Exploring ascomycete diversity in Yunnan, China I: resolving ambiguous taxa in Phaeothecoidiellaceae and investigating conservation implications of fungi
- 1Center of Excellence in Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
- 2Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 3Honghe Center for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Sciences, Honghe, Yunnan, China
- 4CIFOR-ICRAF China Country Program, Kunming, Yunnan, China
- 5Centre for Mountain Futures (CMF), Kunming Institute of Botany, Kunming, Yunnan, China
- 6Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 7Vishnugupta Vishwavidyapeetam, Gokarna, India
- 8Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 9Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 10Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Yunnan, located in southwestern China, is known for its high fungal diversity, and many of which are endemic to the region. As part of our ongoing studies on fungi in Yunnan, we introduce two new genera in Phaeothecoidiellaceae (Mycosphaerellales), to accommodate one Repetophragma-like and another Stomiopeltis-like taxa. Pseudorepetophragma gen. nov. is introduced herein as a monotypic genus to accommodate P. zygopetali comb. nov.(≡ Repetophragma zygopetali), whereas Pseudostomiopeltis gen. nov. is introduced to accommodate Ps. xishuangbannaensis gen. et sp. nov. and Ps. phyllanthi comb. nov.(≡ Stomiopeltis phyllanthi), based on a new collection from Yunnan. In addition, Stomiopeltis sinensis is transferred to Exopassalora as E. sinensis comb. nov. due to its phylogenetic affinity and grouped with E. zambiae, the generic type of Exopassalora. This study provides new insights into the biodiversity of fungal species in this region and adds to our understanding of their ecological roles, as well as the resolution to ambiguous taxa in Phaeothecoidiellaceae.
1 Introduction
China is home to diverse climates and environments containing four of the world’s 36 biodiversity hotspots; of which, three hotspots, the mountain ranges of Southwest China, Eastern Himalaya, and Indo-Burma, intersect with the Yunnan Province (Feng and Yang, 2018; Cai et al., 2019). Yunnan has diverse climate types and environments and is significantly affected by the monsoon rains (abundant rainfall and resultant humid tropical evergreen rainforests). Diverse environments, complex topography and geography, and highly variable plant species allow fungi to specialize and flourish, and these also affect the fungal growth and distribution (Yang et al., 2004; Zhu et al., 2006; Feng and Yang, 2018; Wanasinghe et al., 2020). Furthermore, Yunnan is an agricultural province that cultivates a wide variety of agricultural and horticultural crops such as coffee (Arabica), commercial flowers, fruits (e.g., grapes, passion fruits, bananas, and mangoes), grains (e.g., rice), rubber, sugarcane, tea (Pu’er), tobacco, plantation trees (e.g., Yunnan pine, bamboo, and teak), vegetables as well as wild edible mushrooms (Li et al., 2011; Frayer et al., 2014; Zhang et al., 2014; Shen et al., 2017; ORO Yunnan Commerce, 2023). This also led Yunnan to host a high diversity of fungi that has not been studied so far.
According to Feng and Yang (2018), there may be approximately 104,000 fungal species existing in Yunnan, but only 6,000 are described, including roughly 3,000 species of higher fungi (Ascomycota and Basidiomycota), indicating that fewer than 5% of known species have been described in this province. Two prominent regions of Yunnan have been well documented on fungal diversity, viz., the Eastern Himalayas and Hengduan Mountains in northwestern Yunnan and the tropical region in southern and southwestern Yunnan, while the other parts remain a huge challenge (Feng and Yang, 2018).
Dothideomycetes is the largest and most diverse class of Ascomycota, containing 49 orders, 223 families, 1,765 genera, and over 19,000 species (Hongsanan et al., 2020a; Hongsanan et al., 2020b; Wijayawardene et al., 2022). Dothideomycetes was previously known as Loculoascomycetes (Nannfeldt, 1932; Luttrell, 1955; Barr, 1979; Eriksson, 1981; Barr and Huhndorf, 2001). The taxa in this class are commonly characterized by bitunicate, fissitunicate asci that are also shared with taxa in Arthoniomycetes and Eurotiomycetes (Hyde et al., 2013; Hongsanan et al., 2020a; Hongsanan et al., 2020b; Pem et al., 2021). However, Dothideomycetes can be distinguished from these two classes based on multigene phylogeny, their evolutionary relationships from molecular clock analysis, and ecological niches (Geiser et al., 2015; Schoch and Grube, 2015; Liu et al., 2017; Hongsanan et al., 2020a; Hongsanan et al., 2020b; Wijayawardene et al., 2022). Even though several genera and families of Dothideomycetes have been well-resolved based on molecular phylogeny in the past two decades, there are still over 200 genera that could not be classified into any families or orders due to the lack of informative phylogenetic markers and uncertain morphological features (Thambugala et al., 2014; Phillips et al., 2019; Abdollahzadeh et al., 2020; Haridas et al., 2020; Hongsanan et al., 2020a; Hongsanan et al., 2020b; Pem et al., 2021; Wijayawardene et al., 2022).
One of the Dothideomycetes orders, Mycosphaerellales, comprises eight families, 230 genera (including the doubtful taxa of Mycosphaerellaceae and genera incertae sedis), and over 4,200 species (Abdollahzadeh et al., 2020; Wijayawardene et al., 2022). The order encompassed ecologically and morphologically diverse fungi, including endophytes, epiphytes, lichenicolous fungi, phytopathogens, and saprobes (Abdollahzadeh et al., 2020). Of these, many genera of Mycosphaerellaceae are well-known as important plant pathogens that are quarantine regulated, such as Pseudocercospora angolensis causing fruit and leaf spot disease on citrus, and Septoria malagutii causing angular leaf spot on potato (Quaedvlieg et al., 2012; Videira et al., 2017). Mycosphaerellales was introduced by Kirk et al. (2001) to accommodate Mycosphaerellaceae and is characterized by small perithecial ascomata, often immersed to erumpent through the host surface by black papilla with lysigenous ostiole; the peridium composed of thin pseudoparenchymatous cells; lacking interascal tissue; asci fissitunicate, ovoid to saccate; and ascospores hyaline, septate, lacking mucilaginous sheath. Subsequently, Schoch et al. (2006) and Kirk et al. (2008) placed Mycosphaerellaceae in Capnodiales, and hence Mycosphaerellales was treated as a synonym of Capnodiales. Phylogenetic analyses of a concatenated large subunit (LSU), tef1-α, and rpb2 sequence dataset conducted by Abdollahzadeh et al. (2020) revealed that Mycosphaerellales represents a robust clade, accommodating eight families, viz., Cystocoleaceae, Dissoconiaceae, Extremaceae, Mycosphaerellaceae, Neodevriesiaceae, Phaeothecoidiellaceae, Schizothyriaceae, and Teratosphaeriaceae. Hence, Abdollahzadeh et al. (2020) resurrected the order Mycosphaerellales as distinct from Capnodiales and also provided an amended description for the order.
Family Phaeothecoidiellaceae, a member of Mycosphaerellales, was introduced by Hongsanan et al. (2017) to accommodate the genera Chaetothyrina, Houjia, and Phaeothecoidiella. Members of this family are well-known as sooty blotch/flyspeck fungi which were frequently found as epiphytes or pathogens on fruits, leaves, and stems (Hongsanan et al., 2017; Hongsanan et al., 2020b). Subsequently, Zeng et al. (2018) introduced a new genus Translucidithyrium to this family. Hongsanan et al. (2020b) updated the taxonomic status of Phaeothecoidiellaceae and treated Nowamycetaceae as a synonym of Phaeothecoidiellaceae by transferring Nowamyces to Phaeothecoidiellaceae. Therefore, Hongsanan et al. (2020b) accepted five genera in Phaeothecoidiellaceae, while Wijayawardene et al. (2022) listed nine genera in this family, viz., Chaetothyrina, Exopassalora, Houjia, Neochaetothyrina, Nowamyces, Phaeothecoidiella, Rivilata, Sporidesmajora, and Translucidithyrium.
Exopassalora was established by Videira et al. (2017) to accommodate a single species, previously described as Passalora zambiae (Crous et al., 2004). The genus is characterized by irregularly branched, septate mycelium, composed of brown hyphae, with dark brown chlamydospore-like hyphal swellings; medium brown, smooth, simple or branched conidiophore-bearing terminal and intercalary, subcylindrical, pale to medium brown sympodially proliferating, polyblastic, conidiogenous cells, with darkened, conspicuous conidiogenous loci, and medium brown, smooth, narrowly ellipsoidal, tapering to subtruncate conidia with thickened and darkened hila and produced in simple or branched chains (Videira et al., 2017). According to Videira et al. (2017), Exopassalora zambiae formed a clade with Exopassalora sp. CBS 118964 in Phaeothecoidiellaceae and was phylogenetically distant from other Passalora species in Mycosphaerellaceae. Videira et al. (2017) reassigned most Passalora sensu lata into new genera.
Repetophragma, typified by R. biseptatum (≡Sporidesmium biseptatum), was introduced by Subramanian (1992), to accommodate Sporidesmium-like species with holoblastic, annellidic, percurrently proliferating conidiogenous cells and phragmoseptate conidia and initially included nine species previously known in Sporidesmium. Subsequently, many species were accommodated in Repetophragma based solely on morphological characterizations (McKenzie, 1995; Mena-Portales et al., 2000; Wu and Zhuang, 2005; Castañeda-Ruiz et al., 2006; Marincowitz et al., 2008; Silvera-Simón et al., 2009; Castañeda-Ruiz et al., 2011; Rambelli et al., 2011; Castañeda-Ruiz et al., 2013; Ma et al., 2014; Buyck et al., 2017; Wang et al., 2017; Ai et al., 2019). Even though, there are 40 species epithets available for Repetophragma in Index Fungorum (2023), most of them lack molecular data to clarify their phylogenetic placements. Only four species, namely, R. goidanichii, R. inflatum, R. ontariense, and R. zygopetali, have molecular data available in GenBank. Of these, R. ontariense was treated as a synonym of Vargamyces aquaticus (Amniculicolaceae, Pleosporales) by Hernández-Restrepo et al. (2017), whereas R. goidanichii was placed in Venturiaceae (Venturiales, Dothideomycetes) and R. inflatum was placed in Xylariales incertae sedis (Hernández-Restrepo et al., 2017). This concurs with the phylogenetic studies conducted by Shenoy et al. (2006). Unfortunately, the genus has not yet been clarified in terms of its generic placement based on the molecular data of the type species, R. biseptatum. Hence, the phylogenetic affinity of Repetophragma sensu stricto remains uncertain pending further study.
Stomiopeltis was introduced by Theissen (1914) to initially accommodate a single species S. aspersa which was collected on the leaves of Laurus sp. in India. Subsequently, many species—mostly in the 19th century—that lacked molecular data to clarify their phylogenetic placement were included in the genus (Luttrell, 1946; Müller and von Arx, 1962; Index Fungorum, 2023). Stomiopeltis has been reported as a pathogen causing sooty blotch/flyspeck disease but has also been found as a saprobe on fruits (Mayfield et al., 2013; Ajitomi et al., 2017; Jayasiri et al., 2019; Batzer et al., 2022). The genus is scarcely known, and only a few species have molecular data available in GenBank (Crous et al., 2019; Jayasiri et al., 2019; Renard et al., 2020). Most Stomiopeltis-like isolates were treated as Stomiopeltis sp. (Ajitomi et al., 2017; Batzer et al., 2022). An updated taxonomic treatment of Stomiopeltis was carried out by Zeng et al. (2019) who treated the genus in Capnodiales incertae sedis, and this was followed by Hongsanan et al. (2020b) and Wijayawardene et al. (2022). Whereas Renard et al. (2020) demonstrated that Stomiopeltis is polyphyletic, forming clades within the orders Microthyriales and Venturiales. Since the type species of Stomiopeltis, S. aspersa, has not yet been sequenced, the phylogenetic status of Stomiopeltis remains doubtful and also pending further study.
Several taxonomic studies of Dothideomycetes in Yunnan have been published in the past few years (Tibpromma et al., 2018; Phookamsak et al., 2019; Dong et al., 2020; Hyde et al., 2020; Li et al., 2020; Mortimer et al., 2021; Wanasinghe et al., 2021; Yang et al., 2022). Even though these studies resulted in a substantial increase in the number of described microfungi in Yunnan, there is still a glaring knowledge gap in our understanding of the fungi in this region. The present study aims to introduce two novel genera, one novel species, and three new combinations of Phaeothecoidiellaceae in Yunnan, based on molecular phylogeny coupled with morphological characteristics.
2 Materials and methods
2.1 Sample collection, morphological examination, isolation, and preservation
The sample was collected from Xishuangbanna, Yunnan Province, China in 2021. The sample was stored in a plastic Ziploc bag and returned to the laboratory for observation and examination. Fungal fruiting bodies on host substrates were observed using an Olympus SZ61 series stereomicroscope. Micro-morphologies on squash-mount slides were observed and photographed using a Nikon ECLIPSE Ni-U compound microscope equipped with a Nikon DS-Ri2 camera. Congo red was used to stain the conidiomatal centrum for clarity of conidiophores and conidiogenous cells. Lacto-glycerol was added to preserve important morphological features on permanent slides and edges of the coverslip were sealed with nail polish. All morphological features were measured using Tarosoft (R) Image FrameWork version 0.9.7., and the photographic plate was processed using Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA).
Fungal pure culture was obtained by single spore isolation, according to the methods described in Senanayake et al. (2020). The germinated spores were transferred to freshly sterilized potato dextrose agar (PDA) and incubated under normal light at 20°C–25°C. Culture characteristics were observed and recorded after one- and four-week intervals. The specimen was deposited in the Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), China. Axenic living cultures were preserved at the collection of Rungtiwa Phookamsak housed at Honghe Center for Mountain Futures (RPC) and duplicated in the Culture Collection of the Herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (KUNCC) Kunming, China. Index Fungorum numbers are provided for the newly described taxa.
2.2 DNA extraction, PCR amplification, and sequencing
Fungal genomic DNA was extracted from mycelia that grow on PDA for four weeks by using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, Hangzhou, China) according to the manufacturer’s protocol. The conditions for the polymerase chain reaction (PCR) were determined using the primer pairs LR0R/LR5 (Vilgalys and Hester, 1990) to amplify the 28S large subunit region (LSU), and ITS4/ITS5 (White et al., 1990) to amplify the internal transcribed spacer region (ITS: ITS1-5.8S-ITS2). The amplification of ITS and LSU was carried out with setting times and temperatures for the initialization, denaturation, annealing, and final extension periods following Phookamsak et al. (2017).
The final PCR reaction component was 25 µl, containing 12.5 μl Master Mix (mixture of EasyTaqTM DNA Polymerase, dNTPs, and optimized buffer; Beijing TransGen Biotech Co., Ltd., Chaoyang District, Beijing, China), 8.5 µl of double-distilled water (ddH2O), 1 μl of each forward and reverse primer (10 μm), and 2 μl DNA template. The PCR products were sent to TsingKe Biological Technology, Kunming City, Yunnan Province, China, for purification and Sanger sequencing. The consensus sequences of the newly generated strains are available to the scientific community via submission to GenBank.
2.3 Sequence alignment and phylogenetic analyses
The chromatograms of sequence results were checked, manually edited, trimmed, and assembled into consensus sequences using SeqMan Pro version 11.1.0 (DNASTAR, Inc. Madison, WI, USA). The consensus sequences of the newly generated strains were blasted using the nucleotide BLAST search tool on the NCBI website to search for closely related species in the GenBank database. Sequence data obtained from this study, the closely related species from nucleotide BLAST search, and previous studies were downloaded from GenBank to supplement the datasets (Table 1). The dataset was prepared to generate the phylogenetic trees for taxa in Mycosphaerellales. Each sequence dataset was aligned using MAFFT (Katoh et al., 2019) and checked manually in Bioedit (Hall, 2004). Maximum likelihood (ML) and Bayesian analysis (BI) were conducted based on the individual datasets.
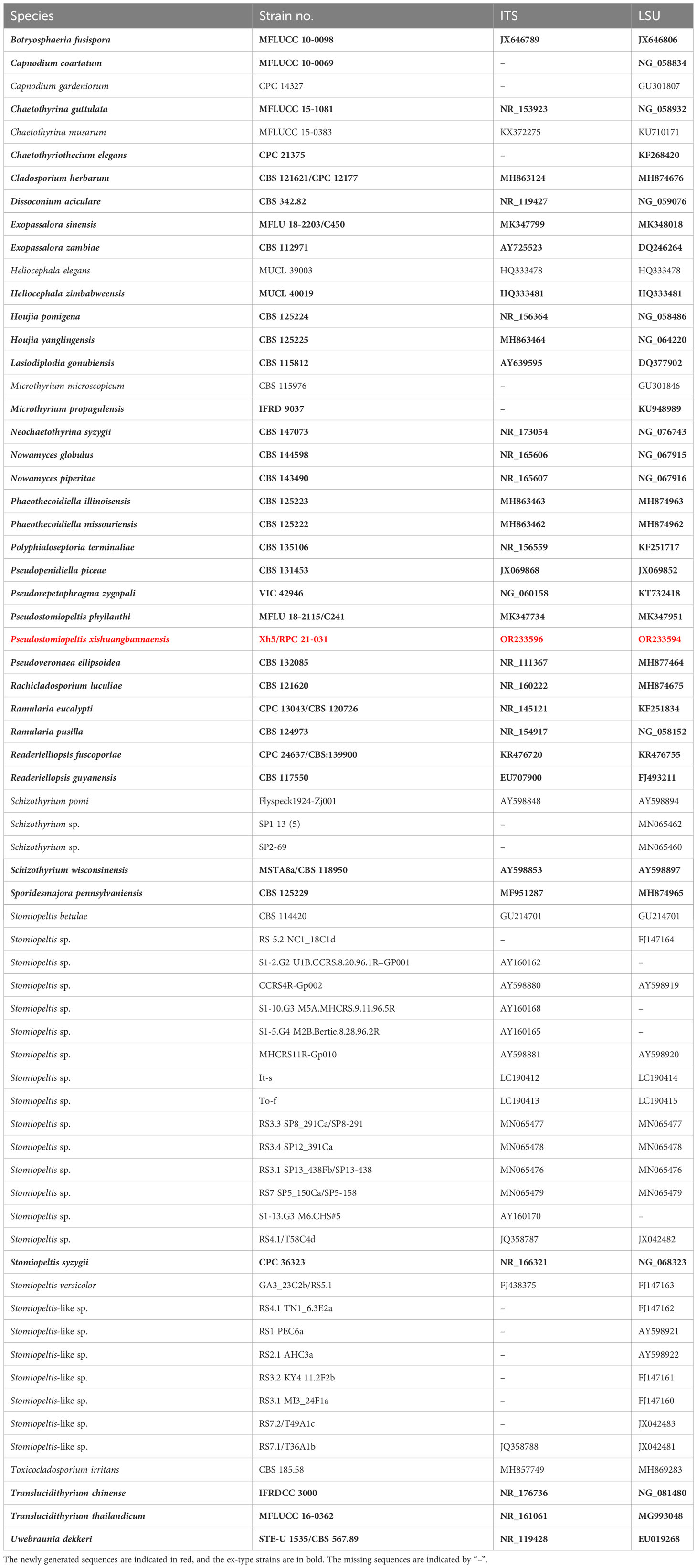
Table 1 Species details and GenBank accession numbers used in the phylogenetic analysis of Phaeothecoidiellaceae (Mycosphaerellales) and other related families and orders.
Maximum likelihood analyses were performed in RAxML-HPC v.8 on the XSEDE (8.2.12) tool in the online web portal CIPRES Science Gateway v. 3.3 using default settings but following adjustments with 1,000 bootstrap replications. The BI analyses were conducted via the same web portal as in ML, with two different runs, and six chains were executed. The initial 25% of sample trees were treated as burn-in and discarded. The trees were visualized using Figtree v. 1.4.0 (Rambaut, 2014) and edited in Adobe Illustrator version 20.0.0.
3 Results
3.1 Molecular phylogeny
The phylogenetic tree which represented novel taxa in Phaeothecoidiellaceae was constructed using sequence data from ITS and LSU genes. A total of 66 strains of taxa in the family Phaeothecoidiellaceae, representative of other related families in Capnodiales, Cladosporiales, Microthyriales, and Mycosphaerellales, were included, with two strains of Botryosphaeria fusispora (MFLUCC 10-0098) and Lasiodiplodia gonubiensis (CBS 115812) as the outgroup. The aligned dataset contained 1,808 characters, including gaps. The best-scoring RAxML tree was selected to represent the relationships among taxa, with a final likelihood value of -17,660.527024. The matrix contained 1,121 distinct alignment patterns, with estimated base frequencies of A = 0.240165, C = 0.248028, G = 0.296115, T = 0.215692; substitution rates AC = 1.209829, AG = 1.862907, AT = 1.368661, CG = 1.131373, CT = 4.503418, GT = 1.000000; and gamma distribution shape parameter α = 0.495210 (Figure 1). For BI analysis, GTR + I + G was selected as the best-fit model by AIC in MrModeltest for each gene (ITS and LSU). Six simultaneous Markov chains were run for 3,000,000 generations, and trees were sampled every 100 generations. The first 25% of trees were discarded as the burn-in phase of the analyses, and the remaining trees were used for calculating posterior probabilities in the majority rule consensus tree (the critical value for the topological convergence diagnostic is 0.01), of which the final average standard deviation of split frequencies at the end of total MCMC generations was 0.009273.
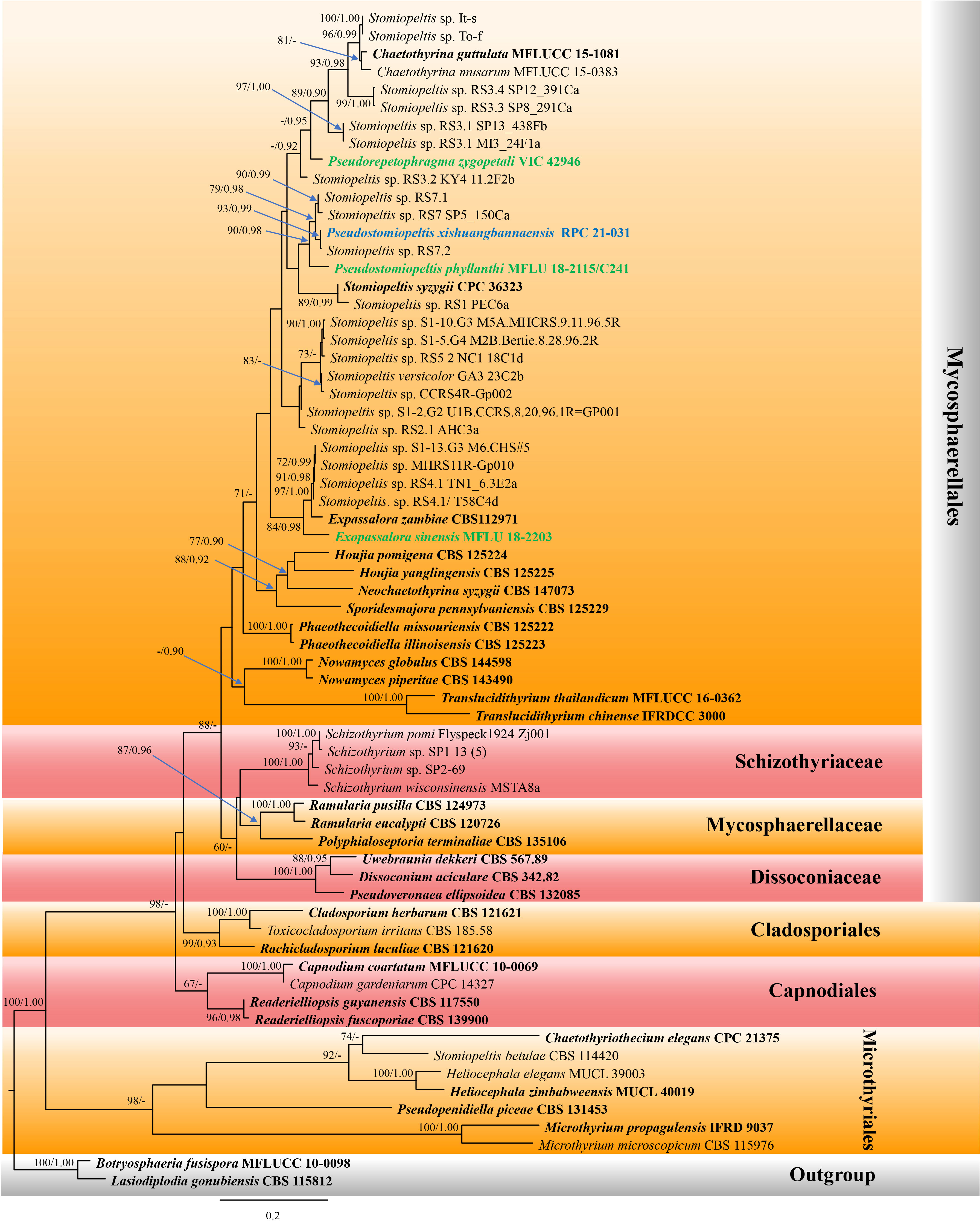
Figure 1 Phylogram of the best-scoring maximum likelihood (ML) consensus tree based on a combined dataset (ITS and LSU) of Pseudostomiopeltis. The new species is indicated in blue, and the new combination species are indicated in green. Isolates from type materials are in bold. The ML ultrafast bootstrap and Bayesian PP values greater than 70% and 0.95 are shown at the nodes. The tree is rooted with Botryosphaeria fusispora (MFLUCC 10-0098) and Lasiodiplodia gonubiensis (CBS 115812).
Our new isolate shared the branch length with Stomiopeltis sp. (RS7.2), with significant support (93% ML, 0.99 BYPP, Figure 1), and formed a well-resolved subclade with Stomiopeltis sp. (strains RS7 SP5_150Ca and RS7.1) within Phaeothecoidiellaceae (Mycosphaerellales). According to Renard et al. (2020), Stomiopeltis is polyphyletic. Moreover, in comparison with the type of Stomiopeltis examined by Zeng et al. (2018), our new isolate is morphologically distinguishable from S. aspersa, the generic type of Stomiopeltis, based on the upper wall arrangement. Hence, the new genus Pseudostomiopeltis is introduced herein to accommodate the new species, Ps. xishuangbannaensis, and Stomiopeltis sensu lato in this subclade. Furthermore, Repetophragma zygopetali (VIC 42946, ex-type strain) formed an independent lineage basal to the genus Chaetothyrina and Stomiopeltis spp. (strains It-s, To-f, RS3.3 SP8_291Ca, RS3.4 SP12_391Ca, RS3.1 SP13_438Fb, and RS3.1 MI3_24F1a). According to Shenoy et al. (2006), Repetophragma has shown to be polyphyletic, affiliating in different families and orders of Dothideomycetes and Sordariomycetes. Moreover, the generic type of Repetophragma, R. biseptatum, lacks molecular data to clarify its phylogenetic placement. Hence, the new genus Pseudorepetophragma is introduced herein to accommodate R. zygopetali as Pseudorepetophragma zygopetali comb. nov. based on phylogenetic evidence and morphological distinction in the conidiogenous proliferations, whereas Stomiopeltis sinensis clustered with Exopassalora zambiae (CBS 125225, ex-type strain) and Stomiopeltis spp. (strains S1-13.G3 M6.CHS#5, MHCRS11R-Gp010, RS4.1 TN1_6.3E2a, and RS4.1/T58C4d) with significant support (84% ML, 0.98 BYPP; Figure 1). Thus, S. sinensis is also transferred to Exopassalora as E. sinensis comb. nov.
3.2 Taxonomy
Exopassalora sinensis (Jayasiri, E.B.G. Jones and K.D. Hyde) Phookamsak and Hongsanan, comb. nov.
Index Fungorum number: IF 900623
≡ Stomiopeltis sinensis Jayasiri, E.B.G. Jones and K.D. Hyde, in Jayasiri, Hyde, Jones, McKenzie, Jeewon, Phillips, Bhat, Wanasinghe, Liu, Lu, Kang, Xu and Karunarathna, Mycosphere 10(1): 129 (2019)
Detailed description: See Jayasiri et al. (2019).
Notes: Exopassalora sinensis was first introduced as a saprobic fungus on decaying fruit pericarp of Harpephyllum in Yunnan, China (Jayasiri et al., 2019). The species is represented by its sexual morph having superficial, rounded thyriothecia, dark brown, textura angularis cell-layered peridium, 4-spored, fissitunicate, oblong to subglobose asci embedded in filiform, unbranched, septate pseudoparaphyses, and hyaline, obovoid to ellipsoid, 1-septate ascospores (Jayasiri et al., 2019). Phylogenetic analyses (Figure 1) demonstrated that the species formed a well-resolved clade with Exopassalora zambiae (CBS 125225, ex-type strain) and unnamed Stomiopeltis spp. within Phaeothecoidiellaceae. Morphologically, the species could not be compared with E. zambiae due to the representative different morphs. Therefore, the species is transferred to Exopassalora herein based on phylogenetic evidence.
Pseudorepetophragma Phookamsak, Bhat and Hongsanan, gen. nov.
Index Fungorum number: IF 900624
Etymology: The generic epithet “Pseudorepetophragma” refers to the genus that is morphologically resembling Repetophragma.
Fungus associated with sooty blotch on living leaves of Zygopetalum mackayi (Orchidaceae). Sexual morph: Undetermined. Asexual morph: Colonies effuse, black, forming a dark mycelial mat. Mycelium superficial, composed of septate, branched, dark brown hyphae. Conidiophores macronematous, mononematous, simple, erect, septate, straight, or slightly narrow toward the apex, percurrently proliferating, septate, dark brown to brown. Conidiogenous cells monoblastic, enteroblastic, integrated, terminal, conspicuously percurrent, with apices remaining wavy or uneven after each conidial secession. Conidia acrogenous, solitary, brown, dark brown, cylindrical to obclavate, with truncate base, septate, thick, and smooth-walled. Conidial secession schizolytic (adapted from Buyck et al., 2017).
Type species: Pseudorepetophragma zygopetali (O.L. Pereira, Meir. Silva and R.F. Castañeda) Phookamsak, Bhat and Hongsanan
Notes: Subramanian (1992) established the genus Repetophragma and accommodated some species previously described as Sporidesmium. The genus is characterized by macronematous, brown, solitary, septate conidiophores, with monoblastic, enteroblastic, integrated, terminal, percurrently proliferating, and distinctly annellidic conidiogenous cells and acrogenous, solitary, dry, euseptate, conidia with a truncate base (Subramanian, 1992; Castañeda-Ruiz et al., 2011). Castañeda-Ruiz et al. (2011) re-illustrated the genus by providing the synopsis table of morphological features and key to the species of Repetophragma. Based on this comprehensive study, Castañeda-Ruiz et al. (2011) introduced a novel species, R. paracambrense, and 12 new combinations in the genus. Of these, most species were previously known in Endophragmiella and Sporidesmium. Considering the species of Repetophragma, most of the accepted species have annellidic, percurrent proliferations of the conidiogenous cells bearing euseptate conidia with apically rounded, well-defined, and without rostrate or appendiculate apical cell (Iturriaga et al., 2008; Silvera-Simón et al., 2009). This led to the inclusion of morphologically diverse species in Repetophragma. The phylogenetic analyses inferred by Shenoy et al. (2006) and Hernández-Restrepo et al. (2017) also revealed the status of some Repetophragma to be polyphyletic. Unfortunately, the phylogenetic affinity of Repetophragma is uncertain due to the lack of molecular data for the type species of Repetophragma. Furthermore, R. zygopetali formed an independent lineage within the Phaeothecoidiellaceae. Morphologically, R. zygopetali is similar to R. biseptatum in having monoblastic, enteroblastic, conspicuously percurrent conidiogenous cells. However, R. zygopetali can be distinguished from the rest of Repetophragma in having integrated, monoblastic, conspicuously percurrent but irregularly distanced conidiogenous cells with wavy or uneven apices after each conidial secession [Figure 29a in Buyck et al. (2017)] rather than equidistantly laid annellidic conidiogenous cells with even apices after each conidial secession (Subramanian, 1992; Seifert et al., 2011). Based on the morphological differences in the conidiogenous cells and phylogenetic analyses, we introduce the new genus Pseudorepetophragma to accommodate R. zygopetali as P. zygopetali comb. nov.
Pseudorepetophragma zygopetali (O.L. Pereira, Meir. Silva and R.F. Castañeda) Phookamsak, Bhat and Hongsanan, comb. nov.
Index Fungorum number: IF 900625
≡ Repetophragma zygopetali O.L. Pereira, Meir. Silva and R.F. Castañeda, in Buyck, Duhem, Das, Jayawardena, Niveiro, Pereira, Prasher, Adhikari, Alberto, Bulgakov, Castañeda-Ruíz, Hembrom, Hyde, Lewis, Michlig, Nuytinck, Parihar, Popoff, Ramirez, Da Silva, Verma and Hofstetter, Cryptog. Mycol. 38(1): 135 (2017)
Detailed description: See Buyck et al. (2017).
Notes: Pseudorepetophragma zygopetali was first introduced as Repetophragma zygopetali by Buyck et al. (2017) due to its morphological resemblance with R. dennisii, but differing in the size of conidiophores and conidia (Castañeda-Ruiz et al., 2011; Buyck et al., 2017). The species was reported as a sooty blotch fungus that occurred on Zygopetalum mackayi in Brazil, while other species of Repetophragma have been mostly reported as hyperparasites or saprobes (Ellis, 1963; Castañeda-Ruiz et al., 2011; Index Fungorun, 2023). Its life mode is fitted well with the genera in Phaeothecoidiellaceae, and this was confirmed by phylogenetic evidence.
Pseudostomiopeltis Phookamsak and Hongsanan, gen. nov.
Index Fungorum number: IF 900626
Etymology: The generic epithet “Pseudostomiopeltis” refers to the genus that resembles Stomiopeltis.
Epiphytic or saprobic on leaves and fruits. Sexual morph: Mycelium absence. Ascomata thyriothecial, black, solitary, gregarious, superficial, rounded, easily removed from the host surface. Upper wall composed of a thin layer of neatly arranged dark cells of textura angularis. Hamathecium lacking pseudoparaphyses. Asci 4-spored, bitunicate, fissitunicate, oblong to subglobose, with a minute pedicel. Ascospores uniseriate, hyaline, asymmetric, obovoid to ellipsoid, 1-septate, constricted at the septum, upper cell slightly broader than the lower cell (Adopted from Jayasiri et al., 2019). Asexual morph: Mycelium absence. Conidiomata thyriothecial, superficial, scattered, or in a small group, hemispherical, dimidiate-scutate, uniloculate, with a central, pore-like ostiole. Upper wall composed of a thin layer, of brown, radiating cells, with loosely irregular lobed cells at the margin. Peridium composed of 1–2 strata of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells arising from the innermost wall cells of the conidiomata, hyaline, enteroblastic, phialidic, lageniform to ampulliform, determinate, discrete, smooth-walled, with minute channel and collarette. Conidia solitary, hyaline, ellipsoidal to oblong, slightly truncate at the base, with obtuse apex, aseptate, smooth-walled, occasionally with attached conidiogenous cell. Sporulation in vitro forming dense, brown, compact hyphae on OA medium, with cream spore masses. Conidiophores hyaline, cylindrical to subcylindrical, septate, branched or unbranched, smooth-walled. Conidiogenous cells hyaline, enteroblastic, phialidic, terminal and intercalary, cylindrical to subcylindrical, aseptate, smooth-walled, with minute channel and collarette. Conidia solitary, hyaline, subglobose to ellipsoidal to oblong, with obtuse ends, aseptate, smooth-walled.
Type species: Pseudostomiopeltis xishuangbannaensis Phookamsak, Hongsanan, Wanas. and Bhat
Notes: Luttrell (1946) re-circumscribed the genus Stomiopeltis based on morphological studies and divided the species of Stomiopeltis into two groups based on the difference in the types of upper wall cell arrangements. The first group included the type species (S. aspersa) that has non-radiating upper wall cells, composed of disorderly arranged, irregularly lobed pseudoparenchymatous cells, while the second group has radiating upper wall cells, somewhat obscured by the curving and twisting of the radiating hyphae and by the irregularly lobed cells, which may be termed as “meandering plectenchyma”. Luttrell (1946) mentioned that the second group should be placed in Microthyriaceae. The phylogenetic analyses conducted by Renard et al. (2020) also showed that Stomiopeltis is polyphyletic due to S. betulae forming a clade within Microthyriales, while two Stomiopeltis-like species formed a clade with Tothia fuscella in Venturiales. The present phylogenetic analyses of a concatenated ITS and LSU sequence dataset demonstrated that our new isolate formed a well-resolved subclade with other Stomiopeltis sensu lato in Phaeothecoidiellaceae, Mycosphaerellales. Besides, the type species of Stomiopeltis, S. aspersa, has not yet been sequenced, and hence the phylogenetic affinity of Stomiopeltis sensu stricto is still uncertain.
Zeng et al. (2018) re-examined the holotype of Stomiopeltis aspersa and provided an updated morphological description that is characterized by superficial, brown, reticulate hyphae, flattened, circular, brown, thyriothecia with an irregular central ostiole. The upper wall comprises brown, meandrous, compact hyphae, lacking a basal plate. Asci are 8-spored, ellipsoidal, short-pedicellate, with an ocular chamber and ascospores are overlapping 2–3-seriate, hyaline, cylindrical, 1-septate, not constricted at the septum, with the upper cell shorter and broader than the lower cell. Morphologically, our new isolate could not be compared with the type of S. aspersa because they form different morphs. However, the new isolate is clearly distinguished from S. aspersa by the absence of superficial, brown, reticulate hyphae that penetrate the host and form hemispherical, dimidiate-scutate thyriothecia. Additionally, the upper wall of the thyriothecia radiates, and the cells at the margin are loosely and irregularly lobed, while S. aspersa has a non-radiating upper wall composed of sinuous, irregularly lobed cells [Figures 20c, d in Zeng et al. (2018)]. Furthermore, S. phyllanthi which formed a clade with our new isolate, is also different from S. aspersa in lacking superficial, reticulate hyphae on the host and pseudoparaphyses (Jayasiri et al., 2019). Based on phylogenetic evidence and morphological distinctiveness with the type of Stomiopeltis, we introduced the new genus Pseudostomiopeltis to accommodate the new species, Ps. xishuangbannaensis, while Stomiopeltis phyllanthi is also transferred to the new genus as Pseudostomiopeltis phyllanthi comb. nov.
Pseudostomiopeltis phyllanthi (Jayasiri, E.B.G. Jones and K.D. Hyde) Phookamsak and Hongsanan, comb. nov.
Index Fungorum number: IF 900639
≡ Stomiopeltis phyllanthi Jayasiri, E.B.G. Jones and K.D. Hyde, in Jayasiri, Hyde, Jones, McKenzie, Jeewon, Phillips, Bhat, Wanasinghe, Liu, Lu, Kang, Xu and Karunarathna, Mycosphere 10(1): 131 (2019)
Detailed description: See Jayasiri et al. (2019).
Notes: Pseudostomiopeltis phyllanthi was first introduced as Stomiopeltis phyllanthi by Jayasiri et al. (2019) which was found as a saprobe on the fruits of Phyllanthus emblica. The species formed a sexual morph and is characterized by black, superficial, rounded thyriothecia, with the upper wall neatly lined by dark cells of textura angularis, lacking pseudoparaphyses, with 4-spored, fissitunicate, oblong to subglobose asci, and hyaline, obovoid to ellipsoid, 1-septate ascospores (Jayasiri et al., 2019). The present phylogenetic analyses of a concatenated ITS and LSU sequence dataset demonstrated that the species formed a separate branch and is basal to the clade Pseudostomiopeltis with significant support (90% ML, 0.98 PP; Figure 1). Therefore, we transferred S. phyllanthi to Pseudostomiopeltis as Ps. phyllanthi comb. nov.
Pseudostomiopeltis xishuangbannaensis Phookamsak, Hongsanan, Wanas. and Bhat, sp. nov., Figure 2

Figure 2 Pseudostomiopeltis xishuangbannaensis (KUN-HKAS 129044, holotype). (A) The appearance of conidiomata on host substrate. (B) Upper view of conidioma. (C) Vertical section of conidioma. (D) Section through the peridium. (E–G) Conidiogenous cells bearing conidia, arising from the inner cavities (note: G = stained with Congo red). (H–L) Conidia. (M) Sporulation on OA with cream conidial masses. (N) Conidiophores and conidiogenous cells in vitro. (O) Conidia sporulated in vitro. Scale bars: (B, C) = 20 μm, (D–L, N, O) = 5 μm.
Index Fungorum number: IF 900640
Etymology: The specific epithet “xishuangbannaensis” refers to the locality, Xishuangbanna Dai Autonomous Prefecture, Yunnan, China, where the holotype was collected.
Holotype: KUN-HKAS 129044
Epiphytic or saprobic on dead leaves of an unidentified dicot, hypophyllous, visible as small, circular, black dots, easily removed from the host surface. Sexual morph: Undetermined. Asexual morph: Conidiomata 85–120 µm high, 70–135 µm, thyriothecial, scattered, or in a small group, hemispherical, dimidiate-scutate, uni-loculate, with a central, pore-like ostiole. Upper wall composed of a thin layer, of brown, radiating cells, with loosely irregular lobed cells at the margin. Peridium 1.5–4 µm wide, composed of 1–2 strata of textura angularis. Conidiophores reduced to the conidiogenous cells. Conidiogenous cells 2.5–5 × 1.5–3 µm (x̄ = 3.9 × 2.3, n = 30), arising from the innermost wall cells of the conidiomata, hyaline, enteroblastic, phialidic, lageniform to ampulliform, determinate, discrete, smooth-walled, with minute channel and collarette. Conidia 7–9 × 2–4 µm (x̄ = 8.1 × 3.2, n = 30), solitary, hyaline, ellipsoidal to oblong, slightly truncate at the base, with obtuse apex, aseptate, smooth-walled, occasionally with attached conidiogenous cell.
Culture characteristics: Conidia germinated on PDA within 24 h. Colonies on OA reaching 25–28 mm in diam. after two weeks at room temperature (15°C–20°C). Colonies dense, circular, flattened to slightly raised, surface smooth with an entire edge, fairly fluffy to floccose; from above gray at the margin, becoming dark gray toward the center, with cream conidial masses; from below dark gray to black at the margin, white-gray at the middle, dark gray to black at the center, radiating. Sporulation in OA after two weeks, forming dense, brown, compact hyphae on OA medium, with cream spore masses. Mycelium 1.5–3 µm thick, brown, branched, septate, with compact hyphae. Conidiophores 10–30 × 1–3 µm (x̄ = 13.9 × 2.2, n = 30), hyaline, cylindrical to subcylindrical, septate, branched or unbranched, smooth-walled. Conidiogenous cells (5–)7–9 × 1–3 µm (x̄ = 7.8 × 1.9, n = 30), hyaline, enteroblastic, phialidic, terminal, cylindrical to subcylindrical, aseptate, smooth-walled, with minute channel and collarette. Conidia 2.5–4 × 1–2 µm (x̄ = 3 × 1.7, n = 30), solitary, hyaline, subglobose to ellipsoidal to oblong, with obtuse ends, aseptate, smooth-walled.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Mengla County, Menglun, Xishuangbanna Tropical Botanical Garden, on dead leaves of an unidentified dicot, 11 January 2021, D.N. Wanasinghe, Xh5 (KUN-HKAS 129044, holotype), ex-type living culture, RPC 21-031 = KUNCC.
Notes: The NCBI nucleotide BLAST search of ITS sequence indicated that Pseudostomiopeltis xishuangbannaensis (RPC 21-031) is similar to Stomiopeltis sp. T49A1c with 99.82% similarity (identities = 557/558 with no gap), Stomiopeltis sp. T36A1b with 97.32% similarity (identities = 545/560 with 5 gaps), Stomiopeltis sp. RS7 with 96.25% similarity (identities = 539/560 with 5 gaps), and S. phyllanthi MFLU 18-2115 with 92.36% similarity (identities = 447/484 with 9 gaps). Similarly, the NCBI nucleotide BLAST search of LSU sequence indicated that Ps. xishuangbannaensis (RPC 21-031) is similar to Cf. Stomiopeltis sp. RS7.2 with 100% similarity (identities = 765/765, with no gap), Cf. Stomiopeltis sp. RS7.1 with 99.87% similarity (identities = 764/765, with no gap), and Stomiopeltis sp. RS7 with 99.76% similarity (identities = 834/836, with 1 gap), while the LSU of S. phyllanthi MFLU 18-2115 is misidentified. The closest hit using the LSU sequence of S. phyllanthi is Meyerozyma guilliermondii culture CBS:8105 with 100% similarity.
The phylogenetic analyses of a concatenated ITS and LSU sequence dataset (Figure 1) revealed that Pseudostomiopeltis xishuangbannaensis (RPC 21-031) has a close relationship with Ps. phyllanthi MFLU 18-2115 (≡ Stomiopeltis phyllanthi). A nucleotide pairwise comparison of ITS sequence indicated that Ps. xishuangbannaensis (RPC 21-031) differs from Ps. phyllanthi (MFLU 18-2115) in 38/485 bp (7.8%). Morphologically, Ps. xishuangbannaensis (RPC 21-031) could not be compared with Ps. phyllanthi as they formed different morphs.
4 Discussion
In the present study, two new genera, one new species, and three new combinations are described and illustrated based on morphology and phylogeny. The new genus, Pseudostomiopeltis, is introduced to accommodate the type species, Ps. xishuangbannaensis sp. nov. and Ps. phyllanthi com. nov. This genus belongs to Phaeothecoidiellaceae (Mycosphaerellales). The members of Pseudostomiopeltis share certain similar characteristics with Stomiopeltis which has been classified as a genus incertae sedis in Capnodiales by Wijayawardene et al. (2022). Stomiopeltis is a polyphyletic genus, and the sequence data of the type species are not available (Renard et al., 2020). It is interesting to note that the morphology of Stomiopeltis shows remarkable similarities to the species in Micropeltidaceae (Micropeltidales, Lecanoromycetes). However, molecular analysis has revealed that the majority of Stomiopeltis strains are classified in Phaeothecoidiellaceae (Mycosphaerellales), and most of these strains have not yet been identified at the species level. In the phylogenetic tree constructed using ITS and LSU sequence data (Figure 1), our new isolate clustered together with Cf. Stomiopeltis sp. RS7.2. Since the strain Cf. Stomiopeltis sp. RS7.2 has not been identified as its morphology is not available, we thus establish our strain as a new species. Whereas, S. phyllanthi is grouped with the clade of Pseudostomiopeltis with significant support (90% ML, 0.98 BYPP; Figure 1). Therefore, we transfer S. phyllanthi to Pseudostomiopeltis based on phylogenetic evidence, although the upper wall structure of S. phyllanthi could not be determined in this study.
Stomiopeltis syzygii is morphologically similar to Pseudostomiopeltis xishuangbannaensis sporulated in vitro in having subcylindrical, septate, branched or unbranched conidiophores, terminal and intercalary, phialidic, subcylindrical, hyaline conidiogenous cells, and hyaline, smooth, aseptate conidia (Crous et al., 2019). However, S. syzygii has slightly larger [(5–)8–10(–12) × 1.5 µm vs. 2.5–4 × 1–2 µm], subcylindrical conidia, whereas Ps. xishuangbannaensis sporulated in vitro has subglobose to ellipsoidal or oblong conidia. The phylogenetic analyses demonstrated that S. syzygii (CPC 36323, ex-type strain) formed a subclade with Stomiopeltis sp. RS1 PEC6a basal to Pseudostomiopeltis with low support. Hence, the species is tentatively excluded from Pseudostomiopeltis until taxon samplings are increased, providing a better phylogenetic resolution of Pseudostomiopeltis with uncertain Stomiopeltis spp. within Phaeothecoidiellaceae. It is notable that S. betulae clustered within Microthyriales. However, the phylogenetic placements of Stomiopeltis and its relationships with other genera remain unclear. Further sequence data and morphological studies are needed to confirm the placement of this genus.
Besides the phylogenetic investigation of Stomiopeltis sensu lato in Phaeothecoidiellaceae, Repetophragma zygopetali formed an independent lineage within Phaeothecoidiellaceae in the present study. Buyck et al. (2017) treated R. zygopetali in Micropeltidaceae (Micropeltidales, Lecanoromycetes) based on phylogenetic evidence that R. zygopetali formed a basal clade with Sporidesmajora pennsylvaniensis (CPC 16112), Houjia pomigena (CPC 16109), and H. yanglingensis (CPC 16110, CPC 16111, CPC 16113, and CPC 16114) in their analysis. Unfortunately, most Repetophragma species were identified based solely on morphological characteristics that distinguished them from Sporidesmium in having conidiophores with terminal annellations, suggestive of repeated percurrent proliferative conidiogenous cells (Subramanian, 1992). Whereas Sporidesmium species have non-hyphopodiate mycelium, simple non-proliferating conidiophores, or with irregularly distanced percurrent proliferations (Subramanian, 1992). Presently, Sporidesmium was accommodated in its own family (Sporidesmiaceae, Sporidesmiales, Sordariomycetes), whereas the phylogenetic affinity of Repetophragma sensu stricto is uncertain. Based solely on morphology, the taxonomy of Repetophragma remains ambiguous. Molecular data of species in Repetophragma is urgently needed to verify their congeneric status within Repetophragma and also clarify the phylogenetic affinity of Repetophragma.
Exopassalora was established to accommodate Passalora zambiae (Crous et al., 2004). The species was isolated from leaf spots of Eucalyptus globulus in Zambia. Crous et al. (2004) classified the species into Passalora due to its being phylogenetically distinct from other Mycosphaerella spp. known from Eucalyptus. Later, Videira et al. (2017) introduced many novel genera to accommodate Passalora sensu lato, including Exopassalora, based on multigene phylogenetic evidence. The sexual morph of E. zambiae is known only for its asci and ascospores, prepared onto the slide (Crous et al., 2004). In the present phylogenetic analyses, Stomiopeltis sinensis formed a separate branch basal to Exopassalora. Hence, the species is transferred to Exopassalora, as E. sinensis comb. nov., based on phylogenetic evidence. Morphologically, E. sinensis could be only compared with E. zambiae in ascospore characters that are similar in having hyaline, 1-septate ascospores (Crous et al., 2004; Jayasiri et al., 2019). Moreover, E. zambiae was found on leaf spots of Eucalyptus globulus, while Stomiopeltis spp. clustered in the subclade of Exopassalora were found as sooty blotch and flyspeck fungi on apples and pears (Batzer, 2005; Ismail et al., 2016). In contrast, E. sinensis was isolated from decaying fruit pericarp of Harpephyllum sp. (wild plum). It is presumable that the species may occur on fresh fruits of Harpephyllum as parasites and continue living on dead fruits as a saprobe, similar to Pseudostomiopeltis xishuangbannaensis. However, the change in life mode may need further study for a better understanding of the pathogenic capabilities of these species.
Most genera of Phaeothecoidiellaceae were known as pathogenic fungi causing sooty blotch and flyspeck or leaf spot diseases on various hosts worldwide (Crous et al., 2004; Batzer, 2005; Yang et al., 2010; Ismail et al., 2016; Buyck et al., 2017; Hongsanan et al., 2017; Zeng et al., 2018; Crous et al., 2019; Hongsanan et al., 2020b; Crous et al., 2021). Species of Chaetothyrina, Houjia, Phaeothecoidiella, Sporidesmajora, Stomiopeltis-like spp., and Translucidithyrium have been reported as sooty blotch and flyspeck fungi, mainly occurring on apples and pears (Batzer, 2005; Yang et al., 2010; Ismail et al., 2016; Hongsanan et al., 2017; Zeng et al., 2018; Hongsanan et al., 2020b; Li et al., 2020), while species of Exopassalora and Nowamyces have been reported as fungi associated with leaf spot diseases (Crous et al., 2004; Crous et al., 2019). Neochaetothyrina, on the other hand, has been reported as a saprobe on Syzygium (Crous et al., 2021). In the present study, Pseudostomiopeltis phyllanthi and Ps. xishuangbannaensis were found as saprobes on fruits and leaves of dicots. However, Stomiopeltis-like spp. clustered with Pseudostomiopeltis were reported as sooty blotch and flyspeck fungi on apples (Mayfield et al., 2013). This leads to questioning that Pseudostomiopeltis may also be capable of causing sooty blotch and flyspeck disease.
Yunnan is a region known for its high biodiversity, serving as a critical habitat for numerous species. However, this region is under threat from habitat loss, climate change, and various human activities. Among these factors, climate change is a major threat to the biodiversity in Yunnan. As global temperatures continue to rise, the region may experience changes in precipitation patterns and temperature regimes that could potentially impact the distribution and survival of numerous species, including fungi. Despite these challenges, there are also opportunities for biodiversity conservation in Yunnan. Scientific research and monitoring play crucial roles in providing valuable information for the biodiversity conservation efforts in the region. The discovery of novel taxa highlights the rich fungal diversity in Yunnan and contributes to our understanding of the ecological roles of fungi in forest ecosystems. The description and documentation of these new taxa not only provide important information on fungi useful in future research but also enhance our knowledge of conservation efforts in the region. By expanding our understanding of the biodiversity of Yunnan, we can better protect and manage its unique ecosystems and ensure the long-term survival of its species.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession numbers OR233594 (LSU) and OR233596 (ITS).
Author contributions
SH and RP: conceptualization. SH and RP: data curation. SH and DS: formal analysis. SL, JX, IP, and NX: funding acquisition. SH, RP, DB, DW, NS, and DS: investigation. SH, RP, and DS: methodology. SH and NS: project administration. SL, JX, IP, and NX: supervision. SH, RP, DW, NS, and DS: writing—original draft. SH, RP, DB, DW, NS, IP, and DS: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research work was partially supported by Chiang Mai University (Grant Number: RG03/2566), and was also supported by the Project on Key Technology for Ecological Restoration and Green Development in Tropical Dry- Hot Valley, under the Yunnan Department of Sciences and Technology of China (grant no. 202202AE090091).
Acknowledgments
SH would like to express gratitude to Chiang Mai University for providing financial support and laboratory facilities. We acknowledge Shenzhen University and the Biology Experimental Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the facilities of molecular laboratory. Dr. Shaun Pennycook at Manaaki Whenua - Landcare Research, is thanked for help in naming the new fungal species. Beinn Purvis at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing. Chun-Fang Liao at Mae Fah Luang University, Thailand is thanked for helping with fungal isolation. Hongbo Jiang and Qinxian Li at Kunming Institute of Botany, Chinese Academy of Sciences are thanked for their general assistance. RP thanks the Yunnan Revitalization Talent Support Program "Young Talent" Project (grant no. YNWR-QNBJ-2020-120) for financial research support. DB gratefully acknowledges the financial support provided under the Distinguished Scientist Fellowship Programme (DSFP), at King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdollahzadeh, J., Groenewald, J. Z., Coetzee, M. P. A., Wingfield, M. J., Crous, P. W. (2020). Evolution of lifestyles in capnodiales. Stud. Mycol. 95, 381–414. doi: 10.1016/j.simyco.2020.02.004
Ai, C. C., Xia, J. W., Zhang, X. G., Ma, L. G. (2019). Repetophragma verrucosum sp. nov. from China. Mycotaxon 134 (3), 443–446. doi: 10.5248/134.443
Ajitomi, A., Takushi, T., Sato, T., Ooshiro, A., Yamashiro, M. (2017). First report of flyspeck of mango caused by Stomiopeltis sp. in Japan. J. Gen. Plant Pathol. 83, 299–303. doi: 10.1007/s10327-017-0726-7
Barr, M. E. (1979). A classification of loculoascomycetes. Mycologia 71, 935–995. doi: 10.1080/00275514.1979.12021099
Barr, M. E., Huhndorf, S. M. (2001). “Loculoascomycetes,” in The mycota VII, part A. Systematics and evolution. Eds. McLaughlin, D. J., McLaughlin, E. G., Lemke, P. A. (Berlin: Springer Verlag), 283–305.
Batzer, J. C. (2005). Sooty blotch and flyspeck on apple: expansion of the fungal complex, post-harvest removal and heterogeneity of apple canopy wetness and its impact on the outcome of a disease-warning system. Retrospec. Theses Dissertat., 1225. doi: 10.31274/RTD-180813-12083
Batzer, J. C., Prado, M. M., Svendsen, J. M., Gleason, M. L. (2022). Diversity of sooty blotch and flyspeck fungi on cider apples in Spain. PhytoFront 2, 289–306. doi: 10.1094/PHYTOFR-11-21-0074-R
Buyck, B., Duhem, B., Das, K., Jayawardena, R. S., Niveiro, N., Pereira, O. L., et al. (2017). Fungal biodiversity profiles 21–30. Cryptogam. Mycol. 38 (1), 101–146. doi: 10.7872/crym/v38.iss1.2017.101
Cai, J., Yu, W. B., Zhang, T., Wang, H., Li, D. Z. (2019) China’s biodiversity hotspots revisited: A treasure chest for plants in Revealing of the plant diversity in China’s biodiversity hotspots Eds. Cai, J., Yu, W.-B., Zhang, T., Li, D.-Z. doi: 10.3897/phytokeys.130.38417
Castañeda-Ruiz, R. F., Gusmão, L. F., Abarca, G. H., Saikawa, M. (2006). Some hyphomycetes from Brazil. Two new species of Brachydesmiella, two new combinations for Repetophragma, and new records. Mycotaxon 95, 261–270.
Castañeda-Ruiz, R. F., Heredia, G., Arias, R. M., McKenzie, E. H. C., Hyde, K. D., Stadler, M., et al. (2011). A new species and re-disposed taxa in Repetophragma. Mycosphere 2 (3), 273–289.
Castañeda-Ruiz, R. F., Hernández-Restrepo, M., Gené, J., Granados, M., Guarro, J. (2013). Two new species of Repetophragma from the Iberian Peninsula. Mycotaxon 125, 209–215. doi: 10.5248/125.209
Crous, P. W., Cowan, D. A., Maggs-Kölling, G., Yilmaz, N., Thangavel, R., Wingfield, M. J., et al. (2021). Fungal Planet description sheets: 1182–1283. Persoonia 46, 313–528. doi: 10.3767/persoonia.2021.46.11
Crous, P. W., Groenewald, J. Z., Mansilla, J. P., Hunter, G. C., Wingfield, M. J. (2004). Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. Stud. Mycol. 50 (1), 195–214. doi: 10.3114/sim.55.1.99
Crous, P. W., Wingfield, M. J., Lombard, L., Roets, F., Swart, W. J., Alvarado, P., et al. (2019). Fungal Planet description sheets: 951–1041. Persoonia 43, 223–425. doi: 10.3767/persoonia.2019.43.06
Dong, W., Wang, B., Hyde, K. D., Raja, H. A., Tanaka, K., Abdel-Wahab, M. A., et al. (2020). Freshwater dothideomycetes. Fungal Divers. 105, 319–575. doi: 10.1007/s13225-020-00463-5
Eriksson, O. E. (1981). The families of bitunicate ascomycetes. Nord. J. Bot. 1, 800. doi: 10.1111/j.1756-1051.1981.tb01167.x
Feng, B., Yang, Z. (2018). Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Divers. 40 (4), 165–171. doi: 10.1016/j.pld.2018.07.001
Frayer, J. M., Sun, Z., Müller, D., Munroe, D. K., Xu, J. (2014). Analyzing the drivers of tree planting in Yunnan, China, with Bayesian networks. Land Use Policy 36, 248–258. doi: 10.1016/j.landusepol.2013.08.005
Geiser, D. M., LoBuglio, K. F., Gueidan, C. (2015). “5 pezizomycotina: eurotiomycetes. Part B,” in Systematics and evolution. The mycota, vol. 7B . Eds. McLaughlin, D., Spatafora, J. (Berlin, Heidelberg: Springer), 121–141. doi: 10.1007/978-3-662-46011-5_5
Hall, T. (2004). BioEdit v. 7.0.1 (North Carolina State University: Department of Microbiology). Available at: www.mbio.ncsu.edu/BioEdit/bioedit.htm.
Haridas, S., Albert, R., Binder, M., Bloem, J., LaButti, K., Salamov, A., et al. (2020). 101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens. Stud. Mycol. 96, 141–153. doi: 10.1016/j.simyco.2020.01.003
Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R. F., Mena-Portales, J., Crous, P. W., Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002
Hongsanan, S., Hyde, K. D., Phookamsak, R., Wanasinghe, D. N., McKenzie, E. H. C., Sarma, V. V., et al. (2020a). Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Divers. 105, 17–318. doi: 10.1007/s13225-020-00462-6
Hongsanan, S., Hyde, K. D., Phookamsak, R., Wanasinghe, D. N., McKenzie, E. H. C., Sarma, V. V., et al. (2020b). Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11 (1), 1553–2107. doi: 10.5943/mycosphere/11/1/13
Hongsanan, S., Zhao, R. L., Hyde, K. D. (2017). A new species of Chaetothyrina on branches of mango, and introducing Phaeothecoidiellaceae fam. nov. Mycosphere. 8 (1), 137–146. doi: 10.5943/mycosphere/8/1/13
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. G. B., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5
Hyde, K. D., Jones, E. B. G., Liu, J. K., Ariyawansa, H., Boehm, E., Boonmee, S., et al. (2013). Families of dothideomycetes. Fungal Divers. 63, 1–313. doi: 10.1007/s13225-013-0263-4
Index Fungorum. Available at: http://www.indexfungorum.org. (Accessed June 2023).
Ismail, S. I., Batzer, J. C., Harrington, T. C., Crous, P. W., Lavrov, D. V., Li, H., et al. (2016). Ancestral state reconstruction infers phytopathogenic origins of sooty blotch and flyspeck fungi on apple. Mycologia 108 (2), 292–302. doi: 10.3852/15-036
Iturriaga, T., Hawksworth, D. L., Crane, J. L. (2008). ‘Sporidesmium’ lichenicola sp. nov., a new lichenicolous fungus on Leptogium from Venezuela. Mycologia 100, 392–396. doi: 10.3852/06-166r
Jayasiri, S. C., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Jeewon, R., Phillips, A. J. L., et al. (2019). Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10 (1), 1–186. doi: 10.5943/mycosphere/10/1/1
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kirk, P. M., Cannon, P. F., David, J. C., Stalpers, J. A. (2001). Ainsworth & Bisby's Dictionary of the fungi. 9th edn (Wallingford: CABI).
Kirk, P. M., Cannon, P. F., Minter, D. W., Stalpers, J. A. (2008). Ainsworth & Bisby’s dictionary of the fungi. 10th edn (Wallingford: CABI).
Li, X. H., Wu, H. X., Li, J. C., Chen, H., Wang, W. (2020). The insights into the evolutionary history of Translucidithyrium: based on a newly-discovered species. MycoKeys 75, 1–16. doi: 10.3897/mycokeys.75.58628
Li, C. Y., Zhang, G. Y., Hammer, K., Yang, C. Y., Long, C. L. (2011). A checklist of the cultivated plants of Yunnan (PR China). Genet. Resour. Crop Evol. 58, 153–164. doi: 10.1007/s10722-010-9638-5
Liu, J. K., Hyde, K. D., Jeewon, R., Phillips, A. J. L., Maharachchikumbura, S. S. N., Ryberg, M., et al. (2017). Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers. 84, 75–99. doi: 10.1007/s13225-017-0385-1
Luttrell, E. S. (1946). The genus stomiopeltis (Hemisphaeriaceae). Mycologia 38 (5), 565–586. doi: 10.1080/00275514.1946.12024079
Luttrell, E. S. (1955). The ascostromatic ascomycetes. Mycologia 47, 511–532. doi: 10.1080/00275514.1955.12024473
Ma, J., Zhang, X. G., Castañeda-Ruíz, R. F. (2014). New species of Acrodictys and Repetophragma from dead branches in China. Mycotaxon 127, 129–134. doi: 10.5248/127.129
Marincowitz, S., Crous., P. W., Groenewaldn, J. Z., Wingfield, M. J. (2008). Microfungi occurring on Proteaceae in the fynbos. CBS Biodiversity Ser. 7, 1–166.
Mayfield, D. A., Karakaya, A., Batzer, J. C., Blaser, J. M., Gleason, M. L. (2013). Diversity of sooty blotch and flyspeck fungi from apples in northeastern Turkey. Eur. J. Plant Pathol. 135, 805–815. doi: 10.1007/s10658-012-0123-1
McKenzie, E. H. C. (1995). Dematiaceous hyphomycetes on Pandanaceae. 5. Sporidesmium sensu lato. Mycotaxon, 56.
Mena-Portales, J., Delgado-Rodríguez, G., Here dia-Abarca, G. (2000). Nuevas combinaciones para especies de Sporidesmium S.L. (Hongos mitospóricos). Bol. Soc Micol. Madrid 25, 265–269.
Mortimer, P. E., Jeewon, R., Xu, J. C., Lumyong, S., Wanasinghe, D. N. (2021). Morpho-phylo taxonomy of novel Dothideomycetous fungi associated with dead woody twigs in Yunnan Province, China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.654683
Müller, E., von Arx, J. A. (1962). Die Gattungen der didymosporen Pyrenomycten. Beitr. Kryptogamenfl. Schweiz 11, 1–992.
Nannfeldt, J. A. (1932). Studien uber die Morphologie und Systematik der nicht–lichenisierten, inoperkulaten Discomyceten. Nova Acta Regiae Societatis Scientiarum Upsaliensis IV 8, 1–368.
ORO Yunnan Commerce (2023) Yunnan agriculture. Available at: https://www.oroyunnanmaldive.com/yunnan-agriculture/ (Accessed May 20, 2023).
Pem, D., Jeewon, R., Chethana, K. W. T., Hongsanan, S., Doilom, M., Suwannarach, N., et al. (2021). Species concepts of Dothideomycetes: classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 109, 283–319. doi: 10.1007/s13225-021-00485-7
Phillips, A. J. L., Hyde, K. D., Alves, A., Liu, J. K. (2019). Families in Botryosphaeriales: a phylogenetic, morphological and evolutionary perspective. Fungal Divers. 94, 1–22. doi: 10.1007/s13225-018-0416-6
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, D. J., Jones, E. G. B., Maharachchikumbura, S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w
Phookamsak, R., Wanasinghe, D. N., Hongsanan, S., Phukhamsakda, C., Huang, S. K., Tennakoon, D., et al. (2017). Towards a natural classification of Ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales). Fungal Divers. 87, 299–339. doi: 10.1007/s13225-017-0393-1
Quaedvlieg, W., Groenewald, J. Z., de Jesús Yáñez-Morales, M., Crous, P. W. (2012). DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia 29 (1), 101–115. doi: 10.3767/003158512X661282
Rambaut, A. (2014) FigTree v1.4: Tree figure drawing tool. Available at: http://treebio.ed.ac.uk/software/figtree/.
Rambelli, A., Ciccarone, C., Tempesta, S., Raimondo, F. M. (2011). Dematiaceous hyphomycetes from Quercus suber litter. Fl. Medit. 21, 325–344.
Renard, L. L., Firmino, A. L., Pereira, O. L., Stockey, R. A., Berbee, M. L. (2020). Character evolution of modern flyspeck fungi and implications for interpreting thyriothecial fossils. Am. J. Bot. 107, 1021–1040. doi: 10.1002/ajb2.1511
Schoch, C., Grube, M. (2015). “6 pezizomycotina: dothideomycetes and arthoniomycetes,” in Systematics and evolution. The Mycota (a comprehensive treatise on fungi as experimental systems for basic and applied research), vol. 7B . Eds. McLaughlin, D., Spatafora, J. (Berlin: Springer).
Schoch, C. L., Shoemaker, R. A., Seifert, K. A., Hambleton, S., Spatafora, J. W., Crous, P. W. (2006). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98, 1041–1052. doi: 10.3852/mycologia.98.6.1041
Seifert, K., Morgan-Jones, G., Gams, W., Kendrick, B. (2011). The genera of hyphomycetes. CBS Biodiversity Series no. 9 (Utrecht, Netherlands: CBS-KNAW Fungal Biodiversity Centre), 1–997.
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Shen, S., Xu, G., Li, D., Clements, D. R., Zhang, F., Jin, G., et al. (2017). Agrobiodiversity and in situ conservation in ethnic minority communities of Xishuangbanna in Yunnan Province, Southwest China. J. Ethnobiol. 13, 28. doi: 10.1186/s13002-017-0158-7
Shenoy, D. B., Jeewon, R., Wu, W. P., Bhat, D. J., Hyde, K. D. (2006). Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 110, 916–928. doi: 10.1016/j.mycres.2006.06.004
Silvera-Simón, C., Mena-Portales, J., Gené, J., Cano, J., Guarro, J. (2009). Repetophragma calongeii sp. nov. and other interesting dematiaceous hyphomycetes from the North of Spain. An. Jard. Bot. Madr. 66 (S1), 33–39. doi: 10.3989/ajbm.2218
Subramanian, C. V. (1992). A reassessment of Sporidesmium (Hyphomycetes) and some related. Taxa. Proc. Indian Acad. Sci. 58 (4), 179v190.
Thambugala, K. M., Ariyawansa, H. A., Li, Y. M., Boonmee, S., Hongsanan, S., Tian, Q., et al. (2014). Dothideales. Fungal Divers. 68, 105–158. doi: 10.1007/s13225-014-0303-8
Tibpromma, S., Hyde, K. D., McKenzie, E. H. C., Bhat, D. J., Phillips, A. J. L., Wanasinghe, D. N., et al. (2018). Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers. 93, 1–160. doi: 10.1007/s13225-018-0408-6
Videira, S. I. R., Groenewald, J. Z., Nakashima, C., Braun, U., Barreto, R. W., de Wit, P. J. G. M., et al. (2017). Mycosphaerellaceae - chaos or clarity? Stud. Mycol. 87, 257–421. doi: 10.1016/j.simyco.2017.09.003
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wanasinghe, D. N., Mortimer, P. E., Xu, J. (2021). Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China). J. Fungi (Basel) 7 (3), 180. doi: 10.3390/jof7030180
Wanasinghe, D. N., Wijayawardene, N. N., Xu, J., Cheewangkoon, R., Mortimer, P. E. (2020). Taxonomic novelties in Magnolia-associated Pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PloS One 15, e0235855. doi: 10.1371/journal.pone.0235855
Wang, X. M., Chen, S. S., Liu, X. M., Zhao, Z. J., Li, H. H., Zhang, X. G., et al. (2017). Repetophragma elegans sp. nov. from Hainan Province, China. Mycotaxon 132, 881–884. doi: 10.5248/132.881
White, T. J., Bruns, T., Lee, S., Taylor, J. W.. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications. Eds. Innes, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (San Diego, CA, USA: Academic Press), 315–322.
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13 (1), 53–453. doi: 10.5943/mycosphere/13/1/2
Wu, W. P., Zhuang, W. (2005). Sporidesmium, Endophragmiella and related genera from China. Fungal Divers. Res. Ser. 15, 1–351.
Yang, H. L., Sun, G. Y., Batzer, J. C., Crous, P. W., Groenewald, J. Z., Gleason, M. L. (2010). Novel fungal genera and species associated with the sooty blotch and flyspeck complex on apple in China and the USA. Persoonia 24, 29–37. doi: 10.3767/003158510X492101
Yang, Y., Tian, K., Hao, J., Pei, S., Yang, Y. X. (2004). Biodiversity and biodiversity conservation in Yunnan, China. Biol. Conserv. 13, 813–826. doi: 10.1023/B:BIOC.0000011728.46362.3c
Yang, E. F., Tibpromma, S., Karunarathna, S. C., Phookamsak, R., Xu, J. C., Zhao, Z. X., et al. (2022). Taxonomy and phylogeny of novel and extant taxa in Pleosporales associated with Mangifera indica from Yunnan, China (Series I). J. Fungi 8 (2), 152. doi: 10.3390/jof8020152
Zeng, X. Y., Hongsanan, S., Hyde, K. D., Putarak, C., Wen, T. C. (2018). Translucidithyrium Thailandicum gen. et sp. nov.: a new genus in Phaeothecoidiellaceae. Mycol. Prog. 17, 1087–1096. doi: 10.1007/s11557-018-1419-0
Zeng, X. Y., Wu, H. X., Hongsanan, S., Jeewon, R., Wen, T. C., Maharachchikumbura, S. S. N., et al. (2019). Taxonomy and the evolutionary history of Micropeltidaceae. Fungal Divers. 97, 393–436. doi: 10.1007/s13225-019-00431-8
Zhang, H., Li, J., Zhou, H., Chen, Z., Song, G., Peng, Z., et al. (2014). “Arabica coffee production in the Yunnan Province of China,” in 24th International Conference on Coffee Science.
Keywords: Dothideomycetes, fungal taxonomy, Mycosphaerellales, novel taxa, sootyblotch/flyspeck fungi
Citation: Hongsanan S, Phookamsak R, Bhat DJ, Wanasinghe DN, Promputtha I, Suwannarach N, Sandamali D, Lumyong S, Xu J and Xie N (2023) Exploring ascomycete diversity in Yunnan, China I: resolving ambiguous taxa in Phaeothecoidiellaceae and investigating conservation implications of fungi. Front. Cell. Infect. Microbiol. 13:1252387. doi: 10.3389/fcimb.2023.1252387
Received: 03 July 2023; Accepted: 02 August 2023;
Published: 04 September 2023.
Edited by:
Yusufjon Gafforov, Academy of Sciences Republic of Uzbekistan (UzAS), UzbekistanReviewed by:
Qing Tian, University of Electronic Science and Technology of China, ChinaHai-Xia Wu, Chinese Academy of Forestry, China
Copyright © 2023 Hongsanan, Phookamsak, Bhat, Wanasinghe, Promputtha, Suwannarach, Sandamali, Lumyong, Xu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rungtiwa Phookamsak, jomjam.rp2@gmail.com
 Sinang Hongsanan
Sinang Hongsanan Rungtiwa Phookamsak
Rungtiwa Phookamsak Darbhe Jayarama Bhat6,7
Darbhe Jayarama Bhat6,7  Dhanushka N. Wanasinghe
Dhanushka N. Wanasinghe Itthayakorn Promputtha
Itthayakorn Promputtha Nakarin Suwannarach
Nakarin Suwannarach Diana Sandamali
Diana Sandamali Saisamorn Lumyong
Saisamorn Lumyong Jianchu Xu
Jianchu Xu