Contribution of TEX15 genetic variants to the risk of developing severe non-obstructive oligozoospermia
- 1Departamento de Genética e Instituto de Biotecnología, Centro de Investigación Biomédica (CIBM), Universidad de Granada, Granada, Spain
- 2Instituto de Investigación Biosanitaria ibs.GRANADA, Granada, Spain
- 3Andrology Laboratory and Sperm Bank, IVIRMA Valencia, Valencia, Spain
- 4IVI Foundation, Health Research Institute La Fe, Valencia, Spain
- 5Servicio de Urología. Hospital Universitari i Politecnic La Fe e Instituto de Investigación Sanitaria La Fe (IIS La Fe), Valencia, Spain
- 6IVI-RMA Lisbon, Lisbon, Portugal
- 7Department of Obstetrics and Gynecology, Faculty of Medicine, University of Lisbon, Lisbon, Portugal
- 8Unidad de Reproducción, UGC Obstetricia y Ginecología, HU Virgen de Las Nieves, Granada, Spain
- 9CEIFER Biobanco—GAMETIA, Granada, Spain
- 10UGC de Urología, HU Virgen de las Nieves, Granada, Spain
- 11UGC de Obstetricia y Ginecología, Complejo Hospitalario de Jaén, Jaén, Spain
- 12Centro de Medicina Reprodutiva, Maternidade Alfredo da Costa, Centro Hospitalar Universitário de Lisboa Central, Lisboa, Portugal
- 13Departamento de Obstetrícia, Ginecologia e Medicina da Reprodução, Hospital de Santa Maria, Centro Hospitalar Universitário de Lisboa Norte, Lisboa, Portugal
- 14Laboratory of Seminology and Embryology, Andrology Service-Fundació Puigvert, Barcelona, Spain
- 15Departamento de Genética Humana, Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisbon, Portugal
- 16i3S—Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto, Portugal
- 17Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP), Porto, Portugal
- 18Serviço de Genética, Departamento de Patologia, Faculdade de Medicina da Universidade do Porto, Porto, Portugal
- 19ToxOmics—Centro de Toxicogenómica e Saúde Humana, Nova Medical School, Lisbon, Portugal
- 20CGPP-IBMC—Centro de Genética Preditiva e Preventiva, Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal
- 21Human Molecular Genetics Group, Bellvitge Biomedical Research Institute (IDIBELL), L’Hospitalet de Llobregat, Barcelona, Spain
- 22Departamento de Bioquímica y Biología Molecular I, Universidad de Granada, Granada, Spain
Background: Severe spermatogenic failure (SPGF) represents one of the most relevant causes of male infertility. This pathological condition can lead to extreme abnormalities in the seminal sperm count, such as severe oligozoospermia (SO) or non-obstructive azoospermia (NOA). Most cases of SPGF have an unknown aetiology, and it is known that this idiopathic form of male infertility represents a complex condition. In this study, we aimed to evaluate whether common genetic variation in TEX15, which encodes a key player in spermatogenesis, is involved in the susceptibility to idiopathic SPGF.
Materials and Methods: We designed a genetic association study comprising a total of 727 SPGF cases (including 527 NOA and 200 SO) and 1,058 unaffected men from the Iberian Peninsula. Following a tagging strategy, three tag single-nucleotide polymorphisms (SNPs) of TEX15 (rs1362912, rs323342, and rs323346) were selected for genotyping using TaqMan probes. Case-control association tests were then performed by logistic regression models. In silico analyses were also carried out to shed light into the putative functional implications of the studied variants.
Results: A significant increase in TEX15-rs1362912 minor allele frequency (MAF) was observed in the group of SO patients (MAF = 0.0842) compared to either the control cohort (MAF = 0.0468, OR = 1.90, p = 7.47E-03) or the NOA group (MAF = 0.0472, OR = 1.83, p = 1.23E-02). The genotype distribution of the SO population was also different from those of both control (p = 1.14E-02) and NOA groups (p = 4.33–02). The analysis of functional annotations of the human genome suggested that the effect of the SO-associated TEX15 variants is likely exerted by alteration of the binding affinity of crucial transcription factors for spermatogenesis.
Conclusion: Our results suggest that common variation in TEX15 is involved in the genetic predisposition to SO, thus supporting the notion of idiopathic SPGF as a complex trait.
Introduction
Infertility is a growing health concern involving over 50–70 million childbearing age couples worldwide, with the male factor contributing in approximately 50% of cases (Fainberg and Kashanian, 2019). Non-obstructive azoospermia (NOA) and severe oligozoospermia (SO) due to spermatogenic failure (SPGF) represent the most severe phenotypes of male factor infertility. However, the causes of such conditions are poorly understood and the aetiology of most affected men is usually defined as being idiopathic (Cannarella et al., 2019; Agarwal et al., 2021). Recent advances clearly point to idiopathic male infertility as a complex trait, in which the combined effect of polymorphic risk variants (causing subtle changes in gene expression) may increase the susceptibility of an individual to suffer from this disorder (Singh and Jaiswal, 2011; Cervan-Martin et al., 2020b; Salas-Huetos and Aston, 2021).
In this regard, common variants in the human genome may increase the susceptibility to develop male infertility by altering key events during spermatogenesis, in which DNA integrity is crucial (Neto et al., 2016). Having such in mind, it is important to stress that alterations in the DNA structure frequently occur in meiosis prophase I during the transition from primary to secondary spermatocytes, due to the generation of double-strand breaks (DSBs) for homologous chromosome pairing and crossing over (CO) (Cannarella et al., 2020). Consequently, DNA repair mechanisms are essential to produce fit mature sperm cells. In fact, the assessment of DNA integrity is a relevant marker to determine the sperm quality in assisted reproductive techniques (ART) (Leduc et al., 2008; Gonzalez-Marin et al., 2012). Thus, while the generation of genetic variability through changes in DNA is the basis for evolution, it is strictly controlled at the individual level (Capilla et al., 2016; Alves et al., 2017).
Consistent with the above, the spermatogenic process shows a very complex molecular and cellular control, involving over 2,000 genes, among which 900 are solely expressed in the male germline (Chalmel et al., 2012; Cervan-Martin et al., 2020b). The so-called Testis expressed (TEX) gene family plays a crucial role in spermatogenesis and some of its members have been strongly associated with male infertility (Cannarella et al., 2019; Bellil et al., 2021). Specifically, TEX15, which is predominantly expressed in spermatogonial and primary spermatocytes, encodes a 2,789 amino acid protein necessary for meiotic recombination and DSB repair in the germ line (Yang et al., 2008). Additionally, TEX15 also has a significant epigenetic function in spermatogenesis by interacting with several PIWI like RNA-mediated gene silencing proteins (namely PIWIL2, PIWIL4), in order to maintain spermatogonial stem cell integrity through the silencing of transposable elements (Schopp et al., 2020; Yang et al., 2020). Indeed, inactivation of Tex15 expression in mutant male mice causes sterility by meiotic arrest and impaired activity of RAD51 and DMC1 recombinases (Yang et al., 2008; Chen et al., 2016).
In this context, high-penetrance pathogenic variants in TEX15 have been widely linked to human male infertility due to spermatogenic impairment in different studies. For instance, non-sense mutations producing truncated TEX15 proteins have been reported to lead to cryptozoospermia (where spermatozoa are apparently absent in fresh semen samples but are recovered in centrifuged pellets) and SPGF in unrelated families (Okutman et al., 2015; Colombo et al., 2017; Wang et al., 2018; Cannarella et al., 2021; Tian et al., 2021). Moreover, mutations in this gene have been associated with NOA due to Sertoli cell-only (SCO) phenotype (characterised by a complete lack of germ cells in the seminiferous tubules) (Araujo et al., 2020).
On the other hand, case-control genetic association studies have attempted to assess the involvement of TEX15 polymorphic variation, such as single nucleotide polymorphisms (SNPs), in the development of SPGF resulting in NOA or SO. The first approach was conducted in a cohort of European ancestry and identified two TEX15 SNPs putatively associated with SPGF (Aston et al., 2010). However, such associations were not replicated in a subsequent study carried out in an independent South-Eastern European population (Plaseski et al., 2012). Similarly, two novel TEX15 variants have been recently proposed to be involved in the genetic predisposition to SPGF in the Iranian population (Ghadirkhomi et al., 2022). Finally, although TEX15 common variation was initially associated with SPGF risk in Han Chinese (Ruan et al., 2012), another report could not confirm this observation in this ethnicity (Zhang et al., 2015). Therefore, no conclusive results are currently available on the possible role of the common TEX15 genetic variation in SPGF development.
In view of the above, we aimed to clarify the involvement of TEX15 common variation in SPGF predisposition in a European genetic background, by analysing a large and phenotypically well-characterised study cohort.
Materials and methods
Patient cohort characterisation
This study was carried out in a large SPGF cohort with European descent. In total, the infertile men group comprised 727 SPGF cases from Spain and Portugal, diagnosed as NOA (n = 527), if a total absence of spermatozoa was observed in the ejaculate, or SO (n = 200), when showing less than 5 million spermatozoa/mL semen. The control set included 1,058 Iberian men, of whom 700 were healthy individuals representative of the general population (most of them with self-reported fatherhood), and 358 men with normal sperm counts, as previously described (Cervan-Martin et al., 2020a; Cervan-Martin et al., 2020c). Cases and controls were matched by age, geographical origin, and ethnicity, and signed an informed written consent in accordance with the Declaration of Helsinki. Each participating centre received ethical approval and complied with the requirements of their local regulatory authorities prior to the study.
SPGF patients were diagnosed after two high-speed centrifugations in two different semen samples in different fertility clinics managed in public and private health Hospitals and centres from Portugal and Spain, based on the guidelines for the management of infertile men by the American Urological Association (AUA)/American Society for Reproductive Medicine (ASRM), the Canadian Urological Association (CUA), and the World Health Organization (WHO) (Cooper et al., 2010; Jarvi et al., 2010; Bjorndahl and Kirkman Brown, 2022). Moreover, the NOA diagnosis was further confirmed by histological examination of testicular biopsies of the NOA patients that decided to undergo testicular sperm extraction (TESE) for use in ART (who represented around half of the NOA cohort). We established stringent selection criteria to include only infertile men due to idiopathic SPGF, which involved an exhaustive medical evaluation, screening for possible karyotype abnormalities and Y-chromosome microdeletions, as well as a thorough revision of the medical records to discard physical testicular disorders (e.g., orchitis and obstruction of vas deferens) and other known causes of male infertility. However, no screening for high-penetrance point mutations was conducted because this procedure is not part of the routine diagnostic workup of infertile men due to SPGF (Krausz et al., 2018).
Study design and single nucleotide polymorphism selection
A candidate gene study was conducted to shed light on the possible association of common genetic variation in the TEX15 locus with idiopathic SPGF risk. During the selection process, we confirmed that, according to publicly available single-cell RNA-seq data in the puberty and adult (Guo et al., 2018; Guo et al., 2020), the highest TEX15 expression was found in spermatogonia and spermatocytes (Supplementary Figure S1).
TEX15 is located in human chromosome 8, spanning an 81.5 kb region that constitutes a single linkage disequilibrium (LD) block in the 1,000 Genomes phase III (1KGPh3) European population (Auton et al., 2015), according to LDlink (Machiela and Chanock, 2015) (Figure 1A). Therefore, we followed a SNP tagging strategy using the European data of the 1KGPh3 and the Haploview V.4.2 software (Barrett, 2009; Auton et al., 2015) to cover most of the genetic variability of the region, prioritising those taggers that tagged, at least, more than five variants. Three taggers representative of three different minor allele frequency (MAF) ranges were selected with this method: rs1362912 (MAF <0.1), and rs323346 (0.1 < MAF <0.2), and rs323342 (MAF >0.2) (Supplementary Table S1).
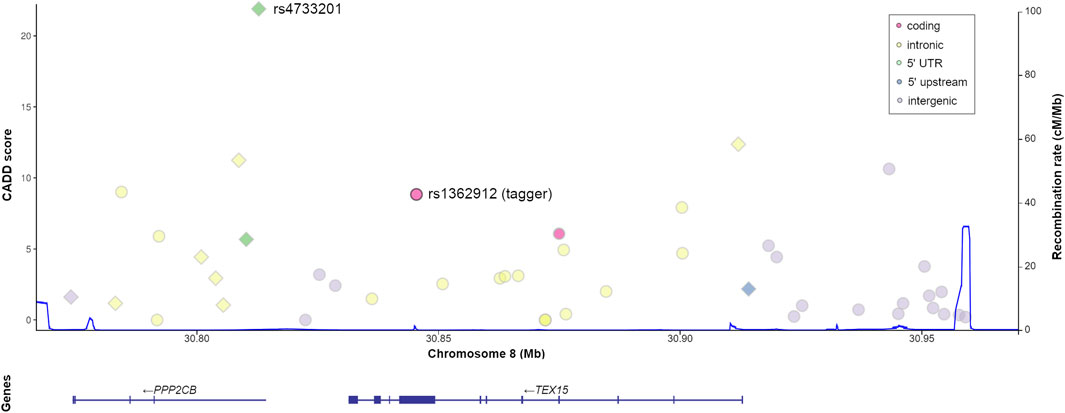
FIGURE 1. Genomic context and predicted functional relevance of the proxy variants tagged by TEX15-rs1362912. Both the CADD scores of each variant (left x-axis) and the recombination rate of the region (right x axis, blue line) are displayed. Variants with functional annotations in testis are represented as diamonds.
Sample preparation and genotyping
For all the recruited individuals, genomic DNA was extracted from peripheral blood mononuclear cells with the QIAamp® DNA Blood Midi/Maxi kit (Qiagen, Hilden, Germany), the MagNA Pure LC-DNA LV Isolation kit I (Roche, Basel, Switzerland), or the Wizard® Genomic DNA Purification Kit Protocol (Promega, Madison, WI, United States), following the manufacturers’ protocols. The genotyping was performed using the TaqMan™ SNP genotyping technology (Applied Biosystems, Foster City, CA, United States). Real-time quantitative polymerase chain reactions (qPCR) were performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, California, United States), using specific predesigned TaqMan™ probes (assay IDs: C___8867446_10; C____622151_10; C____622153_10) and the SDS 2.3 software for allele discrimination (both from Applied Biosystems, Foster City, California, United States).
Case-control statistical association analysis
An estimation of the statistical power of this study was calculated with the CaTS Power Calculator for Genetic Association Studies (Skol et al., 2006) (Supplementary Table S2). Possible deviances from Hardy-Weinberg equilibrium (HWE) were evaluated at the 5% significance level in both case and control groups using a χ2 test.
The statistical analyses were conducted with the software Plink v1.9 (Chang et al., 2015). Case-control comparisons of the allele and genotype frequencies were performed by logistic regression on the genotypes using geographical origin (Spain or Portugal) as a covariate and assuming different association models for genetic risk (including additive, dominant, recessive, and genotypic). p-values, odds ratios (ORs) and their 95% confidence intervals (CIs) were then calculated setting the significance threshold at p-value < 0.05 after correction of possible multiple testing effects by the Benjamini and Hochberg False Discovery Rate (FDR-BH) method (Benjamini et al., 2001).
In silico functional characterisation of risk variants
In an attempt to provide a plausible functional effect of the observed associations and identify possible molecular or cellular mechanisms underlying the pathogenic phenotypes, we implemented a bioinformatic workflow to extract and explore the functional annotation data available in different public databases. In a first step, we extended our functional characterisation to all proxies (genetic variants showing a LD r2 ≥ 0.8) of the selected taggers in the reference European population using the tools for that purpose implemented in LDLink (Machiela and Chanock, 2015). All proxies were equally considered candidates for explaining the observed associations, as described elsewhere (Cervan-Martin et al., 2020a; Cervan-Martin et al., 2021). Briefly, the prioritisation processes were performed with the following resources: GTExPortal (Carithers and Moore, 2015), SNPnexus (Oscanoa et al., 2020), ENCODE (Luo et al., 2020), Haploreg v.4.1. (Ward and Kellis, 2016), RegulomeDB (Dong and Boyle, 2019), SNP2TFBS (Kumar et al., 2017), amongst others. Several scores of deleteriousness were also used, such as CADD, DeepSEA, EIGEN, FATHMM, fitCons, FunSeq2 GWAVA, REMM, and RegulomeDB (Supplementary Tables S3,S4). In addition, we carried out an enrichment analysis of both gene ontology (GO) terms and protein-protein interactions (PPIs), including all transcription factors whose reported binding site (TFBS) sequences overlapped with the SNPs included in the prioritisation analysis, as implemented in STRINGv11.5 (Szklarczyk et al., 2019). Finally, to determine the TEX15 expression at the single cell level, we used the data available at both the Human Testis Atlas Browser (Guo et al., 2018) and the Single Cell Expression Atlas (Papatheodorou et al., 2020).
Results
The genotyping success rate of the three analysed variants was above 98% and none of them showed a significant deviation from HWE either in cases or controls. Additionally, the MAFs in the control group were concordant with those described for both the Iberian subpopulation and the European super population of the 1KGPh3 project (Auton et al., 2015), with no statistically significant differences observed between the population-representative group and the normozoospermic group in either the allele or the genotype frequencies. Furthermore, our study population had an appropriate overall statistical power to identify genetic associations with moderate to high effects, as detailed in Supplementary Table S2.
Common TEX15 gene variation confers susceptibility to severe oligozoospermia
First, we compared the allele and genotype frequencies of the selected taggers between the overall SPFG group and the unaffected control population. These analyses revealed no significant differences under any of the tested models (Table 1).
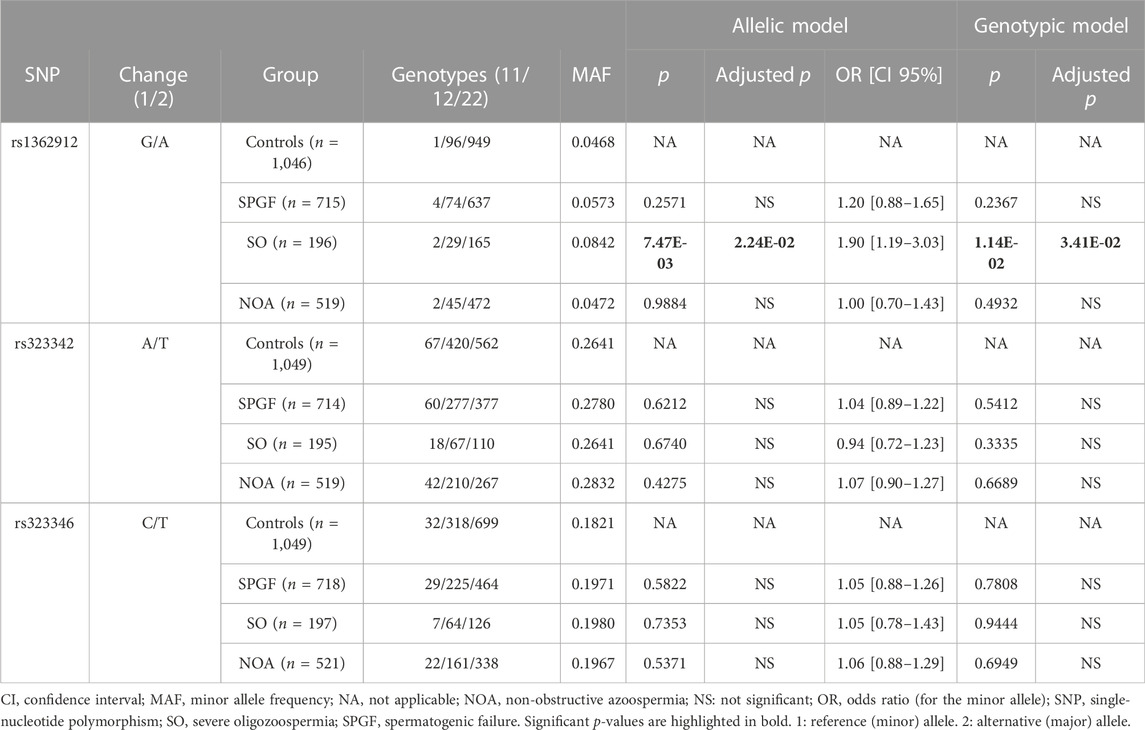
TABLE 1. Analysis of the genotype and allele frequencies of the TEX15 tagger variants comparing groups of male infertility against the unaffected control group.
Subsequently, we compared the groups of cases showing specific phenotypes of male infertility stablished by semen analysis (that is, NOA and SO) against the control cohort. Regarding NOA, no statistically significant differences were observed for any of the three tested taggers. In fact, the MAFs for the NOA group were very similar to those observed for the control group (Table 1).
Conversely, the comparison between the SO group and the control cohort revealed statistically significant differences in the allele/genotype frequencies of rs1362912 under the additive (pADD = 7.47E-03, OR = 1.90, CI 95% = 1.19–3.03) and genotypic (pGENO = 1.14E-02) models (Table 1). Such associations remained significant after multiple testing correction (pADD-FDR = 2.24E-02; pGENO-FDR = 3.41E-02) (Table 1).
To assess the consistency of the rs1362912 association with SO, we decided to eliminate the possible confounding effect of having SPFG in the analysis by using the NOA group as the reference cohort. The comparison between SO and NOA showed a similar risk effect on SO for the rs1362912*G minor allele in the additive model (pADD = 1.23E-02, 1.83, CI 95% = 1.14–2.94). The observed differences in the genotypic test for this SNP were also at the same range (pGENO = 4.33E-02) (Table 2). Interestingly, despite the lower statistical power of this case-case comparison, the association assuming an additive effect of rs1362912*G rameined after multiple testing correction (pADD-FDR = 3.69E-02) (Table 2).
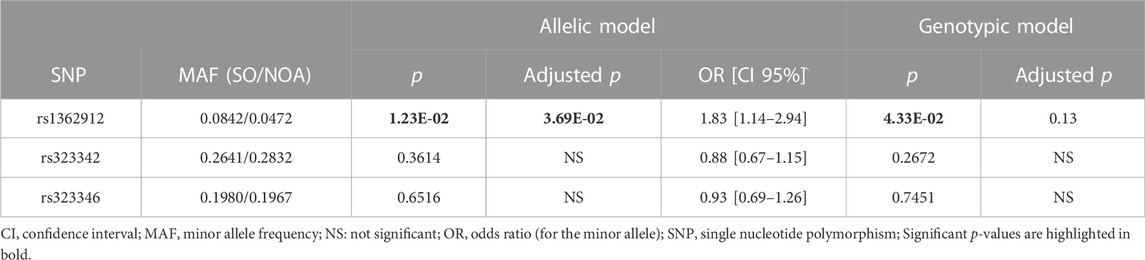
TABLE 2. Analysis of the genotype and allele frequencies of the TEX15 tagger variants comparing the severe oligozoospermia (SO) group against the non-obstructive azoospermia (NOA) group.
No evidence of additional association with the SO group was evident for the remaining selected variants (i.e., rs323346 and rs323342) (Tables 1, 2).
The severe oligozoospermia-associated variants in TEX15 might affect chromatin activity in the testis
The statistical analyses suggested an involvement of low frequency variants, tagged by rs1362912, in the susceptibility to SO. However, our study design relied on a tagging strategy, and the studied SNPs were selected based on their representativeness of different MAFs rather than on their possible functional features. Therefore, to investigate the biological meaning of the observed association between rs1362912 and SO further, we first identified all proxies (r2 ≥ 0.8) of this tagger and, subsequently, we carried out a functional prioritisation to elucidate the putative causal variant/s.
The tagger rs1362912 represents a synonymous SNP in exon eight of the TEX15 gene (Figure 1), whereas most of its proxies are non-coding variants located in both intronic and intergenic regions (Figure 1; Supplementary Table S5). We decided to base our prioritisation mostly on the overlaps with possible regulatory elements in the testis related with changes in gene expression that could affect the spermatogenic process. Amongst the 44 SNPs that were evaluated, the variants rs114435820, rs4733201, and rs1381559038 stood out from the rest using such criterion (Figure 2; Supplementary Table S5). Specifically, the SNP rs114435820 was located in a CpG island, which may indicate a possible effect on nearby gene expression. Interestingly, this proxy overlaps with a CTCF binding site, as well as with histone marks to active enhancers (H3K27ac) and promoters (H3K4me3) in the adult testis, according to ChIP- seq data from ENCODE (Davis et al., 2018) (Figure 2; Supplementary Table S5). Additionally, the position weight matrix (PWM) data extracted from HaploReg (Ward and Kellis, 2016) showed that the SNP rs114435820 could alter the binding affinity of the transcription factor YY1, which plays an important role in DNA repair during spermatogenesis (Wu et al., 2009), amongst others (Figure 2; Supplementary Tables S5, S6). The data of the testis from ENCODE also showed an overlap of the variant rs4733201 with binding sites of CTCF and POLR2A, with the same previously described histone marks, and with a DNAse hypersensitive sites (Figure 2; Supplementary Table S5). Furthermore, this variant was predicted to alter the TFBS of E2F1, which has been previously associated with male infertility due to SPGF (Jorgez et al., 2015; Rocca et al., 2019). Moreover, 31 additional proteins were observed to be bound to this site through ChIP-Seq experiments, being some of them (such as CTCF, YY1, EGR1, TBP, or CCNT2) involved in spermatogenesis (Tourtellotte et al., 2000; Wu et al., 2009; Teng et al., 2011; Hernandez-Hernandez et al., 2016; Ponomarenko et al., 2020) (Figure 2; Supplementary Tables S5, S6). With regards to the scores indicative of functionality, although some proxies (such as rs114433201) showed relevant values, the strongest evidence of deleteriousness was observed for the TEX15 variant rs4733201, which is located in the 5’ untranslated region of the nearby gene PPP2CB and had a considerably high CADD score (score = 21.9) (Figures 1, 2; Supplementary Table S5).
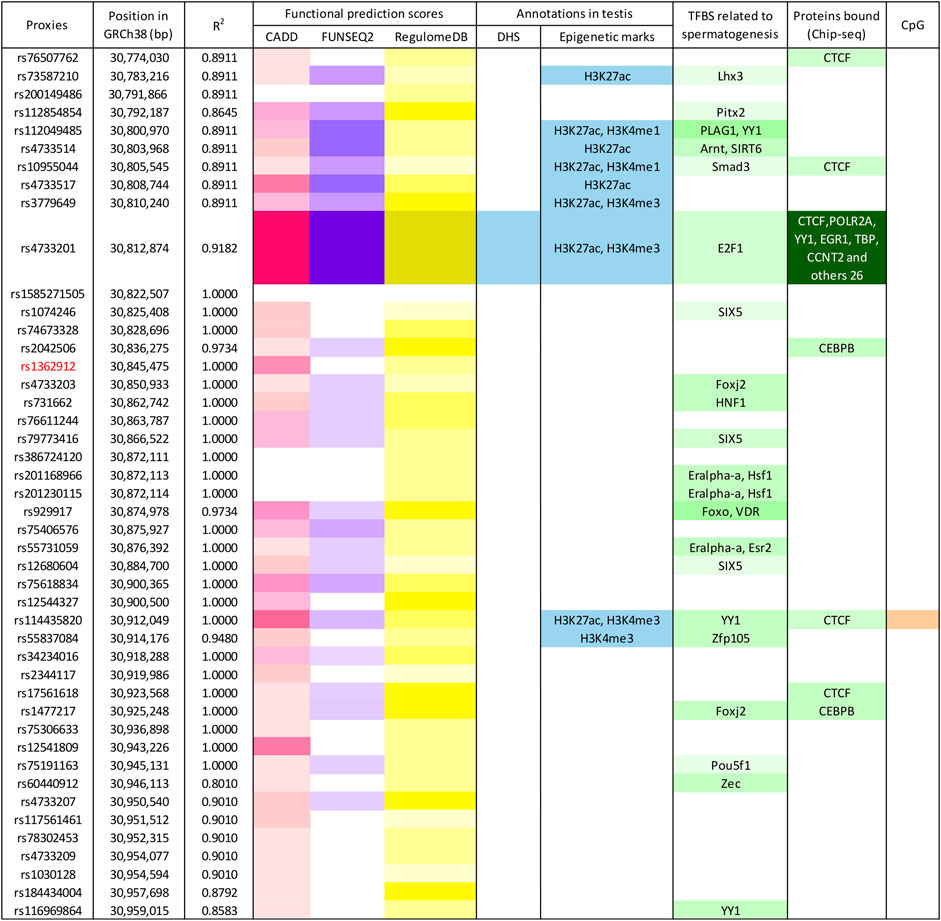
FIGURE 2. Enrichment in functional annotations of the human genome for TEX15-rs1362912 and its proxies. Functional prediction scores, according to different algorithms, are marked in pink (CADD), violet (Funseq2), and yellow (RegulomeDB); blue-coloured cells represent overlap with functional annotations in testis based on the ENCODE and Roadmap Epigenomics projects; variants located within transcription factor binding sites (TFBS) related to spermatogenesis (based on either position weight matrix data or Chip-seq experiments) are shown in green; and variants in CpG sites are highlighted in orange. Colour intensity is correlated with the probability or relevance (dark colours indicate higher probability). Bp, base pairs; GRCh38, Genome Reference Consortium Human Build 38; DHS, DNase I hypersensitive site.
In view of the possible influence on TFBSs of the most likely causal variants of the TEX15 association with SO, we decided to perform an enrichment analysis of both PPIs and biological pathways considering the 112 TFs whose binding affinity to this genetic region was predicted to be altered by the analysed SNPs (Supplementary Tables S5, S6). The PPI network showed significantly more interactions than expected (p = 1.00E-16) (Figure 3). In relation to the functional enrichment of this protein set, “regulation of transcription” and “gene expression” were the GO terms showing the most significant enrichment p-values (p = 3.94E-61 and p = 2.60E-45, respectively). Interestingly, other significantly enriched biological pathways included “reproductive process” (GO:0022414; p = 2.98E-07), “developmental process involved in reproduction” (GO:0003006; p = 1.31E-07), “reproductive structure development” (GO:0048608; p = 3.51E-07), “urogenital system development” (GO:0001655; p = 2.8E-04), “formation of primary germ layer” (GO:0001704; p = 1.70E-03), and “pituitary gland development” (GO:0021983; p = 4.26E-02) (Figure 3; Supplementary Table S7). Furthermore, “male infertility” (WP4673; p = 9.00E-03) and “ovarian infertility” (WP34; p = 4.48E-06) were also highlighted amongst the enriched pathways (Figure 3; Supplementary Table S8).
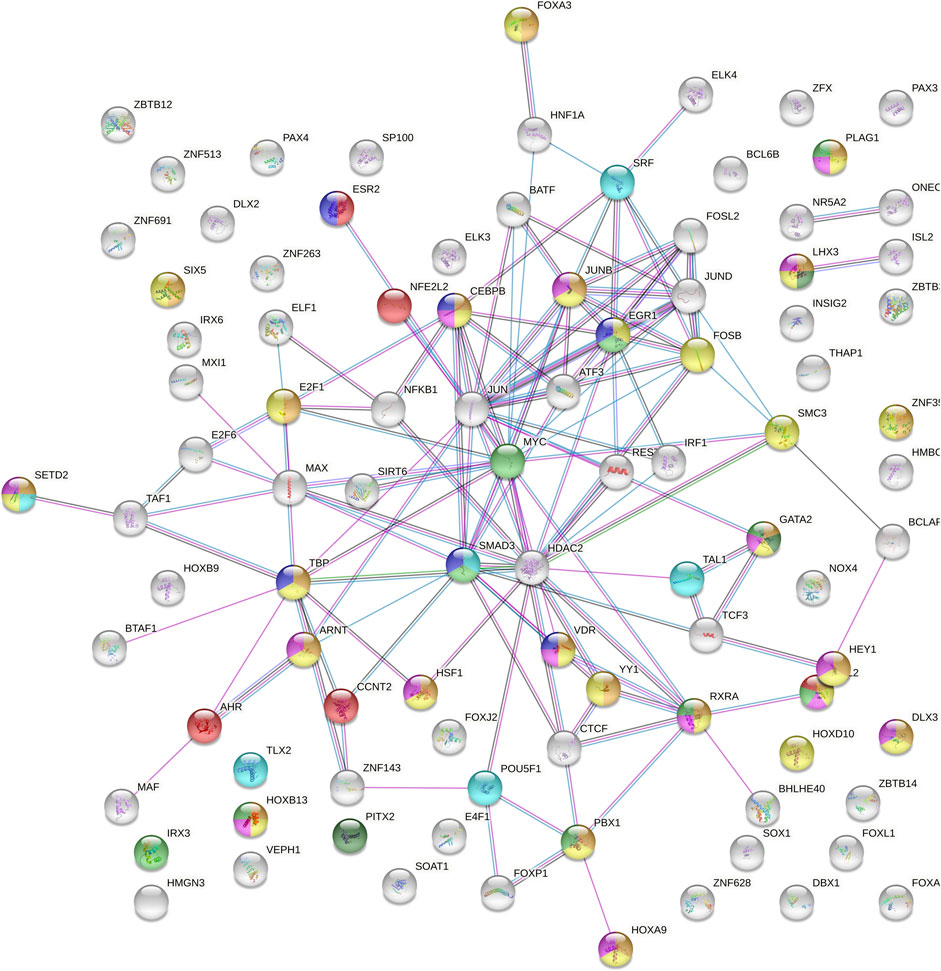
FIGURE 3. Protein-protein interaction network of the 122 transcription factors with predicted binding sites overlapping with TEX15-rs1362912 and its proxies based on ChIP-seq experiments or protein weight matrix data. Blue lines indicate that the interaction is established from curated databases; pink lines indicate experimentally determined connections; green lines link neighbour genes; black lines represent co-expression; and grey lines correspond to proteins with homology. Bubble colour meaning: light blue, formation of primary germ layer; orange, developmental process involved in reproduction; yellow, reproductive process; pink, reproductive structure development; dark green, pituitary gland development; light green, urogenital system development; purple, ovarian infertility and red, male infertility.
Discussion
Understanding the genetic basis of SPGF is a necessary step to improve the clinical management and genetic counselling of infertile patients (Fainberg and Kashanian, 2019; Sharma et al., 2021). In the present study, we conducted a comprehensive evaluation of the possible involvement of TEX15 genetic variation in the predisposition to SPGF, using a large European cohort that ensured a high statistical power.
Our results clearly suggest that TEX15-rs1362912, or any of its tagged SNPs, is directly involved in the pathological mechanisms underlying mild forms of SPGF, i.e., SO, with little or no contribution to extreme patterns, such as NOA. Taking into account the high relevance of TEX15 in the reproductive function and the insight provided by previously published genetic studies on this gene (Aston et al., 2010; Plaseski et al., 2012; Ruan et al., 2012; Okutman et al., 2015; Zhang et al., 2015; Colombo et al., 2017; Wang et al., 2018; Araujo et al., 2020; Cannarella et al., 2021; Ghadirkhomi et al., 2022), we hypothesise that NOA could be mostly influenced by high-penetrance damaging mutations in this locus rather than by common SNPs, which would be responsible for increasing SO predisposition instead. Consistent with this idea, only 27 SO-associated point mutations are currently annotated in both the “Male Infertility Genomic Consortium (IMIGC) database” and the “Infertility Disease Database (IDDB)”, whereas NOA has 138 entries, amongst which TEX15 is included (Houston et al., 2021; Wu et al., 2021). Moreover, a higher proportion of NOA cases present Mendelian causes of their infertility when compared to SO (e.g., AZF microdeletions and chromosomal abnormalities are three times more prevalent in NOA than in SO) (Krausz and Riera-Escamilla, 2018).
Although our data are consistent with this hypothesis, the causality of the TEX15 common variation in SO predisposition remains controversial. Previous genetic association reports in this locus have not been fully replicated likely due to differences in the genetic architecture of the different study groups or, more probably, to type I or II errors caused by the limitations in the statistical power of the studied cohorts to detect low or modest effects. In this regard, the TEX15 SNP rs323346 was not associated with SO neither in our study nor in an independent population of European ancestry previously assessed by Aston et al. (2010). Inconsistent results were obtained in two additional studies performed in Chinese, as Ruan et al. (2012) described a genetic association between this SNP and SO that was not replicated by Zhang et al. (2015) in the same ethnicity. Moreover, there are striking differences between the reported rs323346*C allele frequencies for the human populations included in the 1KGPh3 project (e.g., EUR = 0.17; EAS = 0.11; SAS = 0.33; AFR = 0.80, Supplementary Figure S2) (Auton et al., 2015), which might indicate population-specific causal variants within this region (Aston et al., 2010; Ruan et al., 2012; Ghadirkhomi et al., 2022). Therefore, subsequent studies in independent study cohorts should be performed to clarify further the possible existence of different SO functionally relevant SNPs in this gene.
Remarkably, animal models clearly support the role of the TEX15 locus in SO development (Yang et al., 2008; Schopp et al., 2020; Yang et al., 2020). In fact, TEX15-defficient murine models show reproductive system abnormalities, including male infertility, altered spermatogenesis, arrest of male meiosis, decreased male germ cell number, DSB repair anomalies, and reduced testis weight (Yang et al., 2008; Carvalho-Silva et al., 2019). This gene is mostly expressed in spermatogonia and spermatocytes, in which it has a relevant regulatory role in the control of gene expression by interacting with a vast number of transcription factors related to the reproductive function, as reported in the Open Targets Platform (Carvalho-Silva et al., 2019).
According to the results of our in silico functional analyses, amongst the linked polymorphisms of the tagger rs1362912, the non-coding TEX15 variant rs4733201 may be responsible for an important contribution to the final phenotype. This SNP showed strong evidence of functionality, including high scoring by different functional impact prediction methods as well as an overlap with testis-specific active chromatin epigenetic marks. Interestingly, the sequence surrounding this SNP corresponds to a TFBS for different key transcription factors related with spermatogenesis, including EGR1 (Tourtellotte et al., 2000; Man et al., 2014), CTCF (Hernandez-Hernandez et al., 2016; Rivero-Hinojosa et al., 2021), SMC3 (Eijpe et al., 2000; James et al., 2002), YY1 (Wu et al., 2009; Kim et al., 2016), CCNT2 (Teng et al., 2011), E2F6 (Pohlers et al., 2005; Dahlet et al., 2021), and MAX (Maeda et al., 2013), and it is annotated to be transcriptionally active in the testis (Davis et al., 2018). Moreover, this SNP was predicted to modify the activity of E2F1, which has been associated with human SPGF (Jorgez et al., 2015; Rocca et al., 2019).
Similarly, the results of the biological pathway enrichment analysis of the transcription factors with TFBSs influenced by the analysed variants also support their involvement in SPGF, as some of the enriched pathways included “reproductive process”, “pituitary gland development”, “ovarian infertility”, and “male infertility”. To our knowledge, there are no reported studies in which the possible effects of TEX15 on female infertility were evaluated (Yang et al., 2008; Bellil et al., 2021). However, TEX15 is expressed in the female reproductive tract (Supplementary Figure S1), “ovarian infertility” was amongst the most enriched pathways, and there is evidence of the implications in female fertility of some of the transcription factors with binding sites in the genomic positions of some of the SNPs considered in our functional analysis. Specifically, VDR is associated with female infertility (Djurovic et al., 2020), whereas CEBPB has been described as relevant factor for female reproduction for its role in ovarian follicle development (Fan et al., 2009), together with SMAD3 (Tomic et al., 2004; Li et al., 2008) and ESR2 (Khristi et al., 2018; Chakravarthi et al., 2020). Such is consistent with the assumption that the effect of the common variation in the genome on pathological conditions is not limited to altering the expression or function of a single gene or protein, but rather to unbalance complex molecular networks (Cui et al., 2015).
Regarding the possible limitations of this work, it is important note that our exclusion criteria based on genetic abnormalities considered only karyotype alterations and Y chromosome microdeletions. Although both type of alterations account for the majority of known genetic causes of SPGF (Cioppi et al., 2021), screening of high-penetrance point mutations in reported SPGF genes was not performed. As a consequence, some cases of our study cohort could harbour single-gene mutations that may explain their infertility. However, considering the low frequency of the reported monogenic mutations in SPGF (Cioppi et al., 2021), this limitation is not expected to have a relevant impact in the allele frequencies of our study cohort. The possible statistical noise added by such cases would likely cause a subtle increase in the probability of obtaining type II errors (false negative results), but it would hardly affect the consistency of the observed associations.
Overall, the insight gained in this study supports the notion of idiopathic SPGF as a complex trait, where common genetic variation has a determinant role in disease susceptibility and development. In the case of TEX15, it is likely that the selective pressures have prevented the dissemination of high-penetrance deleterious variants leading to NOA in the human populations, considering the crucial role that this gene has in the reproductive success of an individual (Yang et al., 2008; Chen et al., 2016). Under this assumption, it is reasonable to assume that the common TEX15 polymorphisms affecting male infertility were associated with less extreme manifestations of SPGF, such as SO.
Lisbon clinical group co-authors and IVIRMA group co-authors
Ana Aguiar, (Unidade de Medicina da Reproducao, Hospital de Santa Maria, Centro Hospitalar Lisboa Norte, Lisboa, Portugal); Carlos Calhaz-Jorge, (Unidade de Medicina da Reproducao, Hospital de Santa Maria, Centro Hospitalar Lisboa Norte, Lisboa, Portugal); Joaquim Nunes, (Unidade de Medicina da Reproducao, Hospital de Santa Maria, Centro Hospitalar Lisboa Norte, Lisboa, Portugal); Sandra Sousa (Unidade de Medicina da Reproducao, Hospital de Santa Maria, Centro Hospitalar Lisboa Norte, Lisboa, Portugal), and Sónia Correia (Centro de Medicina Reprodutiva, Maternidade Alfredo da Costa, Centro Hospitalar Lisboa Central, Lisboa, Portugal); Maria Graça Pinto(Centro de Medicina Reprodutiva, Maternidade Alfredo da Costa, Centro Hospitalar Lisboa Central, Lisboa, Portugal). Alberto Pacheco, (IVIRMA Madrid, Spain); Cristina González, (IVIRMA Sevilla, Spain); Susana Gómez, (IVIRMA Lisboa, Portugal); David Amorós, (IVIRMA Barcelona, Spain); Jesús Aguilar, (IVIRMA Vigo, Spain); Fernando Quintana, (IVIRMA Bilbao, Spain).
Data availability
The data generated in this study are either contained in the article file and its Supplementary Material or available upon reasonable request to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee “CEIM/CEI Provincial de Granada” (Andalusia, Spain). The patients/participants provided their written informed consent to participate in this study.
Author contributions
Formal analysis, writing—original draft and review and editing: AG-J and SG-M. Data acquisition, draft review and validation: RR-E, NG, SL, GR, SS-R, and IVIRMA group, Lisbon Clinical group, JC, MG, AC, FV, VM, MP, IP, JN, JS-C, OL-R, IP-C, PM, FC, AB, LB, SS, JG, SL, and AL. Data interpretation and draft review: MC-M, JV-S, MB, and RJ. Conceptualization, draft review and validation: RP-M. Conceptualization, design, supervision, draft review and validation: LB-C and FDC. All authors approved the final version of the manuscript.
Funding
This work was supported by the Spanish Ministry of Science and Innovation through the Spanish National Plan for Scientific and Technical Research and Innovation (PID 2020-120157RB-I00) and the Andalusian Government through the research projects of “Plan Andaluz de Investigacion, Desarrollo e Innovacion (PAIDI 2020)” (ref. PY20_00212) and “Programa Operativo FEDER 2020” (ref. B-CTS-584-UGR20). LB-C was supported by the Spanish Ministry of Science and Innovation through the “Juan de la Cierva Incorporacion” program (Grant ref. IJC 2018-038026-I, funded by MCIN/AEI/10.13039/501100011033), which includes FEDER funds. AG-J was funded by MCIN/AEI/10.13039/501100011033 and FSE “El FSE invierte en tu futuro” (grant ref. FPU20/02926). IPATIMUP integrates the i3S Research Unit, which is partially supported by the Portuguese Foundation for Science and Technology (FCT), financed by the European Social Funds (COMPETE-FEDER) and National Funds (projects PEstC/SAU/LA0003/2013 and POCI-01-0145-FEDER-007274). PM is supported by the FCT post-doctoral fellowship (SFRH/BPD/120777/2016), financed from the Portuguese State Budget of the Ministry for Science, Technology and High Education and from the European Social Fund, available through the Programa Operacional do Capital Humano. ToxOmics—Centre for Toxicogenomics and Human Health, Genetics, Oncology and Human Toxicology, Nova Medical School, Lisbon, is also partially supported by FCT (UID/BIM/00009/2016 and UIDB/00009/2020). SL received support from Instituto de Salud Carlos III (grant: DTS18/00101], co-funded by FEDER funds/European Regional Development Fund (ERDF)-a way to build Europe-), and from “Generalitat de Catalunya” (grant 2017SGR191). SL is sponsored by the “Researchers Consolidation Program” from the SNS-Dpt. Salut Generalitat de Catalunya (Exp. CES09/020). This article is related to the Ph.D. Doctoral Thesis of AG-J.
Acknowledgments
We thank the National DNA Bank Carlos III (University of Salamanca, Spain) for supplying part of the control DNA samples from Spain and all the participants for their essential collaboration.
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1089782/full#supplementary-material
References
Agarwal, A., Baskaran, S., Parekh, N., Cho, C. L., Henkel, R., Vij, S., et al. (2021). Male infertility. Lancet 397, 319–333. doi:10.1016/S0140-6736(20)32667-2
Alves, I., Houle, A. A., Hussin, J. G., and Awadalla, P. (2017). The impact of recombination on human mutation load and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160465. doi:10.1098/rstb.2016.0465
Araujo, T. F., Friedrich, C., Grangeiro, C. H. P., Martelli, L. R., Grzesiuk, J. D., Emich, J., et al. (2020). Sequence analysis of 37 candidate genes for male infertility: Challenges in variant assessment and validating genes. Andrology 8, 434–441. doi:10.1111/andr.12704
Aston, K. I., Krausz, C., Laface, I., Ruiz-Castane, E., and Carrell, D. T. (2010). Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum. Reprod. 25, 1383–1397. doi:10.1093/humrep/deq081
Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., Korbel, J. O., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi:10.1038/nature15393
Barrett, J. C. (2009). Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc. 2009, pdb.ip71. pdb ip71. doi:10.1101/pdb.ip71
Bellil, H., Ghieh, F., Hermel, E., Mandon-Pepin, B., and Vialard, F. (2021). Human testis-expressed (TEX) genes: A review focused on spermatogenesis and male fertility. Basic Clin. Androl. 31, 9. doi:10.1186/s12610-021-00127-7
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N., and Golani, I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284. doi:10.1016/s0166-4328(01)00297-2
Bjorndahl, L., and Kirkman Brown, J.other Editorial Board Members of the WHO Laboratory Manual for the Examination and Processing of Human Semen (2022). The sixth edition of the WHO laboratory manual for the examination and processing of human semen: Ensuring quality and standardization in basic examination of human ejaculates. Fertil. Steril. 117, 246–251. doi:10.1016/j.fertnstert.2021.12.012
Cannarella, R., Condorelli, R. A., Duca, Y., La Vignera, S., and Calogero, A. E. (2019). New insights into the genetics of spermatogenic failure: A review of the literature. Hum. Genet. 138, 125–140. doi:10.1007/s00439-019-01974-1
Cannarella, R., Condorelli, R. A., Mongioi, L. M., La Vignera, S., and Calogero, A. E. (2020). Molecular Biology of spermatogenesis: Novel Targets of apparently idiopathic male infertility. Int. J. Mol. Sci. 21, 1728. doi:10.3390/ijms21051728
Cannarella, R., Condorelli, R. A., Paolacci, S., Barbagallo, F., Guerri, G., Bertelli, M., et al. (2021). Next-generation sequencing: Toward an increase in the diagnostic yield in patients with apparently idiopathic spermatogenic failure. Asian J. Androl. 23, 24–29. doi:10.4103/aja.aja_25_20
Capilla, L., Garcia Caldes, M., and Ruiz-Herrera, A. (2016). Mammalian meiotic recombination: A toolbox for genome evolution. Cytogenet. Genome Res. 150, 1–16. doi:10.1159/000452822
Carithers, L. J., and Moore, H. M. (2015). The genotype-tissue expression (GTEx) project. Biopreserv. Biobank. 13, 307–308. doi:10.1089/bio.2015.29031.hmm
Carvalho-Silva, D., Pierleoni, A., Pignatelli, M., Ong, C., Fumis, L., Karamanis, N., et al. (2019). Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 47, D1056–D1065. doi:10.1093/nar/gky1133
Cervan-Martin, M., Bossini-Castillo, L., Rivera-Egea, R., Garrido, N., Lujan, S., Romeu, G., et al. (2021). Effect and in silico characterization of genetic variants associated with severe spermatogenic disorders in a large Iberian cohort. Andrology 9, 1151–1165. doi:10.1111/andr.13009
Cervan-Martin, M., Bossini-Castillo, L., Rivera-Egea, R., Garrido, N., Lujan, S., Romeu, G., et al. (2020a). Evaluation of male fertility-associated loci in a European population of patients with severe spermatogenic impairment. J. Pers. Med. 11, 22. doi:10.3390/jpm11010022
Cervan-Martin, M., Castilla, J. A., Palomino-Morales, R. J., and Carmona, F. D. (2020b). Genetic landscape of nonobstructive azoospermia and new perspectives for the clinic. J. Clin. Med. 9, 300. doi:10.3390/jcm9020300
Cervan-Martin, M., Suazo-Sanchez, M. I., Rivera-Egea, R., Garrido, N., Lujan, S., Romeu, G., et al. (2020c). Intronic variation of the SOHLH2 gene confers risk to male reproductive impairment. Fertil. Steril. 114, 398–406. doi:10.1016/j.fertnstert.2020.02.115
Chakravarthi, V. P., Ghosh, S., Roby, K. F., Wolfe, M. W., and Rumi, M. a. K. (2020). A gatekeeping role of ESR2 to maintain the primordial follicle reserve. Endocrinology 161, bqaa037. doi:10.1210/endocr/bqaa037
Chalmel, F., Lardenois, A., Evrard, B., Mathieu, R., Feig, C., Demougin, P., et al. (2012). Global human tissue profiling and protein network analysis reveals distinct levels of transcriptional germline-specificity and identifies target genes for male infertility. Hum. Reprod. 27, 3233–3248. doi:10.1093/humrep/des301
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7. doi:10.1186/s13742-015-0047-8
Chen, S. R., Hao, X. X., Zhang, Y., Deng, S. L., Wang, Z. P., Wang, Y. Q., et al. (2016). Androgen receptor in Sertoli cells regulates DNA double-strand break repair and chromosomal synapsis of spermatocytes partially through intercellular EGF-EGFR signaling. Oncotarget 7, 18722–18735. doi:10.18632/oncotarget.7916
Colombo, R., Pontoglio, A., and Bini, M. (2017). Two novel TEX15 mutations in a family with nonobstructive azoospermia. Gynecol. Obstet. Invest. 82, 283–286. doi:10.1159/000468934
Cooper, T. G., Noonan, E., Von Eckardstein, S., Auger, J., Baker, H. W., Behre, H. M., et al. (2010). World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 16, 231–245. doi:10.1093/humupd/dmp048
Cui, H., Dhroso, A., Johnson, N., and Korkin, D. (2015). The variation game: Cracking complex genetic disorders with NGS and omics data. Methods 79-80, 18–31. doi:10.1016/j.ymeth.2015.04.018
Dahlet, T., Truss, M., Frede, U., Al Adhami, H., Bardet, A. F., Dumas, M., et al. (2021). E2F6 initiates stable epigenetic silencing of germline genes during embryonic development. Nat. Commun. 12, 3582. doi:10.1038/s41467-021-23596-w
Davis, C. A., Hitz, B. C., Sloan, C. A., Chan, E. T., Davidson, J. M., Gabdank, I., et al. (2018). The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801. doi:10.1093/nar/gkx1081
Djurovic, J., Stamenkovic, G., Todorovic, J., Aleksic, N., and Stojkovic, O. (2020). Polymorphisms and haplotypes in VDR gene are associated with female idiopathic infertility. Hum. Fertil. 23, 101–110. doi:10.1080/14647273.2018.1515503
Dong, S., and Boyle, A. P. (2019). Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 40, 1292–1298. doi:10.1002/humu.23791
Eijpe, M., Heyting, C., Gross, B., and Jessberger, R. (2000). Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J. Cell Sci. 113, 673–682. doi:10.1242/jcs.113.4.673
Fainberg, J., and Kashanian, J. A. (2019). Recent advances in understanding and managing male infertility. F1000Res 8, 670. doi:10.12688/f1000research.17076.1
Fan, H. Y., Liu, Z., Shimada, M., Sterneck, E., Johnson, P. F., Hedrick, S. M., et al. (2009). MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 324, 938–941. doi:10.1126/science.1171396
Ghadirkhomi, E., Angaji, S. A., Khosravi, M., and Mashayekh, M. R. (2022). Correlation of novel single nucleotide polymorphisms ofUSP26, TEX15, and TNP2 genes with male infertility in north west of Iran. Int. J. Fertil. Steril. 16, 10–16. doi:10.22074/IJFS.2021.521138.1058
Gonzalez-Marin, C., Gosalvez, J., and Roy, R. (2012). Types, causes, detection and repair of DNA fragmentation in animal and human sperm cells. Int. J. Mol. Sci. 13, 14026–14052. doi:10.3390/ijms131114026
Guo, J., Grow, E. J., Mlcochova, H., Maher, G. J., Lindskog, C., Nie, X., et al. (2018). The adult human testis transcriptional cell atlas. Cell Res. 28, 1141–1157. doi:10.1038/s41422-018-0099-2
Guo, J., Nie, X., Giebler, M., Mlcochova, H., Wang, Y., Grow, E. J., et al. (2020). The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell 26, 262–276. doi:10.1016/j.stem.2019.12.005
Hernandez-Hernandez, A., Lilienthal, I., Fukuda, N., Galjart, N., and Hoog, C. (2016). CTCF contributes in a critical way to spermatogenesis and male fertility. Sci. Rep. 6, 28355. doi:10.1038/srep28355
Houston, B. J., Riera-Escamilla, A., Wyrwoll, M. J., Salas-Huetos, A., Xavier, M. J., Nagirnaja, L., et al. (2021). A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum. Reprod. Update 28, 15–29. doi:10.1093/humupd/dmab030
James, R. D., Schmiesing, J. A., Peters, A. H., Yokomori, K., and Disteche, C. M. (2002). Differential association of SMC1alpha and SMC3 proteins with meiotic chromosomes in wild-type and SPO11-deficient male mice. Chromosome Res. 10, 549–560. doi:10.1023/a:1020910601858
Jarvi, K., Lo, K., Fischer, A., Grantmyre, J., Zini, A., Chow, V., et al. (2010). CUA Guideline: The workup of azoospermic males. Can. Urol. Assoc. J. 4, 163–167. doi:10.5489/cuaj.10050
Jorgez, C. J., Wilken, N., Addai, J. B., Newberg, J., Vangapandu, H. V., Pastuszak, A. W., et al. (2015). Genomic and genetic variation in E2F transcription factor-1 in men with nonobstructive azoospermia. Fertil. Steril. 103, 44–52. doi:10.1016/j.fertnstert.2014.09.021
Khristi, V., Chakravarthi, V. P., Singh, P., Ghosh, S., Pramanik, A., Ratri, A., et al. (2018). ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 474, 214–226. doi:10.1016/j.mce.2018.03.012
Kim, J. S., Chae, J. H., Cheon, Y. P., and Kim, C. G. (2016). Reciprocal localization of transcription factors YY1 and CP2c in spermatogonial stem cells and their putative roles during spermatogenesis. Acta Histochem. 118, 685–692. doi:10.1016/j.acthis.2016.08.005
Krausz, C., Cioppi, F., and Riera-Escamilla, A. (2018). Testing for genetic contributions to infertility: Potential clinical impact. Expert Rev. Mol. diagn. 18, 331–346. doi:10.1080/14737159.2018.1453358
Krausz, C., and Riera-Escamilla, A. (2018). Genetics of male infertility. Nat. Rev. Urol. 15, 369–384. doi:10.1038/s41585-018-0003-3
Kumar, S., Ambrosini, G., and Bucher, P. (2017). SNP2TFBS - a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 45, D139–D144. doi:10.1093/nar/gkw1064
Leduc, F., Nkoma, G. B., and Boissonneault, G. (2008). Spermiogenesis and DNA repair: A possible etiology of human infertility and genetic disorders. Syst. Biol. Reprod. Med. 54, 3–10. doi:10.1080/19396360701876823
Li, Q., Pangas, S. A., Jorgez, C. J., Graff, J. M., Weinstein, M., and Matzuk, M. M. (2008). Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol. Cell. Biol. 28, 7001–7011. doi:10.1128/MCB.00732-08
Luo, Y., Hitz, B. C., Gabdank, I., Hilton, J. A., Kagda, M. S., Lam, B., et al. (2020). New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889. doi:10.1093/nar/gkz1062
Machiela, M. J., and Chanock, S. J. (2015). LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. doi:10.1093/bioinformatics/btv402
Maeda, I., Okamura, D., Tokitake, Y., Ikeda, M., Kawaguchi, H., Mise, N., et al. (2013). Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 4, 1754. doi:10.1038/ncomms2780
Man, P. S., Wells, T., and Carter, D. A. (2014). Cellular distribution of Egr1 transcription in the male rat pituitary gland. J. Mol. Endocrinol. 53, 271–280. doi:10.1530/JME-14-0158
Neto, F. T., Bach, P. V., Najari, B. B., Li, P. S., and Goldstein, M. (2016). Spermatogenesis in humans and its affecting factors. Semin. Cell Dev. Biol. 59, 10–26. doi:10.1016/j.semcdb.2016.04.009
Okutman, O., Muller, J., Baert, Y., Serdarogullari, M., Gultomruk, M., Piton, A., et al. (2015). Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum. Mol. Genet. 24, 5581–5588. doi:10.1093/hmg/ddv290
Oscanoa, J., Sivapalan, L., Gadaleta, E., Dayem Ullah, A. Z., Lemoine, N. R., and Chelala, C. (2020). SNPnexus: A web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 48, W185–W192. doi:10.1093/nar/gkaa420
Papatheodorou, I., Moreno, P., Manning, J., Fuentes, A. M., George, N., Fexova, S., et al. (2020). Expression atlas update: From tissues to single cells. Nucleic Acids Res. 48, D77–D83. doi:10.1093/nar/gkz947
Plaseski, T., Noveski, P., Popeska, Z., Efremov, G. D., and Plaseska-Karanfilska, D. (2012). Association study of single-nucleotide polymorphisms in FASLG, JMJDIA, LOC203413, TEX15, BRDT, OR2W3, INSR, and TAS2R38 genes with male infertility. J. Androl. 33, 675–683. doi:10.2164/jandrol.111.013995
Pohlers, M., Truss, M., Frede, U., Scholz, A., Strehle, M., Kuban, R. J., et al. (2005). A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr. Biol. 15, 1051–1057. doi:10.1016/j.cub.2005.04.060
Ponomarenko, M. P., Sharypova, E. B., Drachkova, I. A., Savinkova, L. K., Chadaeva, I. V., Rasskazov, D. A., et al. (2020). Candidate SNP-markers altering TBP binding affinity for promoters of the Y-linked genes CDY2A, SHOX, and ZFY are lowering many indexes of reproductive potential in men. Vavilovskii Zhurnal Genet. Sel. 24, 785–793. doi:10.18699/VJ20.674
Rivero-Hinojosa, S., Pugacheva, E. M., Kang, S., Mendez-Catala, C. F., Kovalchuk, A. L., Strunnikov, A. V., et al. (2021). The combined action of CTCF and its testis-specific paralog BORIS is essential for spermatogenesis. Nat. Commun. 12, 3846. doi:10.1038/s41467-021-24140-6
Rocca, M. S., Di Nisio, A., Sabovic, I., Ghezzi, M., Foresta, C., and Ferlin, A. (2019). E2F1 copy number variations contribute to spermatogenic impairment and cryptorchidism by increasing susceptibility to heat stress. Andrology 7, 251–256. doi:10.1111/andr.12583
Ruan, J., He, X. J., Du, W. D., Chen, G., Zhou, Y., Xu, S., et al. (2012). Genetic variants in TEX15 gene conferred susceptibility to spermatogenic failure in the Chinese Han population. Reprod. Sci. 19, 1190–1196. doi:10.1177/1933719112446076
Salas-Huetos, A., and Aston, K. I. (2021). Defining new genetic etiologies of male infertility: Progress and future prospects. Transl. Androl. Urol. 10, 1486–1498. doi:10.21037/tau.2020.03.43
Schopp, T., Zoch, A., Berrens, R. V., Auchynnikava, T., Kabayama, Y., Vasiliauskaite, L., et al. (2020). TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing. Nat. Commun. 11, 3739. doi:10.1038/s41467-020-17372-5
Sharma, A., Minhas, S., Dhillo, W. S., and Jayasena, C. N. (2021). Male infertility due to testicular disorders. J. Clin. Endocrinol. Metab. 106, e442–e459. doi:10.1210/clinem/dgaa781
Singh, K., and Jaiswal, D. (2011). Human male infertility: A complex multifactorial phenotype. Reprod. Sci. 18, 418–425. doi:10.1177/1933719111398148
Skol, A. D., Scott, L. J., Abecasis, G. R., and Boehnke, M. (2006). Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213. doi:10.1038/ng1706
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Teng, Y., Wang, Y., Fu, J., Cheng, X., Miao, S., and Wang, L. (2011). Cyclin T2: A novel miR-15a target gene involved in early spermatogenesis. FEBS Lett. 585, 2493–2500. doi:10.1016/j.febslet.2011.06.031
Tian, R. H., Huang, Y. H., Chen, H. X., Li, P., Zhi, E. L., Yao, C. C., et al. (2021). Successful microsurgical vasoepididymostomy for a case of cryptozoospermia. Asian J. Androl. 24, 436–437. doi:10.4103/aja202178
Tomic, D., Miller, K. P., Kenny, H. A., Woodruff, T. K., Hoyer, P., and Flaws, J. A. (2004). Ovarian follicle development requires Smad3. Mol. Endocrinol. 18, 2224–2240. doi:10.1210/me.2003-0414
Tourtellotte, W. G., Nagarajan, R., Bartke, A., and Milbrandt, J. (2000). Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol. Cell. Biol. 20, 5261–5268. doi:10.1128/mcb.20.14.5261-5268.2000
Wang, X., Jin, H. R., Cui, Y. Q., Chen, J., Sha, Y. W., and Gao, Z. L. (2018). Case study of a patient with cryptozoospermia associated with a recessive TEX15 nonsense mutation. Asian J. Androl. 20, 101–102. doi:10.4103/1008-682X.194998
Ward, L. D., and Kellis, M. (2016). HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881. doi:10.1093/nar/gkv1340
Wu, S., Hu, Y. C., Liu, H., and Shi, Y. (2009). Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol. Cell. Biol. 29, 6245–6256. doi:10.1128/MCB.00679-09
Wu, J., Li, D., Liu, X., Li, Q., He, X., Wei, J., et al. (2021). Iddb: A comprehensive resource featuring genes, variants and characteristics associated with infertility. Nucleic Acids Res. 49, D1218–D1224. doi:10.1093/nar/gkaa753
Yang, F., Eckardt, S., Leu, N. A., Mclaughlin, K. J., and Wang, P. J. (2008). Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J. Cell Biol. 180, 673–679. doi:10.1083/jcb.200709057
Yang, F., Lan, Y., Pandey, R. R., Homolka, D., Berger, S. L., Pillai, R. S., et al. (2020). TEX15 associates with MILI and silences transposable elements in male germ cells. Genes. Dev. 34, 745–750. doi:10.1101/gad.335489.119
Keywords: oligozoospermia, spermatogenesis, TEX15, polymorphisms, association study
Citation: Guzmán-Jiménez A, González-Muñoz S, Cerván-Martín M, Rivera-Egea R, Garrido N, Luján S, Santos-Ribeiro S, Castilla JA, Gonzalvo MC, Clavero A, Vicente FJ, Maldonado V, Villegas-Salmerón J, Burgos M, Jiménez R, Pinto MG, Pereira I, Nunes J, Sánchez-Curbelo J, López-Rodrigo O, Pereira-Caetano I, Marques PI, Carvalho F, Barros A, Bassas L, Seixas S, Gonçalves J, Lopes AM, Larriba S, Palomino-Morales RJ, Carmona FD, Bossini-Castillo L, IVIRMA Group and Lisbon Clinical Group (2022) Contribution of TEX15 genetic variants to the risk of developing severe non-obstructive oligozoospermia. Front. Cell Dev. Biol. 10:1089782. doi: 10.3389/fcell.2022.1089782
Received: 04 November 2022; Accepted: 28 November 2022;
Published: 15 December 2022.
Edited by:
Mingrong Lv, First Affiliated Hospital of Anhui Medical University, ChinaReviewed by:
Rong Hua, Anhui Medical University, ChinaAminata Toure, Institute for Advanced Biosciences, France
Chunyu Liu, Fudan University, China
Copyright © 2022 Guzmán-Jiménez, González-Muñoz, Cerván-Martín, Rivera-Egea, Garrido, Luján, Santos-Ribeiro, Castilla, Gonzalvo, Clavero, Vicente, Maldonado, Villegas-Salmerón, Burgos, Jiménez, Pinto, Pereira, Nunes, Sánchez-Curbelo, López-Rodrigo, Pereira-Caetano, Marques, Carvalho, Barros, Bassas, Seixas, Gonçalves, Lopes, Larriba, Palomino-Morales, Carmona, Bossini-Castillo, IVIRMA Group and Lisbon Clinical Group. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: F. David Carmona, dcarmona@ugr.es; Lara Bossini-Castillo, lbossinicastillo@ugr.es
†ORCID: F. David Carmona, orcid.org/0000-0002-1427-7639
#IVIRMA Group, and Lisbon Clinical Group authors are listed at the end of the article
‡These authors share first authorship
§These authors share last authorship
 Andrea Guzmán-Jiménez
Andrea Guzmán-Jiménez Sara González-Muñoz1,2‡
Sara González-Muñoz1,2‡  Nicolás Garrido
Nicolás Garrido Samuel Santos-Ribeiro
Samuel Santos-Ribeiro Susana Seixas
Susana Seixas Alexandra M. Lopes
Alexandra M. Lopes Rogelio J. Palomino-Morales
Rogelio J. Palomino-Morales F. David Carmona
F. David Carmona Lara Bossini-Castillo
Lara Bossini-Castillo