Comparative genomic analysis of plasmids harboring blaOXA-48-like genes in Klebsiella pneumoniae
- 1Shandong Provincial Engineering and Technology Research Center for Wild Plant Resources Development and Application of Yellow River Delta, College of Biological and Environmental Engineering, Binzhou University, Binzhou, China
- 2Binzhou Key Laboratory of Chemical Drug R&D and Quality Control (preparation), Binzhou, China
- 3Department of Radiation Oncology, Zhuhai People’s Hospital (Zhuhai hospital affiliated with Jinan University), Zhuhai, China
- 4Department of Stomatology, Zhuhai People’s Hospital (Zhuhai hospital affiliated with Jinan University), Zhuhai, China
- 5Department of Neurosurgery, Zhuhai People’s Hospital (Zhuhai hospital affiliated with Jinan University), Zhuhai, China
- 6Zhuhai Precision Medical Center, Zhuhai People’s Hospital (Zhuhai hospital affiliated with Jinan University), Zhuhai, China
The emergence and spread of carbapenem-resistant Klebsiella pneumoniae (CRKP) is a serious medical problem worldwide. Acquired OXA-48-like carbapenemases encoded by plasmids are important causes of carbapenem resistance in K. pneumoniae. To explore the links between plasmids and blaOXA-48-like genes in K. pneumoniae, we systematically analyzed the variants of blaOXA-48-like plasmid replicon types, phylogenetic patterns, geographic distribution, conjugative transfer regions, and the genetic environments surrounding blaOXA-48-like of 191 blaOXA-48-like-harboring plasmids, which were identified from 4451 plasmids of K. pneumoniae downloaded from GenBank. Our results showed that seven different variants of blaOXA-48-like genes were identified from the 191 blaOXA-48-like-harboring plasmids in K. pneumoniae, with blaOXA-48, blaOXA-232, and blaOXA-181 being highly prevalent. In K. pneumoniae, blaOXA-48 was mainly carried by the composite transposon Tn1999.2 located on IncL/M-type conjugative plasmids, which were mainly geographically distributed in Switzerland, Germany, and China. In K. pneumoniae, the blaOXA-232 gene was mainly carried by 6.1-kb ColKP3-type mobilizable plasmids, which were mainly isolated in India. In K. pneumoniae, blaOXA-181 was mainly carried by a group of 50-kb ColKP3-IncX3 hybrid conjugative plasmids and a group of small ColKP3-type mobilizable plasmids with lengths of 5.9–9.3 kb, the former was sporadically discovered in China, South Korea, India, and Czech Republic, while the latter was almost all isolated in India. In addition, five blaOXA-245-harboring 65.9-kb IncL plasmids of K. pneumoniae isolated in Spain were found to have the genetic context of blaOXA-245 more complicated than that of blaOXA-48-harboring IncL/M-type plasmids, with two copies of IS1R inserted both upstream and downstream of blaOXA-245-lysR. These findings enhance our understanding of the genetic diversity of blaOXA-48-like-harboring plasmids in K. pneumoniae.
1. Introduction
The rapid increase in carbapenemase-producing Enterobacterales has become a threat to public health (Kim et al., 2021). OXA-48-like carbapenemases are important causes of carbapenem resistance in Enterobacterales (Pitout et al., 2019) and consist of 261–265 amino acids (Poirel et al., 2004). Among the various OXA-48-like carbapenemases, OXA-48, OXA-181, OXA-232, OXA-204, OXA-162, and OXA-244, in that order, are the most common. Surveillance studies based on molecular methodologies have indicated that OXA-48-like carbapenemases remain the second or third most common carbapenemases in Enterobacterales globally (de Jonge et al., 2016; Karlowsky et al., 2017; Han et al., 2020). Notably, Enterobacterales producing OXA-48-like carbapenemases are endemic in certain parts of the world (Pitout et al., 2019). OXA-48-like carbapenemases are widely distributed among members of Enterobacterales, mainly Klebsiella pneumoniae isolated from hospital sites (de Jonge et al., 2016; Karlowsky et al., 2017; Han et al., 2020).
K. pneumoniae is a significant cause of both community- and hospital-acquired infections such as pneumonia, urinary tract infections, bloodstream infections, and septicemia (Pitout et al., 2015; Bengoechea and Sa Pessoa, 2019) and is known for its high frequency of antimicrobial resistance (AMR) genes (Navon-Venezia et al., 2017; Wyres and Holt, 2018). K. pneumoniae has been classified into one of the ESKAPE pathogens, which are the leading cause of nosocomial infections (Pandey et al., 2021). The emergence and spread of carbapenem-resistant K. pneumoniae (CRKP) have become a severe medical problem worldwide (Navon-Venezia et al., 2017). AMR in CRKP isolates is frequently encoded by plasmid-borne genes that can disseminate horizontally (Rozwandowicz et al., 2018).
Plasmids are autonomously replicating extrachromosomal DNA molecules in many bacterial strains (Le Roux et al., 2011), which are the key element in horizontal gene transfer in the microbial community (Fang and Zhou, 2020). According to the transferability of plasmids, they can be classified into the non-transferable plasmids, conjugative (self-transferable) plasmids and mobilizable plasmids (Branger et al., 2018). For CRKP isolates, conjugative plasmids are important vehicles for the spread of AMR genes (Smillie et al., 2010; Ravi et al., 2018). Such plasmids typically have a conserved backbone and variable accessory regions (Brown et al., 2013; Sitter et al., 2021). The conserved backbone region includes genes responsible for plasmid-related traits, e.g., replication regulation and conjugation functions. The variable accessory region is loaded with accessory genes, such as AMR genes, usually located on transposons or integrons (Norman et al., 2009; Norberg et al., 2011). For the conjugative plasmids, the transfer regions responsible for conjugation typically comprise four key modules: origin of transfer (oriT) site, relaxase gene, gene encoding type IV coupling protein (T4CP), and gene cluster for type IV secretion system (T4SS) (de la Cruz et al., 2010). Notably, mobilizable plasmids are also important contributors to the dissemination of AMR genes, usually carrying indispensable oriT sites and a limited amount of mob genes, which cannot transfer independently but can be mobilized by conjugative elements (Ramsay and Firth, 2017).
Currently, studies on the comprehensive analysis of blaOXA-48-like-harboring plasmids and their conjugative transfer regions in K. pneumoniae are limited. In this work, we performed in silico characterization and comparative analysis of blaOXA-48-like-harboring plasmids of K. pneumoniae using bacterial plasmids available in the NCBI GenBank database. We systematically analyzed the variants of blaOXA-48-like, replicon type, phylogenetic pattern, conjugative transfer region, and genetic context of blaOXA-48-like-harboring plasmids of K. pneumoniae.
2. Materials and methods
2.1. Plasmid sequences from the NCBI database
A total of 4451 plasmids of K. pneumoniae (Table S1) were downloaded from the GenBank (Benson et al., 2018) genome database (https://www.ncbi.nlm.nih.gov/genome/browse/#!/plasmids/815/) on April 26, 2022. The DNA files in FASTA format were downloaded in batches into our server built on the Linux-CentOS7 operating system using two Bioperl modules: Bio::DB::GenBank and Bio::SeqIO.
2.2. Determination of blaOXA-48-like-harboring plasmids of K. pneumoniae
The acquire antimicrobial resistance genes (ARGs) of plasmids were determined using the software ResFinder standalone version 4.1 (Bortolaia et al., 2020), with a minimum identity of 90%, a minimum coverage of 60%. The term “blaOXA” was used to search in the “Resistance gene” list within the ResFinder results to determine the blaOXA-positive plasmids of K. pneumoniae. The plasmids harboring blaOXA-48-like genes were further determined by mapping the “Resistance gene” to file phenotypes.txt of the ResFinder database (Bortolaia et al., 2020) to extract the information column titles termed “Notes” of the file phenotypes.txt. The variants of blaOXA-48-like genes in some plasmids were not determined using the ResFinder software. They were further analyzed using the CARD database (Alcock et al., 2020) and the beta-lactamase database (Naas et al., 2017).
2.3. Geographic location and origin of the blaOXA-48-like-harboring plasmids of K. pneumoniae
Information about geographic location and origin of the blaOXA-48-like-harboring plasmids in K. pneumoniae were extracted in batches from the section “source” of corresponding genomic files in GenBank format, using the key words “country”, “isolation_source” and “host”. For some plasmids with no relevant records on the geographic location and origin, we tried to search the corresponding information from their published papers according to the PubMed ID (PMID) numbers included in the GenBank files. To ensure accuracy of data, only data collected in strict accordance with the above two standards could be included in our analysis.
2.4. Replicon sequence analysis of the blaOXA-48-like-harboring plasmids of K. pneumoniae
Plasmid replicon typing of blaOXA-48-like-carrying plasmids was executed in the PlasmidFinder software (Carattoli and Hasman, 2020). Based on the database “Enterobacteriales,” the genomic files (FASTA format) of blaOXA-48-like-carrying plasmids were analyzed in batches using the PlasmidFinder software version 2.0.1, with a minimum identity of 95% and a minimum coverage of 60%. The database was updated on November 29, 2021.
2.5. Phylogenetic cladogram of the blaOXA-48-like-harboring plasmids of K. pneumoniae
The files of the blaOXA-48-like-carrying plasmids of K. pneumoniae in GenBank format were downloaded in batches using Bioperl (Bio::DB::GenBank and Bio::SeqIO). Using Bioperl/Bio::SeqIO, genomic files containing protein sequences were extracted from the files in GenBank format. Phylogenetic cladogram according to the presence/absence of orthologous gene families of all blaOXA-48-like-harboring plasmids in K. pneumoniae were analyzed. A binary gene presence/absence matrix was built using OrthoFinder (Emms and Kelly, 2019), and then hierarchical clustering was conducted using PAST3 (Hammer et al., 2001) and eventually displayed using iTOL (Letunic and Bork, 2016).
2.6. In silico characterization of the conjugative transfer regions of blaOXA-48-like-harboring plasmids
The files (GenBank format) of the blaOXA-48-like-harboring plasmids in K. pneumoniae were analyzed in batches using oriTfinder (Li et al., 2018) (standalone version) to identify the presence/absence of oriTs, relaxase genes, T4CP genes, and gene clusters for T4SS. Moreover, the types of oriTs, relaxase genes, T4CP genes, and gene clusters for T4SS of the plasmids were determined based on the oriTDB database (Li et al., 2018). In addition, the types of gene clusters for T4SS were classified based on the SecReT4 database (Bi et al., 2013).
2.7. Analysis of genetic context of the blaOXA-48-like genes
The bacterial insertion sequences of the blaOXA-48-like-harboring plasmids in K. pneumoniae were explored using ISfinder (Siguier et al., 2006). Comparisons among the genetic contexts of blaOXA-48-like genes of the plasmids were carried out using Easyfig (Sullivan et al., 2011) or BLAST Ring Image Generator (Alikhan et al., 2011).
3. Results
3.1. Variants of blaOXA genes in the blaOXA-48-like-harboring plasmids of K. pneumoniae
Using ResFinder, 191 (4.29%) blaOXA-48-like-harboring plasmids (Table S2) were identified from 4451 plasmids of K. pneumoniae downloaded from the GenBank genome database. Among the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae, 197 blaOXA-48-like genes belonging to seven blaOXA-48-like variants were identified. Of these seven variants, blaOXA-48 was the most dominant, followed by blaOXA-232 and blaOXA-181 (Figure 1A). A total of 102 plasmids harboring blaOXA-48 were screened from the 191 blaOXA-48-like-harboring plasmids, including 100 plasmids with only one copy of the blaOXA-48 gene in their genomes and two plasmids containing two copies of blaOXA-48 genes in their genomes (Table S2). Of the 59 blaOXA-232-harboring plasmids, 55 plasmids were found to harbor only one copy of blaOXA-232 in their genomes, and four plasmids were found to carry two copies of blaOXA-232 in their genomes) (Table S2). All 22 blaOXA-181-harboring plasmids harbored one copy of the blaOXA-181 gene in their genomes. In addition, one plasmid harboring blaOXA-204, one carrying blaOXA-244, and five blaOXA-245-harboring plasmids were identified in this study. Meanwhile, one plasmid from K. pneumoniae strain LZK001 harbored one blaOXA gene, similar to blaOXA-48 and blaOXA-244, according to the results of ResFinder.
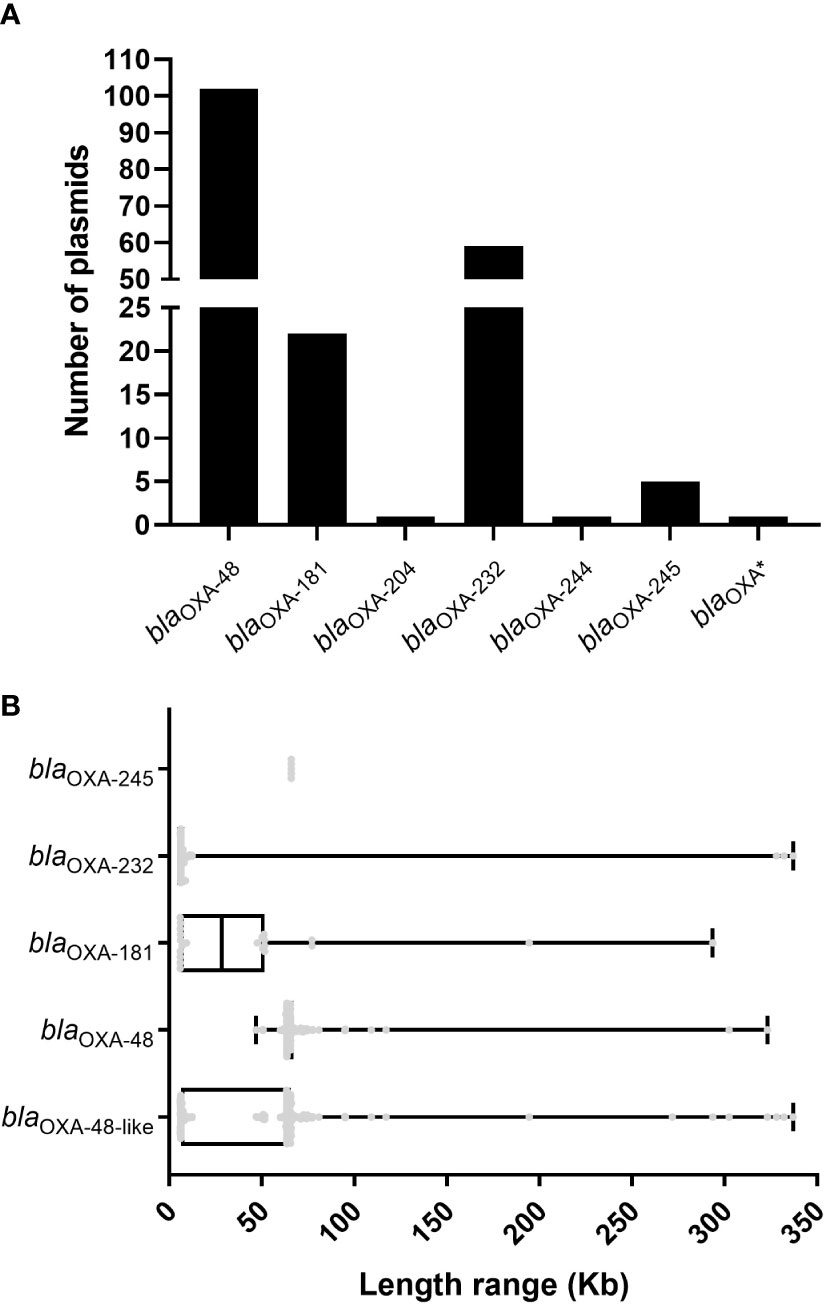
Figure 1 Characteristics of the 191 blaOXA-48-like-harboring plasmids in K. pneumoniae. (A) Histogram of number of variants of blaOXA-48-like genes among the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae. (B) Box plot of the length distribution of the five blaOXA-245-harboring plasmids, 59 blaOXA-232-harboring plasmids, 22 blaOXA-181-harboring plasmids, 102 blaOXA-48-harboring plasmids, and all the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae.
3.2. Lengths of blaOXA-48-like-harboring plasmids of K. pneumoniae
We analyzed and compared the lengths of 191 blaOXA-48-like-harboring plasmids of K. pneumoniae and compared the lengths of plasmids harboring different variants of blaOXA-48-like genes. The lengths of the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae ranged from 5.85 to 337.0 kb, and the 25th percentile, median, and 75th percentile were 6.14 kb, 63.59 kb, and 65.50 kb, respectively (Figure 1B). The lengths of 102 plasmids harboring blaOXA-48 ranged from 46.89 to 323.1 kb (25th percentile, 63.59 kb; 75th percentile, 67.10 kb), with a median size of 63.59 kb (Figure 1B). For the 22 plasmids harboring blaOXA-181, their genome sizes ranged from 5.92 to 293.7 kb. The 25th percentile, median, and 75th percentile were 5.92 kb, 28.47 kb, and 51.48 kb, respectively (Figure 1B). For the 59 plasmids harboring blaOXA-232, their genome sizes ranged from 5.85 to 337 kb, and 42 of these 59 were found to be the small plasmids with a length of 6.14 kb (Figure 1B). In addition, the length of all five blaOXA-245-harboring plasmids was 65.93 kb (Figure 1B).
3.3. Replicon types of blaOXA-48-like-harboring plasmids of K. pneumoniae
Of the 191 blaOXA-48-like-harboring plasmids, the replicon types of 189 were successfully identified, including 167 single-replicon and 22 multi-replicon plasmids (16 plasmids containing two replicons, four plasmids containing three replicons, and two plasmids containing four replicons) (Figure 2).
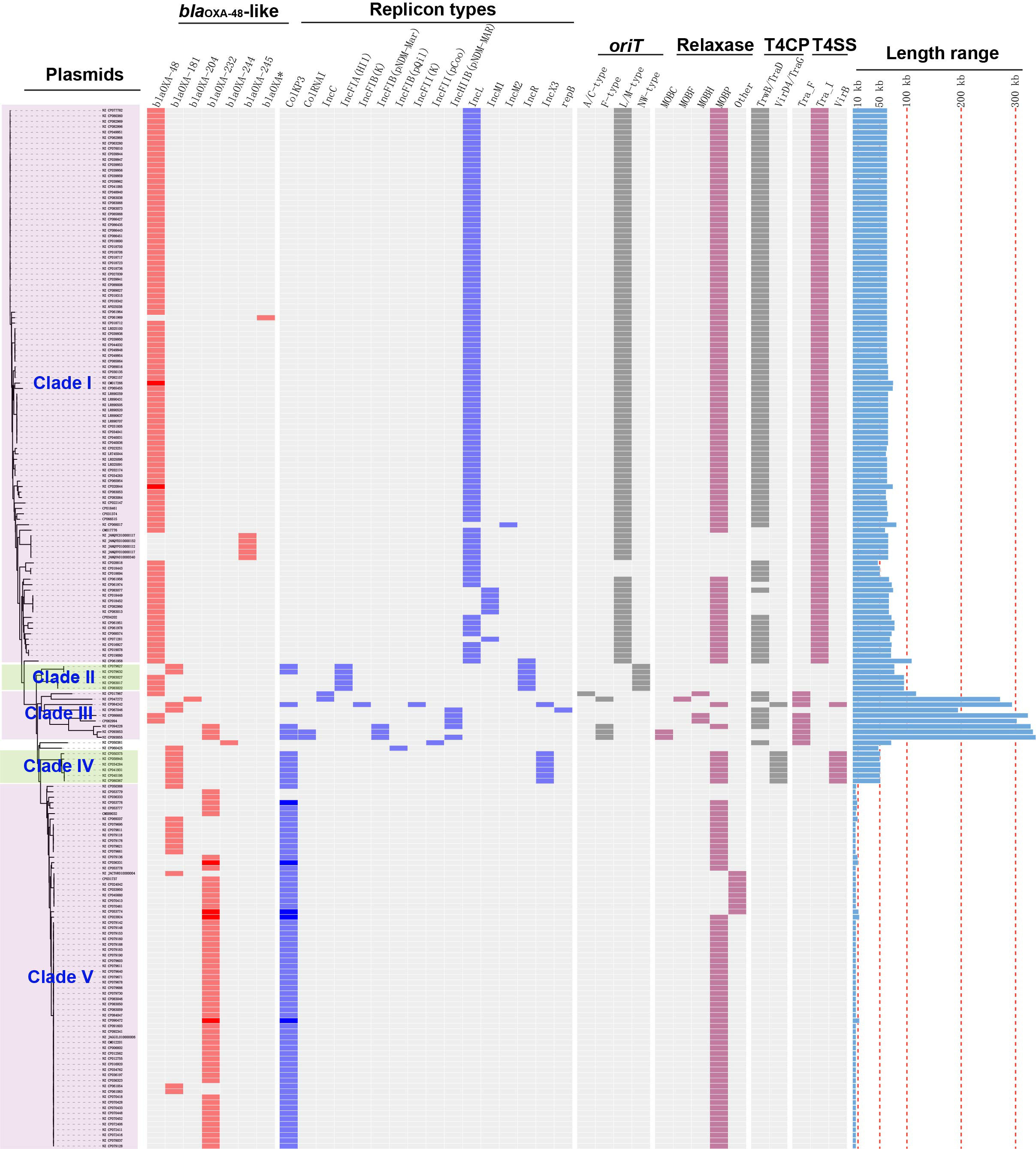
Figure 2 Details of variants of blaOXA-48-like genes, replicon types, conjugative transfer regions, and length distribution of the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae. The four categories of information present in this figure include the phylogenetic tree, variants of blaOXA-48-like, replicon types, conjugative transfer regions (oriT, relaxase, T4CP, and T4SS), and length distribution of the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae.
Among the 167 single-replicon plasmids harboring blaOXA-48-like in K. pneumoniae, plasmids with IncL replicon were the most common, with a total of 94 plasmids (Figure 2). In addition, 60 single-replicon plasmids with ColKP3 replicon were selected from the 167 single-replicon blaOXA-48-like-positive plasmids, which were the second most prevalent single-replicon plasmids harboring blaOXA-48-like genes in K. pneumoniae (Figure 2).
In summary, 77 of the 191 blaOXA-48-like-harboring plasmids of K. pneumoniae were found to carry the ColKP3 replicon, accounting for 40.31% of all blaOXA-48-like-harboring plasmids of K. pneumoniae in this study (Figure 2). Furthermore, 102 plasmids harboring blaOXA-48-like genes were classified as IncL/M-type plasmids, with replicon types including IncL, IncM1, or IncM2 (Figure 2).
3.4. Genetic diversity of the blaOXA-48-like-harboring plasmids in K. pneumoniae
We constructed a phylogenetic cladogram of the 191 blaOXA-48-like-harboring plasmids to obtain a comprehensive overview of blaOXA-48-like-harboring plasmid genes in K. pneumoniae (Figure 2). Based on the phylogenetic cladogram combined with the information on plasmid types, conjugative transfer regions, and genome sizes of the blaOXA-48-like-harboring plasmids, most of the 191 blaOXA-48-like-harboring plasmids were clustered into five main clades (clades I–V), representing five blaOXA-48-like-harboring plasmid patterns in K. pneumoniae.
3.4.1. Clade I: IncL/M-type plasmids harboring blaOXA-48 and blaOXA-245 in K. pneumoniae
A total of 102 IncL/M-type plasmids were classified into the clade I cluster, mainly blaOXA-48-carrying plasmids (Figure 2), accounting for 53.4% of all blaOXA-48-like-harboring plasmids of K. pneumoniae. Most of the 102 IncL/M-type plasmids were single ARG-harboring plasmids (Figure S1). Notably, the five blaOXA-245-harboring plasmids were also classified into the clade I cluster. The most frequent replicon type of blaOXA-48-like-harboring IncL/M-type plasmids was the IncL replicon, followed by IncM1 and IncM2. For the 102 IncL/M-type plasmids harboring blaOXA-48-like genes, their genome sizes ranged from 46.89 to 109.1 kb (25th percentile, 63.59 kb; 75th percentile, 65.68 kb), with a median size of 63.59 kb (Figure S2). For the conjugative transfer regions, most of the plasmids of clade I were found to carry L/M-type oriTs, genes encoding relaxases of the MOBP family characterized by the domain “Relaxase (Pfam: PF03432),” genes encoding T4CPs of TrwB/TraD subfamily characterized by the domain “TrwB_AAD_bind (PF10412),” and Tra_I-like T4SS gene clusters (Figures 2, 3A), inferred as conjugative plasmids. Members of clade I were widely geographically distributed all over the world, mainly in Switzerland (29 plasmids), Germany (16 plasmids), and China (15 plasmids) (Figure 4). Most of the IncL/M-type plasmids were human origins, some were animal origins (collected in Switzerland) and environment origins (also collected in Switzerland) (Figure S1 and Table S3).
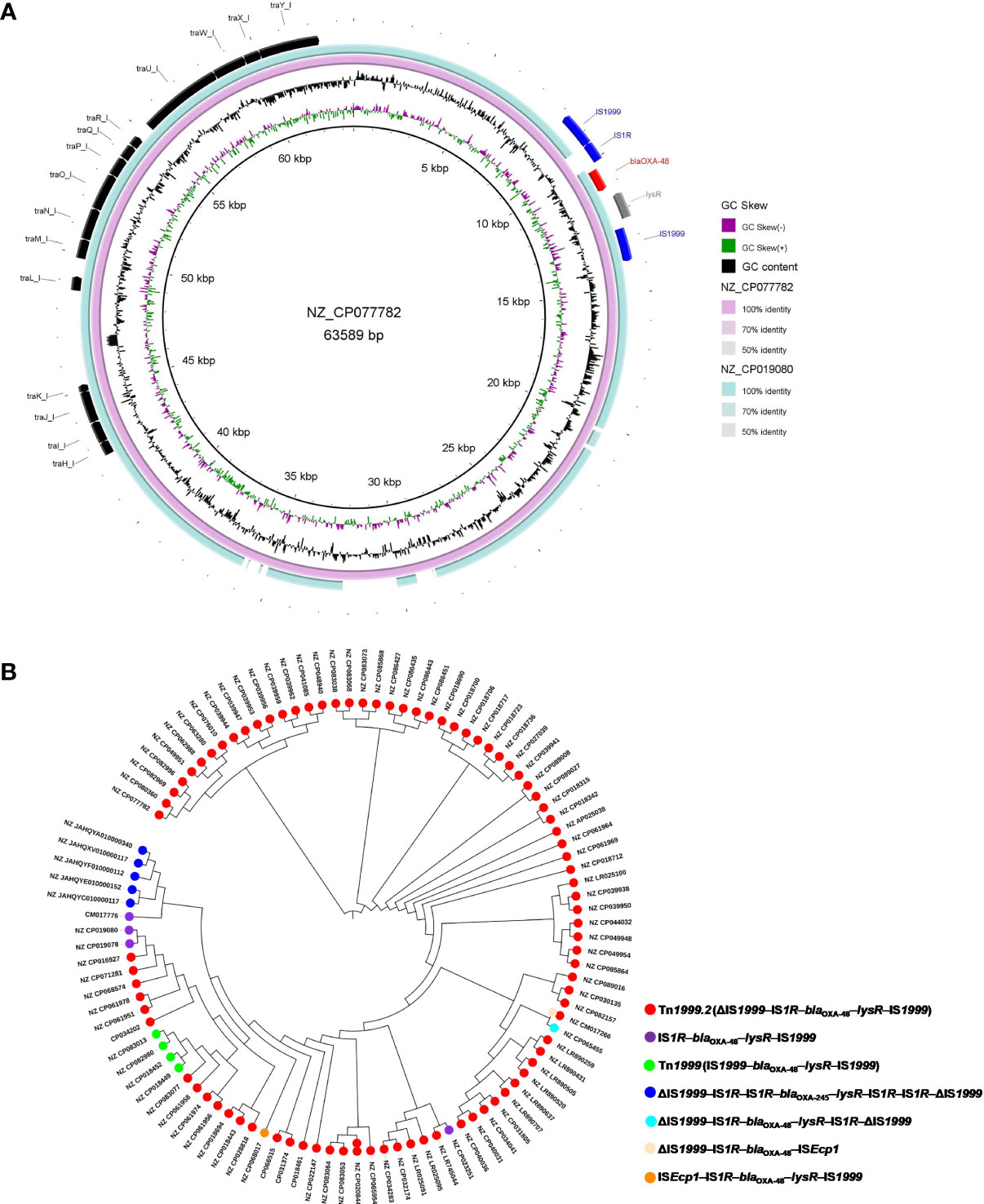
Figure 3 An overview of the conjugative transfer regions and genetic environment surrounding blaOXA-48 or blaOXA-245 carried by the 102 IncL/M-type plasmids classified into the clade I cluster. (A) Details of the Tra_I-like T4SS gene clusters, transposon Tn1999.2, and Tn1999 in the representative plasmids. (B) Distribution of different genetic contexts surrounding blaOXA-48-like genes in the 102 IncL/M-type plasmids.
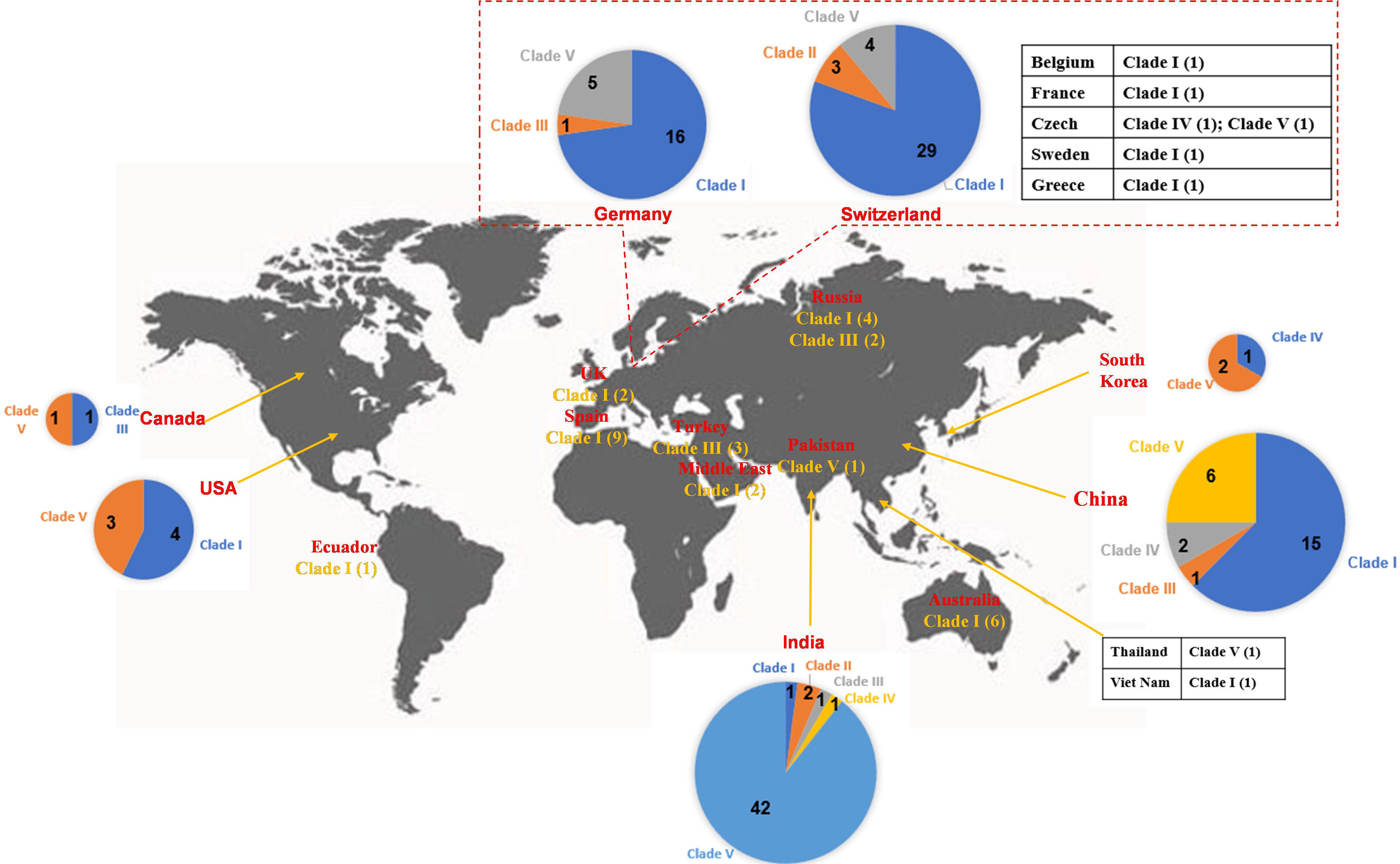
Figure 4 Worldwide distribution of blaOXA-48-like-harboring plasmids in K. pneumoniae. The geographical distribution of the five clades (Clade I–Clade V) from the blaOXA-48-like-harboring plasmids of K. pneumoniae was calculated and displayed by pie chart, text description, as well as tabular form.
We explored the genetic environment surrounding blaOXA-48-like genes carried by the 102 IncL/M-type plasmids in K. pneumoniae. For blaOXA-48-carrying IncL/M-type plasmids in K. pneumoniae, the blaOXA-48 was mainly located on the composite transposon Tn1999.2 (ΔIS1999–IS1R–blaOXA-48–lysR–IS1999), with 87 plasmids carrying intact Tn1999.2 and four plasmids harboring the truncated Tn1999.2 (IS1R–blaOXA-48–lysR–IS1999) (Figure 3A). We also found four blaOXA-48-harboring plasmids of clade I carrying classical Tn1999 (IS1999–blaOXA-48–lysR–IS1999). Notably, several blaOXA-48-harboring plasmids were found to carry specific genetic contexts of blaOXA-48, including K. pneumoniae strain KPN1482 plasmid pKPN1482-3 (NZ_CP020844) with two copies of intact Tn1999.2 (ΔIS1999–IS1R–blaOXA-48–lysR–IS1999), K. pneumoniae strain F64 plasmid pRYC-OXA48 (NZ_CM017266) carrying the intact Tn1999.2 (ΔIS1999–IS1R–blaOXA-48–lysR–IS1999) and the structure “ΔIS1999–IS1R–blaOXA-48–ISEcp1”, K. pneumoniae strain Beach Ranger plasmid pBR_02 (NZ_CP065455) carrying the “ΔIS1999–IS1R–blaOXA-48–lysR–IS1R–ΔIS1999”, as well as the K. pneumoniae strain 2016_49 plasmid pMS3802OXARMA (NZ_CP068017) carrying the structure “ISEcp1–IS1R–blaOXA-48–lysR–IS1999” (Figure S2). In addition, we found that the five blaOXA-245-harboring IncL plasmids, which were all isolated in Spain, carried the structure “ΔIS1999–IS1R–IS1R–blaOXA-245–lysR–IS1R–IS1R–ΔIS1999” (Figure 3B).
3.4.2. Clade II: Mobilizable blaOXA-48-like-harboring plasmids in K. pneumoniae
Five blaOXA-48-like-harboring multidrug-resistant (MDR) plasmids with IncFIA(HI1) and IncR replicons were classified into a small cluster, clade II of the phylogenetic cladogram constructed with the 191 blaOXA-48-like-harboring plasmids, including three 94.9-kb plasmids [IncFIA(HI1):IncR] harboring blaOXA-48 and two 77.1-kb plasmids [ColKP3:IncFIA(HI1):IncR] harboring blaOXA-181 (Figure 2). These five plasmids all carried NW-type oriTs, but not genes encoding relaxase, T4CP, or T4SS in their genomes, indicating that they are mobilizable plasmids (Figure 2). The five plasmids belonging to clade II were found to be distributed in Switzerland and India (Figure 4), which were all human origins (Figure S1 and Table S3).
For the three blaOXA-48-harboring plasmids isolated in Switzerland, blaOXA-48 was situated in the truncated Tn1999.2 (IS1R–blaOXA-48–lysR–IS1999) (Figure S3A). For the two blaOXA-181-harboring plasmids isolated in India, intact IS3000 was located upstream of blaOXA-181 (Figure S3B).
3.4.3. Clade III: Mega blaOXA-48-like-harboring plasmids in K. pneumoniae
One cluster of the phylogenetic cladogram (clade III) composed of nine mega MDR plasmids with lengths ranging from 117.2 to 337.0 kb (25th percentile, 233.1 kb, median, 302.4 kb, and 75th percentile, 330.1 kb) was identified, including blaOXA-48-harboring, blaOXA-181-harboring, blaOXA-204-harboring, and blaOXA-232-harboring plasmids (Figures 2, S2). The plasmids in clade III contained four single-replicon plasmids, one plasmid with two different replicons, two plasmids with three different replicons, and two plasmids with four different replicons (Figure 2). Among the nine mega plasmids, the IncHI1B(pNDM-MAR) and ColKP3 replicons were relatively common, with the IncHI1B(pNDM-MAR) replicon found in five plasmids and the ColKP3 replicon found in four plasmids (Figure 2). Most of the plasmids belonging to clade III had the genes encoding for T4CPs of TrwB/TraD subfamily characterized by the domain “TrwB_AAD_bind (PF10412)” and Tra_F-like T4SS gene clusters (Figure 2). According to the identified conjugative transfer regions, these mega plasmids were inferred as conjugative plasmids. However, they were heterogeneous in the type of relaxase genes, and only five plasmids had their oriTs successfully identified using oriTfinder. The plasmids of clade III were sporadically discovered in Turkey, Russia, China, India, Germany, and Canada (Figure 4), originated from samples of human (Figure S1 and Table S3).
For the three blaOXA-48-harboring plasmids, including one 117.2-kb IncC plasmid and two IncHI1B(pNDM-MAR) plasmids with lengths >300 kb, blaOXA-48 was found to be situated in the truncated Tn1999.2 (IS1R–blaOXA-48–lysR–IS1999). For the two blaOXA-181-harboring plasmids, one 293-kb plasmid carried the genetic environment of blaOXA-181 (IS26–ΔIS3000–ΔISEcp1–blaOXA-181–ΔlysR–Δere–ΔrepA–ISKpn19), and another 194-kb plasmid carried the “ISEcp1–blaOXA-181”. For the blaOXA-204-harboring plasmid and the three blaOXA-232-harboring plasmids, ISEcp1 was inserted upstream of blaOXA-204 or blaOXA-232.
3.4.4. Clade IV: blaOXA-181-harboring plasmids with multi-replicon ColKP3:IncX3 in K. pneumoniae
Six blaOXA-181-harboring plasmids with multi-replicon ColKP3:IncX3 were clustered into clade IV, and their genome sizes ranged from 50.13 to 51.48 kb (Figure 2). Almost all the plasmids of clade IV harbored two ARGs: blaOXA-181 and qnrS1 (Figure S1). They all carried genes encoding relaxases of the MOBP family characterized by the domain “Relaxase (PF03432),” genes encoding T4CPs of the VirD4/TraG subfamily characterized by the domain “T4SS-DNA_transf (PF02534),” and mostly VirB-like T4SS gene clusters (Figures 2, 5A). Although we could not determine the definite oriT sites of the clade IV plasmids, the oriT-like region flanking the relaxase genes was found in all six blaOXA-181-harboring plasmids, characterized by the inverted repeat (IR) sequence (TAACTA.TAGTTA). According to the identified conjugative transfer regions, the plasmids of clade IV should be conjugative plasmids. The plasmids of clade IV (human origins) were sporadically discovered in China, South Korea, India, and Czech Republic (Figures 4, S1; Table S3).
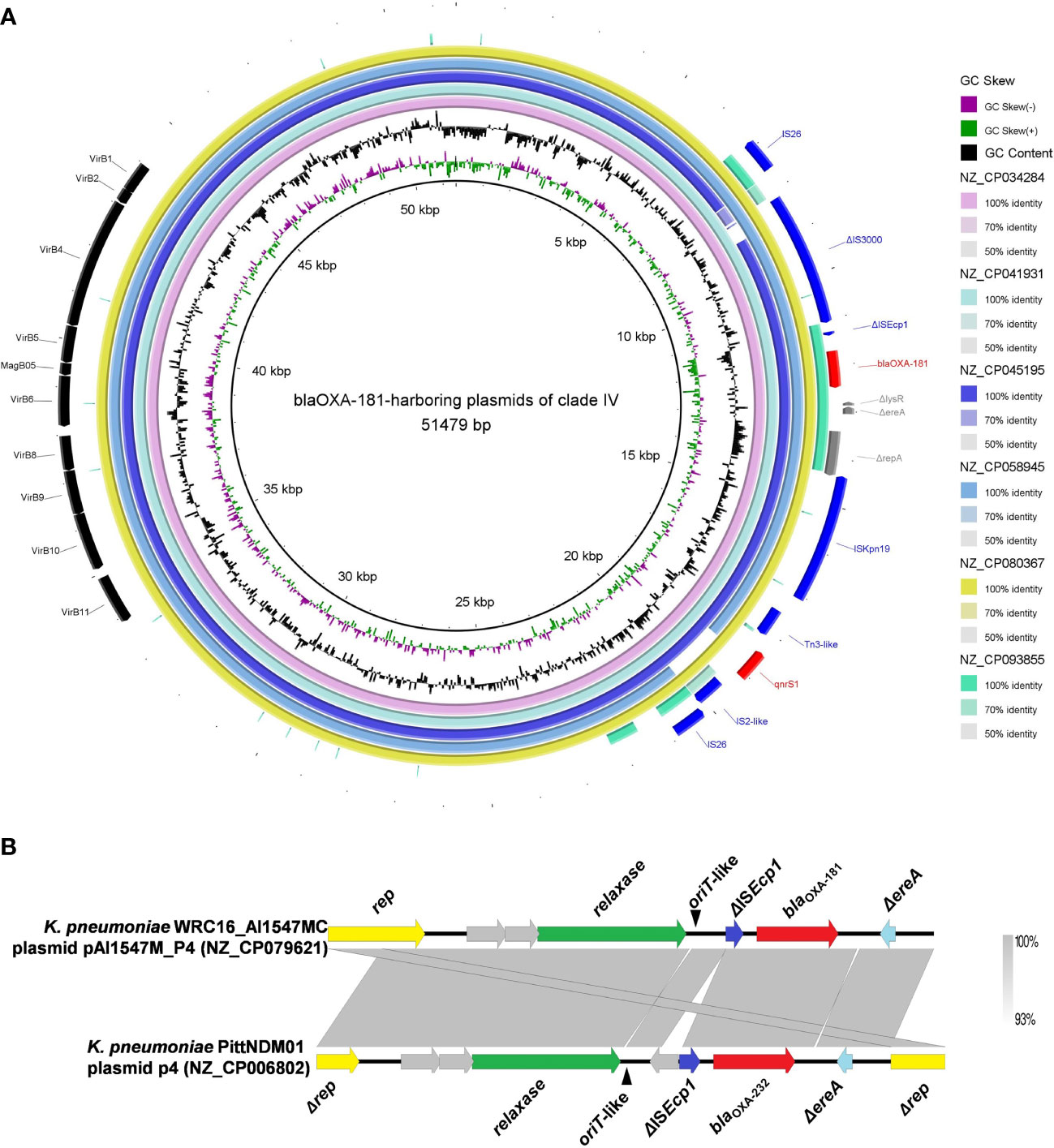
Figure 5 Characteristics of the conjugative transfer regions and genetic environment surrounding blaOXA-48-like carried by the plasmids belonging to clade IV and clade V. (A) Details of the VirB-like T4SS gene clusters and blaOXA-181-associated genetic structures identified among the six blaOXA-181-harboring plasmids clustered into clade IV. (B) The oriT-like regions and the genetic context surrounding the blaOXA-181 or blaOXA-232 genes of the small plasmids belonging to clade V.
For the six ColKP3-IncX3 hybrid plasmids harboring both blaOXA-181 and qnrS1 in their genomes, blaOXA-181 and qnrS1 were found to be located in a composite transposon, which was bracketed by two copies of the insertion sequence IS26 in the same orientation (IS26–ΔIS3000–ΔISEcp1–blaOXA-181–ΔlysR–Δere–ΔrepA–ISKpn19–Tn3-like–qnrS1–IS2-like–IS26) (Figure 5A).
3.4.5. Clade V: Small mobilizable blaOXA-48-like-harboring plasmids in K. pneumoniae
A total of 67 small plasmids with lengths ranging from 5.85 to 12.27 kb (mostly 6.14 kb), including 11 blaOXA-181-harboring plasmids and 56 blaOXA-232-harboring plasmids, were grouped into a large cluster named clade V of the phylogenetic cladogram (Figure 2). Almost all the plasmids classified into clade V were single ARG-harboring plasmids (Figure S1). Most of the 67 small plasmids belonging to clade V were identified as single-replicon plasmids with a ColKP3 replicon. Moreover, most of the plasmids belonging to clade V carried genes encoding relaxases of the MOBP family characterized by the domain “Relaxase (Pfam: PF03432),” but no gene encoding T4CP or T4SS (Figures 2, 5B). Although no oriT was identified in the plasmids of clade V, the oriT-like regions adjacent to relaxase genes were identified with an IR sequence (AAAAGGAAAGTG.CACTTTCCTTTT) (Figure 5B). According to the identified conjugative transfer regions, the plasmids of clade V should be mobilizable plasmids. Among the 67 plasmids belonging to clade V, 42 (62.69%) were geographically found in India (Figure 4). These ColKP3-type mobilizable plasmids were also found in China, Germany, Switzerland, the USA, etc (Figure 4). Notably, the ColKP3-type mobilizable plasmids were found to be human origins (Figure S1 and Table S3).
We explored the genetic context surrounding the blaOXA-181 or blaOXA-232 genes of the 67 small plasmids belonging to clade V. For the 67 small ColKP3-type plasmids belonging to clade V, blaOXA-181 or blaOXA-232 were located downstream of the ΔISEcp1 harbored by the ColKP3-type plasmid (Figure 5B).
4. Discussion
OXA-48-like carbapenemases are important causes of non-susceptibility to carbapenems in Enterobacterales (Pitout et al., 2019). blaOXA-48-like genes are always plasmid-borne, and plasmids make considerable contributions to disseminating blaOXA-48-like genes (de Jonge et al., 2016). To characterize plasmids harboring blaOXA-48-like genes in K. pneumoniae, we systematically analyzed the variants of blaOXA-48-like, replicon types, conjugative transfer regions, and genetic contexts of blaOXA-48-like plasmids among 191 blaOXA-48-like-harboring plasmids, which were selected from 4451 plasmids belonging to K. pneumoniae from the NCBI GenBank database. In our study, seven different variants of blaOXA-48-like genes were identified from 191 blaOXA-48-like-harboring plasmids in K. pneumoniae, with blaOXA-48, blaOXA-232, and blaOXA-181 being highly prevalent.
The blaOXA-48-carrying plasmids were the most prevalent, accounting for 53.40% of the 191 blaOXA-48-like-harboring plasmids in K. pneumoniae. Currently, OXA-48 is the most common OXA-48-like carbapenemase worldwide; it was first reported in 2004 on a 70-kb plasmid of K. pneumoniae isolated in Turkey (Poirel et al., 2004). After the first report, the presence of OXA-48 was reported in many members of Enterobacterales (Pitout et al., 2019). Our results showed that IncL/M-type conjugative plasmids were important carriers of blaOXA-48 in K. pneumoniae, mainly IncL plasmids, followed by IncM1 and IncM2 plasmids. The broad-host-range IncL/M-type plasmids are now frequently found in environmental and clinical strains (Potron et al., 2013; Woerther et al., 2018), which have been demonstrated as contributors to the dissemination of genes encoding broad-spectrum β-lactam resistance, including blaOXA-48 (Carrër et al., 2008) blaNDM-1 (Aubert et al., 2003), and blaCTX-M-3 (Oteo et al., 2015). The spread of blaOXA-48 is largely driven by Tn1999 and its variants, which are situated on pOXA-48a-like IncL/M-type conjugative plasmids (Potron et al., 2013). In our study of blaOXA-48-carrying IncL/M-type plasmids in K. pneumoniae, the blaOXA-48 was mainly located on the composite transposon Tn1999.2, a variant of Tn1999. Tn1999 contains two copies of IS1999; one copy is located upstream of blaOXA-48, and another is situated downstream of blaOXA-48–lysR. IS1999 was first reported in Pseudomonas aeruginosa isolates from Thailand and was inserted into the integron-specific recombination site attI1 upstream of blaVEB-1 (Aubert et al., 2003). Tn1999.2, first described in Turkey from 2006 to 2007, is a Tn1999 variant with an IS1R inserted into IS1999 upstream of blaOXA-48, generating a strong hybrid promoter, resulting in higher enzymatic activity than that of Tn1999 (Carrër et al., 2008).
In our study, blaOXA-232-harboring plasmids were the second most common plasmids carrying blaOXA-48-like genes in K. pneumoniae, accounting for 30.89% of the 191 blaOXA-48-like-harboring plasmids. The variant blaOXA-232 was first found in 2012 in K. pneumoniae and E. coli isolates obtained from French patients who had traveled to India (Potron et al., 2013). In China, the blaOXA-232 was first reported in 2017 in K. pneumoniae (Yin et al., 2017). In K. pneumoniae, the blaOXA-232 gene was mainly carried by 6.1-kb ColKP3-type mobilizable plasmids. These small ColKP3-type mobilizable plasmids harboring blaOXA-232 carried oriT-like regions characterized by the IR sequence (AAAAGGAAAGTG.CACTTTCCTTTT) and relaxases of the MOBP family characterized by the domain “Relaxase (Pfam: PF03432),” with TraI protein encoded by the IncPα plasmid RP4 (Pansegrau et al., 1993) as a representative.
In this study, blaOXA-181 was another common variant in K. pneumoniae, and plasmids harboring blaOXA-181 accounted for 11.52% of the 191 blaOXA-48-like-harboring plasmids. OXA-181 was first identified in CRKP and Enterobacter cloacae strains isolated from Indian hospitals in 2007 (Castanheira et al., 2011). Since then, OXA-181-producing Enterobacterales, mainly K. pneumoniae and Escherichia coli, have been reported in several countries worldwide (Balm et al., 2013; Liu et al., 2015; Rojas et al., 2017; Piazza et al., 2018; Mouftah et al., 2019; Liu et al., 2020). Four plasmid types belonging to ColKP3, IncX3, IncT, and IncN1 replicons have been reported to harbor the OXA-181 gene (blaOXA-181) (Pitout et al., 2019). Our study showed that the blaOXA-181-harboring plasmids mainly included two categories in K. pneumoniae: one was a group of 50-kb ColKP3-IncX3 hybrid conjugative plasmids, and the other was a group of small ColKP3-type mobilizable plasmids with lengths of 5.9–9.3 kb.
Notably, five blaOXA-245-harboring IncL plasmids with a length of 65.9 kb in K. pneumoniae were found in our analysis, which were all isolated from Spain. OXA-245, differing from OXA-48 in a single amino acid substitution (Glu125Tyr), was first identified in 2011 in a K. pneumoniae isolate collected in Spain (Pérez-Vázquez et al., 2016). In Spain, OXA-245 has been closely related to K. pneumoniae ST11 (Oteo et al., 2015) and also exhibited co-production of CTX-M-15 (Pérez-Vázquez et al., 2016). Similar to blaOXA-48, blaOXA-245 was located in the variant Tn1999 and carried by a 60-kb IncL/M-type plasmid. However, the genetic context of blaOXA-245 carried by the five 65.9-kb IncL plasmids was more complicated than that of blaOXA-48-harboring IncL/M-type plasmids, with two copies of IS1R inserted upstream and downstream of blaOXA-245–lysR.
5. Conclusion
In this study, we analyzed the variants of blaOXA-48-like, replicon types, phylogenetic patterns, geographic distribution, conjugative transfer regions, and the genetic environments surrounding blaOXA-48-like of 191 blaOXA-48-like-harboring plasmids, which were identified from 4451 plasmids of K. pneumoniae downloaded from GenBank. Seven variants of blaOXA-48-like were found among the 191 blaOXA-48-like-harboring plasmids, with blaOXA-48, blaOXA-232, and blaOXA-181 as the most dominant. The blaOXA-48 was mainly harbored by the composite transposon Tn1999.2 located on IncL/M-type conjugative plasmids, which were mainly geographically distributed in Switzerland, Germany, and China. The blaOXA-232 was mainly carried by 6.1-kb ColKP3-type mobilizable plasmids, which were mainly geographically distributed in India. The blaOXA-181 was mainly carried by a group of 50-kb ColKP3-IncX3 hybrid conjugative plasmids (sporadically discovered in China, South Korea, India, and Czech Republic) and a group of small ColKP3-type mobilizable plasmids with lengths of 5.9–9.3 kb (mainly isolated in India). In addition, five blaOXA-245-harboring 65.9-kb IncL plasmids in K. pneumoniae (isolated in Spain) were found to have the genetic context of blaOXA-245 more complicated than that of blaOXA-48-harboring IncL/M-type plasmids. This study provides important insights into the phylogeny and evolution of blaOXA-48-like-harboring plasmids in K. pneumoniae and further addresses their role in the acquisition and spread of resistance genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XL and WL conceived and designed the project. WL, HG and XL analysed all the data and wrote the manuscript. YG, XYa, RL, and SL performed data acquisition. CS, WD, and SC provided the technical assistance. PX and WH provided some suggestions for manuscript writing. XL, XYi and JS reviewed and edited the manuscript. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported financially by the National Natural Science Foundation of China (Grant No. 82002170, 82103018 and 32100006), the project ZR2021QH029 supported by Shandong Provincial Natural Science Foundation, the Science and Technology Planning Project of Zhuhai City (Grant No. ZH22036201210105PWC), the Xiangshan Talent Project of Zhuhai People’s Hospital (Grant No. 2020XSYC-02), the Doctor Foundation of Binzhou University (2019Y30), and the Cultivation Project of Zhuhai People’s Hospital (2019PY-19).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1082813/full#supplementary-material
Abbreviations
CRKP, carbapenem-resistant Klebsiella pneumoniae; AMR, antimicrobial resistance; oriT, origin of transfer; T4CP, type IV coupling protein; T4SS, type IV secretion system.
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48 (D1), D517–d525. doi: 10.1093/nar/gkz935
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Aubert, D., Naas, T., Nordmann, P. (2003). IS1999 increases expression of the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. J. Bacteriol. 185 (17), 5314–5319. doi: 10.1128/JB.185.17.5314-5319.2003
Balm, M. N., La, M. V., Krishnan, P., Jureen, R., Lin, R. T., Teo, J. W. (2013). Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin. Microbiol. Infect. 19 (9), E421–E423. doi: 10.1111/1469-0691.12247
Bengoechea, J. A., Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43 (2), 123–144. doi: 10.1093/femsre/fuy043
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., et al. (2018). GenBank. Nucleic Acids Res. 46 (D1), D41–d47. doi: 10.1093/nar/gkx1094
Bi, D., Liu, L., Tai, C., Deng, Z., Rajakumar, K., Ou, H. Y. (2013). SecReT4: A web-based bacterial type IV secretion system resource. Nucleic Acids Res. 41 (Database issue), D660–D665. doi: 10.1093/nar/gks1248
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345
Branger, C., Ledda, A., Billard-Pomares, T., Doublet, B., Fouteau, S., Barbe, V., et al. (2018). Extended-spectrum β-lactamase-encoding genes are spreading on a wide range of Escherichia coli plasmids existing prior to the use of third-generation cephalosporins. Microb. Genom. 4 (9), e000203. doi: 10.1099/mgen.0.000203
Brown, C. J., Sen, D., Yano, H., Bauer, M. L., Rogers, L. M., van der Auwera, G. A., et al. (2013). Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl. Environ. Microbiol. 79 (24), 7684–7695. doi: 10.1128/AEM.02252-13
Carattoli, A., Hasman, H. (2020). PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Carrër, A., Poirel, L., Eraksoy, H., Cagatay, A. A., Badur, S., Nordmann, P. (2008). Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52 (8), 2950–2954. doi: 10.1128/AAC.01672-07
Castanheira, M., Deshpande, L. M., Mathai, D., Bell, J. M., Jones, R. N., Mendes, R. E. (2011). Early dissemination of NDM-1- and OXA-181-producing enterobacteriaceae in Indian hospitals: report from the SENTRY antimicrobial surveillance program 2006-2007. Antimicrob. Agents Chemother. 55 (3), 1274–1278. doi: 10.1128/aac.01497-10
de Jonge, B. L., Karlowsky, J. A., Kazmierczak, K. M., Biedenbach, D. J., Sahm, D. F., Nichols, W. W. (2016). In vitro susceptibility to ceftazidime-avibactam of carbapenem-nonsusceptible enterobacteriaceae isolates collected during the INFORM global surveillance study, (2012 to 2014). Antimicrob. Agents Chemother. 60 (5), 3163–3169. doi: 10.1128/AAC.03042-15
de la Cruz, F., Frost, L. S., Meyer, R. J., Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34 (1), 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Emms, D. M., Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20 (1), 238. doi: 10.1186/s13059-019-1832-y
Fang, Z., Zhou, H. (2020). Identification of the conjugative and mobilizable plasmid fragments in the plasmidome using sequence signatures. Microb. Genom. 6 (11), mgen000459. doi: 10.1099/mgen.0.000459
Hammer, Ø., Harper, D. A., Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4 (1), 9.
Han, R., Shi, Q., Wu, S., Yin, D., Peng, M., Dong, D., et al. (2020). Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant enterobacteriaceae isolated from adult and children patients in China. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00314
Karlowsky, J. A., Lob, S. H., Kazmierczak, K. M., Badal, R. E., Young, K., Motyl, M. R., et al. (2017). In vitro activity of imipenem against carbapenemase-positive enterobacteriaceae isolates collected by the SMART global surveillance program from 2008 to 2014. J. Clin. Microbiol. 55 (6), 1638–1649. doi: 10.1128/jcm.02316-16
Kim, J. S., Yu, J. K., Jeon, S. J., Park, S. H., Han, S., Park, S. H., et al. (2021). Dissemination of an international high-risk clone of escherichia coli ST410 co-producing NDM-5 and OXA-181 carbapenemases in Seoul, republic of Korea. Int. J. Antimicrob. Agents 58 (6), 106448. doi: 10.1016/j.ijantimicag.2021.106448
Le Roux, F., Davis, B. M., Waldor, M. K. (2011). Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 39 (3), 1004–1013. doi: 10.1093/nar/gkq852
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 (W1), W242–W245. doi: 10.1093/nar/gkw290
Liu, C., Fang, Y., Zeng, Y., Lu, J., Sun, Q., Zhou, H., et al. (2020). First report of OXA-181-producing klebsiella pneumoniae in China. Infect. Drug Resist. 13, 995–998. doi: 10.2147/IDR.S237793
Liu, Y., Feng, Y., Wu, W., Xie, Y., Wang, X., Zhang, X., et al. (2015). First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob. Agents Chemother. 59 (8), 5022–5025. doi: 10.1128/aac.00442-15
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46 (W1), W229–w234. doi: 10.1093/nar/gky352
Mouftah, S. F., Pál, T., Darwish, D., Ghazawi, A., Villa, L., Carattoli, A., et al. (2019). Epidemic IncX3 plasmids spreading carbapenemase genes in the united Arab Emirates and worldwide. Infect. Drug Resist. 12, 1729–1742. doi: 10.2147/idr.s210554
Naas, T., Oueslati, S., Bonnin, R. A., Dabos, M. L., Zavala, A., Dortet, L., et al. (2017). Beta-lactamase database (BLDB)–structure and function. J. Enzyme Inhib Med. Chem. 32 (1), 917–919. doi: 10.1080/14756366.2017.1344235
Navon-Venezia, S., Kondratyeva, K., Carattoli, A. (2017). Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41 (3), 252–275. doi: 10.1093/femsre/fux013
Norberg, P., Bergström, M., Jethava, V., Dubhashi, D., Hermansson, M. (2011). The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2 (1), 1–11. doi: 10.1038/ncomms1267
Norman, A., Hansen, L. H., Sørensen, S. J. (2009). Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R Soc. Lond B Biol. Sci. 364 (1527), 2275–2289. doi: 10.1098/rstb.2009.0037
Oteo, J., Ortega, A., Bartolomé, R., Bou, G., Conejo, C., Fernández-Martínez, M., et al. (2015). Prospective multicenter study of carbapenemase-producing enterobacteriaceae from 83 hospitals in Spain reveals high in vitro susceptibility to colistin and meropenem. Antimicrob. Agents Chemother. 59 (6), 3406–3412. doi: 10.1128/AAC.00086-15
Pandey, D., Singhal, N., Kumar, M. (2021). Investigating the OXA variants of ESKAPE pathogens. Antibiotics (Basel) 10 (12), 1539. doi: 10.3390/antibiotics10121539
Pansegrau, W., Schröder, W., Lanka, E. (1993). Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U S A. 90 (7), 2925–2929. doi: 10.1073/pnas.90.7.2925
Pérez-Vázquez, M., Oteo, J., García-Cobos, S., Aracil, B., Harris, S. R., Ortega, A., et al. (2016). Phylogeny, resistome and mobile genetic elements of emergent OXA-48 and OXA-245 Klebsiella pneumoniae clones circulating in Spain. J. Antimicrob. Chemother. 71 (4), 887–896. doi: 10.1093/jac/dkv458
Piazza, A., Comandatore, F., Romeri, F., Pagani, C., Floriano, A. M., Ridolfo, A., et al. (2018). First report of an ST410 OXA-181 and CTX-M-15 coproducing Escherichia coli clone in Italy: A whole-genome sequence characterization. Microb. Drug Resist. 24 (8), 1207–1209. doi: 10.1089/mdr.2017.0366
Pitout, J. D., Nordmann, P., Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59 (10), 5873–5884. doi: 10.1128/AAC.01019-15
Pitout, J. D., Peirano, G., Kock, M. M., Strydom, K.-A., Matsumura, Y. (2019). The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 33 (1), e00102–e00119. doi: 10.1128/CMR.00102-19
Poirel, L., Héritier, C., Tolün, V., Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48 (1), 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Potron, A., Rondinaud, E., Poirel, L., Belmonte, O., Boyer, S., Camiade, S., et al. (2013). Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class d β-lactamase from enterobacteriaceae. Int. J. Antimicrob. Agents 41 (4), 325–329. doi: 10.1016/j.ijantimicag.2012.11.007
Ramsay, J. P., Firth, N. (2017). Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr. Opin. Microbiol. 38, 1–9. doi: 10.1016/j.mib.2017.03.003
Ravi, A., Valdés-Varela, L., Gueimonde, M., Rudi, K. (2018). Transmission and persistence of IncF conjugative plasmids in the gut microbiota of full-term infants. FEMS Microbiol. Ecol. 94 (1), fix158. doi: 10.1093/femsec/fix158
Rojas, L. J., Hujer, A. M., Rudin, S. D., Wright, M. S., Domitrovic, T. N., Marshall, S. H., et al. (2017). NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the americas. Antimicrob. Agents Chemother. 61 (7), e00454-17. doi: 10.1128/aac.00454-17
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in enterobacteriaceae. J. Antimicrob. Chemother. 73 (5), 1121–1137. doi: 10.1093/jac/dkx488
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 (Database issue), D32–D36. doi: 10.1093/nar/gkj014
Sitter, T. L., Vaughan, A. L., Schoof, M., Jackson, S. A., Glare, T. R., Cox, M. P., et al. (2021). Evolution of virulence in a novel family of transmissible mega-plasmids. Environ. Microbiol. 23 (9), 5289–5304. doi: 10.1111/1462-2920.15595
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P., de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74 (3), 434–452. doi: 10.1128/mmbr.00020-10
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 (7), 1009–1010. doi: 10.1093/bioinformatics/btr039
Woerther, P.-L., Jardak, T., Ben Hassine, I., Forget, S., Chachaty, E., Arlet, G., et al. (2018). A long-term study of the diversity of OXA-48-like carbapenemase-producing bacterial strains in infected patients and carriers. Microb. Drug Resist. 24 (2), 181–189. doi: 10.1089/mdr.2017.0060
Wyres, K. L., Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Keywords: Klebsiella pneumoniae, plasmid, blaOXA-48-like, conjugative transfer region, genetic context
Citation: Li W, Guo H, Gao Y, Yang X, Li R, Li S, Sun C, Du W, Chen S, Xu P, Huang W, Shi J, Yi X and Li X (2022) Comparative genomic analysis of plasmids harboring blaOXA-48-like genes in Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 12:1082813. doi: 10.3389/fcimb.2022.1082813
Received: 28 October 2022; Accepted: 05 December 2022;
Published: 20 December 2022.
Edited by:
Biao Tang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Zhihai Liu, Qingdao Agricultural University, ChinaAzer Özad Düzgün, Gumushane University, Turkey
Copyright © 2022 Li, Guo, Gao, Yang, Li, Li, Sun, Du, Chen, Xu, Huang, Shi, Yi and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobin Li, li.xiaobin2009@163.com; Xinfeng Yi, xinfeng8194@sina.com; Jia Shi, shijia117@163.com
†These authors have contributed equally to this work
 Wang Li
Wang Li Hengzhao Guo
Hengzhao Guo Yi Gao1
Yi Gao1  Wen Du
Wen Du Xiaobin Li
Xiaobin Li