Exosomes Mediate APP Dysregulation via APP-miR-185-5p Axis
- 1Department of Anesthesiology, Tongji Hospital Affiliated to Tongji University School of Medicine, Shanghai, China
- 2Center for Translational Neurodegeneration and Regenerative Therapy, Tongji Hospital Affiliated to Tongji University School of Medicine, Shanghai, China
- 3Center for Translational Neurodegeneration and Regenerative Therapy, Shanghai 10th People’s Hospital Affiliated to Tongji University School of Medicine, Shanghai, China
- 4Translational Research Institute of Brain and Brain-Like Intelligence, Shanghai Fourth People’s Hospital Affiliated to Tongji University School of Medicine, Shanghai, China
- 5Collaborative Innovation Center for Brain Science, Tongji University, Shanghai, China
APP misexpression plays a crucial role in triggering a complex pathological cascade, leading to Alzheimer’s disease (AD). But how the expression of APP is regulated in pathological conditions remains poorly understood. In this study, we found that the exosomes isolated from AD mouse brain promoted APP expression in neuronal N2a cells. Moreover, exosomes derived from N2a cells with ectopic expression of APP (APP-EXO) also induced APP dysregulation in normal N2a cells. Surprisingly, the effects of APP-EXO on APP expression in recipient cells were not mediated by the direct transferring of APP gene products. Instead, the effects of APP-EXO were highly likely mediated by the reduction of the expression levels of exosomal miR-185-5p. We found that the 3′UTR of APP transcripts binds to miR-185-5p, therefore inhibiting the sorting of miR-185-5p to exosomes. N2a cell-derived exosomes with less amount of miR-185-5p exert similar roles in APP expression to APP-EXO. Lastly, we demonstrated a significant decline of serum exosomal miR-185-5p in AD patients and AD mice, versus the corresponding controls. Together, our results demonstrate a novel mechanism in the exosome-dependent regulation of APP, implying exosomes and exosomal miRNAs as potential therapeutic targets and biomarkers for AD treatment and diagnosis, respectively.
Introduction
In recent decades, the neurobiology of aging has expanded greatly as a scientific discipline. The abnormal age-related changes in the central nervous system (CNS) have been considered as one key determinant in the pathogenesis of various neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and glaucoma (Jucker and Ingram, 1997; Kalia and Lang, 2015; Mancino et al., 2018). AD is the most common age-related neurodegenerative disease worldwide and the number one cause for dementia in the aged population (Danborg et al., 2014; Reitz and Mayeux, 2014). One key pathological characteristic of AD is the accumulation of insoluble forms of amyloid-β (Aβ) in plaques in extracellular spaces (Masters et al., 2015). In pathological conditions, amyloid precursor protein (APP) is proteolyzed by β- and γ-secretase, generating Aβ peptides of 40 or 42 amino acids (Aβ40 or Aβ42) (Selkoe, 2001). Both peptides, especially Aβ42, are hydrophobic and tend to fold in β-sheet structure which recruits other extracellular proteins to form senile plaques (Masters et al., 2015). The classical Aβ hypothesis of AD claims that the mutant or misexpression of APP generates excessive Aβ (Hardy and Selkoe, 2002), leading to neurotoxicity (O'Brien and Wong, 2011), neuroinflammation (Cai et al., 2014), and regeneration deficiency (Scopa et al., 2019). Due to the key roles of APP in the pathogenesis of AD, APP overexpression has been widely applied in the establishment of AD models in vivo (Webster et al., 2014; Gao et al., 2019) and in vitro (Atluri et al., 2019; Lin et al., 2019). Although APP has been considered as one of the most important risk genes for AD, it remains largely unknown how the expression of APP is regulated under pathological conditions.
Emerging evidence implicates exosomes as a key microenvironment element that contributes to the progression of AD. Exosomes are small bilipid layer-enclosed extracellular vesicles (30–200 nm) that are released by virtually all CNS cells (Xia et al., 2019b). Exosomes have been reported to participate in the spreading of Aβ in the brain (Dinkins et al., 2014). The inhibition of exosome release halts the propagation of tau, an AD-related protein whose phosphorylation and cleavage are induced by Aβ (Hernandez and Avila, 2008; Asai et al., 2015). The functions of exosomes are mediated by their contents such as miRNAs (Xia et al., 2019b). Mounting studies have demonstrated distinct profiles of exosomal miRNAs in various biological fluids collected from AD patients, compared with healthy donors (Liu et al., 2014; Lugli et al., 2015; Yang et al., 2018). Currently, multiple miRNAs including miR-455-3p, miR-193b-3p, and miR-31-5p have been reported to target APP 3′ untranslated region (UTR) and inhibit the expression of APP protein in vivo and in vitro (Liu et al., 2014; Kumar et al., 2019; Barros-Viegas et al., 2020; Kumar et al., 2021; Kumar and Reddy, 2021). Hence, aforementioned literatures suggest the importance to investigate the roles of exosomes, especially exosomal miRNAs, in the pathogenesis of AD. In this study, we firstly demonstrated that exosomes isolated from the AD mouse brains (AD-EXO) significantly elevated APP expression levels in recipient cells. We next collected exosomes from conditioned medium of N2a cells with APP overexpression (APP-N2A), an in vitro AD model, and observed that APP-N2A-derived exosomes (APP-EXO) also enhanced APP expression in recipient cells. Interestingly, the effects of exosomes on APP dysregulation were mediated by reducing the levels of exosomal miR-185-5p, an inhibitory microRNA (miRNA) for APP expression, instead of the direct transferring of APP gene products to recipient cells. We further demonstrated that APP in producer cells regulated the sorting of miR-185-5p to exosomes via the direct interaction of APP 3′UTR and miR-185-5p. At last, we observed a significant reduction of miR-185-5p levels in exosomes isolated from the AD patients and mice sera, compared with the corresponding controls. Together, our study discovered a novel mechanism for APP expression regulation via exosome-mediated delivery of miR-185-5p, indicating the important roles of exosomes and their contents in the pathogenesis of AD.
Materials and Methods
Culture of N2a Cell Line
N2a neuronal cells were purchased from the Chinese Academy of Sciences Cell Bank, and cultured in DMEM (Gibco) containing 10% FBS (Gibco) and 100 U/ml penicillin and streptomycin (ThermoFisher).
Mice
APP/PS1 mice and C57 mice (purchased from Shanghai Model Organisms Center) are housed and bred in the Comparative Medicine animal facilities of Tongji University School of Medicine (TUSM). All procedures were conducted according to protocols approved by the Institutional Animal Care and Use Committee of TUSM. Mouse genotype was validated by PCR.
Study Population
The design of the present study was approved by the ethics committee of Tongji Hospital affiliated to Tongji University School of Medicine (Shanghai, China), and the written informed consents were obtained from all participants. A total of three AD patients (males, mean age 67.3 ± 4.7) and three healthy donors (males, mean age 70.0 ± 7.1) were selected for this study. Peripheral blood was collected from each patient after fasting for 12 h. The serum was separated by centrifugation at 3,000 g for 10 min in room temperature, followed by centrifugation at 12,000 g for 5 min at 4°C.
Collection of Exosomes in Conditioned Medium, Brain Extracellular Spaces, and Serum
Exosomes were isolated from the culture medium of N2a cells as previously described (Ma et al., 2018). Briefly, 3.5 × 106 N2a cells were culture in 10 cm dish. 24 h before collecting supernatants, the culture medium was replaced with FBS free medium containing DMEM (Gibco) and 100 U/ml penicillin and streptomycin (ThermoFisher) only. After conditioned media collection, exosomes were isolated by sequential centrifugation: conditioned media was first centrifuged at 300 g for 10 min to eliminate flowing cells, at 3,000 g for 20 min to eliminate cellular debris, at 10,000 g for 30 min to eliminate intracellular organelles and then at 100,000 g for 2 h to precipitate exosomes. All steps of centrifugation were handled at 4°C. For brain exosome isolation, fresh mouse brains were dissected and treated with 20 units/ml papain (Worthington) in Hibernate E solution (5 ml/brain) for 30 min at 37°C. The brain tissue was gently homogenized in 10 ml cold Hibernate E solution. The brain homogenate was sequentially filtered through a 40 μm mesh filter (BD Biosciences) and a 0.2 μm syringe filter (Thermo Scientific). Exosomes in the filtrate were collected through gradient centrifugation as described above. Exosomes in the serum were isolated using ExoQuick Exosome Precipitation kit (System Biosciences). Exosomes were resuspended in PBS and stored in −80°C until required.
Nanoparticle Tracking Analysis
Nanoparticle Tracking Analysis (NTA) was carried out as previously described (Ma et al., 2019). Briefly, 1 ml of exosome suspension was used for NanoSight analysis. NTA was assessed on NanoSight NS300 system (Malvern Instruments, UK) with a sCMOS camera. The conditions of the measurements were set at 25°C, 1 cP viscosity, 25 s per capture frame, and 60 s measurement time. Three individual measurements were applied for measuring the sizes of exosomes.
Transmission Electron Microscopy
Transmission Electron Microscop (TEM) was carried out as previously described (Ma et al., 2019). Briefly, purified exosomes were negatively stained and then spread on the copper grids. The droplets of exosomes were removed with filter paper and air-dried at room temperature. Images were obtained using transmission electron microscopy (JEM-1230, JEOL Ltd.).
Human APP Full-Length/CDS/3′UTR Plasmids, miR-185-5p Mimics/Inhibitor, and Transfection
The plasmids for the ectopic expression of human APP full-length, APP CDS, and APP 3′UTR sequences were constructed by Generay (Generay Biotech Co., Ltd., Shanghai). Plasmid backbone pcDNA3.1 was purchased from Generay. APP transcript sequences were obtained from UCSC Genome Browser (http://genome.ucsc.edu/) and inserted into pcDNA3.1 using restriction enzymes NheI and NotI for genomic digestion. The mimics control, miR-185-5p mimics, inhibitor control, and miR-185-5p inhibitor were purchased from GenePharma (GenePharma Co., Ltd., Shanghai). Transfection was performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instruction.
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
The messenger RNA (mRNA) and miRNA were isolated from cell samples using RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA was removed and cDNA was synthesized using DNase I digestion kit (Qiagen) and miScript II RT kit (Qiagen), respectively. Transcripts were amplified using specific primer sets (Supplementary Table S1) and SYBR green PCR kit (Qiagen) with the ABI7500 (Applied Biosystems). Reactions were run in triplicates for each sample and no-template blanks were used as negative controls. Values were normalized to the Gapdh (for mRNA) and U6 snRNA (for miRNA).
Western Blotting
Western blotting was performed as previously described (Gao et al., 2019). Exosomes or cells were lysed in RIPA lysis and extraction buffer (ThermoFisher) containing a protease inhibitor cocktail (Sigma). Protein concentrations were determined with a BCA Protein Assay Kit (Pierce). Proteins (20–30 mg) from exosome and cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretic transferred to polyvinylidene fluoride membranes (Millipore and Bio-Rad). Membranes were incubated with primary antibodies for APP (rabbit, Abcam, 1:1,000), CD9 (rabbit, Abcam, 1:2000), Flottlin2 (mouse, BD Biosciences; 1:5,000), overnight at 4°C followed by a secondary anti-rabbit or anti-mouse antibody (Cell Signaling Technologies, 1:10,000) incubation. Antigen-antibody complexes were visualized by Pierce ECL Western Blotting Substrate (ThermoFisher). For data quantification, films were scanned with a CanonScan 9950F scanner; the acquired images were analyzed using ImageJ program.
Small RNA Sequencing and Bioinformatics Analysis
Total RNA was extracted from N2a cell-derived exosomes and 3 μg total RNA per sample was used as input material for the small RNA library. Sequencing libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS (Illumia). After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2,500/2000 platform and 50 bp single-end reads were generated. Raw data (raw reads) of fastq format were firstly processed through custom perl and python scripts for quality control. The small RNA tags were mapped to reference sequence by Bowtie without mismatch to analyze their expression and distribution on the reference. Mapped small RNA tags were used to looking for known miRNA. miRBase 20.0 was used as reference for known miRNA, miRDeep2 and sRNA-tools-cli were used to obtain novel miRNAs and draw the secondary structures, respectively. miRNA expression levels were estimated by TPM (transcript per million) and differentially expressed miRNAs between two groups were determined (p ≤ 0.05). The targets of differentially expressed miRNAs were predicted using TargetScan and miRanda databases according to the scoring criteria of each software. The target genes whose context score percentage is less than 50 or max energy is greater than −10 were removed in targetscan algorithm and Miranda algorithm, respectively. The intersection of predicted targets of both databases was taken as the final target genes of differential expressed miRNAs. Predicted targets of differentially expressed miRNAs were mapped to Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis. GO and KEGG enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.ncifcrf.gov/).
Dual-Luciferase Reporter Assay
Dual-luciferase reporter assay was performed as previously described (Xia et al., 2019a). The APP wild type 3′UTR (APP WT 3′UTR) and miR-185-5p binding site-mutated 3′UTR (APP MU 3′UTR) were synthesized by Genewiz (Genewiz, Suzhou, China) and cloned into the PmeI and SacI site of the pmirGLO vector (Promega, Beijing, China), downstream of the firefly luciferase gene. For the luciferase assay, 3 × 104 293T cells were cultured in 96-well plates with DMEM (Gibco) containing 10% FBS (Gibco) and 100 U/ml penicillin and streptomycin (ThermoFisher). Cells were co-transfected with miR-185-5p mimics and either APP WT 3′UTR or APP MU 3′UTR dual-luciferase reporter vector after reaching ∼70% confluency. Serum-free Opti-MEM (ThermoFisher) was used to prepare the transfection solution, and Lipofectamine 2000 reagent (Invitrogen) was used for transfection. 24 h post transfection, Dual-Luciferase® Reporter Assay System (Promega) was used to determine the luciferase activities on SpectraMax M5 microplate readers (Molecular Devices). The activity of Renilla luciferase was used to normalize that of firefly luciferase.
Statistical Analyses
All results are the means of at least three independent experiments ± s. d. Data from two groups were evaluated statistically by two-tailed, paired or unpaired student t test. Data from multiple groups were evaluated statistically by one-way ANOVA followed by Tukey’s post hoc test. Significance was considered when p < 0.05.
Results
Exosomes From AD Mouse Brains Induce APP Expression in Neuronal N2a Cells
To investigate the effects of exosomes on APP expression, we first isolated exosomes from cortical and hippocampal tissues of 7-month-old APP/PS1 mice (AD-EXO) and control C57 mice (Ctl-EXO). The purity of exosomes was examined by NTA and western blotting. NTA results suggested that the sizes of AD-EXO and Ctl-EXO were between 100 and 200 nm, matching with the typical size distribution of exosomes (Figure 1A). The morphology of exosomes was visualized via TEM, which showed distinct cup-shaped structure of exosomes (Figure 1B). Western blotting detected strong expression of exosome positive markers, Flotillin2, Flotillin1, CD9, TSG101, HSP70, and CD9, in both AD-EXO and Ctl-EXO (Figure 1C). Moreover, the negative markers of exosomes, Apoa1, Apob, and Calnexin, were absent in both AD-EXO and Ctl-EXO, further confirming the purity of exosomes (Figure 1D). To determine the effects of exosomes on APP expression, we co-cultured N2a cells with either 20 μg/ml AD-EXO or Ctl-EXO for 2 days. RT-qPCR analysis showed higher levels of APP transcripts in AD-EXO-treated group, compared with Ctl-EXO group (Figure 1E). Western blotting further revealed a significant increase of the expression levels of APP proteins in AD-EXO-treated cells versus Ctl-EXO group (Figure 1F). Thus, our results suggest an important role of exosomes from AD mouse brains in APP dysregulation in neuronal cells.
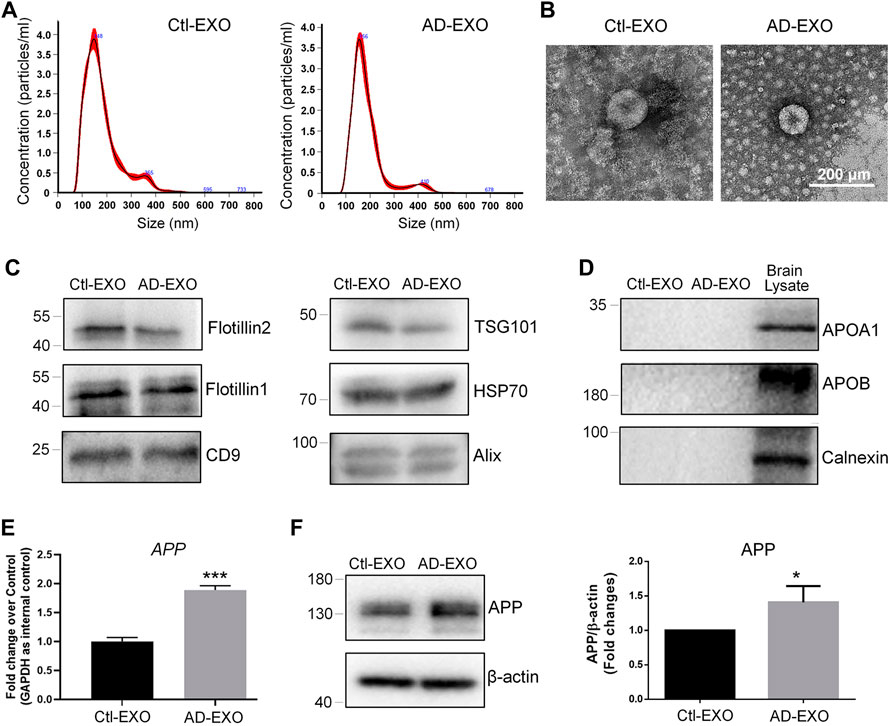
FIGURE 1. AD-EXO promote APP expression in N2a cells. (A) Particle-size distribution and concentration of exosomes were determined by NTA. (B) Ctl-EXO and AD-EXO were visualized under TEM using negative staining. (C) The expression levels of exosome positive markers Flotillin2, Flotillin1, CD9, TSG101, HSP70, and CD9 in protein lysates of exosomes were determined by western blotting. (D) The expression levels of exosome negative markers Apoa1, Apob, and Calnexin in protein lysates of exosomes were determined by western blotting. (E) The APP transcript levels in N2a cells co-cultured with either AD-EXO or Ctl-EXO were determined by RT-qPCR. (F) The APP protein levels in N2a cells co-cultured with either AD-EXO or Ctl-EXO were determined by western blotting. Data were represented as mean ± s. d. from three independent experiments. * and *** denote p < 0.05 and p < 0.001, respectively.
Exosomes Derived From N2a Cells With APP-Overexpression Promote APP Expression in Recipient Cells
To further confirm the exosome-mediated APP dysregulation and investigate the underlying mechanisms, we established an in vitro AD model by ectopically expressing full-length APP mRNAs in N2a cells to mimic the pathological conditions of AD in vivo. The overexpression of APP transcripts and APP proteins in N2a was validated by RT-qPCR (Figure 2A) and western blotting (Figure 2B), respectively. APP-overexpressed (APP OE) cell-derived exosomes (APP-EXO) or empty vector-transfected (control) cell-derived exosomes (EXO) were then collected, visualized, and validated via NTA, TEM, and western blotting assays (Figure 2C–F). To determine the effects of APP-EXO on APP expression, we co-cultured N2a cells with either 20 μg/ml APP-EXO or EXO for 2 days. Both RT-qPCR and western blotting results revealed a significant increase of the expression levels of APP transcripts (Figure 2G) and APP proteins (Figure 2H) in APP-EXO-treated cells versus EXO group, indicating the positive effects of APP-EXO on APP expression in neuronal cells.
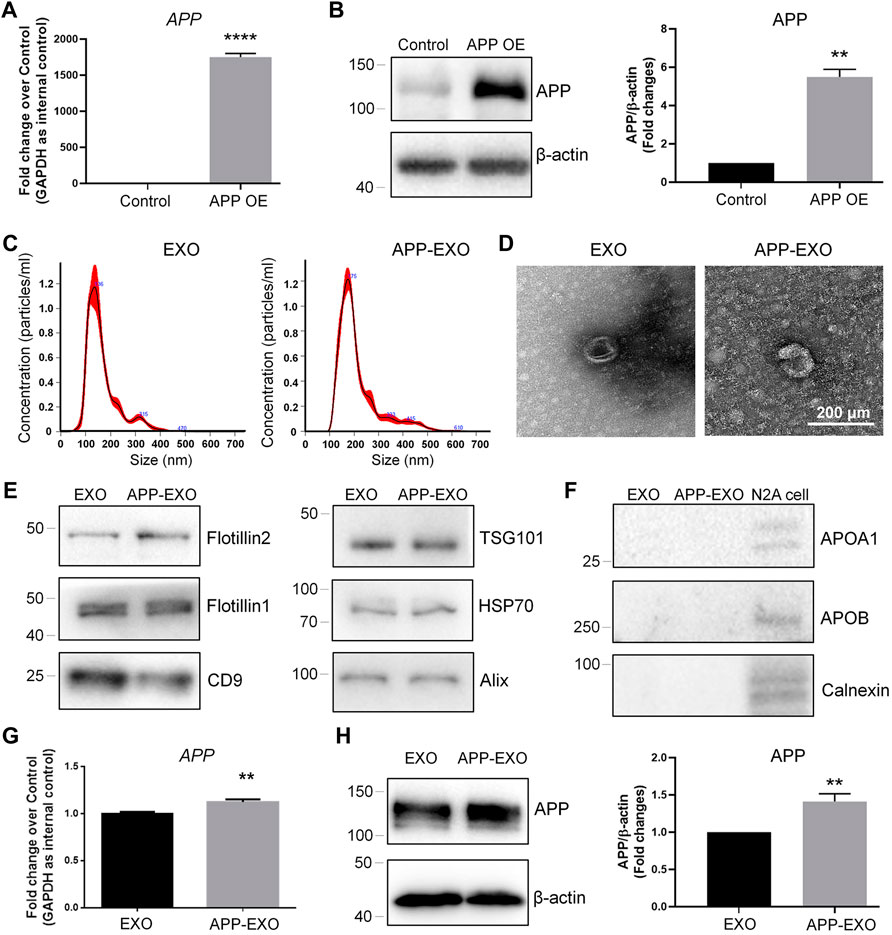
FIGURE 2. APP-EXO promote APP expression in recipient cells. (A,B) N2a cells were transfected with either control vector or APP OE plasmid for 2 days. The expression levels of APP transcripts and APP proteins were determined by RT-qPCR (A) and western blotting (B), respectively. (C) Particle-size distribution and concentration of exosomes were determined by NTA. (D) EXO and APP-EXO were visualized under TEM using negative staining. (E) The expression levels of exosome positive markers Flotillin2, Flotillin1, CD9, TSG101, HSP70, and CD9 in protein lysates of exosomes were determined by western blotting. (F) The expression levels of exosome negative markers Apoa1, Apob, and Calnexin in protein lysates of exosomes were determined by western blotting. (G) The APP transcript levels in N2a cells co-cultured with APP-EXO or EXO were determined by RT-qPCR. (H) The APP protein levels in N2a cells co-cultured either APP-EXO or EXO were determined by western blotting. Data were represented as mean ± s. d. from three independent experiments. ** and **** denote p < 0.01 and p < 0.0001, respectively.
Exosomes Promote APP Expression Independent to the Direct Transferring of APP Gene Products
We next investigated the possible mechanisms of exosome-induced APP dysregulation. The simplest and most obvious one is that exosomes may directly transfer APP transcripts and APP proteins to target cells (Perez-Gonzalez et al., 2012). To test this premise, we first compared the expression levels of APP transcripts and APP proteins in APP-EXO and EXO. Both RT-qPCR and western blotting results indicated that APP-EXO contains higher levels of APP transcripts (Figure 3A) and APP proteins (Figure 3B), compared with EXO. To further confirm our observations, we then transfected N2a cells with plasmid that overexpresses the coding DNA sequence (CDS) region of APP transcript. The empty (control) plasmid and plasmid containing the 3′ UTR of APP transcript were used as controls. Both RT-qPCR and western blotting results suggested a significant up-regulation of APP CDS and APP proteins in APP CDS-overexpressed (CDS OE) N2a cells, compared with APP 3′UTR-overexpressed (3′UTR OE) and control cells (Figures 3C,E). Moreover, APP 3′UTR levels were elevated only in 3′UTR OE group versus CDS OE and control groups (Figure 3D). Our observations suggested that CDS OE has similar promotional effects to APP OE on the expression of APP transcripts and proteins in N2a cells. Based on the results, we proposed that exosomes derived from CDS OE cells (CDS-EXO) should have also similar promotional effects to APP-EXO on the expression of APP transcripts and proteins in recipient cells if exosomes regulated APP expression through the direct transferring of APP gene products among cells.
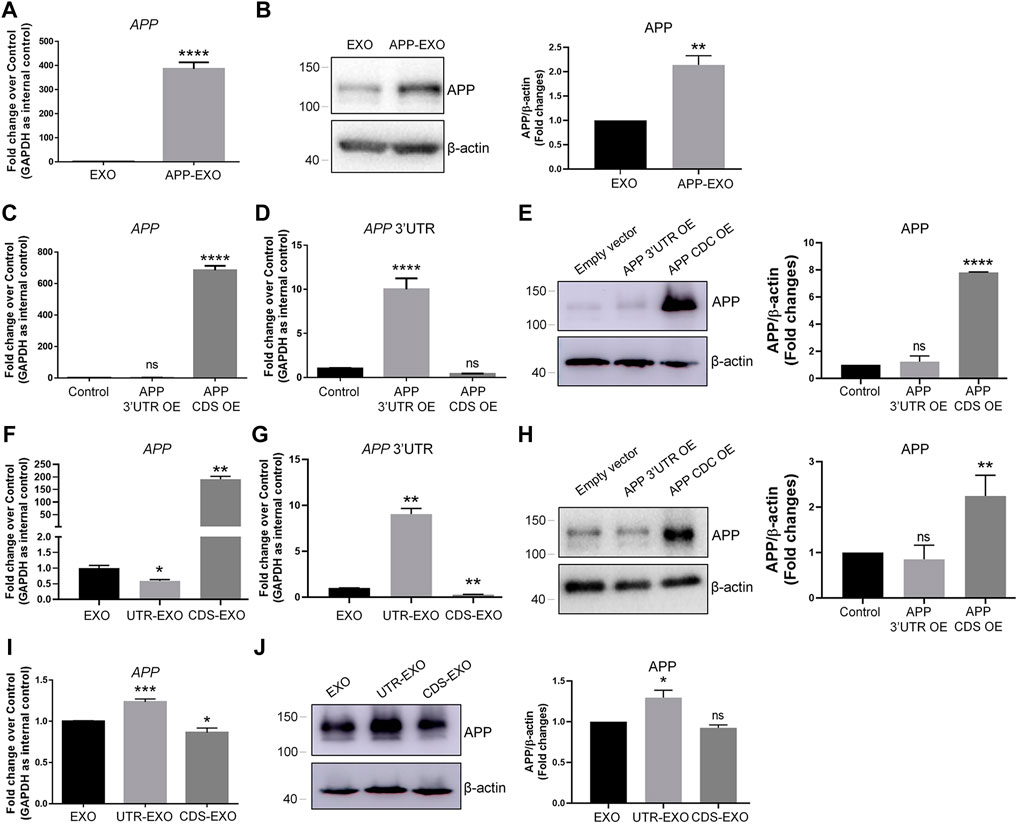
FIGURE 3. APP-EXO promote APP expression in recipient cells independent to the direct delivery of APP gene products. (A,B) The expression levels of APP transcripts and APP proteins in exosomes were determined by RT-qPCR (A) and western blotting (B), respectively. (C,D) N2a cells were transfected with empty control vector, APP 3′UTR OE plasmid, or APP CDS OE plasmid for 2 days. The expression levels of APP transcripts (C) and APP 3′UTR (D) were determined by RT-qPCR. (E) The APP protein levels in control, APP 3′UTR OE, and APP CDS OE groups were determined by western blotting. (F,G) The expression levels of APP transcripts (F) and APP 3′UTR (G) in EXO, UTR-EXO, and CDS-EXO were determined by RT-qPCR. (H,I) N2a cells were co-culture with EXO, UTR-EXO, or CDS-EXO for 2 days. The expression levels of APP transcripts (H) and APP proteins (I) were determined by RT-qPCR and western blotting, respectively. Data were represented as mean ± s. d. from three independent experiments. *, **, and **** denote p < 0.05, p < 0.01, and p < 0.0001, respectively. ns denotes no significance.
We then collected CDS-EXO and exosomes derived from 3′UTR OE cells (UTR-EXO). The sorting of APP gene products into exosomes was confirmed by RT-qPCR and western blotting. The results showed that APP CDS expression levels were significantly higher in CDS-EXO, but not in UTR-EXO, compared with EXO (Figure 3F). APP 3′UTR levels were significantly higher in UTR-EXO, compared with both CDS-EXO and EXO (Figure 3G). Moreover, APP protein levels were significantly elevated in CDS-EXO, but not in UTR-EXO, compared with EXO, confirming the enrichment of APP gene products in CDS-EXO (Figure 3H). Afterwards, we co-cultured N2a cells with either CDS-EXO, UTR-EXO, or EXO for 2 days. Surprisingly, both RT-qPCR and western blotting results showed that CDS-EXO failed to elevate the expression levels of APP transcripts and APP proteins, respectively (Figures 3I,J). In contrast, UTR-EXO significantly increased the expression of APP transcripts and APP proteins in co-cultured cells (Figures 3I,J). Therefore, our results suggested that APP-EXO regulated APP expression in recipient cells independent to the direct transferring of APP gene products.
Exosomes Derived From Neuronal Cells With APP-Overexpression Exhibit Distinct miRNA Profiles
To understand the mechanisms of UTR-EXO-mediated APP regulation, we first carried out small RNA sequencing to determine the miRNA signatures of APP-EXO and EXO, given the importance of mRNA 3′UTR-miRNA interaction in mediating the content sorting and functions of exosomes (Squadrito et al., 2014). 796 miRNAs were detected in either APP-EXO or EXO (Supplementary Table S2). 435 and 56 miRNAs were identified that were significantly up-regulated and down-regulated, respectively, in APP-EXO versus EXO (Figure 4A, Supplementary Table S3). The heatmap that represented the top 60 differentially expressed miRNAs also demonstrated the distinct profiles of exosomal miRNAs between two exosome populations (Figure 4B). GO analysis on biological process revealed that the predicted target genes of differentially expressed miRNAs were strongly associated with response to stimulus, cell differentiation, protein phosphorylation, and apoptotic process that might play roles in the pathogenesis of AD (Figure 4C). More importantly, KEGG analysis of the predicted target genes identified synaptic vesicle cycle, p53 pathway, Pyruvate pathway, glycolysis, and glycosphingolipid biosynthesis that were linked to AD (Figure 4D). Taken together, our study demonstrated distinct miRNA signatures in exosomes derived from N2a cells with and without APP dysregulation, implying an involvement of exosomal miRNAs in the regulation of AD-related pathogenic networks.
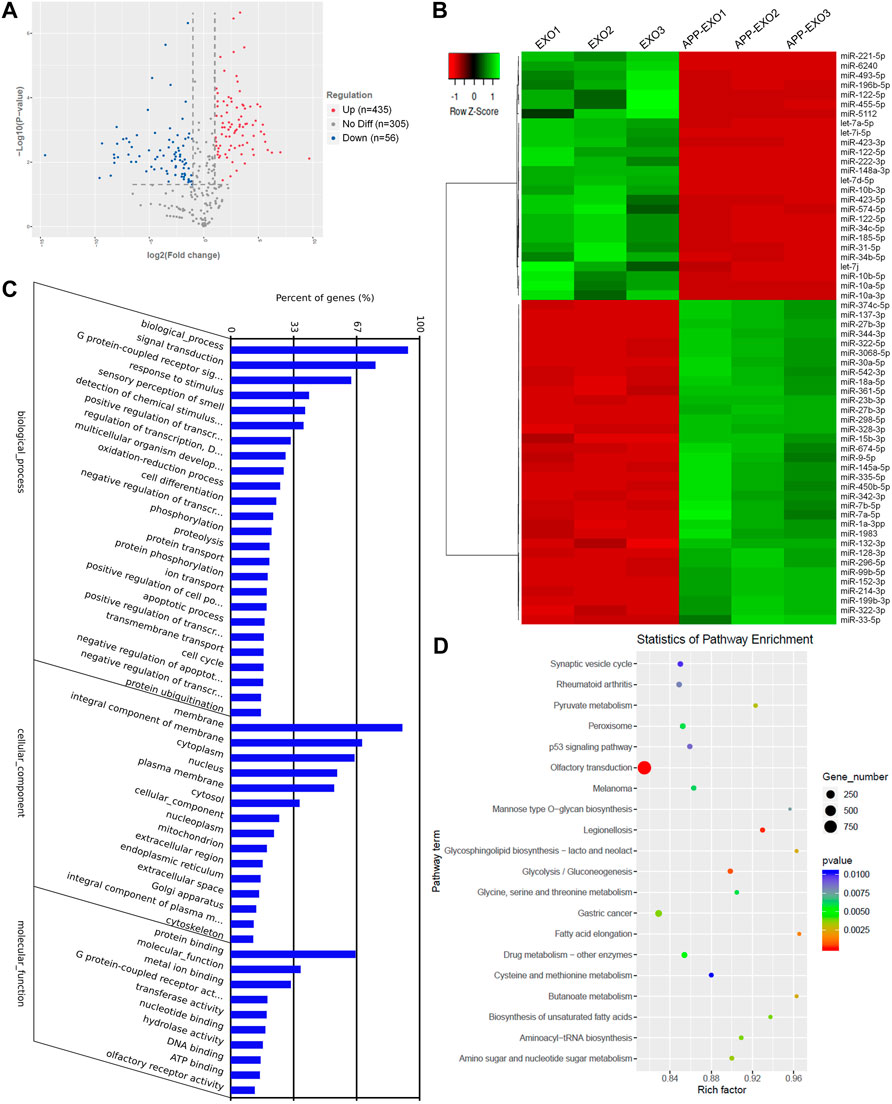
FIGURE 4. APP overexpression significantly alters exosomal miRNA profiles. (A) The volcano plot was generated based on the logarithm of the p-values and the log fold change of all detected miRNAs in APP-EXO and EXO. (B) The top 60 differentially expressed miRNAs between APP-EXO and EXO are represented in the heat map and hierarchical clustering-based dendrograms. (C) The enriched gene ontology (GO) terms in the predicted targets of differentially expressed miRNAs were determined by GO analysis. (D) The enriched signaling pathways in the predicted targets of differentially expressed miRNAs were determined by KEGG analysis.
APP 3′UTR Directly Binds to miR-185-5p and Inhibits miR-185-5p Sorting to Exosomes
To further identify the key miRNAs that mediate the effects of APP-EXO on APP expression in recipient cells, we identified the overlapping miRNAs (miR-185-5p, miR-31-5p, miR-144-3p, miR-369-3p, and miR-15b-5p) between the down-regulated miRNAs in APP-EXO and miRNAs that conservatively bind to APP 3′UTR (Figure 5A). Among these miRNAs in EXOs, miR-185-5p had the largest readcounts (Figure 5B). Furthermore, miR-31a-5p and miR-185-5p were the two with the largest fold change (Figure 5C). Hence, miR-185-5p was selected for following studies. The small RNA sequencing results were corroborated via RT-qPCR which demonstrated a significant decrease in the levels of miR-185-5p in APP-EXO versus EXO (Figure 5D). To make sure that our observations in vitro match with the situation in vivo, we examined the exosomal miR-185-5p levels in AD mouse brains via RT-qPCR and found a significant decline of miR-185-5p expression levels in AD-EXO compared with Ctl-EXO (Figure 5E).
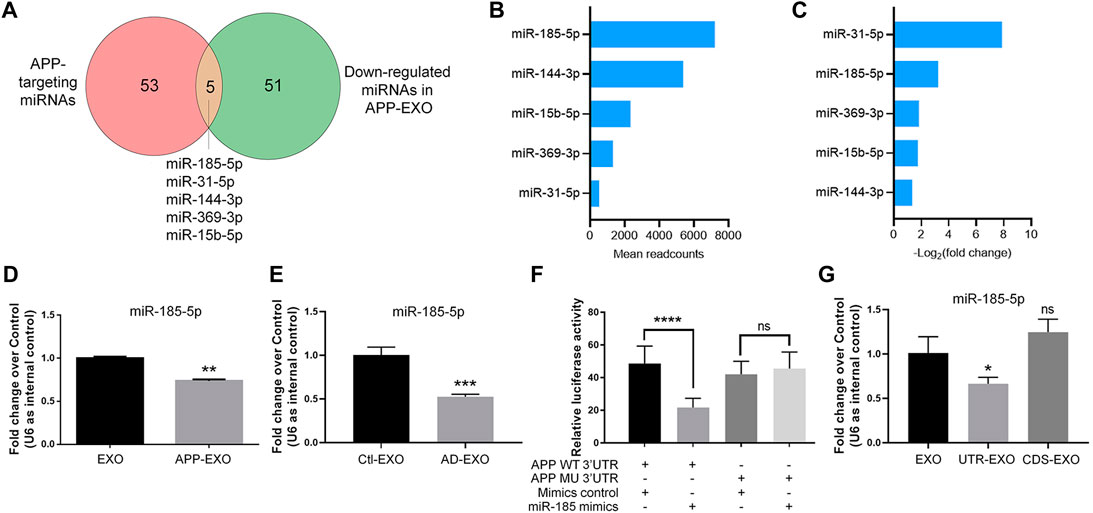
FIGURE 5. APP overexpression inhibits miR-185-5p sorting to exosomes via the direct interaction between APP 3′UTR and miR-185-5p. (A) The numbers of APP-targeting miRNAs and down-regulated miRNAs in APP-EXO were shown in the Venn diagram. The overlapping miRNAs were listed. (B) The readcounts of five overlapping miRNAs in EXO were ranked from more to less. (C) The fold changes of five overlapping miRNAs between APP-EXO and EXO were ranked from more to less. (D) The down-regulation of miR-185-5p in APP-EXO was determined by RT-qPCR. (E) The down-regulation of miR-185-5p in AD-EXO was determined by RT-qPCR. (F) Repression of luciferase activities by the APP 3′UTR were dependent on miR-185-5p. Firefly luciferase activities were normalized to the internal control, Renilla luciferase activities. (G) The down-regulation of miR-185-5p in UTR-EXO, but not CDS-EXO, was confirmed by RT-qPCR. Data were represented as mean ± s. d. from three independent experiments. *, **, ***, and **** denote p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively. ns denotes no significance.
Next, we carried out dual-luciferase assay to validate the direct binding of miR-185-5p with APP 3′UTR (Figure 5F). Co-transfection of miR-185-5p mimics and dual-luciferase reporter constructs containing the wild-type APP 3′UTR, but not miR-185-5p target site-mutated APP 3′UTR, significantly decreased the firefly activity in N2a cells, normalized by the Rellina activity, suggesting that miR-185-5p directly targets APP transcripts. We further tested that whether or not the sorting miR-185-5p in exosomes is regulated by APP 3′UTR. RT-qPCR results showed that miR-185-5p levels was significantly reduced in UTR-EXO, but not in CDS-EXO, compared with EXO, indicating APP 3′UTR but not CDS as the key element in regulating the sorting of miR-185-5p into exosomes (Figure 5G). Taken together, our study identified miR-185-5p as an EXO-enriched miRNA whose loading into exosomes is greatly influenced by endogenous APP 3′UTR.
Neuronal Exosomes With Low miR-185-5p Levels Promote APP Expression in Recipient Cells
To determine whether miR-185-5p mediates the effects of APP-EXO, we first examined the roles of miR-185-5p in regulating APP expression in neuronal cells via miR-185-5p loss-of-function (LOF) and gain-of-function (GOF) approaches. For miR-185-5p LOF, we transfected N2a cells with either miR-185-5p inhibitor (=LOF group) or inhibitor control for 2 days. RT-qPCR analysis showed a significant decrease in the levels of miR-185-5p in miR-185-5p LOF group, validating the efficiency of transfection (Figure 6A). A significant increase of APP transcript levels was observed in LOF group, compared with controls (Figure 6B). RT-qPCR results were confirmed by western blotting, which showed a significant increase of APP protein levels in the cell lysate of miR-185-5p LOF group versus that of control group (Figure 6C). For miR-185-5p GOF, N2a cells were transfected with either miR-185-5p mimics (=GOF group) or mimics control for 2 days. The efficiency of transfection is validated by RT-qPCR (Figure 6D). Moreover, both RT-qPCR and western blotting analyses demonstrated a significant decline of the expression levels of APP transcripts (Figure 6E) and APP protein (Figure 6F), respectively, in miR-185-5p GOF group, compared with control group. Thus, our results suggested that miR-185-5p negatively regulated APP expression in neuronal cells in vitro.
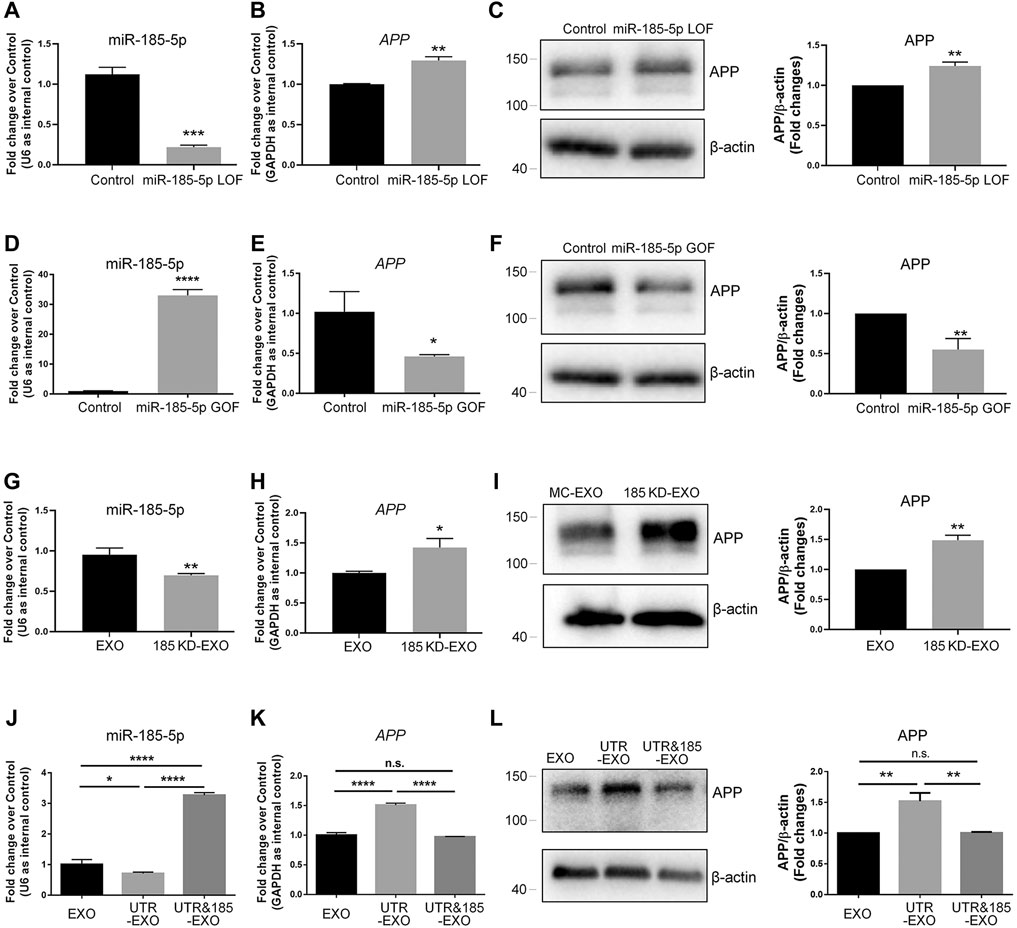
FIGURE 6. Exosomal miR-185-5p is important to inhibit APP expression in recipient cells. (A) N2a cells were transfected with either scrambled control or miR-185-5p inhibitor for 2 days. The expression levels of miR-185-5p were determined by RT-qPCR. (B) The up-regulation of APP transcript expression in miR-185-5p inhibitor-transfected N2a cells were determined by RT-qPCR. (C) The up-regulation of APP protein expression in miR-185-5p inhibitor-transfected N2A cells were determined by western blotting. (D) N2a cells were transfected with either scrambled control or miR-185-5p mimics for 2 days. The expression levels of miR-185-5p were determined by RT-qPCR. (E) The down-regulation of APP transcript expression in miR-185-5p mimics-transfected N2a cells were determined by RT-qPCR. (F) The down-regulation of APP protein expression in miR-185-5p mimics-transfected N2a cells were determined by western blotting. (G) Exosomes were collected from the conditioned medium of N2a cells transfected with either inhibitor control or miR-185-5p inhibitor. The expression levels of exosomal miR-185-5p were determined by RT-qPCR. (H,I) N2a cells were co-cultured with either EXO or 185 KD-EXO for 2 days. The expression levels of APP transcripts and APP protein in exosome-co-cultured N2a cells were determined by RT-qPCR (H) and western blotting (I). (J) Exosomes were collected from the conditioned medium of N2a cells co-transfected with either empty vector + mimics control, APP 3′UTR + mimics control, or APP 3′UTR + miR-185-5p mimics. The expression levels of exosomal miR-185-5p were determined by RT-qPCR. (K,L) N2a cells were co-cultured with either EXO, UTR-EXO, or UTR&185-EXO for 2 days. The expression levels of APP transcripts and APP protein in exosome-co-cultured N2a cells were determined by RT-qPCR (K) and western blotting (L). Data were represented as mean ± s. d. from three independent experiments. *, **, ***, and **** denote p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
Afterwards, we transfected N2a cells with miR-185-5p inhibitor to knockdown (KD) intracellular miR-185-5p and collected exosomes from the conditioned medium of miR-185-5p KD cells (185 KD-EXO). The KD of exosomal miR-185-5p was validated by RT-qPCR (Figure 6G). N2a cells were co-cultured with either EXO control or 185 KD-EXO for 2 days. RT-qPCR results showed a significant increase of APP transcripts in 185 KD-EXO treated cells versus controls (Figure 6H). The RT-qPCR data were further confirmed by western blotting (Figure 6I), indicating miR-185-5p as an essential content of exosomes for inhibiting APP expression in recipient cells.
Since the small RNA sequencing analysis revealed multiple miRNAs whose abundance in exosomes were regulated by APP 3′UTR, we next addressed that whether exosomal miR-185-5p mediates the promoting effects of UTR-EXO on APP expression. Except for EXO and UTR-EXO, we also collected exosomes from the conditioned medium of N2a cells co-transfected with APP 3′UTR and miR-185-5p mimics (UTR&185-EXO). RT-qPCR results revealed that the APP 3′UTR overexpression-induced decline of the levels of exosomal miR-185-5p was abrogated by miR-185-5p GOF in exosome-secreting cells (Figure 6J). N2a cells were then co-cultured with either EXO, UTR-EXO, or UTR&185-EXO for 2 days. RT-qPCR analysis found no significant difference of APP transcripts in UTR&185-EXO group, compared to control group, which indicated that the UTR-EXO-induced excessive expression of APP transcripts in recipient cells was rescued by the overexpression of exosomal miR-185-5p (Figure 6K). Western blotting results also displayed a similar APP protein expression pattern to that of RT-qPCR (Figure 6L). In conclusion, our findings suggest miR-185-5p as an important exosomal cargo in modulating APP expression in recipient cells, which may contribute to the dysregulation of APP metabolism in AD.
miR-185-5p Levels Are Down-Regulated in Exosomes Derived From AD Patients’ and Mouse Sera
As our in vitro observations indicated exosomal miR-185-5p as a key regulator of APP expression that may be involved in AD pathogenesis, we examined the potential translational value of exosomal miR-185-5p in AD diagnosis. We first compared the exosomal miR-185-5p levels in the sera collected from three AD patients and three age-/gender-matched healthy donors. RT-qPCR results suggested that serum exosomal miR-185-5p was significantly down-regulated in AD group versus controls (Figure 7A). We also collected sera from three AD mice and three age-/sex-matched C57 mice. Similarly, RT-qPCR results showed that the expression levels of serum exosomal miR-185-5p were significantly lower in AD mice, compared with controls (Figure 7B). Hence, our results implied a possibility of utilizing serum exosomal miR-185-5p in AD diagnosis, which requires extensive investigations including the introduction of large AD cohort and complicated statistical analyses.
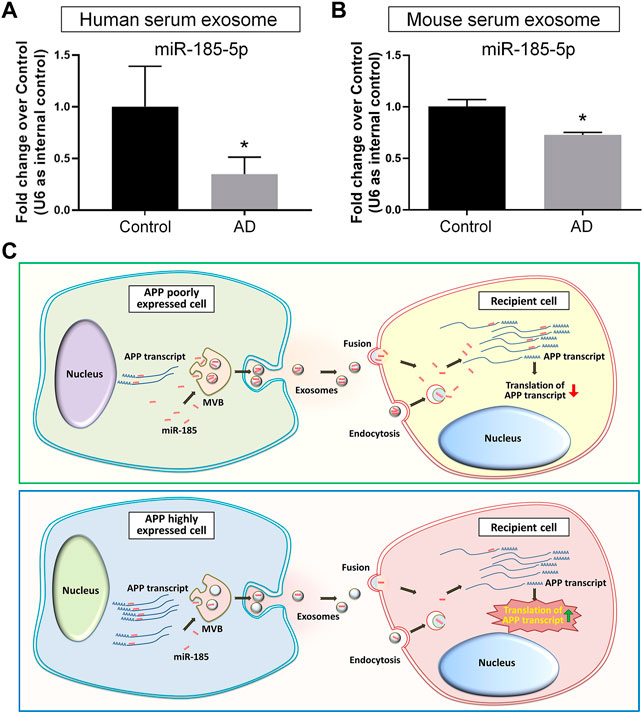
FIGURE 7. Serum exosomal miR-185-5p is lower in AD patients and mice than healthy controls. (A) Human serum exosomes were isolated from three AD patients and three healthy donors. The levels of miR-185-5p in exosomes were determined by RT-qPCR. (B) Mouse serum exosomes were isolated from three APP/PS1 mice and three control mice. The levels of miR-185-5p in exosomes were determined by RT-qPCR. (C) A schematic representation of exosome-mediated APP regulation. Upper panel: in normal conditions, the low levels of endogenous APP transcripts in normal neuronal cells result in high levels of free intracellular miR-185-5p. Consequently, more miR-185-5p can be loaded into exosomes and delivered to recipient cells via exosomes for maintaining APP expression at a low level in recipient cells. Lower panel: in pathological conditions, the expression levels of APP transcripts are abnormally elevated in neuronal cells. Excessive APP transcripts bind to more miR-185-5p, reducing free miR-185-5p intracellularly and leading to less miR-185-5p that can be loaded into exosomes. Exosomes with less miR-185-5p fail to suppress APP expression in recipient cells, causing uncontrolled APP expression. Data were represented as mean ± s. d. * denotes p < 0.05.
Discussion
With the rapid expansion of our knowledge, exosomes have now been considered as a key contributor in the pathogenesis of AD (Malm et al., 2016; Xia et al., 2019b). The inhibition of exosome release has obtained outstanding therapeutic effects including the suppression of glial activation, the reduction of total Aβ42 and plaque burden, the down-regulation of tau phosphorylation, and the enhancement of cognitive functions in animal studies (Dinkins et al., 2014; Dinkins et al., 2016). Since exosomes mediate various physiological processes, the shutdown of exosome biogenesis may also induce adverse reactions such as the impairment of anti-inflammatory responses and cell survival in the CNS (Chen et al., 2019; Yue et al., 2019). Therefore, more attentions have been paid on the identification of AD-related cargos in exosomes. For example, Aβ42 has been identified in neuronal exosomes isolated from AD patients’ peripheral blood and cerebrospinal fluid (Jia et al., 2019). Similarly, hypo-phosphorylated Tau protein can be detected in exosomes derived from primary cortical neurons (Wang et al., 2017). The transmission of exosomal cargos among neurons therefore acts as an essential channel for the spreading of Aβ and Tau in the brain (Jiang et al., 2019). Since current theories deem the formation of Aβ senile plaques and Tau neurofibrillary tangles as clinico-pathological features of AD instead of the causes of AD (Armstrong, 2013), the crosstalk between exosomes and AD-related genes including APP, presenilin (PSEN), and apolipoprotein E (ApoE) has been given with more and more emphasis (Perez-Gonzalez et al., 2012; Fernandes et al., 2018; Fyfe, 2019; Peng et al., 2019). In this study, we have demonstrated that exosomes derived from N2a cells with APP ectopic expression significantly increase the expression of APP in recipient cells. These exosomes achieve their functions highly likely through the delivery of miR-185-5p, instead of the direct transferring of APP gene products to recipient cells. Our study indicated that healthy neurons-derived exosomes, containing high levels of APP inhibiting factors including miR-185-5p, act as a cell-extrinsic sustainer for the inhibitory networks of APP expression. The dysregulation of APP expression confines miR-185-5p to APP 3′UTR, leading to the collapse of the inhibitory networks (Figure 7C).
To date, miR-185-5p has emerged as a key regulator of the pathogenesis of various diseases including tumor (Niu and Tang, 2019). miR-185-5p targets ROCK2 (Niu and Tang, 2019), ELK1 (Fan et al., 2018), ELK3 (Liu et al., 2020), IGF2 (Zhuang et al., 2020), RAGE (Yin et al., 2018), BCL2, BCL2L1 (Ostadrahimi et al., 2018), and many other genes to suppress tumor cell migration and invasion. miR-185-5p dysregulation has also been linked to renal fibrosis (Yuan et al., 2020), intervertebral disc degeneration (Guo et al., 2020), and other diseases (Li et al., 2019). A significant decrease in the levels of serum exosomal miR-185-5p in AD patients has been reported in a comparative study (Lugli et al., 2015). Here, we confirmed this pattern using samples collected from both AD patients and animal models. More importantly, our study is the first one showing the direct interaction between miR-185-5p and APP 3′UTR, which controls the sorting of miR-185-5p into exosomes. Our observations match with the findings of Squadrito et al. that endogenous mRNA down-regulates its targeting miRNAs sorting into exosomes via anchoring free miRNAs to its 3′UTR (Squadrito et al., 2014). Hence, we, in this study, demonstrated the tight association between the abnormal expression of AD-related genes and the shift of exosomal miRNA patterns in the patients’ biological fluids.
Mounting evidence has implicated exosomal miRNAs as biomarkers of AD. Lugli et al. found that plasma exosomal miR-185-5p, together with other six miRNAs, was sufficient to allow highly accurate prediction of AD in individual samples (Lugli et al., 2015). But whether plasma/serum exosomal miR-185-5p alone can be used as an AD biomarker remains unknown. In this study, although we identified a significant reduction in the levels of exosomal miR-185-5p in patients’ serum, the small sample size did not allow us to evaluate the sensitivity and specificity of exosomal miR-185-5p in AD diagnosis. With a larger sample size and more extensive investigations, the potential application of serum exosomal miR-185-5p in the diagnosis of AD may be a near possibility.
In summary, our study has demonstrated that the excessive expression of neuronal APP reduces exosomal miR-185-5p levels through the direct binding of APP 3′UTR to miR-185-5p, therefore releasing APP transcripts in the exosome-receiving cells from exosomal miR-185-5p-mediated translation inhibition. Thus, our study provides a possible mechanism for the intercellular regulation of APP expression via exosomes, implying exosomes and their miRNAs as potential therapeutic targets for AD treatment and biomarkers for AD diagnosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: ArrayExpress: The accession number: E-MTAB-11106.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Tongji Hospital affiliated to Tongji University School of Medicine. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Tongji University School of Medicine.
Author Contributions
XX, JL, and JZ conceptualized the project and designed the experiments. LD, XY, XX, YS, CL, GG, SZ, and SS performed the experiments. LD, XY, XX, YW, YH, and JL analyzed the data. XX prepared the manuscript, YL provided human sample resources. All authors read and approved the final manuscript.
Funding
This work was supported in part by research grants from the National Natural Science Foundation of China (No. 91949204 and No. 81830037 to JZ, No. 81971145 and No. 81901333 to XX, No. 81974155 to JL), Shanghai Pujiang Talent Program (No. 2019PJD049 and No. 82171194 to JL), Shanghai Sailing Program (No. 19YF1451700 to XX), Shanghai Blue Cross Brain Hospital Co., Ltd., and Shanghai Tongji University Education Development Foundation (No. 000000381/2018108 to JZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Yuju Li and Diksha Basnet for excellent technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.793388/full#supplementary-material
Abbreviations
185 KD-EXO, miR-185-5p-knockdown N2a cell-derived exosomes; AD, Alzheimer’s disease; abrAPP-N2A, N2a cells with APP overexpression; ApoE, Apolipoprotein E; APP-EXO, APP-overexpressed N2a cell-derived exosomes; Aβ, Amyloid-β; CDS: Coding DNA sequence; CDS-EXO, APP coding DNA sequence-overexpressed N2a cell-derived exosomes; CNS, Central nervous system; EXO, N2a cell-derived exosomes; GO, Gene ontology; GOF, Gain-of-function; KD, Knockdown; KEGG, Kyoto Encyclopedia of Genes and Genomes; LOF, Loss-of-function; MCI, Mild-cognitive impairment; miRNA, microRNA; mRNA, Messenger RNA; NTA: Nanoparticle tracking; OE, Overexpress; PD, Parkinson’s disease; PSEN, Presenilin; UTR, Untranslated region; UTR-EXO, APP 3′untranslated region-overexpressed N2a cell-derived exosomes.
References
Armstrong, R. A. (2013). Review Article what Causes Alzheimer's Disease? Folia Neuropathol. 3, 169–188. doi:10.5114/fn.2013.37702
Asai, H., Ikezu, S., Tsunoda, S., Medalla, M., Luebke, J., Haydar, T., et al. (2015). Depletion of Microglia and Inhibition of Exosome Synthesis Halt Tau Propagation. Nat. Neurosci. 18, 1584–1593. doi:10.1038/nn.4132
Atluri, V. S. R., Tiwari, S., Rodriguez, M., Kaushik, A., Yndart, A., Kolishetti, N., et al. (2019). Inhibition of Amyloid-Beta Production, Associated Neuroinflammation, and Histone Deacetylase 2-Mediated Epigenetic Modifications Prevent Neuropathology in Alzheimer's Disease In Vitro Model. Front. Aging Neurosci. 11, 342. doi:10.3389/fnagi.2019.00342
Barros-Viegas, A. T., Carmona, V., Ferreiro, E., Guedes, J., Cardoso, A. M., Cunha, P., et al. (2020). miRNA-31 Improves Cognition and Abolishes Amyloid-β Pathology by Targeting APP and BACE1 in an Animal Model of Alzheimer's Disease. Mol. Ther. Nucleic Acids 19, 1219–1236. doi:10.1016/j.omtn.2020.01.010
Cai, Z., Hussain, M. D., and Yan, L.-J. (2014). Microglia, Neuroinflammation, and Beta-Amyloid Protein in Alzheimer's Disease. Int. J. Neurosci. 124, 307–321. doi:10.3109/00207454.2013.833510
Chen, J., Zhou, R., Liang, Y., Fu, X., Wang, D., and Wang, C. (2019). Blockade of lncRNA‐ASLNCS5088-Enriched Exosome Generation in M2 Macrophages by GW4869 Dampens the Effect of M2 Macrophages on Orchestrating Fibroblast Activation. FASEB J. 33, 12200–12212. doi:10.1096/fj.201901610
Danborg, P. B., Simonsen, A. H., Waldemar, G., and Heegaard, N. H. H. (2014). The Potential of microRNAs as Biofluid Markers of Neurodegenerative Diseases - a Systematic Review. Biomarkers 19, 259–268. doi:10.3109/1354750x.2014.904001
Dinkins, M. B., Dasgupta, S., Wang, G., Zhu, G., and Bieberich, E. (2014). Exosome Reduction In Vivo Is Associated with Lower Amyloid Plaque Load in the 5XFAD Mouse Model of Alzheimer's Disease. Neurobiol. Aging 35, 1792–1800. doi:10.1016/j.neurobiolaging.2014.02.012
Dinkins, M. B., Enasko, J., Hernandez, C., Wang, G., Kong, J., Helwa, I., et al. (2016). Neutral Sphingomyelinase-2 Deficiency Ameliorates Alzheimer's Disease Pathology and Improves Cognition in the 5XFAD Mouse. J. Neurosci. 36, 8653–8667. doi:10.1523/jneurosci.1429-16.2016
Fan, H.-X., Feng, Y.-J., Zhao, X.-P., He, Y.-Z., and Tang, H. (2018). MiR-185-5p Suppresses HBV Gene Expression by Targeting ELK1 in Hepatoma Carcinoma Cells. Life Sci. 213, 9–17. doi:10.1016/j.lfs.2018.10.016
Fernandes, A., Ribeiro, A. R., Monteiro, M., Garcia, G., Vaz, A. R., and Brites, D. (2018). Secretome from SH-SY5Y APPSwe Cells Trigger Time-dependent CHME3 Microglia Activation Phenotypes, Ultimately Leading to miR-21 Exosome Shuttling. Biochimie 155, 67–82. doi:10.1016/j.biochi.2018.05.015
Fyfe, I. (2019). APOE*ε4 Links Exosomes to Cognitive Decline. Nat. Rev. Neurol. 15, 60. doi:10.1038/s41582-018-0123-7
Gao, G., Zhao, S., Xia, X., Li, C., Li, C., Ji, C., et al. (2019). Glutaminase C Regulates Microglial Activation and Pro-inflammatory Exosome Release: Relevance to the Pathogenesis of Alzheimer's Disease. Front. Cel. Neurosci. 13, 264. doi:10.3389/fncel.2019.00264
Guo, W., Zhang, B., Sun, C., Duan, H. Q., Liu, W. X., Mu, K., et al. (2020). Circular RNA Derived from TIMP2 Functions as a Competitive Endogenous RNA and Regulates Intervertebral Disc Degeneration by Targeting miR1855p and Matrix Metalloproteinase 2. Int. J. Mol. Med. 46, 621–632. doi:10.3892/ijmm.2020.4621
Hardy, J., and Selkoe, D. J. (2002). The Amyloid Hypothesis of Alzheimer's Disease: Progress and Problems on the Road to Therapeutics. Science 297, 353–356. doi:10.1126/science.1072994
Hernández, F., and Avila, J. (2008). The Role of Glycogen Synthase Kinase 3 in the Early Stages of Alzheimers' Disease. FEBS Lett. 582, 3848–3854. doi:10.1016/j.febslet.2008.10.026
Jia, L., Qiu, Q., Zhang, H., Chu, L., Du, Y., Zhang, J., et al. (2019). Concordance between the Assessment of Aβ42, T-Tau, and P-T181-Tau in Peripheral Blood Neuronal-Derived Exosomes and Cerebrospinal Fluid. Alzheimer's Demen. 15, 1071–1080. doi:10.1016/j.jalz.2019.05.002
Jiang, L., Dong, H., Cao, H., Ji, X., Luan, S., and Liu, J. (2019). Exosomes in Pathogenesis, Diagnosis, and Treatment of Alzheimer's Disease. Med. Sci. Monit. 25, 3329–3335. doi:10.12659/msm.914027
Jucker, M., and Ingram, D. K. (1997). Murine Models of Brain Aging and Age-Related Neurodegenerative Diseases. Behav. Brain Res. 85, 1–25. doi:10.1016/s0166-4328(96)02243-7
Kalia, L. V., and Lang, A. E. (2015). Parkinson's Disease. Lancet 386, 896–912. doi:10.1016/s0140-6736(14)61393-3
Kumar, S., and Reddy, P. H. (2021). Elevated Levels of MicroRNA-455-3p in the Cerebrospinal Fluid of Alzheimer's Patients: A Potential Biomarker for Alzheimer's Disease. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166052. doi:10.1016/j.bbadis.2020.166052
Kumar, S., Reddy, A. P., Yin, X., and Reddy, P. H. (2019). Novel MicroRNA-455-3p and its Protective Effects against Abnormal APP Processing and Amyloid Beta Toxicity in Alzheimer's Disease. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 2428–2440. doi:10.1016/j.bbadis.2019.06.006
Kumar, S., Morton, H., Sawant, N., Orlov, E., Bunquin, L. E., Pradeepkiran, J. A., et al. (2021). MicroRNA-455-3p Improves Synaptic, Cognitive Functions and Extends Lifespan: Relevance to Alzheimer's Disease. Redox Biol. 48, 102182. doi:10.1016/j.redox.2021.102182
Li, C.-C., Qiu, X.-T., Sun, Q., Zhou, J.-P., Yang, H.-J., Wu, W.-Z., et al. (2019). Endogenous Reduction of miR-185 Accelerates Cardiac Function Recovery in Mice Following Myocardial Infarction via Targeting of Cathepsin K. J. Cel Mol. Med. 23, 1164–1173. doi:10.1111/jcmm.14016
Lin, Y., Liang, X., Yao, Y., Xiao, H., Shi, Y., and Yang, J. (2019). Osthole Attenuates APP-Induced Alzheimer's Disease through Up-Regulating miRNA-101a-3p. Life Sci. 225, 117–131. doi:10.1016/j.lfs.2019.04.004
Liu, C.-G., Song, J., Zhang, Y.-Q., and Wang, P.-C. (2014). MicroRNA-193b Is a Regulator of Amyloid Precursor Protein in the Blood and Cerebrospinal Fluid Derived Exosomal microRNA-193b Is a Biomarker of Alzheimer's Disease. Mol. Med. Rep. 10, 2395–2400. doi:10.3892/mmr.2014.2484
Liu, J.-Q., Deng, M., Xue, N.-N., Li, T.-X., Guo, Y.-X., Gao, L., et al. (2020). lncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing miR-185-5p-Targeted KLF3 Inhibition. Mol. Ther. Nucleic Acids 20, 231–241. doi:10.1016/j.omtn.2020.01.020
Lugli, G., Cohen, A. M., Bennett, D. A., Shah, R. C., Fields, C. J., Hernandez, A. G., et al. (2015). Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS One 10, e0139233. doi:10.1371/journal.pone.0139233
Ma, Y., Wang, K., Pan, J., Fan, Z., Tian, C., Deng, X., et al. (2018). Induced Neural Progenitor Cells Abundantly Secrete Extracellular Vesicles and Promote the Proliferation of Neural Progenitors via Extracellular Signal-Regulated Kinase Pathways. Neurobiol. Dis. 124, 322–334. doi:10.1016/j.nbd.2018.12.003
Ma, Y., Li, C., Huang, Y., Wang, Y., Xia, X., and Zheng, J. C. (2019). Exosomes Released from Neural Progenitor Cells and Induced Neural Progenitor Cells Regulate Neurogenesis through miR-21a. Cell Commun. Signal 17, 96. doi:10.1186/s12964-019-0418-3
Malm, T., Loppi, S., and Kanninen, K. M. (2016). Exosomes in Alzheimer's Disease. Neurochem. Int. 97, 193–199. doi:10.1016/j.neuint.2016.04.011
Mancino, R., Martucci, A., Cesareo, M., Giannini, C., Corasaniti, M. T., Bagetta, G., et al. (2018). Glaucoma and Alzheimer Disease: One Age-Related Neurodegenerative Disease of the Brain. Curr. Neuropharmacol. 16, 971–977. doi:10.2174/1570159x16666171206144045
Masters, C. L., Bateman, R., Blennow, K., Rowe, C. C., Sperling, R. A., and Cummings, J. L. (2015). Alzheimer's Disease. Nat. Rev. Dis. Primers 1, 15056. doi:10.1038/nrdp.2015.56
Niu, Y., and Tang, G. (2019). miR-185-5p Targets ROCK2 and Inhibits Cell Migration and Invasion of Hepatocellular Carcinoma. Oncol. Lett. 17, 5087–5093. doi:10.3892/ol.2019.10144
O'Brien, R. J., and Wong, P. C. (2011). Amyloid Precursor Protein Processing and Alzheimer's Disease. Annu. Rev. Neurosci. 34, 185–204. doi:10.1146/annurev-neuro-061010-113613
Ostadrahimi, S., Abedi Valugerdi, M., Hassan, M., Haddad, G., Fayaz, S., Parvizhamidi, M., et al. (2018). miR-1266-5p and miR-185-5p Promote Cell Apoptosis in Human Prostate Cancer Cell Lines. Asian Pac. J. Cancer Prev. 19, 2305–2311. doi:10.22034/APJCP.2018.19.8.2305
Peng, K. Y., Pérez-González, R., Alldred, M. J., Goulbourne, C. N., Morales-Corraliza, J., Saito, M., et al. (2019). Apolipoprotein E4 Genotype Compromises Brain Exosome Production. Brain 142, 163–175. doi:10.1093/brain/awy289
Perez-Gonzalez, R., Gauthier, S. A., Kumar, A., and Levy, E. (2012). The Exosome Secretory Pathway Transports Amyloid Precursor Protein Carboxyl-Terminal Fragments from the Cell into the Brain Extracellular Space. J. Biol. Chem. 287, 43108–43115. doi:10.1074/jbc.m112.404467
Reitz, C., and Mayeux, R. (2014). Alzheimer Disease: Epidemiology, Diagnostic Criteria, Risk Factors and Biomarkers. Biochem. Pharmacol. 88, 640–651. doi:10.1016/j.bcp.2013.12.024
Scopa, C., Marrocco, F., Latina, V., Ruggeri, F., Corvaglia, V., La Regina, F., et al. (2019). Impaired Adult Neurogenesis Is an Early Event in Alzheimer's Disease Neurodegeneration, Mediated by Intracellular Abeta Oligomers. Cell Death Differ. 27, 934. doi:10.1038/s41418-019-0409-3
Selkoe, D. J. (2001). Alzheimer's Disease: Genes, Proteins, and Therapy. Physiol. Rev. 81, 741–766. doi:10.1152/physrev.2001.81.2.741
Squadrito, M. L., Baer, C., Burdet, F., Maderna, C., Gilfillan, G. D., Lyle, R., et al. (2014). Endogenous RNAs Modulate microRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cel Rep. 8, 1432–1446. doi:10.1016/j.celrep.2014.07.035
Wang, Y., Balaji, V., Kaniyappan, S., Krüger, L., Irsen, S., Tepper, K., et al. (2017). The Release and Trans-synaptic Transmission of Tau via Exosomes. Mol. Neurodegen. 12, 5. doi:10.1186/s13024-016-0143-y
Webster, S. J., Bachstetter, A. D., Nelson, P. T., Schmitt, F. A., and Van Eldik, L. J. (2014). Using Mice to Model Alzheimer's Dementia: an Overview of the Clinical Disease and the Preclinical Behavioral Changes in 10 Mouse Models. Front. Genet. 5, 88. doi:10.3389/fgene.2014.00088
Xia, X., Lu, H., Li, C., Huang, Y., Wang, Y., Yang, X., et al. (2019a). miR-106b Regulates the Proliferation and Differentiation of Neural Stem/progenitor Cells through Tp53inp1-Tp53-Cdkn1a axis. Stem Cel Res Ther 10, 282. doi:10.1186/s13287-019-1387-6
Xia, X., Wang, Y., Huang, Y., Zhang, H., Lu, H., and Zheng, J. C. (2019b). Exosomal miRNAs in central Nervous System Diseases: Biomarkers, Pathological Mediators, Protective Factors and Therapeutic Agents. Prog. Neurobiol. 183, 101694. doi:10.1016/j.pneurobio.2019.101694
Yang, T. T., Liu, C. G., Gao, S. C., Zhang, Y., and Wang, P. C. (2018). The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer's Disease Biomarkers. Biomed. Environ. Sci. 31, 87–96. doi:10.3967/bes2018.011
Yin, C., Zhang, G., Sun, R., Pan, X., Wang, X., Li, H., et al. (2018). miR1855p Inhibits Factin Polymerization and Reverses Epithelial Mesenchymal Transition of Human Breast Cancer Cells by Modulating RAGE. Mol. Med. Rep. 18, 2621–2630. doi:10.3892/mmr.2018.9294
Yuan, Q., Xu, T., Chen, Y., Qu, W., Sun, D., Liu, X., et al. (2020). MiR-185-5p Ameliorates Endoplasmic Reticulum Stress and Renal Fibrosis by Downregulation of ATF6. Lab. Invest. 100, 1436–1446. doi:10.1038/s41374-020-0447-y
Yue, K.-Y., Zhang, P.-R., Zheng, M.-H., Cao, X.-L., Cao, Y., Zhang, Y.-Z., et al. (2019). Neurons Can Upregulate Cav-1 to Increase Intake of Endothelial Cells-Derived Extracellular Vesicles that Attenuate Apoptosis via miR-1290. Cell Death Dis. 10, 869. doi:10.1038/s41419-019-2100-5
Keywords: exosome, miR-185-5p, APP, Alzheimer’s disease, brain, extracellular vesicle, microRNA, serum
Citation: Ding L, Yang X, Xia X, Li Y, Wang Y, Li C, Sun Y, Gao G, Zhao S, Sheng S, Liu J and Zheng JC (2022) Exosomes Mediate APP Dysregulation via APP-miR-185-5p Axis. Front. Cell Dev. Biol. 10:793388. doi: 10.3389/fcell.2022.793388
Received: 12 October 2021; Accepted: 18 January 2022;
Published: 11 February 2022.
Edited by:
Allan J. Guimaraes, Fluminense Federal University, BrazilReviewed by:
Richard Killick, King’s College London, United KingdomSubodh Kumar, Texas Tech University Health Sciences Center, United States
Copyright © 2022 Ding, Yang, Xia, Li, Wang, Li, Sun, Gao, Zhao, Sheng, Liu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohuan Xia, xiaohuan_xia1@163.com; Jianhui Liu, jianhuiliu_1246@163.com; Jialin C. Zheng, jialinzheng@tongji.edu.cn
†These authors have contributed equally to this work
 Lu Ding
Lu Ding Xiaoyu Yang
Xiaoyu Yang Xiaohuan Xia
Xiaohuan Xia Yunxia Li
Yunxia Li Yi Wang
Yi Wang Chunhong Li
Chunhong Li Yiyan Sun1
Yiyan Sun1  Ge Gao
Ge Gao Shu Zhao
Shu Zhao Jianhui Liu
Jianhui Liu Jialin C. Zheng
Jialin C. Zheng