Corrigendum: The sole DNA ligase in Entamoeba histolytica is a high-fidelity DNA ligase involved in DNA damage repair
- 1Posgrado en Ciencias Genómicas, Universidad Autónoma de la Ciudad de México, Mexico City, Mexico
- 2Consejo Nacional de Ciencia y Tecnología, Mexico City, Mexico
- 3Departamento de Infectómica y Patogénesis Molecular, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico City, Mexico
- 4Laboratorio Nacional de Genómica para la Biodiversidad, Centro de Investigación y de Estudios Avanzados, Irapuato, Mexico
A Corrigendum on
The soleDNA ligase in Entamoeba histolytica is a high-fidelity DNA ligase involved in DNA damage repair
By Azuara-Liceaga E, Betanzos A, Cardona-Felix CS, Castañeda-Ortiz EJ, Cárdenas H, Cárdenas-Guerra RE, Pastor-Palacios G, García-Rivera G, Hernández-Álvarez D, Trasviña-Arenas CH, Diaz-Quezada C, Orozco E and Brieba LG (2018) Front. Cell. Infect. Microbiol. 8:214. doi: 10.3389/fcimb.2018.00214
Error in Figure/Table
In the published article, there was an error in FIGURE 1B. Assessment of rEhDNAligI fidelity on nicked double stranded DNA mismatches, as published. In FIGURE 1B, part of the figure that corresponds to lanes 10 to 18 (G:T mismatch) was accidentally duplicated in the part of the figure that corresponds to the A:T pair (lanes 19 to 26). The corrected FIGURE 1B and its caption appear below.
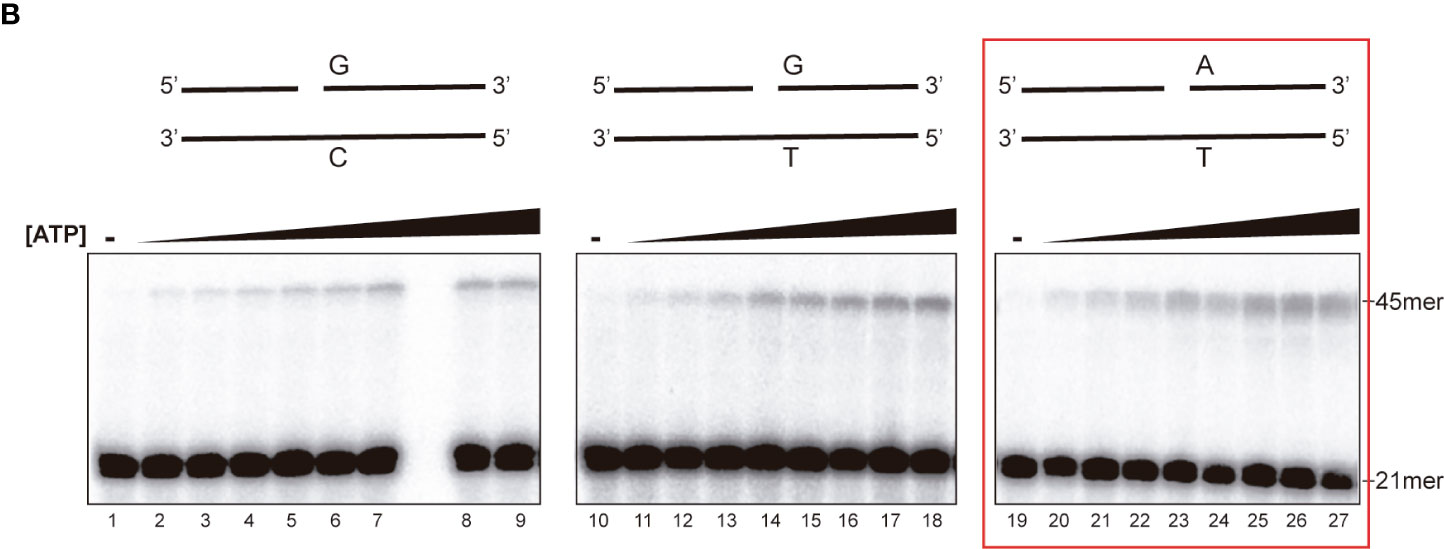
Figure 1B Steady-state kinetics of EhDNAligI on the G:T mismatch at different concentrations of ATP. Lanes 1–9 show EhDNAligI activity at different ATP concentrations in comparison to canonical G:C substrate, lanes 10–18 monitored ligase activity with a mismatched G:T substrate, lanes 19–27 indicate activity with the canonical A:T substrate. The 21mer substrate and the 45mer ligation products are marked with arrows.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: EhDNAligI, protozoan, DNA insults, ligation, repairing, 8-oxoG adduct, NER and BER pathways
Citation: Azuara-Liceaga E, Betanzos A, Cardona-Felix CS, Castañeda-Ortiz EJ, Cárdenas H, Cárdenas-Guerra RE, Pastor-Palacios G, García-Rivera G, Hernández-Álvarez D, Trasviña-Arenas CH, Diaz-Quezada C, Orozco E and Brieba LG (2022) Corrigendum: The sole DNA ligase in Entamoeba histolytica is a high-fidelity DNA ligase involved in DNA damage repair. Front. Cell. Infect. Microbiol. 12:1023564. doi: 10.3389/fcimb.2022.1023564
Received: 19 August 2022; Accepted: 26 September 2022;
Published: 16 December 2022.
Edited and Reviewed by:
Serge Ankri, Technion Israel Institute of Technology, IsraelCopyright © 2022 Azuara-Liceaga, Betanzos, Cardona-Felix, Castañeda-Ortiz, Cárdenas, Cárdenas-Guerra, Pastor-Palacios, García-Rivera, Hernández-Álvarez, Trasviña-Arenas, Diaz-Quezada, Orozco and Brieba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Azuara-Liceaga, elisa.azuara@uacm.edu.mx; Luis G. Brieba, luis.brieba@cinvestav.mx
†Present addresses: Cesar S. Cardona-Felix, Centro Interdisciplinario de Ciencias Marinas de Instituto Politécnico Nacional, La Paz, Mexico
Guillermo Pastor-Palacios, División de Biología Molecular, Instituto Potosino de Investigación Científica y Tecnológica, San Luis Potosí, Mexico
 Elisa Azuara-Liceaga
Elisa Azuara-Liceaga Abigail Betanzos
Abigail Betanzos Cesar S. Cardona-Felix
Cesar S. Cardona-Felix Elizabeth J. Castañeda-Ortiz1
Elizabeth J. Castañeda-Ortiz1  Guillermina García-Rivera
Guillermina García-Rivera Carlos H. Trasviña-Arenas
Carlos H. Trasviña-Arenas Esther Orozco
Esther Orozco Luis G. Brieba
Luis G. Brieba