BAF (mSWI/SNF) complex regulates mediolateral cortical patterning in the developing forebrain
- 1Institute for Neuroanatomy, University Medical Center, Georg-August-University Goettingen, Goettingen, Germany
- 2Faculty of Biotechnology, Thai Nguyen University of Sciences, Thai Nguyen, Vietnam
- 3Department of Human Genetics, Ruhr University Bochum, Bochum, Germany
- 4Department of Anatomy and Molecular Embryology, Ruhr University Bochum, Bochum, Germany
- 5Tata Institute of Fundamental Research, Mumbai, India
- 6Max Planck Institute for Multidisciplinary Sciences, Goettingen, Germany
- 7Harvard Stem Cell Institute, Harvard Medical School, Boston, MA, United States
- 8German Primate Center-Leibniz Institute for Primate Research, Goettingen, Germany
Early forebrain patterning entails the correct regional designation of the neuroepithelium, and appropriate specification, generation, and distribution of neural cells during brain development. Specific signaling and transcription factors are known to tightly regulate patterning of the dorsal telencephalon to afford proper structural/functional cortical arealization and morphogenesis. Nevertheless, whether and how changes of the chromatin structure link to the transcriptional program(s) that control cortical patterning remains elusive. Here, we report that the BAF chromatin remodeling complex regulates the spatiotemporal patterning of the mouse dorsal telencephalon. To determine whether and how the BAF complex regulates cortical patterning, we conditionally deleted the BAF complex scaffolding subunits BAF155 and BAF170 in the mouse dorsal telencephalic neuroepithelium. Morphological and cellular changes in the BAF mutant forebrain were examined using immunohistochemistry and in situ hybridization. RNA sequencing, Co-immunoprecipitation, and mass spectrometry were used to investigate the molecular basis of BAF complex involvement in forebrain patterning. We found that conditional ablation of BAF complex in the dorsal telencephalon neuroepithelium caused expansion of the cortical hem and medial cortex beyond their developmental boundaries. Consequently, the hippocampal primordium is not specified, the mediolateral cortical patterning is compromised, and the cortical identity is disturbed in the absence of BAF complex. The BAF complex was found to interact with the cortical hem suppressor LHX2. The BAF complex suppresses cortical hem fate to permit proper forebrain patterning. We provide evidence that BAF complex modulates mediolateral cortical patterning possibly by interacting with the transcription factor LHX2 to drive the LHX2-dependent transcriptional program essential for dorsal telencephalon patterning. Our data suggest a putative mechanistic synergy between BAF chromatin remodeling complex and LHX2 in regulating forebrain patterning and ontogeny.
Introduction
Telencephalic patterning is crucial for the anatomical and functional designation of various aspects of the forebrain during embryogenesis. The dorsal telencephalon (dTel) is broadly organized into the (cerebral) cortex and the dorsal midline. The cortex is sub-divided into the neocortex and archicortex (the hippocampus), whereas the cortical hem and choroid plexus comprise the dorsal midline [reviewed in (Hébert and Fishell 2008)]. The cortical hem and anti-hem, localized at the border between the dTel and ventral telencephalon (vTel), play opposite instructive roles as telencephalic organizers via production of morphogens for the early molecular patterning of the brain [Reviewed in (Kiecker and Lumsden 2012)]. The commitment of portions of the telencephalic neuroepithelium to generate distinct structural and functional telencephalic regions, such as the neocortex and hippocampus, takes place early in forebrain development, i.e., E10.5–E12.5 in mouse (Levitt et al., 1997; Gitton et al., 1999; Mallamaci et al., 2000). The graded exposure of dTel progenitors to morphogens orchestrate forebrain patterning through their ability to regulate downstream patterning modulators, including transcription factors, that play instrumental roles in positional specification of telencephalic structures (Sur and Rubenstein 2005; Mallamaci and Stoykova 2006; Rash and Grove 2006; O'Leary and Sahara 2008).
The cortical hem is a telencephalic structure flanked by the hippocampal primordium and the choroid plexus. It is a prominent source of patterning signals, including those that belong to the Wingless/Int (WNT) (Grove et al., 1998) and the Bone morphogenetic protein (BMP) families (Furuta et al., 1997). Thus, the cortical hem performs crucial roles in both cortical and hippocampal patterning due to its morphogen enrichment (Grove et al., 1998; Lee et al., 2000; Mangale et al., 2008; Caronia-Brown et al., 2014). Furthermore, the cortical hem generates reelin-producing Cajal-Retzius cells that migrate tangentially to populate the marginal zone of the cortex and perform important roles in neocortical and hippocampal lamination (Rice and Curran 2001; Meyer et al., 2002; Takiguchi-Hayashi et al., 2004; Bielle et al., 2005; Forster et al., 2006).
Precise molecular regulation and proper cytoarchitecture are prerequisites for the establishment of the various cortical domains during dTel patterning [reviewed in (Subramanian and Tole 2009)]. A few factors, such as FOXG1, EMX1/2, PAX6, LHX2, FGF, LMX1A and GLI3 (Grove et al., 1998; Bishop et al., 2000; Mallamaci et al., 2000; Bulchand et al., 2001; Shimogori et al., 2004; Shinozaki et al., 2004; Kimura et al., 2005; Muzio and Mallamaci 2005; Mangale et al., 2008; Chizhikov et al., 2010; Godbole et al., 2018), and components of signaling pathways (WNT, FGF, BMP) (Forster et al., 2006; Hébert and Fishell 2008) are fundamental in directing cortical patterning. Despite the essential role of these factors for correct forebrain patterning, the interconnecting mechanistic network involved is largely unknown.
The Brg1/Brm-associated factor (BAF) is the mammalian SWI/SNF complex which is a multi-subunit ATP-dependent chromatin remodeler. It is capable of re-structuring chromatin through histone mobilization and/or recruitment of epigenetic cofactors to modify the epigenetic landscape of chromatin (Sokpor et al., 2021b), leading to regulation of gene expression during brain development [reviewed in (Sokpor et al., 2017)]. We previously developed mouse models of inactive BAF complex. This was achieved via double conditional knockout (dcKO) of the scaffolding BAF subunits BAF155 and BAF170 in cortical progenitors, which leads to disassembly of other subunits in the BAF complex and their subsequent elimination by the ubiquitin-proteasome machinery (Narayanan et al., 2015; Bachmann et al., 2016; Nguyen et al., 2016; Nguyen et al., 2018). The aberration of the entire BAF complex function in dcKO or partly in single knockouts of BAF155 or BAF170, disturbs several aspects of neural development, including increase in progenitor proliferation and delamination, reduced neural cell differentiation, delayed/stalled neuronal migration, and decrease in oligodendrogenesis (Tuoc et al., 2013; Narayanan et al., 2015; Bachmann et al., 2016; Nguyen et al., 2016; Tuoc et al., 2017; Narayanan et al., 2018; Nguyen et al., 2018; Abbas et al., 2021; Sokpor et al., 2021a).
Interestingly, the BAF complex and the LIM homeodomain transcription factor LHX2 appear to share common developmental functions. Thus, ablation of BAF complex leads to neurodevelopmental phenotypes similar to that of Lhx2 deletion in several neural tissues, including the eye and olfactory neuroepithelia. Disturbance of Lhx2 expression causes misregulation of proliferation and differentiation of the retinal neural progenitors, and defective gliogenesis (Lamba et al., 2008; Tétreault et al., 2009; Balasubramanian et al., 2014; de Melo et al., 2016; de Melo et al., 2018), a phenotype partly recapitulated by BAF complex manipulation in the developing human fetal retina (Lamba et al., 2008). Similarly, the olfactory epithelium of Lhx2 mutant mice have abnormal progenitor differentiation, deprivation of olfactory receptor gene expression, and loss of neuronal identity leading to perturbed development and diversification of olfactory receptor neurons (Hirota and Mombaerts 2004; Kolterud et al., 2004). Likewise, conditional inactivation of the BAF complex in the olfactory epithelium in mice resulted in loss of LHX2-expressing olfactory receptor neurons and overall dysgenesis of the olfactory epithelium (Bachmann et al., 2016). Notably, both BAF complex and LHX2 are known to regulate neurogenesis during forebrain development in a WNT-signaling dependent manner (Hsu et al., 2015; Nguyen et al., 2018). As a result, the respective mutants displayed reduced cortical size due to decreased cortical neurogenesis (Hsu et al., 2015; Nguyen et al., 2018).
In this study, we show that the BAF complex is a determinant of cortical identity and a suppressor of cortical hem enlargement during forebrain development. Interestingly, we found that the BAF complex interacts with LHX2 and is required for normal LHX2 transcriptional activity in the dTel, suggesting their functional interaction to effect correct mediolateral patterning of the developing forebrain. Particularly, loss of BAF complex resulted in an expansion of the cortical hem at the expense of the medial and dorsolateral cortical primordia, leading to distortion of mediolateral cortical patterning. Additionally, we postulate an evolutionary/developmental significance of the differential expression of BAF complex subunits in mouse and the marmoset cortex and hem. These results highlight the neurodevelopmental role of the BAF complex in regulating the anatomical (regional) and functional organization of the mammalian cerebral cortex.
Materials and methods
Animals
Floxed BAF155 (Choi et al., 2012), floxed BAF170 (Tuoc et al., 2013), FoxG1-Cre (Hebert and McConnell 2000), Emx1-Cre (Gorski et al., 2002), and hGFAP-Cre mice (Zhuo et al., 2001) were maintained in a C57BL6/J background (license numbers 14/1636 and 16/2330).
Marmoset monkey (Callithrix jacchus) were obtained from the self-sustaining colony of the German Primate Center (Deutsches Primatenzentrum; DPZ) and housed according to the standard German Primate Center practice for common marmoset monkeys. Embryonic and fetal stages were obtained surgically by hysterotomy, or hysterectomy (license numbers 42502-04-12/0708 and 42502-04-16/2129) as described previously (Aeckerle et al., 2015; Wolff et al., 2019). All surgical work on the monkeys were performed by a veterinarian with several years of experience in handling and operating marmoset monkeys. A detailed description of experiment with marmoset monkey is provided in Supplementary Material. Animals were handled in accordance with the German Animal Protection Law.
Plasmids and antibodies
A list of plasmids and antibodies with detailed descriptions is provided in Supplementary Material.
In utero electroporation, and in vivo (LHX2) and in vitro (LHX2/PAX6) transcriptional activity assay
In vivo and in vitro transcriptional activity assay were performed as previously described (Pinon et al., 2008; Nguyen et al., 2018). Briefly, LHX2 transcriptional activity in vivo was monitored by electroporating brains of E12.5 BAF155fl/fl;BAF170fl/fl embryos with a Cre plasmid (or empty plasmid as a control) and the reporter constructs pGL3-5xLhx2BS-luciferase or pGL3-luciferase (Folgueras et al., 2013) together with pRL-TK constructs at a 5:1:0.3 ratio. LHX2 transcriptional activity was measured in the E14.5 mouse cortex. For in vitro assay, Neuro2A cells at 1×105 per well density were plated into 24-well plates. Cells were transfected with 0.8 μg of shRNA plasmids (shBAF155, shBAF170) along with 50 ng of Pax6-luc (Kammandel et al., 1999) and 10 ng of pRL-TK. Two (2) days post-transfection, cells were then collected for Pax6 transcriptional activity measurement. In all cases, firefly luciferase activity was normalized to that of Renilla luciferase.
CoIP/mass spectrometry, RNA sequencing
Detailed descriptions were provided previously (Narayanan et al., 2015).
Immunohistochemistry
After blocking with 5% goat or donkey normal serum, coronal sections of brain or whole head were incubated overnight with primary antibody at 4°C, and the signal was detected with a fluorescent secondary antibody (Alexa Fluor; 1:400; Invitrogen).
In situ hybridization
ISH was carried out as described previously (Tuoc et al., 2009).
3D reconstruction
3D images of the cortical hem were constructed using Neurolucida software version 11.03. Every third consecutive sections (10 μ each) of BAF mutant and control brains were imaged in rostro-caudal order. Contours were drawn in each section based on the expression of cortical hem-specific markers. The 3D reconstruction was produced from whole-stack contours. The contours were placed into sets for left and right cortical hem as depicted in Supplementary Movie S1. The hem volume estimation was done by using Neurolucida Explorer v. 11.03.
Cell counts and quantitative analysis of IHC signal intensity
Immunostaining in IHC images was quantified using anatomically matched forebrain sections. Nucleus-marker positive cells within the pallium of confocal images were counted for comparison. In most cases, cell counts from six matched sections were averaged from three biological replicates (control/dcKO pallium). In many cases, the number of lineage marker cells was quantified using total marker-positive cells alone, or by normalizing to the total number of DAPI+ (nucleus-stained) cells using the following equation: Normalized number = marker-positive cell number/DAPI + cell number. Statistical analyses of histological data were performed using unpaired Student’s t-test. All bar graphs are plotted as means ± SD. All statistical tests are two-tailed, and p-values are considered to be significant for α = 0.05.
Imaging, quantification, and statistical analyses
All images were acquired with standard (Leica DM 6000) and confocal (Leica TCS SP5) fluorescence microscopes. Images were further analyzed with Adobe Photoshop. IHC and ISH signal intensities were quantified by using ImageJ software. Statistical analyses were done using Student’s t-test. All graphs are plotted as mean ± SD.
Results
Expression of BAF complex in the embryonic mammalian dorsal telencephalon
The expression of BAF complex in the early developing mouse dTel was analyzed by immunohistochemical (IHC) staining of the scaffolding BAF complex subunits BAF155 and BAF170, and an additional BAF complex subunit BAF60a, which together are indicative of BAF complex presence (Narayanan et al., 2015). The examined BAF subunits were observed to be expressed throughout the E11.5 (Figure 1A) and E13.5 control mouse dTel (Supplementary Figure S1A) without an obvious or discernable gradient pattern of expression. The consistency of BAF complex expression is maintained laterally and continues into the vTel. Likewise, the dorsal telencephalic midline structures, cortical hem and choroid plexus, display uniform BAF complex expression (Figure 1A).
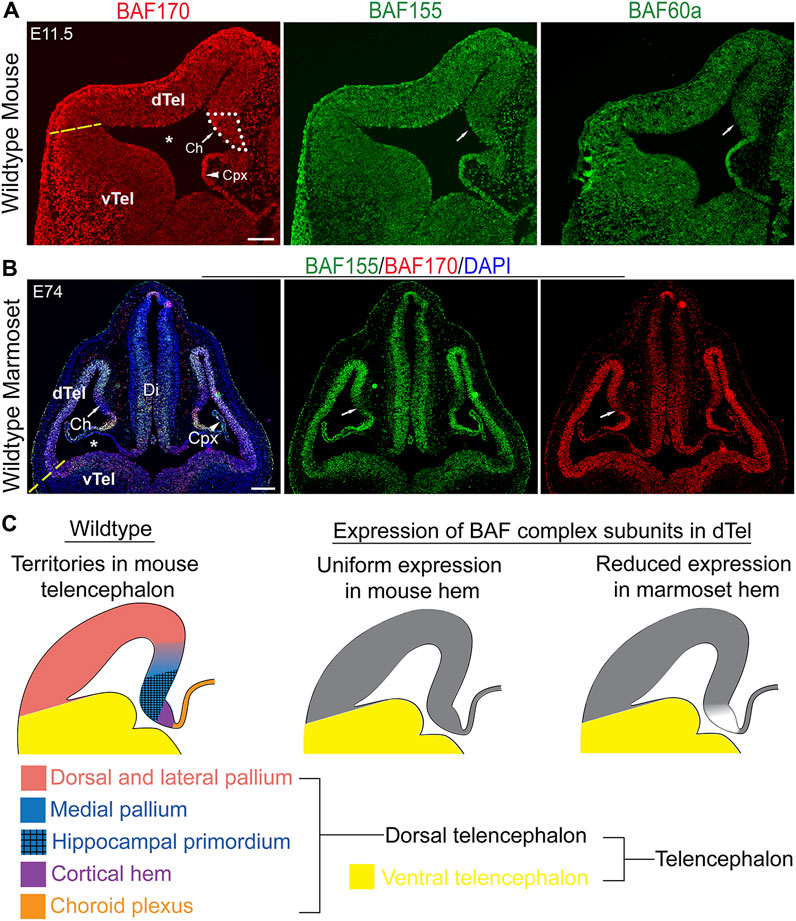
FIGURE 1. Comparative expression analysis suggests a role for BAF complex in mammalian forebrain patterning. (A) Images showing hemi-sections of the E11.5 wild-type mouse telencephalon immunostained for the BAF complex subunits BAF155, BAF170, and BAF60a. An approximation of the border between the dorsal telencephalon and ventral telencephalon is indicated with a yellow dashed line. Medial telencephalic territories, i.e., the cortical hem and choroid plexus, are shown. (B) Images of the marmoset whole head showing immunostaining of the E74 telencephalon with BAF155, BAF170 antibodies. Arrow points to the cortical hem area. The lateral ventricle is indicated by an asterisk. Yellow dashed line indicates an approximation of the dorsal and ventral telencephalon boundary. (C) Schematics showing territorial designation of the mouse dorsal telencephalon (right panel), and the generalized expression of BAF complex subunits in the mouse (middle panel) and marmoset (left panel) dorsal telencephalon. Abbreviations: dorsal telencephalon (dTel), ventral telencephalon (vTel), diencephalon (Di), choroid plexus (Cpx). DAPI counterstaining is shown. Scale bars: 200 µm in (A), 250 µm in (B).
On the other hand, comparative analysis of BAF complex expression in a non-human (primate-like) brain model unveiled demonstrable changes in the expression pattern of BAF complex compared with that of mouse. Notably, we observed intense expression of BAF155 and BAF 170 in the E74 marmoset dTel, which becomes reduced or diffused ventrally (i.e., in the vTel) (Figure 1B). Interestingly, there is a discernable abrupt discontinuation or reduction of BAF complex expression in the region of the cortical hem and its resurgence in the adjoining choroid plexus in the marmoset E74 dTel (Figures 1B,C). The observed lack of BAF complex in the marmoset cortical hem contrasts that seen in the developing mouse cortical hem (Figure 1A vs. 1B; Figure 1C). This observation drew our attention to investigating a possible role for BAF complex in driving cortical hem-mediated dTel patterning or morphogenesis during mammalian brain development.
Loss of BAF155 and BAF170 leads to expansion of the cortical hem
We examined the telencephalic phenotype of the dcKO_Foxg1-Cre mutant brain in which the BAF complex is conditionally abolished in the mouse telencephalon by deleting both BAF155 and BAF170 subunits (Bachmann et al., 2016; Nguyen et al., 2016) with the Foxg1-Cre driver being active from E8.5 on (Hebert and McConnell 2000). At stage E11.5, the GLAST-immuno-delimited cortical hem in the mutant forebrain was markedly expanded beyond its spatial confinement as compared with control (Figures 2A,B; white arrowed, dotted lines). We performed quantitative analysis of the 3D-reconstructed GLAST-stained hems and found that the E11.5 dcKO_Foxg1-Cre cortical hem had undergone about 3-fold volumetric enlargement (Figure 2B; Supplementary Movie S1). Thus, the dcKO_Foxg1-Cre cortical hem extended ectopically beyond its normal tiny location at the end of the medial pallial wall into the neuroepithelial domain of the presumptive cortex (Figure 2A). By applying immunostaining against the cortex selective marker, LHX2, it was evident that the size of LHX2-marked cortical primordium was reduced in the dcKO_Foxg1-Cre dorsal pallium as compared with control (Figure 2A).
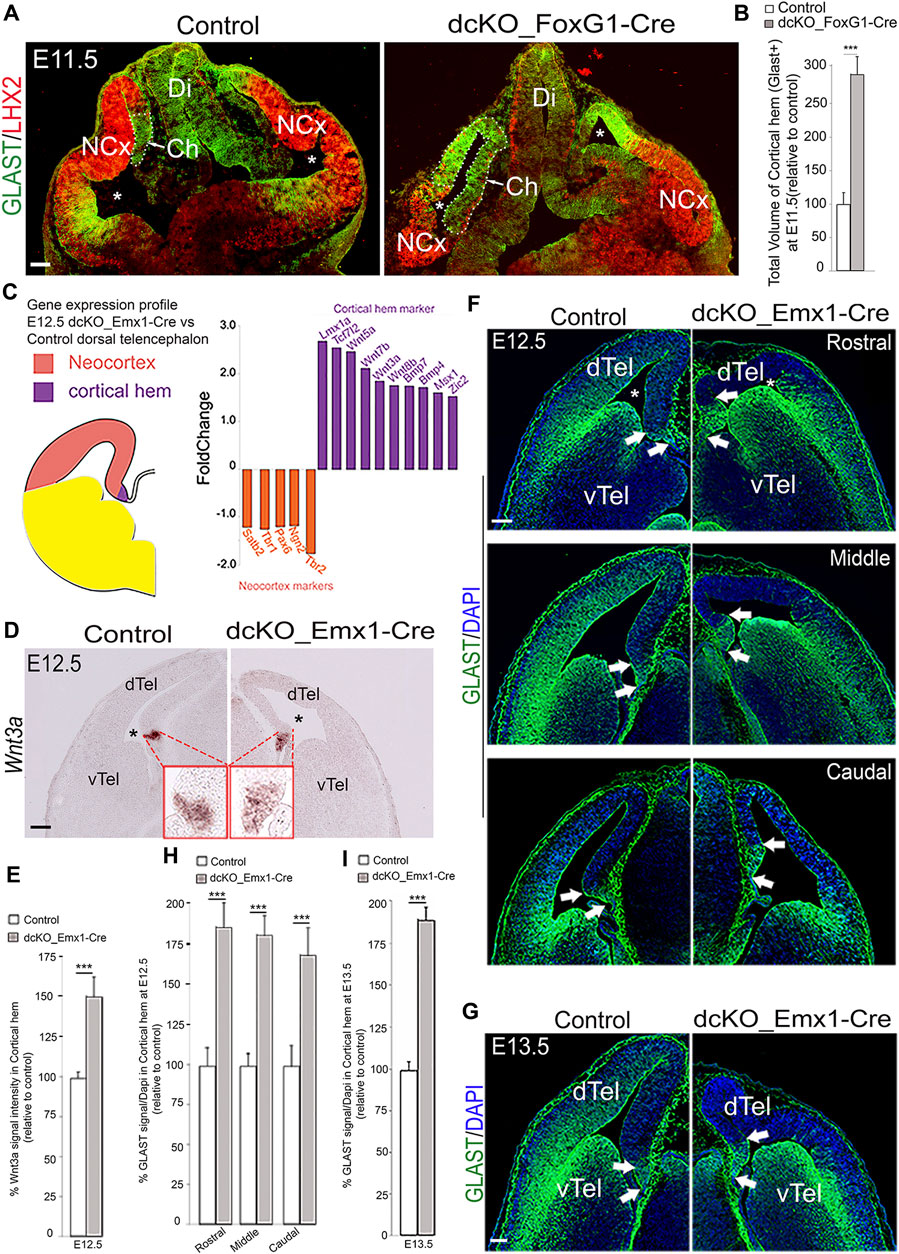
FIGURE 2. BAF complex ablation causes abnormal cortical hem enlargement. (A) Images of the E11.5 mouse whole head sections showing the developing forebrain stained with LHX2 and GLAST antibodies to reveal the presumptive neocortex and the cortical hem (delineated with stippled lines) of the control and dcKO_Foxg1-Cre telencephalon, respectively. (B) Bar chart indicating the statistical difference between the estimated total volume of control and dcKO_Foxg1-Cre cortical hems. (C) Schematic showing demarcation of the neocortex from the cortical hem in hemisection of the mouse telencephalon. Results of RNA sequencing of the E12.5 control and dcKO_Emx1-Cre cortical tissues indicating fold change of downregulated neocortical marker genes (orange bars) and upregulated hem maker genes (purple bars) in the dcKO_Emx1-Cre are graphically represented. (D) Micrographs showing in situ hybridization in the E12.5 control and dcKO_Emx1-Cre dTel with the RNA probe and hem marker Wnt3a. (E) Bar graphs indicating the statistical differences between the estimated size of the control and mutant hem using Wnt3a signal intensity measurement. (F,G) Images showing rostral to caudal sections of the E12.5 (F) and mid-section of the E13.5 (G) control and dcKO_Emx1-Cre brains immmunostained with the GLAST antibody. White arrows point to the boundaries of the cortical hem revealed by intense GLAST staining. (H,I) Bar graphs indicating the statistical differences between the estimated size of the control and mutant hem in the respective micrographs with GLAST staining at E12.5 (rostral–caudal) (F) and E13.5 (G). Where shown, sections are counterstained with DAPI (blue). The lateral ventricle is indicated by an asterisk. Arrows point to the cortical hem. Results are presented as mean ± SD. Unpaired Student’s t-test: ***p < 0.0005; n = 4–6; * denotes level of significance; Scale bars = 100 µm in (A,D,G), 50 µm in (F). Abbreviations: NCx, neocortex; Ch, cortical hem; Di, Diencephalon; dTel, dorsal telencephalon; vTel, ventral telencephalon.
To elucidate BAF complex function during formation of the dTel and specifically in the context of cortical hem-mediated dTel patterning, we crossed mice harboring floxed alleles of BAF155 (Choi et al., 2012) and BAF170 (Tuoc et al., 2013) (i.e., BAF155fl/fl and BAF170fl/fl, respectively) with the dTel neuroepithelium-specific Emx1-Cre to generate dcKO_Emx1-Cre (Narayanan et al., 2015), in which Cre activity is detectable in the presumptive cortex from E10.5 onwards (Gorski et al., 2002). Thus, unlike in the dcKO_Foxg1-Cre telencephalon (Figure 2A), in which BAF complex is broadly deleted at the initial stage (E8.5) of telencephalon formation (Xuan et al., 1995; Martynoga et al., 2005; Bachmann et al., 2016), the Emx1-Cre practically restricted the loss of BAF complex to the dTel at the onset of neurogenesis (Gorski et al., 2002). Indeed, IHC analysis with antibodies against BAF155 and BAF170 revealed overt loss of BAF complex in the dcKO_Emx1-Cre dTel by E11.5 (Supplementary Figure S1).
We proceeded with characterizing the dTel phenotype of the dcKO_Emx1-Cre forebrain by initially reexamining data from our previously performed gene expression profiling of the dcKO_Emx1-Cre dTel containing both neocortex and cortical hem at embryonic stage E12.5 (Figure 2C) [RNA-seq data sheet in (Narayanan et al., 2015); GSE106711]. This analysis revealed that 1,723 transcripts were downregulated and 102 upregulated in the E12.5 dcKO_Emx1-Cre dTel. Notably, many genes important for cortical identity (e.g., Pax6, Ngn1, Ngn2, Tbr1 and Satb2) were downregulated, whereas expression levels of several known cortical hem-specific genes including Wnt3a, Wnt5a and Bmp were upregulated (Figure 2C). We validated our RNA-seq data by examining the expression pattern and level of Wnt3a, which is confined to or concentrated in the cortical hem at early stages of cortical development (Figure 2D; [Lee et al., 2000; Louvi et al., 2007)]. From our ISH analysis, we found that the Wnt3a expression level and area of expression were increased in the mutant dTel as compared with control at E12.5 (Figures 2D,E), thus corroborating the upregulated expression of Wnt3a seen in our RNA-seq data (Figure 2C).
Besides measuring the expression pattern of upregulated genes, we also examined expression of the radial glial markers GLAST and BLBP, which can delimit the cortical hem (Figures 2F,G; Supplementary Figure S2). In early cortical development, between E11.5–E13.5, GLAST expression displays a lateralhigh to mediallow gradient in the neocortex, but with localized high expression level in the cortical hem (Figures 2F,G, demarcated with arrows). Analysis of the 3D-reconstructed E12.5 and E13.5 dcKO_Emx1-Cre cortical hem, as done for dcKO_Foxg1-Cre hem analysis (Figure 2B; Supplementary Movie S1), revealed nearly 2-fold expansion by volume as compared with control (Figures 2H,I). Such expanded dcKO_Emx1-Cre hem was observed at all rostrocaudal levels of the cortical hem system (Figures 2F,H). Thus, our histological analyses suggest that the increased expression of hem marker genes revealed in the RNA-seq data is likely due to cortical hem enlargement in the dcKO_Emx1-Cre forebrain at E12.5. That notwithstanding, the expanded cortical hem and associated dysmorphic cortical phenotypes are less severe in dcKO_Emx1-Cre in comparison to the dcKO_Foxg1-Cre dTel (Figures 2F,G vs. Figures 1A,B). This is consistent with the report of Godbole et al. (2018), in which deletion of Lhx2 at progressively later stages resulted in a corresponding decrease in expansion of the hem.
Of note, Emx1_Cre-mediated single knockout of either BAF155 (BAF155 cKO) or BAF170 (BAF170 cKO) did not elicit an overt expansion of cortical hem (Supplementary Figure S3) as seen in the dcKO_Emx1-Cre mutants (Figures 2D–H). This implies that the function of BAF155 and BAF170 is necessary to orchestrate normal development (suppression) of the cortical hem system. The requirement of BAF complex in temporal regulation of cortical hem size during corticogenesis was suggested by the outcome of the experiment in which BAF complex was ablated later in cortical development as in dcKO_hGFAP-Cre mutants in which Cre is late-active, namely around E13.5 (Zhuo et al., 2001; Nguyen et al., 2018). Notably, we observed no significant change in the size of the E15.5 cortical hem (intensely stained with GLAST antibody, and without PAX6 labeling) in the dcKO_hGFAP-Cre dTel as compared with control (Supplementary Figures S3C–E; unpaired t-test, p-value = 0.926; n = 4). It implies that the modulatory function of the BAF complex in hem morphogenesis is exerted within a limited early developmental time window as demonstrated in the FoxG1-Cre and Emx1-Cre mutants (Figure 2). The temporal regulation of hem size was also reported in previous studies in which hem size was unaffected after later deletion of factors known to regulate hem size at early cortical development stages (Muzio and Mallamaci 2005; Hanashima et al., 2007; Mangale et al., 2008; Godbole et al., 2018).
Taken together, our RNA-seq results and elaborate histological analyses indicate that the loss of BAF complex in the dTel neuroepithelium at the onset of cortical neurogenesis leads to aberrant expansion of the cortical hem. This suggests that the BAF complex may act as a suppressor of hem fate during early forebrain development.
Medial pallium expands and encroaches laterally in the absence of BAF complex
Since loss of BAF complex in the dTel neuroepithelium resulted in abnormal expansion of the cortical hem, we turned our attention to possible impact on mediolateral patterning in the developing BAF mutant telencephalon. We applied ISH analysis to examine the expression pattern of medial dTel (archicortical) markers on serial coronal sections of the E13.5 control and dcKO_Emx1-Cre forebrain (Figure 3). Upon close examination, we observed abnormal patterning of the dorsal midline structures and neocortex in the dcKO_Emx1-Cre mouse dTel. Per Wnt2b and Wnt3a ISH, we noticed mild expansion of the dcKO_Emx1-Cre cortical hem as compared with control (Figures 3A,B). However, the choroid plexus, which is the ventral continuum and derivative of the cortical hem (Louvi et al., 2007), did not present with any noticeable morphological alteration in the dcKO_Emx1-Cre when compared to control, as indicated by ISH staining with the choroid plexus-specific marker transthyretin (Ttr) (Figure 3C; [Herbert et al., 1986; Grove et al., 1998; Monuki et al., 2001)].
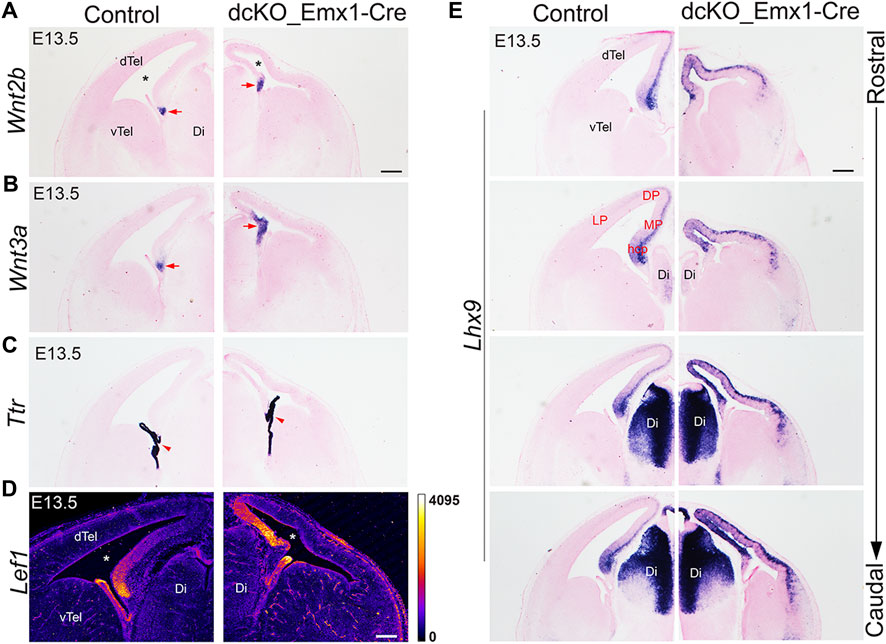
FIGURE 3. Early loss of BAF complex in the developing dorsal telencephalon distorts cortical patterning. (A–E) Images showing in situ hybridization or immuostaining in the control and dcKO_Emx1-Cre E13.5 mouse (dorsal) telencephalon with the riboprobes Wnt2b (A) and Wnt3a (B) to mark the cortical hem, Ttr (transthyretin) to mark the choroid plexus (C), LEF1 antibody to stain the medial cortex (D), and Lhx9 for revealing the cortical hem and medial cortex in rostral to caudal sections (E). The lateral ventricle is indicated by an asterisk. Arrow points to cortical hem. Arrowhead points to choroid plexus Abbreviations: dTel, dorsal telencephalon; vTel, ventral telencephalon Ch, cortical hem; Di, Diencephalon. LP, lateral cortex; DP, dorsal cortex; MP, medial cortex; hcp, hippocampal primordium. Scale bar: 200 µm.
Furthermore, immunostaining for LEF1 and ISH with the RNA probe Lhx9, both of which reveal medial cortex/pallium identity, indicated drastic lateral expansion/extension of the medial cortex at the expense of the dorsolateral cortex in the dcKO_Emx1-Cre dTel (Figures 3D,E). Normally, by E12.5, LHX9 is strongly expressed in the hippocampal primordium and the expression intensity tapers from medial to lateral cortex. Around E13.5, LHX9 is expressed in the ventricular zone of the hippocampal primordium [Figure 3E; (Bulchand et al., 2003)] and in pioneer postmitotic neurons (mainly preplate cells) (Bertuzzi et al., 1999) of the wild-type medial cortex/pallium. However, Lhx9 expression is observed to fill the whole of the medial cortex and is further strongly expressed in the lateral cortex of the dcKO_Emx1-Cre dTel (Figure 3E). This deviation in Lhx9 expression from the aforementioned normal pattern supports the idea of abnormal patterning of the dcKO_Emx1-Cre dTel. It means that the expanded cortical hem and lateral extension of the medial cortex resulted in predominance of the medial pallium in the entire dTel with more severity in the caudal region (Figure 3E; Supplementary Figure S4).
Put together, the data presented here point to a putative function of the BAF chromatin remodeling complex in directing cortical patterning during mouse forebrain development.
BAF complex and hippocampal specification
Existing evidence demonstrates that ectopic hippocampal fields can be induced by an ectopic cortical hem (Mangale et al., 2008), indicating that the hem is an organizing center sufficient in the specification and development of the hippocampal primordium. We previously reported that BAF155 and BAF170 are required for the development of the mouse hippocampus (Nguyen et al., 2018). These findings prompted us to investigate the implication(s) of the expanded cortical hem, as observed in this study, for the specification and/or morphogenesis of the hippocampal anlagen.
Consistent with our findings that deletion of BAF complex components caused progenitor proliferation (Nguyen et al., 2018), the dcKO_Emx1-Cre dTel at E13.5/E14.5 is obviously thinner than controls, with only a few postmitotic neurons. Nonetheless, we examined whether these neurons have been specified to acquire a hippocampal identity. Immunostaining for dentate gyrus marker PROX1 or hippocampal marker ZBTB20 (Xie et al., 2010; Iwano et al., 2012; Rosenthal et al., 2012; Nielsen et al., 2014; Nguyen et al., 2018) revealed no detectable labeling in the vicinity of the cortical hem identified by GLAST staining (Figures 4A,B). Yet, HuCD- and TBR1-expressing postmitotic neurons are present in the E13.5 medial pallium (data not shown). The observed lack of ZBTB20 staining in the BAF complex mutant cortex (Figure 4B) is counterintuitive because ZBTB20 expression is upregulated in the absence of BAF complex (Wang et al., 2021).
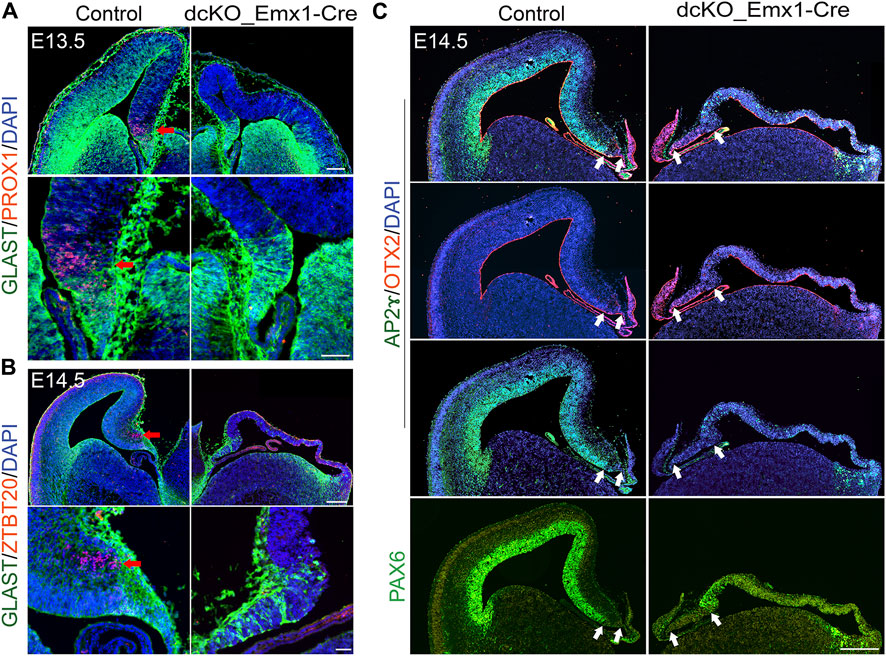
FIGURE 4. Loss of BAF in dTel perturbs hippocampal formation and neocortical patterning. (A) Immunohistochemical micrographs showing hemi-section of the control and dcKO_Emx1-Cre mouse brain at E13.5 stained with GLAST and PROX1 antibodies. Panels of close-up images highlight the presence and absence of the PROX1+ dentate gyrus cells in the control and BAF complex mutant medial pallium, respectively. Red arrow points to PROX1 staining. (B) Micrographs showing hemi-section of the E14.5 mouse brain stained with GLAST and ZBTB20 antibodies in control and dcKO_Emx1-Cre. Panels of close-up images highlight the presence and absence of the ZBTB20 + hippocampal neurons in the control and BAF complex mutant hippocampal primordium, respectively. Red arrow points to ZBTB20 staining. (C) Images showing profound loss of cortical mass and patterning associated with the expanded hem in the E14.5 dcKO_Emx1-Cre dTel as compared with control. The applied antibodies AP2ɣ and PAX6 stain the cortical neuroepithelium/ventricular zone to reveal the unstained cortical hem, while the hem is stained by OTX2. White arrows indicate the size of the cortical hem. Sections are counterstained with DAPI (blue). Scale bars = 250 µm [in upper panel of (A)], 50 µm [in lower panel of (A)], 500 µm [in upper panel of (B)], 50 µm [in lower panel of (B)], 500 µm (C).
To further assess the functional integrity of the expanded dcKO_Emx1-Cre cortical hem, we performed immunostaining for REELIN, which can be used to identify Cajal-Retzius cells. These cells arederivatives of the cortical hem (Rice and Curran 2001; Takiguchi-Hayashi et al., 2004; Bielle et al., 2005; Liu et al., 2018) andcontribute to cortical arealization and laminar patterning via several secreted factors, including REELIN (Frotscher et al., 2009; Causeret et al., 2021). It was observed that the BAF mutant cortical marginal zone was less populated with reelin-expressing cells compared with control, which may partly be due to elevated cell death as indicated by REELIN staining colocalization with cleaved Caspase 3, a marker for apoptosis, in the mutant marginal zone (Supplementary Figure S5). Of note, such REELIN-expressing cells were seen to have accumulated in the region of the prospective hippocampal anlage and in the cortical hem of the dcKO_Emx1-Cre mutant (Supplementary Figure S5), which is indicative of their impaired migration.
Using PAX6 and AP2ɣ (Pinto et al., 2009) as indicators of cortical identity, we observed that loss of BAF complex in dTel engendered massive cortical shrinkage and cortical integrity perturbation by mid-embryonic corticogenesis (E14.5) in the dcKO_Emx1-Cre cortex as compared with control (Figure 4C). The severely dysmorphic BAF mutant cortex is seen to have lost the medial to lateral neuroepithelial patterning revealed by lack of PAX6 or AP2ɣ expression in the ventricular zone, which is normally enriched in the wild-type (control) mouse cortical neuroepithelium (Figure 4C). Meanwhile, the cortical hem, marked by OTX2 and without PAX6 or AP2ɣ staining, remained expanded in the E14.5 dcKO_Emx1-Cre dTel as compared with control (Figure 4C).
Importantly, the non-specification or aplasia of the hippocampus and abnormal alteration of the cortical structure in dcKO_Emx1-Cre forebrain partially phenocopies the Lhx2 null dTel (Bulchand et al., 2001; Chou et al., 2009; Godbole et al., 2018). Taken together, these findings indicate that the BAF complex is necessary for preserving cortical identity and modulates correct formation of the cortical hem needed for hippocampal specification and/or morphogenesis.
BAF155 and BAF170 interact with the cortical hem suppressor LHX2 during early cortical development
The similarities in the hem expansion upon loss of either LHX2 (Bulchand et al., 2001) or BAF155/170 (this study) motivated us to examine whether these factors participate in a common regulatory pathway or interact with each other. We found extensive co-expression (colocalization) of BAF155 and BAF170 with LHX2 in the E11.5 control presumptive cortical neuroepithelium (Figure 5A). However, lack of BAF complex in the early developing dTel did not cause obvious downregulation of LHX2 expression in the cortical neuroepithelium (Figure 5A; Supplementary Figures S2A,C). This indicates that the BAF complex does not, at least in the examined stages of dTel development (E11.5 and E13.5), have a major influence on the expression level of LHX2 in the presumptive cortex.
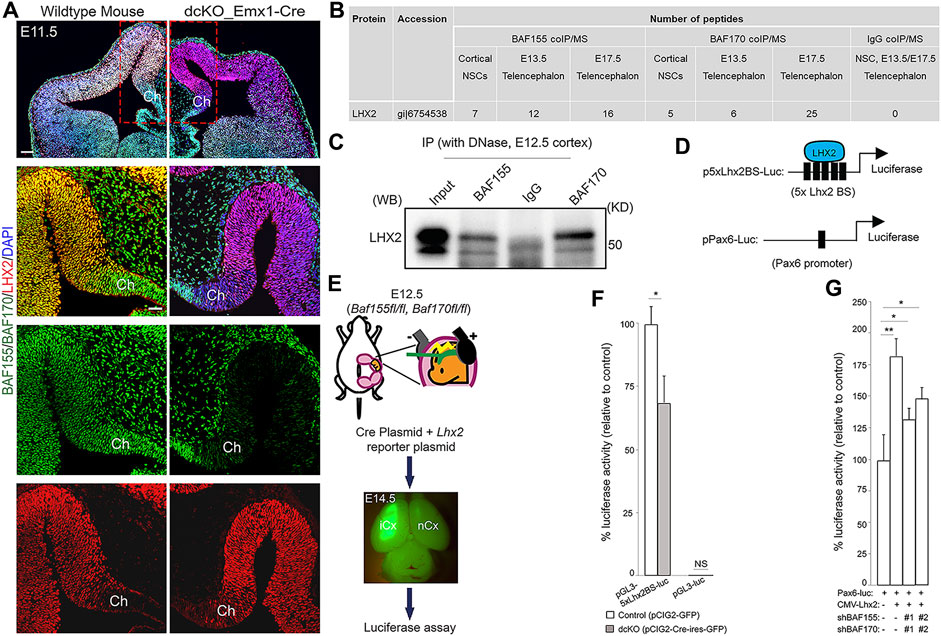
FIGURE 5. BAF complex interacts with LHX2 in the neocortical neuroepithelium. (A) Immunomicrographs showing the E11.5 control and BAF complex mutant mouse telencephalon stained for BAF155, BAF170, and LHX2. Colocalization of BAF complex is indicated by yellow signal. A close-up view of the medial pallium and cortical hem is shown. (B) Image of table indicating the number of LHX2 peptides purified from immunoprecipitates of BAF155 and BAF170 obtained from NS5 cell protein extracts, and from the E13.5 or E17.5 dorsal telencephalon protein extract. IgG coIP/MS was used as control. (C) Interaction of BAF155 and BAF170 with LHX2 was confirmed by coIP/western blot analyses of E12.5 dorsal telencephalic tissue. (D) Illustrations showing the plasmids (p-5xLhx2BS-luciferase and pPax6-Luc) used for the luciferase assay. The multiple (5x) Lhx2 binding sites (BS) for augmenting LHX2-dependent promoter activity is depicted in p-5xLhx2BS-luciferase, whereas pPax6-Luc has one Lhx2 binding sites. (E) Image showing scheme for in vivo luciferase assay. (F,G) Bar charts representing the statistical comparisons of (F) In vivo LHX2-mediated luciferase activity in the control or dcKO_Emx1-Cre cortex, and (G) In vitro PAX6-mediated luciferase activity in culture Neuro2A cells in the presence or absence of BAF155, BAF170 or LHX2 permutationally as indicated. In both cases, two variants of shRNAs (#1 and #2) were used to knockdown BAF155 and BAF170. Unpaired Student’s t-test: *p < 0.05, **p < 0.005; NS stands for not significant; * denotes level of significance; n = 6; Results are presented as mean ± SD. Abbreviations: Ch, cortical hem; iCx, injected cortex; nCx, non-injected cortex.
We explored the molecular interaction partners of the BAF complex that control formation of the cortical hem and modulate hem-dependent establishment of other dTel structures. To this end, we performed co-immmunoprecipitation (coIP) in cell lysates obtained from the E13.5 or E17.5 telencephalic tissue from wild-type mice, and cultured NS-5 neural stem cells (NSC) using antibodies against BAF155 and BAF170 followed by mass spectrometry (MS) (Narayanan et al., 2015; Nguyen et al., 2018). Many factors essential for neural morphogenesis were identified as protein interaction partners of both BAF155 and BAF170 subunits of the BAF complex (data not shown). Strikingly, we found that LHX2, a determinant of cortical identity and suppressor of hem fate (Bulchand et al., 2001; Monuki et al., 2001; Mangale et al., 2008; Roy et al., 2014), was bound to BAF155 and BAF170 in cultured NSCs, and in the early (E13.5) and late (E17.5) developing forebrain as compared with control (IgG coIP/MS) (Figures 5B,C; p score = 1.00).
To examine eventual functional interaction between BAF complex and LHX2, and its neurodevelopmental relevance in the context of hem morphogenesis and cortical formation, we investigated whether the interaction between BAF155/BAF170 and LHX2 influences LHX2-dependent transcriptional activity in the developing mouse cortex. For this purpose, we used an Lhx2-dependent reporter vector (pGL3-5xLhx2BS-luciferase), containing five Lhx2 (5x Lhx2) binding sites (BS) upstream of a luciferase reporter and an empty vector without Lhx2 binding site (pEV-luciferase) as a negative control (Figure 5D) (Folgueras et al., 2013). To eliminate the function of BAF155 and BAF170, the reporter plus Cre-expressing plasmids were in utero co-electroporated into the E12.5 cortex of BAF155fl/fl; BAF170fl/fl embryos (Figure 5E). We then examined isolated tissue samples from the cortex using a luciferase assay. Our analysis indicated that BAF complex knockout in the dTel or pallium significantly diminished the reporter activity of pGL3-5xLhx2BS-luciferase, but not that of the control plasmid lacking the Lhx2 binding motif (Figures 5E,F). This suggests that BAF155 and BAF170 are together required for activation of LHX2-dependent transcriptional activity.
Alternatively, it is possible that BAF complex might cooperate with LHX2 to regulate the expression of genes governing establishment of cortical identity or patterning. In cortical progenitors, LHX2 is reported to directly regulate PAX6 expression by binding to its promoter (Hou et al., 2013; Shetty et al., 2013; Hsu et al., 2015). Given that PAX6 is a key regulator of cortical progenitor identity and patterning (Bishop et al., 2000; Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001; Pinon et al., 2008), we reasoned that its activity may be under the influence of a BAF complex-LHX2 regulatory axis during cortical formation. To verify such a possibility, we first confirmed that Pax6 is downregulated in the dcKO dTel using IHC analyses (Supplementary Figures S2A,C). We then performed luciferase reporter assays to determine how the expression of LHX2 and BAF complex regulate the activity of a Pax6 promoter vector, which contains an Lhx2 binding site (Figure 5D; Kammandel et al., 1999; Hou et al., 2013). The Pax6 promoter vector driving luciferase expression (Pax6-Luc) was transiently transfected into Neuro2A cells together with a combination of Lhx2-expression (CMV-Lhx2) and BAF155/BAF170-silencing vectors (shBAF155 and shBAF170; Figure 5G). After lysate collection and luminescence quantification, the results from the reporter assay revealed that, as compared with control (in Pax6-Luc + CMV-EV condition), LHX2 (in Pax6-Luc + CMV-Lhx2 condition) significantly regulates promoter activity of Pax6 (Figure 5G). However, upon BAF155/BAF170 dual knockdown using short-hairpin RNA (shBAFs) constructs, the level of luciferase activity reduced (Pax6-Luc + CMV-Lhx2 + shBAFs condition), implying that both LHX2 and BAF155/BAF170 are required for maintaining the promoter activity of Pax6 (Figure 5G).
Altogether, the results of our protein-protein interaction analyses and luciferase assay suggest that the telencephalon patterning regulatory factor LHX2 plausibly acts in concert with BAF complex to orchestrate forebrain development. This finding affords description of a possible molecular basis of the involvement of BAF complex in regulating cortical hem fate and/or morphogenesis via functional cooperation with LHX2 and, by extension, its downstream effectors of dorsal telencephalic patterning (Figure 6).

FIGURE 6. Schema depicting how dorsal telencephalon patterning is perturbed by BAF complex mutagenesis. (A) Illustration shows normal regional designation of the dorsal telencephalon (in wild type). The cortical hem, hippocampus, medial cortex/pallium, and neocortex are properly specified and placed in the wild-type dorsal telencephalon. (B) Cartoon depicting disturbance of cortical patterning due to lack of BAF complex in the dorsal telencephalic neuroepithelium (in dcKO_Emx1-Cre). The hem and medial cortex are seen expanded at the expense of the hippocampus and neocortex, making the dcKO_Emx1-Cre dorsal telencephalon have predominance of medial cortex identity. The choroid plexus is not observably affected by BAF complex ablation. (C) A putative mechanism involving the interaction between the scaffolding BAF complex subunits BAF155 and BAF170, and the transcription factor LHX2 is put forward as a previously unknown regulatory system for driving proper cortical hem formation and patterning of the dorsal telencephalon.
Discussion
During development, the dTel is organized into structural domains that correlate with specific functional areas. The cortical hem and anti-hem are embryonic organizers located at the medial and lateral edge of the dTel, respectively. The cortical hem is a hub for morphogen signaling known to regulate areal patterning of the embryonic cortical neuroepithelium [reviewed in (Subramanian et al., 2009)]. Absence or abnormal morphology of the cortical hem or molecular manipulations of related morphogenetic factors therein have been linked to various telencephalic perturbations of the developing brain, including cortical mis-patterning (Xuan et al., 1995; Lee et al., 2000; Martynoga et al., 2005; Yoshida et al., 2006; Mangale et al., 2008; Chizhikov et al., 2010; Caronia-Brown et al., 2014; Godbole et al., 2018). However, our understanding of correlating activity of chromatin regulators to the functionality of specific morphogens that drive forebrain patterning is far from clear.
In this study, we focused on determining the role of the ATP-dependent chromatin remodeler BAF complex in regulating forebrain patterning with emphasis on the forebrain hem. Previous studies identified the BAF (mSWI/SNF) complex to play pivotal roles in many aspects of cortical development, including neural progenitor specification, proliferation and differentiation, and maturation of postmitotic neural cells [Reviewed in (Sokpor et al., 2017)]. To expand our understanding of the molecular regulators that ensure the establishment and maintenance of cortical structures and functions, we spatiotemporally inactivated the multimeric chromatin remodeling BAF complex via deletion of its scaffolding subunits BAF155 and BAF170 by using Foxg1-Cre, Emx1-Cre and hGFAP-Cre. By this means, we demonstrate in this current study that the BAF complex function is instructive for patterning of the mammalian dTel (Figure 6).
BAF complex suppresses expansion of cortical hem
We observed that in vivo loss of BAF complex during early stages of cortical formation (i.e., under Foxg1- and Emx1-Cre drivers) caused abnormal expansion of the cortical hem and medial cortex to the detriment of the hippocampal complex and mediolateral cortical identity. The LHX2+ presumptive cortex, although specified in the BAF complex mutant brain, appeared dysmorphic and lateralized. Cortical patterning was seen to be progressively lost from rostral to caudal levels of the telencephalon due to exaggeration of the medial cortical identity or lopsided medial cortical specification. This outcome points to possible distortion of the blueprint for cortical neuroepithelial patterning due to BAF complex ablation and highlights potential abolishment of the secondary organizer function of the expanded cortical hem. Such secondary patterning activity by hem may be necessary for modulating neocortical arealization to yield the various functional cortical areas.
Given the important role of the cortical hem in dTel patterning, we think the BAF complex likely regulates the extent of the cortical hem to modulate cortical patterning. This notion is consolidated by the observation that although the hem is expanded in the absence of optimal BAF complex function, the size of the choroid plexus, which is derived from the hem (Louvi et al., 2007) is not affected. The demonstrable effect of BAF complex disruption on the cortical hem argues its selective regulatory role therein, and/or in the adjoining neocortex. It is also conceivable that the expanded hem is improperly configured, leading to its incompetence in orchestrating arealization of the adjacent cortical primordium or in executing other hem functions.
Our findings convey the idea that the BAF complex likely suppresses inherent expansion tendencies of dTel midline structures, excluding the choroid plexus, during cortical patterning. However, the said suppressive capacity of the BAF complex is likely exerted at early cortical development stages, perhaps until E12.5, as loss of BAF complex from E13.5 achieved in dcKO_hGFAP-Cre mutants did not lead to an observable hem or medial cortex expansion, albeit hippocampal formation was perturbed in such mutants (Nguyen et al., 2018). The effect of BAF complex deletion on hem expansion is consistent with outcome of other studies in which genetic manipulation of factors such as Foxg1, Lhx2, Lhx2/Pax6 (double deletion), and Nf2 in early development caused abnormal expansion of the mouse dTel midline structures and associated cortical patterning deviations (Bulchand et al., 2001; Monuki et al., 2001; Vyas et al., 2003; Muzio and Mallamaci 2005; Mangale et al., 2008; Lavado et al., 2013; Godbole et al., 2017). It would be interesting to find out the impact on cortical patterning following focal silencing of the BAF complex in the nascent hem using Lmx1a-Cre driver (Chizhikov et al., 2010; Fregoso et al., 2019).
The discernable graded expression of BAF complex in the telencephalon and its distinctive dramatic downregulation in the cortical hem of marmoset, which are not obvious in the mouse cortex, supports a more complex role for BAF complex in driving primate brain morphogenesis. The observations may be adaptive modification or refinement of BAF complex expression that subserves additional functions in the evolved marmoset dTel and hem. In a broader context, these observations may have evolutionary implication for primate cortical patterning and development, especially that the BAF complex has been identified to interact with and augment the transcriptional activity of the well-known cortical patterning regulator LHX2 (Bulchand et al., 2001; Monuki et al., 2001; Mangale et al., 2008).
Implication of BAF complex and LHX2 interaction for suppression of cortical hem expansion
The formation of cortical hem is regulated by the activity of several factors, including the transcription factor LHX2 and FOXG1 (Godbole et al., 2018). Previous studies reported cortical hem expansion with concomitant massive loss of the cortical identity due to complete or constitutive ablation of Lhx2 (Bulchand et al., 2001; Mangale et al., 2008). We show here that LHX2 is an interaction partner of the BAF complex. Furthermore, we found that cortical phenotype caused by silencing of BAF complex (this study) resembles that of Lhx2−/− mutant (Mangale et al., 2008; Roy et al., 2014), suggesting that both factors are co-dependent in directing telencephalic patterning. We infer from our luciferase experiment that the identified interaction may be a potentiating kind to suppress hem expansion and preserve cortical identity. In further support of such a scenario, we found that the BAF complex promotes the LHX2 target gene Pax6, which has a pivotal role in cortical patterning (Bishop et al., 2000; Stoykova et al., 2000; Kimura et al., 2005; Godbole et al., 2017). Moreover, either factor (BAF complex or LHX2) alone is unable to afford sufficient suppression to prevent abnormal expansion of the cortical hem in the absence of the other, hence signifying possible functional cooperation between the BAF complex and LHX2. Together, our new findings disclose the possibility that a correlation between transcription factor activation with specific epigenetic chromatin remodeling machinery act in concert to provide a suppressive function preventing the uncontrolled enlargement of the hem into the normal cortical territory.
The results from the hem phenotype analysis during development in the LHX2- and BAF complex-ablated forebrain, revealed early temporal overlap of their function in effecting hem size restriction. Whereas loss of BAF complex in the dTel under the Emx1-Cre driver, with Cre activity beginning from E10.5 (Guo et al., 2000; Gorski et al., 2002), resulted in cortical hem expansion (this study), loss of Lhx2 at E10.5 or later did not have any impact on hem size compared to control (Godbole et al., 2018). Thus, it appears that the repressive function of the BAF complex on hem fate persists slightly beyond the apparent end point of the LHX2-dependent hem size restriction. In that line of reasoning, we argue that the BAF complex is sufficient in limiting hem expansion and may partly substitute for or complement the function of LHX2 in preventing the uncontrolled expansion of the cortical hem during later or active periods of hem morphogenesis.
Overall, we suggest that the chromatin remodeling BAF complex, partly through interaction with LHX2, engages in the regulatory network that ensures the specification and morphogenesis of the mouse cortical hem, and its placement at the medial edge of the dTel (Figure 6). The pattern of BAF expression in the marmoset telencephalon is suggestive of a similar role in the primate brain. Further investigation is needed to establish the precise role of the BAF complex and its interaction partners necessary for orchestrating development of the highly evolved primate dTel through regulating hem morphogenesis and functionality.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE106711.
Ethics statement
The animal study was reviewed and approved by the Animal Welfare Committees of the University Medical Center Göttingen and local authority (LAVES: Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit) under the license numbers: 14/1636 and 16/2330.
Author contributions
HN and GS characterized the dcKO phenotypes, and drafted and revised the manuscript; AP, NS, and ST provided data in Figure 3/Supplementary Figure S4, and contributed to revising the manuscript; MP and YX performed the protein-protein interaction study; LP, PAU, and JR contributed to histological analyses and animal application; RB provided marmoset tissue; AS, BB-S, HPN, and JFS provided research resources and contributed to discussions; TT conceived, supervised the study, and revised the manuscript.
Funding
This work was supported by the Universitätsmedizin Göttingen (TT), TU432/3, TU432/6 DFG grants (TT), Schram-Stiftung (TT), DFG-CNMPB (TT, JFS), and the RUB/FoRUM/F1008N-20 grant (TT). MP was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (NIH-NIDDK) (K01DK121861). Work in the ST lab was supported by a grant from the Department of Science and Technology [DST/CSRI/2017/202(G)]; by the Canada-Israel Health Research Initiative, jointly funded by the Canadian Institutes of Health Research, the Israel Science Foundation, the International Development Research Centre, Canada and the Azrieli Foundation; intramural funds from TIFR-DAE (12-R&D-TFR-5.10-0100RTI 2001). NS was the recipient of an NPDF award (PDF/2019/002804) from the Science and Engineering Research Board, Government of India.
Acknowledgments
We acknowledge T. Huttanus and H. Fett for their expert animal care and support. We also thank A. Messing, A. P. McMahon, A. Jones, F. Guillemot, and E. Fuchs for providing reagents. We acknowledge support by the DFG Open Access Publication Funds of the Ruhr-Universität Bochum.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1011109/full#supplementary-material
References
Abbas, E., Hassan, M. A., Sokpor, G., Kiszka, K., Pham, L., Kerimoglu, C., et al. (2021). Conditional loss of BAF (mSWI/SNF) scaffolding subunits affects specification and proliferation of oligodendrocyte precursors in developing mouse forebrain. Front. Cell Dev. Biol. 9, 619538. doi:10.3389/fcell.2021.619538
Aeckerle, N., Drummer, C., Debowski, K., Viebahn, C., and Behr, R. (2015). Primordial germ cell development in the marmoset monkey as revealed by pluripotency factor expression: Suggestion of a novel model of embryonic germ cell translocation. Mol. Hum. Reprod. 21, 552. doi:10.1093/molehr/gav016
Bachmann, C., Nguyen, H., Rosenbusch, J., Pham, L., Rabe, T., Patwa, M., et al. (2016). mSWI/SNF (BAF) complexes are indispensable for the neurogenesis and development of embryonic olfactory epithelium. PLoS Genet. 12, e1006274. doi:10.1371/journal.pgen.1006274
Balasubramanian, R., Bui, A., Ding, Q., and Gan, L. (2014). Expression of LIM-homeodomain transcription factors in the developing and mature mouse retina. Gene Expr. Patterns 14, 1–8. doi:10.1016/j.gep.2013.12.001
Bertuzzi, S., Porter, F. D., Pitts, A., Kumar, M., Agulnick, A., Wassif, C., et al. (1999). Characterization of Lhx9, a novel LIM/homeobox gene expressed by the pioneer neurons in the mouse cerebral cortex. Mech. Dev. 81, 193–198. doi:10.1016/S0925-4773(98)00233-0
Bielle, F., Griveau, A., Narboux-Neme, N., Vigneau, S., Sigrist, M., Arber, S., et al. (2005). Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 8, 1002–1012. doi:10.1038/nn1511
Bishop, K. M., Goudreau, G., and O'Leary, D. D. M. (2000). Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science 288, 344–349. doi:10.1126/science.288.5464.344
Bulchand, S., Grove, E. A., Porter, F. D., and Tole, S. (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100, 165–175. doi:10.1016/s0925-4773(00)00515-3
Bulchand, S., Subramanian, L., and Tole, S. (2003). Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev. Dyn. 226, 460–469. doi:10.1002/dvdy.10235
Caronia-Brown, G., Yoshida, M., Gulden, F., Assimacopoulos, S., and Grove, E. A. (2014). The cortical hem regulates the size and patterning of neocortex. Development 141, 2855–2865. doi:10.1242/dev.106914
Causeret, F., Moreau, M. X., Pierani, A., and Blanquie, O. (2021). The multiple facets of Cajal-Retzius neurons. Development 148, dev199409. doi:10.1242/dev.199409
Chizhikov, V. V., Lindgren, A. G., Mishima, Y., Roberts, R. W., Aldinger, K. A., Miesegaes, G. R., et al. (2010). Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc. Natl. Acad. Sci. U. S. A. 107, 10725–10730. doi:10.1073/pnas.0910786107
Choi, J., Ko, M., Jeon, S., Jeon, Y., Park, K., Lee, C., et al. (2012). The SWI/SNF-like BAF complex is essential for early B cell development. J. Immunol. 188, 3791–3803. doi:10.4049/jimmunol.1103390
Chou, S. J., Perez-Garcia, C. G., Kroll, T. T., and O'Leary, D. D. M. (2009). Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 12, 1381–1389. doi:10.1038/nn.2427
de Melo, J., Clark, B. S., and Blackshaw, S. (2016). Multiple intrinsic factors act in concert with Lhx2 to direct retinal gliogenesis. Sci. Rep. 6, 32757. doi:10.1038/srep32757
de Melo, J., Clark, B. S., Venkataraman, A., Shiau, F., Zibetti, C., and Blackshaw, S. (2018). Ldb1- and Rnf12-dependent regulation of Lhx2 controls the relative balance between neurogenesis and gliogenesis in the retina. Development 145, dev159970. doi:10.1242/dev.159970
Folgueras, A. R., Guo, X., Pasolli, H. A., Stokes, N., Polak, L., Zheng, D., et al. (2013). Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell 13, 314–327. doi:10.1016/j.stem.2013.06.018
Forster, E., Zhao, S., and Frotscher, M. (2006). Laminating the hippocampus. Nat. Rev. Neurosci. 7, 259–267. doi:10.1038/nrn1882
Fregoso, S. P., Dwyer, B. E., and Franco, S. J. (2019). Lmx1a drives Cux2 expression in the cortical hem through activation of a conserved intronic enhancer, Development. 146. doi:10.1242/dev.170068
Frotscher, M., Chai, X. J., Bock, H. H., Haas, C. A., Forster, E., and Zhao, S. T. (2009). Role of Reelin in the development and maintenance of cortical lamination. J. Neural Transm. 116, 1451–1455. doi:10.1007/s00702-009-0228-7
Furuta, Y., Piston, D. W., and Hogan, B. L. M. (1997). Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124, 2203–2212. doi:10.1242/dev.124.11.2203
Gitton, Y., Cohen-Tannoudji, M., and Wassef, M. (1999). Specification of somatosensory area identity in cortical explants. J. Neurosci. 19, 4889–4898. doi:10.1523/JNEUROSCI.19-12-04889.1999
Godbole, G., Roy, A., Shetty, A. S., and Tole, S. (2017). Novel functions of LHX2 and PAX6 in the developing telencephalon revealed upon combined loss of both genes. Neural Dev. 12, 19. doi:10.1186/s13064-017-0097-y,
Godbole, G., Shetty, A. S., Roy, A., D'Souza, L., Chen, B., Miyoshi, G., et al. (2018). Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development 145, dev154583. doi:10.1242/dev.154583
Gorski, J. A., Talley, T., Qiu, M. S., Puelles, L., Rubenstein, J. L. R., and Jones, K. R. (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314. doi:10.1523/JNEUROSCI.22-15-06309.2002
Grove, E. A., Tole, S., Limon, J., Yip, L. W., and Ragsdale, C. W. (1998). The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315–2325. doi:10.1242/dev.125.12.2315
Guo, H., Hong, S., Jin, X. L., Chen, R. S., Avasthi, P. P., Tu, Y. T., et al. (2000). Specificity and efficiency of Cre-mediated recombination in Emx1-Cre knock-in mice. Biochem. Biophys. Res. Commun. 273, 661–665. doi:10.1006/bbrc.2000.2870
Hanashima, C., Fernandes, M., Hebert, J. M., and Fishell, G. (2007). The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J. Neurosci. 27, 11103–11111. doi:10.1523/JNEUROSCI.1066-07.2007
Hébert, J. M., and Fishell, G. (2008). The genetics of early telencephalon patterning: Some assembly required. Nat. Rev. Neurosci. 9, 678–685. doi:10.1038/nrn2463
Hebert, J. M., and McConnell, S. K. (2000). Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222, 296–306. doi:10.1006/dbio.2000.9732
Herbert, J., Wilcox, J. N., Pham, K. T., Fremeau, R. T., Zeviani, M., Dwork, A., et al. (1986). Transthyretin: A choroid plexus-specific transport protein in human brain. The 1986 S. Weir mitchell award. Neurology 36, 900–911. doi:10.1212/wnl.36.7.900
Hirota, J., and Mombaerts, P. (2004). The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 101, 8751–8755. doi:10.1073/pnas.0400940101
Hou, P. S., Chuang, C. Y., Kao, C. F., Chou, S. J., Stone, L., Ho, H. N., et al. (2013). LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 41, 7753–7770. doi:10.1093/nar/gkt567
Hsu, L. C., Nam, S., Cui, Y., Chang, C. P., Wang, C. F., Kuo, H. C., et al. (2015). Lhx2 regulates the timing of beta-catenin-dependent cortical neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 112, 12199–12204. doi:10.1073/pnas.1507145112
Iwano, T., Masuda, A., Kiyonari, H., Enomoto, H., and Matsuzaki, F. (2012). Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development 139, 3051–3062. doi:10.1242/dev.080002
Kammandel, B., Chowdhury, K., Stoykova, A., Aparicio, S., Brenner, S., and Gruss, P. (1999). Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 205, 79–97. doi:10.1006/dbio.1998.9128
Kiecker, C., and Lumsden, A. (2012). The role of organizers in patterning the nervous system. Annu. Rev. Neurosci. 35, 347–367. doi:10.1146/annurev-neuro-062111-150543
Kimura, J., Suda, Y., Kurokawa, D., Hossain, Z. M., Nakamura, M., Takahashi, M., et al. (2005). Emx2 and Pax6 function in cooperation with Otx2 and Otx1 to develop caudal forebrain primordium that includes future archipallium. J. Neurosci. 25, 5097–5108. doi:10.1523/Jneurosci.0239-05.2005
Kolterud, A., Alenius, M., Carlsson, L., and Bohm, S. (2004). The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development 131, 5319–5326. doi:10.1242/dev.01416
Lamba, D. A., Hayes, S., Karl, M. O., and Reh, T. (2008). Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev. Dyn. 237, 3016–3023. doi:10.1002/dvdy.21697
Lavado, A., He, Y., Pare, J., Neale, G., Olson, E. N., Giovannini, M., et al. (2013). Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development 140, 3323–3334. doi:10.1242/dev.096537
Lee, S. M., Tole, S., Grove, E., and McMahon, A. P. (2000). A local Wnt-3a signal is required for development of the mammalian hippocampus. Development 127, 457–467. doi:10.1242/dev.127.3.457
Levitt, P., Barbe, M. F., and Eagleson, K. L. (1997). Patterning and specification of the cerebral cortex. Annu. Rev. Neurosci. 20, 1–24. doi:10.1146/annurev.neuro.20.1.1
Liu, B., Xiao, H., and Zhao, C. (2018). Forced expression of Foxg1 in the cortical hem leads to the transformation of cajal-retzius cells into dentate granule neurons. J. Dev. Biol. 6, E16. doi:10.3390/jdb6030016
Louvi, A., Yoshida, M., and Grove, E. A. (2007). The derivatives of the Wnt3a lineage in the central nervous system. J. Comp. Neurol. 504, 550–569. doi:10.1002/cne.21461
Mallamaci, A., Muzio, L., Chan, C. H., Parnavelas, J., and Boncinelli, E. 2000. Area identity shifts in the early cerebral cortex of Emx2-/- mutant mice. Nat. Neurosci. 3: 679–686. doi:10.1038/76630
Mallamaci, A., and Stoykova, A. (2006). Gene networks controlling early cerebral cortex arealization. Eur. J. Neurosci. 23, 847–856. doi:10.1111/j.1460-9568.2006.04634.x
Mangale, V. S., Hirokawa, K. E., Satyaki, P. R., Gokulchandran, N., Chikbire, S., Subramanian, L., et al. (2008). Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 319, 304–309. doi:10.1126/science.1151695
Martynoga, B., Morrison, H., Price, D. J., and Mason, J. O. (2005). Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127. doi:10.1016/j.ydbio.2005.04.005
Meyer, G., Perez-Garcia, C. G., Abraham, H., and Caput, D. (2002). Expression of p73 and Reelin in the developing human cortex. J. Neurosci. 22, 4973–4986. doi:10.1523/Jneurosci.22-12-04973.2002
Monuki, E. S., Porter, F. D., and Walsh, C. A. (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604. doi:10.1016/s0896-6273(01)00504-9
Muzio, L., and Mallamaci, A. (2005). Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J. Neurosci. 25, 4435–4441. doi:10.1523/Jneurosci.4804-04.2005
Narayanan, R., Pham, L., Kerimoglu, C., Watanabe, T., Castro Hernandez, R., Sokpor, G., et al. (2018). Chromatin remodeling BAF155 subunit regulates the Genesis of basal progenitors in developing cortex. iScience 4, 109–126. doi:10.1016/j.isci.2018.05.014
Narayanan, R., Pirouz, M., Kerimoglu, C., Pham, L., Wagener, R. J., Kiszka, K. A., et al. (2015). Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 13, 1842–1854. doi:10.1016/j.celrep.2015.10.046
Nguyen, H., Kerimoglu, C., Pirouz, M., Pham, L., Kiszka, K. A., Sokpor, G., et al. (2018). Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Rep. 10, 1734–1750. doi:10.1016/j.stemcr.2018.04.014
Nguyen, H., Sokpor, G., Pham, L., Rosenbusch, J., Stoykova, A., Staiger, J. F., et al. (2016). Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle 15, 1317–1324. doi:10.1080/15384101.2016.1160984
Nielsen, J. V., Thomassen, M., Mollgard, K., Noraberg, J., and Jensen, N. A. (2014). Zbtb20 defines a hippocampal neuronal identity through direct repression of genes that control projection neuron development in the isocortex. Cereb. Cortex 24, 1216–1229. doi:10.1093/cercor/bhs400
O'Leary, D. D., and Sahara, S. (2008). Genetic regulation of arealization of the neocortex. Curr. Opin. Neurobiol. 18, 90–100. doi:10.1016/j.conb.2008.05.011
Pinon, M. C., Tuoc, T. C., Ashery-Padan, R., Molnar, Z., and Stoykova, A. (2008). Altered molecular regionalization and normal thalamocortical connections in cortex-specific Pax6 knock-out mice. J. Neurosci. 28, 8724–8734. doi:10.1523/JNEUROSCI.2565-08.2008
Pinto, L., Drechsel, D., Schmid, M. T., Ninkovic, J., Irmler, M., Brill, M. S., et al. (2009). AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat. Neurosci. 12, 1229–1237. doi:10.1038/nn.2399
Rash, B. G., and Grove, E. A. (2006). Area and layer patterning in the developing cerebral cortex. Curr. Opin. Neurobiol. 16, 25–34. doi:10.1016/j.conb.2006.01.004
Rice, D. S., and Curran, T. (2001). Role of the Reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci. 24, 1005–1039. doi:10.1146/annurev.neuro.24.1.1005
Rosenthal, E. H., Tonchev, A. B., Stoykova, A., and Chowdhury, K. (2012). Regulation of archicortical arealization by the transcription factor Zbtb20. Hippocampus 22, 2144–2156. doi:10.1002/hipo.22035
Roy, A., Gonzalez-Gomez, M., Pierani, A., Meyer, G., and Tole, S. (2014). Lhx2 regulates the development of the forebrain hem system. Cereb. Cortex 24, 1361–1372. doi:10.1093/cercor/bhs421
Shetty, A. S., Godbole, G., Maheshwari, U., Padmanabhan, H., Chaudhary, R., Muralidharan, B., et al. (2013). Lhx2 regulates a cortex-specific mechanism for barrel formation. Proc. Natl. Acad. Sci. U. S. A. 110, E4913–E4921. doi:10.1073/pnas.1311158110
Shimogori, T., Banuchi, V., Ng, H. Y., Strauss, J. B., and Grove, E. A. (2004). Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development 131, 5639–5647. doi:10.1242/dev.01428
Shinozaki, K., Yoshida, M., Nakamura, M., Aizawa, S., and Suda, Y. (2004). Emx1 and Emx2 cooperate in initial phase of archipallium development. Mech. Dev. 121, 475–489. doi:10.1016/j.mod.2004.03.013
Sokpor, G., Kerimoglu, C., Nguyen, H., Pham, L., Rosenbusch, J., Wagener, R., et al. (2021a). Loss of BAF complex in developing cortex perturbs radial neuronal migration in a WNT signaling-dependent manner. Front. Mol. Neurosci. 14, 687581. doi:10.3389/fnmol.2021.687581
Sokpor, G., Nguyen, H. P., and Tuoc, T. (2021b). Context-specific chromatin remodeling activity of mSWI/SNF complexes depends on the epigenetic landscape. Signal Transduct. Target. Ther. 6, 360. doi:10.1038/s41392-021-00770-6
Sokpor, G., Xie, Y. B., Rosenbusch, J., and Tuoc, T. (2017). Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 10, 243. doi:10.3389/fnmol.2017.00243
Stoykova, A., Treichel, D., Hallonet, M., and Gruss, P. (2000). Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20, 8042–8050. doi:10.1523/JNEUROSCI.20-21-08042.2000
Subramanian, L., Remedios, R., Shetty, A., and Tole, S. (2009). Signals from the edges: The cortical hem and antihem in telencephalic development. Semin. Cell Dev. Biol. 20, 712–718. doi:10.1016/j.semcdb.2009.04.001
Subramanian, L., and Tole, S. (2009). Mechanisms underlying the specification, positional regulation, and function of the cortical hem. Cereb. Cortex 19, I90–I95. doi:10.1093/cercor/bhp031
Sur, M., and Rubenstein, J. L. (2005). Patterning and plasticity of the cerebral cortex. Science 310, 805–810. doi:10.1126/science.1112070
Takiguchi-Hayashi, K., Sekiguchi, M., Ashigaki, S., Takamatsu, M., Hasegawa, H., Suzuki-Migishima, R., et al. (2004). Generation of reelin-positive marginal zone cells from the caudomedial wall of telencephalic vesicles. J. Neurosci. 24, 2286–2295. doi:10.1523/Jneurosci.4671-03.2004
Tétreault, N., Champagne, M-P., and Bernier, G. (2009). The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev. Biol. 327, 541–550. doi:10.1016/j.ydbio.2008.12.022
Toresson, H., Potter, S. S., and Campbell, K. (2000). Genetic control of dorsal-ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Development 127, 4361–4371. doi:10.1242/dev.127.20.4361
Tuoc, T., Dere, E., Radyushkin, K., Pham, L., Nguyen, H., Tonchev, A. B., et al. (2017). Ablation of BAF170 in developing and postnatal dentate gyrus affects neural stem cell proliferation, differentiation, and learning. Mol. Neurobiol. 54, 4618–4635. doi:10.1007/s12035-016-9948-5
Tuoc, T. C., Boretius, S., Sansom, S. N., Pitulescu, M. E., Frahm, J., Livesey, F. J., et al. (2013). Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell 25, 256–269. doi:10.1016/j.devcel.2013.04.005
Tuoc, T. C., Radyushkin, K., Tonchev, A. B., Pinon, M. C., Ashery-Padan, R., Molnar, Z., et al. (2009). Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. J. Neurosci. 29, 8335–8349. doi:10.1523/JNEUROSCI.5669-08.2009
Vyas, A., Saha, B., Lai, E., and Tole, S. (2003). Paleocortex is specified in mice in which dorsal telencephalic patterning is severely disrupted. J. Comp. Neurol. 466, 545–553. doi:10.1002/cne.10900
Wang, A., Wang, J., Tian, K., Huo, D., Ye, H., Li, S., et al. (2021). An epigenetic circuit controls neurogenic programs during neocortex development. Development 148, dev199772. doi:10.1242/dev.199772
Wolff, E., Suplicki, M. M., and Behr, R. (2019). Primordial germ cells do not migrate along nerve fibres in marmoset monkey and mouse embryos. Reproduction 157, 101–109. doi:10.1530/Rep-18-0401
Xie, Z. F., Ma, X. H., Ji, W. L., Zhou, G. D., Lu, Y. Z., Xiang, Z. H., et al. (2010). Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc. Natl. Acad. Sci. U. S. A. 107, 6510–6515. doi:10.1073/pnas.0912315107
Xuan, S., Baptista, C. A., Balas, G., Tao, W., Soares, V. C., and Lai, E. (1995). Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14, 1141–1152. doi:10.1016/0896-6273(95)90262-7
Yoshida, M., Assimacopoulos, S., Jones, K. R., and Grove, E. A. (2006). Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development 133, 537–545. doi:10.1242/dev.02209
Yun, K., Potter, S., and Rubenstein, J. L. (2001). Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205. doi:10.1242/dev.128.2.193
Keywords: cortical development, patterning, dorsal telencephalon, cortical hem, hippocampus, BAF (mSWI/SNF) complex, Lhx2
Citation: Nguyen H, Sokpor G, Parichha A, Pham L, Saikhedkar N, Xie Y, Ulmke PA, Rosenbusch J, Pirouz M, Behr R, Stoykova A, Brand-Saberi B, Nguyen HP, Staiger JF, Tole S and Tuoc T (2022) BAF (mSWI/SNF) complex regulates mediolateral cortical patterning in the developing forebrain. Front. Cell Dev. Biol. 10:1011109. doi: 10.3389/fcell.2022.1011109
Received: 03 August 2022; Accepted: 16 September 2022;
Published: 03 October 2022.
Edited by:
Kunimasa Ohta, Kyushu University, JapanCopyright © 2022 Nguyen, Sokpor, Parichha, Pham, Saikhedkar, Xie, Ulmke, Rosenbusch, Pirouz, Behr, Stoykova, Brand-Saberi, Nguyen, Staiger, Tole and Tuoc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shubha Tole, shubhatole@gmail.com; Tran Tuoc, Tran.Tuoc@ruhr-uni-bochum.de
†These authors have contributed equally to this work and share first authorship
 Huong Nguyen
Huong Nguyen Godwin Sokpor
Godwin Sokpor Arpan Parichha
Arpan Parichha Linh Pham
Linh Pham Nidhi Saikhedkar
Nidhi Saikhedkar Yuanbin Xie
Yuanbin Xie Pauline Antonie Ulmke
Pauline Antonie Ulmke Joachim Rosenbusch
Joachim Rosenbusch Mehdi Pirouz6,7
Mehdi Pirouz6,7  Rüdiger Behr
Rüdiger Behr Anastassia Stoykova
Anastassia Stoykova Beate Brand-Saberi
Beate Brand-Saberi Huu Phuc Nguyen
Huu Phuc Nguyen Jochen F. Staiger
Jochen F. Staiger Shubha Tole
Shubha Tole Tran Tuoc
Tran Tuoc