Co-delivery of Salinomycin and Curcumin for Cancer Stem Cell Treatment by Inhibition of Cell Proliferation, Cell Cycle Arrest, and Epithelial–Mesenchymal Transition
- 1School of Pharmacy, Nantong University, Nantong, China
- 2Danish Cancer Society Research Center, Copenhagen, Denmark
- 3NICM Health Research Institute, Western Sydney University, Sydney, NSW, Australia
Malignant cancer is a devastating disease often associated with a poor clinical prognosis. For decades, modern drug discoveries have attempted to identify potential modulators that can impede tumor growth. Cancer stem cells however are more resistant to therapeutic intervention, which often leads to treatment failure and subsequent disease recurrence. Here in this study, we have developed a specific multi-target drug delivery nanoparticle system against breast cancer stem cells (BCSCs). Therapeutic agents curcumin and salinomycin have complementary functions of limiting therapeutic resistance and eliciting cellular death, respectively. By conjugation of CD44 cell-surface glycoprotein with poly(lactic-co-glycolic acid) (PLGA) nanoparticles that are loaded with curcumin and salinomycin, we investigated the cellular uptake of BCSCs, drug release, and therapeutic efficacy against BCSCs. We determined CD44-targeting co-delivery nanoparticles are highly efficacious against BCSCs by inducing G1 cell cycle arrest and limiting epithelial–mesenchymal transition. This curcumin and salinomycin co-delivery system can be an efficient treatment approach to target malignant cancer without the repercussion of disease recurrence.
Introduction
Resistance to chemotherapy is the major cause of cancer relapse and mortality. Early indications of intrinsic resistance emphasized on the presence of cancer stem cells (CSCs), which are highly mesenchymal in nature and have the ability to self-renew, differentiate, and promote tumorigenesis. They play an important role in chemotherapy and radiotherapy resistance via enhanced anti-apoptotic, DNA repair, and reactive oxygen species (ROS)-protective mechanisms, often leading to tumor recurrence including metastasis. There is now overwhelming clinical evidence to support enriched CSCs as the dominant population in reviving tumor growth despite initial aggressive chemotherapeutic intervention. Therefore, effective elimination of CSCs is essential to impede tumor relapse and drug resistance to improve patients' survival. However, current treatments may harm the normal stem cell pool and result in severe side effects. Therefore, therapeutics to specifically remove CSCs remain a major challenge for anti-cancer treatment.
Salinomycin is a polyether ionophore antibiotic with efficacy as an anti-cancer drug (Naujokat and Steinhart, 2012). Pre-clinical data indicates salinomycin has the potential to reduce tumorigenicity by targeting CSCs and has been effective in several cancer types (Gupta et al., 2009; Lu et al., 2011; Ketola et al., 2012; Lim et al., 2020). The anti-cancer mechanism of salinomycin is associated with dysregulation of metal ions (Paulus et al., 1998), activation of autophagy-mediated cell death, and inhibition of stem cell maintenance (Lu et al., 2011; Yue et al., 2013; Mai et al., 2017). A recent study found that salinomycin and its derivative, ironomycin, exhibited a potent and selective activity against breast cancer stem cells (BCSCs) by accumulating and sequestering iron to induce ferroptosis, which is a recently described form of iron-dependent cell death marked by the oxidative modification of phospholipid membranes (Mai et al., 2017). However, it is challenging to efficiently deliver salinomycin (Sal) to tumor sites due to its hydrophobicity, unfavorable pharmacokinetic profile, and cytotoxicity during systemic drug administration (Naujokat and Steinhart, 2012). Curcumin (Cur) is a bioactive ingredient derived from the root of Curcuma longa L, which have been widely used in the prevention and treatment of a number of diseases, including cancer (Sharma et al., 2005; Zhou et al., 2011; Shanmugam et al., 2015; Gao et al., 2017; Motevalli et al., 2019). Curcumin is a potent tumor suppressor as it can inhibit tumor growth at cellular and in vivo level. Although the mechanism is not fully understood, studies have shown that curcumin can lead to inhibition of cancer cell invasion (Baek et al., 2003; Shao et al., 2017), cell cycle arrest, and induction of autophagic cell death. More recently, researchers have investigated the anti-metastasis potentials of curcumin (Basnet and Skalko-Basnet, 2011; Norris et al., 2013; Teiten et al., 2014; Kotcherlakota et al., 2016) and found it can inhibit CSCs' viability and their stemness via ROS and the signal transducer and activator of transcription 3 (STAT3) signaling pathway (Buhrmann et al., 2020). However, its therapeutic potential is limited due to its rapid metabolism and poor solubility and absorption (Tsai et al., 2011).
Nanomedicines provide new strategies in cancer targeting and treatment due to high drug loading capability, cancer-targeting ability, and even the potential to overcome the efflux pump-mediated drug resistance by delivering a high concentration of intracellular drugs (Cho et al., 2008). Our previous studies showed that conjugation of salinomycin with gold nanoparticles can efficiently induce ferroptotic cell death of BCSCs by increasing the generation of ROS, mitochondrial dysfunction, and lipid oxidation with higher iron accumulation and GPX-4 inactivation (Zhao et al., 2019). Here, the combination of salinomycin and curcumin was used to achieve synergetic effect in treating BCSCs. Salinomycin and curcumin were loaded onto poly(lactic-co-glycolic acid) (PLGA) polymeric nanoparticles via double emulsion method to form nanoparticles (Avgoustakis et al., 2002; Alam et al., 2017). Additionally, hyaluronic acid (HA) was also conjugated to nanoparticles as targeting moiety due to its specific interaction with the CD44 receptor, which is a cell surface glycoprotein and commonly overexpressed on BCSCs (Zöller, 2011; Ganesh et al., 2013; Sanfilippo et al., 2020). The CD44-targeting nanoparticles loaded with salinomycin and curcumin showed improved therapeutic efficiency against BCSCs by inducing G1 cell cycle arrest and limiting epithelial–mesenchymal transition (EMT).
Materials and Methods
Materials
Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin–streptomycin, propidium iodide, Triton X-100, PageRuler Prestained Protein Ladder, and nitrocellulose membrane were purchased from Thermo Fisher. Curcumin, salinomycin, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), and N-hydroxysuccinimide (NHS) were bought from Sigma-Aldrich and used directly without any purification. Mouse anti-vimentin, mouse anti-E-cadherin, and rabbit anti-β-actin primary antibodies purchased from Abcam. Odyssey blocking buffer, IRDye® 800CW goat anti-mouse secondary antibody, and IRDye® 680RD goat anti-rabbit secondary antibody were purchased from LI-COR.
Nanoparticle Synthesis and Surface Functionalization
NH2-PEG-NH2 (MW: 3.4 k), PLGA (MW: 10 k), and PLGA-PEG-NH2 (MW: 18.6 k) co-polymer were bought from Meiluo Tech Company. Curcumin and salinomycin were loaded into the PLGA-PEG-NH2 copolymer via the well-established double emulsion method (Avgoustakis et al., 2002). Briefly, curcumin (1 mg/ml) and salinomycin (molar ratio of Cur and Sal = 1:1) were dissolved in organic phase containing co-polymer (1.75%, w/v), and the mixture was mixed using an ultrasonic homogenizer (LABSONIC M, Sartorius, Germany). The primary particles were added into a PVA solution (10 ml, 2%, w/v), and the mixture was sonicated again. The free drugs were then removed by centrifugation at 20,000 rpm for 15 min and washed with water. The formed nanoparticles were labeled as PEG-PLGA-Cur-Sal.
After drug loading, targeting moiety HA was conjugated onto the polymeric nanoparticles via EDC coupling. Briefly, desalted HA (1 equivalent), EDC (3 equivalents), and NHS (3 equivalents) were dissolved in 2-(N-morpholino)ethanesulfonic acid (MES) buffer and stir in ice bath to activate carboxylic group for half an hour. PEG-PLGA-Cur-Sal was then added and the mixture was stirred for another 4 h at room temperature to form nanoparticles named HA-PEG-PLGA-Cur-Sal. They were purified by using centrifugation at 20,000 rpm for 15 min.
Characterization of the Nanoparticles
1H NMR spectra were acquired on a Bruker AVANCE 400 MHz spectrometer. A gel permeation chromatography—multi-angle laser light scattering (GPC-MALLS) system consisting of a 1,515 isocratic pump (Waters), Styragel HT 6E and Styragel HT 3 columns (Waters), 2,414 differential refractive index detector (Waters), and a DAWN HELEOS laser light scattering detector (Wyatt) with THF as an eluent at a flow rate of 1 ml/min was used to measure the molecular weight and dispersity of the polymers. After drug loading, the size of the nanoparticles was measured via dynamic light scattering (DLS) to measure the hydrodynamic diameter using a Malvern Zetasizer, and a Philips CM100 transmission electron microscope (TEM) was also used to observe their morphology.
Cancer Stem Cell Culture
MCF-7 cells were bought from ATCC (USA) and cultured in DMEM medium containing 10% (v/v) FBS and 1% (v/v) penicillin–streptomycin. They were kept in a humidified 37°C incubator with 5% CO2. After serum-free treatment, BCSC sub-populations in the MCF-7 cells were isolated using MagCellect Human BCSC isolation kit (R&D Systems) according to protocols from the manufacturer. In brief, MCF-7 cells (1 × 107 cells/ml) were prepared in buffer and mixed with anti-human CD24 biotinylated antibody and then Streptavidin Ferrofluid for selection of CD24low cells. Those cells were subsequently incubated with a biotinylated anti-human CD44 antibody and Streptavidin Ferrofluid for selection of CD44high cells. After washing, magnetically tagged CD24low/CD44high BCSCs were isolated through the magnet. The cells were directly treated with drug-loaded nanoparticles and control samples after isolation.
Drug Loading Efficiency and Drug Release Study
Salinomycin loading efficiency was analyzed by using a UV-Vis-NIR spectrophotometer following a published method (Wang et al., 2014). The drug concentration of curcumin was calculated using the linear portion of the calibration curve obtained by the UV spectrophotometer at 427 nm for serial drug concentrations.
Cellular Uptake Assay With Flow Cytometry
Flow cytometry was used to quantify cellular interaction following incubation of the cells with each treatment group. BCSCs were seeded in six-well culture plates (2 × 105 cells/well). Fluorescent dye Cy5.5 was encapsulated in the nanoparticles (NPs) to image cellular uptake. After pre-incubation for 24 h, cells were treated with HA-PEG-PLGA NPs and PEG-PLGA NPs, respectively. After 6 h, the cells were washed with PBS twice and cells of each well were collected and imaged using a FACSCanto A flow cytometer (BD Bioscience, USA). Fluorescent dye coumarin-6 was encapsulated in the NPs, and cellular uptake was monitored using a Leica TCS SP8 CARS confocal microscope (Leica, Germany) after 4-h incubation of untargeted PEG-PLGA NPs and targeted HA-PEG-PLGA NPs.
Proliferation Assay
Cells were seeded at 800 cells per well in 96-well plates and cultured until they reached ~10% confluence. Cells were then treated overnight with various concentrations of drug-loaded nanoparticles or controls. Cell proliferation was monitored using the Incucyte Zoom (Essen, MI, USA). Images were taken at regular time intervals over 6 days, and cell growth curves were plotted with the Incucyte Zoom software.
Cell Cycle G1/S Assessment Using Flow Cytometry
Cells were seeded onto six-well plates and cultured overnight until they reached 80% confluence. After a subsequent 24-h treatment with drug-loaded nanoparticles or the relevant control, the cells were treated with 300 μl of 1 mg/ml propidium iodide, 0.1% (v/v) Triton X-100, and 0.8% (v/v) RNAse A solution for 30 min at room temperature. Cells were washed with PBS and harvested for cell cycle measurements while protected from light. Single cells were identified by measurement of forward and side scatter. Then, doublets were removed using pulse processing. Measurements took place using a B670LP-A filter on a FACSCanto A flow cytometer.
Wound Scratch Assays
Cell migration was assessed using an established wound scratch assay (Fan et al., 2011). Cells were seeded at 64,000 cells per well in 96-well plates and incubated until they reached more than 80% confluence. The plates were placed in an Incucyte wound maker (Essen, MI, USA) to ensure consistency in the wounds made on the cell monolayers in each well. The medium and cell debris were then removed and replaced with 200 μl of fresh medium containing drug-loaded nanoparticles or controls. The plates were placed in the Incucyte Zoom (Essen, MI, USA), and the wound width was monitored over 24 h for later analysis using Incucyte Zoom software.
Cell Attachment
Cells were seeded onto six-well plates and left to attach overnight until over 80% confluence and then treated with drug-loaded nanoparticles or controls for 24 h. The cells were washed with PBS and then detached using versene. They were then seeded 5 × 104 cells per well onto gelatin-precoated 96-well plates for 2 h. After washing with PBS and fixation using 4% PFA for 15 min, the cells were imaged using the Incucyte Zoom (Essen, MI, USA) and the number of cells attached was quantified using Incucyte Zoom software.
Western Blotting
The expression of EMT markers was assessed by western blotting. Cells were seeded onto six-well plates and cultured overnight until they reached over 80% confluence. They were then treated with drug-loaded nanoparticles or controls for 24 h, washed with PBS, and harvested with versene. Ice-cold lysis buffer was added to each cell pellet for 15 min, and protein supernatants were obtained after spinning at 16,000G for 15 min at 4°C. The total protein concentration in each extract was measured using the bicinchoninic acid protein assay.
Protein samples were diluted using Milli-Q water to give a concentration of 50 μg protein/20 μl. Loading buffer (5 μl) was added to each sample, and samples were heated at 75°C for 10 min. Samples were loaded into wells of 15% acrylamide gels with a protein ladder and run at 100 V. The proteins were subsequently transferred onto nitrocellulose membranes at 100 V for 1 h, then blocked with 5 ml of blocking buffer at 4°C overnight. Blots were treated with a mouse anti-vimentin monoclonal antibody (1:1,000) or a mouse anti-E-cadherin monoclonal antibody (1:1,000) in blocking buffer for 1 h at room temperature on a shaker, while a rabbit anti-β-actin monoclonal antibody was used as a loading control (1:1,000). After washing with 1X TBST four times every 15 min for 1 h, the blots were incubated with goat anti-mouse polyclonal secondary antibodies (1:5,000) in the dark for 1 h. After washing for four times and drying, the membranes were scanned with the Odyssey CLx (LI-COR, NE, USA) imaging system and data were analyzed with Image Studio software.
Statistical Analysis
Multiple group comparisons were carried out using T-test, using GraphPad Prism version 7. This enabled significance against DMSO controls to be calculated. All results are shown as mean ± SEM of at least three replicates.
Result and Discussion
Nanoparticle Preparation and Drug Loading
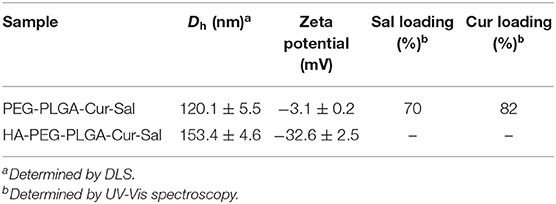
PLGA-PEG-NH2 co-polymer was validated via 1H NMR spectrum (Supplementary Figure 2). PLGA-PEG-NH2 co-polymer showed both the characteristic peak of PLGA (methyl group: 1.44–1.83 ppm and multiple peaks: 5.17–5.40 and 4.66–4.89 ppm) and the typical peak of PEG in which the methane proton is around 3.62 ppm (CH2-CH2-O) and validated the successful conjugation of PLGA-PEG-NH2. PLGA-PEG nanoparticles loaded with salinomycin and curcumin were prepared using the well-established double emulsion solvent evaporation method (Avgoustakis et al., 2002; You et al., 2018). HA was conjugated onto nanoparticles via EDC coupling and confirmed by hexadecyltrimethylammonium bromide (CTAB) turbidimetric method (Chen and Wang, 2009; Song et al., 2009). The hydrodynamic size of the nanoparticles increased from 120.1 ± 5.5 to 153.4 ± 4.6 nm after conjugation with HA, which was measured by dynamic light scattering (Table 1). Nanoparticle morphology was confirmed by TEM (Supplementary Figure 1). The surface charge also exhibited a negative zeta potential value after HA conjugation because of the negative carboxylic moiety of HA, which confirmed the conjugation of HA onto the particles. Approximately 70% of salinomycin and 82% of curcumin were loaded in polymeric nanoparticles, characterized by measuring UV-Vis spectra. Free salinomycin and curcumin molecules were separated from the solution and subsequently removed by centrifugation.
Drug Release Study
The kinetics of the drug release was evaluated over time upon their in vitro incubation in PBS buffer at 37°C, and the results are summarized in Figure 1. The higher hydrophilic moiety of PEG amount in co-polymer contributed to greater drug release. During the initial hours, there was a rapid release that led to a higher loading of salinomycin and curcumin in the vicinity of the surface and faster drug diffusion (Panagi et al., 2001; Wang et al., 2013). After 12 h, levels of salinomycin and curcumin were observed as a sustained curve, possibly due to the slow release of drugs within the polymeric matrix. Post-24 h, ~88% of salinomycin and 90% of curcumin were released from polymeric nanoparticles when pH was 7.4. When pH was reduced to 5.0, ~96% of salinomycin and 94% of curcumin were released. This data suggests both salinomycin and curcumin can be released from the polymeric nanoparticles under a similar condition and is expected when coming in contact with tumor tissues.
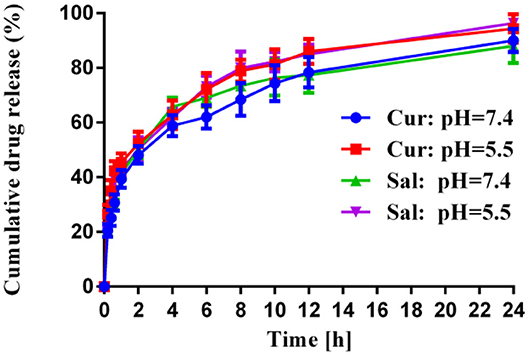
Figure 1. In vitro release profiles of Cur and Sal from HA-PEG-PLGA-Cur-Sal nanoparticles in sodium phosphate buffer at pH 5.0 and pH 7.4 at 37°C, respectively. Error bars show mean ± standard deviation from experiments performed in triplicate.
In vitro CD44-Mediated Cellular Uptake
Flow cytometry analysis was used to determine the internalization of HA-PEG-PLGA NPs by cancer cells and if this correlation is associated with the over-expression of CD44 receptor. BCSCs treated with HA-PEG-PLGA NPs showed a higher fluorescence intensity than BCSCs incubated with PEG-PLGA NPs after 6 h. This is due to a HA receptor that leads to cellular internalization via HA receptor-mediated endocytosis. As shown in Figure 2, the cellular association of HA-PEG-PLGA NPs was approximately three times greater than non-specific PEG-PLGA NPs at 37°C. This suggests HA-PEG-PLGA NPs could enter into the cell membrane and efficiently target cells by binding with CD44 that is overexpressed in BCSCs. HA targeting ability was also confirmed by confocal fluorescence micrograph of MCF-7 cells after 4-h incubation of untargeted PEG-PLGA NPs and targeted HA-PEG-PLGA NPs at the same concentration of coumarin-6. Significant fluorescence signal of coumarin-6 was observed in HA-PEG-PLGA NPs (Supplementary Figure 3).
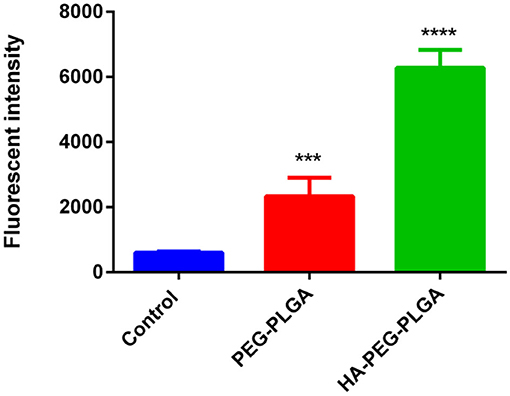
Figure 2. Cell uptake of HA-PEG-PLGA NPs and PEG-PLGA NPs by flow cytometry. The BCSCs were incubated with different formulations for 6 h. Data are mean ± SD (n = 3) (***p < 0.001, ****p < 0.0001).
In vitro Cell Proliferation and Viability
The in vitro cytotoxicity analysis was grouped as follows: HA-PEG-PLGA-Cur-Sal, PEG-PLGA-Cur-Sal, PEG-PLGA-Cur, PEG-PLGA-Sal, PEG-PLGA, Cur, and Sal. These compounds were evaluated against BCSCs by using cell proliferation assay with an incubation period of 144 h. The cytotoxicity data of BCSCs summarized in Figure 3A show the different formulations using a series of eight different concentrations of Sal and each group received the same corresponding Sal concentration. Cur was kept at a constant concentration. From Figure 3A, HA-PEG-PLGA-Cur-Sal, which has a specific affinity with over-expressed CD44 receptor, showed the greatest cytotoxicity as compared to the remaining formulations at the same drug concentration. Additionally, PEG-PLGA-Cur-Sal showed increased cytotoxicity compared to formulations containing either Cur or Sal. ED50 values were derived as described by Tallarida (2011), and synergism was observed in both HA-PEG-PLGA-Cur-Sal and PEG-PLGA-Cur-Sal that exhibited a super-additive effect.
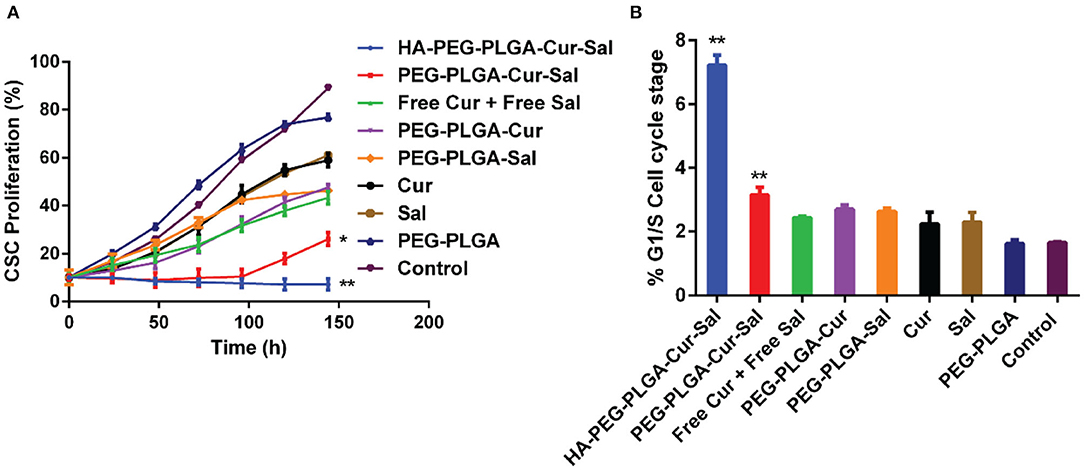
Figure 3. (A) Cell proliferation study of HA-PEG-PLGA-Cur-Sal, PEG-PLGA-Cur-Sal, PEG-PLGA-Cur, PEG-PLGA-Sal, PEG-PLGA, Cur, and Sal against CSCs. (B) Cell cycle arrest study of HA-PEG-PLGA-Cur-Sal, PEG-PLGA-Cur-Sal, PEG-PLGA-Cur, PEG-PLGA-Sal, PEG-PLGA, Cur, and Sal against CSCs. Values are means ± standard deviations (n = 5, S.D.) (*p < 0.05, **p < 0.01).
Cell Cycle Arrest Study
Chemotherapeutic drugs are highly toxic to the progression of cell cycle and thus can promote cellular death. To determine whether the drugs and their formulations cause cell cycle arrest, we investigate cell cycle progression by flow cytometry. The G1 phase of cell cycle is the initial cell cycle phase in which cells progress for subsequent duplication of itself. As shown in Figure 3B, both HA-PEG-PLGA-Cur-Sal and PEG-PLGA-Cur-Sal can significantly prolong the G1/S phase of the cell cycle, while HA-PEG-PLGA-Cur-Sal promoted G1/S arrest to a greater extent than the PEG-PLGA-Cur-Sal group. This suggests that HA-PEG-PLGA-Cur-Sal is the most effective in preventing BCSCs from subsequent progression into S-phase in which the genome duplication occurs. This indicates that co-delivery of salinomycin and curcumin causes extensive G1/S arrest, leading to the subsequent activation of apoptosis.
Cell Migration and EMT
BCSCs play a pivotal role in migration, which is essential for the pathogenesis of cancer during invasion and metastasis. Conversely, lack of cellular migration can lead to a failure in regenerative therapies. To validate cell migration in vitro, we adopted wound-healing assay which is widely used as a model to determine cell migration in vivo. As shown in Figures 4A,B, BCSCs treated with HA-PEG-PLGA-Cur-Sal showed significant reduction of cell migration in both wound length and rate of wound closure when compared to other treated groups. The lack of migration has a direct impact on cancer cell invasion and tumor metastasis. In Figure 4C, data indicated BCSCs that received HA-PEG-PLGA-Cur-Sal treatment had the lowest cellular confluency during cell attachment assay due to its reduced adhesion ability of BCSCs. Figure 4D shows that the immunoblotting of vimentin (mesenchymal marker) and E-cadherin (epithelial maker) confirmed HA-PEG-PLGA-Cur-Sal could reverse the properties of EMT, thereby limiting BCSCs to migrate.
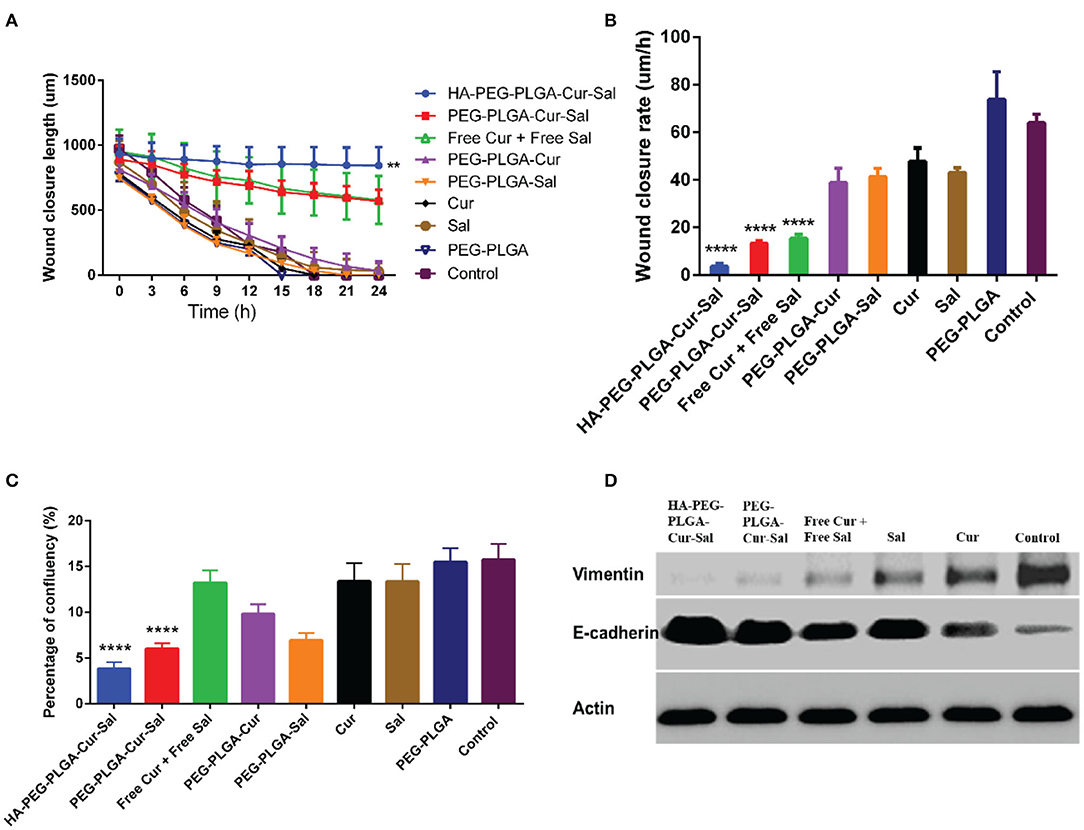
Figure 4. Cell migration behavior after treatment of PEG-PLGA-Cur, PEG-PLGA-Sal, PEG-PLGA, Cur, and Sal against CSCs using wound healing assay: (A) wound closure length, (B) wound closure rate, and (C) percentage of confluency of wound healing assay. (D) Expression of epithelial–mesenchymal transition markers of HA-PEG-PLGA-Cur-Sal, PEG-PLGA-Cur-Sal, free Cur + free Sal, Cur, and Sal. Values are means ± standard deviations (n = 5, S.D.) (**p < 0.01, ****p < 0.0001).
Conclusion
In summary, PLGA nanoparticle delivery system co-loaded with anticancer drug Sal and Cur was successfully synthesized and functionalized with HA as a targeting moiety. BCSCs showed higher sensitivity to this formulation with a combination of Sal and Cur by not only efficiently inducing cell death but also inhibiting cell migration and attachment. Our study revealed HA-PEG-PLGA-Cur-Sal could impede G1 phase cell cycle progression, leading to cell cycle arrest and subsequent induction of induced apoptosis of BCSCs. Collectively, our findings suggest that the combination HA-PEG-PLGA-Cur-Sal is an efficient therapeutic strategy against BCSCs and has the potential of treating tumor metastasis and improve patient outcome.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YZha: methodology and data interpretation and writing—original draft preparation. KW: methodology and data interpretation. YZhe: experiments and data collection. YL: methodology validation and review. TL: conceptualization, supervision, and manuscript editing. XZ: revision of manuscript. All authors contributed to the article and approved the submitted version.
Funding
TL was supported by the National Health and Medical Research Council (NHMRC) Early Career Fellowship (Grant No. 1112258) and the WSU Vice-Chancellor's Research Fellowship. YZha was supported by the National Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 19KJD350002) and the Jiangsu's Mass Entrepreneurship and Innovation Program. XZ was supported by the Natural Science Foundation of Nantong City (JC2018126).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.601649/full#supplementary-material
References
Alam, N., Koul, M., Mintoo, M. J., Khare, V., Gupta, R., Rawat, N., et al. (2017). Development and characterization of hyaluronic acid modified PLGA based nanoparticles for improved efficacy of cisplatin in solid tumor. Biomed. Pharmacother. 95, 856–864. doi: 10.1016/j.biopha.2017.08.108
Avgoustakis, K., Beletsi, A., Panagi, Z., Klepetsanis, P., Karydas, A. G., and Ithakissios, D. S. (2002). PLGA-mPEG nanoparticles of cisplatin: in vitro nanoparticle degradation, in vitro drug release and in vivo drug residence in blood properties. J. Control. Release 79, 123–135. doi: 10.1016/S0168-3659(01)00530-2
Baek, O.-S., Kang, O.-H., Choi, Y.-A., Choi, S.-C., Kim, T.-H., Nah, Y.-H., et al. (2003). Curcumin inhibits protease-activated receptor-2 and−4-mediated mast cell activation. Clin. Chim. Acta 338, 135–141. doi: 10.1016/j.cccn.2003.08.015
Basnet, P., and Skalko-Basnet, N. (2011). Curcumin: an anti-inflammatory molecule from a curry spice on the path to cancer treatment. Molecules 16, 4567–4598. doi: 10.3390/molecules16064567
Buhrmann, C., Yazdi, M., Dezfouli, A. B., Sahraneshin, F. S., and Shayan, P. (2020). Significant decrease in the viability and tumor stem cell marker expression in tumor cell lines treated with curcumin. J. Herb. Med. 22:100339. doi: 10.1016/j.hermed.2020.100339
Chen, Y.-H., and Wang, Q. (2009). Establishment of CTAB turbidimetric method to determine hyaluronic acid content in fermentation broth. Carbohydr. Polym. 78, 178–181. doi: 10.1016/j.carbpol.2009.04.037
Cho, K., Wang, X., Nie, S., Chen, Z., and Shin, D. M. (2008). Therapeutic nanoparticles for drug delivery in cancer. Clin. Cancer Res. 14, 1310–1316. doi: 10.1158/1078-0432.CCR-07-1441
Fan, F., Samuel, S., Gaur, P., Lu, J., Dallas, N. A., Xia, L., et al. (2011). Chronic exposure of colorectal cancer cells to bevacizumab promotes compensatory pathways that mediate tumour cell migration. Br. J. Cancer 104, 1270–1277. doi: 10.1038/bjc.2011.81
Ganesh, S., Iyer, A. K., Morrissey, D. V., and Amiji, M. M. (2013). Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials 34, 3489–3502. doi: 10.1016/j.biomaterials.2013.01.077
Gao, C., Tang, F., Gong, G., Zhang, J., and Wang, R. (2017). pH-Responsive prodrug nanoparticles based on a sodium alginate derivative for selective co-release of doxorubicin and curcumin into tumor cells. Nanoscale 9, 12533–12542. doi: 10.1039/C7NR03611F
Gupta, P. B., Onder, T. T., Jiang, G., Tao, K., Kuperwasser, C., Weinberg, R. A., et al. (2009). Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659. doi: 10.1016/j.cell.2009.06.034
Ketola, K., Hilvo, M., Hyötyläinen, T., Vuoristo, A., Ruskeepää, A. L., Orešič, M., et al. (2012). Salinomycin inhibits prostate cancer growth and migration via induction of oxidative stress. Br. J. Cancer 106, 99–106. doi: 10.1038/bjc.2011.530
Kotcherlakota, R., Barui, A. K., Prashar, S., Fajardo, M., Briones, D., Rodríguez-Diéguez, A., et al. (2016). Curcumin loaded mesoporous silica: an effective drug delivery system for cancer treatment. Biomater. Sci. 4, 448–459. doi: 10.1039/C5BM00552C
Lim, Y. C., Ensbey, K. S., Offenhäuser, C., D'Souza, R., Cullen, J. K., Stringer, B. W., et al. (2020). Simultaneous targeting of DNA replication and homologous recombination in glioblastoma with a polyether ionophore. Neuro Oncol. 22, 216–228. doi: 10.1093/neuonc/noz159
Lu, D., Choi, M. Y., Yu, J., Castro, J. E., Kipps, T. J., and Carson, D. A. (2011). Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 108, 13253–13257. doi: 10.1073/pnas.1110431108
Mai, T. T., Hamai, A., Hienzsch, A., Caneque, T., Muller, S., Wicinski, J., et al. (2017). Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 9, 1025–1033. doi: 10.1038/nchem.2778
Motevalli, S. M., Eltahan, A. S., Liu, L., Magrini, A., Rosato, N., Guo, W., et al. (2019). Co-encapsulation of curcumin and doxorubicin in albumin nanoparticles blocks the adaptive treatment tolerance of cancer cells. Biophys. Rep. 5, 19–30. doi: 10.1007/s41048-018-0079-6
Naujokat, C., and Steinhart, R. (2012). Salinomycin as a drug for targeting human cancer stem cells. J. Biomed. Biotechnol. 2012:950658. doi: 10.1155/2012/950658
Norris, L., Karmokar, A., Howells, L., Steward, W. P., Gescher, A., and Brown, K. (2013). The role of cancer stem cells in the anti-carcinogenicity of curcumin. Mol. Nutr. Food Res. 57, 1630–1637. doi: 10.1002/mnfr.201300120
Panagi, Z., Beletsi, A., Evangelatos, G., Livaniou, E., Ithakissios, D. S., and Avgoustakis, K. (2001). Effect of dose on the biodistribution and pharmacokinetics of PLGA and PLGA–mPEG nanoparticles. Int. J. Pharm. 221, 143–152. doi: 10.1016/S0378-5173(01)00676-7
Paulus, E. F., Kurz, M., Matter, H., and Vértesy, L. (1998). Solid-state and solution structure of the salinomycin–sodium complex: stabilization of different conformers for an ionophore in different environments. J. Am. Chem. Soc. 120, 8209–8221. doi: 10.1021/ja973607x
Sanfilippo, V., Caruso, V. C. L., Cucci, L. M., Inturri, R., Vaccaro, S., and Satriano, C. (2020). Hyaluronan-metal gold nanoparticle hybrids for targeted tumor cell therapy. Int. J. Mol. Sci. 21:3085. doi: 10.3390/ijms21093085
Shanmugam, M. K., Rane, G., Kanchi, M. M., Arfuso, F., Chinnathambi, A., Zayed, M. E., et al. (2015). The multifaceted role of curcumin in cancer prevention and treatment. Molecules 20, 2728–2769. doi: 10.3390/molecules20022728
Shao, N., Jia, H., Li, Y., and Li, J. (2017). Curcumin improves treatment outcome of takayasu arteritis patients by reducing TNF-α: a randomized placebo-controlled double-blind clinical trial. Immunol. Res. 65, 969–974. doi: 10.1007/s12026-017-8917-z
Sharma, R. A., Gescher, A. J., and Steward, W. P. (2005). Curcumin: the story so far. Eur. J. Cancer 41, 1955–1968. doi: 10.1016/j.ejca.2005.05.009
Song, J.-M., Im, J.-H., Kang, J.-H., and Kang, D.-J. (2009). A simple method for hyaluronic acid quantification in culture broth. Carbohydr. Polym. 78, 633–634. doi: 10.1016/j.carbpol.2009.04.033
Tallarida, R. J. (2011). Quantitative methods for assessing drug synergism. Genes Cancer 2, 1003–1008. doi: 10.1177/1947601912440575
Teiten, M. H., Dicato, M., and Diederich, M. (2014). Hybrid curcumin compounds: a new strategy for cancer treatment. Molecules 19, 20839–20863. doi: 10.3390/molecules191220839
Tsai, Y. M., Chien, C. F., Lin, L. C., and Tsai, T. H. (2011). Curcumin and its nano-formulation: the kinetics of tissue distribution and blood-brain barrier penetration. Int. J. Pharm. 416, 331–338. doi: 10.1016/j.ijpharm.2011.06.030
Wang, H., Jia, Y., Hu, W., Jiang, H., Zhang, J., and Zhang, L. (2013). Effect of preparation conditions on the size and encapsulation properties of mPEG-PLGA nanoparticles simultaneously loaded with vincristine sulfate and curcumin. Pharm. Dev. Technol. 18, 694–700. doi: 10.3109/10837450.2012.696267
Wang, Q., Wu, P., Ren, W., Xin, K., Yang, Y., Xie, C., et al. (2014). Comparative studies of salinomycin-loaded nanoparticles prepared by nanoprecipitation and single emulsion method. Nanoscale Res. Lett. 9:351. doi: 10.1186/1556-276X-9-351
You, L., Wang, J., Liu, T., Zhang, Y., Han, X., Wang, T., et al. (2018). Targeted brain delivery of rabies virus glycoprotein 29-modified deferoxamine-loaded nanoparticles reverses functional deficits in parkinsonian mice. ACS Nano 12, 4123–4139. doi: 10.1021/acsnano.7b08172
Yue, W., Hamai, A., Tonelli, G., Bauvy, C., Nicolas, V., Tharinger, H., et al. (2013). Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy 9, 714–729. doi: 10.4161/auto.23997
Zhao, Y., Zhao, W., Lim, Y. C., and Liu, T. (2019). Salinomycin-loaded gold nanoparticles for treating cancer stem cells by ferroptosis-induced cell death. Mol. Pharm. 16, 2532–2539. doi: 10.1021/acs.molpharmaceut.9b00132
Zhou, H., Beevers, C. S., and Huang, S. (2011). The targets of curcumin. Curr. Drug Targets 12, 332–347. doi: 10.2174/138945011794815356
Keywords: nanomedicine, polymeric nanoparticles, curcumin, salinomycin, cancer stem cells
Citation: Zhao Y, Wang K, Zheng Y, Zeng X, Lim YC and Liu T (2021) Co-delivery of Salinomycin and Curcumin for Cancer Stem Cell Treatment by Inhibition of Cell Proliferation, Cell Cycle Arrest, and Epithelial–Mesenchymal Transition. Front. Chem. 8:601649. doi: 10.3389/fchem.2020.601649
Received: 02 September 2020; Accepted: 09 November 2020;
Published: 15 January 2021.
Edited by:
Jie He, University of Connecticut, United StatesReviewed by:
Ruibing Wang, University of Macau, ChinaSeung Rim Hwang, Chosun University, South Korea
Copyright © 2021 Zhao, Wang, Zheng, Zeng, Lim and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianqing Liu, michelle.tianqing.liu@gmail.com
†These authors have contributed equally to this work
 Yongmei Zhao1†
Yongmei Zhao1†  Tianqing Liu
Tianqing Liu