A Novel Species of Penicillium With Inhibitory Effects Against Pyricularia oryzae and Fungal Pathogens Inducing Citrus Diseases
- 1State Key Laboratory of Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
- 2Department of Health Sciences, Faculty of Medicine and Health Sciences, University of Mauritius, Reduit, Mauritius
- 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 4Division of Mycobiology & Neurodegenerative Disease Research, Department of Microbiology, School of Life Sciences, Central University of Tamil Nadu, Thiruvarur, India
A novel species of Penicillium, proposed as P. linzhiense sp.nov was isolated from soil collected in Linzhi Town, Linzhi County, Tibet Autonomous Region, China. DNA sequence analyses from eight different gene regions indicate that the isolate represents a novel species and most closely related to P. janczewskii. The phylogenetic analysis based on a concatenated dataset of three genes, ITS, CaM, and BenA, also confirmed the placement of the novel species within the Canescentia section of the genus Penicillium. Differences in morphology among similar species are detailed and single gene phylogenies based on ITS, CaM and BenA genes as well as a multi-loci gene phylogeny are presented. Cultural studies were performed to study inhibitory activities on plant pathogens. The results reveal a notable antifungal activity against Pyricularia oryzae causing rice blast with an inhibition rate up to 77%, while for other three citrus pathogens, Diaporthe citri, Phyllosticta citrichinaensis, and Colletotrichum gloeosporioides, inhibition rate was 40, 50, and 55% respectively. No noticeable effects were observed for Fusarium graminearum, Botryosphaeria kuwatsukai, and Rhizoctonia solani. Interestingly, unlike other reported members of Canescentia, P. linzhiense showed no antagonistic effect on root rotting fungi. The new taxon isolated here has the potential to be used as a biocontrol agent especially for economically important phytopathogens or emerging pathogens on diseases occurring on citrus or rice.
Introduction
Orange is widely planted and consumed, but its production is threatened by a variety of pathogens during production, resulting in huge economic loss every year. To reduce the loss caused by pathogens, a large amount of pesticides have been applied to protect citrus from plant pathogens. Usage of these pesticides is harmful to the environment and human health (Nicolopoulou-Stamati et al., 2016; Bisht et al., 2019). For example, mancozeb, an effective fungicide applied widely to inhibit D. citri on citrus in China (Chen et al., 2010; Jiang et al., 2012; Liu et al., 2018), is carcinogenic, teratogenic, and mutagenic after degrading to ethylene thiourea (ETU). Besides, with increasing pesticide abuse, pathogens are easily subjected to resistance to pesticides, leading to failure of disease control. Therefore, it is important to screen beneficial fungi for potential biological control leading to the safe production of citrus.
Some fungi are considered as biocontrol agents for inhibiting plant pathogens, but very few are associated with members of the genus Penicillium. Studies have reported that conidia or culture filtrate of P. janczewskii can induce systemic resistance of melon and tobacco to inhibit Rhizoctonia solani (Madi and Katan, 1998; Nicoletti et al., 2007), and one of its main secondary metabolites, pseurotin A, reveals moderate inhibition against Erwinia carotovora and Pseudomonas syringae (Schmeda-Hirschmann et al., 2008).
Penicillium is much more ubiquitous in the environment than other species of fungi and they also exist as endophytes inside plant tissues (Rashmi et al., 2019). Due to a paucity of morphological characteristics, overlap of morphs among different species and largely similar cultural characteristics, traditional morphological classification somehow been somehow unreliable to delineate species in this genus (Houbraken et al., 2014).With the development of modern molecular based phylogeny, the concept of multi-loci sequence typing (MLST) for phylogenetic species was proposed in the classification of Penicillium, thus making identification of strains more precise (Rakeman et al., 2002). MLST is a typing method initially used for distinguishing bacterial mutations by directly determining the nucleotide sequences of multiple housekeeping genes that are highly conservative and usually stably expressed in all cells. It was first applied by Maiden et al. (1998) in typing of the bacterial pathogen (Neisseria meningitidis), gradually implemented to other bacteria and fungi predominantly in epidemiology and taxonomy. Based on multiple gene locus sequence information to identify species, Visagie et al. (2014a) came up with a short standardized gene region namely DNA barcoding which was used in the identification of Penicillium. Up to now, there are DNA barcodes for more than 370 species accepted for Penicillium. Currently, the internal transcribed spacer rDNA regions (ITS) is widely sequenced as an official DNA barcode when discriminating species for fungi (Schoch et al., 2012; Lücking et al., 2020). However, ITS is not robust enough to identify species in Penicillium and alternative barcodes are needed to assist proper identification (Skouboe et al., 1999; Seifert et al., 2007; Visagie et al., 2014a; Nilsson et al., 2018; Tekpinar and Kalmer, 2019). The β-tubulin gene (BenA), the calmodulin gene (CaM), and the RNA polymerase II second largest subunit gene (RPB2) combined with ITS have been successfully employed in species-level identification of Penicillium (Houbraken et al., 2014; Visagie et al., 2016a; Visagie et al., 2016b; Wang et al., 2017; Diao et al., 2019). They, however, have certain limitations sometimes, such as difficulties in amplifying specific gene regions and sequence analyses due to ambiguously aligned sites and these lead to problems in resolving species concepts (Giraud et al., 2010; Houbraken et al., 2012; Chen et al., 2013; Wang and Wang, 2013; Visagie et al., 2014a; Visagie et al., 2016b). In this study, multi gene phylogenetic analyses were performed based on ITS, BenA, and CaM following the recommendations of Visagie et al. (2016b).
During the screening of fungi for potential biocontrol agents on Citrus, a fungal strain, Z863, was isolated from soil by selective medium dilution plate method (Houbraken and Samson, 2011). Morphological examinations and DNA sequence analyses reveals that Z863 is a new species belonging to Penicillium sect. Canescentia. Members of section Canescentia are soil inhabitants and there exist several studies demonstrating their potential antifungal activity predominantly related to the inhibition of soil-borne pathogens (Madi and Katan, 1998; Nicoletti et al., 2007; Schmeda-Hirschmann et al., 2008; Houbraken and Samson, 2011; Urooj et al., 2018). The aim of this paper is to introduce this taxon collected from China as a new species based on morphology supported by phylogenetic analyses of a combined dataset from ITS, BenA, and CaM genetic data. In addition, we also report results based on its potential inhibitory activities on plant pathogens.
Materials and Methods
Sampling and Isolations
Soil samples were collected in Linzhi Town, Linzhi County, Tibet Autonomous Region (29.60146 N, 94.41736 E), China. The fungal samples were separated based on selective medium dilution plate method, in which 10 g soil to 90 ml distilled water, were shaken for 10 min at 120 rpm and diluted twice to 10−1 and 10−2 of the original concentration. Potato dextrose agar (PDA) as an isolation medium was prepared with 200 g potato, 20 g glucose, 18 g agar, 0.3 g chloramphenicol in 1,000 ml ddH2O and sterilized at 121°C for 30 min. Three concentrations of diluent of 0.1 ml were separately pipetted into polystyrene Petri dish with 15 ml coagulated PDA and then the sterile coater was used to homogenize the diluent with three replicates for each concentration. After cultured at 25°C for 3 d, colonies were observed and mycelia (through hyphal typing) were transferred to a new PDA plate and once colonies grow up to 3 cm, they were transferred to new plates again.
Morphological Identification
Macromorphological characters were checked from Czapek’s agar (CZ), Czapek yeast autolysate agar (CYA), malt extract agar (MEA), and yeast extract sucrose agar (YES) media. The medium’s preparation, strain’s inoculation manner, and incubation conditions were performed following the protocols of Visagie et al. (2014b).
After incubation at 25°C for 7 d, plates were checked, observed and colony’s morphology was recorded. The descriptions of color are based on NBS ISCC color name notation. Afterward, 60% lactic acid was used as a floating agent for making slides, and mycelia and conidia were examined under the microscope. Macromorphological and micromorphological details were also examined and recorded by EOS 600D Camera (Canon, China, Beijing) and Leica Microscope DM750 (Leica, China, Shanghai) with an ICC50 Camera and arrangement of photos was done in Adobe Photoshop CC 2018.
DNA Extraction and PCR
DNA of the samples was extracted by Rapid Fungi Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China) as the manufacturer’s instructions.
PCR amplifications of the ITS, BenA, CaM, RPB2, translation elongation factor 1-α (TEF) regions, the large subunit (LSU) and the small subunit (SSU) of ribosomal DNA gene and tubulin gene were performed with corresponding primers listed in Table 1. One amplification reaction consisted of 25 μl Green Taq Mix (Vazyme, Nanjing, China), 2 μl of each primer (10 μM), 2 μl template DNA (30 ng μl−1), and 19 μl ddH2O in reaction system according to manufacturer’s instructions. PCR reactions were performed by an MG96G PCR instrument (LongGene, Hangzhou, China) with the following procedure: pre-denaturing at 94°C for 2 min; subsequent 35 cycles with denaturing at 94°C for 30 s, annealing at 55°C for 40 s, extending at 72°C for 1 min; the final extension at 72°C for 10 min. After PCR reaction, products were detected by 1% agarose gel electrophoresis. Purification of products was conducted by the DNA gel purification kit (Axygen Biotech, Hangzhou, China).
DNA Sequencing and Sequence Analyses
Purified and recovered target DNA fragments were sent to be directly sequenced in an ABI PRISMA377 automatic sequencer (Sangon Biotech, Shanghai, China). Once DNA sequences were obtained, they were verified and then aligned with homologous or similar nucleotide sequences in the GenBank database by BLAST.
Three fragments including ITS, CaM, and BenA were employed for further comprehensive phylogenetic analyses. Three sets of genetic data (ITS, CaM, and BenA) were arranged and corrected by BioEdit Sequence Alignment Editor version 7.2.3, and finally a concatenated DNA sequence dataset was analyzed with Maximum Parsimony (MP) and Maximum Likelihood (ML). Phylogenetic analysis for model selections were performed by Modeltest 3.7 based on the lowest Akaike information criterion (AIC) value. The phylogenetic tree was constructed using PAUP 4.0b10 software under different optimality criteria. The stability of tree branches was evaluated by bootstrap with 1,000 replicates. Trees were processed for publication in Adobe Illustrator CC. Taxa used in the phylogenetic analyses are shown in Table 2.
Cultural Studies and Inhibition Activities on Plant Pathogens
Target strain and each tested pathogen (D. citri, Ph. citrichinaensis, C. gloeosporioides, Py. oryzae, F. graminearum, B. kuwatsukai, and R. solani) were separately inoculated onto the same 9 cm polystyrene Petri dish containing 15 ml PDA, 4 cm between two inoculums. To set up a control group, each pathogen with the same conditions as the test group was individually inoculated in a single medium at the same position in the PDA plate. Then, both groups were cultured at 25°C for 7 d.
A week later, the growth radius was measured (recorded as R) of the control group and the inhibition culture group (recorded as r). Each replicate was measured three times and the average was calculated. The inhibition rate of pathogen radius (abbreviated as IR) was calculated out according to the following formula:
Results
Isolation and Morphology
Twenty isolates with white color colony on PDA medium plate were isolated. The morphology of these isolates was identical when examined under microscope, suggesting that they belong to the same species. One isolate, Z863 was used as representative for further studies. Results of microscopic examination showed that it is characterized by morphologies of the genus Penicillium. Based on phenotypic characters, this taxon belongs to the Canescentia section of the genus Penicillium. The morphological descriptions are provided in the taxonomy section.
Taxonomy
Penicillium linzhiense H-K. Wang & R. Jeewon, sp. nov. –Mycobank MB#838576; Figures 1, 2.
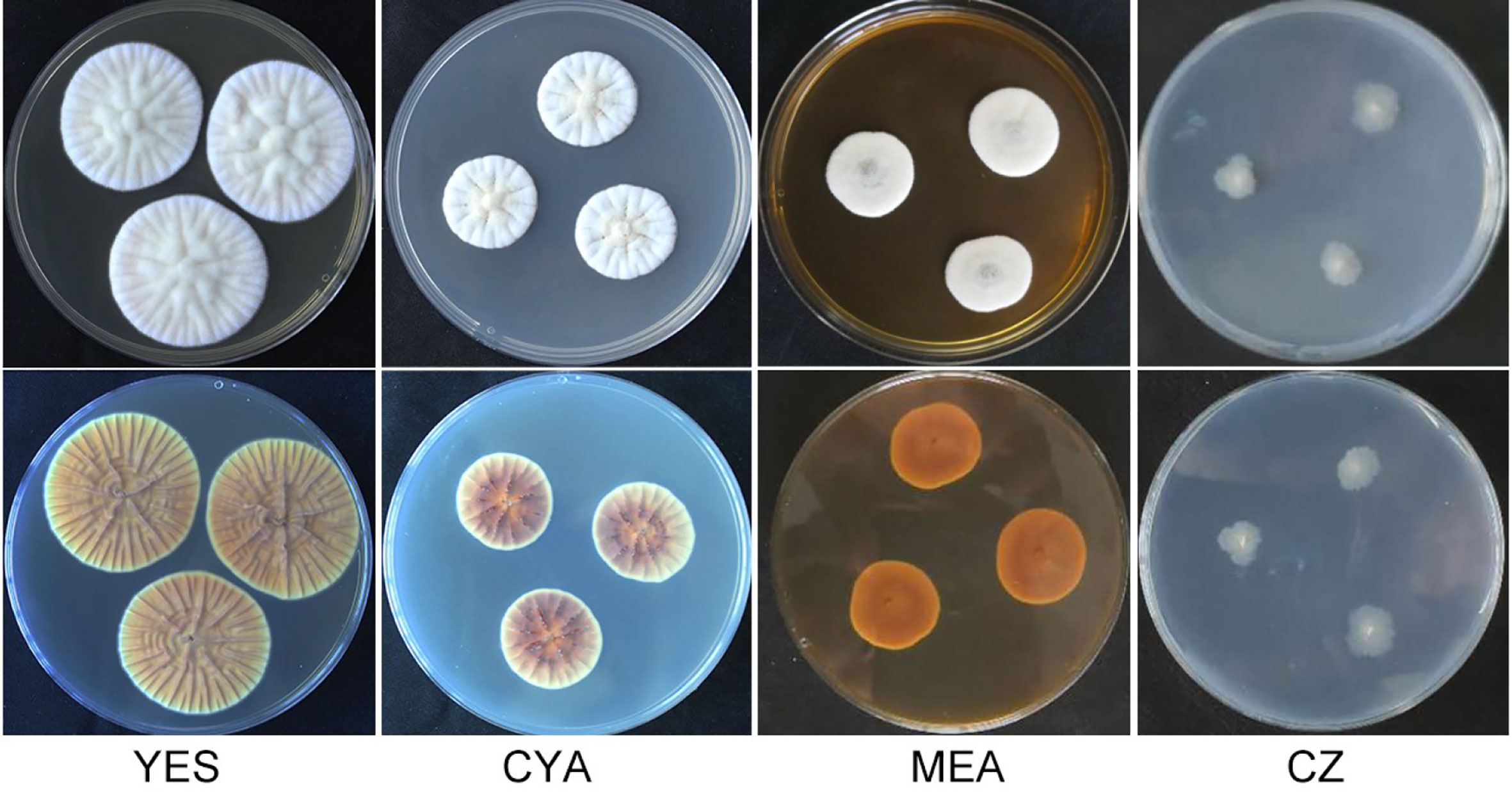
Figure 1 Morphology of Penicillium linzhiense after incubation at 25°C on different medium for 7 days. bottom row: reverse plate.
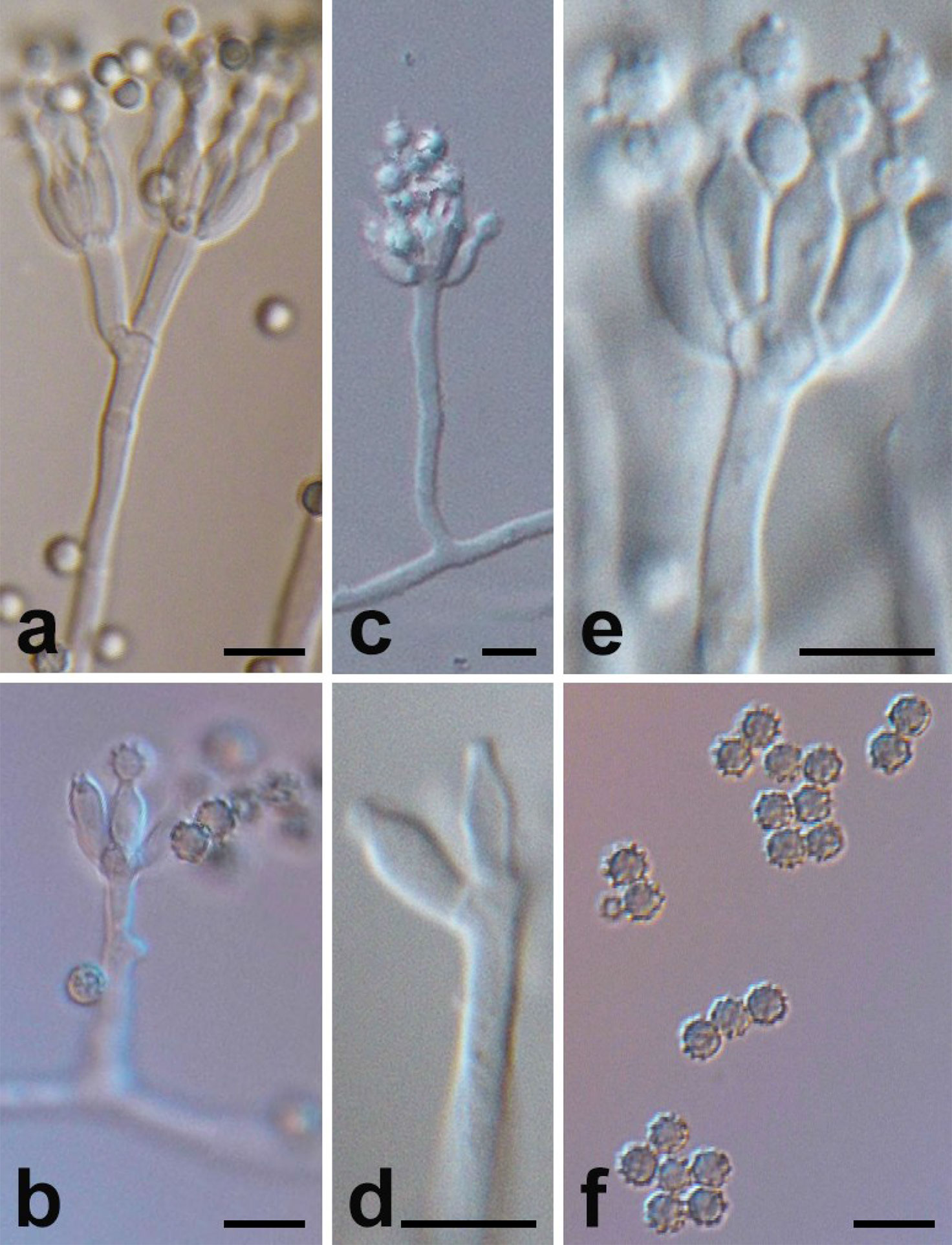
Figure 2 Micromorphology of P. linzhiense. (Scale bars = 5 μm.) (A) Branching status of conidiophore. (B) Conidia’s growth status on the bottle stem. (C) Conidiophore and bottle stem. (D) Morphology of bottle stem. (E) Growth manner of bottle stem on base stem. (F) Morphology of conidia.
In: subgenus Penicillium, section Canescentia.
Barcodes: ITS-MT461156; BenA-MT461157; CaM-MT461162;
Etymology. Latin, linzhiense, named after Linzhi, China, location where the isolates were collected.
Type strain examined: Linzhi Town, Linzhi County, Tibet Autonomous Region (29.60146 N, 94.41736 E), China, 20 Aug. 2016, collected by H-K Wang, CCTCC no: M2019870. Deposited in China typical culture preservation center located in Wuhan University, Wuhan, China.
After incubation at 25°C for 7 d on different medium plates, colonies of P. linzhiense displayed remarkable differences in morphology (Figure 1). Colonies on CYA after 7 d approached 30–32 mm, covered with many radial sulcate, thicker at the center without sulcate; margin entire to somewhat irregular; texture usually flocculent or velvet; mycelia white, moderate yellowish pink (SICC-NBS 29); center white to pinkish white (SICC-NBS 9) with exudate on the surface; soluble pigment absent. Colonies on MEA after 7 d approached 20–30 mm, uneven, navel-like bulge at the center; margin entire to somewhat irregular; mycelia white, texture velutinous; somewhat vivid pink at the center with exudate on the surface; soluble pigment absent. Colonies on CZ after 7 d approached 16–18 mm, low and plane; margin irregular; mycelia (SICC-NBS 184) very pale blue; texture fluffy; exudate and soluble pigment absent. Colonies on YES after 7 d approached 42–44 mm, covered with many radial wrinkles, thicker without radial wrinkles at the center; margin entire; mycelia white, yellowish white (SICC-NBS 92) to pale greenish yellow (SICC-NBS 104); texture mostly flocculent or velvet; exudate and soluble pigment absent.
Microscopic characters were also examined on MEA, conidiophore (Figure 2C), 20–100 × 2–2.5 μm, occurred in aerial or dragging hyphae with smooth walls. Broom branches (Figure 2A) are predominantly single-whorled, with fewer double-whorls and solitary pedicels with enlarged apices. It grew two to eight or more bottle pedicels (6–8 × 2.0–2.5 μm) per whorl, typically flask-shaped, with short and distinct necks (Figures 2D–E). Conidia were spherical or subspherical in shape, 2.6–4.5 μm, markedly spiny and rough (Figure 2F), each in a bottle stem or free (Figure 2B). Conidial chains were loose, nearly cylindrical, or irregular.
Morphology of conidiophore and conidia of P. linzhiense are similar to P. janczewskii. The strain differs from P. janczewskii in that P. linzhiense has a light colony on CYA and is grayish-white without becoming grayish-green within 2 weeks; the conidiophore branching pattern is predominantly monoverticillate, with fewer biverticillate, and conidiophore are solitary. However, colonies of P. janczewskii change from grayish-green to grayish-black on CYA medium; conidiophore branching patterns of P. janczewskii, P. dunedinense, P. echinatum, P. griseoazureum, P. nigricans are mainly biverticillate, with terverticillate, or few monoverticillate, with two to four metula per conidiophore. This novel species can be distinguished from P. corvianum by the spiny conidia.
Sequencing and Phylogenetic Analyses
Eight gene fragments of strain Z863 were obtained using PCR according to the primer pairs in Table 1. All the sequences were uploaded to GenBank with the following accession numbers (ITS: MT461156; BenA: MT461157; LSU: MT461158; SSU: MT461159; TEF: MT461160; RPB2: MT461161; CaM: MT461162; beta-tubulin (Tub): MT461163).
Phylogenetic analysis (Maximum Likelihood, ML) based on a combined ITS, CaM, and BenA dataset of 21 taxa (with 1,238 characters and 130 parsimony informative characters) with P. brevicompactum as outgroup resulted in one tree shown in Figure 3A (TL = 513, CI = 0.789, RI = 0.738, RC = 0.583, HI = 0.211). Phylogeny depicts that Z863 is a new species as it constitutes a strongly supported independent lineage basal to P. janczewskii, P. dunedinense, P. nigricans, P. griseoazureum, and P. echinatum (Figure 3A).
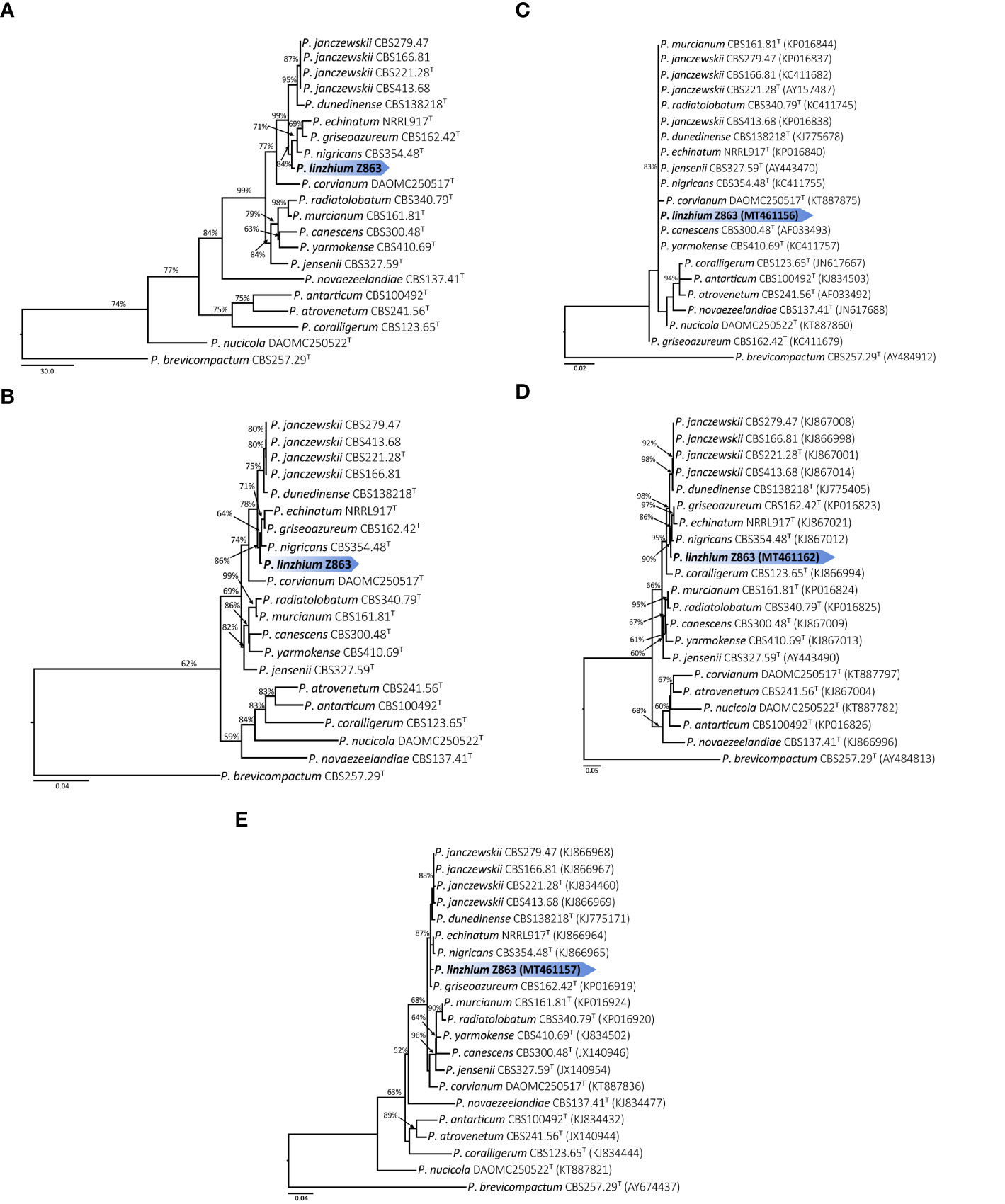
Figure 3 (A) Maximum parsimony tree of the combined sequence of ITS, CaM and BenA of P. linzhiense (T = ex-type; the scale bar shows the number of substitutions and the values above the nodes represent bootstrap support). The new species is marked by blue block. (B) Maximum likelihood tree of the combined sequence of ITS, CaM, and BenA of P. linzhiense (T = ex-type; the scale bar shows the number of substitutions and the values above the nodes represent bootstrap support). The new species is marked by blue block. (C) Maximum likelihood tree derived from DNA sequence analyses of the ITS, gene region. (The scale bar shows the number of substitutions and the values above the nodes represent bootstrap support, but those support lower than 50% are not showed). (D) Maximum likelihood tree derived from DNA sequence analyses of the CaM gene region. (The scale bar shows the number of substitutions and the values above the nodes represent bootstrap support, but those support lower than 50% are not showed). (E) Maximum likelihood tree derived from DNA sequence analyses of the BenA, gene region. (The scale bar shows the number of substitutions and the values above the nodes represent bootstrap support, but those support lower than 50% are not showed).
The Maximum Parsimony (MP) phylogenetic tree derived from the combined dataset shown in Figure 3B is based on 1,238 characters with best model GTR+G+I. The phylogenetic position of Z863 is the same in the ML tree. This result also confirmed that Z863 is a new species in Penicillium sect. Canescentia. During our initial ITS sequence BLAST search in GenBank, the similarity between our strain and P. janczewskii (MK179261), P. arizonense (MH492021), P. canescens (KX359603), P. murcianum (NR_138358), P. janczewskii (KP016839) was 100%. This confirms that our species is undoubtedly a Penicillium species and belongs to Penicillium sect. Canescentia. However, one cannot rely on ITS alone for proper identification and establishing new species, especially for taxonomically complex genera (Jeewon and Hyde, 2016). Even our single gene phylogenetic analyses based on ITS alone also shows that the tree is unresolved with weak branch support and the affinities of P. linzhiense to P. corvianum (KT887875), P. canescens (AF033493), and P. yarmokense (KC411757) are not clear (Figure 3C). However single gene datasets based on CaM and BenA genes provided better resolution (Figures 3D, E).
Inhibition of Penicillium linzhiense on Plant Pathogens
Comparing inhibition culture group and control group, P. linzhiense showed inhibitory effects against Py. oryzae (Figure 4A), D. citri (Figure 4B), Ph. citrichinaensis (Figure 4C), and C. gloeosporioides (Figure 4D). However, the strain did not exhibit significant effect on F. graminearum (Figure 4E) and had no inhibition against B. kuwatsukai (Figure 4F) and R. solani (Figure 4G). The inhibition rate of pathogen radius (IR) is showed in Figure 5.
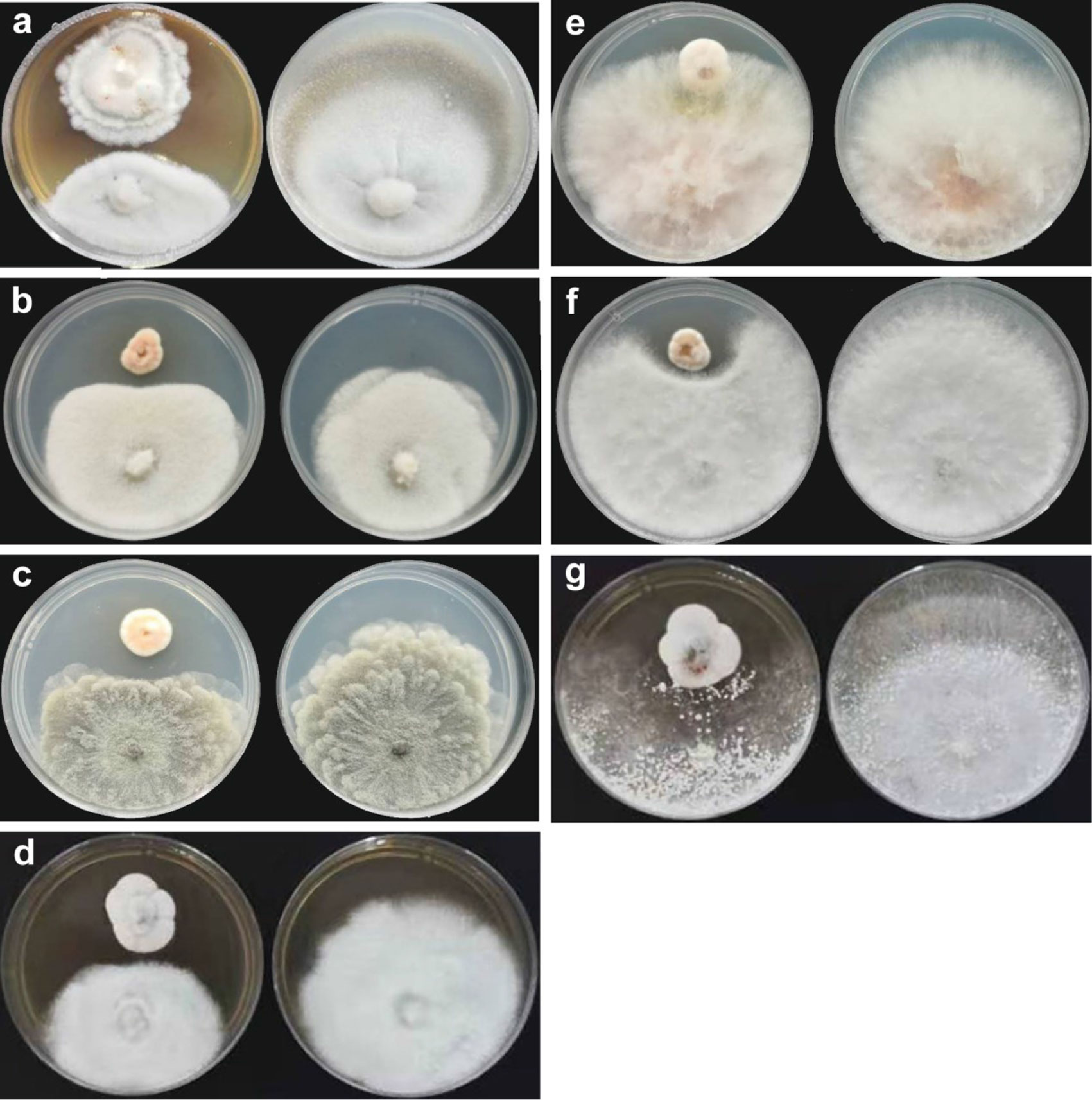
Figure 4 Inhibition of P. linzhiense on plant pathogens. (A) Inhibitory effect of P. linzhiense on Pyricularia oryzae after 14 d; (B) Inhibitory effect of P. linzhiense on Diaporthe citri after 7 d; (C) Inhibitory effect of P. linzhiense on Phyllosticta citrichinaensis after 7 d; (D) Inhibitory effect of P. linzhiense on Colletotrichum gloeosporioides after 7 d; (E) Inhibitory effect of P. linzhiense on Fusarium graminearum after 7 d; (F) Inhibitory effect of P. linzhiense on Botryosphaeria kuwatsukai after 7 d; (G) Inhibitory effect of P. linzhiense on Rhizoctonia solani after 7 d. [(A–G) The left one was an inhibition culture group (above: P. linzhiense; below: the tested pathogen), and the right one was a control group].
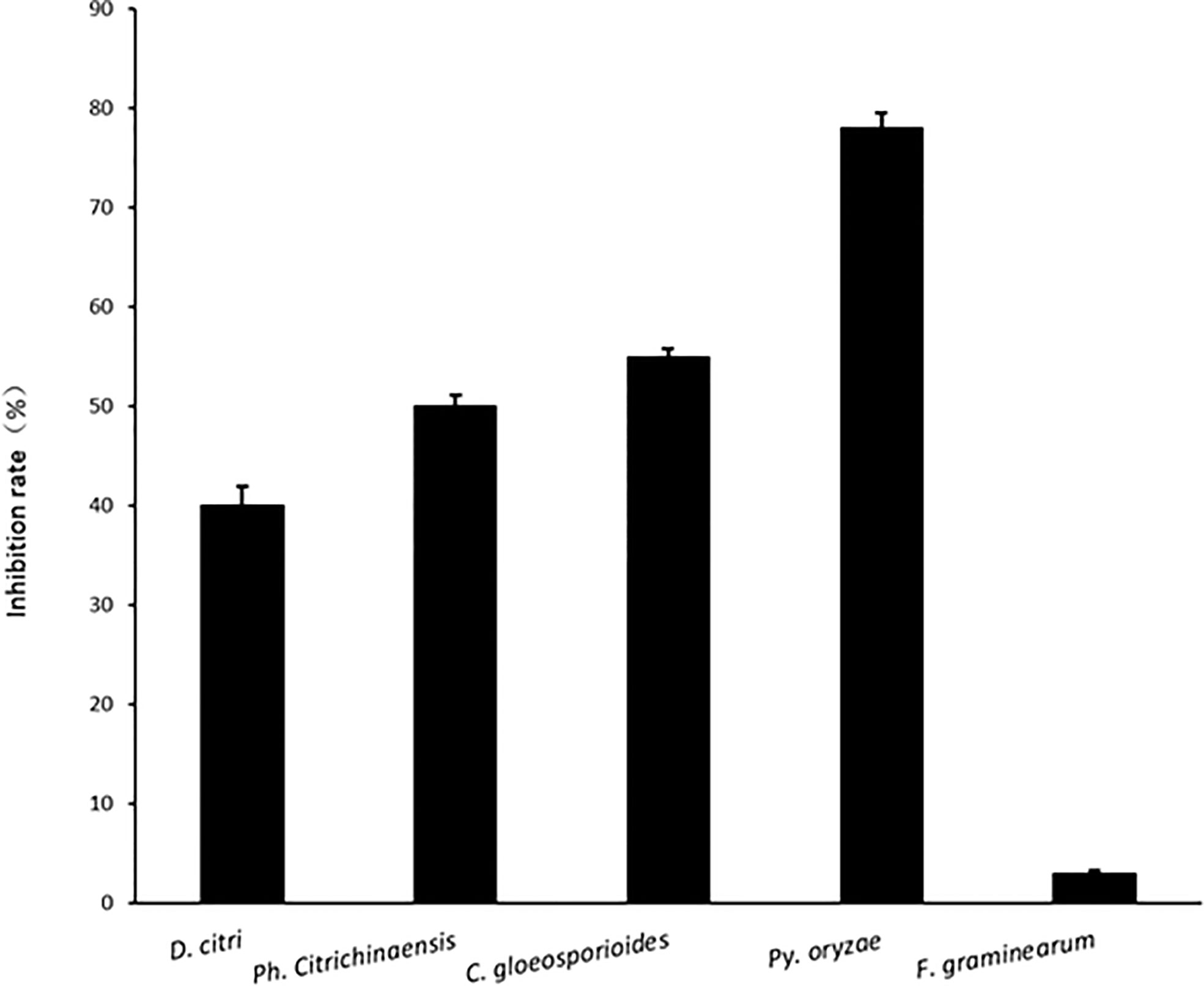
Figure 5 The inhibition rate of pathogen radius (IR) of P. linzhiense against different pathogens in the confrontation culture test.
Discussion
Penicillium is widely distributed in the environment and easily isolated from air and soil. To date many published reports have reported a ubiquitous and high frequency of Penicillium in soil samples from different climatic conditions and geographical regions (Grishkan and Nevo, 2004; Sharma et al., 2011; Cruz et al., 2013; Kalashnikova et al., 2016; Cecchi et al., 2019) and these species are associated with important soil function. Despite their important roles, the traditional morphological delineation of species has always been a taxonomic dilemma and currently DNA based sequence data from a combination of different genes especially ITS, BenA, and CaM should be analyzed for accurate identification. In our study, we isolated a new species, Penicillium linzhiense and its morphological details, ability to restrict growth of fungal pathogens and evolutionary relationships are discussed.
Our multigene phylogeny reveals that P. linzhiense is close to P. janczewskii, P. dunedinense, P. echinata, P. griseoazureum, and P. nigricans. However, P. linzhiense is distinct from species mentioned above particularly in morphs with distinctly thinner mycelium on CZ medium and with single-whorled broom branches mainly accompanied by few double-whorled one. With respect to P. janczewskii, P. linzhiense mainly differs in the color of colony after two weeks’ culture and the broom branches (at the start, there is gray-white and mainly monoverticillate, with fewer biverticillate, with solitary conidiophore, and with time turns grayish green to grayish black and mainly biverticillate, with terverticillate or few monoverticillate, with two to four metula per conidiophore). When the cultural characteristics of P. dunedinense are compared against P. linzhiense at 25°C after 7 d, clear differences can be observed. On CYA medium, wrinkles of P. linzhiense look denser than P. dunedinense; on MEA, P. dunedinense is sulcate but P. linzhiense is not; on YES, P. dunedinense is grayish orange but P. linzhiense is white (Visagie et al., 2014b). As for P. echinata, its conidiophores have diaphragms, conidial chains that are relatively tighter than P. linzhiense and its broom branches are single-whorled (Matsushima, 1972), but conidiophores of P. linzhiense have no diaphragms and there exist few double-whorls of broom branches in P. linzhiense. As is shown in Manual and Atlas of the Penicillia (Ramirez and Martinez, 1982), both P. griseoazureum and P. nigricans on CYA and on MEA do not have any exudate but P. linzhiense possesses yellowish brown exudate on the colonies on CYA and MEA.
Phylogenetic analyses of a combined ITS, CaM, and BenA sequence dataset (Figures 3A, B) in the study show that P. linzhiense forms a distinct lineage, basal to P. nigricans CBS 354.48 with high bootstrap support in ML analysis (84% ML). Based on the recommendations for the establishment of new species proposed by Jeewon and Hyde (2016), we also compared % differences across all genes amplified. Comparison of ITS, CaM, and BenA nucleotides between P. linzhiense and P. nigricans CBS 354.48 reveals 0, 2, and 3 base pair differences, respectively. In the phylogram generated from maximum likelihood analysis based on ITS sequence data, P. linzhiense was observed to be closely related to P. yarmokense (CBS 410.69), P. corvianum (KT887875), and P. canescens (AF033493) (Figure 3C) but this relationship is unstable and unresolved. Furthermore, a comparison of DNA sequences of the ITS regions sequences between P. linzhiense and P. yarmokense shows 0 (0%) base pair differences. Although ITS barcodes play an important role in the taxonomy of Penicillium species, this gene region is not powerful enough to discriminate species due to their low variability (Skouboe et al., 1999; Seifert et al., 2007; Stielow et al., 2015). Upon analysis of the BenA sequences data, P. linzhiense was found to be a sister taxon to P. echinatum (NRRL917) (Figure 3E) and nucleotide comparison reveals 3 (0.8%) base pair differences between these two taxa. Thus, based on the phylogenetic analyses of the concatenated dataset and phenotypic differences and following the guidelines proposed by Jeewon and Hyde (2016), we hereby introduce P. linzhiense as a new species in the genus Penicillium.
In the inhibition studies, as we expected, P. linzhiense shows a distinct inhibitory effect against three important pathogenic fungi causing citrus diseases. Results also show a stronger inhibition against Pyricularia oryzae causing rice blast (Figure 4). It is recommended to perform further pathogenicity studies on P. linzhiense, including field experiments and the effect of metabolites to assess to what extent the latter can be used as a potential biological control agent in integrated disease management strategies. The discovery of P. linzhiense provides one more possibility to control citrus diseases and rice blast.
Members of Penicillium sect. Canescentia are well known as soil-borne fungi (Houbraken and Samson, 2011) and some studies pointed out they possess distinct inhibitory effect against root rotting fungi and even able to promote growth of plants (Madi and Katan, 1998; Nicoletti et al., 2007; Schmeda-Hirschmann et al., 2008; Urooj et al., 2018). Interestingly, as a member of Penicillium sect. Canescentia, P. linzhiense shows little inhibition against common soil-borne pathogens unlike other reported members mentioned above. Instead, P. linzhiense reveals its suppression on pathogens triggering plant disease aboveground. As revealed in this study, results demonstrate that P. linzhiense can be a potential biocontrol agent for Py. oryzae which causes damage to the leaves, stems, and ears of rice; D. citri which causes diseases at the tip of trees, new leaves, and fruits of citrus; Ph. citrichinaensis which is often found associated with fruits of citrus (Baayen et al., 2002); and C. gloeosporioides which is detrimental to leaves, branches, flowers, fruits, and fruit stalks of citrus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT461156; https://www.ncbi.nlm.nih.gov/genbank/, MT461157; https://www.ncbi.nlm.nih.gov/genbank/, MT461158; https://www.ncbi.nlm.nih.gov/genbank/, MT461159; https://www.ncbi.nlm.nih.gov/genbank/, MT461160; https://www.ncbi.nlm.nih.gov/genbank/, MT461161; https://www.ncbi.nlm.nih.gov/genbank/, MT461162; https://www.ncbi.nlm.nih.gov/genbank/, MT461163.
Author Contributions
RJ, HL, F-CL, and H-KW designed the study. LL did the sample collection and laboratory work. LL, RJ, PD, SSKD and H-KW are involved in phylogenetic analyses and initial writing and finalizing drafts. HL, F-CL, and H-KW contributed for the research funds. All authors contributed to the article and approved the submitted version.
Funding
This research is supported by the Key R & D Program of Zhejiang Province (2019C02022), China, and the National Key R & D Program of China (2017YFD0202004). RJ thanks the University of Mauritius for support and the MRC funded project MRC/RUN/1705.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baayen R. P., Bonants P. J. M., Verkley G., Carroll G. C., van der Aa H. A., de Weerdt M., et al. (2002). Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of woody plants, G. mangiferae (Phyllosticta capitalensis). Phytopathology 92, 464–477. doi: 10.1094/PHYTO.2002.92.5.464
Bisht J., Palni L. M. S., Harsh N. S. K. (2019). “Pesticide contamination and human health,” in Handbook of Research on the Adverse Effects of Pesticide Pollution in Aquatic Ecosystems. (IGI Global). 137–149. doi: 10.4018/978-1-5225-6111-8.ch008
Castlebury L. A., Rossman A. Y., Sung G.-H., Hyten A. S., Spatafora J. W. (2004). Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol Res. 108, 864–872. doi: 10.1017/S0953756204000607
Cecchi G., Marescotti P., Di Piazza S., Zappatore S., Zotti M. (2019). Fungal richness in the extreme environments of the Libiola mine (eastern Liguria, Italy): correlations among microfungi, lithology, mineralogy, and contaminants. Environ. Earth Sci. 78, 1–12. doi: 10.1007/s12665-019-8553-0
Chen G., Jiang L., Xu F., Li H. (2010). In vitro and in vivo screening of fungicides for controlling citrus melanose caused by Diaporthe citri. J. Zhejiang Univ. 36, 440–444. doi: 10.3785/j.issn.1008-9209.2010.04.014
Chen A. J., Tang D., Zhou Y. Q., Sun B. D., Li X. J., Wang L. Z., et al. (2013). Identification of ochratoxin A producing fungi associated with fresh and dry liquorice. PLoS One 8, e78285. doi: 10.1371/journal.pone.0078285
Cruz R., Santos C., de Lima J. S., Moreira K. A., de Souza-Motta C. M. (2013). Diversity of Penicillium in soil of Caatinga and Atlantic Forest areas of Pernambuco, Brazil: an ecological approach. Nova Hedwigia 97, 543–556. doi: 10.1127/0029-5035/2013/0127
Diao Y.-Z., Chen Q., Jiang X.-Z., Houbraken J., Barbosa R. N., Cai L., et al. (2019). Penicillium section Lanata-divaricata from acidic soil. Cladistics 35, 514–549. doi: 10.1111/cla.12365
Giraud F., Giraud T., Aguileta G., Fournier E., Samson R., Cruaud C., et al. (2010). Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. Int. J. Food Microbiol. 137, 204–213. doi: 10.1016/j.ijfoodmicro.2009.11.014
Glass N. L., Donaldson G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from Filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995
Grishkan I., Nevo E. (2004). Soil microfungi of Nahal Meitsar, “Evolution Canyon” IV, Golan Heights. Plant Biosyst. 138, 21–26. doi: 10.1080/11263500410001684080
Hong S.-B., Cho H.-S., Shin H.-D., Frisvad J. C., Samson R. A. (2006). Novel Neosartorya species isolated from soil in Korea. Int. J. Syst Evol Microbiol. 56, 477–486. doi: 10.1099/ijs.0.63980-0
Houbraken J., Samson R. A. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol 70, 1–51. doi: 10.3114/sim.2011.70.01
Houbraken J., Frisvad J. C., Seifert K. A., Overy D. P., Tuthill D. M., Valdez J. G., et al. (2012). New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29, 78–100. doi: 10.3767/003158512X660571
Houbraken J., Visagie C. M., Meijer M., Frisvad J. C., Busby P. E., Pitt J. I., et al. (2014). A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud. Mycol 78, 373–451. doi: 10.1016/j.simyco.2014.09.002
Jeewon R., Hyde K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Jiang L., Xu F., Huang Z., Huang F., Chen G., Li H. (2012). Occurrence and control of citrus melanose caused by Diaporthe citri. Acta Agric Zhejiangensis 24, 647–653.
Kalashnikova K. A., Konovalova O. P., Alexandrova A. V. (2016). Soil-inhabiting microfungi of the monsoon dipterocarp forest (the natural reserve Dong Nai, south Vietnam). Mikologiya i Fitopatologiya 50, 97–107.
Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Liu X., Wang M., Mei X., Jiang L., Han G., Li H. (2018). Sensitivity evaluation of Diaporthe citri populations to mancozeb and screening of alternative fungicides for citrus melanose control. Acta Phytophylacica Sin. 45, 373–381.
Lücking R., Aime M. C., Robbertse B., Miller A. N., Ariyawansa H. A., Aoki T., et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11, 1–32. doi: 10.1186/s43008-020-00033-z
Madi L., Katan J. (1998). Penicillium janczewskii and its metabolites, applied to leaves, elicit systemic acquired resistance to stem rot caused by Rhizoctonia solani. Physiol. Mol. Plant Pathol. 53, 163–175. doi: 10.1006/pmpp.1998.0174
Maiden M. C. J., Bygraves J. A., Feil E., Morelli G., Russell J. E., Urwin R., et al. (1998). Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Matsushima T. (1972). Microfungi of the Solomon Islands and Papua-New Guinea. Mycologia 64, 1208–1209. doi: 10.2307/3758096
Nicoletti R., Lopez-Gresa M. P., Manzo E., Carella A., Ciavatta M. L. (2007). Production and fungitoxic activity of Sch 642305, a secondary metabolite of Penicillium canescens. Mycopathologia 163, 295–301. doi: 10.1007/s11046-007-9015-x
Nicolopoulou-Stamati P., Maipas S., Kotampasi C., Stamatis P., Hens L. (2016). Chemical pesticides and human health: The urgent need for a new concept in agriculture. Front. Public Health 4, 148. doi: 10.3389/fpubh.2016.00148
Nilsson R. H., Taylor A. F. S., Adams R. I., Baschien C., Johan B.-P., Cangren P., et al. (2018). Taxonomic annotation of public fungal ITS sequences from the built environment - a report from an April 10-11, 2017 workshop (Aberdeen, UK). Mycokeys 28, 65–82. doi: 10.3897/mycokeys.28.20887
O’Donnell K., Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Rakeman J. L., Bui U., Lafe K., Chen Y. C., Honeycutt R. J., Cookson B. T. (2005). Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J. Clin. Microbiol. 43 (7), 3324–3333. doi: 10.1128/JCM.43.7.3324-3333.2005
Ramirez C., Martinez A. T. (1982). Manual and atlas of the Penicillia (New York; Amsterdam: Elsevier Biomedical Press).
Rashmi M., Kushveer J. S., Sarma V. V. (2019). A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10, 798–1079. doi: 10.5943/mycosphere/10/1/19
Schmeda-Hirschmann G., Hormazabal E., Rodriguez J. A., Theoduloz C. (2008). Cycloaspeptide A and pseurotin A from the endophytic fungus Penicillium janczewskii. Z. Fur Naturforschung - Sec C J. Biosci. 63, 383–388. doi: 10.1515/znc-2008-5-612
Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Seifert K. A., Samson R. A., deWaard J. R., Houbraken J., Levesque C. A., Moncalvo J.-M., et al. (2007). Prospects for fungus identification using C01 DNA barcodes, with Penicillium as a test case. Proc. Natl. Acad. Sci. U. S. A. 104, 3901–3906. doi: 10.1073/pnas.0611691104
Sharma G., Pandey R. R., Singh M. S. (2011). Microfungi associated with surface soil and decaying leaf litter of Quercus serrata in a subtropical natural oak forest and managed plantation in Northeastern India. Afr. J. Microbiol. Res. 5, 777–787. doi: 10.5897/AJMR10.621
Skouboe P., Frisvad J. C., Taylor J. W., Lauritsen D., Boysen M., Rossen L. (1999). Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol Res. 103, 873–881. doi: 10.1017/S0953756298007904
Stielow J. B., Levesque C. A., Seifert K. A., Meyer W., Irinyi L., Smits D., et al. (2015). One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35, 242–263. doi: 10.3767/003158515X689135
Tekpinar A. D., Kalmer A. (2019). Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwigia 109, 187–224. doi: 10.1127/nova_hedwigia/2019/0528
Urooj F., Farhat H., Ali S. A., Ahmed M., Sultana V., Shams Z. I., et al. (2018). Role of endophytic Penicillium species in suppressing the root rotting fungi of sunflower. Pakistan J. Bot. 50, 1621–1628.
Visagie C. M., Houbraken J., Frisvad J. C., Hong S. B., Klaassen C. H. W., Perrone G., et al. (2014a). Identification and nomenclature of the genus Penicillium. Stud. Mycol 78, 343–371. doi: 10.1016/j.simyco.2014.09.001
Visagie C. M., Hirooka Y., Tanney J. B., Whitfield E., Mwange K., Meijer M., et al. (2014b). Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 78, 63–139. doi: 10.1016/j.simyco.2014.07.002
Visagie C. M., Houbraken J., Dijksterhuis J., Seifert K. A., Jacobs K., Samson R. A. (2016a). A taxonomic review of Penicillium species producing conidiophores with solitary phialides, classified in section Torulomyces. Persoonia 36, 134–155. doi: 10.3767/003158516X690952
Visagie C. M., Renaud J. B., Burgess K. M., Malloch D. W., Clark D., Ketch L., et al. (2016b). Fifteen new species of Penicillium. Persoonia 36, 247–280. doi: 10.3767/003158516X691627
Wang B., Wang L. (2013). Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia 105, 1547–1554. doi: 10.3852/13-022
Wang X.-C., Chen K., Zeng Z.-Q., Zhuang W.-Y. (2017). Phylogeny and morphological analyses of Penicillium section Sclerotiora (fungi) lead to the discovery of five new species. Sci. Rep. 7, 8233. doi: 10.1038/s41598-017-08697-1
Keywords: Canescentia, antifungal activity, Pyricularia oryzae, citrus diseases, taxonomy, multigene phylogeny, plant pathology
Citation: Liang L-J, Jeewon R, Dhandevi P, Durairajan SSK, Li H, Lin F-C and Wang H-K (2021) A Novel Species of Penicillium With Inhibitory Effects Against Pyricularia oryzae and Fungal Pathogens Inducing Citrus Diseases. Front. Cell. Infect. Microbiol. 10:604504. doi: 10.3389/fcimb.2020.604504
Received: 09 September 2020; Accepted: 22 December 2020;
Published: 10 February 2021.
Edited by:
Hailing Jin, University of California, Riverside, United StatesReviewed by:
Vishukumar Aimanianda, Institut Pasteur, FranceBanu Metin, Istanbul Sabahattin Zaim University, Turkey
Copyright © 2021 Liang, Jeewon, Dhandevi, Durairajan, Li, Lin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajesh Jeewon, r.jeewon@uom.ac.mu; Hong-Kai Wang, hkwang@zju.edu.cn
 Li-Juan Liang
Li-Juan Liang Rajesh Jeewon
Rajesh Jeewon Pem Dhandevi
Pem Dhandevi Siva Sundara Kumar Durairajan4
Siva Sundara Kumar Durairajan4  Hongye Li
Hongye Li Fu-Cheng Lin
Fu-Cheng Lin