Piscirickettsia salmonis Produces a N-Acetyl-L-Homoserine Lactone as a Bacterial Quorum Sensing System-Related Molecule
- 1Laboratorio de Biopelículas y Microbiología Ambiental, Centro de Biotecnología, Universidad de Concepción, Concepción, Chile
- 2Departamento de Ciencias Biológicas, Facultad de Ciencias de la Vida, Universidad Andres Bello, Talcahuano, Chile
- 3Departamento de Investigación y Desarrollo, Ecombio limitada, Concepción, Chile
- 4Laboratorio de Investigaciones Medioambientales de Zonas Áridas (LIMZA), Depto. Ingeniería Mecánica, Facultad de Ingeniería, Universidad de Tarapacá, Arica, Chile
- 5Departamento de Química Analítica e Inorgánica, Facultad de Ciencias Químicas, Universidad de Concepción, Concepción, Chile
- 6Departamento de Investigación y Desarrollo, Micbiotech Spa, Concepción, Chile
- 7Laboratorio de Inmunología y Estrés de Organismos Acuáticos, Facultad de Ciencias Veterinarias, Instituto de Patología Animal, Valdivia, Chile
- 8Interdisciplinary Center for Aquaculture Research (INCAR), Concepción, Chile
- 9Departamento de Microbiología, Facultad de Ciencias Biológicas, Universidad de Concepción, Concepción, Chile
Piscirickettsia salmonis is the etiological agent of piscirickettsiosis, the most prevalent disease in salmonid species in Chilean salmonids farms. Many bacteria produce N-acyl-homoserine lactones (AHLs) as a quorum-sensing signal molecule to regulate gene expression in a cell density-dependent manner, and thus modulate physiological characteristics and several bacterial mechanisms. In this study, a fluorescent biosensor system method and gas chromatography-tandem mass spectrometry (GC/MS) were combined to detect AHLs produced by P. salmonis. These analyses revealed an emitted fluorescence signal when the biosensor P. putida EL106 (RPL4cep) was co-cultured with both, P. salmonis LF-89 type strain and an EM-90-like strain Ps007, respectively. Furthermore, the production of an AHL-type molecule was confirmed by GC/MS by both P. salmonis strains, which identified the presence of a N-acetyl-L-homoserine Lactone in the supernatant extract. However, It is suggested that an alternate pathway could synthesizes AHLs, which should be address in future experiments in order to elucidate this important bacterial process. To the best of our knowledge, the present report is the first to describe the type of AHLs produced by P. salmonis.
Introduction
Piscirickettsiosis is the main infectious disease affecting Chilean aquaculture (Figueroa et al., 2019). This disease produces high mortality in Atlantic salmon (Salmo salar), Coho salmon (Oncorhynchus kisutch) and rainbow trout (Oncorhynchus mykiss) and causes serious economic losses (Rozas and Enríquez, 2014). Piscirickettsiosis is a systemic disease generated by Piscirickettsia salmonis, a Gram-negative facultative intracellular γ-proteobacterium, which is able to evade the host immune response and establishes a replicative vacuole in the cytoplasm (Ortiz-Severín et al., 2019; Pérez-Stuardo et al., 2020). Current prevention strategies for fish farmed in seawater are based on vaccination; of the 54 registered fish vaccines in Chile, 34 are for Piscirickettsiosis (SAG, 2021). However, protection by vaccination has been difficult to achieve in the long term under field conditions (Figueroa et al., 2019). Additionally, In addition, antibiotics treatments used in aquaculture have not been completely effective in reducing the development of salmonid rickettsial septicemia (SRS) (Miranda et al., 2018), probably due to antibiotic resistance acquired by the pathogen (Henríquez et al., 2016; Sandoval et al., 2016; Cartes et al., 2017) and SRS outbreaks are recurrent in Chile (Sernapesca, 2021). Several studies have described the intracellular life cycle of P. salmonis (McCarthy et al., 2008; Rojas et al., 2009; Pérez-Stuardo et al., 2019; Zúñiga et al., 2020) and even the biofilm formation as a survival strategy of P. salmonis in the seawater (Marshall et al., 2012; Levipan et al., 2020). However, the molecular mechanisms of P. salmonis communication its responses to specific external stimuli are largely unknown.
Bacteria use Quorum Sensing (QS)-mediated communication systems to coordinate physiological functions such as DNA uptake, biofilm formation, defense strategies and secretion of virulence factors (Duddy and Bassler, 2021; Wu and Luo, 2021). The expression of virulence genes via QS includes I- the synthesis of QS signal molecules; II- the release of signaling molecules; III- sensing and binding of the signaling molecules to membrane receptors at high cell density; IV- the retrieval of the receptor-signal complex from the cell and its binding to the promoter region; and V- the transcription of pathogenicity-related genes (Deng et al., 2011; Durán et al., 2016). However, the question of whether the expression of these virulence factors is or not regulated by a QS system in P. salmonis has not yet been studied.
Many γ-proteobacteria orchestrate their behavior in a cell density-dependent manner via N-acyl-homoserine lactones (AHLs) (Miller and Bassler, 2001), which consist of a polar homoserine lactone head group and a nonpolar aliphatic tail (Fuqua and Greenberg, 2002; Whiteley et al., 2017). Most of the described AHLs are fatty acyl AHLs with acyl groups of various lengths (C4 to C18). In addition, an AHL molecule containing a C2- acyl group has recently been reported in Gluconacetobacter strains (Liu et al., 2019). AHLs possess either a hydroxyl, carbonyl, or no substitution on the third carbon and have various degrees of side-chain saturation (Whitehead et al., 2001; Waters and Bassler, 2005). A typical AHL-QS system contains an AHL signal synthase (LuxI) and a transcriptional regulator (LuxR) (Tsai and Winans, 2010; Papenfort and Bassler, 2016). For example, QS in P. aeruginosa is regulated by N-3-oxo-dodecanoyl L-homoserine lactone (3-oxo-C12-AHL) and N-butyryl L-homoserine lactone (C4-AHL), which bind to intracellular LuxR-type receptors and induce the expression of LuxI-type AHL synthases (Brotherton et al., 2018). However, bacteria can contain multiple luxI-luxR homologs and possess orphan or “solo” luxR homologs (Hudaiberdiev et al., 2015). Therefore, many bacteria also contain extra copies of luxR transcriptional regulators that are not proximal to a luxI synthase gene (Gan et al., 2015). LuxI and luxR homologs have been identified in diverse species throughout the Gram-negative Proteobacteria, including phototrophic purple non-sulfur bacteria, marine Vibrios, rhizosphere bacteria, enteric commensals and opportunistic pathogens of plants and animals (Parsek and Greenberg, 2000; Gray and Garey, 2001). It has been reported that strains of P. salmonis have between 2 and 5 copies of luxR (Millar et al., 2018), and LuxR is present in the outer membrane vesicles of P. salmonis LF-89 type strain (Oliver et al., 2017). However, there is no evidence of synthase LuxI gene orthologs or luxI-like genes in P. salmonis.
Recently, Levipán et al. (2021) reported that P. salmonis does not possess a functional QS circuit based on the endogenous production of AHL-type molecules. However, the authors suggest P. salmonis could have a functional QS system in the presence of exogenous AHL-type molecules. Little is known about AHLs-type molecules production in marine pathogens, and AHL production by P. salmonis has not been conclusively documented. However, fluorescence-based biosensors have advantages over other methods due to the fluorescence intensity of single cells can be measured (Rai et al., 2015). Therefore, the selection of the most appropriate fluorescent biosensor bacterium for P. salmonis is important in order to optimize the detection of fluorescence induced by AHL-like molecules. We aimed to determine if P. salmonis can produce active QS AHLs-type molecules by using green fluorescent protein-based biosensor strains and if so, to identify the types of these molecules by Gas-chromatography coupled to mass spectrometry.
Material and Methods
Bacterial Culture
P. salmonis LF-89 (equivalent to ATCC VR-1361) type strain and an EM-90-like isolate (Ps007) were routinely grown on AUSTRAL-TSFe agar plates at 18°C for ten days (Yáñez et al., 2013). For all experiments, P. salmonis colonies were grown in AUSTRAL-SRS broth (ASB) at 18°C for 120 h and 75 rpm (Yañez et al., 2012). The identity of each isolate was confirmed by phenotyping procedures and PCR assays (Karatas et al., 2008; Isla et al., 2021). Bacterial cell density was measured using the BacLight™ LIVE/DEAD Viability Kit (Invitrogen™, USA). To verify the purity of P. salmonis in all the tests performed, before and after the cultures, direct microscopy methods were carried out like Gram staining and/or epifluorescence microscopy using the BacLight™ LIVE/DEAD Viability Kit (SYTO 9/propidium Iodide) (Invitrogen™, USA). In parallel, an aliquot was transferred onto tryptone soy agar plates supplemented with hemoglobin (Austral-TSHem) at 18°C for 10-15 days to validate the purity results by observing the growth of only P. salmonis colonies, and additional Gram staining procedures.
Selection of Bacterial Biosensor for AHLs Detection
The biosensor with the highest sensitivity for N-Octanoyl-L-homoserine lactone (C8-HSL) molecules was selected by testing the Pseudomonas putida EL105 (RPL4las), P. putida EL106 (RPL4cep) and Escherichia coli MT102 (pKR-C12) biosensor strains (Lumjiaktase et al., 2010). Bacterial strains were previously grown overnight at 27°C in Luria Bertani (LB Miller, Merck) agar plates with Kanamycin (100 μg/mL, Sigma-Aldrich, St. Louis, MO) and Rifampicin (50 μg/mL, Sigma-Aldrich, St. Louis, MO) for Pseudomonas strains and only Gentamicin (25 μg/mL, US Biological) for E. coli. Then, single colonies of each strain were inoculated in 25 mL of LB broth (Miller, Merck) with antibiotics and incubated for 12 h as described above. Bacterial cells were then resuspended and washed three times in milli-Q water by centrifugation at 8,000 rpm for 15 min at 4°C. The cell pellet was resuspended in LB broth with four μM C8-HSL (HPLC-grade; Sigma-Aldrich, St. Louis, MO) at a 1x109 cells/mL final density and then incubated for 24 h at 27°C and 120 rpm as previously described (Lumjiaktase et al., 2010). After that, P. putida EL106 (RPL4cep) was grown in the presence of C8-HSL in a concentration range from 10 nM to 1600 nM in order to determine the optimal concentration to use in the experiments. Bacterial cells grown without the addition of AHLs in the medium were used as controls. Fluorescence was measured in 96-wells black plates at 475 nm excitation wavelength and 515 nm emission detection in a microplate reader (TECAN infinite, F200 pro). Measurements were done in triplicate, and green fluorescence values were normalized to 1 mL of bacterial culture and converted into specific green fluorescence by dividing normalized values by the OD600 of the bacterial culture. Data was obtained using i-control™ Microplate Reader software. Subsequently, the biosensor bacterial strain that showed significantly highest fluorescence was also evaluated at 18°C in the same conditions as previously described to evaluate the successful growth and fluorescence emission of biosensor bacterial strains in subsequent assays with P. salmonis. The fluorescence measurements were corrected for autofluorescence.
Induction of AHLs Biosensor by P. salmonis Strains
P. putida EL106 (RPL4cep) strain was grown in LB broth with the corresponding antibiotic. Bacterial cells were pelleted and washed three times in milli-Q water by centrifugation at 8,000 rpm for 15 min and 4°C. Subsequently, P. salmonis LF-89 and Ps007 strains were grown in ASB for 120 h at 18°C and 75 rpm. After that, the bacterial biosensor was resuspended in ASB at 105 and 109 cells/mL and co-cultured with 109 cells/mL of each P. salmonis strain. Cultures were incubated for 24, 96, 144 and 168 h at 18°C and 75 rpm. The bacterial biosensor was resuspended in ASB with 250 nM C8-HSL was used as an inductor response control. This concentration was the highest concentration, within the lowest range of HSL-C8 concentrations used (10 to 250 nM), that emitted the highest fluorescence values, allowing us to optimize the use of the pure inductor and be able to quantify the emitted fluorescence. Fluorescence was measured in triplicate in 96-wells black plates in a microplate reader (TECAN infinite, F200 pro) set at an excitation wavelength of 475 nm and emission detection at 515 nm. Data were obtained using i-control ™ Microplate Reader software. The fluorescence measurements were corrected for autofluorescence.
Extraction of AHLs From P. salmonis Culture Supernatants
AHLs from P. salmonis LF-89 and Ps007 strains culture supernatants were obtained according to the method described by (Shaw et al., 1997) with some modifications. Briefly, each P. salmonis strain was grown in 600 mL of ASB at 18°C for 120 h and 75 rpm. Bacterial cells were centrifuged at 5,000 rpm for 15 min at 4°C, and the supernatant was extracted three times with a ratio of 1:1 of acidified ethyl acetate (0.5% glacial acetic acid). The organic phase obtained was dried with anhydrous MgSO4 (Merck, Darmstadt, Germany), filtered through a cellulose filter (47 mm, 0.22 µm) and concentrated by rotary evaporation at 43 ± 2°C. The dry extract was weighed and resuspended in 5 mL of acetone (GC-ECD/FID grade, Merck, Darmstadt, Germany). The extracts were stored at -20°C until use.
Induction of AHLs Biosensor by P. salmonis Supernatant and Organic Extracts
The filtered supernatant and organic extracts (dissolved in 10% w/v ASB) of each P. salmonis strain were arranged in a 1:1 ratio with the bacterial biosensor P. putida EL106 (RPL4cep) (109 cells/mL), and the emitted fluorescence was evaluated. RFU was measured at 24 and 48 h in triplicate in 96-wells black plates as described above. The fluorescence measurements were corrected for autofluorescence.
Identification of AHLs by GC/MS
The identification of AHLs was performed by GC/MS, similar to Levipan et al. (2021). Briefly, the standard solutions of N-butyryl-DLHomoserine lactone (C4-HSL), N-hexanoyl-DL-homoserine lactone (C6-HSL), N-octanoyl-L-homoserine (C8-HSL), and N-dodecanoyl-L-Homoserine lactone (C12-HSL) (HPLC-grade; Sigma-Aldrich, St. Louis, MO) were prepared in GC-ECD/FID-grade acetone. Analysis was performed using a model 7890A GC system (Agilent, USA) interfaced to a single quadrupole mass selective detector 5975C (triple-axis detector), both of which were controlled by a computer equipped with ChemStation software (Agilent, USA) and an internal NIST Mass Spectral Search Program (NIST v.02, 2009). Sample injection was done in splitless mode into an HP-5MS capillary column, 30 m x 250 mm i.d. and 0.25 mm film thickness (Agilent, USA). Pure helium (99.999%, Linde, Chile) was used as the GC carrier gas at a flow rate of 1.0 mL min-1. The GC injector temperature was set at 250°C. The GC column-oven temperature was set at 60°C holding for 2 min, then up to 270°C and maintained by 5 min with a heating rate of 10°C min−1 (Levipan et al., 2021). One μL of extracted samples and AHLs standards previously diluted with acetone was injected into the GC system for the AHLs identification. The transfer line temperature was adjusted to 280°C. Mass spectrometry conditions were as follows: electron ionization source set to 70 eV, MS Quad 150°C, MS Source 230°C. The mass spectrometer was run in full-scan mode (m/z 15–350) and Selected Ion Monitoring (SIM) at m/z 143.
Statistical Analysis
Graphpad Prism v7 (GraphPad Software Inc., La Jolla, CA, USA) was used for dataset analysis and graphing of all experiments. Differences between experimental conditions were deemed statistically significant at P ≤ 0.01, as established using Student’s t-test and one-way ANOVA with Tukey Multiple Comparison test (Zar, 1999).
Results
Screening of the Fluorescent Bacterial Biosensors for the AHLs Detection
The emitted fluorescence of each biosensor strain at different culture times was determined in order to select the strain with the highest sensitivity for AHLs. Bacterial biosensors were grown in the presence of C8-HSL in a concentration range from 10 nM to 1600 nM. Bacterial cells grown without the addition of AHLs in the medium were used as controls. Thus, C8-HSL at a concentration of 250 nM was selected for the assay (Supplementary Figure 1). We determined that the solvent control (DMSO) for E. coli MT102 (pKR - C12) produced high autofluorescence that prevented the detection of C8-HSL. The emitted fluorescence of P. putida EL105 (RPL4las) and P. putida EL106 (RPL4cep) was significantly (ANOVA, P ≤ 0.01) higher than the control (Figure 1A). Importantly, the emitted fluorescence by P. putida EL106 (RPL4cep) was significantly (ANOVA, P ≤ 0.01) higher than P. putida EL105 (RPL4las). The emitted fluorescence by P. putida EL106 (RPL4cep) in the presence of 250 nM AHL-C8 at 18°C was 7232 ± 27.2 RFU, which was 70% lower than the fluorescence at 27°C (24158 ± 80.3 RFU) (Figure 1B), indicating it was completely functional at the lowest temperature tested and could be used to evaluate the production of AHLs by P. salmonis.
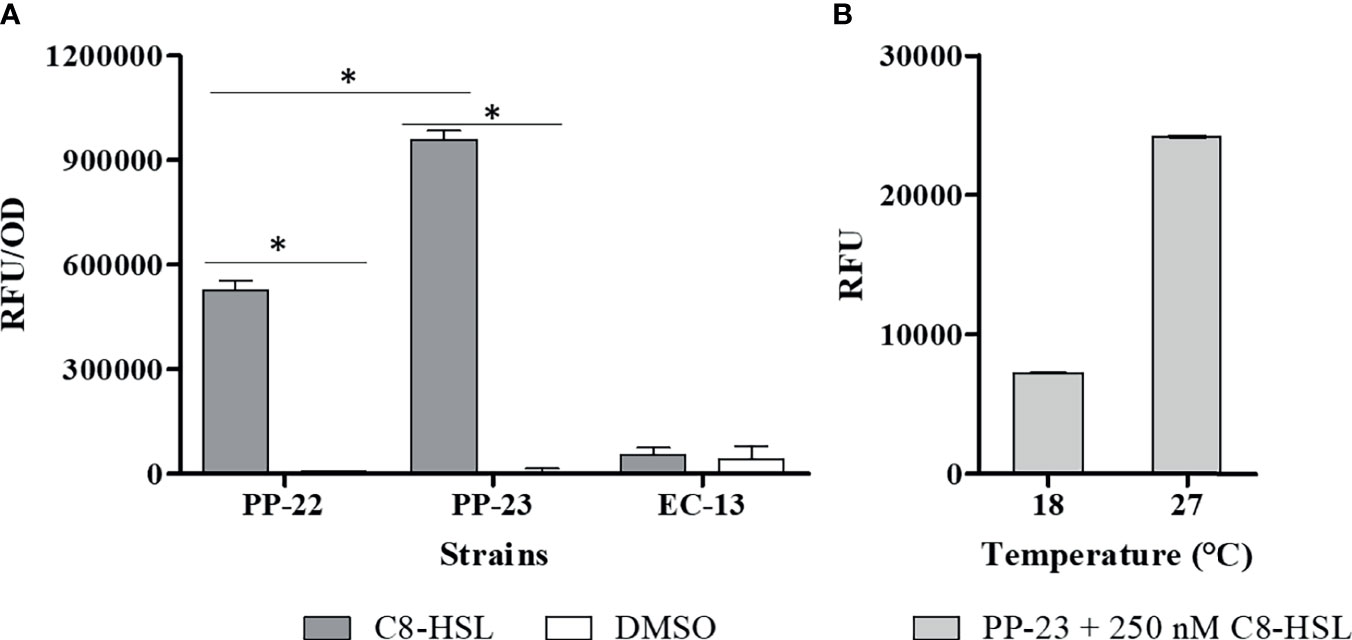
Figure 1 Selection of biosensor strains at the optimal growth conditions of P. salmonis. (A) Response of 4 μM C8-HSL to biosensor strains; PP-22: P. putida EL105 (RPL4las), PP-23: P. putida EL106 (RPL4cep), and EC-12: E. coli MT102 (pKR-C12) at 27°C for 24 h. Control: DMSO (C8-HSL solvent). (B) Response of 250 nM C8-HSL to P. putida EL 106 (RPL4cep) at 18°C and 27°C for 24 h. Emitted fluorescence was recorded in a microtiter plate reader at 515 nm. RFU, relative fluorescence units. Asterisks indicate statistical significance (Student’s t-test P ≤ 0.01, n = 3). Error bars: standard deviation.
P. salmonis Strains Induce an AHL Response in the Bacterial Biosensor
The fluorescence emitted by the selected biosensor P. putida EL106 (RPL4cep) was evaluated in a co-culture with both P. salmonis strains at different concentrations and times. The fluorescent intensity of P. putida EL106 (RPL4cep) was higher in the presence of the two P. salmonis strains and the positive control at 96 h (Figure 2A) at 109 cells/mL. Additionally, the intensity of the fluorescence emitted by the bacterial biosensor increased up to 144 h and 168 h in the presence of the P. salmonis LF-89 and Ps007, respectively, at 105 cell/mL concentration. However, this was also the case up to 96 h in the positive control. Ps007 showed the highest fluorescence intensity in both conditions at 168 h, reaching a maximum of 7825 ± 131 RFU and 2750 ± 79 RFU at a biosensor concentration of 109 CFU/mL and 105 CFU/mL, respectively. LF-89 generated the lowest maximum fluorescence intensity of 203 ± 4 RFU and 997 ± 117 RFU at a biosensor concentration of 109 CFU/mL and 105 CFU/mL, respectively. Based on these results, we decided to use a biosensor concentration of 105 cells/mL and a time of 120 h for our next assay. P. salmonis LF-89 type strain showed an increase in fluorescence intensity (RFU = 2183 ± 380) under these conditions, while the intensity of P. salmonis Ps007 strain was 3209 ± 176 RFU (Figure 2B).
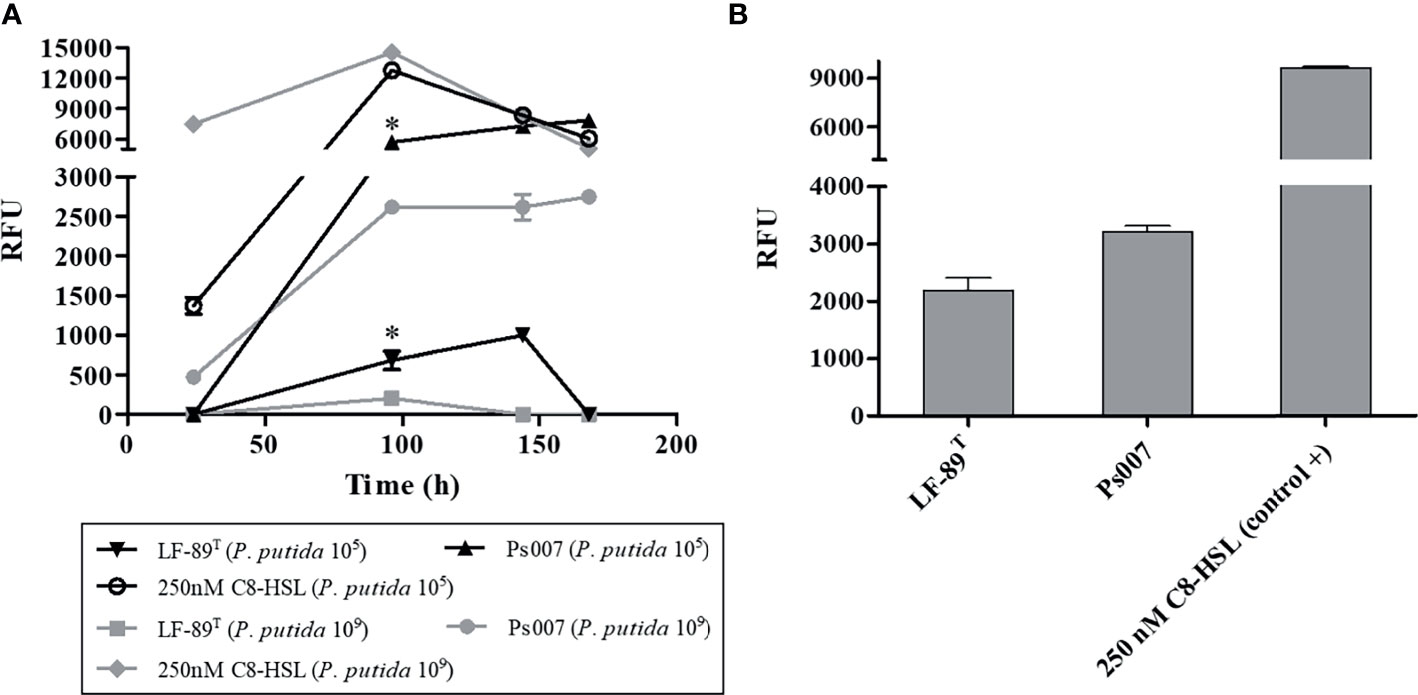
Figure 2 Fluorescence induction in P. putida EL 106 (RPL4cep) in co-culture with P. salmonis LF-89T and EM-90-like (Ps007) strains. (A) P. putida EL 106 (RPL4cep) 105 cell/mL and 109 cell/mL at 24, 96, 144 and 168h. Asterisks indicate statistical significance (ANOVA P ≤ 0.01, n = 3) in P. salmonis Ps007 strain between 105 and 109 cells/mL of the biosensor strain from 96 h to 168 h. In P. salmonis LF-89 strain asterisks indicate statistical significance (ANOVA P ≤ 0.01, n = 3) between 105 and 109 cells/mL of the biosensor strain only at 96 h. (B) P. putida EL 106 (RPL4cep) 105 cell/mL at 120 h. Control: P. putida EL106 (RPL4cep) + 250 nM C8-HSL. Emitted fluorescence was recorded in a microtiter plate reader at 515 nm. RFU, relative fluorescence units. Asterisks indicate statistical significance (ANOVA P ≤ 0.01, n = 3) with the control. Error bars: standard deviation.
P. salmonis Supernatant and Organic Extracts Induce an AHL Response in the Bacterial Biosensor
We only detected fluorescence from the biosensor when evaluating the supernatant from Ps007 at 24 h (Figure 3); however, the supernatants of both strains induced fluorescence at 48 h. We detected fluorescence emission from the organic extracts of both strains at both times. In this sense, the fluorescence intensity induced by both strains was significantly higher at 48h (3864 ± 106 RFU and 3826 ± 55 RFU for P. salmonis LF-89 and Ps007 strains (P < 0.01), respectively) (Figure 3). Furthermore, this emitted fluorescence is significantly (P < 0.01) higher than that induced in the supernatants of the LF-89 and Ps007 strains; 869 ± 50 RFU and 2088 ± 50 RFU, respectively. Therefore, a significant increase in fluorescence measured was observed in the organic extracts with respect to the supernatant for each strain.
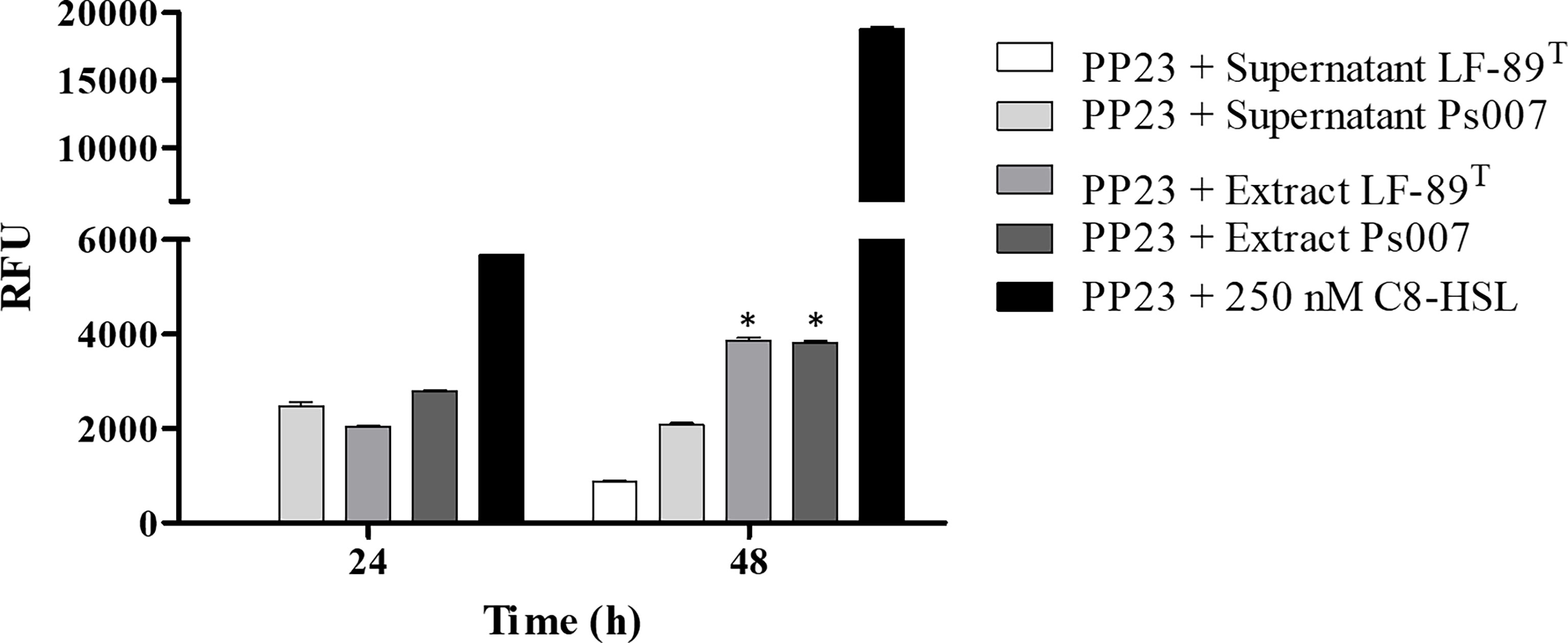
Figure 3 Fluorescence induction in P. putida EL 106 (RPL4cep) of supernatants and organic extracts of P. salmonis at different times. The filtered supernatant and organic extracts (dissolved in 10% w/v ASB) of each P. salmonis strain were arranged in a 1:1 ratio with the bacterial biosensor P. putida EL106 (RPL4cep) (109 cells/mL). Samples and controls were incubated for 24 and 48 h at 27°C. PP23: P. putida EL 106 (RPL4cep); Controls: P. putida EL106 (RPL4cep) (109 cells/mL) + 250 nM C8-HSL. Emitted fluorescence was recorded in a microtiter plate reader at 515 nm. RFU, relative fluorescence units. Asterisks indicate statistical significance (ANOVA P ≤ 0.01, n = 3) between the supernatant and the organic extracts. Error bars: standard deviation.
P. salmonis Produces N-Acetyl-Homoserine Lactone
Four standard solutions were assessed in GC/MS without derivatization using the full-scan acquisition mode: C4-HSL, C6-HSL, C8-HSL, and C12-HSL. We detected and identified the peaks from the respective mass spectra using the retention times of standard samples (Table 1). Similar chromatographic peaks were obtained from organic extracts samples from both strains (Figures 4A, B, respectively). However, most of the peaks did not match the retention times of the assayed standards described in Table 1. Additionally, the C8-HSL and C12-HSL standards matched two peaks in the samples (18.250 min and 22.040 min, respectively). However, the fragmentation pattern in scan mode showed a negligible m/z of 143. The samples were then evaluated using the SIM mode at m/z 143. The SIM chromatogram showed that neither of the possible compounds, C8-HSL or C12-HSL, generated a significant signal at m/z 143. Therefore, none of the available standards were detected in the samples. Nevertheless, the SIM chromatogram showed an intense signal in 10.459 min, where the rest of the peaks had an area at least 1,000 times lower. According to the fragmentation pattern, the compound N-acetyl-homoserine lactone (C2-HSL) (peak SIM at 10.459 min) was the most probable AHL molecule identified in both P. salmonis samples. Nevertheless, the SIM chromatogram showed an intense signal in 10.459 min, where the rest of the spectrum peaks had an area at least 1,000 times lower than m/z 143 (Supplementary Figure 2), becoming in it the molecular ion of this compound (M·+). According to the fragmentation pattern, the compound N-acetyl-homoserine lactone (C2-HSL; C6H9NO3) was the most probable AHL molecule identified in both P. salmonis samples (peak SIM at 10.459 min) due to the m/z fragments detected in this peak match over 80% with the m/z fragments detected for the pure acyl-HSL standards according to NIST Mass Search (Table 1). Additionally, an one-factor ANOVA showed that there not exist differences in the mass fragmentations distribution obtained in the samples when these were compared with the m/z distribution of the pure standards at 95% confidence level (p> 0.05).
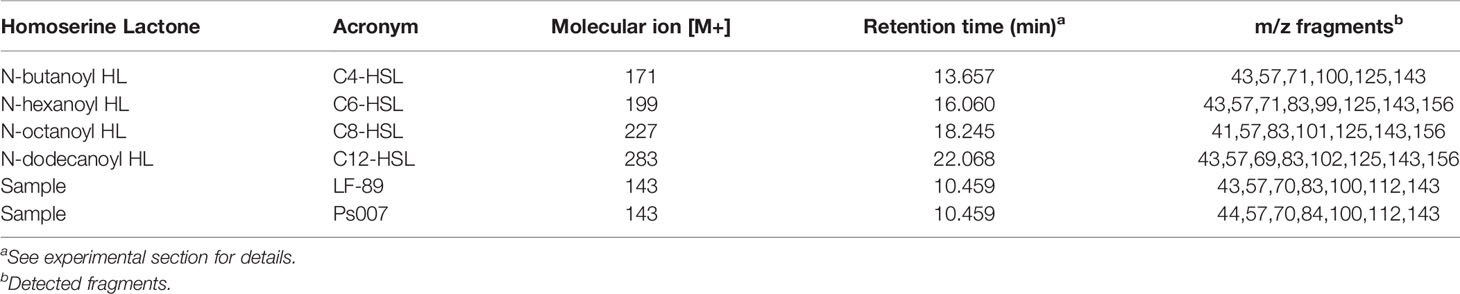
Table 1 Experimental retention times and m/z fragments detected for standards AHLs molecules and samples analyzed.
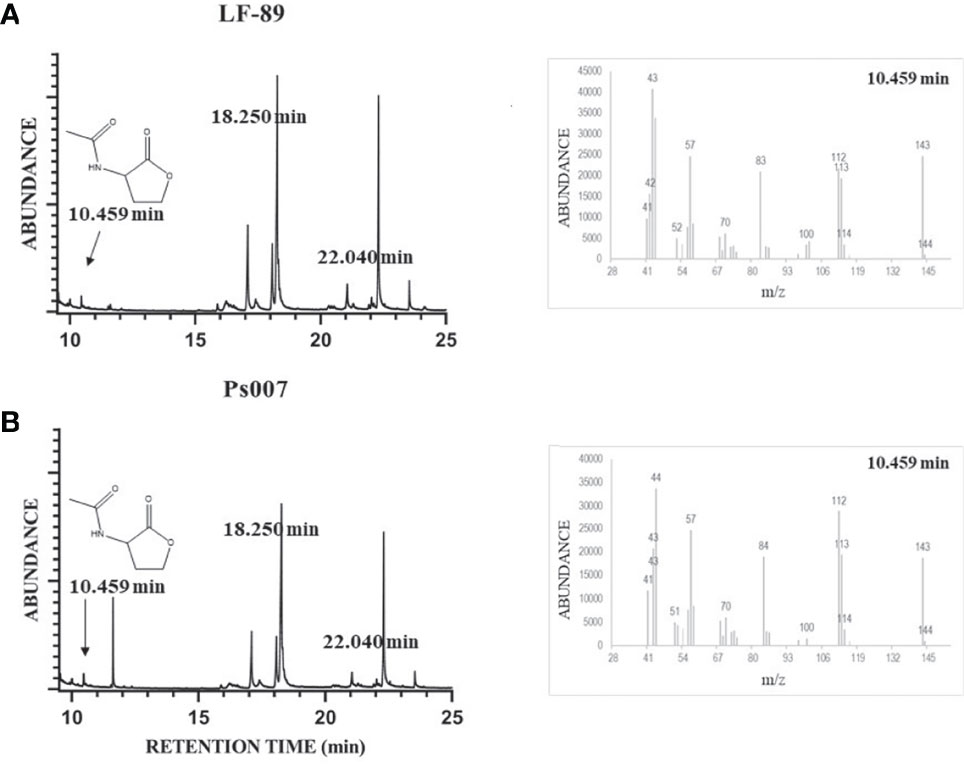
Figure 4 Identification of AHLs molecules in P. salmonis culture organic extract by GC/MS analysis. LF-89T (A) and Ps007 (B) spectrum. Mass spectrum at t=10.459 min was included for both samples. The standard solutions of C4-HSL, C6-HSL, C8-HSL, and C12-HSL were prepared in GC-ECD/FID-grade acetone. MS conditions were as follows: electron ionization source set to 70 eV, MS Quad 150°C, MS Source 230°C. The mass spectrometer was run in full-scan mode (m/z 15–350) and Selected Ion Monitoring (SIM) at m/z 143.The sensor and GC/MS results are representative of three independent experiments.
Discussion
The predominant QS signaling pathway in Gram-negative bacteria is the LuxI/LuxR system that utilizes acyl-homoserine lactones (AHLs) signal molecules (Mok et al., 2003). Several authors described that some salmonid bacterial pathogens, such as Edwardsiella tarda (Morohoshi et al., 2004), Aeromonas hydrophila, A. salmonicida (Swift et al., 1997) and Aliivibrio salmonicida (Hansen et al., 2015), produce AHLs to induce QS-type systems. It has been identified between two and five copies of the luxR gene in P. salmonis but didn’t identify any luxI gene orthologues (Millar et al., 2018). Additionally, LuxR has been reported in the proteome of biologically active membrane vesicles in P. salmonis LF-89 type Strain (Oliver et al., 2017). The known diversity of signals and receptors in microbial communication is still expanding (Chen et al., 2002; Mukherjee and Bassler, 2019). LuxR solo regulators might have evolved by gene duplication and horizontal gene transfer. An increased understanding of the evolutionary roles of QS regulators would be helpful for engineering of cell–cell communication circuits in bacteria (Xu, 2020).
Bacterial biosensors have facilitated the detection of AHLs production via screening-based approaches (Lumjiaktase et al., 2010). Recently, Levipan et al. (2021) did not detect short- and long-chain AHLs-type molecules in two P. salmonis strains via the colorimetric agar plate-based method. This method relies on Chromobacterium violaceum VIR07 and CV026 biosensors strains and C6-HSL and N-hexanoyl-DL-homoserine lactone (C10-HSL) as a positive control. Fluorescence-based biosensors have advantages over other methods because the fluorescence intensity of single cells can be measured (Rai et al., 2015).
We first carried out an experimental standardization focused on the selection of the most appropriate fluorescent biosensor bacterium. Thus, P. putida EL106 (RPL4cep) was selected to evaluate the production of AHLs by P. salmonis. P. putida EL106 (RPL4cep) successfully emitted fluorescent signals when was cultured under the ideal conditions for P. salmonis. This is the first report that suggests this biosensor possesses C2-HSL sensitivity. Nevertheless, the sensor plasmid RP4cep is highly sensitive for AHLs, with acyl side chains ranging from C8 to C12 and has a wide range (C4 to C12) of detection previously reported (Lumjiaktase et al., 2010). We observed the induction of emitted fluorescence by this biosensor strain in supernatant extracts and the organic extracts of P. salmonis LF-89 and Ps007 strains, suggesting that P. salmonis produces AHLs-type molecules. We detected and identified AHLs-type molecules in both P. salmonis strains via GC/MS. The fragmentation pattern observed in each standard coincided with the fragmentation patterns previously described for AHLs molecules (Cataldi et al., 2004). All standards showed the characteristic m/z=143 as one of the main fragments. Other important detected peaks, in the available standards, were m/z 125, 101, 57, and 43, which were similar to the fragments reported by several authors for AHLs (Pearson et al., 1995; Cataldi et al., 2004; Llamas et al., 2005; Cataldi et al., 2007; Rani et al., 2011). When the P. salmonis samples were analyzed it was found that even when retention times of some peaks. Furthermore, even when retention times of some peaks in the P. salmonis samples were close to the standards, the fragmentation patterns did not completely match with those reported for AHLs, and also did not show the characteristic m/z 143 for AHLs-type molecules (Pearson et al., 1995; Levipan et al., 2021). Therefore, this suggests an absence of C4-HSL, C8-HSL, C12-HSL and also C6-HSL, which is in agreement with the previously reported by Levipan et al. (2021) in similar samples. However, this work do not show SIM chromatograms at shorter analysis times, where we have detect a significant signal at 10.459. In this work, the search of m/z 143 was performed in the entire chromatogram time length, finding that exist this small signal in scan mode at 10.459 min that showed a strong signal at SIM mode (m/z=143, Supplementary Figure 2). The fragmentation pattern showed that m/z 57 and 143 were two of the main detected signals, and both are typical for the identification of AHLs in P. salmonis (Pearson et al., 1995; Cataldi et al., 2004; Liu et al., 2019). Additionally, fragments at m/z 43, 84 and 100 were detected, all of which are close to m/z values previously reported for other AHLs (Pearson et al., 1995; Cataldi et al., 2004; Llamas et al., 2005). These outcomes are in agreement with those results reported by Levipan et al., 2021 given by none of the available standards were detected in P. salmonis samples. Nevertheless, at lower retention times this small signal in scan mode at 10.459 min, with a strong signal at SIM mode (Supplementary Figure 2), match in the fragmentation pattern according to the experimental m/z observed for the standards with a coincidence over the 80% in the most important fragments in each sample. Additionally an one-way ANOVA showed that there is not statistical differences between the distribution in the obtained main m/z fragments of each sample compared with the main m/z fragments of the pure standards at 95% confidence level (Supplementary Table 1). Based on these analyses, C2-HSL is AHL-type molecule present in both strains of P. salmonis because the fragmented ion at m/z 143 is typical of carbonyl groups having a hydrogen atom in the γ-position (Supplementary Figure 3). The other fragment ions, i.e., m/z 57, 71 and 43, were probably produced by two major fragmentation processes involving an inductive effect and an α-cleavage (Cataldi et al., 2004). In addition, a peak at m/z 101 was also detected, which could be explained by the α-cleavage of the N-acyclic side chain at the secondary amino group, resulting in the molecular ion formation for de-acylated homoserine lactone (Figure 4). Due to no peaks were detected over m/z 143, this fragment is the molecular ion corresponding to C2-HSL. However, no peaks were detected over m/z 143. Therefore, this fragment would be the molecular ion corresponding to C2-HSL. This QS signaling molecule has been identified by liquid chromatography-tandem mass spectrometry in Gluconacetobacter xylinus CGMCC No. 2955 and Gluconacetobacter sp. strain SX-1, which are acetic acid-producing Gram-negative bacteria and typical bacterial cellulose biosynthesis strains. C2-HSL has been used as a control for QS events because it belongs to a family of cell-to-cell interaction mediators in some bacteria such as Erwinia carotovora ATCC39048, Paraburkholderia sp. BSNB-0670, Gluconacetobacter strains, and others (Chhabra et al., 1993; Liu et al., 2019; Hoang et al., 2020).
In conclusion, this is the first study describing the production of AHL molecules by the fish bacterial pathogen P. salmonis. This valuable information provides a basis for further studies on the QS system of P. salmonis and how it is related to other processes such as pathogenicity, cell-to-cell communication, biofilm formation, antibiotic resistance, and virulence.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
PR, DS, JV, and CO planned most of the experiments and wrote the final version of the manuscript. DS, RR, DC, JB, and CC performed some of the experiments and participated in the writing of the manuscript. AR, NR-T and HU participated in the writing of the manuscript. All authors have approved the final article.
Funding
This work was financially supported by the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) [Grant No. 11180994], Fondo de Financiamiento de Centros de Investigación (FONDAP) Interdisciplinary Center for Aquaculture Research (INCAR) [Grant No. 15110027], FONDAP Solar Energy Research Center (SERC) [Grant No. 15110019], and Vicerrectoría de Investigación, Desarrollo y Creación Artística of the Universidad Austral de Chile (VIDCA-UACh).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are sincerely grateful to Ph.D. Gerardo Cárcamo Oyarce of Research Laboratory Electronics, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA for his technical support and for sharing the biosensor strain Pseudomonas putida EL106 (RPL4cep). The authors also thank Fraunhofer Chile Research for providing the P. salmonis EM-90 like strain used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.755496/full#supplementary-material
References
Brotherton, C. A., Medema, M. H., Greenberg, E. P. (2018). luxR Homolog-Linked Biosynthetic Gene Clusters in Proteobacteria. MSystems 3, e00208–e00217. doi: 10.1128/mSystems.00208-17
Cartes, C., Isla, A., Lagos, F., Castro, D., Muñoz, M., Yañez, A., et al. (2017). Search and Analysis of Genes Involved in Antibiotic Resistance in Chilean Strains of Piscirickettsia Salmonis. J. Fish Dis. 40, 1025–1039. doi: 10.1111/jfd.12579
Cataldi, T., Bianco, G., Frommberger, M., Schmitt-Kopplin, P. (2004). Direct Analysis of Selected N-Acyl-L-Homoserine Lactones by Gas Chromatography/Mass Spectrometry. Rapid Commun. Mass Spectrom 18, 1341–1344. doi: 10.1002/rcm.1480
Cataldi, T. R., Bianco, G., Palazzo, L., Quaranta, V. (2007). Occurrence of N-Acyl-L-Homoserine Lactones in Extracts of Some Gram-Negative Bacteria Evaluated by Gas Chromatography–Mass Spectrometry. Anal. Biochem. 361, 226–235. doi: 10.1016/j.ab.2006.11.037
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural Identification of a Bacterial Quorum-Sensing Signal Containing Boron. Nature 415, 545. doi: 10.1038/415545a
Chhabra, S. R., Stead, P., Bainton, N. J., Salmond, G. P., Stewart, G. S., Williams, P., et al. (1993). Autoregulation of Carbapenem Biosynthesis in Erwinia Carotovora by Analogues of N-(3-Oxohexanoyl)-L-Homoserine Lactone. J. Antibiot Res. 46, 441–454. doi: 10.7164/antibiotics.46.441
Deng, Y., Wu, J. E., Tao, F., Zhang, L.-H. (2011). Listening to a New Language: DSF-Based Quorum Sensing in Gram-Negative Bacteria. Chem. Rev. 111, 160–173. doi: 10.1021/cr100354f
Duddy, O. P., Bassler, B. L. (2021). Quorum Sensing Across Bacterial and Viral Domains. PloS Pathog. 17, e1009074. doi: 10.1371/journal.ppat.1009074
Durán, N., Justo, G. Z., Durán, M., Brocchi, M., Cordi, L., Tasic, L., et al. (2016). Advances in Chromobacterium Violaceum and Properties of Violacein-Its Main Secondary Metabolite: A Review. Biotechnol. Adv. 34, 1030–1045. doi: 10.1016/j.biotechadv.2016.06.003
Figueroa, J., Cárcamo, J., Yañez, A., Olavarria, V., Ruiz, P., Manríquez, R., et al. (2019). Addressing Viral and Bacterial Threats to Salmon Farming in Chile: Historical Contexts and Perspectives for Management and Control. Rev. Aquac 11, 299–324. doi: 10.1111/raq.12333
Fuqua, C., Greenberg, E. P. (2002). Listening in on Bacteria: Acyl-Homoserine Lactone Signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. doi: 10.1038/nrm907
Gan, H. M., Gan, H. Y., Ahmad, N. H., Aziz, N. A., Hudson, A. O., Savka, M. A. (2015). Whole Genome Sequencing and Analysis Reveal Insights Into the Genetic Structure, Diversity and Evolutionary Relatedness of luxI and luxR Homologs in Bacteria Belonging to the Sphingomonadaceae Family. Front. Cell. Infect. Microbiol. 4, 188. doi: 10.3389/fcimb.2014.00188
Gray, K. M., Garey, J. R. (2001). The Evolution of Bacterial LuxI and LuxR Quorum Sensing Regulators. Microbiology 147, 2379–2387. doi: 10.1099/00221287-147-8-2379
Hansen, H., Purohit, A. A., Leiros, H.-K. S., Johansen, J. A., Kellermann, S. J., Bjelland, A. M., et al. (2015). The Autoinducer Synthases LuxI and AinS Are Responsible for Temperature-Dependent AHL Production in the Fish Pathogen Aliivibrio Salmonicida. BMC Microbiol. 15, 1–13. doi: 10.1186/s12866-015-0402-z
Henríquez, P., Kaiser, M., Bohle, H., Bustos, P., Mancilla, M. (2016). Comprehensive Antibiotic Susceptibility Profiling of Chilean Piscirickettsia Salmonis Field Isolates. J. Fish Dis. 39, 441–448. doi: 10.1111/jfd.12427
Hoang, T. P. T., Barthélemy, M., Lami, R., Stien, D., Eparvier, V. R., Touboul, D. (2020). Annotation and Quantification of N-Acyl Homoserine Lactones Implied in Bacterial Quorum Sensing by Supercritical-Fluid Chromatography Coupled With High-Resolution Mass Spectrometry. Anal. Bioanal. Chem. 412, 1–16. doi: 10.1007/s00216-019-02265-4
Hudaiberdiev, S., Choudhary, K. S., Vera Alvarez, R., Gelencsér, Z., Ligeti, B., Lamba, D., et al. (2015). Census of Solo luxR Genes in Prokaryotic Genomes. Front. Cell. Infect. Microbiol. 5, 20. doi: 10.3389/fcimb.2015.00020
Isla, A., Martinez-Hernandez, J. E., Levipan, H. A., Haussmann, D., Figueroa, J., Rauch, M. C., et al. (2021). Development of a Multiplex PCR Assay for Genotyping the Fish Pathogen Piscirickettsia Salmonis Through Comparative Genomics. Front. Microbiol. 12, 673216. doi: 10.3389/fmicb.2021.673216
Karatas, S., Mikalsen, J., Steinum, T., Taksdal, T., Bordevik, M., Colquhoun, D. (2008). Real Time PCR Detection of Piscirickettsia Salmonis From Formalin-Fixed Paraffin-Embedded Tissues. J. Fish Dis. 31, 747–753. doi: 10.1111/j.1365-2761.2008.00948.x
Levipan, H. A., Irgang, R., Yáñez, A., Avendaño-Herrera, R. (2020). Improved Understanding of Biofilm Development by Piscirickettsia Salmonis Reveals Potential Risks for the Persistence and Dissemination of Piscirickettsiosis. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-68990-4
Levipan, H. A., Reyes-Garcia, L., Avendaño-Herrera, R. (2021). Piscirickettsia Salmonis Does Not Evidence Quorum Sensing Based on Acyl-Homoserine Lactones. J. Fish Dis. 44, 1047–1051. doi: 10.1111/jfd.13383
Liu, L.-P., Huang, L.-H., Ding, X.-T., Yan, L., Jia, S.-R., Dai, Y.-J., et al. (2019). Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry. Molecules 24, 2694–2703. doi: 10.3390/molecules24152694
Llamas, I., Quesada, E., Martínez-Cánovas, M. J., Gronquist, M., Eberhard, A., Gonzalez, J. E. (2005). Quorum Sensing in Halophilic Bacteria: Detection of N-Acyl-Homoserine Lactones in the Exopolysaccharide-Producing Species of Halomonas. Extremophiles 9, 333–341. doi: 10.1007/s00792-005-0448-1
Lumjiaktase, P., Aguilar, C., Battin, T., Riedel, K., Eberl, L. (2010). Construction of Self-Transmissible Green Fluorescent Protein-Based Biosensor Plasmids and Their Use for Identification of N-Acyl Homoserine-Producing Bacteria in Lake Sediments. Appl. Environ. Microbiol. 76, 6119–6127. doi: 10.1128/AEM.00677-10
Marshall, S. H., Gómez, F. A., Ramírez, R., Nilo, L., Henríquez, V. (2012). Biofilm Generation by Piscirickettsia Salmonis Under Growth Stress Conditions: A Putative In Vivo Survival/Persistence Strategy in Marine Environments. Res. Microbiol. 163, 557–566. doi: 10.1016/j.resmic.2012.08.002
McCarthy, Ú.M., Bron, J. E., Brown, L., Pourahmad, F., Bricknell, I. R., Thompson, K. D., et al. (2008). Survival and Replication of Piscirickettsia Salmonis in Rainbow Trout Head Kidney Macrophages. Fish Shellfish Immunol. 25, 477–484. doi: 10.1016/j.fsi.2008.07.005
Millar, A. D., Tapia, P., Gómez, F. A., Marshall, S. H., Fuentes, D. E., Valdes, J. H. (2018). Draft Genomes and Reference Transcriptomes Extend the Coding Potential of the Fish Pathogen Piscirickettsia Salmonis. Electron. J. Biotechnol. 33, 36–38. doi: 10.1016/j.ejbt.2018.04.002
Miller, M. B., Bassler, B. L. (2001). Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Miranda, C. D., Godoy, F. A., Lee, M. R. (2018). Current Status of the Use of Antibiotics and the Antimicrobial Resistance in the Chilean Salmon Farms. Front. Microbiol. 9, 1284. doi: 10.3389/fmicb.2018.01284
Mok, K. C., Wingreen, N. S., Bassler, B. L. (2003). Vibrio Harveyi Quorum Sensing: A Coincidence Detector for Two Autoinducers Controls Gene Expression. EMBO J. 22, 870–881. doi: 10.1093/emboj/cdg085
Morohoshi, T., Inaba, T., Kato, N., Kanai, K., Ikeda, T. (2004). Identification of Quorum-Sensing Signal Molecules and the luxRI Homologs in Fish Pathogen Edwardsiella Tarda. J. Biosci. Bioeng 98, 274–281. doi: 10.1016/S1389-1723(04)00281-6
Mukherjee, S., Bassler, B. L. (2019). Bacterial Quorum Sensing in Complex and Dynamically Changing Environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Oliver, C., Hernández, M. A., Tandberg, J. I., Valenzuela, F. K. N., Lagos, L. X., Haro, R. E., et al. (2017). The Proteome of Biologically Active Membrane Vesicles From Piscirickettsia Salmonis LF-89 Type Strain Identifies Plasmid-Encoded Putative Toxins. Front. Cell. Infect. Microbiol. 7, 420. doi: 10.3389/fcimb.2017.00420
Ortiz-Severín, J., Travisany, D., Maass, A., P Chávez, F., Cambiazo, V. (2019). Piscirickettsia Salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors. Pathogens 8:269. doi: 10.3390/pathogens8040269
Papenfort, K., Bassler, B. L. (2016). Quorum Sensing Signal–Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Parsek, M. R., Greenberg, E. P. (2000). Acyl-Homoserine Lactone Quorum Sensing in Gram-Negative Bacteria: A Signaling Mechanism Involved in Associations With Higher Organisms. Proc. Natl. Acad. Sci. U. S. A. 97, 8789–8793. doi: 10.1073/pnas.97.16.8789
Pearson, J. P., Passador, L., Iglewski, B. H., Greenberg, E. P. (1995). A Second N-Acylhomoserine Lactone Signal Produced by Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92, 1490–1494. doi: 10.1073/pnas.92.5.1490
Pérez-Stuardo, D., Espinoza, A., Tapia, S., Morales-Reyes, J., Barrientos, C., Vallejos-Vidal, E., et al. (2020). Non-Specific Antibodies Induce Lysosomal Activation in Atlantic Salmon Macrophages Infected by Piscirickettsia Salmonis. Front. Immunol. 11:544718. doi: 10.3389/fimmu.2020.544718
Pérez-Stuardo, D., Morales-Reyes, J., Tapia, S., Ahumada, D. E., Espinoza, A., Soto-Herrera, V., et al. (2019). Non-Lysosomal Activation in Macrophages of Atlantic Salmon (Salmo Salar) After Infection With Piscirickettsia Salmonis. Front. Immunol. 10, 434. doi: 10.3389/fimmu.2019.00434
Rai, N., Rai, R., Venkatesh, K. (2015). Quorum Sensing Biosensors Quorum Sensing vs. Quorum Quenching: A Battle With No End in Sight, (New Delhi: Springer), 173–183.
Rani, S., Kumar, A., Malik, A. K., Schmitt-Kopplin, P. (2011). Occurrence of N-Acyl Homoserine Lactones in Extracts of Bacterial Strain of Pseudomonas Aeruginosa and in Sputum Sample Evaluated by Gas Chromatography–Mass Spectrometry. Am. J. Analyt Chem. 2, 294. doi: 10.4236/ajac.2011.22037
Rojas, V., Galanti, N., Bols, N. C., Marshall, S. H. (2009). Productive Infection of Piscirickettsia Salmonis in Macrophages and Monocyte-Like Cells From Rainbow Trout, a Possible Survival Strategy. J. Cell. Biochem. 108, 631–637. doi: 10.1002/jcb.22295
Rozas, M., Enríquez, R. (2014). Piscirickettsiosis and Piscirickettsia Salmonis in Fish: A Review. J. Fish Dis. 37, 163–188. doi: 10.1111/jfd.12211
SAG. (2021). Productos Biológicos Inmunológicos Con Registro Provisional. Available at: https://www.sag.gob.cl/sites/default/files/lista_salmonidos_registro_provisional_0.pdf.
Sandoval, R., Oliver, C., Valdivia, S., Valenzuela, K., Haro, R. E., Sánchez, P., et al. (2016). Resistance-Nodulation-Division Efflux Pump acrAB Is Modulated by Florfenicol and Contributes to Drug Resistance in the Fish Pathogen Piscirickettsia Salmonis. FEMS Microbiol. Lett. 363, 1–7. doi: 10.1093/femsle/fnw102
Sernapesca. (2021). Informe Sanitario De La Salmonicultura En Centros Marinos Año 2020. Available at: http://www.sernapesca.cl/sites/default/files/informe_sanitario_salmonicultura_en_centros_marinos_2020v2.pdf.
Shaw, P. D., Ping, G., Daly, S. L., Cha, C., Cronan, J. E., Rinehart, K. L., et al. (1997). Detecting and Characterizing N-Acyl-Homoserine Lactone Signal Molecules by Thin-Layer Chromatography. Proc. Natl. Acad. Sci. U. S. A. 94, 6036–6041. doi: 10.1073/pnas.94.12.6036
Swift, S., Karlyshev, A. V., Fish, L., Durant, E. L., Winson, M. K., Chhabra, S. R., et al. (1997). Quorum Sensing in Aeromonas Hydrophila and Aeromonas Salmonicida: Identification of the luxRI Homologs ahyRI and asaRI and Their Cognate N-Acylhomoserine Lactone Signal Molecules. J. Bacteriol. 179, 5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997
Tsai, C. S., Winans, S. C. (2010). LuxR-Type Quorum-Sensing Regulators That Are Detached From Common Scents. Mol. Microbiol. 77, 1072–1082. doi: 10.1111/j.1365-2958.2010.07279.x
Waters, C. M., Bassler, B. L. (2005). Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J., Salmond, G. P. (2001). Quorum-Sensing in Gram-Negative Bacteria. FEMS Microbiol. Rev. 25, 365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x
Whiteley, M., Diggle, S. P., Greenberg, E. P. (2017). Progress in and Promise of Bacterial Quorum Sensing Research. Nature 551, 313–320. doi: 10.1038/nature24624
Wu, L., Luo, Y. (2021). Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 12, 611413. doi: 10.3389/fmicb.2021.611413
Xu, G. (2020). Evolution of luxR Solos in Bacterial Communication: Receptors and Signals. Biotechnol. Lett. 42, 181–186. doi: 10.1007/s10529-019-02763-6
Yáñez, A., Silva, H., Valenzuela, K., Pontigo, J., Godoy, M., Troncoso, J., et al. (2013). Two Novel Blood-Free Solid Media for the Culture of the Salmonid Pathogen Piscirickettsia Salmonis. J. Fish Dis. 36, 587–591. doi: 10.1111/jfd.12034
Yañez, A., Valenzuela, K., Silva, H., Retamales, J., Romero, A., Enriquez, R., et al. (2012). Broth Medium for the Successful Culture of the Fish Pathogen Piscirickettsia Salmonis. Dis. Aquat Org. 97, 197–205. doi: 10.3354/dao02403
Keywords: Piscirickettsia salmonis, SRS, quorum sensing (QS), N-acetyl-L-homoserine lactone, AHL (N-acyl-homoserine lactone)
Citation: Ruiz P, Sepulveda D, Vidal JM, Romero R, Contreras D, Barros J, Carrasco C, Ruiz-Tagle N, Romero A, Urrutia H and Oliver C (2021) Piscirickettsia salmonis Produces a N-Acetyl-L-Homoserine Lactone as a Bacterial Quorum Sensing System-Related Molecule. Front. Cell. Infect. Microbiol. 11:755496. doi: 10.3389/fcimb.2021.755496
Received: 09 August 2021; Accepted: 11 October 2021;
Published: 25 October 2021.
Edited by:
Matthew L. Nilles, University of North Dakota, United StatesReviewed by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoRodrigo Pulgar Tejo, University of Chile, Chile
Copyright © 2021 Ruiz, Sepulveda, Vidal, Romero, Contreras, Barros, Carrasco, Ruiz-Tagle, Romero, Urrutia and Oliver. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristian Oliver, cristianoliver7@gmail.com; Alex Romero, alexromero@uach.cl; Homero Urrutia, hurrutia@udec.cl
†These authors have contributed equally to this work
 Pamela Ruiz
Pamela Ruiz Daniela Sepulveda
Daniela Sepulveda José Miguel Vidal
José Miguel Vidal Romina Romero
Romina Romero David Contreras5
David Contreras5  Javier Barros
Javier Barros Carlos Carrasco
Carlos Carrasco Nathaly Ruiz-Tagle
Nathaly Ruiz-Tagle Alex Romero
Alex Romero Cristian Oliver
Cristian Oliver