Antimicrobial mechanisms of nanopatterned surfaces—a developing story
- 1Manufacturing Futures Lab, Department of Mechanical Engineering, University College London, London, United Kingdom
- 2Photonic Innovations Lab, Department of Electronic and Electrical Engineering, University College London, London, United Kingdom
Whilst it is now well recognized that some natural surfaces such as seemingly fragile insect wings possess extraordinary antimicrobial properties, a quest to engineer similar nanopatterned surfaces (NPSs) is ongoing. The stake is high as biofouling impacts critical infrastructure leading to massive social and economic burden with an antimicrobial resistance (AMR) issue at the forefront. AMR is one of the most imminent health challenges the world is facing today. Here, in the effort to find more sustainable solutions, the NPSs are proposed as highly promising technology as their antimicrobial activity arises from the topographical features, which could be realized on multiple material surfaces. To fully exploit these potentials however, it is crucial to mechanistically understand the underlying killing pathways. Thus far, several mechanisms have been proposed, yet they all have one thing in common. The antimicrobial process is initiated with bacteria contacting nanopatterns, which then imposes mechanical stress onto bacterial cell wall. Hence, the activity is called “mechano-bactericidal”. From this point on, however, the suggested mechanisms start to diverge partly due to our limited understanding of force interactions at the interface. The aim of this mini review is to analyze the state-of-the-art in proposed killing mechanisms by categorizing them based on the characteristics of their driving force. We also highlight the current gaps and possible future directions in investigating the mechanisms, particularly by shifting towards quantification of forces at play and more elaborated biochemical assays, which can aid validating the current hypotheses.
Introduction
Antimicrobial resistant microorganisms are now one of the top 10 global public health threats according to the World Health Organization. To tackle the AMR challenges, several domestic and global action plans have been developed, largely focused on identifying effective and feasible infection prevention and control measures. Here, bioinspired nanopatterns with antimicrobial properties are considered particularly promising as they can kill bacteria (and other microorganisms) or inhibit their proliferation by virtue of their topography (Linklater et al., 2021; Hawi et al., 2022). The fact that antimicrobial activity stems from the topography arguably enables them to be applied on any surface where biofouling is a problem (by direct surface structuring or application of nanostructured film). Thus, the potential applications of antimicrobial NPSs are widespread spanning from public transportation, mobile displays, water purification systems, packaging, to personal protecting equipment and medical implants (Ganjian et al., 2019; Valiei et al., 2020; Michalska et al., 2021b).
Conventional antimicrobial surfaces rely on diffusion of pre-immobilized biocides, which has certain drawbacks. These include compound degradation, reduced potency over time, uncontrolled release, and, importantly, inefficiency against ever-growing AMR bacteria (Andersson and Hughes, 2014; Cloutier et al., 2015). Though it is noteworthy that antimicrobial NPSs do not comprehensively resolve all these challenges; for example, the reduced efficiency over time and pattern durability remained the major bottlenecks awaiting to be resolved (Wang et al., 2020; Hoque et al., 2023). Nevertheless, the unique topographically triggered antimicrobial activity of these surfaces has drawn significant research interest, and understanding how these surfaces operate is the first research question yet to be fully answered (Lin et al., 2018).
Since the first report on antibacterial properties of natural (Ivanova et al., 2012) and synthetic substrates (Ivanova et al., 2013), researchers have tried to elucidate the underlying killing mechanisms. The prevalent opinion has been that the mechanical interactions and force exchanges between the NPSs and bacteria at the bio-interface compromises the cell wall integrity, which in turn leads to the loss of internal turgor pressure and eventual death. However, the origin of forces acting on the bacterial cell wall and, consequently, its failure mode are still unsettled. For instance, the physical damage was firstly attributed to cell wall uniaxial strain beyond its elastic limit over the suspended region (Pogodin et al., 2013). Alternatively, over the following years, various and often contradictory failure causes have been proposed, whose driving force arises from different sources such as adhesion (Michalska et al., 2018), stored elastic energy (Ivanova et al., 2020), cell motility (Bandara et al., 2017), and capillary forces (Valiei et al., 2020).
Of note, whether rupturing the cell wall is the primary cause of cell death is being additionally questioned. In fact, a recent study on stochastic TiO2 nanopillars (pillars are one example of nanoprotrusions; the notation is used throughout the text for simplicity) reports that bacterial cell wall undergoes localized deformation, which leads to only occasional penetration. Hence, it cannot account for all the observed reduction in viability (Jenkins et al., 2020). This observation alongside complementary biological assays—a recent trend in the field, have prompted a new take on the killing mechanisms. Namely, it postulates that the mechanical stimuli imparted on bacterial envelop could also be transduced to a chemical signal, leading to oxidative stress inside the cells, which eventually can activate apoptosis-like pathways (Zhao et al., 2022; Le et al., 2023). In other words, antimicrobial NPSs may have the capacity to induce gene and proteomic alteration inside the cells. Thus, the antimicrobial properties could also be attributed to a biochemical response to the cell wall perturbances. However, the exact pathways by which the mechanical signal is transduced into a chemical signal that creates redox imbalance inside the cells remains unclear.
The difficulty in reaching a consensus over the killing mechanisms stems from multiple challenges pertinent to small scales of both topography and bacteria, which makes real-time direct observations or force measurements difficult or impossible (Lin et al., 2018; Bandara et al., 2020; Valiei et al., 2020). Lack of universal protocols to evaluate the performance and attention to small but important details like, e.g., presence of air bubbles during testing, further complicates the issue and comparison of the designs (Valiei et al., 2020; Michalska et al., 2021a). Additionally, many highly efficient patterns are random or heterogenous, which makes it harder to delineate the performance boundaries. Overall, it is likely that a combination of mechanisms accounts for the cell death rather than just one. Nevertheless, investigating the mechanisms one at a time, if possible at all, is insightful in terms of determining their contribution in the final bacterial death. Also, establishing the killing mechanism(s) paves the way for further improvements such as increasing the killing efficiency and pattern durability. The latter is equally important as the properties remain as long as the pattern is present, which is, however, beyond the scope of this review. The readers may refer to some recent advances in this regard reported by Yu et al. (2021), Zhang et al. (2021). Here, we firstly review the purely mechanical mechanisms classified by the proposed driving forces, followed by examination of the mechanically triggered mechanisms.
Mechanical mechanisms
To establish that NPSs are “mechano-bactericidal,” it must be proven there are active force exchanges at the bacteria-substrate interface, sufficient to damage the cell wall leading to death either directly or indirectly (mechanical or mechanically triggered mechanisms, respectively). The damage of the cell walls occurs when sufficient mechanical stress, whose minimum magnitude depends on the cell wall mechanical properties, is imparted on the cell wall. To describe mechano-bactericidal mechanisms, the first step is identifying the driving forces that could govern these interactions. Several forces of different origins have been proposed and investigated via simulations and/or experimentally where possible, which can be grouped as follows: 1) non-specific interactions, 2) stored-elastic energy release, and 3) capillary forces.
Non-specific interactions
The “biophysical model” by Pogodin et al. (2013) was the first proposed mechanism to explain the bactericidal action of cicada wings. According to this model, deformational stress distributes non-uniformly across the cell wall at the adsorption sites and the suspended region. It assumes that stress is developed by bacterial cells’ tendency to maximize their adsorption area and minimize the gained free energy through cell wall deformation. Since the nanopillars are more accessible as the adsorption sites, it is more likely that the cell wall ruptures over the suspended region (Figure 1A). Conversely, a recent finite element analysis of a Gram-negative cell on cicada-like surfaces suggests that critical nondevelopable strain occurs at the pillar apex (Figure 1B) (Velic et al., 2021). Likewise, observations from various imaging techniques reveal the critical site of action is rather the protrusions’ top (Linklater et al., 2017; Jenkins et al., 2020).
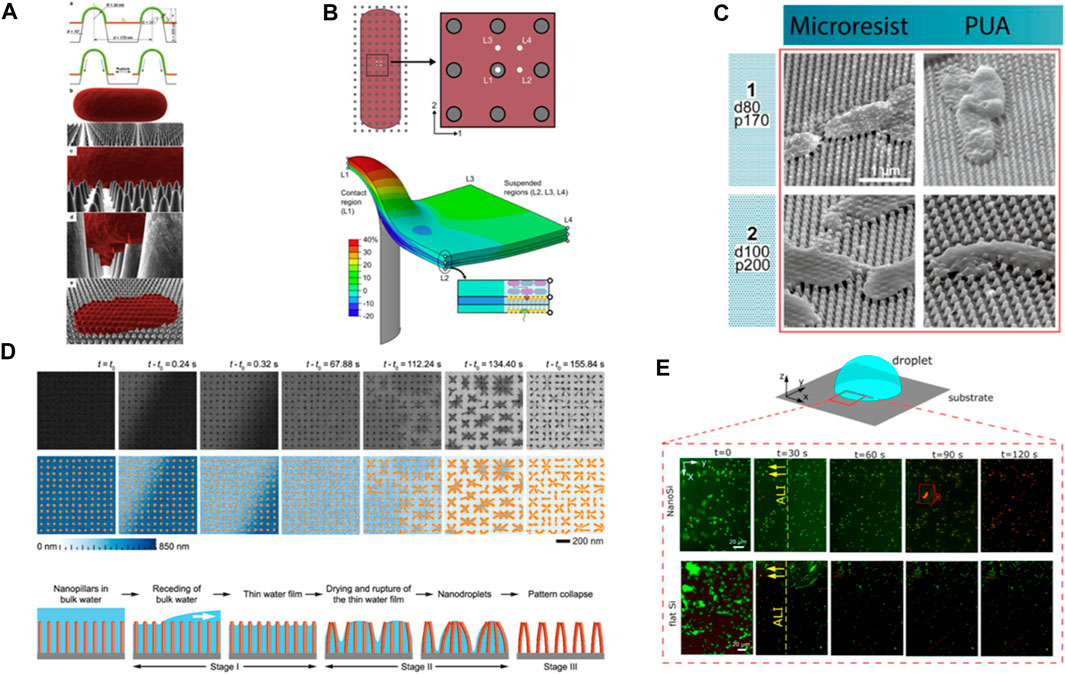
FIGURE 1. (A) Schematic of different stages of bacterial interaction with nanopillars based on biophysical model. The adsorbed layer increases its surface area which leads to the cell wall rupture over suspended region between nanopillars. Reprinted with permission from Pogodin et al. (2013). Copyright 2023 Elsevier. (B) Contour plot illustrates the longitudinal uniaxial strain of a multi-layer bacterial envelop adhered on a nanopillar. The plot reveals a nonuniform strain distribution in different layers and locations, with a critical site of action identified at the apex of the nanopillar. Reprinted with permission from Velic et al. (2021). Copyright 2023 Elsevier. (C) SEM images of E. coli cells interaction with deflected nanopillars. Reprinted with permission from Lohmann et al. (2022). Copyright 2023 American Chemical Society. (D) (Top) Time-resolved in situ TEM image series showing elastocapillary action during water evaporation from high AR silicon nanopillars surfaces. (Bottom) Schematic illustration of different stages of the dewetting process. Reprinted with permission from Vrancken et al. (2020). Copyright 2023 American Chemical Society. (E) P. aeruginosa viability on nanopillars (NanoSi) and control (flat) Si surfaces upon evaporation. Viability of bacteria as a function of time on NanoSi and flat Si that is subject to evaporation (live cells are green and dead cells are red). Reprinted with permission from Valiei et al. (2020). Copyright 2023 American Chemical Society.
The driving force in both models is thought to be nonspecific intermolecular adhesion forces (Van der Waals, hydrogen bonding, electrostatic) with the biophysical model not specifying their magnitude. In the computational model by Velic et al. (2021) on the other hand, the data used to compute adhesion energy lacks an appropriate experimental validation. Here, methods like Single Cell Force Spectroscopy (SCFS) could provide the relevant data. For example, various groups indicate the magnitude of nonspecific interaction forces between Gram-positive/negative bacteria and relatively smooth hydrophilic/hydrophobic silicon substrates to be of the order of a few nN (Potthoff et al., 2015; Beaussart et al., 2020). This value is expected to decrease upon nanostructuring due to the reduction in effective surface area, as reported for Staphylococcus (S.) aureus on nanopillars (Doll et al., 2022). The quantitative approaches in these studies are inspiring indeed for designing future experiments to determine the role of nonspecific forces at the interface.
Stored-elastic energy release
Scanning electron microscopy (SEM) images of bacteria on high aspect ratio (AR) nanopillars from various studies (conducted for 15 min–18 h) revealed that peripheral nanostructures deflect towards the cells (Figure 1C) (Bandara et al., 2020; Ivanova et al., 2020). This observation has prompted researchers to investigate the impact of nanopillars flexural rigidity (i.e., a combination of materials elastic modulus and AR) on killing efficiency of NPSs. For certain patterns, an enhanced killing efficiency is reported and attributed to the release of stored elastic energy of pillars upon deflection that acts as an external force onto bacterial cell wall (Linklater et al., 2018). From these efficient patterns, we learn that depending on nanopillars’ elastic modulus, geometrical features and their deflection, the calculated force needed to deform each pillar varies from ∼10 nN (Ivanova et al., 2020) to 200–500 nN (Lohmann et al., 2022). Despite the thicker geometries in the latter case, the significantly lower elastic modulus of the studied materials when compared to silicon in the former case makes one wonder how realistic these values are. Nevertheless, we consider this range and postulate that is not entirely clear whether bacteria cells can generate such force, to deflect approximately 10 pillars around their cell body. Bearing in mind the challenges to compute these values however, like ambiguity whether elastic modulus changes at nanoscale at all or how nanoscale defects affect the results, we attempt to discuss it in the context of available forces at play. The already described intermolecular adhesion forces act instantaneously and as such, we deem them irrelevant here. However, we consider three probable scenarios including forces generated by 1) migrating bacteria, 2) extracellular polymeric substances (EPS) (Bandara et al., 2017), and 3) capillary action.
We first look at the magnitudes of generated forces during bacteria migrations. To migrate, cells must exert forces, and this is dependent at minimum on the motility type and relevant appendages or their lack. Although little is known about the possibility of bacteria migration on NPSs, the studies in this field yield information about the level of “strong” forces possible to be exerted. For example, Neisseria gonorrhea’s individual type IV pili generate 100 pN, while the pili bundles’ force does not exceed a few nN (Marathe et al., 2014). Single Molecular Force Spectroscopy data suggests similar results for Pseudomonas (P.) aeruginosa’s type IV pili (Dufrêne et al., 2021). In another study, a single cell and collective migration of Myxococcus xanthus were analyzed by Traction Force Microscopy, and revealed for the latter case that the exerted force is below 300 pN (Sabass et al., 2017). Hence, it appears that bacterial force generation apparatus alone may not exert sufficient force to be directly responsible for nanopillars’ deflection.
For the bacterial cells interacting at longer times, the role of secreted extracellular polymeric substance (EPS) cannot be excluded. Here, forces up to 30 nN have been recorded during live imaging of Xylella fastidiosa adhered on protein coated-InP nanowires for 24 h (Sahoo et al., 2016). Considering the surface coating and bacteria dwelling time, it can be argued that the interactions are indeed EPS-mediated. If this is the case however, one may think of EPS-mediated force transfer because the energy stored in pillars may not be in direct contact with the bacterial cells anymore. Of note, the bacteria models above mostly vary from the ones typically used in mechano-bactericidal studies. One of the potential future directions may include closing the gap of force generation capabilities by different strains.
Although trivial at large scales, capillary forces might become dominant at sub-micrometer scale. In fact, capillary forces may deform solid bodies, a phenomenon called elastocapillary. In the field of nanomanufacturing, collapse of high AR nanostructures has been extensively studied and reported for solution-based processing (Chandra and Yang, 2009; Mallavarapu et al., 2020; Michalska et al., 2023). Real-time monitoring of water evaporation from an array of Si nanopillars by using in situ liquid phase Transmission Electron Microscopy (TEM) revealed that as the water evaporates, discrete nanodroplets form, ultimately leading to nanopillars’ collapse (Figure 1D). Depending on whether the interpillar attraction forces are greater than the elastic-stored energy or not, the pillars remain clustered or revert to their initial position (Vrancken et al., 2020). Therefore, capillary forces could be potentially responsible for the deflections when surface tension is high and there is a case for bacteria-NPSs-liquid-air interface. Although we do not know how the presence of adhered bacteria on nanopillars changes the dynamics of capillary forces, SEM images suggest that bacteria may amplify the clustering (Figure 1C). While some of the discussed studies consider the issue and use, e.g., critical point drying during sample preparation for microscopy to exclude it, others do not, which might contribute to an effect. More about the impact of capillary action is described below.
Capillary forces
Capillary forces are present at bacteria cell-substrate interface when the level of liquid falls below the bacterial cell height. Since capillary forces are usually dominant over other surface forces, they are a sensible candidate to suspect as driving force for pulling bacteria towards the nanoprotrusions and damaging the cell wall. In fact, Valiei et al. (2020) reported lack of killing on hydrophilic silicon nanopillars even after 24 h of incubation while cells were still fully immersed in the medium, i.e., no capillary force was present at the interface. However, when the liquid droplet starts to evaporate, a live/dead staining of the cells at the periphery of the moving air-liquid interface revealed remarkable >90% of killing only after 2 min (Figure 1E). The findings in this study indicate that capillary forces are large enough to create downward force and kill bacteria (Valiei et al., 2020). Most likely, harnessing this effect could dramatically increase the antimicrobial property of NPSs, albeit it depends on surface energy and topography. Moreover, at any stage of exposing bacteria to the NPSs, the development of capillary forces may act as an external and dominant force to kill bacteria. Thus, careful experimental operations are suggested if one desires to distinguish between capillary action- or other force-induced killings.
Mechanically triggered mechanisms
The interactions at bacteria-NPSs interface captured by imaging techniques inspired the field to propose new mechanisms or challenge the long-lasting beliefs (Figure 2A). For example, 3D visualization of bacteria-TiO2 nanopillar interactions, reconstructed from electron tomography, suggests that Escherichia (E.) coli’s cell wall can be locally deformed around the pillars as much as 190 nm without being ruptured (Figure 2B). The study indicates that penetration events are infrequent, and they cannot explain the reported reduction in viability assays (Jenkins et al., 2020). These observations, alongside complementary biological assays, have prompted a new take on how NPSs operate—antimicrobial properties of NPSs may not have a purely physical nature. Instead, it has been suggested that the physical signals (mechanical stress) received by bacterial cell wall can be transduced to bio/chemical signals that could impair internal processes such as bacterial growth (Jenkins et al., 2020) or activate apoptosis-like death pathways (Zhao et al., 2022).
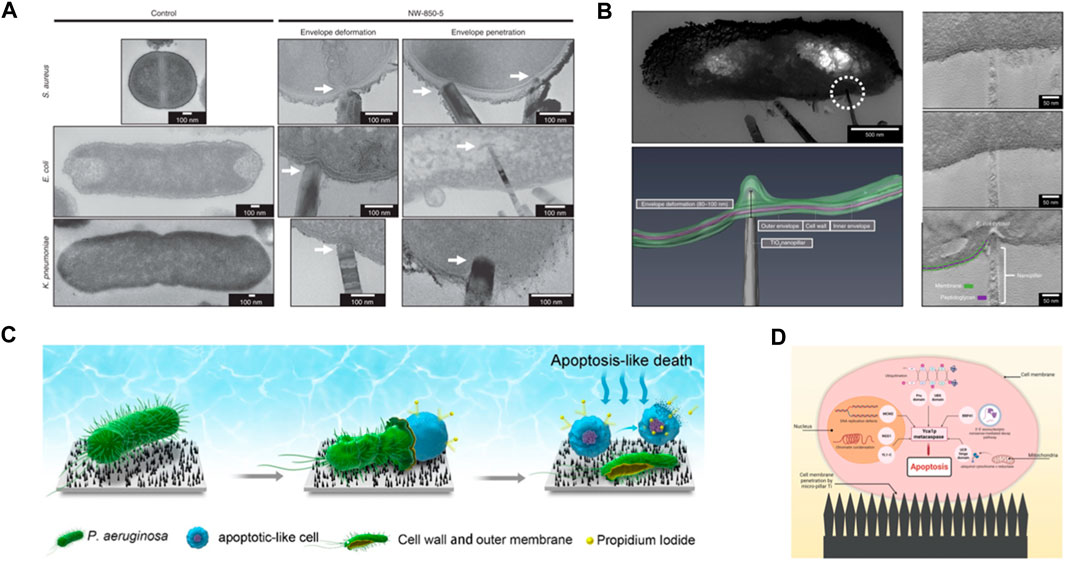
FIGURE 2. (A) TEM micrographs of TiO2 nanopillars-mediated bacterial envelope deformation and penetration in Gram-positive and Gram-negative bacteria. Adapted from Jenkins et al. (2020). (B) Tomographic reconstruction of nanopillar-induced envelop deformation in E. coli. Adapted from Jenkins et al. (2020). (C) Schematic illustration of death process of P. aeruginosa cell in contact with bSi nanospikes. Adapted with permission from Zhao et al. (2022). Copyright 2023 American Chemical Society. (D) Schematic depiction of how mechanically triggered mechanism induces microorganism apoptosis. Mechanical injuries as the result of interactions with nanopillars activate pathways that lead to programmed death. Reprinted from Le et al. (2023).
Analyses of proteomic profiles of E. coli and S. aureus after 24 h incubation on nanopillars show that differentially expressed proteins in both strains are biologically connected, implying a generic response across bacterial strains. Furthermore, the data indicate that some of these proteins are associated with cell response to elevated reactive oxygen species (ROS) production, further confirmed by measuring H2O2 levels—an oxidative stress marker. Therefore, the reduction in bacteria viability here has been attributed to the activation of ROS-dependent lethal pathways (Jenkins et al., 2020).
A recent investigation of mechano-bactericidal property of black silicon (bSi) nanopillars suggests that a combination of mechanically injured cell wall, internal turgor pressure, and a weak coupling between the cell wall and plasma membrane allows the cytoplasmic content of P. aeruginosa to leak out while cells are viable (Figure 2C). Subsequent exposure of the fluid membrane to nanopillars increases the intracellular ROS production, which activates apoptosis-like death (Figure 2D) (Zhao et al., 2022). Even though the mediating biochemical links between each of these stages are not apparent yet, the occurrence of these events is independently supported by electron microscopy images and biological assays.
Discussion
Understanding the mechanism(s) behind the bactericidal action of NPSs underlies the basis for their application. The key to unraveling the role of each mechanism itself is understanding its driving force and, ideally, quantifying it. Simultaneous fluorescence imaging and nanoindentation of E. coli with atomic force microscopy showed that force of 25 nN can puncture the cell wall to kill almost 90% of a cell population (Del Valle et al., 2020). While such quantification is a good reference, one needs to consider the highly localized character of the measured force. Having this in mind, we now attempt to review the magnitude of the proposed driving forces as a quantitative approach in assessing the efficacy of the killing mechanisms. Mechano-microbiological studies indicate that the magnitude of forces generated by bacterial varies between several hundreds of pN (Sabass et al., 2017; Dufrêne et al., 2021) and tens of nN for longer dwelling times in a presence of EPS (Sahoo et al., 2016). On the other hand, studies that investigated the non-specific interactions reported the forces from 1 nN (Doll et al., 2022) to 40 nN (Maikranz et al., 2020) for nanopillars and relatively smooth Si surfaces, respectively. These results imply that the reduction of effective surface area on nanopillars would further undermine the nonspecific interaction role as dominant driving force for cell death.
Capillary action arising from surface tension at the physical boundary of different phases at the bacteria-NPSs-liquid-air interface is suggested to play a critical role in creating downward forces strong enough to damage bacteria cell wall (Valiei et al., 2020). A recent study further confirmed that the morphology of P. aeruginosa cells incubated on superhydrophilic Si nanopillars under continuous wet conditions showed no sign of damage to the cell wall as visualized by SEM, after specimen fixing and critical point drying. When the same samples were prepared for the SEM after water was evaporated, cell morphology was visibly destroyed (Valiei et al., 2022). Considering that capillary forces are only introduced when the liquid level recedes a certain threshold, it is not easy to measure the force experimentally. Nevertheless, there is enough evidence in literature to show that capillary action plays a dominant role at the air/liquid interface of nano/microstructures (Chandra and Yang, 2009; Mallavarapu et al., 2020).
The deflection of nanopillars around bacteria cell body has been widely reported and is believed to contribute to increase the mechano-bactericidal efficiency by exerting the stored elastic energy on the cell wall (Linklater et al., 2018; Ivanova et al., 2020; Lohmann et al., 2022). Here, we argue that forces generated by the cells may not be sufficient to deflect nanopillars within the reported range of 10 nN–500 nN (Lohmann et al., 2022). To the best of our knowledge, currently there is no compelling evidence to support that cells can generate this amount of force on such surfaces. To illuminate the observations, it seems more studies are needed to both quantify the forces exerted by bacteria and attempt to measure and/or simulate with more accuracy the force required to deflect nanopillars. It was also suggested that perhaps we should look at stress instead of absolute force (Lohmann et al., 2022). Perhaps, particularly for the higher reported forces, a careful verification of sample preparation for SEM imaging would be beneficial to exclude a contribution of capillary action. The more extensive discussion around this mechanism in this review stems from the appreciation of complexity to validate its assumptions. While we debate the force magnitude exerted by bacteria under “passive” adhesive conditions, the capillary action is one example of added external force to yield an enhanced killing. Others not discussed here and overall, less explored may include, e.g., shear, acoustic, or magnetic forces. Conversely, under those “passive” conditions, it is plausible that cells experience sublethal damage, which leads us to a discussion of mechanically triggered mechanisms.
Here, both cited groups reveal quite complex mechanisms, which cannot be captured by standard characterization techniques employed in the field, namely, colony counting and live/dead fluorescent staining. The latter technique seems to be the most frequently utilized in the field, often without the former method. These studies hence highlight that interpretation of such results can be grossly misleading, potentially underscoring a real activity/value of some already reported surface designs. In fact, this issue had been already pointed out by Michalska et al. (2021a), where the discrepancies in bactericidal performance were similarly noted when flow cytometry technique was employed, albeit on the loosely attached cells. Importantly for practical applications, while (Zhao et al., 2022), reveal that sublethal injured cells can undergo post-stress ALD when transferred to a non-stressed environment, in contrast, Michalska et al. (2021a) show that some injuries can be repaired. In this aspect, the major differences between the studies are interaction time and “the non-stressed environment,” where phosphate saline buffer and super optimal broth with catabolite repression medium were used, respectively.
Most certainly, post-stress research needs more focus in the future to deepen the mechanistic understanding but also to provide relevant information to practical applications. While quantitative studies on interacting forces are also needed as well as more biochemical investigations, another future challenge appears to be exploring the factors that make one mechanism dominate over the others. These surely include topography, the interfacing mode between bacteria and the surfaces ( ± external force, etc.), and the kind of bacteria species. This would enable true control over the interactions’ mechanics. This, in turn, could be then leveraged to design strategies for more efficient killing as well as harness other untapped opportunities in the field.
Author contributions
AP: Conceptualization, Writing–original draft, Writing–review and editing. IP: Supervision, Writing–review and editing. MM: Conceptualization, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. MM acknowledges the Royal Society RGS\R1\231146 Research Grant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersson, D. I., and Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi:10.1038/nrmicro3270
Bandara, C. D., Ballerin, G., Leppänen, M., Tesfamichael, T., Ostrikov, K. K., and Whitchurch, C. B. (2020). Resolving bio–nano interactions of E. coli bacteria–dragonfly wing interface with helium ion and 3D-structured illumination microscopy to understand bacterial death on nanotopography. ACS Biomater. Sci. Eng. 6, 3925–3932. doi:10.1021/acsbiomaterials.9b01973
Bandara, C. D., Singh, S., Afara, I. O., Wolff, A., Tesfamichael, T., Ostrikov, K., et al. (2017). Bactericidal effects of natural nanotopography of dragonfly wing on Escherichia coli. ACS Appl. Mater Interfaces 9, 6746–6760. doi:10.1021/acsami.6b13666
Beaussart, A., Feuillie, C., and El-Kirat-Chatel, S. (2020). The microbial adhesive arsenal deciphered by atomic force microscopy. Nanoscale 12, 23885–23896. doi:10.1039/d0nr07492f
Chandra, D., and Yang, S. (2009). Capillary-force-induced clustering of micropillar arrays: is it caused by isolated capillary bridges or by the lateral capillary meniscus interaction force? Langmuir 25, 10430–10434. doi:10.1021/la901722g
Cloutier, M., Mantovani, D., and Rosei, F. (2015). Antibacterial coatings: challenges, perspectives, and opportunities. Trends Biotechnol. 33, 637–652. doi:10.1016/j.tibtech.2015.09.002
Del Valle, A., Torra, J., Bondia, P., Tone, C. M., Pedraz, P., Vadillo-Rodriguez, V., et al. (2020). Mechanically induced bacterial death imaged in real time: a simultaneous nanoindentation and fluorescence microscopy study. ACS Appl. Mater Interfaces 12, 31235–31241. doi:10.1021/acsami.0c08184
Doll, P. W., Doll, K., Winkel, A., Thelen, R., Ahrens, R., Stiesch, M., et al. (2022). Influence of the available surface area and cell elasticity on bacterial adhesion forces on highly ordered silicon nanopillars. ACS Omega 7, 17620–17631. doi:10.1021/acsomega.2c00356
Dufrêne, Y. F., Viljoen, A., Mignolet, J., and Mathelié-Guinlet, M. (2021). AFM in cellular and molecular microbiology. Cell Microbiol. 23, e13324. doi:10.1111/cmi.13324
Ganjian, M., Modaresifar, K., Ligeon, M. R. O., Kunkels, L. B., Tümer, N., Angeloni, L., et al. (2019). Nature helps: toward bioinspired bactericidal nanopatterns. Adv. Mater Interfaces 6, 201900640. doi:10.1002/admi.201900640
Hawi, S., Goel, S., Kumar, V., Pearce, O., Ayre, W. N., and Ivanova, E. P. (2022). Critical review of nanopillar-based mechanobactericidal systems. ACS Appl. Nano Mater 5, 1–17. doi:10.1021/acsanm.1c03045
Hoque, M. J., Ma, J., Rabbi, K. F., Yan, X., Singh, B. P., Upot, N. V., et al. (2023). Perspectives on superhydrophobic surface durability. Appl. Phys. Lett. 123, 0164927. doi:10.1063/5.0164927
Ivanova, E. P., Hasan, J., Webb, H. K., Gervinskas, G., Juodkazis, S., Truong, V. K., et al. (2013). Bactericidal activity of black silicon. Nat. Commun. 4, 2838. doi:10.1038/ncomms3838
Ivanova, E. P., Hasan, J., Webb, H. K., Truong, V. K., Watson, G. S., Watson, J. A., et al. (2012). Natural bactericidal surfaces: mechanical rupture of Pseudomonas aeruginosa cells by cicada wings. Small 8, 2489–2494. doi:10.1002/smll.201200528
Ivanova, E. P., Linklater, D. P., Werner, M., Baulin, V. A., Xu, X., Vrancken, N., et al. (2020). The multi-faceted mechano-bactericidal mechanism of nanostructured surfaces. Proc. Natl. Acad. Sci. 117, 12598–12605. doi:10.1073/pnas.1916680117
Jenkins, J., Mantell, J., Neal, C., Gholinia, A., Verkade, P., Nobbs, A. H., et al. (2020). Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 11, 1626. doi:10.1038/s41467-020-15471-x
Le, P. H., Linklater, D. P., Aburto-Medina, A., Nie, S., Williamson, N. A., Crawford, R. J., et al. (2023). Apoptosis of multi-drug resistant Candida species on microstructured titanium surfaces. Adv. Mater Interfaces 10, 202300314. doi:10.1002/admi.202300314
Lin, N., Berton, P., Moraes, C., Rogers, R. D., and Tufenkji, N. (2018). Nanodarts, nanoblades, and nanospikes: mechano-bactericidal nanostructures and where to find them. Adv. Colloid Interface Sci. 252, 55–68. doi:10.1016/j.cis.2017.12.007
Linklater, D. P., Baulin, V. A., Juodkazis, S., Crawford, R. J., Stoodley, P., and Ivanova, E. P. (2021). Mechano-bactericidal actions of nanostructured surfaces. Nat. Rev. Microbiol. 19, 8–22. doi:10.1038/s41579-020-0414-z
Linklater, D. P., De Volder, M., Baulin, V. A., Werner, M., Jessl, S., Golozar, M., et al. (2018). High aspect ratio nanostructures kill bacteria via storage and release of mechanical energy. ACS Nano 12, 6657–6667. doi:10.1021/acsnano.8b01665
Linklater, D. P., Juodkazis, S., Rubanov, S., and Ivanova, E. P. (2017). Comment on “bactericidal effects of natural nanotopography of dragonfly wing on Escherichia coli”. ACS Appl. Mater Interfaces 9, 29387–29393. doi:10.1021/acsami.7b05707
Lohmann, S. C., Tripathy, A., Milionis, A., Keller, A., and Poulikakos, D. (2022). Effect of flexibility and size of nanofabricated topographies on the mechanobactericidal efficacy of polymeric surfaces. ACS Appl. Bio Mater 5, 1564–1575. doi:10.1021/acsabm.1c01318
Maikranz, E., Spengler, C., Thewes, N., Thewes, A., Nolle, F., Jung, P., et al. (2020). Different binding mechanisms of: Staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale 12, 19267–19275. doi:10.1039/d0nr03134h
Mallavarapu, A., Ajay, P., and Sreenivasan, S. V. (2020). Enabling ultrahigh-aspect-ratio silicon nanowires using precise experiments for detecting the onset of collapse. Nano Lett. 20, 7896–7905. doi:10.1021/acs.nanolett.0c02539
Marathe, R., Meel, C., Schmidt, N. C., Dewenter, L., Kurre, R., Greune, L., et al. (2014). Bacterial twitching motility is coordinated by a two-dimensional tug-of-war with directional memory. Nat. Commun. 5, 3759. doi:10.1038/ncomms4759
Michalska, M., Divan, R., Noirot, P., and Laible, P. D. (2021a). Antimicrobial properties of nanostructured surfaces-demonstrating the need for a standard testing methodology. Nanoscale 13, 17603–17614. doi:10.1039/d1nr02953c
Michalska, M., Gambacorta, F., Divan, R., Aranson, I. S., Sokolov, A., Noirot, P., et al. (2018). Tuning antimicrobial properties of biomimetic nanopatterned surfaces. Nanoscale 10, 6639–6650. doi:10.1039/C8NR00439K
Michalska, M., Laney, S. K., Li, T., Portnoi, M., Mordan, N., Allan, E., et al. (2021b). Bioinspired multifunctional glass surfaces through regenerative secondary mask lithography. Adv. Mater. 33, e2102175. doi:10.1002/adma.202102175
Michalska, M., Rossi, A., Kokot, G., Macdonald, C. M., Cipiccia, S., Munro, P. R. T., et al. (2023). Fabrication of high-aspect ratio nanogratings for phase-based X-ray imaging. Adv. Funct. Mater 33, 202212660. doi:10.1002/adfm.202212660
Pogodin, S., Hasan, J., Baulin, V. A., Webb, H. K., Truong, V. K., Phong Nguyen, T. H., et al. (2013). Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces. Biophys. J. 104, 835–840. doi:10.1016/j.bpj.2012.12.046
Potthoff, E., Ossola, D., Zambelli, T., and Vorholt, J. A. (2015). Bacterial adhesion force quantification by fluidic force microscopy. Nanoscale 7, 4070–4079. doi:10.1039/c4nr06495j
Sabass, B., Koch, M. D., Liu, G., Stone, H. A., Shaevitz, J. W., and Schwarz, U. S. (2017). Force generation by groups of migrating bacteria. Proc. Natl. Acad. Sci. U. S. A. 114, 7266–7271. doi:10.1073/pnas.1621469114
Sahoo, P. K., Janissen, R., Monteiro, M. P., Cavalli, A., Murillo, D. M., Merfa, M. V., et al. (2016). Nanowire arrays as cell force sensors to investigate adhesin-enhanced holdfast of single cell bacteria and biofilm stability. Nano Lett. 16, 4656–4664. doi:10.1021/acs.nanolett.6b01998
Valiei, A., Lin, N., Bryche, J.-F., McKay, G., Canva, M., Charette, P. G., et al. (2020). Hydrophilic mechano-bactericidal nanopillars require external forces to rapidly kill bacteria. Nano Lett. 20, 5720–5727. doi:10.1021/acs.nanolett.0c01343
Valiei, A., Lin, N., McKay, G., Nguyen, D., Moraes, C., Hill, R. J., et al. (2022). Surface wettability is a key feature in the mechano-bactericidal activity of nanopillars. ACS Appl. Mater Interfaces 14, 27564–27574. doi:10.1021/acsami.2c03258
Velic, A., Hasan, J., Li, Z., and Yarlagadda, P. K. D. V. (2021). Mechanics of bacterial interaction and death on nanopatterned surfaces. Biophys. J. 120, 217–231. doi:10.1016/j.bpj.2020.12.003
Vrancken, N., Ghosh, T., Anand, U., Aabdin, Z., Chee, S. W., Baraissov, Z., et al. (2020). Nanoscale elastocapillary effect induced by thin-liquid-film instability. J. Phys. Chem. Lett. 11, 2751–2758. doi:10.1021/acs.jpclett.0c00218
Wang, D., Sun, Q., Hokkanen, M. J., Zhang, C., Lin, F.-Y., Liu, Q., et al. (2020). Design of robust superhydrophobic surfaces. Nature 582, 55–59. doi:10.1038/s41586-020-2331-8
Yu, F., Wang, D., Yang, J., Zhang, W., and Deng, X. (2021). Durable super-repellent surfaces: from solid–liquid interaction to applications. Acc. Mater Res. 2, 920–932. doi:10.1021/accountsmr.1c00147
Zhang, W., Wang, D., Sun, Z., Song, J., and Deng, X. (2021). Robust superhydrophobicity: mechanisms and strategies. Chem. Soc. Rev. 50, 4031–4061. doi:10.1039/D0CS00751J
Keywords: bioinspired, biomimetic, biofouling, antimicrobial mechanisms, mechanobactericidal surfaces, nanopatterns, driving force
Citation: Pirouz A, Papakonstantinou I and Michalska M (2024) Antimicrobial mechanisms of nanopatterned surfaces—a developing story. Front. Chem. 12:1354755. doi: 10.3389/fchem.2024.1354755
Received: 12 December 2023; Accepted: 18 January 2024;
Published: 29 January 2024.
Edited by:
Susan Kelleher, Dublin City University, IrelandReviewed by:
Wenluan Zhang, University of Electronic Science and Technology of China, ChinaCopyright © 2024 Pirouz, Papakonstantinou and Michalska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martyna Michalska, m.michalska@ucl.ac.uk
 Arash Pirouz
Arash Pirouz Ioannis Papakonstantinou2
Ioannis Papakonstantinou2  Martyna Michalska
Martyna Michalska