Recent Advances on DNAzyme-Based Biosensors for Detection of Uranyl
- 1Beijing Research Institute of Chemical Engineering and Metallurgy, China National Nuclear Corporation, Beijing, China
- 2School of Environmental and Municipal Engineering, Qingdao University of Technology, Qingdao, China
- 3Intelligent Wearable Engineering Research Center of Qingdao, Research Center for Intelligent and Wearable Technology, College of Textiles and Clothing, State Key Laboratory of Bio-Fibers and Eco-Textiles, Qingdao University, Qingdao, China
Nuclear facilities are widely used in fields such as national defense, industry, scientific research, and medicine, which play a huge role in military and civilian use. However, in the process of widespread application of nuclear technology, uranium and its compounds with high carcinogenic and biologically toxic cause a lot of environmental problems, such as pollutions of water, atmosphere, soil, or ecosystem. Bioensors with sensitivity and specificity for the detection of uranium are highly demand. Nucleic acid enzymes (DNAzyme) with merits of high sensitivity and selectivity for targets as excellent molecular recognition elements are commonly used for uranium sensor development. In this perspective review, we summarize DNAzyme-based biosensors for the quantitative detection of uranyl ions by integrating with diverse signal outputting strategies, such as fluorescent, colorimetry, surface-enhanced Raman scattering, and electrochemistry. Different design methods, limit of detection, and practical applications are fully discussed. Finally, the challenges, potential solutions, and future prospects of such DNAzyme-based sensors are also presented.
Introduction
Uranium, a radioactive metal element, is a significant raw material for the nuclear industry including nuclear power, nuclear weapons, scientific research, and nuclear medicines. Uranium-based nuclear energy effectively reduces global environmental problems such as global warming and energy depletion caused by fossil energy. Economic, efficient, and clean nuclear energy has great development prospects. Whereas, with the development and application of nuclear technology, the radioactive wastes containing uranium will gradually infiltrate environmental media, such as water, soil, and atmosphere, and eventually enter the biosphere system, which will cause great damage to humans and ecosystems. Uranium with strong chemical toxicity and radiotoxicity will cause lasting disturbances and damage to the immune, reproduction, and hematopoietic systems of organisms (Domingo, 2001). The World Health Organization lists uranium as a priority environmental pollutant, stipulating that the concentration limit of uranium in drinking water is 30 ug/L (Ansoborlo et al., 2015). Uranium possesses a lot of forms under different conditions and the uranyl ion (UO22+) is the most stable chemical form in aqueous solution. Many techniques have been developed for UO22+ detection, including X-ray fluorescence spectroscopy, atomic emission spectrometry, inductively coupled plasma mass spectrometry, and high-performance liquid chromatography (Jamali et al., 2006; Jaison et al., 2011; Boulyga et al., 2016; Sanyal et al., 2017). These methods with high selectivity and sensitivity are high cost, requiring sophisticated instrument and tedious pre-treating procedures, which are difficult to achieve the goal of real-time and onsite detection. Therefore, bioensors with high sensitivity and specificity for the detection of uranyl ion have become increasingly necessary.
Nucleic acid enzymes, also called DNAzymes, are isolated through in vitro selection (Liu et al., 2009). DNAzyme typically is composed of a substrate strand and an enzyme strand, and the two DNA strands are partially complementary hybridized by base pairing to form a double-stranded system. The presence of target ion activates the activity of DNAzyme and the ribo-adenosine (rA) on the substrate chain is cleavaged. The released target ion subsequently interacts with another DNAzyme, resulting in a signal-amplifying effect. DNAzymes with high metal-binding affinity and specificity show great promise as molecular tools in the design of diverse biosensors and nanodevices, benefiting from their unique characters, including low nonspecific adsorption, good stability, and easy preparation (Zhou et al., 2017; Jouha and Xiong, 2021). Moreover, the recycling of target molecule properties makes DNAzymes outstanding signal amplifiers for enzyme-free and highly sensitive detection of many different metal ions (Saidur et al., 2017; Jouha and Xiong, 2021; Khan et al., 2021). UO22+-specific DNAzyme was firstly selected by Lu group and a fluorescent sensor was developed simultaneously (Liu et al., 2007). In the past few years, quite a few amplified sensing platforms had been established for the quantitative detection of UO22+ (Farzin et al., 2019; He and Hua, 2019; Wu et al., 2019; Wang et al., 2022). In this review, we only focused on the application of various types of DNAzymes-based methods with diverse signals outputting approaches to achieve quantitative detection of uranyl ion. Design strategies, detection limits, and detection ranges were comprehensively compared. Such DNAzymes based sensors are expected to show great potential in environmental monitoring and nuclear emergency.
Applications of Different Types of Dnazyme-Based Sensors
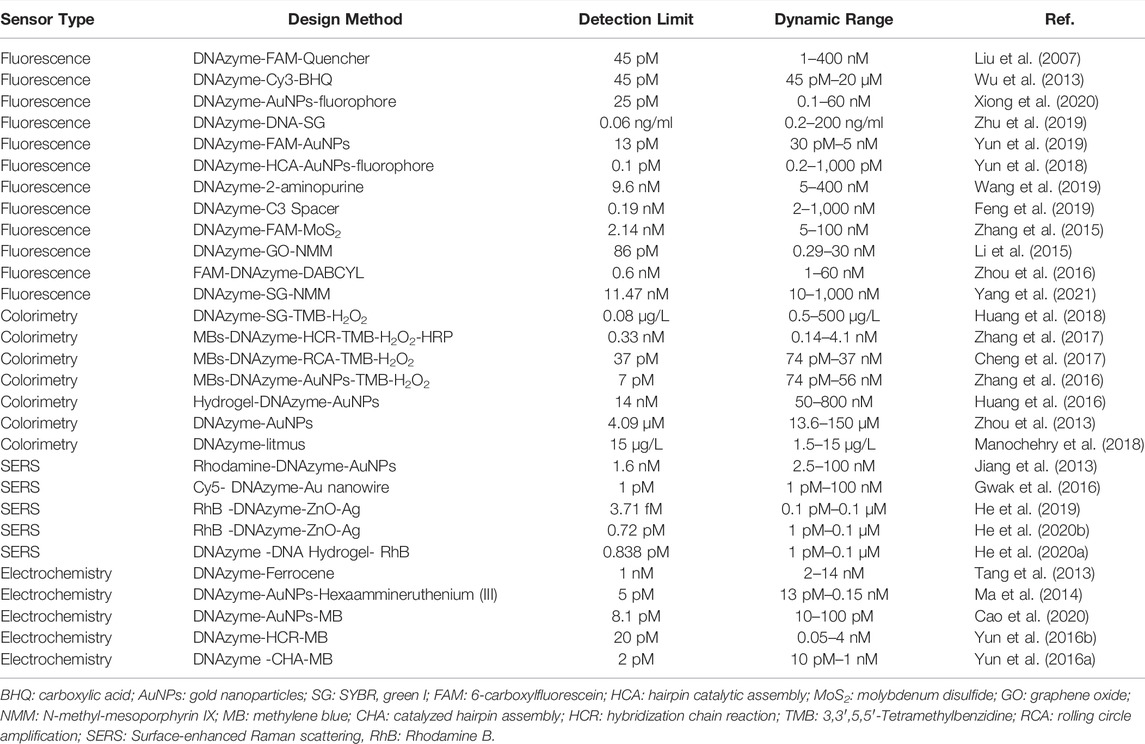
DNAzymes as the recognition element are suitable for the fabrication of uranyl sensors. Target molecules induced huge changes in structure and conformation of DNAzymes to produce diverse signal outputting including fluorescence, electrochemistry, colorimetry, and surface-enhanced Raman scattering (SERS). Table 1 summarized the DNAzyme-based biosensors for the detection of UO22+ by integrating with different signal output types including fluorescent, electrochemistry, colorimetry, and SERS.
DNAzyme-Based Fluorescent Sensors for UO22+ Detection
Fluorescence techniques are commonly used for the detection of diverse targets by measuring the change of fluorescence emission. DNAzyme as affinity ligand was applied in fluorescence sensors. Lu group reported a DNAzyme-based gold nanoparticles (AuNPs) sensor for uranyl ion detection (Wu et al., 2013). AuNPs were functioned with uranyl-specific DNAzyme and the substrate strand was modified with a fluorophore/quencher at 5′ and 3’-end respectively. In the absence of UO22+, the fluorescence of fluorophore was quenched by both quencher and AuNPs. Upon uranyl ion binding, the cleavage of rA resulted in the release of fluorophore and the enhanced fluorescence enabled the sensitive detection of UO22+ in living cells. Xiong et al. presented a DNA tweezer probe for the fluorescent detection of UO22+. DNAzyme catalytic cleavage strategy was used for signal enhancement (Xiong et al., 2020). AuNPs and fluorophore were fixed at the ends of DNA tweezer. In the presence of target UO22+, DNAzyme cleaved substrate linker DNA sequence, the enhanced fluorescent signal allowed the detection of target ions, and the limit of detection was 25 pM UO22+.
Zhu et al. reported a G-quadruplex-assisted enzyme strand recycling strategy-based fluorescent sensor for detecting UO22+ as shown in Figure 1A (Zhu et al., 2019). Such approach contained enzyme strand (E-DNA), cleaved substrate strand (S-DNA), and SYBR green I (SG). In the presence of target UO22+, DNAzyme was activated and further cleaved the S-DNA containing G-quadruplex sequence at the both ends. The formation of G-quadruplex helped the separation between E-DNA and S-DNA, which obviously improved the recycle utilization of E-DNA. The fluorescent signal of SG, a DNA intercalating dye, positively correlated with the amount of target UO22+. The limit of detection was 200 pM UO22+.
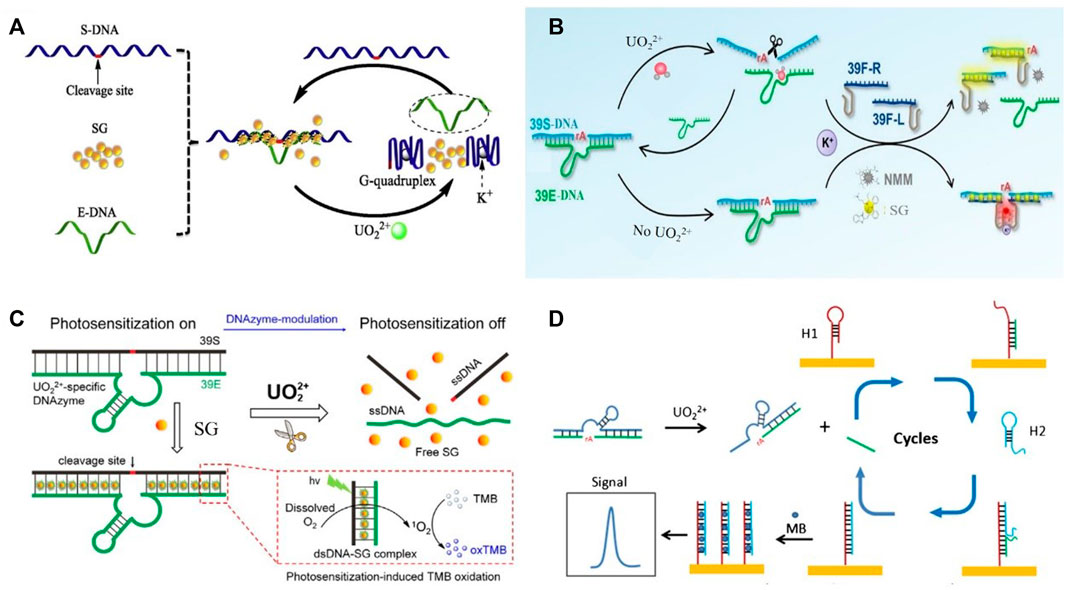
FIGURE 1. DNAzyme-based biosensors for the detection of uranyl ions by integrating with diverse signal outputting strategies in previous reports (A) G-quadruplex-assisted enzyme strand recycling-based fluorescent uranyl ion sensor. Reprinted with permission (Zhu et al., 2019), Copyright 2019 Elsevier B.V. (B) Ratiometric fluorescent biosensor for uranyl detection. Reprinted with permission (Yang et al., 2021). Copyright 2021 Elsevier B.V. (C) Electrochemical biosensor for uranyl detection based on DNAzyme and CHA. Reprinted with permission (Yun et al., 2016a). Copyright 2017, Royal Society of Chemistry (D) DNAzyme-modulated SG photosensitization colorimetric sensor for detection of UO22+. Reprinted with permission (Huang et al., 2018). Copyright 2017, Royal Society of Chemistry.
A variety of signal amplification strategies were also applied to the design of UO22+ sensor based on DNAzyme cleavage. Entropy-driven amplification and DNAzyme circular cleavage amplification-based fluorescent sensor was used for sensitive uranyl ion detection (Yun et al., 2019). The first amplification, entropy-driven amplification, was initiated by a DNA fragment coming from the cleavage of DNAzyme in the addition of target UO22+. Two DNA sequences released from the entropy driven amplification were partly complementary, which was an E-DNA of Mg2-specific DNAzyme. The second amplification, DNAzyme circular cleavage amplification, subsequently was activated. The formation of E-DNA circularly cleaved S-DNA-FAM probes decorated AuNPs. The following recovery of fluorescent signal enabled the sensitive detection of UO22+ and the limit of detection was as low as 13 pM. An enzyme-free dual amplification-based fluorescent sensor for ultra-sensitive detection of UO22+ was reported by Huang et al. (Yun et al., 2018). The hairpin catalytic assembly (HCA) reaction and DNAzyme-strand recycling were used in such strategy. In the absence of UO22+, dye-labeled hairpins absorbed on the surface of AuNPs and aggregation of AuNPs were prohibited. The addition of UO22+ triggered HCA reaction between the three hairpins. The formed rigid DNA triangles with negatively charged released from the negatively charged AuNPs. Turn-on fluorescent signal achieved the sensitive detection of UO22+ and the limit of detection was as low as 0.1 pM.
Wang et al. reported a fluorescent DNAzyme beacon probe for uranyl ion detection by embedding 2-aminopurine into the middle of S-DNA instead of labelling fluorescent dyes at the ends of S-DNA (Wang et al., 2019). 2-aminopurine, a fluorescent analog of adenosine, emitted fluorescence signals in a single-strand DNA (ssDNA); however, it was significantly quenched in the hybridized double-stranded DNA (dsDNA) due to the base-stacking interaction. In the presence of UO22+, fluorescence signal recovered upon the cleavage of DNAzyme. Turn-on sensing process enabled the quantitative detection of UO22+. The original catalytic activity of DNAzyme was hugely enhanced by inserting a C3 spacer. The length of flexible linkers and site of insertion were fully studied (Feng et al., 2019). Such modified DNAzyme was used in a fluorescent sensor and the detection limit was 0.19 nM UO22+.
Nanosheets with merits of great adsorption of DNA probes and excellent fluorescence quenching effect were commonly used for the fabrication of fluorescence sensors. Fu group presented a fluorescent biosensor for the simple and rapid detection of UO22+ in aqueous environment (Zhang et al., 2015). Such an approach used DNAzyme as target recognition element and molybdenum disulfide (MoS2) nanosheets as the fluorescence quencher. In the presence of UO22+, the cleavage occurred and the released FAM-labeled ssDNA adsorbed on the surface of MoS2 nanosheets, resulting in an obvious decreased fluorescence signal. The limit of detection of this turn-on sensor was 2.14 nM. Graphene oxide (GO) was used as a quencher to lower background fluorescence for amplified detection of UO22+ (Li et al., 2015). The presence of targets led to the cleavage of rA in DNAzyme, resulting in the formation of G-quadruplexes, which could interact with N-methyl-mesoporphyrin IX (NMM) to cause an enhanced fluorescence intensity. Free ssDNA and NMM were adsorbed by GO for background reduction. The limit of detection was as low as 86 pM.
DNAzyme nanostructures for UO22+ detection in living cells were developed (Zhou et al., 2016). The fluorescently quenched nanoprobes were decorated by ssDNAs containing the metal ion-dependent enzymatic and substrate sequences. The self-assembly formation nanostructure could specifically recognize target ions to recover fluorescent emissions. Increased fluorescent signals enabled the quantitative detection of uranyl ions. Yang et al. reported a ratiometric fluorescent DNAzyme sensor for UO22+ monitoring (Yang et al., 2021). The fluorescent biosensor contained DNAzyme probes (39E-DNA and 39S-DNA) and the split G-quadruplex probes (39F-R and 39F-L) as shown in Figure 1B. The presence of 39S-DNA leaded the proximity of 39F-R and 39F-L to form G-quadruplex. In the presence of target ions, DNAzyme-induced the cleavage of 39S-DNA splitted 39F-R and 39F-L. The decreased fluorescent of NMM was linear with uranyl ion concentration. Meanwhile, SG was used to monitor the hybridization of G-quadruplex probes and 39S-DNA. The ratiometric signal of NMM and SG enabled the robust detection of UO22+.
DNAzyme-Based Colorimetric Sensors
Colorimetric technique was used to determine the concentration of targets in solution by measuring the absorbance of a specific wavelength, which could be performed by UV-vis spectrophotometry (Priyadarshini and Pradhan, 2017). The sensitivity of colorimetric technique was lower than that of fluorescence; however, the color change was easily captured by naked eyes, making it a facile and convenient method.
DNAzyme-based colorimetric sensors had been developed for UO22+ detection via DNAzyme modulated photosensitization as demonstrated in Figure 1C (Huang et al., 2018). The dsDNA structure allowed SG to be located in, which could activate the photosensitization of SG for TMB (3,3′,5,5′-tetramethylbenzidine) oxidation, and chromogenic reaction occurred subsequently. In the presence of target UO22+, dsDNA structure of DNAzyme was cleaved and SG was released. Therefore, the color was weakened due to the reduced TMB oxidation from SG. This colorimetric sensor offered a detection limit of 0.08 μg/L (UV−vis detection) and 0.5 μg/L (naked eye).
Fu group reported magnetic beads (MBs) and HCR-based colorimetric biosensor for uranyl ion detection (Zhang et al., 2017). The addition of UO22+ cleaved DNAzymes immobilized on MBs surface to release ssDNA. The released ssDNA on MBs surface triggered HCR to capture a large amount of horseradish peroxidase (HRP). Upon the addition of TMB-H2O2 solution, the HRP-DNA-MBs conjugates could catalyze the H2O2-mediated oxidation of TMB, a color change from colorless to blue in solution was observed. This provided a sensitive and selective sensing platform for the visual or colorimetric detection of UO22+. The proposed biosensor has high sensitivity and strong anti-interference capability. In addition, the same group described a UO22+ sensor in combination with rolling circle amplification (RCA) (Cheng et al., 2017). DNAzyme functionalized on MBs was selectively cleaved in the presence of UO22+. The released DNA chains then triggered RCA, which increased the sensitivity of such biosensor. The detection limit was 37 pM. AuNPs-based enzymatic catalysis amplification was applied for UO22+ sensor development (Zhang et al., 2016).
DNAzyme-functionalized MBs was used for UO22+ recognition, released short ssDNA, then fixed HRP-functionalized AuNPs to the surface of MBs. H2O2-mediated oxidation of TMB occurred. The limit detection was 7 pM.
A smart hydrogel sensor was designed and synthesized for rapid, portable, sensitive detection of UO22+ (Huang et al., 2016). DNA-grafted polyacrylamide chains were utilized to crosslink with DNAzyme to form the DNA hydrogel. Colorimetric analysis was achieved by encapsulating AuNPs in the DNAzyme-crosslinked hydrogel. The presence of UO22+ in the sample activated the cleavage of substrate strand from the enzyme strand, thereby decreasing the density of crosslinkers and destabilizing the hydrogel, which then released the encapsulated AuNPs. The dispersion of AuNPs would lead to the change of absorbance. The decreased signal value enabled the quantification of uranyl ion. DNAzyme-functionalized AuNPs were used for UO22+ detection (Zhou et al., 2013). The cleavage of the substrate strand of DNAzyme in the presence of targets resulted in releasing a shorter duplex, leading to the aggregation of AuNPs. The changed signal allowed the sensing of UO22+. A litmus test-based assay for colorimetric uranyl biosensor was developed by Manochehry et al., The addition of target ions produced a pH-increasing enzyme, which was recognized by litmus paper. The changed signal was linear with uranyl ion concentration (Manochehry et al., 2018).
DNAzyme-Based SERS Sensors
SERS technique with advantages of rapid detection speed, high-throughput screening, and high sensitivity had great potential in the high-speed and sensitive detection of diverse targets molecules (Pilot et al., 2019; Fan et al., 2020). Jiang et al. showed a label-free DNAzyme-based SERS method for sensing uranyl ion (Jiang et al., 2013). Such an approach used rhodamine as the Raman signal probe. The addition of UO22+ induced the cleavage of DNAzyme and the released ssDNA was adsorbed on the surface of AuNPs to form a stable conjugate. Subsequently, the combination of rhodamine and AuNPs-ssDNA conjugate gave a strong SERS signal. Gwak et al. presented a DNAzyme-based plasmonic nanowire interstice sensor for uranyl ion detection (Gwak et al., 2016). The DNAzyme reacted with target UO22+ and released Cy5 labeled strand. The plasmonic nanowire interstice sensor sensitively captured the released strands, giving a strong Raman signal and the detection limit was 1pM.
He et al. described a reusable SERS-based microfluidic biosensor for rapid detection of UO22+(He et al., 2019). When target ions were added to the solution, 5′-rhodamine B (RhB)-labeled DNAzymes were cleavaged in the microfluidic chip, ZnO-Ag nanosheet arrays modified with S-DNA, which was sequence-complementary with the RhB-labeled E-DNA. The hybridization of S-DNA and E-DNA fixed RhB close to the surface of ZnO-Ag. The increased Raman signal enabled the sensitive detection of UO22+. In addition, the same group developed a recyclable SERS-microfluidic biosensor for UO22+ detection (He et al., 2020b). ZnO-Ag hybrids arrays were designed as the reaction substrates. In the absence of UO22+, RhB-labeled dsDNA formed a rigid structure and weak Raman signal was detected. Addition of uranyl triggered DNAzyme-cleavage reaction. RhB was dropped down from the surface of SERS substrates, leading to the variation of Raman signals. The detection limit of uranyl was 0.72 pM. Flexible DNAzyme-based hydrogel SERS sensor for the detection of uranyl ions was also developed by Wang group (He et al., 2020a). The presence of UO22+ ions triggered the activity of DNAzyme to cleave the substrate strand; subsequently, the DNA hydrogel structure was destroyed to release RhB, leading to a changed Raman signal. The detection of limit of such approach was 0.84 pM.
DNAzyme-Based Electrochemical Sensors
Electrochemical biosensors had become increasingly popular due to its simplicity, portability, low cost, and high sensitivity (Wongkaew et al., 2019). DNAzymes were employed to achieve target recycling for signal amplification in electrochemical sensors. Tan et al. developed a DNAzyme-based electrochemical sensor for sensitive uranyl ion detection (Tang et al., 2013). A split uranyl-specific DNAzyme decorated with ferrocene unit was immobilized on the surface of a gold electrode. In the presence of uranyl, the cleavage of S-DNA induced the ferrocene release from the electrode. The measurement of changed electrochemical signal enabled the quantitative detection of UO22+.
AuNPs with large surface areas had the ability to absorb large number of DNAzymes and electroactive indicator, which was suitable for electrochemical sensor fabrication. Ma et al. presented a DNAzymes- and AuNPs-based electrochemical biosensor for uranyl detection (Ma et al., 2014). The addition of UO22+ induced the cleavage of DNAzymes and electroactive indicators were removed from the electrode subsequently. Differential current signals were used for sensing uranyl. Cao et al. designed an electrochemical biosensor for UO22+ detection by the integration of DNAzyme and different DNA-modified AuNPs network structure (Cao et al., 2020). Such an approach effectively increased the amount of methylene blue (MB), a commonly used electrochemical indicator. The presence of UO22+ triggered the cleavage of DNAzyme. MB and DNA-AuNPs then released from the gold electrode. The detection of reduced electrochemical response allowed the sensitive uranyl sensing with a low detection limit of 8.1 pM.
Different amplification strategies, such as hybridization chain reaction (HCR) and catalyzed hairpin assembly (CHA), were used for electrochemical biosensor development. Yun et al. reported an ultrasensitive DNAzyme-based electrochemical sensor for uranyl detection (Yun et al., 2016b). DNAzyme was selectively cleaved in the presence of UO22+. The released DNA chains triggered HCR, which increased the sensitivity of monitoring. The detection limit was 20 pM uranyl. An ultrasensitive electrochemical biosensor for uranyl detection based on DNAzyme and CHA was presented by Yun et al. (Yun et al., 2016a). As shown in Figure 1D, DNA was selectively cleaved in the presence of UO22+. The released fragment hybridized with hairpin probe 1 (H1) immobilized on the gold electrode. The unfolding of H1 subsequently induced the hybridization with hairpin 2 (H2). The DNA fragment spontaneously dissociated from the surface and then initiated the next hybridization cycle. MB was added to intercalate into the dsDNA. Double magnification strategy enabled the sensitive detection of UO22+ and the detection limit was 2 pM.
Conclusion and Outlooks
In this review, the recent progresses of sensors functioned with DNAzymes for uranyl ion detection were summarized including the design strategies, limit of detection, and dynamic range. Sensitive sensing for uranyl ion had been achieved with different signal outputting approaches including fluorescent, electrochemistry, colorimetry, and SERS. Despite great progress has already been achieved in this area, there are still many challenges to be overcome. Firstly, even though most DNAzyme-based uranyl sensors immune to the interference of commonly used metal ions, Th4+ can still affect the detection results. Hence, it is an important task in the future to improve the selectivity of such biosensor to tolerance the interfering ions. Secondly, in order to improve the limit of detection, complex construction strategies including using of functional nanomaterials and DNA recycle amplification (e.g., HCR, RCA and CHA) still need to be designed. Low-cost, portability, and onsite sensing platforms are of great promise in future uranyl sensor application. Thirdly, miniaturization and intellectualization are the trend of uranyl sensor in the future. Quantitative detection of uranium based on mobile phones is a very attractive direction (Chen et al., 2018), and chemical hybridization of carbon dots and CdTe QDs were used for the fabrication of ratiometric fluorescent probe. The addition of uranyl ions greatly quenched the red fluorescence of CdTe QDs, whereas the green fluorescence of carbon dots kept constant, leading to an obvious color change. A smartphone was used to analyze the content of uranyl on the basis of captured signals. Onsite and point-of-care detection of uranyl ions was achieved. Therefore, the sensitivity needed to be improved, which was mainly because the amplification strategy was not adopted.
Nowadays, the fabrication of high-quality DNAzyme-based sensors with merits of rapidness, low cost, sensitivity, and selectivity will become more active due to the widely used nuclear energy applications and the combined effects of its radioactive and biological toxicity during the uranium exploitation. Along with the great progress in the field of sensor design, we believe that DNAzyme-based sensors will play a vital role in various applications in environmental monitoring and nuclear environmental protection.
Author Contributions
All authors have read and agreed to the published version of the manuscript. YB collected and analyzed data, completed pictures and spreadsheets, conceived and wrote the paper. HC, LZ and GJ collected and analyzed data; LX and GZ wrote and revised the paper.
Funding
This research was supported by the National Key Research and Development Program of China (No. 2019YFC1907700), the National Natural Science Foundation of China (No. 21801158), the Natural Science Foundation of Shandong Province (No. ZR2020QB092), the China Postdoctoral Science Foundation (No. 2021M691689), and the State Key Laboratory of Bio-Fibers and Eco-Textiles (Qingdao University, Nos. ZKT23, KF2020201, GZRC202025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ansoborlo, E., Lebaron-Jacobs, L., and Prat, O. (2015). Uranium in Drinking-Water: A Unique Case of Guideline Value Increases and Discrepancies between Chemical and Radiochemical Guidelines. Environ. Int. 77, 1–4. doi:10.1016/j.envint.2014.12.011
Boulyga, S. F., Koepf, A., Konegger-Kappel, S., Macsik, Z., and Stadelmann, G. (2016). Uranium Isotope Analysis by MC-ICP-MS in Sub-ng Sized Samples. J. Anal. Spectrom. 31, 2272–2284. doi:10.1039/C6JA00238B
Cao, C., Liu, J., Tang, S., Dai, Z., Xiao, F., Rang, W., et al. (2020). Amplified Electrochemical Determination of UO22+ Based on the Cleavage of the DNAzyme and DNA-Modified Gold Nanoparticle Network Structure. Microchim. Acta 187, 311. doi:10.1007/s00604-020-04263-1
Chen, X., Mei, Q., Yu, L., Ge, H., Yue, J., Zhang, K., et al. (2018). Rapid and On-Site Detection of Uranyl Ions via Ratiometric Fluorescence Signals Based on a Smartphone Platform. ACS Appl. Mater. Inter. 10, 42225–42232. doi:10.1021/acsami.8b13765
Cheng, X., Yu, X., Chen, L., Zhang, H., Wu, Y., and Fu, F. (2017). Visual Detection of Ultra-Trace Levels of Uranyl Ions Using Magnetic Bead-Based Dnazyme Recognition in Combination with Rolling Circle Amplification. Microchim. Acta 184, 4259–4267. doi:10.1007/s00604-017-2472-0
Domingo, J. L. (2001). Reproductive and Developmental Toxicity of Natural and Depleted Uranium: A Review. Reprod. Toxicol. 15, 603–609. doi:10.1016/S0890-6238(01)00181-2
Fan, M., Andrade, G. F. S., and Brolo, A. G. (2020). A Review on Recent Advances in the Applications of Surface-Enhanced Raman Scattering in Analytical Chemistry. Anal. Chim. Acta 1097, 1–29. doi:10.1016/j.aca.2019.11.049
Farzin, L., Shamsipur, M., Sheibani, S., Samandari, L., and Hatami, Z. (2019). A Review on Nanomaterial-Based Electrochemical, Optical, Photoacoustic and Magnetoelastic Methods for Determination of Uranyl Cation. Microchim. Acta 186, 289. doi:10.1007/s00604-019-3426-5
Feng, M., Gu, C., Sun, Y., Zhang, S., Tong, A., and Xiang, Y. (2019). Enhancing Catalytic Activity of Uranyl-Dependent DNAzyme by Flexible Linker Insertion for More Sensitive Detection of Uranyl Ion. Anal. Chem. 91, 6608–6615. doi:10.1021/acs.analchem.9b00490
Gwak, R., Kim, H., Yoo, S. M., Lee, S. Y., Lee, G.-J., Lee, M.-K., et al. (2016). Precisely Determining Ultralow Level UO22+ in Natural Water with Plasmonic Nanowire Interstice Sensor. Sci. Rep. 6, 19646. doi:10.1038/srep19646
He, W., and Hua, D. (2019). Spectrographic Sensors for Uranyl Detection in the Environment. Talanta 201, 317–329. doi:10.1016/j.talanta.2019.04.018
He, X., Wang, S., Liu, Y., and Wang, X. (2019). Ultra-Sensitive Detection of Uranyl Ions with A Specially Designed High-Efficiency SERS-Based Microfluidic Device. Sci. China Chem. 62, 1064–1071. doi:10.1007/s11426-019-9468-x
He, X., Zhou, X., Liu, W., Liu, Y., and Wang, X. (2020a). Flexible DNA Hydrogel SERS Active Biofilms for Conformal Ultrasensitive Detection of Uranyl Ions from Aquatic Products. Langmuir 36, 2930–2936. doi:10.1021/acs.langmuir.9b03845
He, X., Zhou, X., Liu, Y., and Wang, X. (2020b). Ultrasensitive, Recyclable and Portable Microfluidic Surface-Enhanced Raman Scattering (SERS) Biosensor for Uranyl Ions Detection. Sensors Actuators B: Chem. 311, 127676. doi:10.1016/j.snb.2020.127676
Huang, Y., Fang, L., Zhu, Z., Ma, Y., Zhou, L., Chen, X., et al. (2016). Design and Synthesis of Target-Responsive Hydrogel for Portable Visual Quantitative Detection of Uranium with A Microfluidic Distance-Based Readout Device. Biosens. Bioelectron. 85, 496–502. doi:10.1016/j.bios.2016.05.008
Huang, C., Fan, X., Yuan, Q., Zhang, X., Hou, X., and Wu, P. (2018). Colorimetric Determination of Uranyl (UO22+) in Seawater via Dnazyme-Modulated Photosensitization. Talanta 185, 258–263. doi:10.1016/j.talanta.2018.03.079
Jaison, P. G., Telmore, V. M., Kumar, P., and Aggarwal, S. K. (2011). Determination of Uranium in Seawater Samples by Liquid Chromatography Using Mandelic Acid as A Complexing Agent. J. Chromatogr. Sci. 49, 657–664. doi:10.1093/chrsci/49.9.657
Jamali, M. R., Assadi, Y., Shemirani, F., Hosseini, M. R. M., Kozani, R. R., Masteri-Farahani, M., et al. (2006). Synthesis of Salicylaldehyde-Modified Mesoporous Silica and its Application as A New Sorbent for Separation, Preconcentration and Determination of Uranium by Inductively Coupled Plasma Atomic Emission Spectrometry. Anal. Chim. Acta 579, 68–73. doi:10.1016/j.aca.2006.07.006
Jiang, Z., Yao, D., Wen, G., Li, T., Chen, B., and Liang, A. (2013). A Label-Free Nanogold Dnazyme-Cleaved Surface-Enhanced Resonance Raman Scattering Method for Trace UO22+ Using Rhodamine 6G as Probe. Plasmonics 8, 803–810. doi:10.1007/s11468-012-9476-8
Jouha, J., and Xiong, H. (2021). DNAzyme‐Functionalized Nanomaterials: Recent Preparation, Current Applications, and Future Challenges. Small 17, 2105439. doi:10.1002/smll.202105439
Khan, S., Burciu, B., Filipe, C. D. M., Li, Y., Dellinger, K., and Didar, T. F. (2021). DNAzyme-Based Biosensors: Immobilization Strategies, Applications, and Future Prospective. Acs Nano 15, 13943–13969. doi:10.1021/acsnano.1c04327
Li, M.-H., Wang, Y.-S., Cao, J.-X., Chen, S.-H., Tang, X., Wang, X.-F., et al. (2015). Ultrasensitive Detection of Uranyl by Graphene Oxide-Based Background Reduction and Rcdzyme-Based Enzyme Strand Recycling Signal Amplification. Biosens. Bioelectron. 72, 294–299. doi:10.1016/j.bios.2015.05.032
Liu, J., Brown, A. K., Meng, X., Cropek, D. M., Istok, J. D., Watson, D. B., et al. (2007). A Catalytic Beacon Sensor for Uranium with Parts-Per-Trillion Sensitivity and Millionfold Selectivity. Proc. Natl. Acad. Sci. U.S.A. 104, 2056–2061. doi:10.1073/pnas.0607875104
Liu, J., Cao, Z., and Lu, Y. (2009). Functional Nucleic Acid Sensors. Chem. Rev. 109, 1948–1998. doi:10.1021/cr030183i
Ma, D., Yuan, Y., Xiao, X., Gao, Y., Li, Y., Xu, W., et al. (2014). A Label-Free Electrochemical Biosensor for Trace Uranium Based on Dnazymes and Gold Nanoparticles. J. Radioanal. Nucl. Chem. 299, 1911–1919. doi:10.1007/s10967-013-2897-9
Manochehry, S., Mcconnell, E. M., Tram, K. Q., Macri, J., and Li, Y. (2018). Colorimetric Detection of Uranyl Using A Litmus Test. Front. Chem. 6, 332. doi:10.3389/fchem.2018.00332
Pilot, R., Signorini, R., Durante, C., Orian, L., Bhamidipati, M., and Fabris, L. (2019). A Review on Surface-Enhanced Raman Scattering. Biosensors 9, 57. doi:10.3390/bios9020057
Priyadarshini, E., and Pradhan, N. (2017). Gold Nanoparticles as Efficient Sensors in Colorimetric Detection of Toxic Metal Ions: A Review. Sensors Actuators B: Chem. 238, 888–902. doi:10.1016/j.snb.2016.06.081
Saidur, M. R., Aziz, A. R. A., and Basirun, W. J. (2017). Recent Advances in DNA-Based Electrochemical Biosensors for Heavy Metal Ion Detection: A Review. Biosens. Bioelectron. 90, 125–139. doi:10.1016/j.bios.2016.11.039
Sanyal, K., Khooha, A., Das, G., Tiwari, M. K., and Misra, N. L. (2017). Direct Determination of Oxidation States of Uranium in Mixed-Valent Uranium Oxides Using Total Reflection X-Ray Fluorescence X-Ray Absorption Near-Edge Spectroscopy. Anal. Chem. 89, 871–876. doi:10.1021/acs.analchem.6b03945
Tang, Q., Yuan, Y., Xiao, X., Guo, P., Hu, J., Ma, D., et al. (2013). DNAzyme Based Electrochemical Sensors for Trace Uranium. Microchim. Acta 180, 1059–1064. doi:10.1007/s00604-013-1021-8
Wang, X., Zeng, R., Chu, S., Tang, W., Lin, N., Fu, J., et al. (2019). A Quencher-free DNAzyme Beacon for Fluorescently Sensing Uranyl Ions via Embedding 2-Aminopurine. Biosens. Bioelectron. 135, 166–172. doi:10.1016/j.bios.2019.04.021
Wang, S., Jiang, J., He, X., Yang, S., Wu, H., Qin, Z., et al. (2022). Research Progress of SERS on Uranyl Ions and Uranyl Compounds: A Review. J. Mater. Chem. C 10, 4006–4018. doi:10.1039/d1tc05154g
Wongkaew, N., Simsek, M., Griesche, C., and Baeumner, A. J. (2019). Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-On-A-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem. Rev. 119, 120–194. doi:10.1021/acs.chemrev.8b00172
Wu, P., Hwang, K., Lan, T., and Lu, Y. (2013). A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc. 135, 5254–5257. doi:10.1021/ja400150v
Wu, X., Huang, Q., Mao, Y., Wang, X., Wang, Y., Hu, Q., et al. (2019). Sensors for Determination of Uranium: A Review. Trac Trends Anal. Chem. 118, 89–111. doi:10.1016/j.trac.2019.04.026
Xiong, Z., Wang, Q., Zhang, J., Yun, W., Wang, X., Ha, X., et al. (2020). A Simple and Programmed DNA Tweezer Probes for One-step and Amplified Detection of UO22+. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 229, 118017. doi:10.1016/j.saa.2019.118017
Yang, Y., Yang, H., Wan, Y., Zhou, W., Deng, S., He, Y., et al. (2021). Temperature-robust and Ratiometric G-Quadruplex Proximate Dnazyme Assay for Robustly Monitoring of Uranium Pollution and its Microbial Biosorbents Screening. J. Hazard. Mater. 413, 125383. doi:10.1016/j.jhazmat.2021.125383
Yun, W., Cai, D., Jiang, J., Wang, X., Liao, J., Zhang, P., et al. (2016a). An Ultrasensitive Electrochemical Biosensor for Uranyl Detection Based on Dnazyme and Target-Catalyzed Hairpin Assembly. Microchim. Acta 183, 1425–1432. doi:10.1007/s00604-016-1778-7
Yun, W., Jiang, J., Cai, D., Wang, X., Sang, G., Liao, J., et al. (2016b). Ultrasensitive Electrochemical Detection of UO22+ Based on DNAzyme and Isothermal Enzyme-free Amplification. RSC Adv. 6, 3960–3966. doi:10.1039/c5ra22773a
Yun, W., Wu, H., Liu, X., Zhong, H., Fu, M., Yang, L., et al. (2018). Ultra-sensitive Fluorescent and Colorimetric Detection of UO22+ Based on Dual Enzyme-free Amplification Strategies. Sensors Actuators B: Chem. 255, 1920–1926. doi:10.1016/j.snb.2017.08.205
Yun, W., Wu, H., Yang, Z., Wang, R., Wang, C., Yang, L., et al. (2019). A Dynamic, Ultra-sensitive and "Turn-On" Strategy for Fluorescent Detection of Uranyl Based on Dnazyme and Entropy-Driven Amplification Initiated Circular Cleavage Amplification. Anal. Chim. Acta 1068, 104–110. doi:10.1016/j.aca.2019.04.018
Zhang, H., Ruan, Y., Lin, L., Lin, M., Zeng, X., Xi, Z., et al. (2015). A Turn-Off Fluorescent Biosensor for the Rapid and Sensitive Detection of Uranyl Ion Based on Molybdenum Disulfide Nanosheets and Specific DNAzyme. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 146, 1–6. doi:10.1016/j.saa.2015.02.113
Zhang, H., Lin, L., Zeng, X., Ruan, Y., Wu, Y., Lin, M., et al. (2016). Magnetic Beads-Based DNAzyme Recognition and Aunps-Based Enzymatic Catalysis Amplification for Visual Detection of Trace Uranyl Ion in Aqueous Environment. Biosens. Bioelectron. 78, 73–79. doi:10.1016/j.bios.2015.11.024
Zhang, H., Cheng, X., Chen, L., Mo, F., Xu, L., and Fu, F. (2017). Magnetic Beads-Based DNA Hybridization Chain Reaction Amplification and Dnazyme Recognition for Colorimetric Detection of Uranyl Ion in Seafood. Anal. Chim. Acta 956, 63–69. doi:10.1016/j.aca.2016.12.021
Zhou, B., Shi, L.-F., Wang, Y.-S., Yang, H.-X., Xue, J.-H., Liu, L., et al. (2013). Resonance Light Scattering Determination of Uranyl Based on Labeled DNAzyme-Gold Nanoparticle System. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 110, 419–424. doi:10.1016/j.saa.2013.03.036
Zhou, W., Liang, W., Li, D., Yuan, R., and Xiang, Y. (2016). Dual-color Encoded Dnazyme Nanostructures for Multiplexed Detection of Intracellular Metal Ions in Living Cells. Biosens. Bioelectron. 85, 573–579. doi:10.1016/j.bios.2016.05.058
Zhou, W., Saran, R., and Liu, J. (2017). Metal Sensing by DNA. Chem. Rev. 117, 8272–8325. doi:10.1021/acs.chemrev.7b00063
Keywords: uranyl sensor, DNAzyme, environmental monitoring, detection, spectrum, electrochemistry
Citation: Bai Y, Xu L, Chai H, Zhou L, Jiang G and Zhang G (2022) Recent Advances on DNAzyme-Based Biosensors for Detection of Uranyl. Front. Chem. 10:882250. doi: 10.3389/fchem.2022.882250
Received: 23 February 2022; Accepted: 22 March 2022;
Published: 27 April 2022.
Edited by:
Cheng Ma, Yangzhou University, ChinaReviewed by:
Taiping Qing, Xiangtan University, ChinaCopyright © 2022 Bai, Xu, Chai, Zhou, Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lechang Xu, xu_lechang@163.com; Guangyao Zhang, gyzhang@qdu.edu.cn
 Yunlong Bai
Yunlong Bai Lechang Xu1*
Lechang Xu1*  Guangyao Zhang
Guangyao Zhang