The new ceRNA crosstalk between mRNAs and miRNAs in intervertebral disc degeneration
- Department of Spine Surgery, Beijing Jishuitan Hospital, Fourth Clinical College of Peking University, Beijing, China
Degeneration of the intervertebral disc has been linked to lower back pain. To date, pathophysiological mechanisms of intervertebral disc degeneration (IDD) remain unclear; it is meaningful to find effective diagnostic biomarkers and new therapeutic strategies for IDD. This study aimed to reveal the molecular mechanism of IDD pathogenesis from the multidimensional transcriptomics perspective. Here, we acquired IDD bulk omics datasets (GSE67567 and GSE167199) including mRNA, microRNA expression profiles, and single-cell RNA sequencing (GSE199866) from the public Gene Expression Omnibus (GEO) database. Through principal component analysis and Venn analysis, we found different expression patterns in the IDD transcription level and identified 156 common DEGs in both bulk datasets. GO and KEGG functional analyses showed these dysregulators were mostly enriched in the collagen-containing extracellular matrix, cartilage development, chondrocyte differentiation, and immune response pathways. We also constructed a potentially dysregulated competing endogenous RNA (ceRNA) network between mRNAs and miRNAs related to IDD based on microRNA target information and co-expression analysis of RNA profiles and identified 36 ceRNA axes including ZFP36/miR-155-5p/FOS, BTG2/hsa-miR-185-5p/SOCS3, and COL9A2/hsa-miR-664a-5p/IBA57. Finally, in integrating bulk and single-cell transcriptome data analyses, a total of three marker genes, COL2A1, PAX1, and ZFP36L2, were identified. In conclusion, the key genes and the new ceRNA crosstalk we identified in intervertebral disc degeneration may provide new targets for the treatment of IDD.
Introduction
Lower back pain is a leading cause of disability, affecting about 80% of individuals at least once during their life, and may lead to high healthcare costs and poor quality of life (Li et al., 2019; Chen et al., 2020a; Guo et al., 2020; Wang et al., 2021a). Degeneration of the intervertebral disc has been linked to lower back pain (Tang et al., 2020; Huang et al., 2021; Zhang et al., 2021). Intervertebral discs comprise the annulus fibrosus and nucleus pulposus (NP), and the function of the disc is to retain the vertebral stability (Wang et al., 2018a; Tan et al., 2018; Jiang et al., 2020). There are many risk factors for intervertebral disc degeneration (IDD), including genetic susceptibility, aging, smoking, heavy load work, and body weight (Wang et al., 2017a; Chen et al., 2021a). To date, the pathophysiological mechanisms of IDD remain unclear, and it is difficult to find effective diagnostic biomarkers and new therapeutic strategies for IDD.
MicroRNA (miRNA) is a subtype of noncoding, endogenous transcripts with 21–25 nucleotides in length that modulates gene expression in transcription, post-transcription, and epigenetics (Li et al., 2022; Niu et al., 2022; Xiao et al., 2022; Yang et al., 2022). Dysregulated expression of miRNA has been found to participate in many diseases such as neurological diseases, cardiovascular disease, cancer, and other diseases (Lin et al., 2019; Niu et al., 2020; Zhu et al., 2020; Andreucci et al., 2022). Recent studies have demonstrated that miRNAs play important roles in development of orthopedic diseases including IDD (Tan et al., 2021; Yang et al., 2021). Extensive studies indicated transcription RNA products such as lncRNAs and circRNAs can act as natural miRNA sponges through their miRNA response elements (MRE), acting as competing endogenous RNAs (ceRNAs) (Salmena et al., 2011). The ceRNA crosstalk plays a critical role in modulating gene expression (Thomson and Dinger, 2016). Accumulating studies have found several ceRNAs were related to autophagy, apoptosis, and cell cycle in IDD (Xi et al., 2017; Han et al., 2018; Zhan et al., 2020). Previous studies of ceRNA interactions in IDD had focused on lncRNAs and circRNAs as the sponges of miRNAs (Wang et al., 2017a; Xi et al., 2017; Cheng et al., 2018). Also, mRNAs can competitively bind to miRNAs through their MREs, thus acting as ceRNAs to regulate the expression of miRNA–target mRNAs (Tay et al., 2014; Xu et al., 2015). Nevertheless, it lacked a global view of the ceRNA crosstalk between mRNAs and miRNAs in IDD progression. It is important to evaluate the ceRNA regulation network in the type of mRNA–miRNA–mRNA crosstalk to improve our understanding of molecular mechanisms in IDD.
In this study, systematic bioinformatics analysis and expression profiles including microarrays and next-generation sequencing from bulk and single-cell transcriptome analyses were used to identify the key dysregulators in IDD. Then, combining miRNA targeting information and gene expression validation, we constructed the dysregulated ceRNA networks between miRNAs and mRNAs in intervertebral disc degeneration.
Materials and methods
Data acquisition
The dataset GSE67567 profiled by Arraystar Human microarray V2.0 (Agilent-033010) and GSE167199 identified by RNA sequencing (Illumina NovaSeq 6000) were downloaded from the Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo/). GSE67567 and GSE167199 were composed of five and three pairs of intervertebral disc degeneration (IDD), respectively, and normal specimens as control (IDnD). For single-cell transcriptome analysis, GSE199866 was downloaded. Freshly isolated human cells were separately isolated from non-degenerating and degenerating discs of the same individual. Isolated cells were subjected to droplet-based single-cell RNA sequencing using the 10X Genomics platform. The individual cells were profiled from IDnD and IDD discs.
Differential expression analysis
The package in R (version 4.2.1) named “limma” was used to analyze the differential gene expression (Ritchie et al., 2015). The differentially expressed genes (DEGs) between IDD and IDnD tissues were screened with the cutoff value at p-value < 0.05 and |Log2 FoldChange|>1. The differentially expressed mRNAs in both bulk RNA expression datasets were utilized for subsequent analysis. Meanwhile, the differentially expressed miRNAs between IDD and control tissues were also calculated as the aforementioned method.
CeRNA construction between mRNAs and miRNAs in intervertebral disc degeneration
The miRNAs targeting mRNAs pairs were predicted using miRTarbase (Huang et al., 2022) and starBase databases (Li et al., 2014). In order to study the possible cell specific functions of the ceRNA network, we obtained the human miRNA–DEG relationships and selected some eligible pairs with gene expression validation. It requires the following conditions (Xu et al., 2015): 1). The mRNAs that shared miRNA have the positive correlation of expression (p < 0.05). 2).The hypergeometric distribution of the number of shared miRNAs is significant (p < 0.05). 3). The miRNA and target mRNA have the negative correlation of expression (p < 0.05).
Protein–protein interaction network and gene functional analysis
The DEGs were mapped to the STRING database (https://cn.string-db.org/, version 11.5) to construct their PPI network (Szklarczyk et al., 2022; von Mering et al., 2003). The network visualization and analysis was conducted by Cytoscape software (https://cytoscape.org/, version 3.9.1) (Shannon et al., 2003). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were used to analyze the biological pathways (Kanehisa and Goto, 2000). The R package named “clusterProfiler” was applied to acquire the functional enrichment terms of the DEGs (Wu et al., 2021). The annotation clusters with the p-value < 0.05 were treated as statistically significant. All analyses were performed by R software (version 4.2.1). The p-value less than 0.05 was considered statistically significant.
Single-cell transcriptome analysis
The single-cell datasets were integrated using Seurat’s alignment procedure (Hao et al., 2021). Canonical correlation analysis (CCA) was performed to identify shared sources of variation to produce anchors across the datasets, following SCTransform normalization and principal component analysis for feature extraction. An unsupervised nonlinear dimensionality reduction technique was based on the first 30 principal components. Visualization was performed via Uniform Manifold Approximation and Projection (UMAP).
Results
The different transcriptome expression pattern in IDD
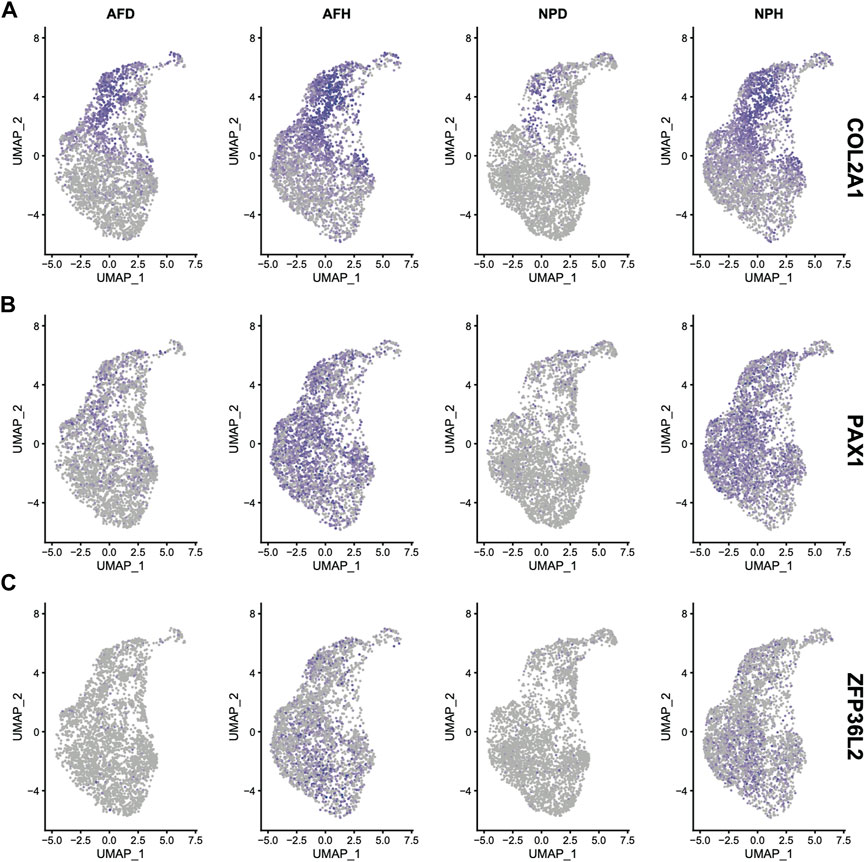
Based on the mRNA expression profiles from bulk datasets GSE67567 and GSE167199, we identified transcription patterns of intervertebral disc degeneration (IDD) samples and the normal controls (intervertebral disc, non-degeneration, and IDnD), respectively. The result of the aforementioned datasets indicated that IDD was different from IDnD in the transcription level using principal component analysis (PCA) (Figures 1A,B) and clustering (Figures 1C,D). Then, the differentially expressed gene analysis (p < 0.05, |Log2 FoldChange|>1) of microarray data GSE67567 showed that a total of 2448 mRNAs were significantly differentially expressed in IDD compared with IDnD. There were 1,864 upregulated mRNAs and 584 downregulated mRNAs (Figure 2A, Supplementary Figure S1A). As for the RNA-seq dataset GSE167199 (Figure 2B, Supplementary Figure S1B), it contained 1890 DEGs (1424 downregulated and 466 upregulated). By Venn analysis, 156 common DEGs were in the same expression trend in both datasets (33 downregulated and 123 upregulated, Figure 2C). The expression heat maps of the common 156 DEGs are visualized in Figure 2D (GSE67567) and Figure 2E (GSE167199). In the same way, we identified a total of six significantly differentially expressed miRNAs including hsa-miR-3131, hsa-miR-3150a-3p, hsa-miR-4731-3p, hsa-miR-4741, hsa-miR-486-5p, and hsa-miR-642b-3p (Table 1).
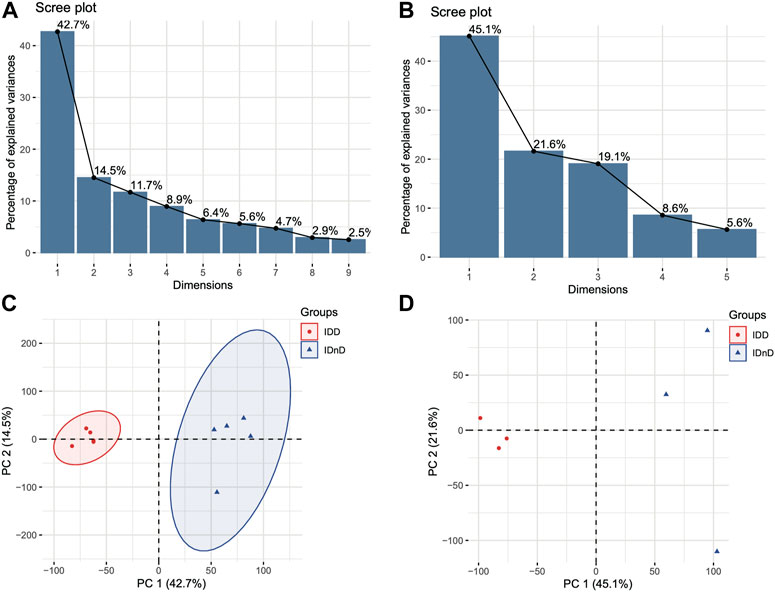
FIGURE 1. Different transcriptome expression patterns between IDD and IDnD. (A) Scree plot of the principal components in the GSE67567 dataset. (B) Scree plot of the principal components in the GSE167199 dataset. (C) PCA and clustering analysis in the GSE67567 dataset. (D) PCA and clustering analysis in the GSE167199 dataset. Axes are principal components 1 and 2.
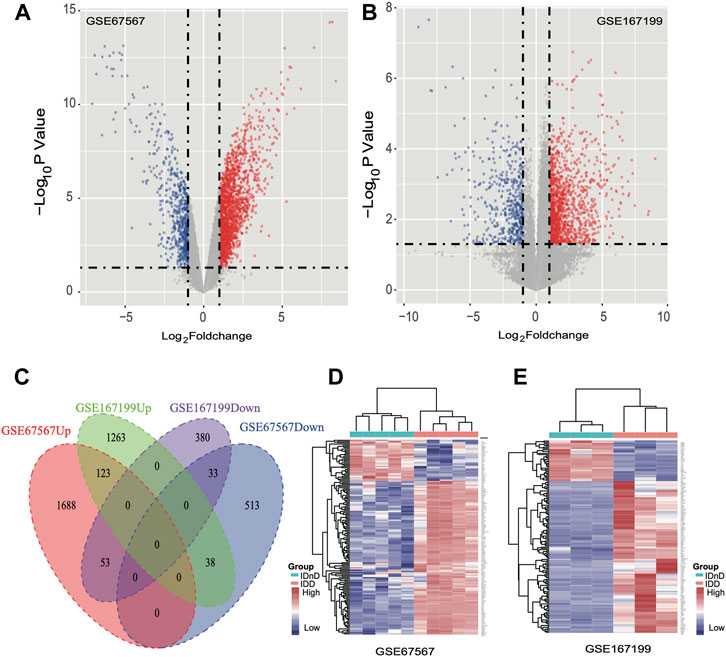
FIGURE 2. Differentially expressed mRNAs between IDD and IDnD. (A) Comparison of differential mRNA volcano plots between nucleus pulposus samples from IDD and IDnD in the GSE67567 dataset. (B) Comparison of DEG volcano plots between nucleus pulposus samples from IDD and IDnD in the GSE167199 dataset. (C) Venn diagram of the DEGs from both datasets. (D) Expression heatmap of 156 common DEGs in the GSE67567 dataset. (E) Expression heatmap of 156 common DEGs in the GSE167199 dataset.
Functional analysis of the DEGs
In GO and KEGG analyses by the R software package clusterProfiler, we identified 73 enriched GO terms and 41 KEGG pathways from the aforementioned 156 common DEGs (p < 0.05, Supplementary Table S1). The top 10 GO terms ranked by the p-value are shown in Figure 3A. The most enriched GO term in BP was the “GO:0030198-extracellular matrix organization” (p < 0:001, n = 18); in CC was the “GO:0062023-collagen-containing extracellular matrix” (p < 0:001, n = 23); and in MF was the “GO:0005201-extracellular matrix structural constituent” (p < 0:001, n = 12). There are more upregulated genes than downregulated genes, suggesting that the GO terms may be activated. The top 10 enriched KEGG pathways of the DEGs are presented in Figure 3B. The most enriched pathways were associated with amoebiasis, rheumatoid arthritis, and the ECM–receptor interaction.
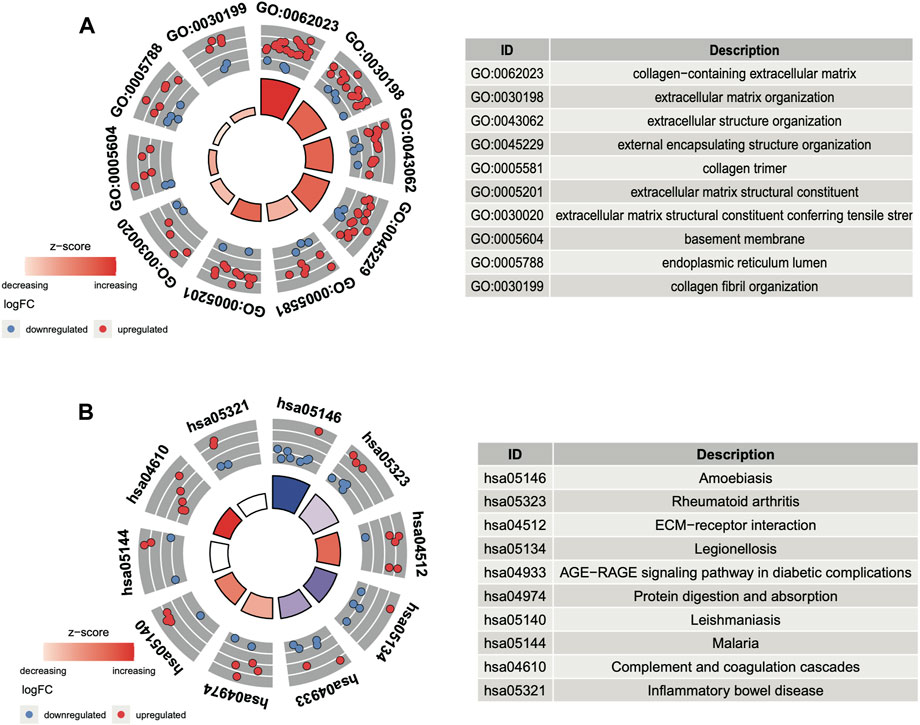
FIGURE 3. Functional analysis of DEGs. (A) Top 10 GO terms ranked by the p-value. (B) Top 10 KEGG pathways ranked by the p-value.
Construction of the PPI network of the DEGs
According to the common 156 DEGs with the same expression trend in both bulk datasets, a PPI network was established including 83 nodes and 167 interacting pairs (Supplementary Table S2). After degree analysis by Cytoscape, the top 10 genes in terms of degree named IL6, FOS, ITGAM, COL3A1, ACAN, HMOX1, COL2A1, TLR2, SOCS3, and CXCL2 were considered hub genes in the network (Figure 4A). It has been reported that they were all involved in progression intervertebral disc degeneration (Osuka et al., 2014; Lv et al., 2016; Johnson et al., 2017; Makino et al., 2017; Lin et al., 2018; Chen et al., 2019; Wu et al., 2019; He et al., 2021; Li et al., 2021). As shown in Figure 4, the hub genes in the top 10 GO terms (Figure 4B) and KEGG pathway (Figure 4C) were closely associated with the collagen-containing extracellular matrix, ECM−receptor interaction, and immune regulation.
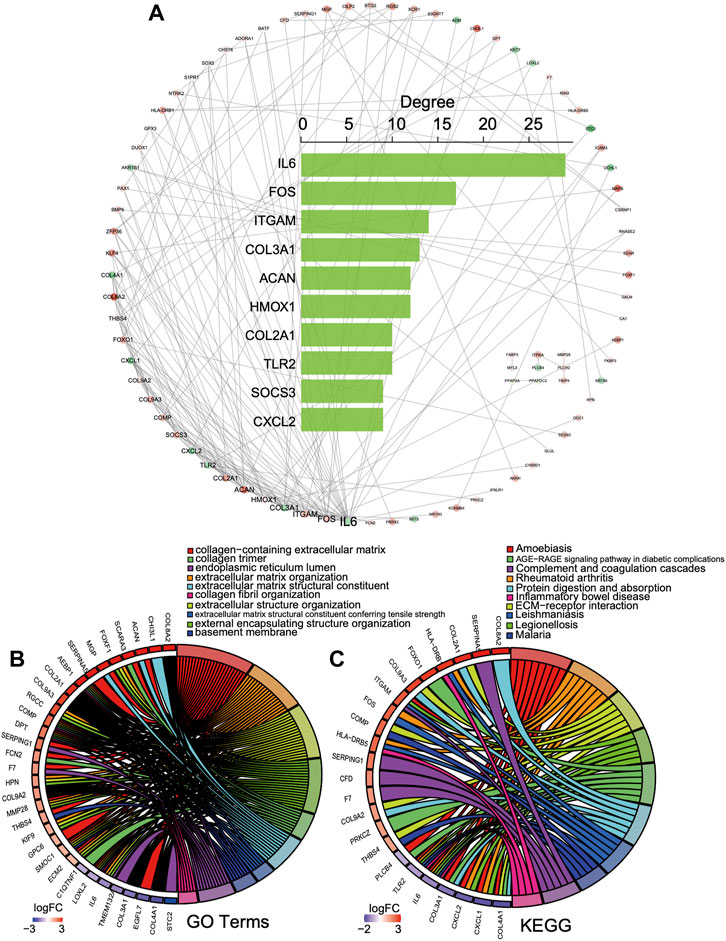
FIGURE 4. PPI network of the DEGs. (A) PPI network and top 10 gene degree distribution: red represents upregulated, and blue represents downregulated. (B, C) Hub gene relations in the top 10 GO terms and KEGG pathways.
Construction of the dysregulated competing endogenous RNA between mRNAs and miRNAs
First, we obtained 5,705 human miRNA–DEG relationships from miRTarbase and starBase based on the 156 common DEGs. Second, the hypergeometric test was used to evaluate the number of shared miRNAs among the DEGs with p < 0.05, and the dysregulated ceRNA network was constructed which contained 308 miRNAs, 133 DEGs, and 1728 interacting pairs (Supplementary Figure S2A). Finally, the expression correlation score of genes was calculated by the Pearson test, and we set the p-value < 0.05, and 36 ceRNA axes were screened (Figure 5A, Supplementary Table S3). We used these qualified relationships to construct the ceRNA network and visualize it by Cytoscape software. This ceRNA network includes 39 edges with 30 DEGs and 12 miRNA nodes. The triangles nodes represent DEGs, ellipse nodes represent miRNAs, and the color gradient indicates the difference in expression (Figure 5A). The DEGs that shared same miRNAs show strong positive correlation of expression in Figure 5B in red color. At the same time, some DEGs are clustered in the specific set, proving that they may have synergistic functions, such as ZFP36, SOCS3, and FOS. On the contrary, the miRNA and target DEGs show strong negative correlation of expression in Figure 5B in blue color. All the aforementioned results can prove the reliability of the ceRNA network and the regulatory function of the miRNA in the target gene directly or indirectly.
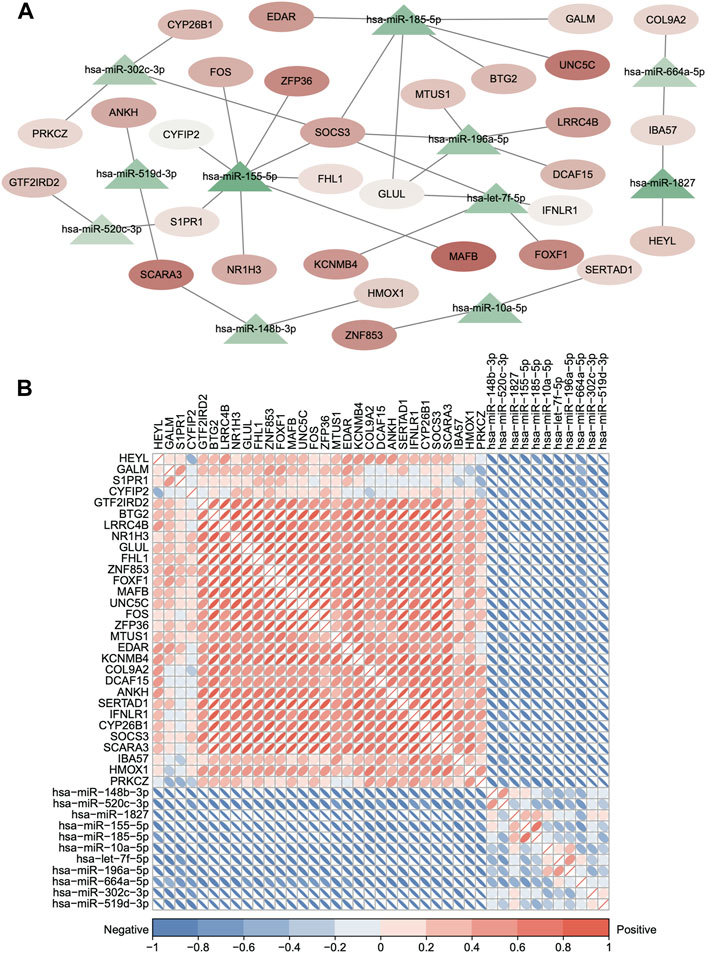
FIGURE 5. Dysregulated ceRNA between mRNAs and miRNAs in IDD. (A) Dysregulated ceRNA network between miRNAs and differentially expressed mRNAs. (hypergeometric test p < 0.05 and Pearson test p < 0.05). Red represents upregulated, and blue represents downregulated. The triangle nodes represent DEGs, and ellipse nodes represent miRNAs; the color gradient indicates the difference in expression. (B) Clustered corrplot of miRNAs and differentially expressed mRNAs in the ceRNA network. The color gradient represents the expression correlation score between genes.
Integrating single-cell and bulk transcriptome analyses
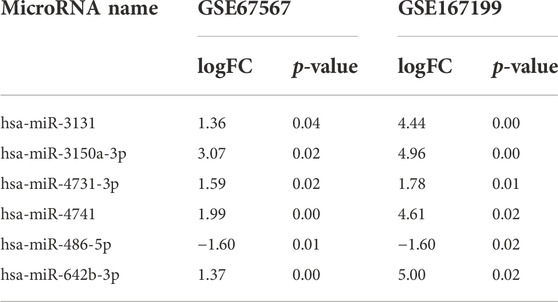
Single-cell clustering in the annulus fibrosus (AF) and NP of patients with IDD is shown in Figure 6A. We obtained 115 marker genes (AF = 30, AFH = 18, NPH = 26, and NPD = 41, Supplementary Table S4) based on 3,142 AF cells from degenerated discs (AFD), 3,226 AF cells from non-degenerated discs (AFH), 3,678 NP cells from degenerated discs (NPD), and 3,955 NP cells from non-degenerated discs (NPH). A heatmap of the top 10 marker gene expressions in each cell group is shown in Figure 6B. Intersection analysis of the marker genes was conducted with the aforementioned 156 bulk DEGs and showed three common differential genes including COL2A1 (Figure 7A), PAX1 (Figure 7B), and ZFP36L2 (Figure 7C).
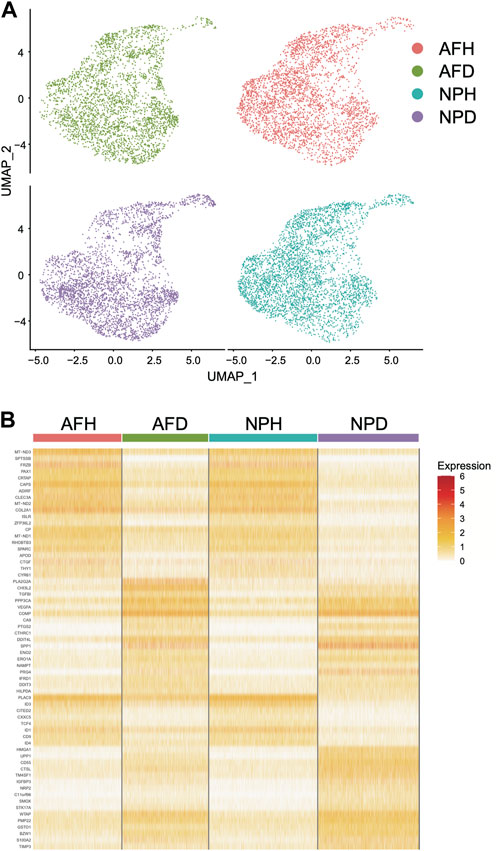
FIGURE 6. Single-cell transcriptome analysis. (A) Single-cell clustering in the annulus fibrosus (AF) and nucleus pulposus (NP) of patients with intervertebral disc degeneration (IDD). (B) Heatmap of marker gene expression in each cell group (top 10).
Discussion
Intervertebral disc degeneration (IDD) is an age-related chronic degeneration with NP cell senescence and imbalance between extracellular matrix (ECM) catabolism and synthesis (Shao et al., 2019; Chen et al., 2021a; Chen et al., 2021b). With the disc degeneration, the synthesis of proteoglycan and collagen I and degradation of the ECM are increased (Chen et al., 2020b; Cui et al., 2020; Gao et al., 2020). Although IDD is an age-induced condition, it is also influenced by other risk factors including genetic susceptibility, aging, smoking, heavy load work, and body weight (Wang et al., 2017b; Wang et al., 2018b; Xiao et al., 2020). Several biotechnologies were shown to abate IDD by inducing ECM regeneration and repair (Wang et al., 2018c; Yang et al., 2020). Thus, it is important to find effective diagnostic biomarkers and new therapeutic strategies for IDD.
Recently, several studies have demonstrated that miRNAs play important roles in many biological processes such as cell apoptosis, ECM degradation, and cell proliferation (Wang et al., 2021b; Yu et al., 2021; Zhou et al., 2021). For example, Zhang et al. (2022) demonstrated that miR-4478 induced apoptosis of NP cells through targeting MTH1. Cui et al. (2022) showed that miR-760 prevented IDD development by targeting the NF-κB signaling pathway and MyD88. Zhou et al. (2022) showed that miR-206 ameliorated IDD through targeting GJA1. Yu et al. (2022) demonstrated that miR-137 suppressed ECM degradation and inflammatory response in lipopolysaccharide-induced NP cells through targeting ACVR1. Chen et al. (2022a) indicated that miR-1260b ameliorated LPS-induced IDD development by targeting TCF7L2. Cao et al. (2021) demonstrated that miR-200c-3p inhibited IDD development through targeting the RAP2C/ERK signaling pathway. Guo et al. (2021) demonstrated that miR-502 inhibited TNF-α-induced NP cell apoptosis through targeting TARF2. These results suggested that miRNAs play important roles in the development of IDD. We found six significantly differentially expressed miRNAs in both datasets, hsa-miR-3131, hsa-miR-3150a-3p, hsa-miR-4731-3p, hsa-miR-4741, hsa-miR-486-5p, and hsa-miR-642b-3p. The role of these miRNAs in IDD has not yet been proven in other studies.
Through Venn analysis, we found 156 DEGs had same expression trends in both bulk datasets. Further GO and KEGG functional analyses showed these dysregulators were mostly enriched in the collagen-containing extracellular matrix, cartilage development, chondrocyte differentiation, and immune response pathways. Moreover, it showed that several dysregulated genes played important roles in the PPI network, including genes that were related to cytokines (IL6, SOCS3, CXCL2, CXCL1, and TLR2), complements (ITGAM), ECM composition including the collagen family, and aggrecan (COL3A1, COL2A1, COL9A2, COL9A3, COL4A1, COL8A2, ACAN, and COMP), as well as cell proliferation or differentiation transcription factors (FOS and FOXO1). In previous studies, collagen and aggrecan turnover in ECM was observed in degenerated discs (Sivan et al., 2014; Trefilova et al., 2021). COMP, the cartilage oligomeric matrix protein, was also found as a marker of IVDD (Qi et al., 2021). Several cytokines including IL1, IL5, IL6, IL7, IL10 TLR4, and TNF-alpha were found to promote extracellular matrix degradation, leading to the degeneration of IVD (Klawitter et al., 2014; Osuka et al., 2014; Risbud and Shapiro, 2014). Macrophage infiltration was also observed in herniated discs, which may cause a variable level of ITGAM expression (Kawaguchi et al., 2001). FoxO1a was found to mediate apoptosis in disc degeneration (Jing et al., 2021). FOS was proved to regulate inflammatory response in IVDD (Makino et al., 2017).
The GO and KEGG analyses in our study also demonstrated multiple pathways related to ECM metabolism and immune response. However, KEGG analysis also demonstrated pathways involving parasites (amoebiasis, malaria, and leishmaniasis) or bacterial infection (legionellosis). The reason was that IVDD and parasite or certain bacterial infection have overlap in pathways that involves cytokine release (Cruz Cubas et al., 1994; Friedman et al., 1998; García-Zepeda et al., 2007; Samant et al., 2021) and ECM degradation (Meza, 2000).
The mRNA–miRNA–mRNA pattern of the ceRNA network has been found in cancers (Tay et al., 2014; Xu et al., 2015); however, it has not been observed in IDD yet. We constructed a potentially dysregulated ceRNA network between mRNAs and miRNAs related to IDD based on microRNA target information and co-expression analysis of RNA profiles and identified 36 new ceRNA axes (Supplementary Table S3 and Figure 5A). Some genes involved in the axes were proved functional in IDD, including ZFP36/miR-155-5p/FOS, BTG2/hsa-miR-185-5p/SOCS3, and COL9A2/hsa-miR-664a-5p/IBA57 (Osuka et al., 2014; Meng et al., 2016; Makino et al., 2017; Xu et al., 2022) by regulating ECM metabolism and immune response; however, the function of more axes still remains unclear.
Single-cell transcriptome sequencing is a new method to delineate heterogeneous tissues at the single-cell level (Chen et al., 2022b; Galuzzi et al., 2022; Muhl et al., 2022). By integrating bulk and single-cell transcriptome data analyses, a total of three marker genes, COL2A1, PAX1, and ZFP36L2, were identified. Pax1 was found to be an important transcription factor in the development of the intervertebral disc (Sivakamasundari et al., 2017; Nakamichi and Asahara, 2020). However, no study was found describing how ZFP36 relates to disc degeneration. ZFP36 was differentially expressed and was a hub gene in GO and KEGG analyses. Its ligand, ZFP36L2, was found differentially expressed in single-cell transcriptome analysis. Zinc finger protein 36 (ZFP36) family proteins are RNA-binding proteins involved in mRNA metabolism pathways (Makita et al., 2021). It plays a significant role in regulating immune responses and inflammatory diseases (Lai et al., 1999) and cancers (Schuschel et al., 2020). This is a pathway worthy of further investigation.
In summary, the key genes and the new ceRNA crosstalk we identified in intervertebral disc degeneration may provide new understanding of the molecular mechanism of IDD pathogenesis from the ceRNA perspective and may provide new targets for treating IDD.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XL, YA, QW, and XH conducted this study. XL wrote the manuscript. YA, QW, and XH gave advice on the interpretation of data and the constitution of the study. XL analyzed the data. All authors have read and approved the final manuscript.
Funding
This study was funded by the Beijing Hospitals Authority Youth Programme (code: QML20200404), the Beijing Nova Program (Z201100006820136), and the Beijing Municipal Health Commission (BJRITO-RDP-2023).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1083983/full#supplementary-material
References
Andreucci, E., Ruzzolini, J., Bianchini, F., Versienti, G., Biagioni, A., Lulli, M., et al. (2022). miR-214-Enriched extracellular vesicles released by acid-adapted melanoma cells promote inflammatory macrophage-dependent tumor trans-endothelial migration. Cancers (Basel) 14 (20), 5090. doi:10.3390/cancers14205090
Cao, J., Jiang, M., Ren, H., and Xu, K. (2021). MicroRNA200c3p suppresses intervertebral disc degeneration by targeting RAP2C/ERK signaling. Mol. Med. Rep. 24 (6), 865. doi:10.3892/mmr.2021.12505
Chen, J., Mei, Z., Huang, B., Zhang, X., Liu, J., Shan, Z., et al. (2019). IL-6/YAP1/β-catenin signaling is involved in intervertebral disc degeneration. J. Cell. Physiol. 234 (5), 5964–5971. doi:10.1002/jcp.27065
Chen, L., Zhang, X., Zhang, Q., Zhang, T., Xie, J., Wei, W., et al. (2022). A necroptosis related prognostic model of pancreatic cancer based on single cell sequencing analysis and transcriptome analysis. Front. Immunol. 13, 1022420. doi:10.3389/fimmu.2022.1022420
Chen, S., Shi, G., Zeng, J., Li, P. H., Peng, Y., Ding, Z., et al. (2022). MiR-1260b protects against LPS-induced degenerative changes in nucleus pulposus cells through targeting TCF7L2. Hum. Cell 35 (3), 779–791. doi:10.1007/s13577-021-00655-4
Chen, W., Li, S., and Zhang, F. (2021). Role of lncRNA XIST/microRNA-19/PTEN network in autophagy of nucleus pulposus cells in intervertebral disc degeneration via the PI3K/Akt signaling pathway. Cell Cycle 20, 1629–1641. doi:10.1080/15384101.2021.1924450
Chen, X., Gu, M., and Zhang, X. (2020). Circular RNAs in compression-induced intervertebral disk degeneration. EBioMedicine 54, 102720. doi:10.1016/j.ebiom.2020.102720
Chen, X., Li, Z., Xu, D., and Li, S. (2020). LINC01121 induced intervertebral disc degeneration via modulating miR-150-5p/MMP16 axis. J. Gene Med. 22 (10), e3231. doi:10.1002/jgm.3231
Chen, Y., Wu, Y., Chen, R., Xu, C., and Chen, Q. (2021). LncRNA LINC00324 is upregulated in intervertebral disk degeneration and upregulates FasL in nucleus pulposus cells. Mol. Cell. Biochem. 476 (5), 1995–2000. doi:10.1007/s11010-021-04058-9
Cheng, X., Zhang, L., Zhang, K., Zhang, G., Hu, Y., Sun, X., et al. (2018). Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 77 (5), 770–779. doi:10.1136/annrheumdis-2017-212056
Cruz Cubas, A. B., Gentilini, M., and Monjour, L. (1994). Cytokines and T-cell response in malaria. Biomed. Pharmacother. 48 (1), 27–33. doi:10.1016/0753-3322(94)90187-2
Cui, S., Liu, Z., Tang, B., Wang, Z., and Li, B. (2020). LncRNA MAGI2-AS3 is down-regulated in intervertebral disc degeneration and participates in the regulation of FasL expression in nucleus pulposus cells. BMC Musculoskelet. Disord. 21 (1), 149. doi:10.1186/s12891-020-3086-y
Cui, X., Li, Y., Bao, J., Wang, K., and Wu, X. (2022). Downregulation of miR-760 causes human intervertebral disc degeneration by targeting the MyD88/nuclear factor-kappa B signaling pathway. Front. Bioeng. Biotechnol. 10, 813070. doi:10.3389/fbioe.2022.813070
Friedman, H., Yamamoto, Y., Newton, C., and Klein, T. (1998). Immunologic response and pathophysiology of Legionella infection. Semin. Respir. Infect. 13 (2), 100–108.
Galuzzi, B. G., Vanoni, M., and Damiani, C. (2022). Combining denoising of RNA-seq data and flux balance analysis for cluster analysis of single cells. BMC Bioinforma. 23, 445. doi:10.1186/s12859-022-04967-6
Gao, D., Hao, L., and Zhao, Z. (2020). Long non-coding RNA PART1 promotes intervertebral disc degeneration through regulating the miR93/MMP2 pathway in nucleus pulposus cells. Int. J. Mol. Med. 46 (1), 289–299. doi:10.3892/ijmm.2020.4580
García-Zepeda, E. A., RojAs-Lopez, A., Esquivel-VelazquezM., , and Ostoa-Saloma, P. (2007). Regulation of the inflammatory immune response by the cytokine/chemokine network in amoebiasis. Parasite Immunol. 29 (12), 679–684. doi:10.1111/j.1365-3024.2007.00990.x
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Li, H. R., et al. (2020). The circular RNA circ-GRB10 participates in the molecular circuitry inhibiting human intervertebral disc degeneration. Cell Death Dis. 11 (8), 612. doi:10.1038/s41419-020-02882-3
Guo, Z., Gao, W. S., Wang, Y. F., Gao, F., Wang, W., and Ding, W. Y. (2021). MiR-502 suppresses TNF-alpha-induced nucleus pulposus cell apoptosis by targeting TARF2. Biomed. Res. Int. 2021, 5558369. doi:10.1155/2021/5558369
Han, Z., Wang, J., Gao, L., Wang, Q., and Wu, J. (2018). Aberrantly expressed messenger RNAs and long noncoding RNAs in degenerative nucleus pulposus cells co-cultured with adipose-derived mesenchymal stem cells. Arthritis Res. Ther. 20 (1), 182. doi:10.1186/s13075-018-1677-x
Hao, Y., Hao, S., Andersen-Nissen, E., Mauck, W. M., Zheng, S., Butler, A., et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184 (13), 3573–3587 e29. doi:10.1016/j.cell.2021.04.048
He, R., Wang, Z., Cui, M., Liu, S., Wu, W., Chen, M., et al. (2021). HIF1A Alleviates compression-induced apoptosis of nucleus pulposus derived stem cells via upregulating autophagy. Autophagy 17 (11), 3338–3360. doi:10.1080/15548627.2021.1872227
Huang, H., Xing, D., Zhang, Q., Li, H., Lin, J., He, Z., et al. (2021). LncRNAs as a new regulator of chronic musculoskeletal disorder. Cell Prolif. 54, e13113. doi:10.1111/cpr.13113
Huang, H. Y., Lin, Y. C. D., Cui, S., Huang, Y., Tang, Y., Xu, J., et al. (2022). miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 50 (D1), D222–D230. doi:10.1093/nar/gkab1079
Jiang, Z., Zeng, Q., Li, D., Ding, L., Lu, W., Bian, M., et al. (2020). Long noncoding RNA MALAT1 promotes high glucoseinduced rat cartilage endplate cell apoptosis via the p38/MAPK signalling pathway. Mol. Med. Rep. 21 (5), 2220–2226. doi:10.3892/mmr.2020.11009
Jing, D., Wu, W., Deng, X., Peng, Y., Yang, W., Huang, D., et al. (2021). FoxO1a mediated cadmium-induced annulus fibrosus cells apoptosis contributes to intervertebral disc degeneration in smoking. J. Cell. Physiol. 236 (1), 677–687. doi:10.1002/jcp.29895
Johnson, Z. I., Doolittle, A. C., Snuggs, J. W., Shapiro, I. M., Le Maitre, C. L., and Risbud, M. V. (2017). TNF-alpha promotes nuclear enrichment of the transcription factor TonEBP/NFAT5 to selectively control inflammatory but not osmoregulatory responses in nucleus pulposus cells. J. Biol. Chem. 292 (42), 17561–17575. doi:10.1074/jbc.M117.790378
Kanehisa, M., and Goto, S. (2000). Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kawaguchi, S., YamashiTa, T., YoKogushi, K., Murakami, T., Ohwada, O., and SatoN., (2001). Immunophenotypic analysis of the inflammatory infiltrates in herniated intervertebral discs. Spine (Phila Pa 1976) 26 (11), 1209–1214. doi:10.1097/00007632-200106010-00008
Klawitter, M., Hakozaki, M., Kobayashi, H., Krupkova, O., Quero, L., Ospelt, C., et al. (2014). Expression and regulation of toll-like receptors (TLRs) in human intervertebral disc cells. Eur. Spine J. 23 (9), 1878–1891. doi:10.1007/s00586-014-3442-4
Lai, W. S., Carballo, E., Strum, J. R., Kennington, E. A., Phillips, R. S., and Blackshear, P. J. (1999). Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19 (6), 4311–4323. doi:10.1128/mcb.19.6.4311
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97. doi:10.1093/nar/gkt1248
Li, N., Liu, T., Guo, F., Yang, J., Shi, Y., Wang, S., et al. (2022). Identification of long non-coding RNA-microRNA-mRNA regulatory modules and their potential roles in drought stress response in wheat (Triticum aestivum L.). Front. Plant Sci. 13, 1011064. doi:10.3389/fpls.2022.1011064
Li, Z., Chen, X., Xu, D., Li, S., Chan, M. T. V., and Wu, W. K. K. (2019). Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 52 (6), e12704. doi:10.1111/cpr.12704
Li, Z., Sun, Y., He, M., and Liu, J. (2021). Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing. Bioengineered 12 (1), 1026–1039. doi:10.1080/21655979.2021.1899533
Lin, Q., Hou, S., Dai, Y., Jiang, N., and Lin, Y. (2019). LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson's disease through RAB3IP. Biol. Chem. 400 (9), 1217–1228. doi:10.1515/hsz-2018-0431
Lin, Y., Jiao, Y., Yuan, Y., Zhou, Z., Zheng, Y., Xiao, J., et al. (2018). Propionibacterium acnes induces intervertebral disc degeneration by promoting nucleus pulposus cell apoptosis via the TLR2/JNK/mitochondrial-mediated pathway. Emerg. Microbes Infect. 7 (1), 1. doi:10.1038/s41426-017-0002-0
Lv, F. J., Peng, Y., Lim, F. L., Sun, Y., LvM., , Zhou, L., et al. (2016). Matrix metalloproteinase 12 is an indicator of intervertebral disc degeneration co-expressed with fibrotic markers. Osteoarthr. Cartil. 24 (10), 1826–1836. doi:10.1016/j.joca.2016.05.012
Makino, H., Seki, S., Yahara, Y., Shiozawa, S., Aikawa, Y., Motomura, H., et al. (2017). A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci. Rep. 7 (1), 16983. doi:10.1038/s41598-017-17289-y
Makita, S., Takatori, H., and Nakajima, H. (2021). Post-transcriptional regulation of immune responses and inflammatory diseases by RNA-binding ZFP36 family proteins. Front. Immunol. 12, 711633. doi:10.3389/fimmu.2021.711633
Meng, T., Ren, Q., Wang, J. M., Shi, H., Zhang, S. T., and Liu, M. T. (2016). Association between COL9A2 Gln326Arg mutations and the development of intervertebral disc disease in a Chinese population. Genet. Mol. Res. 15 (4), 15048958. doi:10.4238/gmr15048958
Meza, I. (2000). Extracellular matrix-induced signaling in entamoeba histolytica: Its role in invasiveness. Parasitol. Today 16 (1), 23–28. doi:10.1016/s0169-4758(99)01586-0
Muhl, L., Mocci, G., Pietila, R., Liu, J., He, L., Genove, G., et al. (2022). A single-cell transcriptomic inventory of murine smooth muscle cells. Dev. Cell 57 (20), 2426–2443 e6. doi:10.1016/j.devcel.2022.09.015
Nakamichi, R., and Asahara, H. (2020). The transcription factors regulating intervertebral disc development. JOR Spine 3 (1), e1081. doi:10.1002/jsp2.1081
Niu, R. Z., Wang, L. Q., Yang, W., Sun, L. Z., Tao, J., Sun, H., et al. (2022). MicroRNA-582-5p targeting Creb1 modulates apoptosis in cardiomyocytes hypoxia/reperfusion-induced injury. Immun. Inflamm. Dis. 10 (11), e708. doi:10.1002/iid3.708
Niu, X., Pu, S., Ling, C., Xu, J., Wang, J., Sun, S., et al. (2020). lncRNA Oip5-as1 attenuates myocardial ischaemia/reperfusion injury by sponging miR-29a to activate the SIRT1/AMPK/PGC1α pathway. Cell Prolif. 53 (6), e12818. doi:10.1111/cpr.12818
Osuka, K., Usuda, N., Aoyama, M., Yamahata, H., Takeuchi, M., Yasuda, M., et al. (2014). Expression of the JAK/STAT3/SOCS3 signaling pathway in herniated lumbar discs. Neurosci. Lett. 569, 55–58. doi:10.1016/j.neulet.2014.03.045
Qi, D. D., Liu, Z. H., Wu, D. S., and Huang, Y. F. (2021). A study on COMP and CTX-II as molecular markers for the diagnosis of intervertebral disc degeneration. Biomed. Res. Int. 2021, 3371091. doi:10.1155/2021/3371091
Risbud, M. V., and Shapiro, I. M. (2014). Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 10 (1), 44–56. doi:10.1038/nrrheum.2013.160
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. doi:10.1093/nar/gkv007
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Samant, M., Sahu, U., Pandey, S. C., and Khare, P. (2021). Role of cytokines in experimental and human visceral Leishmaniasis. Front. Cell. Infect. Microbiol. 11, 624009. doi:10.3389/fcimb.2021.624009
Schuschel, K., Helwig, M., Huttelmaier, S., Heckl, D., Klusmann, J. H., and Hoell, J. I. (2020). RNA-binding proteins in acute leukemias. Int. J. Mol. Sci. 21 (10), E3409. doi:10.3390/ijms21103409
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shao, T., Hu, Y., Tang, W., Shen, H., Yu, Z., and Gu, J. (2019). The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene 715, 144029. doi:10.1016/j.gene.2019.144029
Sivakamasundari, V., Kraus, P., Sun, W., Hu, X., Lim, S. L., Prabhakar, S., et al. (2017). A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol. Open 6 (2), 187–199. doi:10.1242/bio.023218
Sivan, S. S., Wachtel, E., and Roughley, P. (2014). Structure, function, aging and turnover of aggrecan in the intervertebral disc. Biochim. Biophys. Acta 1840 (10), 3181–3189. doi:10.1016/j.bbagen.2014.07.013
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2022). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607. doi:10.1093/nar/gky1131
Tan, H., Zhao, L., Song, R., Liu, Y., and Wang, L. (2018). The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. Am. J. Physiol. Cell Physiol. 315 (1), C21–C27. doi:10.1152/ajpcell.00220.2017
Tan, L., Xie, Y., Yuan, Y., and Hu, K. (2021). LncRNA GAS5 as miR-26a-5p sponge regulates the PTEN/PI3K/akt Axis and affects extracellular matrix synthesis in degenerative nucleus pulposus cells in vitro. Front. Neurol. 12, 653341. doi:10.3389/fneur.2021.653341
Tang, N., Dong, Y., Xiao, T., and Zhao, H. (2020). LncRNA TUG1 promotes the intervertebral disc degeneration and nucleus pulposus cell apoptosis though modulating miR-26a/HMGB1 axis and regulating NF-κB activation. Am. J. Transl. Res. 12 (9), 5449–5464.
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 (7483), 344–352. doi:10.1038/nature12986
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 17 (5), 272–283. doi:10.1038/nrg.2016.20
Trefilova, V. V., Shnayder, N. A., Petrova, M. M., Kaskaeva, D. S., Tutynina, O. V., Petrov, K. V., et al. (2021). The role of polymorphisms in collagen-encoding genes in intervertebral disc degeneration. Biomolecules 11 (9), 1279. doi:10.3390/biom11091279
von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. (2003). String: A database of predicted functional associations between proteins. Nucleic Acids Res. 31 (1), 258–261. doi:10.1093/nar/gkg034
Wang, H., He, P., Pan, H., Long, J., Wang, J., Li, Z., et al. (2018). Circular RNA circ-4099 is induced by TNF-alpha and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 50 (4), 27–14. doi:10.1038/s12276-018-0056-7
Wang, H., Zhu, Y., Cao, L., Guo, Z., Sun, K., Qiu, W., et al. (2021). circARL15 plays a critical role in intervertebral disc degeneration by modulating miR-431-5p/DISC1. Front. Genet. 12, 669598. doi:10.3389/fgene.2021.669598
Wang, K., Song, Y., Liu, W., Wu, X., Zhang, Y., Li, S., et al. (2017). The noncoding RNA linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral disc degeneration through inhibiting ADAMTS5 expression. Clin. Sci. 131 (10), 965–979. doi:10.1042/CS20160918
Wang, L., Tang, R., Zhang, Y., Chen, S., Guo, Y., Wang, X., et al. (2021). PTH-induced EndMT via miR-29a-5p/GSAP/Notch1 pathway contributed to valvular calcification in rats with CKD. Cell Prolif. 54 (6), e13018. doi:10.1111/cpr.13018
Wang, X., Lv, G., Li, J., Wang, B., Zhang, Q., and Lu, C. (2017). LncRNA-RP11-296A18.3/miR-138/HIF1A pathway regulates the proliferation ECM synthesis of human nucleus pulposus cells (HNPCs). J. Cell. Biochem. 118 (12), 4862–4871. doi:10.1002/jcb.26166
Wang, X., Peng, L., Gong, X., Zhang, X., Sun, R., and Du, J. (2018). LncRNA-RMRP promotes nucleus pulposus cell proliferation through regulating miR-206 expression. J. Cell. Mol. Med. 22 (11), 5468–5476. doi:10.1111/jcmm.13817
Wang, X., Wang, B., Zou, M., Li, J., Lu, G., Zhang, Q., et al. (2018). CircSEMA4B targets miR-431 modulating IL-1β-induced degradative changes in nucleus pulposus cells in intervertebral disc degeneration via Wnt pathway. Biochim. Biophys. Acta. Mol. Basis Dis. 1864 (11), 3754–3768. doi:10.1016/j.bbadis.2018.08.033
Wu, Q., Mathers, C., Wang, E. W., Sheng, S., Wenkert, D., and Huang, J. H. (2019). TGF-beta initiates beta-catenin-mediated ctgf secretory pathway in old bovine nucleus pulposus cells: A potential mechanism for intervertebral disc degeneration. JBMR Plus 3 (2), e10069. doi:10.1002/jbm4.10069
Wu, T., Liu, W., Huang, S., Chen, J., He, F., Wang, H., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Mat. Today. Bio 2 (3), 100141. doi:10.1016/j.mtbio.2021.100141
Xi, Y., Jiang, T., Wang, W., Yu, J., Wang, Y., Wu, X., et al. (2017). Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci. Rep. 7 (1), 13234. doi:10.1038/s41598-017-13364-6
Xiao, L., Ding, B., Xu, S., Gao, J., Yang, B., Wang, J., et al. (2020). circRNA_0058097 promotes tension-induced degeneration of endplate chondrocytes by regulating HDAC4 expression through sponge adsorption of miR-365a-5p. J. Cell. Biochem. 121 (1), 418–429. doi:10.1002/jcb.29202
Xiao, M., Yang, S., Zhou, A., Li, T., Liu, J., Chen, Y., et al. (2022). MiR-27a-3p and miR-30b-5p inhibited-vitamin D receptor involved in the progression of tuberculosis. Front. Microbiol. 13, 1020542. doi:10.3389/fmicb.2022.1020542
Xu, H., Dong, R., Zeng, Q., Fang, L., Ge, Q., Xia, C., et al. (2022). Col9a2 gene deletion accelerates the degeneration of intervertebral discs. Exp. Ther. Med. 23 (3), 207. doi:10.3892/etm.2022.11130
Xu, J., Li, Y., Lu, J., Pan, T., Ding, N., Wang, Z., et al. (2015). The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 43 (17), 8169–8182. doi:10.1093/nar/gkv853
Yang, H., Wang, G., Liu, J., Lin, M., Chen, J., Fang, Y., et al. (2021). LncRNA JPX regulates proliferation and apoptosis of nucleus pulposus cells by targeting the miR-18a-5p/HIF-1α/Hippo-YAP pathway. Biochem. Biophys. Res. Commun. 566, 16–23. doi:10.1016/j.bbrc.2021.05.075
Yang, J., Li, C., Wei, H., Du, W., and Hu, Q. (2022). Upregulation of microRNA-762 suppresses the expression of GIPC3 in systemic lupus erythematosus and neuropsychiatric systemic lupus erythematosus. Immun. Inflamm. Dis. 10 (11), e719. doi:10.1002/iid3.719
Yang, P., Gao, R., Zhou, W., and Han, A. (2020). Protective impacts of circular RNA VMA21 on lipopolysaccharide-engendered WI-38 cells injury via mediating microRNA-142-3p. Biofactors 46 (3), 381–390. doi:10.1002/biof.1593
Yu, B., Zhu, Z., Shen, B., Lu, J., Guo, K., Zhao, W., et al. (2022). MicroRNA-137 inhibits the inflammatory response and extracellular matrix degradation in lipopolysaccharide-stimulated human nucleus pulposus cells by targeting activin a receptor type I. Bioengineered 13 (3), 6396–6408. doi:10.1080/21655979.2022.2042987
Yu, H., Wang, K., Liu, P., Luo, P., Zhu, D., Yin, J., et al. (2021). miR-4286 functions in osteogenesis and angiogenesis via targeting histone deacetylase 3 and alleviates alcohol-induced bone loss in mice. Cell Prolif. 54 (6), e13054. doi:10.1111/cpr.13054
Zhan, S., Wang, K., Xiang, Q., Song, Y., Li, S., Liang, H., et al. (2020). lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J. Cell. Physiol. 235 (3), 2195–2208. doi:10.1002/jcp.29129
Zhang, J., Liu, R., Mo, L., Liu, C., and Jiang, J. (2022). MiR-4478 accelerates nucleus pulposus cells apoptosis induced by oxidative stress by targeting MTH1. Spine (Phila Pa 1976). Publish Ahead of Print. doi:10.1097/BRS.0000000000004486
Zhang, Z., Huo, Y., Zhou, Z., Zhang, P., and Hu, J. (2021). Role of lncRNA PART1 in intervertebral disc degeneration and associated underlying mechanism. Exp. Ther. Med. 21 (2), 131. doi:10.3892/etm.2020.9563
Zhou, H., Li, X., Wu, R. X., He, X. T., An, Y., Xu, X. Y., et al. (2021). Periodontitis-compromised dental pulp stem cells secrete extracellular vesicles carrying miRNA-378a promote local angiogenesis by targeting Sufu to activate the Hedgehog/Gli1 signalling. Cell Prolif. 54 (5), e13026. doi:10.1111/cpr.13026
Zhou, P., Xu, P., Yu, W., and Li, H. (2022). MiR-206 improves intervertebral disk degeneration by targeting GJA1. J. Orthop. Surg. Res. 17 (1), 157. doi:10.1186/s13018-022-03044-1
Keywords: intervertebral disc degeneration, miRNAs, ceRNAs, single-cell transcriptome sequencing, mRNAs
Citation: Li X, An Y, Wang Q and Han X (2022) The new ceRNA crosstalk between mRNAs and miRNAs in intervertebral disc degeneration. Front. Cell Dev. Biol. 10:1083983. doi: 10.3389/fcell.2022.1083983
Received: 29 October 2022; Accepted: 18 November 2022;
Published: 02 December 2022.
Edited by:
Shuangfei Ni, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Cheng Huang, China-Japan Friendship Hospital, ChinaDerong Xu, Qingdao University, China
Copyright © 2022 Li, An, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingye Li, xingyeli@yeah.net
 Xingye Li
Xingye Li Yan An
Yan An