Benefits and risks of gene drives for invasive plant management - the case for common tansy
- 1Department of Horticultural Science, University of Minnesota, St. Paul, MN, United States
- 2Foreign Disease-Weed Science Research Unit, United States Department of Agriculture-Agricultural Research Service, Frederick, MD, United States
- 3Department of Agronomy and Plant Genetics, University of Minnesota, St. Paul, MN, United States
Invasive plants cause significant environmental and economic damage, but land managers have few control options. Common tansy (Tanacetum vulgare) is prevalent in many US states and is one of the most reported invasive plants in Minnesota. Controlling common tansy poses a challenge due to its extensive distribution and association with diverse plant communities. A gene drive is being explored as a genetic biocontrol method for the management of several non-native invasives, including common tansy in North America. Gene drives have emerged as a novel biotechnology application with potential to improve public health, promote conservation, and increase agricultural productivity. In common tansy, gene drives could be developed to target genes that would reduce or eliminate female fertility and consequently inhibit common tansy seed production. Using common tansy as an example, we outline risks associated with the use of gene drive technology for invasive plant control and explain how risks may be mitigated. Understanding potential benefits and risks associated with gene drives in the early stages of development is crucial. Mitigating risks, receiving stakeholder input, and navigating the regulatory environment will play an important role in gene drive development and deployment.
1 Introduction
Non-native invasive plants present a critical management problem world-wide (Mashhadi and Radosevich, 2004). A non-native invasive plant is one that has been introduced, either intentionally or by accident, to a region outside its native range, and its spread causes harm to the environment, economy, or public health. Plants are frequently introduced for cultivation or as stowaways on water and land vehicles, and the majority of introduced plants do not cause significant harm. Many non-native plants have provided significant agricultural, economic, and health benefits to the United States (Blackburn et al., 2014; Milanović et al., 2020). However, a small number of introduced plants become invasive and cause immense damage. An estimated 24% of introduced plants become established, and of those, an estimated 18% have detrimental impacts (Jeschke and Pyšek, 2018). Invasive plants quickly dominate an area and outcompete native plants which may lead to the decline of threatened or endangered species (Dueñas et al., 2018). Human health can be impacted by invasive plants like the case of blisters and burns caused by the sap of wild parsnip (Pastinaca sativa) and giant hogweed (Heracleum mantegazzianum) (Averill and DiTommaso, 2007; Cuddington et al., 2022). Plant invasions reduce the recreational value and esthetic appeal of rivers, lakes, and natural areas as is the case of Eurasian watermilfoil (Myriophyllum spicatum) in North America (Westbrooks, 1998; Pimentel et al., 2005; Zhang and Boyle, 2010). Many invasive plants are agricultural weeds and cause economic harm in terms of crop losses or reduced quantity or quality of livestock forage, e.g. non-native invasive Canada thistle (Cirsium arvense) (Guggisberg et al., 2012). Annual crop losses caused by agricultural weeds are estimated to be US$33 billion (Pimentel et al., 2005).
Invasive plant management is challenging for land managers from a logistical and financial standpoint. Herbicides are often one of the most effective options at controlling invasive plants, but herbicides also harm native vegetation. Several target plants have evolved herbicide resistance limiting the effectiveness of this method (Kniss, 2018). Hand-pulling can be highly effective, but it is usually only feasible for small populations and causes soil structure damage. Financial and labor resources often act as a major limiting factor in invasive plant removal (Kettenring and Adams, 2011). Classical biological control has been an effective control method to manage some plants, but it is limited to those with appropriate control agents and often takes many years to assess specificity and risk in order to receive state and federal approval from regulatory agencies for release (Clewley et al., 2012). Classical biological control has risks associated with non-target effects of the biocontrol agent, but those are heavily mitigated by lengthy risk analysis procedures (Scoles et al., 2022; Van Driesche and Center, 2013). All control methods have in some level of risk, and the likelihood of reinvasion is high without effective restoration following any method of removal (Kettenring and Adams, 2011). The challenges associated with traditional management methods have led scientists to explore novel methods of management.
Genetic biocontrol, particularly gene drive technology, is emerging as a novel method of management for many harmful species and diseases. Gene drives work as a mechanism of biased inheritance for a target allele, which can be harnessed to ‘drive’ a desired allele throughout a population (Alphey et al., 2020). Gene drives designed to knock out a reproductive-specific gene would result in sterility, which could lead to population-level decline for an invasive species. While not used to date for the management of any species, gene drives have the potential to become an effective and efficient weed management tool. This review highlights common tansy as a possible species for development and deployment of a gene drive. Many of the principles proposed for common tansy can be applied to other invasive plant species.
2 Background of gene drives
Synthetic gene drives have been developed in insects, mammals, fungi, and bacteria (Bier, 2022) and there is much discussion of their utility in plants (Neve, 2018). Synthetic gene drives are engineered to bias inheritance and spread a desired trait through a population over generations. Conversely, selfish genetic elements occur naturally and are segments of the genome that are able to enhance their own transmission, even if the overall fitness of the organism is reduced (Ågren and Clark, 2018). Meiotic drives are an example of selfish genetic elements achieving drive during meiosis and are often referred to as naturally occurring gene drives (Vergara et al., 2022). Meiotic drive is a method in which a selfish genetic element manipulates its transmission via an interference mechanism and becomes overrepresented in gametes (Ågren and Clark, 2018; McFarlane et al., 2023). Meiotic drives in plants have been studied in maize (Zea mays) chromosome knobs and monkeyflower hybrids (Mimulus) (Buckler et al., 1999; Fishman and Willis, 2005). Design of gene drives may utilize clustered regularly interspaced short palindromic repeats (CRISPR), transposable elements (TEs), zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), maternal-effect dominant embryonic arrest (Medea), or homing endonuclease genes (HEGs) (National Academies of Sciences, Engineering, and Medicine, 2016).
Homing endonuclease genes (HEGs) are a class of selfish genetic elements that facilitate the conversion of a heterozygote to a homozygote. The HEG encodes an enzyme that cleaves the homologous chromosome at the recognition site, and homology directed repair copies the HEG into the homologous chromosome (National Academies of Sciences, Engineering, and Medicine, 2016) Over generations, the gene is expected to have an increased frequency in a population. There is discussion whether the term ‘gene drive’ for naturally occurring selfish genetic elements is appropriate, or if a gene drive should only refer to synthetic gene drives (James et al., 2023). Except when otherwise noted in this paper, gene drives are used to refer to synthetic gene drives, in particular CRISPR-based homing gene drives. CRISPR-based homing gene drives act similarly to homing endonuclease genes.
Harnessing selfish genetic elements was first proposed in the 1960s and revisited in 2003 with the idea of harnessing HEGs (Burt, 2003; Vergara et al., 2022) Scientists harnessed selfish genetic elements to engineer a system of biased inheritance which led to the development of a synthetic homing endonuclease-based gene drive in mosquito (Anopheles gambiae) (Windbichler et al., 2011). Shortly after the introduction of CRISPR as a genome editing tool, a CRISPR/Cas9-based gene drive system, termed a mutagenic chain reaction, was created in Drosophila melanogaster (Gantz and Bier, 2015). Yeast and mosquito gene drives quickly followed in Saccharomyces cerevisiae, Anopheles stephensi, and Anopheles gambiae using CRISPR/Cas9 (DiCarlo et al., 2015; Gantz et al., 2015; Hammond et al., 2016).
CRISPR/Cas9 allows for more targeted, efficient, and cost-effective gene editing and is used to construct an artificial homing endonuclease system. The CRISPR/Cas9 genome editing system utilizes guide RNA to recognize the target sequence, and then the Cas9 nuclease creates a double stranded break near the target sequence that is repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR) mechanisms. Non-homologous end joining repairs the break by joining the DNA fragments. It is prone to small insertions and deletions, which may produce mutations knocking out the targeted gene. Homology directed repair utilizes a DNA template to precisely repair DNA and can be utilized to insert a gene drive element into the genome for development of a CRISPR-based homing drive (Figure 1) (Asmamaw and Zawdie, 2021).
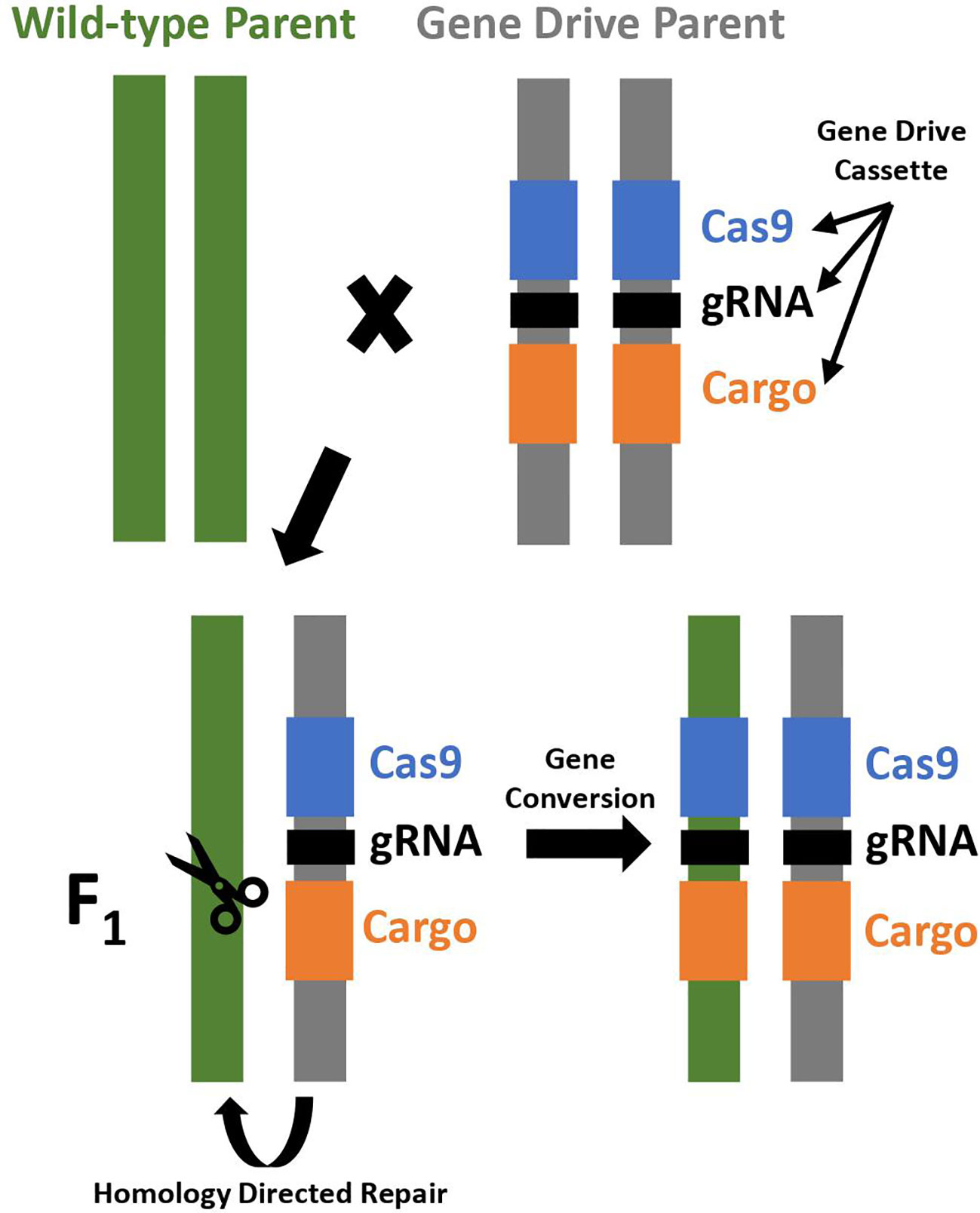
Figure 1 Gene conversion in a CRISPR-based homing drive. A cross between a wild-type parent and a gene drive parent results in F1 progeny that are initially heterozygous, but subsequently converted to homozygous for the gene drive cassette due to gene conversion through homology directed repair. The gene drive cassette expresses a Cas endonuclease (Cas9; shown in blue), guide RNA (gRNA; shown in black), and contains a cargo gene if desired (shown in orange).
There are two strategies for the application of a gene drive to manage invasive plants: population modification and population suppression. Population modification, or a modification drive, spreads a genetic element that alters the population’s genotype. The spread of a genetic element that returns herbicide sensitivity to a population that has developed herbicide resistance would function as a population modification gene drive and is also referred to as a sensitizing drive (Neve, 2018). Population suppression, or a suppression drive, spreads a genetic element, or deleterious allele, that leads to population decline. Population suppression can be achieved by driving reduced fertility into a population (National Academies of Sciences, Engineering, and Medicine, 2016).
An early population model suggests that a sufficient genetic load for population suppression can be reached in fewer than 20 generations (Burt, 2003). Genetic load refers to reduced fitness at the population-level produced by the gene drive. A variety of population models have been created that predict how the gene drives’ fitness cost, conversion rate, and initial release frequency can vary genetic load of a gene drive over time in a population (Unckless et al., 2015; Drury et al., 2017; Rode et al., 2019; Vergara et al., 2022). When evaluated in a cage study of mosquitoes (Anopheles gambiae), Kyrou et al. (2018) demonstrated that the gene drive construct reached 100% prevalence in as few as seven generations (Kyrou et al., 2018).
Gene drives could also prove useful for plant breeding, plant pathogen control, and conservation. Plant breeders could use gene drives to shorten the inbreeding time to produce homozygous parental lines and improve gene-editing in polyploid crops (Siddiqui et al., 2021; Tek and Budak, 2022). The use of gene drives to control pathogens and pests of agricultural crops has also been discussed (Courtier-Orgogozo et al., 2017; Medina, 2018; Gardiner et al., 2020). Conversely, gene drives could be engineered to spread an allele that increases plant fitness, removes deleterious alleles, or promotes the conservation of an endangered plant species (Barrett et al., 2019; Rode et al., 2019).
3 Feasibility of gene drives in plants
Gene drives will not be suitable for all invasive plant management scenarios. Gene drives are transmitted through sexual reproduction and the conversion of heterozygotes to homozygotes. Therefore, development of a gene drive is most suitable in outcrossing plant species that primarily reproduce via seed. Plants that have high selfing rates or primarily spread vegetatively (e.g., rhizomes, tubers, etc.) will be less amenable to control using a gene drive (National Academies of Sciences, Engineering, and Medicine, 2016). Moreover, pollination biology and seed dispersal will influence the range and speed at which a gene drive will spread. The generation time (life history) of the plant species will also factor into the development and feasibility of a gene drive. For example, a perennial species that takes several years to reach reproductive age, such as trees, may require hundreds of years for sufficient generations to spread the gene drive (Rode et al., 2019). Plant species can have extensive seed banks that present a challenge for population control via any method. Dormant seeds in the soil will not be affected by a gene drive until they germinate and reach maturity for gene exchange. Species with low yearly germination rates are expected to require more generations to reach the same genetic load as those with high yearly germination rates (Barrett et al., 2019).
A variety of technical challenges must be considered for the development of a gene drive in a plant species. Genetic transformation is the method of introducing foreign DNA into a host and will be needed to introduce a gene drive construct into an invasive plant host. Agrobacterium-mediated transformation is a genetic transformation method that has been successfully used in many plant species (Gelvin, 2003). However, some species have proven to be recalcitrant to Agrobacterium infection or the subsequent shoot regeneration via tissue culture (Pitzschke, 2013; Armas et al., 2017). Particle bombardment, viral vector-mediated, protoplast transfection, and nanoparticle-mediated DNA delivery are alternative gene drive delivery methods that can be considered (Laforest and Nadakuduti, 2022). Conventional plant genetic transformation methods insert DNA randomly into the host genome. Random integration or multiple copies of transgenes may result in variable expression levels from plants derived from independent transformation events. These variable levels of transgene expression may be the result of position effects or caused by transgene silencing (Gelvin, 2003). In either case, the random insertion necessitates the screening of multiple independent lines for heritability and gene drive expression. Alternatively, site-specific plant transformation methods using CRISPR technology and particle bombardment have been demonstrated (Begemann et al., 2017; Dong et al., 2020). Ultimately, an efficient method of transformation will be necessary for gene drive development and testing in any plant species.
Identifying gene targets and targeting them will be more complex in plant species with an unsequenced or large genome and plants with polyploidy or aneuploidy (Neve, 2018). A gene drive that targets a trait controlled by many genes will pose more challenges than one that targets a trait controlled by a single gene with low sequence diversity within the species and high sequence diversity among species. Suppression gene drives are often proposed to target male or female fertility genes. A number of genes essential to female fertility have been identified in model plant systems such as Arabidopsis, and homologous genes may serve as a starting point for a gene target in other plant species (Siddiqi et al., 2000; Crawford et al., 2007; Johnston et al., 2007; Qin et al., 2009; Trigueros et al., 2009; Zhao et al., 2014). To obtain a high drive efficiency, the guide RNA must correctly find the target site and Cas9 must reliably create a double stranded break near the target sequence. After the double stranded break is made, homology directed repair needs to occur to ensure further propagation of the gene drive element. A high drive efficiency is demonstrated by a high conversion rate of heterozygotes to homozygotes seen in CRISPR-based homing gene drives. There is evidence that homology directed repair does not occur as frequently in plants as non-homologous end joining. (Gorbunova and Levy, 1999). However, the use of specific promoters can increase homology directed repair efficiency (Miki et al., 2018; Wolter et al., 2018). The engineering of maternal effect dominant embryonic arrest (Medea), male gamete killer drives, and CRISPR toxin-antidote drives may be a good choice in species with low homology directed repair efficiency (Barrett et al., 2019; Wang et al., 2022). Medea works by ensuring progeny must inherit Medea in order to survive through the use of a toxin-antidote system. Medea alleles must be associated with a low fitness cost for population-wide spread, so Medea is not suitable for a suppression drive but could be effective for a modification drive (Akbari et al., 2014). Male gamete killer drives also rely on a toxin-antidote system but will have limited sequence specificity (Barrett et al., 2019; Sweigart et al., 2019). CRISPR toxin-antidote drives work similarly to Medea and are discussed in more detail under the Risks section of this manuscript.
Resistance to gene drives has occurred (Hammond et al., 2017), and it is likely that resistance would develop to any gene drive without a design to mitigate resistance development. Resistance alleles are generated when non-homologous end joining occurs in place of homology directed repair, or they can generate from standing genetic variation or de novo mutations (Unckless et al., 2017). Strategies to prevent gene drive resistance include guide RNA multiplexing and designing targets for highly conserved regions of the genome (Champer et al., 2018; Barrett et al., 2019; Price et al., 2020). However, some models have shown that resistance alleles may not become prominent in a population until many generations after the gene drive saturates a population. This lag time will vary depending on the drive’s fitness costs and the rate of resistance allele formation (Unckless et al., 2017). A suppression drive may have a high fitness cost, which can give rise to rapid resistance causing a drive to locally self-extinguish (Noble et al., 2018). Resistance developing unlinked to the drive, or nonallelic, is possible. Gene drive spread has potential to outpace nonallelic resistance evolution through the use of multiple drives with individually moderate effects (Cook et al., 2022).
4 Common tansy as a model for gene drives in plants
Common tansy (Tanacetum vulgare) is an herbaceous perennial native to Western Eurasia. It was introduced to North America as early as the 1600s for its utility as an ornamental, medicinal, and funerary plant (Mitich, 1992; Jacobs, 2008). It is winter hardy to USDA Z3, and spreads both sexually (seed) and asexually (rhizomes). In the state of Minnesota, and five other states, common tansy is designated as a noxious weed meaning efforts must be made to control its spread (Clasen et al., 2011; Chandler, 2013).
Common tansy is a short-lived herbaceous perennial with rhizomes from which shoots emerge in the spring. Rhizomes may persist three to ten years with an average of 5.4 years (Jacobs, 2008). Common tansy plants produce many flowers in their first year of growth, and in the northern U.S. plants can flower from August into early November. Flowerheads on the 0.6 to 1.5 meter tall stems will hold seeds throughout the winter or longer if undisturbed (Jacobs, 2008). Seedling germination can occur in the spring or fall, and rhizome spread is minimal with seed dispersal being the main mode of population establishment (White, 1997).
Common tansy populations have continued to expand and increase over the past 30 years or more, despite widespread management. Common tansy readily forms a monoculture where it supplants native plants. This plant has been shown to invade forests, pastures, and waterways and is very prevalent in the Superior National Forest. It is toxic to livestock and horses, which harms agriculture. It has also been identified as an alternate host to the chrysanthemum stunt virus (CSV) that reduces the commercial potential of infected chrysanthemum plants, (Chrysanthemum xgrandiflorum and Chrysanthemum xhybridum) (White, 1997; Chandler, 2013).
Lake Huron tansy (T. huronense) and camphor tansy (T. camphoratum) are closely related species native to the United States. Lake Huron tansy is state-listed as endangered in Wisconsin and threatened in Michigan (Carlson and Fulkerson, 2017). Feverfew (T. parthenium) is a closely related species that is cultivated and has naturalized in the United States. Feverfew is known to hybridize with common tansy (Brown et al., 1999; Zhou et al., 1999). Hybridization has been shown to increase the invasion risk of some species (Ellstrand and Schierenbeck, 2000). The ability of common tansy to hybridize with Lake Huron tansy or camphor tansy is unknown. Common tansy is also closely related taxonomically to ox-eye daisy (Leucanthemum vulgare), an invasive plant in the Southwestern United States (Jacobs, 2008; United States Department of Agriculture, 2017). Common tansy is widespread throughout North America with a known presence in 45 US states, and 9 Canadian provinces and territories. Minnesota has greater than 38,000 reported occurrences of common tansy (EDDMapS, 2023).
Existing control methods for common tansy have several drawbacks on their effectiveness and ability for safe-use. Herbicides are a commonly used control method for common tansy. However, herbicides are costly and often require repeated applications. Herbicide resistance is a growing concern in weeds, with several weeds developing resistance (Kniss, 2018). In addition, herbicides that are applied broadly over adventitious populations may cause harm to non-target organisms, including insects, animals, and even microbial communities contributing to biodiversity loss (Ruuskanen et al., 2023). Herbicide run-off, leaching, or vapor drift can cause the herbicide to travel further than intended, bringing the harmful effects with it. Control via hand-pulling, mowing, and tillage is labor intensive and only suitable for small populations. Grazing is not a good option for common tansy populations due to livestock toxicity. Mowing and fire-management has limited effectiveness because underground rhizomes allow common tansy to regrow (Chandler, 2013). An international effort to explore biological control of common tansy formally began in 2006 (CABI, 2023) with considerable progress in testing insects as biological control agents for common tansy (Wolf et al., 2012). This form of biological control introduces a host-specific enemy from the species’ area of origin into the invading range to produce population decline. Biological control agents are subject to local and federal approvals and have unique challenges but have been successful in controlling other invasive plants (Carson et al., 2008; Clewley et al., 2012; Van Driesche and Center, 2013).
A gene drive may be a useful tool for controlling invasive common tansy. By using a suppression drive that specifically knocks-out female fertility, it may be possible to significantly reduce the number of plants in common tansy populations over several generations. When female fertility is reduced or eliminated by a gene drive, and male fertility is maintained, pollen containing the gene drive element will spread to non-gene drive plants (wild-type), but the pollen-donor plant will not produce viable seed. Seed will be produced via the hybrid cross between wild-type and gene drive common tansy. However, most of this seed will be female sterile and, thus, unable to produce progeny, yet still capable of spreading the gene drive element via pollen resulting in eventual population decline (Figure 2).
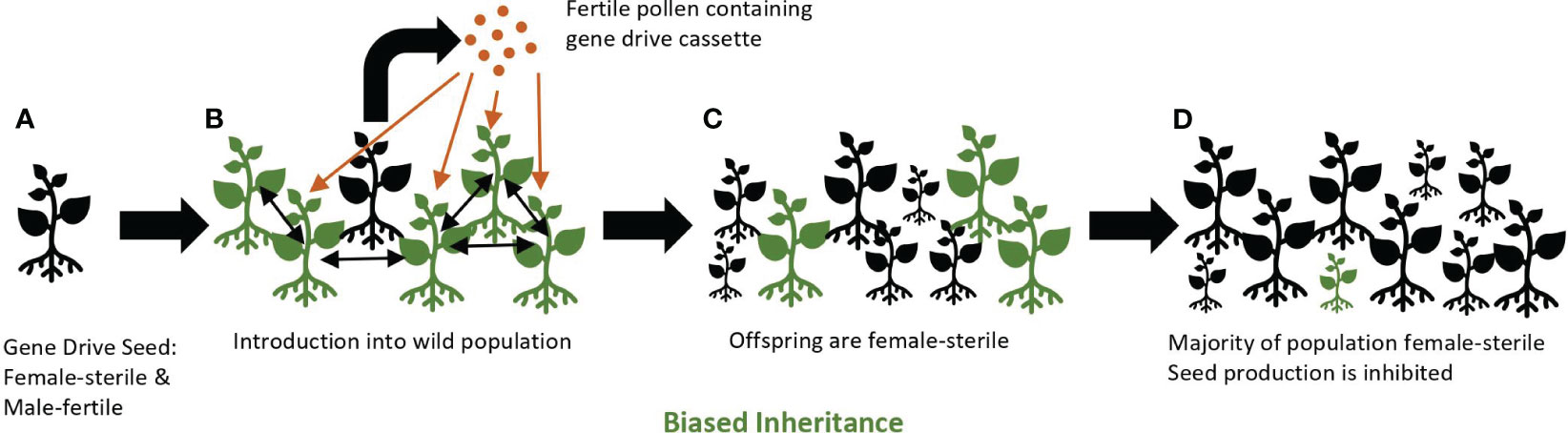
Figure 2 A female-sterile gene drive approach. Gene drive plants (black) containing the gene drive cassette are female-sterile and male-fertile individuals (A). Homozygous gene drive plants are introduced into a population of wild-type plants (green) where fertile pollen from gene drive plants pollinates wild-type plants and produces seeds (B). Seedlings emerge that are homozygous for the gene drive cassette due to gene conversion through biased inheritance (C). Over several generations, a majority of the population becomes female-sterile. Seed production is inhibited and the population declines (D). Wild-type seedlings may still appear in the population due to a persistent seed bank or as offspring of two wild-type plants.
Ensuring heritable site-specific gene targeting requires high rates of homology directed repair in germline cells, egg cells, or during early embryo development (Miki et al., 2018). Additional research is needed to improve gene targeting efficiency, but through tissue-specific Cas9 expression via germline-specific promoters coinciding with site-specific double stranded breaks, heritable site-specific gene targeting is achievable by optimizing the temporal and spatial expression of Cas9 (Miki et al., 2018). Germline differentiation is expected to occur late in development for most plants and occurs shortly before flowering in angiosperms (Lanfear, 2018). In contrast, germline segregation is not well studied, and more research is needed on the timing of germline segregation. One theory proposes a slowly-dividing functional germline (Lanfear, 2018). Taking this into consideration, we expect gene conversion to occur in egg cells or early embryo development prior to germline differentiation and segregation resulting in somatic and germline cells that are homozygous for the gene drive cassette.
Common tansy is a strong candidate for gene drive development and deployment for multiple reasons. Common tansy has high levels of outcrossing and has been reported to be self-incompatible (Clasen et al., 2011). Reproduction produces a high number of seeds, with a single plant capable of producing up to 50,000 seeds (Royer and Dickinson, 1999). While propagation and spread via rhizomes can occur, it is localized and not the primary mode of spread (Chandler, 2013). This ensures the sexual reproduction that is necessary for transmission of the gene drive. By reducing or eliminating seed production in a population through gene drive, the main mode of reproduction and dispersal will be controlled. Diploid organisms with a smaller genome are better for the identification of genetic targets. Common tansy is diploid but does have a large genome at 2C = 8.86 pg (Keskitalo et al., 1998). We are working to sequence the common tansy genome to facilitate target identification in candidate genes. In vitro regeneration has been successfully seen in common tansy, and Agrobacterium-mediated transformation has been optimized in several related species (Keskitalo et al., 1995; Sherman et al., 1998; Patial et al., 2016). We are working to optimize in vitro regeneration as a part of an efficient transformation method for common tansy that will allow for introduction of a gene drive into the genome. Low seed bank persistence and low risk of resistance would be advantageous for gene drive development (Neve, 2018). The seedbank persistence of common tansy and the risk of gene drive resistance developing is not yet known. However, this is true of many plants that may be good gene drive candidates. The challenges in the development of a gene drive for common tansy management are expected to be minimal compared to most invasive plant species (Figure 3).
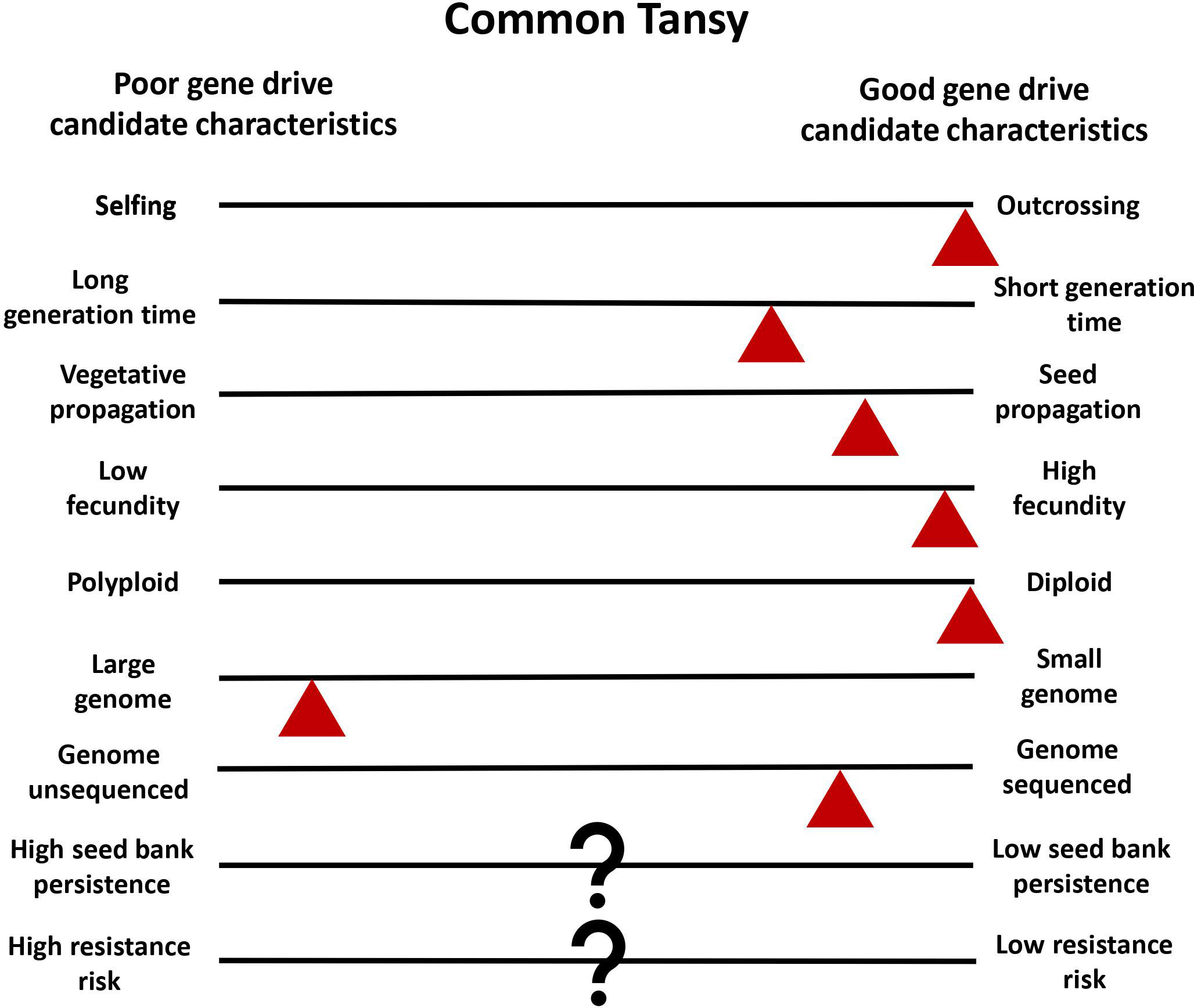
Figure 3 Common tansy as a model for plant gene drives. Characteristics of common tansy are estimated with red triangles in relation to good and poor gene drive candidate characteristics. The triangle placement is used to show where tansy is estimated to fall in comparison to characteristic diversity in all plants. Question marks (?) indicate when information on common tansy is unknown.
5 Benefits
Gene drive technology has been suggested as a possible “silver bullet” to invasive species management (Webber et al., 2015). Theoretically, gene drives have the potential to eliminate invasive species in a precise manner, which could prevent harmful effects to non-target organisms and closely related taxa. A sensitizing drive could be designed to make a species susceptible to a compound that is otherwise non-toxic to other plants, humans, and the environment (Min et al., 2018). In contrast, a suppression drive designed to reduce fertility in a species could prevent continued spread of the invasive plant, such as common tansy. Preventing further spread of the species, and facilitating population decline through a gene drive, is an important step towards restoring natural vegetation in an invaded area (Flory and Clay, 2009). An added benefit of the use of gene drives for control is the lack of disturbances to the soil or environment that can facilitate invasion and reinvasion.
Gene drives would be self-propagating and would not require repeated applications which would be beneficial for control in remote and inaccessible regions. Application of a gene drive via seeds, or in some cases pollen, will allow a large number of gene drive organisms to be released to help reach threshold frequencies in adventitious populations. Since herbicides will not be applied, safety concerns of exposure to harmful chemicals are eliminated (Myers et al., 2016). Once developed, a gene drive may be a more cost-effective tool for invasive plant management and would require minimal human intervention. A more cost-effective option for management can sometimes be the best or only choice for land managers due to financial constraints (Kettenring and Adams, 2011). Overall, gene drives have the potential to minimize environmental harm while still effectively managing invasive plant populations.
6 Risks
Gene drives are capable of spreading to nearby populations and transferring the gene drive cassette throughout a new population. While this may be desirable from an invasive plant control standpoint, the potential for gene drives to spread beyond their intended range is not without concerns. A major concern is the potential for a gene drive to be moved intentionally or unintentionally to the species’ native range. Theoretically, this could result in reduction or elimination of the species in its native range, reducing its ecosystem services. For example, in its native range of Eurasia, common tansy acts as a competitor of the non-native invasive plant, Canada goldenrod (Solidago canadensis) (Schittko and Wurst, 2014). To combat the unintended spread of a gene drive, a reversal drive, or recall drive, has been proposed as a tool that would overwrite genomic changes spread by the initial gene drive (Bier, 2022). The engineering and release of resistant alleles could also result in extinction of an escaped gene drive (Burt, 2003; Bier, 2022). While a reversal drive can function as a safety mechanism, there may still be significant damage caused by a gene drive spreading beyond its intended range (Vergara et al., 2022). A reversal drive is only likely to be introduced after some damage has occurred and that damage may have long-lasting effects (Rode et al., 2019).
Split drives, daisy-chain drives, and sensitizing drives are all types of gene drives that are designed to prevent uncontrolled spread in a gene drive release. Split drives and daisy-chain drives are classified as self-limiting drives. Split drives use molecular confinement for components of the gene drive cassette. This creates a drive that will only work if all necessary parts of the drive are present in an organism. Due to rapid segregation, exponential spread of the drive is prevented. Split drives have been proposed as a safeguarding mechanism for laboratory research and could limit the spread of a drive in the field (Akbari et al., 2015; DiCarlo et al., 2015; Terradas et al., 2021). Daisy-chain drives contain a linear series of unlinked drive elements responsible for driving the next element in the chain. Daisy-chain drives are designed to be self-exhaustive. Earlier elements in the chain will be lost from the population over time until the final element, containing the gene drive cargo, is lost. This stops the spread of the gene drive and can be used to design a gene drive that will only affect a local population (Noble et al., 2019). The use of split drives and daisy-chain drives may allow for safer release of a gene drive, although it may require the re-release of a gene drive after multiple generations. Sensitizing drives spread a neutral allele through a population that produces a sensitivity to a compound. Even if the drive spreads to a non-target population, the effect of the drive is only seen after the drive has spread through the population, and the population is treated with a particular compound, such as glyphosate (Min et al., 2018; Neve, 2018). Split drives, daisy-chain drives, and sensitizing drives will limit uncontrolled spread of a gene drive and minimize harm even if introduced to an invasive plant’s native range.
Confined drives offer another approach to limiting uncontrolled spread of a gene drive release. Toxin-antidote drives, including CRISPR toxin-antidote drives and Medea, can limit gene drive spread by requiring gene drive introduction thresholds. Once a gene drive surpasses its introduction threshold, it is able to increase to fixation in a population (Wang et al., 2022). Medea drives typically only have an introduction threshold if the drive has fitness costs, but CRISPR toxin-antidote drives can be simply designed to have an introduction threshold without a fitness cost (Champer et al., 2021). CRISPR toxin-antidote drives act as a toxin by disrupting a target gene, then it rescues individuals with the drive via a recoded version of the target gene (Champer et al., 2020). Toxin-antidote drives often are not suitable for population suppression like homing drives are. Tethered drive systems can allow homing drives to gain the confinement benefits of other drives by using a confined drive to supply an essential component needed for homing drive function such as guide RNA or the Cas endonuclease (Dhole et al., 2019; Wang et al., 2022). Targeting locally fixed alleles by selecting a fixed target sequence in the desired population that is less prevalent in the non-target population offers another potential for confining a gene drive (Wang et al., 2022).
Horizontal gene transfer is a significant risk of a gene drive release. Horizontal gene transfer occurs when genes move between populations of distinct species (National Academies of Sciences, Engineering, and Medicine, 2016). In gene drives, horizontal gene transfer would result in the transfer of the gene drive cassette to a non-target species. If the gene drive cassette were able to transfer to and spread in a non-target species, the non-target species may experience the same population suppression or modification effects as the target species. In common tansy, horizontal gene transfer to native species such as Lake Huron tansy or camphor tansy is a major concern. Lake Huron tansy is listed as endangered and threatened in some states, and horizontal gene transfer of a deleterious allele would further harm its populations. Non-target naturalized and cultivated species could also be negatively affected by gene drive transfer. Selecting a genetic target that is different from targets in closely related species due to sequence diversity will mitigate its impact after an unintended transfer of the gene drive. Many closely-related species are capable of hybridization (Goulet et al., 2017), so knowledge of the species’ ability to hybridize, and the sequence diversity among related taxa, will be critical for assessing the risks of a gene drive release. Populations with limited gene flow may be able to utilize a precision drive and prevent gene drive spread through non-target populations. A precision drive system would utilize an initial drive spreading a unique sequence that can be quickly followed by a drive spreading the desired cargo gene. The second drive is designed to only recognize and spread the drive to individuals with the unique sequence (Esvelt et al., 2014).
While invasive species negatively impact invaded ecosystems, the complete removal of a widely-prevalent invasive species may produce unintended ecosystem effects. For example, without concentrated restoration efforts, one or more new invaders may establish in place of the removed species. Due to an invasive species’ ability to significantly alter ecosystems, its subsequent removal may harm other members who have now become integrated or dependent (Zavaleta et al., 2001). Since common tansy is insect pollinated (entomophilous), its complete removal may temporarily impact pollinators that have exploited common tansy as an abundant food source. Ecosystem interactions are complex, and restoration efforts must be made to facilitate the recovery of native plant communities (Guido and Pillar, 2017).
The modeling of gene drive population dynamics provides useful information, but these models have many limitations. Much of the research on synthetic gene drives has been done on insects, particularly mosquitoes, and open field trials of gene drive mosquitoes have not yet occurred (Rašić et al., 2021). There is little known about the long term spread and effects of a gene drive. We must assume that some risks of gene drive have yet to be identified and understood. If the release of a gene drive occurs before fully understanding its risks, we may be creating a new problem requiring significant resources to adequately manage.
7 Regulatory environment and public perceptions
The deployment of gene drive technology, even for field testing, may be subject to governance and regulations. The United States Department of Agriculture (USDA), Environmental Protection Agency (EPA), and Food and Drug Administration (FDA) evaluate risks of biotechnology products. The regulation of gene drive organisms in the U.S. will likely fall under the jurisdiction of one or more of these agencies. However, there is concern about gaps in regulatory coverage for gene drives (Rudenko et al., 2018; West et al., 2022). Regulatory agencies take a “science-based” approach to assessing risks and do not explicitly consider ethical and cultural implications (Kuzma et al., 2018). The unique concerns and risks associated with gene drive organisms has led to recommendations to inform the public of risks and benefits to gene drive applications early in their development (Kuzma, 2021). There is broad consensus about the need for community engagement early in development. Community engagement could be evaluating and addressing concerns through public input and education or to obtain community consent for a gene drive release (de Graeff et al., 2023).
The communities in which a gene drive will be released, stakeholders, and the general public will all have concerns and feedback concerning the release and testing of a gene drive (National Academies of Sciences, Engineering, and Medicine, 2016). Diversity among stakeholders will translate to different perceptions and tolerances of risks posed by gene drives compared with their benefits (Rudenko et al., 2018). Oxitec, a private company holding field trials of genetically modified mosquitoes, has faced various hurdles from regulatory decisions to public opinion (Glenza, 2016; Petersen, 2022). We expect similar challenges will exist for field trials and the release of gene drives to control invasive plants, and early community engagement will be necessary to overcome these challenges.
Scientists have developed safeguarding recommendations for responsible gene drive research. Multiple stringent confinement strategies are recommended for laboratory work on gene drive organisms to prevent escape (Akbari et al., 2015). Several scientists have committed to assessing and understanding risks, benefits, efficacy, and safety of gene drive organisms prior to any field trials. These scientists also point to public transparency, community engagement, and strategies for monitoring and mitigating a gene drive release as critical components of responsible trialing of gene drive organisms (Long et al., 2020) These commitments and recommendations promote the ethical research of gene drive organisms and provide safeguards against the potential risks.
8 Conclusions
Gene drives are a novel technology that has promise for invasive plant control that complements or exceeds the capabilities of current control methods. Gene drives allow for a precise, species-specific method of management and minimize collateral damage that is common among traditional control methods. However, this novel technology remains unused for invasive plant control. Factors such as mode of reproduction, fecundity, ploidy, genome size, and seed bank persistence will affect the utility of gene drives for invasive plant control. Common tansy has several characteristics that make it a good candidate for gene drive research. However, caution will be necessary when proceeding with gene drive development and trials. Uncontrolled spread of the gene drive cassette both geographically and to non-target species could cause significant harm to the environment. These risks, and risks that have not yet been identified, need to be fully understood and mitigated prior to field trials. The regulatory atmosphere for gene drive organisms is still in development, which has prompted scientists to make commitments to responsible gene drive research (Akbari et al., 2015; Long et al., 2020). Community engagement is expected to play an important role in gene drive development and release. Gene drive technology has advanced rapidly and has many exciting applications for plants if the benefits are shown to outweigh the risks.
Author contributions
LC: Writing – original draft, Writing – review & editing. AS: Conceptualization, Writing – review & editing. MT: Writing – review & editing. NA: Writing – review & editing. RB: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for this research was supported by the United States Department of Agriculture, Agricultural Research Service appropriated project # 8044-22000-047-00D via the Gene Drive for Tanacetum vulgare Control: A Model for Management agreement # 8044-22000-047-004-A and the Minnesota Agricultural Experiment Station project MIN-21-108.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ågren J. A., Clark A. G. (2018). Selfish genetic elements. PloS Genet. 14 (11), e1007700. doi: 10.1371/journal.pgen.1007700
Akbari O. S., Bellen H. J., Bier E., Bullock S. L., Burt A., Church G. M., et al. (2015). Safeguarding gene drive experiments in the laboratory. Science 349, 927–929. doi: 10.1126/science.aac7932
Akbari O. S., Chen C.-H., Marshall J. M., Huang H., Antoshechkin I., Hay B. A. (2014). Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol. 3, 915–928. doi: 10.1021/sb300079h
Alphey L. S., Crisanti A., Randazzo F. F., Akbari O. S. (2020). Opinion: Standardizing the definition of gene drive. Proc. Natl. Acad. Sci. 117, 30864–30867. doi: 10.1073/pnas.2020417117
Armas I., Pogrebnyak N., Raskin I. (2017). A rapid and efficient in vitro regeneration system for lettuce (Lactuca sativa L.). Plant Methods 13, 58. doi: 10.1186/s13007-017-0208-0
Asmamaw M., Zawdie B. (2021). Mechanism and applications of CRISPR/cas-9-mediated genome editing. Biologics 15, 353–361. doi: 10.2147/BTT.S326422
Averill K. M., DiTommaso A. (2007). Wild parsnip (Pastinaca sativa): A troublesome species of increasing concern. Weed Technol. 21, 279–287. doi: 10.1614/WT-05-186.1
Barrett L. G., Legros M., Kumaran N., Glassop D., Raghu S., Gardiner D. M. (2019). Gene drives in plants: opportunities and challenges for weed control and engineered resilience. Proc. Biol. Sci. 286, 20191515. doi: 10.1098/rspb.2019.1515
Begemann M. B., Gray B. N., January E., Gordon G. C., He Y., Liu H., et al. (2017). Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 7, 11606. doi: 10.1038/s41598-017-11760-6
Bier E. (2022). Gene drives gaining speed. Nat. Rev. Genet. 23, 5–22. doi: 10.1038/s41576-021-00386-0
Blackburn T. M., Essl F., Evans T., Hulme P. E., Jeschke J. M., Kühn I., et al. (2014). A unified classification of alien species based on the magnitude of their environmental impacts. PloS Biol. 12, e1001850. doi: 10.1371/journal.pbio.1001850
Brown A. M. G., Edwards C. M., Hartman T. P. V., Marshall J. A., Smith R. M., Davey M. R., et al. (1999). Sexual hybrids of Tanacetum: biochemical, cytological and pharmacological characterization. J. Exp. Bot. 50, 435–444. doi: 10.1093/jxb/50.333.435
Buckler E.S. ,. 4., Phelps-Durr T. L., Buckler C. S., Dawe R. K., Doebley J. F., Holtsford T. P. (1999). Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153, 415–426. doi: 10.1093/genetics/153.1.415
Burt A. (2003). Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928. doi: 10.1098/rspb.2002.2319
CABI (2023) Tackling common tansy in North America. Available at: https://www.cabi.org/projects/tackling-common-tansy-in-north-america/ (Accessed August 20, 2023).
Carlson M. L., Fulkerson J. R. (2017). Tanacetum camphoratum Less. (syn. = Tanacetum bipinnatum (L.) Sch. Bip. pro parte): Conservation Assessment on the Tongass National Forest, Region 10 (Anchorage, Alaska: Alaska Natural Heritage Program, University of Alaska Anchorage), 41 pp.
Carson W. P., Hovick S. M., Baumert A. J., Bunker D. E., Pendergast T. H. (2008). Evaluating the post-release efficacy of invasive plant biocontrol by insects: a comprehensive approach. Arthropod Plant Interact. 2, 77–86. doi: 10.1007/s11829-008-9036-5
Champer J., Champer S. E., Kim I. K., Clark A. G., Messer P. W. (2021). Design and analysis of CRISPR-based underdominance toxin-antidote gene drives. Evol. Appl. 14, 1052–1069. doi: 10.1111/eva.13180
Champer J., Lee E., Yang E., Liu C., Clark A. G., Messer P. W. (2020). A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082. doi: 10.1038/s41467-020-14960-3
Champer J., Liu J., Oh S. Y., Reeves R., Luthra A., Oakes N., et al. (2018). Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. 115, 5522–5527. doi: 10.1073/pnas.1720354115
Chandler M. (2013). Minnesota Noxious Weed Advisory Committee Risk Assessment Worksheet: Common Tansy (Minnesota Department of Agriculture).
Clasen B., Moss N., Chandler M., Smith A. (2011). A preliminary genetic structure study of the non-native weed, common tansy (Tanacetum vulgare). Can. J. Plant Sci. 91, 717–723. doi: 10.4141/cjps10203
Clewley G. D., Eschen R., Shaw R. H., Wright D. J. (2012). The effectiveness of classical biological control of invasive plants. J. Appl. Ecol. 49, 1287–1295. doi: 10.1111/j.1365-2664.2012.02209.x
Cook F., Bull J. J., Gomulkiewicz R. (2022). Gene drive escape from resistance depends on mechanism and ecology. Evol. Appl. 15, 721–734. doi: 10.1111/eva.13358
Courtier-Orgogozo V., Morizot B., Boëte C. (2017). Agricultural pest control with CRISPR-based gene drive: time for public debate: Should we use gene drive for pest control? EMBO Rep. 18, 878–880. doi: 10.15252/embr.201744205
Crawford B. C. W., Ditta G., Yanofsky M. F. (2007). The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 17, 1101–1108. doi: 10.1016/j.cub.2007.05.079
Cuddington K., Sobek-Swant S., Drake J., Lee W., Brook M. (2022). Risks of giant hogweed (Heracleum mantegazzianum) range increase in North America. Biol. Invasions 24, 299–314. doi: 10.1007/s10530-021-02645-x
de Graeff N., Pirson I., van der Graaf R., Bredenoord A. L., Jongsma K. R. (2023). The boundary problem: Defining and delineating the community in field trials with gene drive organisms. Bioethics. 37, 600–609. doi: 10.1111/bioe.13165
Dhole S., Lloyd A. L., Gould F. (2019). Tethered homing gene drives: A new design for spatially restricted population replacement and suppression. Evol. Appl. 12, 1688–1702. doi: 10.1111/eva.12827
DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., Church G. M. (2015). Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255. doi: 10.1038/nbt.3412
Dong O. X., Yu S., Jain R., Zhang N., Duong P. Q., Butler C., et al. (2020). Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 11, 1178. doi: 10.1038/s41467-020-14981-y
Drury D. W., Dapper A. L., Siniard D. J., Zentner G. E., Wade M. J. (2017). CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv. 3, e1601910. doi: 10.1126/sciadv.1601910
Dueñas M.-A., Ruffhead H. J., Wakefield N. H., Roberts P. D., Hemming D. J., Diaz-Soltero H. (2018). The role played by invasive species in interactions with endangered and threatened species in the United States: a systematic review. Biodivers. Conserv. 27, 3171–3183. doi: 10.1007/s10531-018-1595-x
EDDMapS (2023) Early Detection & Distribution Mapping System (The University of Georgia - Center for Invasive Species and Ecosystem Health). Available at: http://www.eddmaps.org (Accessed July 10, 2023).
Ellstrand N. C., Schierenbeck K. A. (2000). Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl. Acad. Sci. 97, 7043–7050. doi: 10.1073/pnas.97.13.7043
Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M. (2014). Concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401. doi: 10.7554/eLife.03401
Fishman L., Willis J. H. (2005). A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics 169, 347–353. doi: 10.1534/genetics.104.032789
Flory S. L., Clay K. (2009). Invasive plant removal method determines native plant community responses. J. Appl. Ecol. 46, 434–442. doi: 10.1111/j.1365-2664.2009.01610.x
Gantz V. M., Bier E. (2015). Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444. doi: 10.1126/science.aaa5945
Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M., Bier E., et al. (2015). Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. 112, E6736–E6743. doi: 10.1073/pnas.1521077112
Gardiner D. M., Rusu A., Barrett L., Hunter G. C., Kazan K. (2020). Can natural gene drives be part of future fungal pathogen control strategies in plants? New Phytol. 228, 1431–1439. doi: 10.1111/nph.16779
Gelvin S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Glenza J. (2016) Zika virus: Floridians fear “Pandora”s box’ of genetically altered mosquitos (The Guardian). Available at: https://www.theguardian.com/us-news/2016/aug/14/florida-keys-zika-virus-genetically-modified-mosquitoes (Accessed July 15, 2023).
Gorbunova V. V., Levy A. A. (1999). How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4, 263–269. doi: 10.1016/s1360-1385(99)01430-2
Goulet B. E., Roda F., Hopkins R. (2017). Hybridization in plants: old ideas, new techniques. Plant Physiol. 173, 65–78. doi: 10.1104/pp.16.01340
Guggisberg A., Welk E., Sforza R., Horvath D. P., Anderson J. V., Foley M. E., et al. (2012). Invasion history of North American Canada thistle, Cirsium arvense. J. Biogeogr. 39, 1919–1931. doi: 10.1111/j.1365-2699.2012.02746.x
Guido A., Pillar V. D. (2017). Invasive plant removal: assessing community impact and recovery from invasion. J. Appl. Ecol. 54, 1230–1237. doi: 10.1111/1365-2664.12848
Hammond A., Galizi R., Kyrou K., Simoni A., Siniscalchi C., Katsanos D., et al. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles Gambiae. Nat. Biotechnol. 34, 78–83. doi: 10.1038/nbt.3439
Hammond A. M., Kyrou K., Bruttini M., North A., Galizi R., Karlsson X., et al. (2017). The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PloS Genet. 13, e1007039. doi: 10.1371/journal.pgen.1007039
Jacobs J. (2008). Ecology and Management of Common Tansy (Tanacetum vulgare L.). Invasive Species Technical Note No. MT-18 (United States Department of Agriculture).
James S. L., O’Brochta D. A., Randazzo F., Akbari O. S. (2023). A gene drive is a gene drive: the debate over lumping or splitting definitions. Nat. Commun. 14, 1749. doi: 10.1038/s41467-023-37483-z
Jeschke J., Pyšek P. (2018). “Tens rule,” in Invasion Biology: Hypotheses and Evidence. Eds. Jeschke J. M., Heger T. (Wallingford, Oxfordshire, UK: CAB International), 124–134. doi: 10.1079/9781780647647.0124
Johnston A. J., Meier P., Gheyselinck J., Wuest S. E., Federer M., Schlagenhauf E., et al. (2007). Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8, R204. doi: 10.1186/gb-2007-8-10-r204
Keskitalo M., Kanerva T., Pebu F. (1995). Development of in vitro procedures for regeneration of petiole and leaf explants and production of protoplast-derived callus L., in Tanacetum vulgare. Plant Cell Rep. 14, 261–266. doi: 10.1007/BF00233646
Keskitalo M., Lindén A., Valkonen J. P. T. (1998). Genetic and morphological diversity of Finnish tansy (Tanacetum vulgare L., Asteraceae). Züchter Genet. Breed. Res. 96, 1141–1150. doi: 10.1007/s001220050850
Kettenring K. M., Adams C. R. (2011). Lessons learned from invasive plant control experiments: a systematic review and meta-analysis. J. Appl. Ecol. 48, 970–979. doi: 10.1111/j.1365-2664.2011.01979.x
Kniss A. R. (2018). Genetically engineered herbicide-resistant crops and herbicide-resistant weed evolution in the United States. Weed Sci. 66, 260–273. doi: 10.1017/wsc.2017.70
Kuzma J. (2021). Procedurally robust risk assessment framework for novel genetically engineered organisms and gene drives. Regul. gov. 15, 1144–1165. doi: 10.1111/rego.12245
Kuzma J., Gould F., Brown Z., Collins J., Delborne J., Frow E., et al. (2018). A roadmap for gene drives: using institutional analysis and development to frame research needs and governance in a systems context. J. Responsible Innovation 5, S13–S39. doi: 10.1080/23299460.2017.1410344
Kyrou K., Hammond A. M., Galizi R., Kranjc N., Burt A., Beaghton A. K., et al. (2018). A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles Gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066. doi: 10.1038/nbt.4245
Laforest L. C., Nadakuduti S. S. (2022). Advances in delivery mechanisms of CRISPR gene-editing reagents in plants. Front. Genome Ed 4. doi: 10.3389/fgeed.2022.830178
Lanfear R. (2018). Do plants have a segregated germline? PloS Biol. 16, e2005439. doi: 10.1371/journal.pbio.2005439
Long K. C., Alphey L., Annas G. J., Bloss C. S., Campbell K. J., Champer J., et al. (2020). Core commitments for field trials of gene drive organisms. Science 370, 1417–1419. doi: 10.1126/science.abd1908
Mashhadi H. R., Radosevich S. R. (2004). “Invasive plants,” in Weed Biology and Management. Ed. Inderjit (Dordrecht: Springer Netherlands), 1–28. doi: 10.1007/978-94-017-0552-3_1
McFarlane G. R., Whitelaw C. B. A., Lillico S. G. (2023). Gene drive: past, present and future roads to vertebrate biocontrol. Appl. Biosci. 2, 52–70. doi: 10.3390/applbiosci2010006
Medina R. F. (2018). Gene drives and the management of agricultural pests. J. Responsible Innovation 5, S255–S262. doi: 10.1080/23299460.2017.1407913
Miki D., Zhang W., Zeng W., Feng Z., Zhu J.-K. (2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9, 1967. doi: 10.1038/s41467-018-04416-0
Milanović M., Knapp S., Pyšek P., Kühn I. (2020). Linking traits of invasive plants with ecosystem services and disservices. Ecosystem Serv. 42, 101072. doi: 10.1016/j.ecoser.2020.101072
Min J., Smidler A. L., Najjar D., Esvelt K. M. (2018). Harnessing gene drive. J. Responsible Innovation 5, S40–S65. doi: 10.1080/23299460.2017.1415586
Myers J. P., Antoniou M. N., Blumberg B., Carroll L., Colborn T., Everett L. G., et al. (2016). Concerns over use of glyphosate-based herbicides and risks associated with exposures: a consensus statement. Environ. Health 15, 19. doi: 10.1186/s12940-016-0117-0
National Academies of Sciences, Engineering, and Medicine (2016). Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values (Washington D.C.: National Academies Press). doi: 10.17226/23405
Neve P. (2018). Gene drive systems: do they have a place in agricultural weed management? Pest Manage. Sci. 74, 2671–2679. doi: 10.1002/ps.5137
Noble C., Adlam B., Church G. M., Esvelt K. M., Nowak M. A. (2018). Current CRISPR gene drive systems are likely to be highly invasive in wild populations. Elife 7, e33423. doi: 10.7554/eLife.33423
Noble C., Min J., Olejarz J., Buchthal J., Chavez A., Smidler A. L., et al. (2019). Daisy-chain gene drives for the alteration of local populations. Proc. Natl. Acad. Sci. 116, 8275–8282. doi: 10.1073/pnas.1716358116
Patial V., Krishna R., Arya G., Singh V. K., Agarwal M., Goel S., et al. (2016). Development of an efficient, genotype independent plant regeneration and transformation protocol using cotyledonary nodes in safflower (Carthamus tinctorius L.). J. Plant Biochem. Biotechnol. 25, 421–432. doi: 10.1007/s13562-016-0354-x
Petersen M. (2022) In California, an army of genetically engineered mosquitoes awaits release. Will it backfire? Los Angeles Times. Available at: https://www.latimes.com/business/story/2022-04-08/genetically-engineered-mosquitoes-may-soon-fly-in-california (Accessed July 15, 2023).
Pimentel D., Zuniga R., Morrison D. (2005). Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 52, 273–288. doi: 10.1016/j.ecolecon.2004.10.002
Pitzschke A. (2013). Agrobacterium infection and plant defense—transformation success hangs by a thread. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00519
Price T. A. R., Windbichler N., Unckless R. L., Sutter A., Runge J.-N., Ross P. A., et al. (2020). Resistance to natural and synthetic gene drive systems. J. Evol. Biol. 33, 1345–1360. doi: 10.1111/jeb.13693
Qin Y., Leydon A. R., Manziello A., Pandey R., Mount D., Denic S., et al. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PloS Genet. 5, e1000621. doi: 10.1371/journal.pgen.1000621
Rašić G., Lobo N. F., Jeffrey Gutiérrez E. H., Sánchez C H. M., Marshall J. M. (2021). Monitoring needs for gene drive mosquito projects: lessons from vector control field trials and invasive species. Front. Genet. 12. doi: 10.3389/fgene.2021.780327
Rode N. O., Estoup A., Bourguet D., Courtier-Orgogozo V., Débarre F. (2019). Population management using gene drive: molecular design, models of spread dynamics and assessment of ecological risks. Conserv. Genet. 20, 671–690. doi: 10.1007/s10592-019-01165-5
Royer F., Dickinson R. (1999). Weeds of Canada and the Northern United States: A Guide for Identification (Renton, WA: The University of Alberta Press).
Rudenko L., Palmer M. J., Oye K. (2018). Considerations for the governance of gene drive organisms. Pathog. Glob. Health 112, 162–181. doi: 10.1080/20477724.2018.1478776
Ruuskanen S., Fuchs B., Nissinen R., Puigbò P., Rainio M., Saikkonen K., et al. (2023). Ecosystem consequences of herbicides: the role of microbiome. Trends Ecol. Evol. 38, 35–43. doi: 10.1016/j.tree.2022.09.009
Schittko C., Wurst S. (2014). Above- and belowground effects of plant-soil feedback from exotic Solidago canadensis on native Tanacetum vulgare. Biol. Invasions 16, 1465–1479. doi: 10.1007/s10530-013-0584-y
Scoles J., Cuda J. P., Overholt W. A., Bill C. R. (2022). How Scientists Obtain Approval to Release Organisms for Classical Biological Control of Invasive Weeds (University of Florida IFAS).
Sherman J. M., Moyer J. W., Daub M. E. (1998). A regeneration and Agrobacterium-mediated transformation system for genetically diverse chrysanthemum cultivars. J. Am. Soc Hortic. Sci. 123, 189–194. doi: 10.21273/JASHS.123.2.189
Siddiqi I., Ganesh G., Grossniklaus U., Subbiah V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127, 197–207. doi: 10.1242/dev.127.1.197
Siddiqui H. A., Harvey-Samuel T., Mansoor S. (2021). Gene drive: a faster route to plant improvement. Trends Plant Sci. 26, 1204–1206. doi: 10.1016/j.tplants.2021.09.005
Sweigart A. L., Brandvain Y., Fishman L. (2019). Making a murderer: the evolutionary framing of hybrid gamete-killers. Trends Genet. 35, 245–252. doi: 10.1016/j.tig.2019.01.004
Tek M. I., Budak K. (2022). A new approach to develop resistant cultivars against the plant pathogens: CRISPR drives. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.889497
Terradas G., Buchman A. B., Bennett J. B., Shriner I., Marshall J. M., Akbari O. S., et al. (2021). Inherently confinable split-drive systems in Drosophila. Nat. Commun. 12, 1480. doi: 10.1038/s41467-021-21771-7
Trigueros M., Navarrete-Gómez M., Sato S., Christensen S. K., Pelaz S., Weigel D., et al. (2009). The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21, 1394–1409. doi: 10.1105/tpc.109.065508
Unckless R. L., Clark A. G., Messer P. W. (2017). Evolution of resistance against CRISPR/cas9 gene drive. Genetics 205, 827–841. doi: 10.1534/genetics.116.197285
Unckless R. L., Messer P. W., Connallon T., Clark A. G. (2015). Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201, 425–431. doi: 10.1534/genetics.115.177592
United States Department of Agriculture (2017). Field Guide for Managing Oxeye Daisy in the Southwest (United States Department of Agriculture).
Van Driesche R., Center T. (2013). “Biological control of invasive plants in protected areas,” in Plant Invasions in Protected Areas: Patterns, Problems and Challenges. Eds. Foxcroft L. C., Pyšek P., Richardson D. M., Genovesi P. (Dordrecht: Springer Netherlands), 561–597. doi: 10.1007/978-94-007-7750-7_26
Vergara M. M., Labbé J., Tannous J. (2022). Reflection on the challenges, accomplishments, and new frontiers of gene drives. BioDesign Res. 2022:9853416. doi: 10.34133/2022/9853416
Wang G.-H., Du J., Chu C. Y., Madhav M., Hughes G. L., Champer J. (2022). Symbionts and gene drive: two strategies to combat vector-borne disease. Trends Genet. 38, 708–723. doi: 10.1016/j.tig.2022.02.013
Webber B. L., Raghu S., Edwards O. R. (2015). Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc. Natl. Acad. Sci. 112, 10565–10567. doi: 10.1073/pnas.1514258112
West W. F., Buchman L. W., Medina R. F. (2022). Public deliberation and the regulation of gene drive in the USA. Sci. Public Policy 49, 843–852. doi: 10.1093/scipol/scac032
Westbrooks R. (1998). Invasive plants, changing the landscape of America: Fact book (Washington, D.C: Federal Interagency Committee for the Management of Noxious and Exotic Weeds (FICMNEW).
White D. J. (1997). Tanacetum vulgare L: weed potential, biology, response to herbivory, and prospects for classical biological control in Alberta (Edmonton, Alberta: University of Alberta). doi: 10.7939/R3W669G9M
Windbichler N., Menichelli M., Papathanos P. A., Thyme S. B., Li H., Ulge U. Y., et al. (2011). A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473, 212–215. doi: 10.1038/nature09937
Wolf V. C., Gassmann A., Müller C. (2012). Choice behaviour and performance of Cassida stigmatica on various chemotypes of Tanacetum vulgare and implications for biocontrol. Entomol. Exp. Appl. 144, 78–85. doi: 10.1111/j.1570-7458.2012.01242.x
Wolter F., Klemm J., Puchta H. (2018). Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 94, 735–746. doi: 10.1111/tpj.13893
Zavaleta E. S., Hobbs R. J., Mooney H. A. (2001). Viewing invasive species removal in a whole-ecosystem context. Trends Ecol. Evol. 16, 454–459. doi: 10.1016/S0169-5347(01)02194-2
Zhang C., Boyle K. J. (2010). The effect of an aquatic invasive species (Eurasian watermilfoil) on lakefront property values. Ecol. Econ. 70, 394–404. doi: 10.1016/j.ecolecon.2010.09.011
Zhao L., He J., Cai H., Lin H., Li Y., Liu R., et al. (2014). Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 80, 615–628. doi: 10.1111/tpj.12657
Keywords: gene drive, invasives, common tansy, weed management, genetic biocontrol
Citation: Croghan L, Smith AG, Tancos MA, Anderson NO and Becker RL (2023) Benefits and risks of gene drives for invasive plant management - the case for common tansy. Front. Agron. 5:1290781. doi: 10.3389/fagro.2023.1290781
Received: 08 September 2023; Accepted: 09 October 2023;
Published: 20 October 2023.
Edited by:
Hanwen Wu, NSW Government, AustraliaReviewed by:
Jackson Champer, Peking University, ChinaXiaocheng Zhu, Wagga Wagga Agricultural Institute (WWAI), Australia
Copyright © 2023 Croghan, Smith, Tancos, Anderson and Becker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lori Croghan, crogh009@umn.edu
 Lori Croghan
Lori Croghan Alan G. Smith1
Alan G. Smith1  Matthew A. Tancos
Matthew A. Tancos Neil O. Anderson
Neil O. Anderson Roger L. Becker
Roger L. Becker