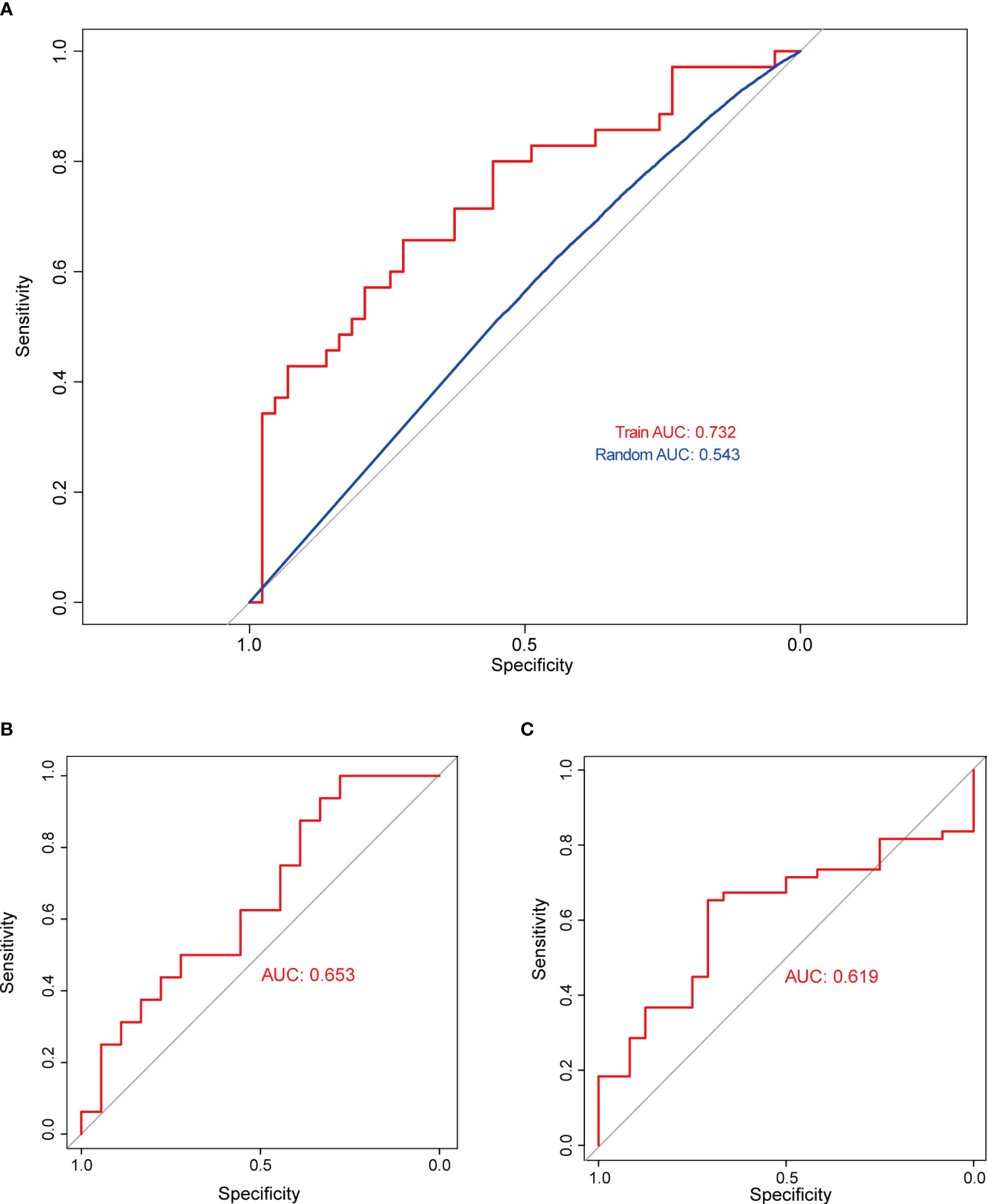
Figure 4 Validation of model classification performance. (A) The ROC curve of the RF model with 20 genes in the training dataset; the AUC reached 0.732. (B) The RF model’s classification performance in the self-test dataset; the AUC reached 0.653. (C) The RF model’s classification performance in the independent test dataset; the AUC reached 0.619.
Eight genes in IICM exhibit better efficacy in pituitary tumors
We then analyzed the function of these 20 key genes. The results suggested that 8 of the 20 genes were enriched in multiple signaling pathways, such as the KRAS signaling pathway and PPAR signaling pathway (Figure 5A). This result suggests that the function of this model is mainly driven by these 8 genes (BMP6, CIB2, FABP5, HOMER2, MAML3, NIN, PRKG2 and SIDT2).
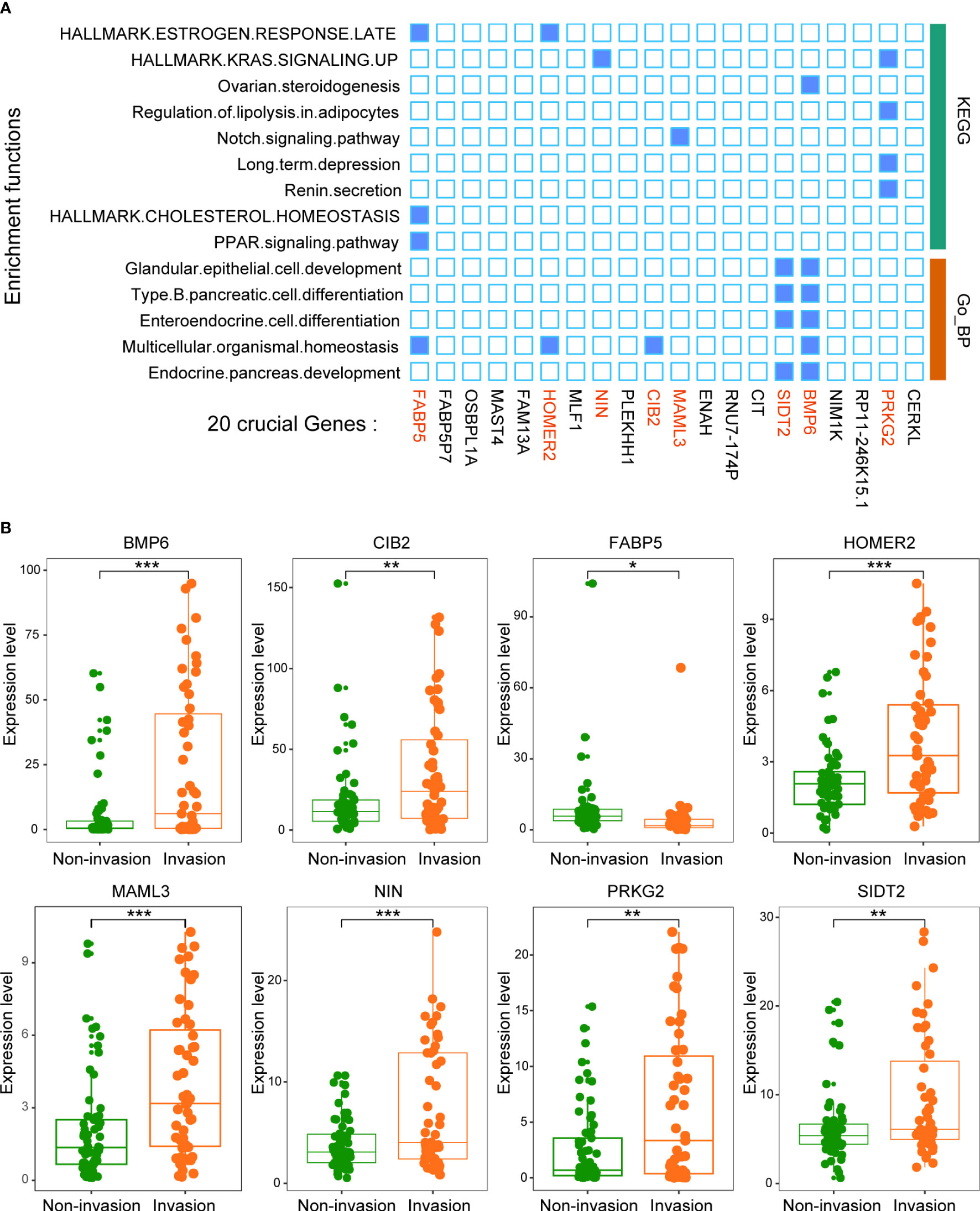
Figure 5 Twenty crucial genes were enriched in multiple pathways. (A) Functional enrichment analysis of the 20 genes. The results showed that 8 genes played an important role in these pathways. (B) The expression levels of the 8 crucial genes between patients with and without invasion. *p < 0.05, **p < 0.01, ***p < 0.001.
All 8 genes were differentially expressed in the invasion and noninvasion groups (Figure 5B), and most were differentially expressed in the high- and low-immune score groups (Figure 6). Subsequently, the consensus clustering analysis suggested that the 8 genes could well divide the patients into two groups (Figures 7A, B). We then constructed a classifier for these 8 genes, and the results showed that the 8-gene classification model showed good classification efficiency in pituitary tumors (AUC=0.671, Figure 7C). Then, qRT-PCR was performed to validate the expression level of these genes between invasive and noninvasive NF-PitNEts (Figure 7D). Consistent with the sequencing data, the mRNA expression of BMP6, CIB2, HOMER2, MAML3, NIN, PRKG2, and SIDT2 was significantly upregulated in invasive NF-PitNEts, while FABP5 was significantly downregulated. Overall, we filtered 8 genes that could predict the invasion status of pituitary tumors and could be used as predictors for further treatment of the tumors.
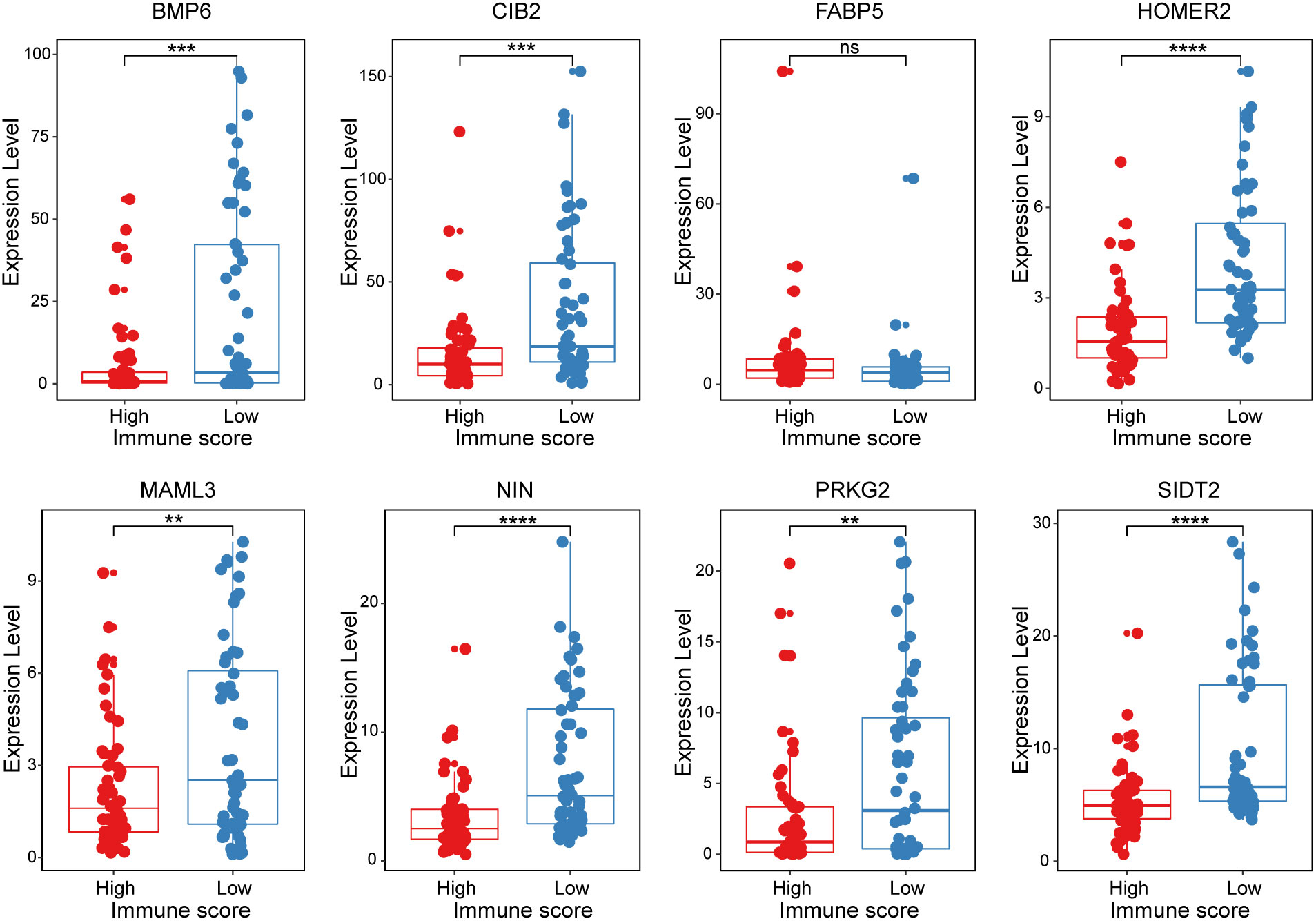
Figure 6 The expression levels of the 8 crucial genes in high Immune-score samples and low Immune-score samples. The results are expressed as the means ± SD (Student’s t test. **p < 0.01, ***p < 0.001, ****p < 0.0001). ns, no significance.
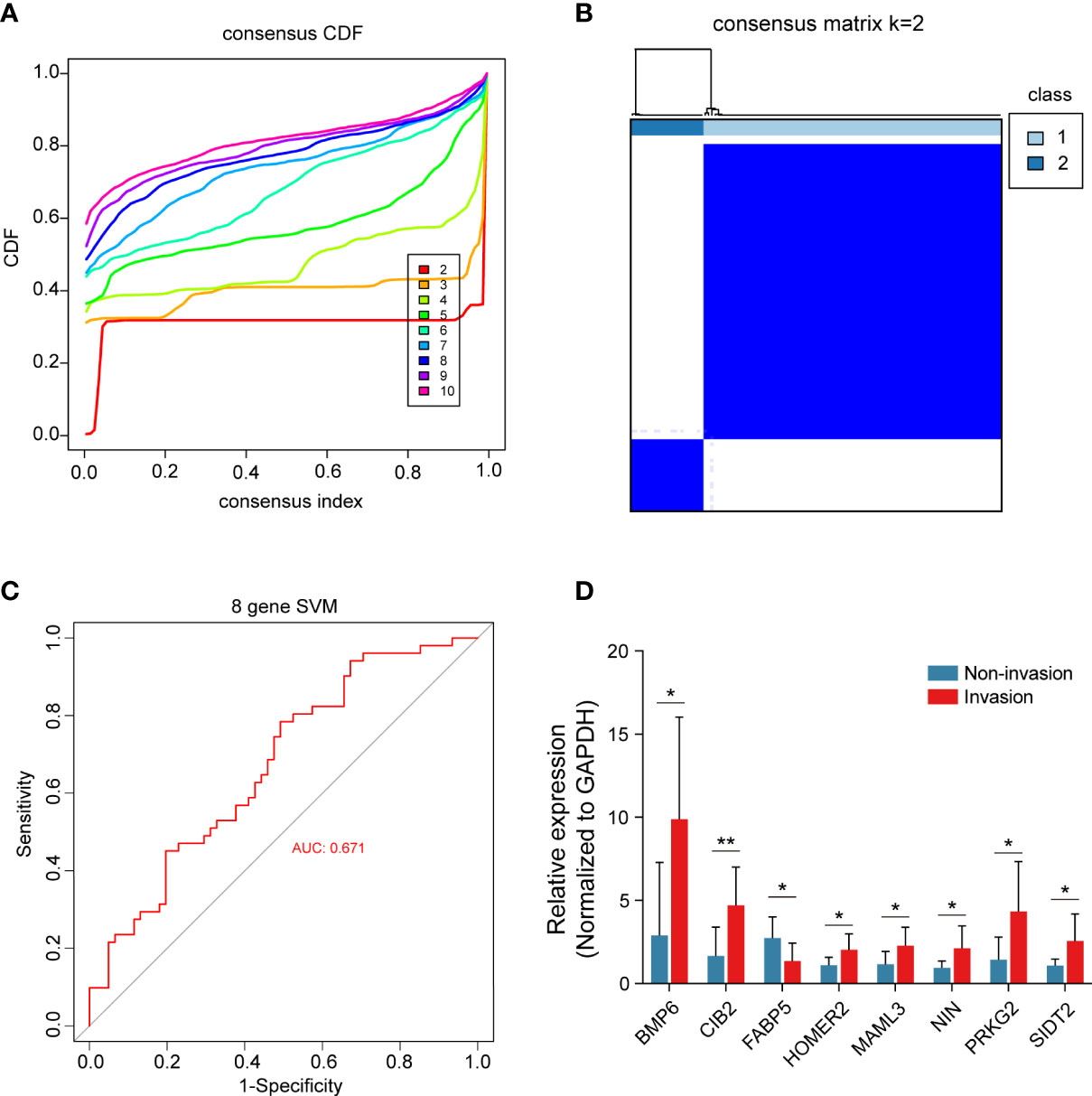
Figure 7 Consensus cluster of the 8 crucial genes. (A) Consensus index of the consensus cluster analysis. The 8 crucial genes could divide the patients into two groups. (B) Heatmap of the two groups divided by the 8 genes. (C) The new model constructed by the 8 crucial genes performed with good classification effectiveness in pituitary tumors. (D) Validation of the 8 crucial genes between 8 CS invasive NF-PitNEts and 8 noninvasive NF-PitNEts. The results are expressed as the means ± SD (Student’s t test. *p < 0.05; **p < 0.01).
Discussion
Although NF-PitNEts are benign neoplasms, they often invade surrounding structures and cannot be cured using standard therapies (29). Moreover, invasion is known as an important prognostic factor for recurrence (30). Recent studies have reported that immune cells infiltrate pituitary adenomas and may play an important role in tumor invasion and progression (31–34). Therefore, understanding the mechanisms involved in immunity with invasive NF-PitNEts could lead to the discovery of new therapeutic targets in the future.
First, 3152 DEGs (662 upregulated and 2490 downregulated) were identified based on the high- and low-immune score groups, which were obtained using the ESTIMATE algorithm. We obtained 525 DEGs between invasive and noninvasive NF-PitNEts. Then, the abundance of immune cells was predicted using the ImmuCellAI database, and Tc, Th2, NKT, DC, B cell, monocyte and neutrophil cells were found to be different between the invasion and noninvasion groups. Huang X et al. (35) found that patients with invasive NF-PitNEts had significantly lower CD3-CD56+ natural killer (NK) cells than patients with noninvasive NF-PitNEts in peripheral blood.
Subsequently, the abundance of these immune cells was used to construct a coexpression network of immune cell-related genes by WGCNA. As a bioinformatics method, WGCNA clustering results (coexpression gene modules) have high biological significance and reliability (36, 37). Next, the 81 interacting genes were identified by immune-related DEGs, invasion-related DEGs and module genes significantly associated with immune cells. To narrow down the number of invasion- and immune-associated genes, random forest analysis was performed. The 20 genes with the highest RF score were selected to constrict the SVM model, and the classification efficiency was verified in the training, self-test, and independent test datasets. This result indicated that these genes play an important role in invasive behavior. Then, functional enrichment analysis was performed on these 20 genes, and 8 were found to be enriched in multiple signaling pathways, such as the KRAS signaling pathway, Notch signaling pathway, and PPAR signaling pathway. Liu et al. (38) found that upregulation of secreted phosphoprotein 1 affects tumor cell proliferation, migration, and invasion via the KRAS/MEK pathway in head and neck cancer. Feng et al. (39) reported that the Notch signaling pathway was associated with the invasion of growth hormone adenomas. Finally, these 8 invasion- and immune-related genes (BMP6, CIB2, FABP5, HOMER2, MAML3, NIN, PRKG2 and SIDT2) were validated by consensus clustering analysis and verified by qRT-PCR between invasive and noninvasive NF-PitNEts.
Bone morphogenetic protein-6 (BMP-6) belongs to the TGF-β superfamily (40). BMP-6 was deemed to be associated with several tumor metastases, such as breast, prostate, rectal, and thyroid carcinomas (41–45). In addition, BMP-6 changes the morphology of macrophages and induces the expression of the cytokine tumor necrosis factor (TNF)-α (46). Calcium and integrin-binding protein 2 (CIB2) is a small EF-hand protein that can bind Mg2+ and Ca2+ ions and participates in basic cellular functions (47). Zhu et al. (48) found that CIB2 is correlated with cell proliferation, migration, and invasion in ovarian cancer. Moreover, Wang et al. (49) reported that CIB2 can cause M2 macrophage death and facilitate tumor microenvironment inflammation. Fatty acid-binding protein 5 (FABP5), which is an intracellular lipid carrier, is correlated with tumor development in multiple human cancers (50–52), including prostate cancer (53), bladder cancer (54), and glioblastoma (55). Liu et al. (56) reported that FABP5 promotes lipid accumulation in monocytes/macrophages and may represent a therapeutic target for tumor-associated monocytes (TAMs) and cancer cells. Homer scaffolding protein 2 (HOMER2) is an adaptor protein that has been reported to be associated with tumor progression in endometrial cancer (57). Mastermind-like 3 (MAML3) is a known transcriptional coactivator of NOTCH (58). Onishi et al. (59) found that the inhibition of MAML3 significantly reduced the proliferation and invasion of tumor cells in small cell lung cancer. SID1 transmembrane family member 2 (SIDT2) is a lysosomal membrane protein that promotes RNA degradation by transporting RNA to lysosomes (60, 61). Yi et al. (62) found that the expression of SIDT2 was associated with the biological behaviors of cancer cells in papillary thyroid carcinoma.
There are still some concepts that could improve our research. First, although we validated the IICM using a self-test dataset and another independent NF-PitNEts cohort, a large-scale multicenter cohort is needed for stronger validation. Second, our study only verified expression levels in NF-PitNEts tissues by qRT-PCR, but the underlying functions and mechanism of these genes still need to be explored in vivo and in vitro.
In summary, we conducted a comprehensive bioinformatic analysis and screened out immune-related genes that were significantly correlated with invasion in patients with NF-PitNEts. The current study may provide a novel potential immunotherapy target for invasive NF-PitNEts.
Data availability statement
The data provided in the study are deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), the accession number is: GSE169498, and another set of data is included in the supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Beijing Tiantan Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
WX, YZ worked on the conception and designed research, and eventually approved the manuscript. QF and YL was contributed to collected and analyzed clinical data of patients. JW and JG were dedicated to data analysis, interpretation, and drafting. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant codes: 82071558, 82071559, 82203174, and 82103028) and the Capital’s Funds for Health Improvement and Research (Grant no.2020-4-1077).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1131693/full#supplementary-material
References
1. Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united states in 2014-2018. Neuro Oncol (2021) 23:iii1–iii105. doi: 10.1093/neuonc/noab200
2. Herrgott GA, Asmaro KP, Wells M, Sabedot TS, Malta TM, Mosella MS, et al. Detection of tumor-specific DNA methylation markers in the blood of patients with pituitary neuroendocrine tumors. Neuro Oncol (2022) 24:1126–39. doi: 10.1093/neuonc/noac050
3. Daly AF, Beckers A. The epidemiology of pituitary adenomas. Endocrinol Metab Clin North Am (2020) 49:347–55. doi: 10.1016/j.ecl.2020.04.002
4. Molitch ME. Diagnosis and treatment of pituitary adenomas: a review. JAMA (2017) 317:516–24. doi: 10.1001/jama.2016.19699
5. Sam AH, Shah S, Saleh K, Joshi J, Roncaroli F, Robinson S, et al. Clinical outcomes in patients with nonfunctioning pituitary adenomas managed conservatively. Clin Endocrinol (Oxf) (2015) 83:861–5. doi: 10.1111/cen.12860
6. Ntali G, Wass JA. Epidemiology, clinical presentation and diagnosis of non-functioning pituitary adenomas. Pituitary (2018) 21:111–8. doi: 10.1007/s11102-018-0869-3
7. Greenman Y, Tordjman K, Kisch E, Razon N, Ouaknine G, Stern N. Relative sparing of anterior pituitary function in patients with growth hormone-secreting macroadenomas: comparison with nonfunctioning macroadenomas. J Clin Endocrinol Metab (1995) 80:1577–83. doi: 10.1210/jcem.80.5.7745003
8. Losa M, Mortini P, Barzaghi R, Ribotto P, Terreni MR, Marzoli SB, et al. Early results of surgery in patients with nonfunctioning pituitary adenoma and analysis of the risk of tumor recurrence. J Neurosurg (2008) 108:525–32. doi: 10.3171/JNS/2008/108/3/0525
9. Nomikos P, Ladar C, Fahlbusch R, Buchfelder M. Impact of primary surgery on pituitary function in patients with non-functioning pituitary adenomas – a study on 721 patients. Acta Neurochir (Wien) (2004) 146:27–35. doi: 10.1007/s00701-003-0174-3
10. Penn DL, Burke WT, Laws ER. Management of non-functioning pituitary adenomas: surgery. Pituitary (2018) 21:145–53. doi: 10.1007/s11102-017-0854-2
11. Hansen TM, Batra S, Lim M, Gallia GL, Burger PC, Salvatori R, et al. Invasive adenoma and pituitary carcinoma: a SEER database analysis. Neurosurg Rev (2014) 37:279–85. doi: 10.1007/s10143-014-0525-y
12. Chen Y, Wang CD, Su ZP, Chen YX, Cai L, Zhuge QC, et al. Natural history of postoperative nonfunctioning pituitary adenomas: a systematic review and meta-analysis. Neuroendocrinology (2012) 96:333–42. doi: 10.1159/000339823
13. Raverot G, Vasiljevic A, Jouanneau E. Prognostic factors of regrowth in nonfunctioning pituitary tumors. Pituitary (2018) 21:176–82. doi: 10.1007/s11102-017-0861-3
14. Mercado M, Melgar V, Salame L, Cuenca D. Clinically non-functioning pituitary adenomas: pathogenic, diagnostic and therapeutic aspects. Endocrinol Diabetes Nutr (2017) 64:384–95. doi: 10.1016/j.endinu.2017.05.009
15. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
16. Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett (2021) 500:1–10. doi: 10.1016/j.canlet.2020.12.024
17. Lazarus J, Maj T, Smith JJ, Perusina Lanfranca M, Rao A, D'Angelica MI, et al. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight (2018) 3:e121932. doi: 10.1172/jci.insight.121932
18. Marques P, Barry S, Carlsen E, Collier D, Ronaldson A, Awad S, et al. Chemokines modulate the tumour microenvironment in pituitary neuroendocrine tumours. Acta Neuropathol Commun (2019) 7:172. doi: 10.1186/s40478-019-0830-3
19. Principe M, Chanal M, Ilie MD, Ziverec A, Vasiljevic A, Jouanneau E, et al. Immune landscape of pituitary tumors reveals association between macrophages and gonadotroph tumor invasion. J Clin Endocrinol Metab (2020) 105:dgaa520. doi: 10.1210/clinem/dgaa520
20. Zhou W, Zhang C, Zhang D, Peng J, Ma S, Wang X, et al. Comprehensive analysis of the immunological landscape of pituitary adenomas: implications of immunotherapy for pituitary adenomas. J Neurooncol (2020) 149:473–87. doi: 10.1007/s11060-020-03636-z
21. Lupi I, Manetti L, Caturegli P, Menicagli M, Cosottini M, Iannelli A, et al. Tumor infiltrating lymphocytes but not serum pituitary antibodies are associated with poor clinical outcome after surgery in patients with pituitary adenoma. J Clin Endocrinol Metab (2010) 95:289–96. doi: 10.1210/jc.2009-1583
22. Yagnik G, Rutowski MJ, Shah SS, Aghi MK. Stratifying nonfunctional pituitary adenomas into two groups distinguished by macrophage subtypes. Oncotarget (2019) 10:2212–23. doi: 10.18632/oncotarget.26775
23. Knosp E, Steiner E, Kitz K, Matula C. Pituitary adenomas with invasion of the cavernous sinus space: a magnetic resonance imaging classification compared with surgical findings. Neurosurgery (1993) 33:610–7. doi: 10.1227/00006123-199310000-00008
24. Guo J, Fang Q, Liu Y, Xie W, Li C, Zhang Y. Screening and identification of key microenvironment-related genes in non-functioning pituitary adenoma. Front Genet (2021) 12:627117. doi: 10.3389/fgene.2021.627117
25. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
26. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
27. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
28. Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) (2020) 7:1902880. doi: 10.1002/advs.201902880
29. Asa SL, Casar-Borota O, Chanson P, Delgrange E, Earls P, Ezzat S, et al. From pituitary adenoma to pituitary neuroendocrine tumor (PitNET): an international pituitary pathology club proposal. Endocr Relat Cancer (2017) 24:C5–8. doi: 10.1530/ERC-17-0004
30. Maletkovic J, Dabbagh A, Zhang D, Zahid A, Bergsneider M, Wang MB, et al. Residual tumor confers a 10-fold increased risk of regrowth in clinically nonfunctioning pituitary tumors. J Endocr Soc (2019) 3:1931–41. doi: 10.1210/js.2019-00163
31. Barry S, Carlsen E, Marques P, Stiles CE, Gadaleta E, Berney DM, et al. Tumor microenvironment defines the invasive phenotype of AIP-mutation-positive pituitary tumors. Oncogene (2019) 38:5381–95. doi: 10.1038/s41388-019-0779-5
32. Iacovazzo D, Chiloiro S, Carlsen E, Bianchi A, Giampietro A, Tartaglione T, et al. Tumour-infiltrating cytotoxic T lymphocytes in somatotroph pituitary neuroendocrine tumours. Endocrine (2020) 67:651–8. doi: 10.1007/s12020-019-02145-y
33. Marques P, Silva AL, Lopez-Presa D, Faria C, Bugalho MJ. The microenvironment of pituitary adenomas: biological, clinical and therapeutical implications. Pituitary (2022) 25:363–82. doi: 10.1007/s11102-022-01211-5
34. Suteau V, Collin A, Menei P, Rodien P, Rousselet MC, Briet C. Expression of programmed death-ligand 1 (PD-L1) in human pituitary neuroendocrine tumor. Cancer Immunol Immunother (2020) 69:2053–61. doi: 10.1007/s00262-020-02611-x
35. Huang X, Xu J, Wu Y, Sheng L, Li Y, Zha B, et al. Alterations in CD8(+) tregs, CD56(+) natural killer cells and IL-10 are associated with invasiveness of nonfunctioning pituitary adenomas (NFPAs). Pathol Oncol Res (2021) 27:598887. doi: 10.3389/pore.2021.598887
36. Bian Y, Huang J, Zeng Z, Yao H, Tu J, Wang B, et al. Construction of survival-related co-expression modules and identification of potential prognostic biomarkers of osteosarcoma using WGCNA. Ann Transl Med (2022) 10:296. doi: 10.21037/atm-22-399
37. Langfelder P, Horvath S. WGCNA: an r package for weighted correlation network analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
38. Liu K, Hu H, Jiang H, Liu C, Zhang H, Gong S, et al. Upregulation of secreted phosphoprotein 1 affects malignant progression, prognosis, and resistance to cetuximab via the KRAS/MEK pathway in head and neck cancer. Mol Carcinog (2020) 59:1147–58. doi: 10.1002/mc.23245
39. Feng J, Wang J, Liu Q, Li J, Zhang Q, Zhuang Z, et al. DAPT, a gamma-secretase inhibitor, suppresses tumorigenesis, and progression of growth hormone-producing adenomas by targeting notch signaling. Front Oncol (2019) 9:809. doi: 10.3389/fonc.2019.00809
40. Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney Int (2000) 57:2207–14. doi: 10.1046/j.1523-1755.2000.00081.x
41. Clement JH, Sanger J, Hoffken K. Expression of bone morphogenetic protein 6 in normal mammary tissue and breast cancer cell lines and its regulation by epidermal growth factor. Int J Cancer (1999) 80:250–6. doi: 10.1002/(SICI)1097-0215(19990118)80:2<250::AID-IJC14>3.0.CO;2-D
42. Dai J, Keller J, Zhang J, Lu Y, Yao Z, Keller ET. Bone morphogenetic protein-6 promotes osteoblastic prostate cancer bone metastases through a dual mechanism. Cancer Res (2005) 65:8274–85. doi: 10.1158/0008-5472.CAN-05-1891
43. Hatakeyama S, Gao YH, Ohara-Nemoto Y, Kataoka H, Satoh M. Expression of bone morphogenetic proteins of human neoplastic epithelial cells. Biochem Mol Biol Int (1997) 42:497–505. doi: 10.1080/15216549700202901
44. Kawabata A, Okano K, Uchida K, Yamaguchi R, Hayashi T, Tateyama S. Co-Localization of chondromodulin-I (ChM-I) and bone morphogenetic protein-6 (BMP-6) in myoepithelial cells of canine mammary tumors. J Vet Med Sci (2005) 67:1097–102. doi: 10.1292/jvms.67.1097
45. Zhang M, Yan JD, Zhang L, Wang Q, Lü SJ, Zhang J, et al. Activation of bone morphogenetic protein-6 gene transcription in MCF-7 cells by estrogen. Chin Med J (Engl) (2005) 118:1629–36. doi: 10.1016/j.mcm.2008.08.009
46. Hong JH, Lee GT, Lee JH, Kwon SJ, Park SH, Kim SJ, et al. Effect of bone morphogenetic protein-6 on macrophages. Immunology (2009) 128:e442–450. doi: 10.1111/j.1365-2567.2008.02998.x
47. Seki N, Hattori A, Hayashi A, Kozuma S, Ohira M, Hori T, et al. Structure, expression profile and chromosomal location of an isolog of DNA-PKcs interacting protein (KIP) gene. Biochim Biophys Acta (1999) 1444:143–7. doi: 10.1016/S0167-4781(98)00253-X
48. Zhu W, Jarman KE, Lokman NA, Neubauer HA, Davies LT, Gliddon BL, et al. CIB2 negatively regulates oncogenic signaling in ovarian cancer via sphingosine kinase 1. Cancer Res (2017) 77:4823–34. doi: 10.1158/0008-5472.CAN-17-0025
49. Wang X, Yang Y, Cai WQ, Lu Y. The relationship of sphingosine kinase 1 with pyroptosis provides a new strategy for tumor therapy. Front Immunol (2020) 11:574990. doi: 10.3389/fimmu.2020.574990
50. Campos B, Centner FS, Bermejo JL, Ali R, Dorsch K, Wan F, et al. Aberrant expression of retinoic acid signaling molecules influences patient survival in astrocytic gliomas. Am J Pathol (2011) 178:1953–64. doi: 10.1016/j.ajpath.2011.01.051
51. Han J, Kioi M, Chu WS, Kasperbauer JL, Strome SE, Puri RK. Identification of potential therapeutic targets in human head & neck squamous cell carcinoma. Head Neck Oncol (2009) 1:27. doi: 10.1186/1758-3284-1-27
52. Ogawa R, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, et al. Identification of candidate genes involved in the radiosensitivity of esophageal cancer cells by microarray analysis. Dis Esophagus (2008) 21:288–97. doi: 10.1111/j.1442-2050.2007.00759.x
53. Pang J, Liu WP, Liu XP, Li LY, Fang YQ, Sun QP, et al. Profiling protein markers associated with lymph node metastasis in prostate cancer by DIGE-based proteomics analysis. J Proteome Res (2010) 9:216–26. doi: 10.1021/pr900953s
54. Chen R, Feng C, Xu Y. Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J Int Med Res (2011) 39:533–40. doi: 10.1177/147323001103900222
55. Barbus S, Tews B, Karra D, Hahn M, Radlwimmer B, Delhomme N, et al. Differential retinoic acid signaling in tumors of long- and short-term glioblastoma survivors. J Natl Cancer Inst (2011) 103:598–606. doi: 10.1093/jnci/djr036
56. Liu J, Sun B, Guo K, Yang Z, Zhao Y, Gao M, et al. Lipid-related FABP5 activation of tumor-associated monocytes fosters immune privilege via PD-L1 expression on treg cells in hepatocellular carcinoma. Cancer Gene Ther (2022) 29:1951–60. doi: 10.1038/s41417-022-00510-0
57. Mhawech-Fauceglia P, Walia S, Yessaian A, Machida H, Matsuo K, Lawrenson K. Overexpression of HOMER2 predicts better outcome in low-grade endometrioid endometrial adenocarcinoma. Pathology (2018) 50:499–503. doi: 10.1016/j.pathol.2018.03.004
58. Heynen GJ, Nevedomskaya E, Palit S, Jagalur Basheer N, Lieftink C, Schlicker A, et al. Mastermind-like 3 controls proliferation and differentiation in neuroblastoma. Mol Cancer Res (2016) 14:411–22. doi: 10.1158/1541-7786.MCR-15-0291-T
59. Onishi H, Ichimiya S, Yanai K, Umebayashi M, Nakamura K, Yamasaki A, et al. RBPJ and MAML3: potential therapeutic targets for small cell lung cancer. Anticancer Res (2018) 38:4543–7. doi: 10.21873/anticanres.12758
60. Aizawa S, Fujiwara Y, Contu VR, Hase K, Takahashi M, Kikuchi H, et al. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy (2016) 12:565–78. doi: 10.1080/15548627.2016.1145325
61. Hase K, Contu VR, Kabuta C, Sakai R, Takahashi M, Kataoka N, et al. Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes. Autophagy (2020) 16:1974–88. doi: 10.1080/15548627.2020.1712109
Keywords: nonfunctioning pituitary neuroendocrine tumors (NF-PitNEts), invasive, immune microenvironment, WGCNA, biomarkers
Citation: Wu J, Guo J, Fang Q, Liu Y, Li C, Xie W and Zhang Y (2023) Identification of biomarkers associated with the invasion of nonfunctional pituitary neuroendocrine tumors based on the immune microenvironment. Front. Endocrinol. 14:1131693. doi: 10.3389/fendo.2023.1131693
Received: 05 January 2023; Accepted: 15 June 2023;
Published: 14 July 2023.
Edited by:
Jesper Krogh, Rigshospitalet, DenmarkReviewed by:
Liuguan Bian, Shanghai Jiao Tong University, ChinaKe Li, Chongqing Medical University, China
Copyright © 2023 Wu, Guo, Fang, Liu, Li, Xie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yazhuo Zhang, zyztxzz@126.com, ; Weiyan Xie, weiyanxie@ccmu.edu.cn
†These authors have contributed equally to this work
 Jiangping Wu1,2†
Jiangping Wu1,2† Jing Guo
Jing Guo Chuzhong Li
Chuzhong Li Yazhuo Zhang
Yazhuo Zhang